- 1School of Pharmaceutical Sciences, South-Central University for Nationalities, Wuhan, China
- 2College of Chemistry and Material Sciences, South-Central University for Nationalities, Wuhan, China
- 3Guangxi International Zhuang Medical Hospital, Nanning, China
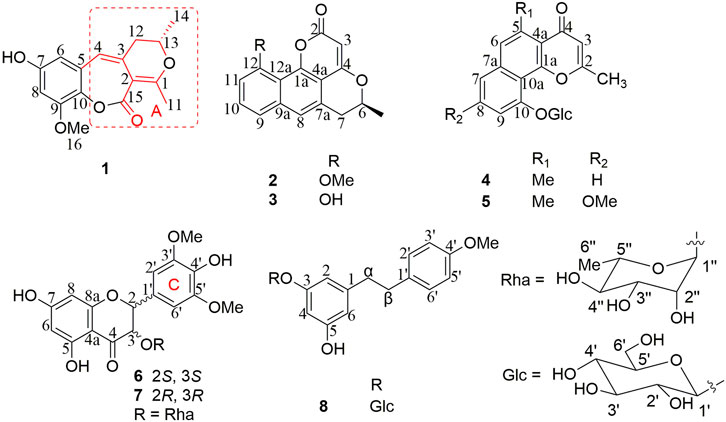
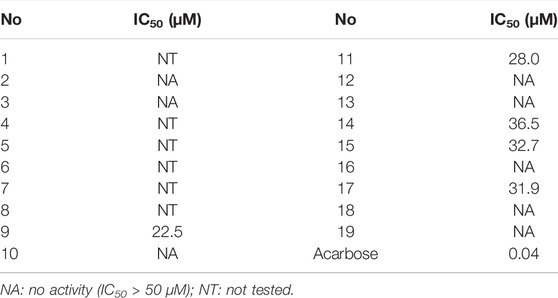
Eight new phenolic compounds, named bercheminols A-H (1–8), and eleven known analogues were isolated from the stems and leaves of Berchemia lineata (L.) DC. Their structures including the absolute configurations were elucidated by extensive spectroscopic analysis, chemical method, and quantum chemical calculations. Compound 1 possesses an unprecedented 3,4-dihydro-11H-benzo[b]pyrano[4,3-e] oxepin-11-one skeleton. The other new compounds belong to three structural types of natural products, including naphthopyrones (2–5), flavonoids (6–7), and bibenzyl (8). The α-glucosidase inhibitory activities of the isolated compounds were assayed. As a result, vittarin-B (9), rubrofusarin-6-O-β-D-glucopyranoside (11), quercetin (14), kaempferol (15), and dihydrokaempferol (17) showed moderate inhibitory activities against α-glucosidase with IC50 values of 22.5, 28.0, 36.5, 32.7, and 31.9 μM, respectively.
Introduction
Berchemia lineata (L.) DC. belongs to the genus of Berchemia, defined as the plant origin of “Tiebaojin” in Guangxi Traditional Chinese Medicine Standard. “Tiebaojin” is an important ethnic medicine commonly used in Guangxi Zhuang nationality and southwest minority areas of China. It can be used for the treatment of pulmonary tuberculosis hemoptysis, icteric hepatitis, abdominal pain, traumatic injury, snake bite, etc. Through investigation, it is found that the actual source of the medicinal materials of “Tiebaojin” commonly used clinically in the Zhuang region of Guangxi mainly include B. lineata, B. floribunda, B. polyphylla Wall. ex Laws., B. polyphylla var. leioclada (Zhang et al., 2011; Jing et al., 2017). Previous phytochemical studies on B. floribunda and B. polyphylla showed that they mainly include flavonoids, glycosides, lignans, quinones, and terpenoids (Chen et al., 2006). However, there are few studies on the chemical constituents of B. lineata, only some report chromones, flavonoids, and lignans (Shen et al., 2010a; Shen, et al., 2010b; Li et al., 2016; Jiang et al., 2019). To further search for new active compounds from B. lineata, phytochemical investigations of an extract of the stems and leaves of this plant afforded eight new phenolic compounds and eleven known analogues (Figure 1). This study reported the isolation, structure identification, and biological activity of these compounds.
Materials and Methods
General Experimental Procedures
The optical rotation was measured in MeOH by an Autopol IV polarimeter (Rudolph Research Analytical, Hackettstown, NJ, United States). UV spectra were obtained by a UH5300 UV–VIS double beam spectrophotometer (Hitachi Co., Tokyo, Japan). 1D and 2D NMR spectra were obtained by a Bruker AVANCE IIITM 500 and 600 MHz spectrometers (Bruker, Ettlingen, Germany) in methanol-d4, DMSO-d6 using TMS as internal standard. HR-ESI-MS data was obtained on a Thermo Scientific Q Exactive Orbitrap LC-MS/MS System (Thermo Scientific, Waltham, MA, United States). An Ultimate 3000 HPLC system (Dionex Co., Sunnyvale, CA, United States) with an UltiMate 3000 pump and UltiMate 3000 variable wavelength detector was employed to carry out semi-preparative HPLC, with a Nacalai Tesque 5C18-MS-II column (250 × 10 mm, 5 μm). Silica gel (200–300 mesh and 300–400 mesh) for open column chromatography (CC) was purchased from Qingdao Haiyang Chemical Group Co., Ltd. (Qingdao, China). The organic solvents were purchased from Sinopharm Chemical Reagent Co., Ltd. (Shanghai, China). Reagents used for α-glucosidase inhibitory assay (α-glucosidase, 4-nitrophenyl-α-D-glucopyranoside, and acarbose) were purchased from Shanghai Yuanye Biology Co., Ltd. (Shanghai, China), and the absorbance was measured by a full-wavelength microplate reader (Thermo Fisher Scientific Shier Technology Co., Ltd.).
Plant Material
The stem and leaves of Berchemia lineata (L.) DC. were purchased from Nanning, Guangxi Zhuang autonomous Region, P. R. China, and were identified by Prof. Hongli Teng, Guangxi Zhuang Medicine International Hospital. The voucher specimen was deposited in the herbarium of the School of Pharmaceutical Sciences, South Central University for Nationalities.
Extraction and Isolation
The stem and leaves of Berchemia lineata (L.) DC. (10 kg) were powdered and extracted using 70% EtOH three times for 24 h to obtain ethanol extract (700 g), which was then successively extracted with petroleum ether (PE), EtOAc, and n-butanol three times to obtain PE extract (57.31 g), EtOAc extract (100 g), and n-butanol extract (300 g). The n-butanol extract was chromatographed on a D-101 macroporous resin column, eluted successively with H2O-EtOH (7:3 to 1:9), and obtained 3 fractions (Fr.I-III). Fr.II (90 g) was chromatographed on a silica gel column, eluted successively with CH2Cl2-MeOH gradient (20:1, 9:1, 8:2, 7:3, 1:1) to obtain 12 fractions (Fr.II- A ∼ L). Fr.II-C was repeatedly prepared by semi-preparative HPLC to obtain 9 (CH3CN: H2O = 32: 68, tR 50.1 min, 3 mg), and compound 12 (7 mg) was obtained from Fr.II-H by recrystallization. Fr.II-F was subjected to ODS column chromatography with a gradient of H2O-MeOH (7:3 to 0:1) to obtain 11 fractions (Fr.II-F.1∼F.11). Fr.II-F.2 was repeatedly prepared by semi-preparative HPLC (CH3CN: H2O = 19 : 81) to yield 6 (tR 31.0 min, 1.5 mg) and 7 (tR 34.9 min, 1.3 mg). Compounds 10 (4.5 mg) and 11 (4 mg) were obtained after recrystallization from Fr.II-F.3 and F.4, respectively. The mother liquid of Fr.II-F.3 was subjected to Sephadex LH-20 column chromatography eluting with methanol, and then repeatedly prepared using semi-preparative HPLC (CH3CN: H2O = 19 : 81) to obtain compounds 4 (1 mg, tR 34.0 min), 5 (tR 46.7 min, 1.1 mg), and 8 (tR 25.6 min, 1 mg). Fr.II-J was subjected to ODS with a gradient of H2O-MeOH (7:3 to 0:1) and then repeatedly prepared by HPLC (CH3CN:H2O = 18:82) to obtain compound 13 (tR 42.1 min, 3.89 mg). The EtOAc extract was chromatographed on a silica gel column chromatography, eluted successively with PE/EtOAc gradient (15:1 to 1:1) to obtain 16 fractions (Fr.1∼Fr.16), and compound 14 (4.5 mg) was obtained from Fr.15 by recrystallization. Fr.9 was subjected to a silica gel column chromatography, eluted successively with PE/EtOAc gradient (20:1 to 1:1), and then repeatedly prepared by semi-preparative HPLC to obtain 19 (CH3CN:H2O = 50:50, tR 36.9 min, 4.9 mg). Fr.14 was subjected to ODS column chromatography with a gradient of H2O-MeOH (7:3 to 0:1) to obtain Fr.14.1∼Fr.14.14, and then repeatedly prepared by semi-preparative HPLC to obtain 1 (CH3CN:H2O = 30:70, tR 67.2 min, 1 mg), 2 (CH3CN:H2O = 40:60, tR 40.5 min, 4.5 mg), 3 (CH3CN:H2O = 40:60, tR 33.5 min, 2 mg), 15 (CH3CN:H2O = 40:60, tR 14.5 min, 12 mg), 16 (CH3CN:H2O = 35:65, tR 21.7 min, 6 mg), 17 (CH3CN:H2O = 25:75, tR 43.9 min, 3 mg), and 18 (CH3CN: H2O = 40:60, tR 22.9 min, 3.2 mg).
Spectroscopic Data
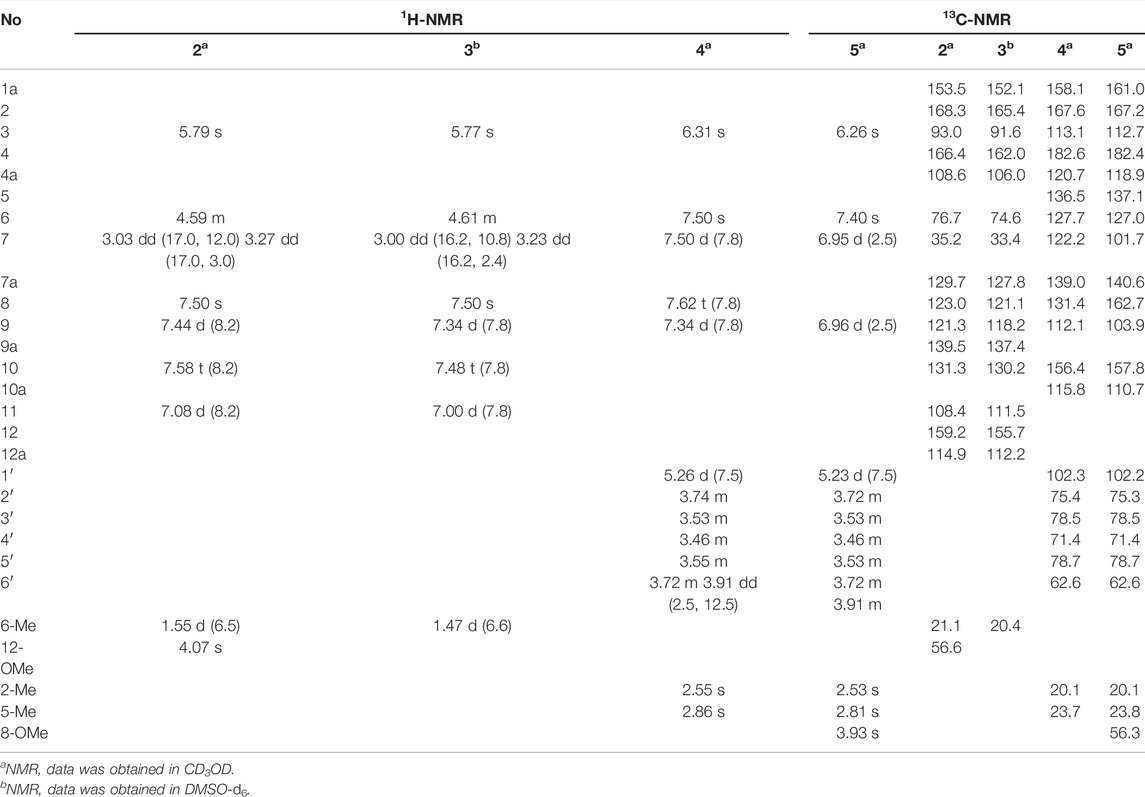
Bercheminol (1): brown amorphous powder; [α]20D -27.3 (c 0.05, MeOH); UV (MeOH) λmax (log ε) 210 (2.87), 295 (2.63) nm; ECD (MeOH) λ (θ) 227 (+14.58), 259 (+7.72), 307 (−11.78) nm; 1H NMR (500 MHz, CD3OD) δH 6.07 (1H, s, H-4), 6.15 (1H, d, J = 2.5 Hz, H-6), 6.40 (1H, d, J = 2.5 Hz, H-8), 2.31 (3H, s, CH3-11), 2.64 (1H, dd, J = 13.5, 3.5 Hz, H-12a), 2.35 (1H, dd, J = 13.5, 8.5, H-12b), 4.45 (1H, m, H-13), 1.34 (3H, d, J =6.5, CH3-14), 3.81 (3H, s, OMe); 13C NMR (125 MHz, CD3OD) δC 173.1 (C-1), 105.1 (C-2), 132.0 (C-3), 122.2 (C-4), 132.4 (C-5), 106.5 (C-6), 156.2 (C-7), 100.8 (C-8), 153.6 (C-9), 132.8 (C-10), 21.9 (C-11), 39.1 (C-12), 77.3 (C-13), 20.7 (C-14), 171.9 (C-15), 56.7 (OMe); HRESIMS m/z 289.10695 [M + H]+ (calcd for C16H17O5, 289.10705).
Bercheminol B (2): brown amorphous powder; [α]20D +15.6 (c 0.05, MeOH); UV (MeOH) λmax (log ε) 220 (2.87), 280 (2.61), 365 (2.10) nm; ECD (MeOH) λ (θ) 212 (+26.79), 228 (−14.00), 247 (+1.39), 286 (−5.44) nm; 1H and 13C NMR see Table 1; HRESIMS m/z 283.09594 [M + H]+ (calcd for C17H15O4, 283.09649).
Bercheminol C (3): brown amorphous powder; [α]20D +9.6 (c 0.05, MeOH); UV (MeOH) λmax (log ε) 220 (2.64), 285 (2.35) nm; ECD (MeOH) λ (θ) 212 (+2.65), 231 (-1.96) nm; 1H and 13C NMR see Table 1; HRESIMS m/z 269.08072 [M + H]+ (calcd for C16H13O4, 269.08084).
Bercheminol D (4): white amorphous powder; [α]20D -8.3 (c 0.02, MeOH); UV (MeOH) λmax (log ε) 225 (3.11), 255 (2.98), 360 (2.37) nm; 1H and 13C NMR (MeOH) Table 1; HRESIMS m/z 403.13870 [M + H]+ (calcd for C21H23O8, 403.13874).
Bercheminol E (5): white amorphous powder; [α]20D -1.1 (c 0.05, MeOH); UV (MeOH) λmax (log ε) 235 (2.36), 270 (2.49), 350 (1.90) nm; 1H and 13C NMR see Table 1; HRESIMS m/z 433.14941 [M + H]+ (calcd for C22H25O9, 433.14931).
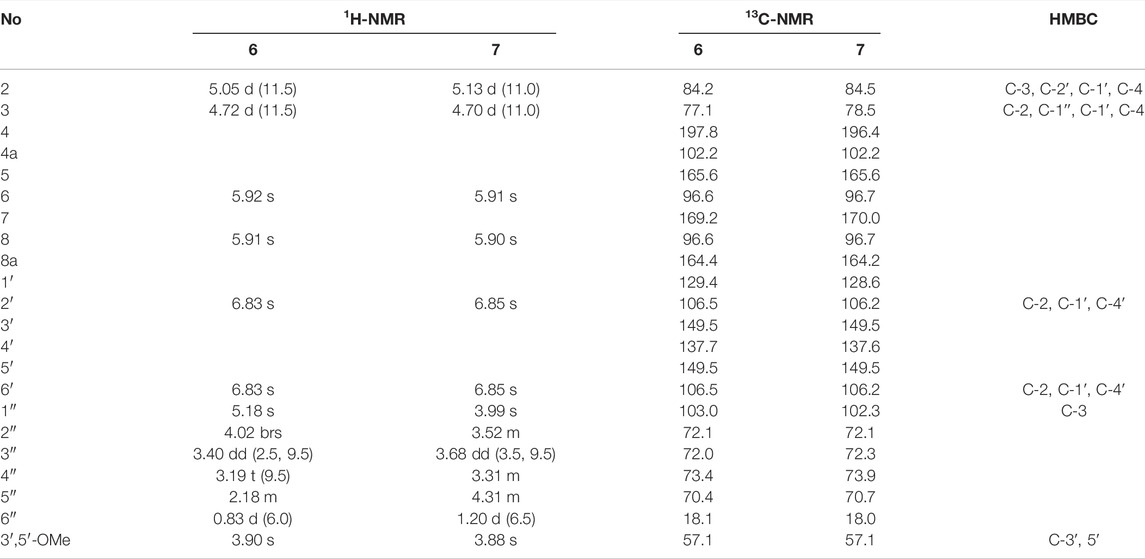
Bercheminol F (6): brown amorphous powder; [α]20D-65.0 (c 0.02, MeOH); UV (MeOH) λmax (log ε) 230 (3.84), 290 (3.79) nm; ECD (MeOH) λ (θ) 207 (+8.93), 220 (-16.13), 245 (+0.41), 260 (-2.24), 294 (+10.27), 329 (-3.36) nm; 1H and 13C NMR see Table 2; HRESIMS m/z 493.13760 [M-H]- (calcd for C23H25O12, 493.13515).
Bercheminol G (7): brown amorphous powder; [α]20D +17.8 (c 0.02, MeOH); UV (MeOH) λmax (log ε) 230 (3.81), 295 (3.70) nm; ECD (MeOH) λ (θ) 208 (-7.38), 229 (+9.17), 295 (-4.61), 321 (+2.54) nm; 1H and 13C NMR see Table 2; HRESIMS m/z 493.13589 [M-H]- (calcd for C23H25O12, 493.13515).
Bercheminol H (8): yellow amorphous powder; [α]20D +6.0 (c 0.05, MeOH); UV (MeOH) λmax (log ε) 210 (3.04), 275 (2.19) nm; 1H NMR (500 MHz, CD3OD) δH 6.39 (1H, br s, H-2), 6.37 (1H, t, J = 2.0 Hz, H-4), 6.29 (1H, br s, H-6), 7.06 (2H, d, J = 8.5 Hz, H-2′, 6′), 6.79 d (2H, d, J = 8.5 Hz, H-3′, 5′), 4.78 (1H, d, J = 7.5 Hz, H-1″), 3.40 (2H, m, H-2″, 6″), 3.41 (2H, m, H-3″, 5″), 3.38 (1H, m, H-4″), 3.88 (1H, d, J = 12.5 Hz, H2-6″a), 3.69 (1H, dd, J =5.0, 12.0, H2-6″b), 2.76 (4H, m H2-α, H2-β); 13C NMR (125 MHz, CD3OD) δC 145.6 (C-1), 110.9 (C-2), 159.4 (C-3), 102.8 (C-4), 160.3 (C-5), 109.4 (C-6), 135.2 (C-1′), 130.6 (C-2′), 114.8 (C-3′), 159.4 (C-4′), 114.8 (C-5′), 130.6 (C-6′), 102.4 (C-1″), 75.0 (C-2″), 78.2 (C-3″), 71.5 (C-4″), 78.2 (C-5″), 62.6 (C-6″), 39.6 (C-α), 37.0 (C-β), 55.8 (OMe); HRESIMS m/z 407.17007 [M + H]+ (calcd for C21H27O8, 407.17004).
α-Glucosidase Inhibitory Assay
According to the literature method (Özgünseven et al., 2021) with some modifications, 20 μL compounds at different concentrations reacted with 20 μL of 4-nitrophenyl-α-D-glucopyranoside (20 mM) and 20 μL of α-glucosidase (0.5 U/mL) in a 96-well plate at 37°C for 30 min. Na2CO3 (0.2%, 80 μL) was then added to terminate the reaction and the absorbance value was measured at 405 nm using a microplate reader. IC50 values were calculated from the graph of inhibition percentage against the logarithm of the concentrations of compounds.
Determination of Sugar
Compounds 4, 5, 6, 7, and 8 (0.5 mg)were refluxed with 2 ml of 4N HCl–dioxane (1 : 1) for 2 h, and then cooled to room temperature. The mixtures were extracted three times with 2 ml EtOAc. The aqueous layers were dried and refluxed with 0.5 ml pyridine and 1 mg L-cysteine methyl ester hydrochloride at 60 C for 1 h. 5 μL 2-methylphenyl isothiocyanate was added to the reaction mixtures and continued to reflux at 60 C for 1 h (Min et al., 2003; Tanaka, T., 2007). These dithiocarbamate derivatives of D-glucose and L-rhamnose were prepared in the same way. The reaction mixtures were analyzed by HPLC (column: YMC-Pack ODS-A 250 × 4.6 mm I.D.; CH3CN/H2O = 25: 75, wavelength: 250 nm; flow rate: 0.8 ml/min). The retention time of three sugar fractions of compounds 4, 5, and 8 were detected at 22.56, 22.40, and 22.43 min respectively, which were almost the same as that of D-glucose (22.61 min). The retention time of two sugar fractions of compounds 6 and 7 were detected at 25.52 and 25.49 min, respectively, which were almost the same as that of L-rhamnose (25.56 min). The results showed that compounds 4, 5, and 8 contained D-glucose, and compounds 6 and 7 contained L-rhamnose.
Results and Discussion
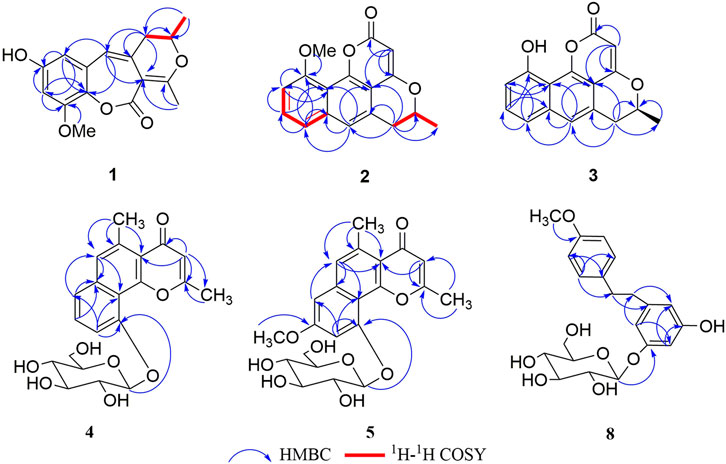
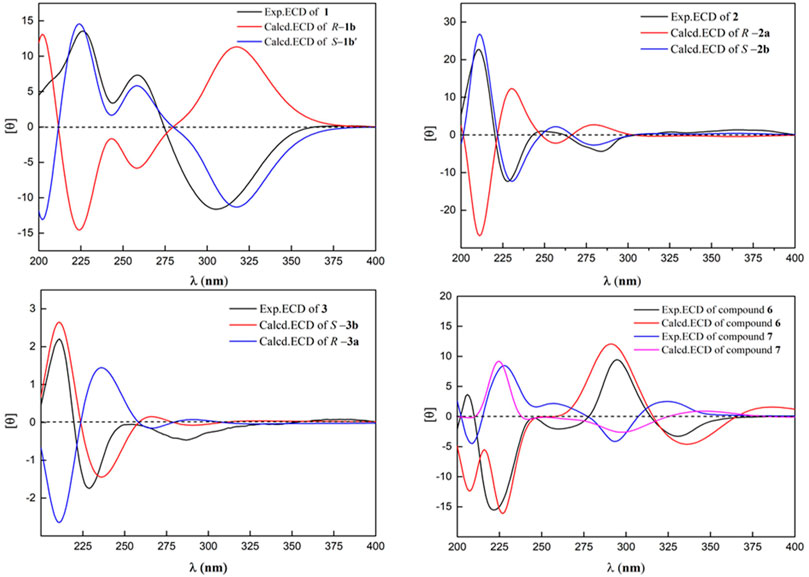
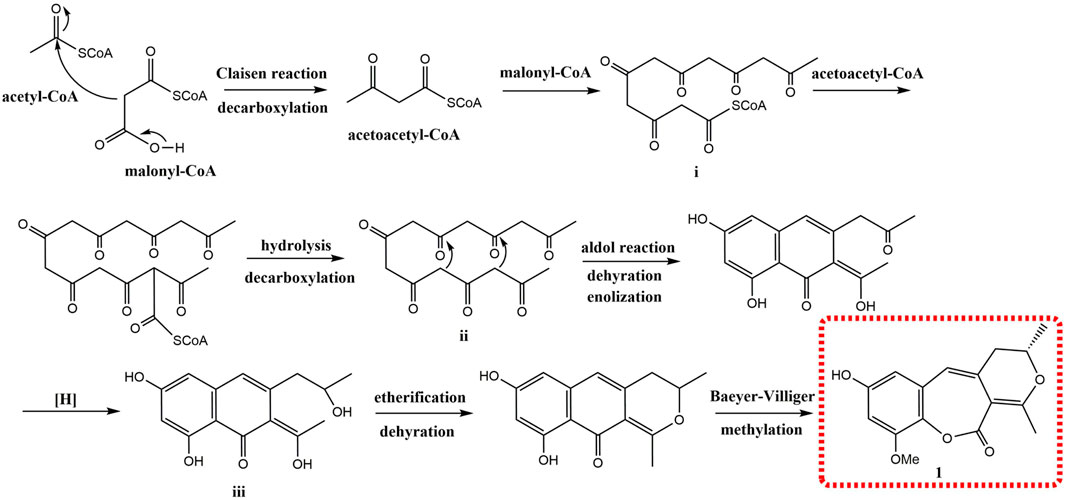
Compound 1 was obtained as a brown amorphous powder. According to the pseudo molecular ion peak at m/z 289.10695 [M + H]+ (calcd 289.10705) of HR-ESI-MS, the molecular formula was determined as C16H16O5, suggesting nine degrees of unsaturation (DOUs). The 1H-NMR spectrum of 1 indicated that 1 contained a pair of meta-coupled aromatic proton signals [δH 6.15 (1H, d, J = 2.5 Hz, H-6), 6.40 (1H, d, J = 2.5 Hz, H-8)], an isolated olefin proton signal [δH 6.07 (1H, s, H-4)], a pair of methylene signals [δH 2.64 (1H, dd, J = 13.5, 3.5 Hz), 2.35 (1H, dd, J = 13.5, 8.5 Hz), an oxygenated methine [δH 4.45 (1H, m)], two methyls [δH 2.31 (3H, s), 1.34 (3H, d, J = 6.5 Hz)], and a methoxy [δH 3.81 (3H, s)]. The 13C-NMR spectra that combined with HSQC and HMBC spectrum indicated the presence of 16 carbon signals, including one 1, 2, 3, 5-tetrasubstituted phenyl group [δC 132.4 (s), 106.5 (d), 156.2 (s), 100.8 (d), 153.6 (s), 132.8 (s)], one trisubstituted double bond [δC 122.2 (d), 132.0 (s)], an enolized double bond [δC 173.1 (s), 105.1 (s)], one ester carbonyl carbon [δC 171.9 (s)], an oxygenated methine [δC 77.3 (d)], two methyls [δC 21.9 (q), 20.7 (q)], one methoxy [δC 56.7 (q)], and one methylene [δC 39.1 (t)]. According to 1H–1H COSY and HSQC spectra, the structural fragment CH3(14)CH(O) (13)CH2(12)- was deduced. HMBC correlations (Figure 2) from H2-12 to δC 105.1 (s, C-2), 132.0 (s, C-3) and 122.2 (s, C-4), from H-4 to δC 105.1 (s, C-2), from CH3-11 [δH 2.31 (s)] to δC 105.1 (s, C-2), and δC 173.1 (s, C-1) suggested that the trisubstituted double bond was connected to C-12, and the trisubstituted and enolized double bonds were connected through C-3 to C-2. The remaining ester carbonyl carbon should be connected to C-2. Therefore, substructure A was established as depicted in Figure 1. Furthermore, HMBC correlations from H-4 to δC 106.5 (d, C-6) and 132.8 (s, C-10) suggested that substructure A was connected to C-5. ROESY correlation of MeO to H-8 and HMBC correlation of MeO to δC 153.6 (s, C-9) indicated that the methoxy group was connected to C-9. Except for the presence of two double bonds, one carbonyl and one phenyl, two DOUs were needed to satisfy the molecular formula of 1. Therefore, compound 1 still had two more rings. Owing to insufficient HMBC correlations, the connection positions of the two rings could not be determined, which ring was formed through ether bond, and which ring was formed by an ester bond. Therefore, there were two possible structures for 1. The ether bond was formed between C-1 and C-10, and C-2 was connected to C-13 through the carbonyl C-15 to form the ester bond in the candidate structure 1a. In contrast, C-2 was connected to C-10 through the carbonyl C-15 to form the ester bond, and the ether bond was formed between C-1 and C-13 in the candidate structure 1b. To further assign the connection positions of the two rings, NMR calculations with DP4+ analysis for two possible isomers were carried out. As a result, 1b was the most likely structure based on DP4+ probability with 100% (Figure 3). Ultimately, its absolute configuration was confirmed as (13S) by ECD calculations (Figure 4). Therefore, the structure of 1 was defined and named bercheminol A, which possessed an unprecedented 3, 4-Dihydro-11H-benzo [b]pyrano [4,3-e] oxepin-11-one skeleton. From the biogenic analysis, compound 1 comes from the acetate-malonate (AA-MA) pathway. The reasonable biosynthetic pathway is shown in Scheme 1. First, one acetyl coenzyme A (CoA) and five malonyl-CoA undergo Claisen condensation and decarboxylation to obtain polyketone i. Then, i react with acetoacetyl-CoA through a series of reactions, including Claisen condensation, hydrolysis and decarboxylation to obtain the key intermediate ii. Starting from ii, phenolic compound iii is obtained through aldol condensation, dehydration, enolization, and reduction. Finally, iii is etherified, oxidized, and methylated to give compound 1.
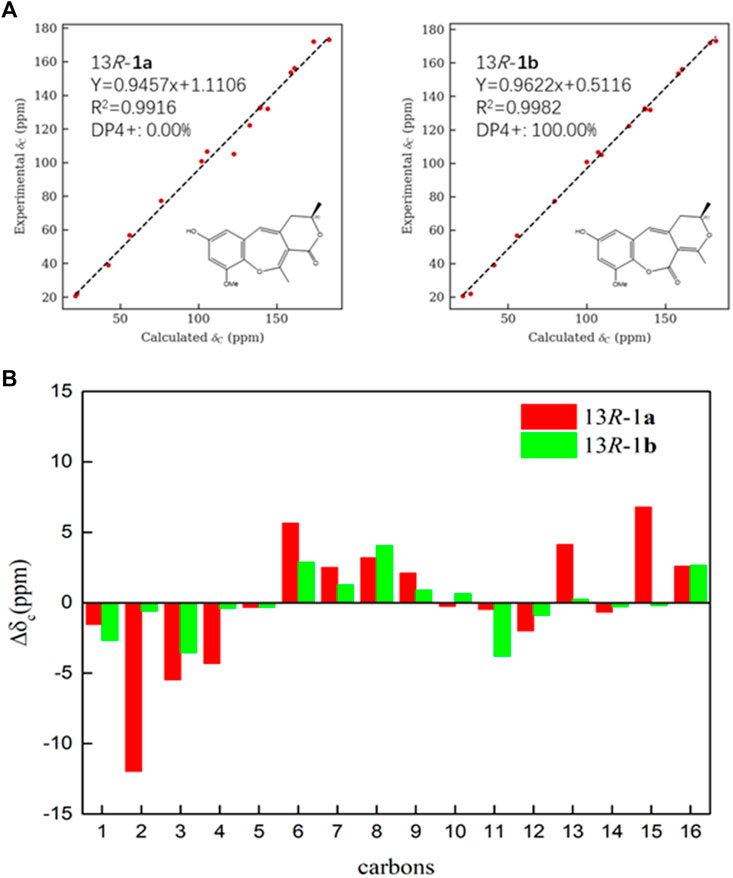
FIGURE 3. (A) Linear regression fitting of calculated 13C chemical shifts of two possible isomers of 1a and 1b with the experimental values (B) Deviation between calculated 13C chemical shifts of two possible isomers of 1a and 1b with the experimental values.
Compound 2 was obtained as a brown amorphous powder. The molecular formula was determined as C17H14O4 based on the HR-ESI-MS of the protonated molecular ion peak at m/z 283.09594 [M + H]+ (calcd 283.09649), suggesting 11 DOU. The 1H-NMR spectrum of 2 indicated the presence of a set of 1, 2, 3-trisubstituted benzene ring [δH 7.44 (1H, d, J =8.2 Hz, H-9), 7.58 (1H, t, J =8.2 Hz, H-10), 7.08(1H, d, J =8.2 Hz, H-11)], an isolated aromatic proton signal [δH 7.50 (1H, s, H-8)], an olefin proton signal [δH 5.79 (1H, s, H-3)], a pair of double doublets [δH 3.03 (1H, dd, J =12.0, 17.0 Hz, H-7), 3.27 (1H, dd, J =3.0, 17.0 Hz, H-7), and an oxygenated methine [δH 4.59 (1H, m, H-6)]. The 13C-NMR and DEPT spectra exhibited 17 carbon signals, including 5 sp2 methines, 1 methyl, 1 methylene, 1 methoxy, and 4 sp2 quaternary carbons, 3 oxygenated sp2 tertiary carbons, one oxygenated methine, and one ester carbonyl carbon. The above NMR data suggested that 2 might belong to naphthopyranone. By comparison of the NMR data of 2 with those of pannorin B (Kaur et al., 2015) suggested that their structures closely resembled except for two major differences. First, the signal of 1, 2, 3-trisubstituted benzene ring in 2 replaced a pair of meta-coupled aromatic protons in pannorin B, suggesting that the methoxy and hydroxyl groups at C-10 and C-12 in the latter were replaced by the hydrogen and methoxy group, respectively, which was further supported by HMBC spectrum. Second, the chemical shifts of C-6 have shifted upfield 24.7 ppm, respectively, compared to pannorin B, suggesting that the hemiacetal carbon at C-6 in pannorin B was replaced by an oxygenated methine which was further confirmed by 1H–1H COSY correlation of CH3CH(O)CH2- and HMBC spectrum. Ultimately, its absolute configuration was established as (6S) by ECD calculations (Figure 4). Therefore, the structure of 2 was defined and named bercheminol B.
Compound 3 was obtained as a brown amorphous powder. The molecular formula was determined as C16H12O4 by the HR-ESI-MS of the protonated molecular ion peak at m/z269.08072 [M + H]+ (calcd 269.08084), suggesting 14 mass units less than 2. Compared with NMR data of 2, 3 had one less methoxy signal, suggesting that 3 was demethylated of 2 which was further confirmed by HMBC spectrum and ECD calculations. Thus, the structure of 3 was defined and named bercheminol C.
Compound 4 was obtained as a white amorphous powder. The molecular formula was determined as C21H22O8 by the HR-ESI-MS of the protonated molecular ion peak at m/z 403.13870 [M + H]+ (calcd 403.13874). Acid hydrolysis of 4 and HPLC analysis of dithiocarbamate derivative of sugar provided D-glucose. The configuration of the D-glucose was determined β by the coupling constant of 7.5 Hz of an anomeric proton (Cai et al., 2021). In addition to the NMR signal of glucose, the 1H-NMR spectrum of the aglycone of 4 contained an isolated aromatic proton signal [δH 7.50(1H, s, H-6)], a set of signals of 1, 2, 3-trisubstituted benzene ring [δH 7.62 (1H, t, J = 7.8 Hz, H-8), 7.34 (1H, d, J = 7.8 Hz, H-9), 7.50 (1H, d, J = 7.8 Hz, H-7)], an olefinic proton [δH 6.31 (1H, s, H-3)], and two tertiary methyls [δH 2.55, 2.86 (each 3H, s)]. The 13C-NMR and DEPT spectra of the aglycone of 4 revealed the presence of five sp2 methines, two methyls, and eight non-protonated carbons, including four sp2 quaternary carbons, a conjugated carbonyl carbon [δC 182.6 (C-4)], three oxygenated tertiary carbons [δC 167.6 (C-2), 158.1(C-1a), 156.4 (C-10)]. The above NMR data suggested that 4 might belong to angular naphthopyrone (Shen et al., 2007; Chovolou et al., 2011). By careful comparison of NMR data of 4 with those of pleuropyrone A, their structures resembled each other. The major difference between them was the presence of signals of a 1, 2, 3-trisubstituted benzene ring in 4, instead of a pair of meta-coupled aromatic proton signals in pleuropyrone A (Min et al., 2003). This deduction was further confirmed by HMBC correlations and H-7/H-8/H-9 of 1H–1H COSY. Therefore, the structure of 4 was defined and named bercheminol D.
Compound 5 was obtained as a white amorphous powder. The molecular formula was determined as C22H24O9 by the HR-ESI-MS of the protonated molecular ion peak at m/z 433.14941 [M + H]+ (calcd 433.14931) suggesting 14 mass units more than pleuropyrone A. By careful comparison of NMR data of 5 with those of pleuropyrone A, it was found that the only difference was the presence of an additional methoxy signal. It was suggested that 5 was an 8-O-methylated derivative of pleuropyrone A, which was further confirmed by the HMBC correlation from the methoxy group to C-8. Therefore, the structure of 5 was defined and named bercheminol E.
Compounds 6 and 7 were obtained as brown amorphous powders. The negative mode of HR-ESI-MS of 6 and 7 showed their quasimolecular ions at m/z 493.13760 and 493.13589 [M-H]- respectively, suggesting the molecular formula C23H26O12. The detailed analyses of their NMR data indicated that they also possessed identical planar structures and belonged to dihydroflavonol glycoside containing L-rhamnopyranosyl moiety, which was further supported by acid hydrolysis of 6 and 7. By comparison of the NMR data of 6 and 7 with those of neoastilbin (De Britto et al., 1995), it was found that they are different from neoastilbin in the substituent pattern of the C ring. The ring C of 3,4-dihydroxyphenyl in neoastilbin was replaced by 3, 5-dimethoxy-4-hydroxyphenyl in 6 and 7. HMBC correlations from H-2′ and 6′ to C-2, C-1′ and C-4′, and MeO to C-3′ and C-5′ confirmed these findings. Careful analyses of the NMR data of 6 and 7 indicated that the difference between 6 and 7 were the absolute configurations of C-2 and C-3, which were the same as those of neoastilbin and astilbin. Compared to a (2R, 3R) configuration, the chemical shifts of H-1″ and H-2″ of the O-rhamnopyranosyl at C-3 in the (2S, 3S) configuration were located downfield, the chemical shifts of H-5″ and CH3-6″ were located upfield (De Britto et al., 1995). According to this rule, the absolute configurations of C-2 and C-3 of 6 and 7 were determined as (2S, 3S) and (2R, 3R), respectively. The presence of negative and positive cotton effect at about 330 nm in the CD spectrum of 6 and 7, respectively, and ECD calculations confirmed these findings (Figure 4). Therefore, the structures of 6 and 7 were defined and named as bercheminols F and G.
Compound 8 was obtained as a yellow amorphous powder. The molecular formula was determined as C21H26O8 by the HR-ESI-MS of the protonated molecular ion peak at m/z 407.17007 (calcd 407.17004). Acid hydrolysis of 8 and HPLC analysis of dithiocarbamate derivative of sugar provided D-glucose. The configuration of D-glucose was determined as β based on the coupling constant of 7.5 Hz of an anomeric proton. Detailed analyses of its NMR data indicated that the aglycone of 8 was determined as vittarin A (Wu et al., 2005). Therefore, 8 was 3-O-glucosylated derivative of vittarin A, which was further confirmed by the HMBC correlation from the anomeric proton of glycopyranosyl to C-3. Therefore, the structure of 8 was defined and named bercheminol H.
By comparing the spectral data of these compounds with those reported in the literature, the structures of 11 known compounds are identified as vittarin-B (9) (Wu et al., 2005), demethylflavasperone-10-O-β-D-glucopyranoside (10) (Xiong et al., 2019), rubrofusarin-6-O-β-D-glucopyranoside (11) (Messana et al., 1991), rubrofusarin-6-O-α-L-rhamnosyl-(1→6)-O-β-D-glucopyranside (12) (Shen et al., 2007), kaempferol-3-O-α-L-rhamnopyranosyl-(1→6)-β-D-glucopyranoside (13) (Thissera et al., 2020), quercetin (14) (Xiong et al., 2019), kaempferol (15) (Li et al., 2016), naringenin (16) (Gao et al., 2005), dihydrokaempferol (17) (Lu et al., 2015), 2-hydroxyemodin 1-methyl ether (18) (Lin et al., 2001), emodin (19) (Cohen et al., 1995).
The study of the chemical constituents of this plant led to the isolation of five types of phenolic compounds. Compound 1 possesses an unprecedented carbon skeleton with 3, 4-dihydro-11H-benzo [b]pyrano [4,3-e]oxepin-11-one. In addition to this new skeleton, four known types of phenolic compounds were also isolated, including naphthopyrones (2-5, 10-12), flavonoids (6-7, 13-17), bibenzyls (8-9), and anthraquinones (18–19). Furthermore, naphthopyrones may be categorized into two types: naphtho-α-pyrones, such as 2-3, and naphtho-γ-pyrones, such as 4-5, 10 which belonged to angular ones, and 11-12 which belonged to linear ones. Because of the insufficient amount of compounds 1 and 4–8, their biological activities were not tested. The α-glucosidase inhibitory activities of the remaining compounds were assayed (Table 3). As a result, vittarin-B (9), rubrofusarin-6-O-β-D-glucopyranoside (11), quercetin (14), kaempferol (15), and dihydrokaempferol (17) showed moderateα-glucosidase inhibitory activities with IC50 values of 22.5, 28.0, 36.5, 32.7, and 31.9 μM, respectively.
Conclusion
In conclusion, 19 structurally diverse phenolic compounds were isolated from the stem and leaves of Berchemia lineata (L.) DC., among which compounds 1–8 were previously undescribed. Most of isolated compounds were evaluated for their α-glucosidase inhibitory activities. As a result, bibenzyl (9), linear naphtho-γ-pyrone (11), and flavonoids (14–15, 17) displayed moderate inhibitory activities against α-glucosidase. Therefore, those compounds might be accountable for the antihyperglycemic effect of this herb.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
Author Contributions
HT and GY conceived, designed the experiments, and revised the manuscript. YL carried out the isolation of compounds and wrote the original draft. YC carried out structure elucidation. WX carried out NMR and ECD calculations. XL, GM, and JX carried out the experiments and data analyses. XZ collected the plant material. All authors have read and approved the published version of the manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (No. 81960762), the Research and Development Program of Hubei Province (No. 2021ACB003), and the Major Scientific and Technological Project of Hubei Province (No. 2020ACA007).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fchem.2022.889441/full#supplementary-material
References
Cai, W., Xu, S., Ma, T., Zhang, X., Liu, B., and Xu, F. (2021). Five Novel Triterpenoid Saponins from Hovenia Dulcis and Their Nrf2 Inhibitory Activities. Arabian J. Chem. 14, 103292–103299. doi:10.1016/j.arabjc.2021.103292
Chen, L., and Dong, J. X. (2006). Advances in Studies on Chemical Constituents from Plants of Berchemia Neck and Their Bioactivities. Chin. Tradit Herb Drugs 37, 627–630. doi:10.7501/j.issn.0253-2670.2006.4.259
Chovolou, Y., Ebada, S. S., Wätjen, W., and Proksch, P. (2011). Identification of Angular Naphthopyrones from the Philippine Echinoderm Comanthus Species as Inhibitors of the NF-Κb Signaling Pathway. Eur. J. Pharmacol. 657, 26–34. doi:10.1016/j.ejphar.2011.01.039
Cohen, P. A., and Neil Towers, G. H. (1995). The Anthraquinones of Heterodermia Obscurata. Phytochemistry 40, 911–915. doi:10.1016/0031-9422(95)00407-X
De Britto, J., Manickam, V. S., Gopalakrishnan, S., Ushioda, T., and Tanaka, N. (1995). Chemical and Chemotaxonomical Studies of Ferns. Part LXXXVI. Determination of Aglycone Chirality in Dihydroflavonol 3-O-.ALPHA.-L-Rhamnosides by 1H-NMR Spectroscopy. Chem. Pharm. Bull. 43, 338–339. doi:10.1248/cpb.43.338
Gao, S., Fu, G.-M., Fan, L.-H., Yu, S.-S., and Yu, D.-Q. (2005). Flavonoids fromLysidice rhodostegiaHance. J. Integr. Plant Biol. 47, 759–763. doi:10.1111/j.1744-7909.2005.00063.x
Jiang, X., Chen, S., Guo, Z., Gu, D., and Liang, X. (2019). Components Characterisation of Berchemia Lineata (L.) DC. By UPLC-QTOF-MS/MS and its Metabolism with Human Liver Microsomes. Nat. Product. Res. 35, 521–524. doi:10.1080/14786419.2019.1637871
Jing, Y. S., Xie, G. Y., Gu, W. W., Shi, L., and Qin, M. J. (2017). Research Advances in Chemical Constituents and Pharmacological Activities of Tiebaojin Medicine. Chin. Wild Plant Resour. 36, 49–53. doi:10.3969/j.issn.1006-9690.2017.01.014
Kaur, A., Raja, H. A., Deep, G., Agarwal, R., and Oberlies, N. H. (2015). Pannorin B, a New Naphthopyrone from an Endophytic Fungal Isolate ofPenicilliumsp. Magn. Reson. Chem. 54, 164–167. doi:10.1002/mrc.4324
Li, J., Deng, G.-R., Cheng, W., He, B., Zhang, G.-L., Huang, B.-S., et al. (2016). Chemical Constituents of Berchemia Lineata. Med. Biopharm. Proc. Int. Conf. 1140–1148. doi:10.1142/9789814719810_0146
Lin, L.-C., Chou, C.-J., and Kuo, Y.-C. (2001). Cytotoxic Principles from Ventilago leiocarpa. J. Nat. Prod. 64, 674–676. doi:10.1021/np000569d
Lu, C.-l., Zhu, W., Wang, D.-m., Chen, W.-l., Hu, M.-m., Wang, M., et al. (2015). Inhibitory Effects of Chemical Compounds Isolated from the Rhizome ofSmilax Glabraon Nitric Oxide and Tumor Necrosis Factor-αProduction in Lipopolysaccharide-Induced RAW264.7 Cell. Evidence-Based Complement. Altern. Med. 2015, 1–9. doi:10.1155/2015/602425
Messana, I., Ferrari, F., Cavalcanti, M. S. B., and Morace, G. (1991). An Anthraquinone and Three Naphthopyrone Derivatives from Cassia Pudibunda. Phytochemistry 30, 708–710. doi:10.1016/0031-9422(91)83762-A
Min, B.-S., Lee, J.-P., Na, M.-K., An, R.-B., Lee, S.-M., Lee, H.-K., et al. (2003). A New Naphthopyrone from the Root of Pleuropterus Ciliinervis. Chem. Pharm. Bull. 51, 1322–1324. doi:10.1002/chin.20041618210.1248/cpb.51.1322
Özgünseven, A., Barut, B., Šoral, M., Sari, S., Akaydın, G., Özel, A., et al. (2021). Alpha-glucosidase and Tyrosinase Inhibiton of Polyphenols Isolated from Potentilla Speciosa Var. Speciosa: In Vitro and In Silico Perspectives. Ind. Crops Prod. 170, 113806–113812. doi:10.1016/j.indcrop.2021.113806
Shen, J.-W., Jiang, J.-S., Zhang, X.-F., Zheng, C.-F., and Zhang, P.-C. (2007). Two New Benzochromone Glycosides from the Stem of Berchemia Racemosa. J. Asian Nat. Prod. Res. 9, 499–503. doi:10.1080/10286020600782074
Shen, Y. X., Teng, H. L., Yang, G. Z., Mei, Z. N., and Chen, X. L. (2010b). A New Chromone Derivative from Berchemia Lineata. Yao Xue Xue Bao 45, 1139–1143. doi:10.16438/j.0513-4870.2010.09.004
Shen, Y. X., Teng, H. L., Chen, X. L., Yang, G. Z., and Mei, Z. N. (2010a). Studies on the Chemical Constituents of the Roots of Berchemia Lineata (L.) DC. Chin. Tradit Herb Drugs 41, 1955–1957.
Tanaka, T., Nakashima, T., Ueda, T., Tomii, K., and Kouno, I. (2007). Facile Discrimination of Aldose Enantiomers by Reversed-phase HPLC. Chem. Pharm. Bull. 55, 899–901. doi:10.1248/cpb.55.899
Thissera, B., Visvanathan, R., Khanfar, M. A., Qader, M. M., Hassan, M. H. A., Hassan, H. M., et al. (2020). Sesbania Grandiflora L. Poir Leaves: A Dietary Supplement to Alleviate Type 2 Diabetes through Metabolic Enzymes Inhibition. South Afr. J. Bot. 130, 282–299. doi:10.1016/j.sajb.2020.01.011
Wu, P.-L., Hsu, Y.-L., Zao, C.-W., Damu, A. G., and Wu, T.-S. (2005). Constituents of Vittaria Anguste-elongata and Their Biological Activities. J. Nat. Prod. 68, 1180–1184. doi:10.1021/np050060o
Xiong, Y., Du, C. X., Duan, Y. S., Yuan, C. M., Huang, L. J., Gu, W., et al. (2019). Chemical Constituents and Pharmacological Activities of Sedum Aizoon Form Guizhou Province. Chin. Tradit Herb Drugs 50, 5404–5409. doi:10.7501/j.issn.0253-2670.2019.22.004
Keywords: Berchemia lineata, Berchemia, phenolic compounds, antihyperglycemic effect, α-glucosidase inhibitory activity
Citation: Li Y, Chen Y, Xie W, Li X, Mei G, Xu J, Zhao X, Teng H and Yang G (2022) Phenolic Compounds From the Stems and Leaves of Berchemia lineata (L.) DC. Front. Chem. 10:889441. doi: 10.3389/fchem.2022.889441
Received: 04 March 2022; Accepted: 16 March 2022;
Published: 14 April 2022.
Edited by:
Xiaoxiao Huang, Shenyang Pharmaceutical University, ChinaReviewed by:
Shifang Li, Shenyang Pharmaceutical University, ChinaYu-Xi Wang, Institute of Applied Ecology (CAS), China
Copyright © 2022 Li, Chen, Xie, Li, Mei, Xu, Zhao, Teng and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hongli Teng, NTY0OTg4MTc3QHFxLmNvbQ==; Guangzhong Yang, eWFuZ2d6ODg4QDEyNi5jb20=
†These authors have contributed equally to this work and share first authorship
 Yitong Li
Yitong Li Yu Chen
Yu Chen Wenli Xie
Wenli Xie Xueni Li
Xueni Li Gui Mei
Gui Mei Jing Xu
Jing Xu Xiangpei Zhao3
Xiangpei Zhao3 Guangzhong Yang
Guangzhong Yang