
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
BRIEF RESEARCH REPORT article
Front. Cell. Infect. Microbiol., 19 January 2022
Sec. Clinical Microbiology
Volume 11 - 2021 | https://doi.org/10.3389/fcimb.2021.810887
This article is part of the Research TopicWomen in Clinical Microbiology 2021View all 10 articles
Cryptosporidium spp. are responsible for moderate to severe diarrhea, mainly in children and immunocompromised patients. Using ELISA, the recognition of synthetic peptides generated from the sequences of the Cryptosporidium parvum gp40 and gp15 proteins by serum IgM and IgG antibodies from patients infected (cases) with Cryptosporidium hominis, C. parvum, and Cryptosporidium canis, and uninfected individuals (controls) was evaluated. A statistically significant difference (p = 0.0025) was found in terms of the recognition of peptides A133 and A32 between cases and controls. Additional studies are necessary to understand the potential of these peptides as vaccine candidates.
Cryptosporidium spp. of the phylum Apicomplexa are protozoan parasites of medical and veterinary importance worldwide because they are causal agents of moderate to severe diarrhea, with higher prevalence in developing countries (Chalmers and Giles, 2010; Bouzid et al., 2013). Children under 5 years old, and immunocompromised people, are the most susceptible to cryptosporidiosis (Sarkar et al., 2012). In immunocompetent people, the disease can occur asymptomatically; if there are symptoms, they are self-limited with a duration of 1–2 weeks (Widerström et al., 2014). The Food and Drug Administration (FDA) has approved nitazoxanide as a drug against cryptosporidiosis with an effectiveness of 56%–96% in immunocompetent people (Checkley et al., 2015).
Recent efforts to develop a vaccine against Cryptosporidium spp. have focused on the identification and characterization of immunogenic proteins (GP900, 27-kD, gp15/CP15, gp40, CSL) involved in the adhesion and invasion of Cryptosporidium merozoites and sporozoites (Frost et al., 2005; Ajjampur et al., 2011; Allison et al., 2011; Avendaño et al., 2018). One of the most studied proteins is GP60 or gp40/15, which is located in the apical complex of the parasite (Cevallos et al., 2000; Borad and Ward, 2010). Anti-gp15 and anti-gp40 antibodies are associated with protection in humans (Ajjampur et al., 2011; Allison et al., 2011). Synthetic peptide-based vaccines versus attenuated pathogen vaccines or DNA vaccines are less difficult to manufacture and require less stringent storage conditions (Avendaño et al., 2018); therefore, synthetic peptides with potential as vaccine candidates are of great interest. The objective of this study was to evaluate humoral immunity, as demonstrated in the recognition of synthetic peptides of the Cryptosporidium parvum gp15 and gp40 proteins by serum IgM and IgG antibodies from patients infected (cases) with Cryptosporidium hominis, C. parvum, and Cryptosporidium canis, and uninfected individuals (controls).
The protocol was registered in the Secretaría de Salud del Estado de Sonora (project No. 196) and approved by the Ethics Committee of the Hospital Infantil del Estado de Sonora (HIES) and the Centro Médico Dr. Ignacio Chávez (CMDICH) (CEI-09-2015). Informed consent was obtained from each patient after the objectives of the study were clearly explained; in the case of minors, informed consent was also obtained from the parent or guardian. The clinical characteristics of the patients were obtained with prior authorization from the patients’ clinical files at each hospital.
The inclusion criteria of the patients (case) for the study were being positive for Cryptosporidium by Kinyoun staining, male or female, of any age and with or without clinical symptoms of acute gastroenteritis (AGE). The group of cases consisted of 39 children from 5 months to 9 years of age (51.3% male). Patients in this group were carriers of Cryptosporidium spp., as diagnosed by microscopic observation of oocysts (Henriksen and Pohlenz, 1981) and molecular characterization via nested analysis (PCR sequencing) of small subunit rRNA (SSU rRNA) and GP60 (Valenzuela et al., 2014; Gonzalez-Diaz et al., 2016; Urrea-Quezada et al., 2018); 89.7% of the cryptosporidiosis cases (35/39) were recruited from the HIES (Supplementary Table 1). The most frequent admission diagnoses (87.2%) were acute gastroenteritis (AGE), malnutrition (25.6%), and dehydration (15.4%); however, three patients had an admission diagnosis that was unrelated to a gastrointestinal disorder: acute otitis, pulmonary tuberculosis, pneumonia, and bronchiolitis. Ten percent of the cases (4/39) were recruited from the CMDICH. The admission diagnosis of each of them was different; one case went to the doctor for a general check-up, two for AGE, and the last for a urinary tract infection (UTI) (Table 1).
The inclusion criterion for the controls was being negative for Cryptosporidium as determined microscopically and via molecular techniques. This group includes male or female individuals of any age with or without clinical symptoms of acute gastroenteritis (AGE) and others symptoms not related to AGE. The control group comprised 90 subjects from 1 month to 65 years old who were Cryptosporidium negative, 47.8% of whom were female (Supplementary Table 1). All of the included participants reside in urban areas of the state of Sonora. Thirty-four of the subjects were recruited from the HIES; all of the subjects were under 15 years of age, and 88% of them (30/34) had admission diagnoses of various infections: respiratory tract (16/30) and GEA (6/30) (Supplementary Table 1). Thirty-one of the controls were recruited from the CMDICH (1–65 years), and the admission diagnosis was not determined in any of them. In addition, a group of 21-year-old volunteers were included in the study, 15 of whom were apparently healthy and 10 of whom presented with clinical manifestations (diarrhea and abdominal pain).
Serum and fecal samples of each participant were sent to the Laboratory of Parasitology of the Departamento de Ciencias Químico Biológicas of the Universidad de Sonora. Feces was used for the microscopic diagnosis of intestinal parasites and molecular characterization of Cryptosporidium; serum was used for the determination of whether there was humoral recognition (IgM and IgG) of the peptides using ELISA. The fecal samples were stored at 4°C, and the serum samples were stored at −20°C until further analysis.
Sera from four apparently healthy patients (two toddlers and two adults) without cryptosporidiosis who did not show recognition for the recombinant proteins (rCpGP15, rCpGP40, and rChGP40) and peptides (A109, A133, V30, A33, and R61) were used as negative controls. For the positive controls, we used two groups: a group composed of three children infected with C. parvum and a group composed of three children infected with C. hominis; all the positive controls had strong recognition for all the peptides and recombinant proteins (data not shown). All the patient samples were tested in duplicate. A sample was considered positive when the absorbance was above the cutoff, which consisted of the mean plus two standard deviations of the negative control.
Human fecal samples were collected from children and adults from Hermosillo, Sonora, México. Oocysts were detected by the modified Ziehl–Neelsen (MZN) method. The oocysts were stained pink on a pale green background, appeared round or spherical in shape, and were 4–6 µm in diameter (Henriksen and Pohlenz, 1981).
DNA extraction from the stool samples was performed with the ZR Fecal DNA MiniPrep Extraction Kit (Zymo Research Corp., Irvine, CA) according to the manufacturer’s instructions after five freeze (−20°C) and thaw (95°C) cycles, and the extracted DNA was stored at −20°C until further processing (Valenzuela et al., 2014).
Molecular characterization was performed by analyzing the sequences of the nested PCR products for the SSU rRNA (826–864 bp) and GP60 (880–900 bp) genes (Valenzuela et al., 2014). To identify patients with possible coinfections of different Cryptosporidium species, real-time PCR (qPCR) analysis was performed using the Chos-1 and Cops-2 genes of C. hominis and C. parvum, respectively (Bouzid et al., 2016).
The peptides were designed from the sequences of the C. parvum gp15 protein and gp40 protein using the Chromas Lite version 2.1 program. The Clustalw program was used to align multiple sequences and detect differences and similarities among the different DNA sequences of the samples obtained. Finally, to compare the sequences identified in this work with others reported worldwide, the Blastn program was used. The sequences were synthesized by GenScript (Piscataway, NJ, USA) to crude purity and adjusted to a concentration of 1 mg/ml in water.
The serum samples were used to detect peptide-specific antibodies using enzyme-linked immunosorbent assay (ELISA) modified from Priest (Priest et al., 2005). Briefly, 96-well clear flat bottom microplates (Corning, Inc., New York, NY, USA) were coated with 0.5 µg of each peptide in carbonate buffer 0.1 M, pH 9.6. The 96-well plates were coated overnight at 4°C. Excess antigen was removed with three washes with 0.05% Tween 20–PBS [pH 7.4; phosphate-buffered saline (PBS)]; nonspecific binding was blocked with a 5% nonfat dry milk solution for 4 h at room temperature. After three washes with Tween 20–PBS solution, the wells were incubated with serum diluted 1:100 in PBS for 1 h at room temperature. After washing, goat anti-human IgG and IgM with peroxidase (dilutions of 1:1,000 and 1:500, respectively) (Southern Biotech Associates, Inc., Birmingham, AL, USA) were added and incubated for 1 h at room temperature. After being washed, the wells were incubated with the TMB substrate solution (Sigma, St. Louis, MO, USA) at room temperature in the dark, and the reaction was stopped after 5 min with H2S04. Absorbance was read on a microplate absorbance reader at 450 nm (iMark Bio-Rad).
Data were analyzed using GraphPad Prism 5 for Windows (GraphPad Software, Inc., San Diego, CA). The Mann–Whitney U-test was used to determine differences between the patient group and the control group.
Of the 39 cases with cryptosporidiosis included in this study, C. parvum (19/39) was identified in 48.7%, C. hominis (13/39) in 33.3%, and C. canis in 2.6% (1/39) by analyzing the sequence of the SSU rRNA gene. In 7.7% of the cases (3/39), a coinfection of C. parvum and C. hominis was determined by qPCR, and in 2.6% of the cases (1/39), a coinfection of C. canis and C. hominis was determined (Table 1).
Five peptides from C. parvum, four peptides of the gp15 protein (A109, A133, A32, and R61), and one peptide of the gp40 protein (V30) were designed and used in this study (Supplementary Table 2).
Table 2 shows the seroprevalence result of the IgG response of infected patients (cases) and noninfected individuals (controls) to peptides generated from gp15 sequences (A109, A133, A32, and R61) and a gp40 sequence (V30) from C. parvum. The seroprevalences of the cases for peptides A133 and A32 were 84.6% and 74.4%, respectively. The peptides A133 and A32 showed a statistically significant difference (p <0.003) in the recognition of sera between cases and controls, with stronger recognition by the cases (Figures 1A, B). The recognition of peptides R61, A109, and V30 did not show a statistically significant difference (p >0.05) between cases and controls (Figures 1C–E).
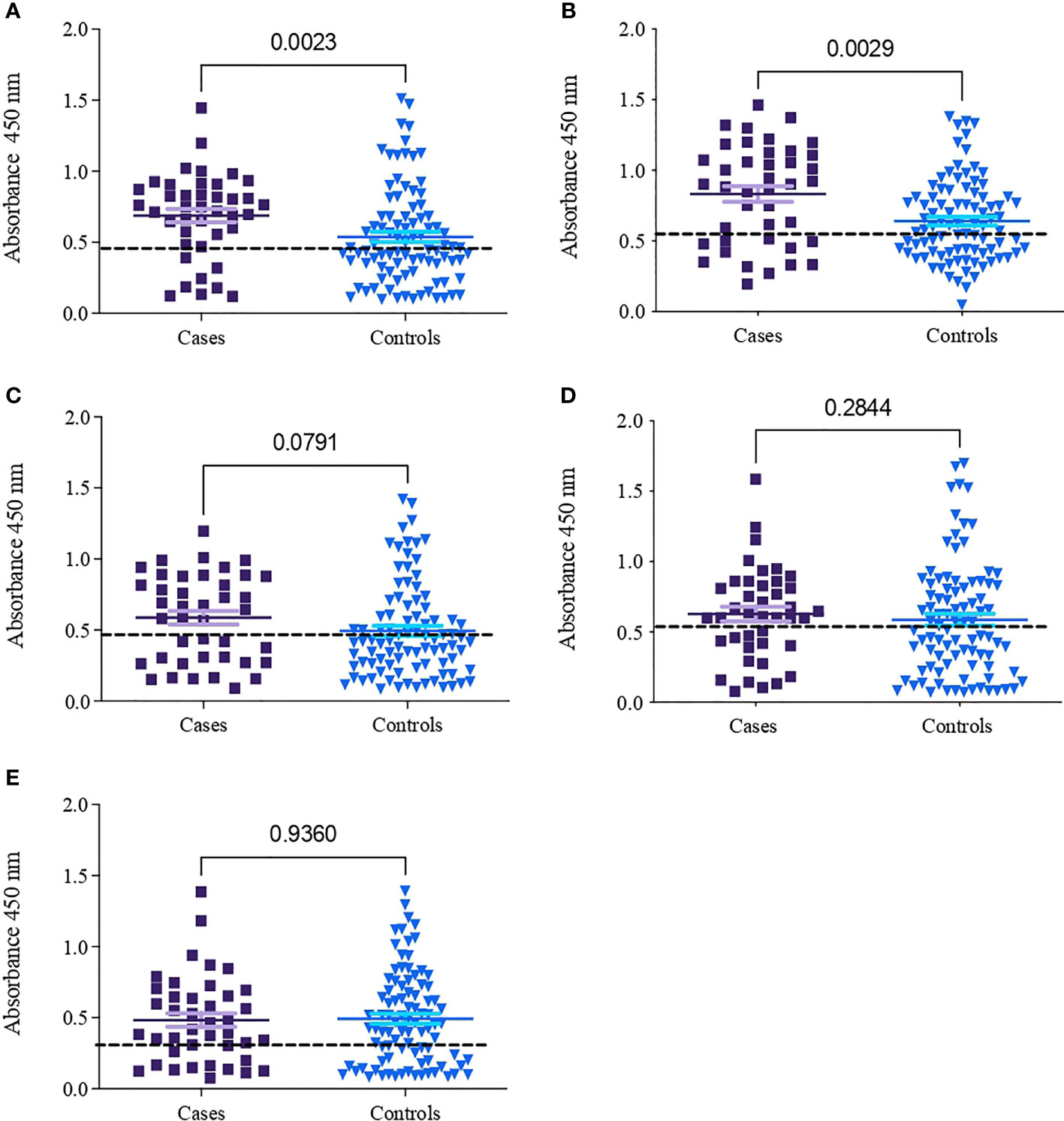
Figure 1 Absorbances obtained between cases and controls for four peptides of the Cryptosporidium parvum proteins gp15 and gp40. (A) A133; cutoff line (0.46). (B) A32; cutoff line (0.57). (C) R61; cutoff line (0.47). (D) A109; cutoff line (0.55). (E) V30; cutoff line (0.33).
In the control group, two age groups were observed: those who were younger than 15 years old and those who were older than 19 years old (Table 2). Statistically significant differences were not found for the recognition of peptides A133 and A32 by age group (p = 0.862, p = 0.082).
The serum recognition (IgG) of the five peptides tested in this study by infecting genotype/species (C. parvum, C. hominis, and C. canis) is shown in Supplementary Figure 1, and no statistically significant difference was found (p > 0.4).
Supplementary Figures 2A, B show a statistically significant difference between the IgM and IgG antibody titers both in cases (p < 0.0001) and controls (p < 0.0001) for the recognition of the A133 peptide. Similarly, after determining the cutoff (0.5), it was identified that 79.5% of the cases (31/39) (Supplementary Figure 2C) and 43.3% of the controls (39/90) were above the cutoff (Supplementary Figure 2D).
Supplementary Figures 3A, B show that there was a statistically significant difference between the IgM and IgG antibody titers in both the cases (p = 0.0001) and controls (p = 0.0001) for the recognition of the A32 peptide; after determining the cohort line (0.55), it was identified that 74.4% of the cases (29/39) (Supplementary Figure 3C) and 54.4% of the controls (49/90) were above the cutoff (Supplementary Figure 3D).
The A32 and A133 peptides described here have been submitted to the Instituto Mexicano de la Propiedad Industrial to obtain their patents (MX/a/2019/013932 and MX/a/2019/013933).
Despite the design of the peptides tested, A109, A133, A32, and R61 were generated from the sequence of C. parvum gp15; after alignment with other reported sequences, peptides A109, A32, and R61 were found in C. hominis gp15. Therefore, these peptides are present in the gp15 region of both species (conserved epitopes), and no statistically significant differences were found according to the genotype/infecting species. However, for the A133 and V30 peptides, whose sequences were only aligned in isolates of C. parvum, there was no difference in recognition by cases according to the genotype/infecting species. In previous studies, the recognition of recombinant proteins of gp15 (rgp15) of C. hominis (Allison et al., 2011) and rgp40 of C. parvum (Ajjampur et al., 2011) by serum IgG has been observed. Additionally, statistically significant differences in the recognition of the antigens between cases and controls, and cross-reactivity between epitopes (peptides) of these antigens (rgp15Ch, rgp40Cp), have been observed (Ajjampur et al., 2011; Allison et al., 2011).
The recognition of these peptides determines whether the patient is or was infected by Cryptosporidium. Based on the way the peptides were designed, it was expected to find specific genotype/species recognition; however, in the cases included in the study, specific genotype/species recognition was not observed, perhaps because both the cases and controls had a previous encounter with Cryptosporidium. Additionally, the serum IgG titer towards three of the peptides (A109, V30, and R61) tested did not show differences between the cases and controls. In previous studies, it has been shown that multiple encounters with a parasite are required to generate a significant IgG titer (DuPont et al., 1995). Despite this, in this study, it was managed to generate two peptides, A133 and A32, from gp15 and gp40 of C. parvum, respectively; however, A133 and A32 were similar in terms of their amino acid sequences. In addition, it was determined that the recognition of A133 and A32 was significantly different between the cases and controls (p = 0.0023 and p = 0.0029, respectively); however, it was not possible to determine statistically significant differences (p > 0.05) in the recognition of the peptides by infecting species.
Using the peptides A133 and A32, an analysis of the serum IgM antibody was performed to determine whether cases with lower absorbance of IgG had higher absorbance of IgM based on the fact that those with an acute infection have higher IgM titers that decrease over the course of a few weeks, which is opposite of the trend of IgG. It was found that there were statistically significant differences (p < 0.0001) between the serum IgM and IgG antibodies in the cases and controls, which led it to suggest that, in most of these cases, the patients were not infected for the first time or had been infected for several weeks. It was observed that the controls that were above the cut-off had high levels of the IgG antibody; one explanation for this result is memory antibodies.
Our study reveals the ability of two peptides (A133 and A32) of the C. parvum gp15 protein to identify and discriminate between infected (cases) and uninfected (controls) individuals. These results agree with a previous report by Avendaño et al.; this study evaluated five synthetic peptides of the gp15 (Cp15) and CSL proteins. The results showed that all peptides can stimulate antibody production, but only two peptides neutralize parasites in vitro (Avendaño et al., 2018). In addition, another report showed that antibodies against anti-gp40/15 and anti-gp40 antibodies block C. parvum infection in vitro (Cui et al., 2020). These data suggest that regions on the gp15 protein of C. parvum induce a strong antibody response in infected individuals. These regions probably can induce antibodies able to neutralize the Cryptosporidium. These results can support these peptides in future research to develop a Cryptosporidium vaccine. Other studies evaluating the cellular immune response are required to understand better the immune response and the participation of these peptides.
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
The studies involving human participants were reviewed and approved by Ethics Committee of the Hospital Infantil del Estado de Sonora (HIES). Written informed consent to participate in this study was provided by the participants’ legal guardian/next of kin.
OV: conception, project administration, and supervision. AU-Q and VO: writing original draft. JH and OV: visualization. AU-Q, RB-B, AG, JH, and OV: investigation and data analysis. OV, JH, and AG: validation. AU-Q and RB-B: methodology. OV: resources. OV: funding acquisition. All authors contributed to the article and approved the submitted version.
This work was supported by the Consejo Nacional de Ciencia y Tecnología (CONACyT), Fondo Sectorial de Investigación para la Educación (grant number 258454).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We thank the administrative and medical staff of the hospitals. The participation of volunteers is also acknowledged.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2021.810887/full#supplementary-material
Supplementary Table 1 | Distribution of cases and controls by hospital of origin.
Supplementary Table 2 | Peptide antigens of the Cryptosporidium parvum gp15 and gp40 proteins.
Supplementary Figure 1 | Recognition of peptides among infecting Cryptosporidium species. The line is cut-off (average plus two standard deviations) for each peptide obtained from the average of the absorbances of the controls. Cp, C. parvum; Ch, C. hominis; Cc, C. canis. (A) Peptide A109. (B) Peptide A133. (C) Peptide V30. (D) Peptide A32. (E) Peptide R61.
Supplementary Figure 2 | IgM and IgG anti-Cryptosporidium A133 peptide. (A) Cases. (B) Controls. (C) Cases with IgG above the cutoff line (0.5). (D) Controls with IgG above the cut-off line (0.5). (E) Cases with IgG below the cutoff line (0.5). (F) Controls with IgG below the cutoff line.
Supplementary Figure 3 | IgM and IgG anti-Cryptosporidium A32 peptide. (A) Cases. (B) Controls. (C) Cases with IgG above the cutoff line (0.556). (D) Controls with IgG above the cut-off line (0.05). (E) Cases with IgG below the cutoff line (0.05). (F) Controls with IgG below the cut-off line.
Ajjampur, S. S., Sarkar, R., Allison, G., Banda, K., Kane, A., Muliyil, J., et al. (2011). Serum IgG Response to Cryptosporidium Immunodominant Antigen Gp15 and Polymorphic Antigen Gp40 in Children With Cryptosporidiosis in South India. Clin. Vaccine Immunol. 18, 633–639. doi: 10.1128/CVI.00464-10
Allison, G. M., Rogers, K. A., Borad, A., Ahmed, S., Karim, M. M., Kane, A. V., et al. (2011). Antibody Responses to the Immunodominant Cryptosporidium Gp15 Antigen and Gp15 Polymorphisms in a Case-Control Study of Cryptosporidiosis in Children in Bangladesh. Am. J. Trop. Med. Hyg. 85, 97–104. doi: 10.4269/ajtmh.2011.11-0043
Avendaño, C., Jenkins, M., Méndez-Callejas, G., Oviedo, J., Guzmán, F., Patarroyo, M. A., et al. (2018). Cryptosporidium Spp. CP15 and CSL Protein-Derived Synthetic Peptides’ Immunogenicity and In Vitro Seroneutralisation Capability. Vaccine 36, 6703–6710. doi: 10.1016/j.vaccine.2018.09.044
Borad, A., Ward, H. (2010). Human Immune Responses in Cryptosporidiosis. Future Microbiol. 5, 507–519. doi: 10.2217/fmb.09.128
Bouzid, M., Elwin, K., Nader, J. L., Chalmers, R. M., Hunter, P. R., Tyler, K. M. (2016). Novel Real-Time PCR Assays for the Specific Detection of Human Infective Cryptosporidium Species. Virulence 7, 395–399. doi: 10.1080/21505594.2016.1149670
Bouzid, M., Hunter, P. R., Chalmers, R. M., Tyler, K. M. (2013). Cryptosporidium Pathogenicity and Virulence. Clin. Microbiol. Rev. 26, 115–134. doi: 10.1128/CMR.00076-12
Cevallos, A. M., Zhang, X., Waldor, M. K., Jaison, S., Zhou, X., Tzipori, S., et al. (2000). Molecular Cloning and Expression of a Gene Encoding Cryptosporidium Parvum Glycoproteins Gp40 and Gp15. Infect. Immun. 68, 4108–4116. doi: 10.1128/IAI.68.7.4108-4116.2000
Chalmers, R. M., Giles, M. (2010). Zoonotic Cryptosporidiosis in the UK - Challenges for Control. J. Appl. Microbiol. 109, 1487–1497. doi: 10.1111/j.1365-2672.2010.04764.x
Checkley, W., White, A. C., Jaganath, D., Arrowood, M. J., Chalmers, R. M., Chen, X. M., et al. (2015). A Review of the Global Burden, Novel Diagnostics, Therapeutics, and Vaccine Targets for Cryptosporidium. Lancet Infect. Dis. 15, 85–94. doi: 10.1016/S1473-3099(14)70772-8
Cui, Z., Wang, L., Wang, Y., Li, J., Wang, R., Sun, M., et al. (2020). Cryptosporidium Parvum Gp40/15 Is Associated With the Parasitophorous Vacuole Membrane and Is a Potential Vaccine Target. Microorganisms 8, 1–11. doi: 10.3390/microorganisms8030363
DuPont, H. L., Chappell, C. L., Sterling, C. R., Okhuysen, P. C., Rose, J. B., Jakubowski, W. (1995). The Infectivity of Cryptosporidium Parvum in Healthy Volunteers. N. Engl. J. Med. 332, 855–859. doi: 10.1056/NEJM199503303321304
Frost, F. J., Tollestrup, K., Craun, G. F., Fairley, C. K., Sinclair, M. I., Kunde, T. R. (2005). Protective Immunity Associated With a Strong Serological Response to a Cryptosporidium-Specific Antigen Group, in HIV-Infected Individuals. J. Infect. Dis. 192, 618–621. doi: 10.1086/431681
Gonzalez-Diaz, M., Urrea-Quezada, A., Villegas-Gomez, I., Durazo, M., Garibay-Escobar, A., Hernandez, J., et al. (2016). Cryptosporidium Canis in Two Mexican Toddlers. Pediatr. Infect. Dis. J. 35, 1265–1266. doi: 10.1097/INF.0000000000001287
Henriksen, S. A., Pohlenz, J. F. (1981). Staining of Cryptosporidia by a Modified Ziehl-Neelsen Technique. Acta Vet. Scand. 22, 594–596. doi: 10.1186/BF03548684
Priest, J. W., Bern, C., Roberts, J. M., Kwon, J. P., Lescano, A. G., Checkley, W., et al. (2005). Changes in Serum Immunoglobulin G Levels as a Marker for Cryptosporidium Sp. Infection in Peruvian Children. J. Clin. Microbiol. 43, 5298–5300. doi: 10.1128/JCM.43.10.5298-5300.2005
Sarkar, R., Ajjampur, S. S., Muliyil, J., Ward, H., Naumova, E. N., Kang, G. (2012). Serum IgG Responses and Seroconversion Patterns to Cryptosporidium Gp15 Among Children in a Birth Cohort in South India. Clin. Vaccine Immunol. 19, 849–854. doi: 10.1128/CVI.00051-12
Urrea-Quezada, A., González-Díaz, M., Villegas-Gómez, I., Durazo, M., Hernández, J., Xiao, L., et al. (2018). Clinical Manifestations of Cryptosporidiosis and Identification of a New Cryptosporidium Subtype in Patients From Sonora, Mexico. Pediatr. Infect. Dis. J. 37, e136–e138. doi: 10.1097/INF.0000000000001762
Valenzuela, O., González-Díaz, M., Garibay-Escobar, A., Burgara-Estrella, A., Cano, M., Durazo, M., et al. (2014). Molecular Characterization of Cryptosporidium Spp. In Children From Mexico. PloS One 9, e96128. doi: 10.1371/journal.pone.0096128
Keywords: human cryptosporidiosis, emerging parasites, humoral response, synthetic peptides, IgG
Citation: Urrea-Quezada A, Balmaceda-Baca R, Garibay A, Hernández J and Valenzuela O (2022) Serum IgG Responses to gp15 and gp40 Protein-Derived Synthetic Peptides From Cryptosporidium parvum. Front. Cell. Infect. Microbiol. 11:810887. doi: 10.3389/fcimb.2021.810887
Received: 08 November 2021; Accepted: 16 December 2021;
Published: 19 January 2022.
Edited by:
Krisztina M. Papp-Wallace, Louis Stokes Cleveland VA Medical Center, United StatesReviewed by:
Cristina Cabanacan Salibay, De La Salle University—Dasmariñas, PhilippinesCopyright © 2022 Urrea-Quezada, Balmaceda-Baca, Garibay, Hernández and Valenzuela. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Olivia Valenzuela, b2xpdmlhLnZhbGVuenVlbGFAdW5pc29uLm14; dmFsZW56dWVsYS5vQGdtYWlsLmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.