
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell. Infect. Microbiol. , 22 February 2017
Sec. Molecular Bacterial Pathogenesis
Volume 7 - 2017 | https://doi.org/10.3389/fcimb.2017.00045
This article is part of the Research Topic Pseudomonas and Acinetobacter: From Drug Resistance to Pathogenesis View all 13 articles
 Xiaoting Hua1,2
Xiaoting Hua1,2 Lilin Liu1,2
Lilin Liu1,2 Youhong Fang3
Youhong Fang3 Qiucheng Shi1,2
Qiucheng Shi1,2 Xi Li4
Xi Li4 Qiong Chen5
Qiong Chen5 Keren Shi1,2
Keren Shi1,2 Yan Jiang1,2
Yan Jiang1,2 Hua Zhou6
Hua Zhou6 Yunsong Yu1,2,7*
Yunsong Yu1,2,7*Acinetobacter baumannii has emerged as an important opportunistic pathogen due to its ability to acquire resistance to most currently available antibiotics. Colistin is often considered as the last line of therapy for infections caused by multidrug-resistant A. baumannii (MDRAB). However, colistin-resistant A. baumannii strain has recently been reported. To explore how multiple drug-resistant A. baumannii responded to colistin resistance, we compared the genomic, transcriptional and proteomic profile of A. baumannii MDR-ZJ06 to the induced colistin-resistant strain ZJ06-200P5-1. Genomic analysis showed that lpxC was inactivated by ISAba1 insertion, leading to LPS loss. Transcriptional analysis demonstrated that the colistin-resistant strain regulated its metabolism. Proteomic analysis suggested increased expression of the RND efflux pump system and down-regulation of FabZ and β-lactamase. These alterations were believed to be response to LPS loss. In summary, the lpxC mutation not only established colistin resistance but also altered global gene expression.
Acinetobacter baumannii has emerged as an important opportunistic pathogen due to its ability to acquire resistance to most currently available antibiotics (Peleg et al., 2008; Howard et al., 2012; Antunes et al., 2014). Since current treatment options for multi-drug resistant (MDR) A. baumannii are extremely limited, colistin is often considered as the last line of the therapy for infections caused by MDR A. baumannii (Bae et al., 2016; Cheah et al., 2016b). However, colistin-resistant A. baumannii strain has recently been reported (Cai et al., 2012).
Colistin is a polycationic antimicrobial peptide that targets the polyanionic bacterial lipopolysaccharide (LPS) of Gram-negative bacteria. Two different colistin resistance mechanisms have previously been reported (Beceiro et al., 2014). The first mechanism inactivates the lipid A biosynthesis pathway, leading to the complete loss of surface LPS. Mutations in lpxC, lpxA, or lpxD are involved in the first mechanism. The pmrAB two-component system mediates the second resistance mechanism. Mutations in pmrA and pmrB induce the activity of pmrC, which adds phosphoethanolamine (PEtn) to the hepta-acylated form of lipid A (Beceiro et al., 2011). Further mutations in vacJ, pldA, ttg2C, pheS and a conserved hypothetical protein were reported to involve in reduced colistin susceptibility through novel resistance mechanisms (Thi Khanh Nhu et al., 2016). Four putative colistin resistant genes: A1S_1983, hepA, A1S_3026, and rsfS were also identified in our previous study (Mu et al., 2016).
The response to LPS alteration has been investigated via transcriptional analysis. In response to LPS alteration, A. baumannii alters the expression of critical transport and biosynthesis systems associated with modulating the composition and structure of the bacterial surface (lpxA; Henry et al., 2012) or alters the expression of genes associated with outer membrane structure and biogenesis (pmrB; Cheah et al., 2016a). Moreover, the response to colistin is highly similar to the transcriptional alteration observed in an LPS-deficient strain (Henry et al., 2015). Colistin resistance was also explored using proteomic methods. There were 35 differentially expressed proteins. Most differentially expressed proteins were down-regulated in the colistin resistant strain, including outer membrane proteins, chaperones, protein biosynthesis factors, and metabolic enzymes (Fernandez-Reyes et al., 2009). However, the combination of genomic, transcriptomic, and proteomic methods to examine the colistin resistance mechanism in A. baumannii has rarely been reported. Furthermore, the strain used in this study was an MDR strain, but not laboratory strains (ATCC 19606, ATCC 17978) that do not represent clonal lineages in a clinical environment. Here, we used genome, transcriptome, and proteome to elucidate the colistin resistance mechanism in MDR A. baumannii. There was an ISAba1 insertion in lpxC (ABZJ_03720) in ZJ06-200P5-1 compared with the genome sequence of MDR-ZJ06, where lpxC encoded an UDP-3-O-acyl-N-acetylglucosamine deacetylase.
Restriction enzymes, T4 ligase, and Taq DNA polymerase were purchased from TaKaRa (Otsu, Shiga, Japan). The A. baumannii strain MDR-ZJ06 was isolated from the bloodstream of a patient in Hangzhou, China, in 2006. All A. baumannii cultures were grown at 37 °C in Mueller-Hinton (MH) agar and cation-adjusted MH broth (CAMHB) (Oxoid, Basingstoke, UK). Colistin was purchased from Sigma (Shanghai, China).
A colistin-resistant mutant was generated in A. baumannii MDR-ZJ06 by a previously described method (Li et al., 2006). Briefly, first, MDR-ZJ06 was cultured in CAMHB containing colistin at 8 × minimum inhibitory concentration (MIC). After overnight incubation, the culture was diluted 1:1000 with CAMHB containing colistin at 64 × MIC and then incubated at 37 °C overnight. Finally, the culture was diluted 1:100 with CAMHB containing colistin at 200 × MIC. After overnight incubation, the culture was plated on plates containing 10 μg of colistin at an appropriate dilution, and then one of colistin resistant colonies was collected for further experiments and designated as ZJ06-200P5-1. MICs for colistin and tigecycline were determined by E-test (bioMérieux, France) on MH agar, and the antimicrobial activities of the other antimicrobial agents were detected by disk diffusion. The results were interpreted according to CLSI or EUCAST breakpoints.
ZJ06-200P5-1 cells were cultured from a single colony overnight at 37 °C in MH broth. The genomic DNA was extracted via a QIAamp DNA minikit (Qiagen, Valencia, CA) following the manufacturer's protocol. Agarose gel and a NanoDrop spectrophotometer were used to determine the quality and quantity of extracted genomic DNA. The 300 bp library for Illumina paired-end sequencing was constructed from 5 μg of genome DNA of ZJ06-200P5-1 by staff at Zhejiang Tianke (Hangzhou, China). Mapping and SNP detection were performed via Breseq (Deatherage and Barrick, 2014). The regions containing the detected SNPs were amplified by PCR. The PCR products were sent to Biosune (Biosune, Hangzhou, China) for Sanger sequencing.
A. baumannii MDR-ZJ06 and ZJ06-200P5-1 were grown overnight at 37 °C in LB broth. Strains were subcultured 1/100 into fresh LB broth and grown at 37 °C for 2 h (OD600: 0.29 ± 0.02 for MDR-ZJ06, 0.26 ± 0.02 for ZJ06-200P5-1). The cells were collected at 4 °C, and the RNA was extracted using TRIZOL Reagent (Invitrogen, Carlsbad, CA, USA) after liquid nitrogen grinding. For RNA sequencing, wild type and mutants were sampled in triplicate. The subsequent RNA extraction, bacteria mRNA sequence library construction, transcriptome analysis and real-time quantitative PCR verification were performed by staff at Zhejiang Tianke (Hangzhou, China) as described previously in reference (Hua et al., 2014). Sequenced reads were mapped to the MDR-ZJ06 genome (CP001937-8) using Rockhopper (McClure et al., 2013). The output data was analyzed by edgeR (McCarthy et al., 2012). Data generated by RNA sequencing were deposited to the NCBI Sequence Read Archive with accession number SRR5234544 (the wild type) and SRR5234545 (the colistin resistant strain).
A. baumannii MDR-ZJ06 and ZJ06-200P5-1 were grown overnight at 37 °C in LB broth. Strains were subcultured 1/100 into fresh LB broth and grown at 37 °C for 2 h (OD600: 0.29 ± 0.02 for MDR-ZJ06, 0.26 ± 0.02 for ZJ06-200P5-1). The cells were collected at 4 °C and sent to Shanghai Applied Protein Technology Co. Ltd. The cell pellets were washed twice with PBS, and 500 μl SDT lysis buffer (4% SDS, 100 mM Tris-HCl, 1 mM DTT, pH 7.6) was added. After being sonicated for 2 mins on ice, the cells were centrifuged at 14,000 × g for 30 min at 4 °C. The protein concentration in the supernatant was determined by the BCA method.
In brief, 300 μg protein was added to 200 μl UA buffer (8 M urea, 150 mM Tris-HCl pH 8.0) and ultrafiltered (Sartorius, 10 kD) with UA buffer. To block reduced cysteine residues, 100 μl iodoacetamide (IAA) buffer (50 mM IAA in UA buffer) was added, centrifuged at 600 rpm for 1 min, and incubated for 30 min in the dark. The filter was washed twice with 100 μl UA buffer and twice with 100 μl Dissolution buffer (50 mM triethylammonium bicarbonate, pH 8.5). Finally, the proteins were digested with 2 μg trypsin (Promega) in 40 μl Dissolution buffer at 37 °C for 16–18 h. The peptides were collected as a filtrate, and its content was estimated at OD280.
For iTRAQ labeling, the peptides were labeled with the 4-plex iTRAQ reagent following the manufacturer's instructions (AB SCIEX). The peptides from MDR-ZJ06 were labeled with 114 and 116 isobaric reagents, and the peptides from ZJ06-200P5-1 were labeled with 115 and 117 isobaric reagents.
RP-HPCL online-coupled to MS/MS (LC-MS/MS) analysis of the iTRAQ-labeled peptides was performed on an EASY-nLC nanoflow LC system (Thermo Fisher Scientific) connected to an Orbitrap Elite hybrid mass spectrometer (Thermo Fisher Scientific). After the samples were reconstituted and acidified with buffer A (0.1% (v/v) formic acid in water), a set-up involving a pre-column and analytical column was used. The pre-column was a 2 cm EASY-column (100, 5 μm C18; Thermo Fisher Scientific), while the analytical column was a 10 cm EASY-column (75, 3 μm, C18; Thermo Fisher Scientific). The 120 min linear gradient from 0 to 100% buffer B (0.1% (v/v) formic acid and 80% acetonitrile) at a constant flow rate of 250 nl/min was as follows: 0–100 min, 0–35% buffer B; 100–108 min, 35–100% buffer B; 108–120 min, 100% buffer B. MS data were acquired using a data-dependent top 10 method, dynamically choosing the most abundant precursor ions from the survey scan (300–180 m/z) for HCD fragmentation. The Dynamic exclusion was set to a repeat count of 1 with a 30 s duration. Survey scans were acquired at a resolution of 30,000 at m/z 200, and the resolution for HCD spectra was set to 15,000 at m/z 200. The normalized collision energy was 35 eV, and the underfill ratio was defined as 0.1%.
The MS/MS spectra were searched using the MASCOT engine (Matrix Science, London, UK; version 2.2) against the A. baumannii MDR-ZJ06 FASTA database. False discovery rates (FDR) were calculated via running all spectra against the FASTA database using the MASCOT software. The following options were used to identify proteins: peptide mass tolerance = 20 ppm, fragment mass tolerance = 0.1 Da, Enzyme = Trypsin, Max missed cleavages = 2, Fixed modification: Carbamidomethyl (C), iTRAQ 4plex (N-term), iTRAQ 4plex (K), Variable modification: Oxidation (M). Quantification was performed based on the peak intensities of the reporter ions in the MS/MS spectra. The proteins were considered overexpressed when the iTRAQ ratio was above 1.5 and underexpressed when the iTRAQ ratio was lower than 0.67 (Wang et al., 2016). Functional classification of differentially expression genes were annotated using the KEGG databases. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (Vizcaino et al., 2016) partner repository with the dataset identifier PXD005265 and 10.6019/PXD005265. Reviewer account details: Username: cmV2aWV3ZXI1NDI0MkBlYmkuYWMudWs=; Password: zR8mE9wu.
Four independent cultures per strain were grown overnight, diluted to 1:1000 in MH and aliquots placed into a flat-bottom 100-well plate in four replicates. The plate was incubated at 37 °C with agitation. The OD600 of each culture was determined every 5 min for 16 h using a Bioscreen C MBR machine (Oy Growth Curves Ab Ltd., Finland). The growth rate was estimated based on OD600 curves using an R script (Fang et al., 2016).
The colistin-resistant mutant ZJ06-200P5-1 generated from the culture in CAMHB containing colistin was sent for whole genome sequencing. There was an ISAba1 insertion in lpxC in ZJ06-200P5-1 compared with the genome sequence of MDR-ZJ06 (Figure 1). The MIC of MDR-ZJ06 and ZJ06-200P5-1 were detected and listed in Table 1. The MIC for colistin increased from 0.38 mg/L (MDR-ZJ06) to >256 mg/L (ZJ06-200P5-1). However, ZJ06-200P5-1 showed higher sensitivity to multiple antibiotics: β-lactams, carbapenem, tetracycline, and ciprofloxacin, but not aminoglycosides. Furthermore, ZJ06-200P5-1 showed a lower growth rate (0.81 ± 0.05) than wild type.
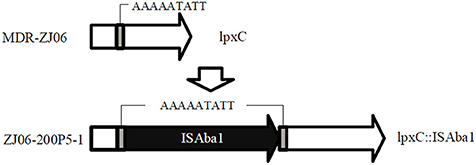
Figure 1. Whole genome sequencing revealed the colistin-resistance mechanism in A. baumannii ZJ06-200P5-1. The gene lpxC was intact in MDR-ZJ06, while in ZJ06-200P5-1, lpxC was inactivated by the insertion sequence ISAba1.
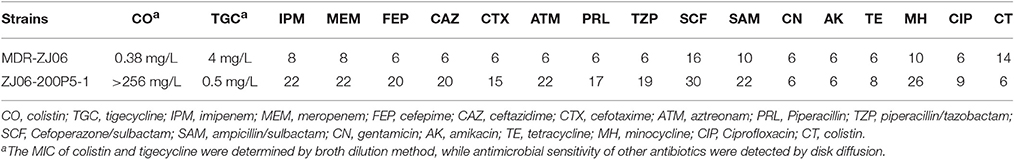
Table 1. Antibiotic susceptibility of A. baumannii MDR-ZJ06 and its colistin resistant mutant ZJ06-200P5-1.
The transcriptome analysis of ZJ06-200P5-1 and MDR-ZJ06 was performed by Illumina RNA deep sequencing technology. Cells of the two strains were collected in the early exponential phase. A total of 137 genes showed significant differential expression [log2(FoldChange) > 1 or log2(FoldChange) < −1], among which 48 genes were upregulated and 89 were downregulated (Table 2). Sixteen selected genes, three up-regulated and thirteen down-regulated genes, were well-validated by RT-qPCR (Figure 2). After mapping the differentially expressed genes into the KEGG pathway, we observed that genes involved in Energy metabolism and Amino acid metabolism were down-regulated, while Carbohydrate metabolism was up-regulated.
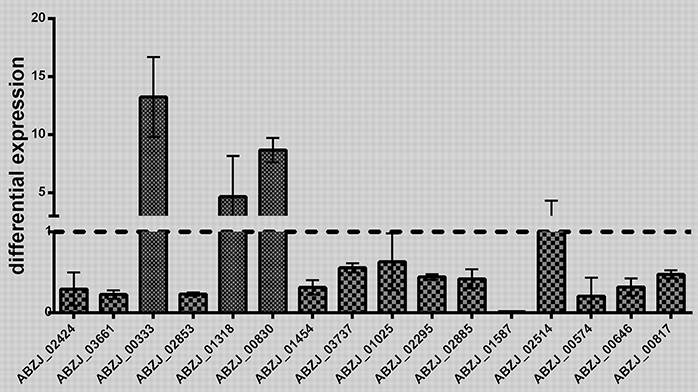
Figure 2. Validation of the RNA sequencing results. The transcriptomic results obtained by RNA-seq were validated by quantitative RT-PCR analysis. The differential expression of 16 genes was detected in this study. Three biology replicates were used in this experiment. The results were presented as expression in ZJ06-200P5-1, relative to MDR-ZJ06. The reference gene rpoB was used for inter-sample normalization. Error bars denote standard deviation.
A total of 1582 proteins were identified in the iTRAQ experiment. A protein ratio >1.5 or <0.67 (p <0.05) was considered to be differentially expressed. After filtration, 82 differentially expressed proteins were identified between ZJ06-200P5-1 and MDR-ZJ06. The detailed information is shown in Table 3.
The expression of AdeABC was up-regulated in the LPS-loss ZJ06-200P5-1 strain. The AdeABC efflux pump confers resistance to various antibiotics classes. The expression of AdeABC genes was increased approximately two-fold in ZJ06-200P5-1 (Figure 3A). However, ZJ06-200P5-1 showed higher susceptibility to multiple antibiotics than MDR-ZJ06 (Table 1).
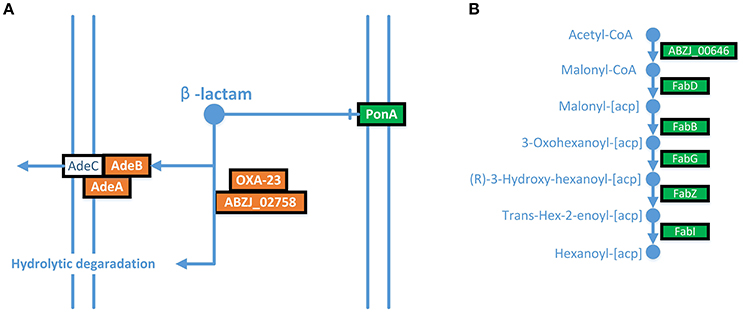
Figure 3. ITRAQ analysis showed that AdeABC were up-regulated, and the fatty acid biosynthesis pathway was down-regulated in ZJ06-200P5-1. (A) AdeABC efflux pump, (B) fatty acid biosynthesis pathway. Green shows genes with significantly reduced expression levels, and red shows genes with significantly increased expression levels.
The fatty acid biosynthesis pathway was down-regulated in the ZJ06-200P5-1 strain (Figure 3B). The expression of FabZ was decreased by approximately two-fold in ZJ06-200P5-1. The β-lactamases blaOXA−23 and blaADC−25 were down-regulated in ZJ06-200P5-1 strain. The expression levels of blaOXA−23 and blaADC−25 were decreased two- to four-fold in ZJ06-200P5-1.
A total of 15 differentially expressed genes (or proteins) were identified in both transcriptome and proteome (Table 4). Among them, three genes were both up-regulated, and nine genes were both down-regulated. Although there was correlation between transcriptome and proteome data, the absolute expression difference values in transcriptome data was higher than those in proteome data. In addition, the result of three gene/proteins were contradictory (highlighted in red letters in Table 4). The contradictory result might be caused by post-transcriptional regulation.
Due to the limitation of antimicrobial agents in clinical use, it is urgent to extend our understanding of the emergence of colistin resistance in A. baumannii. A. baumannii MDR-ZJ06, a multidrug-resistant clinical strain isolated from bloodstream, has been sequenced and was considered an ideal strain for examining the colistin-resistant mechanism in A. baumannii (Zhou et al., 2011). In this study, colistin-resistant strain was rapidly obtained, and its resistance mechanism was LPS loss caused by ISAba1 insertion in lpxC. This result confirmed a previous finding (Moffatt et al., 2010). The rapid isolation of colistin-resistant mutant from multiple drug-resistant A. baumannii indicated a high risk of A. baumannii evolving resistance to colistin in clinical use.
We successfully detected the whole transcriptional profile of A. baumannii strain MDR-ZJ06 and its colistin-resistant mutant ZJ06-200P5-1 via Illumina RNA-sequencing. In another transcriptome study (Henry et al., 2012), A. baumannii ATCC 19606 and its lpxA mutant were used. Although both the lpxC and lpxA mutation lead to LPS loss, the different transcriptional response may be due to differences in the strain genetic background and the resistant mutation. In transcriptional analysis, we observed that genes involved in Energy metabolism and Amino acid metabolism were down-regulated, while Carbohydrate metabolism was up-regulated.
The expression of AdeABC was up-regulated in the LPS-loss ZJ06-200P5-1 strain. Similar results were also observed in all polymyxin-treated samples (Cheah et al., 2016a). In addition, the expression levels of adeIJK and macAB-tolC were up-regulated in the LPS loss mutant (Henry et al., 2012). Increased expression of the RND efflux pump system (AdeABC) was a common finding across all experiments in colistin exposure. The up-regulation of AdeABC indicated the diminished integrity and barrier function of the outer membrane in colistin-resistant A. baumannii (Henry et al., 2015; Cheah et al., 2016a). However, ZJ06-200P5-1 showed higher susceptibility to multiple antibiotics than MDR-ZJ06. The higher susceptibility might result from the higher outer membrane permeability of ZJ06-200P5-1 due to LPS-loss. The increased expression of the efflux pump was thought to be a response to toxic substances that accumulated in the cells due to the increased membrane permeability (Henry et al., 2012).
The fatty acid biosynthesis pathway was down-regulated in the ZJ06-200P5-1 strain. In E. coli, it is important to balance LPS and fatty acid biosynthesis to maintain cell integrity. FabZ, which dehydrates R-3-hydroxymyristoyl-acyl carrier protein in fatty acid biosynthesis, plays an important role in rebalancing lipid A and fatty acid homeostasis (Bojkovic et al., 2016). The decrease in FabZ was considered to be a response to LPS-loss in ZJ06-200P5-1. The β-lactamases blaOXA−23 and blaADC−25 were down-regulated in the ZJ06-200P5-1 strain. Decreased expression levels of blaOXA−23 and blaADC−25 were also observed in A. baumannii MDR-ZJ06 under a subinhibitory concentration of tigecycline (Hua et al., 2014). Meanwhile, the strain under tigecycline stress showed a lower MIC of ceftazidime (Hua et al., 2014). The decrease in blaOXA−23 and blaADC−25 might contribute to the increased sensitivity to β-lactam antimicrobial agents.
A multi-omics approach was adopted to obtain a more global view of colistin-resistant A. baumannii. Genomic analysis showed that lpxC was inactivated by ISAba1 insertion, leading to LPS loss. Transcriptional analysis demonstrated that the colistin-resistant strain regulated its metabolism. Metabolic change and LPS loss were concomitant. Proteomic analysis suggested increased expression of the RND efflux pump system and the down-regulation of FabZ and β-lactamase. These alterations are believed to be responses to LPS loss. Together, the lpxC mutation not only confirmed colistin resistance but also altered global gene expression.
The whole-genome shotgun sequencing results for A. baumannii ZJ06-200P5-1 have been deposited at DDBJ/EMBL/GenBank under the accession number MIFW00000000.
XH and YY conceived and designed the study. XH, LL, YF, QS, XL, QC, KS, YJ, and HZ performed the experiments. XH and YY performed data analysis and drafted the manuscript. All authors reviewed and approved the final manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
This work was supported by the National Natural Science Foundation of China (81230039, 31670135, 81378158), the 973 Preliminary Research Program (2014CB560707), the Natural Science Foundation of Zhejiang province, China (LY15H190004, Y16H190013) and the Zhejiang Province Medical Platform Backbone Talent Plan (2016DTA003).
Antunes, L. C., Visca, P., and Towner, K. J. (2014). Acinetobacter baumannii: evolution of a global pathogen. Pathog. Dis. 71, 292–301. doi: 10.1111/2049-632X.12125
Bae, S., Kim, M. C., Park, S. J., Kim, H. S., Sung, H., Kim, M. N., et al. (2016). In vitro synergistic activity of antimicrobial agents in combination against clinical isolates of colistin-resistant Acinetobacter baumannii. Antimicrob Agents Chemother. 60, 6774–6779. doi: 10.1128/AAC.00839-16
Beceiro, A., Llobet, E., Aranda, J., Bengoechea, J. A., Doumith, M., Hornsey, M., et al. (2011). Phosphoethanolamine modification of lipid A in colistin-resistant variants of Acinetobacter baumannii mediated by the pmrAB two-component regulatory system. Antimicrob. Agents Chemother. 55, 3370–3379. doi: 10.1128/AAC.00079-11
Beceiro, A., Moreno, A., Fernandez, N., Vallejo, J. A., Aranda, J., Adler, B., et al. (2014). Biological cost of different mechanisms of colistin resistance and their impact on virulence in Acinetobacter baumannii. Antimicrob. Agents Chemother. 58, 518–526. doi: 10.1128/AAC.01597-13
Bojkovic, J., Richie, D. L., Six, D. A., Rath, C. M., Sawyer, W. S., Hu, Q., et al. (2016). Characterization of an Acinetobacter baumannii lptD deletion strain: permeability defects and response to inhibition of lipopolysaccharide and fatty acid biosynthesis. J. Bacteriol. 198, 731–741. doi: 10.1128/JB.00639-15
Cai, Y., Chai, D., Wang, R., Liang, B., and Bai, N. (2012). Colistin resistance of Acinetobacter baumannii: clinical reports, mechanisms and antimicrobial strategies. J. Antimicrob. Chemother. 67, 1607–1615. doi: 10.1093/jac/dks084
Cheah, S. E., Johnson, M. D., Zhu, Y., Tsuji, B. T., Forrest, A., Bulitta, J. B., et al. (2016a). Polymyxin resistance in Acinetobacter baumannii: genetic mutations and transcriptomic changes in response to clinically relevant dosage regimens. Sci. Rep. 6:26233. doi: 10.1038/srep26233
Cheah, S. E., Li, J., Tsuji, B. T., Forrest, A., Bulitta, J. B., and Nation, R. L. (2016b). Colistin and polymyxin B dosage regimens against Acinetobacter baumannii: differences in activity and the emergence of resistance. Antimicrob. Agents Chemother. 60, 3921–3933. doi: 10.1128/AAC.02927-15
Deatherage, D. E., and Barrick, J. E. (2014). Identification of mutations in laboratory-evolved microbes from next-generation sequencing data using breseq. Methods Mol. Biol. 1151, 165–188. doi: 10.1007/978-1-4939-0554-6_12
Fang, L., Chen, Q., Shi, K., Li, X., Shi, Q., He, F., et al. (2016). Step-Wise increase in tigecycline resistance in klebsiella pneumoniae associated with Mutations in ramR, lon and rpsJ. PLoS ONE 11:e0165019. doi: 10.1371/journal.pone.0165019
Fernandez-Reyes, M., Rodriguez-Falcon, M., Chiva, C., Pachon, J., Andreu, D., and Rivas, L. (2009). The cost of resistance to colistin in Acinetobacter baumannii: a proteomic perspective. Proteomics 9, 1632–1645. doi: 10.1002/pmic.200800434
Henry, R., Crane, B., Powell, D., Deveson Lucas, D., Li, Z., Aranda, J., et al. (2015). The transcriptomic response of Acinetobacter baumannii to colistin and doripenem alone and in combination in an in vitro pharmacokinetics/pharmacodynamics model. J. Antimicrob. Chemother. 70, 1303–1313. doi: 10.1093/jac/dku536
Henry, R., Vithanage, N., Harrison, P., Seemann, T., Coutts, S., Moffatt, J. H., et al. (2012). Colistin-resistant, lipopolysaccharide-deficient Acinetobacter baumannii responds to lipopolysaccharide loss through increased expression of genes involved in the synthesis and transport of lipoproteins, phospholipids, and poly-beta-1,6-N-acetylglucosamine. Antimicrob. Agents Chemother. 56, 59–69. doi: 10.1128/AAC.05191-11
Howard, A., O'Donoghue, M., Feeney, A., and Sleator, R. D. (2012). Acinetobacter baumannii: an emerging opportunistic pathogen. Virulence 3, 243–250. doi: 10.4161/viru.19700
Hua, X., Chen, Q., Li, X., and Yu, Y. (2014). Global transcriptional response of Acinetobacter baumannii to a subinhibitory concentration of tigecycline. Int. J. Antimicrob. Agents 44, 337–344. doi: 10.1016/j.ijantimicag.2014.06.015
Li, J., Rayner, C. R., Nation, R. L., Owen, R. J., Spelman, D., Tan, K. E., et al. (2006). Heteroresistance to colistin in multidrug-resistant Acinetobacter baumannii. Antimicrob. Agents Chemother. 50, 2946–2950. doi: 10.1128/AAC.00103-06
McCarthy, D. J., Chen, Y., and Smyth, G. K. (2012). Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 40, 4288–4297. doi: 10.1093/nar/gks042
McClure, R., Balasubramanian, D., Sun, Y., Bobrovskyy, M., Sumby, P., Genco, C. A., et al. (2013). Computational analysis of bacterial RNA-Seq data. Nucleic Acids Res. 41:e140. doi: 10.1093/nar/gkt444
Moffatt, J. H., Harper, M., Harrison, P., Hale, J. D., Vinogradov, E., Seemann, T., et al. (2010). Colistin resistance in Acinetobacter baumannii is mediated by complete loss of lipopolysaccharide production. Antimicrob. Agents Chemother. 54, 4971–4977. doi: 10.1128/AAC.00834-10
Mu, X., Wang, N., Li, X., Shi, K., Zhou, Z., Yu, Y., et al. (2016). The Effect of Colistin Resistance-Associated Mutations on the Fitness of Acinetobacter baumannii. Front. Microbiol. 7:1715. doi: 10.3389/fmicb.2016.01715
Peleg, A. Y., Seifert, H., and Paterson, D. L. (2008). Acinetobacter baumannii: emergence of a successful pathogen. Clin. Microbiol. Rev. 21, 538–582. doi: 10.1128/CMR.00058-07
Thi Khanh Nhu, N., Riordan, D. W., Do Hoang Nhu, T., Thanh, D. P., Thwaites, G., Huong Lan, N. P., et al. (2016). The induction and identification of novel Colistin resistance mutations in Acinetobacter baumannii and their implications. Sci. Rep. 6:28291. doi: 10.1038/srep28291
Vizcaino, J. A., Csordas, A., Del-Toro, N., Dianes, J. A., Griss, J., Lavidas, I., et al. (2016). 2016 update of the PRIDE database and its related tools. Nucleic Acids Res. 44, D447–D456. doi: 10.1093/nar/gkw880
Wang, S., Yang, Y., Zhao, Y., Zhao, H., Bai, J., Chen, J., et al. (2016). Sub-MIC tylosin inhibits Streptococcus suis biofilm formation and results in differential protein expression. Front. Microbiol. 7:384. doi: 10.3389/fmicb.2016.00384
Keywords: Acinetobacter baumannii, colistin, whole-genome sequencing, transcriptome, proteome
Citation: Hua X, Liu L, Fang Y, Shi Q, Li X, Chen Q, Shi K, Jiang Y, Zhou H and Yu Y (2017) Colistin Resistance in Acinetobacter baumannii MDR-ZJ06 Revealed by a Multiomics Approach. Front. Cell. Infect. Microbiol. 7:45. doi: 10.3389/fcimb.2017.00045
Received: 10 November 2016; Accepted: 07 February 2017;
Published: 22 February 2017.
Edited by:
Ghassan M. Matar, American University of Beirut, LebanonReviewed by:
Rahul Raghavan, Portland State University, USACopyright © 2017 Hua, Liu, Fang, Shi, Li, Chen, Shi, Jiang, Zhou and Yu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yunsong Yu, eXZ5czExOUAxNjMuY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.