
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Cell Dev. Biol., 04 July 2022
Sec. Signaling
Volume 10 - 2022 | https://doi.org/10.3389/fcell.2022.935224
This article is part of the Research TopicProtein-RNA Interplay-Regulated Signaling in Stem Cells and CancerView all 10 articles
N6-Methyladenosine (m6A) is the most abundant modification on eukaryote messenger RNA and plays a key role in posttranscriptional regulation of RNA metabolism including splicing, intracellular transport, degradation, and translation. m6A is dynamically regulated by methyltransferases (writers), RNA-binding proteins (readers), and demethylases (erasers). Recent studies demonstrate that perturbation of m6A regulators remarkably influences cell fate transitions through rewiring various biological processes, such as growth, differentiation, and survival. Moreover, aberrant m6A modification is implicated in a variety of diseases, in particular cancer. In this review, we describe the functional linkage of m6A modifications to cellular reprogramming and cancer stemness properties.
N6-Methyladenosine modification (m6A) refers to the methylation of the adenosine base at the nitrogen-6 position and tends to occur in a consensus sequence RRACH. It was originally discovered in 1970s and now recognized as the most abundant modification present in eukaryotic messenger RNA (mRNA) (Desrosiers et al., 1974; Adams and Cory, 1975; Lavi and Shatkin, 1975; Wei et al., 1975). m6A modification is present in different types of RNAs including mRNAs, transfer RNAs (tRNAs), ribosomal RNAs (rRNAs), circular RNAs (circRNAs), micro RNAs (miRNA), and long non-coding RNAs (lncRNAs) (Liu and Pan, 2016). The process of m6A modification is reversible and regulated by methyltransferases (writers), demethylases (erasers), and RNA-binding proteins (readers). Methyltransferase complex consisting of methyltransferase 3 (METTL3) (Bokar et al., 1997), methyltransferase 14 (METTL14) (Liu et al., 2014), and WT1-associated protein (WTAP) (Ping et al., 2014) catalyzes m6A formation. Other m6A writers such as RNA-binding motif protein 15/15B (RBM15/15B) (Patil et al., 2016), vir-like M6A methyltransferase associated (VIRMA) (Yue et al., 2018), and zinc finger CCCH-type containing 13 (ZC3H13) (Wen et al., 2018) have been identified to facilitate the function of the methyltransferase complex. On the other hand, fat mass and obesity-associated protein (FTO) (Jia et al., 2011) and AlkB homolog H5 (ALKBH5) (Zheng et al., 2013; Alemu et al., 2016), two key demethylases, demethylate m6A modification. Besides, m6A readers, e.g., YTH domain-containing proteins (YTHDF1-3 (Wang et al., 2014a; Wang et al., 2015; Shi et al., 2017) and YTHDC1-2 (Xiao et al., 2016; Wojtas et al., 2017)) and insulin-like growth factor-2 mRNA-binding proteins (IGF2BP1/2/3) (Huang et al., 2018), target m6A marks of transcripts and trigger RNA processing and metabolism such as alternative splicing, intracellular transport, degradation, and translation.
Self-renewal and differentiation are two unique properties of stem cells with the former referring to the capability of stem cells to make more stem cells and maintain the undifferentiated state, while the latter indicating the change of stem cells to a more specialized cell type. Notably, the processes of self-renewal and differentiation are controlled at a transcriptional level wherein epigenetic and epitranscriptomic regulation play critical roles. To date, m6A is proven to be a mark of transcriptome flexibility involved in regulating the function of stem cells. Emerging evidence have demonstrated that m6A modifications are involved in the process of mouse embryonic development (Geula et al., 2015), stem cell self-renewal (Batista et al., 2014; Wang et al., 2014b), spermatogenesis (Zheng et al., 2013), and so on. However, the origins and functions of m6A marks in reprogramming stemness properties are still largely unclear.
Perturbation of m6A regulators strongly affects gene expression patterns and biological functions of cells, leading to a variety of diseases including cancer. Recent evidence reveals a subpopulation of tumor cells, named cancer stem cells (CSCs), responsible for tumor initiation, metastasis, and relapse. The roles of cancer stem cells have been reported in both solid (Visvader and Lindeman, 2008) and hematological cancers (Zagozdzon and Golab, 2015), although the origin of the CSCs remains elusive. They may derive from differentiated cells or tissue-resident stem cells upon tumor initiation. Intriguingly, genes critical for self-renewal of normal stem cells also function as cancer-related genes, e.g., Bmi-1 (Siddique and Saleem, 2012), Nanog (Gawlik-Rzemieniewska and Bednarek, 2016), Notch (Ranganathan et al., 2011), Sox2 (Novak et al., 2020), and Wnt (Zhan et al., 2017). Given that m6A modifications regulate the expression of stemness-related genes, it is not surprising that they also play an important role in CSCs (Zhang et al., 2016; Li et al., 2017; Zhang et al., 2017; Chen et al., 2021).
In this review, we discuss recent studies that underscore the multifaceted role of m6A modifications in controlling gene expression, highlighting key findings that m6A modifications are essential in stem cells reprogramming and cancer stemness properties regulation.
m6A controls almost every step of RNA metabolism including alternative splicing, intracellular transport, degradation, and translation (Figure 1). In this part, we describe the influences of m6A writers, erasers, and readers on RNA metabolic process through dynamic regulation of m6A.
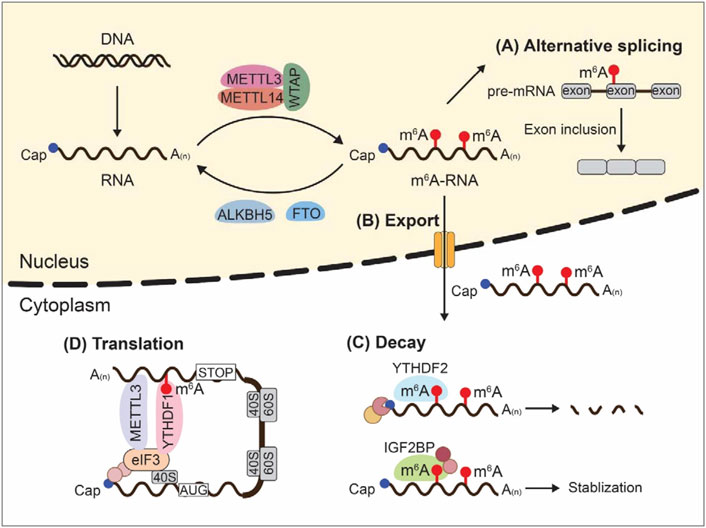
FIGURE 1. m6A modifications and RNA metabolism. Dynamic m6A modifications inside cells influence almost every step of RNA metabolism including alternative splicing, intracellular transport, degradation, and translation.
Alternative splicing (AS) is the process of making messenger RNA (mRNA) from messenger RNA precursor (pre-mRNA) by selecting different combinations of splice sites in pre-mRNA, thus allowing a single gene to code for multiple proteins. AS is essential for generating functional diversity given the limited gene number in eukaryotic organisms. Emerging evidence shows that m6A writers, METTL3, METTL14, and WTAP, and m6A erasers, ALKBH5 and FTO, are located in nuclear speckles where the AS occurs, indicating a potential role of m6A for controlling pre-mRNA processing. In support of this, treatment of S-adenosylmethionine (SAM) synthesis inhibitors, neplanocin A or cycloleucine, reduced RNA m6A methylation and resulted in nuclear accumulation of unspliced transcripts (Stoltzfus and Dane, 1982; Carroll et al., 1990). Consistently, Dominissini et al. observed a correlation between m6A methylation of multi-isoform genes and isoform switching by analyzing human and mouse transcriptome-wide m6A profiling (Dominissini et al., 2012).
METTL3 was found to colocalize with spliceosomal protein U2 small nuclear ribonucleoprotein B'' (U2B′)' in nuclear speckles (Bokar et al., 1997). Silencing of METTL3 could affect AS patterns and gene expressions (Dominissini et al., 2012). WTAP is a regulatory subunit of RNA m6A methyltransferase complex. Localization of METTL3 and METTL14 into nuclear speckles requires interaction with WTAP. Yang et al. showed that WTAP promoted METTL3 and METTL14 accumulation in nuclear speckles and regulated AS of targeted genes (Ping et al., 2014). Knockdown of METTL3 or WTAP led to a remarkable change of transcriptional isoform numbers (Ping et al., 2014). ALKBH5 also colocalized with mRNA-processing factors, including phosphorylated serine/arginine-rich splicing factor 2 (SC35), smith antigen (SM), and alternative splicing factor/splicing factor 2 (ASF/SF2) in nuclear speckles (Zheng et al., 2013). Similarly, FTO was present in nucleoplasm and partially colocalized with splicing factors U4/U6.U5 tri-snRNP-associated protein 1 (SART1), serine/arginine-rich splicing factor 2 (SC35), and RNA polymerase II phosphorylated at Ser2 (Pol II-S2P) (Jia et al., 2011). These two key m6A erasers are both capable of controlling mRNA splicing. ALKBH5 was found to regulate assembly of mRNA processing factors (Zheng et al., 2013); on the other hand, FTO depletion increased the m6A level of target genes, thereby raising the binding capacity of serine/arginine-rich splicing factor 2 (SRSF2) which subsequently increased inclusion of target exons (Zhao et al., 2014). m6A reader YTHDC1 is also engaged in the AS process. YTHDC1 was present in YT bodies near nuclear speckles. Wild type YTHDC1, but not m6A-binding-defective YTHDC1, could recruit pre-mRNA splicing factor SRSF3 (SRp20) but block the binding of SRSF10 (SRp38) to targeted mRNAs in the nucleus, thus promoting exon inclusion (Xiao et al., 2016). All these data indicate an essential regulatory role of m6A in mRNA splicing.
RNAs produced in the nucleus are exported to the cytoplasm through nuclear pore complexes. This is a fundamental step in gene expression process. TREX complex is important for mRNA export. Recent work identified the interactions between TREX subunits (ALYREF, UAP56, THOC5, and CHTOP) and m6A methyltransferases (METTL3, METTL14, WTAP, and KIAA1429) (Lesbirel et al., 2018). The m6A methyltransferase complex could recruit TREX to m6A-modified mRNAs to facilitate their export (Lesbirel et al., 2018). Moreover, depletion of KIAA1429 and WTAP led to an export block for m6A-modified mRNAs (Lesbirel et al., 2018). m6A eraser ALKBH5 also affects mRNA export dependent on its demethylation activity. Cytoplasmic mRNA level was significantly increased after silencing of ALKBH5 because of accelerated nuclear RNA export; re-expression of wild type ALKBH5, but not catalytic inactive mutant H204A, could rescue this phenomenon (Zheng et al., 2013). Binding of YTHDC1 to m6A-modified genes is important for mRNA export. Knockdown of YTHDC1 induced an export block for nuclear m6A-modified mRNA, resulting in accumulation of transcripts in the nucleus (Roundtree et al., 2017). Mechanistically, YTHDC1 interacted with SRSF3, an mRNA export adaptor, to increase RNA binding to SRSF3 (Roundtree et al., 2017).
m6A modifications also participate in circular RNA nuclear export. Depletion of m6A writer METTL3 induced circNSUN2 accumulation in the nucleus, and re-expression of METTL3 could rescue this phenomenon (Chen et al., 2019). Moreover, the m6A reader YTHDC1 was capable of binding to m6A marks of circNSUN2 in the backsplicing junction sites to facilitate the export process (Chen et al., 2019). Together, m6A modifications regulate RNA export.
RNA decay is the process whereby RNA is enzymatically degraded. RNA decay is important for effective mRNA surveillance and turnover. Accumulating evidence suggest m6A modifications affect RNA stability through dynamic interplays with RNA-binding proteins. In mouse embryonic stem cells, m6A level was found to be negatively correlated with mRNA stability (Wang et al., 2014b). m6A writers METTL3 and METTL14 could form a stable heterodimer to catalyze m6A deposition on RNA. Downregulation of METTL3 and METTL14 reduced the m6A level of mRNA, resulting in more binding of human antigen R (HuR) to mRNA which in turn promoted mRNA stability (Wang et al., 2014b). In line with these findings, depletion of METTL3 in both human and mouse cells led to m6A erasure and prolonged half-life of targeted mRNAs (Batista et al., 2014). Although WTAP lacks m6A catalytic activity, it binds to METTL3-METTL14 complex to enhance m6A deposition. As such, WTAP-mediated m6A modifications were negatively correlated with mRNA stability (Schwartz et al., 2014). Furthermore, silencing of METTL3, METTL14, or WTAP reduced global m6A methylation and increased the lifetime of nascent RNAs (Liu et al., 2014). Therefore, m6A modifications affect RNA stability.
Recent reports state that YTHDF2 is the major decay-inducing reader protein that binds to m6A-modified mRNAs to facilitate RNA degradation (Du et al., 2016; Park et al., 2019). Two distinct mechanisms of YTHDF2-induced mRNA degradation have been identified: RNase P/MRP-mediated endoribonucleolytic-cleavage pathway and carbon catabolite repression 4 (CCR4)-negative on TATA-less (NOT)-mediated deadenylation pathway, depending on whether messenger ribonucleoprotein (mRNP) has heat-responsive protein 12 (HRSP12)-binding site or not (Lee et al., 2020). Showed that m6A-modified RNAs underwent endoribonucleolytic cleavage via YTHDF2, HRSP12, and RNase P/MRP, of which HRSP12 acted as an adaptor to connect YTHDF2 and RNase P/MRP Park et al. (2019). In this case, HRSP12-binding site and RNase P/MRP-directed cleavage site were identified upstream and downstream of YTHDF2-binding site, respectively (Park et al., 2019). Of note, m6A-modified circular RNA could also be degraded through YTHDF2-HRSP12-RNase P/MRP-mediated endoribonucleolytic cleavage (Park et al., 2019). On the other hand, Du et al. reported that YTHDF2 directly recruited CCR4/NOT deadenylase complex to m6A-modified mRNAs, leading to deadenylation of mRNAs (Du et al., 2016). Besides, YTHDF3 was identified to regulate the RNA accessibility of YTHDF2 and enhanced YTHDF2-mediated mRNA decay (Shi et al., 2017). In contrast to YTHDF2-mediated mRNA decay, a recent study revealed that IGF2BP1-3 could recognize m6A markers through their KH domains to stabilize m6A-modified RNA (Huang et al., 2018). Intriguingly, although YTHDF2 and IGF2BP1-3 were all proved to bind to m6A markers, their transcriptome-wide binding sites were distinct (Huang et al., 2018). Therefore, m6A modifications can either enhance or inhibit mRNA stability depending on the binding of specific m6A readers.
Translation is the decoding of mRNA by ribosomes to produce polypeptide which later forms a functional protein inside the cells. Recent studies demonstrate that m6A modifications modulate mRNA translation efficiency through different mechanisms. YTHDF1 is known to promote the translation of m6A-modified mRNA. Mechanistically, YTHDF1 could promote ribosome occupancy of targeted mRNA in the cytoplasm by recruiting the initiation factor eukaryotic initiation factor 3 (eIF3) (Wang et al., 2015). In addition, YTHDF3 was reported to facilitate YTHDF1-promoted translation (Shi et al., 2017). METTL3 also enhances mRNA translation. Barbieri et al. found that the transcription factor, CEBPZ, recruited METTL3 to the promoters of select active gene to catalyze m6A methylation in the coding region (CDS) of targeted mRNA, resulting in enhanced translation by relieving ribosome stalling (Barbieri et al., 2017). Consistently, knockdown of METTL3 decreased translational efficiency of m6A-modified transcripts in both human myeloid leukemia and HeLa cell lines (Vu et al., 2017). Surprisingly, METTL3-promoted translation could be independent of m6A catalytic activity (Lin et al., 2016). Gregory and others showed that tethering a wild type or catalytically inactive METTL3 to the 3′UTR of a reporter mRNA exhibited similar translation enhancement (Lin et al., 2016). They further identified a direct physical and functional interaction between METTL3 at 3′UTR near the stop codon and eIF3h at the 5′ untranslated region (5′ UTR) of the mRNA and that METTL3-eIF3h loop may promote translation through ribosome recycling (Choe et al., 2018). Intriguingly, depletion of YTHDF1 did not influence the expression of METTL3 targets (Choe et al., 2018). Thus, METTL3 promotes mRNA translation through diverse mechanisms. It is worth noting that mouse embryonic stem cells (mESCs) with METTL3 knockout exhibited a modest increased translation efficiency (TE) compared to wild type (WT) cells, although this effect was observed for both methylated and unmethylated transcripts with higher GC content (Geula et al., 2015). In this study, loss of m6A could directly enhance mRNA stability of m6A-marked transcripts while indirectly favoring translation of GC-rich transcripts (Geula et al., 2015). Intriguingly, Slobodin et al. reported that transcription rate positively affected the efficiency of mRNA translation which was mediated by m6A modification (Slobodin et al., 2017). Therefore, mRNA m6A could mediate the communication between transcription and translation.
Qian and Jaffrey’s team suggested that m6A could enable mRNA translation in a cap- and IRES-independent manner (Meyer et al., 2015; Zhou et al., 2015; Coots et al., 2017; Zhou et al., 2018). They showed that heat shock stress promoted nuclear localization of YTHDF2 which in turn increased 5′ UTR m6A of stress-inducible mRNAs through competing with FTO in preserving m6A modification, leading to enhanced cap-independent translation initiation (Zhou et al., 2015). In addition, eIF3 could bind to 5′ UTR m6A and recruit the 43S complex to initiate translation without the cap-binding factor eIF4E under stress (Meyer et al., 2015). Furthermore, depletion of METTL3 selectively inhibited translation of mRNAs with 5′ UTR m6A, but not mRNAs with 5′ terminal oligopyrimidine (TOP) elements (Coots et al., 2017). Notably, ABCF1 was identified to coordinate with METTL3 in promoting translation of m6A-modified mRNA (Coots et al., 2017). Thus, 5′ UTR m6A facilitates cap-independent translation under stress.
m6A is also thought to facilitate efficient translation of circular RNA (circRNA) (Yang et al., 2017). Initiation factor eIF4G2 and YTHDF3 were identified to be required for m6A-driven circRNAs translation, which were enhanced by METTL3/14-mediated methylation or suppressed by FTO-mediated demethylation (Yang et al., 2017). Consistently, Bozzoni et al. demonstrated that METTL3 and YTHDC1 could direct the back-splicing reaction of circRNAs, and recognition of m6A marks by YTHDF3 and eIF4G2 modulate circRNAs translation (Di Timoteo et al., 2020).
Mammalian development is thought to be continuous and unidirectional in which stem cells give rise to specialized differentiated cells through a series of cellular changes. However, recent studies have shown that it is possible to modify cell identity by somatic cell nuclear transfer (SCNT) (Matoba and Zhang, 2018), forced expression of specific transcription factors (Takahashi and Yamanaka, 2016) or micro-RNAs (Judson et al., 2009), and using small signaling molecules (Hou et al., 2013). In 2006, Kazutoshi Takahashi and Shinya Yamanaka successfully reprogrammed mouse embryonic fibroblasts (MEF) and adult mouse tail-tip fibroblasts to generate induced pluripotent stem cells (iPSCs) by ectopic expression of four transcription factors, namely Oct3/4, Sox2, c-Myc, and Klf4 (Takahashi and Yamanaka, 2006). In 2007, they further demonstrated the generation of iPSC from adult human dermal fibroblasts with the same four factors (Takahashi et al., 2007). The fact that terminally differentiated somatic cells can be reprogrammed to generate iPSCs has opened new gateways for therapeutics research. Recent evidence has revealed epigenetic profile changes during the process of cell differentiation and reprogramming and that epigenetic perturbations could affect the efficiency of reprogramming iPSCs (Young, 2011; Liang and Zhang, 2013; Hochedlinger and Jaenisch, 2015; Xu and Xie, 2018). In this part, we describe the influences of m6A modifications on stemness and reprogramming.
To maintain self-renewal and pluripotency, stem cells need to stably express pluripotency genes; however, they are also capable of rapidly altering gene expression programming for differentiation. m6A is involved in cell fate determination and is now considered as a mark of transcriptome flexibility required by stem cells. Zhao and others identified that depletion of METTL3 or METTL14 in mESCs suppressed m6A methylation and self-renewal capability (Wang et al., 2014b). Mechanistically, m6A marks blocked the binding of RNA stabilizer protein HuR and protected mRNA from degradation induced by RNA-induced silencing complex (RISC) (Wang et al., 2014b). Consequently, developmental regulators were more enriched than pluripotency genes upon METTL3 or METTL14 knockdown (Wang et al., 2014b). Thus, METTL3/METTL14-mediated m6A modification is required to maintain the pluripotency of ES cells. However, Batista et al. reported that m6A loss promoted ESC self-renewal and hindered differentiation (Batista et al., 2014). In this study, they profiled m6A methylome in mouse and human ESCs, revealing extensive m6A modification of ESC genes, including core pluripotency regulators such as Nanog, Klf4, Myc, Lin28, Med1, Jarid2, and Eed (Batista et al., 2014). They considered m6A as a mark for RNA turnover over in a timely fashion, and knockout of METTL3 improved mESCs self-renewal without affecting cell viability (Batista et al., 2014). The differences in phenotypes between these two studies may partially be explained by the methodology used (RNAi and CRISPR) which may affect downstream m6A-modified RNAs pattern. Another possibility is that the mESCs used in these two studies were at different states. TGFβ signaling is essential for human pluripotent stem cells (hPSCs) to maintain pluripotency (James et al., 2005). Vallier et al. identified a functional interaction between SMAD2/3 transcription factors and METTL3-METTL14-WTAP complex (Bertero et al., 2018). SMAD2/3 could promote the binding of METTL3-METTL14-WTAP to specific SMAD2/3 transcriptional targets involved in early cell fate decisions, e.g., pluripotency factor gene NANOG, leading to increased m6A methylations that facilitate mRNA degradation (Bertero et al., 2018). Consequently, m6A-mediated rapid downregulation of SMAD2/3-targeted genes facilitated timely shut down of pluripotency on differentiation (Bertero et al., 2018). Intriguingly, Filipczyk and others reported that depletion of m6A could both support pluripotency maintenance and exit through activating pAkt and pErk signaling, respectively (Jin et al., 2021).
m6A modification is required for embryo development. Knockout of METTL3 or METTL14 led to early embryonic lethality (Geula et al., 2015; Meng et al., 2019). In Mettl3−/− mice, preimplantation epiblasts and naïve embryonic stem cells with loss of m6A were still viable; however, they failed to terminate the naïve state toward lineage differentiation, resulting in early embryonic lethality (Geula et al., 2015). The abnormal expression and location of NANOG caused by METTL3 ablation was regarded as the leading cause (Geula et al., 2015). Meanwhile, METTL14 is indispensable for postimplantation embryonic development. Silencing of METTL14 contributed to abnormal embryo development since embryonic day 6.5 (E6.5), mainly due to resistance to differentiation (Meng et al., 2019). Mechanistically, METTL14 depletion caused dysregulation of genes associated with embryo development pathways (Meng et al., 2019). The m6A readers YTHDF2 and YTHDC1 are also important for mammalian development (Ivanova et al., 2017; Kasowitz et al., 2018). Maternal RNA degradation, which was mediated by YTHDF2, facilitated oocyte maturation; oocytes with YTHDF2 deficiency failed to change metaphase II (MII) transcriptome, leading to female-specific infertility in mice (Ivanova et al., 2017). On the other hand, knockout of YTHDC1 caused massive alternative splicing defects in oocytes, resulting in a block at the primary stage of folliculogenesis (Kasowitz et al., 2018).
The epigenetic modifications could lock cells into a differentiated state during cell differentiation; therefore, targeting repressive epigenetic marks in differentiated cells improve the efficiency of iPSC formation (Huangfu et al., 2008; Shi et al., 2008). Recent studies also pinpoint m6A as an important player during cellular reprogramming. Chen et al. reported that m6A formation facilitated cell reprogramming to pluripotency (Chen et al., 2015). In this study, lots of cell-type specific markers are m6A-modifed, such as Oct4, Nanog, and DPPA2 for ESCs and iPSCs; POU3F2 and ROBO2 for neural stem cells; and DHH and Sox8 for testicular sertoli cells (Chen et al., 2015). These genes are critical for stem cell maintenance and developmental regulation. Intriguingly, miRNAs could target m6A marks by base pairing and modulate the binding of METTL3, thus leading to the change of cellular m6A abundance (Chen et al., 2015). Deletion of Dicer, an essential endonuclease for producing mature miRNAs, remarkably inhibited the RNA m6A level; in contrast, overexpression of miRNAs increased the binding of METTL3 on mRNAs and enhanced m6A abundance (Chen et al., 2015). To investigate the role of m6A in cell reprogramming, manipulation of METTL3 was conducted in MEFs transduced with four Yamanaka transcription factors. The result indicated that ectopic expression of METTL3 increased colonies of iPSC, enhanced expressions of key pluripotent factors (Oct4, Sox2, and Nanog), and promoted the reprogramming of MEFs to pluripotent stem cells; conversely, depletion of METTL3 reduced m6A and led to impeded reprogramming (Chen et al., 2015).
The crosstalk between epigenetic and epitranscriptomic networks is important to cellular reprogramming. Aguilo et al. reported that chromatin-associated zinc finger protein 217 (ZFP217) coordinated epigenetic and epitranscriptomic regulation to ensure ESC self-renewal and somatic cell reprogramming (Aguilo et al., 2015). They identified gradually increased ZFP217 expression along with decreased METTL3 expression during somatic reprogramming (Aguilo et al., 2015). ZFP217 could induce transcription of core reprogramming factors and repress m6A deposition of pluripotency genes by sequestering METTL3 (Aguilo et al., 2015). Depletion of ZFP217 in MEFs increased the m6A level of Nanog, Sox2, Klf4, and c-Myc mRNAs, promoting their degradation and leading to diminished iPSC colonies formation; this phenomenon could be partially rescued by METTL3 knockdown (Aguilo et al., 2015). Therefore, m6A modifications may be a barrier for ZFP217-meidiated somatic cell reprogramming. In support of these, Song et al. demonstrated that ZFP217 suppressed m6A mRNA methylation by promoting FTO expression (Song et al., 2019). Silencing of ZFP217 decreased FTO expression to enhance m6A levels, resulting in retarded adipogenic differentiation (Song et al., 2019).
So, how to understand the conflicting phenomena regarding the role of m6A on somatic cell reprogramming? One possible explanation is that m6A on cell fate choice is context dependent. Geula et al. reported that depletion of METTL3 exerted a divergent effect on naïve and primed PSCs (Geula et al., 2015). In naïve PSCs, pluripotency genes were highly expressed, and silencing of METTL3 could further enhance their expression to boost naïve circuitry stability; by contrast, the expression of pluripotency genes was downregulated while lineage commitment markers were upregulated in primed cells; thus silencing of METTL3 exerted a minor effect on expression of pluripotency genes while it remarkably increased the expression of lineage commitment markers, making the cells tend toward differentiation (Geula et al., 2015). Therefore, epigenetics and epitranscriptomics can form a complex network to regulate stem cell pluripotency and differentiation.
CSCs or tumor-initiating cells (TICs) are a small subpopulation of cancer cells which could give rise to tumors through processes of self-renewal and differentiation, just like normal stem cell (Figure 2). Tumor development and iPSC generation share striking similarities on gene expression programming, implying a potential link between pluripotency and cancer (Wong et al., 2008). Furthermore, recent evidence state that cancer cells could be reprogrammed to retrieve benign cell functions or differentiate into other unrelated cell types by re-expression of lineage-specific genes, opening a new avenue for cancer treatment (Bussard et al., 2010; Pezzolo et al., 2011). Understanding the molecular drivers of CSCs will advance the development of anticancer therapeutics. In this section, we summarize the key findings on how m6A modifications modulate cancer stemness (Table 1).
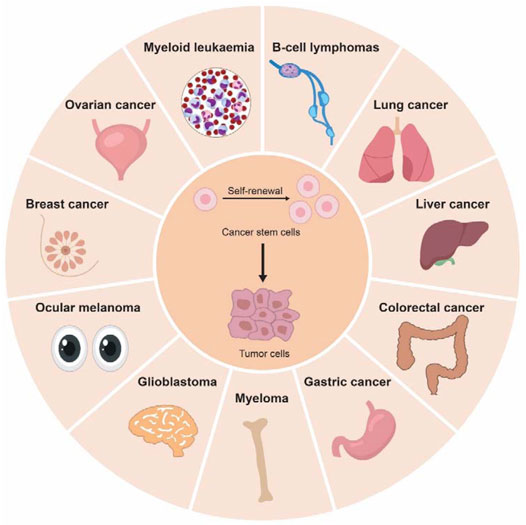
FIGURE 2. Cancer stem cells are key drivers to tumor initiation and progression. Cancer stem cells (CSCs) have been identified in different type of solid cancers (colorectal cancer, gastric cancer, liver cancer, glioblastoma, melanoma, breast cancer, lung cancer, and ovarian cancer) and hematological cancers (myeloid leukemia, lymphoma, and myeloma).
Solid tumors refer to an abnormal mass of tissue in “solid” organs. Gastrointestinal (GI) cancer is one of them, referring to cancers that affect the digestive system, e.g., colorectal cancer (CRC), gastric cancer (GC), and liver cancer. Several m6A regulators have been reported to play important roles in GI cancer. Our team recently identified the novel oncogenic epitranscriptome axis of METTL3-m6A-GLUT1-mTORC1 (Chen et al., 2021) and YTHDF1-m6A-ARHGEF2 (Wang et al., 2022) in promoting CRC tumorigenesis. In the former study, METTL3 was found to promote GLUT1 translation in an m6A-dependent manner by integrative m6A sequencing, RNA sequencing, and ribosome profiling analyses, resulting in increased glucose uptake and lactate production which subsequently activated mTORC1 signaling; consequently, depletion of METTL3 impaired the self-renewal capacity of colon cancer-initiating cells (Chen et al., 2021). As to the latter study, knockdown of YTHDF1 suppressed CRC organoids and decreased cell growth; mechanistically, YTHDF1 bind to m6A marks of ARHGEF2 mRNA and enhanced ARHGEF2 translation by multiomic analysis of m6A sequencing, RNA sequencing, YTHDF1 RNA immunoprecipitation sequencing and proteomics (Wang et al., 2022). In line with our findings, Han et al. reported that high expression of YTHDF1 was induced by Wnt signaling in intestinal stem cells (ISCs) which in turn promoted translation of TCF7L2/TCF4, leading to enhanced β-catenin activity that promoted stemness of ISCs (Han et al., 2020). YTHDF1 also activates Wnt/β-catenin signaling in GC. Pi et al. revealed that YTHDF1 increased translation of frizzled7 (FZD7), a key Wnt receptor, in an m6A-dependent manner; consequently, hyperactivation of the Wnt/β-catenin was induced that facilitated GC tumorigenesis (Pi et al., 2021). Intriguingly, long non-coding RNAs (lncRNAs) could influence the m6A modification process. LNC942 was identified to induce GC stemness and chemoresistance by stabilizing Musashi2 (MSI2), a member of RNA-binding proteins (RBPs); MSI2 then bind to m6A sites of c-Myc mRNA to increase mRNA stability (Zhu et al., 2022).
Current evidence also pinpoints a pivotal role of m6A modifications in liver cancer stem cells (Zhang et al., 2020; Zhou et al., 2020; Bian et al., 2021; Wang et al., 2021). In purified CD133+ liver cancer stem cells, knockdown of YTHDF2 impaired tumor-initiating ability; in contrast, overexpression of YTHDF2 exerted the opposite effect (Zhang et al., 2020). YTHDF2 was capable of binding to m6A sites in the 5′UTR of OCT4 mRNA to promote its translation as determined by luciferase activity assay and polysome profiling assay (Zhang et al., 2020). Meanwhile, METTL14 induced m6A methylation of hepatocyte nuclear factor 3γ (HNF3γ) mRNA, a hepatocyte nuclear factor, leading to reduced HNF3γ expression in hepatocellular carcinoma (HCC) (Zhou et al., 2020). Notably, enforced HNF3γ expression promoted differentiation of HCC cells and liver CSCs, resulting in retarded growth of HCC (Zhou et al., 2020). In addition, HNF3γ expression rendered sensitivity of HCC cells to sorafenib treatment, implying the potential of HNF3γ as a therapeutic target for HCC (Zhou et al., 2020). RALY RNA-binding protein-like (RALYL), a liver progenitor specific gene, was also related with HCC differentiation (Wang et al., 2021). Overexpression of RALYL suppressed the m6A level of TGF-β2 mRNA to enhance its mRNA stability, leading to subsequent activation of TGF-β signaling that contributed to HCC self-renewal and chemoresistance (Wang et al., 2021). In this study, FTO was found to bind to RALYL and thought to be responsible for m6A demethylation of TGF-β2 mRNA (Wang et al., 2021). Furthermore, FTO-mediated RNA demethylation was also involved in S-adenosylmethionine decarboxylase proenzyme (AMD1)-induced cancer stemness in HCC (Bian et al., 2021). AMD1 was capable of stabilizing the interaction between Ras GTPase-activating-like protein 1 (IQGAP1) and FTO, leading to enhanced FTO expression which in turn promoted HCC stemness (Bian et al., 2021). Together, m6A modifications are critical for self-renewal and differentiation of CSCs in GI cancer.
Dysregulated m6A modifications play an important role in lung cancer. Yin et al. identified an lncRNA named RNA Component of Mitochondrial RNA Processing Endoribonuclease (RMRP) which exhibited enriched m6A modifications and increased RNA stability in non-small cell lung cancer (NSCLC) (Yin et al., 2021). Both in vitro and in vivo experiments in this study revealed that RMRP induced TGFBR1/SMAD2/SMAD3 axis and promoted the cancer stem cell properties of NSCLC (Yin et al., 2021). However, how m6A modifications regulate RMRP stability warrants further investigation. On the other hand, Liu et al. (2022) recently reported that ALKBH5 was highly expressed in CSCs isolated from NSCLC. They revealed that depletion of ALKBH5 increased the global m6A level, suppressed expression of Nanog and Oct4, two essential transcription factors for self-renewal and pluripotency of ESCs, and inhibited stemness of CSCs (Liu et al., 2022). Intriguingly, p53 was reported to regulate malignancies of CSCs partially through transactivating ALKBH5 expression (Liu et al., 2022).
Glioblastoma is a prevalent and malignant cancer that occurs in the brain or spinal cord. m6A modifications could regulate gene expression and cell fate in glioblastoma stem-like cells (GSCs). Compared to normal neural stem cells (NSCs), GSCs preferentially expressed YTHDF2 which was essential for GSCs maintenance (Dixit et al., 2021). Intriguingly, instead of destabilizing mRNAs, YTHDF2 was found to increase MYC and VEGFA mRNA stability in an m6A-dependent manner in GSCs, although the mRNA-stabilizing function of YTHDF2 was unclear (Dixit et al., 2021). YTHDF2-MYC-IGFBP3 axis was further identified to promote glioblastoma growth both in vitro and in vivo (Dixit et al., 2021). Importantly, administration of linsitinib, an IGF1/IGF1R inhibitor, exerted potent inhibitory effect against YTHDF2-expressing GSCs without affecting NSCs (Dixit et al., 2021). ALKBH5 was also found highly expressed in GSCs, and silencing of ALKBH5 inhibited the growth of patient-derived GSCs (Zhang et al., 2017). Mechanistically, ALKBH5 reduced the m6A level of FOXM1 mRNA, resulting in enhanced FOXM1 expression which in turn promoted GSC tumorigenesis (Zhang et al., 2017). m6A modifications are critical for self-renewal of GSCs. Knockdown of METTL3 or METTL14 promoted growth, self-renewal, and tumorigenesis of human GSC; conversely, overexpression of METTL3 or inhibition of FTO exerted the opposite effect (Cui et al., 2017). ADAM19 was a downstream target of METTL3/METTL14 that exerted critical biological functions in GSCs (Cui et al., 2017). m6A modifications could influence nonsense-mediated mRNA decay (NMD) in GSCs. Li et al. reported that METTL3 regulated the NMD of splicing factors and AS process in glioblastoma (Li et al., 2019). Depletion of METTL3 inhibited the m6A levels of serine- and arginine-rich splicing factors (SRSF), leading to NMD of SRSF which was mediated by YTHDC1 (Li et al., 2019). Subsequently, downregulated SRSFs significantly changed alternative splicing events of several genes including BCL-X and NCOR2, contributing to suppression of GSCs self-renewal (Li et al., 2019). All these findings establish a critical role of m6A modifications in GSCs.
Breast cancer and ovarian cancer are common cancers in women. m6A modifications exert profound and diverse functions in breast cancer stem cells and ovarian cancer stem cells. In response to hypoxia, hypoxia-inducible factor (HIF)-1α and HIF-2α were stimulated to promote ALKBH5 expression in breast cancer cells; subsequently, ALKBH5 inhibited the m6A level in the 3′UTR of Nanog mRNA and increased NANOG expression, resulting in enhanced breast cancer stem cell phenotype (Zhang et al., 2016). Conversely, ALKBH5 knockdown in human breast cancer cells suppressed tumor initiation capacity (Zhang et al., 2016). Therefore, ALKBH5-mediated m6A modifications play a pivotal role in maintaining breast cancer stemness in the hypoxic environment. Aurora kinase A (AURKA) is a member of serine/threonine kinases family and was reported to stabilize METTL14 protein by preventing its ubiquitylation in breast cancer stem-like cells (Peng et al., 2021). Subsequently, upregulated METTL14 expression induced the m6A level of DROSHA, a Class 2 ribonuclease III enzyme, to stabilize DROSHA mRNA which was meditated by m6A reader IGF2BP2 (Peng et al., 2021). Intriguingly, AURKA could strengthen the binding of IGF2BP2 to DROSHA mRNA, thus promoting DROSHA expression (Peng et al., 2021). Furthermore, DROSHA interacted with β-catenin to transactivate STC1, resulting in enhanced stemness of breast cancer (Peng et al., 2021). In ovarian cancer, FTO is suggested to suppress self-renewal of ovarian CSCs. Huang et al. revealed reduced FTO expression in ovarian tumors and ovarian CSCs (Huang et al., 2020). In this study, ectopic expression of FTO in ovarian cancer cells inhibited the m6A level in the 3′UTR of two phosphodiesterase genes, PDE1C and PDE4B, and reduced their mRNA stability, leading to activation of second messenger 3′, 5′-cyclic adenosine monophosphate (cAMP) signaling and suppression of stemness features (Huang et al., 2020). Furthermore, FTO could suppress self-renewal of ovarian CSCs in vivo in an m6A-dependent manner (Huang et al., 2020). All these studies unveil a key role of m6A modifications in regulating stemness phenotype of breast cancer and ovarian cancer.
Hematologic malignancies comprise three main types: leukemia, lymphoma, and multiple myeloma (MM). In acute myeloid leukemia (AML), a subpopulation of AML cells, called leukemia stem cells (LSCs), exert self-renewal capacity and is responsible for the maintenance of the AML phenotype. There have been numerous studies reporting the functional importance of m6A modifications in AML. Li et al. revealed increased expression of FTO in AML (Li et al., 2017). High FTO expression suppressed the m6A levels of ankyrin repeat and SOCS box protein 2 (ASB2) and retinoic acid receptor α (RARA), leading to reduced mRNA stability of these two genes (Li et al., 2017). However, future study is required to identify m6A readers that are responsible for stabilizing FTO target transcripts, such as ASB2 and RARA. Consequently, FTO promoted leukemogenesis and inhibited Tretinoin-induced AML cell differentiation (Li et al., 2017). Given the functional significance of FTO in AML, several FTO inhibitors have been developed. In a subsequent study, Su et al. reported that R-2-hydroxyglutarate (R-2HG), originally thought to be an oncometabolite, strongly inhibited FTO activity, thereby increasing global m6A modifications, resulting in reduced mRNA stability of MYC/CEBPA in R-2HG-sensitive leukemia cells (Su et al., 2018). Of note, R-2HG treatment also increased ASB2 and RARA expressions in R-2HG-sensitive cells, but not in the resistant cells (Su et al., 2018). Importantly, R-2HG exhibited a potent anti-tumor effect against leukemia with high FTO expression by targeting FTO-m6A-MYC/CEBPA axis (Su et al., 2018). However, whether and how R-2HG exerted its effect on cancer metabolism in leukemia was unclear. Accordingly, Qing et al. showed that R-2HG could effectively inhibit aerobic glycolysis in R-2HG-sensitive leukemia cells, but not in normal CD34+ hematopoietic stem/progenitor cells (Qing et al., 2021). Aerobic glycolysis, termed Warburg effect, converts glucose to lactate even without oxygen, thereby providing the energy required by the cancer cells. R-2HG inhibited FTO activity and increased the m6A level of phosphofructokinase platelet (PFKP) and lactate dehydrogenase B (LDHB), two critical glycolytic genes, thereby reducing their mRNA stability which was mediated by YTHDF2 (Qing et al., 2021). Notably, FTO, PFKP, or LDHB depletion recapitulated R-2HG-induced glycolytic inhibition and suppressed leukemogenesis in vivo (Qing et al., 2021). Using structure-based rational design, Huang et al. recently developed two FTO inhibitors, FB23 and FB23-2 (derivatives of meclofenamic acid), which could directly bind to FTO and suppress its demethylase activity (Huang et al., 2019). FB23-2 strongly inhibited cell proliferation but induced differentiation/apoptosis of human AML cells both in vitro and in vivo; moreover, FB23-2 exhibited a promising therapeutic efficacy in patient-derived xeno-transplantation AML mouse model (Huang et al., 2019). Notably, FB23-2 treatment could significantly eliminate LSCs in these mice models, thereby disrupting AML maintenance (Huang et al., 2019). However, the half-maximal inhibitory concentration (IC50) values of FB23 and FB23-2 in suppressing AML are still high: >20 μM and >1 μM for FB23 and FB23-2, respectively (Huang et al., 2019). To develop efficacious inhibitors against FTO, Chen’s team conducted a structure-based virtual screening of the 260,000 compounds and validation assays, leading to the identification of two compounds, CS1 and CS2, which displayed strong inhibitory effects against FTO activity and AML cell viability with 10- to 30-fold lower IC50 (Su et al., 2020). FTO was frequently overexpressed in LSCs, and pharmacological inhibition of FTO by CS1 and CS2 suppressed self-renewal of LSCs (Su et al., 2020). In addition, targeting FTO decreased the expression of immune checkpoints, such as PD-L1, PD-L2, and LILRB, to reverse immune evasion of leukemia cells (Su et al., 2020), highlighting the potential of FTO inhibitors for cancer therapy. Nevertheless, there remains some limitations for small-molecule FTO inhibitors, e.g., toxic side effects, the sensitivity and specificity of inhibitors against LSCs. As such, Cao et al. developed FTO inhibitor-loaded GSH-bioimprinted nanocomposites (GNPIPP12MA) of synergistic FTO inhibition and GSH depletion (Cao et al., 2022). Notably, GNPIPP12MA not only selectively targeted LSCs but also enhanced the efficacy of the PD-L1 blockade, thereby suppressing leukemogenesis (Cao et al., 2022). Other m6A regulators, such as METTL3 (Barbieri et al., 2017; Vu et al., 2017), METTL14 (Weng et al., 2018), YTHDF2 (Paris et al., 2019), YTHDC1 (Cheng et al., 2021; Sheng et al., 2021), and ALKBH5 (Shen et al., 2020; Wang et al., 2020), have also been demonstrated to regulate LSCs features and contribute to leukemogenesis. It is worth noting that Yankova et al. recently developed a highly potent and selective METTL3 inhibitor, named STM2457, that posed a strong effect in suppressing growth while increasing differentiation and apoptosis of AML (Yankova et al., 2021). Together, all these studies suggest that targeting m6A regulators is a potential therapeutic strategy against AML.
Myeloma is a blood cancer of plasma cells derived from bone marrow. Recent evidence implies a functional role of m6A in MM pathogenesis. Upregulated isocitrate dehydrogenase 2 (IDH2) in CD138+ MM cells reduced global RNA m6A modification through activating FTO (Song et al., 2021). The m6A level of WNT7B mRNA was decreased by IDH2, leading to increased WNT7B expression and subsequent activation of Wnt pathway which eventually facilitated tumorigenesis and progression of MM (Song et al., 2021). Consistently, FTO was highly expressed in plasma cells from MM patients, concomitant with decreased RNA m6A level (Xu et al., 2022). FTO inhibited m6A modifications of heat shock factor 1 (HSF1), thereby increasing its mRNA stability in a YTHDF2-dependent manner (Xu et al., 2022). Importantly, FTO-m6A-HSF1 promoted MM cells growth and metastasis (Xu et al., 2022). Similarly, ALKBH5 was overexpressed in MM and promoted MM tumorigenesis (Qu et al., 2022). ALKBH5 inhibited m6A modifications in 3′UTR of TNF receptor-associated factor 1 (TRAF1) and enhanced its mRNA stability, leading to activation of NF-κB and MAPK signaling pathways (Qu et al., 2022).
Lymphoma is cancer of lymphocytes from lymph nodes, spleen, thymus, or bone marrow. Han et al. reported that PIWI-interacting RNAs (piRNAs)-30473 upregulated WTAP and increased the global m6A level in diffuse large B-cell lymphoma (DLBCL) (Han et al., 2021). Hexokinase 2 (HK2) was further identified as the downstream target of piRNA-30473-WTAP-m6A, and upregulated HK2 by piRNA-30473 contributed to DLBCL tumorigenesis (Han et al., 2021). On the other hand, proto-oncogene MYC was found to transcriptionally activate ALKBH5 and FTO and inhibit m6A levels of SPI1 and PHF12 transcripts, thereby suppressing their mRNA translation which was mediated by YTHDF3 (Wu et al., 2021). Furthermore, depletion of ALKBH5 effectively reduced growth of B-cell lymphomas with deregulated MYC expression (Wu et al., 2021).
To date, great efforts have been made to explore the roles of RNA m6A modifications in different biological processes, and improvements have been achieved to advance our understanding of m6A-mediated epitranscriptomic regulation and its potential as therapeutic targets for cancer patients. However, many questions remain elusive: 1) the origins and functions of m6A marks at different stages of human development are still largely unclear; 2) the contribution of m6A modifications in iPSC pluripotency should be further clarified; 3) m6A writers (e.g., METTL3) and erasers (e.g., FTO) both play an oncogenic role in several cancer types (e.g., AML). Thus, m6A regulators likely target different groups of transcripts and regulate different biological processes; 4) the position of m6A sites (e.g., 5ʹUTR, CDS, or 3ʹUTR) in transcripts likely influence the recognition of m6A and the subsequent RNA metabolism; 5) m6A readers could exhibit opposite functions. YTHDF2 promotes RNA degradation of m6A-modified mRNAs while IGF2BP1-3 stabilizes them, although they target different transcripts. Besides, more and more m6A readers are being discovered, adding to the complexity of m6A epitranscriptome; and 6) the crosstalk or competition among m6A writers, readers and erasers should be further explored. Although the functions of m6A regulators are context dependent, targeting m6A offers great potential for cancer treatment. Future studies on understanding the context-dependent role of m6A modification in cellular reprogramming and cancer stemness is of utmost importance.
HC and YW wrote the manuscript. HC and JY supervised the study and revised the paper. The remaining authors assisted in editing.
This project was supported by the National Natural Science Foundation of China (NSFC; 82103245, 81972576); RGC-CRF Hong Kong (C4039-19GF); RGC-GRF Hong Kong (14107321, 14110819); Heath and Medical Research Fund (HMRF) (18190951); and CUHK Direct Grant for Research (2020.006).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer YD declared a shared affiliation with the authors to the handling editor at the time of the review.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Adams, J. M., and Cory, S. (1975). Modified Nucleosides and Bizarre 5′-termini in Mouse Myeloma mRNA. Nature 255 (5503), 28–33. doi:10.1038/255028a0
Aguilo, F., Zhang, F., Sancho, A., Fidalgo, M., Di Cecilia, S., Vashisht, A., et al. (2015). Coordination of M 6 A mRNA Methylation and Gene Transcription by ZFP217 Regulates Pluripotency and Reprogramming. Cell Stem Cell 17 (6), 689–704. doi:10.1016/j.stem.2015.09.005
Alemu, E. A., He, C., and Klungland, A. (2016). ALKBHs-Facilitated RNA Modifications and De-modifications. DNA Repair (Amst) 44, 87–91. doi:10.1016/j.dnarep.2016.05.026
Barbieri, I., Tzelepis, K., Pandolfini, L., Shi, J., Millán-Zambrano, G., Robson, S. C., et al. (2017). Promoter-bound METTL3 Maintains Myeloid Leukaemia by m6A-dependent Translation Control. Nature 552 (7683), 126–131. doi:10.1038/nature24678
Batista, P. J., Molinie, B., Wang, J., Qu, K., Zhang, J., Li, L., et al. (2014). m6A RNA Modification Controls Cell Fate Transition in Mammalian Embryonic Stem Cells. Cell Stem Cell 15 (6), 707–719. doi:10.1016/j.stem.2014.09.019
Bertero, A., Brown, S., Madrigal, P., Osnato, A., Ortmann, D., Yiangou, L., et al. (2018). The SMAD2/3 Interactome Reveals that TGFβ Controls m6A mRNA Methylation in Pluripotency. Nature 555 (7695), 256–259. doi:10.1038/nature25784
Bian, X., Shi, D., Xing, K., Zhou, H., Lu, L., Yu, D., et al. (2021). AMD1 Upregulates Hepatocellular Carcinoma Cells Stemness by FTO Mediated mRNA Demethylation. Clin. Transl. Med. 11 (3), e352. doi:10.1002/ctm2.352
Bokar, J. A., Shambaugh, M. E., Polayes, D., Matera, A. G., and Rottman, F. M. (1997). Purification and cDNA Cloning of the AdoMet-Binding Subunit of the Human mRNA (N6-Adenosine)-Methyltransferase. RNA 3 (11), 1233–1247.
Bussard, K. M., Boulanger, C. A., Booth, B. W., Bruno, R. D., and Smith, G. H. (2010). Reprogramming Human Cancer Cells in the Mouse Mammary Gland. Cancer Res. 70 (15), 6336–6343. doi:10.1158/0008-5472.can-10-0591
Cao, K., Du, Y., Bao, X., Han, M., Su, R., Pang, J., et al. (2022). Glutathione-Bioimprinted Nanoparticles Targeting of N6-Methyladenosine FTO Demethylase as a Strategy against Leukemic Stem Cells. Small 18 (13), e2106558. doi:10.1002/smll.202106558
Carroll, S. M., Narayan, P., and Rottman, F. M. (1990). N6-methyladenosine Residues in an Intron-specific Region of Prolactin Pre-mRNA. Mol. Cell Biol. 10 (9), 4456–4465. doi:10.1128/mcb.10.9.4456-4465.1990
Chen, H., Gao, S., Liu, W., Wong, C.-C., Wu, J., Wu, J., et al. (2021). RNA N6-Methyladenosine Methyltransferase METTL3 Facilitates Colorectal Cancer by Activating the m6A-GLUT1-mTORC1 Axis and Is a Therapeutic Target. Gastroenterology 160 (4), 1284–1300 e16. doi:10.1053/j.gastro.2020.11.013
Chen, R.-X., Chen, X., Xia, L.-P., Zhang, J.-X., Pan, Z.-Z., Ma, X.-D., et al. (2019). N6-methyladenosine Modification of circNSUN2 Facilitates Cytoplasmic Export and Stabilizes HMGA2 to Promote Colorectal Liver Metastasis. Nat. Commun. 10 (1), 4695. doi:10.1038/s41467-019-12651-2
Chen, T., Hao, Y.-J., Zhang, Y., Li, M.-M., Wang, M., Han, W., et al. (2015). m6A RNA Methylation Is Regulated by MicroRNAs and Promotes Reprogramming to Pluripotency. Cell Stem Cell 16 (3), 289–301. doi:10.1016/j.stem.2015.01.016
Cheng, Y., Xie, W., Pickering, B. F., Chu, K. L., Savino, A. M., Yang, X., et al. (2021). N6-Methyladenosine on mRNA Facilitates a Phase-Separated Nuclear Body that Suppresses Myeloid Leukemic Differentiation. Cancer Cell 39 (7), 958–972 e8. doi:10.1016/j.ccell.2021.04.017
Choe, J., Lin, S., Zhang, W., Liu, Q., Wang, L., Ramirez-Moya, J., et al. (2018). mRNA Circularization by METTL3-eIF3h Enhances Translation and Promotes Oncogenesis. Nature 561 (7724), 556–560. doi:10.1038/s41586-018-0538-8
Coots, R. A., Liu, X.-M., Mao, Y., Dong, L., Zhou, J., Wan, J., et al. (2017). m6A Facilitates eIF4F-independent mRNA Translation. Mol. Cell 68 (3), 504–514 e7. doi:10.1016/j.molcel.2017.10.002
Cui, Q., Shi, H., Ye, P., Li, L., Qu, Q., Sun, G., et al. (2017). m 6 A RNA Methylation Regulates the Self-Renewal and Tumorigenesis of Glioblastoma Stem Cells. Cell Rep. 18 (11), 2622–2634. doi:10.1016/j.celrep.2017.02.059
Desrosiers, R., Friderici, K., and Rottman, F. (1974). Identification of Methylated Nucleosides in Messenger RNA from Novikoff Hepatoma Cells. Proc. Natl. Acad. Sci. U.S.A. 71 (10), 3971–3975. doi:10.1073/pnas.71.10.3971
Di Timoteo, G., Dattilo, D., Centrón-Broco, A., Colantoni, A., Guarnacci, M., Rossi, F., et al. (2020). Modulation of circRNA Metabolism by m6A Modification. Cell Rep. 31 (6), 107641. doi:10.1016/j.celrep.2020.107641
Dixit, D., Prager, B. C., Gimple, R. C., Poh, H. X., Wang, Y., Wu, Q., et al. (2021). The RNA m6A Reader YTHDF2 Maintains Oncogene Expression and Is a Targetable Dependency in Glioblastoma Stem Cells. Cancer Discov. 11 (2), 480–499. doi:10.1158/2159-8290.cd-20-0331
Dominissini, D., Moshitch-Moshkovitz, S., Schwartz, S., Salmon-Divon, M., Ungar, L., Osenberg, S., et al. (2012). Topology of the Human and Mouse m6A RNA Methylomes Revealed by m6A-Seq. Nature 485 (7397), 201–206. doi:10.1038/nature11112
Du, H., Zhao, Y., He, J., Zhang, Y., Xi, H., Liu, M., et al. (2016). YTHDF2 Destabilizes m6A-Containing RNA through Direct Recruitment of the CCR4-Not Deadenylase Complex. Nat. Commun. 7, 12626. doi:10.1038/ncomms12626
Gawlik-Rzemieniewska, N., and Bednarek, I. (2016). The Role of NANOG Transcriptional Factor in the Development of Malignant Phenotype of Cancer Cells. Cancer Biol. Ther. 17 (1), 1–10. doi:10.1080/15384047.2015.1121348
Geula, S., Moshitch-Moshkovitz, S., Dominissini, D., Mansour, A. A., Kol, N., Salmon-Divon, M., et al. (2015). m 6 A mRNA Methylation Facilitates Resolution of Naïve Pluripotency toward Differentiation. Science 347 (6225), 1002–1006. doi:10.1126/science.1261417
Han, B., Yan, S., Wei, S., Xiang, J., Liu, K., Chen, Z., et al. (2020). YTHDF1-mediated Translation Amplifies Wnt-Driven Intestinal Stemness. EMBO Rep. 21 (4), e49229. doi:10.15252/embr.201949229
Han, H., Fan, G., Song, S., Jiang, Y., Qian, C. a., Zhang, W., et al. (2021). piRNA-30473 Contributes to Tumorigenesis and Poor Prognosis by Regulating m6A RNA Methylation in DLBCL. Blood 137 (12), 1603–1614. doi:10.1182/blood.2019003764
Hochedlinger, K., and Jaenisch, R. (2015). Induced Pluripotency and Epigenetic Reprogramming. Cold Spring Harb. Perspect. Biol. 7 (12), a019448. doi:10.1101/cshperspect.a019448()
Hou, P., Li, Y., Zhang, X., Liu, C., Guan, J., Li, H., et al. (2013). Pluripotent Stem Cells Induced from Mouse Somatic Cells by Small-Molecule Compounds. Science 341 (6146), 651–654. doi:10.1126/science.1239278
Huang, H., Wang, Y., Kandpal, M., Zhao, G., Cardenas, H., Ji, Y., et al. (2020). FTO-dependent N6-Methyladenosine Modifications Inhibit Ovarian Cancer Stem Cell Self-Renewal by Blocking cAMP Signaling. Cancer Res. 80 (16), 3200–3214. doi:10.1158/0008-5472.can-19-4044
Huang, H., Weng, H., Sun, W., Qin, X., Shi, H., Wu, H., et al. (2018). Recognition of RNA N6-Methyladenosine by IGF2BP Proteins Enhances mRNA Stability and Translation. Nat. Cell Biol. 20 (3), 285–295. doi:10.1038/s41556-018-0045-z
Huang, Y., Su, R., Sheng, Y., Dong, L., Dong, Z., Xu, H., et al. (2019). Small-Molecule Targeting of Oncogenic FTO Demethylase in Acute Myeloid Leukemia. Cancer Cell 35 (4), 677–691 e10. doi:10.1016/j.ccell.2019.03.006
Huangfu, D., Maehr, R., Guo, W., Eijkelenboom, A., Snitow, M., Chen, A. E., et al. (2008). Induction of Pluripotent Stem Cells by Defined Factors Is Greatly Improved by Small-Molecule Compounds. Nat. Biotechnol. 26 (7), 795–797. doi:10.1038/nbt1418
Ivanova, I., Much, C., Di Giacomo, M., Azzi, C., Morgan, M., Moreira, P. N., et al. (2017). The RNA M 6 A Reader YTHDF2 Is Essential for the Post-transcriptional Regulation of the Maternal Transcriptome and Oocyte Competence. Mol. Cell 67 (6), 1059–1067 e4. doi:10.1016/j.molcel.2017.08.003
James, D., Levine, A. J., Besser, D., and Hemmati-Brivanlou, A. (2005). TGFβ/Activin/Nodal Signaling Is Necessary for the Maintenance of Pluripotency in Human Embryonic Stem Cells. Development 132 (6), 1273–1282. doi:10.1242/dev.01706
Jia, G., Fu, Y., Zhao, X., Dai, Q., Zheng, G., Yang, Y., et al. (2011). N6-methyladenosine in Nuclear RNA Is a Major Substrate of the Obesity-Associated FTO. Nat. Chem. Biol. 7 (12), 885–887. doi:10.1038/nchembio.687
Jin, K. X., Zuo, R., Anastassiadis, K., Klungland, A., Marr, C., and Filipczyk, A. (2021). N6-methyladenosine (m(6)A) Depletion Regulates Pluripotency Exit by Activating Signaling Pathways in Embryonic Stem Cells. Proc. Natl. Acad. Sci. U. S. A. 118 (51), e2105192118. doi:10.1073/pnas.2105192118
Judson, R. L., Babiarz, J. E., Venere, M., and Blelloch, R. (2009). Embryonic Stem Cell-specific microRNAs Promote Induced Pluripotency. Nat. Biotechnol. 27 (5), 459–461. doi:10.1038/nbt.1535
Kasowitz, S. D., Ma, J., Anderson, S. J., Leu, N. A., Xu, Y., Gregory, B. D., et al. (2018). Nuclear m6A Reader YTHDC1 Regulates Alternative Polyadenylation and Splicing during Mouse Oocyte Development. PLoS Genet. 14 (5), e1007412. doi:10.1371/journal.pgen.1007412
Lavi, S., and Shatkin, A. J. (1975). Methylated Simian Virus 40-specific RNA from Nuclei and Cytoplasm of Infected BSC-1 Cells. Proc. Natl. Acad. Sci. U.S.A. 72 (6), 2012–2016. doi:10.1073/pnas.72.6.2012
Lee, Y., Choe, J., Park, O. H., and Kim, Y. K. (2020). Molecular Mechanisms Driving mRNA Degradation by m6A Modification. Trends Genet. 36 (3), 177–188. doi:10.1016/j.tig.2019.12.007
Lesbirel, S., Viphakone, N., Parker, M., Parker, J., Heath, C., Sudbery, I., et al. (2018). The m6A-Methylase Complex Recruits TREX and Regulates mRNA Export. Sci. Rep. 8 (1), 13827. doi:10.1038/s41598-018-32310-8
Li, F., Yi, Y., Miao, Y., Long, W., Long, T., Chen, S., et al. (2019). N6-Methyladenosine Modulates Nonsense-Mediated mRNA Decay in Human Glioblastoma. Cancer Res. 79 (22), 5785–5798. doi:10.1158/0008-5472.can-18-2868
Li, Z., Weng, H., Su, R., Weng, X., Zuo, Z., Li, C., et al. (2017). FTO Plays an Oncogenic Role in Acute Myeloid Leukemia as a N 6 -Methyladenosine RNA Demethylase. Cancer Cell 31 (1), 127–141. doi:10.1016/j.ccell.2016.11.017
Liang, G., and Zhang, Y. (2013). Embryonic Stem Cell and Induced Pluripotent Stem Cell: an Epigenetic Perspective. Cell Res. 23 (1), 49–69. doi:10.1038/cr.2012.175
Lin, S., Choe, J., Du, P., Triboulet, R., and Gregory, R. I. (2016). The M 6 A Methyltransferase METTL3 Promotes Translation in Human Cancer Cells. Mol. Cell 62 (3), 335–345. doi:10.1016/j.molcel.2016.03.021
Liu, J., Yue, Y., Han, D., Wang, X., Fu, Y., Zhang, L., et al. (2014). A METTL3-METTL14 Complex Mediates Mammalian Nuclear RNA N6-Adenosine Methylation. Nat. Chem. Biol. 10 (2), 93–95. doi:10.1038/nchembio.1432
Liu, N., and Pan, T. (2016). N6-methyladenosine-encoded Epitranscriptomics. Nat. Struct. Mol. Biol. 23 (2), 98–102. doi:10.1038/nsmb.3162
Liu, X., Wang, Z., Yang, Q., Hu, X., Fu, Q., Zhang, X., et al. (2022). RNA Demethylase ALKBH5 Prevents Lung Cancer Progression by Regulating EMT and Stemness via Regulating P53. Front. Oncol. 12, 858694. doi:10.3389/fonc.2022.858694
Matoba, S., and Zhang, Y. (2018). Somatic Cell Nuclear Transfer Reprogramming: Mechanisms and Applications. Cell Stem Cell 23 (4), 471–485. doi:10.1016/j.stem.2018.06.018
Meng, T.-G., Lu, X., Guo, L., Hou, G.-M., Ma, X.-S., Li, Q.-N., et al. (2019). Mettl14 Is Required for Mouse Postimplantation Development by Facilitating Epiblast Maturation. FASEB J. 33 (1), 1179–1187. doi:10.1096/fj.201800719r
Meyer, K. D., Patil, D. P., Zhou, J., Zinoviev, A., Skabkin, M. A., Elemento, O., et al. (2015). 5′ UTR m6A Promotes Cap-independent Translation. Cell 163 (4), 999–1010. doi:10.1016/j.cell.2015.10.012
Novak, D., Hüser, L., Elton, J. J., Umansky, V., Altevogt, P., and Utikal, J. (2020). SOX2 in Development and Cancer Biology. Seminars Cancer Biol. 67 (Pt 1), 74–82. doi:10.1016/j.semcancer.2019.08.007
Paris, J., Morgan, M., Campos, J., Spencer, G. J., Shmakova, A., Ivanova, I., et al. (2019). Targeting the RNA m6A Reader YTHDF2 Selectively Compromises Cancer Stem Cells in Acute Myeloid Leukemia. Cell Stem Cell 25 (1), 137–148 e6. doi:10.1016/j.stem.2019.03.021
Park, O. H., Ha, H., Lee, Y., Boo, S. H., Kwon, D. H., Song, H. K., et al. (2019). Endoribonucleolytic Cleavage of m6A-Containing RNAs by RNase P/MRP Complex. Mol. Cell 74 (3), 494–507 e8. doi:10.1016/j.molcel.2019.02.034
Patil, D. P., Chen, C.-K., Pickering, B. F., Chow, A., Jackson, C., Guttman, M., et al. (2016). m6A RNA Methylation Promotes XIST-Mediated Transcriptional Repression. Nature 537 (7620), 369–373. doi:10.1038/nature19342
Peng, F., Xu, J., Cui, B., Liang, Q., Zeng, S., He, B., et al. (2021). Oncogenic AURKA-Enhanced N6-Methyladenosine Modification Increases DROSHA mRNA Stability to Transactivate STC1 in Breast Cancer Stem-like Cells. Cell Res. 31 (3), 345–361. doi:10.1038/s41422-020-00397-2
Pezzolo, A., Parodi, F., Marimpietri, D., Raffaghello, L., Cocco, C., Pistorio, A., et al. (2011). Oct-4+/Tenascin C+ Neuroblastoma Cells Serve as Progenitors of Tumor-Derived Endothelial Cells. Cell Res. 21 (10), 1470–1486. doi:10.1038/cr.2011.38
Pi, J., Wang, W., Ji, M., Wang, X., Wei, X., Jin, J., et al. (2021). YTHDF1 Promotes Gastric Carcinogenesis by Controlling Translation of FZD7. Cancer Res. 81 (10), 2651–2665. doi:10.1158/0008-5472.can-20-0066
Ping, X.-L., Sun, B.-F., Wang, L., Xiao, W., Yang, X., Wang, W.-J., et al. (2014). Mammalian WTAP Is a Regulatory Subunit of the RNA N6-Methyladenosine Methyltransferase. Cell Res. 24 (2), 177–189. doi:10.1038/cr.2014.3
Qing, Y., Dong, L., Gao, L., Li, C., Li, Y., Han, L., et al. (2021). R-2-hydroxyglutarate Attenuates Aerobic Glycolysis in Leukemia by Targeting the FTO/m6A/PFKP/LDHB axis. Mol. Cell 81 (5), 922–939 e9. doi:10.1016/j.molcel.2020.12.026
Qu, J., Hou, Y., Chen, Q., Chen, J., Li, Y., Zhang, E., et al. (2022). RNA Demethylase ALKBH5 Promotes Tumorigenesis in Multiple Myeloma via TRAF1-Mediated Activation of NF-κB and MAPK Signaling Pathways. Oncogene 41 (3), 400–413. doi:10.1038/s41388-021-02095-8
Ranganathan, P., Weaver, K. L., and Capobianco, A. J. (2011). Notch Signalling in Solid Tumours: a Little Bit of Everything but Not All the Time. Nat. Rev. Cancer 11 (5), 338–351. doi:10.1038/nrc3035
Roundtree, I. A., Luo, G. Z., Zhang, Z., Wang, X., Zhou, T., Cui, Y., et al. (2017). YTHDC1 Mediates Nuclear Export of N6-Methyladenosine Methylated mRNAs. Elife 6, 6. doi:10.7554/eLife.31311
Schwartz, S., Mumbach, M. R., Jovanovic, M., Wang, T., Maciag, K., Bushkin, G. G., et al. (2014). Perturbation of m6A Writers Reveals Two Distinct Classes of mRNA Methylation at Internal and 5′ Sites. Cell Rep. 8 (1), 284–296. doi:10.1016/j.celrep.2014.05.048
Shen, C., Sheng, Y., Zhu, A. C., Robinson, S., Jiang, X., Dong, L., et al. (2020). RNA Demethylase ALKBH5 Selectively Promotes Tumorigenesis and Cancer Stem Cell Self-Renewal in Acute Myeloid Leukemia. Cell Stem Cell 27 (1), 64–80 e9. doi:10.1016/j.stem.2020.04.009
Sheng, Y., Wei, J., Yu, F., Xu, H., Yu, C., Wu, Q., et al. (2021). A Critical Role of Nuclear m6A Reader YTHDC1 in Leukemogenesis by Regulating MCM Complex-Mediated DNA Replication. Blood 138 (26), 2838–2852. doi:10.1182/blood.2021011707
Shi, H., Wang, X., Lu, Z., Zhao, B. S., Ma, H., Hsu, P. J., et al. (2017). YTHDF3 Facilitates Translation and Decay of N6-Methyladenosine-Modified RNA. Cell Res. 27 (3), 315–328. doi:10.1038/cr.2017.15
Shi, Y., Tae Do, J., Desponts, C., Hahm, H. S., Schöler, H. R., and Ding, S. (2008). A Combined Chemical and Genetic Approach for the Generation of Induced Pluripotent Stem Cells. Cell Stem Cell 2 (6), 525–528. doi:10.1016/j.stem.2008.05.011
Siddique, H. R., and Saleem, M. (2012). Role of BMI1, a Stem Cell Factor, in Cancer Recurrence and Chemoresistance: Preclinical and Clinical Evidences. Stem Cells 30 (3), 372–378. doi:10.1002/stem.1035
Slobodin, B., Han, R., Calderone, V., Vrielink, J. A. F. O., Loayza-Puch, F., Elkon, R., et al. (2017). Transcription Impacts the Efficiency of mRNA Translation via Co-transcriptional N6-Adenosine Methylation. Cell 169 (2), 326–337 e12. doi:10.1016/j.cell.2017.03.031
Song, S., Fan, G., Li, Q., Su, Q., Zhang, X., Xue, X., et al. (2021). IDH2 Contributes to Tumorigenesis and Poor Prognosis by Regulating m6A RNA Methylation in Multiple Myeloma. Oncogene 40 (35), 5393–5402. doi:10.1038/s41388-021-01939-7
Song, T., Yang, Y., Wei, H., Xie, X., Lu, J., Zeng, Q., et al. (2019). Zfp217 Mediates m6A mRNA Methylation to Orchestrate Transcriptional and Post-transcriptional Regulation to Promote Adipogenic Differentiation. Nucleic Acids Res. 47 (12), 6130–6144. doi:10.1093/nar/gkz312
Stoltzfus, C. M., and Dane, R. W. (1982). Accumulation of Spliced Avian Retrovirus mRNA Is Inhibited in S -Adenosylmethionine-Depleted Chicken Embryo Fibroblasts. J. Virol. 42 (3), 918–931. doi:10.1128/jvi.42.3.918-931.1982
Su, R., Dong, L., Li, C., Nachtergaele, S., Wunderlich, M., Qing, Y., et al. (2018). R-2HG Exhibits Anti-tumor Activity by Targeting FTO/m6A/MYC/CEBPA Signaling. Cell 172 (1-2), 90–e23. doi:10.1016/j.cell.2017.11.031
Su, R., Dong, L., Li, Y., Gao, M., Han, L., Wunderlich, M., et al. (2020). Targeting FTO Suppresses Cancer Stem Cell Maintenance and Immune Evasion. Cancer Cell 38 (1), 79–96 e11. doi:10.1016/j.ccell.2020.04.017
Takahashi, K., Tanabe, K., Ohnuki, M., Narita, M., Ichisaka, T., Tomoda, K., et al. (2007). Induction of Pluripotent Stem Cells from Adult Human Fibroblasts by Defined Factors. Cell 131 (5), 861–872. doi:10.1016/j.cell.2007.11.019
Takahashi, K., and Yamanaka, S. (2016). A Decade of Transcription Factor-Mediated Reprogramming to Pluripotency. Nat. Rev. Mol. Cell Biol. 17 (3), 183–193. doi:10.1038/nrm.2016.8
Takahashi, K., and Yamanaka, S. (2006). Induction of Pluripotent Stem Cells from Mouse Embryonic and Adult Fibroblast Cultures by Defined Factors. Cell 126 (4), 663–676. doi:10.1016/j.cell.2006.07.024
Visvader, J. E., and Lindeman, G. J. (2008). Cancer Stem Cells in Solid Tumours: Accumulating Evidence and Unresolved Questions. Nat. Rev. Cancer 8 (10), 755–768. doi:10.1038/nrc2499
Vu, L. P., Pickering, B. F., Cheng, Y., Zaccara, S., Nguyen, D., Minuesa, G., et al. (2017). The N6-Methyladenosine (m6A)-Forming Enzyme METTL3 Controls Myeloid Differentiation of Normal Hematopoietic and Leukemia Cells. Nat. Med. 23 (11), 1369–1376. doi:10.1038/nm.4416
Wang, J., Li, Y., Wang, P., Han, G., Zhang, T., Chang, J., et al. (2020). Leukemogenic Chromatin Alterations Promote AML Leukemia Stem Cells via a KDM4C-ALKBH5-AXL Signaling Axis. Cell Stem Cell 27 (1), 81–97 e8. doi:10.1016/j.stem.2020.04.001
Wang, S., Gao, S., Zeng, Y., Zhu, L., Mo, Y., Wong, C. C., et al. (2022). N6-Methyladenosine Reader YTHDF1 Promotes ARHGEF2 Translation and RhoA Signaling in Colorectal Cancer. Gastroenterology 162 (4), 1183–1196. doi:10.1053/j.gastro.2021.12.269
Wang, X., Lu, Z., Gomez, A., Hon, G. C., Yue, Y., Han, D., et al. (2014). N6-methyladenosine-dependent Regulation of Messenger RNA Stability. Nature 505 (7481), 117–120. doi:10.1038/nature12730
Wang, X., Wang, J., Tsui, Y.-M., Shi, C., Wang, Y., Zhang, X., et al. (2021). RALYL Increases Hepatocellular Carcinoma Stemness by Sustaining the mRNA Stability of TGF-β2. Nat. Commun. 12 (1), 1518. doi:10.1038/s41467-021-21828-7
Wang, X., Zhao, B. S., Roundtree, I. A., Lu, Z., Han, D., Ma, H., et al. (2015). N6-methyladenosine Modulates Messenger RNA Translation Efficiency. Cell 161 (6), 1388–1399. doi:10.1016/j.cell.2015.05.014
Wang, Y., Li, Y., Toth, J. I., Petroski, M. D., Zhang, Z., and Zhao, J. C. (2014). N6-methyladenosine Modification Destabilizes Developmental Regulators in Embryonic Stem Cells. Nat. Cell Biol. 16 (2), 191–198. doi:10.1038/ncb2902
Wei, C.-M., Gershowitz, A., and Moss, B. (1975). Methylated Nucleotides Block 5′ Terminus of HeLa Cell Messenger RNA. Cell 4 (4), 379–386. doi:10.1016/0092-8674(75)90158-0
Wen, J., Lv, R., Ma, H., Shen, H., He, C., Wang, J., et al. (2018). Zc3h13 Regulates Nuclear RNA m6A Methylation and Mouse Embryonic Stem Cell Self-Renewal. Mol. Cell 69 (6), 1028–1038 e6. doi:10.1016/j.molcel.2018.02.015
Weng, H., Huang, H., Wu, H., Qin, X., Zhao, B. S., Dong, L., et al. (2018). METTL14 Inhibits Hematopoietic Stem/Progenitor Differentiation and Promotes Leukemogenesis via mRNA m6A Modification. Cell Stem Cell 22 (2), 191–205 e9. doi:10.1016/j.stem.2017.11.016
Wojtas, M. N., Pandey, R. R., Mendel, M., Homolka, D., Sachidanandam, R., and Pillai, R. S. (2017). Regulation of M(6)A Transcripts by the 3'-->5' RNA Helicase YTHDC2 Is Essential for a Successful Meiotic Program in the Mammalian Germline. Mol. Cell 68 (2), 374–387 e12. doi:10.1016/j.molcel.2017.09.021
Wong, D. J., Liu, H., Ridky, T. W., Cassarino, D., Segal, E., and Chang, H. Y. (2008). Module Map of Stem Cell Genes Guides Creation of Epithelial Cancer Stem Cells. Cell Stem Cell 2 (4), 333–344. doi:10.1016/j.stem.2008.02.009
Wu, G., Suo, C., Yang, Y., Shen, S., Sun, L., Li, S. T., et al. (2021). MYC Promotes Cancer Progression by Modulating M6 A Modifications to Suppress Target Gene Translation. EMBO Rep. 22 (3), e51519. doi:10.15252/embr.202051519
Xiao, W., Adhikari, S., Dahal, U., Chen, Y.-S., Hao, Y.-J., Sun, B.-F., et al. (2016). Nuclear M 6 A Reader YTHDC1 Regulates mRNA Splicing. Mol. Cell 61 (4), 507–519. doi:10.1016/j.molcel.2016.01.012
Xu, A., Zhang, J., Zuo, L., Yan, H., Chen, L., Zhao, F., et al. (2022). FTO Promotes Multiple Myeloma Progression by Posttranscriptional Activation of HSF1 in an m6A-YTHDF2-dependent Manner. Mol. Ther. 30 (3), 1104–1118. doi:10.1016/j.ymthe.2021.12.012
Xu, Q., and Xie, W. (2018). Epigenome in Early Mammalian Development: Inheritance, Reprogramming and Establishment. Trends Cell Biol. 28 (3), 237–253. doi:10.1016/j.tcb.2017.10.008
Yang, Y., Fan, X., Mao, M., Song, X., Wu, P., Zhang, Y., et al. (2017). Extensive Translation of Circular RNAs Driven by N6-Methyladenosine. Cell Res. 27 (5), 626–641. doi:10.1038/cr.2017.31
Yankova, E., Blackaby, W., Albertella, M., Rak, J., De Braekeleer, E., Tsagkogeorga, G., et al. (2021). Small-molecule Inhibition of METTL3 as a Strategy against Myeloid Leukaemia. Nature 593 (7860), 597–601. doi:10.1038/s41586-021-03536-w
Yin, H., Chen, L., Piao, S., Wang, Y., Li, Z., Lin, Y., et al. (2021). M6A RNA Methylation-Mediated RMRP Stability Renders Proliferation and Progression of Non-small Cell Lung Cancer through Regulating TGFBR1/SMAD2/SMAD3 Pathway. Cell Death Differ. doi:10.1038/s41418-021-00888-8 Online ahead of print
Young, R. A. (2011). Control of the Embryonic Stem Cell State. Cell 144 (6), 940–954. doi:10.1016/j.cell.2011.01.032
Yue, Y., Liu, J., Cui, X., Cao, J., Luo, G., Zhang, Z., et al. (2018). VIRMA Mediates Preferential m6A mRNA Methylation in 3′UTR and Near Stop Codon and Associates with Alternative Polyadenylation. Cell Discov. 4, 10. doi:10.1038/s41421-018-0019-0
Zagozdzon, R., and Golab, J. (2015). Cancer Stem Cells in Haematological Malignancies. Contemp. Oncol. Pozn. 19 (1A), A1–A6. doi:10.5114/wo.2014.47127
Zhan, T., Rindtorff, N., and Boutros, M. (2017). Wnt Signaling in Cancer. Oncogene 36 (11), 1461–1473. doi:10.1038/onc.2016.304
Zhang, C., Samanta, D., Lu, H., Bullen, J. W., Zhang, H., Chen, I., et al. (2016). Hypoxia Induces the Breast Cancer Stem Cell Phenotype by HIF-dependent and ALKBH5-Mediated m⁶A-Demethylation of NANOG mRNA. Proc. Natl. Acad. Sci. U. S. A. 113 (14), E2047–E2056. doi:10.1073/pnas.1602883113
Zhang, C., Huang, S., Zhuang, H., Ruan, S., Zhou, Z., Huang, K., et al. (2020). YTHDF2 Promotes the Liver Cancer Stem Cell Phenotype and Cancer Metastasis by Regulating OCT4 Expression via m6A RNA Methylation. Oncogene 39 (23), 4507–4518. doi:10.1038/s41388-020-1303-7
Zhang, S., Zhao, B. S., Zhou, A., Lin, K., Zheng, S., Lu, Z., et al. (2017). m 6 A Demethylase ALKBH5 Maintains Tumorigenicity of Glioblastoma Stem-like Cells by Sustaining FOXM1 Expression and Cell Proliferation Program. Cancer Cell 31 (4), 591–606 e6. doi:10.1016/j.ccell.2017.02.013
Zhao, X., Yang, Y., Sun, B.-F., Shi, Y., Yang, X., Xiao, W., et al. (2014). FTO-dependent Demethylation of N6-Methyladenosine Regulates mRNA Splicing and Is Required for Adipogenesis. Cell Res. 24 (12), 1403–1419. doi:10.1038/cr.2014.151
Zheng, G., Dahl, J. A., Niu, Y., Fedorcsak, P., Huang, C.-M., Li, C. J., et al. (2013). ALKBH5 Is a Mammalian RNA Demethylase that Impacts RNA Metabolism and Mouse Fertility. Mol. Cell 49 (1), 18–29. doi:10.1016/j.molcel.2012.10.015
Zhou, J., Wan, J., Gao, X., Zhang, X., Jaffrey, S. R., and Qian, S.-B. (2015). Dynamic m6A mRNA Methylation Directs Translational Control of Heat Shock Response. Nature 526 (7574), 591–594. doi:10.1038/nature15377
Zhou, J., Wan, J., Shu, X. E., Mao, Y., Liu, X.-M., Yuan, X., et al. (2018). N6-Methyladenosine Guides mRNA Alternative Translation during Integrated Stress Response. Mol. Cell 69 (4), 636–647 e7. doi:10.1016/j.molcel.2018.01.019
Zhou, T., Li, S., Xiang, D., Liu, J., Sun, W., Cui, X., et al. (2020). m6A RNA Methylation-Mediated HNF3γ Reduction Renders Hepatocellular Carcinoma Dedifferentiation and Sorafenib Resistance. Sig Transduct. Target Ther. 5 (1), 296. doi:10.1038/s41392-020-00299-0
Keywords: RNA metabolism, stemness, tumorigenesis, N6-methyladenose, reprogramming
Citation: Chen H, Wang Y, Su H, Zhang X, Chen H and Yu J (2022) RNA N6-Methyladenine Modification, Cellular Reprogramming, and Cancer Stemness. Front. Cell Dev. Biol. 10:935224. doi: 10.3389/fcell.2022.935224
Received: 03 May 2022; Accepted: 26 May 2022;
Published: 04 July 2022.
Edited by:
Yanquan Zhang, University of Kentucky, United StatesReviewed by:
Liu Dabin, Harbin Medical University, ChinaCopyright © 2022 Chen, Wang, Su, Zhang, Chen and Yu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Huarong Chen, aGNoZW4yQGN1aGsuZWR1Lmhr; Jun Yu, anVueXVAY3Voay5lZHUuaGs=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.