
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell Dev. Biol. , 20 February 2025
Sec. Morphogenesis and Patterning
Volume 13 - 2025 | https://doi.org/10.3389/fcell.2025.1552716
 Jessica C. Britton1
Jessica C. Britton1 Anett Somogyi-Leatigaga1
Anett Somogyi-Leatigaga1 Billy A. Watson1
Billy A. Watson1 Endika Haro1
Endika Haro1 Cassidy G. Mulder1
Cassidy G. Mulder1 Kari D. Kennedy1
Kari D. Kennedy1 Allen M. Cooper1
Allen M. Cooper1 Kristen L. Whitley1
Kristen L. Whitley1 Ruth-Love Yeboah1
Ruth-Love Yeboah1 Jeanyoung Kim1
Jeanyoung Kim1 Micah C. Yu1,2
Micah C. Yu1,2 Jairo D. Campos1,2
Jairo D. Campos1,2 Japhet Amoah1,2
Japhet Amoah1,2 Shimako Kawauchi3
Shimako Kawauchi3 Eunyoung Kim3
Eunyoung Kim3 Charmaine U. Pira1
Charmaine U. Pira1 Kerby C. Oberg1*
Kerby C. Oberg1*Introduction: In vertebrate limb morphogenesis, wingless-related integration site (Wnt) proteins and fibroblast growth factors (Fgfs) secreted from the apical ectodermal ridge (AER) coordinate proximodistal outgrowth. Fgfs also sustain sonic hedgehog (Shh) in the zone of polarizing activity (ZPA). Shh directs anteroposterior patterning and expansion and regulates AER-Fgfs, establishing a positive regulatory feedback loop that is vital in sustaining limb outgrowth. The transcription factor LIM homeodomain 2 (Lhx2) is expressed in the distal mesoderm and coordinates AER and ZPA signals that control cellular proliferation, differentiation, and shaping of the developing limb. Yet how Lhx2 is transcriptionally regulated to support such functions has only been partially characterized.
Methods/Results: We have identified two limb-specific cis-regulatory modules (CRMs) active within the Lhx2 expression domain in the limb. Chromatin conformation analysis of the Lhx2 locus in mouse embryonic limb bud cells predicted CRMs-Lhx2 promoter interactions. Single-cell RNA-sequencing analysis of limb bud cells revealed co-expression of several Fgf-related Ets and Wnt-related Tcf/Lef transcripts in Lhx2-expressing cells. Additionally, disruption of Ets and Tcf/Lef binding sites resulted in loss of reporter-driven CRM activity. Finally, binding of β-catenin to both Lhx2-associated CRMs supports the associated binding of Tcf/Lef transcription factors.
Discussion: These results suggest a role for Ets and Tcf/Lef transcription factors in the regulation of Lhx2 expression through these limb-specific Lhx2-associated CRMs. Moreover, these CRMs provide a mechanism for Fgf and Wnt signaling to localize and maintain distal Lhx2 expression during vertebrate limb development.
LIM homeodomain 2 (Lhx2) is a transcription factor critical for regulating the development of the brain, eye, skin, and limb. In the regulation of the limb, Lhx2 is expressed in the distal mesoderm and plays an essential role in maintaining cells in a responsive state as axis-specific signals coordinate patterned limb outgrowth (Tzchori et al., 2009; Watson et al., 2018). In Drosophila, apterous regulates dorsal fates and is required for limb outgrowth (Blair et al., 1994; Williams et al., 1991). LHX2 is considered the vertebrate homologue of apterous controlling limb outgrowth (Rodriguez-Esteban et al., 1998). Knockdown of LHX2 during limb outgrowth in chicken causes severe limb truncations (Rodriguez-Esteban et al., 1998). In mouse, another family member, Lhx9, is also expressed in the distal limb mesoderm and exhibits redundant functions of Lhx2. Knockout of both Lhx2 and Lhx9 disrupts limb outgrowth similar to chicken (Tzchori et al., 2009).
The loss of Lhx2/9 function disrupts several molecular pathways critical for limb patterning and outgrowth. Fgf10 is expressed in the mesoderm of the emerging limb bud and induces the epithelium at the distal tip to thicken and form the apical ectodermal ridge (AER) (Ohuchi et al., 1997; Ohuchi et al., 1999). The AER, through Wnt proteins, secretes several fibroblast growth factors (Fgfs) that maintain Fgf10 in the underlying mesoderm, establishing a reciprocal positive feedback loop that promotes proximodistal limb outgrowth and patterning (Ohuchi et al., 1997). In mice lacking Lhx2/9 function, Fgf10 expression is greatly reduced (Tzchori et al., 2009).
Lhx2 also regulates sonic hedgehog (Shh) which mediates anteroposterior patterning and outgrowth. Shh is secreted from the zone of polarizing activity (ZPA), a collection of mesodermal cells in the distal posterior limb bud (Yang et al., 1997; Zhu et al., 2008). Shh also supports AER Fgfs expression, and in turn, AER-Fgfs maintain the ZPA’s expression of Shh in another reciprocal positive feedback loop (Niswander et al., 1994). In the absence of Lhx2/9 function, Shh expression is markedly decreased (Tzchori et al., 2009).
Although Lhx2 is critical for proximodistal and anteroposterior patterning, the mechanisms that regulate Lhx2 for these important roles have not been characterized. Cis-regulatory modules (CRMs) control spatiotemporal gene expression (Kolovos et al., 2012; Lewis et al., 2019) and thus were the focus of this investigation. In a previous study, Lee and colleagues (Lee et al., 2011) identified four CRMs labeled as conserved non-coding DNA elements (CNEs) within the Lhx2 locus that were active in the central nervous system overlapping Lhx2 expression in mice. However, none of the CNEs showed activity in the limb.
In this study, we used vertebrate conservation combined with mouse limb chromatin modification data to examine and identify potential limb-associated CRMs within the Lhx2 locus. We utilized the chicken enhancer bioassay (Oberg et al., 2002; Pira et al., 2008; Uchikawa et al., 2003) to screen and localize functional CRMs in the limb in ovo and confirmed their pattern of activity in transgenic mice. We analyzed the co-expression of candidate transcription factors in Lhx2-expressing murine limb cells and identified transcription factor binding site sequences important for CRM activity. Our collective results suggest a mechanism by which key transcription factors linked to Fgf and Wnt signaling regulate Lhx2 expression through limb-specific CRMs during limb development.
Conserved non-coding DNA sequences 1 megabase (Mb) both upstream and downstream of the Lhx2 gene were identified by pairwise alignment using VISTA Genome browser (http://genome.lbl.gov/vista/), UCSC Genome browser (https://genome.ucsc.edu), and the enhancer prediction database Enhancer Atlas (http://www.enhanceratlas.org/). Sequences exhibiting at least 70% homology between vertebrate species and possessing binding sites for limb-associated regulated transcription factors were selected. We excluded conserved regions described by Lee et al. (2011) since none of these were found to be active in limbs. Available embryonic day 11.5 (E11.5) and E12.5 mouse limb ChIP-seq data were used to determine enhancer-related chromatin modification proteins coincident with predicted conserved regions (Gene Expression Omnibus database (GEO; http://www.ncbi.nlm.nih.gov/geo/) under accession numbers: GSE42413 for H3K27ac (Cotney et al., 2013); GSE42237 for H3K4me2 and H3K27me3 (DeMare et al., 2013); GSE13845 for p300 (Visel et al., 2009); RNAP2 and Med12 (Berlivet et al., 2013)).
Conserved regions were scored based on the number of aligned marks, and regions associated with 3 or more enhancer-related chromatin marks were chosen as candidates for potential cis-regulatory modules (PCRMs) (Supplementary Table S1). Regions characterized by two or less active regulatory marks or by repressive regulatory marks were not selected for further analysis. PCRMs were between 150 and 2,100 base pairs in length and were isolated from chicken genomic DNA by PCR using the primers listed in Supplementary Table S2. PCRM DNA sequences were cloned into pCRII TOPO vector (Thermofisher Scientific, Waltham, MA; Catalog #K450002) and then sub-cloned into a thymidine kinase (tk) promoter-driven enhanced GFP (eGFP) reporter construct (generous gift of Masanori Uchikawa) (Uchikawa et al., 2003) for assessment in ovo.
Merged hic reads from E11.5 mouse limb Hi-C data ((Kraft et al., 2019); under accession number GSE116794) were converted into the cooler (hic.cool) format using hic2cool converter package (https://github.com/4dn-dcic/hic2cool). Using the Hi-C Explorer software (http://hicexplorer.readthedocs.io/) (Ramírez et al., 2018; Wolff et al., 2018), TAD calling was performed using the hicFindTAD command with the following parameters: threshold value of 0.05 and delta value of 0.01 and a min and max depth of 3,000 and 31,500, respectively. The output file (domains.bed) was used to plot TAD boundaries (Supplementary Table S3) and visualized using pyGenome tracks (Lopez-Delisle et al., 2021). E11.5 mouse limb CTCF ChIP-seq data was obtained from DeMare and colleagues ((DeMare et al., 2013); under accession GSE42237), and visualized using UCSC Genome Browser (https://genome.ucsc.edu). Accessibility of the Lhx2 locus was evaluated using E11.5 mouse ATAC-seq data reported by Jhanwar and coworkers ((Jhanwar et al., 2021); under accession number GSE164736).
Chicken eggs (Chino Valley Ranchers, Colton, CA) were incubated in a humidified chamber according to Hamburger and Hamilton (HH) stages (Hamburger and Hamilton, 1951). To screen for potential CRM activity, we performed confined microelectroporation (CMEP) to transfect the distal chicken limb bud mesoderm at HH23 modified from the technique described by Oberg et al. (2002). A cocktail consisting of 2 μg/μL of PCRM-eGFP reporter constructs, 0.2 μg/μL pCAGGS-RFP, 0.25% fast green, and Tris-EDTA (TE) buffer was injected into the limb mesoderm ∼50 μm from distal tip. The pCAGGS-RFP is a β-actin promoter-driven RFP plasmid that was used to assess transfection efficiency (kind gift from Cheryll Tickle). The insulated anode needle was inserted into the limb mesoderm anterior to the DNA injection site, whereas the blunt cathode was placed posterior to the injection site, touching the tip of the distal limb bud. Electroporation was performed using the CUY21 Electroporator (Protech International, Boerne, TX) with the following parameters: 10 pulses of 30 V at a duration of 25 ms and with 50 ms intervals. Embryos were incubated in a humidified chamber at 37°C for 24 h and then harvested. Fluorescence was visualized and digital images were acquired using the Sony DKC-5000 or the Leica MZ 10F dissecting microscope camera. Images were compiled in Adobe Photoshop Version 2024.
To determine the pattern of CRM activity, we performed targeted regional electroporation (TREP) to transfect CRM-eGFP constructs into the presumptive limb bud at HH14, as described by Pira et al. (2008). Embryos were incubated in a humidified chamber at 37°C for 48 h and then harvested. Fluorescence was visualized and digital images were acquired as described above.
The mouse DNA sequence of CRM (−8)/LADLRM and CRM (−9)/LASARM were cloned into the PCR4-Shh-lacZ-H11 reporter (Addgene plasmid # 139098; http://n2t.net/addgene:139098) via Gibson Assembly (New England Biolabs, Ipswich, MA; Catalog# E2611S). The constructs were used to generate transgenic mice embryos (University of California Irvine Transgenic Mouse Facility, Irvine, CA). Embryos were harvested at E11.5 and processed for detection of LacZ activity as described by Shen et al. (2017). CRMs that showed reproducible expression patterns in at least three embryos were designated enhancers.
For each image, red and green fluorescence channels of images were separated and converted to grayscale (16 bit) using the image analysis software FIJI (Schindelin et al., 2012; Shihan et al., 2021). The region of interest was outlined based on the selected threshold and the mean fluorescence intensity calculated from the measured mean of both RFP and GFP images. GFP fluorescence intensity was normalized to that of RFP. One-way ANOVA followed by Dunnett’s multiple comparison test with alpha = 0.05 was performed using GraphPad Prism (version 10.0) to determine difference in the mean fluorescence intensity. Data were summarized using box and whisker plots showing the interquartile range, range, and median values, with p-value format as: *: p < 0.05, ns: non-significant.
Chicken cDNA LHX2 and SHH were sub-cloned into pCRII-TOPO vector following the manufacturer’s instructions and used to generate digoxigenin-labeled mRNA probes (Yamada et al., 1999). Whole mount in situ hybridization was performed on HH23 chicken embryos as described (Watson et al., 2018). Embryos were fixed in MEMFA (0.1 M MOPS, 2 mM EGTA, 1 mM MgSO4, 3.7% Formaldehyde) for 90 min at room temperature or overnight at 4°C and then dehydrated in 90% methanol. Proteinase K treatment was 10 μg/mL with an incubation time of 7 min at room temperature. Hybridization and post-hybridization washes were carried out at 60°C and 63°C, respectively. At least 5 embryos per gene were analyzed.
Putative transcription factor binding sites with prediction binding scores of p < 10−2 were identified and visualized using the JASPAR Transcription Factor Binding Site 2024 Database track on UCSC Genome browser (https://genome.ucsc.edu). Primers to the core nucleotide sequences of binding sites were designed to introduce a restriction enzyme to disrupt the sequence (Supplementary Table S4). The primers containing the mutation core sequences were introduced into the chicken CRM (-8)/LADLRM, CRM (-9)/LASARM-eGFP, and ZRS-eGFP reporter constructs via the QuickChange Lightning Site-Directed Mutagenesis Kit (Agilent Technologies, Santa Clara, CA, Catalog# 210513) following manufacturer recommendations. The mutated constructs were sequence-validated and rechecked on Ciiider, a tool for predicting transcription factor binding sites, to determine if new putative transcription factor binding sites were generated by the mutation (Supplementary Figure S1).
E11 mouse forelimb mesoderm single-cell RNA sequencing (scRNA-seq) data reported by He and colleagues (He et al., 2020) was evaluated for cells expressing Lhx2. Using Partek® Flow® software, v10.0.23.0214 (RRID:SCR_011860), filtered h5 matrices were imported and processed as described (Yeboah et al., 2023). In brief, batch correction was used to minimize cross-sample variation and differential expression between Lhx2-expressing (Lhx2+) and Lhx2 non-expressing (Lhx2-) cells (normalized expression lower than 0.5) using ANOVA and Hurdle. Principal component analysis (PCA) was conducted for dimensionality reduction and visualization of relationships among sequenced cells. The number of cells expressing each transcript of interest was recorded (Supplementary Table S5), and t-SNE plots were generated to visualize and analyze co-segregation of transcripts of interest with Lhx2+ cells. We also confirmed the mesodermal enrichment of cell data analyzed (Supplementary Figure S2).
In silico analysis of the Lhx2 locus revealed a total of 49 conserved non-coding DNA sequences with 70% or more sequence homology between vertebrate species both upstream and downstream of the Lhx2 promoter. These 49 conserved regions also possessed binding sites for limb-associated transcription factors (Supplementary Table S1). Twelve out of the 49 conserved regions were associated with three or more enhancer-related chromatin marks: H3K27ac, H3K4me2, p300, RNAP2, and Med12 (Figure 1), indicating potential cis-regulatory function. Ten of the potential cis-regulatory modules (PCRMs) are located upstream of the Lhx2 promoter (PCRM (−1) through (−10), while two are located downstream (PCRM 1 and 2) (Figure 1).
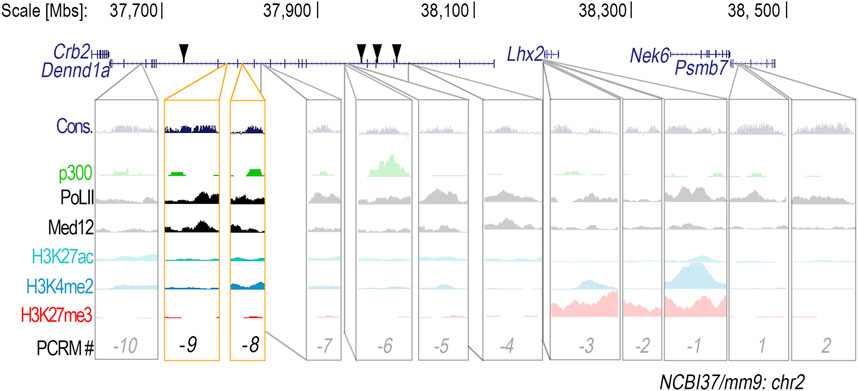
Figure 1. Twelve conserved non-coding DNA regions within the Lhx2 locus are suitable candidates for potential cis-regulatory modules. Conserved (Cons.) non-coding DNA regions in the Lhx2 locus were found to be associated with enhancer-related chromatin modification marks–H3K4me2, H3K27ac, p300, RNAP2, and Med12 – and thus potential cis-regulatory modules (PCRMs). PCRMs located upstream of the Lhx2 promoter were given a negative number and PCRMs located downstream were given a positive number. Location of active CNEs described by Lee and colleagues (Lee et al., 2011) are marked by black triangles. CRM (-8) and CRM (-9), located within the Dennd1a gene, are highlighted in yellow since they showed enhancer activity in the distal limb.
Regulation of gene expression by long-range CRMs (>5 kb from the promoter) requires chromatin folding that facilitates CRM-promoter interactions (Kraft et al., 2019; Long et al., 2022). Frequently interacting chromatin regions called topologically associated domains (TADs) are bordered by sequences that bind CCCTC-binding factors (CTCF) that define TAD boundaries within which chromatin interactions are most likely to occur (Kraft et al., 2019; Long et al., 2022). In addition, accessible chromatin is needed for CRMs to be functionally active (Buenrostro et al., 2013; Buenrostro et al., 2015). Therefore, we analyzed the chromatin organization and accessibility of the Lhx2 locus using published Hi-C data from embryonic day 11.5 (E11.5) limbs (Kraft et al., 2019), CTCF ChIP-seq (DeMare et al., 2013), and ATAC-seq data (Jhanwar et al., 2021).
Lhx2 and the Dennd1a gene, wherein many of the upstream PCRMs reside, are flanked by two CTCF binding sites signifying that they are both within the same TAD (Figure 2). Hi-C data identified strong interactions between the Lhx2 promoter and several PCRMs including −5, −7, −8, −9, and −10. ATAC-seq in the E11.5 limbs demonstrated accessibility of the PCRMs -7, -8, -9, and -10 (Figure 2) consistent with functional activity.

Figure 2. CRM (-8) and CRM (-9) are predicted to interact with the Lhx2 promoter and are accessible for gene regulation. Hi-C heatmap of mouse E11.5 limb buds depicts regions of interaction within the Lhx2 locus. Color bar represents the log of interaction frequency (log(IF). Black triangles highlight the border of topologically associated domain (TAD) interactions, and TAD boundaries are defined by peak calling from CTCF ChIP-seq data aligned beneath the Hi-C data. CRM (-8) and CRM (-9) are located in intron 11 and 12, respectively, of the Dennd1a gene (gray highlight) and are predicted to interact with the Lhx2 promoter (black arrow) within the same TAD. The pink arrow highlights the Lhx2 promoter. Aligned ATAC-seq data shows that the chromatin within this TAD is in an open state for transcription.
To screen the PCRMs for enhancer activity, we isolated the chicken sequences using PCR, constructed CRM-eGFP reporters, and then used confined microelectroporation (CMEP) (Oberg et al., 2002) to focally transfect the CRM constructs into the distal limb mesoderm of Hamburger Hamilton stage 23 (HH23) chicken wing buds. Using this chicken limb bioassay, two of the twelve PCRMs we demonstrated were actual CRMs, (CRM (-8) (191 bp sequence) and (-9) (684 bp sequence)). They showed robust, consistent activity in the distal limb mesoderm overlapping the LHX2 expression domain (Figures 3, 4). CRM (-8) was active in a broad distal band of limb mesoderm, extending about 500 μm from the tip and was present in both the dorsal and ventral mesoderm (Figure 3A). CRM (-9) had activity confined to the distal 100 μm of the mesenchyme abutting the apical ectodermal ridge (AER) (Figure 4A), with symmetrical dorsal and ventral activity. CRM (-7) exhibited weak, inconsistent activity in the distal limb bud (n = 10/45, Supplementary Figure S3A) and was not evaluated further.
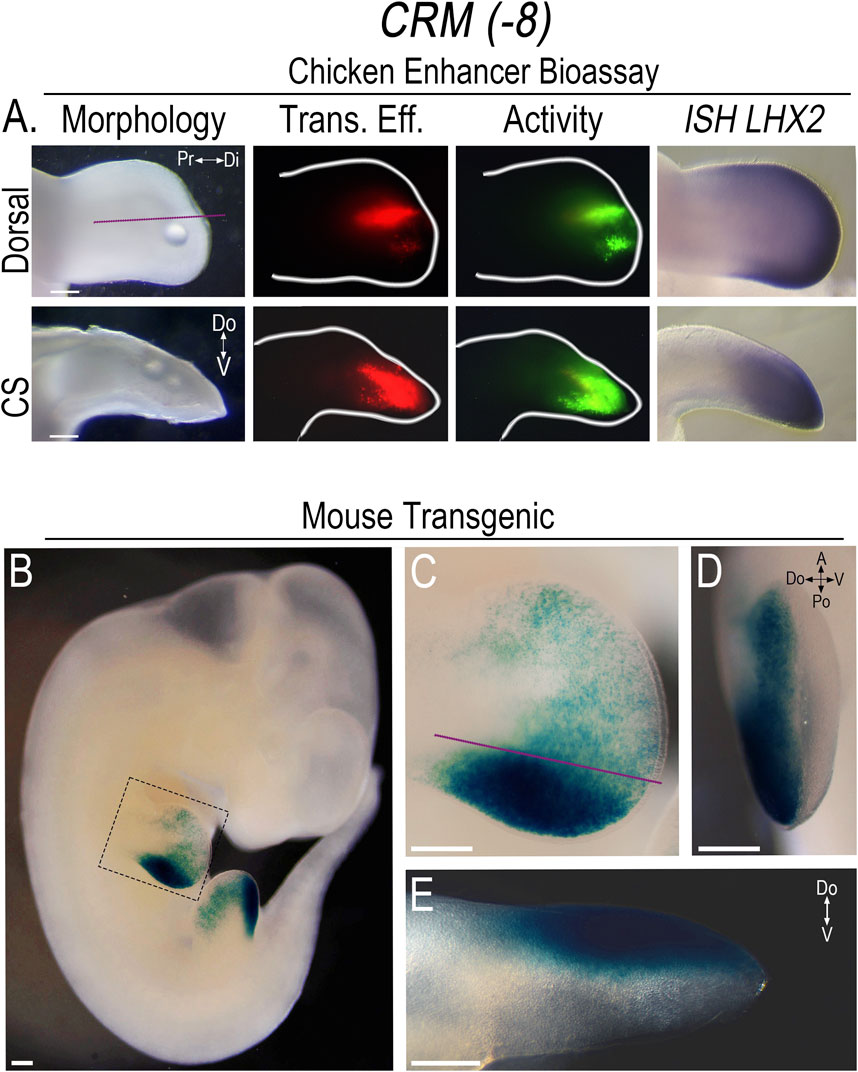
Figure 3. CRM (-8) shows robust activity in the distal limb overlapping Lhx2 expression. (A) Dorsal view of limb bud shows the activity (Activity) of CRM (-8) (n = 31) in the distal limb mesoderm overlapping LHX2 expression (ISH LHX2), using the chicken enhancer bioassay. Purple lines indicate longitudinal cross-sections (CS) of the limb bud shown in the bottom row. Note: CRM (−8) exhibits activity in both the dorsal and ventral mesoderm. Transfection efficiency (Trans. Eff) was determined with a β-actin promoter-driven RFP plasmid (the small bubble present in each limb bud indicates mineral oil used to seal the DNA cocktail into the mesoderm at the injection site). (B) LacZ staining showing the activity pattern of CRM (-8) (n = 5) in E11.5 transgenic mouse embryos. (C) Close-up of the forelimb (highlighted by the dotted box in “(B)”) showing the broad distal limb activity with accentuated activity over the ZPA. (D) End-on view of the forelimb showing activity restricted to the dorsal mesoderm. (E) Longitudinal cross-section of the forelimb in “(C)” demonstrating the dorsally restricted activity. Scale bars: 250 µm. Abbreviations used: A (Anterior), Po (Posterior), Pr (Proximal), Di (Distal), V (Ventral), Do (Dorsal).
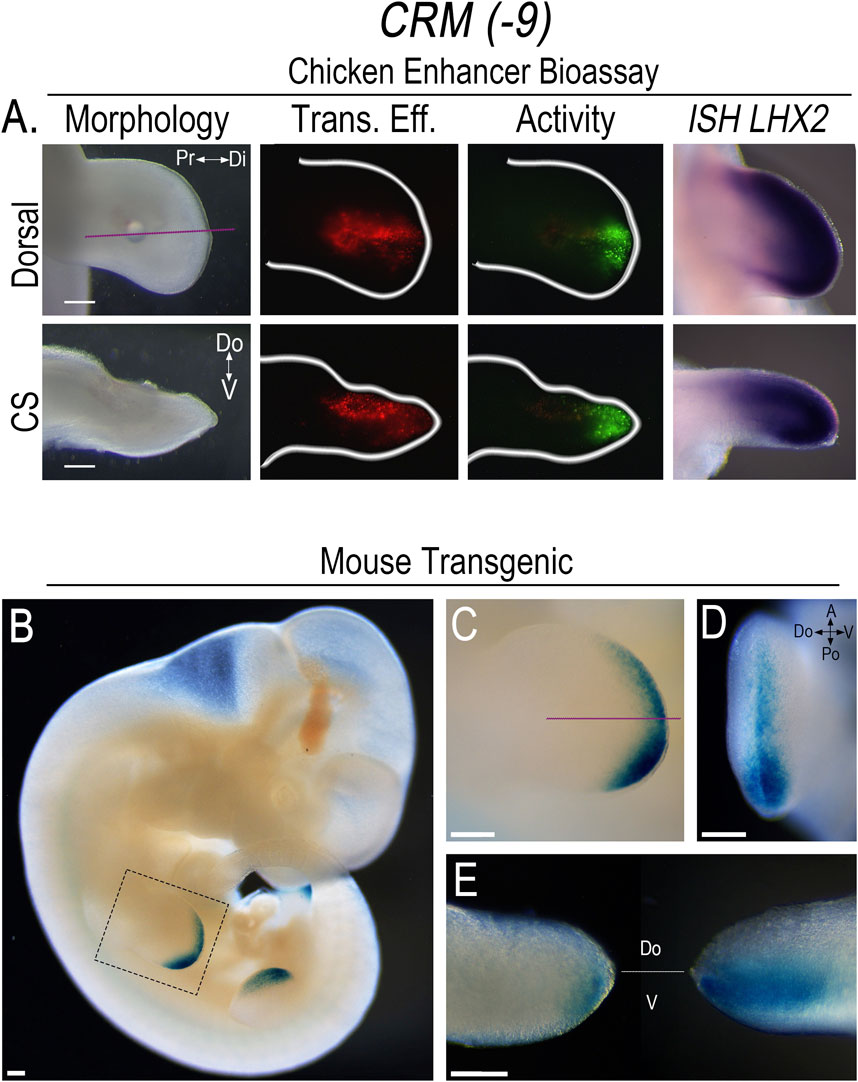
Figure 4. CRM (-9) exhibits activity in the distal sub-AER mesoderm overlapping the domain of Lhx2 expression. (A) Dorsal view of limb bud shows the activity (Activity) of CRM (-9) (n = 30) in the distal sub-AER mesoderm overlapping LHX2 expression (ISH LHX2), using the chicken enhancer bioassay. Purple lines indicate longitudinal cross-sections (CS) of the limb bud shown in the bottom row. Transfection efficiency (Trans. Eff) was determined with a β-actin promoter-driven RFP plasmid. The small bubble present in each limb bud indicates mineral oil used to seal the DNA cocktail into the mesoderm at the injection site. (B) LacZ staining showing the activity pattern of CRM (-9) (n = 11) in E11.5 transgenic mouse embryos. (C) Close-up of the forelimb (highlighted by the dotted box in “(B)”) showing the distal sub-AER activity that is accentuated posteriorly. (D) End-on view of the forelimb showing activity in both the dorsal and ventral mesoderm. (E) Longitudinal cross-section of the forelimb in “(C)” viewed towards the anterior aspect (left) shows activity accentuated in the ventral sub-AER mesoderm, and the posterior aspect (right) shows activity in both the dorsal and ventral mesoderm. Scale bars: 250 µm. Abbreviations used: A (Anterior), Po (Posterior), Pr (Proximal), Di (Distal), V (Ventral), Do (Dorsal).
The corresponding murine sequences for CRM (−8) and CRM (-9) were also isolated and screened in the chicken limb bioassay using CMEP to confirm that they exhibited the same distribution of activity. To better demonstrate the pattern of CRM activity, we used the murine sequences to generate CRM-Shh-lacZ-H11 reporter constructs for transient transfection.
In embryonic day 11.5 (E11.5) transgenic mice, the activity pattern of CRM (-8) showed a broad rim of activity that extended about 500 μm from the distal tip covering much of the distal mesoderm. However, the intensity of activity varied, with scattered, punctate activity in the anterior limb mesoderm and markedly enhanced activity in the posterior mesoderm overlapping the zone of polarizing activity (ZPA) domain (Figures 3B, C). A noted difference in the activity pattern of CRM (-8) in the transgenic mice was the sharp restriction of activity to just the dorsal compartment (Figures 3D, E). To determine whether the same pattern of activity was also present in the chicken model, we performed targeted regional electroporation (TREP) of the murine CRM (-8)-eGFP reporter into the presumptive limb mesoderm prior to limb bud formation, at HH14. The murine CRM (-8) showed the same anteroposterior pattern of activity in the chicken limb as seen in the mouse with accentuated activity in the posterior limb mesoderm. However, in contrast to the mouse model, activity was present in both dorsal and ventral mesoderm (Supplementary Figure S3B). These data indicate that the chicken ventral limb mesoderm can activate murine CRM (-8), while the mouse either lacks this capacity or is suppressing activity. Further work is needed to investigate this species-specific difference in ventral limb mesoderm.
The pattern of CRM (-9) activity in E11.5 transgenic mouse embryos was confined to the distal sub-AER mesoderm, with expanded activity in the posterior distal sub-AER region (Figures 4B–E). The posteriorly expanded CRM (-9) activity was present in both dorsal and ventral mesoderm, but was increased in the ventral mesoderm. In the anterior half, activity was even more accentuated ventrally (Figure 4E). This data, like that of CRM (-8), also shows a dorsoventral asymmetry that was not present in the chicken limb model (Figure 4A) and worthy of further investigation.
Neither CRM (-8) nor (-9) exhibited activity in other Lhx2 expression domains in the transgenic mice indicating that the activity was limb-specific. With extended staining times, some weak reporter staining was observed in the basal plate of the developing spinal cord (Supplementary Figure S3C); however, Lhx2 is not expressed in this region (Lee et al., 2011). Thus, we considered this as non-specific staining or low-level construct-related activation. Since the activity of both CRMs are limb-specific, overlap Lhx2 expression, and are predicted to interact with the Lhx2 promoter, we renamed these CRMs based on their pattern of activity as follows: CRM (-8) as Lhx2-associated distal limb regulatory module (LADLRM) and CRM (-9) as Lhx2-associated sub-AER regulatory module (LASARM).
Fgfs and Wnts are known to work together to coordinate limb outgrowth and AER initiation and formation (ten Berge et al., 2008). Ets transcription factor proteins relay FGF signaling via the Ras/MAPK pathway to regulate limb outgrowth and patterning (proximodistal and anteroposterior) (Sharrocks, 2001; Yordy and Muise-Helmericks, 2000; Mao et al., 2009; Yamamoto-Shiraishi et al., 2014). In addition, Tcf/Lef transcription factor proteins are downstream effectors of Wnt/β-catenin signaling and regulate limb morphogenesis, patterning, and AER maturation (Galceran et al., 1999; Kardon et al., 2003). Such roles for Ets and Tcf/Lef proteins during limb development suggest that these transcription factors may regulate Lhx2 expression.
Several Ets and Tcf/Lef transcripts are expressed in mouse limbs (Galceran et al., 1999; Lettice et al., 2012; Mathew et al., 2011; Wu et al., 2012; Yamamoto-Shiraishi et al., 2014). To determine whether Ets and Tcf/Lef transcription factors are present in Lhx2-expressing (Lhx2+) cells and thus capable of its regulation, we analyzed published mouse forelimb mesoderm single-cell RNA-sequencing (scRNA-seq) data (He et al., 2020). Msx1 and Wnt5a were used as makers for the distal sub-AER cells, and Meis2 was used as a marker for proximal limb mesoderm (Galceran et al., 1999; Zhang et al., 2023).
Principal component analysis (PCA), displayed as t-SNE plots, demonstrated co-segregation of Lhx2+ cells with Msx1+ and Wnt5a+ cells consistent with distal limb mesenchyme. In contrast, most Meis2+ cells marking the proximal limb mesoderm do not co-segregate with Lhx2+ cells. A fraction of the Meis2+ cells, however, do overlap with Lhx2+ cells, congruent with an early stage of limb development before limb elongation fully separates proximal from distal.
Multiple Ets transcripts (Ets1, Ets2, Elk3, Elk4, Etv1, Etv2, Etv4, and Etv5) are co-expressed in Lhx2+ murine (Figure 5) and human limb cells (Supplementary Figure S4). Ets2, Elk3, Ets1, Elk4, and Etv1 are ubiquitously expressed in limb cells (Figure 5). However, there are more cells expressing Ets2, Elk3, and Ets1 transcripts than Elk4 and Etv1 (Supplementary Table S5). A portion of the cells that segregate with Lhx2+ cells by PCA also co-express Lhx2. Etv4, Etv5, and Etv2 expressing cells share a closer principal component segregation pattern with Lhx2+ cells (Figure 5), and most of those cells co-express Lhx2 (Supplementary Table S5).
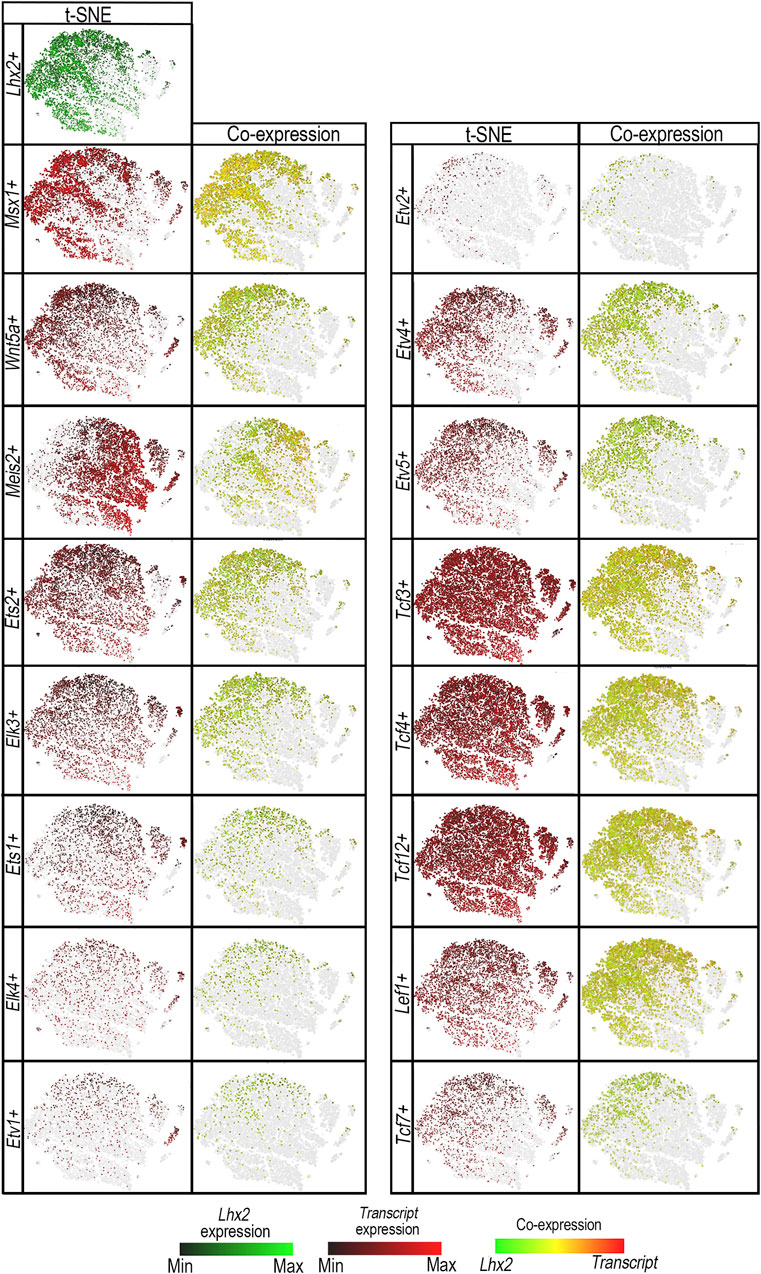
Figure 5. ETS and TCF/LEF transcription factor proteins are co-expressed in Lhx2+ cells. t-distributed stochastic neighbor embedding (t-SNE) plots shows segregation of E11.0 limb bud mesodermal cell populations by principal component analysis in Lhx2-expressing (Lhx2+) cells (green) vs. Ets transcripts: Ets1+, Ets2+, Elk3+, Elk4+, Etv1+, Etv2+, Etv4+, or Etv5+ cells (red), and Tcf/Lef transcripts: Tcf3+, Tcf4 +, Tcf12+, Tcf7+, or Lef1+ cells (red) in scRNA-seq E11 mouse limb data. The levels of Lhx2 expression range from bright green (high) to black (low), while candidate transcription factor expression levels range from red (high) to black (low). Cells expressing both Lhx2 and the candidate transcript are illustrated on a range from green to red, with yellow indicating similar levels of expression. The gray represents cells that do not express candidate transcripts, but are shown to preserve the segregation clusters and shape of the plot. Msx1+ and Wnt5a + cells were used as markers for the distal mesoderm and showed co-expression with Lhx2+ cells (yellow/orange) and co-segregation in the t-SNE plots with Lhx2+ cell populations. Meis2+ cells indicate the proximal mesoderm and do not co-segregate with Lhx2+ cells. Ets1+, Ets2+, Elk3+, Elk4+, and Etv1+ cells are ubiquitous throughout the limb bud with some cells segregating with Lhx2+ cells in the distal limb mesoderm and co-expressing Lhx2 (yellow/orange). Similarly, Tcf3+, Tcf4 +, Tcf12+, and Lef1+ cells are present in most limb bud cells, but cells that do co-segregate with Lhx2+ cells also co-express Lhx2. Etv2+, Etv4+, Etv5+, and Tcf7+ cells display a similar segregation pattern with Lhx2+ cells with most cells co-expressing Lhx2.
Similarly, multiple Tcf/Lef transcripts (Tcf3, Tcf4, Tcf7, Tcf12, and Lef1) are co-expressed in Lhx2+ murine (Figure 5) and human limb cells (Supplementary Figure S4). Cells expressing Tcf3, Tcf4, Tcf12, and Lef1 transcripts segregate ubiquitously throughout limb cells with a fraction of them co-expressing Lhx2. Tcf7+ cells appear to segregate with Lhx2+ cells, and although Tcf7 is expressed in fewer cells (Supplementary Table S5) than other Tcf/Lef1 transcripts (Tcf3, Tcf4, Tcf12, Lef1), most Tcf7+ cells co-express Lhx2 (Figure 5).
Since Etv4+, Etv5+, Etv2+, Tcf7+, and Lef1+ cells co-express Lhx2 and demonstrate co-segregation patterns most similar to Lhx2+ cells, we conclude that these transcription factors are the best candidates for regulating Lhx2 expression via LADLRM and LASARM.
JASPAR transcription factor binding site prediction database identified one putative Ets and one Tcf/Lef binding site within LADLRM that is conserved among human, mouse, and chicken with prediction binding scores of p < 10−2 (Figure 6; Supplementary Figure S5). To determine whether these binding sites are important for enhancer activity, we performed site-directed mutagenesis of the core Ets nucleotide sequence 5′-GGAA-3′ (Cooper et al., 2015; Sharrocks, 2001) and Tcf/Lef core nucleotide sequence 5′-SCTTTGATS-3′ (Cadigan and Waterman, 2012) in the chicken LADLRM sequence. Disruption of both ETS and TCF/LEF binding sites simultaneously (ΔETS/TCF) abolished enhancer activity. Mutation of the ETS binding site (ΔETS) significantly reduced enhancer activity (Figure 6) compared to wildtype, but was insufficient to remove activity. On the other hand, mutation of TCF/LEF binding site (ΔTCF) was sufficient to eliminate activity to negative control levels (empty reporter vector) (graph to negative control in Supplementary Figure S6A), demonstrating that this TCF/LEF binding site is necessary for LADLRM activity (Figure 6A).
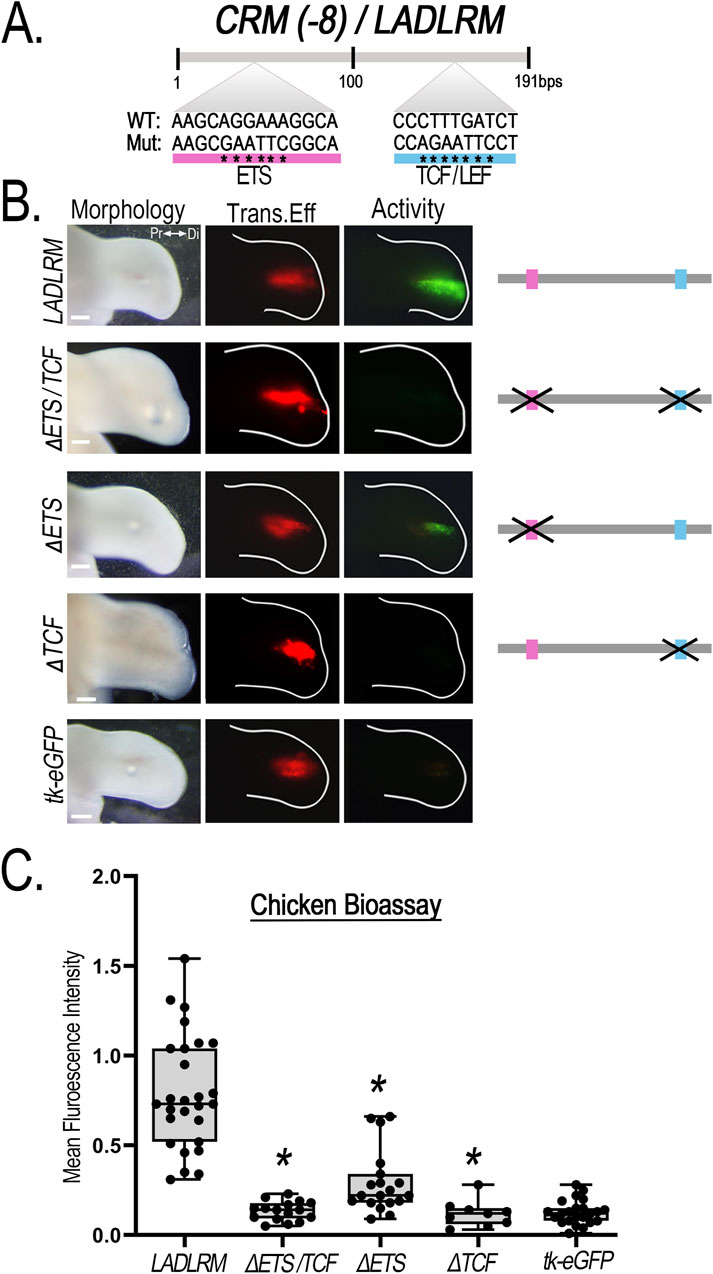
Figure 6. Disruption of a single putative TCF/LEF binding site removes CRM (-8)/LADLRM enhancer activity. (A) Schematic representation of the relative location of the putative ETS (pink) and TCF/LEF (blue) transcription factor binding site sequence in LADLRM. Asterisks (*) indicate the nucleotide base pair substitutions performed to generate mutated (Mut) sequences compared to the wild type (WT). (B) Simultaneous disruption of putative ETS and TCF binding site eliminates LADLRM activity. Alteration of predicted ETS binding site (ΔETS, n = 19) significantly reduces activity, while simultaneous disruption of the ETS and TCF/LEF binding site (ΔETS/TCF, n = 16) or loss of the TCF/LEF binding site (ΔTCF, n = 9) alone removes LADLRM activity to the level of the empty reporter vector (tk-eGFP, n = 15). (C) Boxplots depicting the mean fluorescence intensity of the CRM-GFP (normalized to RFP) using Fiji software. One-way ANOVA followed by Dunnett’s multiple comparison test was performed using LADLRM (n = 28) as the positive control *p < 0.05. Abbreviations used: Pr (proximal), Di (distal). Scale bars: 250 µm.
LASARM possesses two conserved putative ETS binding sites, 155 bp apart, and one putative TCF/LEF binding site (Figure 7). Mutation of both ETS binding sites (ΔETS [1/2]) eliminated activity (Figure 7). Mutation of ETS binding site 1 (ΔETS[1]) significantly reduced activity (Figure 7) when compared to wildtype, suggesting that this site contributes to, but is not necessary for, LASARM activity. Disruption of either the ETS binding site 2 (ΔETS[2]) or the TCF/LEF binding site (ΔTCF) was sufficient to remove activity (Figure 7) as statistical analysis revealed activity was not significantly different from negative control (empty reporter vector, Supplementary Figure S6B). These results indicate that both the ETS binding site 2 and the TCF/LEF binding site are required for LASARM activity.
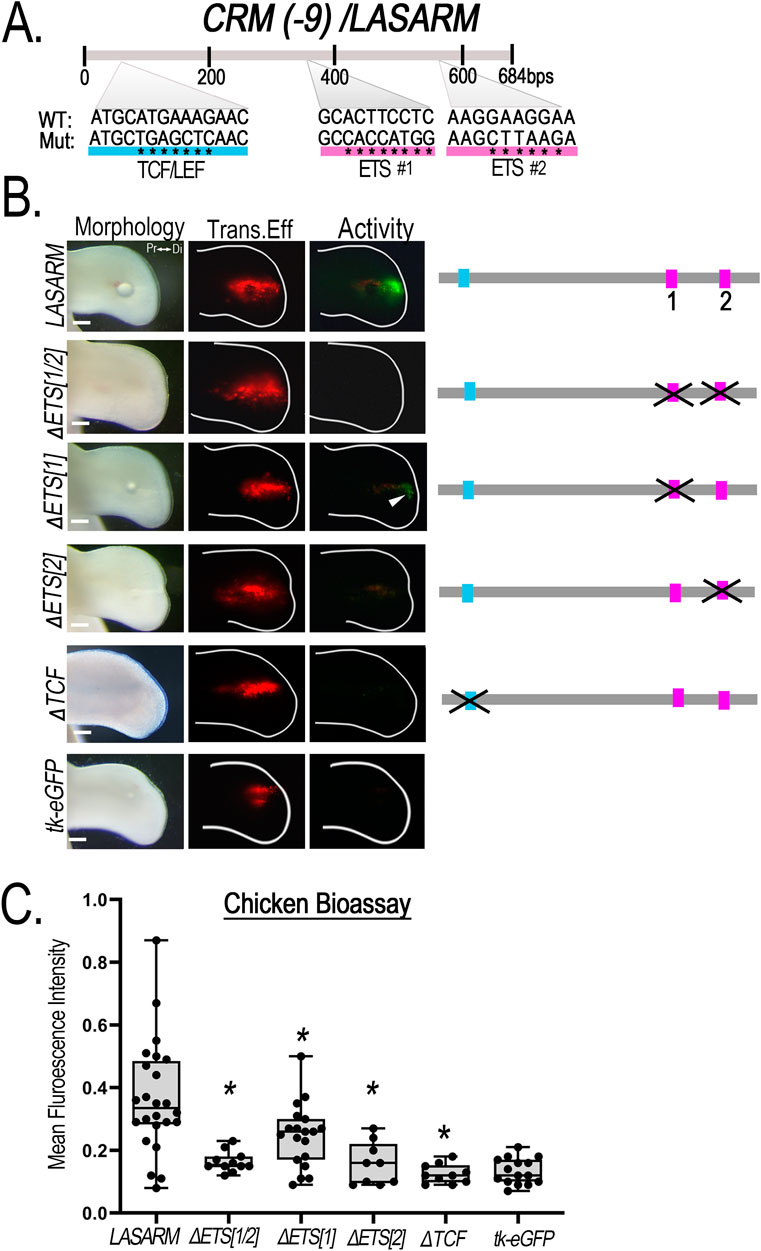
Figure 7. Disruption of putative ETS and TCF/LEF binding sites alters CRM (-9)/LASARM activity. (A) Schematic representation of the relative location of the two putative ETS (pink) and single TCF/LEF (blue) binding sites sequence in LASARM. Asterisks (*) indicate the nucleotide base pair substitutions performed to generate mutated (Mut) sequences compared to the wild type (WT). (B) Mutation of both ETS binding sites (ΔETS [1/2], n = 11) together abolishes LASARM activity. Disruption of ETS binding site 1 (ΔETS [1], n = 19) significantly reduces activity, while disruption of the single ETS binding site 2 (ΔETS[2], n = 9) or TCF/LEF binding site (ΔTCF, n = 10) abolishes LASARM activity to the level of the empty reporter vector (tk-eGFP, n = 15). (C) Boxplots depicting mean fluorescence intensity of the CRM-GFP (normalized to RFP) using Fiji software. One-way ANOVA followed by Dunnett’s multiple comparison test was performed using LASARM (n = 24) as the control, *p < 0.05. Abbreviations used: Pr (proximal), Di (distal). Scale bars: 250 µm.
Binding of Tcf/Lef to both LADLRM and LASARM is supported by a preliminary report by Malkmus et al. (2024) showing associated binding of β-catenin (a co-factor in Tcf/Lef binding) by ChIP-seq to these CRMs (Supplementary Figure S6C). Unfortunately, we were unable to find published ChIP-seq data for embryonic mouse limbs for any of the ETS transcription factors to confirm binding to LADLRM or LASARM.
In Lhx2/Lhx9 mutant mice, Shh expression is markedly reduced in the ZPA with mice exhibiting distal limb truncations (Tzchori et al., 2009). In chicken limbs, inhibition of LHX2 function and morpholino knockdown of LHX2 in the ZPA reduce SHH expression and aborts limb outgrowth (Rodriguez-Esteban et al., 1998; Watson et al., 2018). Furthermore, LADLRM and LASARM show intense activity in the distal posterior limb mesoderm overlapping the ZPA domain (Figures 3, 4), supporting a role for Lhx2 in the expression and maintenance of Shh.
The limb-specific ZRS enhancer regulates Shh expression (Lettice et al., 2002; Lettice et al., 2017). Lhx2 expression overlaps Shh and ZRS activity in the ZPA (Figure 8A). The ZRS sequence contains two conserved putative Lhx2 binding sites that possess the core 5′-TAATTA-3′motif (Roberson et al., 1994), located 139 bp pairs from each other (Figure 8B). This provides a potential mechanism for direct regulation of Shh by Lhx2. To determine whether these two Lhx2 binding sites are required for ZRS activity, we simultaneously mutated both LHX2 binding sites (ΔLHX2) in the chicken. Surprisingly, the ZRS with mutant binding sites had no significant loss of activity (Figures 8C, D) indicating that Lhx2 does not regulate Shh expression through these two binding sites.
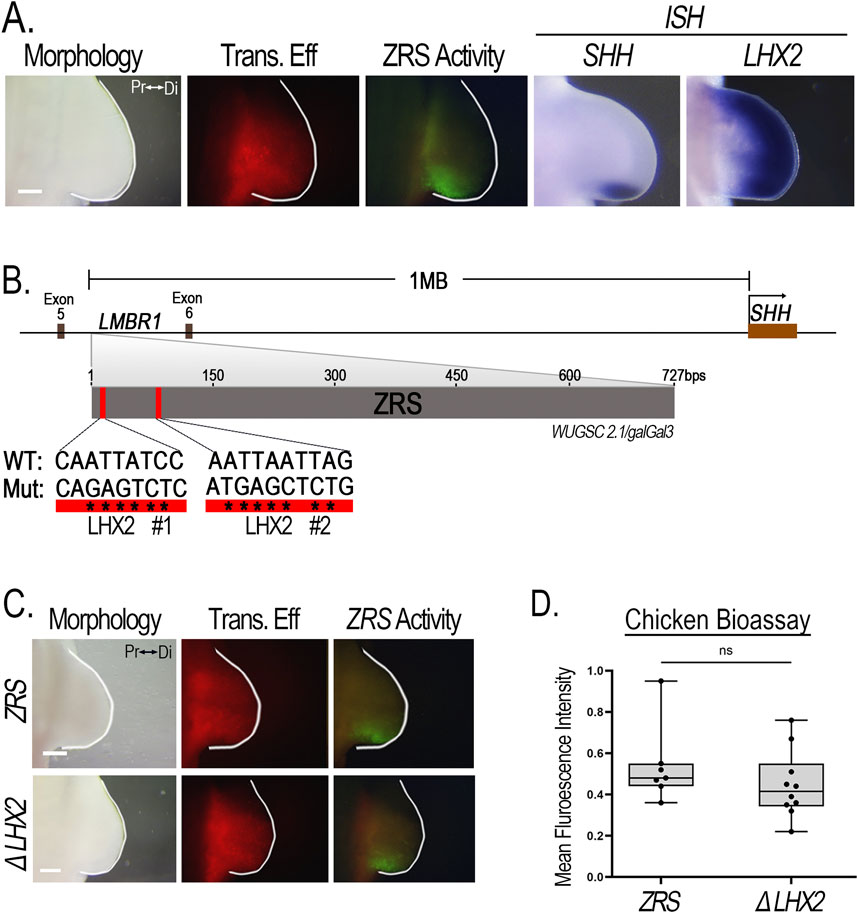
Figure 8. ZRS activity is not affected when LHX2 binding sites are disrupted. (A) LHX2 expression overlaps the activity of the ZRS and SHH expression posteriorly in chicken limb buds. (B) Schematic diagram showing the ZRS situated between Exon 5 and 6 of the LMBR1 gene, 1 Megabase (Mb) upstream of the SHH promoter. The ZRS contains two putative LHX2 binding sites (red rectangles). Asterisks (*) indicate the nucleotide base pair substitutions performed to generate mutated (Mut) sequences compared to the wild type (WT). (C) Disruption of the two LHX2 binding sites (ΔLHX2, n = 10) yields no change in ZRS activity compared to wildtype (ZRS, n = 7). (D) Boxplots depicting the mean fluorescence intensity of the ZRS-GFP-reporter (normalized to RFP) using Fiji software. Transfection efficiency (Trans. Eff) was assessed with a β-actin promoter-driven RFP plasmid. Student’s t-test (two-tailed, p < 0.05) was performed comparing the WT ZRS to the mutant. Abbreviations used: Pr (proximal), Di (distal), ns (non-significant). Scale bars: 250 µm.
Lhx2 modulates the delicate switch between the proliferation and differentiation of progenitor cells into their respective lineages by regulating the expression of pathway-specific genes during embryogenesis (Hsu et al., 2015; Li et al., 2022; Zibetti et al., 2019). For example, during brain development, Lhx2 via the Wnt/B-catenin pathway supports the proliferation and subsequent neurogenesis of cortical progenitors needed for appropriate cortex growth and differentiation; loss of Lhx2 results in precocious initiation of neurogenesis and a smaller cortex (Bulchand et al., 2001; Hsu et al., 2015). In addition, Lhx2 exhibits pioneering transcription factor activity, establishing competence for gene expression by increasing chromatin accessibility and recruiting transcriptional proteins to developmental gene loci that regulate cell fate (Suresh et al., 2023; Zibetti et al., 2019). For instance, in the retina, Lhx2 remodels the chromatin of retinal progenitor cells in preparation for development and sustains their proliferation, with loss of Lhx2 resulting in premature differentiation (Gordon et al., 2013; Zibetti et al., 2019).
In the limb, Lhx2 is expressed in the distal mesoderm sub-adjacent to the apical ectodermal ridge (AER) and overlapping the zone of polarizing activity (ZPA) (Rodriguez-Esteban et al., 1998; Tzchori et al., 2009; Watson et al., 2018; Yokoyama et al., 2017). This sub-AER mesoderm is a region of undifferentiated progenitor cells that contributes to patterned limb outgrowth (Savage et al., 1993; Hara et al., 1998). Tzchori et al. (2009) suggested that Lhx2 (and Lhx9) function to integrate AER-Fgfs and ZPA-Shh signals that sustain progenitor cell populations in the distal limb during patterning and outgrowth. However, the mechanisms that induce and maintain Lhx2 to support these functions during limb development are unknown.
In this study, we identified two cis-regulatory modules (CRMs) within the Lhx2 locus with limb-specific activity in the distal mesoderm coincident with Lhx2 expression (Figures 3, 4). Functional enhancers of Lhx2 have been identified for the midbrain and hindbrain (Lee et al., 2011); however, our study is the first to identify limb-specific Lhx2 CRMs active in the limb. We named the CRMs based on their activity pattern, with Lhx2-associated distal Iimb regulatory module (LADLRM) exhibiting more broad activity extending about 500 μm from the distal tip with accentuated activity over the ZPA. The other CRM, Lhx2-associated sub-AER regulatory module (LASARM), has activity within the cells just beneath the AER (sub-AER). The intense activity of both LADLRM and LASARM in the posterior ZPA-related domain implies these CRMs work to ensure that Lhx2-expressing cells are kept responsive to AER and ZPA signals during proximodistal limb segment differentiation and anteroposterior patterning.
The AER expresses multiple Fgfs (Fgf4, Fgf8, Fgf9, and Fgf17) (Sun et al., 2002) and Wnts (Wnt3, Wnt4, Wnt6, and Wnt7b) (Geetha-Loganathan et al., 2008) that contribute to AER formation and maintenance. These factors also participate in the feedback loops that control limb outgrowth and patterning. Ets and Tcf/Lef transcription factors are part of the Fgf and Wnt signaling pathways, respectively. Ets transcription factors are recognized as regulators of proximodistal and anteroposterior limb patterning (Koyano-Nakagawa et al., 2022; Mao et al., 2009; Yamamoto-Shiraishi et al., 2014). For example, data suggest that the Etv4/5 transcription factors regulate anteroposterior patterning by localizing Shh expression to the ZPA. In their absence, ectopic anterior Shh is expressed, and radial polydactyly develops (Koyano-Nakagawa et al., 2022; Lettice et al., 2012; Mao et al., 2009; Zhang et al., 2009). Tcf/Lef transcription factors are also important regulators of AER development, limb outgrowth, and proximodistal patterning (Kawakami et al., 2001; Kengaku et al., 1998; ten Berge et al., 2008). For instance, Lef1−/−Tcf1−/− mutant mice fail to develop a functional AER and limb development is arrested (Galceran et al., 1999). The co-expression of several Ets and Tcf/Lef transcripts in Lhx2-expressing cells, the associated binding of β-catenin to both CRMs, and the importance of their predicted binding sites for both LADLRM and LASARM activity suggest that Fgfs and Wnts participate in the maintenance of Lhx2 expression.
An unexpected finding in this study is the differential activity pattern of LADLRM and LASARM between the chicken and mouse. In chicken, LADLRM derived from either chicken or mouse is active in both the dorsal and ventral mesoderm, whereas in mice, the murine LADLRM is active only in the dorsal limb mesenchyme. These data suggest that mouse ventral mesoderm, but not chicken ventral mesoderm, is expressing a regulatory transcription factor that is inhibiting LADLRM activity. Similarly, the chicken LASARM has no dorsal-ventral bias in the chicken. However, the activity of the murine LASARM sequence in transgenic mice is accentuated ventrally, while posteriorly, adjacent to the ZPA, the activity is also expressed in the dorsal mesoderm. The suspected regulatory transcription factor in the mouse ventral limb mesoderm could also act to ventrally accentuate LASARM activity. Further work is needed to clarify the transcriptional differences between murine and chicken ventral limb bud mesoderm.
A probable explanation is the species-specific (chicken-mouse) differences in Lhx2 and Lhx9 expression. In chicken, LHX9 is expressed only in the anterior mesoderm and does not play a role in the regulation of SHH (Watson et al., 2018). However, LHX2 is expressed throughout the distal mesenchyme, extending the full anterior and posterior length of the limb, and is necessary for the maintenance of SHH expression (Watson et al., 2018). Moreover, repression of chicken LHX2 expression stops limb outgrowth and halts AER formation (Rodriguez-Esteban et al., 1998). This suggests that in chicken LHX2 may be the main LIM homeodomain transcription factor in the distal sub-AER mesoderm to regulate patterning and outgrowth, requiring LADLRM activity in both the dorsal and ventral mesoderm. In contrast in mice, Lhx9 expression extends beyond the anterior mesoderm at the distal tip and into the posterior limb domain overlapping the ZPA. The redundant, overlapping Lhx9 expression in the Lhx2 domain may lessen the constraints for Lhx2 to have both dorsal and ventral expression. Alternatively, mice may harbor another ventrally-active CRM to regulate the ventral expression of Lhx2.
Although Lhx2 is required for Shh activity, and both LADLRM and LASARM show accentuated posterior activity associated with the ZPA, our data indicate that predicted LHX2 binding sites are not necessary for the activity of the ZRS, the Shh enhancer, in an isolated reporter construct. Similarly, preliminary data from Bower et al. (2024) found that disruption of Lhx2 binding sites within a ZRS-reporter construct did not alter activity; however, when these same sites are removed from the endogenous ZRS in mice, limb truncation defects occurred indistinguishable from the complete loss of Shh. They suggest that these binding sites are necessary for mediating long-range enhancer activation (Bower et al., 2024). Collectively these data suggest a role for Lhx2 in tethering the ZRS to its promoter region (or promoting their interaction) and a rationale for the accentuated ZPA-related activity of the Lhx2 CRMs. Alternatively, accentuated ZPA-related Lhx2 expression could increase chromatin accessibility (Zibetti et al., 2019) and accentuate ZPA-related Shh transcription compared to other sites lacking Lhx2.
Lhx2 could also regulate Shh expression independent of the ZRS. Shh co-receptors Cdon and Gas1 mediate Shh signaling and regulate limb patterning and digit specification, as mutants display zeugopod and autopod defects (Allen et al., 2007; Allen et al., 2011; Echevarría-andino et al., 2023). In mouse retinal progenitor cells, Lhx2 binds to the Cdon and Gas1 loci to support activation of the Shh (Li et al., 2022) and likewise may do so in the limb.
In summary, we discovered two Lhx2-associated cis-regulatory modules, LADLRM and LASARM, within the Lhx2 locus that have limb-specific activity overlapping Lhx2 expression. Several Ets and Tcf/Lef members co-express with Lhx2. Moreover, Ets transcription factor binding sites contribute to LADLRM activity and are necessary for LASARM activity. Additionally, Tcf/Lef transcription factor binding sites are essential for both the activity of LADLRM and LASARM. Together, these data suggest that Fgf and Wnt signaling pathways participate in regulating Lhx2 expression in the distal limb mesoderm.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
The animal study was approved by Loma Linda Institutional Animal Care and Utilization Committe. The study was conducted in accordance with the local legislation and institutional requirements.
JB: Conceptualization, Data curation, Formal Analysis, Investigation, Methodology, Writing–original draft, Writing–review and editing, Software, Supervision, Visualization. AS-L: Conceptualization, Data curation, Investigation, Visualization, Writing–review and editing, Methodology, Formal Analysis. BW: Conceptualization, Data curation, Formal Analysis, Investigation, Methodology, Visualization, Writing–review and editing. EH: Formal Analysis, Methodology, Visualization, Writing–review and editing, Data curation. CM: Data curation, Formal Analysis, Methodology, Writing–review and editing. KK: Data curation, Formal Analysis, Methodology, Writing–review and editing. AC: Data curation, Formal Analysis, Visualization, Writing–review and editing, Methodology. KW: Data curation, Formal Analysis, Methodology, Visualization, Writing–review and editing. R-LY: Methodology, Writing–review and editing, Formal Analysis. JK: Data curation, Methodology, Writing–review and editing, Formal Analysis. MY: Data curation, Writing–review and editing, Formal Analysis, Visualization. JC: Data curation, Writing–review and editing. JA: Data curation, Writing–review and editing. SK: Methodology, Writing–review and editing. EK: Methodology, Writing–review and editing. CP: Data curation, Methodology, Writing–review and editing, Investigation, Project administration, Supervision, Writing–original draft. KO: Conceptualization, Formal Analysis, Funding acquisition, Methodology, Project administration, Resources, Supervision, Visualization, Writing–original draft, Writing–review and editing, Data curation, Investigation.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This research was supported in part by the Pathology Research Endowment, Loma Linda University.
The authors acknowledge the support of the Chao Family Comprehensive Cancer Center Transgenic Mouse Facility Shared Resource, supported by the National Cancer Institute of the National Institutes of Health under award number P30CA062203 (SK, EK). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors declare that no Generative AI was used in the creation of this manuscript.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2025.1552716/full#supplementary-material
Allen, B. L., Song, J. Y., Izzi, L., Althaus, I. W., Kang, J., Krauss, R. S., et al. (2011). Overlapping roles and collective requirement for the coreceptors GAS1, CDO, and BOC in SHH pathway function. Cell Press 20 (6), 775–787. doi:10.1016/j.devcel.2011.04.018
Allen, B. L., Tenzen, T., and Mcmahon, A. P. (2007). The Hedgehog-binding proteins Gas1 and Cdo cooperate to positively regulate Shh signaling during mouse development. Genes and Dev. 21 (10), 1244–1257. doi:10.1101/gad.1543607
Berlivet, S., Paquette, D., Dumouchel, A., Langlais, D., Dostie, J., and Kmita, M. (2013). Clustering of tissue-specific sub-TADs accompanies the regulation of HoxA genes in developing limbs. PLoS Genet. 9 (12), e1004018. doi:10.1371/journal.pgen.1004018
Blair, S. S., Brower, D. L., Thomas, J. B., and Zavortink, M. (1994). The role of apterous in the control of dorsoventral compartmentalization and PS integrin gene expression in the developing wing of Drosophila. Development 120 (7), 1805–1815. doi:10.1242/dev.120.7.1805
Bower, G., Hollingsworth, E. W., Jacinto, S., Clock, B., Cao, K., Liu, M., et al. (2024). Conserved cis-acting range extender element mediates extreme long-range enhancer activity in mammals. BioRxiv, 2024.05.26.595809. Available at: https://www.biorxiv.org/content/10.1101/2024.05.26.595809v1.
Buenrostro, J. D., Giresi, P. G., Zaba, L. C., Chang, H. Y., and Greenleaf, W. J. (2013). Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods 10 (12), 1213–1218. doi:10.1038/nmeth.2688
Buenrostro, J. D., Wu, B., Litzenburger, U. M., Ruff, D., Gonzales, M. L., Snyder, M. P., et al. (2015). Single-cell chromatin accessibility reveals principles of regulatory variation. Nature 523 (7561), 486–490. doi:10.1038/nature14590
Bulchand, S., Grove, E. A., Porter, F. D., and Tole, S. (2001). LIM-homeodomain gene Lhx2 regulates the formation of the cortical hem. Mech. Dev. 100 (2), 165–175. doi:10.1016/S0925-4773(00)00515-3
Cadigan, K. M., and Waterman, M. L. (2012). TCF/LEFs and Wnt signaling in the nucleus. Cold Spring Harb. Perspect. Biol. 4 (11), a007906. doi:10.1101/cshperspect.a007906
Cooper, C. D. O., Newman, J. A., Aitkenhead, H., Allerston, C. K., and Gileadi, O. (2015). Structures of the Ets protein DNA-binding domains of transcription factors Etv1, Etv4, Etv5, and Fev. DETERMINANTS OF DNA BINDING AND REDOX REGULATION BY DISULFIDE BOND FORMATION. Journal of Biological Chemistry 290 (22), 13692–13709. doi:10.1074/jbc.M115.646737
Cotney, J., Leng, J., Yin, J., Reilly, S. K., Demare, L. E., Emera, D., et al. (2013). The evolution of lineage-specific regulatory activities in the human embryonic limb. Cell 154 (1), 185–196. doi:10.1016/j.cell.2013.05.056
DeMare, L. E., Leng, J., Cotney, J., Reilly, S. K., Yin, J., Sarro, R., et al. (2013). The genomic landscape of cohesin-Associated chromatin interactions. Genome Res. 23 (8), 1224–1234. doi:10.1101/gr.156570.113
Echevarría-andino, M. L., Franks, N. E., Schrader, H. E., Hong, M., Krauss, R. S., and Allen, B. L. (2023). CDON contributes to Hedgehog-dependent patterning and growth of the developing limb. Dev. Biol. 493 (2022), 1–11. doi:10.1016/j.ydbio.2022.09.011
Galceran, J., Fariñas, I., Depew, M. J., Farin, I., Clevers, H., and Grosschedl, R. (1999). Wnt3a −/−-like phenotype and limb deficiency in Lef1−/− Tcf1 −/− mice. Genes and Dev. 13 (6), 709–717. doi:10.1101/gad.13.6.709
Geetha-Loganathan, P., Nimmagadda, S., and Scaal, M. (2008). Wnt signaling in limb organogenesis. Organogenesis 4 (2), 109–115. doi:10.4161/org.4.2.5857
Gordon, P. J., Yun, S., Clark, A. M., Monuki, E. S., Murtaugh, L. C., and Levine, E. M. (2013). Lhx2 balances progenitor maintenance with neurogenic output and promotes competence state progression in the developing retina. J. Neurosci. 33 (30), 12197–12207. doi:10.1523/JNEUROSCI.1494-13.2013
Hamburger, V., and Hamilton, H. L. (1951). A series of normal stages in the development of the chick embryo. J. Morphol. 4, 231–272. doi:10.1002/aja.1001950404
Hara, K., Kimura, J., and Ide, H. (1998). Effects of FGFs on the morphogenic potency and AER-maintenance activity of cultured progress zone cells of chick limb bud. Int. J. Dev. Biol. 42 (4), 591–599.
He, P., Williams, B. A., Trout, D., Marinov, G. K., Amrhein, H., Berghella, L., et al. (2020). The changing mouse embryo transcriptome at whole tissue and single-cell resolution. Nature 583 (7818), 760–767. doi:10.1038/s41586-020-2536-x
Hsu, L. C. L., Nam, S., Cui, Y., Chang, C., Wang, C., Kuo, H., et al. (2015). Lhx2 regulates the timing of β-catenin-dependent cortical neurogenesis. Proc. Natl. Acad. Sci. U. S. A. 112 (39), 12199–12204. doi:10.1073/pnas.1507145112
Jhanwar, S., Malkmus, J., Stolte, J., Romashkina, O., Zuniga, A., and Zeller, R. (2021). Conserved and species-specific chromatin remodeling and regulatory dynamics during mouse and chicken limb bud development. Nat. Commun. 12 (1), 5685. doi:10.1038/s41467-021-25935-3
Kardon, G., Harfe, B. D., and Tabin, C. J. (2003). A Tcf4-positive mesodermal population provides a prepattern for vertebrate limb muscle patterning. Dev. Cell 5 (6), 937–944. doi:10.1016/S1534-5807(03)00360-5
Kawakami, Y., Capdevila, J., Büscher, D., Itoh, T., Esteban, C. R., and Belmonte, J. C. I. (2001). WNT signals control FGF-dependent limb initiation and AER induction in the chick embryo. Cell 104 (6), 891–900. doi:10.1016/S0092-8674(01)00285-9
Kengaku, M., Capdevila, J., Rodriguez-Esteban, C., De La Peña, J., Johnson, R. L., Belmonte, J. C. I., et al. (1998). Distinct WNT pathways regulating AER formation and dorsoventral polarity in the chick limb bud. Science 280 (5367), 1274–1277. doi:10.1126/science.280.5367.1274
Kolovos, P., Knoch, T. A., Grosveld, F. G., Cook, P. R., and Papantonis, A. (2012). Enhancers and silencers: an integrated and simple model for their function. Epigenetics Chromatin 5 (1), 1. doi:10.1186/1756-8935-5-1
Koyano-Nakagawa, N., Gong, W., Das, S., Theisen, J. W. M., Swanholm, T. B., Van Ly, D., et al. (2022). Etv2 regulates enhancer chromatin status to initiate Shh expression in the limb bud. Nat. Commun. 13 (1), 4221–4238. doi:10.1038/s41467-022-31848-6
Kraft, K., Magg, A., Heinrich, V., Riemenschneider, C., Schöpflin, R., Markowski, J., et al. (2019). Serial genomic inversions induce tissue-specific architectural stripes, gene misexpression and congenital malformations. Nat. Cell Biol. 21 (3), 305–310. doi:10.1038/s41556-019-0273-x
Lee, A. P., Brenner, S., and Venkatesh, B. (2011). Mouse transgenesis identifies conserved functional enhancers and cis-regulatory motif in the vertebrate LIM homeobox gene Lhx2 locus. PLoS ONE 6 (5), e20088. doi:10.1371/journal.pone.0020088
Lettice, L. A., Devenney, P., De Angelis, C., and Hill, R. E. (2017). The conserved sonic hedgehog limb enhancer consists of discrete functional elements that regulate precise spatial expression. Cell Rep. 20 (6), 1396–1408. doi:10.1016/j.celrep.2017.07.037
Lettice, L. A., Horikoshi, T., Heaney, S. J. H., Van Baren, M. J., Van Der Linde, H. C., Breedveld, G. J., et al. (2002). Disruption of a long-range cis-acting regulator for Shh causes preaxial polydactyly. Proc. Natl. Acad. Sci. U. S. A. 99 (11), 7548–7553. doi:10.1073/pnas.112212199
Lettice, L. A., Williamson, I., Wiltshire, J. H., Peluso, S., Devenney, P. S., Hill, A. E., et al. (2012). Opposing functions of the ETS factor family define Shh spatial expression in limb buds and underlie polydactyly. Dev. Cell 22 (2), 459–467. doi:10.1016/j.devcel.2011.12.010
Lewis, M. W., Li, S., and Franco, H. L. (2019). Transcriptional control by enhancers and enhancer RNAs. Transcription 10 (4–5), 171–186. doi:10.1080/21541264.2019.1695492
Li, X., Gordon, P. J., Gaynes, J. A., Fuller, A. W., Ringuette, R., Santiago, C. P., et al. (2022). Lhx2 is a progenitor-intrinsic modulator of Sonic Hedgehog signaling during early retinal neurogenesis. ELife 11, 783422–e78435. doi:10.7554/ELIFE.78342
Long, H. S., Greenaway, S., Powell, G., Mallon, A. M., Lindgren, C. M., and Simon, M. M. (2022). Making sense of the linear genome, gene function and TADs. Epigenetics Chromatin 15 (1), 4–19. doi:10.1186/s13072-022-00436-9
Lopez-Delisle, L., Rabbani, L., Wolff, J., Bhardwaj, V., Backofen, R., Grüning, B., et al. (2021). pyGenomeTracks: reproducible plots for multivariate genomic datasets. Bioinformatics 37 (3), 422–423. doi:10.1093/bioinformatics/btaa692
Malkmus, J., Morabito, A., Lopez-Delisle, L., Esteban, L. A., Mayran, A., Zuniga, A., et al. (2024). WNT signaling coordinately controls mouse limb bud outgrowth and establishment of the digit-interdigit pattern. BioRxiv 12 (24), 629665. doi:10.1101/2024.12.25.629665
Mao, J., McGlinn, E., Huang, P., Tabin, C. J., and McMahon, A. P. (2009). Fgf-Dependent Etv4/5 activity is required for posterior restriction of sonic hedgehog and promoting outgrowth of the vertebrate limb. Dev. Cell 16 (4), 600–606. doi:10.1016/j.devcel.2009.02.005
Mathew, S. J., Hansen, J. M., Merrell, A. J., Murphy, M. M., Lawson, J. A., Hutcheson, D. A., et al. (2011). Connective tissue fibroblasts and Tcf4 regulate myogenesis. Development 138 (2), 371–384. doi:10.1242/dev.057463
Niswander, L., Jeffrey, S., Martin, G. R., and Tickle, C. (1994). A positive feedback loop coordinates growth and patterning in the vertebrate limb. Nature 371 (6498), 609–612. doi:10.1038/371609a0
Oberg, K. C., Pira, C. U., Revelli, J. P., Ratz, B., Aguilar-Cordova, E., and Eichele, G. (2002). Efficient ectopic gene expression targeting chick mesoderm. Dev. Dyn. 224 (3), 291–302. doi:10.1002/dvdy.10104
Ohuchi, H., Nakagawa, T., Itoh, N., and Noji, S. (1999). FGF10 can induce Fgf8 expression concomitantly with En1 and R-fng expression in chick limb ectoderm, independent of its dorsoventral specification. Dev. Growth Differ. 41 (6), 665–673. doi:10.1046/j.1440-169X.1999.00466.x
Ohuchi, H., Nakagawa, T., Yamamoto, A., Araga, A., Ohata, T., Ishimaru, Y., et al. (1997). The mesenchymal factor, FGF10, initiates and maintains the outgrowth of the chick limb bud through interaction with FGF8, an apical ectodermal factor. Development 124 (11), 2235–2244. doi:10.1242/dev.124.11.2235
Pira, C. U., Caltharp, S. A., Kanaya, K., Manu, S. K., Greer, L. F., and Oberg, K. C. (2008). “Identification of developmental enhancers using targeted regional electroporation (Trep) of evolutionarily conserved regions,” in Bioluminescence and chemiluminescence, 319–322. doi:10.1142/9789812839589_0073
Ramírez, F., Bhardwaj, V., Arrigoni, L., Lam, K. C., Grüning, B. A., Villaveces, J., et al. (2018). High-resolution TADs reveal DNA sequences underlying genome organization in flies. Nat. Commun. 9 (1), 189. doi:10.1038/s41467-017-02525-w
Roberson, M. S., Schoderbek, W. E., Tremml, G., and Maurer, R. A. (1994). Activation of the glycoprotein hormone alpha-subunit promoter by a LIM-homeodomain transcription factor. Mol. Cell. Biol. 14 (5), 2985–2993. doi:10.1128/mcb.14.5.2985
Rodriguez-Esteban, C., Schwabe, J. W. R., De La Peña, J., Rincon-Limas, D. E., Magallón, J., Botas, J., et al. (1998). Lhx2, a vertebrate homologue of apterous, regulates vertebrate limb outgrowth. Development 125 (20), 3925–3934. doi:10.1242/dev.125.20.3925
Savage, M. P., Fallon, J. F., Hart, C. E., Riley, B. B., Sasse, J., and Olwin, B. B. (1993). Distribution of FGF-2 suggests it has a role in chick limb bud growth. Dev. Dyn. 198 (3), 159–170. doi:10.1002/aja.1001980302
Schindelin, J., Arganda-Carreras, I., Frise, E., Kaynig, V., Longair, M., Pietzsch, T., et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat. Methods 9 (7), 676–682. doi:10.1038/nmeth.2019
Sharrocks, A. D. (2001). The ETS-domain transcription factor family. Nat. Rev. Mol. Cell Biol. 2 (11), 827–837. doi:10.1038/35099076
Shen, X., Bao, W., Yu, W., Liang, R., Nguyen, B., and Liu, Y. (2017). An improved method with high sensitivity and low background in detecting low β-galactosidase expression in mouse embryos. PLoS ONE 12 (5), 01769155–e177011. doi:10.1371/journal.pone.0176915
Shihan, M. H., Novo, S. G., Le Marchand, S. J., Wang, Y., and Duncan, M. K. (2021). A simple method for quantitating confocal fluorescent images. Biochem. Biophysics Rep. 25, 100916. doi:10.1016/j.bbrep.2021.100916
Sun, X., Mariani, F. V., and Martin, G. R. (2002). Functions of FGF signalling from the apical ectodermal ridge in limb development. Nature 418 (6897), 501–508. doi:10.1038/nature00902
Suresh, V., Muralidharan, B., Pradhan, S. J., Bose, M., D’Souza, L., Parichha, A., et al. (2023). Regulation of chromatin accessibility and gene expression in the developing hippocampal primordium by LIM-HD transcription factor LHX2. PLoS Genet. 19 (8 August), 10108744–1010922. doi:10.1371/journal.pgen.1010874
ten Berge, D., Brugmann, S. A., Helms, J. A., and Nusse, R. (2008). Wnt and FGF signals interact to coordinate growth with cell fate specification during limb development. Development 135 (19), 3247–3257. doi:10.1242/dev.023176
Tzchori, I., Day, T. F., Carolan, P. J., Zhao, Y., Wassif, C. A., Li, L. Q., et al. (2009). LIM homeobox transcription factors integrate signaling events that control three-dimensional limb patterning and growth. Development 136 (8), 1375–1385. doi:10.1242/dev.026476
Uchikawa, M., Ishida, Y., Takemoto, T., Kamachi, Y., Kondoh, H., and Embryos, C. (2003). Functional analysis of chicken Sox2 enhancers highlights an array of diverse regulatory elements that are conserved in mammals. Dev. Cell 4 (4), 509–519. doi:10.1016/s1534-5807(03)00088-1
Visel, A., Blow, M. J., Li, Z., Zhang, T., Akiyama, J. A., Holt, A., et al. (2009). ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature 457 (7231), 854–858. doi:10.1038/nature07730
Watson, B. A., Feenstra, J. M., Van Arsdale, J. M., Rai-Bhatti, K. S., Kim, D. J. H., Coggins, A. S., et al. (2018). LHX2 mediates the FGF-to-SHH regulatory loop during limb development. J. Dev. Biol. 6 (2), 13–19. doi:10.3390/JDB6020013
Williams, J. A., Bell, J. B., and Carroll, S. B. (1991). Control of Drosophila wing and haltere development by the nuclear vestigial gene product. Genes Dev. 5 (12), 2481–2495. doi:10.1101/gad.5.12b.2481
Wolff, J., Bhardwaj, V., Nothjunge, S., Richard, G., Renschler, G., Gilsbach, R., et al. (2018). Galaxy HiCExplorer: a web server for reproducible Hi-C data analysis, quality control and visualization. Nucleic Acids Res. 46 (W1), W11–W16. doi:10.1093/nar/gky504
Wu, C. I., Hoffman, J. A., Shy, B. R., Ford, E. M., Fuchs, E., Nguyen, H., et al. (2012). Function of wnt/β-catenin in counteracting Tcf3 repression through the tcf3-β-catenin interaction. Dev. Camb. 139 (12), 2118–2129. doi:10.1242/dev.076067
Yamada, M., Szendro, P. I., Prokscha, A., Schwartz, R. J., and Eichele, G. (1999). Evidence for a role of Smad6 in chick cardiac development. Dev. Biol. 215 (1), 48–61. doi:10.1006/dbio.1999.9419
Yamamoto-Shiraishi, Y., Higuchi, H., Yamamoto, S., Hirano, M., and Kuroiwa, A. (2014). Etv1 and Ewsr1 cooperatively regulate limb mesenchymal Fgf10 expression in response to apical ectodermal ridge-derived fibroblast growth factor signal. Dev. Biol. 394 (1), 181–190. doi:10.1016/j.ydbio.2014.07.022
Yang, Y., Drossopoulou, G., Chuang, P., Duprez, D., Marti, E., Bumcrot, D., et al. (1997). Relationship between dose, distance and time in Sonic Hedgehog-mediated regulation of anteroposterior polarity in the chick limb. Development 4404 (124), 4393–4404. doi:10.1242/dev.124.21.4393
Yeboah, R. L., Pira, C. U., Shankel, M., Cooper, A. M., Haro, E., Ly, V. D., et al. (2023). Sox, Fox, and Lmx1b binding sites differentially regulate a Gdf5-Associated regulatory region during elbow development. Front. Cell Dev. Biol. 11, 1215406. doi:10.3389/fcell.2023.1215406
Yokoyama, S., Furukawa, S., Kitada, S., Mori, M., Saito, T., Kawakami, K., et al. (2017). Analysis of transcription factors expressed at the anterior mouse limb bud. PLoS ONE 12 (5), e0175673. doi:10.1371/journal.pone.0175673
Yordy, J. S., and Muise-Helmericks, R. C. (2000). Signal transduction and the Ets family of transcription factors. Oncogene 19 (55), 6503–6513. doi:10.1038/sj.onc.1204036
Zhang, B., He, P., Lawrence, J. E. G., Wang, S., Tuck, E., Williams, B. A., et al. (2023). A human embryonic limb cell atlas resolved in space and time. Nature 635 (809), 668–678. doi:10.1038/s41586-023-06806-x
Zhang, Z., Verheyden, J. M., Hassell, J. A., and Sun, X. (2009). FGF-regulated Etv genes are essential for repressing Shh expression in mouse limb buds. Dev. Cell 16 (4), 607–613. doi:10.1016/j.devcel.2009.02.008
Zhu, J., Nakamura, E., Nguyen, M. T., Bao, X., Akiyama, H., and Mackem, S. (2008). Uncoupling sonic hedgehog control of pattern and expansion of the developing limb bud. Dev. Cell 14 (4), 624–632. doi:10.1016/j.devcel.2008.01.008
Keywords: Lhx2, vertebrate limb development, cis-regulatory module, gene regulation, FGFs, Wnt, ETS, Tcf/Lef
Citation: Britton JC, Somogyi-Leatigaga A, Watson BA, Haro E, Mulder CG, Kennedy KD, Cooper AM, Whitley KL, Yeboah R-L, Kim J, Yu MC, Campos JD, Amoah J, Kawauchi S, Kim E, Pira CU and Oberg KC (2025) Evidence for Fgf and Wnt regulation of Lhx2 during limb development via two limb-specific Lhx2-associated cis-regulatory modules. Front. Cell Dev. Biol. 13:1552716. doi: 10.3389/fcell.2025.1552716
Received: 29 December 2024; Accepted: 22 January 2025;
Published: 20 February 2025.
Edited by:
Carlos Estella, Centre for Molecular Biology Severo Ochoa, Spanish National Research Council (CSIC), SpainReviewed by:
Alberto Rosello-Diez, University of Cambridge, United KingdomCopyright © 2025 Britton, Somogyi-Leatigaga, Watson, Haro, Mulder, Kennedy, Cooper, Whitley, Yeboah, Kim, Yu, Campos, Amoah, Kawauchi, Kim, Pira and Oberg. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kerby C. Oberg, koberg@llu.edu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.