
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell. Infect. Microbiol., 24 February 2021
Sec. Clinical Microbiology
Volume 11 - 2021 | https://doi.org/10.3389/fcimb.2021.625430
This article is part of the Research TopicProgress in Pathogen Identification Based on Mass SpectrometryView all 17 articles
Acinetobacter baumannii is one of the main causes of nosocomial infections. Increasing numbers of multidrug-resistant Acinetobacter baumannii cases have been reported in recent years, but its antibiotic resistance mechanism remains unclear. We studied 9 multidrug-resistant (MDR) and 10 drug-susceptible Acinetobacter baumannii clinical isolates using Label free, TMT labeling approach and glycoproteomics analysis to identify proteins related to drug resistance. Our results showed that 164 proteins exhibited different expressions between MDR and drug-susceptible isolates. These differential proteins can be classified into six groups: a. proteins related to antibiotic resistance, b. membrane proteins, membrane transporters and proteins related to membrane formation, c. Stress response-related proteins, d. proteins related to gene expression and protein translation, e. metabolism-related proteins, f. proteins with unknown function or other functions containing biofilm formation and virulence. In addition, we verified seven proteins at the transcription level in eight clinical isolates by using quantitative RT-PCR. Results showed that four of the selected proteins have positive correlations with the protein level. This study provided an insight into the mechanism of antibiotic resistance of multidrug-resistant Acinetobacter baumannii.
1) 164 proteins exhibited different expressions between drug-resistant isolates and drug-susceptible isolates.
2) Classifying six groups of differential expressed proteins between drug-resistant isolates and drug-susceptible isolates.
3) A specifically expressed polysaccharide on the S21 site of adenylate kinase was found in most MDR strains.
Nosocomial infections caused by multidrug-resistant (MDR) bacteria strains are a serious problem worldwide in decades. Acinetobacter baumannii has become one of the most common species that cause nosocomial infections and healthcare-associated infections such as bacteremia, pneumonia, meningitis, skin and wound infections, and urinary tract infection due to its strong biofilm formation ability and the ability to resist nutrient deprivation and antibiotics (Dijkshoorn et al., 2007; Shin and Park, 2015; Mujawar et al., 2019). The traditional first-line treatment of A. baumannii uses carbapenem antibiotics such as imipenem, but since the early 1990s, there have been reports of outbreaks caused by imipenem-resistant A. baumannii (Tankovic et al., 1994). Other therapeutic antibiotics include aminoglycosides, sulbactam, polymyxin and tigecycline, etc., and combined antibiotics are also being used. However, studies have shown that MDR strains which are resistant to different antibiotics are reported commonly (Li et al., 2006; Peleg et al., 2007; Göttig et al., 2014; Al-Kadmy et al., 2020). Whether for developing new drugs or making full use of old drugs to treat infections caused by MDR strains, it is necessary to fully understand the antibiotic resistance mechanism. A full understanding of the resistance mechanism is critical to improve the eradication rate of A. baumannii. Studies have shown that the antibiotics resistant mechanism mainly includes the modification of the target site, inactivation or modification of the drugs by producing enzymes such as β-lactamases, the activation of the efflux pump and the changes of the membrane structure and permeability of bacteria (Dijkshoorn et al., 2007; Zarrilli et al., 2013; Lee et al., 2017; Ramirez et al., 2020).
Genomics and proteomics studies can explore the expression of genes or proteins under various conditions thus to help understand the different mechanisms of bacteria drug resistance. At present, there have been extensive researches on A. baumannii through proteomic analysis to explore the relevant mechanisms of drug resistance, drought tolerance, biofilm formation, virulence, and nutrient regulation (Kwon et al., 2009; Shin et al., 2009; Nwugo et al., 2011; Long et al., 2013; Gayoso et al., 2014; Scribano et al., 2019). Researches on drug resistance of A. baumannii have studied the differences of a single strain pre and after the induction of resistance (Fernández-Reyes et al., 2009; Hood et al., 2010; Hua et al., 2017) or under different culture conditions (Yun et al., 2011; De Silva et al., 2018), or the difference between susceptible strains and resistant strains (Siroy et al., 2006; Vashist et al., 2010; Li et al., 2015; Wang et al., 2016). However, the researches usually analyze only one or two strains and focused only on a certain antibiotic. Different proteomics methods have their advantages and disadvantages. Using more than one proteomics method can complement each other and enhance the reproducibility and effectiveness of proteomics (Tiwari and Tiwari, 2014). In addition, protein glycosylation is an important means of protein modification, and different glycosylation modifications are critical to protein function (Ahmad Izaham and Scott, 2020). In this study, we intend to use the approach of label free and tandem mass tag (TMT) labeling-based proteomics and glycoproteomes to analyze the different proteins between MDR and drug-susceptible A. baumannii to fully clarify the mechanism of the antibiotic resistance.
Nineteen A. baumannii clinical strains (9 MDR and 10 susceptible isolates) were isolated from different patients in the second affiliated hospital of Nanchang University, China, during 2011–2019. The isolates were identified by VITEK-2 compact system and 16S ribosomal DNA identification. The 16S ribosomal DNA were amplified with the primer (16s-PCRF and 16s-PCRR) showed in Supplementary Table 1. The fragments were sequenced and blasted in NCBI non-redundant database for species identification. Antibiotic susceptibility of the following antibiotics were tested by Kirby–Bauer test (KB-test): β-lactam antibiotics [piperacillin, ceftazidime, ceftriaxone, cefotaxime, cefepime, imipenem, Unasyn (ampicillin/sulbactam), and Tazocilin (piperacillin/tazobactam)], aminoglycoside (gentamicin and tobramycin), tetracyclines (minocycline and tigecycline), polymyxin B, fluoroquinolone (levofloxacin, Ciprofloxacin), and paediatric compound sulfamethoxazole tablets. Data of these isolates are shown in Supplementary Table 2.
The experiment process is shown in Figure 1. All the isolates were cultured in LB broth at 37°C with shaking until OD600 nm of 0.7–0.8 reached. The cells were collected and lysed by 8M urea in 50 mM triethyl ammonium bicarbonate (TEAB) and ultrasonic breakage for 20 s. The protein samples were collected via centrifugation at 16,000×g for 10 min at 4°C. The protein concentration of the supernatant was determined using BCA Protein Assay Kit (Thermo-Fisher Scientific).
The proteins were reduced by incubation with TCEP (200 mM) at 55°C for 1 h and alkylated by incubation with iodoacetamide (IAA, 375 mM, Thermo Scientific) for 30 min in dark at room temperature. TEAB (100 mM) was used to adjust the urea concentration of less than 1M in all the protein samples, and then the proteins were digested to peptides using trypsin (Promega) at a trypsin/protein ratio of 1:50 (w/w) overnight at 37°C. The generated tryptic peptides were dried by speed vacuum at 4°C and desalted with C18 Spin column. For TMT labeling, the resultant peptide mixture of 10 isolates were labeled using TMT reagent kit (Thermo Scientific, USA) as resistant strain-127 isobaric tag and susceptible strain-126 isobaric tag (Chen et al., 2014). The combinations are as follows: A151 and A9, A159 and A11, A160 and A21, A161 and A90, A162 and A132.
The samples were reconstituted in 0.1% formic acid (FA) and separated on a NanoAcquity Ultra Performance Liquid Chromatography (UPLC) system (EASY-nLC 1000, Thermo Scientific). Afterward, the samples were fitted with a nanoAcquity Symmetry C18 trap column (100 μm × 2 cm, NanoViper C18, 5 μm, 100Å) and an analytical column (75 μm × 15 cm, NanoViper C18, 3 μm, 100Å). The mobile phase A was 100:0.1 HPLC grade water/FA, and mobile phase B was 100:0.1 ACN/FA. Each sample was loaded on the trapping column with a flow rate of 2.0 μl/min, followed by separation on the analytical column using a 100 min 3%–35% mobile phase B linear gradient at a flow rate of 0.8 μl/min. Retention Time Calibration Mixture (Thermo Scientific) was used to optimize LC and MS parameters and was used to monitor the stability of the system.
The analytical column was coupled to a high-resolution Q-Exactive Plus mass spectrometer (Thermo Fisher Scientific, San Jose, CA) using a nano-electrospray ion source, which was operated in positive ion mode. The source was operated at 2.0 kV with transfer-capillary temperature maintained at 250°C and S-Lens RF level set at 60. MS spectra were obtained by scanning over the range m/z 350–2000. Mass spectra were acquired in the Orbitrap mass analyzer with 1 microscan per spectrum for both MS and MS/MS. Resolving power for MS and MS/MS were set at 70,000 and 17,500, respectively. Tandem MS data were acquired in parallel with MS, on the top 20 most abundant multiply charged precursors, with higher energy collisional dissociation (HCD) at normalized collision energy of 30V. Precursors were isolated using a 2.0 m/z window and dynamic exclusion of 60 s was enabled during precursor selection. The data were determined twice.
For TMT labeling, Proteome Discoverer (version 1.4, Thermo Scientific, USA) was used to search the UniProtKB/Swiss-Prot database. The parameters were set as follows: integration tolerance, 20 ppm; precursor mass tolerance, 10 ppm; fragment mass tolerance, 0.02 Da. Dynamic modification Oxidation/+15.99 Da and carbamidomethyl/+57.02 Da) were set as dynamic and static modifications. Proteins that were differentially expressed were determined by peptide identifications with 95% confidence interval. Meanwhile, TMT signal analyses showed at least two-fold change in abundance, and its P value was <0.05 in unpaired Student’s t-test.
For Label free, peptide identification and label free relative quantification analysis were carried out using Peaks Studio 8.5 software (Bioinformatics Solutions Inc., Waterloo, ON, Canada). Using A. baumannii UniProtKB database (326,258 sequences, downloaded in June 2019). Input parameters: 20 ppm precursor mass tolerance, 0.02 Da fragment mass tolerance. The false discovery rates for protein and peptides were set at a maximum of 1%.
Only those protein groups which passed the filter are displayed in the protein profile heatmap. The relative protein abundance is represented as a heat map of the representative proteins of each protein group. The representative proteins are clustered if they exhibit a similar expression trend across the samples.
Glycopeptides in the samples were enriched by HILIC (The Nest Group, Inc.). Briefly, the tryptic and desalted peptides were resuspended in 80% ACN. The appropriate amounts of HILIC particle in 80% ACN were placed in Pierce spin columns (Thermo Scientific) and equilibrated three times using 80% ACN, which was followed by sample loading three times and washing two times with 80% ACN. Then, glycopeptides bound to the HILIC column were eluted three times with 100 μl of 0.1% TFA. The samples were dried by a SpeedVac and stored at −80°C until analysis.
The analytical column was coupled to a high-resolution Q-Exactive Plus mass spectrometer (Thermo Fisher Scientific, San Jose, CA) with a nano electrospray ion source operated in positive ion mode. The source was operated at 2.0 kV with the transfer capillary temperature maintained at 250°C and the S-lens RF level set at 60. MS spectra were obtained by scanning over an m/z range of 350–2000. Mass spectra in both MS and MS/MS were acquired in an Orbitrap mass analyzer with 1 microscan per spectrum. The resolving power for MS and MS/MS was set at 70,000 and 17,500, respectively. Tandem MS data on the top 20 most abundant multiply charged precursors were acquired in parallel with MS data, with higher energy collisional dissociation (HCD) at a normalized collision energy of 30 V. Precursors were isolated using a 2.0 m/z window, and dynamic exclusion of 60 s was enabled during precursor selection.
Database searches were performed using Byonic software (v2.13.17, Protein Metrics, Inc.). The following parameters were set for the search: cleavage sites, RK; cleavage side, C-terminal; digestion specificity, fully specific; missed cleavages, 2; precursor mass tolerance, 10 ppm; fragmentation type, QTOF/HCD; fragment mass tolerance, 0.02 Da, and protein false discovery rate (FDR), 1% FDR (or 20 reverse counts). All the other settings were set at their default values.
Byonic scores reflect the absolute quality of the peptide-spectrum match and not the relative quality compared to other candidate peptides. The Byonic score ranges from 0 to approximately 1,000, with 300 being a good score, 400 a very good score, and peptide-spectrum matches with scores over 500 almost certainly correct. The DeltaMod value indicates whether modifications are confidently localized; DeltaMod values over 10 indicate a high likelihood that all modification placements are correct. Therefore, a score over 300, a DeltaMod value over 10, a q-value < 0.05, and an FDR < 0.1% were set as thresholds in this study. Systematic and comprehensive analyses of specific glycopeptides, glycoforms, and glycosylation sites related to our samples from all the proteins identified by Byonic were carried out.
To further understand the functions of differential expressed proteins (DEP) between drug-resistant and drug-susceptible A. baumannii isolates, The DEPs were further submitted to NCBI (National Center for Biotechnology Information) and Uniprot (https://www.uniprot.org/) for GO enrichment analysis (statistically significant differences of GO terms were defined by P < 0.05), KOBAS 2.0 (KEGG Orthology Based Annotation System; http://kobas.cbi.pku.edu.cn/home.do) for KEGG pathway analysis and STRING database (https://string-db.org/) for protein-protein interaction (PPI) analysis. The subcellular localization and the specific information of the DEPs were identified by pSORTb version 3.0.2 (https://www.psort.org/psortb/) and Pubmed, respectively. The potential glycosylation sites were output from NetOGlyc 4.0 Server (http://www.cbs.dtu.dk/services/NetOGlyc/).
Eight A. baumannii clinical isolates (four resistant strains and four susceptible strains) from the same hospital (data shown in Supplementary Table 3) were used to determine the transcription of seven kinds of DEPs: Aminoglycoside (3’) phosphotransferase AphA1 or APH (3’)-Ia (AFV53106), Beta-lactamase AmpC (AFA35105/AFA35107), Outer membrane protein assembly factor BamD (WP_000056810), MFS transporter (RSR57702), ABC transporter (AXV52620), HlyD membrane-fusion protein of T1SS (ENV25944), and Elongation factor Tu (KLT84190). Primers are listed in Supplementary Table 1. All isolates were grown overnight at 37°C in LB broth, and sub-cultured 1/100 into fresh LB broth for 4 h. RNA extraction was performed using RNAprep Pure Kit (TianGen). The extracted RNA was reversed to cDNA using the All-in-One™ First-Strand cDNA Synthesis Kit (TaKara). Then qRT-PCR were performed using the 2*SYBR Green qPCR Master Mix (Low Rox). The CT value was obtained by using the 7500 Fast DX instrument, rpoB was used as the internal parameter. The normalized relative expression levels of the target genes were calculated by the comparative cycle threshold (2-ΔΔCT). The data obtained were analyzed and plotted with Graphpad prism version 5.0. Error bars represent the SDs. Significant differences were defined by P < 0.05 (*), P < 0.01(**), and P < 0.001(***).
Proteomics analysis uses non-targeted research to directly detect the expression of a large number of proteins. In this study, we used TMT labeling-based proteomics, label-free proteomics, and glycoproteomics to analyze the differentially expressed proteins between 9 MDR and 10 drug-susceptible A.baumannii isolates. All MDR strains are resistant to Cefepime, imipenem, gentamicin, tobramycin, levofloxacin, ciprofloxacin, and paediatric compound sulfamethoxazole. Drug-susceptible isolates are only non-susceptible to penicillins and cephalosporins of the eight antimicrobial categories listed in Supplementary Table 2.
For the TMT labeling-based proteomics, we randomly selected 10 isolates (5 MDR and 5 drug-susceptible) and made them into 5 drug-resistant and drug-susceptible pairs. Each pair was subjected to two biological replicates. The MDR isolates and drug-susceptible isolates were labelled by 127 and 126 reagent, respectively. In the first pair, 2,270 and 2,730 proteins were found in two biological iterations, and a total of 3,884 proteins were identified. In the second pair, 2,050 and 2,061 proteins were found respectively, and a total of 3,150 proteins were found. In the third pair, 2,695 and 2,200 proteins were found, and a total of 3,782 proteins were found. In the fourth pair, 1,953 and 1,874 proteins were found, and a total of 2,977 proteins were identified. In the fifth pair, 2,021 and 1,927 proteins were found, and a total of 3,051 proteins were found. In order to conduct an overall analysis, we finally selected 127/126 ≥ 2 and ≤0.5 data for analysis. As seen in Supplementary Table 4, a total of 70 proteins were obtained with the same expression trend in more than 4 pairs, among which there were 23 kinds of proteins with the same expression trend in 5 pairs. The relative molecular mass of the protein is between 6 and 118 kda, a larger proportion is between 10 and 50kda, and the isoelectric point is between 4.42 and 11.12. 58 up-regulated proteins (127/126 ≥ 2) and 12 down-regulated proteins (127/126 ≤ 0.5) were expressed in MDR isolates. Gene Ontology (GO) analysis can classify genes to different groups according to their functions. Based on the GO annotation analysis, the proteins were classified into three categories: molecular function, cellular component, and biological process. GO analysis with the largest number of proteins involved were shown in Figure 2, up-regulated proteins are classified into 20 molecular function related proteins, 11 cellular component related proteins, and 14 biological process related proteins; downregulated proteins are classified into 7 molecular function related proteins, 3 cellular component related proteins, and 5 biological process related proteins. The DEPs of MDR and drug-susceptible isolates focused on catalytic activity and binding. There are 22 KEGG pathways involved in the down-regulated proteins, most of which are involved in metabolic pathways; 39 KEGG pathways are involved in the up-regulated proteins, which are mostly involved in metabolic pathways, carbon metabolism, and biosynthesis of amino acids (Supplementary Figure 1). The PPI net showed the interactions of the 59 proteins. The average node degree is 2.78 and the interaction of ribosomal-related proteins is relatively dense (Supplementary Figure 2).
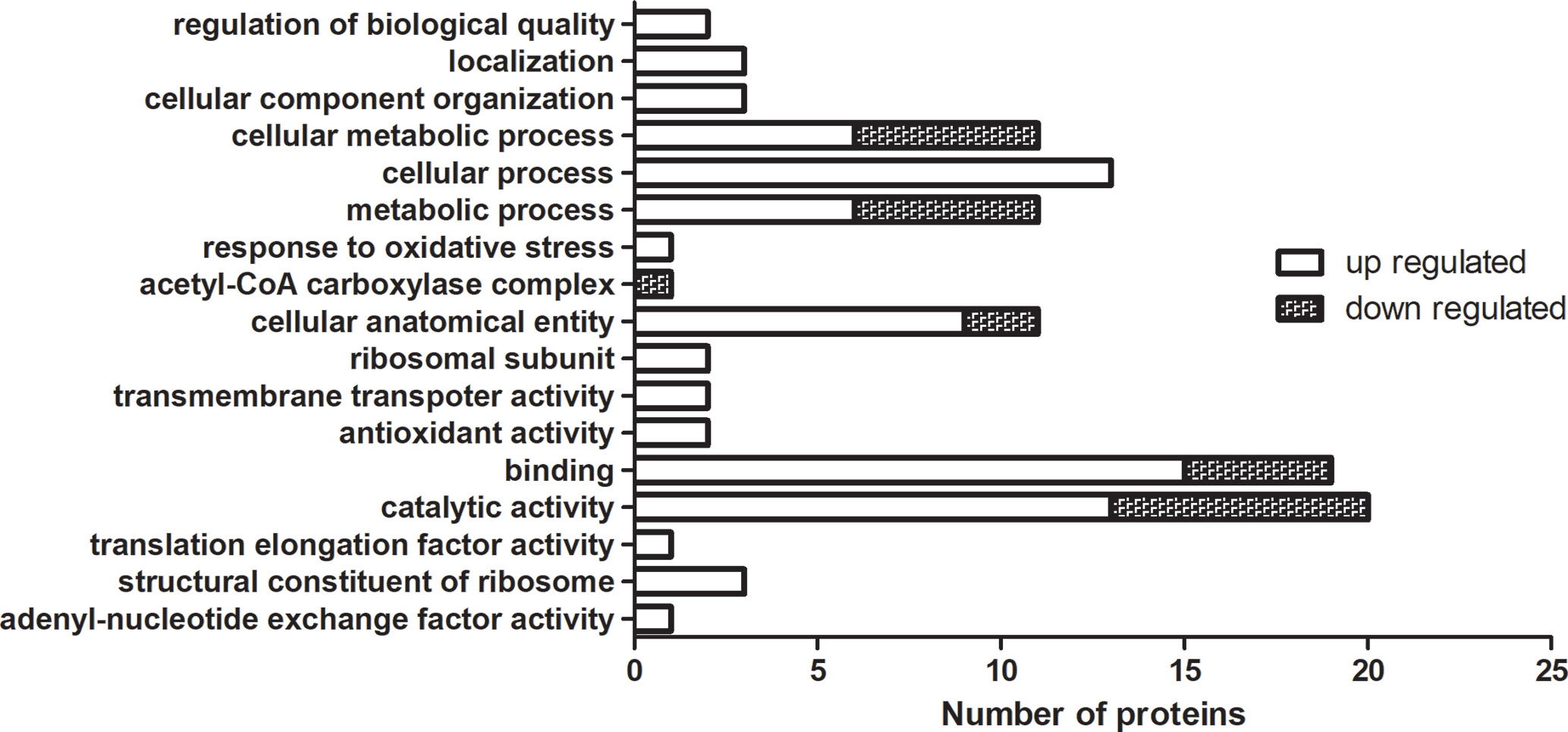
Figure 2 GO enrichment analysis of DEPs in TMT-labeling proteomics. The MDR isolates and drug-susceptible isolates were labelled by 127 and 126 reagent. Up-regulated and down-regulated means 127/126 ≥ 2 and ≤0.5, respectively. Each column represented the number of proteins involved in GO annotation analysis of DEPs in TMT-labeling proteomics. Statistically significant differences of GO terms were defined by P < 0.05.
We analyzed 10 drug-susceptible isolates and 9 MDR isolates by label free proteomics and obtained 102 proteins that are all present in more than 8 resistant isolates and only in 1 or less susceptible isolate (Supplementary Table 5). The number of proteins involved in molecular function, cellular component, and biological process are 52, 20, and 33, respectively (Figure 3). DEPs mainly focused on catalytic activity, binding, cellular process, metabolic process, and cellular anatomical entity. A total of 48 KEGG pathways (input numbers) were involved in Metabolic pathways (28), Carbon metabolism (11), Biosynthesis of amino acids (10), Glycine, serine and threonine metabolism (7), Valine, leucine and isoleucine degradation (6), Cysteine and methionine metabolism (6), Glyoxylate and dicarboxylate metabolism (5), etc. (Supplementary Figure 3). The PPI network diagram showed a total of 87 protein interactions, with an average node degree of 0.437 (Supplementary Figure 4). The heat map of similar expressed proteins showed differences in the expression of resistant isolates and susceptible isolates, differences in strains would also lead to different expression levels of some proteins in different bacteria (Figure 4).
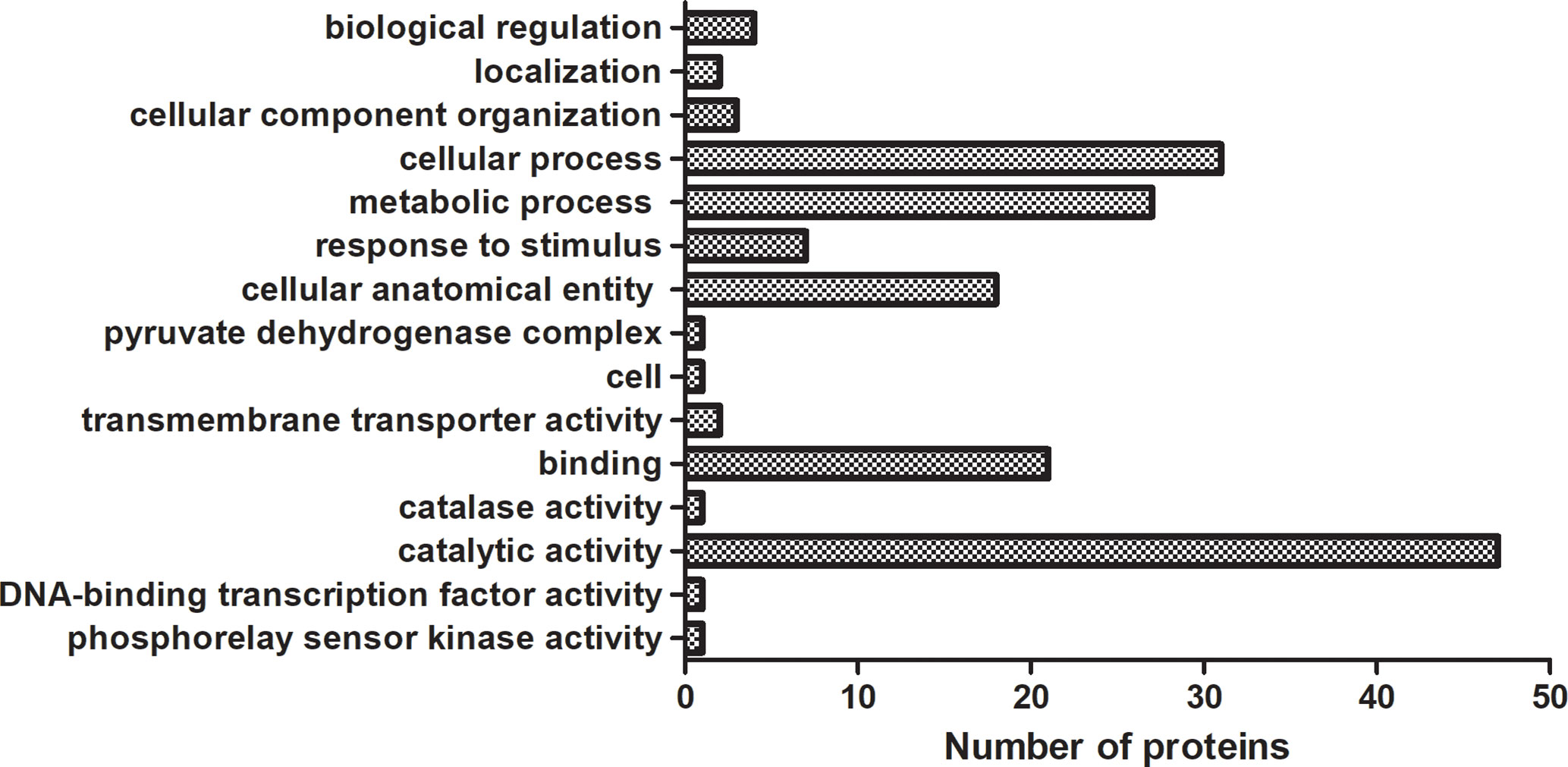
Figure 3 GO enrichment analysis of DEPs in label free proteomics. The proteins in label free proteomics are all present in more than 8 resistant isolates and only in 1 or less susceptible isolate. Each column represented the number of proteins involved in GO annotation analysis of DEPs in label free proteomics. Statistically significant differences of GO terms were defined by P < 0.05.
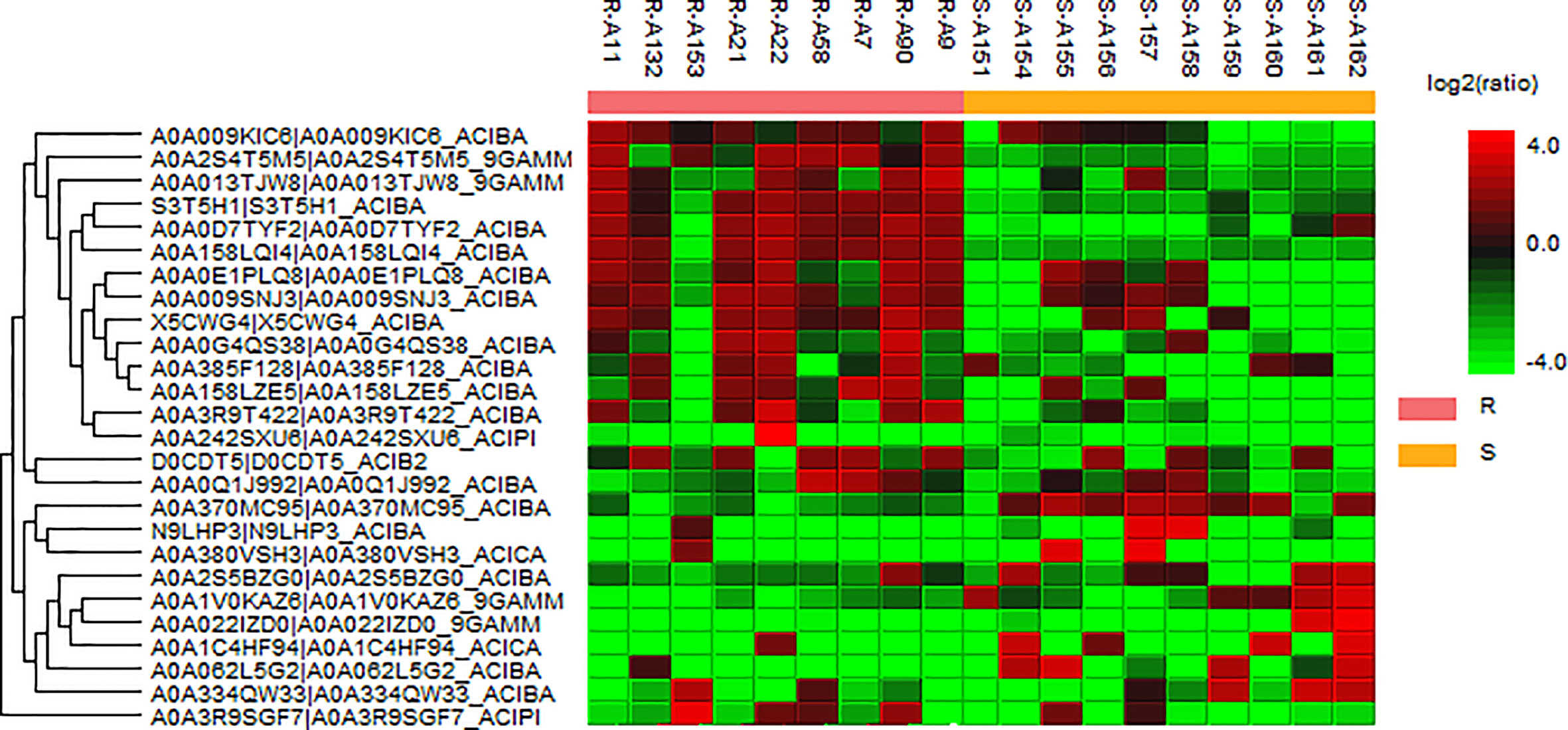
Figure 4 Heat map of the representative proteins of each isolate. “R” means drug-resistant isolates and “S” means drug-susceptible isolates. The representative proteins identified from label free proteomics are clustered if they exhibit a similar expression trend across the samples. The hierarchical clustering is generated using neighbor joining algorithm with a Euclidean distance similarity measurement of the log2 ratios of the abundance of each sample relative to the average abundance.
The results from TMT labeling-based proteomics and label free proteomics were combined and we found a total of 164 DEPs. The subcellular localizations of these DEPs were mainly in the cytoplasm, the proteins up-regulated or identified by label free proteomics mainly on the periplasmic or outer membrane of the cell (Figure 5). We further classified the proteins into six groups with different functions. According to this classification, there were 12, 20, 22, 14, 60, and 37 differentially expressed proteins of A, B, C, D, E, and F group, respectively. As following:
In this study, we found 12 proteins more abundant in the MDR isolates classified to group a (Table 1). These 12 proteins belong to a variety of known antibiotic resistance proteins, such as beta-lactamases and aminoglycoside phosphotransferase. The resistance of Acinetobacter to β-lactam is mainly due to the synthesis and enzymatic degradation of the species-specific β-lactamase, all four types of β-lactamases have been identified in A. baumannii (Lee et al., 2017), and the analysis of 23 MDR A. baumannii clinical isolates in Taiwan has shown that all A. baumannii can encode AmpC cephalosporins (Lin et al., 2011). OXA-23 and OXA-72 belong to class D β-lactamases (Donald et al., 2000; Evans and Amyes, 2014). Metal-dependent hydrolases such as metal β-lactamase (MBL) are zinc-dependent hydrolases that can cleave the β-lactam bond of most β-lactam antibiotics (Parimelzaghan et al., 2016; Kwapien et al., 2017). In this study, both metal-dependent hydrolase-related proteins and β-lactamase-related proteins were up-regulated in MDR isolates. The resistance of Acinetobacter to aminoglycoside antibiotics is mainly through N-acetylation, adenylation or O-phosphorylation modification to inactivate aminoglycosides (Seward et al., 1998; Shakil et al., 2008). AphA1b is one of the modifying enzymes involved in aminoglycoside resistance (Nigro et al., 2011). In this study, it is uniquely expressed in MDR isolates, which can explain its prevalence to tobramycin, gentamicin, and other aminoglycoside antibiotics. DacD (D-alanyl-D-alanine carboxypeptidase) belongs to penicillin binding proteins (PBPs) (Spidlova et al., 2018), also called PBP6b in A. baumannii, which is involved in the metabolism of peptidoglycans (Cayô et al., 2011). In addition, studies have found that Cell division protein ZapA is related to the resistance of β-lactam antibiotics (Knight et al., 2016). These proteins are all up-regulated in the drug-resistant isolates in this study.
The second category contains of 20 DEPs (Table 2) including 1 down-regulated protein and 19 up-regulated proteins. Thioredoxin, which is a member of the thioredoxin superfamily, is involved in the virulence of bacteria and also related to the expression of genes related to the ABC transport system (May et al., 2019). This protein is shown to be down-regulated in our results which are consistent with the literature. The upregulated proteins in this group include proteins that play a key role in the formation of the outer membrane, proteins related to maintaining the integrity of the outer membrane, pump proteins, and outer membrane proteins: β Barrel Outer Membrane Protein (BAM) is related to bacterial nutrient acquisition, protein secretion, signal transduction and bacterial survival, and drug resistance (Sikora et al., 2018). Porin has been shown to play an important role in the resistance mechanism (Lee et al., 2017). Lipoprotein can act as a fusion protein to promote the complete biogenesis of the cell membrane (Melly et al., 2019). Studies have found that lipoproteins are upregulated in MDR A. baumannii strains (Chopra et al., 2013). SurA is a periplasmic chaperone protein involved in the folding of outer membrane porins, and is closely related to the integrity of the outer membrane (Bell et al., 2018). The alpha/beta hydrolase folding superfamily is a class of hydrolase enzymes involved in lipid metabolism, cell membrane maintenance, virulence, efflux, and metabolism of cell (Johnson, 2017). Outer membrane protein A (OmpA) is the outer membrane protein of bacteria, which is related to the efflux pump and drug resistance of bacteria (Kwon et al., 2017). The expression of OmpW is down-regulated in carbapenem-resistant strains, and its down-regulation can make PBPs unavailable (Tiwari et al., 2012). Study have also found that OmpW has a higher expressed in MDR strains (Chopra et al., 2013). The outer membrane protein CarO is associated with carbapenem drug resistance (Xiao et al., 2016). The multicomponent efflux pump system is widely present in bacteria and it can make bacteria resistant to antibiotics by pumping out antibiotics. Six superfamily of resistant pumps have been identified in A. baumannii: major facilitator superfamily (MFS), resistance nodulation division (RND), multidrug and toxic compound extrusion (MATE), small multidrug resistance (SMR), ATP-binding cassette (ABC), and proteobacterial antimicrobial compound efflux (PACE) (Pérez-Varela et al., 2019). In our study, we found that both ABC transporter, MFS transporter, and RND transporter were up-regulated. MFS efflux pump and ABC transporter have been found to be associated with quinolone resistance and beta lactam resistance (Correia et al., 2016; Xiao et al., 2016; Lari et al., 2018; Pérez-Varela et al., 2018). T1SS is formed by HlyB (ABC transporter), HlyD (membrane fusion protein), and TolC (outer membrane). Its C-terminal can carry a secretion signal, and the deletion of the C-terminal will cause secretion blocked (Holland et al., 2016). In our study, both the C-terminal target domain and the hypothetical protein F962_01862 encoding HlyD were up-regulated.
We found there were 22 stress response-related proteins differentially expressed between MDR and drug-susceptible isolates (Table 3), among them, the expression of Antibiotic biosynthesis monooxygenase which can oxidize and inactivate antibiotics (Minerdi et al., 2016; Koteva et al., 2018) is interestingly down-regulated. The expression of heat shock proteins and acid shock proteins were upregulated. Heat shock protein is generally used as a molecular chaperone or protease to repair damaged proteins, and its expression increases during stress response such as antibiotic induction. Bacteria with heat shock proteins induced are more resistant to antibiotic environments (Cardoso et al., 2010). Acid shock protein can improve the acid resistance of bacteria (Villarreal et al., 2000). Other proteins related to the stress response and resistance to environment were also upregulated as following: Trigger factor (TF) can play a key role as a molecular chaperone, also related to the resistance to the external environment (Lee et al., 2009). Heavy metal-associated (HMA) domain proteins can give bacteria the ability to resist high metal environments (Maynaud et al., 2014). Rhodanese-Like Domain (RHD) Protein participates in biological processes such as sulfur metabolism and environmental adaptability (Cipollone et al., 2007). Superoxide dismutase (SOD) can effectively catalyze the conversion of superoxide free radicals and protect bacteria from reactive oxygen. It has been shown to be related to the oxidative stress response of Acinetobacter baumannii and its resistance to antibiotics (Heindorf et al., 2014). Catalase catalyzes the degradation of hydrogen peroxide and is closely related to the defense of bacteria against related environments (Sun et al., 2016). Universal stress protein (Usp) helps bacteria adapt to oxidative stress, high temperature, pH, etc. (Elhosseiny et al., 2015). Aldehyde dehydrogenase (AldA) is related to a variety of metabolic processes such as redox regulation of bacteria, and can participate in environmental stress defense such as hypochlorite stress (Imber et al., 2018). Cysteine synthase CysK can be used to synthesize cysteine (Bogicevic et al., 2016), and cysteine related products are important molecules required for the oxidative stress response of bacteria (Hicks and Mullholland, 2018). NAD(P)H-quinone oxidoreductase participates in quinone detoxification and helps bacteria to survive under adverse conditions (Ryan et al., 2014), which is also related to resistance to oxidative stress (Kishko et al., 2012). Bacterial proteases play an important role in the survival, stress response, and pathogenicity of bacteria (Culp and Wright, 2017). Isochorismatase family protein is related to serum resistance in A. baumannii (Jacobs et al., 2010). Response regulators are related to the tolerance to dehydration and resistance to oxidative stress (Farrow et al., 2018). The toxin antitoxin system also regulates the response of SOS stress (Fernández-García et al., 2016).These proteins all showed upregulated or unique expressed in MDR isolates in this study. The upregulated expression of these stress proteins that resist the external environment may promote the resistance of bacteria by making the resistance of bacteria more stable.
The forth group is proteins that have important functions for gene expression and protein translation or modification (Table 4). The expression of Serine hydroxymethyltransferase in this class is down-regulated. This protein is an iron inhibitory protein and can bind to mRNA to control gene expression and participate in the overall bacterial response (Nwugo et al., 2011). Up-regulated proteins includes enolase, DNA breaking-rejoining elements, ribosomal proteins, elongation factor Tu (EF-Tu), ribonuclease E (RNase), Valyl-tRNA synthetase, NusA, and long-chain fatty acid transport proteins. Enolase can play a central role in RNA processing (Krucinska et al., 2019). DNA breaking-rejoining enzymes play an important role in the transmission of genetic elements (Van Houdt et al., 2012). The ribosomal protein S4 RpsD is related to the assembly of ribosomes (Olsson and Isaksson, 1979). The 50S ribosomal proteins L15 and L16 are important translation proteins (McNicholas et al., 2001; Dutton et al., 2016). EF-Tu is also related to protein translation and can interact with a variety of proteins to perform different biological functions (Premkumar et al., 2014). EF-Tu and ribosomal protein can help the production of bacterial protein, and some study have found them upregulated in carbapenem-resistant strains (Tiwari et al., 2012). RNase can perform different processing on RNA to regulate gene expression (Mardle et al., 2019). Valyl-tRNA synthetase is responsible for the aminoacylation of tRNA (Heck and Hatfield, 1988). Nucleoid-associated proteins play an important role in concentrating DNA and regulating gene expression (Lee and Marians, 2013). Transcription termination/antitermination protein NusA can bind to RNA polymerase or nascent RNA to influence transcription (Qayyum et al., 2016). Long-chain fatty acid transport proteins are involved in the transport of fatty acids and can affect intracellular signal transduction and gene expression (Dirusso and Black, 2004).These proteins may help the expression of drug resistance-related proteins by influencing the progress of gene expression or protein translation.
The largest category contains 60 DEPs expressed differently between the MDR isolates and drug-susceptible isolates. These proteins are mainly related to metabolism (Supplementary Table 6). This group includes seven downregulated proteins, which are Serine hydroxymethyltransferase, NADH-quinone oxidoreductase, Malate dehydrogenase, Non-heme chloroperoxidase, 3,4-dihydroxy-2-butanone 4-phosphate synthase, Ketol-acid reductoisomerase [NADP (+)] and Acetyl-coenzyme A carboxylase carboxyl transferase subunit beta. They participate in the biosynthesis of serine, tetrahydrofolate, nitropyrrolidin, branched-chain amino acids, and fatty acids which are related to cellular processes such as bacterial respiration and TCA cycle (Shin et al., 2009; Nwugo et al., 2011; Deris et al., 2014; Reddy et al., 2014; Krishna et al., 2019). Among them, Malate dehydrogenase, which is upregulated in bacterial biofilm state (Shin et al., 2009), and the expression of it in carbapenem-resistant A.baumannii was up-regulated, the researcher hypothesis that it contributes to energy production and can improve the survival rate of bacteria (Tiwari et al., 2012). In our results, the protein is down-regulated. We speculate this may be due to strain differences. There are 53 up-regulated or uniquely expressed proteins in MDR strains, which are involved in lipid metabolism (Jang et al., 2008; Gu et al., 2019), amino acid metabolism (Stancik et al., 2002; Bezsudnova et al., 2017), TCA cycle (Corregido et al., 2019), purine anabolic metabolism (Spurr et al., 2012), pyruvate metabolism (Song et al., 2010), fatty acid metabolism (Nishimaki-Mogami et al., 1987), intracellular electron transfer of bacteria (López Rivero et al., 2019), nutrition and energy acquisition (Drewke et al., 1996), and other metabolic processes. In addition, the heavy metal associated (HMA) domain protein which is closely related to the utilization and metabolism of metal ions such as copper ions and zinc ions (Furukawa et al., 2018) is also up-regulated.
The final category F includes proteins with unknown functions or other functions besides the other five groups such as virulence or biofilm formation (Supplementary Table 7). Studies have found that the resistance of bacteria to disinfectants and antibacterial agents will be greatly increased after the formation of biofilms (Høiby et al., 2010) and the resistance of A. baumannii that produces biofilms is significantly higher than that of bacteria that cannot produce biofilms (Gurung et al., 2013). The mechanisms of biofilm formation causing resistance include delaying the penetration of antibacterial agents into bacteria, causing changes in the growth rate of membrane-forming microorganisms, and upregulating efflux pumps and other physiological metabolic differences (Donlan and Costerton, 2002; Kentache et al., 2017). Our study also found the proteins related to biofilm formation expressed more in drug-resistance isolates. For example, flagellin which is involved in the formation of bacterial flagella and the fimbriae assembly protein FilF are both related to bacterial biofilm formation (Karatan and Watnick, 2009). Histidine kinase and esterase members are also involved in the formation of biofilms (Chen et al., 2017; Larsen and Johnson, 2019). They both are upregulated in our study. A comparison of drug-resistant clinical strains and susceptible clinical strains found that drug-resistant clinical strains contain more virulence factors such as FilF, GroEL, and hemagglutinin-like protein (Li et al., 2015). In our study another type of up-regulated protein is mainly related to the virulence of bacteria. For example, hemolysin is one of the virulence factors of bacteria and is closely related to the pathogenicity of bacteria (Bhakdi et al., 1988). Cupin family protein is a superfamily of proteins with multiple functions such as metalloenzymes, sugar binding, and pathogenicity (Sim et al., 2016). Several hypothetical proteins are also up-regulated in MDR strains. Among them, putative septicolysin, cholesterol-dependent cytolysin family, and related proteins are generally virulence factors produced by bacteria (Lukoyanova et al., 2016). Other uncharacterized proteins are also upregulated. It is worth noting that an undefined protein is a member of LysM domain/BON superfamily protein. The unknown functional protein of LysM domain/BON superfamily protein was detected in both upregulated and downregulated proteins. A previous study found that a 16 kDa protein of LysM domain/BON superfamily protein detected in the outer membrane protein of susceptible Klebsiella pneumoniae (Kádár et al., 2017). Another study found that it may be related to the stress response of Klebsiella pneumoniae to Carbapenem (Khan et al., 2017). However, its specific function is unknown and deserves further study.
Studies have found that O-glycosylation mechanism is widespread in A.baumannii, and it is closely related to the virulence and biofilm formation ability of the bacteria (Iwashkiw et al., 2012; Kinsella et al., 2015), but its association with drug resistance was rarely reported. Other post-translational modifications such as phosphorylation and acetylation have been shown to be related to drug resistance (Kentache et al., 2016). Our study aimed to identify the different glycosylation between the MDR and drug-susceptible strains. A total of 77 glycoproteins were found in the 9 MDR isolates and 97 glycoproteins were found in the 10 drug-susceptible isolates. MDR strains had 10 specifically expressed polysaccharides, and drug-susceptible strains had 30 specifically expressed polysaccharides (Supplementary Table 8). The specifically expressed polysaccharides found in MDR strain contain chaperone protein DnaK and phosphoenolpyruvate carboxykinase which both are essential for metabolism and survival (Supplementary Table 9). By further analysis, we found a polysaccharide form of HexNAc(2)Hex(2)Fuc(1) on the S21 (OGlycan/876.3223) site of adenylate kinase (the product of adk) is present in six MDR isolates and not exist in any drug-susceptible isolates. NetOGlyc predicts an additional glycosylation site at site 129 that is more likely to carry O-GalNAc modifications in this protein. The adenylate kinase is related to energy metabolism (Shin and Park, 2015). There is a study showed that the main mechanism of multidrug resistance is the increased activity of adenosine triphosphate (ATP)-dependent drug efflux transporters (Wen et al., 2018). Therefore, we speculate that glycosylation of adenylate kinase is closely related to the metabolism of bacteria, which may enhance the bacteria’s metabolic ability and efflux ability to enhance their drug resistance.
To verify the results from proteomics, we randomly chose another eight A.baumannii isolates from the same hospital to identify the transcription level of 7 DEPs: Aminoglycoside (3’) phosphotransferase AphA1 or APH (3’)-Ia (AFV53106), Beta-lactamase AmpC (AFA35105/AFA35107), Outer membrane protein assembly factor BamD (WP_000056810), MFS transporter (RSR57702), ABC transporter (AXV52620), HlyD membrane-fusion protein of T1SS (ENV25944), and Elongation factor Tu (KLT84190). Our results (Figure 6) showed that the transcription levels of AFV53106, AF135105, AXV52620, and ENV25944 in MDR isolates have a higher trend than that of drug-susceptible isolates, which is consistent with the results of proteomics. However, the transcription levels of WP_000056810, RSR57702 and KLT84190 in MDR isolates and susceptible isolates showed no difference. Previous studies have also found disparity between the protein levels and transcription levels of certain genes. This may be due to protein expression and post-translational modifications. In general, the verification of other MDR strains and drug-sensitive strains showed consistency with the proteomic results, indicating that the protein obtained in our results is closely related to the resistance mechanism of A.baumannii.
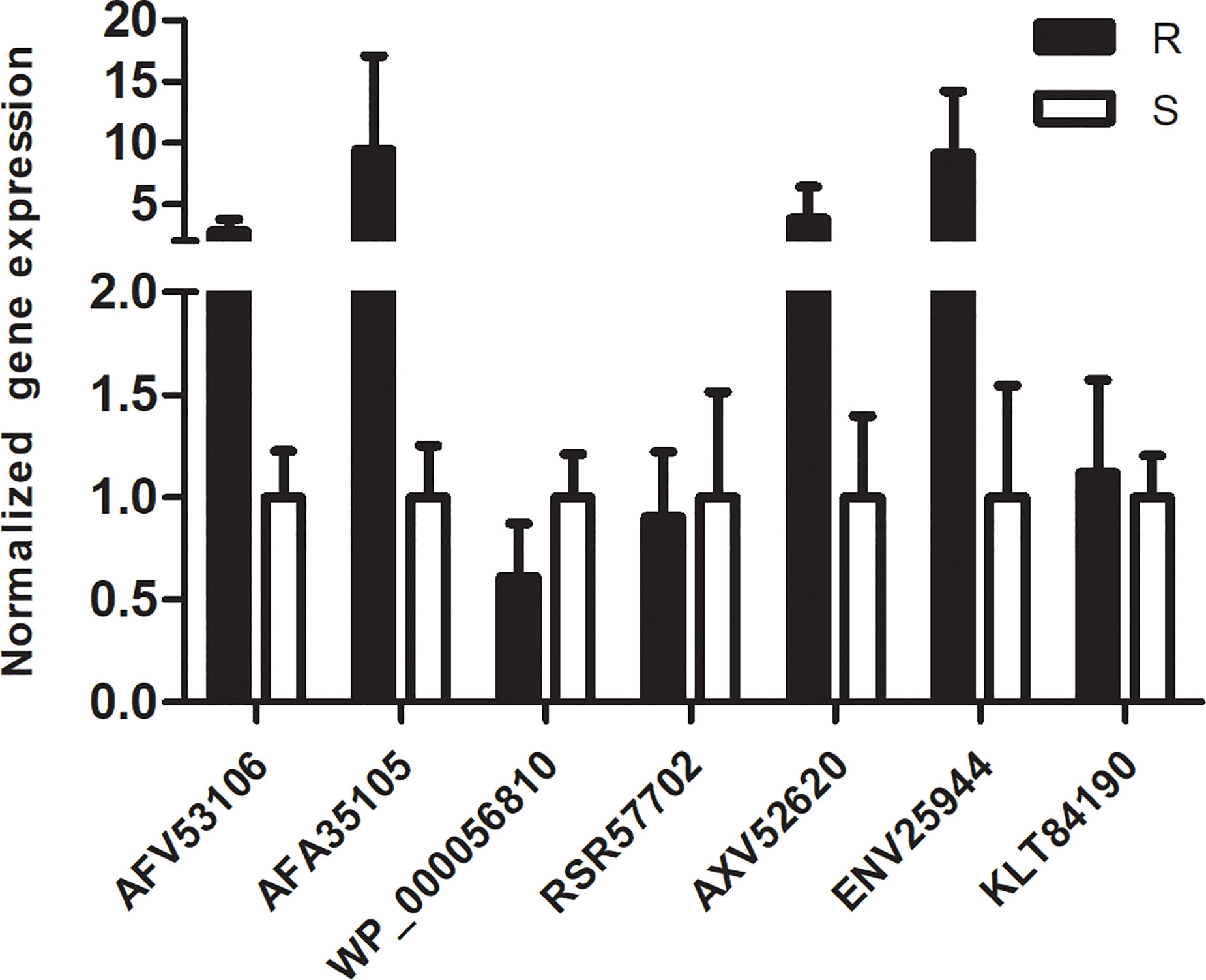
Figure 6 qRT-PCR analysis of 7 representative proteins: The normalized expression level (2-ΔΔct) of 7 DEPs genes of the resistant isolates and susceptible isolates were tested by qRT-PCR. rpoB was used as the internal parameter. Error bars represent the SDs.
In conclusion, our study found that in MDR strains, a large number of membrane proteins and membrane formation and efflux-related proteins, metabolism-related proteins, stress response-related proteins, and proteins involved in gene expression regulation and protein translation are all upregulated, and glycosylation of adenosine triphosphate is unique in MDR strains. Through the study of the mechanism of multidrug resistance of A.baumannii, treatment can be adopted for its resistance mechanism to improve the success rate of treatment of A.baumannii infection, such as using engineered endolysin to degrade bacterial peptidoglycan to replace carbapenem drugs (Lee et al., 2017) or add certain compounds that can increase the energy production of bacteria and enhance the permeability of their cell membranes during antibiotic treatment to promote the therapeutic effect of antibiotics (Shin and Park, 2015). The development of vaccines against drug-resistant-related proteins is also another effective strategy to solve drug-resistant bacterial infections (Mujawar et al., 2019). This study uses plenty of isolates for comparative and comprehensive analysis, however, because these isolates were isolated from the same hospital and might have similar genetic phenotypes, we still need further study like expanding sample sources or sequencing these isolates to found the genetic mechanism. In addition, the specific mechanism of how DEPs-related genes influence drug resistance still needs to be further studied and explored using auxiliary methods such as gene knockout to support the results of proteomics.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
DX and Q-HZ conceived and designed the study. PW and R-QL analyzed the data. LW and W-TY performed experiments. PW and Q-HZ wrote the manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by The Major Infectious Diseases AIDS and Viral Hepatitis Prevention and Control Technology Major Projects (Grant No. 2018ZX10712001-006012 and Grand No. 2018ZX10733402-003002).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2021.625430/full#supplementary-material
MDR, multidrug-resistant; TMT, tandem mass tag; TEAB, triethyl ammonium bicarbonate; HCD, higher energy collisional dissociation; PPI, protein-protein interaction; DEP, differential expressed proteins; GO, Gene Ontology; MBL, metal β-lactamase; PBP, penicillin binding proteins; BAM, β Barrel Outer Membrane Protein; Omp, Outer membrane protein; RHD, Rhodanese-Like Domain; SOD, Superoxide dismutase; Usp, Universal stress protein.
Ahmad Izaham A. R., Scott N. E. (2020). Open Database Searching Enables the Identification and Comparison of Bacterial Glycoproteomes without Defining Glycan Compositions Prior to Searching. Mol. Cell Proteomics 19 (9), 1561–1574. doi: 10.1074/mcp.TIR120.002100
Al-Kadmy I. M. S., Ibrahim S. A., Al-Saryi N., Aziz S. N., Besinis A., Hetta H. F. (2020). Prevalence of Genes Involved in Colistin Resistance in Acinetobacter baumannii: First Report from Iraq. Microb. Drug Resist. 26 (6), 616–622. doi: 10.1089/mdr.2019.0243
Bell E. W., Zheng E. J., Ryno L. M. (2018). Identification of inhibitors of the E. coli chaperone SurA using in silico and in vitro techniques. Bioorg. Med. Chem. Lett. 28 (22), 3540–3548. doi: 10.1016/j.bmcl.2018.09.034
Bezsudnova E. Y., Boyko K. M., Popov V. O. (2017). Properties of Bacterial and Archaeal Branched-Chain Amino Acid Aminotransferases. Biochemistry (Mosc) 82 (13), 1572–1591. doi: 10.1134/s0006297917130028
Bhakdi S., Mackman N., Menestrina G., Gray L., Hugo F., Seeger W., et al. (1988). The hemolysin of Escherichia coli. Eur. J. Epidemiol. 4 (2), 135–143. doi: 10.1007/bf00144740
Bogicevic B., Berthoud H., Portmann R., Bavan T., Meile L., Irmler S. (2016). Cysteine biosynthesis in Lactobacillus casei: identification and characterization of a serine acetyltransferase. FEMS Microbiol. Lett. 363 (4), fnw012. doi: 10.1093/femsle/fnw012
Cardoso K., Gandra R. F., Wisniewski E. S., Osaku C. A., Kadowaki M. K., Felipach-Neto V., et al. (2010). DnaK and GroEL are induced in response to antibiotic and heat shock in Acinetobacter baumannii. J. Med. Microbiol. 59 (Pt 9), 1061–1068. doi: 10.1099/jmm.0.020339-0
Cayô R., Rodríguez M. C., Espinal P., Fernández-Cuenca F., Ocampo-Sosa A. A., Pascual A., et al. (2011). Analysis of genes encoding penicillin-binding proteins in clinical isolates of Acinetobacter baumannii. Antimicrob. Agents Chemother. 55 (12), 5907–5913. doi: 10.1128/aac.00459-11
Chen C., Xiao D., Zhou W., Shi Q., Zhang H. F., Zhang J., et al. (2014). Global protein differential expression profiling of cerebrospinal fluid samples pooled from Chinese sporadic CJD and non-CJD patients. Mol. Neurobiol. 49 (1), 290–302. doi: 10.1007/s12035-013-8519-2
Chen R., Lv R., Xiao L., Wang M., Du Z., Tan Y., et al. (2017). A1S_2811, a CheA/Y-like hybrid two-component regulator from Acinetobacter baumannii ATCC17978, is involved in surface motility and biofilm formation in this bacterium. Microbiologyopen 6 (5), e510. doi: 10.1002/mbo3.510
Chopra S., Ramkissoon K., Anderson D. C. (2013). A systematic quantitative proteomic examination of multidrug resistance in Acinetobacter baumannii. J. Proteomics 84, 17–39. doi: 10.1016/j.jprot.2013.03.008
Cipollone R., Ascenzi P., Visca P. (2007). Common themes and variations in the rhodanese superfamily. IUBMB Life 59 (2), 51–59. doi: 10.1080/15216540701206859
Corregido M. C., Asención Diez M. D., Iglesias A., Piattoni C. V. (2019). New pieces to the carbon metabolism puzzle of Nitrosomonas europaea: Kinetic characterization of glyceraldehyde-3 phosphate and succinate semialdehyde dehydrogenases. Biochimie 158, 238–245. doi: 10.1016/j.biochi.2019.01.013
Correia S., Hébraud M., Chafsey I., Chambon C., Viala D., Torres C., et al. (2016). Impacts of experimentally induced and clinically acquired quinolone resistance on the membrane and intracellular subproteomes of Salmonella Typhimurium DT104B. J. Proteomics 145, 46–59. doi: 10.1016/j.jprot.2016.04.001
Culp E., Wright G. D. (2017). Bacterial proteases, untapped antimicrobial drug targets. J. Antibiot. (Tokyo) 70 (4), 366–377. doi: 10.1038/ja.2016.138
De Silva P. M., Chong P., Fernando D. M., Westmacott G., Kumar A. (2018). Effect of Incubation Temperature on Antibiotic Resistance and Virulence Factors of Acinetobacter baumannii ATCC 17978. Antimicrob. Agents Chemother. 62 (1), e01514-17. doi: 10.1128/aac.01514-17
Deris Z. Z., Akter J., Sivanesan S., Roberts K. D., Thompson P. E., Nation R. L., et al. (2014). A secondary mode of action of polymyxins against Gram-negative bacteria involves the inhibition of NADH-quinone oxidoreductase activity. J. Antibiot. (Tokyo) 67 (2), 147–151. doi: 10.1038/ja.2013.111
Dijkshoorn L., Nemec A., Seifert H. (2007). An increasing threat in hospitals: multidrug-resistant Acinetobacter baumannii. Nat. Rev. Microbiol. 5 (12), 939–951. doi: 10.1038/nrmicro1789
Dirusso C. C., Black P. N. (2004). Bacterial long chain fatty acid transport: gateway to a fatty acid-responsive signaling system. J. Biol. Chem. 279 (48), 49563–49566. doi: 10.1074/jbc.R400026200
Donald H. M., Scaife W., Amyes S. G., Young H. K. (2000). Sequence analysis of ARI-1, a novel OXA beta-lactamase, responsible for imipenem resistance in Acinetobacter baumannii 6B92. Antimicrob. Agents Chemother. 44 (1), 196–199. doi: 10.1128/aac.44.1.196-199.2000
Donlan R. M., Costerton J. W. (2002). Biofilms: survival mechanisms of clinically relevant microorganisms. Clin. Microbiol. Rev. 15 (2), 167–193. doi: 10.1128/cmr.15.2.167-193.2002
Drewke C., Klein M., Clade D., Arenz A., Müller R., Leistner E. (1996). 4-O-phosphoryl-L-threonine, a substrate of the pdxC(serC) gene product involved in vitamin B6 biosynthesis. FEBS Lett. 390 (2), 179–182. doi: 10.1016/0014-5793(96)00652-7
Dutton L. C., Paszkiewicz K. H., Silverman R. J., Splatt P. R., Shaw S., Nobbs A. H., et al. (2016). Transcriptional landscape of trans-kingdom communication between Candida albicans and Streptococcus gordonii. Mol. Oral. Microbiol. 31 (2), 136–161. doi: 10.1111/omi.12111
Elhosseiny N. M., Amin M. A., Yassin A. S., Attia A. S. (2015). Acinetobacter baumannii universal stress protein A plays a pivotal role in stress response and is essential for pneumonia and sepsis pathogenesis. Int. J. Med. Microbiol. 305 (1), 114–123. doi: 10.1016/j.ijmm.2014.11.008
Evans B. A., Amyes S. G. (2014). OXA β-lactamases. Clin. Microbiol. Rev. 27 (2), 241–263. doi: 10.1128/cmr.00117-13
Farrow J. M. III., Wells G., Pesci E. C. (2018). Desiccation tolerance in Acinetobacter baumannii is mediated by the two-component response regulator BfmR. PloS One 13 (10), e0205638. doi: 10.1371/journal.pone.0205638
Fernández-García L., Blasco L., Lopez M., Bou G., García-Contreras R., Wood T., et al. (2016). Toxin-Antitoxin Systems in Clinical Pathogens. Toxins (Basel) 8 (7), 227. doi: 10.3390/toxins8070227
Fernández-Reyes M., Rodríguez-Falcón M., Chiva C., Pachón J., Andreu D., Rivas L. (2009). The cost of resistance to colistin in Acinetobacter baumannii: a proteomic perspective. Proteomics 9 (6), 1632–1645. doi: 10.1002/pmic.200800434
Furukawa Y., Lim C., Tosha T., Yoshida K., Hagai T., Akiyama S., et al. (2018). Identification of a novel zinc-binding protein, C1orf123, as an interactor with a heavy metal-associated domain. PloS One 13 (9), e0204355. doi: 10.1371/journal.pone.0204355
Gayoso C. M., Mateos J., Méndez J. A., Fernández-Puente P., Rumbo C., Tomás M., et al. (2014). Molecular mechanisms involved in the response to desiccation stress and persistence in Acinetobacter baumannii. J. Proteome Res. 13 (2), 460–476. doi: 10.1021/pr400603f
Göttig S., Gruber T. M., Higgins P. G., Wachsmuth M., Seifert H., Kempf V. A. (2014). Detection of pan drug-resistant Acinetobacter baumannii in Germany. J. Antimicrob. Chemother. 69 (9), 2578–2579. doi: 10.1093/jac/dku170
Gu Q., Yuan Q., Zhao D., Huang J., Hsiang T., Wei Y., et al. (2019). Acetyl-coenzyme A synthetase gene ChAcs1 is essential for lipid metabolism, carbon utilization and virulence of the hemibiotrophic fungus Colletotrichum higginsianum. Mol. Plant Pathol. 20 (1), 107–123. doi: 10.1111/mpp.12743
Gurung J., Khyriem A. B., Banik A., Lyngdoh W. V., Choudhury B., Bhattacharyya P. (2013). Association of biofilm production with multidrug resistance among clinical isolates of Acinetobacter baumannii and Pseudomonas aeruginosa from intensive care unit. Indian J. Crit. Care Med. 17 (4), 214–218. doi: 10.4103/0972-5229.118416
Heck J. D., Hatfield G. W. (1988). Valyl-tRNA synthetase gene of Escherichia coli K12. Primary structure and homology within a family of aminoacyl-TRNA synthetases. J. Biol. Chem. 263 (2), 868–877. doi: 10.1016/S0021-9258(19)35434-1
Heindorf M., Kadari M., Heider C., Skiebe E., Wilharm G. (2014). Impact of Acinetobacter baumannii superoxide dismutase on motility, virulence, oxidative stress resistance and susceptibility to antibiotics. PloS One 9 (7), e101033. doi: 10.1371/journal.pone.0101033
Hicks J. L., Mullholland C. V. (2018). Cysteine biosynthesis in Neisseria species. Microbiology (Reading) 164 (12), 1471–1480. doi: 10.1099/mic.0.000728
Høiby N., Bjarnsholt T., Givskov M., Molin S., Ciofu O. (2010). Antibiotic resistance of bacterial biofilms. Int. J. Antimicrob. Agents 35 (4), 322–332. doi: 10.1016/j.ijantimicag.2009.12.011
Holland I. B., Peherstorfer S., Kanonenberg K., Lenders M., Reimann S., Schmitt L. (2016). Type I Protein Secretion-Deceptively Simple yet with a Wide Range of Mechanistic Variability across the Family. EcoSal Plus 7 (1). doi: 10.1128/ecosalplus.ESP-0019-2015
Hood M. I., Jacobs A. C., Sayood K., Dunman P. M., Skaar E. P. (2010). Acinetobacter baumannii increases tolerance to antibiotics in response to monovalent cations. Antimicrob. Agents Chemother. 54 (3), 1029–1041. doi: 10.1128/aac.00963-09
Hua X., Liu L., Fang Y., Shi Q., Li X., Chen Q., et al. (2017). Colistin Resistance in Acinetobacter baumannii MDR-ZJ06 Revealed by a Multiomics Approach. Front. Cell Infect. Microbiol. 7:45:45. doi: 10.3389/fcimb.2017.00045
Imber M., Loi V. V., Reznikov S., Fritsch V. N., Pietrzyk-Brzezinska A. J., Prehn J., et al. (2018). The aldehyde dehydrogenase AldA contributes to the hypochlorite defense and is redox-controlled by protein S-bacillithiolation in Staphylococcus aureus. Redox Biol. 15, 557–568. doi: 10.1016/j.redox.2018.02.001
Iwashkiw J. A., Seper A., Weber B. S., Scott N. E., Vinogradov E., Stratilo C., et al. (2012). Identification of a general O-linked protein glycosylation system in Acinetobacter baumannii and its role in virulence and biofilm formation. PloS Pathog. 8 (6), e1002758. doi: 10.1371/journal.ppat.1002758
Jacobs A. C., Hood I., Boyd K. L., Olson P. D., Morrison J. M., Carson S., et al. (2010). Inactivation of phospholipase D diminishes Acinetobacter baumannii pathogenesis. Infect. Immun. 78 (5), 1952–1962. doi: 10.1128/iai.00889-09
Jang H. J., Chang M. W., Toghrol F., Bentley W. E. (2008). Microarray analysis of toxicogenomic effects of triclosan on Staphylococcus aureus. Appl. Microbiol. Biotechnol. 78 (4), 695–707. doi: 10.1007/s00253-008-1349-x
Johnson G. (2017). The α/β Hydrolase Fold Proteins of Mycobacterium tuberculosis, with Reference to their Contribution to Virulence. Curr. Protein Pept. Sci. 18 (3), 190–210. doi: 10.2174/1389203717666160729093515
Kádár B., Kocsis B., Tóth Á., Kristóf K., Felső P., Kocsis B., et al. (2017). Colistin resistance associated with outer membrane protein change in Klebsiella pneumoniae and Enterobacter asburiae. Acta Microbiol. Immunol. Hung. 64 (2), 217–227. doi: 10.1556/030.64.2017.017
Karatan E., Watnick P. (2009). Signals, regulatory networks, and materials that build and break bacterial biofilms. Microbiol. Mol. Biol. Rev. 73 (2), 310–347. doi: 10.1128/mmbr.00041-08
Kentache T., Jouenne T., Dé E., Hardouin J. (2016). Proteomic characterization of Nα- and Nϵ-acetylation in Acinetobacter baumannii. J. Proteomics 144, 148–158. doi: 10.1016/j.jprot.2016.05.021
Kentache T., Ben Abdelkrim A., Jouenne T., Dé E., Hardouin J. (2017). Global Dynamic Proteome Study of a Pellicle-forming Acinetobacter baumannii Strain. Mol. Cell Proteomics 16 (1), 100–112. doi: 10.1074/mcp.M116.061044
Khan A., Sharma D., Faheem M., Bisht D., Khan A. U. (2017). Proteomic analysis of a carbapenem-resistant Klebsiella pneumoniae strain in response to meropenem stress. J. Glob. Antimicrob. Resist. 8, 172–178. doi: 10.1016/j.jgar.2016.12.010
Kinsella R. L., Scott N. E., Feldman M. F. (2015). Clinical implications of glycoproteomics for Acinetobacter baumannii. Expert Rev. Proteomics 12 (1), 1–3. doi: 10.1586/14789450.2015.987756
Kishko I., Harish B., Zayats V., Reha D., Tenner B., Beri D., et al. (2012). Biphasic kinetic behavior of E. coli WrbA, an FMN-dependent NAD(P)H:quinone oxidoreductase. PloS One 7 (8), e43902. doi: 10.1371/journal.pone.0043902
Knight D., Dimitrova D. D., Rudin S. D., Bonomo R. A., Rather P. N. (2016). Mutations Decreasing Intrinsic β-Lactam Resistance Are Linked to Cell Division in the Nosocomial Pathogen Acinetobacter baumannii. Antimicrob. Agents Chemother. 60 (6), 3751–3758. doi: 10.1128/aac.00361-16
Koteva K., Cox G., Kelso J. K., Surette M. D., Zubyk H. L., Ejim L., et al. (2018). Rox, a Rifamycin Resistance Enzyme with an Unprecedented Mechanism of Action. Cell Chem. Biol. 25 (4), 403–412.e405. doi: 10.1016/j.chembiol.2018.01.009
Krishna V. S., Zheng S., Rekha E. M., Guddat L. W., Sriram D. (2019). Discovery and evaluation of novel Mycobacterium tuberculosis ketol-acid reductoisomerase inhibitors as therapeutic drug leads. J. Comput. Aided Mol. Des. 33 (3), 357–366. doi: 10.1007/s10822-019-00184-1
Krucinska J., Falcone E., Erlandsen H., Hazeen A., Lombardo M. N., Estrada A., et al. (2019). Structural and Functional Studies of Bacterial Enolase, a Potential Target against Gram-Negative Pathogens. Biochemistry 58 (9), 1188–1197. doi: 10.1021/acs.biochem.8b01298
Kwapien K., Damergi M., Nader S., El Khoury L., Hobaika Z., Maroun R. G., et al. (2017). Calibration of 1,2,4-Triazole-3-Thione, an Original Zn-Binding Group of Metallo-β-Lactamase Inhibitors. Validation of a Polarizable MM/MD Potential by Quantum Chemistry. J. Phys. Chem. B 121 (26), 6295–6312. doi: 10.1021/acs.jpcb.7b01053
Kwon S. O., Gho Y. S., Lee J. C., Kim S. I. (2009). Proteome analysis of outer membrane vesicles from a clinical Acinetobacter baumannii isolate. FEMS Microbiol. Lett. 297 (2), 150–156. doi: 10.1111/j.1574-6968.2009.01669.x
Kwon H. I., Kim S., Oh M. H., Na S. H., Kim Y. J., Jeon Y. H., et al. (2017). Outer membrane protein A contributes to antimicrobial resistance of Acinetobacter baumannii through the OmpA-like domain. J. Antimicrob. Chemother. 72 (11), 3012–3015. doi: 10.1093/jac/dkx257
Lari A. R., Ardebili A., Hashemi A. (2018). AdeR-AdeS mutations & overexpression of the AdeABC efflux system in ciprofloxacin-resistant Acinetobacter baumannii clinical isolates. Indian J. Med. Res. 147 (4), 413–421. doi: 10.4103/ijmr.IJMR_644_16
Larsen E. M., Johnson R. J. (2019). Microbial esterases and ester prodrugs: An unlikely marriage for combating antibiotic resistance. Drug Dev. Res. 80 (1), 33–47. doi: 10.1002/ddr.21468
Lee C., Marians K. J. (2013). Characterization of the nucleoid-associated protein YejK. J. Biol. Chem. 288 (44), 31503–31516. doi: 10.1074/jbc.M113.494237
Lee K., Choi H., Im H. (2009). Identification and expression of the tig gene coding for trigger factor from psychrophilic bacteria with no information of genome sequence available. Curr. Microbiol. 59 (2), 160–166. doi: 10.1007/s00284-009-9412-0
Lee C. R., Lee J. H., Park M., Park K. S., Bae I. K., Kim Y. B., et al. (2017). Biology of Acinetobacter baumannii: Pathogenesis, Antibiotic Resistance Mechanisms, and Prospective Treatment Options. Front. Cell Infect. Microbiol. 7:55:55. doi: 10.3389/fcimb.2017.00055
Li J., Rayner C. R., Nation R. L., Owen R. J., Spelman D., Tan K. E., et al. (2006). Heteroresistance to colistin in multidrug-resistant Acinetobacter baumannii. Antimicrob. Agents Chemother. 50 (9), 2946–2950. doi: 10.1128/aac.00103-06
Li Z. T., Zhang R. L., Bi X. G., Xu L., Fan M., Xie D., et al. (2015). Outer membrane vesicles isolated from two clinical Acinetobacter baumannii strains exhibit different toxicity and proteome characteristics. Microb. Pathog. 81, 46–52. doi: 10.1016/j.micpath.2015.03.009
Lin M. F., Chang K. C., Lan C. Y., Chou J., Kuo J. W., Chang C. K., et al. (2011). Molecular epidemiology and antimicrobial resistance determinants of multidrug-resistant Acinetobacter baumannii in five proximal hospitals in Taiwan. Jpn. J. Infect. Dis. 64 (3), 222–227. doi: 10.1258/ijsa.2010.010389
Long Q., Huang C., Liao P., Xie J. (2013). Proteomic insights into Acinetobacter baumannii drug resistance and pathogenesis. Crit. Rev. Eukaryot. Gene Expr. 23 (3), 227–255. doi: 10.1615/critreveukaryotgeneexpr.2013007266
López Rivero A. S., Rossi M. A., Ceccarelli E. A., Catalano-Dupuy D. L. (2019). A bacterial 2[4Fe4S] ferredoxin as redox partner of the plastidic-type ferredoxin-NADP(+) reductase from Leptospira interrogans. Biochim. Biophys. Acta Gen. Subj. 1863 (4), 651–660. doi: 10.1016/j.bbagen.2019.01.004
Lukoyanova N., Hoogenboom B. W., Saibil H. R. (2016). The membrane attack complex, perforin and cholesterol-dependent cytolysin superfamily of pore-forming proteins. J. Cell Sci. 129 (11), 2125–2133. doi: 10.1242/jcs.182741
Mardle C. E., Shakespeare T. J., Butt L. E., Goddard L. R., Gowers D. M., Atkins H. S., et al. (2019). A structural and biochemical comparison of Ribonuclease E homologues from pathogenic bacteria highlights species-specific properties. Sci. Rep. 9 (1), 7952. doi: 10.1038/s41598-019-44385-y
May H. C., Yu J. J., Zhang H., Wang Y., Cap A. P., Chambers J. P., et al. (2019). Thioredoxin-A is a virulence factor and mediator of the type IV pilus system in Acinetobacter baumannii. PloS One 14 (7), e0218505. doi: 10.1371/journal.pone.0218505
Maynaud G., Brunel B., Yashiro E., Mergeay M., Cleyet-Marel J. C., Le Quéré A. (2014). CadA of Mesorhizobium metallidurans isolated from a zinc-rich mining soil is a P(IB-2)-type ATPase involved in cadmium and zinc resistance. Res. Microbiol. 165 (3), 175–189. doi: 10.1016/j.resmic.2014.02.001
McNicholas P. M., Mann P. A., Najarian D. J., Miesel L., Hare R. S., Black T. A. (2001). Effects of mutations in ribosomal protein L16 on susceptibility and accumulation of evernimicin. Antimicrob. Agents Chemother. 45 (1), 79–83. doi: 10.1128/aac.45.1.79-83.2001
Melly G. C., Stokas H., Dunaj J. L., Hsu F. F., Rajavel M., Su C. C., et al. (2019). Structural and functional evidence that lipoprotein LpqN supports cell envelope biogenesis in Mycobacterium tuberculosis. J. Biol. Chem. 294 (43), 15711–15723. doi: 10.1074/jbc.RA119.008781
Minerdi D., Zgrablic I., Castrignanò S., Catucci G., Medana C., Terlizzi M. E., et al. (2016). Escherichia coli Overexpressing a Baeyer-Villiger Monooxygenase from Acinetobacter radioresistens Becomes Resistant to Imipenem. Antimicrob. Agents Chemother. 60 (1), 64–74. doi: 10.1128/aac.01088-15
Mujawar S., Mishra R., Pawar S., Gatherer D., Lahiri C. (2019). Delineating the Plausible Molecular Vaccine Candidates and Drug Targets of Multidrug-Resistant Acinetobacter baumannii. Front. Cell Infect. Microbiol. 9:203:203. doi: 10.3389/fcimb.2019.00203
Nigro S. J., Post V., Hall R. M. (2011). Aminoglycoside resistance in multiply antibiotic-resistant Acinetobacter baumannii belonging to global clone 2 from Australian hospitals. J. Antimicrob. Chemother. 66 (7), 1504–1509. doi: 10.1093/jac/dkr163
Nishimaki-Mogami T., Yamanaka H., Mizugaki M. (1987). Involvement of the fatty acid oxidation complex in acetyl-CoA-dependent chain elongation of fatty acids in Escherichia coli. J. Biochem. 102 (2), 427–432. doi: 10.1093/oxfordjournals.jbchem.a122070
Nwugo C. C., Gaddy J. A., Zimbler D. L., Actis L. A. (2011). Deciphering the iron response in Acinetobacter baumannii: A proteomics approach. J. Proteomics 74 (1), 44–58. doi: 10.1016/j.jprot.2010.07.010
Olsson M. O., Isaksson L. A. (1979). Analysis of rpsD mutations in Escherichia coli. I. Comparison of mutants with various alterations in ribosomal protein S4. Mol. Gen. Genet. 169 (3), 251–257. doi: 10.1007/bf00382271
Parimelzaghan A., Anbarasu A., Ramaiah S. (2016). Gene Network Analysis of Metallo Beta Lactamase Family Proteins Indicates the Role of Gene Partners in Antibiotic Resistance and Reveals Important Drug Targets. J. Cell Biochem. 117 (6), 1330–1339. doi: 10.1002/jcb.25422
Peleg A. Y., Potoski B. A., Rea R., Adams J., Sethi J., Capitano B., et al. (2007). Acinetobacter baumannii bloodstream infection while receiving tigecycline: a cautionary report. J. Antimicrob. Chemother. 59 (1), 128–131. doi: 10.1093/jac/dkl441
Pérez-Varela M., Corral J., Aranda J., Barbé J. (2018). Functional Characterization of AbaQ, a Novel Efflux Pump Mediating Quinolone Resistance in Acinetobacter baumannii. Antimicrob. Agents Chemother. 62 (9), e00906-18. doi: 10.1128/aac.00906-18
Pérez-Varela M., Corral J., Aranda J., Barbé J. (2019). Roles of Efflux Pumps from Different Superfamilies in the Surface-Associated Motility and Virulence of Acinetobacter baumannii ATCC 17978. Antimicrob. Agents Chemother. 63 (3), e02190-18. doi: 10.1128/aac.02190-18
Premkumar L., Kurth F., Duprez W., Grøftehauge M. K., King G. J., Halili M. A., et al. (2014). Structure of the Acinetobacter baumannii dithiol oxidase DsbA bound to elongation factor EF-Tu reveals a novel protein interaction site. J. Biol. Chem. 289 (29), 19869–19880. doi: 10.1074/jbc.M114.571737
Qayyum M. Z., Dey D., Sen R. (2016). Transcription Elongation Factor NusA Is a General Antagonist of Rho-dependent Termination in Escherichia coli. J. Biol. Chem. 291 (15), 8090–8108. doi: 10.1074/jbc.M115.701268
Ramirez M. S., Bonomo R. A., Tolmasky M. E. (2020). Carbapenemases: Transforming Acinetobacter baumannii into a Yet More Dangerous Menace. Biomolecules 10 (5), 720. doi: 10.3390/biom10050720
Reddy M. C., Breda A., Bruning J. B., Sherekar M., Valluru S., Thurman C., et al. (2014). Structure, activity, and inhibition of the Carboxyltransferase β-subunit of acetyl coenzyme A carboxylase (AccD6) from Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 58 (10), 6122–6132. doi: 10.1128/aac.02574-13
Ryan A., Kaplan E., Nebel J. C., Polycarpou E., Crescente V., Lowe E., et al. (2014). Identification of NAD(P)H quinone oxidoreductase activity in azoreductases from P. aeruginosa: azoreductases and NAD(P)H quinone oxidoreductases belong to the same FMN-dependent superfamily of enzymes. PloS One 9 (6), e98551. doi: 10.1371/journal.pone.0098551
Scribano D., Marzano V., Levi Mortera S., Sarshar M., Vernocchi P., Zagaglia C., et al. (2019). Insights into the Periplasmic Proteins of Acinetobacter baumannii AB5075 and the Impact of Imipenem Exposure: A Proteomic Approach. Int. J. Mol. Sci. 20 (14), 3451. doi: 10.3390/ijms20143451
Seward R. J., Lambert T., Towner K. J. (1998). Molecular epidemiology of aminoglycoside resistance in Acinetobacter spp. J. Med. Microbiol. 47 (5), 455–462. doi: 10.1099/00222615-47-5-455
Shakil S., Khan R., Zarrilli R., Khan A. U. (2008). Aminoglycosides versus bacteria–a description of the action, resistance mechanism, and nosocomial battleground. J. BioMed. Sci. 15 (1), 5–14. doi: 10.1007/s11373-007-9194-y
Shin B., Park W. (2015). Synergistic Effect of Oleanolic Acid on Aminoglycoside Antibiotics against Acinetobacter baumannii. PloS One 10 (9), e0137751. doi: 10.1371/journal.pone.0137751
Shin J. H., Lee H. W., Kim S. M., Kim J. (2009). Proteomic analysis of Acinetobacter baumannii in biofilm and planktonic growth mode. J. Microbiol. 47 (6), 728–735. doi: 10.1007/s12275-009-0158-y
Sikora A. E., Wierzbicki I. H., Zielke R. A., Ryner R. F., Korotkov K. V., Buchanan S. K., et al. (2018). Structural and functional insights into the role of BamD and BamE within the β-barrel assembly machinery in Neisseria gonorrhoeae. J. Biol. Chem. 293 (4), 1106–1119. doi: 10.1074/jbc.RA117.000437
Sim D. W., Kim J. H., Kim H. Y., Jang J. H., Lee W. C., Kim E. H., et al. (2016). Structural identification of the lipopolysaccharide-binding capability of a cupin-family protein from Helicobacter pylori. FEBS Lett. 590 (17), 2997–3004. doi: 10.1002/1873-3468.12332
Siroy A., Cosette P., Seyer D., Lemaître-Guillier C., Vallenet D., Van Dorsselaer A., et al. (2006). Global comparison of the membrane subproteomes between a multidrug-resistant Acinetobacter baumannii strain and a reference strain. J. Proteome Res. 5 (12), 3385–3398. doi: 10.1021/pr060372s
Song J., Park Y. H., Nemeria N. S., Kale S., Kakalis L., Jordan F. (2010). Nuclear magnetic resonance evidence for the role of the flexible regions of the E1 component of the pyruvate dehydrogenase complex from gram-negative bacteria. J. Biol. Chem. 285 (7), 4680–4694. doi: 10.1074/jbc.M109.082842
Spidlova P., Stojkova P., Dankova V., Senitkova I., Santic M., Pinkas D., et al. (2018). Francisella tularensis D-Ala D-Ala Carboxypeptidase DacD Is Involved in Intracellular Replication and It Is Necessary for Bacterial Cell Wall Integrity. Front. Cell Infect. Microbiol. 8:111:111. doi: 10.3389/fcimb.2018.00111
Spurr I. B., Birts C. N., Cuda F., Benkovic S. J., Blaydes J. P., Tavassoli A. (2012). Targeting tumour proliferation with a small-molecule inhibitor of AICAR transformylase homodimerization. Chembiochem 13 (11), 1628–1634. doi: 10.1002/cbic.201200279
Stancik L. M., Stancik D. M., Schmidt B., Barnhart D. M., Yoncheva Y. N., Slonczewski J. L. (2002). pH-dependent expression of periplasmic proteins and amino acid catabolism in Escherichia coli. J. Bacteriol. 184 (15), 4246–4258. doi: 10.1128/jb.184.15.4246-4258.2002
Sun D., Crowell S. A., Harding C. M., De Silva P. M., Harrison A., Fernando D. M., et al. (2016). KatG and KatE confer Acinetobacter resistance to hydrogen peroxide but sensitize bacteria to killing by phagocytic respiratory burst. Life Sci. 148, 31–40. doi: 10.1016/j.lfs.2016.02.015
Tankovic J., Legrand P., De Gatines G., Chemineau V., Brun-Buisson C., Duval J. (1994). Characterization of a hospital outbreak of imipenem-resistant Acinetobacter baumannii by phenotypic and genotypic typing methods. J. Clin. Microbiol. 32 (11), 2677–2681. doi: 10.1128/jcm.32.11.2677-2681.1994
Tiwari V., Tiwari M. (2014). Quantitative proteomics to study carbapenem resistance in Acinetobacter baumannii. Front. Microbiol. 5:512:512. doi: 10.3389/fmicb.2014.00512
Tiwari V., Vashistt J., Kapil A., Moganty R. R. (2012). Comparative proteomics of inner membrane fraction from carbapenem-resistant Acinetobacter baumannii with a reference strain. PloS One 7 (6), e39451. doi: 10.1371/journal.pone.0039451
Van Houdt R., Leplae R., Lima-Mendez G., Mergeay M., Toussaint A. (2012). Towards a more accurate annotation of tyrosine-based site-specific recombinases in bacterial genomes. Mob. DNA 3 (1):6. doi: 10.1186/1759-8753-3-6
Vashist J., Tiwari V., Kapil A., Rajeswari M. R. (2010). Quantitative profiling and identification of outer membrane proteins of beta-lactam resistant strain of Acinetobacter baumannii. J. Proteome Res. 9 (2), 1121–1128. doi: 10.1021/pr9011188
Villarreal L., Heredia N. L., García S. (2000). Changes in protein synthesis and acid tolerance in Clostridium perfringens type A in response to acid shock. Int. Microbiol. 3 (2), 113–116.
Wang J., Zhang J., Fu Q., Guo S., Ta L., Sun P. (2016). Proteomic Analyses Uncover the Mechanisms Underlying Antibiotic Resistance Differences among Three Acinetobacter baumannii Isolates. J. Mol. Microbiol. Biotechnol. 26 (6), 401–409. doi: 10.1159/000447454
Wen S. H., Su S. C., Liou B. H., Lin C. H., Lee K. R. (2018). Sulbactam-enhanced cytotoxicity of doxorubicin in breast cancer cells. Cancer Cell Int. 18, 128. doi: 10.1186/s12935-018-0625-9
Xiao S. Z., Chu H. Q., Han L. Z., Zhang Z. M., Li B., Zhao L., et al. (2016). Resistant mechanisms and molecular epidemiology of imipenem-resistant Acinetobacter baumannii. Mol. Med. Rep. 14 (3), 2483–2488. doi: 10.3892/mmr.2016.5538
Yun S. H., Choi C. W., Kwon S. O., Park G. W., Cho K., Kwon K. H., et al. (2011). Quantitative proteomic analysis of cell wall and plasma membrane fractions from multidrug-resistant Acinetobacter baumannii. J. Proteome Res. 10 (2), 459–469. doi: 10.1021/pr101012s
Keywords: Acinetobacter baumannii, antibiotic resistance, proteomic, TMT, label free, glycopeptides
Citation: Wang P, Li R-Q, Wang L, Yang W-T, Zou Q-H and Xiao D (2021) Proteomic Analyses of Acinetobacter baumannii Clinical Isolates to Identify Drug Resistant Mechanism. Front. Cell. Infect. Microbiol. 11:625430. doi: 10.3389/fcimb.2021.625430
Received: 03 November 2020; Accepted: 11 January 2021;
Published: 24 February 2021.
Edited by:
Yang Zhang, University of Pennsylvania, United StatesReviewed by:
Israa M. S. Al-Kadmy, Al-Mustansiriya University, IraqCopyright © 2021 Wang, Li, Wang, Yang, Zou and Xiao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Di Xiao, eGlhb2RpQGljZGMuY24=; Qing-Hua Zou, em91cWluZ2h1YUBiam11LmVkdS5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.