- 1Department of Pathogeny Biology, College of Basic Medical Sciences, Jilin University, Changchun, China
- 2Signature Research Program in Emerging Infectious Diseases, Duke – National University of Singapore (NUS) Graduate Medical School, Singapore, Singapore
- 3School of Medicine and Public Health, The University of Newcastle, Newcastle, NSW, Australia
- 4Viruses, Infections/Immunity, Vaccines and Asthma, Hunter Medical Research Institute, Newcastle, NSW, Australia
Humanity has experienced four major pandemics since the twentieth century, with the 1918 Spanish flu, the 2002 severe acute respiratory syndrome (SARS), the 2009 swine flu, and the 2019 coronavirus disease (COVID)-19 pandemics having the most important impact in human health. The 1918 Spanish flu caused unprecedented catastrophes in the recorded human history, with an estimated death toll between 50 – 100 million. While the 2002 SARS and 2009 swine flu pandemics caused approximately 780 and 280,000 deaths, respectively, the current COVID-19 pandemic has resulted in > 6 million deaths globally at the time of writing. COVID-19, instigated by the SARS – coronavirus-2 (SARS-CoV-2), causes unprecedented challenges in all facets of our lives, and never before brought scientists of all fields together to focus on this singular topic. While for the past 50 years research have been heavily focused on viruses themselves, we now understand that the host immune responses are just as important in determining the pathogenesis and outcomes of infection. Research in innate immune mechanisms is crucial in understanding all aspects of host antiviral programmes and the mechanisms underpinning virus-host interactions, which can be translated to the development of effective therapeutic avenues. This review summarizes what is known and what remains to be explored in the innate immune responses to influenza viruses and SARS-CoVs, and virus-host interactions in driving disease pathogenesis. This hopefully will encourage discussions and research on the unanswered questions, new paradigms, and antiviral strategies against these emerging infectious pathogens before the next pandemic occurs.
Introduction
Influenza A viruses (IAVs) and severe acute respiratory syndrome – coronaviruses (SARS-CoVs) are both regarded as the most important respiratory infectious pathogens in human population. Their ever-changing nature constantly escapes from our current therapeutics, while emerging into novel viruses with unpredictable lethality. We have little immunity to these new viruses, and when they evolve and if they become pandemic, the consequences would be unthinkable. The 2009 H1N1 pandemic caused devastating effects, although by sheer luck the virus was not a lethal virus. The International Health Regulations Committee (World Health Organization) concluded in their 2010 report that “the world is ill-prepared to respond to a severe influenza pandemic or to any similarly global, sustained and threatening public health emergency.” (WHO, 2011). Ten years later, the unexpected appearance of the novel SARS-CoV-2 and CoV disease 2019 (COVID-19) pandemic has again emphasised our lack of preparedness and apart from costly public health measures a complete absence of effective antiviral therapeutics. Despite serious concerns of vaccine mismatch and escape mutants (Australian Government, D.o.H., 2017; Paules et al., 2018), vaccination remains the most important preventative measure against IAVs and SARS-CoV-2. Nevertheless, there is also a need to develop immune-targeting therapeutics that can be immediately deployed at the initial phase of the pandemic before vaccines become available and accessible to all nations.
Human innate immunity is an ancient immune architecture that has evolved to protect the host from microbial insults. The host pattern recognition receptors (PRRs) recognise specific genetic features common to viruses, and these factors are essential barriers to most respiratory viruses and cross-species viral transmission.
IAVs and SARS-CoVs, in agreement with the Red Queen Hypothesis, have also evolved to withstand the selective pressure from the host immune responses (Brockhurst et al., 2014). These viruses produce not only essential proteins for viral replication but also factors that effectively weaken the host antiviral responses and enhances their virulence and survival. These virus-host interactions are the primary drivers of severe diseases in the host. While rapid advances in knowledge in the host innate antiviral responses and viral infections has been achieved by fundamental discoveries, how virus-host interactions mediate dysregulated immune responses and severe diseases remain to be further defined. Understanding these mechanisms will provide important insights in the development of effective antiviral therapeutics against these infectious pathogens.
In this Review, we summarize what the host antiviral networks have taught us, how the viruses compromise our defensive responses, and current unsolved mysteries in infection and innate immunity.
Intracellular Sentinels and the Enablers of the Antiviral Responses
IAVs and CoVs primarily infect human airway epithelial cells for viral replication. Human IAVs specifically bind with host cell surface glycoproteins carrying terminal α2,6 linked sialic acid residues, whereas avian IAVs bind to that with α2,3 linked sialic acid residues (Ito et al., 1997; Matrosovich et al., 2004). In contrast, SARS-CoVs gain cell entry by binding to angiotensin-converting enzyme (ACE)2 on airways epithelial cells (Wang Q et al., 2020; Scialo et al., 2020). IAVs and SARS-CoVs also infect alveolar type II pneumocytes and cause viral pneumonia that often requires hospitalization (Weinheimer et al., 2012; Wahl et al., 2021). Host innate immune responses initiated by these airway epithelial cells provide critical first line of defence against viral infections.
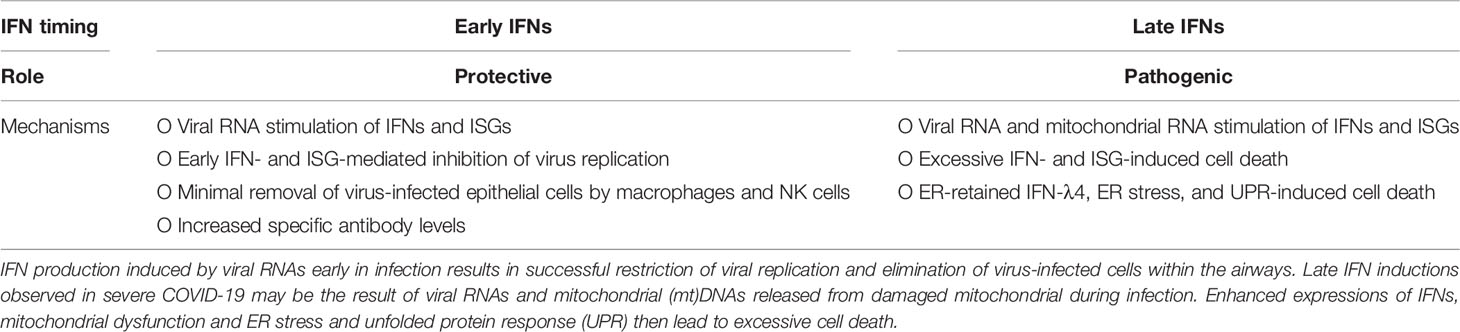
Human innate immune system has developed a suite of intracellular PRRs to detect viral RNAs and trigger innate immune responses that restrict viral replication (Figure 1). There are two major classes of PRRs that facilitate the host antiviral responses, retinoic acid-inducible gene I (RIG-I)-like receptors (RLRs), endosomal Toll-like receptors (TLRs). RLRs include RIG-I and melanoma differentiation–associated gene 5 (MDA5). RIG-I specifically recognises 5′ triphosphate RNAs (5′pppRNAs) that are produced by the IAVs, whereas MDA5 and TLR3/7 binds with virus double stranded (ds)RNAs. Two recent studies independently showed that RIG-I (Thorne et al., 2021) and MDA5 (Yin et al., 2021) was crucial in inducing antiviral responses to SARS-CoV-2, however the importance of the former remains controversial as SARS-CoV-2 has not been shown to produce 5′pppRNAs. Endosomal TLR3/7 also recognise IAV RNAs and are predicted to also detect that of SARS-CoVs, and mediate both antiviral and inflammatory responses.
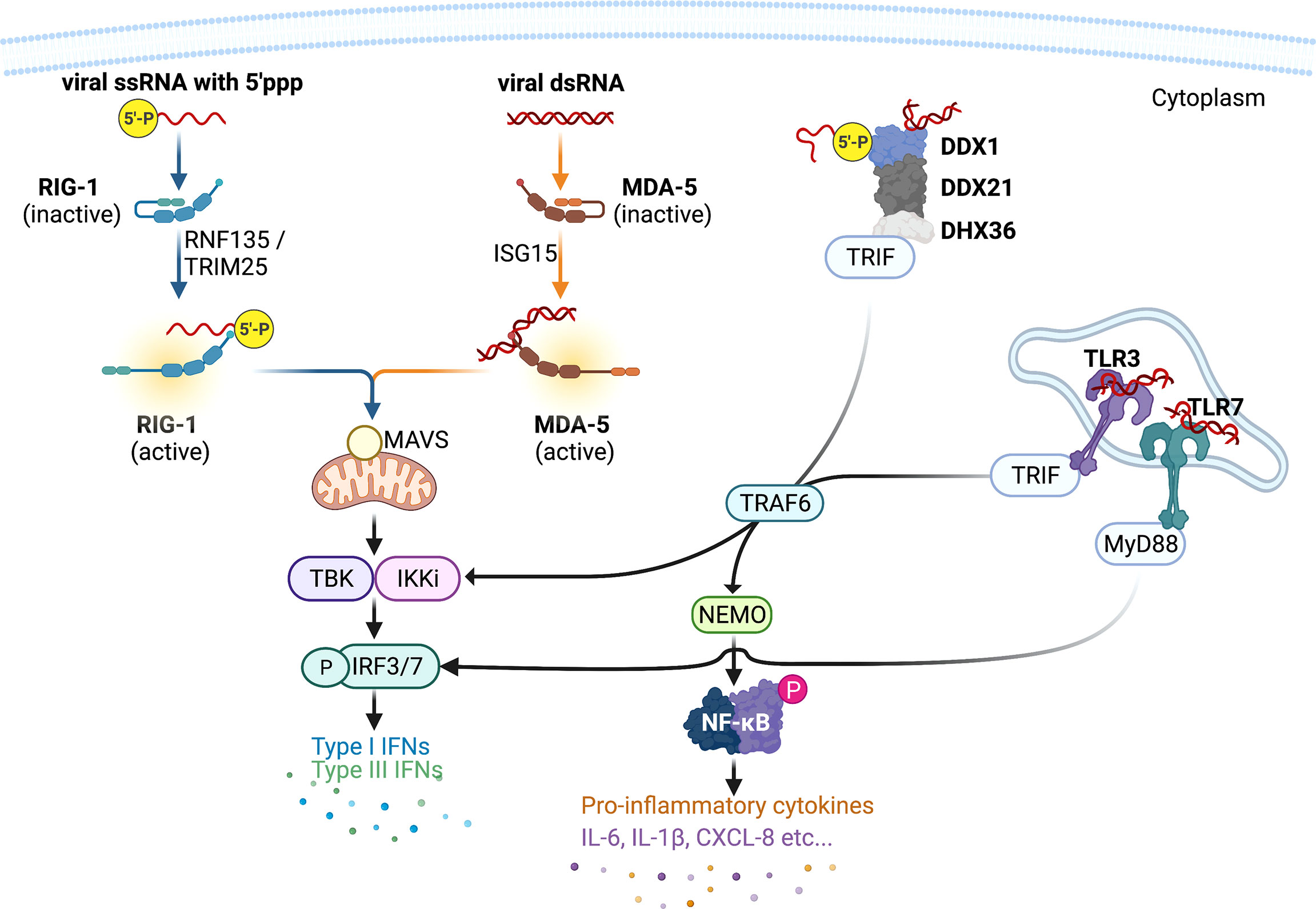
Figure 1 Host intracellular viral RNA recognition and innate immune signalling network. Pattern recognition receptor RIG-I recognises influenza virus ssRNAs with 5` triphosphate group, whereas MDA5, DDX1-DDX21-DHX36 complex, and TLR3 recognises viral dsRNAs. Once engaged with viral RNAs, RIG-I and MDA5 are activated by adaptor proteins RIPLET and TRIM25 and ISG15, respectively, and then initiates the production of type I and III interferons (IFNs) via the mitochondrial antiviral (MAVS) adaptor protein and transcription factor IFN-regulatory factor (IRF)3. DDX1-DDX21-DHX36 complex and TLR3 induces the expression of both type I and III IFNs and pro-inflammatory cytokines [interleukin (IL)-6, IL-1β, etc…] via the adaptor TRIF and the transcription factors IRF3/7 and nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB). This figure was created with BioRender.com.
Once engaged with viral RNAs, RIG-I is activated by adaptor proteins RING Finger (RNF)135 (also known as RIPLET) and Tripartite Motif Containing 5 (TRIM25) (Gack et al., 2007; Hayman et al., 2019), and MDA-5 activation is dependent on interferon-inducible gene (ISG)15 (Liu et al., 2021). Activated RIG-I and MDA-5 interacts with mitochondrial antiviral signalling (MAVS) adaptor protein. MAVS forms fibrillar oligomeric complexes when activated and drives downstream antiviral cytokine production. MAVS recruits tumor-necrosis-factor (TNF)-receptor associated factor (TRAF)3, which activates TANK binding kinase (TBK1) – IκB kinase (IKK)ϵ – IKKγ complex and mediates activations of interferon (IFN) regulatory factor 3/7 (Paz et al., 2011). Activated IRFs then translocate into nucleus where they facilitate the production of antiviral proteins including type I IFNs [IFN-ɑ/-β/-κ/-ϵ (epsilon)/-ω (omega)] and type III IFNs IFN-λ1 [interleukin-29 (IL-29)], IFN-λ2 (IL-28A), IFN-λ3 (IL-28B), and IFN-λ4. The released IFNs act on the same/neighbouring cells to induce the expression of over 300 IFN-stimulated genes (ISGs) via the transcription factor signal transducer and activator of transcription (STAT)1. Notable ISGs that have been well characterised include protein kinase R (PKR) that inhibits viral replication and induces apoptosis (Stark et al., 1998; Fitzgerald et al., 2003; Seth et al., 2005; Hsu et al., 2012; Hsu et al., 2016), and IFN-inducible transmembrane (IFITM) 1, 2, and 3 proteins that entrap infecting virions within endosome and subsequent elimination in the endolysomes (Brass et al., 2009; Feeley et al., 2011; Compton et al., 2016).
In addition to RLRs and TLR3/, Asp-Glu-Ala-Asp (DEAD) box (DDX)1 protein is another PRR that has been shown to be important in IFN responses. Both RIG-I (aka. DDX58) and DD1 belongs to the same DEAD protein family. DDX1 forms a biomolecular condensate with DDX21 and Asp-Glu-Ala-His (DEAH) box (DHX)36 proteins (Figure 1) and induces type I IFNs via the adaptor toll-interleukin receptor (TIR)-containing adaptor molecule-1 (TICAM-1)/TIR domain-containing adaptor-induced IFN-β (TRIF) (Fuller-Pace, 2006; Zhang et al., 2011).
Endosomal TLR3 also employs TRIF as its adaptor protein and facilitates the activation of IKKɑ/β/γ complex (aka. NEMO) and nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB). NF-κB then drives the production of type I IFNs and pro-inflammatory cytokines and chemokines including interleukin (IL)-6, IL-1β, CXCL-8, and TNF-ɑ (Vallabhapurapu and Karin, 2009). These cytokines recruit immune cells such as macrophages, neutrophils, and natural killer (NK) cells to the site of infection and remove virus-infected cells (Biron et al., 1999; Mandelboim et al., 2001; Hsu et al., 2016). TLR7 and its adaptor myeloid differentiation primary response (MyD)88 mediates activation of IRF7 and subsequent transcription of type I IFNs.
IAV infection can lead to heightened inflammation (aka. cytokine storm) that promotes tissue destruction and pneumonia (Hsu et al., 2017; Scharenberg et al., 2019; Vanders et al., 2019; Gu et al., 2019). Excessive IL-6, IL-1β, and TNFα levels along with increased recruitment of immune cells in the airways contribute to epithelial damage and acute lung injury, driving the development of acute respiratory distress syndrome (ARDS), leading to severe vascular leakage and pulmonary edema (Short et al., 2014). Local inflammation can spill over into the systemic circulation, causing systemic inflammation, sepsis, and death.
Similar to IAV infection, excessive airway inflammation and epithelial damage also occurs in severe COVID-19. In addition to elevated IL-1β and TNF-ɑ productions (Sheridan et al., 1997; Kolb et al., 2001; Kimura et al., 2009; Al-Sadi et al., 2013; Kimura et al., 2013), chemokine CCL2 that is typically released by damaged cells and recruits monocytes has also been shown to be increased in COVID-19 patients (Huang et al., 2020; Li et al., 2020). These uncontrolled inflammation and tissue damage compromises the integrity of epithelial-endothelial (air-blood) barrier, leading to ARDS that is evident in severe COVID-19 (Huang et al., 2020; McGonagle et al., 2020; Mehta et al., 2020) as well as in highly pathogenic avian IAV H5N1 infection (Chotpitayasunondh et al., 2005; Tisoncik et al., 2012; Gu et al., 2019).
Constitutively Protective or Pathogenic IFNs
Protective, constitutive, and Inducible IFNs
The innate immune cytokines are thought to be only induced upon virus infection, reports have demonstrated that type I IFNs were constitutively expressed at low levels at resting state and primed the epithelial cells for a more robust antiviral responses to infection (Figure 2). This “revving-up” model has been observed with both IFN-ɑ and IFN-β in IAV and hepatitis C virus (HCV) infection (Hsu et al., 2012; Tsugawa et al., 2014), and may be driven by the constitutive expression of IRF3. This however is unlikely as IRF3 dimerization is not evident at resting state (Yoo et al., 2014; Liu et al., 2015), and IRF3 deficiency had no effect on the production of constitutive IFN-β (Hata et al., 2001). Alternative mechanisms are possibly involved in the constitutive IFN responses. Indeed, transcriptional factors NF-κB and activating protein-1 (AP-1) appears to be essential in constitutive IFN-β production (Gough et al., 2010; Balachandran and Beg, 2011; Basagoudanavar et al., 2011), as subunits of these factors (RelA and c-Jun, respectively) have been reported to bind to the IFN-β promoter region at resting state, although they have been shown to be dispensable in virus-induced IFN-β expression in airway epithelial cells. Deeper elucidation is needed to decipher the exact mechanisms by which constitutive IFNs and ISGs are maintained, particularly the interactions amongst transcriptional factors such as IRFs, NF-κB, AP-1, and other factors.
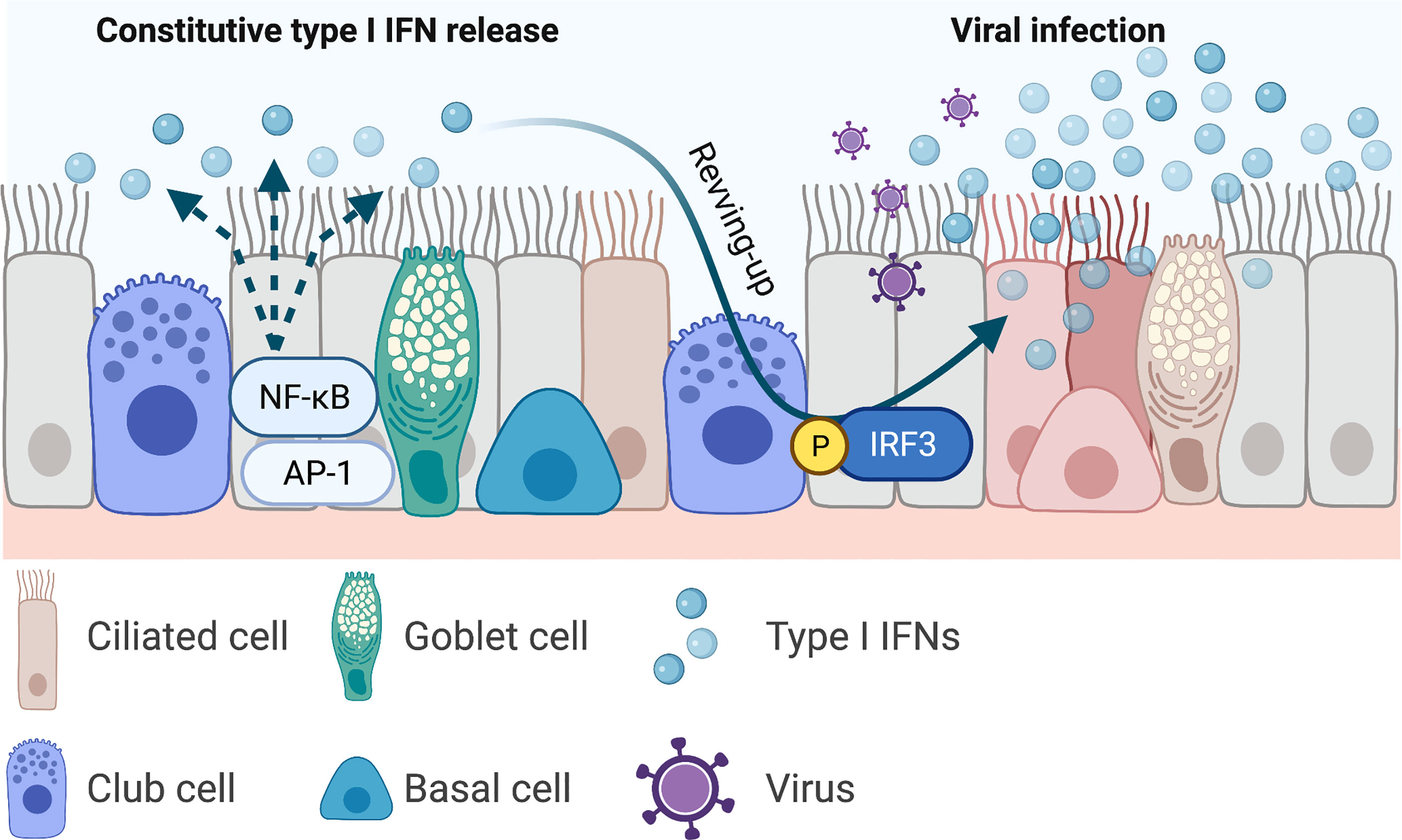
Figure 2 Constitutive type I interferons and the revving-up model. Airway epithelial-derived type I interferons (IFNs) are constitutively expressed and primes the local microenvironment at resting state. This priming mediates a more robust antiviral responses upon viral infection. Current evidence shows that the constitutively expressed type I IFNs are driven by the transcription factors nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB) and activating protein (AP)1. Infection induced type I IFNs are mediated by the transcription factor IFN regulatory factor (IRF)3. Increased production of type I IFNs restrict viral replication. This figure was created with BioRender.com.
Pathogenic IFNs
Type I and III IFNs have consistently been reported to inhibit IAV viral replication in both in vitro (Hsu et al., 2011), in vivo animal models (Mordstein et al., 2008; Davidson et al., 2015), and human clinical trials (Bennett et al., 2013). However, the same protective force appears to be detrimental in severe SARS-CoV infections. Longitudinal studies revealed that while most patients with SARS-CoV infection had early IFN expression and the disease was resolved as the SARS-CoV-specific antibodies increased, those with severe diseases had initially delayed but later persistent IFN responses that were associated with reduced levels of SARS-CoV-specific antibodies and poor clinical outcomes (Cameron et al., 2007). Similarly, patients with mild-to-moderate COVID-19 also showed early IFN responses (Galani et al., 2021) (Figure 3A), and those with severe diseases and those deceased had increased and prolonged IFN expressions in the blood (Lucas et al., 2020) (Figure 3B). Interestingly, human seasonal CoV OC43 has also been shown to induce a delayed but productive viral replication with minimal type I and III IFNs as well as ISGs expressions in differentiated human primary bronchial epithelial cell model (Loo et al., 2020). The same pathological roles of IFNs have also been recapitulated in in vivo models. Infection with SARS-CoV or with SARS-CoV-2 showed delayed but enhanced levels of type I IFN expressions (after peak viral replication) and increased infiltration of inflammatory cytokines and macrophages, vascular leakage, and increased disease severity (Channappanavar et al., 2016; Bessiere et al., 2021). Prophylactic IFN-α treatment (1 day before infection) or early treatment post infection (1 day post inoculation) in experimental SARS-CoV-2 infection in hamster led to reduced mortality. Late IFN-α treatment (3 days post inoculation) had no protective effects (Bessiere et al., 2021) (Figure 3C). The protracted type I and possibly type III IFNs may be a common innate immunological feature in beta-CoV infections (including OC43 and SARS-CoVs), and this may be driven by continuous stimulation of ISG expressions. ISGs including STATs, RLRs, and IRFs further amplify type I IFN productions, which increases the recruitment and activation of immune cells into the lung. IFN-β-treated, and SARS-CoV-infected mice showed increased infiltration of macrophages in the lung compared with non-IFN treated and infected group, and IFN-β-mediated pathogenesis was reduced when monocytes were depleted (Channappanavar et al., 2016).
Figure 3 Protective interferons and interferonopathy. (A) Mild SARS-CoV infection is associated with early productions of type I interferons (IFNs) and resolution of viral infection. (B) Severe infection is accompanied with delayed but sustained type I IFN expressions and is associated with more severe diseases with increased mortality. (C) Early exogenous type I IFN treatment results in early resolution of COVID-19, whereas late treatment fails to show protective effects. This figure was created with BioRender.com.
Possible Pathogenic Mechanisms by Late IFN Productions
Severe SARS-CoV-2 infection has been characterized with high levels of circulating mitochondrial (mt)DNA in the peripheral blood (Scozzi et al., 2021), strongly indicating compromised mitochondrial membrane integrity and function. mtDNAs stimulate the productions of type I IFNs expressions via cGAS-cGAMP-mediated STING pathway (Yang et al., 2018; Yu et al., 2020). Viral RNAs and host mtDNAs may likely trigger excessive expression of type I and III IFNs (Table 1).
In addition to enhanced clearance of virus-infected epithelial cells by macrophages and NK cells, exaggerated IFN levels may directly instigate sudden epithelial death. IAV infections have been shown to induce epithelial cell necroptosis (Shubina et al., 2020), and constitutive type I IFNs have recently been implicated in the induction of necroptosis via a key effector Mixed Lineage Kinase Domain Like Pseudokinase (MLKL) (Sarhan et al., 2019). ISGs including PKR, 2’5’A oligoadenylate synthetase (OAS), TNF-alpha related apoptosis inducing ligand (TRAIL), and galectin 9 have also been implicated in apoptosis induction (Chawla-Sarkar et al., 2003; Haw et al., 2016). While the exact ISGs and mechanisms that participate in IFN-mediated pathogenesis require further investigation, these signals enable an extensive suicidal response that clears virus-infected cells but at the expense of lung tissue integrity.
This late persistent IFN signature appears to be unique to severe COVID-19 and have not been observed with severe IAV infections (Wong et al., 2018; Turianova et al., 2019). This “interferonopathy” in COVID-19 may not be solely driven by type I IFNs. Type I and III IFNs have distinct homology and receptor utilisation but appear to induce similar ISG profiles (Kotenko et al., 2003; Sheppard et al., 2003; Prokunina-Olsson et al., 2013). While their functional differences and redundancies remain to be further defined, type I IFN-β has been reported to mediate acute ISG signatures, while type III IFN-λ induced more sustained ISG responses (Levy et al., 2011; Lin et al., 2016). IFN-λ has also been shown to be the primary effector of the antiviral defences against viral infection at the musical barrier such as the gastrointestinal tract (Pott et al., 2011; Li et al., 2019). Surprisingly, IFN-λ4, a member of the type III IFNs, was recently demonstrated to be expressed but mostly retained intracellularly in HCV and Sendai virus infection, instead of being secreted like other IFNs (Obajemu et al., 2017; Onabajo et al., 2021). Intracellular IFN-λ4 caused endoplasmic reticulum (ER) stress and the associated unfolded protein response (UPR), indicating that the translated IFN-λ4 were likely misfolded, and that the prolonged ER stress and UPR could lead to apoptotic death and tissue damage. It remains to be defined if IAV and CoV infections also cause accumulation of misfolded IFN-λ4, albeit ER stress and UPR has been demonstrated in SARS-CoV-2 infection (Koseler et al., 2020; Pathinayake et al., 2020; Bartolini et al., 2022).
The timing and magnitudes of IFN inductions, as well as the mechanisms of action of ISGs is therefore crucial in determining the molecular equipoise of protective and pathogenic IFNs. While the precise mechanisms require further investigation, it is tempting to speculate that IFN-mediated pathogenesis in COVID-19 is also attributed to viral factors.
Counterstrike by the IAVs and SARS-CoVs
Both IAVs and CoVs encode virulence factors that subvert host innate antiviral systems. IAV produces non-structural (NS)1 and polymerase basic (PB)1-F2 proteins that target multiple components of the IFN system (Hsu, 2018). NS1 is a small multifunctional protein that provides stealth within the cells and impairs the host antiviral responses. The RNA-binding domain of the NS1 recognises and shields viral nucleic acids from the host PRR system (Hatada and Fukuda, 1992; Qian et al., 1995; Chien et al., 2004). The non-structured effector domain stabilizes the RNA-binding domain but also interacts and interferes with host proteins including RIPLET, TRIM25, and PKR (Figure 4). RIPLET and TRIM25 facilitates activation of RIG-I and NS1 directly binds with these adaptors and inhibits RIG-I-mediated type I and III IFN expression (Wang et al., 2002; Bornholdt and Prasad, 2008; Rajsbaum et al., 2012). NS1 also impedes PKR from binding to viral RNAs and prevents PKR-mediated apoptosis. NS1 inhibits viral RNA recognition by PKR via the NS1 RNA-binding domain, and also directly interacts with PKR and suppresses PKR-mediated apoptosis through its effector domain (Bergmann et al., 2000; Dauber et al., 2006; Li et al., 2006; Min et al., 2007). Host PI3K signalling pathway is an important modulator of cellular proliferation and apoptosis. IAV has been shown to utilize this pathway as an alternative entry into cells in addition to the sialic acid residues-bearing glycoproteins (Hsu et al., 2011; Hsu et al., 2015). NS1 effector domain directly interacts with PI3K subunit p85β, further enhancing the rate of virus internalization, and this interaction also led to inhibition of PI3K-induced apoptosis (Hale et al., 2006; Shin et al., 2007; Li et al., 2008; Kedzierski et al., 2017). In addition to subverting host antiviral responses, NS1 also stalls host mRNA processing. NS1 binds with the pre-mRNA cleavage and polyadenylation specificity factor 30 (CPSF30) and poly A binding II (PABII) proteins, shutting down the host protein synthesis during infection (Nemeroff et al., 1998; Chen et al., 1999; Noah et al., 2003) (Figure 4).
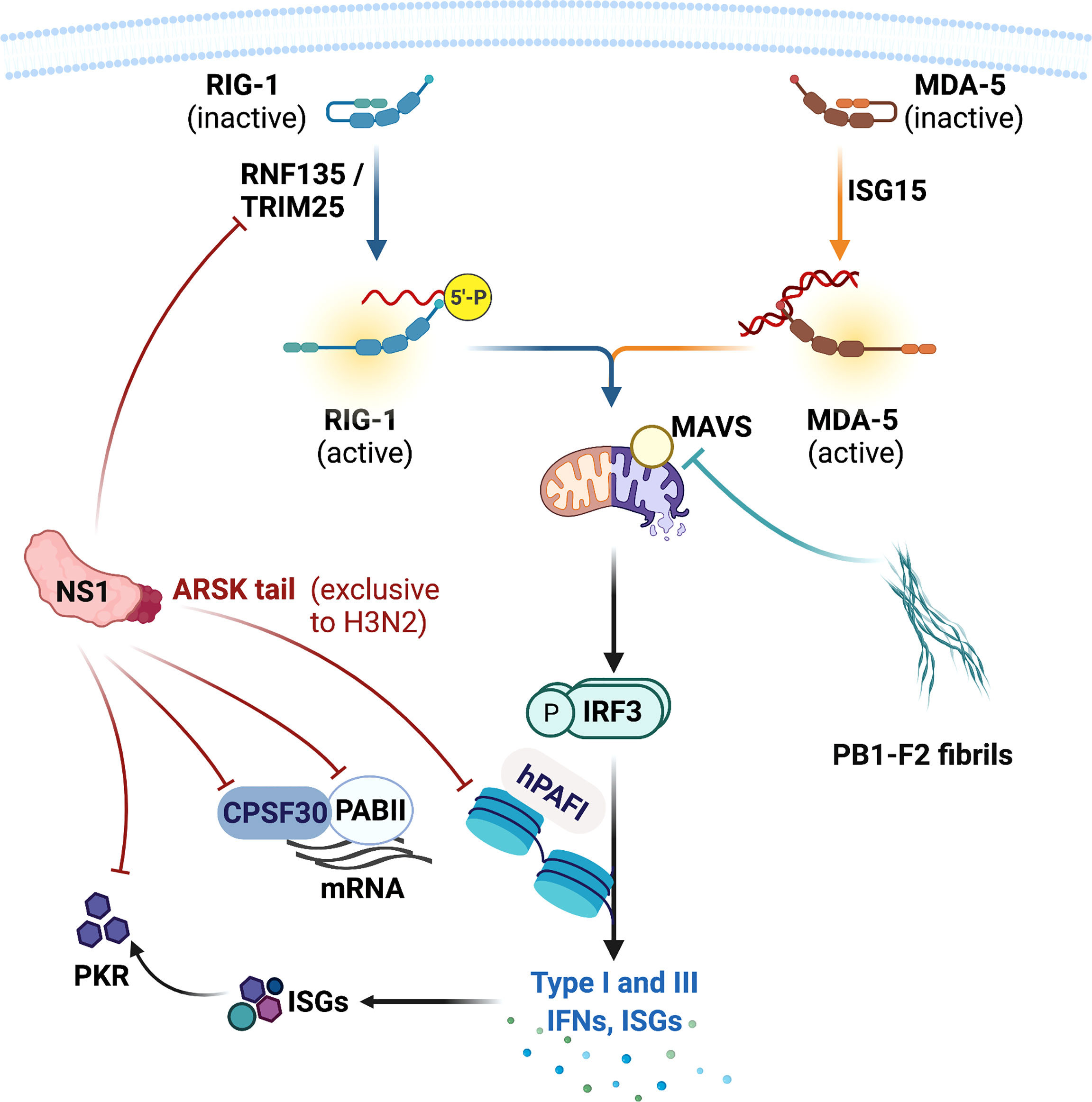
Figure 4 Influenza A virus virulence factors and their targets in the host. Influenza A viruses produce non-structural (NS)1 and polymerase basic 1 (PB1)-F2 proteins that inhibit host antiviral signalling and responses. NS1 protein impairs the production of type I and III IFNs by directly targeting RIG-I activators RIPLET and TRIM25, and many IFN-stimulated genes (ISGs) including protein kinase R (PKR). NS1 also inhibits host protein synthesis by targeting pre-mRNA cleavage and polyadenylation specificity factor 30 (CPSF30) and poly A binding II (PABII) proteins. NS1 also possesses a ARSK tail that inhibits human polymerase II-associated factor 1 (hPAFI) function and host protein transcription. PB1-F2 forms a fibrillar higher-molecular weight aggregate that mainly targets mitochondria and its proteins. PB1-F2 binds with mitochondrial antiviral (MAVS) protein and other mitochondrial membrane proteins, leading to inhibition of type I and III IFN productions and mitochondrial dysfunction. This also results in apoptotic cell death. This figure was created with BioRender.com.
Human H3N2 NS1 possesses a Ala-Arg-Ser-Lys (ARSK) tail at the carboxyl terminus (amino acid residues 226 – 229), which is analogous to the ART(Thr)K motif found on the lysine 4 of histone H3 (H3K4) in the host cell nucleus (Marazzi et al., 2012). Like H3K4 ARTK, NS1 ARSK tail acts as a molecular mimic that directly interacts with human polymerase II-associated factor 1 (hPAFI) and inhibits host protein synthesis (Figure 4). Interestingly, this ARSK tail is exclusively found in the human H3N2 and appears to be lost in other IAV subtypes, including highly pathogenic viruses H5N1, H7N9, and the pandemic H1N1 2009. The reason for this loss of a seemingly beneficial motif in other subtypes remains a mystery.
PB1-F2 is an accessory protein translated from an alternative open reading frame of PB1. PB1-F2 promotes inflammatory cytokine storm during IAV infection (McAuley et al., 2013) and also modulate antiviral responses. PB1-F2 contains a carboxyl-terminal mitochondrial targeting sequence and directly binds with MAVS and disrupts RIG-I docking (Varga et al., 2011; Varga et al., 2012) (Figure 5A). PB1-F2 also targets inner and outer mitochondrial membrane transport proteins adenine nucleotide translocator 3 (ANT3) and voltage-dependent anion channel 1 (VDAC1), impairing mitochondrial membrane integrity and inducing apoptosis (Yoshizumi et al., 2014). Remarkably PB1-F2 has been shown to form a fibrillar aggregate structure that is dependent on the α-helical oligomerization domain at the carboxyl-terminus. This amyloid-like fibres have been hypothesized to form membrane pores, indicating that the PB1-F2 fibrillar structure may directly impair mitochondrial function (Chanturiya et al., 2004; Bruns et al., 2007; McAuley et al., 2013). Indeed, PB1-F2 has been demonstrated to translocate into the innate mitochondrial space via mitochondrial import receptor TOM40 protein (Yoshizumi et al., 2014), and avian IAV H7N9 PB1-F2 enhanced production of mitochondrial reactive oxygen species and calcium efflux, leading to mitochondrial dysfunction, inflammation and apoptosis (Pinar et al., 2017). Interestingly, full length PB1-F2 is mostly expressed by the highly pathogenic viruses including H5N1 and 1918 H1N1, as well as the human seasonal H3N2 virus. In contrast most seasonal H1N1 viruses express PB1-F2 without the carboxyl-terminus, which did not translocate into mitochondrial membrane (Yoshizumi et al., 2014). The truncated PB1-F2 of the 2009 pandemic H1N1 is thought to contribute to the reduced severity of the 2009 pandemic H1N1. This then raises an interesting question. Have the PB1-F2 of seasonal human IAVs lost all of its IFN antagonistic, inflammation- and death-inducing capabilities and what evolutionary advantages do this loss serve for the IAVs that adapt in humans? What does the truncated PB1-F2 do in the host cells? It is possible that PB1-F2 progressively loses the carboxyl-terminus as newly emerged viruses adapt in human, evolving into variants that replicate efficiently in human hosts without extensive apoptosis.
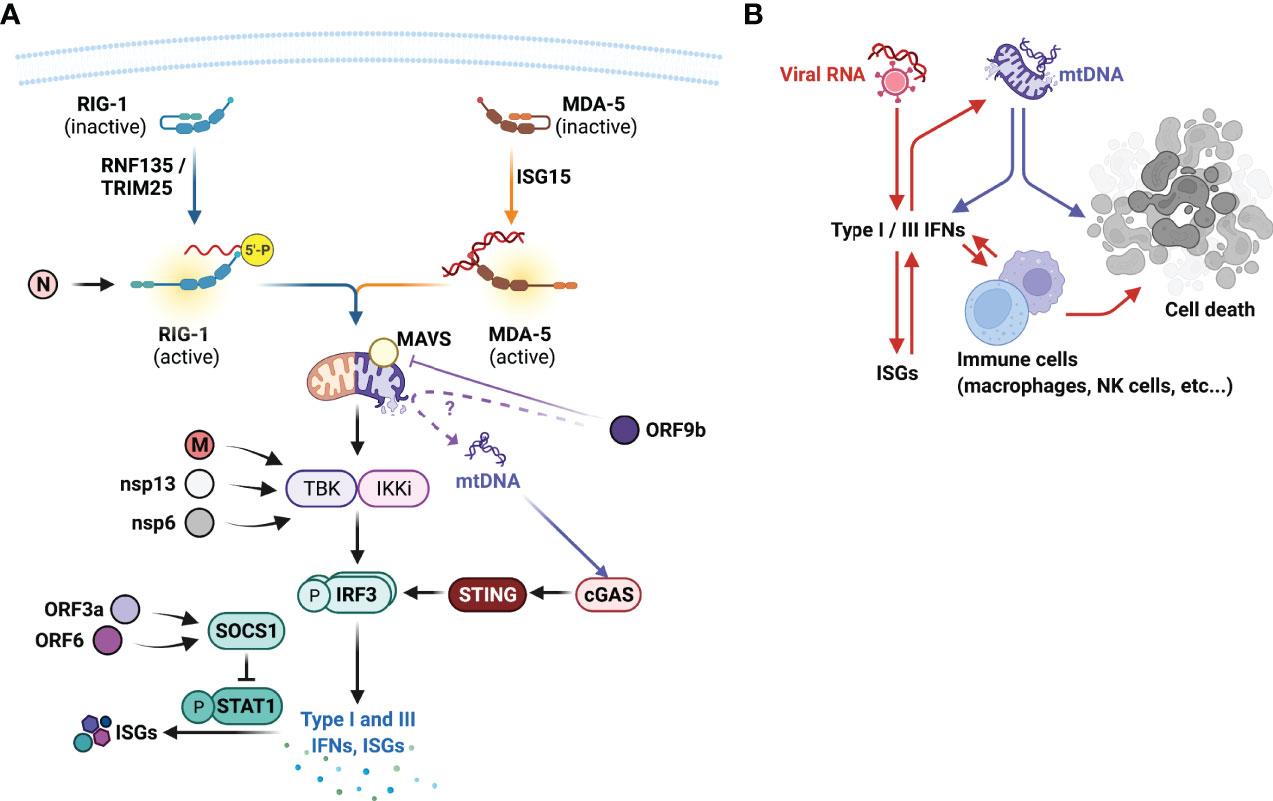
Figure 5 SARS-CoV virulence factors, their targets in the host and type I interferonopathies. (A) SARS-CoVs encode numerous viral factors that directly reduce the hosts antiviral responses. Nucleocapsid (N) protein inhibits RIG-I activation, matrix (M), non-structural protein (nsp)13, and nsp6 impairs TANK binding kinase (TBK1) activation, thus impairing the production of type I and III IFN expressions. Open-reading frame (ORF)3a and ORF6 targets suppressor of cytokine signalling (SOCS)1 that negatively regulates signal transducer and activator of transcription (STAT)1 activation. ORF9b inhibits MAVS activity, suppressing RIG-I signal transduction. Does ORF9b compromise mitochondrial membrane integrity and promote mitochondrial (mt)DNAs escape? mtDNAs further drive the production of type I IFNs via cytoplasmic DNA sensor cGAS and STING and contribute to the type I interferonopathies observed in severe COVID-19. (B) Type I interferonopathies is initially facilitated by viral RNAs that induce the expression of type I and III IFNs and IFN-stimulated genes (ISGs), which further promote the productions of these IFNs. Excessive IFNs also promote the recruitment and activation of innate immune cells including macrophages and natural killer (NK) cells that destroy virus-infected epithelial cells. Intrinsically viral proteins cause mitochondrial membrane rupture, leading to the release of mtDNAs that not only further promote IFN productions, but also cell death. This figure was created with BioRender.com.
There also appears to be two opposing signals by the two most important IAV virulence factors, NS1 (anti-apoptotic) and PB1-F2 (pro-apoptotic). The molecular equipoise of apoptosis is currently unknown, although for human viruses the lost PB1-F2 carboxyl-terminus may indicate a more anti-apoptotic signals for the host cells. An inefficient virus kills it host, and a clever virus stays with it.
For SARS-CoVs, despite the protracted type I IFN signatures observed in severe COVID-19, SARS-CoVs express numerous molecules that inhibit host antiviral responses. Recent in vitro screening assays have indicated several SARS-CoV-2 factors that appeared to modulate host IFN responses, including structural N and M proteins, open reading frames (ORFs) 3a, 6, 7b, and 9b, and non-structural proteins (nsp) 1, 6, and 13 (Lei et al., 2020; Xia et al., 2020) (Figure 5A). Ectopically expressed SARS-CoV-2 N protein has been shown to bind and impede RIG-I-mediated IFN responses upon sendai virus infection (Chen et al., 2020). Whether this indicates SARS-CoV-2 viral RNA recognition by RIG-I require further investigation, but this demonstrates an interesting targeting choice by the N protein, and may indicate non-viral RNA binding functions of RIG-I.
M protein, nsp6 and nsp13 has been shown to directly interfere with TBK1 activation and inhibit IRF3 activation and IFN productions (Xia et al., 2020; Sui et al., 2021; Sui et al., 2022). ORF3a and ORF6 inhibits type I IFN responses by upregulating the suppressor of cytokine signalling (SOCS) 1 expression, a negative regulator of signal transducer and activator of transcription (STAT)1 activation (Lei et al., 2020; Wang et al., 2021) (Figure 5A). ORF9b has been shown to localize to the mitochondria and impair type I IFN productions via its interactions with MAVS and TOM70 (Shi et al., 2014; Bojkova et al., 2020; Jiang et al., 2020). This may thus explain the presence of high circulating mtDNAs in those with severe COVID-19 (Scozzi et al., 2021) and COVID-19 interferonopathies (Figure 5A).
It is remarkable for SARS-CoVs to encode multiple IFN-antagonistic factors, could this, along with higher levels of initial exposure to virus and viral replication, explain the delayed type I IFN and type III IFN responses in those with severe COVID-19? Expression of these viral antagonistic factors in the early phase of infection restricts early IFN responses and maximize viral replication, whilst eliciting strong pro-inflammatory responses in the airways. High levels of viral RNAs and mtDNAs from damaged mitochondria then continuously drive excessive production of IFNs, which potentially become self-sustained through positive feed-back loops (Figure 5B). Integrated spatial and temporal transcriptomic, proteomic, and interactomic analyses of viral and host proteins will thus provide critical insights in the dynamic expressions and interactions of viral proteins and host antiviral responses, and to reveal optimal intervention strategies.
Concluding Perspectives
The arms race between the host and the virus dictates the outcomes of the infection. While advanced genetic screening technologies have accelerated the discovery of host intrinsic restriction factors involved in host antiviral immunity and viral infection, their detailed molecular mechanisms of actions require further investigation. This includes constitutive IFN revving-up system and how they are produced and maintained at the mucosal sites and the transcription mechanisms involved. Are they constitutively expressed at low levels and released or are they stored within the cells such as IFN-λ4 in the ER? Despite the shared Jak-STAT pathway activated by type I and III IFNs, how are they differentially and transcriptionally tuned and whether they induce unique ISG signatures also deserve further clarification. These are fundamental but underexplored areas in innate antiviral immunity, identifying the diversity and functional insights into IFN and ISG biology will provide a wealth of advances in conserved and novel mechanisms of protection from viral pathogens.
Furthermore, there are many important but unanswered questions central to the inception of interferonopathies. This includes the identities of the stimulating moieties (viral RNAs, self-DNAs (mtDNAs), and/or other unidentified molecules) that induce exaggerated type I and perhaps type III IFNs, their signalling kinetics and specific ISG repertoires and precise mechanisms that control viral infection or drive interferonopathies.
The continuous viral mutations give the viruses a huge evolutionary advantage in subverting host innate antiviral systems for survival. IAVs and SARS-CoVs virulence factors strategically compromise key PRR and IFN signalling transduction, host gene transcription, as well as ISG effector functions. As more intrinsic antiviral and cell biological factors are continually being characterised, more of which have been shown to be targeted by viral virulence factors. It is critical that we understand the full spectrum of interacting partners of the IAV and CoV virulence factors and the mutations that render the viruses more infectious or virulent. This may contribute to the surveillance of not just emerging viruses but also their virulence potentials in human.
As we head into the third year of the COVID-19 pandemic and eased restrictions, SARS-CoV-2 is likely to become epidemic and co-circulate with seasonal IAVs in the community. Several studies have reported incidences of IAV and SARS-CoV-2 co-infection amongst those recovered from severe COVID-19 and in those deceased (Wang D et al., 2020; Khodamoradi et al., 2020; Konala et al., 2020; Wu et al., 2020; Cuadrado-Payan et al., 2020; Khorramdelazad et al., 2021). Whether and how co-infection leads to worsened clinical outcomes need to be elucidated, although a recent study showed IAV and SARS-CoV-2 co-infection was significantly associated with increased risk of death (Swets et al., 2022). More importantly, IAV infection has also been shown to increase angiotensin-converting enzyme (ACE)2 expression, the host surface receptor required for SARS-CoV-2 entry into host cells, which may lead to enhanced diseases (Schweitzer et al., 2021). Is SARS-influenza or “flurona” winter coming or is it here already?
It is also important to understand how inborn error of type I IFN signalling impacts overall antiviral responses in severe viral infections and immune-targeted therapeutics such as inhaled IFN-β (Djukanović et al., 2014). In particular, autosomal-recessive deficiencies in IRF7 and IFNAR1 and autosomal-dominant deficiencies in TLR3, TBK1, IRF3, IRF7, IFNAR1 and IFNAR2 genes have been found in a small percentage of patients with severe influenza and COVID-19 (Ciancanelli et al., 2015; Lim et al., 2019; Thomsen et al., 2019). This indicates that not all would benefit from IFN treatment and alternative immune-targeting therapeutic options need to be developed. Regardless, emerging respiratory infectious viruses will always pose a threat to human and our way of life, we need to continually invest in and support research in virology, immunology, and therapeutic strategies before the next pandemic occurs.
In summary, there has been a major explosion of knowledge in PRRs and IFNs, and this will provide important understandings in critical antiviral elements in controlling viral infection. We look forward to further exciting fundamental discoveries and translational research, and how these fields might develop into potential therapeutics against emerging respiratory infectious viruses in the future.
Author Contributions
A-YH conceptualized and wrote the manuscript with GW, YG, CW, and FW. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Al-Sadi, R., Guo, S., Ye, D., Dokladny, K., Alhmoud, T., Ereifej, L., et al. (2013). Mechanism of IL-1beta Modulation of Intestinal Epithelial Barrier Involves P38 Kinase and Activating Transcription Factor-2 Activation. J. Immunol. 190 (12), 6596–6606. doi: 10.4049/jimmunol.1201876
Australian Government, D.o.H (2017). Australian Influenza Surveillance Report and Activity Updates. (Victoria, Australia: D.o. Health)
Balachandran, S., Beg, A. A. (2011). Defining Emerging Roles for NF-kappaB in Antivirus Responses: Revisiting the Interferon-Beta Enhanceosome Paradigm. PLoS Pathog. 7 (10), e1002165. doi: 10.1371/journal.ppat.1002165
Bartolini, D., Stabile, A. M., Vacca, C., Pistilli, A., Rende, M., Gioiello, A., et al. (2022). Endoplasmic Reticulum Stress and NF-kB Activation in SARS-CoV-2 Infected Cells and Their Response to Antiviral Therapy. IUBMB Life 74 (1), 93–100. doi: 10.1002/iub.2537
Basagoudanavar, S. H., Thapa, R. J., Nogusa, S., Wang, J., Beg, A. A., Balachandran, S. (2011). Distinct Roles for the NF-Kappa B RelA Subunit During Antiviral Innate Immune Responses. J. Virol. 85 (6), 2599–610. doi: 10.1128/JVI.02213-10
Bennett, A. L., Smith, D. W., Cummins, M. J., Jacoby, P. A., Cummins, J. M., Beilharz, M. W. (2013). Low-Dose Oral Interferon Alpha as Prophylaxis Against Viral Respiratory Illness: A Double-Blind, Parallel Controlled Trial During an Influenza Pandemic Year. Influenza Other Respir. Viruses 7 (5), 854–862. doi: 10.1111/irv.12094
Bergmann, M., Garcia-Sastre, A., Carnero, E., Pehamberger, H., Wolff, K., Palese, P., et al. (2000). Influenza Virus NS1 Protein Counteracts PKR-Mediated Inhibition of Replication. J. Virol. 74 (13), 6203–6. doi: 10.1128/JVI.74.13.6203-6206.2000
Bessiere, P., Wasniewski, M., Picard-Meyer, E., Servat, A., Figueroa, T., Foret-Lucas, C., et al. (2021). Intranasal Type I Interferon Treatment is Beneficial Only When Administered Before Clinical Signs Onset in the SARS-CoV-2 Hamster Model. PLoS Pathog. 17 (8), e1009427. doi: 10.1371/journal.ppat.1009427
Biron, C. A., Nguyen, K. B., Pien, G. C., Cousens, L. P., Salazar-Mather, T. P. (1999). Natural Killer Cells in Antiviral Defense: Function and Regulation by Innate Cytokines. Annu. Rev. Immunol. 17, 189–220. doi: 10.1146/annurev.immunol.17.1.189
Bojkova, D., Klann, K., Koch, B., Widera, M., Krause, D., Ciesek, S., et al. (2020). Proteomics of SARS-CoV-2-Infected Host Cells Reveals Therapy Targets. Nature 583 (7816), 469–472. doi: 10.1038/s41586-020-233207
Bornholdt, Z. A., Prasad, B. V. (2008). X-Ray Structure of NS1 From a Highly Pathogenic H5N1 Influenza Virus. Nature 456 (7224), 985–8. doi: 10.1038/nature07444
Brass, A. L., Huang, I. C., Benita, Y., John, S. P., Krishnan, M. N., Feeley, E. M., et al. (2009). The IFITM Proteins Mediate Cellular Resistance to Influenza A H1N1 Virus, West Nile Virus, and Dengue Virus. Cell 139 (7), 1243–54. doi: 10.1016/j.cell.2009.12.017
Brockhurst, M. A., Chapman, T., King, K. C., Mank, J. E., Paterson, S., Hurst, G. D. (2014). Running With the Red Queen: The Role of Biotic Conflicts in Evolution. Proc. Biol. Sci. 281 (1797). doi: 10.1098/rspb.2014.1382
Bruns, K., Studtrucker, N., Sharma, A., Fossen, T., Mitzner, D., Eissmann, A., et al. (2007). Structural Characterization and Oligomerization of PB1-F2, a Proapoptotic Influenza A Virus Protein. J. Biol. Chem. 282 (1), 353–363. doi: 10.1074/jbc.M606494200
Cameron, M. J., Ran, L., Xu, L., Danesh, A., Bermejo-Martin, J. F., Cameron, C. M., et al. (2007). Interferon-Mediated Immunopathological Events are Associated With Atypical Innate and Adaptive Immune Responses in Patients With Severe Acute Respiratory Syndrome. J. Virol. 81 (16), 8692–8706. doi: 10.1128/JVI.00527-07
Channappanavar, R., Fehr, A. R., Vijay, R., Mack, M., Zhao, J., Meyerholz, D. K., et al. (2016). Dysregulated Type I Interferon and Inflammatory Monocyte-Macrophage Responses Cause Lethal Pneumonia in SARS-CoV-Infected Mice. Cell Host Microbe 19 (2), 181–193. doi: 10.1016/j.chom.2016.01.007
Chanturiya, A. N., Basanez, G., Schubert, U., Henklein, P., Yewdell, J. W., Zimmerberg, J. (2004). PB1-F2, an Influenza A Virus-Encoded Proapoptotic Mitochondrial Protein, Creates Variably Sized Pores in Planar Lipid Membranes. J. Virol. 78 (12), 6304–12. doi: 10.1128/JVI.78.12.6304-6312.2004
Chawla-Sarkar, M., Lindner, D. J., Liu, Y. F., Williams, B. R., Sen, G. C., Silverman, R. H., et al. (2003). Apoptosis and Interferons: Role of Interferon-Stimulated Genes as Mediators of Apoptosis. Apoptosis 8 (3), 237–249. doi: 10.1023/A:1023668705040
Chen, K., Xiao, F., Hu, D., Ge, W., Tian, M., Wang, W., et al. (2020). SARS-CoV-2 Nucleocapsid Protein Interacts With RIG-I and Represses RIG-Mediated IFN-Beta Production. Viruses 13 (1), 47. doi: 10.3390/v13010047
Chen, Z., Li, Y., Krug, R. M. (1999). Influenza A Virus NS1 Protein Targets Poly(A)-Binding Protein II of the Cellular 3'-End Processing Machinery. EMBO J. 18 (8), 2273–83. doi: 10.1093/emboj/18.8.2273
Chien, C. Y., Xu, Y., Xiao, R., Aramini, J. M., Sahasrabudhe, P. V., Krug, R. M., et al. (2004). Biophysical Characterization of the Complex Between Double-Stranded RNA and the N-Terminal Domain of the NS1 Protein From Influenza A Virus: Evidence for a Novel RNA-Binding Mode. Biochemistry 43 (7), 1950–62. doi: 10.1021/bi030176o
Chotpitayasunondh, T., Ungchusak, K., Hanshaoworakul, W., Chunsuthiwat, S., Sawanpanyalert, P., Kijphati, R., et al. (2005). Human Disease From Influenza A (H5N1), Thailand, 2004. Emerg. Infect. Dis. 11 (2), 201–9. doi: 10.3201/eid1102.041061
Ciancanelli, M. J., Huang, S.X., Luthra, P., Garner, H., Itan, Y., Volpi, S., et al. (2015). Infectious Disease. Life-Threatening Influenza and Impaired Interferon Amplification in Human IRF7 Deficiency. Science 348 (6233), 448–453. doi: 10.1126/science.aaa1578
Compton, A. A., Roy, N., Porrot, F., Billet, A., Casartelli, N., Yount, J. S., et al. (2016). Natural Mutations in IFITM3 Modulate Post-Translational Regulation and Toggle Antiviral Specificity. EMBO Rep. 17 (11), 1657–1671. doi: 10.15252/embr.201642771
Cuadrado-Payan, E., Montagud-Marrahi, E., Torres-Elorza, M., Bodro, M., Blasco, M., Poch, E., et al. (2020). SARS-CoV-2 and Influenza Virus Co-Infection. Lancet 395 (10236), e84. doi: 10.1016/S0140-6736(20)31052-7
Dauber, B., Schneider, J., Wolff, T. (2006). Double-Stranded RNA Binding of Influenza B Virus Nonstructural NS1 Protein Inhibits Protein Kinase R But is Not Essential to Antagonize Production of Alpha/Beta Interferon. J. Virol. 80 (23), 11667–77. doi: 10.1128/JVI.01142-06
Davidson, S., Maini, M. K., Wack, A. (2015). Disease-Promoting Effects of Type I Interferons in Viral, Bacterial, and Coinfections. J. Interferon Cytokine Res. 35 (4), 252–264. doi: 10.1089/jir.2014.0227
Djukanović, R., Harrison, T., Johnston, S. L., Gabbay, F., Wark, P., Thomson, N. C., et al. (2014). The Effect of Inhaled IFN-β on Worsening of Asthma Symptoms Caused by Viral Infections. A Randomized Trial. Am. J. Respir. Crit. Care Med. 190 (2), 145–154. doi: 10.1164/rccm.201312-2235OC
Feeley, E. M., Sims, J. S., John, S. P., Chin, C. R., Pertel, T., Chen, L. M., et al. (2011). IFITM3 Inhibits Influenza A Virus Infection by Preventing Cytosolic Entry. PLoS Pathog. 7 (10), e1002337. doi: 10.1371/journal.ppat.1002337
Fitzgerald, K. A., McWhirter, S. M., Faia, K. L., Rowe, D. C., Latz, E., Golenbock, D. T., et al. (2003). IKKepsilon and TBK1 are Essential Components of the IRF3 Signaling Pathway. Nat. Immunol. 4 (5), 491–6. doi: 10.1038/ni921
Fuller-Pace, F. V. (2006). DExD/H Box RNA Helicases: Multifunctional Proteins With Important Roles in Transcriptional Regulation. Nucleic Acids Res. 34 (15), 4206–15. doi: 10.1093/nar/gkl460
Gack, M. U., Shin, Y. C., Joo, C. H., Urano, T., Liang, C., Sun, L., et al. (2007). TRIM25 RING-Finger E3 Ubiquitin Ligase is Essential for RIG-I-Mediated Antiviral Activity. Nature 446 (7138), 916–20. doi: 10.1038/nature05732
Galani, I. E., Rovina, N., Lampropoulou, V., Triantafyllia, V., Manioudaki, M., Pavlos, E., et al. (2021). Untuned Antiviral Immunity in COVID-19 Revealed by Temporal Type I/III Interferon Patterns and Flu Comparison. Nat. Immunol. 22 (1), 32–40. doi: 10.1038/s41590-020-00840-x
Gough, D. J., Messina, N. L., Hii, L., Gould, J. A., Sabapathy, K., Robertson, A. P., et al. (2010). Functional Crosstalk Between Type I and II Interferon Through the Regulated Expression of STAT1. PLoS Biol. 8 (4), e1000361. doi: 10.1371/journal.pbio.1000361
Gu, Y., Hsu, A. C., Pang, Z., Pan, H., Zuo, X., Wang, G., et al. (2019). Role of the Innate Cytokine Storm Induced by the Influenza A Virus. Viral Immunol. 32 (6), 244–51. doi: 10.1089/vim.2019.0032
Hale, B. G., Jackson, D., Chen, Y. H., Lamb, R. A., Randall, R. E. (2006). Influenza A Virus NS1 Protein Binds P85beta and Activates Phosphatidylinositol-3-Kinase Signaling. Proc. Natl. Acad. Sci. U.S.A. 103 (38), 14194–9. doi: 10.1073/pnas.0606109103
Hatada, E., Fukuda, R. (1992). Binding of Influenza A Virus NS1 Protein to dsRNA In Vitro. J. Gen. Virol. 73 (Pt 12), 3325–29. doi: 10.1099/0022-1317-73-12-3325
Hata, N., Sato, M., Takaoka, A., Asagiri, M., Tanaka, N., Taniguchi, T., et al. (2001). Constitutive IFN-Alpha/Beta Signal for Efficient IFN-Alpha/Beta Gene Induction by Virus. Biochem. Biophys. Res. Commun. 285 (2), 518–525. doi: 10.1006/bbrc.2001.5159
Haw, T. J., Starkey, M. R., Nair, P. M., Pavlidis, S., Liu, G., Nguyen, D. H., et al. (2016). A Pathogenic Role for Tumor Necrosis Factor-Related Apoptosis-Inducing Ligand in Chronic Obstructive Pulmonary Disease. Mucosal Immunol. 9 (4), 859–72. doi: 10.1038/mi.2015.111
Hayman, T. J., Hsu, A. C., Kolesnik, T. B., Dagley, L. F., Willemsen, J., Tate, M. D., et al. (2019). RIPLET, and Not TRIM25, is Required for Endogenous RIG-I-Dependent Antiviral Responses. Immunol. Cell Biol. 97 (9), 840–52. doi: 10.1111/imcb.12284
Hsu, A. C. (2018). Influenza Virus: A Master Tactician in Innate Immune Evasion and Novel Therapeutic Interventions. Front. Immunol. 9 (743), 743. doi: 10.3389/fimmu.2018.00743
Hsu, A. C. Y., Barr, I., Hansbro, P. M., Wark, P. A. (2011). Human Influenza Is More Effective Than Avian Influenza at Antiviral Suppression in Airway Cells. Am. J. Respir. Cell Mol. Biol. 44 (6), 906–913. doi: 10.1165/rcmb.2010-0157OC
Hsu, A. C., Parsons, K., Barr, I., Lowther, S., Middleton, D., Hansbro, P. M., et al. (2012). Critical Role of Constitutive Type I Interferon Response in Bronchial Epithelial Cell to Influenza Infection. PloS One 7 (3), e32947. doi: 10.1371/journal.pone.0032947
Hsu, A. C., Starkey, M. R., Hanish, I., Parsons, K., Haw, T. J., Howland, L. J., et al. (2015). Targeting PI3K-P110alpha Suppresses Influenza Virus Infection in Chronic Obstructive Pulmonary Disease. Am. J. Respir. Crit. Care Med. 191 (9), 1012–1023. doi: 10.1164/rccm.201501-0188OC
Hsu, A. C., Parsons, K., Moheimani, F., Knight, D. A., Hansbro, P. M., Fujita, T., et al. (2016). Impaired Antiviral Stress Granule and IFN-Beta Enhanceosome Formation Enhances Susceptibility to Influenza Infection in Chronic Obstructive Pulmonary Disease Epithelium. Am. J. Respir. Cell Mol. Biol. 55 (1), 117–127. doi: 10.1165/rcmb.2015-0306OC
Hsu, A. C., Dua, K., Starkey, M. R., Haw, T. J., Nair, P. M., Nichol, K., et al. (2017). MicroRNA-125a and -B Inhibit A20 and MAVS to Promote Inflammation and Impair Antiviral Response in COPD. JCI Insight 2 (7), e90443. doi: 10.1172/jci.insight.90443
Huang, C., Wang, Y., Li, X., Ren, L., Zhao, J., Hu, Y., et al. (2020). Clinical Features of Patients Infected With 2019 Novel Coronavirus in Wuhan, China. Lancet 395 (10223), 497–506. doi: 10.1016/S0140-6736(20)30183-5
Ito, T., Suzuki, Y., Mitnaul, L., Vines, A., Kida, H., Kawaoka, Y. (1997). Receptor Specificity of Influenza A Viruses Correlates With the Agglutination of Erythrocytes From Different Animal Species. Virology 227 (2), 493–499. doi: 10.1006/viro.1996.8323
Jiang, H. W., Zhang, H. N., Meng, Q. F., Xie, J., Li, Y., Chen, H., et al. (2020). SARS-CoV-2 Orf9b Suppresses Type I Interferon Responses by Targeting TOM70. Cell Mol. Immunol. 17 (9), 998–1000. doi: 10.1038/s41423-020-0514-8
Kedzierski, L., Tate, M. D., Hsu, A. C., Kolesnik, T. B., Linossi, E. M., Dagley, L., et al. (2017). Suppressor of Cytokine Signaling (SOCS)5 Ameliorates Influenza Infection via Inhibition of EGFR Signaling. Elife 6:e20444. doi: 10.7554/eLife.20444.025
Khodamoradi, Z., Moghadami, M., Lotfi, M. (2020). Co-Infection of Coronavirus Disease 2019 and Influenza A: A Report From Iran. Arch. Iran Med. 23 (4), 239–43. doi: 10.34172/aim.2020.04
Khorramdelazad, H., Kazemi, M. H., Najafi, A., Keykhaee, M., Zolfaghari Emameh, R., Falak, R (2021). Immunopathological Similarities Between COVID-19 and Influenza: Investigating the Consequences of Co-Infection. Microb. Pathog. 152, 104554. doi: 10.1016/j.micpath.2020.104554
Kimura, K., Morita, Y., Orita, T., Haruta, J., Takeji, Y., Sonoda, K. H. (2013). Protection of Human Corneal Epithelial Cells From TNF-Alpha-Induced Disruption of Barrier Function by Rebamipide. Invest. Ophthalmol. Vis. Sci. 54 (4), 2572–2760.
Kimura, K., Teranishi, S., Nishida, T. (2009). Interleukin-1beta-Induced Disruption of Barrier Function in Cultured Human Corneal Epithelial Cells. Invest. Ophthalmol. Vis. Sci. 50 (2), 597–603. doi: 10.1167/iovs.08-2606
Kolb, M., Margetts, P. J., Anthony, D. C., Pitossi, F., Gauldie, J. (2001). Transient Expression of IL-1beta Induces Acute Lung Injury and Chronic Repair Leading to Pulmonary Fibrosis. J. Clin. Invest. 107 (12), 1529–36. doi: 10.1172/JCI12568
Konala, V. M., Adapa, S., Gayam, V., Naramala, S., Daggubati, S. R., Kammari, C. B., et al. (2020). Co-Infection With Influenza A and COVID-19. Eur. J. Case Rep. Intern. Med. 7 (5), 001656. doi: 10.12890/2020_001656
Koseler, A., Sabirli, R., Goren, T., Turkcuer, I., Kurt, O. (2020). Endoplasmic Reticulum Stress Markers in SARS-COV-2 Infection and Pneumonia: Case-Control Study. In Vivo 34 (3 Suppl), 1645–1650. doi: 10.21873/invivo.11956
Kotenko, S. V., Gallagher, G., Baurin, V. V., Lewis-Antes, A., Shen, M., Shah, N. K., et al. (2003). IFN-Lambdas Mediate Antiviral Protection Through a Distinct Class II Cytokine Receptor Complex. Nat. Immunol. 4 (1), 69–77. doi: 10.1038/ni875
Lei, X., Dong, X., Ma, R., Wang, W., Xiao, X., Tian, Z., et al. (2020). Activation and Evasion of Type I Interferon Responses by SARS-CoV-2. Nat. Commun. 11 (1), 3810. doi: 10.1038/s41467-020-17665-9
Levy, D. E., Marie, I. J., Durbin, J. E. (2011). Induction and Function of Type I and III Interferon in Response to Viral Infection. Curr. Opin. Virol. 1 (6), 476–86. doi: 10.1016/j.coviro.2011.11.001
Li, S., Min, J. Y., Krug, R. M., Sen, G. C. (2006). Binding of the Influenza A Virus NS1 Protein to PKR Mediates the Inhibition of its Activation by Either PACT or Double-Stranded RNA. Virology 349 (1), 13–21. doi: 10.1016/j.virol.2006.01.005
Li, Y., Anderson, D. H., Liu, Q., Zhou, Y. (2008). Mechanism of Influenza A Virus NS1 Protein Interaction With the P85beta, But Not the P85alpha, Subunit of Phosphatidylinositol 3-Kinase (PI3K) and Up-Regulation of PI3K Activity. J. Biol. Chem. 283 (34), 23397–409. doi: 10.1074/jbc.M802737200
Li, L., Xue, M., Fu, F., Yin, L., Feng, L., Liu, P. (2019). IFN-Lambda 3 Mediates Antiviral Protection Against Porcine Epidemic Diarrhea Virus by Inducing a Distinct Antiviral Transcript Profile in Porcine Intestinal Epithelia. Front. Immunol. 10, 2394. doi: 10.3389/fimmu.2019.02394
Li, H., Liu, L., Zhang, D., Xu, J., Dai, H., Tang, N., et al. (2020). SARS-CoV-2 and Viral Sepsis: Observations and Hypotheses. Lancet 395 (10235), 1517–20. doi: 10.1016/S0140-6736(20)30920-X
Lim, H. K., Huang, S. X. L., Chen, J., Kerner, G., Gilliaux, O., Bastard, P., et al. (2019). Severe Influenza Pneumonitis in Children With Inherited TLR3 Deficiency. J. Exp. Med. 216 (9), 2038–56. doi: 10.1084/jem.20181621
Lin, J. D., Feng, N., Sen, A., Balan, M., Tseng, H. C., McElrath, C., et al. (2016). Distinct Roles of Type I and Type III Interferons in Intestinal Immunity to Homologous and Heterologous Rotavirus Infections. PLoS Pathog. 12 (4), e1005600. doi: 10.1371/journal.ppat.1005600
Liu, S., Cai, X., Wu, J., Cong, Q., Chen, X., Li, T., et al. (2015). Phosphorylation of Innate Immune Adaptor Proteins MAVS, STING, and TRIF Induces IRF3 Activation. Science 347 (6227), aaa2630. doi: 10.1126/science.aaa2630
Liu, G., Lee, J. H., Parker, Z. M., Acharya, D., Chiang, J. J., van Gent, M., et al. (2021). ISG15-Dependent Activation of the Sensor MDA5 is Antagonized by the SARS-CoV-2 Papain-Like Protease to Evade Host Innate Immunity. Nat. Microbiol. 6 (4), 467–478. doi: 10.1038/s41564-021-00884-1
Loo, S. L., Wark, P. A. B., Esneau, C., Nichol, K. S., Hsu, A. C., Bartlett, N. W. (2020). Human Coronaviruses 229E and OC43 Replicate and Induce Distinct Anti-Viral Responses in Differentiated Primary Human Bronchial Epithelial Cells. Am. J. Physiol. Lung Cell Mol. Physiol. 319(6):L926–L931. doi: 10.1152/ajplung.00374.2020
Lucas, C., Wong, P., Klein, J., Castro, T. B. R., Silva, J., Sundaram, M., et al. (2020). Longitudinal Analyses Reveal Immunological Misfiring in Severe COVID-19. Nature 584 (7821), 463–469. doi: 10.1038/s41586-020-2588-y
Mandelboim, O., Lieberman, N., Lev, M., Paul, L., Arnon, T. I., Bushkin, Y., et al. (2001). Recognition of Haemagglutinins on Virus-Infected Cells by NKp46 Activates Lysis by Human NK Cells. Nature 409 (6823), 1055–1060.
Marazzi, I., Ho, J. S., Kim, J., Manicassamy, B., Dewell, S., Albrecht, R. A., et al. (2012). Suppression of the Antiviral Response by an Influenza Histone Mimic. Nature 483 (7390), 428–433. doi: 10.1038/nature10892
Matrosovich, M. N., Matrosovich, T. Y., Gray, T., Roberts, N. A., Klenk, H. D. (2004). Human and Avian Influenza Viruses Target Different Cell Types in Cultures of Human Airway Epithelium. Proc. Natl. Acad. Sci. U.S.A. 101 (13), 4620–24. doi: 10.1073/pnas.0308001101
McAuley, J. L., Tate, M. D., MacKenzie-Kludas, C. J., Pinar, A., Zeng, W., Stutz, A., et al. (2013). Activation of the NLRP3 Inflammasome by IAV Virulence Protein PB1-F2 Contributes to Severe Pathophysiology and Disease. PLoS Pathog. 9 (5), e1003392. doi: 10.1371/journal.ppat.1003392
McGonagle, D., Sharif, K., O'Regan, A., Bridgewood, C. (2020). The Role of Cytokines Including Interleukin-6 in COVID-19 Induced Pneumonia and Macrophage Activation Syndrome-Like Disease. Autoimmun Rev., 102537. doi: 10.1016/j.autrev.2020.102537
Mehta, P., McAuley, D. F., Brown, M., Sanchez, E., Tattersall, R. S., Manson, J. J., et al. (2020). COVID-19: Consider Cytokine Storm Syndromes and Immunosuppression. Lancet 395 (10229), 1033–1034. doi: 10.1016/S0140-6736(20)30628-0
Min, J. Y., Li, S., Sen, G. C., Krug, R. M. (2007). A Site on the Influenza A Virus NS1 Protein Mediates Both Inhibition of PKR Activation and Temporal Regulation of Viral RNA Synthesis. Virology 363 (1), 236–43. doi: 10.1016/j.virol.2007.01.038
Mordstein, M., Kochs, G., Dumoutier, L., Renauld, J. C., Paludan, S. R., Klucher, K., et al. (2008). Interferon-Lambda Contributes to Innate Immunity of Mice Against Influenza A Virus But Not Against Hepatotropic Viruses. PLoS Pathog. 4 (9), e1000151. doi: 10.1371/journal.ppat.1000151
Nemeroff, M. E., Barabino, S. M., Li, Y., Keller, W., Krug, R. M. (1998). Influenza Virus NS1 Protein Interacts With the Cellular 30 kDa Subunit of CPSF and Inhibits 3'end Formation of Cellular pre-mRNAs. Mol. Cell 1 (7), 991–1000. doi: 10.1016/S1097-2765(00)80099-4
Noah, D. L., Twu, K. Y., Krug, R. M. (2003). Cellular Antiviral Responses Against Influenza A Virus are Countered at the Posttranscriptional Level by the Viral NS1A Protein via its Binding to a Cellular Protein Required for the 3' End Processing of Cellular pre-mRNAS. Virology 307 (2), 386–395. doi: 10.1016/S0042-6822(02)00127-7
Obajemu, A. A., Rao, N., Dilley, K. A., Vargas, J. M., Sheikh, F., Donnelly, R. P., et al. (2017). IFN-Lambda4 Attenuates Antiviral Responses by Enhancing Negative Regulation of IFN Signaling. J. Immunol. 199 (11), 3808–20. doi: 10.4049/jimmunol.1700807
Onabajo, O. O., Wang, F., Lee, M. H., Florez-Vargas, O., Obajemu, A., Tanikawa, C., et al. (2021). Intracellular Accumulation of IFN-Lambda4 Induces ER Stress and Results in Anti-Cirrhotic But Pro-HCV Effects. Front. Immunol. 12, 692263. doi: 10.3389/fimmu.2021.692263
Pathinayake, P. S., Hsu, A. C., Wark, P. A. B. (2020). PAT in the ER for Transmembrane Protein Folding. Trends Biochem. Sci. 45 (12), 1007–8. doi: 10.1016/j.tibs.2020.10.001
Paules, C. I., Sullivan, S. G., Subbarao, K., Fauci, A. S. (2018). Chasing Seasonal Influenza - The Need for a Universal Influenza Vaccine. N Engl. J. Med. 378 (1), 7–9. doi: 10.1056/NEJMp1714916
Paz, S., Vilasco, M., Werden, S. J., Arguello, M., Joseph-Pillai, D., Zhao, T., et al. (2011). A Functional C-Terminal TRAF3-Binding Site in MAVS Participates in Positive and Negative Regulation of the IFN Antiviral Response. Cell Res. 21 (6), 895–910. doi: 10.1038/cr.2011.2
Pinar, A., Dowling, J. K., Bitto, N. J., Robertson, A. A., Latz, E., Stewart, C. R., et al. (2017). PB1-F2 Peptide Derived From Avian Influenza A Virus H7N9 Induces Inflammation via Activation of the NLRP3 Inflammasome. J. Biol. Chem. 292 (3), 826–36. doi: 10.1074/jbc.M116.756379
Pott, J., Mahlakoiv, T., Mordstein, M., Duerr, C. U., Michiels, T., Stockinger, S., et al. (2011). IFN-Lambda Determines the Intestinal Epithelial Antiviral Host Defense. Proc. Natl. Acad. Sci. U.S.A. 108 (19), 7944–9. doi: 10.1073/pnas.1100552108
Prokunina-Olsson, L., Muchmore, B., Tang, W., Pfeiffer, R. M., Park, H., Dickensheets, H., et al. (2013). A Variant Upstream of IFNL3 (IL28B) Creating a New Interferon Gene IFNL4 is Associated With Impaired Clearance of Hepatitis C Virus. Nat. Genet. 45 (2), 164–171. doi: 10.1038/ng.2521
Qian, X. Y., Chien, C. Y., Lu, Y., Montelione, G. T., Krug, R. M. (1995). An Amino-Terminal Polypeptide Fragment of the Influenza Virus NS1 Protein Possesses Specific RNA-Binding Activity and Largely Helical Backbone Structure. RNA 1 (9), 948–56.
Rajsbaum, R., Albrecht, R. A., Wang, M. K., Maharaj, N. P., Versteeg, G. A., Nistal-Villan, E., et al. (2012). Species-Specific Inhibition of RIG-I Ubiquitination and IFN Induction by the Influenza A Virus NS1 Protein. PLoS Pathog. 8 (11), e1003059. doi: 10.1371/journal.ppat.1003059
Sarhan, J., Liu, B. C., Muendlein, H. I., Weindel, C. G., Smirnova, I., Tang, A. Y., et al. (2019). Constitutive Interferon Signaling Maintains Critical Threshold of MLKL Expression to License Necroptosis. Cell Death Differ 26 (2), 332–47. doi: 10.1038/s41418-018-0122-7
Scharenberg, M., Vangeti, S., Kekalainen, E., Bergman, P., Al-Ameri, M., Johansson, N., et al. (2019). Influenza A Virus Infection Induces Hyperresponsiveness in Human Lung Tissue-Resident and Peripheral Blood NK Cells. Front. Immunol. 10, 1116. doi: 10.3389/fimmu.2019.01116
Schweitzer, K. S., Crue, T., Nall, J. M., Foster, D., Sajuthi, S., Correll, K. A., et al. (2021). Influenza Virus Infection Increases ACE2 Expression and Shedding in Human Small Airway Epithelial Cells. Eur. Respir. J. 58 (1):2003988. doi: 10.1183/13993003.03988-2020
Scialo, F., Daniele, A., Amato, F., Pastore, L., Matera, M. G., Cazzola, M., et al. (2020). ACE2: The Major Cell Entry Receptor for SARS-CoV-2. Lung 198 (6), 867–77. doi: 10.1007/s00408-020-00408-4
Scozzi, D., Cano, M., Ma, L., Zhou, D., Zhu, J. H., O'Halloran, J. A., et al. (2021). Circulating Mitochondrial DNA is an Early Indicator of Severe Illness and Mortality From COVID-19. JCI Insight 6 (4):e143299. doi: 10.1172/jci.insight.143299
Seth, R. B., Sun, L., Ea, C. K., Chen, Z. J. (2005). Identification and Characterization of MAVS, a Mitochondrial Antiviral Signaling Protein That Activates NF-kappaB and IRF 3. Cell 122 (5), 669–682. doi: 10.1016/j.cell.2005.08.012
Sheppard, P., Kindsvogel, W., Xu, W., Henderson, K., Schlutsmeyer, S., Whitmore, T. E., et al. (2003). IL-28, IL-29 and Their Class II Cytokine Receptor IL-28r. Nat. Immunol. 4 (1), 63–8. doi: 10.1038/ni873
Sheridan, B. C., McIntyre, R. C., Meldrum, D. R., Fullerton, D. A. (1997). Pentoxifylline Treatment Attenuates Pulmonary Vasomotor Dysfunction in Acute Lung Injury. J. Surg. Res. 71 (2), 150–4. doi: 10.1006/jsre.1997.5144
Shi, C. S., Qi, H. Y., Boularan, C., Huang, N. N., Abu-Asab, M., Shelhamer, J. H., et al. (2014). SARS-Coronavirus Open Reading Frame-9b Suppresses Innate Immunity by Targeting Mitochondria and the MAVS/TRAF3/TRAF6 Signalosome. J. Immunol. 193 (6), 3080–9. doi: 10.4049/jimmunol.1303196
Shin, Y. K., Li, Y., Liu, Q., Anderson, D. H., Babiuk, L. A., Zhou, Y. (2007). SH3 Binding Motif 1 in Influenza A Virus NS1 Protein is Essential for PI3K/Akt Signaling Pathway Activation. J. Virol. 81 (23), 12730–9. doi: 10.1128/JVI.01427-07
Short, K. R., Kroeze, E., Fouchier, R. A. M., Kuiken, T. (2014). Pathogenesis of Influenza-Induced Acute Respiratory Distress Syndrome. Lancet Infect. Dis. 14 (1), 57–69. doi: 10.1016/S1473-3099(13)70286-X
Shubina, M., Tummers, B., Boyd, D. F., Zhang, T., Yin, C., Gautam, A., et al. (2020). Necroptosis Restricts Influenza A Virus as a Stand-Alone Cell Death Mechanism. J. Exp. Med. 217 (11):e20191259. doi: 10.1084/jem.20191259
Stark, G. R., Kerr, I. M., Williams, B. R., Silverman, R. H., Schreiber, R. D. (1998). How Cells Respond to Interferons. Annu. Rev. Biochem. 67, 227–264. doi: 10.1146/annurev.biochem.67.1.227
Sui, L., Zhao, Y., Wang, W., Wu, P., Wang, Z., Yu, Y., et al. (2021). SARS-CoV-2 Membrane Protein Inhibits Type I Interferon Production Through Ubiquitin-Mediated Degradation of TBK1. Front. Immunol. 12, 662989. doi: 10.3389/fimmu.2021.662989
Sui, C., Xiao, T., Zhang, S., Zeng, H., Zheng, Y., Liu, B., et al. (2022). SARS-CoV-2 NSP13 Inhibits Type I IFN Production by Degradation of TBK1 via P62-Dependent Selective Autophagy. J. Immunol. 208 (3), 753–61. doi: 10.4049/jimmunol.2100684
Swets, M. C., Russell, C. D., Harrison, E. M., Docherty, A. B., Lone, N., Girvan, M., et al. (2022). SARS-CoV-2 Co-Infection With Influenza Viruses, Respiratory Syncytial Virus, or Adenoviruses. Lancet. 399(10334), 1463–64. doi: 10.1016/S0140-6736(22)00383-X
Thomsen, M. M., Jorgensen, S. E., Gad, H. H., Storgaard, M., Gjedsted, J., Christiansen, M., et al. (2019). Defective Interferon Priming and Impaired Antiviral Responses in a Patient With an IRF7 Variant and Severe Influenza. Med. Microbiol. Immunol. 208 (6), 869–76. doi: 10.1007/s00430-019-00623-8
Thorne, L. G., Reuschl, A. K., Zuliani-Alvarez, L., Whelan, M. V. X., Turner, J., Noursadeghi, M., et al. (2021). SARS-CoV-2 Sensing by RIG-I and MDA5 Links Epithelial Infection to Macrophage Inflammation. EMBO J. 40 (15), e107826. doi: 10.15252/embj.2021107826
Tisoncik, J. R., Korth, M. J., Simmons, C. P., Farrar, J., Martin, T. R., Katze, M. G. (2012). Into the Eye of the Cytokine Storm. Microbiol. Mol. Biol. Rev. 76 (1), 16–32. doi: 10.1128/MMBR.05015-11
Tsugawa, Y., Kato, H., Fujita, T., Shimotohno, K., Hijikata, M. (2014). Critical Role of Interferon-α Constitutively Produced in Human Hepatocytes in Response to RNA Virus Infection. PLoS One 9 (2), e89869. doi: 10.1371/journal.pone.0089869
Turianova, L., Lachova, V., Svetlikova, D., Kostrabova, A., Betakova, T. (2019). Comparison of Cytokine Profiles Induced by Nonlethal and Lethal Doses of Influenza A Virus in Mice. Exp. Ther. Med. 18 (6), 4397–405. doi: 10.3892/etm.2019.8096
Vallabhapurapu, S., Karin, M. (2009). Regulation and Function of NF-kappaB Transcription Factors in the Immune System. Annu. Rev. Immunol. 27, 693–733. doi: 10.1146/annurev.immunol.021908.132641
Vanders, R. L., Hsu, A., Gibson, P. G., Murphy, V. E., Wark, P. A. B. (2019). Nasal Epithelial Cells to Assess In Vitro Immune Responses to Respiratory Virus Infection in Pregnant Women With Asthma. Respir. Res. 20 (1), 259. doi: 10.1186/s12931-019-1225-5
Varga, Z. T., Ramos, I., Hai, R., Schmolke, M., Garcia-Sastre, A., Fernandez-Sesma, A., et al. (2011). The Influenza Virus Protein PB1-F2 Inhibits the Induction of Type I Interferon at the Level of the MAVS Adaptor Protein. PLoS Pathog. 7 (6), e1002067. doi: 10.1371/journal.ppat.1002067
Varga, Z. T., Grant, A., Manicassamy, B., Palese, P. (2012). Influenza Virus Protein PB1-F2 Inhibits the Induction of Type I Interferon by Binding to MAVS and Decreasing Mitochondrial Membrane Potential. J. Virol. 86 (16), 8359–66. doi: 10.1128/JVI.01122-12
Wahl, A., Gralinski, L. E., Johnson, C. E., Yao, W., Kovarova, M., Dinnon, K. H., 3rd, et al. (2021). SARS-CoV-2 Infection is Effectively Treated and Prevented by EIDD-2801. Nature 591 (7850), 451–457.
Wang, X., Basler, C. F., Williams, B. R., Silverman, R. H., Palese, P., Garcia-Sastre, A. (2002). Functional Replacement of the Carboxy-Terminal Two-Thirds of the Influenza A Virus NS1 Protein With Short Heterologous Dimerization Domains. J. Virol. 76 (24), 12951–62. doi: 10.1128/JVI.76.24.12951-12962.2002
Wang, Q., Zhang, Y., Wu, L., Niu, S., Song, C., Zhang, Z., et al. (2020). Structural and Functional Basis of SARS-CoV-2 Entry by Using Human Ace2. Cell 181 (4), 894–904 e9. doi: 10.1016/j.cell.2020.03.045
Wang, D., Hu, B., Hu, C., Zhu, F., Liu, X., Zhang, J., et al. (2020). Clinical Characteristics of 138 Hospitalized Patients With 2019 Novel Coronavirus-Infected Pneumonia in Wuhan, China. JAMA 323 (11), 1061–69. doi: 10.1001/jama.2020.1585
Wang, R., Yang, X., Chang, M., Xue, Z., Wang, W., Bai, L., et al. (2021). ORF3a Protein of Severe Acute Respiratory Syndrome Coronavirus 2 Inhibits Interferon-Activated Janus Kinase/Signal Transducer and Activator of Transcription Signaling via Elevating Suppressor of Cytokine Signaling 1. Front. Microbiol. 12, 752597. doi: 10.3389/fmicb.2021.752597
Weinheimer, V. K., Becher, A., Tonnies, M., Holland, G., Knepper, J., Bauer, T. T., et al. (2012). Influenza A Viruses Target Type II Pneumocytes in the Human Lung. J. Infect. Dis. 206 (11), 1685–94. doi: 10.1093/infdis/jis455
WHO (2011). Report of the Review Committee on the Functioning of the International Health Regulations (2005) in Relation to Pandemic (H1N1) 2009 (World Health Organization).
Wong, S. S., Oshansky, C. M., Guo, X. J., Ralston, J., Wood, T., Seeds, R., et al. (2018). Severe Influenza Is Characterized by Prolonged Immune Activation: Results From the SHIVERS Cohort Study. J. Infect. Dis. 217 (2), 245–256. doi: 10.1093/infdis/jix571
Wu, Q., Xing, Y., Shi, L., Li, W., Gao, Y., Pan, S., et al. (2020). Coinfection and Other Clinical Characteristics of COVID-19 in Children. Pediatrics 146 (1):e20200961. doi: 10.1542/peds.2020-0961
Xia, H., Cao, Z., Xie, X., Zhang, X., Chen, J. Y., Wang, H., et al. (2020). Evasion of Type I Interferon by SARS-CoV-2. Cell Rep. 33 (1), 108234. doi: 10.1016/j.celrep.2020.108234
Yang, K., Huang, R., Fujihira, H., Suzuki, T., Yan, N. (2018). N-Glycanase NGLY1 Regulates Mitochondrial Homeostasis and Inflammation Through NRF1. J. Exp. Med. 215 (10), 2600–16. doi: 10.1084/jem.20180783
Yin, X., Riva, L., Pu, Y., Martin-Sancho, L., Kanamune, J., Yamamoto, Y., et al. (2021). MDA5 Governs the Innate Immune Response to SARS-CoV-2 in Lung Epithelial Cells. Cell Rep. 34 (2), 108628. doi: 10.1016/j.celrep.2020.108628
Yoo, J. S., Takahasi, K., Ng, C. S., Ouda, R., Onomoto, K., Yoneyama, M., et al. (2014). DHX36 Enhances RIG-I Signaling by Facilitating PKR-Mediated Antiviral Stress Granule Formation. PLoS Pathog. 10 (3), e1004012. doi: 10.1371/journal.ppat.1004012
Yoshizumi, T., Ichinohe, T., Sasaki, O., Otera, H., Kawabata, S., Mihara, K., et al. (2014). Influenza A Virus Protein PB1-F2 Translocates Into Mitochondria via Tom40 Channels and Impairs Innate Immunity. Nat. Commun. 5, 4713. doi: 10.1038/ncomms5713
Yu, C. H., Davidson, S., Harapas, C. R., Hilton, J. B., Mlodzianoski, M. J., Laohamonthonkul, P., et al. (2020). TDP-43 Triggers Mitochondrial DNA Release via mPTP to Activate cGAS/STING in ALS. Cell 183 (3), 636–649 e18. doi: 10.1016/j.cell.2020.09.020
Keywords: influenza A virus, coronavirus, coronavirus – COVID-19, virus infection, innate immunity, innate immune system
Citation: Wang GQ, Gu Y, Wang C, Wang F and Hsu AC-Y (2022) A Game of Infection – Song of Respiratory Viruses and Interferons. Front. Cell. Infect. Microbiol. 12:937460. doi: 10.3389/fcimb.2022.937460
Received: 06 May 2022; Accepted: 25 May 2022;
Published: 29 June 2022.
Edited by:
Rameez Raja, Cleveland Clinic, United StatesReviewed by:
Showkat A. Dar, National Institutes of Health (NIH), United StatesReyaz Ur Rasool, University of Pennsylvania, United States
Copyright © 2022 Wang, Gu, Wang, Wang and Hsu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Fang Wang, d2ZAamx1LmVkdS5jbg==; Alan Chen-Yu Hsu, QWxhbi5Ic3VAZHVrZS1udXMuZWR1LnNn
 Guo Qiang Wang
Guo Qiang Wang Yinuo Gu1
Yinuo Gu1 Chao Wang
Chao Wang Fang Wang
Fang Wang Alan Chen-Yu Hsu
Alan Chen-Yu Hsu