- Infectious Diseases and Immunology Division, Indian Institute of Chemical Biology, Kolkata, West Bengal, India
Leishmania is an intracellular, zoonotic, kinetoplastid eukaryote with more than 1.2 million cases all over the world. The leishmanial chromosomes are divided into polymorphic chromosomal ends, conserved central domains, and antigen-encoding genes found in telomere-proximal regions. The genome flexibility of chromosomal ends of the leishmanial parasite is known to cause drug resistance and intracellular survival through the evasion of host defense mechanisms. Therefore, in this review, we discuss the plasticity of Leishmania genome organization which is the primary cause of drug resistance and parasite survival. Moreover, we have not only elucidated the causes of such genome plasticity which includes aneuploidy, epigenetic factors, copy number variation (CNV), and post-translation modification (PTM) but also highlighted their impact on drug resistance and parasite survival.
1 Introduction
Leishmaniasis primarily a zoonotic poverty-driven neglected tropical disease largely overlooked by the pharmaceutical industry (Bray, 1974), is usually transmitted to the human host by the bite of a sandfly. Leishmania amastigotes enter the gut of the sandfly after the bloodmeal wherein they develop into pro-cyclic promastigotes. These promastigotes divide and replicate to form the metacyclic promastigotes after 4-7 days. During this stage, they migrate to the pharynx of the sandfly and after its bite, the promastigotes get inside the host body to get phagocytized by the macrophage. Once inside the phagosome, they differentiated into amastigotes. When the sandfly withdraws blood from the mammalian host the parasite cells get inside its gut where it again gets transformed into the promastigote form and thus the cycle continues (Bates, 2018).
Even though leishmaniasis is present all over the world, the disease burden in humans is concentrated in certain parts only. Visceral leishmaniasis (VL) is mainly relevant in India, Brazil, Sudan, Somalia and Kenya. Despite its endemicity in these countries, new cases tend to arise and approximately 500,000 have been reported in rural parts of South Asia, Brazil, and parts of Africa. VL, like all leishmaniases, affects mostly the rural populations. For decades VL has caused epidemics in Bihar, one of the poorest and most densely populated states in India (Bi et al., 2018). Cutaneous leishmaniasis (CL) is mainly relevant in Iran, Syria, Pakistan, Saudi Arabia, Algeria and Peru (Roatt et al., 2014). Kabul is considered to be the epicenter of CL in the world with 67,000 new cases per year (Burza et al., 2018). Currently, there is no approved vaccine against this disease, and the majority of control strategies involve therapeutics (Handman, 2001; Murray et al., 2005; Palatnik-De-Sousa, 2008). In India, antimonial based medications for treating the disease is disused due to the development of resistance against these formulations. However, pentavalent antimony (Sb) the standard treatment for the last 70 years remains the backbone in many regions of Leishmania endemicity. The polyene antibiotic amphotericin B (AMB), which has been proven to be 95% effective against VL in India is another first-line therapy (Sundar et al., 2010).
Even though Liposomal AMB requires intravenous delivery, still it has become a common treatment in many countries. However, there were geographical disparities in the response of liposomal AMB, with VL cases in India responding better than those in East Africa or South America (Berman et al., 1998; Saravolatz et al., 2006). Clinical AMB resistance is uncommon, and parasites were found to be sensitive even after numerous treatments in the same patient (Lachaud et al., 2009). Miltefosine (MTF) is an alkyl-lysophospholipid analog administered orally to treat leishmanial parasites (Croft et al., 1987; Jha et al., 1999). Since its introduction in India in 2002, miltefosine has been successfully used to treat VL but the relapse rates have increased owing to MTF resistance (Sundar et al., 2002; Zuckerman et al., 2012; Burza et al., 2013). Aminoglycoside paromomycin (PMM) has been licensed for the treatment of VL in India (Jha et al., 1998; Sundar et al., 2007). So far, the restricted use of PMM has prevented the establishment of resistance. However, there have been geographical differences in PMM efficacy against VL between East Africa and India (Hailu et al., 2010; Musa et al., 2010). Finally, due to severe toxicity, pentamidine (PTD) has been discontinued for the treatment of VL and is now only used in the treatment of CL in South America (Soto et al., 1994; Lai a Fat et al., 2002; Roussel et al., 2006; Bhattacharyya et al., 2022). The mode of action of antimonials is unknown despite its use for more than six decades. It has been reported to cause the generation of reactive oxygen species, trypanothione depletion, and apoptosis-like death, although an exact mechanism is yet to be determined (Sereno et al., 2001; Lee et al., 2002; Sudhandiran and Shaha, 2003; Wyllie et al., 2004; Mehta and Shaha, 2006; Mookerjee Basu et al., 2006; Mandal et al., 2007; Vergnes et al., 2007; Moreira et al., 2011). Thus, there is a considerable need for new anti-leishmanial drugs. Further understanding of genome organization would aid in designing of targeted therapies against drug resistance in Leishmania.
The Leishmania sp. uses various strategies to adapt to changing environment while shifting between different hosts. These include a shift in the collection of expressed genes, mosaic aneuploidy, and copy number variation. The copy number variation (CNV) causes variation in host-parasite interaction for cellular invasion and immune evasion mechanisms causing genomic and phenotypic heterogeneity (Reis-Cunha et al., 2018). Furthermore, CNV produces extrachromosomal circular or linear amplified DNAs by amplification of chromosomes, affecting entire chromosomes or specific genomic areas for mosaic aneuploidy. Mosaic aneuploidy may be viewed as a potent intracellular parasite survival strategy causing enhanced genomic variability and a population clearance of heterozygous cells without altering genetic heterogeneity (Sterkers et al., 2012). Moreover, the changes in environmental variables cause phenotypic variability which leads to drug resistance. Thus, the capacity of leishmanial parasites to respond to the pharmacological inhibition via CNVs and mosaic aneuploidy led to the development of genomic screens like Cos-Seq, and FISH to speed up drug targets for novel prospective therapeutic candidates (Grünebast and Clos, 2020).
Thus, in this review, we have discussed the regulation of genome plasticity via CNV and its impact on drug resistance and intracellular parasite survival (Figure 1). Moreover, we not only highlighted the influence of PTMs on genome plasticity for parasite survival through environmental factors but also elucidated the role of epigenetics in parasite growth.
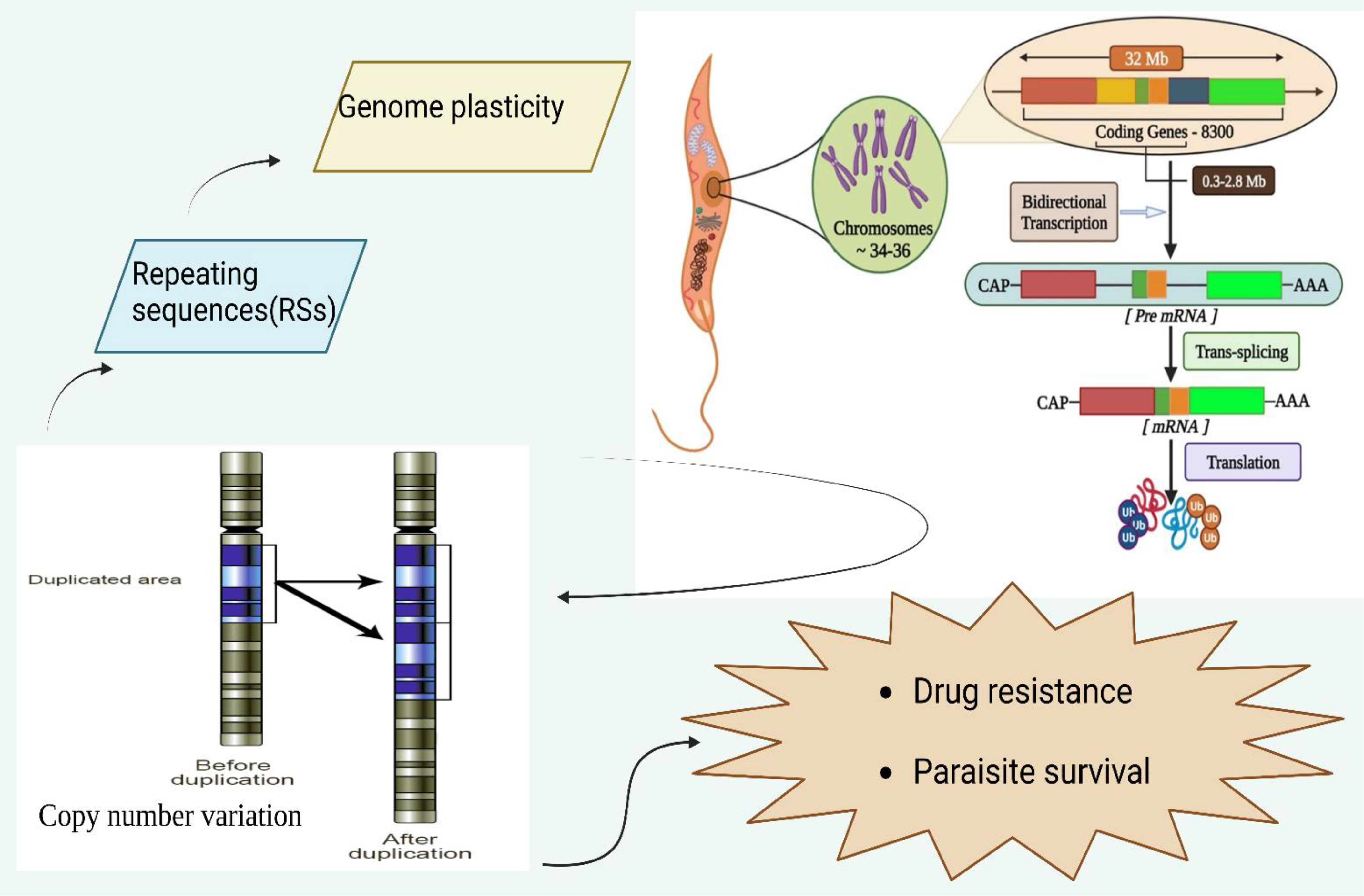
Figure 1 Genomic organization of leishmanial parasites and its regulation of Copy number variation to modulate and influence drug resistance and parasite survival. The leishmanial species contains 34-36 chromosomes where a vast polycistronic unit spans through the protein-coding genes with a size from 0.3 to 2.8 mb. The transcription of these lengthy polycistronic units occurs bi-directionally from the start sites situated at switch regions of the strand.
2 Genome organization
The Leishmania sp genome is approximately 32 mb in size containing around 8,300 coding genes (Ivens et al., 2005; Peacock et al., 2007). More than 99% of genes in the Leishmania genus are conserved with only a few species-specific genes found altogether in L. major, L. infantum, and L. braziliensis (Peacock et al., 2007). The leishmanial species contains 34-36 chromosomes where a vast polycistronic unit spans through the protein-coding genes with a size from 0.3 to 2.8 mb (Wincker et al., 1996; Britto et al., 1998; Martínez-Calvillo et al., 2003; El-Sayed et al., 2005; Ivens et al., 2005; Peacock et al., 2007; Raymond et al., 2012). The transcription of these lengthy polycistronic units occurs bi-directionally from the start sites situated at switch regions of the strand in the absence of RNA polymerase-II promoters (Martínez-Calvillo et al., 2004). The attachment occurs through the trans-splicing of nucleotides at the long leader RNA of the 5’end of each mRNA with simultaneous polyadenylation at the 3’end for processing of mRNA (Matthews et al., 1994; Haile and Papadopoulou, 2007). The mRNA stability and translational rate of the leishmanial parasite depends on 3′-untranslated regions (3′-UTRs) to regulate gene expression via altered environmental factors for its development due to its lack of transcriptional control (Bringaud et al., 2007; Dupé et al., 2014). Leishmania frequently uses DNA copy number variations (CNVs) (aneuploidy, gene amplification, or gene deletion) to regulate the expression of drug targets, drug transporters, or other determinants of resistance to overcome drug pressure (Gordon et al., 2012; Pfau and Amon, 2012; Shor and Perlin, 2015; Rutledge and Cimini, 2016). Single-nucleotide polymorphisms (SNPs) are caused due to CNVs in pharmacological targets or transporters leading to drug resistance without affecting gene expression (Alwi, 2005).
2.1 Regulation of genome plasticity
Leishmanial parasites were previously thought to be diploid but recent research has revealed their aneuploidy (Ubeda et al., 2008a; Leprohon et al., 2009; Downing et al., 2011; Rogers et al., 2011; Sterkers et al., 2011; Mannaert et al., 2012; Sterkers et al., 2012). Aneuploidy patterns were seen among populations of leishmanial parasites at various individual cells. Using mosaic aneuploidy, it was discovered that an average diploid population of a few parasites shared the same ploidy for individual chromosomes when the cumulative ploidy was obtained from next-generation DNA sequencing data (Ubeda et al., 2008a; Sterkers et al., 2011; Lachaud et al., 2014). Variations in chromosomal size were identified across different strains of the related leishmanial parasites (Minning et al., 2011; Reis-Cunha et al., 2015). However, links between increased or decreased chromosome number and drug resistance are circumstantial. This demonstrates that a particular set of genes on the variant chromosomes coordinate to develop resistance. But aneuploidy is a survival mechanism for leishmanial parasites through genomic stability and enhanced virulence in recent years (Sterkers et al., 2012; Sterkers et al., 2014).
The CNV serves as a mechanism for controlling the expression of conditional factors in the absence of transcription initiation and thus establishing a strong link between chromosome ploidy and nucleic acid expression levels (Ubeda et al., 2008a; Leprohon et al., 2009). The mis-aggregation of allele would lead to enhanced chromosomal number leading to overexpression of genes via increased copy numbers leading to aneuploidy in the leishmanial parasite. However, the Leishmania genome (32 mb) is scattered across 34 to 36 chromosomes and decreased co-expression of genes would cause the parasite to amplify or delete certain smaller portions of DNA as extrachromosomal elements via recombination rearrangements at the level of homologous repeating sequences (RSs) (Ubeda et al., 2014).
In a recent study, the full set of RSs in several Leishmania species was highlighted, and the entire leishmanial genome was found to possess the ability to alter RSs at the RNA level, causing parasite survival via aneuploidy through the generation of extrachromosomal elements (Ubeda et al., 2014). Moreover, LmSIDER1 and LmSIDER2 are two families of SIDER found in L. major causing plasticity via CNV for parasite survival. SIDER2 destabilizes mRNA and causes mRNA decay, whereas the SIDER1 family regulates mRNA translation in a stage-specific manner (Boucher et al., 2002; Bringaud et al., 2007; Azizi et al., 2017).
3 Intracellular host-parasite survival via CNV
The genome of L. infantum constitutes 2000 RSs for the synthesis of over 3,000 extrachromosomal DNA elements (Ubeda et al., 2014). Short interspersed degenerate retrotransposons (SIDERs), small degenerated retrotransposons, having no specificity for site-specific integration, reside mostly in the vicinity of intergenic regions of directional gene cluster and majority of these elements are located within 3′ UTRs. SIDER elements possess two functions, they either regulate gene expression or act as a structural backbone to facilitate gene rearrangements for CNV of DNA regions (Ouellette and Papadopoulou, 2009; Smith et al., 2009). When leishmanial parasites are exposed to drugs, the extrachromosomal DNA amplifications occur causing amplicons via episomes (Ubeda et al., 2008a; Leprohon et al., 2009; Downing et al., 2011; Rogers et al., 2011; Sterkers et al., 2011; Mannaert et al., 2012; Sterkers et al., 2012). The episomes are produced by rearrangements of direct or inverted homologous RSs and are amplified as either circular or linear extrachromosomal DNA (Ubeda et al., 2014). The amplicon production varies with increased drug resistance that allows the parasites to adjust to changes in their environment for parasite survival within the host (Ubeda et al., 2014). Since gene rearrangements are caused by RSs in response to drug resistance, the revelation of newer recombinase proteins involved in rearrangements would lead to preventing drug resistance.
Several drugs cause indirect or direct DNA damage as DSB. The RAD51 is a DNA repair protein that plays a crucial role in homologous recombination (HR) in the intracellular parasite. The double-strand breaks (DSBs) were found to promote RAD51 expression leading to drug resistance in the leishmanial parasite (Mckean et al., 2001; Genois et al., 2012). The inactivation of RAD51 would prevent parasites to synthesize circular extrachromosomal elements but linear amplicons production remained unchanged leading to drug resistance and parasite survival (Ubeda et al., 2014). There are three DNA-repair proteins, that are present in the leishmanial parasite as RAD51 paralogs (RAD51-3, RAD51-4, and RAD51-6) that showed to cooperate for the promotion of homologous recombination (HR) by stimulating RAD51 activity (Genois et al., 2015). The inactivation of RAD51-4 leads to the suppression of the generation of circular amplicons by RSs in an inverted orientation but not linear amplicons in L. infantum, when exposed to drugs (Laffitte et al., 2014).
Similarly, MRE11 is a DNA repair nuclease that forms MRN (meiotic recombination) complex with RAD50 as well as NBS1 and is critical for DSB repair by homologous recombination (HR) or non-homologous end joining (Mimitou and Symington, 2011; Genois et al., 2014; Shibata et al., 2014). L. infantum constitutes linear amplicons upon treated with drug via inverted RSs-causing inactivation of MRE11 for intracellular parasite survival (Laffitte et al., 2014). Furthermore, parasites expressing DNA-binding-proficient but nucleotide-deficient MRE11 only produce circular amplicons during drug selection, indicating that a fully functional MRE11 is required for linear amplification. The inactivation of MRE11 alone or in combination with RAD50 causes significant chromosomal translocation in L. infantum, indicating that the MRE11/RAD50 complex is critical for genomic integrity and gene rearrangements (Laffitte et al., 2016a).
Based on the aforementioned studies it could be inferred that modulation of RSs causes the DNA repair protein to promote intracellular host-parasite survival via DSBs. Thus, drugs or inhibitory molecules targeting DNA repair proteins could serve as therapeutic interventions to cause regression of leishmanial infection. However, further understanding of other DNA repair proteins modulated by RSs of CNV needs to be done before reaching a definitive conclusion on this aspect. Even though there is a plethora of studies discussing drug resistance and parasite survival, studies focusing on a definite pattern based on genome plasticity are limited and need to be focused on to get a detailed mechanistic overview of intracellular host-parasite survival.
4 Intracellular host-parasite survival via PTM and epigenetics
4.1 Gene expression and translational efficiency
The post-transcriptional regulation of gene expression in the leishmanial parasite is mediated through external environmental stimuli (Brandau et al., 1995; Quijada et al., 1997; Clayton and Shapira, 2007; Grünebast and Clos, 2020). Leishmanial genes are constitutively expressed with no gene-specific transcription control and verified by nuclear run-on and microarray investigations. When axenic amastigotes were compared to promastigotes, a downregulation of RNA production for parasite survival was observed (Leifso et al., 2007; Alcolea et al., 2010). The process of stage differentiation causes enhanced amastigote-specific protein synthesis inside the host body for leishmanial survival (Rosenzweig et al., 2008b; Lahav et al., 2011).
A system-wide RNA abundance study indicated that Leishmania gene expression is regulated at levels other than RNA synthesis, such as RNA processing and stability (Clayton and Shapira, 2007). However, changes in mRNA abundance do not always correspond to changes in protein synthesis rates or abundance, indicating the significance of translation efficiency and protein half-life as regulatory targets (Charest et al., 1996; Yao et al., 2002; Decuypere et al., 2012; Azevedo et al., 2015; Bifeld et al., 2018). The RNA half-life poses resistance to antimony as a post-transcriptional mechanism. It can be correlated with the stability of mRNA coding for AQP1 in a 3’-UTR-dependent manner with non-long terminal repeat retrotransposons causing parasite survival and drug resistance via plasticity in the leishmanial parasite (Mandal et al., 2015).
4.2 Post-translational modifications (PTMs)
Various PTMs including phosphorylation, acetylation, methylation, and glycosylation influence axenic differentiation into amastigotes through modified proteins (Morales et al., 2008; Rosenzweig et al., 2008a; Morales et al., 2010). MAPKs are serine/threonine-specific protein kinases found in all eukaryotes for signal transduction and regulation of gene expression profiles. MAPKs regulate important cellular activities like cell viability, life cycle, morphology, and drug resistance in leishmanial parasites by phosphorylating their substrates (Wiese, 1998; Wiese et al., 2003; Bengs et al., 2005; Kuhn and Wiese, 2005; Wang et al., 2005; Mccarter, 2006; Von Freyend et al., 2010; Cayla et al., 2014). The stability of heat shock proteins and their roles during the life cycle is affected by phosphorylation of HSP70 and HSP90 in MAP kinase 1 of L. donovani (Kaur et al., 2017). Moreover, when stationary phase cells were compared with rapidly expanding cells, acetylation levels were enhanced in rapidly growing cells. The acetylation of the H3 variant in the switch region of the telomeric divergent strand causes parasite survival via chromatin modification in L. major promastigotes (Thomas et al., 2009).
Furthermore, the leishmanial telomeric repeat sequence (GGGTTA) contains glycosylated thymine, known as “Base J” or beta-D-glucosyl hydroxy methyl uracil (Van Luenen et al., 2012; Olinski et al., 2016; Reynolds et al., 2016) replacing the thymine in nuclear DNA, for transcriptional regulation and termination in kinetoplastid protozoans such as Leishmania major, Leishmania donovani and Trypanosoma cruzi facilitating intracellular parasite survival (Van Leeuwen et al., 1998). 5-hydroxymethyluracil is another epigenetic modification of thymine base alteration, which has been recently linked to the Leishmania genome. However, its epigenetic role in parasite survival has yet to be determined (Kawasaki et al., 2017).
Besides, a recent investigation pointed out a novel PTMs via bromodomain (BDs) complexes, mainly BD5. BD5 has emerged as a pivotal modulating factor in transcriptional factor of polymerase II in leishmanial parasite. Thus, BD5 can be used as an potential target to inhibit leishmanial infection by targeting the aforementioned mechanism to regulate transcriptional regulation in kinetoplastids (Jones et al., 2022). A plethora of investigation needs to be done in this direction to get an in-depth analysis of BDs based leishmanial regulation.
4.3 Epigenetics
The epigenetic markers such as H3K27ac, H3K9me3, H3K23ac H3K14ac promote transcriptional activation of ribosomal RNA (rRNA) genes in the promoter region of L. major, whereas H4K20me3 epigenetic marks in the coding region causes transcriptional silence (Vizuet-De-Rueda et al., 2016). H2A.Z and H2B.V histone variants are epigenetic markers found to be critical for L. major survival (Afrin et al., 2019). Histone acetyltransferase (HAT)4 acetylates H4K14 in L. donovani, promoting euchromatin maintenance (Yadav et al., 2016). L. donovani maintains its survival through epigenetic modulation via acetylation of H410 and H414 through HAT2 and HAT3 respectively (Chandra et al., 2017). To successfully establish itself inside the host, L. donovani utilizes epigenetic programming of genes in a histone lysine methyltransferase as well as demethylase dependent manner resulting in macrophage polarization in both J774 macrophages and BALB/c mice (Parmar et al., 2020).
In the promastigote and amastigote stages, epigenetic markers are regulated differently. Some histone deacetylases (HDAC) are preferentially increased in the logarithmic phase of promastigotes in L. infantum compared to intracellular amastigotes, allowing the amastigotes to adapt to the intra-phagolysosomal environment (Alcolea et al., 2010). Recent reports have indicated that NAD dependent HDACs sirtuins results in parasite persistence through inhibition of apoptosis. However, sirtinol causes apoptosis in axenic amastigotes of L.infantum (Vergnes et al., 2002; Vergnes et al., 2005).
A subset of non-coding RNAs is identified specifically in L. infantum and L. donovani amastigotes as a critical element for parasite survival within macrophages by causing modulation of epigenetics markers (Dumas et al., 2006). L. donovani causes epigenetic changes in host macrophages, irreversibly suppressing innate immune defenses (Marr et al., 2014). Upon L. donovani infection, several genes of signaling pathways such as JAK/STAT, calcium, MAPK, notch, mTOR and cell adhesion proteins (beta1 integrin) are differentially methylated at the CpG sites of the DNA resulting in increased oxidative phosphorylation (Lecoeur et al., 2022). Moreover, oxidative phosphorylation within the host increases the survival of the parasite by downregulation of innate immune response (Marr et al., 2014). The matrix metalloproteinase 1 (MMP1) gene which is involved in tissue repair gets downregulated by L. braziliensis causing lesions that facilitate the infection and ultimately lead to parasite survival (Wang et al., 2006; Almeida et al., 2015). Furthermore, epigenetic regulation by DNA methylation at the CpG promoters of the LOX gene upon activation by IL-6 in a homocysteine-dependent manner would lead to parasite survival (Thaler et al., 2011; Castellucci et al., 2014). The downregulation of iNOS by L. amazonensis in infected macrophages via histone deacetylase causes parasite survival (Calegari-Silva et al., 2018).
During L. donovani infection upregulation of miRNA-30A-3p causes parasite survival by targeting host autophagy machinery (Singh et al., 2016). The miRNA-3620 was found to influence iron homeostasis and hypoxia, while miRNA-3473f was associated with autophagy suppression in L. donovani-infected macrophages. The upregulation of miRNA-763, miRNA-1264, and miRNA-3473f causes drug resistance due to the overexpression of efflux pumps via ABC transporters (Singh et al., 2016; Tiwari et al., 2017). Thus, to establish a protective niche for its survival the leishmanial parasite causes a hypoxic environment within macrophages to regulate miRNA-210 in a HIF-1-dependent manner causing a pro-inflammatory response via regulating the NF-kB pathway (Kumar et al., 2018).
The leishmanial parasite influences the ncRNA (non-coding RNA) network via manipulation of the transcriptional arrest of the protein-coding genes of the macrophages for its survival (Rana et al., 2011). Downregulation of signal recognition particle RNA through ncRNA for parasitic survival inside the host is found in M2 macrophages by inhibition of RNA pol III (Misra et al., 2005). Additionally, L. donovani drives BRD4-p300-RNA-polII dependent unique super-enhancer element mediated miR146-5p transcription to promote parasite-supportive M2 macrophage niche (Das et al., 2021; Gupta et al., 2022). Gp63 and LPG are surface glycoproteins and glycolipids of Leishmania, shown to downregulate ncRNAs in M2 macrophages, thus helping in parasite persistence (Farrow et al., 2011). The overexpression of the host transcription factor c-myc leads to the downregulation of 19 miRNAs in infected macrophages with L. donovani. This would act as a prognostic marker for M2 macrophages for gene suppression causing parasite survival and proliferation. miRNA-34a which regulates the expression of c-myc is reciprocally downregulated in Leishmania-infected cells (Colineau et al., 2018).
4.4 RNA profiling and translational inducer
The level of proteins or translation rates can be assessed to study the effect of environmental stimuli on parasite gene expression (Bente et al., 2003; Rosenzweig et al., 2008a; Lahav et al., 2011), while the latter can be accomplished through ribosome profiling analysis or a combination of metabolic labeling/mass spectrometry method (Ingolia et al., 2009; Kalesh and Denny, 2019). The combination of ribosome profiling and RNA-Seq allows researchers to examine not only the relationship between mRNA abundance and translation but also the relative translational efficiency of mRNAs to environmental stimuli. Ribosome profiling paired with RNA-Seq revealed the inhibition of HSP90 to alter steady-state levels for mRNAs and cause increased or decreased protein synthesis rates for around 10% of the proteome in L. donovani (Bifeld et al., 2018). The early amastigote differentiation is enhanced when HSP90 is inhibited causing morphological alterations in the parasite (Lahav et al., 2011; Mandal et al., 2015; Bifeld et al., 2018).
Axenic amastigote differentiation is caused by decreased overall translation and phosphorylation of the translation factor eIF2a (Cloutier et al., 2012). Protein synthesis reduced in amastigotes causes reduced proliferation in the intracellular stage of the leishmanial life cycle (Jara et al., 2019). Under the influence of stress, there is a switch from cap-dependent to cap-independent translation in the leishmanial parasite to modify their translation machinery to environmental stress in a cap4, eIF2-alpha, eIF4E, and Leish4E-IP dependent manner (Zinoviev et al., 2011). Besides these discoveries, the involvement of translation factors in Leishmania’s inducible gene expression is unknown for their survival and drug resistance. Further studies in this direction needs to be done to get a proper understanding of the impact of translational inducer of leishmanial parasite survival.
5 Drug resistance in leishmanial parasite
5.1 Copy number variations (CNVs)
Even though CNVs are the most common form of resistance in the leishmanial parasite, SNPs are minor nucleotide insertions or deletions that cause drug resistance. Leishmania frequently uses DNA CNVs (aneuploidy, gene amplification, or gene deletion) to regulate the expression of drug targets, drug transporters, or drug resistance (Gordon et al., 2012; Pfau and Amon, 2012; Shor and Perlin, 2015; Rutledge and Cimini, 2016). Single-nucleotide polymorphisms (SNPs) caused due to CNVs in pharmacological targets or transporters lead to drug resistance without affecting gene expression (Alwi, 2005). SNP causes changes in the amino acid via substitution or non-sense mutations in the MTF transporter (MT) or its Ros3 subunit (Vasudevan et al., 2001; Pérez-Victoria et al., 2003; Pérez-Victoria et al., 2006; Laffitte et al., 2016c). This was further validated using whole-genome sequencing of MT (Coelho et al., 2012; Laffitte et al., 2016b; Mondelaers et al., 2016).
The drug resistance occurred primarily due to mutations in the MT gene of L. infantum isolates obtained from MTF-treated patients (Cojean et al., 2012). A genetic polymorphism in L. donovani isolates from the Indian subcontinent exhibited antimonial resistance via SNP (Imamura et al., 2016). Surprisingly, a group of highly resistant isolates clustering together were found to share genetic characteristics for drug resistance. The genetic characteristics for this drug resistance would include a high copy number of H-locus coding for ABC transporter MRPA and a homozygous two-base-pair insertion in the aquaglyceroporin 1 (AQP1) gene are involved for drug resistance in a Sb dependent manner (Légaré et al., 2001). Moreover, MRPA and AQP1 are involved in Sb uptake and their downregulation causes drug resistance both in vivo and in vitro. Thus, these Sb resistances may serve as a marker for the treatment of SNPs based on plasticity on a diverse set of isolates (Gourbal et al., 2004; Decuypere et al., 2005; Marquis et al., 2005; Uzcategui et al., 2008; Mandal et al., 2010; Mandal et al., 2015; Monte-Neto et al., 2015; Plourde et al., 2015).
5.2 Aneuploidy
The individual chromosomal ploidy is highly varied as it can occur between one to six sister chromosomes within leishmanial populations (Mannaert et al., 2012; Lachaud et al., 2014; Sterkers et al., 2014). Intermediate fold values of the NGS read alignment density can signal mosaic aneuploidy, such as the occurrence of population variations or incomplete chromosomal duplication occurrences (Imamura and Dujardin, 2019). Drug resistance via aneuploidy has been intensively investigated and due to decades of use of pentavalent antimony-based drugs in endemic areas, resistance has developed, making these drugs nearly obsolete in some locations (Dube et al., 2005; Croft et al., 2006; Singh et al., 2006; Ostyn et al., 2014). Advances are occurring in systems biology methodologies in Leishmania spp. for antimony resistance to aneuploidy patterns peculiar to resistant parasites (Leprohon et al., 2009; Laffitte et al., 2016c; Dumetz et al., 2017).
Mosaic aneuploidy plays a pivotal in vector and host adaptation owing to their diverse patterns of ploidy identified for vector and host-derived leishmanial cells grown in vitro. Ploidy changes occur in vitro, which produces clones with a ploidy pattern that differs from the parent (Sterkers et al., 2011; Sterkers et al., 2012; Dumetz et al., 2017). A recent study comparing ploidy and somy patterns of L. donovani strains was acquired through whole-genome sequencing (WGS) for parasite development in in vitro and karyotypic diversification. While cultured parasites possess a wide range of somy patterns, ploidies for bone-marrow-derived parasite genomes were strikingly identical (Domagalska et al., 2019), indicating similar selective pressures within human hosts. Despite this, Leishmania appears to be able to cope with genomic plasticity. Aneuploidy is easily reversed after the benefits of a supernumerary chromosome have worn off and when the overexpression owing to extra gene copies appears to be limited (Kröber-Boncardo et al., 2020). Regulated translation could thus help to alleviate the impacts of aneuploidy. Chromosomal amplification resulted in enhanced copy number of the gene which modulated the expression of the genes through host dependent environmental factor. The modulation in host dependent environmental factor prompted genetic tropism in species specific genes of leismanial parasites for genetic tropism as seen in species-specific genes 2, 14, 19, and 67 in L. major, L. infantum, and L. braziliensis (Rogers et al., 2011). This shows that aneuploidy is inherently unstable within a strain, fluctuating to changes in environmental factors causing plasticity for parasite survival (Rogers et al., 2011). Furthermore, extensive variation in the copy number of the chromosome for ploidy variability was concluded based on the WGS of varied strains of L.donovani. Thus, phylogenetically close strains (BPK173/Ocl3 and BPK275/Ocl18) exhibit a different somy for chromosomes 2, 8, 11, 12, 14, 20, and 33 owing to their heterogeneity of aneuploidy making it unstable over time. This ploidy instability explains the differences in the ploidy of the L. major Friedlin strain (2011) (Downing et al., 2011).
6 Future perspective and outlook
Even though drugs are introduced to the clinic without knowing more about their mode of action, a comprehensive understanding of the molecular targets for drug discovery is required. Target identification of possible resistance mechanism due to CNVs should be considered for drug use (Laffitte et al., 2016a; Paananen and Fortino, 2020). Genetic techniques experimentally mimicking CNVs for causing drug resistance and functional cloning were utilized for the gain-of-function screening method wherein leishmanial cosmid libraries were electroporated into Leishmania sp and transfectants for a specified phenotype (Ryan et al., 1993). The high copy number and gene expression of cosmids and selection of the phenotype were made possible for the selection of drug resistance or susceptibility. This method favors cosmids with dominant phenotypes while excluding less enriched cosmids (Vasudevan et al., 1998; Cotrim et al., 1999; Kündig et al., 1999; Carter et al., 2000; Coelho et al., 2003).
Amplification itself can produce drug resistance through different direct and indirect mechanisms. Interestingly, the artificial expression of cosmid-based functional screening coupled with a next-generation sequencing technique known as Cos-Seq has emerged as a novel technology that aids in the identification of target genes and their mechanism of drug resistance (Tejera Nevado et al., 2016) (Gazanion et al., 2016). Even though Cos-Seq identified an extraordinary number of resistance genes both known and novel but none appeared as an obvious target candidate. Thus, it is still unclear whether non-protein targets represent another possibility as these would not be detected by Cos-Seq and hence further research in this direction needs to be done.
The loss-of-function mutations in aquaglyceroporin AQP1 or MT transporter genes cannot be isolated using the Cos-Seq approach and require high-throughput dominant-negative screening methods through inducible RNA interference target sequencing (RIT-Seq) (Alsford et al., 2012). RNA interference is found to be absent in most of the leishmanial parasites but presents in Leishmania (Viannia) subgenus (Lye et al., 2010). Moreover, the lack of inducible expression in Leishmania (Viannia) was a drawback of this technique (Kraeva et al., 2014; Duncan et al., 2016). RNA-guided nuclease systems based on CRISPR has been developed as an alternative RIT-Seq for accomplishing effective specific genomic changes in a variety of organisms (Sander and Joung, 2014; Selle and Barrangou, 2015; Wright et al., 2016). The CRISPR/Cas9 system, derived from Streptococcus pyogenes, was used to disrupt genes in intracellular parasites such as L. major and L. donovani, and could be used to generate whole-genome Cas9-mediated gene deletion libraries (Sollelis et al., 2015; Zhang and Matlashewski, 2015). Thus, future research in this direction needs to be done to optimize this novel technology to be utilized against drug resistance to solve the issue of CNV in Leishmania.
7 Possible genomic variability mechanism
The understanding of genome plasticity at the molecular level in Leishmania opens a pathway to utilize such processes in combating drug resistance (Laffitte et al., 2016a) (Figure 2). Defective chromosomal replication through post-translational modification of replicative sequences as well as mis-segregation of the chromosome during mitosis results in mosaic aneuploidy as seen in these parasites (Zhu et al., 2018). Parasites amplify/delete gene copy number and such amplification of genes occurs via homologous recombination. DNA amplification occurs mainly when extrachromosomal elements are formed due to the rearrangement of direct or inverted homologous repeated sequences (Ubeda et al., 2008b). Genomic amplification of virulence factors such as GP63, Amastin, etc. increases the chance of survival of these parasites inside the host (Samarasinghe et al., 2018). Leishmania possesses unique replication machinery as there is a single polycistronic transcription unit with a single origin of replication in each chromosome which ultimately helps the parasite survive even under drug pressure (Downing et al., 2011). Leishmanial genes are constitutively expressed in these parasites by RNA polymerase II. In Leishmania, the promotor sequence is not yet deciphered properly, so the mechanism of transcription initiation is still under investigation (Grünebast and Clos, 2020b). Epigenetics mechanism seems to be involved by influencing DNA accessibility, which is an active area of research. It may be speculated that post-translational regulation of protein occurs through RNA degradation as there is a lack of proper transcriptional control. However, such studies show promise to decipher more about these mechanisms in the future.
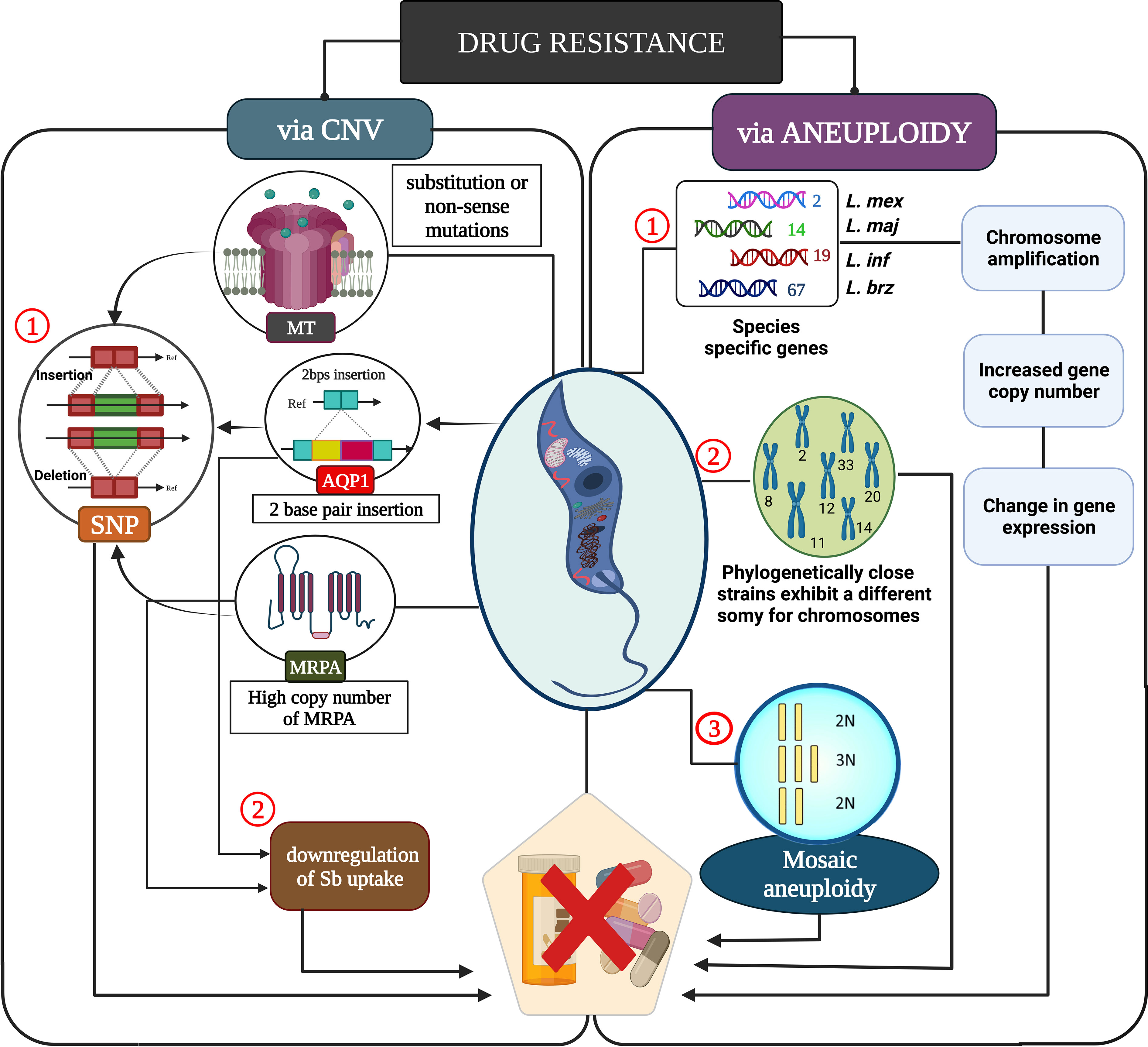
Figure 2 A molecular understanding of drug resistance caused by leishmanial parasite. Copy number variation influences drug resistance through single nucleotide polymorphism which affects the Sb uptake, ultimately leading to drug resistance. Mosaic aneuploidy causes increased copy number variation which leads to chromosomal amplification thus modulating the gene expression in several members of leishmanial parasites for drug resistance.
8 Conclusion and summary
Successful treatment strategies for major public concerns like leishmaniasis are mainly jeopardized due to arising cases of resistant parasites. To survive in wide geographic real and different host species, Leishmania adapts its genome to be highly plastic. The genome plasticity is mainly maintained by aneuploidy, transposons, copy number variations, RNA trans-splicing of mono- and poly-cistronic mRNAs, and post-translational modifications. Hence, it is quite difficult to extrapolate and predict the role of essential proteins from general -omics data of Leishmania. The enormous plasticity of the genome not only helps parasites sabotage host machinery but also supports its survival under extreme drug pressure making new drug discovery quite difficult. Due to this, upon CNVs and PTMs prediction of promastigotes and amastigotes transcriptomics should be done cautiously as promastigotes may contain different sets of RNA expressions which can get modified and altered when they differentiate into amastigotes. Additionally, genome plasticity has a solid impact on drug metabolism and severely affects the outcome of advanced genome editing tools like RNAi, DiCre, plasmid shuffling, and CRISPR/Cas9 to understand their role in drug resistance. Our review has concisely described the set of proteins and epigenetic modifiers of parasites that contribute to CNVs and aneuploidy. Moreover, how such mediators can significantly bring about the unexpected outcomes of treatment cases with new anti-leishmanial agents and result in resistance generation can be an interesting area to explore. In a nutshell, our illustration depicts that an integrated understanding of the parasite proteins which mainly maintain the genome integrity and plasticity will assist in achieving successful chemotherapy against parasites like Leishmania.
Author contributions
The manuscript was conceptualized by MK, RB, and NA. It was written and drafted by MK and RB. SD and SM helped in collecting and analyzing the data and proofreading the manuscript. NA guided the whole project and helped in funding acquisitions. All authors contributed to the article and approved the submitted version.
Acknowledgments
The authors would also like to appreciate Biorender.com for their software. We would like to thank our funding agencies such as Global Challenges Research Fund under the grant agreement, ‘A Global Network for Neglected Tropical Diseases.’ Grant number MR/P027989/1, India, Indian Council of Medical Research and Council of Scientific and Industrial Research, India, and Sir J.C. Bose Fellowship grant number SB/S2/JCB-21/2015, India.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abbreviations
VL, Visceral Leishmaniasis; CL, Cutaneous leishmaniasis; Sb, pentavalent antimony; AMB, Amphotericin B; MTF, Miltefosine; PMM, Paromomycin; PTD, Pentamidine; CNV, Copy number variation; RSs, repeating sequences; SIDERs, Short interspersed degenerate retrotransposons; DSBs, double-strand breaks; HR, homologous recombination; MRN, meiotic recombination; PTMs, Post Translation Modification; HAT, Histone acetyltransferase; HDAC, histone deacetylases; MMP1, matrix metalloproteinase 1; ncRNA, non-coding RNA; SNPs, Single-nucleotide polymorphisms; WGS, whole-genome sequencing; RIT-Seq, RNA interference target sequencing
References
Afrin, F., Khan, I., Hemeg, H. A. (2019). Leishmania-host interactions-an epigenetic paradigm. Front. Immunol. 10, 492. doi: 10.3389/fimmu.2019.00492
Alcolea, P. J., Alonso, A., Gómez, M. J., Moreno, I., Domínguez, M., Parro, V., et al. (2010). Transcriptomics throughout the life cycle of leishmania infantum: high down-regulation rate in the amastigote stage. Int. J. Parasitol. 40, 1497–1516. doi: 10.1016/j.ijpara.2010.05.013
Almeida, L., Oliveira, J., Guimarães, L. H., Carvalho, E. M., Blackwell, J. M., Castellucci, L. (2015). Wound healing genes and susceptibility to cutaneous leishmaniasis in Brazil: Role of COL1A1. Infection Genet. Evol. 30, 225–229. doi: 10.1016/j.meegid.2014.12.034
Alsford, S., Eckert, S., Baker, N., Glover, L., Sanchez-Flores, A., Leung, K. F., et al. (2012). High-throughput decoding of antitrypanosomal drug efficacy and resistance. Nature 482, 232–236. doi: 10.1038/nature10771
Azevedo, A., Toledo, J. S., Defina, T., Pedrosa, A. L., Cruz, A. K. (2015). Leishmania major phosphoglycerate kinase transcript and protein stability contributes to differences in isoform expression levels. Exp. Parasitol. 159, 222–226. doi: 10.1016/j.exppara.2015.09.008
Azizi, H., Dumas, C., Papadopoulou, B. (2017). The pumilio-domain protein PUF6 contributes to SIDER2 retroposon-mediated mRNA decay in leishmania. Rna 23, 1874–1885. doi: 10.1261/rna.062950.117
Bates, P. A. (2018). Revising leishmania’s life cycle. Nat. Microbiol. 3, 529–530. doi: 10.1038/s41564-018-0154-2
Bengs, F., Scholz, A., Kuhn, D., Wiese, M. (2005). LmxMPK9, a mitogen-activated protein kinase homologue affects flagellar length in leishmania mexicana. Mol. Microbiol. 55, 1606–1615. doi: 10.1111/j.1365-2958.2005.04498.x
Bente, M., Harder, S., Wiesgigl, M., Heukeshoven, J., Gelhaus, C., Krause, E., et al. (2003). Developmentally induced changes of the proteome in the protozoan parasite leishmania donovani. Proteomics 3, 1811–1829. doi: 10.1002/pmic.200300462
Berman, J., Badaro, R., Thakur, C., Wasunna, K., Behbehani, K., Davidson, R., et al. (1998). Efficacy and safety of liposomal amphotericin b (AmBisome) for visceral leishmaniasis in endemic developing countries. Bull. World Health Organ. 76, 25.
Bhattacharyya, A., Kamran, M., Ejazi, S. A., Das, S., Didwania, N., Bhattacharjee, R., et al. (2022). Revealing a novel antigen repressor of differentiation kinase 2 for diagnosis of human visceral leishmaniasis in India. Pathogens 11. doi: 10.3390/pathogens11020120
Bi, K., Chen, Y., Zhao, S., Kuang, Y., John Wu, C.-H. (2018). Current visceral leishmaniasis research: a research review to inspire future study. BioMed. Res. Int. 2018. doi: 10.1155/2018/9872095
Bifeld, E., Lorenzen, S., Bartsch, K., Vasquez, J.-J., Siegel, T. N., Clos, J. (2018). Ribosome profiling reveals HSP90 inhibitor effects on stage-specific protein synthesis in leishmania donovani. Msystems 3, e00214–e00218. doi: 10.1128/mSystems.00214-18
Boucher, N., Wu, Y., Dumas, C., Dubé, M., Sereno, D., Breton, M., et al. (2002). A common mechanism of stage-regulated gene expression in leishmania mediated by a conserved 3′-untranslated region element. J. Biol. Chem. 277, 19511–19520. doi: 10.1074/jbc.M200500200
Brandau, S., Dresel, A., Clos, J. (1995). High constitutive levels of heat-shock proteins in human-pathogenic parasites of the genus leishmania. Biochem. J. 310, 225–232. doi: 10.1042/bj3100225
Bray, R. (1974). Leishmania. Annu. Rev. Microbiol. 28, 189–217. doi: 10.1146/annurev.mi.28.100174.001201
Bringaud, F., Müller, M., Cerqueira, G. C., Smith, M., Rochette, A., El-Sayed, N. M. A., et al. (2007). Members of a large retroposon family are determinants of post-transcriptional gene expression in leishmania. PloS Pathog. 3, e136. doi: 10.1371/journal.ppat.0030136
Britto, C., Ravel, C., Bastien, P., Blaineau, C., Pagès, M., Dedet, J.-P., et al. (1998). Conserved linkage groups associated with large-scale chromosomal rearrangements between old world and new world leishmania genomes. Gene 222, 107–117. doi: 10.1016/S0378-1119(98)00472-7
Burza, S., Croft, S., Boelaert, M. (2018). Leishmaniasis. Lancet 392 (10151), 951–970. doi: 10.1016/S0140-6736(18)31204-2
Burza, S., Nabi, E., Mahajan, R., Mitra, G., Lima, M. A. (2013). One-year follow-up of immunocompetent male patients treated with miltefosine for primary visceral leishmaniasis in bihar, India. Clin. Infect. Dis. 57, 1363–1364. doi: 10.1093/cid/cit508
Calegari-Silva, T. C., Vivarini, Á.C., Pereira, R. D. M., Dias-Teixeira, K. L., Rath, C. T., Pacheco, A. S., et al. (2018). Leishmania amazonensis downregulates macrophage iNOS expression via histone deacetylase 1 (HDAC1): a novel parasite evasion mechanism. Eur. J. Immunol. 48, 1188–1198. doi: 10.1002/eji.201747257
Carter, N. S., Drew, M. E., Sanchez, M., Vasudevan, G., Landfear, S. M., Ullman, B. (2000). Cloning of a novel inosine-guanosine transporter gene fromLeishmania donovani by functional rescue of a transport-deficient mutant. J. Biol. Chem. 275, 20935–20941. doi: 10.1074/jbc.M002418200
Castellucci, L. C., Almeida, L. F. D., Jamieson, S. E., Fakiola, M., Carvalho, E. M. D., Blackwell, J. M. (2014). Host genetic factors in American cutaneous leishmaniasis: A critical appraisal of studies conducted in an endemic area of Brazil. Memorias do Instituto Oswaldo Cruz 109, 279–288. doi: 10.1590/0074-0276140028
Cayla, M., Rachidi, N., Leclercq, O., Schmidt-Arras, D., Rosenqvist, H., Wiese, M., et al. (2014). Transgenic analysis of the leishmania MAP kinase MPK10 reveals an auto-inhibitory mechanism crucial for stage-regulated activity and parasite viability. PloS Pathog. 10, e1004347. doi: 10.1371/journal.ppat.1004347
Chandra, U., Yadav, A., Kumar, D., Saha, S. (2017). Cell cycle stage-specific transcriptional activation of cyclins mediated by HAT2-dependent H4K10 acetylation of promoters in leishmania donovani. PloS Pathog. 13, e1006615. doi: 10.1371/journal.ppat.1006615
Charest, H., Zhang, W.-W., Matlashewski, G. (1996). The developmental expression of leishmania donovani A2 amastigote-specific genes is post-transcriptionally mediated and involves elements located in the 3′-untranslated region. J. Biol. Chem. 271, 17081–17090. doi: 10.1074/jbc.271.29.17081
Clayton, C., Shapira, M. (2007). Post-transcriptional regulation of gene expression in trypanosomes and leishmanias. Mol. Biochem. Parasitol. 156, 93–101. doi: 10.1016/j.molbiopara.2007.07.007
Cloutier, S., Laverdière, M., Chou, M.-N., Boilard, N., Chow, C., Papadopoulou, B. (2012). Translational control through eIF2alpha phosphorylation during the leishmania differentiation process. PloS One 7, e35085. doi: 10.1371/journal.pone.0035085
Coelho, A. C., Beverley, S. M., Cotrim, P. C. (2003). Functional genetic identification of PRP1, an ABC transporter superfamily member conferring pentamidine resistance in leishmania major. Mol. Biochem. Parasitol. 130, 83–90. doi: 10.1016/S0166-6851(03)00162-2
Coelho, A. C., Boisvert, S., Mukherjee, A., Leprohon, P., Corbeil, J., Ouellette, M. (2012). Multiple mutations in heterogeneous miltefosine-resistant leishmania major population as determined by whole genome sequencing. PloS Negl. Trop. Dis. 6, e1512. doi: 10.1371/journal.pntd.0001512
Cojean, S., Houzé, S., Haouchine, D., Huteau, F., Lariven, S., Hubert, V., et al. (2012). Leishmania resistance to miltefosine associated with genetic marker. Emerging Infect. Dis. 18, 704. doi: 10.3201/eid1804.110841
Colineau, L., Lambertz, U., Fornes, O., Wasserman, W. W., Reiner, N. E. (2018). C-myc is a novel leishmania virulence factor by proxy that targets the host miRNA system and is essential for survival in human macrophages. J. Biol. Chem. 293, 12805–12819. doi: 10.1074/jbc.RA118.002462
Cotrim, P. C., Garrity, L. K., Beverley, S. M. (1999). Isolation of genes mediating resistance to inhibitors of nucleoside and ergosterol metabolism in leishmania by overexpression/selection. J. Biol. Chem. 274, 37723–37730. doi: 10.1074/jbc.274.53.37723
Croft, S., Neal, R., Pendergast, W., Chan, J. (1987). The activity of alkyl phosphorylcholines and related derivatives against leishmania donovani. Biochem. Pharmacol. 36, 2633–2636. doi: 10.1016/0006-2952(87)90543-0
Croft, S. L., Sundar, S., Fairlamb, A. H. (2006). Drug resistance in leishmaniasis. Clin. Microbiol. Rev. 19, 111–126. doi: 10.1128/CMR.19.1.111-126.2006
Das, S., Mukherjee, S., Ali, N. (2021). Super enhancer-mediated transcription of miR146a-5p drives M2 polarization during leishmania donovani infection. PloS Pathog. 17, e1009343–e1009343. doi: 10.1371/journal.ppat.1009343
Decuypere, S., Rijal, S., Yardley, V., De Doncker, S., Laurent, T., Khanal, B., et al. (2005). Gene expression analysis of the mechanism of natural Sb (V) resistance in leishmania donovani isolates from Nepal. Antimicrobial Agents Chemotherapy 49, 4616–4621. doi: 10.1128/AAC.49.11.4616-4621.2005
Decuypere, S., Vanaerschot, M., Brunker, K., Imamura, H., Müller, S., Khanal, B., et al. (2012). Molecular mechanisms of drug resistance in natural leishmania populations vary with genetic background. PloS Negl. Trop. Dis. 6, e1514. doi: 10.1371/journal.pntd.0001514
Domagalska, M. A., Imamura, H., Sanders, M., Van Den Broeck, F., Bhattarai, N. R., Vanaerschot, M., et al. (2019). Genomes of leishmania parasites directly sequenced from patients with visceral leishmaniasis in the Indian subcontinent. PloS Negl. Trop. Dis. 13, e0007900. doi: 10.1371/journal.pntd.0007900
Downing, T., Imamura, H., Decuypere, S., Clark, T. G., Coombs, G. H., Cotton, J. A., et al. (2011). Whole genome sequencing of multiple leishmania donovani clinical isolates provides insights into population structure and mechanisms of drug resistance. Genome Res. 21, 2143–2156. doi: 10.1101/gr.123430.111
Dube, A., Singh, N., Sundar, S., Singh, N. (2005). Refractoriness to the treatment of sodium stibogluconate in Indian kala-azar field isolates persist in in vitro and in vivo experimental models. Parasitol. Res. 96, 216–223. doi: 10.1007/s00436-005-1339-1
Dumas, C., Chow, C., MüLler, M., Papadopoulou, B. (2006). A novel class of developmentally regulated noncoding RNAs in leishmania. Eukaryotic Cell 5, 2033–2046. doi: 10.1128/EC.00147-06
Dumetz, F., Imamura, H., Sanders, M., Seblova, V., Myskova, J., Pescher, P., et al. (2017). Modulation of aneuploidy in leishmania donovani during adaptation to different in vitro and in vivo environments and its impact on gene expression. MBio 8, e00599–e00517. doi: 10.1128/mBio.00599-17
Duncan, S. M., Myburgh, E., Philipon, C., Brown, E., Meissner, M., Brewer, J., et al. (2016). Conditional gene deletion with DiCre demonstrates an essential role for CRK3 in l eishmania mexicana cell cycle regulation. Mol. Microbiol. 100, 931–944. doi: 10.1111/mmi.13375
Dupé, A., Dumas, C., Papadopoulou, B. (2014). An alba-domain protein contributes to the stage-regulated stability of amastin transcripts in l eishmania. Mol. Microbiol. 91, 548–561. doi: 10.1111/mmi.12478
El-Sayed, N. M., Myler, P. J., Blandin, G., Berriman, M., Crabtree, J., Aggarwal, G., et al. (2005). Comparative genomics of trypanosomatid parasitic protozoa. Science 309, 404–409. doi: 10.1126/science.1112181
Farrow, A. L., Rana, T., Mittal, M. K., Misra, S., Chaudhuri, G. (2011). Leishmania-induced repression of selected non-coding RNA genes containing b-box element at their promoters in alternatively polarized M2 macrophages. Mol. Cell. Biochem. 350, 47–57. doi: 10.1007/s11010-010-0681-5
Gazanion, É., Fernández-Prada, C., Papadopoulou, B., Leprohon, P., Ouellette, M. (2016). Cos-seq for high-throughput identification of drug target and resistance mechanisms in the protozoan parasite leishmania. Proc. Natl. Acad. Sci. 113, E3012–E3021. doi: 10.1073/pnas.1520693113
Genois, M.-M., Mukherjee, A., Ubeda, J.-M., Buisson, R., Paquet, E., Roy, G., et al. (2012). Interactions between BRCA2 and RAD51 for promoting homologous recombination in leishmania infantum. Nucleic Acids Res. 40, 6570–6584. doi: 10.1093/nar/gks306
Genois, M.-M., Paquet, E. R., Laffitte, M.-C. N., Maity, R., Rodrigue, A., Ouellette, M., et al. (2014). DNA Repair pathways in trypanosomatids: from DNA repair to drug resistance. Microbiol. Mol. Biol. Rev. 78, 40–73. doi: 10.1128/MMBR.00045-13
Genois, M.-M., Plourde, M., Ethier, C., Roy, G., Poirier, G. G., Ouellette, M., et al. (2015). Roles of Rad51 paralogs for promoting homologous recombination in leishmania infantum. Nucleic Acids Res. 43, 2701–2715. doi: 10.1093/nar/gkv118
Gordon, D. J., Resio, B., Pellman, D. (2012). Causes and consequences of aneuploidy in cancer. Nat. Rev. Genet. 13, 189–203. doi: 10.1038/nrg3123
Gourbal, B., Sonuc, N., Bhattacharjee, H., Legare, D., Sundar, S., Ouellette, M., et al. (2004). Drug uptake and modulation of drug resistance in leishmania by an aquaglyceroporin. J. Biol. Chem. 279, 31010–31017. doi: 10.1074/jbc.M403959200
Grünebast, J., Clos, J. (2020). Leishmania: Responding to environmental signals and challenges without regulated transcription. Comput. Struct. Biotechnol. J. 18, 4016–4023. doi: 10.1016/j.csbj.2020.11.058
Gupta, A. K., Das, S., Kamran, M., Ejazi, S. A., Ali, N. (2022). The pathogenicity and virulence of leishmania - interplay of virulence factors with host defenses. Virulence 13, 903–935. doi: 10.1080/21505594.2022.2074130
Haile, S., Papadopoulou, B. (2007). Developmental regulation of gene expression in trypanosomatid parasitic protozoa. Curr. Opin. Microbiol. 10, 569–577. doi: 10.1016/j.mib.2007.10.001
Hailu, A., Musa, A., Wasunna, M., Balasegaram, M., Yifru, S., Mengistu, G., et al. (2010). Geographical variation in the response of visceral leishmaniasis to paromomycin in East Africa: a multicentre, open-label, randomized trial. PloS Negl. Trop. Dis. 4, e709. doi: 10.1371/journal.pntd.0000709
Handman, E. (2001). Leishmaniasis: current status of vaccine development. Clin. Microbiol. Rev. 14, 229–243. doi: 10.1128/CMR.14.2.229-243.2001
Imamura, H., Downing, T., Van Den Broeck, F., Sanders, M. J., Rijal, S., Sundar, S., et al. (2016). Evolutionary genomics of epidemic visceral leishmaniasis in the Indian subcontinent. Elife 5, e12613. doi: 10.7554/eLife.12613.031
Imamura, H., Dujardin, J.-C. (2019). “A guide to next generation sequence analysis of leishmania genomes,” in Leishmania (New York, NY: Humana Press), 69–94.
Ingolia, N. T., Ghaemmaghami, S., Newman, J. R., Weissman, J. S. (2009). Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. science 324, 218–223. doi: 10.1126/science.1168978
Ivens, A. C., Peacock, C. S., Worthey, E. A., Murphy, L., Aggarwal, G., Berriman, M., et al. (2005). The genome of the kinetoplastid parasite, leishmania major. Science 309, 436–442. doi: 10.1126/science.1112680
Jara, M., Maes, I., Imamura, H., Domagalska, M. A., Dujardin, J. C., Arevalo, J. (2019). Tracking of quiescence in leishmania by quantifying the expression of GFP in the ribosomal DNA locus. Sci. Rep. 9, 1–12. doi: 10.1038/s41598-019-55486-z
Jha, T., Lockwood, D. N., Olliaro, P., Thakur, C., Kanyok, T., Singhania, B., et al. (1998). Randomised controlled trial of aminosidine (paromomycin) v sodium stibogluconate for treating visceral leishmaniasis in north bihar, IndiaCommentary: Some good news for treatment of visceral leishmaniasis in bihar. Bmj 316, 1200–1205. doi: 10.1136/bmj.316.7139.1200
Jha, T., Sundar, S., Thakur, C., Bachmann, P., Karbwang, J., Fischer, C., et al. (1999). Miltefosine, an oral agent, for the treatment of Indian visceral leishmaniasis. New Engl. J. Med. 341, 1795–1800. doi: 10.1056/NEJM199912093412403
Jones, N. G., Geoghegan, V., Moore, G., Carnielli, J. B. T., Newling, K., Calderón, F., et al. (2022). Bromodomain factor 5 is an essential regulator of transcription in leishmania. Nat. Commun. 13, 4071. doi: 10.1038/s41467-022-31742-1
Kalesh, K., Denny, P. W. (2019). A BONCAT-iTRAQ method enables temporally resolved quantitative profiling of newly synthesised proteins in leishmania mexicana parasites during starvation. PloS Negl. Trop. Dis. 13, e0007651. doi: 10.1371/journal.pntd.0007651
Kaur, P., Garg, M., Hombach-Barrigah, A., Clos, J., Goyal, N. (2017). MAPK1 of leishmania donovani interacts and phosphorylates HSP70 and HSP90 subunits of foldosome complex. Sci. Rep. 7, 1–11. doi: 10.1038/s41598-017-09725-w
Kawasaki, F., Beraldi, D., Hardisty, R. E., Mcinroy, G. R., Van Delft, P., Balasubramanian, S. (2017). Genome-wide mapping of 5-hydroxymethyluracil in the eukaryote parasite leishmania. Genome Biol. 18, 1–8. doi: 10.1186/s13059-017-1150-1
Kraeva, N., Ishemgulova, A., Lukeš, J., Yurchenko, V. (2014). Tetracycline-inducible gene expression system in leishmania mexicana. Mol. Biochem. Parasitol. 198, 11–13. doi: 10.1016/j.molbiopara.2014.11.002
Kröber-Boncardo, C., Lorenzen, S., Brinker, C., Clos, J. (2020). Casein kinase 1.2 over expression restores stress resistance to leishmania donovani HSP23 null mutants. Sci. Rep. 10, 1–16. doi: 10.1038/s41598-020-72724-x
Kuhn, D., Wiese, M. (2005). LmxPK4, a mitogen-activated protein kinase kinase homologue of leishmania mexicana with a potential role in parasite differentiation. Mol. Microbiol. 56, 1169–1182. doi: 10.1111/j.1365-2958.2005.04614.x
Kumar, V., Kumar, A., Das, S., Kumar, A., Abhishek, K., Verma, S., et al. (2018). Leishmania donovani activates hypoxia inducible factor-1α and miR-210 for survival in macrophages by downregulation of NF-κB mediated pro-inflammatory immune response. Front. Microbiol. 9, 385. doi: 10.3389/fmicb.2018.00385
Kündig, C., Haimeur, A., Légaré, D., Papadopoulou, B., Ouellette, M. (1999). Increased transport of pteridines compensates for mutations in the high affinity folate transporter and contributes to methotrexate resistance in the protozoan parasite leishmania tarentolae. EMBO J. 18, 2342–2351. doi: 10.1093/emboj/18.9.2342
Lachaud, L., Bourgeois, N., Kuk, N., Morelle, C., Crobu, L., Merlin, G., et al. (2014). Constitutive mosaic aneuploidy is a unique genetic feature widespread in the leishmania genus. Microbes infection 16, 61–66. doi: 10.1016/j.micinf.2013.09.005
Lachaud, L., Bourgeois, N., Plourd, M., Leproho, P., Bastien, P., Ouellette, M. (2009). Parasite susceptibility to amphotericin b in failures of treatment for visceral leishmaniasis in patients coinfected with HIV type 1 and leishmania infantum. Clin. Infect. Dis. 48, e16–e22. doi: 10.1086/595710
Laffitte, M.-C. N., Genois, M.-M., Mukherjee, A., Légaré, D., Masson, J.-Y., Ouellette, M. (2014). Formation of linear amplicons with inverted duplications in leishmania requires the MRE11 nuclease. PloS Genet. 10, e1004805. doi: 10.1371/journal.pgen.1004805
Laffitte, M.-C. N., Leprohon, P., Hainse, M., Légaré, D., Masson, J.-Y., Ouellette, M. (2016a). Chromosomal translocations in the parasite leishmania by a MRE11/RAD50-independent microhomology-mediated end joining mechanism. PloS Genet. 12, e1006117. doi: 10.1371/journal.pgen.1006117
Laffitte, M.-C. N., Leprohon, P., Légaré, D., Ouellette, M. (2016b). Deep-sequencing revealing mutation dynamics in the miltefosine transporter gene in leishmania infantum selected for miltefosine resistance. Parasitol. Res. 115, 3699–3703. doi: 10.1007/s00436-016-5195-y
Laffitte, M.-C. N., Leprohon, P., Papadopoulou, B., Ouellette, M. (2016c). Plasticity of the leishmania genome leading to gene copy number variations and drug resistance. F1000Research 5. doi: 10.12688/f1000research.9218.1
Lahav, T., Sivam, D., Volpin, H., Ronen, M., Tsigankov, P., Green, A., et al. (2011). Multiple levels of gene regulation mediate differentiation of the intracellular pathogen leishmania. FASEB J. 25, 515–525. doi: 10.1096/fj.10-157529
Lai a Fat, E. J., Vrede, M. A., Soetosenojo, R. M., Lai a Fat, R. F. (2002). Pentamidine, the drug of choice for the treatment of cutaneous leishmaniasis in Surinam. Int. J. Dermatol. 41, 796–800. doi: 10.1046/j.1365-4362.2002.01633.x
Lecoeur, H., Prina, E., Gutiérrez-Sanchez, M., Späth, G. F. (2022). Going ballistic: Leishmania nuclear subversion of host cell plasticity. Trends Parasitol. 38, 205–216. doi: 10.1016/j.pt.2021.09.009
Lee, N., Bertholet, S., Debrabant, A., Muller, J., Duncan, R., Nakhasi, H. (2002). Programmed cell death in the unicellular protozoan parasite leishmania. Cell Death Differentiation 9, 53–64. doi: 10.1038/sj.cdd.4400952
Légaré, D., Richard, D., Mukhopadhyay, R., Stierhof, Y.-D., Rosen, B. P., Haimeur, A., et al. (2001). The leishmania ATP-binding cassette protein PGPA is an intracellular metal-thiol transporter ATPase. J. Biol. Chem. 276, 26301–26307. doi: 10.1074/jbc.M102351200
Leifso, K., Cohen-Freue, G., Dogra, N., Murray, A., Mcmaster, W. R. (2007). Genomic and proteomic expression analysis of leishmania promastigote and amastigote life stages: The leishmania genome is constitutively expressed. Mol. Biochem. Parasitol. 152, 35–46. doi: 10.1016/j.molbiopara.2006.11.009
Leprohon, P., Legare, D., Raymond, F., Madore, E., Hardiman, G., Corbeil, J., et al. (2009). Gene expression modulation is associated with gene amplification, supernumerary chromosomes and chromosome loss in antimony-resistant leishmania infantum. Nucleic Acids Res. 37, 1387–1399. doi: 10.1093/nar/gkn1069
Lye, L.-F., Owens, K., Shi, H., Murta, S. M., Vieira, A. C., Turco, S. J., et al. (2010). Retention and loss of RNA interference pathways in trypanosomatid protozoans. PloS Pathog. 6, e1001161. doi: 10.1371/journal.ppat.1001161
Mandal, S., Maharjan, M., Singh, S., Chatterjee, M., Madhubala, R. (2010). Assessing aquaglyceroporin gene status and expression profile in antimony-susceptible and-resistant clinical isolates of leishmania donovani from India. J. Antimicrobial Chemotherapy 65, 496–507. doi: 10.1093/jac/dkp468
Mandal, G., Mandal, S., Sharma, M., Charret, K. S., Papadopoulou, B., Bhattacharjee, H., et al. (2015). Species-specific antimonial sensitivity in leishmania is driven by post-transcriptional regulation of AQP1. PloS Negl. Trop. Dis. 9, e0003500. doi: 10.1371/journal.pntd.0003500
Mandal, G., Wyllie, S., Singh, N., Sundar, S., Fairlamb, A., Chatterjee, M. (2007). Increased levels of thiols protect antimony unresponsive leishmania donovani field isolates against reactive oxygen species generated by trivalent antimony. Parasitology 134, 1679–1687. doi: 10.1017/S0031182007003150
Mannaert, A., Downing, T., Imamura, H., Dujardin, J.-C. (2012). Adaptive mechanisms in pathogens: universal aneuploidy in leishmania. Trends Parasitol. 28, 370–376. doi: 10.1016/j.pt.2012.06.003
Marquis, N., Gourbal, B., Rosen, B. P., Mukhopadhyay, R., Ouellette, M. (2005). Modulation in aquaglyceroporin AQP1 gene transcript levels in drug-resistant leishmania. Mol. Microbiol. 57, 1690–1699. doi: 10.1111/j.1365-2958.2005.04782.x
Marr, A. K., Macisaac, J. L., Jiang, R., Airo, A. M., Kobor, M. S., Mcmaster, W. R. (2014). Leishmania donovani infection causes distinct epigenetic DNA methylation changes in host macrophages. PloS Pathog. 10, e1004419. doi: 10.1371/journal.ppat.1004419
Martínez-Calvillo, S., Nguyen, D., Stuart, K., Myler, P. J. (2004). Transcription initiation and termination on leishmania major chromosome 3. Eukaryotic Cell 3, 506–517. doi: 10.1128/EC.3.2.506-517.2004
Martínez-Calvillo, S., Yan, S., Nguyen, D., Fox, M., Stuart, K., Myler, P. J. (2003). Transcription of leishmania major friedlin chromosome 1 initiates in both directions within a single region. Mol. Cell 11, 1291–1299. doi: 10.1016/S1097-2765(03)00143-6
Matthews, K. R., Tschudi, C., Ullu, E. (1994). A common pyrimidine-rich motif governs trans-splicing and polyadenylation of tubulin polycistronic pre-mRNA in trypanosomes. Genes Dev. 8, 491–501. doi: 10.1101/gad.8.4.491
Mccarter, L. L. (2006). Regulation of flagella. Curr. Opin. Microbiol. 9, 180–186. doi: 10.1016/j.mib.2006.02.001
Mckean, P. G., Keen, J. K., Smith, D. F., Benson, F. E. (2001). Identification and characterisation of a RAD51 gene from leishmania major. Mol. Biochem. Parasitol. 115, 209–216. doi: 10.1016/S0166-6851(01)00288-2
Mehta, A., Shaha, C. (2006). Mechanism of metalloid-induced death in leishmania spp.: role of iron, reactive oxygen species, Ca2+, and glutathione. Free Radical Biol. Med. 40, 1857–1868. doi: 10.1016/j.freeradbiomed.2006.01.024
Mimitou, E. P., Symington, L. S. (2011). DNA End resection–unraveling the tail. DNA Repair 10, 344–348. doi: 10.1016/j.dnarep.2010.12.004
Minning, T. A., Weatherly, D. B., Flibotte, S., Tarleton, R. L. (2011). Widespread, focal copy number variations (CNV) and whole chromosome aneuploidies in trypanosoma cruzi strains revealed by array comparative genomic hybridization. BMC Genomics 12, 1–11. doi: 10.1186/1471-2164-12-139
Misra, S., Tripathi, M. K., Chaudhuri, G. (2005). Down-regulation of 7SL RNA expression and impairment of vesicular protein transport pathways by leishmania infection of macrophages. J. Biol. Chem. 280, 29364–29373. doi: 10.1074/jbc.M504162200
Mondelaers, A., Sanchez-Cañete, M. P., Hendrickx, S., Eberhardt, E., Garcia-Hernandez, R., Lachaud, L., et al. (2016). Genomic and molecular characterization of miltefosine resistance in leishmania infantum strains with either natural or acquired resistance through experimental selection of intracellular amastigotes. PloS One 11, e0154101. doi: 10.1371/journal.pone.0154101
Monte-Neto, R., Laffitte, M.-C. N., Leprohon, P., Reis, P., Frézard, F., Ouellette, M. (2015). Intrachromosomal amplification, locus deletion and point mutation in the aquaglyceroporin AQP1 gene in antimony resistant leishmania (Viannia) guyanensis. PloS Negl. Trop. Dis. 9, e0003476. doi: 10.1371/journal.pntd.0003476
Mookerjee Basu, J., Mookerjee, A., Sen, P., Bhaumik, S., Sen, P., Banerjee, S., et al. (2006). Sodium antimony gluconate induces generation of reactive oxygen species and nitric oxide via phosphoinositide 3-kinase and mitogen-activated protein kinase activation in leishmania donovani-infected macrophages. Antimicrobial Agents chemotherapy 50, 1788–1797. doi: 10.1128/AAC.50.5.1788-1797.2006
Morales, M. A., Watanabe, R., Dacher, M., Chafey, P., Y Fortéa, J. O., Scott, D. A., et al. (2010). Phosphoproteome dynamics reveal heat-shock protein complexes specific to the leishmania donovani infectious stage. Proc. Natl. Acad. Sci. 107, 8381–8386. doi: 10.1073/pnas.0914768107
Morales, M. A., Watanabe, R., Laurent, C., Lenormand, P., Rousselle, J. C., Namane, A., et al. (2008). Phosphoproteomic analysis of leishmania donovani pro-and amastigote stages. Proteomics 8, 350–363. doi: 10.1002/pmic.200700697
Moreira, W., Leprohon, P., Ouellette, M. (2011). Tolerance to drug-induced cell death favours the acquisition of multidrug resistance in leishmania. Cell Death Dis. 2, e201–e201. doi: 10.1038/cddis.2011.83
Murray, H. W., Berman, J. D., Davies, C. R., Saravia, N. G. (2005). Advances in leishmaniasis. Lancet 366, 1561–1577. doi: 10.1016/S0140-6736(05)67629-5
Musa, A. M., Younis, B., Fadlalla, A., Royce, C., Balasegaram, M., Wasunna, M., et al. (2010). Paromomycin for the treatment of visceral leishmaniasis in Sudan: A randomized, open-label, dose-finding study. PloS Negl. Trop. Dis. 4, e855. doi: 10.1371/journal.pntd.0000855
Olinski, R., Starczak, M., Gackowski, D. (2016). Enigmatic 5-hydroxymethyluracil: Oxidatively modified base, epigenetic mark or both? Mutat. Research/Reviews Mutat. Res. 767, 59–66. doi: 10.1016/j.mrrev.2016.02.001
Ostyn, B., Hasker, E., Dorlo, T. P., Rijal, S., Sundar, S., Dujardin, J.-C., et al. (2014). Failure of miltefosine treatment for visceral leishmaniasis in children and men in south-East Asia. PloS One 9, e100220. doi: 10.1371/journal.pone.0100220
Ouellette, M., Papadopoulou, B. (2009). Coordinated gene expression by post-transcriptional regulons in African trypanosomes. J. Biol. 8, 1–4. doi: 10.1186/jbiol203
Paananen, J., Fortino, V. (2020). An omics perspective on drug target discovery platforms. Brief Bioinform. 21, 1937–1953. doi: 10.1093/bib/bbz122
Palatnik-De-Sousa, C. B. (2008). Vaccines for leishmaniasis in the fore coming 25 years. Vaccine 26, 1709–1724. doi: 10.1016/j.vaccine.2008.01.023
Parmar, N., Chandrakar, P., Kar, S. (2020). Leishmania donovani subverts host immune response by epigenetic reprogramming of macrophage m (Lipopolysaccharides+ IFN-γ)/M (IL-10) polarization. J. Immunol. 204, 2762–2778. doi: 10.4049/jimmunol.1900251
Peacock, C. S., Seeger, K., Harris, D., Murphy, L., Ruiz, J. C., Quail, M. A., et al. (2007). Comparative genomic analysis of three leishmania species that cause diverse human disease. Nat. Genet. 39, 839–847. doi: 10.1038/ng2053
Pérez-Victoria, F. J., Gamarro, F., Ouellette, M., Castanys, S. (2003). Functional cloning of the miltefosine transporter: a novel p-type phospholipid translocase from leishmania involved in drug resistance. J. Biol. Chem. 278, 49965–49971. doi: 10.1074/jbc.M308352200
Pérez-Victoria, F. J., Sánchez-Cañete, M. P., Castanys, S., Gamarro, F. (2006). Phospholipid translocation and miltefosine potency require both l. donovani miltefosine transporter and the new protein LdRos3 in leishmania parasites. J. Biol. Chem. 281, 23766–23775. doi: 10.1074/jbc.M605214200
Pfau, S. J., Amon, A. (2012). Chromosomal instability and aneuploidy in cancer: from yeast to man: ‘Exploring aneuploidy: the significance of chromosomal imbalance’review series. EMBO Rep. 13, 515–527. doi: 10.1038/embor.2012.65
Plourde, M., Ubeda, J.-M., Mandal, G., Do Monte-Neto, R. L., Mukhopadhyay, R., Ouellette, M. (2015). Generation of an aquaglyceroporin AQP1 null mutant in leishmania major. Mol. Biochem. Parasitol. 201, 108–111. doi: 10.1016/j.molbiopara.2015.07.003
Quijada, L., Soto, M., Alonso, C., Requena, J. M. (1997). Analysis of post-transcriptional regulation operating on transcription products of the tandemly linked leishmania infantum hsp70 genes. J. Biol. Chem. 272, 4493–4499. doi: 10.1074/jbc.272.7.4493
Rana, T., Misra, S., Mittal, M. K., Farrow, A. L., Wilson, K. T., Linton, M. F., et al. (2011). Mechanism of down-regulation of RNA polymerase III-transcribed non-coding RNA genes in macrophages by leishmania. J. Biol. Chem. 286, 6614–6626. doi: 10.1074/jbc.M110.181735
Raymond, F., Boisvert, S., Roy, G., Ritt, J.-F., Legare, D., Isnard, A., et al. (2012). Genome sequencing of the lizard parasite leishmania tarentolae reveals loss of genes associated to the intracellular stage of human pathogenic species. Nucleic Acids Res. 40, 1131–1147. doi: 10.1093/nar/gkr834
Reis-Cunha, J. L., Rodrigues-Luiz, G. F., Valdivia, H. O., Baptista, R. P., Mendes, T. A., De Morais, G. L., et al. (2015). Chromosomal copy number variation reveals differential levels of genomic plasticity in distinct trypanosoma cruzi strains. BMC Genomics 16, 1–15. doi: 10.1186/s12864-015-1680-4
Reis-Cunha, J. L., Valdivia, H. O., Bartholomeu, D. C. (2018). Gene and chromosomal copy number variations as an adaptive mechanism towards a parasitic lifestyle in trypanosomatids. Curr. Genomics 19, 87–97. doi: 10.2174/1389202918666170911161311
Reynolds, D. L., Hofmeister, B. T., Cliffe, L., Siegel, T. N., Anderson, B. A., Beverley, S. M., et al. (2016). Base J represses genes at the end of polycistronic gene clusters in leishmania major by promoting RNAP II termination. Mol. Microbiol. 101, 559–574. doi: 10.1111/mmi.13408
Roatt, B. M., Aguiar-Soares, R. D. D. O., Coura-Vital, W., Ker, H. G., Moreira, N. D. D., Vitoriano-Souza, J., et al. (2014). Immunotherapy and immunochemotherapy in visceral leishmaniasis: Promising treatments for this neglected disease. Front. Immunol. 5, 272. doi: 10.3389/fimmu.2014.00272
Rogers, M. B., Hilley, J. D., Dickens, N. J., Wilkes, J., Bates, P. A., Depledge, D. P., et al. (2011). Chromosome and gene copy number variation allow major structural change between species and strains of leishmania. Genome Res. 21, 2129–2142. doi: 10.1101/gr.122945.111
Rosenzweig, D., Smith, D., Myler, P. J., Olafson, R. W., Zilberstein, D. (2008a). Post-translational modification of cellular proteins during leishmania donovani differentiation. Proteomics 8, 1843–1850. doi: 10.1002/pmic.200701043
Rosenzweig, D., Smith, D., Opperdoes, F., Stern, S., Olafson, R. W., Zilberstein, D. (2008b). Retooling leishmania metabolism: From sand fly gut to human macrophage. FASEB J. 22, 590–602. doi: 10.1096/fj.07-9254com
Roussel, M., Nacher, M., Fremont, G., Rotureau, B., Clyti, E., Sainte-Marie, D., et al. (2006). Comparison between one and two injections of pentamidine isethionate, at 7 mg/kg in each injection, in the treatment of cutaneous leishmaniasis in French Guiana. Ann. Trop. Med. Parasitol. 100, 307–314. doi: 10.1179/136485906X105561
Rutledge, S. D., Cimini, D. (2016). Consequences of aneuploidy in sickness and in health. Curr. Opin. Cell Biol. 40, 41–46. doi: 10.1016/j.ceb.2016.02.003
Ryan, K. A., Garraway, L. A., Descoteaux, A., Turco, S. J., Beverley, S. M. (1993). Isolation of virulence genes directing surface glycosyl-phosphatidylinositol synthesis by functional complementation of leishmania. Proc. Natl. Acad. Sci. 90, 8609–8613. doi: 10.1073/pnas.90.18.8609
Samarasinghe, S. R., Samaranayake, N., Kariyawasam, U. L., Siriwardana, Y. D., Imamura, H., Karunaweera, N. D. (2018). Genomic insights into virulence mechanisms of leishmania donovani: Evidence from an atypical strain. BMC Genomics 19, 843. doi: 10.1186/s12864-018-5271-z
Sander, J. D., Joung, J. K. (2014). CRISPR-cas systems for editing, regulating and targeting genomes. Nat. Biotechnol. 32, 347–355. doi: 10.1038/nbt.2842
Saravolatz, L. D., Bern, C., Adler-Moore, J., Berenguer, J., Boelaert, M., Den Boer, M., et al. (2006). Liposomal amphotericin b for the treatment of visceral leishmaniasis. Clin. Infect. Dis. 43, 917–924. doi: 10.1086/507530
Selle, K., Barrangou, R. (2015). Harnessing CRISPR–cas systems for bacterial genome editing. Trends Microbiol. 23, 225–232. doi: 10.1016/j.tim.2015.01.008
Sereno, D., Holzmuller, P., Mangot, I., Cuny, G., Ouaissi, A., Lemesre, J.-L. (2001). Antimonial-mediated DNA fragmentation in leishmania infantum amastigotes. Antimicrobial Agents chemotherapy 45, 2064–2069. doi: 10.1128/AAC.45.7.2064-2069.2001
Shibata, A., Moiani, D., Arvai, A. S., Perry, J., Harding, S. M., Genois, M.-M., et al. (2014). DNA Double-strand break repair pathway choice is directed by distinct MRE11 nuclease activities. Mol. Cell 53, 7–18. doi: 10.1016/j.molcel.2013.11.003
Shor, E., Perlin, D. S. (2015). Coping with stress and the emergence of multidrug resistance in fungi. PloS Pathog. 11, e1004668. doi: 10.1371/journal.ppat.1004668
Singh, A. K., Pandey, R. K., Shaha, C., Madhubala, R. (2016). MicroRNA expression profiling of leishmania donovani-infected host cells uncovers the regulatory role of MIR30A-3p in host autophagy. Autophagy 12, 1817–1831. doi: 10.1080/15548627.2016.1203500
Singh, R., Pandey, H., Sundar, S. (2006). Visceral leishmaniasis (kala-azar): challenges ahead. Indian J. Med. Res. 123, 331.
Smith, M., Bringaud, F., Papadopoulou, B. (2009). Organization and evolution of two SIDER retroposon subfamilies and their impact on the leishmania genome. BMC Genomics 10, 1–14. doi: 10.1186/1471-2164-10-240
Sollelis, L., Ghorbal, M., Macpherson, C. R., Martins, R. M., Kuk, N., Crobu, L., et al. (2015). First efficient CRISPR-c as9-mediated genome editing in l eishmania parasites. Cell. Microbiol. 17, 1405–1412. doi: 10.1111/cmi.12456
Soto, J., Buffet, P., Grogl, M., Berman, J. (1994). Successful treatment of Colombian cutaneous leishmaniasis with four injections of pentamidine. Am. J. Trop. Med. hygiene 50, 107–111. doi: 10.4269/ajtmh.1994.50.107
Sterkers, Y., Crobu, L., Lachaud, L., Pagès, M., Bastien, P. (2014). Parasexuality and mosaic aneuploidy in leishmania: alternative genetics. Trends Parasitol. 30, 429–435. doi: 10.1016/j.pt.2014.07.002
Sterkers, Y., Lachaud, L., Bourgeois, N., Crobu, L., Bastien, P., Pagès, M. (2012). Novel insights into genome plasticity in eukaryotes: mosaic aneuploidy in leishmania. Mol. Microbiol. 86, 15–23. doi: 10.1111/j.1365-2958.2012.08185.x
Sterkers, Y., Lachaud, L., Crobu, L., Bastien, P., Pagès, M. (2011). FISH analysis reveals aneuploidy and continual generation of chromosomal mosaicism in leishmania major. Cell. Microbiol. 13, 274–283. doi: 10.1111/j.1462-5822.2010.01534.x
Sudhandiran, G., Shaha, C. (2003). Antimonial-induced increase in intracellular Ca2+ through non-selective cation channels in the host and the parasite is responsible for apoptosis of intracellular leishmania donovani amastigotes. J. Biol. Chem. 278, 25120–25132. doi: 10.1074/jbc.M301975200
Sundar, S., Chakravarty, J., Agarwal, D., Rai, M., Murray, H. W. (2010). Single-dose liposomal amphotericin b for visceral leishmaniasis in India. New Engl. J. Med. 362, 504–512. doi: 10.1056/NEJMoa0903627
Sundar, S., Jha, T., Thakur, C., Engel, J., Sindermann, H., Fischer, C., et al. (2002). Oral miltefosine for Indian visceral leishmaniasis. New Engl. J. Med. 347, 1739–1746. doi: 10.1056/NEJMoa021556
Sundar, S., Jha, T., Thakur, C. P., Sinha, P. K., Bhattacharya, S. K. (2007). Injectable paromomycin for visceral leishmaniasis in India. New Engl. J. Med. 356, 2571–2581. doi: 10.1056/NEJMoa066536
Tejera Nevado, P., Bifeld, E., Höhn, K., Clos, J. (2016). A telomeric cluster of antimony resistance genes on chromosome 34 of leishmania infantum. Antimicrobial Agents chemotherapy 60, 5262–5275. doi: 10.1128/AAC.00544-16
Thaler, R., Agsten, M., Spitzer, S., Paschalis, E. P., Karlic, H., Klaushofer, K., et al. (2011). Homocysteine suppresses the expression of the collagen cross-linker lysyl oxidase involving IL-6, Fli1, and epigenetic DNA methylation. J. Biol. Chem. 286, 5578–5588. doi: 10.1074/jbc.M110.166181
Thomas, S., Green, A., Sturm, N. R., Campbell, D. A., Myler, P. J. (2009). Histone acetylations mark origins of polycistronic transcription in leishmania major. BMC Genomics 10, 1–15. doi: 10.1186/1471-2164-10-152
Tiwari, N., Kumar, V., Gedda, M. R., Singh, A. K., Singh, V. K., Singh, S. P., et al. (2017). Identification and characterization of miRNAs in response to leishmania donovani infection: delineation of their roles in macrophage dysfunction. Front. Microbiol. 8, 314. doi: 10.3389/fmicb.2017.00314
Ubeda, J.-M., Légaré, D., Raymond, F., Ouameur, A. A., Boisvert, S., Rigault, P., et al. (2008a). Modulation of gene expression in drug resistant leishmania is associated with gene amplification, gene deletion and chromosome aneuploidy. Genome Biol. 9, 1–16. doi: 10.1186/gb-2008-9-7-r115
Ubeda, J. M., Légaré, D., Raymond, F., Ouameur, A. A., Boisvert, S., Rigault, P., et al. (2008b). Modulation of gene expression in drug resistant leishmania is associated with gene amplification, gene deletion and chromosome aneuploidy. Genome Biol. 9, R115. doi: 10.1186/gb-2008-9-7-r115
Ubeda, J.-M., Raymond, F., Mukherjee, A., Plourde, M., Gingras, H., Roy, G., et al. (2014). Genome-wide stochastic adaptive DNA amplification at direct and inverted DNA repeats in the parasite leishmania. PloS Biol. 12, e1001868. doi: 10.1371/journal.pbio.1001868
Uzcategui, N. L., Zhou, Y., Figarella, K., Ye, J., Mukhopadhyay, R., Bhattacharjee, H. (2008). Alteration in glycerol and metalloid permeability by a single mutation in the extracellular c-loop of leishmania major aquaglyceroporin LmAQP1. Mol. Microbiol. 70, 1477–1486. doi: 10.1111/j.1365-2958.2008.06494.x
Van Leeuwen, F., Taylor, M. C., Mondragon, A., Moreau, H., Gibson, W., Kieft, R., et al. (1998). β-d-glucosyl-hydroxymethyluracil is a conserved DNA modification in kinetoplastid protozoans and is abundant in their telomeres. Proc. Natl. Acad. Sci. 95, 2366–2371. doi: 10.1073/pnas.95.5.2366
Van Luenen, H. G., Farris, C., Jan, S., Genest, P.-A., Tripathi, P., Velds, A., et al. (2012). Glucosylated hydroxymethyluracil, DNA base J, prevents transcriptional readthrough in leishmania. Cell 150, 909–921. doi: 10.1016/j.cell.2012.07.030
Vasudevan, G., Carter, N. S., Drew, M. E., Beverley, S. M., Sanchez, M. A., Seyfang, A., et al. (1998). Cloning of leishmania nucleoside transporter genes by rescue of a transport-deficient mutant. Proc. Natl. Acad. Sci. 95, 9873–9878. doi: 10.1073/pnas.95.17.9873
Vasudevan, G., Ullman, B., Landfear, S. M. (2001). Point mutations in a nucleoside transporter gene from leishmania donovani confer drug resistance and alter substrate selectivity. Proc. Natl. Acad. Sci. 98, 6092–6097. doi: 10.1073/pnas.101537298
Vergnes, B., Gourbal, B., Girard, I., Sundar, S., Drummelsmith, J., Ouellette, M. (2007). A proteomics screen implicates HSP83 and a small kinetoplastid calpain-related protein in drug resistance in leishmania donovani clinical field isolates by modulating drug-induced programmed cell death. Mol. Cell. Proteomics 6, 88–101. doi: 10.1074/mcp.M600319-MCP200
Vergnes, B., Sereno, D., Madjidian-Sereno, N., Lemesre, J.-L., Ouaissi, A. (2002). Cytoplasmic SIR2 homologue overexpression promotes survival of leishmania parasites by preventing programmed cell death. Gene 296, 139–150. doi: 10.1016/S0378-1119(02)00842-9
Vergnes, B., Vanhille, L., Ouaissi, A., Sereno, D. (2005). Stage-specific antileishmanial activity of an inhibitor of SIR2 histone deacetylase. Acta tropica 94, 107–115. doi: 10.1016/j.actatropica.2005.03.004
Vizuet-De-Rueda, J. C., Florencio-Martínez, L. E., Padilla-Mejía, N. E., Manning-Cela, R., Hernández-Rivas, R., Martínez-Calvillo, S. (2016). Ribosomal RNA genes in the protozoan parasite leishmania major possess a nucleosomal structure. Protist 167, 121–135. doi: 10.1016/j.protis.2016.02.001
Von Freyend, S. J., Rosenqvist, H., Fink, A., Melzer, I. M., Clos, J., Jensen, O. N., et al. (2010). LmxMPK4, an essential mitogen-activated protein kinase of leishmania mexicana is phosphorylated and activated by the STE7-like protein kinase LmxMKK5. Int. J. Parasitol. 40, 969–978. doi: 10.1016/j.ijpara.2010.02.004
Wang, Y., Fan, P. S., Kahaleh, B. (2006). Association between enhanced type I collagen expression and epigenetic repression of the FLI1 gene in scleroderma fibroblasts. Arthritis Rheumatism: Off. J. Am. Coll. Rheumatol. 54, 2271–2279. doi: 10.1002/art.21948
Wang, Q., Melzer, I. M., Kruse, M., Sander-Juelch, C., Wiese, M. (2005). LmxMPK4, a mitogen-activated protein (MAP) kinase homologue essential for promastigotes and amastigotes of leishmania mexicana. Kinetoplastid Biol. Dis. 4, 1–9. doi: 10.1186/1475-9292-4-6
Wiese, M. (1998). A mitogen-activated protein (MAP) kinase homologue of leishmania mexicana is essential for parasite survival in the infected host. EMBO J. 17, 2619–2628. doi: 10.1093/emboj/17.9.2619
Wiese, M., Kuhn, D., Grünfelder, C. G. (2003). Protein kinase involved in flagellar-length control. Eukaryot Cell 2, 769–777. doi: 10.1128/EC.2.4.769-777.2003
Wincker, P., Ravel, C., Blaineau, C., Pages, M., Jauffret, Y., Dedet, J.-P., et al. (1996). The leishmania genome comprises 36 chromosomes conserved across widely divergent human pathogenic species. Nucleic Acids Res. 24, 1688–1694. doi: 10.1093/nar/24.9.1688
Wright, A. V., Nuñez, J. K., Doudna, J. A. (2016). Biology and applications of CRISPR systems: harnessing nature’s toolbox for genome engineering. Cell 164, 29–44. doi: 10.1016/j.cell.2015.12.035
Wyllie, S., Cunningham, M. L., Fairlamb, A. H. (2004). Dual action of antimonial drugs on thiol redox metabolism in the human pathogen leishmania donovani. J. Biol. Chem. 279, 39925–39932. doi: 10.1074/jbc.M405635200
Yadav, A., Chandra, U., Saha, S. (2016). Histone acetyltransferase HAT4 modulates navigation across G2/M and re-entry into G1 in leishmania donovani. Sci. Rep. 6, 27510. doi: 10.1038/srep27510
Yao, C., Leidal, K. G., Brittingham, A., Tarr, D. E., Donelson, J. E., Wilson, M. E. (2002). Biosynthesis of the major surface protease GP63 of leishmania chagasi. Mol. Biochem. Parasitol. 121, 119–128. doi: 10.1016/S0166-6851(02)00030-0
Zhang, W.-W., Matlashewski, G. (2015). CRISPR-Cas9-mediated genome editing in leishmania donovani. MBio 6, e00861–e00815. doi: 10.1128/mBio.00861-15
Zhu, J., Tsai, H. J., Gordon, M. R., Li, R. (2018). Cellular stress associated with aneuploidy. Dev. Cell 44, 420–431. doi: 10.1016/j.devcel.2018.02.002
Zinoviev, A., Leger, M., Wagner, G., Shapira, M. (2011). A novel 4E-interacting protein in leishmania is involved in stage-specific translation pathways. Nucleic Acids Res. 39, 8404–8415. doi: 10.1093/nar/gkr555
Keywords: leishmania, plasticity, parasite survival, drug resistance, genome organization
Citation: Kamran M, Bhattacharjee R, Das S, Mukherjee S and Ali N (2023) The paradigm of intracellular parasite survival and drug resistance in leishmanial parasite through genome plasticity and epigenetics: Perception and future perspective. Front. Cell. Infect. Microbiol. 13:1001973. doi: 10.3389/fcimb.2023.1001973
Received: 24 July 2022; Accepted: 16 January 2023;
Published: 06 February 2023.
Edited by:
Junaid Jawed, Presidency University, IndiaReviewed by:
Budhaditya Mukherjee, Indian Institute of Technology Kharagpur, IndiaSergio Schenkman, Federal University of São Paulo, Brazil
Santanu Kar Mahapatra, Midnapore City College, India
Copyright © 2023 Kamran, Bhattacharjee, Das, Mukherjee and Ali. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Nahid Ali, bmFsaUBpaWNiLnJlcy5pbg==
†These authors have contributed equally to this work
 Mohd. Kamran
Mohd. Kamran Rahul Bhattacharjee
Rahul Bhattacharjee Sonali Das
Sonali Das Sohitri Mukherjee
Sohitri Mukherjee Nahid Ali
Nahid Ali