- 1Department of Clinical Laboratory, The Children’s Hospital, Zhejiang University School of Medicine, National Clinical Research Center for Child Health, Hangzhou, China
- 2Microbiological Laboratory, Zhejiang Provincial Center for Disease Control and Prevention, Hangzhou, China
Objective: This study aims to analyze the molecular epidemiology, resistance, and pathogenicity of Salmonella enterica subsp. diarizonae isolated from children.
Methods: Whole genome sequencing was carried out, and molecular serotypes, sequence types, resistance genes, and virulence genes of S. enterica subsp. diarizonae isolates were analyzed. Antimicrobial susceptibility test was determined by commercialized microdilution method.
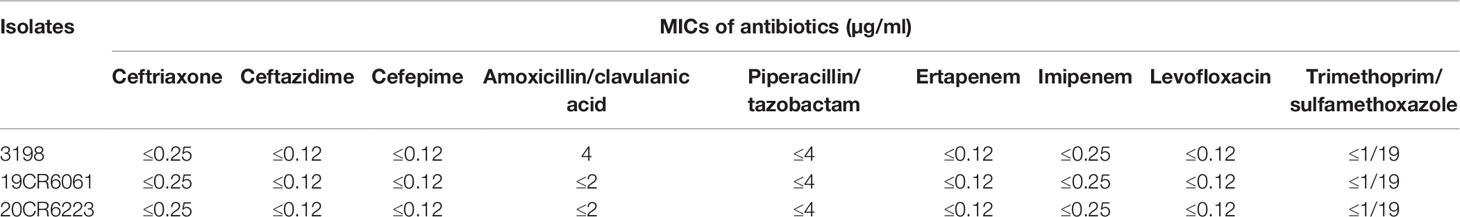
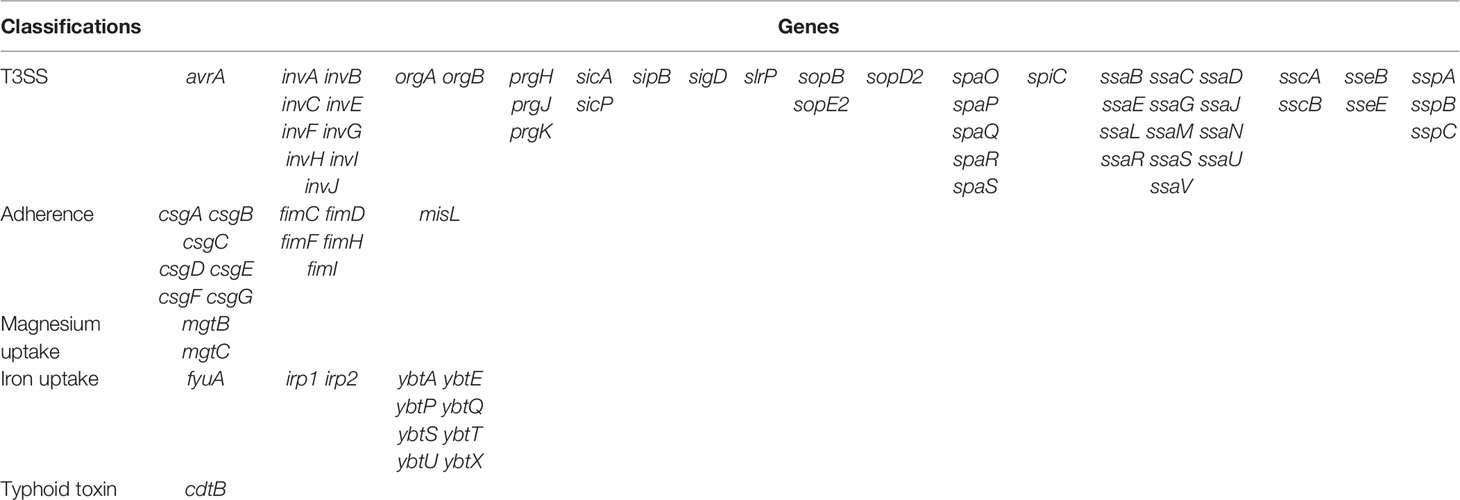
Results: A total of three isolates of S. enterica subsp. diarizonae were isolated during 2015 to 2020. The molecular serotypes of the three strains were 61:c:z35, 61:l,v:1,5,7:[z57], and 65:k:z, respectively, and the sequence types were ST1845, ST233, and ST1263. All the three isolates were susceptible to ceftriaxone, ceftazidime, cefepime, amoxycillin/clavulanic acid, piperacillin/tazobactam, ertapenem, imipenem, levofloxacin, and trimethoprim/sulfamethoxazole. No other resistant gene was detected except aac(6’)-Iaa. There were no resistant plasmids detected in all the three isolates. A total of 76 genes were present in all isolates, containing 49 genes of Type III Secretion System (T3SS) mediated by SPI-1and SPI-2, 13 genes of adherence (type 1 fimbriae, Agf, and MisL-related genes), 11 genes of iron uptake (Yersiniabactin), two genes of magnesium uptake, and one gene of typhoid toxin(cdtB).
Conclusion: The serotypes and sequence types of S. enterica subsp. diarizonae isolates were rarely reported in children; all the S. enterica subsp. diarizonae isolates were susceptible to detected antibiotics; T3SS, adherence, iron uptake, magnesium uptake, and typhoid toxin were responsible for pathogenicity of the S. enterica subsp. diarizonae isolates in children.
Introduction
Salmonella is the predominant bacteria causing diarrhea in children, especially in infancy and early childhood, which seriously threatens the lives and health of children, and previous study has stated that there were over 93 million cases of gastroenteritis and 155,000 deaths caused by nontyphoidal Salmonella per year (Majowicz et al., 2010). The genus Salmonella includes only two species, Salmonella enterica and Salmonella bongori. S. enterica is further subdivided into six subspecies: enterica, salamae, arizonae, diarizonae, houtenae, and indica (Guibourdenche et al., 2010). Virtually all of Salmonella infections in humans were caused by the strain of S. enterica subsp. enterica (McClelland et al., 2001). S. enterica subsp. diarizonae is known to cause infections in ectothermic animals (Schroter et al., 2004). However, S. enterica subsp. diarizonae related cases in human have been gradually reported (Chong et al., 1991; Gerlach et al., 2017; Giner-Lamia et al., 2019; Uelze et al., 2020; Pan et al., 2021). Additionally, limited study has been reported in the area of molecular epidemiology, resistance, and pathogenicity of S. enterica subsp. diarizonae in children (Gerlach et al., 2017; Giner-Lamia et al., 2019). Whole genome sequencing (WGS) provided information regarding multilocus sequence typing (MLST), pathogenicity genes, and other contents (Kozyreva et al., 2016), which have rendered the systematic study of bacterial pathogens at the molecular level efficient and convenient. Accordingly, this study set out using WGS to determine the molecular epidemiology, resistance, and pathogenicity of S. enterica subsp. diarizonae in children from a tertiary university children’s hospital in China during 2015 to 2020.
Methods
Strain Collection and Identification
Three clinical S. enterica subsp. diarizonae isolates were collected at The Children’s Hospital, Zhejiang University School of Medicine from January 2015 to December 2020. First, fecal specimens were inoculated on SS medium (Comagal, Shanghai, China) and incubated for 18–24 h, while other specimens were inoculated on Columbia blood agar (Bioivd, Zhengzhou, China). The suspicious colonies of Salmonella were then identified using matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF MS, Bruker, Germany). Salmonella isolates were serotyped according to the modified Kauffmann-White scheme using commercial antisera (SSI, Copenhagen, Denmark) (Guibourdenche et al., 2010). Salmonella typhimurium ATCC14028 was used for quality control.
Antimicrobial Susceptibility Test
Antimicrobial susceptibility test was determined by commercialized microdilution method (VITEK COMPACT, BioMérieux, Marcy-l’Étoile, France), and the results were interpreted according to the Clinical Laboratory Standards Institute (CLSI) guidelines M100-S30. Ceftriaxone, ceftazidime, cefepime, amoxycillin/clavulanic acid, piperacillin/tazobactam, ertapenem, imipenem, levofloxacin, and trimethoprim/sulfamethoxazole were detected. Escherichia coli ATCC25922 was used for quality control.
DNA Extraction and Sequencing
Genomic DNA was extracted using QIAGEN DNA miniprep kit (QIAGEN, Hilden, Germany). The concentration and purity of the samples were detected by BioDrop μLite+ (BioDrop, Cambridge, UK). The extracted DNA was sent to Hangzhou Digital-Micro Biotechnology Co., Ltd. for sequencing. After library construction, WGS was performed on Illumina Hiseq xTen platform using a 2 × 150-bp paired end (PE) configuration. Sequencing reads were trimmed and de novo assembled into contigs using the Shovill pipeline (https://github.com/tseemann/shovill).
Analysis of WGS Data
MLST, resistance genes, and plasmids were conducted in the Center for Genomic Epidemiology (CGE) by uploading the contigs files obtained from the de novo assembly of the WGS data (Larsen et al., 2012). The molecular serotypes of S. enterica subsp. diarizonae were obtained on the pathogenwatch online (Yoshida et al., 2016). Virulence factors were identified using ABRicate software (V.0.8.10) (https://github.com/tseemann/abricate) by aligned against the Virulence Factors Database (VFDB) (Chen et al., 2016).
Results
Source of Isolates and Molecular Epidemiology
In total, three isolates of S. enterica subsp. diarizonae were isolated during 2015 to 2020; two strains were isolated from 2019 and one from 2020. All of the three strains were derived from fecal specimens. Of these, one isolate was from male and two were from female, and the three patients were all pediatric patients under 1 year old. The serotypes of the three strains were assigned to 61:c:z35, 61:l,v:1,5,7:[z57], and 65:k:z, respectively, while the MLST types were ST1845, ST233, and ST1263, respectively (Table 1).
Antimicrobial Susceptibility Test and Resistance Genes
Antibiotic resistance profiles and resistant genes of three isolates of S. enterica subsp. diarizonae were entirely consistent. All the three isolates were susceptible to ceftriaxone, ceftazidime, cefepime, amoxycillin/clavulanic acid, piperacillin/tazobactam, ertapenem, imipenem, levofloxacin, and trimethoprim/sulfamethoxazole (Table 2). No other resistant gene was detected except for aac(6’)-Iaa. No gene in any of the resistant plasmids was found in this study.
Virulence Genes
The distribution of virulence genes of three isolates of S. enterica subsp. diarizonae was identical. A total of 76 genes were detected, containing 49 genes of Type III Secretion System (T3SS) mediated by SPI-1and SPI-2, 13 genes of adherence (type 1 fimbriae, Agf, and MisL-related genes), 11 genes of iron uptake (Yersiniabactin), two genes of magnesium uptake, and one gene of typhoid toxin (Table 3).
Discussion
Salmonella causes a variety of symptoms ranging from a mild intestinal infection to life-threatening systemic infections (Chanana et al., 2007), including enteric fever, gastroenteritis, bacteraemia, and systemic infection. All isolates analyzed in this study were derived from fecal specimens, and all the three patients were under 1 year old, which means that S. enterica subsp. diarizonae mainly causes gastroenteritis, especially in infants. S. enterica subsp. diarizonae serotypes 43:g,t:- and 8:r:z have been reported in human stools (Guibourdenche et al., 2010), while 48:i:z reported in endocervical tissue and cerebrospinal fluid (Giner-Lamia et al., 2019) and 60:r:z reported in a patient suffering from diarrhea and sepsis (Gerlach et al., 2017). Unlike the studies mentioned above, our current data demonstrated different serotypes (61:c:z35, 61:l,v:1,5,7:[z57] and 65:k:z) of S. enterica subsp. diarizonae in children’s stools, which were rarely reported in children. However, S. enterica subsp. diarizonae serotype IIIb_61:I,v:1,5 (Giner-Lamia et al., 2019), strains were reported in wheat grains (Shridhar et al., 2021). MLST is considered to be a well-adopted genotyping method for bacterial epidemiological studies; however, WGS can be used to confirm unusual or unexpected genotyping results, especially in highly recombinogenic pathogens (Aziz et al., 2017). Using WGS, all the three isolates belonged to distinct STs in this study, suggesting that these isolates were not a cluster of epidemic strain. Previous study has reported ST1256 diarizonae strains isolated from endocervical tissue and cerebrospinal fluid; other STs of S. enterica subsp. diarizonae also related to human infections, including ST233, ST430, and ST432 (Giner-Lamia et al., 2019). However, the STs of S. enterica subsp. diarizonae were ST1845, ST233, and ST1263 in this study. These differences of STs may depend on the geographic location and infection site in which the strains were obtained.
All the three isolates of S. enterica subsp. diarizonae were susceptible to all the detected antibiotics, and no resistant plasmids were detected in this study. Similarly, two diarizonae strains with sequence type ST1256 were found to be susceptible to trimethoprim-sulfametoxazol, quinonoles, aminoglycosides, and most common beta-lactams (Giner-Lamia et al., 2019); a S. enterica subsp. diarizonae serotype 61:k:1,5 (Giner-Lamia et al., 2019) isolate from urine sample was susceptible to all tested antibiotics (Uelze et al., 2020). However, recent study reported a multidrug-resistant clinical isolate of S. diarizonae with a conjugative IncHI2A plasmid (Pan et al., 2021). Therefore, the resistance of S. enterica subsp. diarizonae needs ongoing attention. In this study, WGS analysis revealed that the resistance gene to aminoglycosides (aac (6’)-laa) was present in all isolates; however, the aac(6′)-Iaa gene does not appear to encode for resistance (Leon et al., 2018).
It is known that the critical factor for Salmonella survival and establishment of disease in a host is entering into host cells. First, Salmonella enters host cells by host invasion pathways, where Salmonella adhere to host cells; then, Salmonella enters cell invasion pathways, when T3SS secrets a large number of virulence factors (Velge et al., 2012). The most well-studied Salmonella pathogenicity islands (SPI) are SPI-1, SPI-2, SPI-3, SPI-4, and SPI-5 (Hensel, 2004). SPI-1 presented in virtually all Salmonella isolates, encoded T3SS-1, and related to Salmonella invasion of eukaryotic cells; T3SS-2, which was encoded by SPI-2, is required for intracellular proliferation and survival (Coombes et al., 2004). Up to 49 virulence genes of T3SS (mediated by SPI-1and SPI-2) and 13 genes of adherence were identified in the evaluated genomes of the S. enterica subsp. diarizonae isolates in this study, suggesting that adherence and T3SS played an important role in pathogenicity of S. enterica subsp. diarizonae. MgtCB can maintain the growth in low Mg2+ and the survival of Salmonella in macrophages (Smith et al., 1998). In the present study, mgtB and mgtC, which are located in SPI-3, were present in all three S. enterica subsp. diarizonae strains, which indicated that MgtCB contributes to the pathogenicity of S. enterica subsp. diarizonae. Additionally, misL in SPI-3 region and sopB in SPI-5 region were present in all S. enterica subsp. diarizonae strains in this study. Overall, virulence genes of SPI-1, SPI-2, SPI-3, and SPI-5 regions were present in S. enterica subsp. diarizonae in this study. Distinctly, other studies revealed that diarizonae isolates harbor more SPIs, containing SPIs of SPI-1~5, SPI-9, SPI-11, SPI-13, SPI-18, and SPI-21 (Giner-Lamia et al., 2019) and SPIs of SPI-1~5, SPI-9, SPI-12, SPI-13, and SPI-18 (Gerlach et al., 2017). Yersiniabactin is an iron carrier that helps bacteria gain the ability to chelate iron from infected host cells (Perry and Fetherston, 2011). In the present study, the Yersiniabactin genes were present in all isolates, which may pose challenges for clinical treatment. It was known that cdtB is strongly associated with persistent and chronic infections of Salmonella typhi (Del Bel Belluz et al., 2016). Here, cdtB were detected in all three strains of S. enterica subsp. diarizonae in this study. This finding is in accordance with the study by Gaballa et al. (2021), who revealed the presence of cdtB in S. enterica subsp. enterica, S. enterica subsp. arizonae and S. enterica subsp. diarizonae strains. Overall, S. enterica subsp. diarizonae strains appear to have complex virulence genes, which indicate that much still remains to be learned about the pathogenicity.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: NCBI; PRJNA740099, SAMN19819870, SAMN19819871, SAMN19819872.
Ethics Statement
The studies involving human participants were reviewed and approved by The Research and Ethics committee of The Children’s Hospital, Zhejiang University School of Medicine (2021-IRB-031). Written informed consent from the participants’ legal guardian/next of kin was not required to participate in this study in accordance with the national legislation and the institutional requirements.
Author Contributions
MZ designed the study and drafted the manuscript. QS contributed to the analysis and interpretation of data for the work and revised and critically reviewed the manuscript. XZ performed DNA extraction. LM performed serotyping. YY collected the data. CF carried out antimicrobial susceptibility test. SS designed the study and reviewed the manuscript. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Aziz, A., Sarovich, D. S., Harris, T. M., Kaestli, M., McRobb, E., Mayo, M., et al. (2017). Suspected Cases of Intracontinental Burkholderia Pseudomallei Sequence Type Homoplasy Resolved Using Whole-Genome Sequencing. Microbial Genomics 3. doi: 10.1099/mgen.0.000139
Chanana, V., Ray, P., Rishi, D. B., Rishi, P. (2007). Reactive Nitrogen Intermediates and Monokines Induce Caspase-3 Mediated Macrophage Apoptosis by Anaerobically Stressed Salmonella Typhi. Clin. Exp. Immunol. 150, 368–374. doi: 10.1111/j.1365-2249.2007.03503.x
Chen, L., Zheng, D., Liu, B., Yang, J., Jin, Q. (2016). VFDB 2016: Hierarchical and Refined Dataset for Big Data Analysis-10 Years on. Nucleic Acids Res. 44, D694–D697. doi: 10.1093/nar/gkv1239
Chong, Y., Kwon, O. H., Lee, S. Y., Chung, K. S., Shimada, T. (1991). Salmonella Enterica Subspecies Diarizonae Bacteremia in an Infant With Enteritis-a Case Report. Yonsei Med. J. 32, 275–278. doi: 10.3349/ymj.1991.32.3.275
Coombes, B. K., Brown, N. F., Valdez, Y., Brumell, J. H., Finlay, B. B. (2004). Expression and Secretion of Salmonella Pathogenicity Island-2 Virulence Genes in Response to Acidification Exhibit Differential Requirements of a Functional Type III Secretion Apparatus and Ssal. J. Biol. Chem. 279, 49804–49815. doi: 10.1074/jbc.M404299200
Del Bel Belluz, L., Guidi, R., Pateras, I. S., Levi, L., Mihaljevic, B., Rouf, S. F., et al. (2016). The Typhoid Toxin Promotes Host Survival and the Establishment of a Persistent Asymptomatic Infection. PloS Pathogens 12, e1005528. doi: 10.1371/journal.ppat.1005528
Gaballa, A., Cheng, R. A., Harrand, A. S., Cohn, A. R., Wiedmann, M. (2021). The Majority of Typhoid Toxin-Positive Salmonella Serovars Encode ArtB, an Alternate Binding Subunit. mSphere 6, e01255–20. doi: 10.1128/mSphere.01255-20
Gerlach, R. G., Walter, S., McClelland, M., Schmidt, C., Steglich, M., Prager, R., et al. (2017). Comparative Whole Genome Analysis of Three Consecutive Salmonella Diarizonae Isolates. Int. J. Med. Microbiol: IJMM. 307, 542–551. doi: 10.1016/j.ijmm.2017.09.001
Giner-Lamia, J., Vinuesa, P., Betancor, L., Silva, C., Bisio, J., Soleto, L., et al. (2019). Genome Analysis of Salmonella Enterica Subsp. Diarizonae Isolates From Invasive Human Infections Reveals Enrichment of Virulence-Related Functions in Lineage ST1256. BMC Genomics 20, 99. doi: 10.1186/s12864-018-5352-z
Guibourdenche, M., Roggentin, P., Mikoleit, M., Fields, P. I., Bockemuhl, J., Grimont, P. A., et al. (2010). Supplement 2003-2007 (No. 47) to the White-Kauffmann-Le Minor Scheme. Res. Microbiol. 161, 26–29. doi: 10.1016/j.resmic.2009.10.002
Hensel, M. (2004). Evolution of Pathogenicity Islands of Salmonella Enterica. Int. J. Med. Microbiology: IJMM. 294, 95–102. doi: 10.1016/j.ijmm.2004.06.025
Kozyreva, V. K., Crandall, J., Sabol, A., Poe, A., Zhang, P., Concepcion-Acevedo, J., et al. (2016). Laboratory Investigation of Salmonella Enterica Serovar Poona Outbreak in California: Comparison of Pulsed-Field Gel Electrophoresis (PFGE) and Whole Genome Sequencing (WGS) Results. PloS Curr. 8. doi: 10.1371/currents.outbreaks.1bb3e36e74bd5779bc43ac3a8dae52e6
Larsen, M. V., Cosentino, S., Rasmussen, S., Friis, C., Hasman, H., Marvig, R. L., et al. (2012). Multilocus Sequence Typing of Total-Genome-Sequenced Bacteria. J. Clin. Microbiol. 50, 1355–1361. doi: 10.1128/JCM.06094-11
Leon, I. M., Lawhon, S. D., Norman, K. N., Threadgill, D. S., Ohta, N., Vinasco, J., et al. (2018). Serotype Diversity and Antimicrobial Resistance Among Salmonella Enterica Isolates From Patients at an Equine Referral Hospital. Appl. Environ. Microbiol. 84, e02829–17. doi: 10.1128/AEM.02829-17
Majowicz, S. E., Musto, J., Scallan, E., Angulo, F. J., Kirk, M., O’Brien, S. J., et al. (2010). The Global Burden of Nontyphoidal Salmonella Gastroenteritis. Clin. Infect. Dis. 50, 882–889. doi: 10.1086/650733
McClelland, M., Sanderson, K. E., Spieth, J., Clifton, S. W., Latreille, P., Courtney, L., et al. (2001). Complete Genome Sequence of. Salmonella enterica serovar Typhimurium LT2. Nature 413, 852–856. doi: 10.1038/35101614
Pan, Y., Fang, Y., Song, X., Lyu, N., Chen, L., Feng, Y., et al. (2021). Co-Occurrence of Mcr-9, Extended Spectrum Beta-Lactamase (ESBL) and Ampc Genes in a Conjugative Inchi2a Plasmid From a Multidrug-Resistant Clinical Isolate of Salmonella Diarizonae. J. infection. 82, 84–123. doi: 10.1016/j.jinf.2020.11.008
Perry, R. D., Fetherston, J. D. (2011). Yersiniabactin Iron Uptake: Mechanisms and Role in Yersinia Pestis Pathogenesis. Microbes Infect. 13, 808–817. doi: 10.1016/j.micinf.2011.04.008
Schroter, M., Roggentin, P., Hofmann, J., Speicher, A., Laufs, R., Mack, D. (2004). Pet Snakes as a Reservoir for Salmonella Enterica Subsp. Diarizonae (Serogroup Iiib): A Prospective Study. Appl. Environ. Microbiol. 70, 613–615. doi: 10.1128/AEM.70.1.613-615.2004
Shridhar, P. B., Amachawadi, R. G., Atobatele, M., Shi, X., Adams, P. A., Phebus, R. K., et al. (2021). Draft Genome Sequences of Salmonella Enterica Subsp. Diarizonae Serotype Iiib_61:I,V:1,5,(7) Strains Isolated From Wheat Grains. Microbiol. Res. Ann. 10, e00035–21. doi: 10.1128/MRA.00035-21
Smith, R. L., Kaczmarek, M. T., Kucharski, L. M., Maguire, M. E. (1998). Magnesium Transport in Salmonella Typhimurium: Regulation of Mgta and Mgtcb During Invasion of Epithelial and Macrophage Cells. Microbiol. (Reading) 144 (Pt 7), 1835–1843. doi: 10.1099/00221287-144-7-1835
Uelze, L., Borowiak, M., Flieger, A., Simon, S., Tausch, S. H., Malorny, B. (2020). Complete Genome Sequence of Salmonella Enterica Subsp. Diarizonae Serovar 61:K:1,5,(7) Strain 14-SA00836-0, Isolated From Human Urine. Microbiol. Res Ann. 9, e00683–20. doi: 10.1128/MRA.00683-20
Velge, P., Wiedemann, A., Rosselin, M., Abed, N., Boumart, Z., Chausse, A. M., et al. (2012). Multiplicity of Salmonella Entry Mechanisms, a New Paradigm for Salmonella Pathogenesis. Microbiologyopen 1, 243–258. doi: 10.1002/mbo3.28
Yoshida, C. E., Kruczkiewicz, P., Laing, C. R., Lingohr, E. J., Gannon, V. P., Nash, J. H., et al. (2016). The Salmonella in Silico Typing Resource (SISTR): An Open Web-Accessible Tool for Rapidly Typing and Subtyping Draft Salmonella Genome Assemblies. PloS One 11, e0147101. doi: 10.1371/journal.pone.0147101
Keywords: Salmonella enterica, diarizonae, molecular epidemiology, resistance, pathogenicity, children, WGS
Citation: Zhou M, Shi Q, Zhang X, Mei L, Ye Y, Fang C and Shang S (2021) Salmonella enterica subsp. diarizonae Harboring ST233, ST1263, and ST1845 in Children. Front. Cell. Infect. Microbiol. 11:727811. doi: 10.3389/fcimb.2021.727811
Received: 02 July 2021; Accepted: 02 August 2021;
Published: 19 August 2021.
Edited by:
Ghassan M. Matar, American University of Beirut, LebanonReviewed by:
Graciela Castro Escarpulli, Instituto Politécnico Nacional de México (IPN), MexicoGisela Di Venanzio, Washington University School of Medicine in St. Louis, United States
Copyright © 2021 Zhou, Shi, Zhang, Mei, Ye, Fang and Shang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shiqiang Shang, c2hhbmdzcUB6anUuZWR1LmNu
 Mingming Zhou
Mingming Zhou Qiucheng Shi
Qiucheng Shi Xiucai Zhang
Xiucai Zhang Lingling Mei
Lingling Mei Yihua Ye
Yihua Ye Chao Fang
Chao Fang Shiqiang Shang
Shiqiang Shang