- 1Department of Biology, Trent University, Peterborough, ON, Canada
- 2Centre for Biomedical Sciences, School of Biological Sciences, Royal Holloway University of London, Egham, United Kingdom
- 3Department of Cell Biology (Anatomy III), Biomedical Center (BMC), LMU Munich, Munich, Germany
Editorial on the Research Topic
Dictyostelium: A Tractable Cell and Developmental Model in Biomedical Research
For almost a century, the social amoeba Dictyostelium discoideum has been used as an inexpensive and high-throughput model system for studying a variety of fundamental cellular and developmental processes including cell movement, chemotaxis, differentiation, and autophagy (Müller-Taubenberger et al., 2013; Mathavarajah et al., 2017). The life cycle of Dictyostelium is comprised of a unicellular growth phase and a 24-h multicellular developmental phase with distinct stages (Figure 1A). Dictyostelium development shares many common features with metazoan development but occurs in a much shorter time frame, which allows for the rapid detection of developmental phenotypes. The fully sequenced, low redundancy genome of Dictyostelium provides a less complex system to work with, whilst still maintaining many genes and related signalling pathways found in more complex eukaryotes (Eichinger et al., 2005). The Dictyostelium genome is haploid, which allows researchers to introduce one or multiple gene disruptions with relative ease, and gene function can be studied in a true multicellular organism with measurable phenotypic outcomes (Kuspa, 2006; Faix et al., 2013; Friedrich et al., 2015; Sekine et al., 2018). In addition, insertional mutant libraries facilitate pharmacogenetics screens that have enhanced our understanding of the function of bioactive compounds at a cellular level (Damstra Oddy et al., 2021; Warren et al., 2020). Finally, a variety of expression constructs are available that enable studies on protein localization and function in Dictyostelium (Levi et al., 2000; Veltman et al., 2009; Müller-Taubenberger and Ishikawa-Ankerhold, 2013).
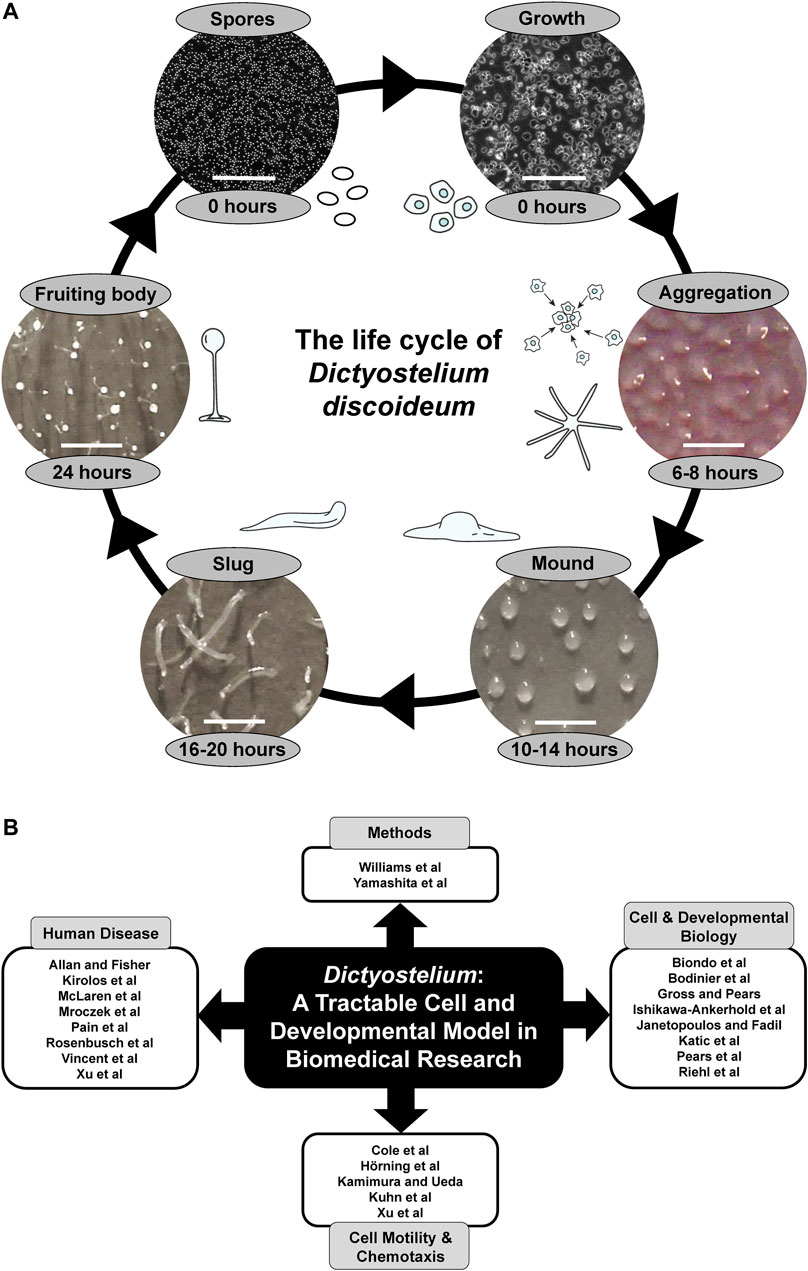
FIGURE 1. (A) The life cycle of Dictyostelium discoideum. During the growth phase, haploid amoebae consume nutrients and divide mitotically. Starvation initiates a 24-h developmental program that begins with the chemotactic aggregation of cells to form multicellular mounds, which is followed by a series of morphological events that generate motile slugs. The final phase of development involves terminal differentiation of cells and the formation of fruiting bodies composed of a mass of viable spores that rest atop a slender stalk. When nutrients become available, the spores germinate to restart the life cycle. White scale bars: 0.2 mm (spores and growth), 1 mm (aggregation, mound, slug, fruiting body). Some of the illustrations and microscopy images depicting the different life cycle stages were previously published in Huber, 2021 (permission provided by CC-BY license). (B) Themes of this Research Topic. Articles within this Research Topic are grouped into four main categories: Methods, Cell and Developmental Biology, Cell Motility and Chemotaxis, and Human Disease.
More recently, Dictyostelium has emerged as a valuable biomedical model system for studying several human diseases. The genome encodes orthologs of genes associated with human disease and the signalling pathways that regulate the behaviour of Dictyostelium cells are remarkably similar to those observed in mammalian cells, which has allowed findings from Dictyostelium to be successfully translated to mammalian systems (Alexander and Alexander, 2011; Chang et al., 2012). As a result, Dictyostelium has, and will continue to offer, excellent opportunities to advance biomedical research.
This Research Topic contains 23 articles that showcase the use of Dictyostelium as a tractable cell, molecular, and developmental model system in biomedical research, and includes two methods articles that enhance the biomedical applications of this valuable model organism (Figure 1B). Yamashita et al. describe the application of CRISPR-based gene disruption in Dictyostelium, while Williams et al. report the development of a new positive selection high throughput genetic screen.
The use of Dictyostelium as a model system for studying fundamental cellular and developmental processes is well established and this Research Topic contains several articles describing new findings on conserved processes in Dictyostelium with biomedical relevance (Figure 1B). In their original research article, Bodinier et al. reveal a mechanism regulated by the leucine-rich repeat kinase LrrkA that facilitates the sensing, phagocytosis, and killing of bacteria by Dictyostelium amoebae. In addition, Biondo et al. describe how Dictyostelium can be used as a model system for studying aerotaxis, Riehl et al. propose a role for UBX domain-containing protein nine in protein homeostasis in Dictyostelium, and Ishikawa-Ankerhold et al. reveal the role of pH in cytoplasmic rod formation in Dictyostelium, which has implications for human diseases caused by actin-cofilin rod formation. In review articles, Katic et al. summarize the roles of dynamin superfamily proteins in regulating vesicular trafficking and host-pathogen interactions in Dictyostelium, Janetopoulos and Fadil review the role of PIP2 in regulating the localization and exocytosis of the contractile vacuole system in migrating Dictyostelium amoebae, and Pears et al. describe DNA repair mechanisms in Dictyostelium, and how further study of DNA repair in Dictyostelium can help us better understand how this process is dysregulated in cancer. Finally, in their Hypothesis and Theory article, Gross and Pears discuss how Dictyostelium can be used to study the roles of the nutrient and energy sensors, mTORC1 and AMPK, which are linked to several human diseases including Alzheimer’s disease, cancer, obesity, and type 2 diabetes.
Historically, Dictyostelium has been used as a model system to elucidate the signalling pathways regulating eukaryotic cell motility and chemotaxis, which has furthered our understanding of the mechanisms regulating cancer cell movement. This Research Topic includes several exciting articles highlighting recent advancements in this area (Figure 1B). For example, in their original research article, Hörning et al. describe the dynamics of actin polymerization and PIP3 activity in amoeboid cells. In addition, Xu et al. reveal the molecular mechanism and biological function of C2GAP1 membrane targeting for chemotaxis, and Cole et al. show the roles of actin-binding proteins in sensing and transmitting mechanical stimuli that drive directed cell migration. To supplement these original research articles, Kamimura and Ueda review studies in Dictyostelium that have helped elucidate the role of G protein-coupled receptor signalling in regulating eurkaryotic chemotaxis, and Kuhn et al. summarize recent advances in imaging, synthetic biology, and computational analysis that have allowed researchers to tune the activity of individual molecules in cells and precisely measure the effects on cellular motility and signalling.
A central theme that emerges from this Research Topic is the use of Dictyostelium as a model system for studying specific human diseases (Figure 1B). In their original research article, McLaren et al. knock out the Dictyostelium ortholog of human ceroid lipofuscinosis neuronal 5 (CLN5) and show that loss of the gene impacts growth and multicellular development by affecting autophagy. In humans, mutations in CLN5 cause a subtype of Batten disease, the most common form of childhood neurodegeneration. This Research Topic also reports the ability of Dictyostelium to further our understanding of Parkinson’s disease. For example, in their original research article, Rosenbusch et al. report findings from Dictyostelium that link mutations in Parkinson’s disease-associated genes to aberrant mitochondrial activity. In addition, Mroczek et al. present data that improve our understanding of the interactions and cytotoxicity of tau and alpha-synuclein, both of which are linked to Parkinson’s disease. Further supporting the use of Dictyostelium as a model system for studying neurological disease, Vincent et al. review work in Dictyostelium and yeast that has provided insight into the roles of WIPI proteins in neurodegeneration, and Allan and Fisher characterize a new Dictyostelium model for the lysosomal storage disease, mucolipidosis type IV. This Research Topic also contains articles related to other human diseases. In their review, Pain et al. describe work in Dictyostelium that studied the role of decanoic acid in ketogenic diets, which have been used in the treatment of epilepsy, bipolar disorder, cancer, and diabetes. In addition, Kirolos et al. review work in Dictyostelium that has enhanced our understanding of the pathology underlying acute respiratory distress syndrome, and Xu et al. summarize studies in Dictyostelium that have provided insight into how phagocytes use chemotaxis and phagocytosis to detect and kill pathogens.
In total, this Research Topic showcases exciting new research in Dictyostelium biology that strengthen its position as a tractable cellular and developmental model system in biomedical research. However, we offer here only a sample of the many facets of medically relevant research currently performed using Dictyostelium. Therefore, we encourage readers to seek out additional literature from the field that has used Dictyostelium as a biomedical model system to learn more about its valuable contributions to human disease research.
Author Contributions
RH: writing—original draft, review and editing, Figure 1A and Figure 1B preparation. RW and AM-T, writing—review and editing. All authors approved the final draft of this Editorial.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The Editors would like to thank all the authors who contributed articles to this Research Topic and the reviewers who evaluated the submitted manuscripts. They would also like to thank the dictyBase team (dictybase.org) for their continuing support of Dictyostelium researchers worldwide.
References
Alexander, S., and Alexander, H. (2011). Lead Genetic Studies in Dictyostelium discoideum and Translational Studies in Human Cells Demonstrate that Sphingolipids Are Key Regulators of Sensitivity to Cisplatin and Other Anticancer Drugs. Semin. Cel Dev. Biol. 22, 97–104. doi:10.1016/j.semcdb.2010.10.005
Chang, P., Orabi, B., Deranieh, R. M., Dham, M., Hoeller, O., Shimshoni, J. A., et al. (2012). The Antiepileptic Drug Valproic Acid and Other Medium-Chain Fatty Acids Acutely Reduce Phosphoinositide Levels Independently of Inositol in Dictyostelium. Dis. Model. Mech. 5 (1), 115–124. doi:10.1242/dmm.008029
Damstra-Oddy, J. L., Warren, E. C., Perry, C. J., Desfougères, Y., Fitzpatrick, J. K., Schaf, J., et al. (2021). Phytocannabinoid-dependent mTORC1 Regulation Is Dependent upon Inositol Polyphosphate Multikinase Activity. Br. J. Pharmacol. 178, 1149–1163. doi:10.1111/bph.15351
Eichinger, L., Pachebat, J. A., Glöckner, G., Rajandream, M. A., Sucgang, R., Berriman, M., et al. (2005). The Genome of the Social Amoeba Dictyostelium discoideum. Nature 435, 43–57. doi:10.1038/nature03481
Faix, J., Linkner, J., Nordholz, B., Platt, J. L., Liao, X.-H., and Kimmel, A. R. (2013). The Application of the Cre-loxP System for Generating Multiple Knock-Out and Knock-In Targeted Loci. Methods Mol. Biol. 983, 249–267. doi:10.1007/978-1-62703-302-2_13
Friedrich, M., Meier, D., Schuster, I., and Nellen, W. (2015). A Simple Retroelement Based Knock-Down System in Dictyostelium: Further Insights into RNA Interference Mechanisms. PLoS One 10, e0131271. doi:10.1371/journal.pone.0131271
Huber, R. J. (2021). Altered Protein Secretion in Batten Disease. Dis. Model. Mech. 14, dmm049152. doi:10.1242/dmm.049152
Kuspa, A. (2006). Restriction Enzyme-Mediated Integration (REMI) Mutagenesis. Methods Mol. Biol. 346, 201–209. doi:10.1385/1-59745-144-4:201
Levi, S., Polyakov, M., and Egelhoff, T. T. (2000). Green Fluorescent Protein and Epitope Tag Fusion Vectors for Dictyostelium discoideum. Plasmid 44, 231–238. doi:10.1006/plas.2000.1487
Mathavarajah, S., Flores, A., and Huber, R. J. (2017). Dictyostelium discoideum: a Model System for Cell and Developmental Biology. Curr. Protoc. Essent. Lab. Tech. 15, 14. doi:10.1002/cpet.15
Müller-Taubenberger, A., and Ishikawa-Ankerhold, H. C. (2013). Fluorescent Reporters and Methods to Analyze Fluorescent Signals. Methods Mol. Biol. 983, 93–112.
Müller-Taubenberger, A., Kortholt, A., and Eichinger, L. (2013). Simple System - Substantial Share: the Use of Dictyostelium in Cell Biology and Molecular Medicine. Eur. J. Cel Biol. 92, 45–53.
Sekine, R., Kawata, T., and Muramoto, T. (2018). CRISPR/Cas9 Mediated Targeting of Multiple Genes in Dictyostelium. Sci Rep. 8 (1), 8471.
Veltman, D. M., Akar, G., Bosgraaf, L., and Van Haastert, P. J. M. (2009). A New Set of Small, Extrachromosomal Expression Vectors for Dictyostelium discoideum. Plasmid 61, 110–118. doi:10.1016/j.plasmid.2008.11.003
Keywords: cancer, cell biology, cell motility and chemotaxis, cell signalling, development, model system, neurological disease, pharmacogenetics screens
Citation: Huber RJ, Williams RSB and Müller-Taubenberger A (2022) Editorial: Dictyostelium: A Tractable Cell and Developmental Model in Biomedical Research. Front. Cell Dev. Biol. 10:909619. doi: 10.3389/fcell.2022.909619
Received: 31 March 2022; Accepted: 07 April 2022;
Published: 26 April 2022.
Edited and reviewed by:
Ramani Ramchandran, Medical College of Wisconsin, United StatesCopyright © 2022 Huber, Williams and Müller-Taubenberger. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Robert J. Huber, roberthuber@trentu.ca; Robin SB Williams, robin.williams@rhul.ac.uk; Annette Müller-Taubenberger, amueller@bmc.med.lmu.de
 Robert J. Huber
Robert J. Huber