- Department of Biomedical Sciences, Unit of Biology and Genetics, University of Cagliari, Cagliari, Italy
DNA methylation is an epigenetic signature consisting of a methyl group at the 5’ cytosine of CpG dinucleotides. Modifications in DNA methylation pattern have been detected in cancer and infectious diseases and may be associated with gene expression changes. In cancer development DNA methylation aberrations are early events whereas in infectious diseases these epigenetic changes may be due to host/pathogen interaction. In particular, in leishmaniasis, a parasitic disease caused by the protozoan Leishmania, DNA methylation alterations have been detected in macrophages upon infection with Leishmania donovani and in skin lesions from patients with cutaneous leishmaniasis. Interestingly, different types of cancers, such as cutaneous malignant lesions, lymphoma and hepatocellular carcinoma, have been diagnosed in patients with a history of leishmaniasis. In fact, it is known that there exists an association between cancer and infectious diseases. Leishmania infection may increase susceptibility to develop cancer, but the mechanisms involved are not entirely clear. Considering these aspects, in this review we discuss the hypothesis that DNA methylation alterations induced by Leishmania may trigger tumorigenesis in long term infection since these epigenetic modifications may enhance and accumulate during chronic leishmaniasis.
Introduction
Several works reported a possible association between infectious diseases and cancer development but the mechanisms leading to malignant transformation are not well elucidated (Kopterides et al., 2007; Al-Kamel, 2017; van Tong et al., 2017; Yasunaga and Matsuoka, 2018). Epigenetics is an interesting research area whose study is increasing in pathogen diseases while it is widely investigated in cancer. It refers to heritable and reversible changes that affects the genetic material packaging and expression without altering the DNA sequence. The epigenetic regulatory network mainly includes DNA methylation, histone modifications and non-coding RNAs (Gal-Yam et al., 2008; Saavedra et al., 2012). Pathogens hijack the epigenome of host cells generating a suitable environment for their survival and replication. DNA methylation, an epigenetic sign, is modulated by pathogens during their interaction with host cells. The DNA methylation profile is relevant for gene expression regulation and chromosome stability. Bacteria, virus and parasites may alter the host methylome and the enzymes or cofactors involved in DNA methylation modulation (Silmon de Monerri and Kim, 2014). In particular, several DNA methylation aberrations have been detected in macrophages upon infection with the protozoan Leishmania spp., responsible for leishmaniasis, a neglected tropical disease (Marr et al., 2014).
Alterations in DNA methylation pattern are also early events in cancerogenesis, and they are tumour-specific signs. Therefore, they are considered excellent diagnostic and useful prognostic biomarkers (Gal-Yam et al., 2008). These aberrations frequently lead to downregulation of tumour suppressor genes and upregulation of oncogenes (Skvortsova et al., 2019). Pathogen infections such as leishmaniasis can be considered tumour promoters since they generate an environment prone to malignancy development employing host colonization strategies similar to cancer cells. In fact, cancer cases have been reported in patients with a history of Leishmania infection (Kopterides et al., 2007; Schwing et al., 2019). The novelty of the present review resides in the description of a possible association between Leishmania infection and cancer onset due to changes in the DNA methylation profile.
DNA methylation
DNA methylation is an epigenetic signature consisting of a methyl group at the 5’ cytosine of CpG dinucleotides. CpG sites are distributed across the genome, in regulatory regions, at DNA repetitive elements and gene bodies. Regions with high CpG content termed CpG islands (CGI) are often located at gene promoter regions. DNA methylation maintains genome stability and regulates gene expression, in particular when methylated CpG sites are located in promoter regions, i.e. a high content of methylated cytosines is generally associated with transcriptional downregulation whereas a hypomethylated state is frequently linked to gene expression upregulation. The addition of the methyl group can be maintained during cell division but it is also a reversible modification (Gal-Yam et al., 2008; Ambrosi et al., 2017). Two protein families, DNA methyltransferases (DNMTs) and Ten-eleven translocation (TET), together with protein partners, are relevant players of the DNA methylation pathway. DNMTs include three enzymes; DNMT1 restores the methylation pattern of the new DNA strand during replication, whereas DNMT3A and DNMT3B with their cofactor DNMT3L are involved in de novo DNA methylation. DNMT1 and DNMT3 mainly differ at the N-terminal domain mediating their specific activity at genomic sites. DNMTs interact with several accessory molecules such as DNMT3L, UHRF1 and RNAs. DNMT3L acts during genomic imprinting establishment regulating DNMT3-target specificity and enzyme activity, while UHRF1 recruits DNMT1 to hemimethylated DNA during replication (Lee et al., 2014; Ambrosi et al., 2017). On the other hand, DNA demethylation is catalysed by TET enzymes which are responsible for 5-methylcytosine oxidation whose products are removed by the DNA repair mechanism. TET family consists of three members which contain or interact with CXXC domains, key sequences in CpG regions recognition (Schubeler, 2015; Ambrosi et al., 2017). The expression of DNMT and TET enzymes varies during development leading to DNA methylation pattern changes that, with other epigenetic mechanisms, participate in the silencing or activation of pathways for cellular stemness, differentiation, proliferation, among others. For instance, TET1 and TET2 maintain stemness phenotype by interaction with NANOG, a pluripotency protein (Lee et al., 2014; Ambrosi et al., 2017).
DNA methylation alterations in cancer and infectious diseases
DNA methylation alterations have been reported in different pathologies, including cancer and infectious diseases. Normal cells present global hypermethylation and unmethylated CGIs at promoter regions, whereas cancer cells undergo a global hypomethylation and hypermethylation at the 5’ gene regulatory regions (Gal-Yam et al., 2008). High content of methylated CpGs at promoter regions, frequently belonging to tumour suppressor genes, is generally associated with gene expression downregulation (Fadda et al., 2018; Loi et al., 2019; Skvortsova et al., 2019; Vega-Benedetti et al., 2020; Vega-Benedetti et al., 2022) (Figure 1). Methylated cytosines can affect transcription factor (TF) binding and thus the recruitment of the RNA polymerase to the transcription start site. For example, Sp1 cannot bind to its consensus site at the Retinoblastoma (RB) gene promoter whether it contains methylated CpGs (Clark et al., 1997; Heberle and Bardet, 2019). Gain of methylation at enhancer regions could also disrupt normal TF binding contributing to tumour suppressor gene downregulation (Schubeler, 2015; Skvortsova et al., 2019) (Figure 1). DNA methylation differently influences TF binding and thus its function (Yin et al., 2017). Instead, in several cancers, such as Wilms tumour, ovarian and breast carcinomas, hypomethylation in satellite DNA could predispose to chromosomal translocations (Feinberg and Tycko, 2004; Gal-Yam et al., 2008). Loss of methylation at retrotranspons may reactivate them leading to possible dysregulation of normal gene expression (Suter et al., 2004). Demethylation in cancer can also occur at promoter regions of oncogenes increasing or reactivating their transcription. DNA methylation works in combination with histone modifications and nucleosome organization to regulate gene expression (Skvortsova et al., 2019). Several experimental studies have been performed to demonstrate the role of DNA methylation changes in tumorigenesis. For instance, in mice models induced hypomethylation led to chromosome instability and tumour promotion, whereas DNMT inhibition prevented malignancy development (Gal-Yam et al., 2008; Kulis and Esteller, 2010). DNA methylation alterations are cancer-specific early events in tumorigenesis resulting in promising diagnostic biomarkers. These aberrations have been found in preneoplastic and histologically normal tissues of people susceptible to develop cancer (Feinberg and Tycko, 2004; Gal-Yam et al., 2008; Saavedra et al., 2012; Luo et al., 2014; Wong Doo et al., 2016; Micevic et al., 2017; Fadda et al., 2018; Loi et al., 2019). To note, methylated-based biomarkers can be traced in cell-free DNA from several matrices including plasma, stool, urine and bile (Gal-Yam et al., 2008; Fadda et al., 2018; Vega-Benedetti et al., 2020; Loi et al., 2022). Moreover, DNA methylation alterations are useful prognostic biomarkers in several cancers including cutaneous melanoma, lung adenocarcinoma, ovarian, breast and cervical cancer (Fleischer et al., 2014; Guo et al., 2018; Guo et al., 2019; Wang et al., 2019; Yang et al., 2020).
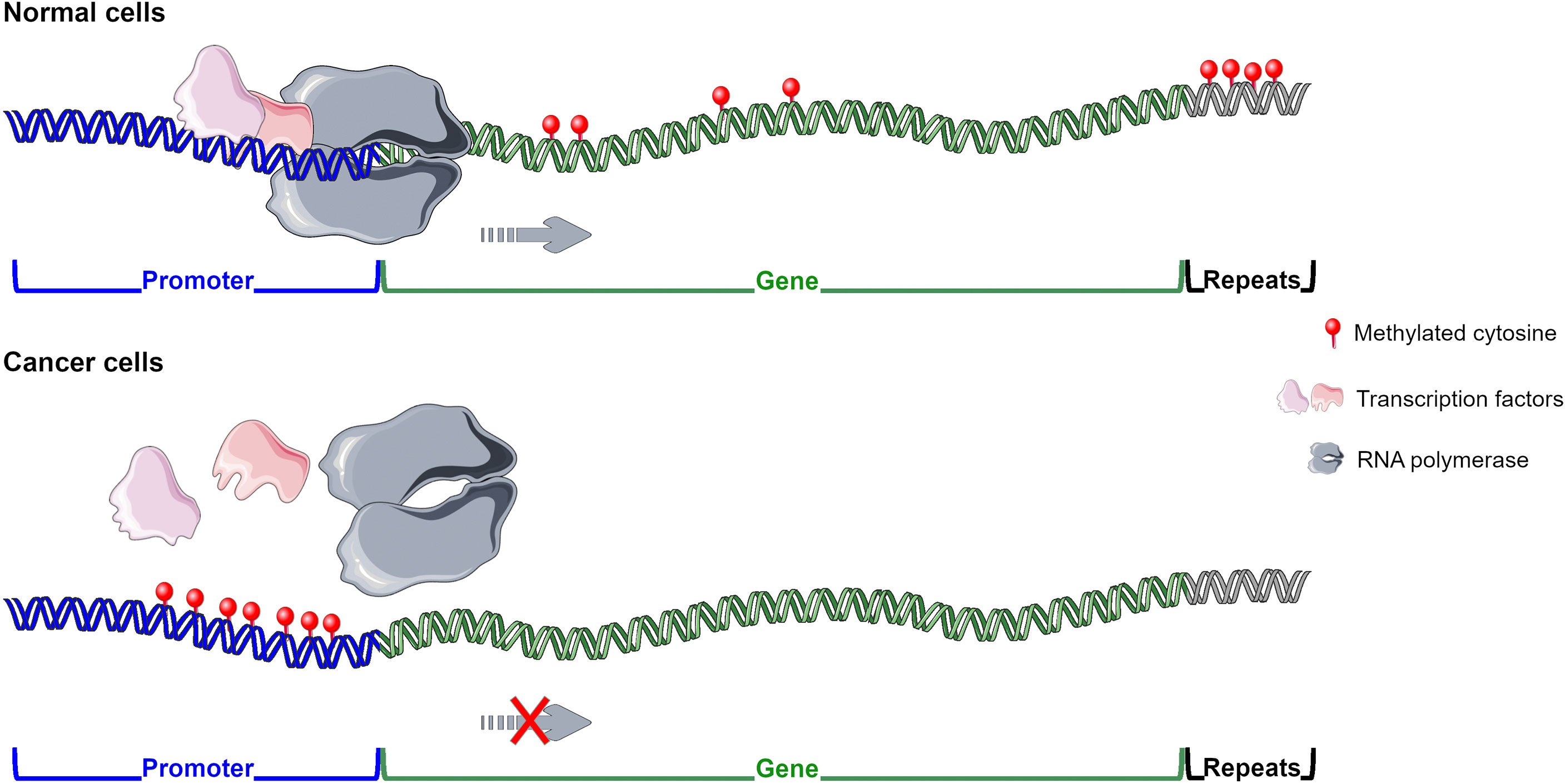
Figure 1 DNA methylation pattern in normal and cancer cells. In normal cells methyl group is widely distributed except for CGIs located at promoters and enhancers of tumour suppressor genes enabling their transcription. In cancer cells hypermethylation is generally observed at regulatory regions of tumour suppressor genes inhibiting their expression, whereas global hypomethylation is detected. Figure created using Servier Medical Art images, licensed under a Creative Commons Attribution 3.0 Unported License; https://smart.servier.com/.
On the other hand, in infectious diseases microbes hijack the epigenome of host cells, including DNA methylation, to elude host defensive mechanisms and promote their survival. Previous evidence showed that virus, bacteria and parasites manipulate the transcription of host defence genes leading to immunosuppression (Paschos and Allday, 2010; Gómez-Díaz et al., 2012; Silmon de Monerri and Kim, 2014). To achieve this aim different strategies are employed, for instance viral DNA integration in host genome induces DNA methylation changes in flanking regions, enabling viral latency. Modulation of host enzymes, such as the DNA methyltransferases, promotes modifications in DNA methylation pattern during hepatitis B (HBV) and Epstein-Barr virus (EBV) infections (Silmon de Monerri and Kim, 2014). Helicobacter pylori infection induces epigenome modification of host cells such as hypermethylation of FOXD3 promoter, a key participant in apoptosis. The bacteria Mycobacterium leprae facilitates its dissemination in the host through DNA demethylation at promoters of genes involved in the epithelial-mesenchymal transition (Silmon de Monerri and Kim, 2014). Regarding parasites, they may regulate host DNA methylation pattern during cell invasion or through protein secretion delivered by vesicles (Silmon de Monerri and Kim, 2014). In vivo and in vitro assays showed DNA methylation modifications upon Toxoplasma gondii and Leishmania donovani infection (Hari Dass and Vyas, 2014; Marr et al., 2014). In agreement with pathogen modulation of host defence system, many altered genes upon Leishmania infection participate in host immune response including the following mechanisms: cytotoxicity mediated by Natural killer cells, interaction between cytokines and their receptors, chemokine/adipocytokine signalling and leukocyte migration (Marr et al., 2014).
Leishmania infection: Host/parasite interplay
Leishmaniasis is a neglected tropical disease caused by different species of the protozoan Leishmania such as L. braziliensis/amazonensis, responsible for the development of the cutaneous and muco-cutaneous clinical forms, and L. infantum/donovani, causative agents of visceral leishmaniasis (Afrin et al., 2019; WHO, 2020). This parasite has a complex life cycle, alternating two stages: the flagellated promastigote and the amastigote forms. Once it is injected in the host by the bite of a female sandfly (Lutzomyia species), promastigotes enter macrophages where they replicate as amastigotes. Leishmania parasites successfully colonize macrophages due to their capability to avoid host defence mechanisms by modulating their surface molecules and host immune response including macrophage activation and antigen processing (Robert McMaster et al., 2016; Afrin et al., 2019). These strategies favour protozoan survival and establishment within the host (Marr et al., 2014; Robert McMaster et al., 2016; Al-Kamel, 2017). Interestingly, parasites manipulate host epigenome. i.e. DNA methylation, histone modifications and non-coding RNA, altering gene expression and thus signalling pathways (Parmar et al., 2020; Roy et al., 2020). Marr et al. reported several differentially methylated CpG sites in macrophages upon infection with L. donovani (Marr et al., 2014). Genes affected by these altered features are implicated in the following signalling pathways: JAK/STAT, MAPK, Notch and Wnt signalling, focal adhesion, among others (Marr et al., 2014). Moreover, low methylation level at FLI1 promoter was observed in cutaneous lesions from patients with L. braziliensis infection and in IL-6 treated macrophages infected with the same Leishmania sp. (Almeida et al., 2017). As observed in the experiments performed by Almeida et al., IL-6 affects methylation of CpGs in the FLI1 promoter suggesting that cellular communication and the surrounding environment are important variables during parasite/host interaction (Almeida et al., 2017).
Although macrophages are the main host cells for Leishmania spp., dendritic cells responsible for antigen presentation also phagocytose parasites and migrate to lymph nodes. Leishmania infection progression is mainly regulated by a complex interplay between macrophages and dendritic cells. IL-12 produced by infected dendritic cells triggers a cascade of events leading to macrophage classical activation involved in parasiticidal activity, whereas IL-10 and TGF-β released by infected macrophages result in alternative macrophage activation and thus in parasite survival (Liu and Uzonna, 2012). However, some Leishmania species such as L. infantum present high tropism to hepatocytes establishing a strong interaction with their membrane and regulating gene expression although parasite internalization has not been confirmed (Kausalya et al., 1993; Rodrigues et al., 2019). Little is known about the factors that guide Leishmania tissue tropism (Seblova et al., 2015). Few genes that may contribute to the disease tropism have been identified in different Leishmania species (Peacock et al., 2007). Tropism may also depend on host susceptibility including immunity and genetics, and on parasite virulence (Chang and McGwire, 2002; Reithinger et al., 2002).
This evidence suggests that Leishmania is able to hijack the epigenome, in particular DNA methylation, and regulate the transcriptional machinery not only of antigen-presenting cells but also of other cell types located at the infection site, such as fibroblasts and hepatocytes (Almeida et al., 2017; Rodrigues et al., 2019). It is not clear whether these changes occur due to molecules, including methyltransferase inhibitors, activators, and ncRNAs, released by Leishmania spp. and delivered via exosomes and/or microvesicles before parasite internalization and/or only when the pathogen is already inside macrophages (Silverman et al., 2010a; Coakley et al., 2015; Lambertz et al., 2015; Robert McMaster et al., 2016). Since parasite internalization is not reported in some immune cells, such as T cells, and in non-immune cells, extracellular vesicles communication may have a key role in cell epigenome modification. It has been reported that extracellular vesicles released by L. major target T cells leading to an increase production of IL-4 (Silverman et al., 2010b; Coakley et al., 2015). The interaction between the molecules released by parasites and the host epigenome is not fully established. Of note, the glycoprotein GP63 and the elongation factor-1α (EF-1α) secreted by Leishmania spp. have been reported to alter host pathways regulating kinases and phosphatases activity (Lambertz et al., 2012; Robert McMaster et al., 2016). Other released molecules, LmS3a, a ribosomal protein, and lipophosphoglycan glycoconjugates (LPP) regulate T-cell activation, iNOS gene expression and nitric oxide production (Ouaissi and Ouaissi, 2005). Parasites, including Leishmania spp. and Trypanosoma cruzi, also alter exosomes content and release from the infected cells. In this way, they are able to regulate the crosstalk between host cells and thus the host response against infection. This evidence supports vesicles as an useful signal transmission mechanism between parasites, parasites and host cells, and from host cells to the surrounding environment (Silverman et al., 2010a; Hassani and Olivier, 2013; Coakley et al., 2015). Therefore, in leishmaniasis exosomes may be an alternative system for molecules delivery to different cell types, such as T cells, fibroblasts and hepatocytes, possibly leading to their function dysregulation.
Association between infectious diseases and cancer
Previous works reported a casual association between cancer and infectious disease including bacteria, virus and parasite infection (Machicado and Marcos, 2016; van Tong et al., 2017; Yasunaga and Matsuoka, 2018; Schwing et al., 2019). Pathological conditions due to infectious diseases contribute to tumour onset and on the other hand cancer cells and surrounding environment lead to a susceptible landscape for pathogen infection (Schwing et al., 2019). Pathogens promote several carcinogenic mechanisms including chronic inflammation, immune response modulation and a series of other events that may trigger further cell function alterations (Figure 2).
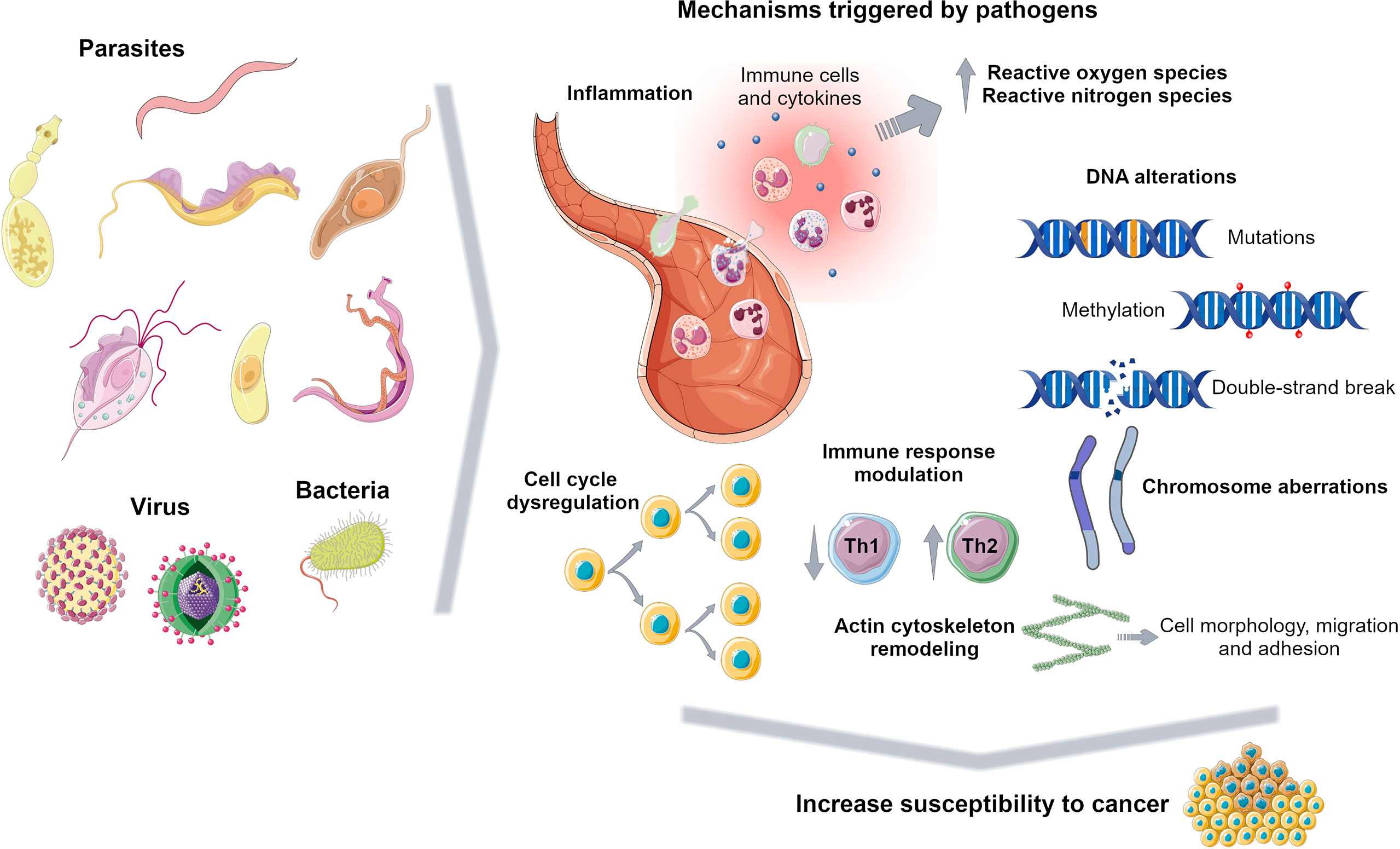
Figure 2 Carcinogenic mechanisms triggered by pathogens. Pathogens promote several events to survive in the host including inflammation, DNA alterations, immune response dysregulation and cell cycle modulation, increasing susceptibility to tumorigenesis. Figure created using Servier Medical Art images, licensed under a Creative Commons Attribution 3.0 Unported License; https://smart.servier.com/.
Infection-associated inflammation
Inflammation is a biological response to cellular damage due to injury or infection and its chronicity increases cancer risk (Shalapour and Karin, 2015). It enhances reactive oxygen and nitrogen species production leading to DNA damage (Machicado and Marcos, 2016; Schwing et al., 2019). For instance, it has been reported that liver fluke infection results in an overproduction of free radicals due to the inflammatory environment consisting of eosinophils, macrophages, neutrophils and the released cytokines. On one hand, a persistent oxidative stress condition triggers lipid peroxidation whose toxic products together with free radicals may induce DNA damage and dysregulate cell homeostasis (Andrade et al., 2012; Bahrami et al., 2014; Kim et al., 2016). On the other hand, Clonorchis sinensis-associated prolonged inflammation maintains elevated cytokines and NF-kB levels inducing further proinflammatory responses such as nitric oxide production, responsible for the DNA repair inhibition and the induction of COX-2 expression, involved in cell growth modulation (Kim et al., 2016). This persistent environment promotes cholangiocarcinoma development (Kim et al., 2016). Trichomonas vaginalis infection increases susceptibility to develop prostate cancer through a secreted protein triggering cell proliferation and inflammation, possibly contributing to angiogenesis (Twu et al., 2014). In leishmaniasis chronic inflammation due to parasite persistence orchestrates a tumour microenvironment characterized by hypoxia, altered expression of COX-2 and NF-kB-target genes (Gregory et al., 2008; Charpentier et al., 2016; Al-Kamel, 2017). However, Leishmania upregulates anti-inflammatory response reducing NF-Kb and inflammasome activation and increasing IL-10 to generate a safe niche for its survival (Mahanta et al., 2018; Afrin et al., 2019; Lecoeur et al., 2020; Parmar et al., 2020). Leishmania may disturb oxygen supply to the tissue lesions contributing to a complex scenario including proinflammatory and anti-inflammatory macrophages (Charpentier et al., 2016; Schatz et al., 2018; Saunders et al., 2021).
Genomic instability induced by infections, a malignancy promoter
Genomic instability, a carcinogenic mechanism, including point mutations, structural chromosomal aberrations and DNA strand breaks, can be caused by pathogen infections (van Tong et al., 2017). In fact, an association between parasitic infections and lymphoid neoplasia has been reported. Burkitt’s lymphoma (BL) cases are common in endemic malaria regions with high Plasmodium falciparum transmission. P. falciparum may induce C-Myc translocation in B cells predisposing patients to develop BL (Torgbor et al., 2014; van Tong et al., 2017). Previous evidence shows that missense mutations at TP53 gene were more frequent in patients with Schistosoma japonicum-induced rectal cancer than in non-schistosomiasis rectal cancer, whereas Schistosoma mansoni targets p53 altering its expression and may cause somatic mutations in BCL2 and C-Myc oncogenes inducing colorectal cancer (van Tong et al., 2017). In leishmaniasis ROS and RNS, produced as a host defence mechanism, may lead to DNA strand breaks and increase cancer risk. Mononuclear leukocytes from infected patients presented increase DNA damage compared to healthy subjects (Oliveira and Cecchini, 2000; Kocyigit et al., 2005; Almeida et al., 2013). These results suggest that pathogen-induced DNA damage play a key role in cellular transformation (van Tong et al., 2017).
Cell cycle regulation in pathogen infection and cancer progression
Cell cycle dysregulation, i.e. constant cell proliferation and apoptosis inhibition, upon pathogen infection is a frequent process that promotes cancer onset (Machicado and Marcos, 2016). Function of p53, a key player in cell cycle progression, is altered during Theileria spp. and Cryptosporidium parvum infections possibly due to its cellular mislocalization or continuous degradation favouring cell malignant transformation (Hayashida et al., 2013; Benamrouz et al., 2014). Alterations in APC and β-catenin genes, involved in Wnt signalling pathway, are altered in C. parvum infection affecting actin cytoskeleton organization and other cellular processes that might result in neoplasia development (Benamrouz et al., 2014). This cell cycle dysregulation is also observed in viral infection through the production of oncogenic factors. For example, Human T-cell leukaemia virus type 1 synthesizes Tax, a viral replication protein, which is responsible for the activation of Wnt and NF-Kb pathways and the inhibition of DNA repair resulting in cell malignant transformation. (Yasunaga and Matsuoka, 2018). In leishmaniasis the parasite modulates host cell survival and death; Leishmania through released factors or surface proteins, such as the lipophosphoglycan, hijack the apoptosis machinery leading to its inhibition in infected macrophages (Ouaissi and Ouaissi, 2005), whereas elevated oxidative stress during infection leads to the death of other immune cell types (Almeida et al., 2013).
Immune response evasion, a common survival strategy of pathogens and cancer cells
An immunological association between pathogen infection and cancer has been suggested (Ouaissi and Ouaissi, 2005; Silmon de Monerri and Kim, 2014; Machicado and Marcos, 2016; Schwing et al., 2019). Similar strategies are orchestrated by pathogens and tumour cells to evade host immune response enabling their survival and proliferation (Ouaissi and Ouaissi, 2005). Leishmania spp. immune-modulate host system activating Th2 response and reducing Th1 protective defence. In visceral leishmaniasis caused by L. chagasi this altered balance between Th1 and Th2 may cooperate to acute leukaemia development (de Vasconcelos et al., 2014). This immune environment favours infection progression avoiding pathogen killing and it increases susceptibility to tumour formation as well (Ouaissi and Ouaissi, 2005; Machicado and Marcos, 2016; Schwing et al., 2019). Moreover, previous evidence shows that T. cruzi and Leishmania spp. manage to kill immune cells and inhibit apoptosis of host cells. Alteration of apoptotic pathways is a strategy employed also by tumours to prevent their clearance by immune cells (Ouaissi and Ouaissi, 2005).
Pathogen coinfections
Association between infectious diseases and cancer development become more complex when simultaneous pathogen infections occur, possibly cooperating to tumorigenesis by triggering cell proliferation, inflammation and inducing genomic/epigenomic alterations (van Tong et al., 2017; Yasunaga and Matsuoka, 2018). For example, Strongiloides stercolaris, the pathogen agent of a chronic gastrointestinal parasitic infection, promotes proliferation of human T-cell leukaemia virus type 1-infected cells increasing the risk to develop adult T-cell leukaemia/lymphoma (Gabet et al., 2000; Yasunaga and Matsuoka, 2018). P. falciparum and EBV are associated with BL, a frequent children tumour in tropical Africa. BL has been diagnosed in hyperendemic malaria regions and EBV was first discovered in BL tumour biopsies (Torgbor et al., 2014). P. falciparum interacts with B cells and impairs their function leading to an uncontrolled proliferation including EBV-infected B cells and on the other hand the parasite inhibits EBV-specific T cell immunity reactivating virus cycle and promoting chromosome aberrations. This environment may lead to the onset and expansion of malignant B-cells and consequent BL development (van Tong et al., 2017). BL has been also reported in a human immunodeficiency virus (HIV)-infected subject presenting leishmaniasis (Boutros et al., 2006; Schwing et al., 2019). HIV-mediated immunosuppression facilitates parasite replication and dissemination, whereas Leishmania interferes with monocytes and macrophages normal function increasing dNTP levels, essential for efficient HIV replication and thus favouring HIV progression (Mock et al., 2012; Zijlstra, 2014). Moreover, few cases of Kaposi sarcoma and Leishmania infection in HIV-positive patients have been reported, all these patients presented typical lesions of Kaposi sarcoma with high Leishmania parasitaemia (Kopterides et al., 2007). No association with malignant transformation has been reported in cases of coinfection of Leishmania and other pathogens, such as P. falciparum and T. cruzi (Martinez et al., 2018).
DNA methylation, an epigenetic sign, linking infectious disease and cancer onset
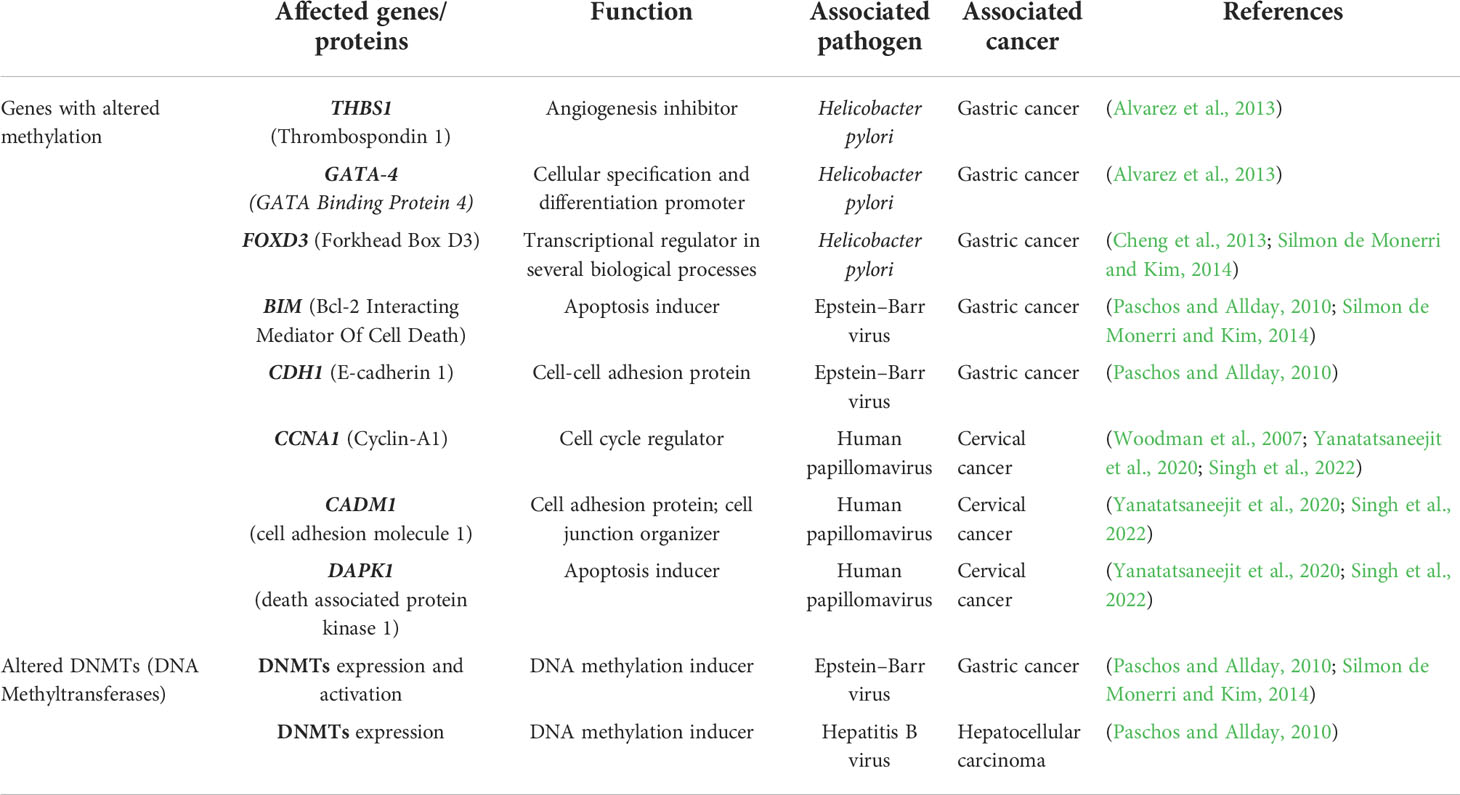
DNA methylation is a fascinating landscape that needs to be further explored in infectious diseases. Few works focused their attention on DNA methylation alterations upon pathogen infection highlighting their importance during host cell colonization and suggesting an increased predisposition to develop cancer. These aberrations may result in transcriptional dysregulation affecting normal cell function and the surrounding environment (Silmon de Monerri and Kim, 2014; Robert McMaster et al., 2016). H. pylori infection induces changes in DNA methylation at the promoter regions of THBS1, GATA-4 and FOXD3, associated with angiogenesis, cell differentiation and apoptosis, promoting gastric cancer development. Interestingly, some of these aberrations are long-lasting modifications that persist after eradication of the bacteria (Alvarez et al., 2013; Cheng et al., 2013; Silmon de Monerri and Kim, 2014). In EBV infection, activation of DNA methyltransferases and transcriptional regulation of BIM, an apoptosis inducer, and CDH1, a cell-cell adhesion protein, may also play a key role in gastric cancer onset (Paschos and Allday, 2010; Silmon de Monerri and Kim, 2014). Another correlation has been indicated between the presence of HBV in hepatocellular carcinoma and aberrant DNA methylation in host cells. In fact, a nuclear viral protein regulates the expression of DNMT1 and DNMT3A enzymes, relevant players in mechanisms that modulate the methylome (Paschos and Allday, 2010). Human papillomavirus (HPV) infection is considered an important risk factor for cervical cancer development. Two viral oncoproteins, E6 and E7, alter cellular DNA methylation of tumour suppressor genes, such as CCNA1, CADM1 and DAPK1, through the formation of a complex with DNMT1 and probably with transcription factors (Woodman et al., 2007; Yanatatsaneejit et al., 2020; Singh et al., 2022). HPV-linked methylation aberrations are useful for prognosis and identification of cervical cancer subtypes (Yang et al., 2020). As mentioned previously, it is well known the involvement of DNA methylation alterations, despite their aetiology, in tumours. DNA methylation alterations may dysregulate gene expression and affect a cascade of events promoting tumorigenesis (Feinberg and Tycko, 2004) and they are also considered specific tumour signatures useful for prevention and diagnosis since they precede mutations (Kulis and Esteller, 2010). However, cancer development is the result of the accumulation of genetic and epigenetic alterations (Kulis and Esteller, 2010). Table 1 summarizes the DNA methylation-related alterations detected in pathogen infection-associated cancer.
May DNA methylation alterations in leishmaniasis trigger tumorigenesis?
Long-term Leishmania infection causes chronic inflammation favouring accumulation of epigenetic and genetic mutations that can promote cancer development. During acute and chronic infection, amastigotes ensure their survival within phagolysosomes developing semi-quiescent and dormant stages, i.e. reducing or shutting down expensive metabolic processes and maintaining a slow or very slow growth (Saunders et al., 2021). Leishmania chronic infection and cancer lead to a susceptible environment allowing the progression of both diseases (Al-Kamel, 2017; Schwing et al., 2019). It is known that many cancers arise from infection sites (Al-Kamel, 2017). The pathophysiologic contribution of Leishmania spp. in cancer onset is not well elucidated but many patients with prior Leishmania spp. infection developed tumours (Al-Kamel, 2017). To note, it is believed that approximately 120 million people have a chronic infection (Saunders et al., 2021), thus monitoring infection parameters achieve great relevance to prevent and early diagnose cancer in endemic regions. The tumour cases reported in patients with leishmaniasis included basal and squamous cell carcinoma, lymphoma, leukaemia, hemangiosarcoma and hepatocellular carcinoma, summarized in Al-Kamel (Al-Kamel, 2017).
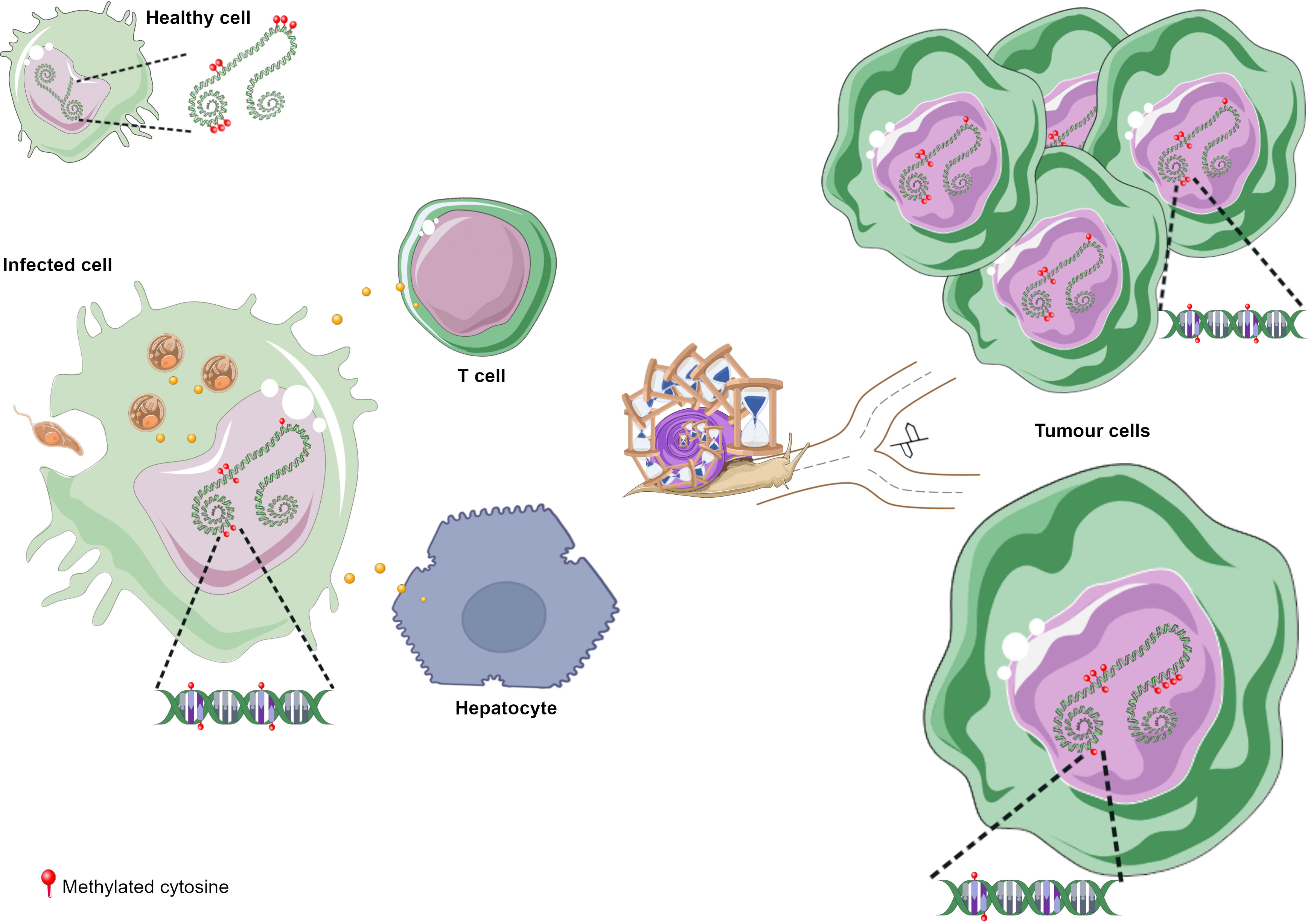
DNA methylation aberrations induced by Leishmania spp. infection may be a possible factor that trigger cell malignant transformation since DNA methylation alterations are frequent early events in cancer. It has been reported that a macrophage cell line infected with L. donovani displays DNA methylation changes in loci linked to pathways frequently altered in cancer (Marr et al., 2014). The affected genes were CTBP1, CTBP2, RASSF5, AXIN1, DVL2, STAT3 and WNT5A (Table 2) (Marr et al., 2014). Some of these gene-related pathways have been above described in pathogen infection-associated cancer. AXIN1, DVL2 and WNT5A are involved in Wnt signalling that regulates several processes such as proliferation, differentiation and adhesion. Wnt5A has been related to cancer-associated inflammation through macrophage recruitment. CTBPs corepressors are involved in cell cycle and WNT gene family regulation (Stankiewicz et al., 2014; Blevins et al., 2017). As aforementioned, Wnt pathway abnormal activation and mutations or mislocalization of its members are frequently observed in infectious diseases and are considered possible tumour promoters (Benamrouz et al., 2014; Mazzoni and Fearon, 2014; Asem et al., 2016; Yasunaga and Matsuoka, 2018; Mei et al., 2020). RASSF5 may be implicated in p53 activity regulation, a key cell cycle guardian frequently altered in infectious diseases leading to cancer onset (Hayashida et al., 2013; Benamrouz et al., 2014; Li et al., 2018). DNA methylation aberrations may explain the altered expression of these genes and thus signalling pathways dysregulation. In fact, these altered loci showed a significant differential methylation with a range from 3% to 9% between infected and non-infected cells (Marr et al., 2014). Notably, although these alterations are located in the gene body and not in regulatory regions, they may also modify gene expression. Long-term consequences of chronic infection in the epigenetic modulation may lead to further methylome alterations in the target cells such as T cells, hepatocytes and fibroblasts, and an increased number of infected cells than in previous phases of infection that may induce aberrations in neighbour cells, probably contributing to the occurrence of malignancy (Figure 3). Additional DNA methylation aberrations in target cells may be caused by Leishmania released molecules delivered by extracellular vesicles, whereas a higher number of infected macrophages may enhance parasite dissemination to visceral organs, resulting in amastigotes and/or exosome release towards other target cells. In agreement with this statement, few works reported cancer-specific methylation alterations several years before mature B-cell neoplasm (MBCN) diagnosis (Wong Doo et al., 2016; Georgiadis et al., 2017; Loi et al., 2019). In fact, in a prospective MBCN cohort analysis Loi et al. found a hypermethylation event at a CGI associated with SHANK1 gene (differential methylation of 3%) in blood samples collected around 10 years prior to diagnosis (Loi et al., 2019). This evidence supports the hypothesis that weak DNA methylation changes observed upon Leishmania spp. infection may enhance during long-term infection predisposing tumorigenesis (Figure 3). To further support the possible association between Leishmania spp. infection and cancer onset susceptibility, DNA methylation alterations at FLI1 promoter region, a gene dysregulated in melanoma, have been detected in macrophages infected with L. braziliensis in vitro and in fibroblasts from cutaneous lesion biopsies as well (Almeida et al., 2017; Ramani et al., 2017). To note, it is believed that epigenetic modulation, including DNA methylation, has a main role in fibroblasts transformation into cancer-associated fibroblasts (CAFs) and this process can be mediated by exosomes, a frequent vehicle used in parasite/host communication. CAFs are an essential component of tumour microenvironment and they can also infiltrate and metastasize together with tumour-specific cells (Ping et al., 2021). Cancer onset in leishmaniasis wounds could arise even several years after apparent healing reinforcing the relevance of chronic infections and the persistence of parasite-associated alterations in host cells. Some cases of basal cell carcinoma in patients with Leishmania infection have been reported. Therefore, cutaneous leishmaniasis can be considered a predisposing factor for skin malignancies (Suster and Ronnen, 1988; Morsy et al., 1992; Yavuzer et al., 2001; Mangoud et al., 2005; Asilian et al., 2012; Morsy, 2013; Schwing et al., 2019). Another type of tumour reported in patients with leishmaniasis is the T-cell lymphoma that may arise in consequence of chronic antigenic stimulation and immunosuppression due to leishmaniasis (Al-Kamel, 2017). Interestingly, Leishmania vesicles target T cells altering their function (Silverman et al., 2010b; Coakley et al., 2015). In 2003, a case report of a patient with hepatocellular carcinoma with pre-existing visceral leishmaniasis and chronic hepatitis C has been published (Precone et al., 2003; Kopterides et al., 2007). As aforementioned, it has been reported Leishmania/hepatocyte membrane interaction and gene expression alterations in these target cells (Rodrigues et al., 2019). Therefore, it is reasonable to wonder whether this interplay induces host DNA methylation changes leading to gene dysregulation, cell dysfunction and probably cell malignant transformation in long-term infections. Further studies are needed to elucidate a possible association between visceral leishmaniasis and cancer onset.
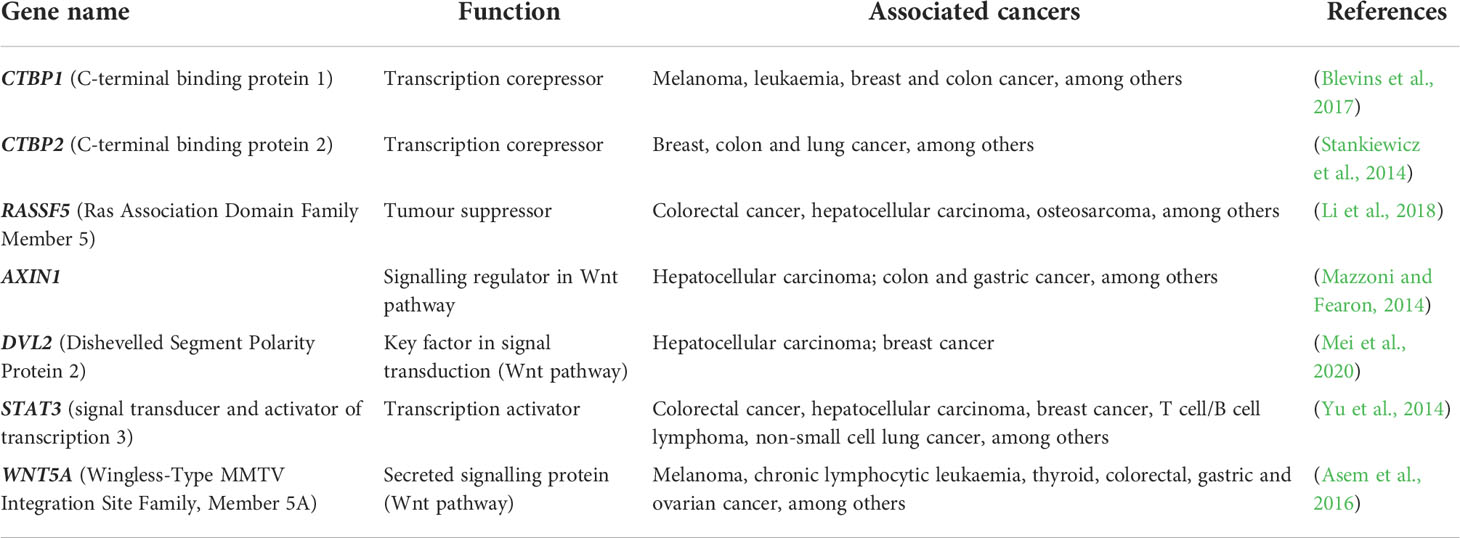
Table 2 Cancer pathway’s genes, associated with altered CpG sites methylation upon infection with L. donovani.
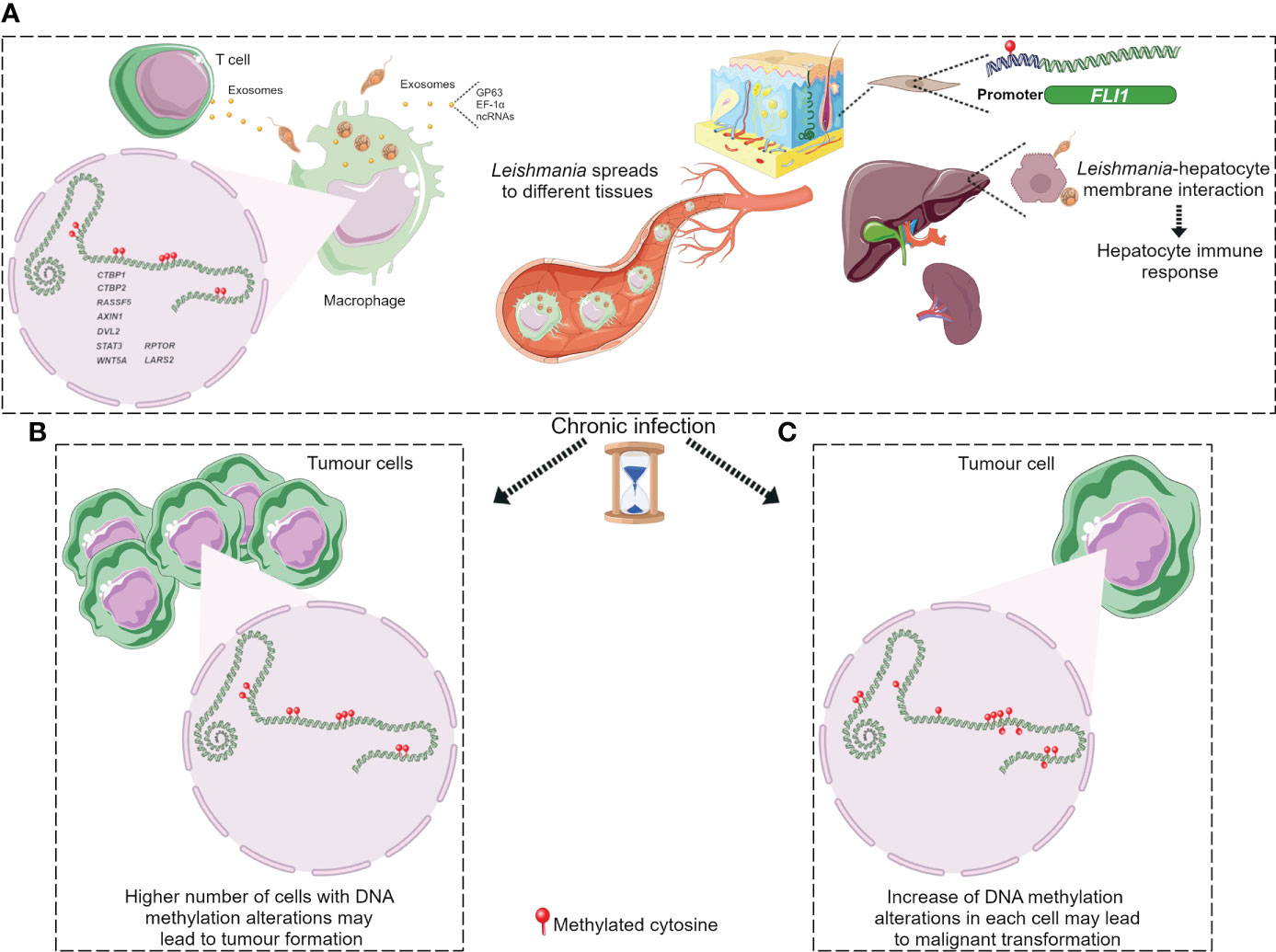
Figure 3 DNA methylation alterations upon Leishmania infection possibly leading to tumorigenesis. (A) Leishmania molecules, possibly delivered by exosomes, induce DNA methylation changes in macrophages and maybe in T cells. Parasite dissemination to different tissues may induce methylome alterations in other cell types, such as fibroblasts and hepatocytes, through Leishmania/host cell interaction. Cancer development may arise through different ways during chronic leishmaniasis: (B) increased number of cells with DNA methylation alterations; (C) additional methylome alterations in target cells. Figure created using Servier Medical Art images, licensed under a Creative Commons Attribution 3.0 Unported License; https://smart.servier.com/.
DNA methylation alterations induced by Leishmania spp. may result in an immune response dysregulation that leads to a non-correct surveillance and enables eventual malignant cells at the leishmanial lesion to elude immune detection (Al-Kamel, 2017). Leishmania infection activates mTOR pathway triggering cell proliferation and altering the balance of M1/M2 macrophages polarization, a biological process regulated by DNA methylation modifications and other mechanisms (Li et al., 2020). The increased M2 macrophage polarization represents an appropriate niche for Leishmania survival and on another hand it has a key role in angiogenesis, tumour formation and progression (Kumar et al., 2018; Yao et al., 2019; Zou et al., 2020). However, it is not elucidated how mTOR signalling is activated in leishmaniasis (Kumar et al., 2018). Marr et al. reported DNA methylation alterations in CpG sites associated with LARS2 and RPTOR genes, involved in the mTOR pathway (Marr et al., 2014). These authors also reported methylation alterations at IL-10, an M2 macrophage marker, frequently altered in inflammation processes and secreted by many tumour types to suppress T cells response (Ouaissi and Ouaissi, 2005; Marr et al., 2014). Figure 3 depicts the hypothesis described in this section regarding cancer onset in patients with leishmaniasis.
Discussion
In this review we propose a possible mechanism for cancer onset during leishmaniasis. The protozoan Leishmania manipulates the epigenome, including DNA methylation, of host cells to enable its survival and replication (Robert McMaster et al., 2016). Changes in the DNA methylation profile may lead to gene expression dysregulation and cell dysfunction. Several DNA methylation modifications due to Leishmania infection are associated with genes involved in cancer pathways such as Wnt signalling and cell cycle regulation, and in immune mechanisms including cytokines signalling and Natural killer cells-mediated cytotoxicity (Marr et al., 2014). Case reports describe cancer onset in patients with long term infection suggesting the persistence or even worsening of the cell and tissue alterations caused by Leishmania (Kopterides et al., 2007; Schwing et al., 2019). DNA methylation alterations are also early events during tumorigenesis and few events of DNA methylation changes can be found even 10 years before cancer onset (Wong Doo et al., 2016; Georgiadis et al., 2017; Loi et al., 2019). Considering these characteristics, it is plausible that the association between parasitic infection and cancer may reside in these epigenetic changes. Long-term Leishmania infection may lead to a high number of altered cells and further increase DNA methylation aberrations in host cells enhancing patient susceptibility to cancer onset. An increased number of infected macrophages may allow parasite dissemination and induce methylome alterations in neighbour cells. These alterations are possibly induced by Leishmania molecules delivered by exosomes to different cell types including T cells, fibroblasts and hepatocytes resulting in cell dysfunction. Further studies are necessary to explore parasite/host cells interplay, including non-immune cells, to elucidate the consequences of DNA methylation changes during infection. Since a correlation between host DNA methylation alterations and cancer risk has been reported in other pathogen diseases such as H. pylori, EBV, HBV and HPV infection, it can be expected a similar mechanism in leishmaniasis. The comprehension of these molecular aberrations and the association between leishmaniasis and cancer could be useful to monitor infected patients in constant follow up and thus the progression of both pathologies.
Data availability statement
The original contributions presented in the study are included in the article/supplementary material. Further inquiries can be directed to the corresponding author.
Author contributions
Conceptualization: AFV-B and PZ; literature search, AFV-B; writing original draft preparation: AFV-B; writing, review and editing: AFV-B, EL, and PZ; visualization: AFV-B; funding acquisition: AFV-B. All authors contributed to the article and approved the submitted version.
Funding
This study has been funded by a Research Grant [2021] from the European Society of Clinical Microbiology and Infectious Diseases (ESCMID) to AV-B.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Afrin, F., Khan, I., Hemeg, H. A. (2019). Leishmania-host interactions-an epigenetic paradigm. Front. Immunol. 10. doi: 10.3389/fimmu.2019.00492
Al-Kamel, M. A. N. (2017). Leishmaniasis and malignancy: A review and perspective. Clin. Skin Cancer 2 (1), 54–58. doi: 10.1016/j.clsc.2017.10.003
Almeida, B. F., Narciso, L. G., Melo, L. M., Preve, P. P., Bosco, A. M., Lima, V. M., et al. (2013). Leishmaniasis causes oxidative stress and alteration of oxidative metabolism and viability of neutrophils in dogs. Vet. J. 198 (3), 599–605. doi: 10.1016/j.tvjl.2013.08.024
Almeida, L., Silva, J. A., Andrade, V. M., Machado, P., Jamieson, S. E., Carvalho, E. M., et al. (2017). Analysis of expression of FLI1 and MMP1 in American cutaneous leishmaniasis caused by leishmania braziliensis infection. Infect. Genet. Evol. 49, 212–220. doi: 10.1016/j.meegid.2017.01.018
Alvarez, M. C., Ladeira, M. S., Scaletsky, I. C., Pedrazzoli, J., Jr., Ribeiro, M. L. (2013). Methylation pattern of THBS1, GATA-4, and HIC1 in pediatric and adult patients infected with helicobacter pylori. Dig Dis. Sci. 58 (10), 2850–2857. doi: 10.1007/s10620-013-2742-6
Ambrosi, C., Manzo, M., Baubec, T. (2017). Dynamics and context-dependent roles of DNA methylation. J. Mol. Biol. 429 (10), 1459–1475. doi: 10.1016/j.jmb.2017.02.008
Andrade, R. L., Dantas, A. F., Pimentel, L. A., Galiza, G. J., Carvalho, F. K., Costa, V. M., et al. (2012). Platynosomum fastosum-induced cholangiocarcinomas in cats. Vet. Parasitol. 190 (1-2), 277–280. doi: 10.1016/j.vetpar.2012.04.015
Asem, M. S., Buechler, S., Wates, R. B., Miller, D. L., Stack, M. S. (2016). Wnt5a signaling in cancer. Cancers (Basel) 8 (9). doi: 10.3390/cancers8090079
Asilian, A., Momeni, I., Khosravani, P. (2012). Basal cell carcinoma superimposed on a cutaneous leishmaniasis lesion in an immunocompromised patient. J. Res. Med. Sci. 17 (1), 108–110.
Bahrami, S., Esmaeilzadeh, S., Oryan, A. (2014). Role of oxidative stress in concomitant occurrence of fasciola gigantica and leiomyoma in cattle. Vet. Parasitol. 203 (1-2), 43–50. doi: 10.1016/j.vetpar.2014.02.047
Benamrouz, S., Conseil, V., Chabe, M., Praet, M., Audebert, C., Blervaque, R., et al. (2014). Cryptosporidium parvum-induced ileo-caecal adenocarcinoma and wnt signaling in a mouse model. Dis. Model. Mech. 7 (6), 693–700. doi: 10.1242/dmm.013292
Blevins, M. A., Huang, M., Zhao, R. (2017). The role of CtBP1 in oncogenic processes and its potential as a therapeutic target. Mol. Cancer Ther. 16 (6), 981–990. doi: 10.1158/1535-7163.MCT-16-0592
Boutros, N., Hawkins, D., Nelson, M., Lampert, I. A., Naresh, K. N. (2006). Burkitt lymphoma and leishmaniasis in the same tissue sample in an AIDS patient. Histopathology 48 (7), 880–881. doi: 10.1111/j.1365-2559.2006.02434.x
Chang, K. P., McGwire, B. S. (2002). Molecular determinants and regulation of leishmania virulence. Kinetoplastid Biol. Dis. 1 (1), 1. doi: 10.1186/1475-9292-1-1
Charpentier, T., Hammami, A., Stager, S. (2016). Hypoxia inducible factor 1alpha: A critical factor for the immune response to pathogens and leishmania. Cell Immunol. 309, 42–49. doi: 10.1016/j.cellimm.2016.06.002
Cheng, A. S., Li, M. S., Kang, W., Cheng, V. Y., Chou, J. L., Lau, S. S., et al. (2013). Helicobacter pylori causes epigenetic dysregulation of FOXD3 to promote gastric carcinogenesis. Gastroenterology 144 (1), 122–133.e129. doi: 10.1053/j.gastro.2012.10.002
Clark, S. J., Harrison, J., Molloy, P. L. (1997). Sp1 binding is inhibited by (m)Cp(m)CpG methylation. Gene 195 (1), 67–71. doi: 10.1016/s0378-1119(97)00164-9
Coakley, G., Maizels, R. M., Buck, A. H. (2015). Exosomes and other extracellular vesicles: The new communicators in parasite infections. Trends Parasitol. 31 (10), 477–489. doi: 10.1016/j.pt.2015.06.009
de Vasconcelos, G. M., Azevedo-Silva, F., Dos Santos Thuler, L. C., Pina, E. T., Souza, C. S., Calabrese, K., et al. (2014). The concurrent occurrence of leishmania chagasi infection and childhood acute leukemia in Brazil. Rev. Bras. Hematol. Hemoter 36 (5), 356–362. doi: 10.1016/j.bjhh.2014.07.013
Fadda, A., Gentilini, D., Moi, L., Barault, L., Leoni, V. P., Sulas, P., et al. (2018). Colorectal cancer early methylation alterations affect the crosstalk between cell and surrounding environment, tracing a biomarker signature specific for this tumor. Int. J. Cancer 143 (4), 907–920. doi: 10.1002/ijc.31380
Feinberg, A. P., Tycko, B. (2004). The history of cancer epigenetics. Nat. Rev. Cancer 4 (2), 143–153. doi: 10.1038/nrc1279
Fleischer, T., Frigessi, A., Johnson, K. C., Edvardsen, H., Touleimat, N., Klajic, J., et al. (2014). Genome-wide DNA methylation profiles in progression to in situ and invasive carcinoma of the breast with impact on gene transcription and prognosis. Genome Biol. 15 (8), 435. doi: 10.1186/PREACCEPT-2333349012841587
Gabet, A. S., Mortreux, F., Talarmin, A., Plumelle, Y., Leclercq, I., Leroy, A., et al. (2000). High circulating proviral load with oligoclonal expansion of HTLV-1 bearing T cells in HTLV-1 carriers with strongyloidiasis. Oncogene 19 (43), 4954–4960. doi: 10.1038/sj.onc.1203870
Gal-Yam, E. N., Saito, Y., Egger, G., Jones, P. A. (2008). Cancer epigenetics: modifications, screening, and therapy. Annu. Rev. Med. 59, 267–280. doi: 10.1146/annurev.med.59.061606.095816
Georgiadis, P., Liampa, I., Hebels, D. G., Krauskopf, J., Chatziioannou, A., Valavanis, I., et al. (2017). Evolving DNA methylation and gene expression markers of b-cell chronic lymphocytic leukemia are present in pre-diagnostic blood samples more than 10 years prior to diagnosis. BMC Genomics 18 (1), 728. doi: 10.1186/s12864-017-4117-4
Gómez-Díaz, E., Jordà, M., Peinado, M., Rivero, A. (2012). Epigenetics of host–pathogen interactions: The road ahead and the road behind. PloS Pathog. 8, e1003007. doi: 10.1371/journal.ppat.1003007
Gregory, D. J., Sladek, R., Olivier, M., Matlashewski, G. (2008). Comparison of the effects of leishmania major or leishmania donovani infection on macrophage gene expression. Infect. Immun. 76 (3), 1186–1192. doi: 10.1128/IAI.01320-07
Guo, W., Zhu, L., Yu, M., Zhu, R., Chen, Q., Wang, Q. (2018). A five-DNA methylation signature act as a novel prognostic biomarker in patients with ovarian serous cystadenocarcinoma. Clin. Epigenet. 10 (1), 142. doi: 10.1186/s13148-018-0574-0
Guo, W., Zhu, L., Zhu, R., Chen, Q., Wang, Q., Chen, J. Q. (2019). A four-DNA methylation biomarker is a superior predictor of survival of patients with cutaneous melanoma. Elife 8. doi: 10.7554/eLife.44310
Hari Dass, S. A., Vyas, A. (2014). Toxoplasma gondii infection reduces predator aversion in rats through epigenetic modulation in the host medial amygdala. Mol. Ecol. 23 (24), 6114–6122. doi: 10.1111/mec.12888
Hassani, K., Olivier, M. (2013). Immunomodulatory impact of leishmania-induced macrophage exosomes: a comparative proteomic and functional analysis. PloS Negl. Trop. Dis. 7 (5), e2185. doi: 10.1371/journal.pntd.0002185
Hayashida, K., Kajino, K., Hattori, M., Wallace, M., Morrison, I., Greene, M. I., et al. (2013). MDM2 regulates a novel form of incomplete neoplastic transformation of theileria parva infected lymphocytes. Exp. Mol. Pathol. 94 (1), 228–238. doi: 10.1016/j.yexmp.2012.08.008
Heberle, E., Bardet, A. F. (2019). Sensitivity of transcription factors to DNA methylation. Essays Biochem. 63 (6), 727–741. doi: 10.1042/EBC20190033
Kausalya, S., Malla, N., Ganguly, N. K., Mahajan, R. C. (1993). Leishmania donovani: in vitro evidence of hepatocyte damage by kupffer cells and immigrant macrophages in a murine model. Exp. Parasitol. 77 (3), 326–333. doi: 10.1006/expr.1993.1090
Kim, T. S., Pak, J. H., Kim, J. B., Bahk, Y. Y. (2016). Clonorchis sinensis, an oriental liver fluke, as a human biological agent of cholangiocarcinoma: a brief review. BMB Rep. 49 (11), 590–597. doi: 10.5483/bmbrep.2016.49.11.109
Kocyigit, A., Keles, H., Selek, S., Guzel, S., Celik, H., Erel, O. (2005). Increased DNA damage and oxidative stress in patients with cutaneous leishmaniasis. Mutat. Res. 585 (1-2), 71–78. doi: 10.1016/j.mrgentox.2005.04.012
Kopterides, P., Mourtzoukou, E. G., Skopelitis, E., Tsavaris, N., Falagas, M. E. (2007). Aspects of the association between leishmaniasis and malignant disorders. Trans. R Soc. Trop. Med. Hyg. 101 (12), 1181–1189. doi: 10.1016/j.trstmh.2007.08.003
Kulis, M., Esteller, M. (2010). DNA Methylation and cancer. Adv. Genet. 70, 27–56. doi: 10.1016/B978-0-12-380866-0.60002-2
Kumar, A., Das, S., Mandal, A., Verma, S., Abhishek, K., Kumar, A., et al. (2018). Leishmania infection activates host mTOR for its survival by M2 macrophage polarization. Parasite Immunol. 40 (11), e12586. doi: 10.1111/pim.12586
Lambertz, U., Oviedo Ovando, M. E., Vasconcelos, E. J., Unrau, P. J., Myler, P. J., Reiner, N. E. (2015). Small RNAs derived from tRNAs and rRNAs are highly enriched in exosomes from both old and new world leishmania providing evidence for conserved exosomal RNA packaging. BMC Genomics 16, 151. doi: 10.1186/s12864-015-1260-7
Lambertz, U., Silverman, J. M., Nandan, D., McMaster, W. R., Clos, J., Foster, L. J., et al. (2012). Secreted virulence factors and immune evasion in visceral leishmaniasis. J. Leukoc. Biol. 91 (6), 887–899. doi: 10.1189/jlb.0611326
Lecoeur, H., Prina, E., Rosazza, T., Kokou, K., N'Diaye, P., Aulner, N., et al. (2020). Targeting macrophage histone H3 modification as a leishmania strategy to dampen the NF-kappaB/NLRP3-Mediated inflammatory response. Cell Rep. 30 (6), 1870–1882.e1874. doi: 10.1016/j.celrep.2020.01.030
Lee, H. J., Hore, T. A., Reik, W. (2014). Reprogramming the methylome: erasing memory and creating diversity. Cell Stem Cell 14 (6), 710–719. doi: 10.1016/j.stem.2014.05.008
Li, S., Teng, J., Li, H., Chen, F., Zheng, J. (2018). The emerging roles of RASSF5 in human malignancy. Anticancer Agents Med. Chem. 18 (3), 314–322. doi: 10.2174/1871520617666170327120747
Liu, D., Uzonna, J. E. (2012). The early interaction of leishmania with macrophages and dendritic cells and its influence on the host immune response. Front. Cell Infect. Microbiol. 2. doi: 10.3389/fcimb.2012.00083
Li, X., Zhang, Y., Pei, W., Zhang, M., Yang, H., Zhong, M., et al. (2020). LncRNA Dnmt3aos regulates Dnmt3a expression leading to aberrant DNA methylation in macrophage polarization. FASEB J. 34 (4), 5077–5091. doi: 10.1096/fj.201902379R
Loi, E., Moi, L., Fadda, A., Satta, G., Zucca, M., Sanna, S., et al. (2019). Methylation alteration of SHANK1 as a predictive, diagnostic and prognostic biomarker for chronic lymphocytic leukemia. Oncotarget 10 (48), 4987–5002. doi: 10.18632/oncotarget.27080
Loi, E., Zavattari, C., Tommasi, A., Moi, L., Canale, M., Po, A., et al. (2022). HOXD8 hypermethylation as a fully sensitive and specific biomarker for biliary tract cancer detectable in tissue and bile samples. Br. J. Cancer 126, 1783–1794. doi: 10.1038/s41416-022-01738-1
Luo, Y., Wong, C. J., Kaz, A. M., Dzieciatkowski, S., Carter, K. T., Morris, S. M., et al. (2014). Differences in DNA methylation signatures reveal multiple pathways of progression from adenoma to colorectal cancer. Gastroenterology 147 (2), 418–429.e418. doi: 10.1053/j.gastro.2014.04.039
Machicado, C., Marcos, L. A. (2016). Carcinogenesis associated with parasites other than schistosoma, opisthorchis and clonorchis: A systematic review. Int. J. Cancer 138 (12), 2915–2921. doi: 10.1002/ijc.30028
Mahanta, A., Ganguli, P., Barah, P., Sarkar, R. R., Sarmah, N., Phukan, S., et al. (2018). Integrative approaches to understand the mastery in manipulation of host cytokine networks by protozoan parasites with emphasis on plasmodium and leishmania species. Front. Immunol. 9. doi: 10.3389/fimmu.2018.00296
Mangoud, A. M., Sanad, E. M., Fouad, M. A., Morsy, T. A. (2005). Proliferative changes of epidermal cells in lesions of cutaneous leishmaniasis. J. Egypt Soc. Parasitol. 35 (3), 761–772.
Marr, A. K., MacIsaac, J. L., Jiang, R., Airo, A. M., Kobor, M. S., McMaster, W. R. (2014). Leishmania donovani infection causes distinct epigenetic DNA methylation changes in host macrophages. PloS Pathog. 10 (10), e1004419. doi: 10.1371/journal.ppat.1004419
Martinez, D. Y., Verdonck, K., Kaye, P. M., Adaui, V., Polman, K., Llanos-Cuentas, A., et al. (2018). Tegumentary leishmaniasis and coinfections other than HIV. PloS Negl. Trop. Dis. 12 (3), e0006125. doi: 10.1371/journal.pntd.0006125
Mazzoni, S. M., Fearon, E. R. (2014). AXIN1 and AXIN2 variants in gastrointestinal cancers. Cancer Lett. 355 (1), 1–8. doi: 10.1016/j.canlet.2014.09.018
Mei, J., Yang, X., Xia, D., Zhou, W., Gu, D., Wang, H., et al. (2020). Systematic summarization of the expression profiles and prognostic roles of the dishevelled gene family in hepatocellular carcinoma. Mol. Genet. Genomic Med. 8 (9), e1384. doi: 10.1002/mgg3.1384
Micevic, G., Theodosakis, N., Bosenberg, M. (2017). Aberrant DNA methylation in melanoma: biomarker and therapeutic opportunities. Clin. Epigenet. 9, 34. doi: 10.1186/s13148-017-0332-8
Mock, D. J., Hollenbaugh, J. A., Daddacha, W., Overstreet, M. G., Lazarski, C. A., Fowell, D. J., et al. (2012). Leishmania induces survival, proliferation and elevated cellular dNTP levels in human monocytes promoting acceleration of HIV co-infection. PLoS Pathog. 8 (4), e1002635. doi: 10.1371/journal.ppat.1002635
Morsy, T. A. (2013). Cutaneous leishmaniasis predisposing to human skin cancer: forty years local and regional studies. J. Egypt Soc. Parasitol. 43 (3), 629–648. doi: 10.12816/0006420
Morsy, T. A., Mangoud, A. M., el-Sebai, M. M., al Seghayer, S. M. (1992). Cutaneous leishmaniasis as a possible predisposing factor for skin malignancy. J. Egypt Soc. Parasitol. 22 (3), 599–602.
Oliveira, F. J., Cecchini, R. (2000). Oxidative stress of liver in hamsters infected with leishmania (L.) chagasi. J. Parasitol. 86 (5), 1067–1072. doi: 10.1645/0022-3395(2000)086[1067:OSOLIH]2.0.CO;2
Ouaissi, A., Ouaissi, M. (2005). Molecular basis of trypanosoma cruzi and leishmania interaction with their host(s): exploitation of immune and defense mechanisms by the parasite leading to persistence and chronicity, features reminiscent of immune system evasion strategies in cancer diseases. Arch. Immunol. Ther. Exp. (Warsz) 53 (2), 102–114.
Parmar, N., Chandrakar, P., Kar, S. (2020). Leishmania donovani subverts host immune response by epigenetic reprogramming of macrophage M(Lipopolysaccharides + IFN-gamma)/M(IL-10) polarization. J. Immunol. 204 (10), 2762–2778. doi: 10.4049/jimmunol.1900251
Paschos, K., Allday, M. J. (2010). Epigenetic reprogramming of host genes in viral and microbial pathogenesis. Trends Microbiol. 18 (10), 439–447. doi: 10.1016/j.tim.2010.07.003
Peacock, C. S., Seeger, K., Harris, D., Murphy, L., Ruiz, J. C., Quail, M. A., et al. (2007). Comparative genomic analysis of three leishmania species that cause diverse human disease. Nat. Genet. 39 (7), 839–847. doi: 10.1038/ng2053
Ping, Q., Yan, R., Cheng, X., Wang, W., Zhong, Y., Hou, Z., et al. (2021). Cancer-associated fibroblasts: overview, progress, challenges, and directions. Cancer Gene Ther. 28 (9), 984–999. doi: 10.1038/s41417-021-00318-4
Precone, D. F., Stornaiuolo, G., Galante, D., Amato, A., Gradoni, L., Gaeta, G. B. (2003). Case report: effect of antileishmanial treatment on hepatitis c viraemia in a visceral leishmaniasis patient with chronic hepatitis c. Trans. R Soc. Trop. Med. Hyg 97 (5), 559–560. doi: 10.1016/s0035-9203(03)80028-7
Ramani, N., Aung, P. P., Hwu, W. J., Nagarajan, P., Tetzlaff, M. T., Curry, J. L., et al. (2017). Aberrant expression of FLI-1 in melanoma. J. Cutan Pathol. 44 (9), 790–793. doi: 10.1111/cup.12979
Reithinger, R., Lambson, B. E., Barker, D. C., Counihan, H., Espinoza, C. J., Gonzalez, J. S., et al. (2002). Leishmania (Viannia) spp. dissemination and tissue tropism in naturally infected dogs (Canis familiaris). Trans. R Soc. Trop. Med. Hyg 96 (1), 76–78. doi: 10.1016/s0035-9203(02)90249-x
Robert McMaster, W., Morrison, C. J., Kobor, M. S. (2016). Epigenetics: A new model for intracellular parasite-host cell regulation. Trends Parasitol. 32 (7), 515–521. doi: 10.1016/j.pt.2016.04.002
Rodrigues, A., Alexandre-Pires, G., Valerio-Bolas, A., Santos-Mateus, D., Rafael-Fernandes, M., Pereira, M. A., et al. (2019). Dog hepatocytes are key effector cells in the liver innate immune response to leishmania infantum. Parasitology 146 (6), 753–764. doi: 10.1017/S0031182018002068
Roy, G., Brar, H. K., Muthuswami, R., Madhubala, R. (2020). Epigenetic regulation of defense genes by histone deacetylase1 in human cell line-derived macrophages promotes intracellular survival of leishmania donovani. PloS Negl. Trop. Dis. 14 (4), e0008167. doi: 10.1371/journal.pntd.0008167
Saavedra, K. P., Brebi, P. M., Roa, J. C. (2012). Epigenetic alterations in preneoplastic and neoplastic lesions of the cervix. Clin. Epigenet. 4 (1), 13. doi: 10.1186/1868-7083-4-13
Saunders, E. C., Sernee, M. F., Ralton, J. E., McConville, M. J. (2021). Metabolic stringent response in intracellular stages of leishmania. Curr. Opin. Microbiol. 63, 126–132. doi: 10.1016/j.mib.2021.07.007
Schatz, V., Neubert, P., Rieger, F., Jantsch, J. (2018). Hypoxia, hypoxia-inducible factor-1alpha, and innate antileishmanial immune responses. Front. Immunol. 9. doi: 10.3389/fimmu.2018.00216
Schubeler, D. (2015). Function and information content of DNA methylation. Nature 517 (7534), 321–326. doi: 10.1038/nature14192
Schwing, A., Pomares, C., Majoor, A., Boyer, L., Marty, P., Michel, G. (2019). Leishmania infection: Misdiagnosis as cancer and tumor-promoting potential. Acta Trop. 197, 104855. doi: 10.1016/j.actatropica.2018.12.010
Seblova, V., Myskova, J., Hlavacova, J., Votypka, J., Antoniou, M., Volf, P. (2015). Natural hybrid of leishmania infantum/L. donovani: development in phlebotomus tobbi, p. perniciosus and lutzomyia longipalpis and comparison with non-hybrid strains differing in tissue tropism. Parasit Vectors 8, 605. doi: 10.1186/s13071-015-1217-3
Shalapour, S., Karin, M. (2015). Immunity, inflammation, and cancer: an eternal fight between good and evil. J. Clin. Invest. 125 (9), 3347–3355. doi: 10.1172/JCI80007
Silmon de Monerri, N. C., Kim, K. (2014). Pathogens hijack the epigenome: A new twist on host-pathogen interactions. Am. J. Pathol. 184 (4), 897–911. doi: 10.1016/j.ajpath.2013.12.022
Silverman, J. M., Clos, J., de'Oliveira, C. C., Shirvani, O., Fang, Y., Wang, C., et al. (2010a). An exosome-based secretion pathway is responsible for protein export from leishmania and communication with macrophages. J. Cell Sci. 123 (Pt 6), 842–852. doi: 10.1242/jcs.056465
Silverman, J. M., Clos, J., Horakova, E., Wang, A. Y., Wiesgigl, M., Kelly, I., et al. (2010b). Leishmania exosomes modulate innate and adaptive immune responses through effects on monocytes and dendritic cells. J. Immunol. 185 (9), 5011–5022. doi: 10.4049/jimmunol.1000541
Singh, P., Chalertpet, K., Sukbhattee, J., Wongmanee, N., Suwannakart, P., Yanatatsaneejit, P. (2022). Association between promoter methylation and gene expression of CGB3 and NOP56 in HPV-infected cervical cancer cells. BioMed. Rep. 16 (1), 1. doi: 10.3892/br.2021.1484
Skvortsova, K., Stirzaker, C., Taberlay, P. (2019). The DNA methylation landscape in cancer. Essays Biochem. 63 (6), 797–811. doi: 10.1042/EBC20190037
Stankiewicz, T. R., Gray, J. J., Winter, A. N., Linseman, D. A. (2014). C-terminal binding proteins: central players in development and disease. Biomol Concepts 5 (6), 489–511. doi: 10.1515/bmc-2014-0027
Suster, S., Ronnen, M. (1988). Basal cell carcinoma arising in a leishmania scar. Int. J. Dermatol. 27 (3), 175–176. doi: 10.1111/j.1365-4362.1988.tb04924.x
Suter, C. M., Martin, D. I., Ward, R. L. (2004). Hypomethylation of L1 retrotransposons in colorectal cancer and adjacent normal tissue. Int. J. Colorectal Dis. 19 (2), 95–101. doi: 10.1007/s00384-003-0539-3
Torgbor, C., Awuah, P., Deitsch, K., Kalantari, P., Duca, K. A., Thorley-Lawson, D. A. (2014). A multifactorial role for p. falciparum malaria in endemic burkitt's lymphoma pathogenesis. PloS Pathog. 10 (5), e1004170. doi: 10.1371/journal.ppat.1004170
Twu, O., Dessi, D., Vu, A., Mercer, F., Stevens, G. C., de Miguel, N., et al. (2014). Trichomonas vaginalis homolog of macrophage migration inhibitory factor induces prostate cell growth, invasiveness, and inflammatory responses. Proc. Natl. Acad. Sci. U.S.A. 111 (22), 8179–8184. doi: 10.1073/pnas.1321884111
van Tong, H., Brindley, P. J., Meyer, C. G., Velavan, T. P. (2017). Parasite infection, carcinogenesis and human malignancy. EBioMedicine 15, 12–23. doi: 10.1016/j.ebiom.2016.11.034
Vega-Benedetti, A. F., Loi, E., Moi, L., Orru, S., Ziranu, P., Pretta, A., et al. (2020). Colorectal cancer early detection in stool samples tracing CpG islands methylation alterations affecting gene expression. Int. J. Mol. Sci. 21 (12), 1–16. doi: 10.3390/ijms21124494
Vega-Benedetti, A. F., Loi, E., Moi, L., Restivo, A., Cabras, F., Deidda, S., et al. (2022). Colorectal cancer promoter methylation alteration affects the expression of glutamate ionotropic receptor AMPA type subunit 4 alternative isoforms potentially relevant in colon tissue. Hum. Cell. 35 (1), 310–319. doi: 10.1007/s13577-021-00640-x
Wang, Y., Deng, H., Xin, S., Zhang, K., Shi, R., Bao, X. (2019). Prognostic and predictive value of three DNA methylation signatures in lung adenocarcinoma. Front. Genet. 10. doi: 10.3389/fgene.2019.00349
WHO (2020). Leishmaniasis (Geneva, Switzerland: World Health Organization). Available at: https://www.who.int/news-room/fact-sheets/detail/leishmaniasis. Geneva, Switzerland
Wong Doo, N., Makalic, E., Joo, J. E., Vajdic, C. M., Schmidt, D. F., Wong, E. M., et al. (2016). Global measures of peripheral blood-derived DNA methylation as a risk factor in the development of mature b-cell neoplasms. Epigenomics 8 (1), 55–66. doi: 10.2217/epi.15.97
Woodman, C. B., Collins, S. I., Young, L. S. (2007). The natural history of cervical HPV infection: unresolved issues. Nat. Rev. Cancer 7 (1), 11–22. doi: 10.1038/nrc2050
Yanatatsaneejit, P., Chalertpet, K., Sukbhattee, J., Nuchcharoen, I., Phumcharoen, P., Mutirangura, A. (2020). Promoter methylation of tumor suppressor genes induced by human papillomavirus in cervical cancer. Oncol. Lett. 20 (1), 955–961. doi: 10.3892/ol.2020.11625
Yang, S., Wu, Y., Wang, S., Xu, P., Deng, Y., Wang, M., et al. (2020). HPV-related methylation-based reclassification and risk stratification of cervical cancer. Mol. Oncol. 14 (9), 2124–2141. doi: 10.1002/1878-0261.12709
Yao, Y., Xu, X. H., Jin, L. (2019). Macrophage polarization in physiological and pathological pregnancy. Front. Immunol. 10. doi: 10.3389/fimmu.2019.00792
Yasunaga, J. I., Matsuoka, M. (2018). Oncogenic spiral by infectious pathogens: Cooperation of multiple factors in cancer development. Cancer Sci. 109 (1), 24–32. doi: 10.1111/cas.13443
Yavuzer, R., Akyurek, N., Ozmen, S., Demirtas, Y., Ataoglu, O. (2001). Leishmania cutis with b-cell cutaneous hyperplasia. Plast. Reconstr Surg. 108 (7), 2177–2178. doi: 10.1097/00006534-200112000-00087
Yin, Y., Morgunova, E., Jolma, A., Kaasinen, E., Sahu, B., Khund-Sayeed, S., et al. (2017). Impact of cytosine methylation on DNA binding specificities of human transcription factors. Science 356 (6337). doi: 10.1126/science.aaj2239
Yu, H., Lee, H., Herrmann, A., Buettner, R., Jove, R. (2014). Revisiting STAT3 signalling in cancer: New and unexpected biological functions. Nat. Rev. Cancer 14 (11), 736–746. doi: 10.1038/nrc3818
Zijlstra, E. E. (2014). PKDL and other dermal lesions in HIV co-infected patients with leishmaniasis: Review of clinical presentation in relation to immune responses. PLoS Negl. Trop. Dis. 8 (11), e3258. doi: 10.1371/journal.pntd.0003258
Keywords: DNA methylation alterations, CpG sites, Leishmania infection, chronic infection, cancer onset, pathogen-associated cancer, Leishmania/host cell interaction
Citation: Vega-Benedetti AF, Loi E and Zavattari P (2022) DNA methylation alterations caused by Leishmania infection may generate a microenvironment prone to tumour development. Front. Cell. Infect. Microbiol. 12:984134. doi: 10.3389/fcimb.2022.984134
Received: 01 July 2022; Accepted: 10 August 2022;
Published: 29 August 2022.
Edited by:
Xing-Quan Zhu, Shanxi Agricultural University, ChinaReviewed by:
Jun-Jun He, Chinese Academy of Agricultural Sciences (CAAS), ChinaShuai Wang, Xinxiang Medical University, China
Zhao-Rong Lun, Sun Yat-sen University, China
Copyright © 2022 Vega-Benedetti, Loi and Zavattari. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ana Florencia Vega-Benedetti, YW5hZi52ZWdhYkB1bmljYS5pdA==
 Ana Florencia Vega-Benedetti
Ana Florencia Vega-Benedetti Eleonora Loi
Eleonora Loi Patrizia Zavattari
Patrizia Zavattari