- 1Guangdong Provincial Key Laboratory of Immune Regulation and Immunotherapy, School of Laboratory Medicine and Biotechnology, Southern Medical University, Guangzhou, China
- 2Dermatology Hospital, Southern Medical University, Guangzhou, China
- 3Institute of Immunology, The People’s Liberation Army (PLA), Third Military Medical University, Chongqing, China
Upon acute viral infection, virus-specific CD4+ T cells differentiate into either TH1 cells or follicular helper T (TFH) cells. The molecular pathways governing such bimodal cell fate commitment remain elusive. Additionally, effector virus-specific TFH cells further differentiate into corresponding memory population, which confer long-term protection against re-infection of same viruses by providing immediate help to virus-specific memory B cells. Currently, the molecular mechanisms underlying the long-term maintenance of memory TFH cells are largely unknown. In this review, we discuss current understanding of early differentiation of virus-specific effector TFH cells and long-term maintenance of virus-specific memory TFH cells in mouse models of viral infection and patients of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection.
Introduction
During viral infection, the orchestration of CD4+ T cells, CD8+ T cells and B cells constitutes the core events of host adaptive immunity, which confers specialized and long-term cellular and humoral immune protection. As “helper” cells, CD4+ T cells not only optimize the cytotoxic function and memory generation of CD8+ T cells, but also play indispensable roles in both efficient neutralizing antibody production and antibody-producing long-lived plasma cells as well as memory B cells development (Seder and Ahmed, 2003; Kurosaki et al., 2015). Regulated by specific cytokine milieu and transcriptional factors, activated CD4+ T cells have the potential to differentiate into various cellular subsets, including TH1, TH2, TH9, TH17, TH22, TH25, follicular helper T (TFH), and induced regulatory T (iTREG) cells (Figure 1), to deal with different types of infection or non-infection situations (O’Shea John and Paul William, 2010; Caza and Landas, 2015; Das et al., 2017; Umar et al., 2020). During viral infection, virus-specific CD4+ T cells mainly differentiate into T helper type 1 (TH1) cells and follicular helper T (TFH) cells (Crotty, 2014; Xu et al., 2015; Huang et al., 2019). TFH cell subset was first identified in human tonsils and peripheral blood, characterized by the expression of C-X-C chemokine receptor type 5 (CXCR5) and inducible costimulator (ICOS), in which the former facilitates TFH cells to interact with cognate B cells and further drives B cells homing to follicles and sustains T-B interaction (Breitfeld et al., 2000; Schaerli et al., 2000).
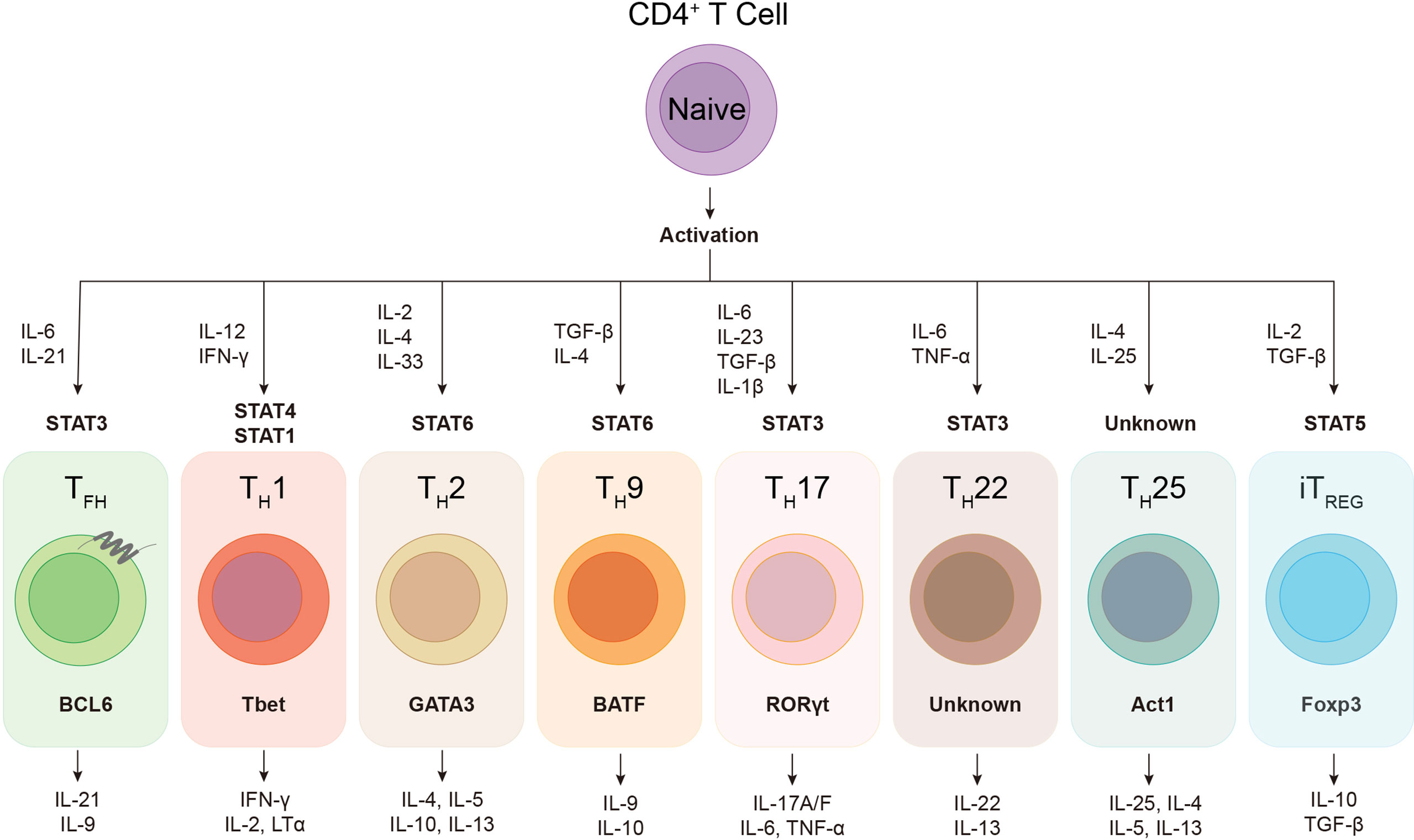
Figure 1 The plasticity of helper CD4+ T cells. Upon activation, naïve CD4+ T cell can differentiate into various subsets of T helper lineages, regulated by certain cytokines and activated signal transducers and activators of transcription (STATs). Each CD4+ helper lineage has the specific lineage-defining transcription factors, e.g. BCL6 in TFH cell, and the characteristic profile of cytokine production, e.g. IL-21 and IL-9 in TFH cell.
Canonical TFH cells usually locate in the intra-follicle germinal center (GC) of secondary lymphoid organs (SLO), such as lymph nodes, spleen, and tonsils, in which TFH cells frequently wander out and in different GC regions, keeping close interaction with the cognate B cells (Shulman et al., 2013). GC is a highly dynamic structure where the high-affinity mutants of B cells are generated via somatic hypermutation (SHM) and affinity-based selection implemented by TFH cells (Victora and Nussenzweig, 2012). In addition, a fraction of TFH cells lingering in T-B border helps cognate B cells ahead to the extrafollicular pathways of antibody generation, which provides immediate protection against invading viruses at the early time of infection (Lee et al., 2011; Di Niro et al., 2015). In addition to SLO, TFH cells have also been witnessed functional in the inducible bronchus-associated lymphoid tissues (iBALT) in lung (Tan et al., 2019), and the tertiary lymphoid structures (TLS) in tumor (Garaud et al., 2022), and the periphery circulation (Morita et al., 2011).
TFH cells are featured as the key players to facilitate high-affinity antibody production of B cells via guaranteeing efficient SHM of immunoglobulin genes and the selective processes in GC during viral infection. After primed by dendritic cells (DCs) through engagement of viral peptide-major histocompatibility complex class II molecules (p-MHCII) complex and virus-specific TCR, TFH-committed CD4+ helper T cells initiate GC responses by moving to the T-B border and interact with cognate B cells to elicit B cell proliferation. During this process, B cells circulate between the light zone (LZ), where follicular dendritic cells (FDCs) deposit antigens and TFH cells recognize the p-MHCII complexes on cognate B cells, and the dark zone (DZ), where B cells extensively proliferate after receiving “help” signals from TFH cells. In DZ, GC B cells undergo rapid proliferation accompanied by SHM, allowing generation of mutated BCRs with diverse affinities to antigens. When back into LZ, mutated GC B cells with higher affinity are selected by TFH cells for another circulation of proliferation and mutation (Victora and Nussenzweig, 2012; Shulman et al., 2013; Crotty, 2014). In addition to directly providing costimulatory signaling to cognate B cells via ICOS, CD40L, and SAP (Qi et al., 2008; Crotty, 2014), TFH cells produce high levels of IL-21, which is essential for B cell survival, proliferation, plasma cell differentiation, and isotype switching (Chtanova et al., 2004; Kuchen et al., 2007; Linterman et al., 2010). In addition to IL-21, TFH cell-derived IL-9 also promotes the development of memory B cells in GC (Wang et al., 2017). It was long and widely believed that TH1 cells rather than TFH cells primarily contribute to promote killing function of CD8+ T cells. Of late, however, Cui et al. revealed that IL-21 produced by tumor-specific TFH cells directly promotes the anti-tumor capacity of CD8+ T cell (Cui et al., 2021). Meanwhile, Zander et al. demonstrated that TFH-derived IL-21 promotes the development and antiviral immunity of CD8+ T cells during chronic viral infection (Zander et al., 2022). Since IL-21 promotes the formation of stem-like/memory CD8+ T cells (Tian and Zajac, 2016), it is possible that the help from CD4+ T cells to CD8+ T cell memory may be mediated by TFH cells. Moreover, CXCR5+ CD4+ TFH cells locating in perifollicular areas of iBALT act to enhance the homing and fitness of CD8+ T cells through IL-21 and IFN-γ production during influenza A virus infection (Pruner and Pepper, 2021).
Overall, TFH cells bridge the cellular and humoral immunity in host, thus playing an essential role in adaptive immune responses. Here we firstly focus on the current understanding of the generation and longevity of virus-specific TFH cells during viral infection, including the fate commitment, lineage differentiation, memory formation, and long-term maintenance (Figure 2). Then, we also discuss the role of SARS-CoV-2-specific TFH cells during currently still ongoing pandemic coronavirus disease 2019 (COVID-19) (Figure 3).
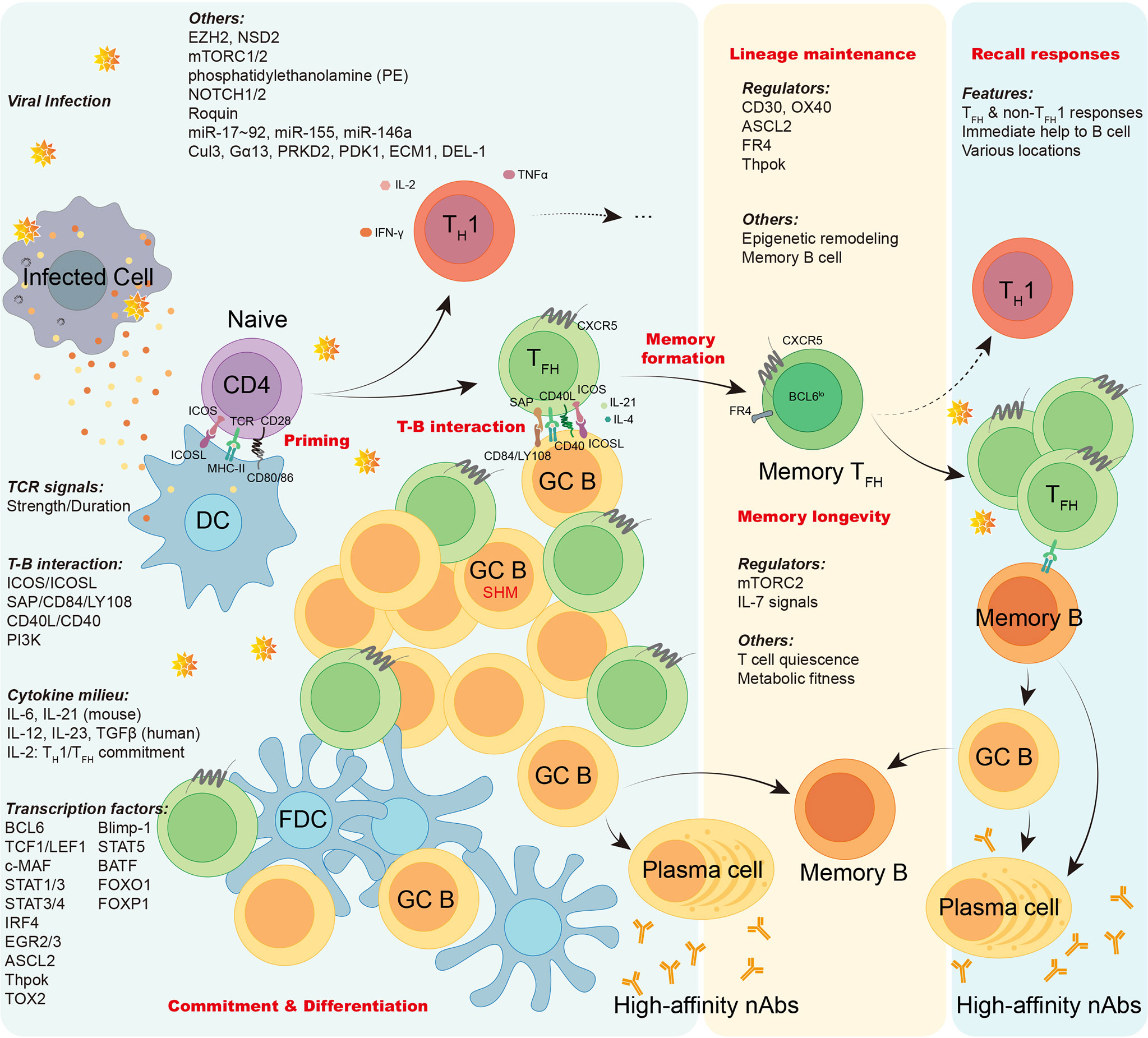
Figure 2 The generation, maintenance, and functions of virus-specific TFH cells. In viral infection, antigen presenting cells like dendritic cells (DCs) conduct antigen uptake, processing, and presentation to prime naïve virus-specific CD4+ T cells, inducing T cell activation. Upon activation, virus-specific CD4+ T cells are mainly committed into either TH1 cells or TFH cells. The lineage commitment and differentiation of TH1/TFH cell are orchestrated under the regulation of TCR signals, T-B interaction, cytokine milieu, transcription factors, and other factors as illustrated. Virus-specific effector TFH cells work on initiating GC responses, facilitating the cognate B cell maturation, promoting generation of long-lived memory B cells and high-affinity neutralizing antibodies. Memory virus-specific TFH cells are developed and sustained after viral clearance under the regulation of a variety of regulators, epigenetic remodeling, and cognate memory B cells. The longevity of memory TFH cells are guaranteed by T cell quiescence and viability conferred by mTORC2 signaling. Upon re-challenge of encountered viruses, memory virus-specific TFH cells provide immediate help to cognate memory B cells, promoting robust cellular and humoral recall responses.
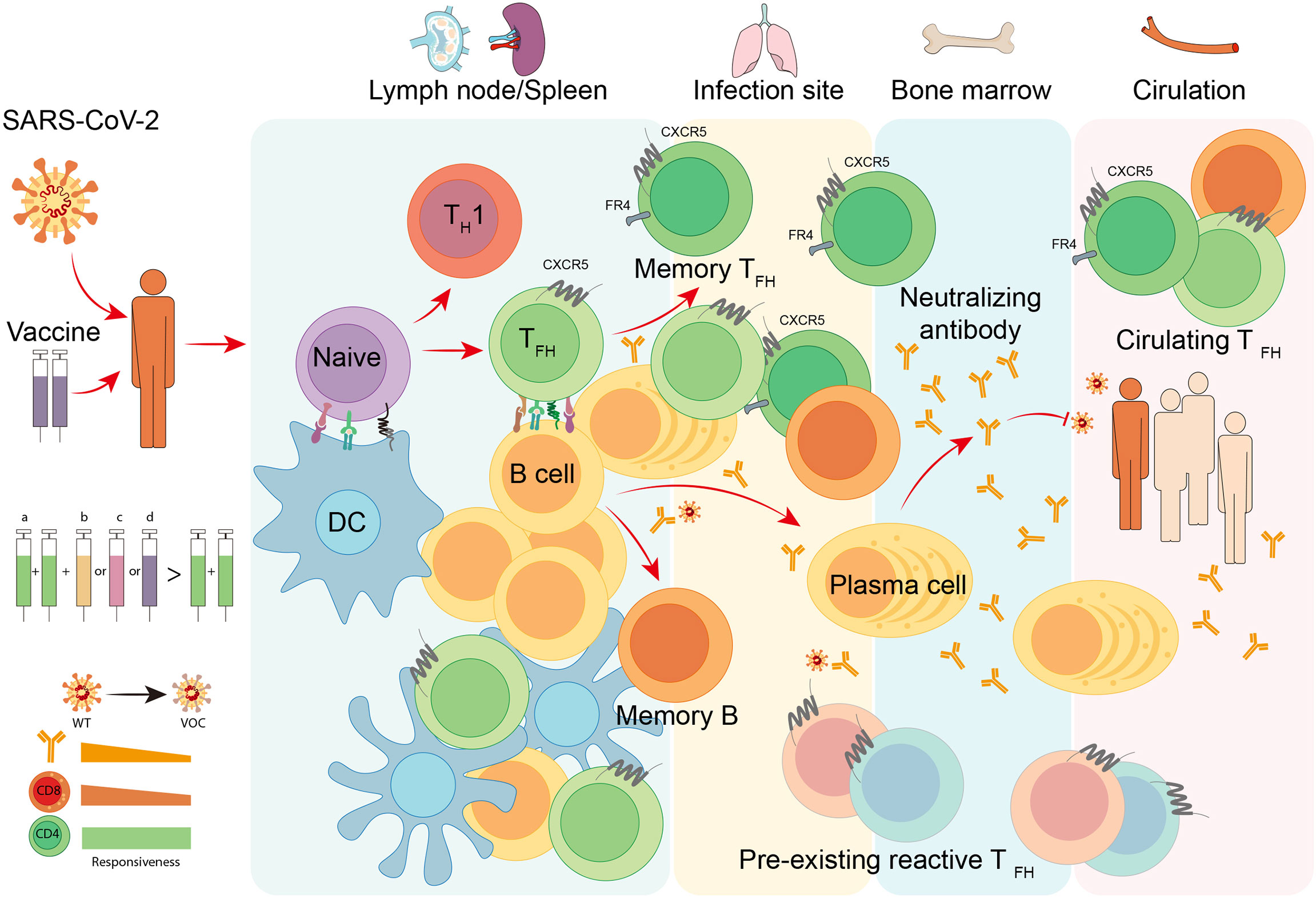
Figure 3 TFH cell responses to COVID-19. Infection or vaccination of SARS-CoV-2 induces differentiation of virus-specific TFH cells and activation of cognate B cells in secondary lymphoid organs like lymph nodes and spleen. With the help of TFH cells, antigen-specific B cells develop into plasma cells to produce neutralizing antibodies with increased affinity, and meanwhile, generate long-lived plasma cells and memory B cells that traffic to the bone marrow and provide long-term protection. Neutralizing antibodies are capable of blocking the attachment and entry of SARS-CoV-2 to prevent COVID-19. In infection site and circulation, the pre-existing cross-reactive TFH cells and circulating TFH cells are proved to be beneficial for the immune protection of natural infection and vaccination. To date, multiple vaccine platforms are utilized to develop SARS-CoV-2 vaccines, including (a) inactivated virus vaccines, (b) adjuvanted recombinant protein vaccines, (c) adenoviral vector-based vaccines, and (d) mRNA vaccines. The heterologous sequential immunization provides a superior effectiveness to protect vaccines from variant of concern (VOC) of SARS-CoV-2 compared to homologous vaccination. The superiority of this vaccination strategy may attribute to the competent responsiveness of memory CD4+ T cells to SARS-CoV-2 VOCs.
Virus-Specific TFH Cell Differentiation
Signals from T Cell Receptor Elicit TFH Commitment
TFH cell differentiation is a multistep and multifactorial process. Naïve CD4+ T cells are primed by TCR recognition of p-MHCII presented on DCs to initiate the activation and lineage differentiation (Goenka et al., 2011). TFH cell program starts at a very early stage after TCR activation. After immunization, antigen-specific CD4+ T cells in draining lymph nodes upregulate expression of BCL6, which is the master transcriptional factor for TFH cells, within 48 hours (Baumjohann et al., 2011). Meanwhile, using lymphocytic choriomeningitis virus- (LCMV) specific TCR transgenic CD4+ T (SMARTA) cells, Choi et al. showed an early development of virus-specific TFH cells at day 3 post-infection (Choi et al., 2011). Previous studies demonstrated that DCs are necessary and sufficient to induce CXCR5+ BCL6+ TFH generation (Goenka et al., 2011), while the late B cell interactions are required for complete differentiation of TFH cells (Barnett et al., 2014; Hao et al., 2018). However, in some scenarios, TFH cells are generated in DC-depleted mice as long as cognate T-B interactions are available (Dahlgren et al., 2015; Arroyo and Pepper, 2019). The strength and duration of TCR signaling are considered to affect the bimodal fate commitment of TFH/TH1 cell during viral infection. By adoptive transfer of TCR transgenic T cells with different TCR affinities, Fazilleau et al. demonstrated that CD4+ helper T cells with higher specific binding of p-MHCII and more restricted TCR junctional diversities tend to commit to TFH cell differentiation (Fazilleau et al., 2009). Further investigations suggested that the interaction between TCR and p-MHCII with long duration favors TFH cell commitment (Baumjohann et al., 2013b; Tubo et al., 2013). This mechanism of TFH cell commitment echoes the observation of the accumulation of TFH cells during chronic viral infection, in which persistent antigen induces sustained TCR stimulation with a long dwell time (Fahey et al., 2011; Vella et al., 2017). However, there are controversial views about the facilitation of stronger TCR signals in determining TFH cell commitment and differentiation (Keck et al., 2014; Snook Jeremy et al., 2018; Kotov et al., 2019). Both Keck et al. and Kotov et al. utilized the Listeria monocytogenes expressing peptides with different TCR affinities and corresponding TCR transgenic mice to demonstrate that TCRs with higher affinity promote TH1 cell formation, whereas TCRs with lower affinity poised to the TFH-biased differentiation of naïve CD4+ T cells (Keck et al., 2014; Kotov et al., 2019). In addition, Jeremy P. Snook et al. confirmed ectopically enhanced TCR signaling via knockdown SHP-1, which is a tyrosine phosphatase that suppresses early TCR signaling events, increases the differentiation of TH1 cells rather than TFH cells (Snook Jeremy et al., 2018). These discrepancies warrant to be reconciled by future studies.
ICOS, SAP, and CD40L Maintain TFH Differentiation
In addition to the interaction of TCR and cognate p-MHCII on DCs, molecules associated with T-B cell conjugation, like ICOS, SAP and CD40L, are also important regulators of TFH cell differentiation during viral infection (Crotty, 2011). With a high expression of CXCR5 and a low level of CCR7, TFH cells are capable of migrating to the T-B border (Breitfeld et al., 2000; Schaerli et al., 2000), where the B cell-dependent TFH cell differentiation occurs. ICOSL expressed on B cells is essential for the responses of TFH cells (Akiba et al., 2005; Bossaller et al., 2006; Gigoux et al., 2009; Xu et al., 2013; Liu et al., 2015), in both CD28-dependent and CD28-independent pathways (Tan et al., 2006; Linterman et al., 2009). ICOSL expression on B cell is subject to the negative feedback regulation of ICOS-ICOSL interaction (Watanabe et al., 2008), while ICOS expression on TFH cells seems under the negative regulation of transcriptional suppressor FOXP1 (Wang et al., 2014). ICOS signaling inactivates FOXO1, which strongly inhibits TFH cell development via negatively regulating BCL6 expression (Stone et al., 2015). ICOS-ICOSL interaction is also required for the persistence of TFH cells and GC responses by down-regulating KLF2, which serves to inhibit TFH cell differentiation (Weber et al., 2015). In addition to conduct TCR signaling, PI3K signaling acts to mediate the TFH-promoting function of ICOS (Gigoux et al., 2009). Rolf et al. showed that the number of TFH cells, GC B cells, and high-affinity antibody-secreting cells is correlated with the magnitude of PI3K signaling (Rolf et al., 2010). SAP expression on TFH cell is critical for the formation of T-B interaction (Qi et al., 2008; Cannons et al., 2010a), which is indispensable for GC TFH cell differentiation. Moreover, SAP actively participates in the modulation of TCR signaling in TFH cells (Cannons et al., 2004; Cannons et al., 2010b). In addition, CD40L expressed on TFH cells is essential for the GC B cells survival and GC maintenance as well as the function of TFH cells (Elgueta et al., 2009; Crotty, 2011; Vinuesa et al., 2016).
Cytokines Shape TFH Lineage Differentiation
Cytokine milieu is pivotal to the lineage fate determination of CD4+ helper T cells. Unlike other subsets of CD4+ T cells which have the default cytokine-driving paradigm of lineage differentiation, for examples, IFN-γ and IL-12 promote TH1 differentiation, whereas IL-4 facilitates TH2 generation, TFH cells manifest no default cytokine-driving differentiation pathway. Though without determining cytokines, TFH cells can be shaped by multiple cytokine types (Pawlak et al., 2020).
IL-6 and IL-21 are important cytokines for TFH cell differentiation in mice (Karnowski et al., 2012), whereas IL-12, IL-23, and TGF-β play prominent roles in human TFH cell differentiation (Ma et al., 2009; Sweet et al., 2012; Schmitt et al., 2014). IL-21 is also an essential effector molecular of TFH cells, though it can be mainly expressed by both TH17 and TFH cells (Chtanova et al., 2004). It is generally acknowledged that IL-21 signaling is critical for the maintenance of GC in a B cell intrinsic mechanism (Linterman et al., 2010; Zotos et al., 2010). IL-6 produced by DCs induces early up-regulation of BCL6 in mouse TFH cells (Eto et al., 2011), as well as promotes the maintenance of TFH cells during chronic viral infection (Harker James et al., 2011). IL-21 also functions in an autocrine manner to support TFH cell responses (Nurieva et al., 2008). In human, plasmacytes-derived IL-6 induces the differentiation of circulating TFH cells (Chavele et al., 2015). Different with murine CD4+ T cells, human CD4+ T cells differentiate into IL-21 producing TFH cells with increased expression of CXCR5, ICOS, and BCL6 under the regulation of IL-12 rather than IL-6 (Ma et al., 2009; Schmitt et al., 2009). Moreover, TGF-β, which substantially inhibits TFH cell differentiation in mice, induces human TFH cell development via activating STAT3-STAT4 signaling (Schmitt et al., 2014).
Recently, an important role of IL-2 in controlling TFH/non-TFH cell commitment was revealed. By using IL-2 reporter mice, DiToro et al. demonstrated that naïve CD4+ T cells receiving highest TCR signals and producing IL-2 will differentiate into TFH cells, whereas IL-2 non-producers will differentiate into non-TFH cells (DiToro et al., 2018). Given abovementioned discrepant roles between certain human and mouse cytokines in regulating TFH cell differentiation, whether IL-2 signaling exerts the same or similar function on mouse and human TH1/TFH commitment needs further investigation.
Transcription Factors Regulate TFH Commitment and Differentiation
Intrinsic programs for TFH cell commitment and differentiation initiate very early upon activation (Baumjohann et al., 2011; Choi et al., 2011). BCL6 is required for TFH cell differentiation by inhibiting Blimp-1, which drives CD4+ T cells developing into non-TFH lineages (Johnston Robert et al., 2009; Nurieva Roza et al., 2009; Yu et al., 2009). BCL6 expression is associated with upregulation of CXCR5 and downregulation of CCR7 and PSGL1, allowing the migration of TFH cells to T-B border and GC (Hatzi et al., 2015). KLF2, another target of BCL6, impedes TFH cell differentiation via inducing expression of Prdm1. Tbx21, and Gata3, and meanwhile repressing Cxcr5 transcription via directly binding to its genomic region (Hatzi et al., 2015; Lee et al., 2015; Weber et al., 2015). Recent studies further elucidated that BCL6 also negatively regulates ID2 to facilitate TFH cell differentiation (Shaw et al., 2016) and positively regulates TOX2 to promote chromatin accessibility of TFH-associated genes (Xu et al., 2019).
Since BCL6 was identified to be the master transcription factor in TFH cell differentiation, plenty of transcription factors have been discovered to regulate TFH cell differentiation via directly or indirectly affecting BCL6 expression and function (Vinuesa et al., 2016; Choi et al., 2020; Schroeder et al., 2021). TCF1 and LEF1 initiate and promote TFH cell differentiation by ensuring the early expression of BCL6 and the repression of Blimp-1 (Choi et al., 2015; Wu et al., 2015; Xu et al., 2015). TCF1 is also involved in suppression of IL-2Rα (Wu et al., 2015), which together with Blimp-1 forms a negative feedback loop of TCF1/IL-2R/Blimp-1 regulating the TFH responses during viral infection. In addition, signal transducers and activator (STAT) 1 and 3 both contribute to TFH differentiation via IL-21 and IL-6 signaling (Nurieva et al., 2008). STAT3 and STAT4, in response to IL-12 and IL-23, cooperatively with TGF-β promote human but not mouse TFH cell differentiation by promoting TFH cell associated molecules (CXCR5, ICOS, IL-21, Bcl-6, etc.) expression and repressing Blimp-1 expression (Schmitt et al., 2014). IRF4 promotes TFH cell differentiation also through signals mediated by STATs (Nurieva et al., 2008) or other transcription factors (Huber and Lohoff, 2014). However, STAT5, in collaboration with Blimp-1 and IL-2 signals, is a potent negative regulator of TFH cell differentiation (Johnston et al., 2012). Also, BATF directly induces transcription of BCL6 and c-MAF in TFH cell to promote the TFH cell differentiation (Betz et al., 2010). Not surprisingly, absence of c-MAF decreases the amount of TFH cells and IL-21 production (Bauquet et al., 2009; Andris et al., 2017). Transcription factors EGR2/3 are also required for TFH cell differentiation and GC formation by regulating BCL6 (Ogbe et al., 2015). Moreover, ASCL2, which has multiple binding sites in Cxcr5 locus, directs the migration of TFH cells towards B cell follicles, and is essential for early TFH cell differentiation (Liu et al., 2014). Recently, Vacchio et al. revealed that Thpok promotes BCL6 and MAF to facilitate virus-specific TFH cell differentiation and GC formation in LCMV infection (Vacchio et al., 2019).
Two forkhead box proteins, FOXO1 and FOXP1, are demonstrated to negatively regulate TFH cell differentiation (Wang et al., 2014; Stone et al., 2015). FOXO1 closely binds to the region of Bcl6 locus, limiting the BCL6 expression and TFH cell development (Stone et al., 2015). FOXP1 directly binds to the Il21 promoter region to suppress IL-21 expression. In addition, FOXP1-deficient CD4+ T cells upregulate expression of ICOS during T cell activation (Wang et al., 2014), indicating a repressive role of FOXP1 on ICOS. In addition, a recent investigation showed that TOX2 acts to bind to and facilitate the chromatin accessibility of gene loci associated with TFH cell differentiation and function, including BCL6 (Xu et al., 2019).
Other Factors Regulating TFH Cell Differentiation
Epigenetic modulation is also involved in TFH cell differentiation. Besides abovementioned chromatin remodeling of TFH cell associated genes via BCL6-TOX2 (Xu et al., 2019), the histone methyltransferase EZH2 also plays an important role in epigenetic regulation of TFH cell differentiation. Using assay for transposase-accessible chromatin with high-throughput sequencing (ATAC-seq), Chen et al. demonstrated that EZH2 is essential for chromatin accessibility remodeling of TFH-associated genes at the early commitment of TFH cells (Chen et al., 2020a). Li et al. revealed that abundant EZH2 binding peaks overlapped with TCF1 peaks, explaining the defective differentiation of TFH cell with EZH2 deficiency (Li et al., 2018). Another histone methyltransferase, NSD2, which is induced by CD28 stimulation and sustained by ICOS signaling, is also required for the early expression of BCL6 and late maintenance of TFH cells (Long et al., 2020).
Moreover, T cell activation and differentiation always manifest substantial re-programming of cellular metabolism (MacIver et al., 2013; Chapman et al., 2020). The serine/threonine kinase mammalian target of rapamycin (mTOR) is a potent regulator of T cell response via sensing and integrating inputs from nutrients, growth factors, energy, and cellular stress (Chi, 2012; Yang and Chi, 2012; Huang et al., 2020). By down-regulating mTOR, Myr-Akt, and/or CD25 signals in LCMV-specific CD4+ T cells, Ray et al. demonstrated that IL-2/mTORC1 axis orchestrates the reciprocal balance between TFH and TH1 cell differentiation during viral infection (Ray et al., 2015). Further studies revealed the discrete regulatory roles of two different mTOR complexes, mTORC1 and mTORC2 (Yang et al., 2016; Zeng et al., 2016; Hao et al., 2018). Deficiency of mTORC1 substantially impairs cell proliferation and TFH cell differentiation, whereas mTORC2 is needed for TFH cell differentiation by promoting Akt activation and TCF1 expression without impacting cell proliferation (Yang et al., 2016). Hao et al. further demonstrated that mTORC2 signals induced by TCR and ICOS stimulation participates in cell migration, late differentiation and maturation of TFH cells (Hao et al., 2018). Recently, using in vivo CRISPR-Cas9 screening and functional validation in mice, Fu et al. revealed a direct regulatory function of de novo synthesis of phosphatidylethanolamine (PE) on TFH cell development via controlling surface expression of CXCR5 (Fu et al., 2021). Taken together, those findings highlight the metabolic control of TFH cell differentiation.
In addition, these are many other factors involved in regulating TFH cell commitment and differentiation. For example, RNA-binding protein Roquin exerts negative post-transcriptional regulation on TFH cells via binding to the TFH-associated genes like Icos, Ox40 (Vinuesa et al., 2005). Some microRNAs also play roles in post-transcriptional regulation of TFH cell differentiation. miR-17~92 promotes TFH cell differentiation by enhancing PI3K signaling as well as repressing non-TFH genes (Baumjohann et al., 2013a). miR-155 promotes TFH cell differentiation during chronic inflammation in which miR-155-knockout diminished the accumulation of TFH cells (Hu et al., 2014). In addition, miR-146a acts as a post-transcriptional repressor to ICOS-ICOSL signaling and the subsequent TFH cell differentiation and GC responses (Pratama et al., 2015). Also, NOTCH1/2 deficiency in CD4+ T cell reduces the expression of BCL6, IL-21, and CXCR5, but increases Blimp-1 expression, resulting in impairment of the development and function of TFH cells. Additional factors have been verified in regulating TFH cell differentiation, including but not limited to the E3 ligase cullin 3 (Cul3) (Mathew et al., 2014), heterotrimeric G protein Gα13 (Kuen et al., 2021), kinase PRKD2 (Misawa et al., 2020), kinase PDK1 (Sun et al., 2021), extracellular matrix protein 1 (ECM1) (He et al., 2018), and stromal cell-derived DEL-1 (Wang et al., 2021).
Memory Virus-Specific TFH Cells
Identification of Virus-Specific Memory TFH Cells
In acute viral infection or vaccination, a small proportion of the antigen-experienced CD4+ T cells survive after antigen clearance, subsequently become the memory CD4+ helper T cells. In addition to survival capacity and homeostatic proliferation without antigenic stimulation, a memory CD4+ helper T cell also need to preserve the lineage features during resting and recall responses (MacLeod et al., 2009; Hale and Ahmed, 2015). In the first report describing the existence of antigen-specific memory TFH cells, Fazilleau et al. found a group of antigen-specific CXCR5+ICOSlo TFH cells in the memory phase of protein vaccination (Fazilleau et al., 2007). It is worth noting that those CXCR5+ICOSlo cells were retained along with persistent peptide-MHCII (Fazilleau et al., 2007), which raises a question about the true memory property of TFH cells. By adoptive transfer of TCR transgenic antigen-specific CD4+ T cells or TFH/non-TFH cells into second recipients, MacLeod et al. further confirmed the existence of antigen-specific memory TFH cells after protein immunization (MacLeod et al., 2011). Accumulating studies further demonstrated the validity and characteristics of virus-specific memory TFH cells during viral infection and vaccination (Weber et al., 2012; Bentebibel et al., 2013; Hale et al., 2013; Locci et al., 2013).
Memory TFH cells are usually marked by co-expression of CXCR5, CCR7, CD62L, and FR4, along with downregulated expression of PD1, ICOS, Ly6c, and BCL6 (Iyer et al., 2013; Hale and Ahmed, 2015). Memory TFH cells exert a superior help function on naïve B cells than primary responding TFH cells (MacLeod et al., 2011). In recall responses, virus-specific TFH cells provide immediate help to virus-specific memory B cells (He et al., 2013; Locci et al., 2013; Phan and Tangye, 2017). In addition, local memory TFH cells colocalized with B cells within the parenchymal lung tissues are critical for the production of virus-specific B cells and antibodies (Son Young et al., 2021).
Formation of Virus-Specific Memory TFH Cells
A consensus is that virus-specific memory CD4+ T cells are progenies of corresponding effector CD4+ T cells, the so-called memory precursors generated during effector phase of acute viral infection (Hale and Ahmed, 2015). But the high plasticity (Zhou et al., 2009) and non-default differentiation pathway (Choi et al., 2020) of TFH cells make it hard to track a destined memory precursor TFH cell at early effector phase. Some studies suggest formation of memory TFH cells can be prior to GC development (He et al., 2013; Tsai and Yu, 2014). However, given the antigen retention in follicle and a rather long time of persistent GC reaction, another view is that since TFH cells can shuttle between different GCs, and when they emigrate into follicles where no presented antigens exist, they acquire less activated phenotypes, resultantly, these TFH cells gradually differentiate into memory cells with a resting state (Kitano et al., 2011; Choi et al., 2013).
Since TFH cells are prone to stay at GC while TH1 cells migrate into infected location, TFH cells are regarded to be more likely to differentiate into central memory TFH cells (Zhu et al., 2010). Nevertheless, distribution of memory TFH cells is not necessarily limited in GCs. Circulating HIV-specific effector memory TFH cells are potent players for immune surveillance in situ (Locci et al., 2013). Moreover, iBALT-resident memory TFH cells in lung are essential for the robust recall humoral responses and provide help to local CD8+ TRM cells (Pruner and Pepper, 2021; Son Young et al., 2021).
Early study showed that CD30 and OX40 signals are needed to form CD4+ T cell memory (Gaspal et al., 2005). Further investigations revealed that memory TFH cells down-regulate BCL6 while retain the surface expression of CXCR5 (Hale and Ahmed, 2015). ASCL2 may be important for the expression of CXCR5 in memory TFH cells, because it binds to the conserved non-coding sequence regions of Cxcr5 locus to promote Cxcr5 transcription without inducing BCL6 expression (Liu et al., 2014). FR4, highly expressed by naïve CD4+ T cells, is down-regulated upon activation and re-expressed on TFH cells (Iyer et al., 2013), and is maintained on memory TFH cells as CXCR5 does (Hale and Ahmed, 2015). What’s more, recent study shows that long-lived TFH cells persisted for over 400 days after infection are marked by high expression of FR4 (Künzli et al., 2020). Yet the mechanism driving the FR4 expression is still unknown. In addition, Ciucci et al. recently showed that Thpok is required for the signatures and emergence of memory CD4+ T cell via antagonizing the expression of Blimp-1 and Runx3 (Ciucci et al., 2019). Overall, mechanisms underlying the formation of virus-specific memory TFH cells remain largely unknown. The imprinting effects from effector phase, the various features induced by different niches (GC in SLO, peripheral residence, or circulation), and driving force of the continuous expression of CXCR5, are parts of the unsolved issues about TFH cell memory formation.
Lineage Maintenance Versus Plasticity
CD4+ helper T cells are featured with high plasticity. Several studies showed that adoptively transferred memory TFH cells eventually differentiate into both TFH and non-TFH cells in second recipients upon rechallenge (Pepper et al., 2011; Lüthje et al., 2012; Künzli et al., 2020). The plasticity of TFH cells is highlighted by the work of Lu et al, in which the authors found that TFH cells harboring chromatin accessibility of Tbx21, Gata3, and Rorc genes that could drive CD4+ Helper T cells to differentiate into TH1, TH2, and TH17 cells under the respective polarizing conditions (Lu et al., 2011).
Given the extrinsic factors, like antigens and effector cytokine milieu (O’Shea John and Paul William, 2010), which promote T helper cell differentiation, are absent after viral clearance, it is reasonable to consider that intrinsic programs play a dominant role in sustaining memory virus-specific TFH cell lineage during memory phase. However, the master transcription factor of TFH cells like BCL6, and other factors constraining T helper cell lineages are down-regulated in memory CD4+ T cells (Hale and Ahmed, 2015). How do memory T cells retain lineage commitment? It is still an unsolved question. Transcriptional and epigenetic profiling suggest that epigenetic remodeling during effector phase may play an important role in retaining lineage characteristics in memory TFH cells (Wilson et al., 2009; Josefowicz, 2013; Youngblood et al., 2013). In addition, memory B cells conduct immediate antigen presentation to memory TFH cells, inducing rapid re-expression of BCL6 in reactivated TFH cells (Ise et al., 2014), which safeguards the TFH-oriented recall responses.
Longevity of Virus-Specific Memory TFH Cells
Following the clearance of virus in acute viral infection, virus-specific memory CD8+ T cells can persist for a very long period at a stable level, whereas virus-specific memory CD4+ T cells gradually decay over time (Homann et al., 2001). However, there is much less knowledge about how memory CD4+ T cells sustain longevity compared to that of memory CD8+ T cells. By transcriptional profiling, Song et al. reveal multiple genetic programs contributed to the longevity of antigen-specific memory CD4+ T cells by maintaining T cell quiescence (Song et al., 2020). In a mouse model of acute infection with LCMV, Wang et al. demonstrated that the mTORC2-Akt-GSK3β axis is critical for the longevity of virus-specific memory TFH and TH1 cells by protecting these memory cells from ferroptosis, however this signaling axis seems to be non-essential for memory CD8+ T cells (Wang et al., 2022c). The tonic mTORC2 activity in virus-specific memory CD4+ T cells is sustained by the IL-7 signaling at memory phase, which suggests an important role of IL-7 signaling in memory TFH cell maintenance (Wang et al., 2022c). Given the essential functions of mTOR signaling pathway in regulating cellular metabolism and the divergent role of mTORC2 in memory CD4+ T cells and CD8+ T cells (Pollizzi et al., 2015), it is reasonable to speculate that there are certain differences either in metabolic features or/and redox homeostasis between virus-specific memory CD4+ and CD8+ T cells. Paired comparison analysis of memory CD4+ and CD8+ T cells may provide valuable clues in further understanding memory CD4+ T cell longevity.
Virus-Specific TFH Cells in COVID-19
TFH Cell Responses in SARS-CoV-2-Infection and Vaccination
Severe Acute Respiratory Syndrome Coronavirus (SARS-CoV-2) is an enveloped single-stranded positive-sense RNA virus accounting for the ongoing pandemic of COVID-19. SARS-CoV-2 genome encodes 4 structural proteins including surface spike (S) glycoprotein, membrane protein (M), envelop protein (E), and nucleoprotein (N). The S protein of SARS-CoV-2 facilitates viral attachment and entry through the engagement with its cognate receptor, angiotensin converting enzyme-2 (ACE2), which is mediated by the receptor binding domain (RBD) within the S1 subunit of S protein (V'Kovski et al., 2021). SARS-CoV-2 infection induces both humoral and cellular immune responses in hosts (Post et al., 2020; Sette and Crotty, 2021; Shrotri et al., 2021). Commonly, neutralizing antibodies are vital for sterilizing immunity of viral infection. Antibodies targeting S protein and RBD are capable of blocking the attachment and entry of SARS-CoV-2 into host cells to prevent COVID-19 infection (Robbiani et al., 2020; Gupta et al., 2021; Wang et al., 2022a; Wang et al., 2022b). The production of neutralizing antibodies targeting SARS-CoV-2 with high magnitude and durability requires potent B cell responses with the help from virus-specific TFH cells. Evidences from non-human primate studies validated that natural infection of SARS-CoV-2 protects rhesus macaques from reinfection, indicating the occurrence of protective immunological memory post SARS-CoV-2 infection (Chandrashekar et al., 2020; Deng et al., 2020). Both two studies verified viral S protein-specific memory CD4+ T cells in rhesus macaques after infection of SARS-CoV-2; moreover, Deng et al. further detected the increased viral S protein-specific central memory CD4+ T cells in lymph nodes of rhesus macaques post rechallenge of SARS-CoV-2. Given the bimodal differentiation of virus-specific TFH and TH1 cells in acute infection, these studies highlight the importance of virus-specific TFH cells in SARS-CoV-2 natural immunity and vaccine-induced immunological protection.
Vaccine candidates that can induce sufficient antibodies targeting S protein and RBD are reckoned to be protective against SARS-CoV-2 infection. Therefore, investigations on SARS-CoV-2-specifc T cell responses, especially the virus-specific TFH cell’s help to cognate B cell, help to accelerate the vaccine testing pipeline and contribute to vaccine development under the pandemic COVID-19. Currently, a plethora of vaccines are applied to fight against COVID-19, including mRNA vaccines, adenoviral vector-based vaccines, recombinant protein vaccines, and inactivated virus vaccines (Grigoryan and Pulendran, 2020). According to the results from clinical trials, all vaccines induce detectable antibodies against SARS-CoV-2. To now, the most efficient vaccine platform is mRNA/LNP, which delivers mRNA encoding S protein of SARS-CoV-2 by lipid nanoparticle (LNP) to host cells and induces robust immune responses towards the S antigen (Laczkó et al., 2020; Painter et al., 2021; Mudd et al., 2022). Immunization with SARS-CoV-2 mRNA vaccines foster potent antigen-specific GC responses (Laczkó et al., 2020) and virus-specific CD4+ T cells (Painter et al., 2021) to generate robust neutralizing antibody responses. By analyzing T cells from samples of lymph nodes acquired by fine-needle aspiration from donors who received mRNA vaccines, Mudd et al. further underscored the vital role of virus-specific TFH cell responses in mRNA vaccine-induced robust and durable immunological protection against SARS-CoV-2 (Mudd et al., 2022).
The Functions of SARS-CoV-2-Specific TFH Cells
In infected individuals, the durable neutralization and memory B cells can be predicted by prompt CD4+ T cell responses, especially the strong circulating TFH (cTFH) cell responses (Gong et al., 2020; Boppana et al., 2021; Narowski et al., 2022). Nevertheless, the imbalanced humoral and cellular immunity were often observed in COVID-19 patients (Oja et al., 2020; Gao et al., 2021). Among convalescents, stronger antibody responses were observed in individuals experienced a severe COVID-19, compared to those got moderate symptoms or asymptomatic individuals (Chen et al., 2020b; Gudbjartsson et al., 2020; Röltgen et al., 2020). In some severe sick patients, remarkably strong virus-specific IgG responses were observed, along with decreased CD4+ T cell responses (Oja et al., 2020). By sequencing the B cell receptor repertoires, Schultheiß et al. further found that individuals who have much severer clinical course got a markedly lower percentage of B cells carrying un-mutated BCRs (Schultheiß et al., 2020), which indicates a profound TFH cell-mediated SHM-and-selection have occurred. Above data suggest a gradually compromise of CD4+ T cell responses during COVID-19 progression. Impaired GC reaction was also observed in some cases, of which Naoki Kaneko et al. showed that GC and BCL6+ TFH and B cells were absent in lymph nodes and spleens from severely SARS-CoV-2 infected patients who eventually succumbed after admission (Kaneko et al., 2020). Altogether, these findings emphasize the essential role of virus-specific TFH cells in natural immunity to control COVID-19.
Evidences of robust responses of SARS-CoV-2-specific TFH cell responses upon administration of mRNA vaccines suggest that virus-specific TFH cell responses contribute to the successful immunization of this preeminent vaccine platform (Painter et al., 2021; Mudd et al., 2022). Moreover, using TFH cell-deletion (TFH-DTR) mice, Cavazzoni et al. showed that reduction of TFH cells results in compromised GC responses and decreased production of anti-S and anti-RBD IgG upon SARS-CoV-2 protein vaccination (Cavazzoni et al., 2022), suggesting that virus-specific TFH cell response is also essential to establish an optimized immune protection in traditional immunization strategy.
On the other hand, TFH cells may also play a pathogenic role in certain circumstances. Wang et al. showed that the hyper-functional CD8+ and CD4+ T cells were associated with the pathogenesis of extremely severe COVID-19 patients (Wang et al., 2020). In another study conducted by Meckiff et al., the authors found compared to non-hospitalized COVID-19 patients, increased cytotoxic TFH cells which manifest high production of IFN-γ, IL-2, and TNF-α were observed in hospitalized patients with severe illness (Meckiff et al., 2020). In addition, Fenoglio et al. revealed the pathogenic roles of CCR4+ and CCR6+ TFH cells in COVID-19 patients (Fenoglio et al., 2021). Moreover, it is necessary to pay additional attentions to COVID-19 patients who also suffer from other diseases associated with TFH cell responses, like HIV-infection, autoimmune diseases and cancers treated with immune checkpoint blockade (ICB) therapies (Picchianti-Diamanti et al., 2021; Riou et al., 2021; Salomé and Horowitz, 2021).
Memory SARS-Cov-2-Specific TFH Cells
Bacher et al. suggested that there are pre-existing memory T cells with low avidity and a cross-reactivity to SARS-CoV-2 in unexposed individuals (Bacher et al., 2020). Given the observation that excessive but low-avidity T cell response to SARS-CoV-2 features the severe COVID-19 but not the mild disease, Bacher et al. questions the protective role of pre-existing cross-reactive memory T cells in anti-SARS-CoV-2 immunity (Bacher et al., 2020). However, other investigations targeting T cell responses to SARS-CoV-2 in unexposed individuals revealed that cross-reactive memory TFH cells could trigger a rapid and superior antibody response to SARS-CoV-2, which might exert better viral control in upper respiratory tract and lung (Grifoni et al., 2020; Lipsitch et al., 2020). The latter view was appreciated by Bonifacius et al., who showed that COVID-19 patients with pre-existing anti-human coronavirus CD4+ and CD8+ T cells with cross-reactivity of endemic coronaviruses manifested higher frequency of SARS-CoV-2 S protein-specific T cells (Bonifacius et al., 2021). In addition, Mateus et al. observed a significantly higher frequency of viral S protein-specific CD4+ T cells and stronger neutralizing antibody responses in vaccinated individuals who present pre-existing SARS-CoV-2 S protein-specific CD4+ T cells than subjects with no cross-reactive memory (Mateus et al., 2021). Therefore, besides the classical functions of virus-specific memory TFH cells, the pre-existing virus-specific TFH cells also function to favor T cell and antibody responses against SARS-CoV-2.
Following SARS-CoV-2 infection, substantial virus-specific T cell memory responses are induced in convalescent individuals, whereas the breadth and magnitude were positively correlated to the disease severity of COVID-19 (Peng et al., 2020). The longitude study of T/B cell and antibody responses to COVID-19 revealed a long duration of CD8+ and CD4+ T cell memory and neutralizing antibodies against SARS-CoV-2 (Dan et al., 2021). In general, accruing evidences suggest a profound protection of nature infection of COVID-19 and vaccination. The emerging incompetent immune protection from vaccination or immunological memory of convalescent individuals is more associated with the rapid evolution of SARS-CoV-2 variants (Thakur et al., 2022; Woldemeskel et al., 2022). SARS-CoV-2 continuously undergoes genetic mutations or viral recombination, resulting in variants with possible differences in transmissibility, clinical manifestation, and immunogenicity. To date, the variant of concern (VOC) is Omicron variant (B.1.1.529), which harbors as many as 36 substitutions in viral S protein and total 59 mutations in whole genome compared with SARS-CoV-2 ancestral strain, leading to immune evasion from neutralization by vaccination- and infection-induced antibodies (Woldemeskel et al., 2022). Although Omicron evade a large fraction of antibodies, its neutralizing antibodies are still represented in a portion of memory B cell repertoire induced by mRNA vaccines (Sokal et al., 2022).
Multiple studies revealed that despite the decay of protective serologic components and decreased effectiveness against infection, vaccines developed based on ancestral strains still efficiently protect individuals from SARS-CoV-2 variants-induced hospitalization and/or severe diseases (Nanduri et al., 2021; Rosenberg et al., 2021; Tenforde et al., 2021). This phenomenon highlights the relative impervious function of T cell immunity against SARS-CoV-2 variants, given that virus-specific T cells mainly act to eliminate infected cells (CD8+ T cell) and help B/CD8+ T cells responses after activation (CD4+ T cell), rather than to directly prevent infection like neutralizing antibodies do. In addition, a recent study demonstrated that the preserved T cell reactivity to variant Omicron variant in most infected and vaccinated individuals can be enhanced shortly after booster vaccination (Naranbhai et al., 2022). In this study, Naranbhai et al. identified about 79% individuals with a preserved T cell reactivity to the viral S protein of Omicron. Moreover, the effector T cell responses to SARS-CoV-2, including both wild type and Omicron strain, were enhanced after additional booster vaccine, accompanied with proliferative memory viral S protein-specific CD4+ T cell responses but reduced CD8+ T cell responses to Omicron. This evidence indicates that virus-specific memory T cell, especially virus-specific memory CD4+ T cell, is a silver lining to the plight of controlling circulating SARS-CoV-2 variants. Notably, heterologous vaccination was used in the abovementioned infection/vaccination-booster stratagem, providing corroborative evidence to the effectiveness of the heterologous sequential vaccination strategy against mutant VOCs of SARS-CoV-2. Compared to homologous vaccination, multiple kinds of heterologous sequential immunization were proved to be superior to induce broad neutralization against VOCs, including combination of inactivated vaccine followed by heterologous mutant RBD vaccine (Song et al., 2022), adenoviral vectored vaccine followed by mRNA vaccine (Pozzetto et al., 2021), inactivated vaccine followed by mRNA vaccine (Zuo et al., 2022), and inactivated vaccine followed by adenoviral vectored vaccine (Li et al., 2022). Given the barely affected memory CD4+ T cell responses to peptide pool of Omicron S protein (Naranbhai et al., 2022), it is thus clear that virus-specific memory TFH cells play an important role in the generation of potent and broad neutralizing antibodies to VOCs induced by the heterologous sequential vaccination strategy.
Concluding Remarks
Since the initial seminal description of TFH cells in 2000 (Breitfeld et al., 2000; Schaerli et al., 2000), many characteristics, functions, and underlying mechanisms of TFH cells have been uncovered over the past two decades. We illustrate the multiple lines and underpinnings of the TFH cell differentiation and maintenance during viral infection in Figure 1. But still a lot of puzzles remain to be solved. For example, the extrinsic and intrinsic factors that determine the fate commitment of antigen-specific TFH cells are still unknown. Maybe the differentiation of TFH cells is not a “default” pathway (Choi et al., 2020), and deciphering the networks regulating TFH cell differentiation needs more intense investigations. In addition, how memory TFH cells retain lineage features and prolong over time is fascinating and merits further studies. Better understanding of virus-specific TFH cells will be of great importance for optimizing anti-viral vaccine development, including SARS-CoV-2 vaccines.
Author Contributions
YW and QT drafted and revised the manuscript with LY. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by grants from the National Key Research Development Plan (2021YFC2300502), and the National Natural Science Foundation of China (No. 32030041 to LY, No. 31800748 to YW, and No. 31800742 to QT).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Akiba, H., Takeda, K., Kojima, Y., Usui, Y., Harada, N., Yamazaki, T., et al. (2005). The Role of ICOS in the CXCR5+ Follicular B Helper T Cell Maintenance In Vivo. J. Immunol. 175 (4), 2340. doi: 10.4049/jimmunol.175.4.2340
Andris, F., Denanglaire, S., Anciaux, M., Hercor, M., Hussein, H., Leo, O. (2017). The Transcription Factor C-Maf Promotes the Differentiation of Follicular Helper T Cells. Front. Immunol. 8. doi: 10.3389/fimmu.2017.00480
Arroyo, E. N., Pepper, M. (2019). B Cells are Sufficient to Prime the Dominant CD4+ Tfh Response to Plasmodium Infection. J. Exp. Med. 217 (2), e20190849. doi: 10.1084/jem.20190849
Bacher, P., Rosati, E., Esser, D., Martini, G. R., Saggau, C., Schiminsky, E., et al. (2020). Low-Avidity CD4(+) T Cell Responses to SARS-CoV-2 in Unexposed Individuals and Humans With Severe COVID-19. Immunity 53 (6), 1258–1271.e1255. doi: 10.1016/j.immuni.2020.11.016
Barnett, L. G., Simkins, H. M. A., Barnett, B. E., Korn, L. L., Johnson, A. L., Wherry, E. J., et al. (2014). B Cell Antigen Presentation in the Initiation of Follicular Helper T Cell and Germinal Center Differentiation. J. Immunol. (Baltimore Md. 1950) 192 (8), 3607–3617. doi: 10.4049/jimmunol.1301284
Baumjohann, D., Kageyama, R., Clingan, J. M., Morar, M. M., Patel, S., de Kouchkovsky, D., et al. (2013a). The microRNA Cluster miR-17∼92 Promotes TFH Cell Differentiation and Represses Subset-Inappropriate Gene Expression. Nat. Immunol. 14 (8), 840–848. doi: 10.1038/ni.2642
Baumjohann, D., Okada, T., Ansel, K. M. (2011). Cutting Edge: Distinct Waves of BCL6 Expression During T Follicular Helper Cell Development. J. Immunol. 187 (5), 2089. doi: 10.4049/jimmunol.1101393
Baumjohann, D., Preite, S., Reboldi, A., Ronchi, F., Ansel, K. M., Lanzavecchia, A., et al. (2013b). Persistent Antigen and Germinal Center B Cells Sustain T Follicular Helper Cell Responses and Phenotype. Immunity 38 (3), 596–605. doi: 10.1016/j.immuni.2012.11.020
Bauquet, A. T., Jin, H., Paterson, A. M., Mitsdoerffer, M., Ho, I. C., Sharpe, A. H., et al. (2009). The Costimulatory Molecule ICOS Regulates the Expression of C-Maf and IL-21 in the Development of Follicular T Helper Cells and TH-17 Cells. Nat. Immunol. 10 (2), 167–175. doi: 10.1038/ni.1690
Bentebibel, S.-E., Lopez, S., Obermoser, G., Schmitt, N., Mueller, C., Harrod, C., et al. (2013). Induction of ICOS+CXCR3+CXCR5+ TH Cells Correlates With Antibody Responses to Influenza Vaccination. Sci. Trans. Med. 5 (176), 176ra132–176ra132. doi: 10.1126/scitranslmed.3005191
Betz, B. C., Jordan-Williams, K. L., Wang, C., Kang, S. G., Liao, J., Logan, M. R., et al. (2010). Batf Coordinates Multiple Aspects of B and T Cell Function Required for Normal Antibody Responses. J. Exp. Med. 207 (5), 933–942. doi: 10.1084/jem.20091548
Bonifacius, A., Tischer-Zimmermann, S., Dragon, A. C., Gussarow, D., Vogel, A., Krettek, U., et al. (2021). COVID-19 Immune Signatures Reveal Stable Antiviral T Cell Function Despite Declining Humoral Responses. Immunity 54 (2), 340–354.e346. doi: 10.1016/j.immuni.2021.01.008
Boppana, S., Qin, K., Files, J. K., Russell, R. M., Stoltz, R., Bibollet-Ruche, F., et al. (2021). SARS-CoV-2-Specific Circulating T Follicular Helper Cells Correlate With Neutralizing Antibodies and Increase During Early Convalescence. PloS Pathog. 17 (7), e1009761–e1009761. doi: 10.1371/journal.ppat.1009761
Bossaller, L., Burger, J., Draeger, R., Grimbacher, B., Knoth, R., Plebani, A., et al. (2006). ICOS Deficiency Is Associated With a Severe Reduction of CXCR5+ CD4 Germinal Center Th Cells. J. Immunol. 177 (7), 4927. doi: 10.4049/jimmunol.177.7.4927
Breitfeld, D., Ohl, L., Kremmer, E., Ellwart, J., Sallusto, F., Lipp, M., et al. (2000). Follicular B Helper T Cells Express Cxc Chemokine Receptor 5, Localize to B Cell Follicles, and Support Immunoglobulin Production. J. Exp. Med. 192 (11), 1545–1552. doi: 10.1084/jem.192.11.1545
Cannons, J. L., Qi, H., Lu, K. T., Dutta, M., Gomez-Rodriguez, J., Cheng, J., et al. (2010a). Optimal Germinal Center Responses Require a Multistage T Cell:B Cell Adhesion Process Involving Integrins, SLAM-Associated Protein, and CD84. Immunity 32 (2), 253–265. doi: 10.1016/j.immuni.2010.01.010
Cannons, J. L., Wu, J. Z., Gomez-Rodriguez, J., Zhang, J., Dong, B., Liu, Y., et al. (2010b). Biochemical and Genetic Evidence for a SAP-PKC-θ Interaction Contributing to IL-4 Regulation. J. Immunol. 185 (5), 2819. doi: 10.4049/jimmunol.0902182
Cannons, J. L., Yu, L. J., Hill, B., Mijares, L. A., Dombroski, D., Nichols, K. E., et al. (2004). SAP Regulates TH2 Differentiation and Pkcθ-Mediated Activation of NF-κb1. Immunity 21 (5), 693–706. doi: 10.1016/j.immuni.2004.09.012
Cavazzoni, C. B., Hanson, B. L., Podestà, M. A., Bechu, E. D., Clement, R. L., Zhang, H., et al. (2022). Follicular T Cells Optimize the Germinal Center Response to SARS-CoV-2 Protein Vaccination in Mice. Cell Rep. 38 (8), 110399–110399. doi: 10.1016/j.celrep.2022.110399
Caza, T., Landas, S. (2015). Functional and Phenotypic Plasticity of CD4+ T Cell Subsets. BioMed. Res. Int. 2015, 521957. doi: 10.1155/2015/521957
Chandrashekar, A., Liu, J., Martinot, A. J., McMahan, K., Mercado, N. B., Peter, L., et al. (2020). SARS-CoV-2 Infection Protects Against Rechallenge in Rhesus Macaques. Sci. (New York N.Y.) 369 (6505), 812–817. doi: 10.1126/science.abc4776
Chapman, N. M., Boothby, M. R., Chi, H. (2020). Metabolic Coordination of T Cell Quiescence and Activation. Nat. Rev. Immunol. 20 (1), 55–70. doi: 10.1038/s41577-019-0203-y
Chavele, K.-M., Merry, E., Ehrenstein, M. R. (2015). Cutting Edge: Circulating Plasmablasts Induce the Differentiation of Human T Follicular Helper Cells via IL-6 Production. J. Immunol. 194 (6), 2482. doi: 10.4049/jimmunol.1401190
Chen, X., Cao, G., Wu, J., Wang, X., Pan, Z., Gao, J., et al. (2020a). The Histone Methyltransferase EZH2 Primes the Early Differentiation of Follicular Helper T Cells During Acute Viral Infection. Cell. Mol. Immunol. 17 (3), 247–260. doi: 10.1038/s41423-019-0219-z
Chen, X., Pan, Z., Yue, S., Yu, F., Zhang, J., Yang, Y., et al. (2020b). Disease Severity Dictates SARS-CoV-2-Specific Neutralizing Antibody Responses in COVID-19. Signal Transduction Targeted Ther. 5 (1), 180. doi: 10.1038/s41392-020-00301-9
Chi, H. (2012). Regulation and Function of mTOR Signalling in T Cell Fate Decisions. Nat. Rev. Immunol. 12 (5), 325–338. doi: 10.1038/nri3198
Choi, J., Diao, H., Faliti, C. E., Truong, J., Rossi, M., Bélanger, S., et al. (2020). Bcl-6 is the Nexus Transcription Factor of T Follicular Helper Cells via Repressor-of-Repressor Circuits. Nat. Immunol. 21 (7), 777–789. doi: 10.1038/s41590-020-0706-5
Choi, Y. S., Gullicksrud, J. A., Xing, S., Zeng, Z., Shan, Q., Li, F., et al. (2015). LEF-1 and TCF-1 Orchestrate T(FH) Differentiation by Regulating Differentiation Circuits Upstream of the Transcriptional Repressor Bcl6. Nat. Immunol. 16 (9), 980–990. doi: 10.1038/ni.3226
Choi, Y. S., Kageyama, R., Eto, D., Tania, C., Robert, J., Monticelli, L., et al. (2011). ICOS Receptor Instructs T Follicular Helper Cell Versus Effector Cell Differentiation via Induction of the Transcriptional Repressor Bcl6. Immunity 34 (6), 932–946. doi: 10.1016/j.immuni.2011.03.023
Choi, Y. S., Yang, J. A., Yusuf, I., Johnston, R. J., Greenbaum, J., Peters, B., et al. (2013). Bcl6 Expressing Follicular Helper CD4 T Cells are Fate Committed Early and Have the Capacity to Form Memory. J. Immunol. (Baltimore Md. 1950) 190 (8), 4014–4026. doi: 10.4049/jimmunol.1202963
Chtanova, T., Tangye, S. G., Newton, R., Frank, N., Hodge, M. R., Rolph, M. S., et al. (2004). T Follicular Helper Cells Express a Distinctive Transcriptional Profile, Reflecting Their Role as Non-Th1/Th2 Effector Cells That Provide Help for B Cells. J. Immunol. 173 (1), 68. doi: 10.4049/jimmunol.173.1.68
Ciucci, T., Vacchio, M. S., Gao, Y., Tomassoni Ardori, F., Candia, J., Mehta, M., et al. (2019). The Emergence and Functional Fitness of Memory CD4(+) T Cells Require the Transcription Factor Thpok. Immunity 50 (1), 91–105.e104. doi: 10.1016/j.immuni.2018.12.019
Crotty, S. (2011). Follicular Helper CD4 T Cells (TFH). Annu. Rev. Immunol. 29 (1), 621–663. doi: 10.1146/annurev-immunol-031210-101400
Crotty, S. (2014). T Follicular Helper Cell Differentiation, Function, and Roles in Disease. Immunity 41 (4), 529–542. doi: 10.1016/j.immuni.2014.10.004
Cui, C., Wang, J., Fagerberg, E., Chen, P.-M., Connolly, K. A., Damo, M., et al. (2021). Neoantigen-Driven B Cell and CD4 T Follicular Helper Cell Collaboration Promotes Anti-Tumor CD8 T Cell Responses. Cell 184 (25), 6101–6118.e6113. doi: 10.1016/j.cell.2021.11.007
Dahlgren, M. W., Gustafsson-Hedberg, T., Livingston, M., Cucak, H., Alsén, S., Yrlid, U., et al. (2015). T Follicular Helper, But Not Th1, Cell Differentiation in the Absence of Conventional Dendritic Cells. J. Immunol. 194 (11), 5187–5199. doi: 10.4049/jimmunol.1401938
Dan, J. M., Mateus, J., Kato, Y., Hastie, K. M., Yu, E. D., Faliti, C. E., et al. (2021). Immunological Memory to SARS-CoV-2 Assessed for Up to 8 Months After Infection. Sci. (New York N.Y.) 371 (6529), eabf4063. doi: 10.1126/science.abf4063
Das, A., Ranganathan, V., Umar, D., Thukral, S., George, A., Rath, S., et al. (2017). Effector/memory CD4 T Cells Making Either Th1 or Th2 Cytokines Commonly Co-Express T-Bet and GATA-3. PloS One 12 (10), e0185932–e0185932. doi: 10.1371/journal.pone.0185932
Deng, W., Bao, L., Liu, J., Xiao, C., Liu, J., Xue, J., et al. (2020). Primary Exposure to SARS-CoV-2 Protects Against Reinfection in Rhesus Macaques. Sci. (New York N.Y.) 369 (6505), 818–823. doi: 10.1126/science.abc5343
Di Niro, R., Lee, S.-J., Heiden, V., Jason, A., Rebecca, A., Trivedi, N., et al. (2015). Salmonella Infection Drives Promiscuous B Cell Activation Followed by Extrafollicular Affinity Maturation. Immunity 43 (1), 120–131. doi: 10.1016/j.immuni.2015.06.013
DiToro, D., Winstead Colleen, J., Pham, D., Witte, S., Andargachew, R., Singer Jeffrey, R., et al. (2018). Differential IL-2 Expression Defines Developmental Fates of Follicular Versus Nonfollicular Helper T Cells. Science 361 (6407), eaao2933. doi: 10.1126/science.aao2933
Elgueta, R., Benson, M. J., De Vries, V. C., Wasiuk, A., Guo, Y., Noelle, R. J. (2009). Molecular Mechanism and Function of CD40/CD40L Engagement in the Immune System. Immunol. Rev. 229 (1), 152–172. doi: 10.1111/j.1600-065X.2009.00782.x
Eto, D., Lao, C., DiToro, D., Barnett, B., Escobar, T. C., Kageyama, R., et al. (2011). IL-21 and IL-6 are Critical for Different Aspects of B Cell Immunity and Redundantly Induce Optimal Follicular Helper CD4 T Cell (Tfh) Differentiation. PloS One 6 (3), e17739–e17739. doi: 10.1371/journal.pone.0017739
Fahey, L. M., Wilson, E. B., Elsaesser, H., Fistonich, C. D., McGavern, D. B., Brooks, D. G. (2011). Viral Persistence Redirects CD4 T Cell Differentiation Toward T Follicular Helper Cells. J. Exp. Med. 208 (5), 987–999. doi: 10.1084/jem.20101773
Fazilleau, N., Eisenbraun, M. D., Malherbe, L., Ebright, J. N., Pogue-Caley, R. R., McHeyzer-Williams, L. J., et al. (2007). Lymphoid Reservoirs of Antigen-Specific Memory T Helper Cells. Nat. Immunol. 8 (7), 753–761. doi: 10.1038/ni1472
Fazilleau, N., McHeyzer-Williams, L. J., Rosen, H., McHeyzer-Williams, M. G. (2009). The Function of Follicular Helper T Cells is Regulated by the Strength of T Cell Antigen Receptor Binding. Nat. Immunol. 10 (4), 375–384. doi: 10.1038/ni.1704
Fenoglio, D., Dentone, C., Parodi, A., Di Biagio, A., Bozzano, F., Vena, A., et al. (2021). Characterization of T Lymphocytes in Severe COVID-19 Patients. J. Med. Virol. 93 (9), 5608–5613. doi: 10.1002/jmv.27037
Fu, G., Guy, C. S., Chapman, N. M., Palacios, G., Wei, J., Zhou, P., et al. (2021). Metabolic Control of TFH Cells and Humoral Immunity by Phosphatidylethanolamine. Nature 595 (7869), 724–729. doi: 10.1038/s41586-021-03692-z
Gao, L., Zhou, J., Yang, S., Wang, L., Chen, X., Yang, Y., et al. (2021). The Dichotomous and Incomplete Adaptive Immunity in COVID-19 Patients With Different Disease Severity. Signal Transduction Targeted Ther. 6 (1), 113. doi: 10.1038/s41392-021-00525-3
Garaud, S., Dieu-Nosjean, M.-C., Willard-Gallo, K. (2022). T Follicular Helper and B Cell Crosstalk in Tertiary Lymphoid Structures and Cancer Immunotherapy. Nat. Commun. 13 (1), 2259. doi: 10.1038/s41467-022-29753-z
Gaspal, F. M. C., Kim, M.-Y., McConnell, F. M., Raykundalia, C., Bekiaris, V., Lane, P. J. L. (2005). Mice Deficient in OX40 and CD30 Signals Lack Memory Antibody Responses Because of Deficient CD4 T Cell Memory. J. Immunol. 174 (7), 3891–3896. doi: 10.4049/jimmunol.174.7.3891
Gigoux, M., Shang, J., Pak, Y., Xu, M., Choe, J., Mak Tak, W., et al. (2009). Inducible Costimulator Promotes Helper T-Cell Differentiation Through Phosphoinositide 3-Kinase. Proc. Natl. Acad. Sci. 106 (48), 20371–20376. doi: 10.1073/pnas.0911573106
Goenka, R., Barnett, L. G., Silver, J. S., O’Neill, P. J., Hunter, C. A., Cancro, M. P., et al. (2011). Cutting Edge: Dendritic Cell-Restricted Antigen Presentation Initiates the Follicular Helper T Cell Program But Cannot Complete Ultimate Effector Differentiation. J. Immunol. 187 (3), 1091. doi: 10.4049/jimmunol.1100853
Gong, F., Dai, Y., Zheng, T., Cheng, L., Zhao, D., Wang, H., et al. (2020). Peripheral CD4+ T Cell Subsets and Antibody Response in COVID-19 Convalescent Individuals. J. Clin. Invest. 130 (12), 6588–6599. doi: 10.1172/JCI141054
Grifoni, A., Weiskopf, D., Ramirez, S. I., Mateus, J., Dan, J. M., Moderbacher, C. R., et al. (2020). Targets of T Cell Responses to SARS-CoV-2 Coronavirus in Humans With COVID-19 Disease and Unexposed Individuals. Cell 181 (7), 1489–1501.e1415. doi: 10.1016/j.cell.2020.05.015
Grigoryan, L., Pulendran, B. (2020). The Immunology of SARS-CoV-2 Infections and Vaccines. Semin. Immunol. 50, 101422–101422. doi: 10.1016/j.smim.2020.101422
Gudbjartsson, D. F., Norddahl, G. L., Melsted, P., Gunnarsdottir, K., Holm, H., Eythorsson, E., et al. (2020). Humoral Immune Response to SARS-CoV-2 in Iceland. New Engl. J. Med. 383 (18), 1724–1734. doi: 10.1056/NEJMoa2026116
Gupta, A., Gonzalez-Rojas, Y., Juarez, E., Crespo Casal, M., Moya, J., Falci, D. R., et al. (2021). Early Treatment for Covid-19 With SARS-CoV-2 Neutralizing Antibody Sotrovimab. New Engl. J. Med. 385 (21), 1941–1950. doi: 10.1056/NEJMoa2107934
Hale, J. S., Ahmed, R. (2015). Memory T Follicular Helper CD4 T Cells. Front. Immunol. 6. doi: 10.3389/fimmu.2015.00016
Hale, J. S., Youngblood, B., Latner, D. R., Mohammed, A. U. R., Ye, L., Akondy, R. S., et al. (2013). Distinct Memory CD4+ T Cells With Commitment to T Follicular Helper- and T Helper 1-Cell Lineages are Generated After Acute Viral Infection. Immunity 38 (4), 805–817. doi: 10.1016/j.immuni.2013.02.020
Hao, Y., Wang, Y., Liu, X., Yang, X., Wang, P., Tian, Q., et al. (2018). The Kinase Complex mTOR Complex 2 Promotes the Follicular Migration and Functional Maturation of Differentiated Follicular Helper CD4(+) T Cells During Viral Infection. Front. Immunol. 9. doi: 10.3389/fimmu.2018.01127
Harker James, A., Lewis Gavin, M., Mack, L., Zuniga Elina, I. (2011). Late Interleukin-6 Escalates T Follicular Helper Cell Responses and Controls a Chronic Viral Infection. Science 334 (6057), 825–829. doi: 10.1126/science.1208421
Hatzi, K., Nance, J. P., Kroenke, M. A., Bothwell, M., Haddad, E. K., Melnick, A., et al. (2015). BCL6 Orchestrates Tfh Cell Differentiation via Multiple Distinct Mechanisms. J. Exp. Med. 212 (4), 539–553. doi: 10.1084/jem.20141380
He, L., Gu, W., Wang, M., Chang, X., Sun, X., Zhang, Y., et al. (2018). Extracellular Matrix Protein 1 Promotes Follicular Helper T Cell Differentiation and Antibody Production. Proc. Natl. Acad. Sci. U States America 115 (34), 8621–8626. doi: 10.1073/pnas.1801196115
He, J., Louis, M., Yew, A., Hu, X., Cindy, S., Chevalier, N., et al. (2013). Circulating Precursor CCR7loPD-1hi CXCR5+ CD4+ T Cells Indicate Tfh Cell Activity and Promote Antibody Responses Upon Antigen Reexposure. Immunity 39 (4), 770–781. doi: 10.1016/j.immuni.2013.09.007
Homann, D., Teyton, L., Oldstone, M. B. A. (2001). Differential Regulation of Antiviral T-Cell Immunity Results in Stable CD8+ But Declining CD4+ T-Cell Memory. Nat. Med. 7 (8), 913–919. doi: 10.1038/90950
Huang, Q., Hu, J., Tang, J., Xu, L., Ye, L. (2019). Molecular Basis of the Differentiation and Function of Virus Specific Follicular Helper CD4+ T Cells. Front. Immunol. 10. doi: 10.3389/fimmu.2019.00249
Huang, H., Long, L., Zhou, P., Chapman, N. M., Chi, H. (2020). mTOR Signaling at the Crossroads of Environmental Signals and T-Cell Fate Decisions. Immunol. Rev. 295 (1), 15–38. doi: 10.1111/imr.12845
Huber, M., Lohoff, M. (2014). IRF4 at the Crossroads of Effector T-Cell Fate Decision. Eur. J. Immunol. 44 (7), 1886–1895. doi: 10.1002/eji.201344279
Hu, R., Kagele, D. A., Huffaker, T. B., Runtsch, M. C., Alexander, M., Liu, J., et al. (2014). miR-155 Promotes T Follicular Helper Cell Accumulation During Chronic, Low-Grade Inflammation. Immunity 41 (4), 605–619. doi: 10.1016/j.immuni.2014.09.015
Ise, W., Inoue, T., McLachlan James, B., Kometani, K., Kubo, M., Okada, T., et al. (2014). Memory B Cells Contribute to Rapid Bcl6 Expression by Memory Follicular Helper T Cells. Proc. Natl. Acad. Sci. 111 (32), 11792–11797. doi: 10.1073/pnas.1404671111
Iyer, S. S., Latner, D. R., Zilliox, M. J., McCausland, M., Akondy, R. S., Penaloza-Macmaster, P., et al. (2013). Identification of Novel Markers for Mouse CD4(+) T Follicular Helper Cells. Eur. J. Immunol. 43 (12), 3219–3232. doi: 10.1002/eji.201343469
Johnston, R. J., Choi, Y. S., Diamond, J. A., Yang, J. A., Crotty, S. (2012). STAT5 is a Potent Negative Regulator of TFH Cell Differentiation. J. Exp. Med. 209 (2), 243–250. doi: 10.1084/jem.20111174
Johnston Robert, J., Poholek Amanda, C., DiToro, D., Yusuf, I., Eto, D., Barnett, B., et al. (2009). Bcl6 and Blimp-1 Are Reciprocal and Antagonistic Regulators of T Follicular Helper Cell Differentiation. Science 325 (5943), 1006–1010. doi: 10.1126/science.1175870
Josefowicz, S. Z. (2013). Regulators of Chromatin State and Transcription in CD4 T-Cell Polarization. Immunology 139 (3), 299–308. doi: 10.1111/imm.12115
Kaneko, N., Kuo, H.-H., Boucau, J., Farmer, J. R., Allard-Chamard, H., Mahajan, V. S., et al. (2020). Loss of Bcl-6-Expressing T Follicular Helper Cells and Germinal Centers in COVID-19. Cell 183 (1), 143–157.e113. doi: 10.1016/j.cell.2020.08.025
Karnowski, A., Chevrier, S., Belz, G. T., Mount, A., Emslie, D., D’Costa, K., et al. (2012). B and T Cells Collaborate in Antiviral Responses via IL-6, IL-21, and Transcriptional Activator and Coactivator, Oct2 and OBF-1. J. Exp. Med. 209 (11), 2049–2064. doi: 10.1084/jem.20111504
Keck, S., Schmaler, M., Ganter, S., Wyss, L., Oberle, S., Huseby Eric, S., et al. (2014). Antigen Affinity and Antigen Dose Exert Distinct Influences on CD4 T-Cell Differentiation. Proc. Natl. Acad. Sci. 111 (41), 14852–14857. doi: 10.1073/pnas.1403271111
Kitano, M., Moriyama, S., Ando, Y., Hikida, M., Mori, Y., Kurosaki, T., et al. (2011). Bcl6 Protein Expression Shapes Pre-Germinal Center B Cell Dynamics and Follicular Helper T Cell Heterogeneity. Immunity 34 (6), 961–972. doi: 10.1016/j.immuni.2011.03.025
Kotov, D. I., Mitchell, J. S., Pengo, T., Ruedl, C., Way, S. S., Langlois, R. A., et al. (2019). TCR Affinity Biases Th Cell Differentiation by Regulating CD25, Eef1e1, and Gbp2. J. Immunol. 202 (9), 2535. doi: 10.4049/jimmunol.1801609
Kuchen, S., Robbins, R., Sims, G. P., Sheng, C., Phillips, T. M., Lipsky, P. E., et al. (2007). Essential Role of IL-21 in B Cell Activation, Expansion, and Plasma Cell Generation During CD4+ T Cell-B Cell Collaboration. J. Immunol. 179 (9), 5886–5896. doi: 10.4049/jimmunol.179.9.5886
Kuen, D.-S., Park, M., Ryu, H., Choi, G., Moon, Y.-H., Kim, J.-O., et al. (2021). Critical Regulation of Follicular Helper T Cell Differentiation and Function by Gα13 Signaling. Proc. Natl. Acad. Sci. 118 (43), e2108376118. doi: 10.1073/pnas.2108376118
Künzli, M., Schreiner, D., Pereboom Tamara, C., Swarnalekha, N., Litzler Ludivine, C., Lötscher, J., et al. (2020). Long-Lived T Follicular Helper Cells Retain Plasticity and Help Sustain Humoral Immunity. Sci. Immunol. 5 (45), eaay5552. doi: 10.1126/sciimmunol.aay5552
Kurosaki, T., Kometani, K., Ise, W. (2015). Memory B Cells. Nat. Rev. Immunol. 15 (3), 149–159. doi: 10.1038/nri3802
Laczkó, D., Hogan, M. J., Toulmin, S. A., Hicks, P., Lederer, K., Gaudette, B. T., et al. (2020). A Single Immunization With Nucleoside-Modified mRNA Vaccines Elicits Strong Cellular and Humoral Immune Responses Against SARS-CoV-2 in Mice. Immunity 53 (4), 724–732.e727. doi: 10.1016/j.immuni.2020.07.019
Lee, S. K., Rigby, R. J., Zotos, D., Tsai, L. M., Kawamoto, S., Marshall, J. L., et al. (2011). B Cell Priming for Extrafollicular Antibody Responses Requires Bcl-6 Expression by T Cells. J. Exp. Med. 208 (7), 1377–1388. doi: 10.1084/jem.20102065
Lee, J.-Y., Skon, C. N., Lee, Y. J., Oh, S., Taylor, J. J., Malhotra, D., et al. (2015). The Transcription Factor KLF2 Restrains CD4+ T Follicular Helper Cell Differentiation. Immunity 42 (2), 252–264. doi: 10.1016/j.immuni.2015.01.013
Linterman, M. A., Beaton, L., Yu, D., Ramiscal, R. R., Srivastava, M., Hogan, J. J., et al. (2010). IL-21 Acts Directly on B Cells to Regulate Bcl-6 Expression and Germinal Center Responses. J. Exp. Med. 207 (2), 353–363. doi: 10.1084/jem.20091738
Linterman, M. A., Rigby, R. J., Wong, R., Silva, D., Withers, D., Anderson, G., et al. (2009). Roquin Differentiates the Specialized Functions of Duplicated T Cell Costimulatory Receptor Genes Cd28 and Icos. Immunity 30 (2), 228–241. doi: 10.1016/j.immuni.2008.12.015
Lipsitch, M., Grad, Y. H., Sette, A., Crotty, S. (2020). Cross-Reactive Memory T Cells and Herd Immunity to SARS-CoV-2. Nat. Rev. Immunol. 20 (11), 709–713. doi: 10.1038/s41577-020-00460-4
Liu, X., Chen, X., Zhong, B., Wang, A., Wang, X., Chu, F., et al. (2014). Transcription Factor Achaete-Scute Homologue 2 Initiates Follicular T-Helper-Cell Development. Nature 507 (7493), 513–518. doi: 10.1038/nature12910
Liu, D., Xu, H., Shih, C., Wan, Z., Ma, X., Ma, W., et al. (2015). T–B-Cell Entanglement and ICOSL-Driven Feed-Forward Regulation of Germinal Centre Reaction. Nature 517 (7533), 214–218. doi: 10.1038/nature13803
Li, J.-X., Wu, S.-P., Guo, X.-L., Tang, R., Huang, B.-Y., Chen, X.-Q., et al. (2022). Safety and Immunogenicity of Heterologous Boost Immunisation With an Orally Administered Aerosolised Ad5-Ncov After Two-Dose Priming With an Inactivated SARS-CoV-2 Vaccine in Chinese Adults: A Randomised, Open-Label, Single-Centre Trial. Lancet Respir. Med. S2213-2600 (22), 00087-X. doi: 10.1016/S2213-2600(22)00087-X
Li, F., Zeng, Z., Xing, S., Gullicksrud, J. A., Shan, Q., Choi, J., et al. (2018). Ezh2 Programs T(FH) Differentiation by Integrating Phosphorylation-Dependent Activation of Bcl6 and Polycomb-Dependent Repression of p19Arf. Nat. Commun. 9 (1), 5452–5452. doi: 10.1038/s41467-018-07853-z
Locci, M., Havenar-Daughton, C., Landais, E., Wu, J., Kroenke, M. A., Arlehamn, C. L., et al. (2013). Human Circulating PD-1+CXCR3-CXCR5+ Memory Tfh Cells are Highly Functional and Correlate With Broadly Neutralizing HIV Antibody Responses. Immunity 39 (4), 758–769. doi: 10.1016/j.immuni.2013.08.031
Long, X., Zhang, L., Zhang, Y., Min, M., Lin, B., Chen, J., et al. (2020). Histone Methyltransferase Nsd2 is Required for Follicular Helper T Cell Differentiation. J. Exp. Med. 217 (1), e20190832. doi: 10.1084/jem.20190832
Lu, K. T., Kanno, Y., Jennifer, L., Handon, R., Bible, P., Abdel, G., et al. (2011). Functional and Epigenetic Studies Reveal Multistep Differentiation and Plasticity of In Vitro-Generated and In Vivo-Derived Follicular T Helper Cells. Immunity 35 (4), 622–632. doi: 10.1016/j.immuni.2011.07.015
Lüthje, K., Kallies, A., Shimohakamada, Y., Belz, G. T., Light, A., Tarlinton, D. M., et al. (2012). The Development and Fate of Follicular Helper T Cells Defined by an IL-21 Reporter Mouse. Nat. Immunol. 13 (5), 491–498. doi: 10.1038/ni.2261
MacIver, N. J., Michalek, R. D., Rathmell, J. C. (2013). Metabolic Regulation of T Lymphocytes. Annu. Rev. Immunol. 31, 259–283. doi: 10.1146/annurev-immunol-032712-095956
MacLeod, M. K. L., Clambey, E. T., Kappler, J. W., Marrack, P. (2009). CD4 Memory T Cells: What are They and What can They do? Semin. Immunol. 21 (2), 53–61. doi: 10.1016/j.smim.2009.02.006
MacLeod, M. K. L., David, A., McKee, A. S., Crawford, F., Kappler, J. W., Marrack, P. (2011). Memory CD4 T Cells That Express CXCR5 Provide Accelerated Help to B Cells. J. Immunol. (Baltimore Md. 1950) 186 (5), 2889–2896. doi: 10.4049/jimmunol.1002955
Ma, C. S., Suryani, S., Avery, D. T., Chan, A., Nanan, R., Santner-Nanan, B., et al. (2009). Early Commitment of Naïve Human CD4(+) T Cells to the T Follicular Helper (T(FH)) Cell Lineage is Induced by IL-12. Immunol. Cell Biol. 87 (8), 590–600. doi: 10.1038/icb.2009.64
Mateus, J., Dan, J. M., Zhang, Z., Rydyznski Moderbacher, C., Lammers, M., Goodwin, B., et al. (2021). Low-Dose mRNA-1273 COVID-19 Vaccine Generates Durable Memory Enhanced by Cross-Reactive T Cells. Sci. (New York N.Y.) 374 (6566), eabj9853–eabj9853. doi: 10.1126/science.abj9853
Mathew, R., Mao, A., Chiang, A. H., Bertozzi-Villa, C., Bunker, J. J., Scanlon, S. T., et al. (2014). A Negative Feedback Loop Mediated by the Bcl6–cullin 3 Complex Limits Tfh Cell Differentiation. J. Exp. Med. 211 (6), 1137–1151. doi: 10.1084/jem.20132267
Meckiff, B. J., Ramírez-Suástegui, C., Fajardo, V., Chee, S. J., Kusnadi, A., Simon, H., et al. (2020). Imbalance of Regulatory and Cytotoxic SARS-CoV-2-Reactive CD4+ T Cells in COVID-19. Cell 183 (5), 1340–1353.e1316. doi: 10.1016/j.cell.2020.10.001
Misawa, T., SoRelle, J. A., Choi, J. H., Yue, T., Wang, K.-W., McAlpine, W., et al. (2020). Mutual Inhibition Between Prkd2 and Bcl6 Controls T Follicular Helper Cell Differentiation. Sci. Immunol. 5 (43), eaaz0085. doi: 10.1126/sciimmunol.aaz0085
Morita, R., Schmitt, N., Bentebibel, S.-E., Ranganathan, R., Bourdery, L., Zurawski, G., et al. (2011). Human Blood CXCR5+CD4+ T Cells Are Counterparts of T Follicular Cells and Contain Specific Subsets That Differentially Support Antibody Secretion. Immunity 34 (1), 108–121. doi: 10.1016/j.immuni.2010.12.012
Mudd, P. A., Minervina, A. A., Pogorelyy, M. V., Turner, J. S., Kim, W., Kalaidina, E., et al. (2022). SARS-CoV-2 mRNA Vaccination Elicits a Robust and Persistent T Follicular Helper Cell Response in Humans. Cell 185 (4), 603–613.e615. doi: 10.1016/j.cell.2021.12.026
Nanduri, S., Pilishvili, T., Derado, G., Soe, M. M., Dollard, P., Wu, H., et al. (2021). Effectiveness of Pfizer-BioNTech and Moderna Vaccines in Preventing SARS-CoV-2 Infection Among Nursing Home Residents Before and During Widespread Circulation of the SARS-CoV-2 B.1.617.2 (Delta) Variant - National Healthcare Safety Network, March 1-August 1, 2021. MMWR. Morbid mortal weekly Rep. 70 (34), 1163–1166. doi: 10.15585/mmwr.mm7034e3
Naranbhai, V., Nathan, A., Kaseke, C., Berrios, C., Khatri, A., Choi, S., et al. (2022). T Cell Reactivity to the SARS-CoV-2 Omicron Variant is Preserved in Most But Not All Individuals. Cell 185 (6), 1041–1051.e1046. doi: 10.1016/j.cell.2022.01.029
Narowski, T. M., Raphel, K., Adams, L. E., Huang, J., Vielot, N. A., Jadi, R., et al. (2022). SARS-CoV-2 mRNA Vaccine Induces Robust Specific and Cross-Reactive IgG and Unequal Neutralizing Antibodies in Naive and Previously Infected People. Cell Rep. 38 (5), 110336–110336. doi: 10.1016/j.celrep.2022.110336
Nurieva, R. I., Chung, Y., Hwang, D., Yang, X. O., Kang, H. S., Ma, L., et al. (2008). Generation of T Follicular Helper Cells Is Mediated by Interleukin-21 But Independent of T Helper 1, 2, or 17 Cell Lineages. Immunity 29 (1), 138–149. doi: 10.1016/j.immuni.2008.05.009
Nurieva Roza, I., Chung, Y., Martinez Gustavo, J., Yang Xuexian, O., Tanaka, S., Matskevitch Tatyana, D., et al. (2009). Bcl6 Mediates the Development of T Follicular Helper Cells. Science 325 (5943), 1001–1005. doi: 10.1126/science.1176676
Ogbe, A., Miao, T., Symonds, A. L. J., Omodho, B., Singh, R., Bhullar, P., et al. (2015). Early Growth Response Genes 2 and 3 Regulate the Expression of Bcl6 and Differentiation of T Follicular Helper Cells *. J. Biol. Chem. 290 (33), 20455–20465. doi: 10.1074/jbc.M114.634816
Oja, A. E., Saris, A., Ghandour, C. A., Kragten, N. A. M., Hogema, B. M., Nossent, E. J., et al. (2020). Divergent SARS-CoV-2-Specific T- and B-Cell Responses in Severe But Not Mild COVID-19 Patients. Eur. J. Immunol. 50 (12), 1998–2012. doi: 10.1002/eji.202048908
O’Shea John, J., Paul William, E. (2010). Mechanisms Underlying Lineage Commitment and Plasticity of Helper CD4+ T Cells. Science 327 (5969), 1098–1102. doi: 10.1126/science.1178334
Painter, M. M., Mathew, D., Goel, R. R., Apostolidis, S. A., Pattekar, A., Kuthuru, O., et al. (2021). Rapid Induction of Antigen-Specific CD4+ T Cells is Associated With Coordinated Humoral and Cellular Immunity to SARS-CoV-2 mRNA Vaccination. Immunity 54 (9), 2133–2142.e2133. doi: 10.1016/j.immuni.2021.08.001
Pawlak, M., Ho, A. W., Kuchroo, V. K. (2020). Cytokines and Transcription Factors in the Differentiation of CD4+ T Helper Cell Subsets and Induction of Tissue Inflammation and Autoimmunity. Curr. Opin. Immunol. 67, 57–67. doi: 10.1016/j.coi.2020.09.001
Peng, Y., Mentzer, A. J., Liu, G., Yao, X., Yin, Z., Dong, D., et al. (2020). Broad and Strong Memory CD4(+) and CD8(+) T Cells Induced by SARS-CoV-2 in UK Convalescent Individuals Following COVID-19. Nat. Immunol. 21 (11), 1336–1345. doi: 10.1038/s41590-020-0782-6
Pepper, M., Pagán, Antonio, J., Igyártó, Botond, Z., Taylor, Justin, J., Jenkins, Marc, K. (2011). Opposing Signals From the Bcl6 Transcription Factor and the Interleukin-2 Receptor Generate T Helper 1 Central and Effector Memory Cells. Immunity 35 (4), 583–595. doi: 10.1016/j.immuni.2011.09.009
Phan, T. G., Tangye, S. G. (2017). Memory B Cells: Total Recall. Curr. Opin. Immunol. 45, 132–140. doi: 10.1016/j.coi.2017.03.005
Picchianti-Diamanti, A., Aiello, A., Laganà, B., Agrati, C., Castilletti, C., Meschi, S., et al. (2021). ImmunosuppressiveTherapies Differently Modulate Humoral- and T-Cell-Specific Responses to COVID-19 mRNA Vaccine in Rheumatoid Arthritis Patients. Front. Immunol. 12. doi: 10.3389/fimmu.2021.740249
Pollizzi, K. N., Patel, C. H., Sun, I.-H., Oh, M.-H., Waickman, A. T., Wen, J., et al. (2015). Mtorc1 and Mtorc2 Selectively Regulate CD8+ T Cell Differentiation. J. Clin. Invest. 125 (5), 2090–2108. doi: 10.1172/JCI77746
Post, N., Eddy, D., Huntley, C., van Schalkwyk, M. C. I., Shrotri, M., Leeman, D., et al. (2020). Antibody Response to SARS-CoV-2 Infection in Humans: A Systematic Review. PloS One 15 (12), e0244126–e0244126. doi: 10.1371/journal.pone.0244126
Pozzetto, B., Legros, V., Djebali, S., Barateau, V., Trouillet-Assant, S. (2021). Immunogenicity and Efficacy of Heterologous ChadOx1/BNT162b2 Vaccination. Nature 600 (7890), 6. doi: 10.1038/s41586-021-04120-y
Pratama, A., Srivastava, M., Williams, N. J., Papa, I., Lee, S. K., Dinh, X. T., et al. (2015). MicroRNA-146a Regulates ICOS–ICOSL Signalling to Limit Accumulation of T Follicular Helper Cells and Germinal Centres. Nat. Commun. 6 (1), 6436. doi: 10.1038/ncomms7436
Pruner, K. B., Pepper, M. (2021). Local Memory CD4 T Cell Niches in Respiratory Viral Infection. J. Exp. Med. 218 (8), e20201733. doi: 10.1084/jem.20201733
Qi, H., Cannons, J. L., Klauschen, F., Schwartzberg, P. L., Germain, R. N. (2008). SAP-Controlled T-B Cell Interactions Underlie Germinal Centre Formation. Nature 455 (7214), 764–769. doi: 10.1038/nature07345
Ray, J. P., Staron, Matthew, M., Shyer, Justin, A., Ho, P.-C., Marshall, Heather, D., Simon, M., et al. (2015). The Interleukin-2-Mtorc1 Kinase Axis Defines the Signaling, Differentiation, and Metabolism of T Helper 1 and Follicular B Helper T Cells. Immunity 43 (4), 690–702. doi: 10.1016/j.immuni.2015.08.017
Riou, C., du Bruyn, E., Stek, C., Daroowala, R., Goliath, R. T., Abrahams, F., et al. (2021). Relationship of SARS-CoV-2-Specific CD4 Response to COVID-19 Severity and Impact of HIV-1 and Tuberculosis Coinfection. J. Clin. Invest. 131 (12), e149125. doi: 10.1172/JCI149125
Robbiani, D. F., Gaebler, C., Muecksch, F., Lorenzi, J. C. C., Wang, Z., Cho, A., et al. (2020). Convergent Antibody Responses to SARS-CoV-2 in Convalescent Individuals. Nature 584 (7821), 437–442. doi: 10.1038/s41586-020-2456-9
Rolf, J., Bell, S. E., Kovesdi, D., Janas, M. L., Soond, D. R., Webb, L. M. C., et al. (2010). Phosphoinositide 3-Kinase Activity in T Cells Regulates the Magnitude of the Germinal Center Reaction. J. Immunol. 185 (7), 4042. doi: 10.4049/jimmunol.1001730
Röltgen, K., Powell, A. E., Wirz, O. F., Stevens, B. A., Hogan, C. A., Najeeb, J., et al. (2020). Defining the Features and Duration of Antibody Responses to SARS-CoV-2 Infection Associated With Disease Severity and Outcome. Sci. Immunol. 5 (54), eabe0240. doi: 10.1126/sciimmunol.abe0240
Rosenberg, E. S., Holtgrave, D., Dorabawila, V., et al. (2021). New COVID-19 Cases and Hospitalizations Among Adults, by Vaccination Status_New York, May 3-July 25. MMWR Morb Mortal Wkly Rep. 2021 (70), 6. doi: 10.15585/mmwr.mm7037a7
Salomé, B., Horowitz, A. (2021). Impaired CD4 T-Cell Response to SARS-CoV-2: Rationale for PD-1 Blockade in Patients With Cancer and COVID-19? Cancer Discovery 11 (8), 1877–1878. doi: 10.1158/2159-8290.CD-21-0613
Schaerli, P., Willimann, K., Lang, A. B., Lipp, M., Loetscher, P., Moser, B. (2000). Cxc Chemokine Receptor 5 Expression Defines Follicular Homing T Cells With B Cell Helper Function. J. Exp. Med. 192 (11), 1553–1562. doi: 10.1084/jem.192.11.1553
Schmitt, N., Liu, Y., Bentebibel, S.-E., Munagala, I., Bourdery, L., Venuprasad, K., et al. (2014). The Cytokine TGF-β Co-Opts Signaling via STAT3-STAT4 to Promote the Differentiation of Human TFH Cells. Nat. Immunol. 15 (9), 856–865. doi: 10.1038/ni.2947
Schmitt, N., Morita, R., Bourdery, L., Bentebibel, S. E., Zurawski, S. M., Banchereau, J., et al. (2009). Human Dendritic Cells Induce the Differentiation of Interleukin-21-Producing T Follicular Helper-Like Cells Through Interleukin-12. Immunity 31 (1), 158–169. doi: 10.1016/j.immuni.2009.04.016
Schroeder, A. R., Zhu, F., Hu, H. (2021). Stepwise Tfh Cell Differentiation Revisited: New Advances and Long-Standing Questions. Faculty Rev. 10, 3. doi: 10.12703/r/10-3
Schultheiß, C., Paschold, L., Simnica, D., Mohme, M., Willscher, E., von Wenserski, L., et al. (2020). Next-Generation Sequencing of T and B Cell Receptor Repertoires From COVID-19 Patients Showed Signatures Associated With Severity of Disease. Immunity 53 (2), 442–455.e444. doi: 10.1016/j.immuni.2020.06.024
Seder, R. A., Ahmed, R. (2003). Similarities and Differences in CD4+ and CD8+ Effector and Memory T Cell Generation. Nat. Immunol. 4 (9), 835–842. doi: 10.1038/ni969
Sette, A., Crotty, S. (2021). Adaptive Immunity to SARS-CoV-2 and COVID-19. Cell 184 (4), 861–880. doi: 10.1016/j.cell.2021.01.007
Shaw, L. A., Bélanger, S., Omilusik, K. D., Cho, S., Scott-Browne, J. P., Nance, J. P., et al. (2016). Id2 Reinforces TH1 Differentiation and Inhibits E2A to Repress TFH Differentiation. Nat. Immunol. 17 (7), 834–843. doi: 10.1038/ni.3461
Shrotri, M., van Schalkwyk, M. C. I., Post, N., Eddy, D., Huntley, C., Leeman, D., et al. (2021). T Cell Response to SARS-CoV-2 Infection in Humans: A Systematic Review. PloS One 16 (1), e0245532–e0245532. doi: 10.1371/journal.pone.0245532
Shulman, Z., Gitlin, A. D., Targ, S., Jankovic, M., Pasqual, G., Nussenzweig, M. C., et al. (2013). T Follicular Helper Cell Dynamics in Germinal Centers. Science 341 (6146), 673–677. doi: 10.1126/science.1241680
Snook Jeremy, P., Kim, C., Williams Matthew, A. (2018). TCR Signal Strength Controls the Differentiation of CD4+ Effector and Memory T Cells. Sci. Immunol. 3 (25), eaas9103. doi: 10.1126/sciimmunol.aas9103
Sokal, A., Broketa, M., Barba-Spaeth, G., Meola, A., Fernández, I., Fourati, S., et al. (2022). Analysis of mRNA Vaccination-Elicited RBD-Specific Memory B Cells Reveals Strong But Incomplete Immune Escape of the SARS-CoV-2 Omicron Variant. Immunity. 55 (6), 1096–1104 doi: 10.1016/j.immuni.2022.04.002
Song, N., Sengupta, S., Khoruzhenko, S., Welsh, R. A., Kim, A., Kumar, M. R., et al. (2020). Multiple Genetic Programs Contribute to CD4 T Cell Memory Differentiation and Longevity by Maintaining T Cell Quiescence. Cell. Immunol. 357, 104210. doi: 10.1016/j.cellimm.2020.104210
Song, S., Zhou, B., Cheng, L., Liu, W., Fan, Q., Ge, X., et al. (2022). Sequential Immunization With SARS-CoV-2 RBD Vaccine Induces Potent and Broad Neutralization Against Variants in Mice. Virol. J. 19 (1), 2–2. doi: 10.1186/s12985-021-01737-3
Son Young, M., Cheon In, S., Wu, Y., Li, C., Wang, Z., Gao, X., et al. (2021). Tissue-Resident CD4+ T Helper Cells Assist the Development of Protective Respiratory B and CD8+ T Cell Memory Responses. Sci. Immunol. 6 (55), eabb6852. doi: 10.1126/sciimmunol.abb6852
Stone, E. L., Pepper, M., Katayama, C. D., Kerdiles, Y. M., Lai, C.-Y., Emslie, E., et al. (2015). ICOS Coreceptor Signaling Inactivates the Transcription Factor FOXO1 to Promote Tfh Cell Differentiation. Immunity 42 (2), 239–251. doi: 10.1016/j.immuni.2015.01.017
Sun, Z., Yao, Y., You, M., Liu, J., Guo, W., Qi, Z., et al. (2021). The Kinase PDK1 is Critical for Promoting T Follicular Helper Cell Differentiation. eLife 10, e61406. doi: 10.7554/eLife.61406
Sweet, R. A., Lee, S. K., Vinuesa, C. G. (2012). Developing Connections Amongst Key Cytokines and Dysregulated Germinal Centers in Autoimmunity. Curr. Opin. Immunol. 24 (6), 658–664. doi: 10.1016/j.coi.2012.10.003
Tan, H.-X., Esterbauer, R., Vanderven, H. A., Juno, J. A., Kent, S. J., Wheatley, A. K. (2019). Inducible Bronchus-Associated Lymphoid Tissues (iBALT) Serve as Sites of B Cell Selection and Maturation Following Influenza Infection in Mice. Front. Immunol. 10. doi: 10.3389/fimmu.2019.00611
Tan, A. H.-M., Wong, S.-C., Lam, K.-P. (2006). Regulation of Mouse Inducible Costimulator (ICOS) Expression by Fyn-NFATc2 and ERK Signaling in T Cells *. J. Biol. Chem. 281 (39), 28666–28678. doi: 10.1074/jbc.M604081200
Tenforde, M. W., Self, W. H., Naioti, E. A., Ginde, A. A., Douin, D. J., Olson, S. M., et al. (2021). Sustained Effectiveness of Pfizer-BioNTech and Moderna Vaccines Against COVID-19 Associated Hospitalizations Among Adults - United States, March-July 2021. MMWR. Morbid mortal weekly Rep. 70 (34), 1156–1162. doi: 10.15585/mmwr.mm7034e2
Thakur, V., Bhola, S., Thakur, P., Patel, S. K. S., Kulshrestha, S., Ratho, R. K., et al. (2022). Waves and Variants of SARS-CoV-2: Understanding the Causes and Effect of the COVID-19 Catastrophe. Infection 50 (2), 309–325. doi: 10.1007/s15010-021-01734-2
Tian, Y., Zajac, A. J. (2016). IL-21 and T Cell Differentiation: Consider the Context. Trends Immunol. 37 (8), 557–568. doi: 10.1016/j.it.2016.06.001
Tsai, L. M., Yu, D. (2014). Follicular Helper T-Cell Memory: Establishing New Frontiers During Antibody Response. Immunol. Cell Biol. 92 (1), 57–63. doi: 10.1038/icb.2013.68
Tubo, N. J., Pagán, A. J., Taylor, J. J., Nelson, R. W., Linehan, J. L., Ertelt, J. M., et al. (2013). Single Naive CD4+ T Cells From a Diverse Repertoire Produce Different Effector Cell Types During Infection. Cell 153 (4), 785–796. doi: 10.1016/j.cell.2013.04.007
Umar, D., Das, A., Gupta, S., Chattopadhyay, S., Sarkar, D., Mirji, G., et al. (2020). Febrile Temperature Change Modulates CD4 T Cell Differentiation via a TRPV Channel-Regulated Notch-Dependent Pathway. Proc. Natl. Acad. Sci. 117 (36), 22357–22366. doi: 10.1073/pnas.1922683117
V'Kovski, P., Kratzel, A., Steiner, S., Stalder, H., Thiel, V. (2021). Coronavirus Biology and Replication: Implications for SARS-CoV-2. Nat. Rev. Microbiol. 19 (3), 155–170. doi: 10.1038/s41579-020-00468-6
Vacchio, M. S., Ciucci, T., Gao, Y., Watanabe, M., Balmaceno-Criss, M., McGinty, M. T., et al. (2019). A Thpok-Directed Transcriptional Circuitry Promotes Bcl6 and Maf Expression to Orchestrate T Follicular Helper Differentiation. Immunity 51 (3), 465–478.e466. doi: 10.1016/j.immuni.2019.06.023
Vella, L. A., Herati, R. S., Wherry, E. J. (2017). CD4+ T Cell Differentiation in Chronic Viral Infections: The Tfh Perspective. Trends Mol. Med. 23 (12), 1072–1087. doi: 10.1016/j.molmed.2017.10.001
Victora, G. D., Nussenzweig, M. C. (2012). Germinal Centers. Annu. Rev. Immunol. 30 (1), 429–457. doi: 10.1146/annurev-immunol-020711-075032
Vinuesa, C. G., Cook, M. C., Angelucci, C., Athanasopoulos, V., Rui, L., Hill, K. M., et al. (2005). A RING-Type Ubiquitin Ligase Family Member Required to Repress Follicular Helper T Cells and Autoimmunity. Nature 435 (7041), 452–458. doi: 10.1038/nature03555
Vinuesa, C. G., Linterman, M. A., Yu, D., MacLennan, I. C. M. (2016). Follicular Helper T Cells. Annu. Rev. Immunol. 34 (1), 335–368. doi: 10.1146/annurev-immunol-041015-055605
Wang, X., Chen, X., Tan, J., Yue, S., Zhou, R., Xu, Y., et al. (2022a). 35B5 Antibody Potently Neutralizes SARS-CoV-2 Omicron by Disrupting the N-Glycan Switch via a Conserved Spike Epitope. Cell Host Microbe S1931-3128 (1922), 00165–2. doi: 10.1016/j.chom.2022.03.035
Wang, H., Geng, J., Wen, X., Bi, E., Kossenkov, A. V., Wolf, A. I., et al. (2014). The Transcription Factor Foxp1 is a Critical Negative Regulator of the Differentiation of Follicular Helper T Cells. Nat. Immunol. 15 (7), 667–675. doi: 10.1038/ni.2890
Wang, F., Hou, H., Luo, Y., Tang, G., Wu, S., Huang, M., et al. (2020). The Laboratory Tests and Host Immunity of COVID-19 Patients With Different Severity of Illness. JCI Insight 5 (10), e137799. doi: 10.1172/jci.insight.137799
Wang, X., Hu, A., Chen, X., Zhang, Y., Yu, F., Yue, S., et al. (2022b). A Potent Human Monoclonal Antibody With Pan-Neutralizing Activities Directly Dislocates S Trimer of SARS-CoV-2 Through Binding Both Up and Down Forms of RBD. Signal transduction targeted Ther. 7 (1), 114–114. doi: 10.1038/s41392-022-00954-8
Wang, H., Li, X., Kajikawa, T., Shin, J., Lim, J.-H., Kourtzelis, I., et al. (2021). Stromal Cell-Derived DEL-1 Inhibits Tfh Cell Activation and Inflammatory Arthritis. J. Clin. Invest. 131 (19), e150578. doi: 10.1172/JCI150578
Wang, Y., Shi, J., Yan, J., Xiao, Z., Hou, X., Lu, P., et al. (2017). Germinal-Center Development of Memory B Cells Driven by IL-9 From Follicular Helper T Cells. Nat. Immunol. 18 (8), 921–930. doi: 10.1038/ni.3788
Wang, Y., Tian, Q., Hao, Y., Yao, W., Lu, J., Chen, C., et al. (2022c). The Kinase Complex Mtorc2 Promotes the Longevity of Virus-Specific Memory CD4+ T Cells by Preventing Ferroptosis. Nat. Immunol. 23 (2), 303–317. doi: 10.1038/s41590-021-01090-1
Watanabe, M., Takagi, Y., Kotani, M., Hara, Y., Inamine, A., Hayashi, K., et al. (2008). Down-Regulation of ICOS Ligand by Interaction With ICOS Functions as a Regulatory Mechanism for Immune Responses. J. Immunol. (Baltimore Md. 1950) 180 (8), 5222–5234. doi: 10.4049/jimmunol.180.8.5222
Weber, J. P., Fuhrmann, F., Feist, R. K., Lahmann, A., Al Baz, M. S., Gentz, L.-J., et al. (2015). ICOS Maintains the T Follicular Helper Cell Phenotype by Down-Regulating Krüppel-Like Factor 2. J. Exp. Med. 212 (2), 217–233. doi: 10.1084/jem.20141432
Weber, J. P., Fuhrmann, F., Hutloff, A. (2012). T-Follicular Helper Cells Survive as Long-Term Memory Cells. Eur. J. Immunol. 42 (8), 1981–1988. doi: 10.1002/eji.201242540
Wilson, C. B., Rowell, E., Sekimata, M. (2009). Epigenetic Control of T-Helper-Cell Differentiation. Nat. Rev. Immunol. 9 (2), 91–105. doi: 10.1038/nri2487
Woldemeskel, B. A., Garliss, C. C., Aytenfisu, T. Y., Johnston, T. S., Beck, E. J., Dykema, A. G., et al. (2022). SARS-CoV-2 -Specific Immune Responses in Boosted Vaccine Recipients With Breakthrough Infections During the Omicron Variant Surge. JCI Insight. 7 (10), e159474. doi: 10.1172/jci.insight.159474
Wu, T., Shin, H. M., Moseman, E. A., Ji, Y., Huang, B., Harly, C., et al. (2015). TCF1 Is Required for the T Follicular Helper Cell Response to Viral Infection. Cell Rep. 12 (12), 2099–2110. doi: 10.1016/j.celrep.2015.08.049
Xu, L., Cao, Y., Xie, Z., Huang, Q., Bai, Q., Yang, X., et al. (2015). The Transcription Factor TCF-1 Initiates the Differentiation of TFH Cells During Acute Viral Infection. Nat. Immunol. 16 (9), 991–999. doi: 10.1038/ni.3229
Xu, H., Li, X., Liu, D., Li, J., Zhang, X., Chen, X., et al. (2013). Follicular T-Helper Cell Recruitment Governed by Bystander B Cells and ICOS-Driven Motility. Nature 496 (7446), 523–527. doi: 10.1038/nature12058
Xu, W., Zhao, X., Wang, X., Feng, H., Gou, M., Jin, W., et al. (2019). The Transcription Factor Tox2 Drives T Follicular Helper Cell Development via Regulating Chromatin Accessibility. Immunity 51 (5), 826–839.e825. doi: 10.1016/j.immuni.2019.10.006
Yang, K., Chi, H. (2012). mTOR and Metabolic Pathways in T Cell Quiescence and Functional Activation. Semin. Immunol. 24 (6), 421–428. doi: 10.1016/j.smim.2012.12.004
Yang, J., Lin, X., Pan, Y., Wang, J., Chen, P., Huang, H., et al. (2016). Critical Roles of mTOR Complex 1 and 2 for T Follicular Helper Cell Differentiation and Germinal Center Responses. eLife 5, e17936. doi: 10.7554/eLife.17936
Youngblood, B., Hale, J. S., Ahmed, R. (2013). T-Cell Memory Differentiation: Insights From Transcriptional Signatures and Epigenetics. Immunology 139 (3), 277–284. doi: 10.1111/imm.12074
Yu, D., Rao, S., Tsai, L. M., Lee, S. K., He, Y., Sutcliffe, E. L., et al. (2009). The Transcriptional Repressor Bcl-6 Directs T Follicular Helper Cell Lineage Commitment. Immunity 31 (3), 457–468. doi: 10.1016/j.immuni.2009.07.002
Zander, R., Kasmani, M. Y., Chen, Y., Topchyan, P., Shen, J., Zheng, S., et al. (2022). Tfh-Cell-Derived Interleukin 21 Sustains Effector CD8+ T Cell Responses During Chronic Viral Infection. Immunity 55 (3), 475–493.e475. doi: 10.1016/j.immuni.2022.01.018
Zeng, H., Cohen, S., Guy, C., Shrestha, S., Neale, G., Brown, S. A., et al. (2016). Mtorc1 and Mtorc2 Kinase Signaling and Glucose Metabolism Drive Follicular Helper T Cell Differentiation. Immunity 45 (3), 540–554. doi: 10.1016/j.immuni.2016.08.017
Zhou, L., Chong, M. M. W., Littman, D. R. (2009). Plasticity of CD4+ T Cell Lineage Differentiation. Immunity 30 (5), 646–655. doi: 10.1016/j.immuni.2009.05.001
Zhu, J., Yamane, H., Paul, W. E. (2010). Differentiation of Effector CD4 T Cell Populations (*). Annu. Rev. Immunol. 28, 445–489. doi: 10.1146/annurev-immunol-030409-101212
Zotos, D., Coquet, J. M., Zhang, Y., Light, A., D'Costa, K., Kallies, A., et al. (2010). IL-21 Regulates Germinal Center B Cell Differentiation and Proliferation Through a B Cell–Intrinsic Mechanism. J. Exp. Med. 207 (2), 365–378. doi: 10.1084/jem.20091777
Keywords: CD4+ T cell, follicular helper T cell, Viral infection, COVID-19, SARS-CoV-2.
Citation: Wang Y, Tian Q and Ye L (2022) The Differentiation and Maintenance of SARS-CoV-2-Specific Follicular Helper T Cells. Front. Cell. Infect. Microbiol. 12:953022. doi: 10.3389/fcimb.2022.953022
Received: 25 May 2022; Accepted: 20 June 2022;
Published: 14 July 2022.
Edited by:
Rameez Raja, Cleveland Clinic, United StatesReviewed by:
Showkat A Dar, National Institutes of Health (NIH), United StatesMuhammad Bilal Latif, Emory University, United States
Arundhoti Das, National Institutes of Health (NIH), United States
Copyright © 2022 Wang, Tian and Ye. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lilin Ye, eWVsaWxpbmxjbXZAdG1tdS5lZHUuY24=
†These authors have contributed equally to this work and share first authorship
 Yifei Wang
Yifei Wang Qin Tian
Qin Tian Lilin Ye1,3*
Lilin Ye1,3*