- 1State Key Laboratory of Quality Research in Chinese Medicine, School of Pharmacy, Macau University of Science and Technology, Taipa, Macau, China
- 2Department of Clinical Pharmacy, School of Pharmacy, Second Military University, Shanghai, China
- 3School of Biosciences, University of Nottingham, Nottingham, United Kingdom
- 4Shanghai Key Laboratory of Bioactive Small Molecules, Department of Pharmacology, School of Pharmacy, Fudan University, Shanghai, China
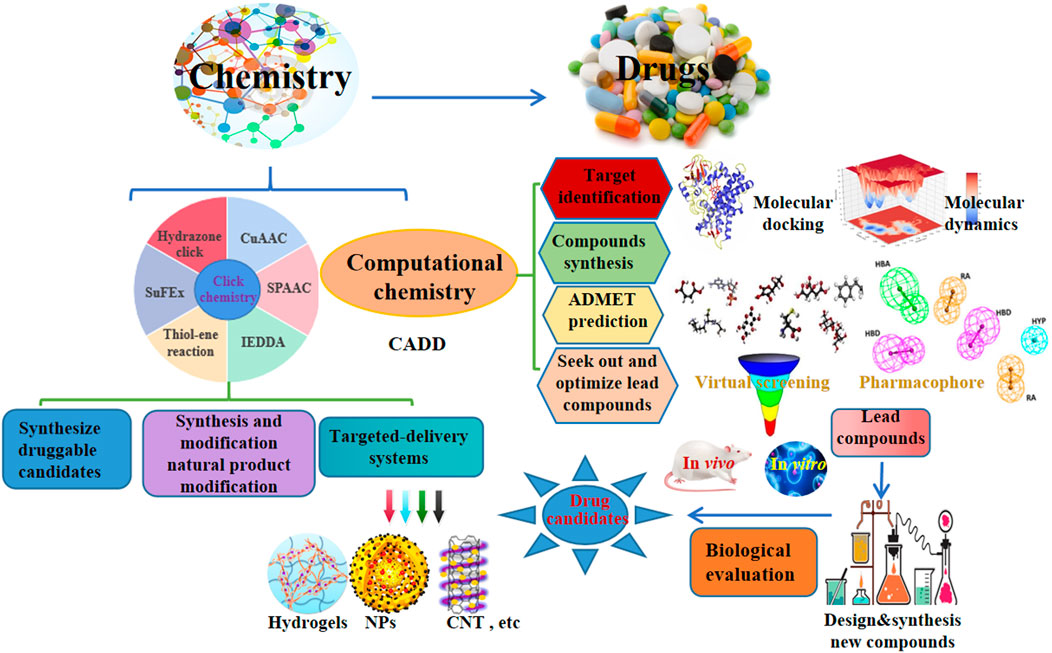
Drug discovery is a costly and time-consuming process with a very high failure rate. Recently, click chemistry and computer-aided drug design (CADD) represent popular areas for new drug development. Herein, we summarized the recent updates in click and computational chemistry for drug discovery and development including clicking to effectively synthesize druggable candidates, synthesis and modification of natural products, targeted delivery systems, and computer-aided drug discovery for target identification, seeking out and optimizing lead compounds, ADMET prediction as well as compounds synthesis, hopefully, inspires new ideas for novel drug development in the future.
Introduction
Click chemistry, an efficient chemo-selective synthesis method for coupling molecular fragments under mild reaction conditions, mainly includes Cu-catalyzed azide-alkyne cycloaddition reaction (CuAAC), strain-promoted azide-alkyne cycloaddition reaction (SPAAC), thiol-ene reaction, inverse electron demand Diels–Alder reaction (IEDDA), hydrazone click chemistry and the newly emerging sulfur fluoride exchange (SuFEx) reaction, has been a hot research topic in the field of chemistry since it was first reported in 2001 (Zhang et al., 2021a; Ashe, 2022). Computer-aided drug design (CADD) has attracted a lot of attention for its potential to accelerate and reduce the cost of the drug development process (Wu et al., 2020). In addition, natural products provide a variety of lead compounds and novel drugs, are worthy of further development. Furthermore, early and late-stage development of new drugs may be slowed down by problems such as poor target selectivity or side effects, toxicity, resistance, inappropriate physicochemical and pharmacokinetic properties. Therefore, we summarized the recent applications of click and computational chemistry in drug development such as click to effectively synthesize druggable candidates, synthesis and modification of natural products, targeted delivery systems including hydrogels, nanoparticles (NPs), carbon nanotubes (CNT), etc, and computer-aided drug discovery including molecular docking and molecular dynamics to identify target, virtual screening (VS.) and pharmacophore to found and optimize lead compounds, ADMET prediction as well as compounds synthesis, which are making a splash in new drug development, hopefully, providing new insights for the discovery of new drug from click and computational chemistry.
Click chemistry
Click to efficiently synthesize druggable candidates
The transformation of the active compound skeleton is a magic weapon for researchers to break through patent restrictions and improve the activity of compounds in the development of new drugs. Copper-catalyzed 1,3-dipolar cycloaddition (CuAAC) to form 1,2,3-triazoles is the most popular reaction in click chemistry. Recently, 1,2,3-triazole backbones with hydrogen bonds, moderate dipole moments and enhanced water solubility had been widely used to generate drug candidates of anti-tumor (Brown et al., 2022; Elganzory et al., 2022; Mohammed et al., 2022; Oekchuae et al., 2022; Oliveira et al., 2022; Mironov et al., 2023), anti-seizure (Bhattacherjee et al., 2022), anti-diabetic (Dhameja et al., 2022), anti-parasitic (Aljohani et al., 2022), anti-bacterial (Daher et al., 2022; Mokariya et al., 2022; Nsira et al., 2022) and anti-viral (Kutkat et al., 2022; Tatarinov et al., 2022) via CuAAC click chemistry (Figure 1A).
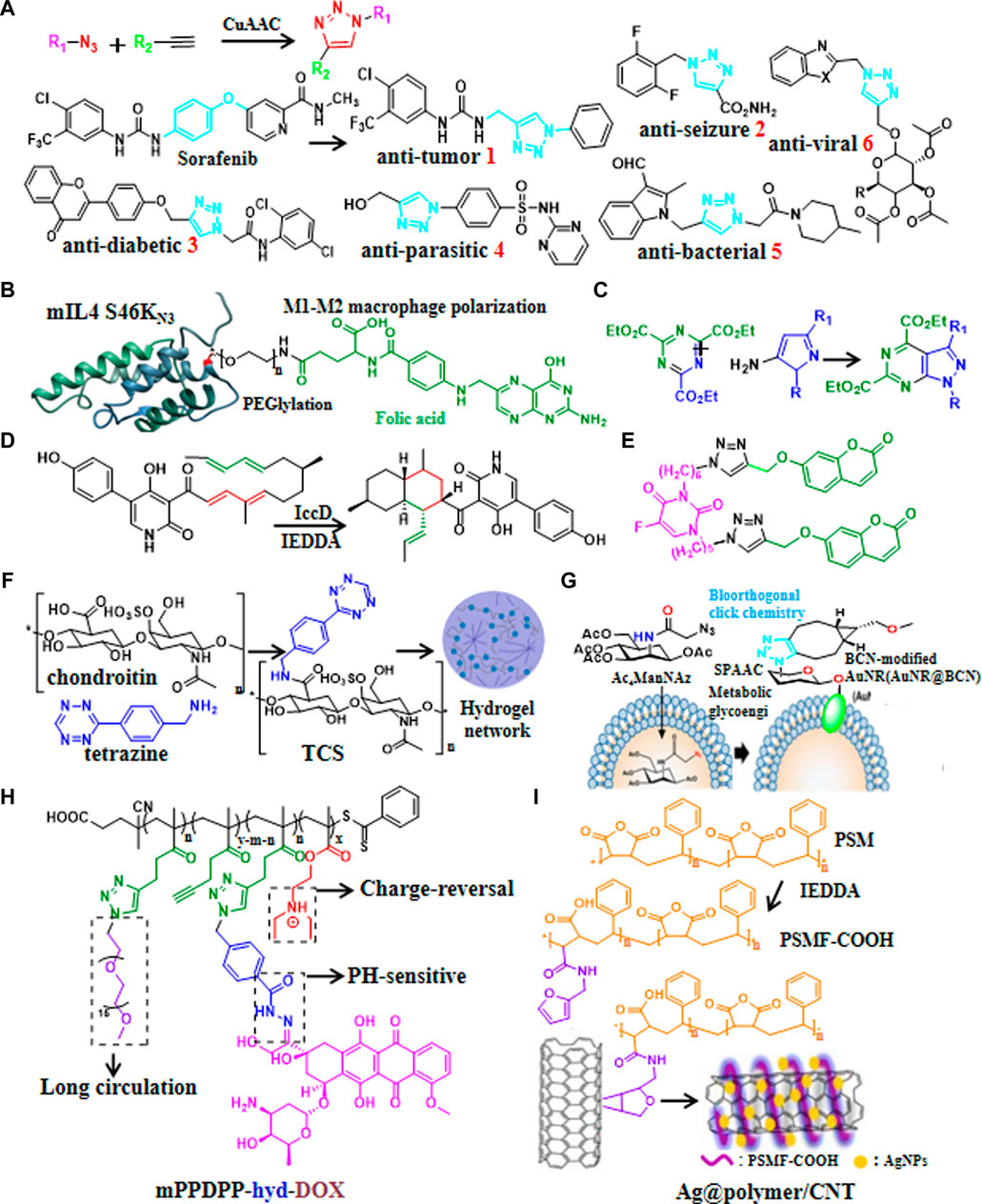
FIGURE 1. Recent updates in click chemistry for drug discovery and development. (A) Reaction formula of CuAAC and some recent applications of CuAAC for developing drug candidates containing 1,2,3-triazoles ring. (B) An example of natural product modification to improve half-life and target IL-4 to arthritic joint via SPAAC. (C) An example of introduction aromatic heterocycles via IEDDA. (D) An example of efficient synthesis of natural products via IEDDA. (E) An example of the generation of anti-cancer drug candidate by modification of the natural product coumarin via CuAAC. (F) Catalyst-free click chemistry to generate chondroitin sulfate-multiarmed PEG hydrogels for skin tissue engineering. (G) An example of the generation of MSCs-mediated deep tumor delivery of gold nanorod for anti-tumor therapy via SPAAC. (H) An example of polymer nanomicelle platform for cancer treatment via CuAAC. (I) An example of the generation of silver nanoparticle-supported polymer-encapsulated carbon nanotubes (CNTs) via IEDDA for nonenzymatic glucose sensing and antimicrobial activity applications.
Synthesis and modification of natural products
Natural products have provide abundant resources for drug discovery. Recently, click chemistry had been adopted for synthesis and modification of natural products, for instances, SPAAC was used to modularly generate Bcl-xL inhibitor (Brauer et al., 2022), adjust PEG chain length and targeting moiety to further improve half-life as well as targeting IL-4 to arthritic joint (Figure 1B) (Spieler et al., 2020). It was reported that poly (globalide-co-ε-caprolactone) could be functionalized with N-acetylcysteine side chains via thiol-ene reaction (Guindani et al., 2019). Furthermore, IEDDA could be used to introduce aromatic heterocycles (Figure 1C) (Xu et al., 2020) and triazines (Zhang et al., 2021b). Similarly, the synthetic efficiency of biosynthesis of anti-fungal drug candidate Ilicicolin H increased 3 × 105 times via IEDDA (Figure 1D) (Zhang et al., 2019). Moreover, 5-fluorouracil-coumarin conjugation (Figure 1E) as anti-cancer drug candidate (ópez et al., 2022) and pH responsive doxorubicin delivery polymers nano-particles (Wallat et al., 2018)for treatment of breast and ovarian cancer were generated by modification of natural products via CuAAC. In addition, quercetin-gold quantum dots for adenocarcinoma treatment (Pansare et al., 2022) and chondroitin sulfate-multiarmed PEG hydrogels for skin tissue engineering (Sousa et al., 2022) had been developed by modification of natural products (Figure 1F).
Targeted delivery systems
Existing drugs may have dis-advantages such as low selectivity, long synthetic routes, poor stability and side effects, thence the development of targeted delivery systems make great sense. Recently click-generated hydrogels had broad applications in the fields of anti-tumor (Ali et al., 2022; Bonardd et al., 2022), wound repair (Basurto et al., 2022) and long term regeneration therapy (Jang et al., 2021) via IEDDA, CuAAC,thiol-ene reaction, and SuFEx, respectively. Biomimetic stiffening of cell-laden hydrogels via sequential thiol-ene and hydrazone click reactions (Chang et al., 2021). Furthermore, nanoscale covalent organic frameworks (COFs) (Guan et al., 2022), Nisin-shelled nanoemulsion (Hashad et al., 2022), and MSCs-mediated deep tumor delivery of gold nanorod (Figure 1G) (Yun et al., 2022) had been synthesized for anti-tumor therapy via thiol-ene reactions, SPAAC, and SPAAC, respectively. Moreover, pH-sensitive polysaccharide-gold nanorod conjugate (Hou et al., 2019) and polymer nanomicelle platform (Figure 1H) (Liao et al., 2021) were reported to treat cancers via hydrazone click reaction and CuAAC, respectively. In addition, silver nanoparticle-supported polymer-wrapped carbon nanotubes (CNT) (Cao et al., 2022) for non-enzymatic glucose sensing and antimicrobial applications (Figure 1I), COF-based nanoreactors for click-activated pro-drug delivery and precise anti-vascular therapy (Wang et al., 2022) had been synthesized via IEDDA, these click chemistry-based targeting strategies may find widespread application in drug delivery in the future.
Computational chemistry in drug discovery
To effectively and efficiently design and develop new drugs, computational methods had been applied for drug design including target identification, seeking out and optimizing lead compounds prediction of pharmacokinetic and toxicological properties as well as compound synthesis by molecular docking and molecular dynamics, virtual screening, pharmacophore and ADMET prediction. Novel quinazoline derivative 1 as tubulin polymerization inhibitor (Dwivedi et al., 2022), PARP-1 inhibitor 2 (Syam et al., 2022), CDK2 inhibitor 3 (Qayed et al., 2022), HDAC-1-3 inhibitor 4 (Cheshmazar et al., 2022), VEGFR-2 inhibitor 5 (Taghour et al., 2022) were identified for cancer therapies. Furthermore, AChE inhibitor 6 (Macedo Vaz et al., 2022) for treatment of Alzheimer’s disease and Mtb RNAP inhibitor 7 (Mekonnen Sanka et al., 2022) for antitubercular and antimicrobial treatment were deserve further study. Moreover, a lead compound 8 of DDP4 inhibitor (Maslov et al., 2022) and acetamide derivative 9 (Zhou et al., 2022) as P2Y14R antagonist were considered as drug candidates for treating type 2 diabetes and gout, respectively. Additionally, potential SARS-CoV-2 main protease inhibitor 10 (Dong et al., 2023) and carbazole alkaloids from Murraya koenigii (Wadanambi et al., 2023) were identified as a promising drug candidates for inhibiting coronavirus infection. Surprisingly, it had been reported a computationally guided asymmetric total synthesis of resveratrol dimers, which possessed a wide range of biological activities such as antioxidant, anti-tumor and cardiovascular activities (Nakajima et al., 2022), suggesting that computationally guided organic synthesis may be a powerful strategy to advance the chemistry of natural products (Figure 2).
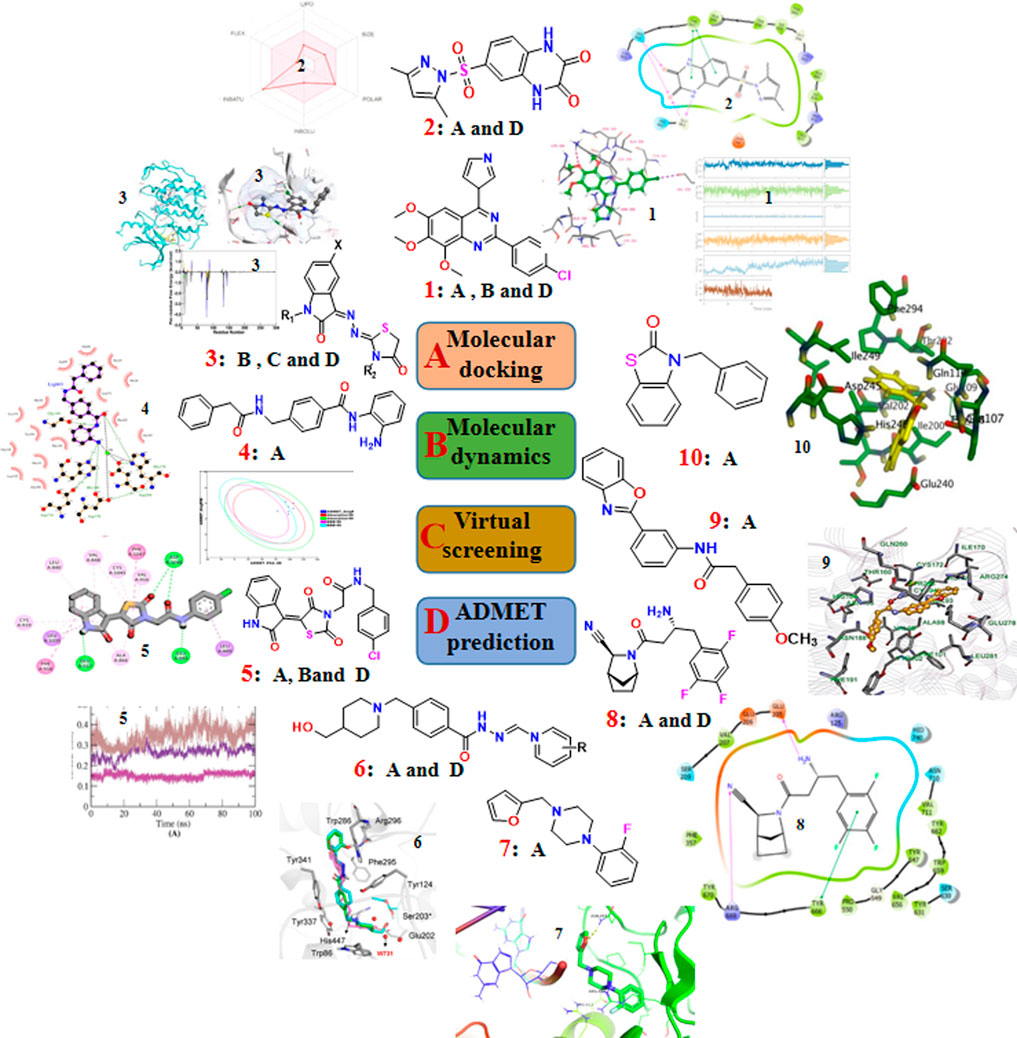
FIGURE 2. The recent updates of computational chemistry in target identification, lead compound discovery and ADMET prediction for drug development.
Conclusion and prospects
In the review, we summarized recent updates in click chemistry for drug discovery and development, including chemical click synthesis of druggable candidates, synthesis and modification of natural products, targeted delivery systems. In addition, we introduced updated computational chemistry in drug discovery for target identification, discovery and optimization of lead compounds, compounds synthesis and prediction of pharmacokinetic and toxicological properties. Click chemistry is a very powerful tool in drug discovery, in which the synthesis of 1,2,3-triazole ring as a pharmacophore, bioisostere via CuAAC has great potential in the drug design for a variety of diseases, however, 1,2,3-triazole ring itself is not a commonly used pharmacophore, and it is rare in marketed drugs, indicating that the use of 1,2,3-triazole as drug molecules still has certain limitations. Furthermore, the CuAAC reaction introduces copper species into biological systems and organisms, leading to potential toxicity issues while many Cu chelation sites may inhibit catalyst activity. Moreover, Copper-free cycloaddition SPAAC reaction and IEDDA reaction have their own issues: for example, they are susceptible to side reactions with nucleophilic residues (e.g., thiol residues in glutathione), and the reactive (electrophilic) nature of the requisite cyclic alkynes/alkenes may result in poor regiospecificity. Although computer molecular docking and molecular dynamics have important applications for target identification, however, the protein used for molecular docking may have a huge unknown difference from the protein in the pathological state due to site mutation. Additionally, computational chemistry needs to be combined with more biological activity test and mechanism exploration. In a word, although click and computational chemistry have shortcomings, which still hold a great and unnegligible potential for drug discovery and development, hopefully, this review can stimulate new ideas for the development of drugs with high selectivity, low toxicity, good stability and their clinical application in the near future.
Author contributions
JC and XZ: Writing-original draft. XL and YZ: proof-reading and editing. XTL and others: data collection, the article was approved for submission by all authors.
Funding
This study was supported by the grants received from the Macau Science and Technology Development fund (FDCT (file no.0021/2020/AGJ, 0011/2020/A1, 0012/2021/AMJ, 003/2022/ALC, 0092/2022/A2). The National Natural Science Foundation of China (Nos. 81973320).
Acknowledgments
The authors are grateful to the Macau University of Science and Technology and the State Key Laboratory of Quality Research in Chinese Medicine (Macau, China) for support.
Conflict of interest
The authors declared that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Ali, I., Gulfam, M., Jo, S. H., Seo, J. W., Rizwan, A., Park, S. H., et al. (2022). Reduction-responsive and bioorthogonal carboxymethyl cellulose based soft hydrogels cross-linked via IEDDA click chemistry for cancer therapy application. Int. J. Biol. Macromol. 219, 109–120. doi:10.1016/j.ijbiomac.2022.07.229
Aljohani, F. S., Rezki, N., Aouad, M. R., Elwakil, B. H., Hagar, M., Sheta, E., et al. (2022). Synthesis, characterization and nanoformulation of novel sulfonamide-1,2,3-triazole molecular conjugates as potent antiparasitic agents. Int. J. Mol. Sci. 23 (8), 4241. doi:10.3390/ijms23084241
Ashe, K. (2022). Chemistry just a click away. Nat. Chem. 14 (12), 1341. doi:10.1038/s41557-022-01108-7
Basurto, I. M., Passipieri, J. A., Gardner, G. M., Smith, K. K., Amacher, A. R., Hansrisuk, A. I., et al. (2022). Photoreactive hydrogel stiffness influences volumetric muscle loss repair. Tissue Eng. Part A 28 (7-8), 312–329. doi:10.1089/ten.TEA.2021.0137
Bhattacherjee, D., Kovalev, I. S., Kopchuk, D. S., Rahman, M., Santra, S., Zyryanov, G. V., et al. (2022). Mechanochemical approach towards multi-functionalized 1,2,3-triazoles and anti-seizure drug rufinamide analogs using copper beads. Molecules 27 (22), 7784. doi:10.3390/molecules27227784
Bonardd, S., Maiti, B., Grijalvo, S., Rodriguez, J., Enshaei, H., Kortaberria, G., et al. (2022). Biomass-derived isosorbide-based thermoresponsive hydrogel for drug delivery. Soft Matter 18 (26), 4963–4972. doi:10.1039/d2sm00623e
Brauer, J., Mötzing, M., Gröst, C., Hoffmann, R., and Berg, T. (2022). Templated generation of a bcl-xL inhibitor by isomer-free SPAAC based on azacyclonon-5-yne. Chemistry 28 (66), e202202259. doi:10.1002/chem.202202259
Brown, T., Cao, M., and Zheng, Y. G. (2022). Synthesis and activity of triazole-adenosine analogs as protein arginine methyltransferase 5 inhibitors. Molecules 27 (12), 3779. doi:10.3390/molecules27123779
Cao, X. T., Ngan Tran, T. Q., Ngo, D. H., Tai, D. C., and Kumar, S. (2022). Click-chemistry-mediated synthesis of silver nanoparticle-supported polymer-wrapped carbon nanotubes: Glucose sensor and antibacterial material. ACS Omega 7 (42), 37095–37102. doi:10.1021/acsomega.2c02832
Chang, C. Y., Johnson, H. C., Babb, O., Fishel, M. L., and Lin, C. C. (2021). Biomimetic stiffening of cell-laden hydrogels via sequential thiol-ene and hydrazone click reactions. Acta Biomater. 130, 161–171. doi:10.1016/j.actbio.2021.05.054
Cheshmazar, N., Hemmati, S., Hamzeh-Mivehroud, M., Sokouti, B., Zessin, M., Schutkowski, M., et al. (2022). Development of new inhibitors of HDAC1-3 enzymes aided by in silico design strategies. J. Chem. Inf. Model 62 (10), 2387–2397. doi:10.1021/acs.jcim.1c01557
Daher, S. S., Lee, M., Jin, X., Teijaro, C. N., Barnett, P. R., Freundlich, J. S., et al. (2022). Alternative approaches utilizing click chemistry to develop next-generation analogs of solithromycin. Eur. J. Med. Chem. 233, 114213. doi:10.1016/j.ejmech.2022.114213
Dhameja, M., Kumar, H., Kurella, S., Uma, A., and Gupta, P. (2022). Flavone-1,2,3-triazole derivatives as potential α-glucosidase inhibitors: Synthesis, enzyme inhibition, kinetic analysis and molecular docking study. Bioorg Chem. 127, 106028. doi:10.1016/j.bioorg.2022.106028
Dong, J., Varbanov, M., Philippot, S., Vreken, F., Zeng, W. B., and Blay, V. (2023). Ligand-based discovery of coronavirus main protease inhibitors using MACAW molecular embeddings. J. Enzyme Inhib. Med. Chem. 38 (1), 24–35. doi:10.1080/14756366.2022.2132486
Dwivedi, A. R., Rawat, S. S., Kumar, V., Kumar, N., Anand, P., Yadav, R. P., et al. (2022). Synthesis and screening of novel 4-N-heterocyclic-2-aryl-6,7,8-trimethoxyquinazolines as antiproliferative and tubulin polymerization inhibitors. Bioorg Med. Chem. 72, 116976. doi:10.1016/j.bmc.2022.116976
Elganzory, H. H., Alminderej, F. M., El-Bayaa, M. N., Awad, H. M., Nossier, E. S., and El-Sayed, W. A. (2022). Design, synthesis, anticancer activity and molecular docking of new 1,2,3-triazole-based glycosides bearing 1,3,4-thiadiazolyl, indolyl and arylacetamide scaffolds. Molecules 27 (20), 6960. doi:10.3390/molecules27206960
Guan, Q., Zhou, L. L., Zhou, W., and Dong, Y. B. (2022). A vinyl-decorated covalent organic framework for ferroptotic cancer therapy via visible-light-triggered cysteine depletion. J. Mater Chem. B 10 (43), 8894–8909. doi:10.1039/d2tb01815b
Guindani, C., Dozoretz, P., Araújo, P. H. H., Ferreira, S. R. S., and de Oliveira, D. (2019). N-acetylcysteine side-chain functionalization of poly(globalide-co-ε-caprolactone) through thiol-ene reaction. Mater Sci. Eng. C Mater Biol. Appl. 94, 477–483. doi:10.1016/j.msec.2018.09.060
Hashad, R. A., Singla, R., Kaur Bhangu, S., Jap, E., Zhu, H., Peleg, A. Y., et al. (2022). Chemoenzymatic surface decoration of Nisin-shelled nanoemulsions: Novel targeted drug-nanocarriers for cancer applications. Ultrason. Sonochem 90, 106183. doi:10.1016/j.ultsonch.2022.106183
Hou, G., Qian, J., Xu, W., Sun, T., Wang, Y., Wang, J., et al. (2019). A novel pH-sensitive targeting polysaccharide-gold nanorod conjugate for combined photothermal-chemotherapy of breast cancer. Carbohydr. Polym. 212, 334–344. doi:10.1016/j.carbpol.2019.02.045
Jang, K. J., Lee, W. S., Park, S., Han, J., Kim, J. E., Kim, B. M., et al. (2021). Sulfur(VI) fluoride exchange (SuFEx)-Mediated synthesis of the chitosan-PEG conjugate and its supramolecular hydrogels for protein delivery. Nanomater. (Basel) 11 (2), 318. doi:10.3390/nano11020318
Kutkat, O., Kandeil, A., Moatasim, Y., Elshaier, Y. A. M. M., El-Sayed, W. A., Gaballah, S. T., et al. (2022). In vitro and in vivo antiviral studies of new heteroannulated 1,2,3-triazole glycosides targeting the neuraminidase of influenza A viruses. Pharm. (Basel) 15 (3), 351. doi:10.3390/ph15030351
Liao, J., Peng, H., Liu, C., Li, D., Yin, Y., Lu, B., et al. (2021). Dual pH-responsive-charge-reversal micelle platform for enhanced anticancer therapy. Mater Sci. Eng. C Mater Biol. Appl. 118, 111527. doi:10.1016/j.msec.2020.111527
Macedo Vaz, S., de Freitas Silva, M., Dos Reis Rosa Franco, G., Jorge R. Guimaraes, M., Motta R. da Silva, F., Goncalves Castro, N., et al. (2022). Synthesis and biological evaluation of 4-hydroxy-methylpiperidinyl-N-benzyl-acylarylhydrazone hybrids designed as novel multifunctional drug candidates for Alzheimer's disease. Bioorg Med. Chem. 71, 116952. doi:10.1016/j.bmc.2022.116952
Maslov, I. O., Zinevich, T. V., Kirichenko, O. G., Trukhan, M. V., Shorshnev, S. V., Tuaeva, N. O., et al. (2022). Design, synthesis and biological evaluation of neogliptin, a novel 2-azabicyclo[2.2.1]heptane-based inhibitor of dipeptidyl peptidase-4 (DPP-4). Pharm. (Basel) 15 (3), 273. doi:10.3390/ph15030273
Mekonnen Sanka, B., Mamo Tadesse, D., Teju Bedada, E., Mengesha, E. T., and Babu, G. N. (2022). Design, synthesis, biological screening and molecular docking studies of novel multifunctional 1,4-di (aryl/heteroaryl) substituted piperazine derivatives as potential antitubercular and antimicrobial agents. Bioorg Chem. 119, 105568. doi:10.1016/j.bioorg.2021.105568
Mironov, M. E., Rybalova, T. V., Pokrovskii, M. A., Emaminia, F., Gandalipov, E. R., Pokrovskii, A. J., et al. (2023). Synthesis of fully functionalized spirostanic 1,2,3-triazoles by the three component reaction of diosgenin azides with acetophenones and aryl aldehydes and their biological evaluation as antiproliferative agents. Steroids 190, 109133. doi:10.1016/j.steroids.2022.109133
Mohammed, H. H. H., Abd El-Hafeez, A. A., Ebeid, K., Mekkawy, A. I., Abourehab, M. A. S., Wafa, E. I., et al. (2022). New 1,2,3-triazole linked ciprofloxacin-chalcones induce DNA damage by inhibiting human topoisomerase I& II and tubulin polymerization. J. Enzyme Inhib. Med. Chem. 37 (1), 1346–1363. doi:10.1080/14756366.2022.2072308
Mokariya, J. A., Kalola, A. G., Prasad, P., and Patel, M. P. (2022). Simultaneous ultrasound- and microwave-assisted one-pot 'click' synthesis of 3-formyl-indole clubbed 1,2,3-triazole derivatives and their biological evaluation. Mol. Divers 26 (2), 963–979. doi:10.1007/s11030-021-10212-8
Nakajima, M., Adachi, Y., and Nemoto, T. (2022). Computation-guided asymmetric total syntheses of resveratrol dimers [published correction appears in Nat Commun. 2022 Apr 27;13(1):2418. Nat. Commun. 13 (1), 152. doi:10.1038/s41467-021-27546-4
Nsira, A., Mtiraoui, H., Chniti, S., Al-Ghulikah, H., Gharbi, R., and Msaddek, M. (2022). Regioselective one-pot synthesis, biological activity and molecular docking studies of novel conjugates N-(p-Aryltriazolyl)-1,5-benzodiazepin-2-ones as potent antibacterial and antifungal agents. Molecules 27 (13), 4015. doi:10.3390/molecules27134015
Oekchuae, S., Sirirak, J., Charoensuksai, P., Wongprayoon, P., Chuaypen, N., Boonsombat, J., et al. (2022). The design and synthesis of a new series of 1,2,3-triazole-cored structures tethering aryl urea and their highly selective cytotoxicity toward HepG2. Pharm. (Basel) 15 (5), 504. doi:10.3390/ph15050504
Oliveira, A., Moura, S., Pimentel, L., Neto, J., Dantas, R., Silva-Jr, F., et al. (2022). New imatinib derivatives with antiproliferative activity against A549 and K562 cancer cells. Molecules 27 (3), 750. doi:10.3390/molecules27030750
ópez, S., Gracia, I., Plaza-Pedroche, R., Rodriguez, J. F., Perez-Ortiz, J. M., Rodriguez-Lopez, J., et al. (2022). In vitro antioxidant and pancreatic anticancer activity of novel 5-fluorouracil-coumarin conjugates. Pharmaceutics 14 (10), 2152. doi:10.3390/pharmaceutics14102152
Pansare, A. V., Pansare, P. V., Shedge, A. A., Pansare, S. V., Patil, V. R., Terrasi, G. P., et al. (2022). Click gold quantum dots biosynthesis with conjugation of quercetin for adenocarcinoma exertion. RSC Adv. 12 (29), 18425–18430. doi:10.1039/d2ra02529a
Qayed, W. S., Hassan, M. A., El-Sayed, W. M., Rogério A Silva, J., and Aboul-Fadl, T. (2022). Novel azine linked hybrids of 2-indolinone and thiazolodinone scaffolds as CDK2 inhibitors with potential anticancer activity: In silico design, synthesis, biological, molecular dynamics and binding free energy studies. Bioorg Chem. 126, 105884. doi:10.1016/j.bioorg.2022.105884
Sousa, G. F., Afewerki, S., Dittz, D., Santos, F. E. P., Gontijo, D. O., Scalzo, S. R. A., et al. (2022). Catalyst-free click chemistry for engineering chondroitin sulfate-multiarmed PEG hydrogels for skin tissue engineering. J. Funct. Biomater. 13 (2), 45. doi:10.3390/jfb13020045
Spieler, V., Ludwig, M. G., Dawson, J., Tigani, B., Littlewood-Evans, A., Safina, C., et al. (2020). Targeting interleukin-4 to the arthritic joint. J. Control Release 326, 172–180. doi:10.1016/j.jconrel.2020.07.005
Syam, Y. M., Anwar, M. M., Abd El-Karim, S. S., Elokely, K. M., and Abdelwahed, S. H. (2022). New quinoxaline-based derivatives as PARP-1 inhibitors: Design, synthesis, antiproliferative, and computational studies. Molecules 27 (15), 4924. doi:10.3390/molecules27154924
Taghour, M. S., Elkady, H., Eldehna, W. M., El-Deeb, N., Kenawy, A. M., Elkaeed, E. B., et al. (2022). Design, synthesis, anti-proliferative evaluation, docking, and MD simulations studies of new thiazolidine-2,4-diones targeting VEGFR-2 and apoptosis pathway. PLoS One 17 (9), e0272362. doi:10.1371/journal.pone.0272362
Tatarinov, D. A., Garifullin, B. F., Belenok, M. G., Andreeva, O. V., Strobykina, I. Y., Shepelina, A. V., et al. (2022). The first 5'-phosphorylated 1,2,3-triazolyl nucleoside analogues with uracil and quinazoline-2,4-dione moieties: A synthesis and antiviral evaluation. Molecules 27 (19), 6214. doi:10.3390/molecules27196214
Wadanambi, P. M., Jayathilaka, N., and Seneviratne, K. N. (2023). A computational study of carbazole alkaloids from Murraya koenigii as potential SARS-CoV-2 main protease inhibitors. Appl. Biochem. Biotechnol. 195 (1), 573–596. doi:10.1007/s12010-022-04138-6
Wallat, J. D., Harrison, J. K., and Pokorski, J. K. (2018). pH responsive doxorubicin delivery by fluorous polymers for cancer treatment. Mol. Pharm. 15 (8), 2954–2962. doi:10.1021/acs.molpharmaceut.7b01046
Wang, P., Li, M., Zhou, F., Yang, Y., Yin, X., Zhang, X. B., et al. (2022). COF-based nanoreactors for click-activated prodrug delivery and precise anti-vascular therapy. Chem. Commun. (Camb). 58 (79), 11107–11110. doi:10.1039/d2cc03931a
Wu, F., Zhou, Y., Li, L., Shen, X., Chen, G., Wang, X., et al. (2020). Computational approaches in preclinical studies on drug discovery and development. Front. Chem. 8, 726. doi:10.3389/fchem.2020.00726
Xu, G., Bai, X., and Dang, Q. (2020). Aromatic heterocycles as productive dienophiles in the inverse electron-demand diels-alder reactions of 1,3,5-triazines. Acc. Chem. Res. 53 (4), 773–781. doi:10.1021/acs.accounts.9b00604
Yun, W. S., Shim, M. K., Lim, S., Song, S., Kim, J., Yang, S., et al. (2022). Mesenchymal stem cell-mediated deep tumor delivery of gold nanorod for photothermal therapy. Nanomater. (Basel). 12 (19), 3410. doi:10.3390/nano12193410
Zhang, F. G., Chen, Z., Tang, X., and Ma, J. A. (2021). Triazines: Syntheses and inverse electron-demand diels-alder reactions. Chem. Rev. 121 (23), 14555–14593. doi:10.1021/acs.chemrev.1c00611
Zhang, X., Zhang, S., Zhao, S., Wang, X., Liu, B., and Xu, H. (2021). Click chemistry in natural product modification. Front. Chem. 9, 774977. doi:10.3389/fchem.2021.774977
Zhang, Z., Jamieson, C. S., Zhao, Y. L., Li, D., Ohashi, M., Houk, K. N., et al. (2019). Enzyme-catalyzed inverse-electron demand diels–alder reaction in the biosynthesis of antifungal Ilicicolin H. J. Am. Chem. Soc. 141 (14), 5659–5663. doi:10.1021/jacs.9b02204
Keywords: click chemistry, computational chemistry, CADD, druggable candidates, drug development
Citation: Cai JH, Zhu XZ, Guo PY, Rose P, Liu XT, Liu X and Zhu YZ (2023) Recent updates in click and computational chemistry for drug discovery and development. Front. Chem. 11:1114970. doi: 10.3389/fchem.2023.1114970
Received: 03 December 2022; Accepted: 27 January 2023;
Published: 07 February 2023.
Edited by:
Lhassane Ismaili, Université Bourgogne Franche-Comté, FranceReviewed by:
Mohamed Benchekroun, Aelis Farma, FranceCopyright © 2023 Cai, Zhu, Guo, Rose, Liu, Liu and Zhu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yi Zhun Zhu, eXp6aHVAbXVzdC5lZHUubW8=
†These authors have contributed equallyto this work
 Jiang Hong Cai
Jiang Hong Cai Xuan Zhe Zhu1†
Xuan Zhe Zhu1† Peter Rose
Peter Rose Xia Liu
Xia Liu Yi Zhun Zhu
Yi Zhun Zhu