- 1Faculty of Medicine, University of Toronto, Toronto, ON, Canada
- 2Division of Neurosurgery, Hospital for Sick Children, Department of Surgery, University of Toronto, Toronto, ON, Canada
- 3Institute of Biomedical Engineering, University of Toronto, Toronto, ON, Canada
- 4Faculty of Medicine, University of Ottawa, Ottawa, ON, Canada
- 5Program in Neuroscience and Mental Health, Hospital for Sick Children Research Institute, Toronto, ON, Canada
- 6Vector Institute for Artificial Intelligence, MaRS Centre, Toronto, ON, Canada
- 7Department of Computer Science, University of Toronto, Toronto, ON, Canada
- 8Hospital for Sick Children, Toronto, ON, Canada
- 9Institute of Medical Science, University of Toronto, Toronto, ON, Canada
Advances in intracranial electroencephalography (iEEG) and neurophysiology have enabled the study of previously inaccessible brain regions with high fidelity temporal and spatial resolution. Studies of iEEG have revealed a rich neural code subserving healthy brain function and which fails in disease states. Machine learning (ML), a form of artificial intelligence, is a modern tool that may be able to better decode complex neural signals and enhance interpretation of these data. To date, a number of publications have applied ML to iEEG, but clinician awareness of these techniques and their relevance to neurosurgery, has been limited. The present work presents a review of existing applications of ML techniques in iEEG data, discusses the relative merits and limitations of the various approaches, and examines potential avenues for clinical translation in neurosurgery. One-hundred-seven articles examining artificial intelligence applications to iEEG were identified from 3 databases. Clinical applications of ML from these articles were categorized into 4 domains: i) seizure analysis, ii) motor tasks, iii) cognitive assessment, and iv) sleep staging. The review revealed that supervised algorithms were most commonly used across studies and often leveraged publicly available timeseries datasets. We conclude with recommendations for future work and potential clinical applications.
Introduction
Intracranial electroencephalography (iEEG) provides exquisite detail with which to study neural activity. Stereotactic EEG (sEEG), strips, grids and depth electrodes are among the most common iEEG modalities. Modern diagnostic and treatment strategies for neurological conditions such as epilepsy rely heavily on interpreting iEEG signals to extract meaningful information that can impact clinical decision making (Lachaux et al., 2003). Furthermore, the study of these signals can serve to uncover the neural syntax associated with both healthy and diseased states by using explainable ML algorithms to shed light on important features that differentiate these states.
Machine learning (ML) is a rapidly growing field that has shown significant potential in complex biomedical applications (Rajkomar et al., 2019). Simply, ML refers to algorithms that can make seemingly intelligent decisions after identifying and learning from hidden patterns in large pre-existing data. Training these algorithms on large biologic datasets for example has enabled the identification of complex patterns not apparent to human experts and informed intelligent decision-making without explicit programming (Kotsiantis et al., 2007). Examples of success in these endeavors include automated diagnostic algorithms in radiology (Syeda-Mahmood, 2018) and pathology (Bera et al., 2019).
ML algorithms can be dichotomized into “supervised” and “unsupervised” learning approaches. An overview of these methodologies is illustrated in Figure 1. Supervised learning involves the use of labeled data during training such that the algorithm learns to associate certain patterns with a predefined label (for example, data associated with seizure vs. non-seizure states) (Kotsiantis et al., 2007). Unsupervised learning involves unlabelled data where the algorithm is provided a large dataset and learns to group certain signals together without a priori knowledge of classification. Each method possesses respective advantages and pitfalls. Deep learning approaches, which were initially designed based on a simplified model of interconnected nodes that mimic neuronal connections, also warrant special note. Each node is analogous to a cell body and the connections (“synapses”) between nodes are assigned weights during training analogous to the strength of synaptic connections between neurons. A primary advantage of deep learning compared to standard ML is that these algorithms can learn from the raw data and automatically extract meaningful information thereby bypassing human-intensive feature selection steps. However, it is important to consider that deep networks can take significantly longer to train and often require larger sample sizes than commonly available in neurosurgical populations.
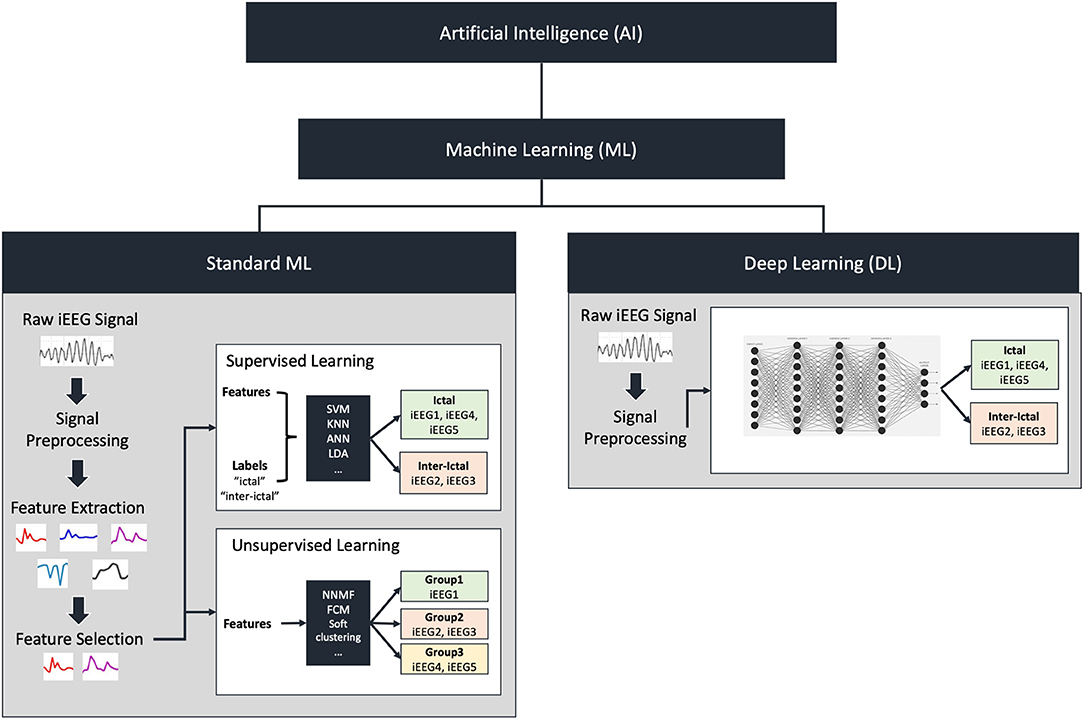
Figure 1. Overview of artificial intelligence and machine learning. Artificial intelligence includes machine learning (ML) which can be divided into standard machine learning or deep learning. In standard ML, raw iEEG signal is preprocessed followed by feature extraction and selection. Feature extraction involves breaking down the raw signal into individual quantifiable components such as power at various frequencies. Feature selection on the other hand involves selecting a subset of these components to be used to train a model. These features can either be fed to a supervised learning algorithm in addition to group labels, or an unsupervised learning algorithm that does not contain labels. Examples of supervised algorithms include support vector machines (SVM), K-nearest neighbors (KNN), artificial neural networks (ANN) or linear discriminant analysis (LDA). These classify the features into the group labels provided as inputs. Examples of unsupervised algorithms include non-negative matrix factorization (NNMF), fuzzy-c-means (FCM), and soft clustering. These will classify the features into groups based on similarity. Deep learning does not require feature extraction and selection. The processed or unprocessed signal can be fed directly into the deep learning model to classify the signals.
Despite the potential for application of ML to iEEG data, evidence to support the utilization of different ML techniques is limited, and considerable heterogeneity has been reported in the literature. As such, the present systematic review aims to: 1) assess the current landscape of ML utilization in iEEG and 2) examine the relative benefits and limitations of ML approaches as they relate to specific neurosurgical problems.
Methods
Search Strategy
In order to identify published, peer-reviewed articles that employ ML in the context of iEEG data, we performed a systematic search of articles using 3 commonly accessed medicine and engineering databases: OVID MEDLINE, IEEE, and Web of Science. We selected these databases as we aimed to capture the breadth of applications of this technology in the fields of science, medicine, and engineering. The search was conducted on August 18th, 2020. Our inclusion criteria comprised of two categories of keywords which must appear in either the article title, abstract or article keywords. The first category comprised of machine learning keywords (“machine learning”, “deep learning”, “artificial intelligence”, “neural network$”), while the second category comprised of keywords relevant to intracranial EEG (“stereoelectroencephalography”, “stereotactic EEG”, “sEEG”, “electrocorticography”, “ECoG”, “intracranial EEG”, “intracranial electroencephalography”, “iEEG”). Relevant articles were required to contain at least one keyword from the first category and at least one from the second category. A detailed summary of our search strategy and yield is provided in the Supplementary Material 1.
Article Filtering
The PRISMA guidelines (Moher et al., 2009) were followed to filter through articles in a systematic and transparent manner (Figure 2). Following our database search, 674 articles were found (OVID MEDLINE: 159, IEEE: 225, Web of Science: 290). One-hundred-sixty-five duplicates were removed leaving 509 for title and abstract filtering. Three-hundred-seventy-three articles were excluded following this first step of filtering as they did not fit our inclusion criteria for the following reasons: reviews, editorials, case reports, conference proceedings, book sections, animal studies, not relevant to intracranial EEG or machine learning. Two independent reviewers (NM, NW) then filtered the full text of 136 remaining articles. Disagreements were resolved by discussion and, when required, a third adjudicator (GMI). Twenty-nine articles were excluded for the following reasons: preliminary results, not clinical application, brief book series, not machine learning, animal studies, not intracranial EEG, not relevant, abstract only. Following this step, 107 articles were selected for this review.
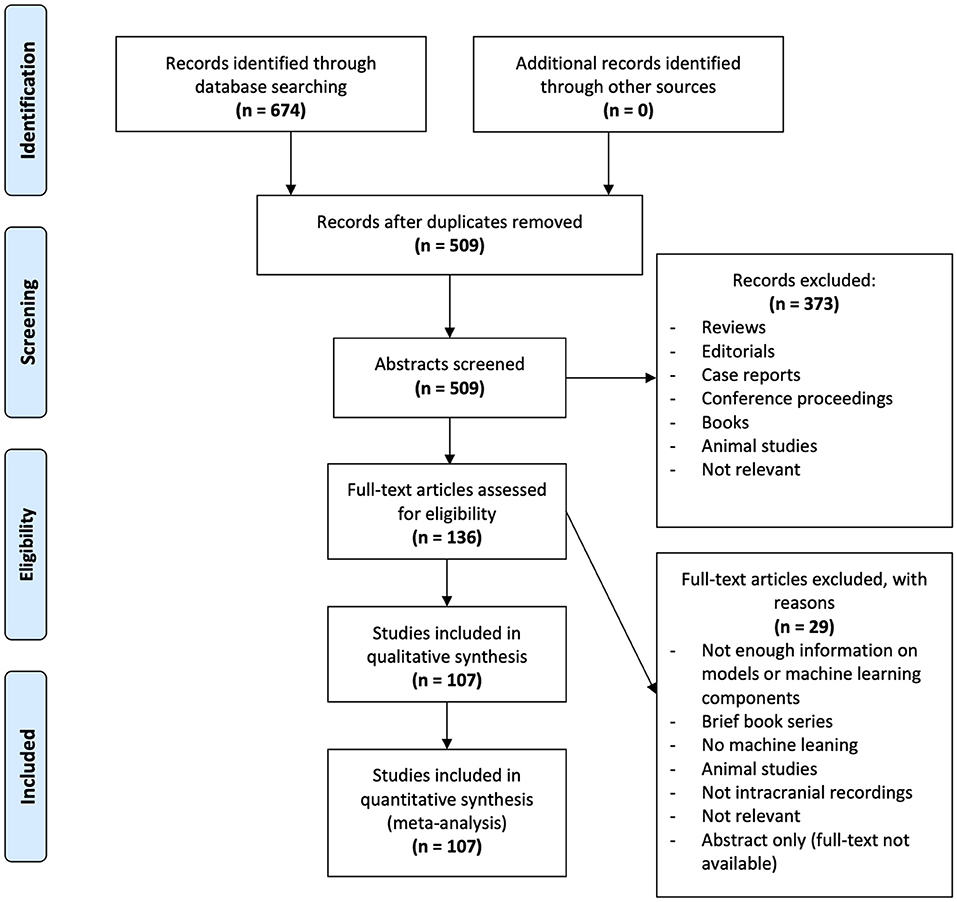
Figure 2. PRISMA analysis of articles searched, filtered and included in the systematic review. Six-hundred-seventy-four articles were identified through database searching in OVID MEDLINE, IEEE, and Web of Science. Duplicates were removed leaving 509 abstracts for screening. Three-hundred-seventy-three articles were excluded as they did not fit out inclusion criteria. From the remaining 136 articles, 29 were excluded following full-text screening. The remaining 107 articles were analyzed in this study.
Data Collection
Each full-text article was reviewed and data regarding demographics, data acquisition, machine learning application, and types of features were gathered into a spreadsheet.
Demographics
Demographics data was composed of year of publication, country of origin, name of the journal, journal category (medical, engineering/computer science, or multidisciplinary). The country of origin was defined by the affiliation of the first author.
Data Acquisition
Data acquisition comprised of intracranial recording method. This includes sEEG, and electrocorticographic (ECoG) strips or grids, or depth electrodes. A fourth option was available if the studies did not specify the type of recording method used. The source of EEG data was also compiled as free-text entries to track whether studies used their own data, or data obtained from online databases.
Machine Learning
The application of machine learning algorithms was classified into four broad categories. Due to the heterogeneity of studies in these fields, studies in the seizure and motor categories were further subdivided as follows:
1. Seizure: seizure prediction/detection, surgical outcome prediction, pathological tissue detection, high-frequency oscillation (HFO) detection/classification, seizure onset zone (SOZ)/epileptic focus localization, bad channel detection, spike detection, tumor tissue detection.
2. Motor: movement classification, motor imagery, speech production.
3. Cognitive tasks
4. Sleep Staging.
The type of ML algorithm was categorized into four groups: Standard supervised ML includes popular algorithms such as support vector machines (SVM), K-nearest neighbors (KNN), single-layered artificial neural networks (ANN), linear discriminant analysis (LDA) and any others that did not fit the definition of deep learning. Deep learning algorithms include recurrent neural networks (RNN), convolutional neural networks (CNN) and multi-layered neural networks (LeCun et al., 2015). The third option applied to studies that employed both standard ML and deep learning methods. The last option included unsupervised machine learning algorithms such as non-negative matrix factorization (NNMF) amongst others. We also recorded whether a study employed one or multiple types of algorithm for purposes of comparing classification performance. Studies which compared their algorithm performance with previously published algorithms trained on the same data were considered one algorithm as they did not themselves train multiple algorithms.
A summary of relevant feature selection steps was also collected in order to understand the data extraction approach utilized by each study. For purposes of comparison, feature selection approaches were split into two main categories: 1) higher order feature selection algorithms; and 2) physically interpretable features (raw or hand-crafted). Studies that used higher-order feature-selection algorithms were defined as those in which the feature selection/pre-processing workflows contained one or more of: i) non-standard time-frequency transforms; ii) dimensionality reduction of feature space; iii) abstract feature selection algorithms; or iv) abstract features (entropy, fractal dimension, etc.).
Performance metrics of the best final algorithms from each paper were recorded to gain insight on relative performance based on algorithmic and feature selection decisions. As algorithm performance can only be fairly compared between studies where both studies use the same dataset, this step was only done in studies employing the same publicly available datasets further discussed in Section Intracranial EEG Datasets. Although accuracy may be considered a sufficient measure of performance, it holds many caveats whereby it may give a false impression of performance in highly unbalanced data. As such, we opted to record all performance metrics presented within each study to provide a more objective picture. These include accuracy, area-under-ROC-curve, sensitivity, specificity, precision, positive predictive value, and/or negative predictive value. As model specificity was most commonly reported in all included studies, this metric is used to compare performance of algorithms between studies. If this metric was not provided, accuracy is used for comparison. Where studies solely reported cross-validation performance, a single mean performance metric was tabulated. In others that split their data into validation and testing sets, both the validation and testing performance were compiled and the testing performance was used for comparison.
Results
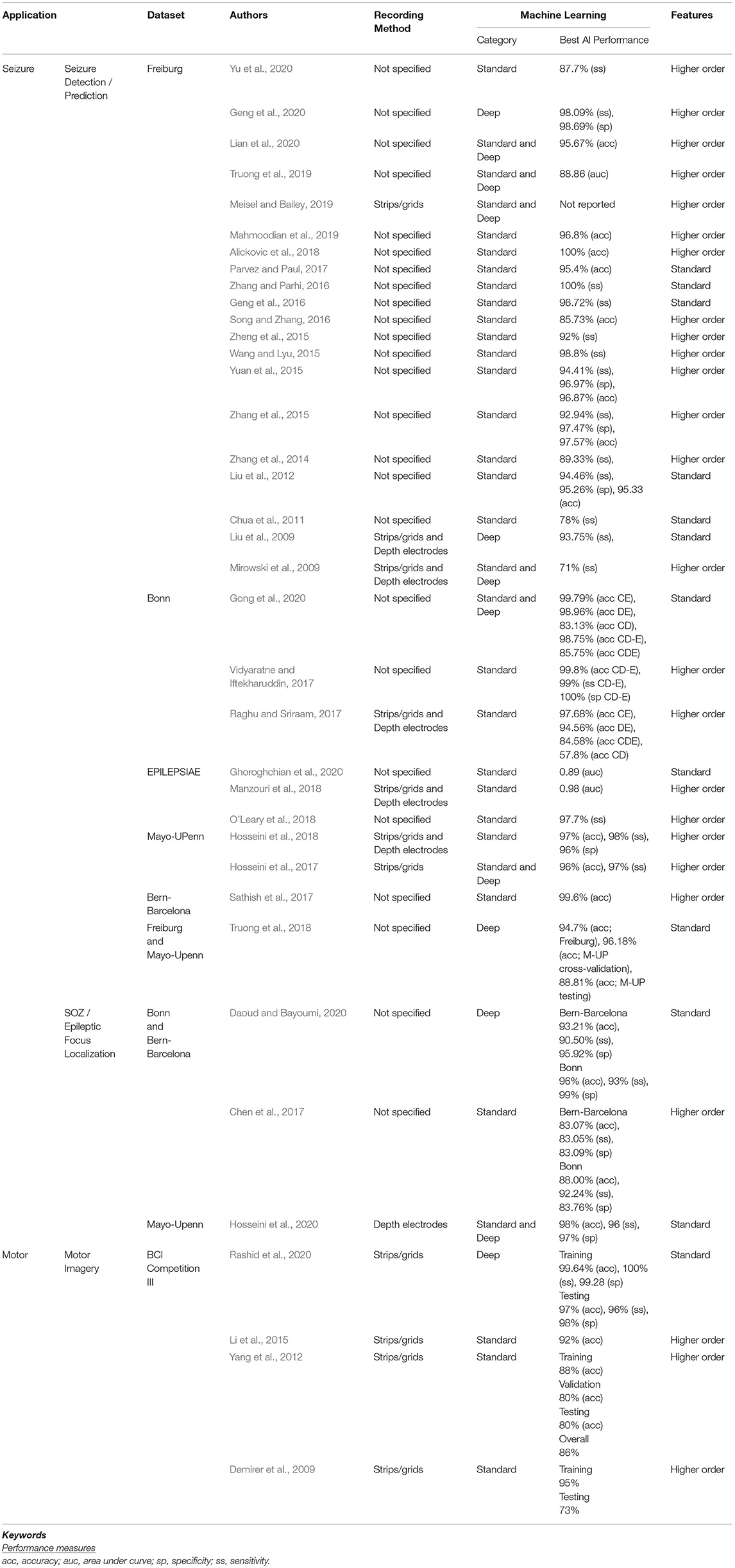
One-hundred-seven articles were included in the systematic review (Hellmann, 1999; Petrosian et al., 2000; D'Alessandro et al., 2005; Hill et al., 2006; Firpi et al., 2007a,b; Chan et al., 2008; Shenoy et al., 2008; Demirer et al., 2009; Liu et al., 2009, 2012; Mirowski et al., 2009; Scherer et al., 2009; Yanagisawa et al., 2009; Ayala et al., 2011; Chua et al., 2011; Kharbouch et al., 2011; Benz et al., 2012; Yang et al., 2012; Ikeda et al., 2014; McMullen et al., 2014; Zhang et al., 2014, 2015; Combrisson and Jerbi, 2015; Li et al., 2015; Memarian et al., 2015; Wang and Lyu, 2015; Yuan et al., 2015; Zheng et al., 2015; Boussen et al., 2016; Geng et al., 2016, 2020; Schrouff et al., 2016; Song and Zhang, 2016; Zhang and Parhi, 2016; Andrade et al., 2017; Antoniades et al., 2017; Chen et al., 2017; Combrisson et al., 2017; Elahian et al., 2017; Hosseini et al., 2017, 2018, 2020; Jrad et al., 2017; Khambhati et al., 2017; Kragel et al., 2017; Kremen et al., 2017, 2019; Parvez and Paul, 2017; Raghu and Sriraam, 2017; Sathish et al., 2017; Tomlinson et al., 2017; Vidyaratne and Iftekharuddin, 2017; Alickovic et al., 2018; Arora et al., 2018; Baud et al., 2018; Derner et al., 2018; Grinenko et al., 2018; Hermiz et al., 2018; Kiral-Kornek et al., 2018; Manzouri et al., 2018; Muller et al., 2018; O'Leary et al., 2018; Pan et al., 2018; Ramsey et al., 2018; Rutigliano et al., 2018; Shoaran et al., 2018; Truong et al., 2018, 2019; Tuyisenge et al., 2018; Varatharajah et al., 2018; Wu et al., 2018; Xie et al., 2018; Angrick et al., 2019a,b; Cimbalnik et al., 2019; Klimes et al., 2019; Lai et al., 2019, 2020; Livezey et al., 2019; Mahmoodian et al., 2019; Medvedev et al., 2019; Meisel and Bailey, 2019; Nejedly et al., 2019a,b; Pailla et al., 2019; Principe et al., 2019; Saboo et al., 2019; Sumsky and Santaniello, 2019; Thomas et al., 2019; Weidemann et al., 2019; Abou Jaoude et al., 2020; Akter et al., 2020; Burrello et al., 2020; Daoud and Bayoumi, 2020; Ghoroghchian et al., 2020; Gong et al., 2020; Karthick et al., 2020; Lian et al., 2020; Makaram et al., 2020; Makin et al., 2020; Rashid et al., 2020; RaviPrakash et al., 2020; Sciaraffa et al., 2020; Yu et al., 2020; Zhao et al., 2020; Zhu et al., 2020). Information regarding each article was collected into four categories: demographics, data acquisition, machine learning model, types of features. A succinct summary of studies employing publicly available datasets and reporting best AI model performance is presented in Table 1 while a more extensive table containing all studies is available in Supplementary Material 2.
Demographics
The number of publications per year is illustrated in Figure 3A. A dramatic increase in the number of publications was observed from 2017 onwards. The majority of articles before 2017 utilized standard machine learning algorithms as opposed to deep learning or unsupervised learning. For comparison, the use of deep learning or both (standard ML and deep learning) made up only 20% of articles published in 2017 compared to 68% of those published in 2020.
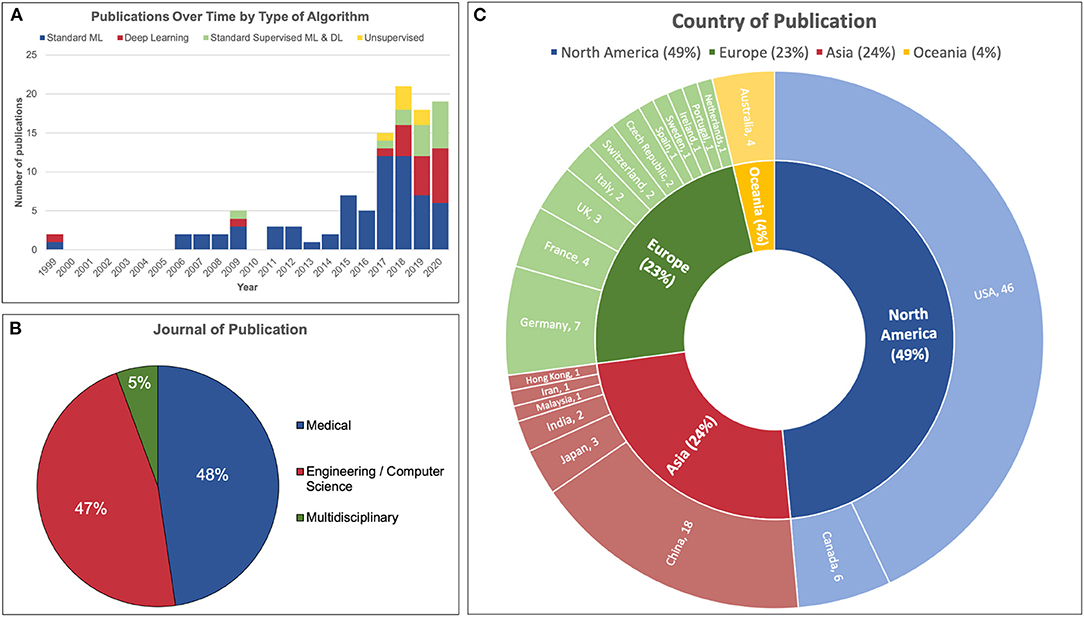
Figure 3. Demographics of 107 articles reviewed. (A) The number of publications in the field of machine learning and iEEG dramatically increased in 2017 incorporating more deep learning and unsupervised learning models. (B) Just under half of articles reviewed were published in a medical journal and a similar proportion were published in an engineering or computer science journal. A minority was published in a multidisciplinary journal. (C) Almost half of the studies were conducted in North America. The United States published the most papers, followed by China, Germany and Canada.
Fifty-one articles were published in medical journals while 50 were published in engineering or computer science journals. Six articles were published in multidisciplinary journals. This distribution is illustrated in Figure 3B.
The distribution of country of origin for each article is displayed in Figure 3C. The majority of articles originated from the United States of America (46/107, 42%), followed by China (18/107, 17%), Germany (7/107, 7%) and Canada (6/107, 6%).
Data Acquisition
The types of intracranial recording varied across each study. Some articles employed only one type of intracranial recording, while others used two methods. Articles which did not specify their method of intracranial EEG recording were simply labeled as unspecified iEEG. The distribution of intracranial recording method is illustrated in Figure 4A. Of those which specified a recording technique, ECoG strips or grids were used most frequently (53/107 articles), followed by depth electrodes (23/107) and sEEG (9/107). Thirty-seven articles were labeled as unspecified iEEG. Some articles used more than one method, hence this sum is greater than the number of articles.
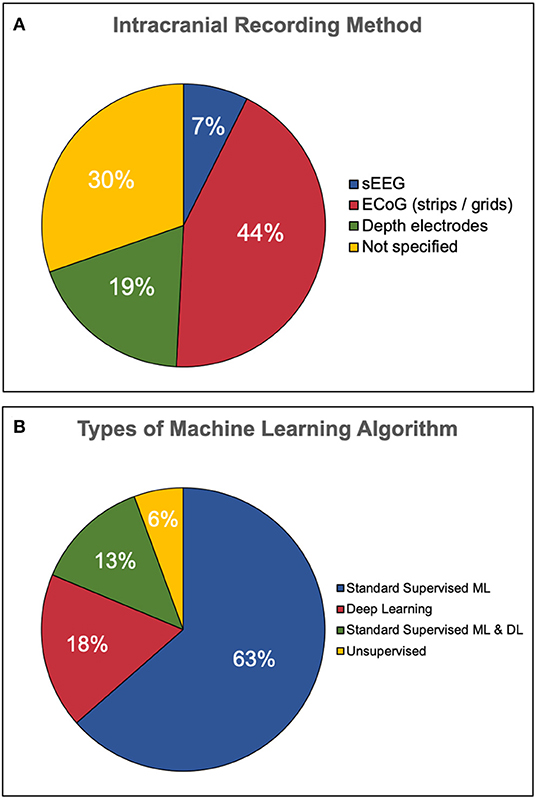
Figure 4. Types of iEEG recording and classification algorithm employed. (A) Less than half of the publications employed electrocorticography techniques such as strips and grids. The second most popular recording method was depth electrodes followed by a minority of stereotactic EEG. One third of the articles did not specify the type of recording method. (B) The majority of studies employ standard supervised machine learning (ML) only, while only 18% used deep learning (DL) only. Thirteen percent used both standard and deep learning while a minority used unsupervised learning.
Just over half of the articles analyzed in this review reported using their own datasets (57/107, 53%). The 50 remaining reported the use of at least one pre-existing dataset, either from their own research group or a freely available dataset available online. Some publications use more than one source of data.
Intracranial EEG Datasets
Pre-existing iEEG datasets are utilized in a number of these papers. The most popular of these include the Freiburg (Andrzejak et al., 2012) (21/50, 42%), the UBonn (Andrzejak et al., 2001) dataset (6/50, 12%), BCI Competition III (Blankertz et al., 2006) (4/50, 8%), Mayo Clinic (Stead et al., 2010) (3/50, 6%), Bern-Barcelona (3/50, 6%), and EPILEPSIAE (3/50, 6%). The Freiburg EEG database is composed of grid, strip and depth electrode recordings from 21 patients suffering from medically intractable focal epilepsy. The UBonn dataset is composed of strips and depth electrodes recorded from epilepsy patients. It is divided into 5 sets where A and B are scalp EEG and C, D and E are iEEG recordings from patients with temporal lobe epilepsy. Sets C and D are composed of inter-ictal intervals while set E shows ictal activity. The BCI Competition III includes 8 scalp and intracranial EEG datasets involving motor imagery and P300 speller paradigms. Specifically, only dataset 1 includes ECoG grids, while the remaining solely include scalp EEG. The Mayo Clinic dataset is composed of strips, grids and depth electrode recordings from 11 epilepsy patients. The Bern-Barcelona dataset comprises of strips and depth electrodes from 5 pharmacoresistant temporal lobe patients. Finally, the EPILEPSIAE database is the product of a collaboration between epilepsy centers in Portugal, France and Germany. It contains both scalp and intracranial EEG recordings including grids and depth electrodes.
Machine Learning
The ML applications for each article were grouped into 4 categories, all illustrated in Figure 5. Four articles employed machine learning for more than one application, thereby leading to 111 counts for 107 articles. The vast majority of applications relate to seizures (74/111, 67%), followed by motor (25/111, 23%), cognitive tasks (8/111, 7%), and sleep staging (4/111, 4%).
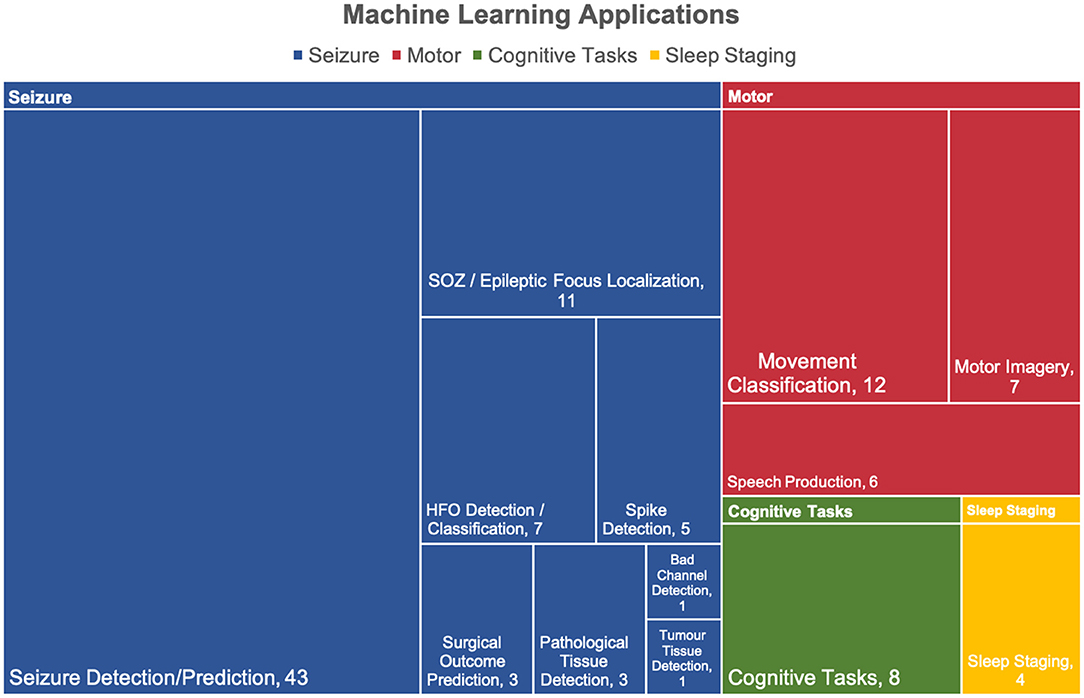
Figure 5. Applications of machine learning in intracranial EEG. The applications were divided into 4 categories: seizure, motor, cognitive tasks. Over half of the articles were relevant to seizures. Four studies had more than one application thereby leading to a total of 111 applicants for 107 articles.
Applications
The seizure category is divided into 8 subcategories in which seizure detection/prediction (43/74, 58%) and SOZ/epileptic focus localization (11/74, 15%) were the most popular. The motor category is composed of 3 subcategories where movement classification makes up half of the applications (12/25, 48%).
Algorithm
The majority of articles reviewed in this study employed standard supervised ML methods only (68/107, 63%), followed by DL only (19/107, 18%). Fourteen articles employed both standard ML and DL in order to compare algorithm performance. Unsupervised learning made up a minority of articles found (6/107, 6%). An illustration of this distribution is presented in Figure 4B. The majority of studies presented results from a single algorithm (78/107, 73%) rather than multiple algorithms (29/107, 27%).
Comparing Algorithmic Performance
Performance metrics for each study employing a publicly available dataset are summarized in Table 1. Five public datasets were identified amongst the studies employing ML for seizure detection or prediction. Amongst studies utilizing the same dataset, a variety of different metrics were reported. It is also important to note that while some studies employ all patient data from public datasets, some may only select a subgroup of participant data from a dataset. Therefore, Table 1 provides the specific recording method as reported by the authors.
Amongst the studies utilizing the Freiburg dataset, 11 reported metrics on the entire set of 21 patients (Liu et al., 2012; Zhang et al., 2014, 2015; Yuan et al., 2015; Geng et al., 2016, 2020; Parvez and Paul, 2017; Alickovic et al., 2018; Mahmoodian et al., 2019; Meisel and Bailey, 2019; Yu et al., 2020). Within this group, deep learning methods achieved significantly higher sensitivity for seizure detection compared to standard methods (mean 0.922 vs. 0.980, p = 0.019 by Student's t-test). Few of these studies, however, reported specificity thereby making a performance comparison based on this metric more difficult. In those using the Freiburg dataset and exclusively investigating standard machine learning (Chua et al., 2011; Liu et al., 2012; Zhang et al., 2014, 2015; Wang and Lyu, 2015; Yuan et al., 2015; Zheng et al., 2015; Geng et al., 2016; Song and Zhang, 2016; Zhang and Parhi, 2016; Parvez and Paul, 2017; Alickovic et al., 2018; Mahmoodian et al., 2019; Yu et al., 2020), SVM was the most popular. Fewer studies employed the Bonn dataset, where deep learning and a vector machine algorithm achieved greater accuracies compared to a standard multilayer perceptron (MLP) with a single hidden layer (Raghu and Sriraam, 2017; Vidyaratne and Iftekharuddin, 2017; Gong et al., 2020). Studies using EPILEPSIAE data all employed standard ML but specificity metrics were not reported for comparison (Manzouri et al., 2018; O'Leary et al., 2018; Ghoroghchian et al., 2020). Those using the Mayo-UPenn show similar accuracies with deep learning and a combination classifier of standard supervised algorithms (Hosseini et al., 2017, 2018; Truong et al., 2018).
Three datasets were employed by studies for SOZ or epileptic focus localization. Two studies used the Bonn and Bern-Barcelona datasets where a deep MLP (more than one hidden layer) (Daoud and Bayoumi, 2020) outperformed a standard SVM (Chen et al., 2017) by achieving greater specificity. The only study that employed the Mayo-UPenn dataset in this context revealed a greater specificity with an LSTM-SVM in comparison to an LSTM alone (Hosseini et al., 2020). This is particularly interesting as it shows that combining a deep learning approach for feature extraction and selection with a standard SVM model may further outperform deep learning alone.
A single publicly available dataset was found amongst the motor studies in the context of motor imagery classification. Three of the 4 studies employing the BCI Competition III dataset split their data into a training and testing group (Demirer et al., 2009; Yang et al., 2012; Rashid et al., 2020) while one only reported their validation accuracy (Li et al., 2015). Amongst the standard ML methods, SVM (Demirer et al., 2009) achieved a greater accuracy than LVQ (Li et al., 2015) and ANN (Yang et al., 2012) in validation sets, but the ANN performed better in testing. This may indicate that although the SVM may performed well in training, it appears to overfit the training data and did not generalize as well as the ANN in this context. However, the most recent study using this dataset employed deep learning and achieved significantly greater accuracies compared to all previous standard ML methods in both training and testing (Rashid et al., 2020).
All studies involved in cognitive tasks or sleep staging used unique datasets and therefore cannot be effectively compared based on classification performance. Amongst the cognitive task studies, two compared different types of algorithm within their own dataset. Though detailed statistical comparison is not possible, deep learning appeared to outperform linear regressions and SVM (Arora et al., 2018). A second study revealed that a combination of LSTM and CNN outperformed CNN alone thereby further probing the potential of combined approach (RaviPrakash et al., 2020). No sleep staging studies were reported to compare different algorithms.
Influence of Feature Selection
In general, studies focusing on seizure detection and prediction applied time-frequency representations of raw EEG signals. This was accomplished with bandpass filtering or signal decomposition using Fourier or wavelet transforms. Less commonly other signal decomposition methods were also used for this purpose, such as the Stockwell transform (Geng et al., 2020), harmonic wavelets (Vidyaratne and Iftekharuddin, 2017) or empirical mode decomposition (EMD) (Zheng et al., 2015). These transforms yield frequency-domain representations of the signal, namely the amplitudes of various frequency ranges in a given signal segment or epoch. These amplitudes were most often used to calculate statistical features such as the mean, variance, and standard deviation (Alickovic et al., 2018; Meisel and Bailey, 2019), as well as higher-order statistical features including skew and kurtosis (Mirowski et al., 2009; Hosseini et al., 2017). Amplitude at various frequencies were also be used to calculate the envelope (Wang and Lyu, 2015; Meisel and Bailey, 2019) (a function enclosing the peaks of the signal, or instantaneous amplitude), or line length (Baud et al., 2018) to capture spikes. Several algorithms in our dataset used bivariate or multivariate (i.e., features calculated from pairs or groups of electrodes) features including cross-correlation and phase-locking value between pairs of signals, as well as higher-order measures of synchrony such as non-linear interdependence, dynamical entrainment, and entropy in difference of phases (Mirowski et al., 2009; O'Leary et al., 2018).
Signal entropy is a measure of the predictability of a signal–that is, disordered and unpredictable signals have high entropy and vice versa. Studies reviewed herein used entropy of various time series, in particular the raw EEG signal, frequency and power spectra, phase, as well as multi-electrode signals (Hosseini et al., 2017; Vidyaratne and Iftekharuddin, 2017; Mahmoodian et al., 2019). A closely related measure to entropy is fractal dimension, in that the latter measures the “roughness” of a signal at different scales. The fractal dimension of frequency- or time-domain signals can be estimated using box-counting methods, as well as the more formal Hausdorff dimension.
SOZ localization depends on the detection and analysis of signal phenomena such as HFOs, phase-amplitude coupling (PAC), and interictal epileptiform discharges (IED). In particular, HFOs have been proposed as epileptogenic biomarkers with emerging prognostic utility in cortical resection outcomes (Jacobs et al., 2010; Frauscher et al., 2017). These events are first detected in raw EEG signals using well-validated detectors in the literature, such as the line-length and Cimbalnik-Stead HFO detectors. The rate of these events in specific electrodes are used to localize the epileptic focus, either as the sole feature (Sumsky and Santaniello, 2019) or in an ensemble with other first-order or abstract features (Klimes et al., 2019).
Features for speech production, motor imagery, and cognitive tasks differed in key respects with those for seizure prediction. While high-frequency oscillations in high-gamma (70-135Hz) and ripple (135-200Hz) bands were most useful in seizure prediction (Pan et al., 2018), features sources from lower frequencies, such as theta (Livezey et al., 2019), alpha (Andrade et al., 2017; Hosseini et al., 2020), and beta bands (Yang et al., 2012) were most useful for speech, motor and cognitive tasks. In addition, features for these three categories generally focused on amplitude, spectral power and its transformations rather than measures of disorder like entropy and fractal dimension (Pan et al., 2018; Xie et al., 2018; Pailla et al., 2019; Thomas et al., 2019). In particular, the normalized (or Z-transformed) log-spectral power obtained from a morlet wavelet transform is commonly used in speech and cognitive tasks (Ikeda et al., 2014; Schrouff et al., 2016; Kragel et al., 2017; Arora et al., 2018; Weidemann et al., 2019).
Hand-selected feature sets through these various methods may be ensemble-optimized to reduce dimensionality and redundancy, thereby making downstream classification more accurate and computationally efficient. Parallel arrays of time-varying features may be subject to dimensionality reduction using principal component analysis (PCA) or its variants as a data pre-processing step (Tomlinson et al., 2017; O'Leary et al., 2018; Yu et al., 2020). Some algorithms rely on feature selection using some optimization algorithm or neural network (Firpi et al., 2007b; Antoniades et al., 2017; Abou Jaoude et al., 2020), or even generating artificial features de novo using a genetic algorithm (Firpi et al., 2007a; Yang et al., 2012; Sathish et al., 2017).
Measures such as fractal dimension and higher-order moments have unintuitive physical correlates and may not correlate with underlying biological signals. Thus, we compared “higher-order” feature selection algorithms with approaches with physically interpretable features to determine if differences in model performance arise. Studies that used higher-order feature-selection algorithms were defined as those in which the feature selection/pre-processing workflows contained one or more of: i) non-standard time-frequency transforms; ii) dimensionality reduction of feature space; iii) abstract feature selection algorithms; or iv) abstract features (entropy, fractal dimension, etc.). When comparing seizure prediction/detection algorithms that were tested on all 21 patients from the Freiburg iEEG dataset, “standard” and “higher-order” feature selection approaches did not differ in overall sensitivity (mean 0.966 vs. 0.930, p = 0.1696 by Student's t-test). This suggests that while higher-order input features have few biological and physical correlates, this penalty to interpretability does not necessarily translate into better model performance.
Discussion
The study of iEEG provides unprecedented insight into the neural code, revealing the underpinnings of a variety of physiological and pathological brain processes. At present, the gold standard of iEEG analysis involves extraction of features through digital signal processing and analyses by human experts. However, ML is rapidly becoming a viable tool that may facilitate better understanding of intracranial data. In the current review, we describe the landscape of ML applied to iEEG. We also compare and contrast various methods and provide a synopsis of relative advantages and pitfalls.
Seizure
Machine learning applications to iEEG predominantly relate to epilepsy. The majority of studies reviewed herein focused on classification of signals as seizures or non-seizure events, while others employed ML for seizure onset prediction. In the latter, the algorithm identifies patterns in the pre-ictal signal in order to pre-emptively predict seizure onset. This is a particularly challenging task for humans to accomplish and therefore an ideal opportunity for the implementation of AI tools (Assi et al., 2019).
Deep learning algorithms demonstrate the best performance in the classification of seizures and SOZ localization. Unlike an ML-informed approach, current practices of seizure detection are susceptible to inter-rater variability. A recent study studying seizure detection from ECoG signal revealed an interrater agreement of only 79% (Quigg et al., 2015). This supports the potential role of using ML-powered algorithms to better inform experts in complex cases thereby reducing variability.
Another benefit of ML-based tools is the ability to integrate multiple data streams including EEG, electromyography (EMG), accelerometry (ACM), electrocardiography (ECG), near-infrared spectroscopy (NIRS), video, and electrodermal activity (EDA) inputs beyond what can be achieved by a human interpreter (Ulate-Campos et al., 2016). This type of approach has also been applied for other applications relevant to neurology such as early detection of Parkinson's disease (Prashanth et al., 2016), falls (Nahiduzzaman et al., 2020) and identification of cortical dysplasia (Mo et al., 2019). One of the studies reviewed utilized this multidimensional approach and achieved an accuracy of 95% for surgical outcome prediction in epilepsy patients, albeit on an in-house dataset (Memarian et al., 2015).
Enhanced SOZ localization is an emerging area for increased ML utilization. Indeed, in medically-refractory epilepsy, post-operative seizure freedom rates remain in the range of 50–70% despite multimodal analytic tools to guide localization and resection of the putative epileptogenic zone (Englot et al., 2011, 2012). Accurate localization of the SOZ also remains the strongest predictor of positive surgical outcome (Jacobs et al., 2010). Through network-based analytic approaches, ML algorithms can provide more detailed localizations of seizure onset zones based on higher-order patterns not immediately apparent to experts, with the potential to improve post-operative outcomes. Studies reviewed herein demonstrate accuracies in excess of 90% using deep learning and 80% with standard ML (Chen et al., 2017; Daoud and Bayoumi, 2020; Hosseini et al., 2020). A growing body of literature also supports the use of ML in identifying spectral patterns such as high-frequency oscillations that may further refine SOZ localization (Jacobs et al., 2010; Weiss et al., 2019; Si, 2020).
Beyond epilepsy, the breadth of ML use in neurosurgical outcome prediction spans multiple fields from neoplasms to spinal disease to arteriovenous malformations, with promising success. (Senders et al., 2018) Such applications of ML may not only guide the decision-making process of neurosurgical teams when selecting surgical candidates, but may also reveal patterns previously unknown to influence surgical outcome.
Despite the significant potential of deep learning, there are several challenges to its clinical deployment. First, deep learning generally requires a significantly larger pool of data compared with standard ML algorithms, and datasets of this size may be clinically unavailable. A second limitation of deep learning relates to its status as a “blackbox” algorithm whereby the underlying decision-making process of the algorithm cannot be readily understood. A great deal of contemporary literature has examined this subject and is producing algorithmic approaches to explain such “blackbox” models. Examples include game theoretic “Shapley” values and permutation importance which provide insight through backpropagation of altered to retrospectively determined feature importance (Wong et al., 2019; Sundararajan and Najmi, 2020). Conversely, standard ML remains more transparent without the use of these complex middle-ground interpreters. Given the reported performance advantages of deep learning, we suggest that an area for substantial future research should relate to the development of clinical tools that may improve inherent sample sizes (such as the development of large, centralized databases). We also recommend that future work should also focus on deeper development of backpropagation tools to explain model predictions.
Interestingly, the realm of neuro-oncology may also be impacted by these advanced AI technologies. A study reviewed herein presented a novel proof of concept using ML to differentiate healthy tissue from brain tumor tissue during awake craniotomy (Boussen et al., 2016). Although the gold standard of intraoperative mapping remains direct cortical stimulation, Boussen et al. were able to discriminate tumoral tissue from eloquent brain using an ANN based on spectral data from ECoG electrodes with a classification accuracy of 93.6%. The finding of profound spectral alteration within tumoral tissue when compared to healthy brain may serve as a useful intraoperative adjunct in awake surgery.
Motor
A significant number of machine learning applications to iEEG relate to motor tasks. Our analysis of the model performances in this context provided a reminder on the importance of model generalizability. Although Demirer et al.'s SVM (Demirer et al., 2009) showed better validation metrics for motor imagery classification with the BCI Competition III dataset compared to Yang et al.'s ANN (Yang et al., 2012), the final testing accuracy was significantly lower. This finding poses a critical reminder with regards to overfitting. When a validation set is used to explicitly optimize a model, the metrics must be interpreted with caution as they may provide an overly optimistic estimate of model performance, as seen in the Demirer study. Therefore, whenever possible, performance should be evaluated on an unseen testing set once the model has been finalized to accurately judge generalization performance. As it relates to the classification of motors tasks, given the small number of studies for comparison, it is not possible to conclude whether an ANN or SVM would be superior in terms of motor classification tasks. However, it does underscore the importance of using unseen data to generate an unbiased estimate of model performance.
The majority of movement classification studies employ algorithms to differentiate motor tasks such as upper limb movement (Yanagisawa et al., 2009; McMullen et al., 2014; Thomas et al., 2019) and hand or finger gesturing (Scherer et al., 2009; Pan et al., 2018; Xie et al., 2018; Pailla et al., 2019; Zhu et al., 2020) based on electrophysiological recordings. One study used a similar paradigm but trained their algorithm to differentiate rest, movement intention and movement execution (Combrisson et al., 2017). By distinguishing the vital steps of motor preparation based on neural correlates, ML in these cases can provide insight on the importance of specific features such as oscillatory phase and amplitude in understanding motor function. On the other hand, motor imagery tasks require participants to imagine performing a specific action. In these studies (Hill et al., 2006; Shenoy et al., 2008; Demirer et al., 2009; Yang et al., 2012; Li et al., 2015; Andrade et al., 2017; Rashid et al., 2020), algorithms were trained to identify when a participant imagined a specific action. Within the examined studies, we see that modern ML approaches can achieve a significant degree of accuracy–up to 98%-in these classification tasks.
The relevance of combining iEEG signals with ML in these domains can relate to better tailoring of neuroprostheses to individual patient dynamics. Whereas, current myoelectric prostheses follow a “one-size-fits-all” approach, the adaptability of ML-based neuroprotheses could learn from the patient's individual movements and adapt to become more responsive and precise over time. Indeed, a recent survey of upper limb prothesis users reveals a desire for adaptable grip strength and individual finger and thumb movements (Cordella et al., 2016). Just as humans learn to adapt these composites of motor movements, a brain-computer interface (BCI) powered by machine learning may be able to learn to adapt as the user interacts with the environment. Furthermore, recent studies indicate that BCI prosthetics may play a role in reducing phantom limb pain (Yanagisawa et al., 2020). The underlying mechanism for this change has yet to be elucidated although it is believed to be related to neuroplasticity in the sensoricortex.
The last sub-category of motor tasks relates to speech production (Ramsey et al., 2018; Livezey et al., 2019). Here, participants were asked to articulate sounds, phenomes or words presented on a screen. Algorithms were trained to differentiate these sounds based on iEEG. Unlike the traditional myoelectric or keyboard-based voice prothesis, neuroprostheses involve a BCI that can extract EEG signals to reconstruct the patient's speech. In this review, a single study (Makin et al., 2020) employed ML to reconstruct sentences from iEEG signal in a small number of participants, demonstrating a word error rate (measure of incorrect predictions) of 3% compared to the professional level of 5% (Xiong et al., 2017). Though these applications are clearly within their infancy, speech prostheses powered by ML may similarly learn from each individual patient's speech patterns to improve speech reconstruction performance with use.
Cognitive Tasks
The study of cognitive performance is unique in that the neural circuitry and activity patterns remain poorly understood. This has limited the development of neuromodulatory interventions, such as deep brain or closed-loop stimulation that could potentially treat deficits in subdomains of cognition. A key role for ML therefore lies in its potential to identify activity distributed across neural assemblies related to specific tasks.
Within the studies we report, memory was the most common cognitive task studied (Kragel et al., 2017; Arora et al., 2018; Derner et al., 2018; Saboo et al., 2019; Weidemann et al., 2019). Three studies employed standard ML including SVM and LR although no comparisons were made between different types of algorithm. Derner et al. (2018) obtained average classification accuracies for memory recall in the range of 60%; similarly, Kragel et al. (2017) presented AUCs of 0.59 and 0.68 with a classifier able to estimate memory task success during retrieval and encoding, respectively. Weidemann et al. (2019) developed LR models that performed better than chance while also noticing that their memory performance prediction classifiers could generalize to unrelated lists. This supports that their classifier is trained to distinguish episodic memory without discriminating for sematic content (Weidemann et al., 2019). This has significant implications for the future development of closed-loop stimulation powered by ML which may be able to predict memory performance across a breadth of contexts. Arora et al. (2018) is the only study that employed deep learning for memory task performance classification and showed that LSTM (mean AUC 0.72) modestly outperformed SVM (mean AUC 0.68) which itself significantly outperformed LR (mean AUC 0.59). Hence, there remains debate whether a modest improvement in performance outweighs the lack of transparency of LSTM in comparison with SVM.
The complexity of neural activity during cognitive tasks is reflected in the relatively modest improvements in algorithm classification performance when compared to chance. However, despite these challenges, these models have shown practical utility in closed-loop stimulation for memory (Ezzyat et al., 2018).
Other studies in the cognitive realm employed machine learning for unique purposes. Saboo et al. (2019) employed a novel approach by comparing an unsupervised clustering model with LDA and SVM to identify active electrodes during a verbal memory task. Their unsupervised method significantly outperformed standard supervised ML reaching a sensitivity of 97% and specificity of 92.9% (Saboo et al., 2019). However, as the ground truth for these labels was defined by expert reviewers, the authors were unable to compare their performance with human experts as the algorithm aimed to recreate the human decisions. Nonetheless, this method provides a much faster alternative to the highly time-consuming and subjective process of human expert labeling (Saboo et al., 2019).
In another, SVMs were used to differentiate whether participants were performing a mathematical or a memory task and achieved best accuracies of 89.51, 87.18 and 76.80% for 3 participants (Schrouff et al., 2016). Another study by Hermiz et al. (2018) employed numerous approaches with a logistic regression to differentiate different types of auditory and visual stimuli achieving accuracies in the 80 to 95% range. Lastly, RaviPrakash et al. (2020) developed a deep learning language comprehension algorithm able to differentiate whether participants are actively listening to a story, or listening to broadband noise. The authors achieved a greater performance with a combination algorithm of LSTM and CNN as opposed to CNN alone, with a classification accuracy of 83%. Although several studies in the field of cognition revealed a better performance of DL algorithms, the small sample size limits definitive conclusions.
Sleep Staging
Studies identified in this review employed supervised (Kremen et al., 2017; Rutigliano et al., 2018; Principe et al., 2019) or unsupervised (Kremen et al., 2019) machine learning to differentiate sleep stages in patients diagnosed with epilepsy. Kremen et al. (2017) employed SVM to classify awake states and slow wave sleep with an accuracy of 97.8% and 89.4% in healthy and epileptic tissue, respectively. A more recent study by this group (Kremen et al., 2019) obtained a 94% accuracy with unsupervised learning for more complex classification of three groups: awake states, N2 and N3 sleep. Rutigliano et al. (2018) employed an ANN with a single hidden layer to classify wakefulness and non-REM sleep with an accuracy of 98.57%. As the labels were defined by a human neurologist, a comparison to expert performance cannot be established.
The primary advantage of ML in this context is the speed of classification and accuracy of sleep-stage detection which would be of importance to a variety of neuromodulatory therapeutics that function during specific stages of sleep (Kremen et al., 2019). An example of such an approach includes closed-loop auditory stimulation during sleep for epilepsy (Fattinger et al., 2019). Furthermore, such tools can lessen the healthcare burden related to long waiting periods for patients awaiting formal sleep studies (Rotenberg et al., 2010).
Limitations of ML Algorithms
The literature search conducted herein revealed that multiple different types of ML algorithms can be employed in the context of iEEG analysis and classification for variety of applications. However, each algorithm faces its own set of limitations. Generally, standard algorithms including SVM, KNN, and decisions trees may not be suitable for very large datasets. Training an SVM also involves numerous parameters which can be a lengthy process to optimize, and they can be also be biased if the number of features provided is greater than the number of samples in the training set. KNNs also perform poorly when using a large number of features and they are particularly sensitive to outliers or missing values. Decision trees can become hypersensitive to its input data whereby small changes in the input can have drastic effects on the output classification. As an ensemble of decision trees, RFs are can be limited by their interpretability and are often considered a black box. Similarly, deep learning models are generally criticized for their lack of interpretability, although novel methods attempting to elucidate how these complex multilayered models make decisions are discussed in the literature (Montavon et al., 2018). A thorough understanding of the dataset, as well as the goals of the ML algorithm are critical to select appropriate models that are best suited to data.
Future Recommendations
Exploring the current ML literature, we identified three recommendations which may be useful for future work in ML and iEEG. First, we believe that future studies should aim to not only report performance metrics but provide a specific reference point for comparison–whether the best previous ML performance or contemporary expert performance. Such comparisons can help ground clinical interpretation of the data and identify the potential practical utilizations of ML in routine clinical practice. Second, large datasets representative of the target population may be advantageous over small datasets as these are more likely to generalize well when applied to new patients. This reinforces the importance of internal and external validation to ensure that algorithms can perform equally well with previously unseen data. Third, it may be advantageous to employ explainable ML algorithms rather than “black box” models in order to shed light on algorithms' decision-making processes. More transparent models may not only offer new knowledge on important features of disease but may also improve the trust from the medical community to employ these models clinically. A study centered around classification performance may employ a more convoluted algorithm while sacrificing interpretability while one aiming to provide new knowledge on the classification process may sacrifice some performance for interpretability.
Conclusions
Artificial intelligence is expected to have a significant impact on the future practice of neurosurgery. By identifying complex and hidden patterns that may be inconspicuous to human counterparts, ML algorithms may not only offer more accurate and more rapid tools to assist decision making but may also elucidate the underlying mechanisms involved in a number of neurological disorders.
This study achieved both of our aims: to assess the current landscape of ML in iEEG, and to examine the relative benefits and limitations of ML approached as they related to neurosurgical problems. We conducted a systematic search across three databases encompassing the fields of medicine, science and engineering and identified 107 articles. We nonetheless recognize that some articles may have been missed within our systematic search considering the rapid rate at which papers within the realm of AI are currently being published.
The studies reviewed herein reveal a promising future of AI in iEEG by shedding light on the variety of ML algorithms that can be employed to classify data in the context of seizures, motor tasks, cognitive tasks and sleep. Of these, supervised algorithms were most commonly employed with most articles published in the realm of seizures and epilepsy. The merits of using ML to tackle neurosurgical problems are broad including the development of more personalized neuroprostheses, standardizing seizure detection compared to human analyses, predicting surgical outcomes, and improving the speed and classification of sleep stages.
As AI becomes increasingly utilized in medicine and surgery, we intend for this work to serve as a resource to aid in the interoperation and development of relevant technologies employing ML in the context of iEEG analysis.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
Author Contributions
NM and NW led the study and conducted the literature search, review and analyses. FZ conducted the statistical analyses and contributed to extracting content from the literature. SW and LE provided extensive input on machine learning. SW, HS, KM, LE, and GI assisted and provided input throughout the review process. NM, NW, and FZ wrote the manuscript with input and feedback from all authors. GI was in charge of the overall direction and planning. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fnhum.2022.913777/full#supplementary-material
References
Abou Jaoude, M., Jing, J., Sun, H., Jacobs, C. S., Pellerin, K. R., Westover, M. B., et al. (2020). Detection of mesial temporal lobe epileptiform discharges on intracranial electrodes using deep learning. Clin. Neurophysiol. 131, 133–141. doi: 10.1016/j.clinph.2019.09.031
Akter, M. S., Islam, M. R., Iimura, Y., Sugano, H., Fukumori, K., Wang, D., et al. (2020). Multiband entropy-based feature-extraction method for automatic identification of epileptic focus based on high-frequency components in interictal iEEG. Sci. Rep. 10, 7044. doi: 10.1038/s41598-020-62967-z
Alickovic, E., Kevric, J., and Subasi, A. (2018). Performance evaluation of empirical mode decomposition, discrete wavelet transform, and wavelet packed decomposition for automated epileptic seizure detection and prediction. Biomed. Signal Process. Control 39, 94–102. doi: 10.1016/j.bspc.2017.07.022
Andrade, J., Cecilio, J., Simoes, M., Sales, F., and Castelo-Branco, M. (2017). Separability of motor imagery of the self from interpretation of motor intentions of others at the single trial level: an EEG study. J. Neuroeng. Rehabil. 14, 63. doi: 10.1186/s12984-017-0276-4
Andrzejak, R. G., Lehnertz, K., Mormann, F., Rieke, C., David, P., and Elger, C. E. (2001). Indications of nonlinear deterministic and finite-dimensional structures in time series of brain electrical activity: Dependence on recording region and brain state. Phys. Rev. E. 64, 61907. doi: 10.1103/PhysRevE.64.061907
Andrzejak, R. G., Schindler, K., and Rummel, C. (2012). Nonrandomness, nonlinear dependence, and nonstationarity of electroencephalographic recordings from epilepsy patients. Phys. Rev. E. 86, 46206. doi: 10.1103/PhysRevE.86.046206
Angrick, M., Herff, C., Johnson, G., Shih, J., Krusienski, D., and Schultz, T. (2019a). Interpretation of convolutional neural networks for speech spectrogram regression from intracranial recordings. Neurocomputing. 342, 145–151. doi: 10.1016/j.neucom.2018.10.080
Angrick, M., Herff, C., Mugler, E., Tate, M. C., Slutzky, M. W., Krusienski, D. J., et al. (2019b). Speech synthesis from ECoG using densely connected 3D convolutional neural networks. J. Neural Eng. 16, 036019. doi: 10.1101/478644
Antoniades, A., Spyrou, L., Martin-Lopez, D., Valentin, A., Alarcon, G., Sanei, S., et al. (2017). Detection of interictal discharges with convolutional neural networks using discrete ordered multichannel intracranial EEG. IEEE Trans. Neural Syst. Rehabil. Eng. 25, 2285–2294. doi: 10.1109/TNSRE.2017.2755770
Arora, A., Lin, J.-J., Gasperian, A., Maldjian, J., Stein, J., Kahana, M., et al. (2018). Comparison of logistic regression, support vector machines, and deep learning classifiers for predicting memory encoding success using human intracranial EEG recordings. J. Neural Eng. 15, 066028. doi: 10.1088/1741-2552/aae131
Assi, E. B., Rihana, S., Nguyen, D. K., and Sawan, M. (2019). Effective connectivity analysis of iEEG and accurate localization of the epileptogenic focus at the onset of operculo-insular seizures. Epilepsy Res. 152, 42–51. doi: 10.1016/j.eplepsyres.2019.02.006
Ayala, M., Cabrerizo, M., Jayakar, P., and Adjouadi, M. (2011). Subdural EEG classification into seizure and nonseizure files using neural networks in the gamma frequency band. J. Clin. Neurophysiol. 28, 20–29. doi: 10.1097/WNP.0b013e31820512ee
Baud, M. O., Kleen, J. K., Anumanchipalli, G. K., Hamilton, L. S., Tan, Y.-L., Knowlton, R., et al. (2018). Unsupervised learning of spatiotemporal interictal discharges in focal epilepsy. Neurosurgery. 83, 683–691. doi: 10.1093/neuros/nyx480
Benz, H. L., Zhang, H., Bezerianos, A., Acharya, S., Crone, N. E., Zheng, X., et al. (2012). Connectivity analysis as a novel approach to motor decoding for prosthesis control. IEEE Trans. Neural Syst. Rehabil. Eng. 20, 143–152. doi: 10.1109/TNSRE.2011.2175309
Bera, K., Schalper, K. A., Rimm, D. L., Velcheti, V., and Madabhushi, A. (2019). Artificial intelligence in digital pathology—new tools for diagnosis and precision oncology. Nat. Rev. Clin. Oncol. 16, 703–715. doi: 10.1038/s41571-019-0252-y
Blankertz, B., Muller, K.-R., Krusienski, D. J., Schalk, G., Wolpaw, J. R., Schlogl, A., et al. (2006). The BCI competition III: validating alternative approaches to actual BCI problems. IEEE Trans. Neural Syst. Rehabil. Eng. 14, 153–159. doi: 10.1109/TNSRE.2006.875642
Boussen, S., Velly, L., Benar, C., Metellus, P., Bruder, N., and Trebuchon, A. (2016). In vivo tumour mapping using electrocorticography alterations during awake brain surgery: a pilot study. Brain Topogr. 29, 766–782. doi: 10.1007/s10548-016-0502-6
Burrello, A., Schindler, K., Benini, L., and Rahimi, A. (2020). Hyperdimensional computing with local binary patterns: one-shot learning of seizure onset and identification of ictogenic brain regions using short-time iEEG recordings. IEEE Trans. Biomed. Eng. 67, 601–613. doi: 10.1109/TBME.2019.2919137
Chan, A. M., Sun, F. T., Boto, E. H., and Wingeier, B. M. (2008). Automated seizure onset detection for accurate onset time determination in intracranial EEG. Clin. Neurophysiol. 119, 2687–2696. doi: 10.1016/j.clinph.2008.08.025
Chen, D., Wan, S., and Bao, F. S. (2017). Epileptic focus localization using discrete wavelet transform based on interictal intracranial EEG. IEEE Trans. Neural Syst. Rehabil. Eng. 25, 413–425. doi: 10.1109/TNSRE.2016.2604393
Chua, E. C.-P., Patel, K., Fitzsimons, M., and Bleakley, C. J. (2011). Improved patient specific seizure detection during pre-surgical evaluation. Clin. Neurophysiol. 122, 672–679. doi: 10.1016/j.clinph.2010.10.002
Cimbalnik, J., Klimes, P., Sladky, V., Nejedly, P., Jurak, P., Pail, M., et al. (2019). Multi-feature localization of epileptic foci from interictal, intracranial EEG. Clin. Neurophysiol. 130, 1945–1953. doi: 10.1016/j.clinph.2019.07.024
Combrisson, E., and Jerbi, K. (2015). Exceeding chance level by chance: the caveat of theoretical chance levels in brain signal classification and statistical assessment of decoding accuracy. J. Neurosci. Methods 250, 126–136. doi: 10.1016/j.jneumeth.2015.01.010
Combrisson, E., Perrone-Bertolotti, M., Soto, J. L. P., Alamian, G., Kahane, P., Lachaux, J.-P., et al. (2017). From intentions to actions: neural oscillations encode motor processes through phase, amplitude and phase-amplitude coupling. Neuroimage. 147, 473–487. doi: 10.1016/j.neuroimage.2016.11.042
Cordella, F., Ciancio, A. L., Sacchetti, R., Davalli, A., Cutti, A. G., Guglielmelli, E., et al. (2016). Literature review on needs of upper limb prosthesis users. Front. Neurosci. 10, 209. doi: 10.3389/fnins.2016.00209
D'Alessandro, M., Vachtsevanos, G., Esteller, R., Echauz, J., Cranstoun, S., Worrell, G., et al. (2005). A multi-feature and multi-channel univariate selection process for seizure prediction. Clin. Neurophysiol. 116, 506–516. doi: 10.1016/j.clinph.2004.11.014
Daoud, H., and Bayoumi, M. (2020). Deep Learning Approach for Epileptic Focus Localization. IEEE Trans. Biomed. Circuits Syst. 14, 209–220. doi: 10.1109/TBCAS.2019.2957087
Demirer, R. M., Ozerdem, M. S., and Bayrak, C. (2009). Classification of imaginary movements in ECoG with a hybrid approach based on multi-dimensional Hilbert-SVM solution. J. Neurosci. Methods 178, 214–218. doi: 10.1016/j.jneumeth.2008.11.011
Derner, M., Jahanbekam, A., Bauckhage, C., Axmacher, N., and Fell, J. (2018). Prediction of memory formation based on absolute electroencephalographic phases in rhinal cortex and hippocampus outperforms prediction based on stimulus-related phase shifts. Eur. J. Neurosci. 47, 824–831. doi: 10.1111/ejn.13878
Elahian, B., Yeasin, M., Mudigoudar, B., Wheless, J. W., and Babajani-Feremi, A. (2017). Identifying seizure onset zone from electrocorticographic recordings: a machine learning approach based on phase locking value. Seizure. 51, 35–42. doi: 10.1016/j.seizure.2017.07.010
Englot, D. J., Han, S. J., Lawton, M. T., and Chang, E. F. (2011). Predictors of seizure freedom in the surgical treatment of supratentorial cavernous malformations. J. Neurosurg. 115, 1169–1174. doi: 10.3171/2011.7.JNS11536
Englot, D. J., Wang, D. D., Rolston, J. D., Shih, T. T., and Chang, E. F. (2012). Rates and predictors of long-term seizure freedom after frontal lobe epilepsy surgery: a systematic review and meta-analysis. J. Neurosurg. 116, 1042–1048. doi: 10.3171/2012.1.JNS111620
Ezzyat, Y., Wanda, P. A., Levy, D. F., Kadel, A., Aka, A., Pedisich, I., et al. (2018). Closed-loop stimulation of temporal cortex rescues functional networks and improves memory. Nat. Commun. 9, 1–8. doi: 10.1038/s41467-017-02753-0
Fattinger, S., Heinzle, B. B., Ramantani, G., Abela, L., Schmitt, B., and Huber, R. (2019). Closed-loop acoustic stimulation during sleep in children with epilepsy: a hypothesis-driven novel approach to interact with spike-wave activity and pilot data assessing feasibility. Front. Hum. Neurosci. 13, 166. doi: 10.3389/fnhum.2019.00166
Firpi, H., Goodman, E. D., and Echauz, J. (2007a). Epileptic Seizure Detection Using Genetically Programmed Artificial Features. IEEE Trans. Biomed. Eng. 54, 212–224. doi: 10.1109/TBME.2006.886936
Firpi, H., Smart, O., Worrell, G., Marsh, E., Dlugos, D., and Litt, B. (2007b). High-frequency oscillations detected in epileptic networks using swarmed neural-network features. Ann. Biomed. Eng. 35, 1573–1584. doi: 10.1007/s10439-007-9333-7
Frauscher, B., Bartolomei, F., Kobayashi, K., Cimbalnik, J., van ‘t Klooster, M. A., Rampp, S., et al. (2017). High-frequency oscillations: the state of clinical research. Epilepsia. 58, 1316–1329. doi: 10.1111/epi.13829
Geng, D., Zhou, W., Zhang, Y., and Geng, S. (2016). Epileptic seizure detection based on improved wavelet neural networks in long-term intracranial EEG. Biocybern. Biomed. Eng. 36, 375–384. doi: 10.1016/j.bbe.2016.03.001
Geng, M., Zhou, W., Liu, G., Li, C., and Zhang, Y. (2020). Epileptic seizure detection based on stockwell transform and bidirectional long short-term memory. IEEE Trans. NEURAL Syst. Rehabil. Eng. 28, 573–580. doi: 10.1109/TNSRE.2020.2966290
Ghoroghchian, N., Groppe, D. M., Genov, R., Valiante, T. A., and Draper, S. C. (2020). Node-centric graph learning from data for brain state identification. IEEE Trans. Signal Inf. Process. over Networks. 6, 120–132. doi: 10.1109/TSIPN.2020.2964230
Gong, C., Zhang, X., and Niu, Y. (2020). Identification of epilepsy from intracranial EEG signals by using different neural network models. Comput. Biol. Chem. 87, 107310. doi: 10.1016/j.compbiolchem.2020.107310
Grinenko, O., Li, J., Mosher, J. C., Wang, I. Z., Bulacio, J. C., Gonzalez-Martinez, J., et al. (2018). A fingerprint of the epileptogenic zone in human epilepsies. Brain. 141, 117–131. doi: 10.1093/brain/awx306
Hellmann, G. (1999). Multifold features determine linear equation for automatic spike detection applying neural nin interictal ECoG. Clin. Neurophysiol. 110, 887–894. doi: 10.1016/S1388-2457(99)00040-1
Hermiz, J., Rogers, N., Kaestner, E., Ganji, M., Cleary, D. R., Carter, B. S., et al. (2018). Sub-millimeter ECoG pitch in human enables higher fidelity cognitive neural state estimation. Neuroimage. 176, 454–464. doi: 10.1016/j.neuroimage.2018.04.027
Hill, N. J., Lal, T. N., Schroder, M., Hinterberger, T., Wilhelm, B., Nijboer, F., et al. (2006). Classifying EEG and ECoG signals without subject training for fast BCI implementation: comparison of nonparalyzed and completely paralyzed subjects. IEEE Trans. Neural Syst. Rehabil. Eng. 14, 183–186. doi: 10.1109/TNSRE.2006.875548
Hosseini, M., Pompili, D., Elisevich, K., and Soltanian-Zadeh, H. (2017). Optimized deep learning for EEG big data and seizure prediction BCI via Internet of Things. IEEE Trans. Big Data 3, 392–404. doi: 10.1109/TBDATA.2017.2769670
Hosseini, M.-P., Pompili, D., Elisevich, K., and Soltanian-Zadeh, H. (2018). Random ensemble learning for EEG classification. Artif. Intell. Med. 84, 146–158. doi: 10.1016/j.artmed.2017.12.004
Hosseini, M.-P., Tran, T. X., Pompili, D., Elisevich, K., and Soltanian-Zadeh, H. (2020). Multimodal data analysis of epileptic EEG and rs-fMRI via deep learning and edge computing. Artif. Intell. Med. 104, 101813. doi: 10.1016/j.artmed.2020.101813
Ikeda, S., Shibata, T., Nakano, N., Okada, R., Tsuyuguchi, N., Ikeda, K., et al. (2014). Neural decoding of single vowels during covert articulation using electrocorticography. Front. Hum. Neurosci. 8, 125. doi: 10.3389/fnhum.2014.00125
Jacobs, J., Zijlmans, M., Zelmann, R., Chatillon, C. É., Hall, J., Olivier, A., et al. (2010). High-frequency electroencephalographic oscillations correlate with outcome of epilepsy surgery. Ann. Neurol. 67, 209–220. doi: 10.1002/ana.21847
Jrad, N., Kachenoura, A., Merlet, I., Bartolomei, F., Nica, A., Biraben, A., et al. (2017). Automatic detection and classification of high-frequency oscillations in depth-EEG signals. IEEE Trans. Biomed. Eng. 64, 2230–2240. doi: 10.1109/TBME.2016.2633391
Karthick, P. A., Tanaka, H., Khoo, H. M., and Gotman, J. (2020). Could we have missed out the seizure onset: A study based on intracranial EEG. Clin. Neurophysiol. 131, 114–126. doi: 10.1016/j.clinph.2019.10.011
Khambhati, A. N., Bassett, D. S., Oommen, B. S., Chen, S. H., Lucas, T. H., Davis, K. A., et al. (2017). Recurring functional interactions predict network architecture of interictal and ictal states in neocortical epilepsy. eNeuro. 4, ENEURO.0091-16.2017. doi: 10.1523/ENEURO.0091-16.2017
Kharbouch, A., Shoeb, A., Guttag, J., and Cash, S. S. (2011). An algorithm for seizure onset detection using intracranial EEG. EPILEPSY Behav. 22, S29–S35. doi: 10.1016/j.yebeh.2011.08.031
Kiral-Kornek, I., Roy, S., Nurse, E., Mashford, B., Karoly, P., Carroll, T., et al. (2018). Epileptic seizure prediction using big data and deep learning: toward a mobile system. EBIOMEDICINE. 27, 103–111. doi: 10.1016/j.ebiom.2017.11.032
Klimes, P., Cimbalnik, J., Brazdil, M., Hall, J., Dubeau, F., Gotman, J., et al. (2019). NREM sleep is the state of vigilance that best identifies the epileptogenic zone in the interictal electroencephalogram. Epilepsia. 60, 2404–2415. doi: 10.1111/epi.16377
Kotsiantis, S. B., Zaharakis, I., and Pintelas, P. (2007). Supervised machine learning: a review of classification techniques. Emerg. Artif. Intell. Appl. Comput. Eng. 160, 3–24. doi: 10.1007/s10462-007-9052-3
Kragel, J. E., Ezzyat, Y., Sperling, M. R., Gorniak, R., Worrell, G. A., Berry, B. M., et al. (2017). Similar patterns of neural activity predict memory function during encoding and retrieval. Neuroimage. 155, 60–71. doi: 10.1016/j.neuroimage.2017.03.042
Kremen, V., Brinkmann, B. H., Van Gompel, J. J., Stead, M., St Louis, E. K., and Worrell, G. A. (2019). Automated unsupervised behavioral state classification using intracranial electrophysiology. J. Neural Eng. 16, 026004. doi: 10.1088/1741-2552/aae5ab
Kremen, V., Duque, J. J., Brinkmann, B. H., Berry, B. M., Kucewicz, M. T., Khadjevand, F., et al. (2017). Behavioral state classification in epileptic brain using intracranial electrophysiology. J. Neural Eng. 14, 26001. doi: 10.1088/1741-2552/aa5688
Lachaux, J. P., Rudrauf, D., and Kahane, P. (2003). Intracranial EEG and human brain mapping. J. Physiol. 97, 613–628. doi: 10.1016/j.jphysparis.2004.01.018
Lai, D., Zhang, X., Chen, W., Zhang, H., Kang, T., Yuan, H., et al. (2020). Channel-wise characterization of high frequency oscillations for automated identification of the seizure onset zone. IEEE Access. 8, 45531–45543. doi: 10.1109/ACCESS.2020.2978290
Lai, D., Zhang, X., Ma, K., Chen, Z., Chen, W., Zhang, H., et al. (2019). Automated detection of high frequency oscillations in intracranial EEG using the combination of short-time energy and convolutional neural networks. IEEE Access. 7, 82501–82511. doi: 10.1109/ACCESS.2019.2923281
LeCun, Y., Bengio, Y., and Hinton, G. (2015). Deep learning. Nature. 521, 436–444. doi: 10.1038/nature14539
Li, M., Cui, Y., Hao, D., and Yang, J. (2015). An adaptive feature extraction method in BCI-based rehabilitation. J. Intell. FUZZY Syst. 28, 525–535. doi: 10.3233/IFS-141329
Lian, Q., Qi, Y., Pan, G., and Wang, Y. (2020). Learning graph in graph convolutional neural networks for robust seizure prediction. J. Neural Eng. 17, 035004. doi: 10.1088/1741-2552/ab909d
Liu, D., Pang, Z., and Wang, Z. (2009). Epileptic seizure prediction by a system of particle filter associated with a neural network. EURASIP J. Adv. Signal Process. 2009, 638534. doi: 10.1155/2009/638534
Liu, Y., Zhou, W., Yuan, Q., and Chen, S. (2012). Automatic seizure detection using wavelet transform and SVM in long-term intracranial EEG. IEEE Trans. Neural Syst. Rehabil. Eng. 20, 749–755. doi: 10.1109/TNSRE.2012.2206054
Livezey, J. A., Bouchard, K. E., and Chang, E. F. (2019). Deep learning as a tool for neural data analysis: Speech classification and cross-frequency coupling in human sensorimotor cortex. PLoS Comput. Biol. 15, e1007091. doi: 10.1371/journal.pcbi.1007091
Mahmoodian, N., Boese, A., Friebe, M., and Haddadnia, J. (2019). Epileptic seizure detection using cross-bispectrum of electroencephalogram signal. Seizure. 66, 4–11. doi: 10.1016/j.seizure.2019.02.001
Makaram, N., von Ellenrieder, N., Tanaka, H., and Gotman, J. (2020). Automated classification of five seizure onset patterns from intracranial electroencephalogram signals. Clin. Neurophysiol. 131, 1210–1218. doi: 10.1016/j.clinph.2020.02.011
Makin, J. G., Moses, D. A., and Chang, E. F. (2020). Machine translation of cortical activity to text with an encoder-decoder framework. Nat. Neurosci. 23, 575–582. doi: 10.1038/s41593-020-0608-8
Manzouri, F., Heller, S., Duempelmann, M., Woias, P., and Schulze-Bonhage, A. (2018). A comparison of machine learning classifiers for energy-efficient implementation of seizure detection. Front. Syst. Neurosci. 12, 43. doi: 10.3389/fnsys.2018.00043
McMullen, D. P., Hotson, G., Katyal, K. D., Wester, B. A., Fifer, M. S., McGee, T. G., et al. (2014). Demonstration of a semi-autonomous hybrid brain–machine interface using human intracranial EEG, eye tracking, and computer vision to control a robotic upper limb prosthetic. IEEE Trans. Neural Syst. Rehabil. Eng. 22, 784–796. doi: 10.1109/TNSRE.2013.2294685
Medvedev, V. A., Agoureeva, I., and Murro, A. M. (2019). A Long Short-Term Memory neural network for the detection of epileptiform spikes and high frequency oscillations. Sci. Rep. 9, 19374. doi: 10.1038/s41598-019-55861-w
Meisel, C., and Bailey, K. A. (2019). Identifying signal-dependent information about the preictal state: A comparison across ECoG, EEG and EKG using deep learning. EBIOMEDICINE. 45, 422–431. doi: 10.1016/j.ebiom.2019.07.001
Memarian, N., Kim, S., Dewar, S., Engel, J. J., and Staba, R. J. (2015). Multimodal data and machine learning for surgery outcome prediction in complicated cases of mesial temporal lobe epilepsy. Comput. Biol. Med. 64, 67–78. doi: 10.1016/j.compbiomed.2015.06.008
Mirowski, P., Madhavan, D., LeCun, Y., and Kuzniecky, R. (2009). Classification of patterns of EEG synchronization for seizure prediction. Clin. Neurophysiol. 120, 1927–1940. doi: 10.1016/j.clinph.2009.09.002
Mo, J.-J., Zhang, J.-G., Li, W.-L., Chen, C., Zhou, N.-J., Hu, W.-H., et al. (2019). Clinical value of machine learning in the automated detection of focal cortical dysplasia using quantitative multimodal surface-based features. Front. Neurosci. 12, 1008. doi: 10.3389/fnins.2018.01008
Moher, D., Liberati, A., Tetzlaff, J., Altman, D. G., Altman, D., Antes, G., et al. (2009). Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. PLoS Med. 6, e1000097. doi: 10.1371/journal.pmed.1000097
Montavon, G., Samek, W., and Müller, K.-R. (2018). Methods for interpreting and understanding deep neural networks. Digit. Signal Process. 73, 1–15. doi: 10.1016/j.dsp.2017.10.011
Muller, M., Schindler, K., Goodfellow, M., Pollo, C., Rummel, C., and Steimer, A. (2018). Evaluating resective surgery targets in epilepsy patients: A comparison of quantitative EEG methods. J. Neurosci. Methods. 305, 54–66. doi: 10.1016/j.jneumeth.2018.04.021
Nahiduzzaman, M., Tasnim, M., Newaz, N. T., Kaiser, M. S., and Mahmud, M. (2020). “Machine learning based early fall detection for elderly people with neurological disorder using multimodal data fusion,” in International Conference on Brain Informatics (Padua, Italy: Springer), 204–214. doi: 10.1007/978-3-030-59277-6_19
Nejedly, P., Cimbalnik, J., Klimes, P., Plesinger, F., Halamek, J., Kremen, V., et al. (2019b). Intracerebral EEG Artifact Identification Using Convolutional Neural Networks. Neuroinformatics. 17, 225–234. doi: 10.1007/s12021-018-9397-6
Nejedly, P., Kremen, V., Sladky, V., Cimbalnik, J., Klimes, P., Plesinger, F., et al. (2019a). Exploiting graphoelements and convolutional neural networks with long short term memory for classification of the human electroencephalogram. Sci. Rep. 9, 11383. doi: 10.1038/s41598-019-47854-6
O'Leary, G., Groppe, D. M., Valiante, T. A., Verma, N., and Genov, R. (2018). NURIP: neural interface processor for brain-state classification and programmable-waveform neurostimulation. IEEE J. Solid-State Circuits 53, 3150–3162. doi: 10.1109/JSSC.2018.2869579
Pailla, T., Miller, K. J., and Gilja, V. (2019). Autoencoders for learning template spectrograms in electrocorticographic signals. J. Neural Eng. 16, 016025. doi: 10.1088/1741-2552/aaf13f
Pan, G., Li, J.-J., Qi, Y., Yu, H., Zhu, J.-M., Zheng, X.-X., et al. (2018). Rapid decoding of hand gestures in electrocorticography using recurrent neural networks. Front. Neurosci. 12, 555. doi: 10.3389/fnins.2018.00555
Parvez, M. Z., and Paul, M. (2017). Seizure prediction using undulated global and local features. IEEE Trans. Biomed. Eng. 64, 208–217. doi: 10.1109/TBME.2016.2553131
Petrosian, A., Prokhorov, D., Homan, R., Dasheiff, R., and Wunsch, D. (2000). Recurrent neural network based prediction of epileptic seizures in intra- and extracranial EEG. Neurocomputing. 30, 201–218. doi: 10.1016/S0925-2312(99)00126-5
Prashanth, R., Roy, S. D., Mandal, P. K., and Ghosh, S. (2016). High-accuracy detection of early Parkinson's disease through multimodal features and machine learning. Int. J. Med. Inform. 90, 13–21. doi: 10.1016/j.ijmedinf.2016.03.001
Principe, A., Ley, M., Conesa, G., and Rocamora, R. (2019). Prediction error connectivity: A new method for EEG state analysis. Neuroimage. 188, 261–273. doi: 10.1016/j.neuroimage.2018.11.052
Quigg, M., Sun, F., Fountain, N. B., Jobst, B. C., Wong, V. S. S., Mirro, E., et al. (2015). Interrater reliability in interpretation of electrocorticographic seizure detections of the responsive neurostimulator. Epilepsia. 56, 968–971. doi: 10.1111/epi.12998
Raghu, S., and Sriraam, N. (2017). Optimal configuration of multilayer perceptron neural network classifier for recognition of intracranial epileptic seizures. Expert Syst. Appl. 89, 205–221. doi: 10.1016/j.eswa.2017.07.029
Rajkomar, A., Dean, J., and Kohane, I. (2019). Machine learning in medicine. N. Engl. J. Med. 380, 1347–1358. doi: 10.1056/NEJMra1814259
Ramsey, N. F., Salari, E., Aarnoutse, E. J., Vansteensel, M. J., Bleichner, M. G., and Freudenburg, Z. V. (2018). Decoding spoken phonemes from sensorimotor cortex with high-density ECoG grids. Neuroimage. 180, 301–311. doi: 10.1016/j.neuroimage.2017.10.011
Rashid, M., Islam, M., Sulaiman, N., Bari, B. S., Saha, R. K., and Hasan, M. J. (2020). Electrocorticography based motor imagery movements classification using long short-term memory (LSTM) based on deep learning approach. SN Appl. Sci. 2, 211. doi: 10.1007/s42452-020-2023-x
RaviPrakash, H., Korostenskaja, M., Castillo, E. M., Lee, K. H., Salinas, C. M., Baumgartner, J., et al. (2020). Deep learning provides exceptional accuracy to ecog-based functional language mapping for epilepsy surgery. Front. Neurosci. 14, 409. doi: 10.3389/fnins.2020.00409
Rotenberg, B. W., George, C. F., Sullivan, K. M., and Wong, E. (2010). Wait times for sleep apnea care in Ontario: a multidisciplinary assessment. Can. Respir. J. 17, 170–4. doi: 10.1155/2010/420275
Rutigliano, T., Rivolta, M. W., Pizzi, R., and Sassi, R. (2018). Composition of feature extraction methods shows interesting performances in discriminating wakefulness and NREM sleep. IEEE Signal Process. Lett. 25, 204–208. doi: 10.1109/LSP.2017.2777919
Saboo, K. V., Varatharajah, Y., Berry, B. M., Kremen, V., Sperling, M. R., Davis, K. A., et al. (2019). Unsupervised machine-learning classification of electrophysiologically active electrodes during human cognitive task performance. Sci. Rep. 9, 17390. doi: 10.1038/s41598-019-53925-5
Sathish, E., Sivakumaran, N., Simon, S. P., and Raghavan, S. (2017). Genetic algorithm based feature selection for classification of focal and non-focal intracranial electroencephalographic signals. J. Sci. Ind. Res. 76, 614–619.
Scherer, R., Zanos, S. P., Miller, K. J., Rao, R. P. N., and Ojemann, J. G. (2009). Classification of contralateral and ipsilateral finger movements for electrocorticographic brain-computer interfaces. Neurosurg. Focus. 27, E12. doi: 10.3171/2009.4.FOCUS0981
Schrouff, J., Mourao-Miranda, J., Phillips, C., and Parvizi, J. (2016). Decoding intracranial EEG data with multiple kernel learning method. J. Neurosci. Methods. 261, 19–28. doi: 10.1016/j.jneumeth.2015.11.028
Sciaraffa, N., Klados, M. A., Borghini, G., Di Flumeri, G., Babiloni, F., and Arico, P. (2020). Double-step machine learning based procedure for HFOs detection and classification. BRAIN Sci. 10, 220. doi: 10.3390/brainsci10040220
Senders, J. T., Staples, P. C., Karhade, A. V., Zaki, M. M., Gormley, W. B., Broekman, M. L. D., et al. (2018). Machine learning and neurosurgical outcome prediction: a systematic review. World Neurosurg. 109, 476–486. doi: 10.1016/j.wneu.2017.09.149
Shenoy, P., Miller, K. J., Ojemann, J. G., and Rao, R. P. N. (2008). Generalized features for electrocorticographic BCIs. IEEE Trans. Biomed. Eng. 55, 273–280. doi: 10.1109/TBME.2007.903528
Shoaran, M., Haghi, B. A., Taghavi, M., Farivar, M., and Emami-Neyestanak, A. (2018). Energy-efficient classification for resource-constrained biomedical applications. IEEE J. Emerg. Sel. Top. Circuits Syst. 8, 693–707. doi: 10.1109/JETCAS.2018.2844733
Si, Y. (2020). Machine learning applications for electroencephalograph signals in epilepsy: a quick review. Acta Epileptol. 2, 5. doi: 10.1186/s42494-020-00014-0
Song, Y., and Zhang, J. (2016). Discriminating preictal and interictal brain states in intracranial EEG by sample entropy and extreme learning machine. J. Neurosci. Methods. 257, 45–54. doi: 10.1016/j.jneumeth.2015.08.026
Stead, M., Bower, M., Brinkmann, B. H., Lee, K., Marsh, W. R., Meyer, F. B., et al. (2010). Microseizures and the spatiotemporal scales of human partial epilepsy. Brain. 133, 2789–2797. doi: 10.1093/brain/awq190
Sumsky, S. L., and Santaniello, S. (2019). Decision support system for seizure onset zone localization based on channel ranking and high-frequency EEG activity. IEEE J. Biomed. Heal. Informatics. 23, 1535–1545. doi: 10.1109/JBHI.2018.2867875
Sundararajan, M., and Najmi, A. (2020). “The many Shapley values for model explanation.” In: International Conference on Machine Learning. PMLR, 9269–9278.
Syeda-Mahmood, T. (2018). Role of big data and machine learning in diagnostic decision support in radiology. J. Am. Coll. Radiol. 15, 569–576. doi: 10.1016/j.jacr.2018.01.028
Thomas, T. M., Candrea, D. N., Fifer, M. S., McMullen, D. P., Anderson, W. S., Thakor, N. V., et al. (2019). Decoding native cortical representations for flexion and extension at upper limb joints using electrocorticography. IEEE Trans. Neural Syst. Rehabil. Eng. 27, 293–303. doi: 10.1109/TNSRE.2019.2891362
Tomlinson, S. B., Porter, B. E., and Marsh, E. D. (2017). Interictal network synchrony and local heterogeneity predict epilepsy surgery outcome among pediatric patients. Epilepsia. 58, 402–411. doi: 10.1111/epi.13657
Truong, N. D., Kuhlmann, L., Bonyadi, M. R., Querlioz, D., Zhou, L., and Kavehei, O. (2019). Epileptic seizure forecasting with generative adversarial networks. IEEE Access. 7, 143999–144009. doi: 10.1109/ACCESS.2019.2944691
Truong, N. D., Nguyen, A. D., Kuhlmann, L., Bonyadi, M. R., Yang, J., Ippolito, S., et al. (2018). Integer convolutional neural network for seizure detection. IEEE J. Emerg. Sel. Top. CIRCUITS Syst. 8, 849–857. doi: 10.1109/JETCAS.2018.2842761
Tuyisenge, V., Trebaul, L., Bhattacharjee, M., Chanteloup-Foret, B., Saubat-Guigui, C., Mindruta, I., et al. (2018). Automatic bad channel detection in intracranial electroencephalographic recordings using ensemble machine learning. Clin. Neurophysiol. 129, 548–554. doi: 10.1016/j.clinph.2017.12.013
Ulate-Campos, A., Coughlin, F., Gaínza-Lein, M., Fernández, I. S., Pearl, P. L., and Loddenkemper, T. (2016). Automated seizure detection systems and their effectiveness for each type of seizure. Seizure. 40, 88–101. doi: 10.1016/j.seizure.2016.06.008
Varatharajah, Y., Berry, B., Cimbalnik, J., Kremen, V., Van Gompel, J., Stead, M., et al. (2018). Integrating artificial intelligence with real-time intracranial EEG monitoring to automate interictal identification of seizure onset zones in focal epilepsy. J. Neural Eng. 15, 046035. doi: 10.1088/1741-2552/aac960
Vidyaratne, L. S., and Iftekharuddin, K. M. (2017). Real-Time Epileptic Seizure Detection Using EEG. IEEE Trans. Neural Syst. Rehabil. Eng. 25, 2146–2156. doi: 10.1109/TNSRE.2017.2697920
Wang, N., and Lyu, M. R. (2015). Extracting and selecting distinctive EEG features for efficient epileptic seizure prediction. IEEE J. Biomed. Heal. Informatics. 19, 1648–1659. doi: 10.1109/JBHI.2014.2358640
Weidemann, C. T., Kragel, J. E., Lega, B. C., Worrell, G. A., Sperling, M. R., Sharan, A. D., et al. (2019). Neural activity reveals interactions between episodic and semantic memory systems during retrieval. J. Exp. Psychol. Gen., [Erratum in: J Exp Psychol Gen. (2019)0.148:782] 148, 1–12. doi: 10.1037/xge0000604
Weiss, S. A., Waldman, Z., Raimondo, F., Slezak, D., Donmez, M., Worrell, G., et al. (2019). Localizing epileptogenic regions using high-frequency oscillations and machine learning. Biomark. Med. 13, 409–418. doi: 10.2217/bmm-2018-0335
Wong, K. K., Rostomily, R., and Wong, S. T. C. (2019). Prognostic gene discovery in glioblastoma patients using deep learning. Cancers. 11, 53. doi: 10.3390/cancers11010053
Wu, M., Wan, T., Ding, M., Wan, X., Du, Y., and She, J. (2018). A new unsupervised detector of high-frequency oscillations in accurate localization of epileptic seizure onset zones. IEEE Trans. Neural Syst. Rehabil. Eng. 26, 2280–2289. doi: 10.1109/TNSRE.2018.2877820
Xie, Z., Schwartz, O., and Prasad, A. (2018). Decoding of finger trajectory from ECoG using deep learning. J. Neural Eng. 15, 036009. doi: 10.1088/1741-2552/aa9dbe
Xiong, W., Droppo, J., Huang, X., Seide, F., Seltzer, M. L., Stolcke, A., et al. (2017). Toward human parity in conversational speech recognition. IEEE/ACM Trans. Audio, Speech, Lang. Process. 25, 2410–2423. doi: 10.1109/TASLP.2017.2756440
Yanagisawa, T., Fukuma, R., Seymour, B., Tanaka, M., Hosomi, K., Yamashita, O., et al. (2020). BCI training to move a virtual hand reduces phantom limb pain: a randomized crossover trial. Neurology. 95, e417–e426. doi: 10.1212/WNL.0000000000009858
Yanagisawa, T., Hirata, M., Saitoh, Y., Kato, A., Shibuya, D., Kamitani, Y., et al. (2009). Neural decoding using gyral and intrasulcal electrocorticograms. Neuroimage. 45, 1099–1106. doi: 10.1016/j.neuroimage.2008.12.069
Yang, J., Singh, H., Hines, E. L., Schlaghecken, F., Iliescu, D. D., Leeson, M. S., et al. (2012). Channel selection and classification of electroencephalogram signals: an artificial neural network and genetic algorithm-based approach. Artif. Intell. Med. 55, 117–126. doi: 10.1016/j.artmed.2012.02.001
Yu, Z., Nie, W., Zhou, W., Xu, F., Yuan, S., Leng, Y., et al. (2020). Epileptic seizure prediction based on local mean decomposition and deep convolutional neural network. J. Supercomput. 76, 3462–3476. doi: 10.1007/s11227-018-2600-6
Yuan, S., Zhou, W., Yuan, Q., Li, X., Wu, Q., Zhao, X., et al. (2015). Kernel collaborative representation-based automatic seizure detection in intracranial EEG. Int. J. Neural Syst. 25, 1550003. doi: 10.1142/S0129065715500033
Zhang, Y., Zhou, W., Yuan, Q., and Wu, Q. (2014). A low computation cost method for seizure prediction. Epilepsy Res. 108, 1357–1366. doi: 10.1016/j.eplepsyres.2014.06.007
Zhang, Y., Zhou, W., and Yuan, S. (2015). Multifractal analysis and relevance vector machine-based automatic seizure detection in intracranial EEG. Int. J. Neural Syst. 25, 1550020. doi: 10.1142/S0129065715500203
Zhang, Z., and Parhi, K. K. (2016). Low-complexity seizure prediction from iEEG/sEEG using spectral power and ratios of spectral power. IEEE Trans. Biomed. Circuits Syst. 10, 693–706. doi: 10.1109/TBCAS.2015.2477264
Zhao, B., Hu, W., Zhang, C., Wang, X., Wang, Y., Liu, C., et al. (2020). Integrated automatic detection, classification and imaging of high frequency oscillations with stereoelectroencephalography. Front. Neurosci. 14, 546. doi: 10.3389/fnins.2020.00546
Zheng, Y., Zhu, J., Qi, Y., Zheng, X., and Zhang, J. (2015). An automatic patient-specific seizure onset detection method using intracranial electroencephalography. Neuromodulation. 18, 79–84. doi: 10.1111/ner.12214
Keywords: intracranial EEG (iEEG), seizure, epilepsy, neurorecording, machine learning, artificial intelligence, deep learning
Citation: Mirchi N, Warsi NM, Zhang F, Wong SM, Suresh H, Mithani K, Erdman L and Ibrahim GM (2022) Decoding Intracranial EEG With Machine Learning: A Systematic Review. Front. Hum. Neurosci. 16:913777. doi: 10.3389/fnhum.2022.913777
Received: 06 April 2022; Accepted: 31 May 2022;
Published: 27 June 2022.
Edited by:
Mark Stecker, Self-Employed, Fresno, United StatesReviewed by:
Waldemar Karwowski, University of Central Florida, United StatesElliot H. Smith, The University of Utah, United States
Copyright © 2022 Mirchi, Warsi, Zhang, Wong, Suresh, Mithani, Erdman and Ibrahim. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Nykan Mirchi, bnlrYW4ubWlyY2hpQG1haWwudXRvcm9udG8uY2E=
 Nykan Mirchi
Nykan Mirchi Nebras M. Warsi2,3
Nebras M. Warsi2,3 Simeon M. Wong
Simeon M. Wong Lauren Erdman
Lauren Erdman George M. Ibrahim
George M. Ibrahim