- 1Department of Gynecology and Obstetrics, West China Second University Hospital, Sichuan University, Chengdu, China
- 2Key Laboratory of Birth Defects and Related Diseases of Women and Children, Sichuan University, Ministry of Education, Chengdu, China
- 3Qingbaijiang Maternal and Child Health Hospital, Chengdu, China
The maternal-fetal interface is an essential environment for embryonic growth and development, and a successful pregnancy depends on the dynamic balance of the microenvironment at the maternal-fetal interface. Single-cell sequencing, which unlike bulk sequencing that provides averaged data, is a robust method for interpreting the cellular and molecular landscape at single-cell resolution. With the support of single-cell sequencing, the issue of maternal-fetal interface heterogeneity during pregnancy has been more deeply elaborated and understood, which is important for a deeper understanding of physiological and pathological pregnancy. In this paper, we analyze the recent studies of single-cell transcriptomics in the maternal-fetal interface, and provide new directions for understanding and treating various pathological pregnancies.
Introduction
The maternal-fetal interface consists of maternal-derived decidua and fetal-derived placenta (Ander et al., 2019), and includes embryonic-derived trophoblast cells, maternal-derived decidual stromal cells, and decidual immune cells (Yang et al., 2019). Trophoblast cells carry human lymphocyte antigen (HLA) inherited from the paternal lineage, which invade the maternal decidua to anchor the blastocyst to the meconium and participate in the formation of the placenta (Ander et al., 2019). Decidual stromal cells synthesize components of extracellular matrix components, hormones, peptides, cytokines, growth factors, etc., and accumulate glycogen, lipids and proteins to support embryonic growth in early pregnancy (Dey et al., 2004). The decidual immune cells include natural killer (NK) cells, macrophages, dendritic cells, T cells, innate lymphocytes and B cells (Faas and de Vos, 2017; Liu et al., 2017; Vazquez et al., 2019), and play a key role in the immune microenvironment at the maternal-fetal interface and in the establishment of maternal-fetal immune tolerance (PrabhuDas et al., 2015; Liu et al., 2017; Ander et al., 2019). The fact that the fetus acts as a homozygous semi-allograft yet is not rejected by the maternal immune system suggests that complex immune regulatory mechanisms exist between mother and fetus during gestation to maintain homeostasis among the various cell populations as well as within the cells. The imbalance between these cells may lead to pathological pregnancies such as RSA, preeclampsia, preterm delivery and congenital infection (Hoo et al., 2020; Yuan et al., 2020; van 't Hof et al., 2021; Wang et al., 2021b). However, the interactions between cells at the maternal-fetal interface and the dynamics of immune cells are not yet fully described.
Single-cell sequencing is an emerging technology based on the level of individual cell isolation to explore the dynamic differentiation among cell populations and within cells, mainly including Single-cell genomics, transcriptomics, proteomics, epigenomics and interactomics sequencing (Lei et al., 2021). Unlike previous high-throughput sequencing technologies, single-cell genomics not only provides in-depth analysis of individual cell subpopulations measured and the variability of gene expression levels, but also helps to further explore the relationship between cell lineages and cell differentiation trajectories, and to construct the interplay network among cell subpopulations. Currently, research related to single-cell sequencing has gained breakthroughs in several disciplines, including tumor cell heterogeneity, embryonic development, neural activity, immune cell population detection, and plant growth and development (Baslan and Hicks, 2017; Zhao et al., 2018a; Potter, 2018; Shulse et al., 2019; Armand et al., 2021). With the growing development of single-cell sequencing technology, single-cell RNA sequencing (scRNA-seq) has been increasingly applied in the study of pregnancy, and the analysis of various cell types, cell subpopulations and factors such as the variability of gene expression in maternal-fetal interface tissues at the individual cell level has helped to reveal the mechanisms of inter- and intracellular interactive dialogues and thus to better understand cell heterogeneity. This not only promotes the study of cell differentiation and development and cell fate at the maternal-fetal interface, but also advances the research of maternal-fetal medicine. By analyzing the recent studies related to scRNA-seq in maternal-fetal interface heterogeneity in pregnancy, this review aims to deeply explore the heterogeneity of maternal-fetal interface tissues and further understand the potential mechanisms of maternal-fetal immune tolerance to provide guidance direction for the treatment and research of clinical diseases.
Single-cell RNA sequencing
scRNA-seq, based on the principle that trace amounts of mRNA in isolated individual cells are efficiently amplified and then sequenced at high throughput, is a new technique for high-throughput sequencing of the cellular transcriptome at the level of individual cells. The general procedures of scRNA-seq include the isolation of a single cell, RNA extraction, reverse transcription, preamplification and detection (Kolodziejczyk et al., 2015). In contrast to conventional sequencing, scRNA-seq requires that cell populations in tissues or body fluids are first separated into individual cells, which is precisely where the difficulty lies. The isolation of viable, individual cells from fresh tissues is clearly the most crucial, which determine the accuracy and amount of the amplified dissociation (Lei et al., 2021). The main techniques include micromanipulation, limiting dilution, laser capture microdissection (LCM), fluorescence-activated cell sorting (FACS), magnetic associated cell separation (MACS), immunomagnetic separation and microfluidics (Li et al., 2020; Yasen et al., 2020).
The core strategies include two types: one is to isolate individual cells and construct sequencing libraries independently; the other is labeling (like Barcode) based single cell identification. With strategy one, single cells are isolated one by one and then sequenced separately, which is not only limited by cost but also very low throughput in the face of tens of thousands of cells in one tissue sample. To overcome this limitation, the clever application of droplet-based microfluidics, such as Drop-sep and 10x Genomics has improved the efficiency of single cell identification (Macosko et al., 2015; Zheng et al., 2017): instead of isolating and extracting individual cells one by one, each cell can be tagged with a unique nucleic acid sequence and then sequenced and analyzed. This principle is applied in microfluidics to improve cell throughput with fast cycle times, low cost, and much higher cell capture efficiency.
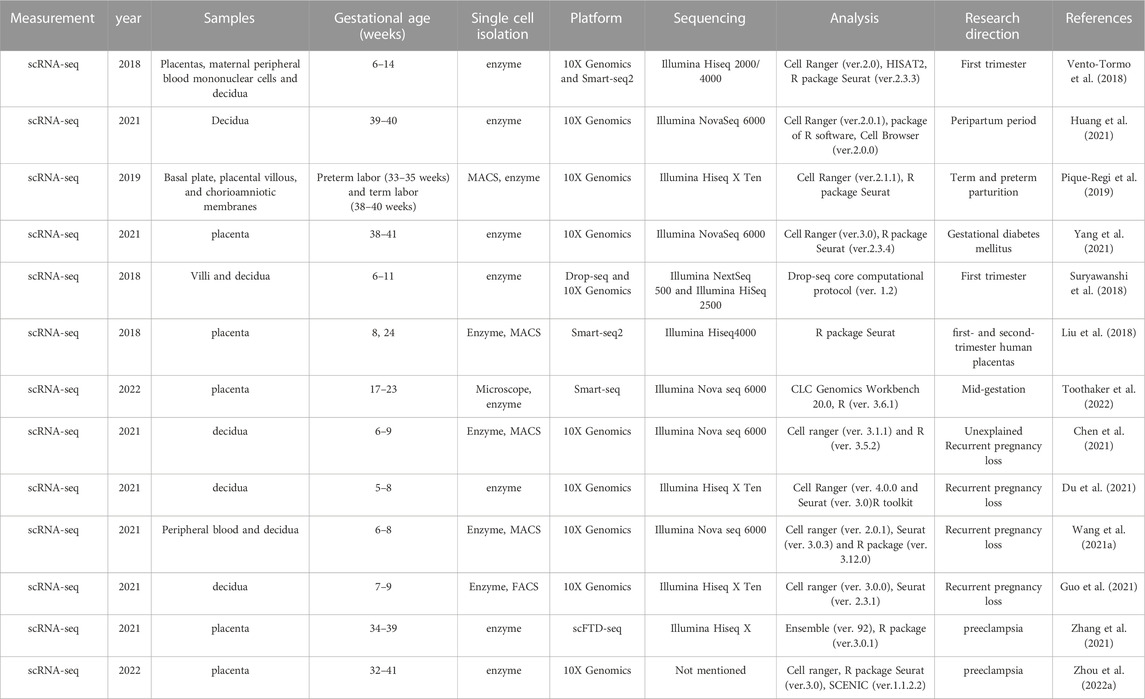
Methods for scRNA-seq differ in how they tag transcripts for their cell-of-origin and generate libraries for sequencing (Ding et al., 2020), which include Cell expression by linear amplification and sequencing (CEL-seq) (Hashimshony et al., 2012), Smart-seq (Ramskold et al., 2012), Smart-seq2 (Picelli et al., 2014), Drop-seq (Macosko et al., 2015), inDrop-sep (Klein et al., 2015), massively parallel RNA single-cell sequencing (MARS-seq) (Jaitin et al., 2014), 10x Chromium (Zheng et al., 2017), Seq-Well (Gierahn et al., 2017), single-cell combinatorial indexiong RNA sequencing (sci-RNA-seq) (Cao et al., 2017) and so on. Ding (Ding et al., 2020) compared seven methods for single-cell sequencing, including two low-throughput plate-based methods (Smart-seq and CEL-Seq2) and five high-throughput methods (10x Chromium, Drop-seq, Seq-Well, inDrops, and sci-RNA-seq), and found that for the low-throughput methods, Smart-seq2 and CEL-Seq2 performed similarly. Among the high-throughput methods, 10x Chromium was the best-performing method (Ding et al., 2020). Currently, scRNA-seq is widely used to analyze the transcriptome of individual cells at the maternal-fetal interface, while the droplet-based 10X Genomics Chromium is the commonly used platform. As presented in Table 1.
The 10X genomics Chromium platform builds on the Gem Code technology, which has been frequently used for Single cell transcriptome research in human placenta and decidua (Vento-Tormo et al., 2018; Wang et al., 2021a; Du et al., 2021; Huang et al., 2021; Yang et al., 2021; Zhou et al., 2022a). The core of the technology is a Gel bead in Emulsion (GEM) (Zheng et al., 2017). Each gel bead if functionalized with barcoded oligonucleotides, which are composed of the following parts: (i) sequencing adapters and primers, (ii) a 14 bp barcode drawn from B750,000 designed sequences to index GEMs, (iii) a 10 bp randomer to index molecules (unique molecular identififier, UMI) and (iv) an anchored 30 bp oligo-dT to prime polyadenylated RNA transcripts (Zheng et al., 2017). In the 8-channel microfluidic “double-cross” crossover system of the 10X Genomics Chromium, Gel Beads bind to the cell/enzyme mixture at the first crossover, and then at the second crossover, oil coats the Gel Beads with the cell/enzyme mixture, forming an oil-in-water structure, or GEM (Gel Beads-in-emulsion). After the formation of GEMs, the cells are lysed and the Gel Beads automatically lyse to release a large number of barcode sequences, which are combined with mRNA and subsequently reverse transcribed by mRNA to produce cDNA; the obtained cDNA is purified and enriched for subsequent construction of a standard sequencing library. The cDNA sequences from the same cell carry the same barcode tag specific to the gel microbeads, and each cDNA molecule also carries a specific UMI tag (Kivioja et al., 2011; Zheng et al., 2017; Zheng et al., 2020). In summary, the Gem Code technology enables the separation of individual cells, Barcoding technology distinguishes between different cells and different transcripts of the same gene in the same cell. After the oil droplets are broken, PCR amplification is performed using cDNA with 10× barcode and UMI as a template. Ultimately, mRNA reverse transcription produces cDNA with 10× barcode and UMI information to construct a standard sequencing library (Kivioja et al., 2011; Zheng et al., 2020).
These sequencing libraries are then sequenced on lllumina sequencer and the data is analyzed after sequencing is completed.
Single-cell RNA sequencing in trophoblast cells
The blastocyst consists of the trophoblast ectoderm, the outer layer of the blastocyst and the inner cell mass (Nakashima et al., 2021), in which the trophoblast proliferates and differentiates into proliferating cytotrophoblast cells (CTBs), which form a cell mass at the tip of the chorion that can differentiate in two directions: the syncytial trophoblasts (STBs) and the extravillous trophoblasts (ETVs) on the fetal side (Saito and Nakashima, 2013). the STB, formed by the fusion of the CTB, is the outer layer of the placental villi that It is in direct contact with maternal glandular secretions and participates in the exchange of nutrients, gases and waste products between maternal and fetal blood, preventing direct contact between maternal and fetal blood (Turco and Moffett, 2019; Nakashima et al., 2021). The STB is the main site of steroid and peptide hormone synthesis and secretion (Burton and Fowden, 2015), and because it does not express MHC class I and II molecules and is not recognized by CD8+ T cells, it also acts as a protective immune barrier (Turco and Moffett, 2019).
The invasive effect of EVTs on the metaphase is the initiating and key factor in the formation of the maternal-fetal interface (Pollheimer et al., 2018). Vento-Tormo et al. (2018) use single-cell transcriptomics from first-trimester placentas with matched maternal blood and decidual cells to reveal the upregulation of receptors involved in immunomodulation, cellular adhesion and invasion of EVT. Suryawanshi et al. (2018) performed a scRNA-seq study of early pregnancy placenta and metaphase tissue and found that EVT specifically expresses the extracellular matrix (ECM) glycoprotein MFAP5, a component of microfibrils that induces cell motility and cancer cell invasion, demonstrating the invasive and mobile role of EVT (Leung et al., 2014; Suryawanshi et al., 2018). EVT interacts with maternal immune cells and plays an important role in the maternal-fetal tolerance process, such as expressing HLA-C, HLA-E and HLA-G (but not HLA-A and HLA-B) to avoid maternal immune rejection (Moffett and Loke, 2006). Interaction with CD4+ T cells leads to an increase in Treg cells and increases the expression of the Treg-specific transcription factor FOXP3 to increase immune tolerance (Tilburgs et al., 2015). Meanwhile, HLA-G expressed by EVT promotes the secretion of large amounts of growth-promoting factors by decidual natural killer cells through the ILT2-KIR2DL4 axis, which promotes fetal growth and development (Fu et al., 2017).
Liu et al. (2018) revealed 14 subtypes of placental cells from first- (8 weeks of gestation) and second-trimester (24 weeks of gestation) human placentas by scRNA-seq. They identified three CTB subtybes (CTB-8W-1, CTB-8W-2 and CTB-8W-3) from the first-trimester placenta. CTB-8W-3 cells highly express cell cycle-related genes, including RRM2, CCNB1 and CDK1, exhibit the highest cell proliferative activity and may act as trophoblast stem cells. CTB-8W-1 has the lowest proliferative capability, but high expression of Syncytin-2 gene, promotes cell fusion and is the progenitor cell of STB. CTB-8W-2 cell function is not known. EVT in early pregnancy is divided into three subtypes (Liu et al., 2018), which are classified as EVT-8W-1, 2 and 3. EVT-8W-1 highly expresses RRM2, has cell proliferative potential and is located in the proximal segment of the cell column, EVT-8W-3 cells are associated with receptor activity regulation and immune response and highly express Tachykinin-3 (TAC3), fibrinogen activator inhibitor-1 (SERPINE1), PRG2 and JAM2, located distal to the cell column. EVT aggregates in mid-gestation to form 2 subtypes, EVT-24W-1 and EVT-24W-2. type 1 is associated with negative regulatory responses to trauma, digestion and the immune system, whereas type 2 is associated with growth regulation, gonadotropin secretion and pregnancy. An ordered pattern of trophoblast differentiation from CTB-8W-2 to CTB-8W-3 and then to EVT-8W and EVT-24W cells was predicted by pseudo-temporal analysis (Liu et al., 2018). The results of Vento-Tormo et al. (2018) et al. also showed that EVT is at the end of the CTB differentiation trajectory. Huang et al. (2021) found that EVT-specific expression of KRT7, PERP and HLA-G in late pregnancy was divided into five clusters by single-cell transcriptome analysis of metaphase before and after delivery, while there was high heterogeneity in gene expression between each cluster, driving different functions.
Preeclampsia is one of the most serious obstetrical complications, affecting 5%–8% of pregnant women (Hutcheon et al., 2011; Than et al., 2018), and is a principal cause of maternal and perinatal morbidity and mortality (Sarno et al., 2015; Ozimek et al., 2016). Despite the severity of the problem, there is a lack of insight into the molecular pathways that were disturbed earlier (Than et al., 2018). PE is a syndrome caused by multiple factors, mechanisms and pathways, and trophoblast dysfunction in the placenta is a central aspect of PE pathogenesis. A progressive body of evidence supports the conclusion that dysplasia and/or dysfunction of different trophoblast lineages of the placenta play a central role in the pathogenesis of preeclampsia. In the preclinical phase, EVT development may be impaired, resulting in EVT dysfunction, shallow trophoblast invasion, failure of physiological transformation of maternal spiral arteries, abnormal placental blood flow, and histological changes consistent with poor maternal vascular perfusion (Redman and Sargent, 2010; Chaiworapongsa et al., 2014). Zhang et al. (2021) revealed the difference between the preeclampsia and healthy pregnancy groups by analyzing the transcripts of 11,518 cells at the single cell level and found that GO terms enriched in EVT from patients with preeclampsia were mainly associated with immune responses, suggesting altered placental immune function in preeclampsia. Zhou et al. (2022b) classified EVT into four subtypes and found that pro-inflammatory, immune, and oxidative stress-related pathways in preeclampsia EVT subtypes were activated. Further analysis revealed that the transcription factors CEBPB, ATF3, and GTF2B were significantly reduced in expression in preeclamptic EVT, and CEBPB and GTF2B were selected for the next functional study, which revealed that knockdown of these two molecules significantly reduced cell viability and invasiveness, and were hypothesized to be involved in the pathogenesis of Pre-eclampsia (Zhou et al., 2022b). Overall, scRNA-seq provides a fresh molecular theoretical basis for preeclampsia trophoblast dysfunction, which may help in the diagnosis and treatment of this disease.
Single-cell RNA sequencing in decidual stromal cells
Decidual stromal cells (DSC) are a major component of the decidua and secrete a variety of factors that regulate the microenvironment at the maternal-fetal interface in early pregnancy, including IGFBP1, PRL, growth factors, cytokines, and interleukins (Gellersen and Brosens, 2014; James-Allan et al., 2018). Suryawanshi et al. (2018) analyzed the gene expression in more than 20,000 cells from villi and decidua samples by scRNA-seq analysis of human first-trimester (6–11 weeks of gestation) placental and decidual cells using 10x Genomics and Drop-seq platforms. DSC expressed genes involved in PRL, IGFBP1, APOA1, CHI3L2, SEROINA3, IL1B, and PROk1. To address stromal cell differentiation, Suryawanshi et al. (2018) placed 1524 stromal cells of decidua in pseudotemporal order and observed the trajectories originating from the fibroblast population toward DSCs. Vento-Tormo et al. (2018) analyzed early gestational (6–14 weeks of gestation) DSC by scRNA-seq and classified them into three cell subpopulations (labeled DS1, DS2, and DS3). The DS1 cell subpopulation expressed ACTA2 and TAGLN, and lacked expression of the classical molecular markers of metaphase, IGFBP1 and PRL. the DS2 and DS3 cell subpopulations expressed IGFBP1, IGFBP2, and IGFBP6, and the DS3 cell subpopulation expressed PRL and genes involved in steroid biosynthesis at different locations. They used immunohistochemistry and multiplexed single molecule fluorescent in situ hybridization for selected markers on serial section of decidua parietails to demonstrate that DS1 cells are present between glands in the decidua spongiosa, while DS2 and DS3 cells are located in decidua compacta. In addition, they developed a repository of ligand-receptor interacting pairs called CellPhone to identify interactions between decidual NK cells and invading fetal EVTs, maternal immunity, and stromal cells. Using this tool, they predicted that DS2 and DS3 express high levels of LGALS9 and CLEC2D, which could interact with their receptors TIM3 and KLRB1-both expressed by subsets of dNKs-enabling the stroma to suppress the inflammatory response in the decidua (Vento-Tormo et al., 2018). Huang et al. (2021) investigated the transcriptome of 29231 decidual cells before and after delivery (39–40 weeks of gestation) by scRNA-seq and found that the main function of DSC at peripartum period included extracellular matrix organization, protein processing and cell-substrate adhesion.
Recurrent spontaneous abortion (RSA), which is to be defined as the loss of two or more consecutive pregnancies, affects up to 5% of women who are attempting to conceive (Practice Committee of the American Society for Reproductive Medicine. Electronic address, 2020). The known etiologies of RPL include genetic abnormalities, endocrine disorders, uterine malformations, Vitamin D deficiency and other influencing factors such as thrombosis and maternal infections (Garrido-Gimenez and Alijotas-Reig, 2015; Tamblyn et al., 2022). Up to 50% of cases the etiology is not known (Pillarisetty and Mahdy, 2022). Du et al. (2021) analysed a total of 66,078 single cells from decidua samples isolated from patients with RSA (5–8 weeks of gestation) and healthy controls (5–8 weeks of gestation) by scRNA-seq and revealed that RSA samples were accompanied by abnormal decidualization and marked impairment of communication between stromal cells and other cell types, such as abnormal activation of macrophages and NK cells. They identified five clusters of DSCs (DS1-DS5) according to their gene expression profile and found a significant reduction in the number of DSC in RSA decidua and that the genes upregulated in RSA DSC were mainly associated with cellular senescence, wire mesh, apoptosis, endocytosis and autophagy. Meanwhile, a new cell population DS5 appeared in RSA decidua, and DS5 highly expressed genes related to apoptosis and senescence pathways, indicating that DSC underwent abnormal development in RSA decidua. This study confirmed new molecular and cellular mechanisms associated with RSA development.
Single-cell RNA sequencing in NK cells
Most studies show that the proportion of NK cells in the endometrium of non-pregnant women is low during the follicular and early secretory phases and gradually increases after ovulation (Lee et al., 2010; Drury et al., 2018). NK cells reach their highest proportion in early gestation and are the most abundant leukocytes in maternal-fetal immunity, accounting for about 70% of metaphase leukocytes (Bulmer et al., 1991), and then their number gradually decreases (Williams et al., 2009; Bulmer et al., 2010). The typical phenotype of NK cells is CD3−CD56+, which can differentiate into different phenotypes in peripheral blood, where about 90% of peripheral blood NK cells (pNK) have a phenotype of CD56dimCD16+ and can mediate natural and antibody-dependent killing and exhibit high cytotoxicity, while the other 10% of NK cells have a CD56bright CD16-phenotype, produce a variety of cytokines and are less cytotoxic (Koopman et al., 2003; De Maria et al., 2011; Co et al., 2013; Faas and de Vos, 2017; Le Bouteiller and Bensussan, 2017). Decidual NK cells (dNK) have a similar phenotype to adult peripheral blood CD56brightCD16−CD3−. On the other hand, like the CD56dimCD16+ pNK, dNK cells are rich in granules, including granzyme/granulysin and perforin, but their cytotoxicity is rare and they have a weak ability to kill target cells (Koopman et al., 2003; Le Bouteiller and Bensussan, 2017). dNK cells play an important role in trophoblast invasion, uterine spiral artery remodeling and placenta formation, and promote embryonic development, and maintain maternal tolerance to the embryo (Yang et al., 2019). However, dNK cell imbalance can lead to a variety of adverse pregnancy outcomes, such as recurrent miscarriage, preeclampsia, intrauterine growth restriction, preterm delivery, and intrauterine infection (Zhao et al., 2018b; Faas and De Vos, 2018; Jabrane-Ferrat, 2019).
Vento-Tormo et al. (2018) identified three major dNK populations, namely dNK1, dNK2, and dNK3, which have distinct molecular characteristics and functions. dNK1 cells expressed CD39, CYP26A1, and B4GALNT1. dNK2 cells expressed ANXA1 and ITGB2, and dNK3 markers expressed ITGB2, CD160, KLRB1 and CD103. Further analysis revealed that dNK1 cells KIR genes (KIR2DS1, KIR2DS4, KIR2DL1, KIR2DL2 and KIR2DL3), and cytoplasmic granule proteins (PREF1, GNLY, GZMA and GZMB) were highly expressed and glycolytic metabolism was active. When KIR2DL on dNK cells interacted with HLA-G on EVT, dNK cell activity was inhibited (Tilburgs et al., 2015). It is hypothesized that dNK1 cells interact specifically with EVT to regulate maternal uterine spiral artery remodeling to ensure adequate blood supply for fetal development (Hiby et al., 2004; Li et al., 2009; Hiby et al., 2010; Hiby et al., 2014). dNK3 is highly expressed in CCL5, an inflammatory mediator produced by immune cells that has a role in regulating migration and invasion (Yu-Ju Wu et al., 2020), and EVT expresses CCR1, a receptor for CCL5, suggesting a role for dNK3 in regulating EVT invasion (Vento-Tormo et al., 2018).
Studies suggest that abnormalities in the metaplastic immune microenvironment may be associated with the development of RSA (Triggianese et al., 2016). However, the underlying mechanisms by which dysregulation of the meconium immune microenvironment leads to RSA are not known. Guo et al. (2021) analyzed a total of 18,646 CD45+ single transcriptomes from 24 human first-trimester (7–9 weeks of gestation) decidua samples by scRNA-seq. They identified three known subsets of dNK cells (dNK1-3) (Vento-Tormo et al., 2018) and a group of proliferating natural killer cells (dNKp) and found that dNK1 cells express LILRB1, which has pro-embryonic growth activity, as well as high expression of angiopoietin receptor Tie-2-mediated signaling pathway genes. dNK2 and dNK3 cells readily secrete cytokines, with dNK3 highly expressing immunomodulatory IFNG. Further analysis revealed a significant decrease in dNK1 cells and a significant increase in dNK3 cells in RSA patients. It is hypothesized that in RSA decidua, the presence of dNK has a diminished angiogenic capacity, accompanied by an enhanced pro-inflammatory capacity. Wang et al. (2021a) analyzed a total of 56,758 CD45+ single transcriptomes from six human first-trimester (6–8 weeks of gestation) peripheral blood and decidual samples by scRNA-seq and identified five major dNK clusters, named as dNKp, dNK1, dNK2, dNK3, and dNK4. dNKp cells were highly expressed in MCM5, STMN1, and PCNA and had a significant proliferative capacity. dNK4 cells highly expressed LILRB1, a marker gene for memory NK cells, indicating that dNK4 cells are involved in recurrent adverse pregnancy outcomes in recurrent miscarriage. Further comparison of differential dNK cell gene expression between recurrent miscarriage and normal pregnancy revealed that inflammation-related genes were increased in recurrent miscarriage (Wang et al., 2021a). Guo et al. (2021) practical high-resolution pseudo-temporal prediction algorithm Palantir (Setty et al., 2019) postulated that dNKp differentiates into dNK1, dNK2 and dNK3 through three developmental branches, with dNKp differentiation into dNK3 being unique to RSA. It was also determined that dNK2 has a clear tendency to convert to dNK1. Chen et al. (2021) analyzed the transcriptomes of 13,953 CD45+cells from six human first-trimester (6–9 weeks of gestation) decidual samples by scRNA-seq and identified a group of CSF+CD59+KIRs dNK cells predominantly present in the normal decidua, a subpopulation of cells that performs anti-inflammatory and regulates the function, infiltration and phenotype of multiple immune cells by expressing CD39 and CD59 (Blom, 2017; Zeng et al., 2020). While the proportion of CSF+CD59+KIRs dNK cells was decreased in RSA (Chen et al., 2021). In summary, the above studies promote our understanding of maternal-fetal interface heterogeneity in disease states from a single-cell perspective and provide new perspectives for clinical immunological studies of RSA.
Single-cell RNA sequencing in macrophages
Decidual macrophages (dMΦ), phenotypically CD14+CD206+, account for 20%–30% of maternal immune cells at the site of placental implantation (Heikkinen et al., 2003), constitute the second largest population of metaphase leukocytes in early pregnancy and remain relatively constant in number throughout pregnancy (Vince et al., 1990). dMΦ plays an important role in the establishment and maintenance of normal pregnancy, promoting spiral artery remodeling and trophoblast invasion and maintaining immune tolerance at the maternal-fetal interface by downregulating the inflammatory response and inducing tolerance to embryonic antigens (Petroff et al., 2002; Abrahams et al., 2004; Smith et al., 2009; Faas et al., 2014). Macrophages in tissues are usually divided into two subpopulations: pro-inflammatory M1 and anti-inflammatory M2 cell types (Zhang et al., 2017). dMΦ presents an M1 phenotype before blastocyst implantation, and as pregnancy progresses, after placenta formation and helical artery remodeling, the M2 subpopulation becomes the major macrophage population in the meconium, maintaining maternal-fetal tolerance during pregnancy (Sun et al., 2021).
Yang et al. (2021) analyzed the transcriptomes of 27,220 cells from four freshly placental tissues (two GDM and two normal controls) by scRNA-seq. As presented in Table 1. In their research, 4,306 MΦ were detected, and most of them were M2-polarized. Roger (Pique-Regi et al., 2019) analyzed the transcriptomes of 79,906 cells from three placental compartments: basal plate, placental villous, and chorioamniotic membranes collected from nine women in three study group: term no labor, term in labor, and preterm labor. They found higher NFKB1 expression in maternal macrophages in women who delivered at term compared to the non-delivery group, and this change was more pronounced in preterm delivery. Differential gene expression in macrophages associated with delivery was mainly involved in the activation of immune responses and regulation of pro-inflammatory cytokine production, suggesting that decidua macrophages undergo M1-type macrophage polarization during full-term and non-full-term deliveries. Wang et al. (2021a) found increased expression of CXCL8, TNF and IFIT2 in RSA, while angiogenic factor VEGFA and immunosuppressive molecule-encoding gene LAGLAS1 expression was decreased, presenting a pro-inflammatory state and possibly failing to regulate uterine spiral artery remodeling. Guo et al. (2021) classified dMΦ cells into two cell subtypes, mac1 and mac2, at the single cell level, which were enriched for M1 and M2 genes, respectively, and found that mac1 cells were elevated and mac2 cells were significantly reduced in the RSA, while genes with T Cell tropism were found to be elevated in the expression of dM cellsΦ in the RSA. This once again demonstrated decidual immune microenvironment disorder of patients with RSA.
Single-cell RNA sequencing in T cells
Decidual T cells are involved in maintaining immune tolerance at the maternal-fetal interface and consist mainly of CD4+ T cells, CD8+ T cells, and FOXP3+ regulatory T cells (Treg) (Nancy and Erlebacher, 2014). Toothaker et al. (2022) researched the mid-gestation (17–23 weeks of gestation) human placenta villi using single-cell analysis and found that decidua had a higher proportion of CD4T cells compared with either of the fetal layers, and that most of the T cells in placenta villi were of memory phenotypes that could help to limit T cell activation. Chen et al. (2021) revealed that the regulatory role of CD4+ T cells and the cytotoxic role of CD8+ T cells in early gestation were coordinated to maintain the balance of the decidual immune microenvironment. Guo et al. (2021) studied the heterogeneity of T cells in the recurrent miscarriage and healthy individuals and found enhanced cytokine-mediated signaling pathways and pro-inflammatory properties of various subsets of T cells in the recurrent miscarriage population. It was reported that the normal gestational decidual immune microenvironment is biased toward helper T cells (Th) 2 type, immunity, whereas Th1 immunity may lead to pregnancy failure (Raghupathy et al., 2000), whereas the decidual microenvironment in the recurrent miscarriage population predominantly exhibits Th1 type (Guo et al., 2021). Wang et al. (2021a) similarly confirmed the presence of highly immune activating properties in meconium T cells from RSA patients. Immune factors play an important role in the development of RSA, and correcting the immune imbalance of Th1/Th2 cells may be an important way to reduce the incidence of RSA.
Single-cell RNA sequencing in dendritic cells
Dendritic cells (DCs) account for about 1%–2% of metaphase free cells and are key antigen presenting cells during pregnancy. DCs can be divided into CD83+ mature dendritic cells (mDC), CD209+ (DC-SIGN), immature dendritic cells (imDC) and intermediate state DEC-205 + DCs according to their maturity, with DC-SIGN + DCs predominating in early gestation. Chen et al. (2021) found that most DC cells were in a resting state in early pregnancy, and these resting state dDCs may induce CD8+ T cell tolerance in the metaphase microenvironment, while activated state dDCs were found in the recurrent miscarriage population, which may be related to the pathogenesis of RSA.
Conclusion
Pregnancy is a dynamic and changing process, and the cellular composition in the placenta and decidua may change at each period. In recent years, single-cell genomics technologies have advanced our understanding of tissue heterogeneity at the maternal-fetal interface during pregnancy, advancing the field of maternal-fetal medicine research and providing powerful technical support for studying the physiological and pathological processes of pregnancy. However, we should recognize the limitations of single-cell sequencing. Firstly, single-cell sequencing studies must take into consideration temporal, spatial and individual differences, such as the location of the material, different gestational week, specimen handling procedures and sample size. In addition, single-cell sequencing requires fresh samples and a high percentage of live cells after dissociation, which makes it more difficult to obtain specimens and increases the bias of experimental batches. Finally, scRNA-seq alone cannot link genotype and phenotype, and we still need in-depth multi-omics studies of the maternal-fetal interface supported by single-cell high-resolution technology to ultimately reduce the incidence of maternal-fetal disease during pregnancy and protect maternal-fetal health.
Author contributions
QC, DS, and YH conceived and designed the manuscript. YX, XL, YW, QC, LZ, and RD reviewed the literature, QC wrote the original draft; DS and YH reviewed and revised the manuscript. YH acquired funding. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by Sichuan Provincial Department of Science and Technology (2022YFS0043), Sichuan University Zigong City School-land Science and Technology Cooperative R&D Project (00402153A2153).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2022.1079961/full#supplementary-material
References
Abrahams, V. M., Kim, Y. M., Straszewski, S. L., Romero, R., and Mor, G. (2004). Macrophages and apoptotic cell clearance during pregnancy. Am. J. Reprod. Immunol. 51 (4), 275–282. doi:10.1111/j.1600-0897.2004.00156.x
Ander, S. E., Diamond, M. S., and Coyne, C. B. (2019). Immune responses at the maternal-fetal interface. Sci. Immunol. 4 (31), eaat6114. doi:10.1126/sciimmunol.aat6114
Armand, E. J., Li, J., Xie, F., Luo, C., and Mukamel, E. A. (2021). Single-cell sequencing of brain cell transcriptomes and epigenomes. Neuron 109 (1), 11–26. doi:10.1016/j.neuron.2020.12.010
Baslan, T., and Hicks, J. (2017). Unravelling biology and shifting paradigms in cancer with single-cell sequencing. Nat. Rev. Cancer 17 (9), 557–569. doi:10.1038/nrc.2017.58
Blom, A. M. (2017). The role of complement inhibitors beyond controlling inflammation. J. Intern Med. 282 (2), 116–128. doi:10.1111/joim.12606
Bulmer, J. N., Morrison, L., Longfellow, M., Ritson, A., and Pace, D. (1991). Granulated lymphocytes in human endometrium: Histochemical and immunohistochemical studies. Hum. Reprod. 6 (6), 791–798. doi:10.1093/oxfordjournals.humrep.a137430
Bulmer, J. N., Williams, P. J., and Lash, G. E. (2010). Immune cells in the placental bed. Int. J. Dev. Biol. 54 (2-3), 281–294. doi:10.1387/ijdb.082763jb
Burton, G. J., and Fowden, A. L. (2015). The placenta: A multifaceted, transient organ. Philos. Trans. R. Soc. Lond B Biol. Sci. 370 (1663), 20140066. doi:10.1098/rstb.2014.0066
Cao, J., Packer, J. S., Ramani, V., Cusanovich, D. A., Huynh, C., Daza, R., et al. (2017). Comprehensive single-cell transcriptional profiling of a multicellular organism. Science 357 (6352), 661–667. doi:10.1126/science.aam8940
Chaiworapongsa, T., Chaemsaithong, P., Yeo, L., and Romero, R. (2014). Pre-eclampsia part 1: Current understanding of its pathophysiology. Nat. Rev. Nephrol. 10 (8), 466–480. doi:10.1038/nrneph.2014.102
Chen, P., Zhou, L., Chen, J., Lu, Y., Cao, C., Lv, S., et al. (2021). The immune atlas of human deciduas with unexplained recurrent pregnancy loss. Front. Immunol. 12, 689019. doi:10.3389/fimmu.2021.689019
Co, E. C., Gormley, M., Kapidzic, M., Rosen, D. B., Scott, M. A., Stolp, H. A., et al. (2013). Maternal decidual macrophages inhibit NK cell killing of invasive cytotrophoblasts during human pregnancy. Biol. Reprod. 88 (6), 155. doi:10.1095/biolreprod.112.099465
De Maria, A., Bozzano, F., Cantoni, C., and Moretta, L. (2011). Revisiting human natural killer cell subset function revealed cytolytic CD56(dim)CD16+ NK cells as rapid producers of abundant IFN-gamma on activation. Proc. Natl. Acad. Sci. U. S. A. 108 (2), 728–732. doi:10.1073/pnas.1012356108
Dey, S. K., Lim, H., Das, S. K., Reese, J., Paria, B. C., Daikoku, T., et al. (2004). Molecular cues to implantation. Endocr. Rev. 25 (3), 341–373. doi:10.1210/er.2003-0020
Ding, J., Adiconis, X., Simmons, S. K., Kowalczyk, M. S., Hession, C. C., Marjanovic, N. D., et al. (2020). Systematic comparison of single-cell and single-nucleus RNA-sequencing methods. Nat. Biotechnol. 38 (6), 737–746. doi:10.1038/s41587-020-0465-8
Drury, J. A., Parkin, K. L., Coyne, L., Giuliani, E., Fazleabas, A. T., and Hapangama, D. K. (2018). The dynamic changes in the number of uterine natural killer cells are specific to the eutopic but not to the ectopic endometrium in women and in a baboon model of endometriosis. Reprod. Biol. Endocrinol. 16 (1), 67. doi:10.1186/s12958-018-0385-3
Du, L., Deng, W., Zeng, S., Xu, P., Huang, L., Liang, Y., et al. (2021). Single-cell transcriptome analysis reveals defective decidua stromal niche attributes to recurrent spontaneous abortion. Cell Prolif. 54 (11), e13125. doi:10.1111/cpr.13125
Faas, M. M., and De Vos, P. (2018). Innate immune cells in the placental bed in healthy pregnancy and preeclampsia. Placenta 69, 125–133. doi:10.1016/j.placenta.2018.04.012
Faas, M. M., and de Vos, P. (2017). Uterine NK cells and macrophages in pregnancy. Placenta 56, 44–52. doi:10.1016/j.placenta.2017.03.001
Faas, M. M., Spaans, F., and De Vos, P. (2014). Monocytes and macrophages in pregnancy and pre-eclampsia. Front. Immunol. 5, 298. doi:10.3389/fimmu.2014.00298
Fu, B., Zhou, Y., Ni, X., Tong, X., Xu, X., Dong, Z., et al. (2017). Natural killer cells promote fetal development through the secretion of growth-promoting factors. Immunity 47 (6), 1100–1113 e1106. doi:10.1016/j.immuni.2017.11.018
Garrido-Gimenez, C., and Alijotas-Reig, J. (2015). Recurrent miscarriage: Causes, evaluation and management. Postgrad. Med. J. 91 (1073), 151–162. doi:10.1136/postgradmedj-2014-132672
Gellersen, B., and Brosens, J. J. (2014). Cyclic decidualization of the human endometrium in reproductive health and failure. Endocr. Rev. 35 (6), 851–905. doi:10.1210/er.2014-1045
Gierahn, T. M., Wadsworth, M. H., Hughes, T. K., Bryson, B. D., Butler, A., Satija, R., et al. (2017). Seq-well: Portable, low-cost RNA sequencing of single cells at high throughput. Nat. Methods 14 (4), 395–398. doi:10.1038/nmeth.4179
Guo, C., Cai, P., Jin, L., Sha, Q., Yu, Q., Zhang, W., et al. (2021). Single-cell profiling of the human decidual immune microenvironment in patients with recurrent pregnancy loss. Cell Discov. 7 (1), 1. doi:10.1038/s41421-020-00236-z
Hashimshony, T., Wagner, F., Sher, N., and Yanai, I. (2012). CEL-seq: Single-cell RNA-seq by multiplexed linear amplification. Cell Rep. 2 (3), 666–673. doi:10.1016/j.celrep.2012.08.003
Heikkinen, J., Mottonen, M., Komi, J., Alanen, A., and Lassila, O. (2003). Phenotypic characterization of human decidual macrophages. Clin. Exp. Immunol. 131 (3), 498–505. doi:10.1046/j.1365-2249.2003.02092.x
Hiby, S. E., Apps, R., Chazara, O., Farrell, L. E., Magnus, P., Trogstad, L., et al. (2014). Maternal KIR in combination with paternal HLA-C2 regulate human birth weight. J. Immunol. 192 (11), 5069–5073. doi:10.4049/jimmunol.1400577
Hiby, S. E., Apps, R., Sharkey, A. M., Farrell, L. E., Gardner, L., Mulder, A., et al. (2010). Maternal activating KIRs protect against human reproductive failure mediated by fetal HLA-C2. J. Clin. Invest 120 (11), 4102–4110. doi:10.1172/JCI43998
Hiby, S. E., Walker, J. J., O'Shaughnessy K, M., Redman, C. W., Carrington, M., Trowsdale, J., et al. (2004). Combinations of maternal KIR and fetal HLA-C genes influence the risk of preeclampsia and reproductive success. J. Exp. Med. 200 (8), 957–965. doi:10.1084/jem.20041214
Hoo, R., Nakimuli, A., and Vento-Tormo, R. (2020). Innate immune mechanisms to protect against infection at the human decidual-placental interface. Front. Immunol. 11, 2070. doi:10.3389/fimmu.2020.02070
Huang, J., Li, Q., Peng, Q., Xie, Y., Wang, W., Pei, C., et al. (2021). Single-cell RNA sequencing reveals heterogeneity and differential expression of decidual tissues during the peripartum period. Cell Prolif. 54 (2), e12967. doi:10.1111/cpr.12967
Hutcheon, J. A., Lisonkova, S., and Joseph, K. S. (2011). Epidemiology of pre-eclampsia and the other hypertensive disorders of pregnancy. Best. Pract. Res. Clin. Obstet. Gynaecol. 25 (4), 391–403. doi:10.1016/j.bpobgyn.2011.01.006
Jabrane-Ferrat, N. (2019). Features of human decidual NK cells in healthy pregnancy and during viral infection. Front. Immunol. 10, 1397. doi:10.3389/fimmu.2019.01397
Jaitin, D. A., Kenigsberg, E., Keren-Shaul, H., Elefant, N., Paul, F., Zaretsky, I., et al. (2014). Massively parallel single-cell RNA-seq for marker-free decomposition of tissues into cell types. Science 343 (6172), 776–779. doi:10.1126/science.1247651
James-Allan, L. B., Whitley, G. S., Leslie, K., Wallace, A. E., and Cartwright, J. E. (2018). Decidual cell regulation of trophoblast is altered in pregnancies at risk of pre-eclampsia. J. Mol. Endocrinol. 60 (3), 239–246. doi:10.1530/JME-17-0243
Kivioja, T., Vaharautio, A., Karlsson, K., Bonke, M., Enge, M., Linnarsson, S., et al. (2011). Counting absolute numbers of molecules using unique molecular identifiers. Nat. Methods 9 (1), 72–74. doi:10.1038/nmeth.1778
Klein, A. M., Mazutis, L., Akartuna, I., Tallapragada, N., Veres, A., Li, V., et al. (2015). Droplet barcoding for single-cell transcriptomics applied to embryonic stem cells. Cell 161 (5), 1187–1201. doi:10.1016/j.cell.2015.04.044
Kolodziejczyk, A. A., Kim, J. K., Svensson, V., Marioni, J. C., and Teichmann, S. A. (2015). The technology and biology of single-cell RNA sequencing. Mol. Cell 58 (4), 610–620. doi:10.1016/j.molcel.2015.04.005
Koopman, L. A., Kopcow, H. D., Rybalov, B., Boyson, J. E., Orange, J. S., Schatz, F., et al. (2003). Human decidual natural killer cells are a unique NK cell subset with immunomodulatory potential. J. Exp. Med. 198 (8), 1201–1212. doi:10.1084/jem.20030305
Le Bouteiller, P., and Bensussan, A. (2017). Up-and-down immunity of pregnancy in humans. F1000Res 6, 1216. doi:10.12688/f1000research.11690.1
Lee, S., Kim, J., Jang, B., Hur, S., Jung, U., Kil, K., et al. (2010). Fluctuation of peripheral blood T, B, and NK cells during a menstrual cycle of normal healthy women. J. Immunol. 185 (1), 756–762. doi:10.4049/jimmunol.0904192
Lei, Y., Tang, R., Xu, J., Wang, W., Zhang, B., Liu, J., et al. (2021). Applications of single-cell sequencing in cancer research: Progress and perspectives. J. Hematol. Oncol. 14 (1), 91. doi:10.1186/s13045-021-01105-2
Leung, C. S., Yeung, T. L., Yip, K. P., Pradeep, S., Balasubramanian, L., Liu, J., et al. (2014). Calcium-dependent FAK/CREB/TNNC1 signalling mediates the effect of stromal MFAP5 on ovarian cancer metastatic potential. Nat. Commun. 5, 5092. doi:10.1038/ncomms6092
Li, C., Houser, B. L., Nicotra, M. L., and Strominger, J. L. (2009). HLA-G homodimer-induced cytokine secretion through HLA-G receptors on human decidual macrophages and natural killer cells. Proc. Natl. Acad. Sci. U. S. A. 106 (14), 5767–5772. doi:10.1073/pnas.0901173106
Li, H., Huang, Q., Liu, Y., and Garmire, L. X. (2020). Single cell transcriptome research in human placenta. Reproduction 160 (6), R155–R167. doi:10.1530/REP-20-0231
Liu, S., Diao, L., Huang, C., Li, Y., Zeng, Y., and Kwak-Kim, J. Y. H. (2017). The role of decidual immune cells on human pregnancy. J. Reprod. Immunol. 124, 44–53. doi:10.1016/j.jri.2017.10.045
Liu, Y., Fan, X., Wang, R., Lu, X., Dang, Y. L., Wang, H., et al. (2018). Single-cell RNA-seq reveals the diversity of trophoblast subtypes and patterns of differentiation in the human placenta. Cell Res. 28 (8), 819–832. doi:10.1038/s41422-018-0066-y
Macosko, E. Z., Basu, A., Satija, R., Nemesh, J., Shekhar, K., Goldman, M., et al. (2015). Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell 161 (5), 1202–1214. doi:10.1016/j.cell.2015.05.002
Moffett, A., and Loke, C. (2006). Immunology of placentation in eutherian mammals. Nat. Rev. Immunol. 6 (8), 584–594. doi:10.1038/nri1897
Nakashima, A., Shima, T., Aoki, A., Kawaguchi, M., Yasuda, I., Tsuda, S., et al. (2021). Molecular and immunological developments in placentas. Hum. Immunol. 82 (5), 317–324. doi:10.1016/j.humimm.2021.01.012
Nancy, P., and Erlebacher, A. (2014). T cell behavior at the maternal-fetal interface. Int. J. Dev. Biol. 58 (2-4), 189–198. doi:10.1387/ijdb.140054ae
Ozimek, J. A., Eddins, R. M., Greene, N., Karagyozyan, D., Pak, S., Wong, M., et al. (2016). Opportunities for improvement in care among women with severe maternal morbidity. Am. J. Obstet. Gynecol. 215 (4), e1–e6. doi:10.1016/j.ajog.2016.05.022
Petroff, M. G., Sedlmayr, P., Azzola, D., and Hunt, J. S. (2002). Decidual macrophages are potentially susceptible to inhibition by class Ia and class Ib HLA molecules. J. Reprod. Immunol. 56 (1-2), 3–17. doi:10.1016/s0165-0378(02)00024-4
Picelli, S., Faridani, O. R., Bjorklund, A. K., Winberg, G., Sagasser, S., and Sandberg, R. (2014). Full-length RNA-seq from single cells using Smart-seq2. Nat. Protoc. 9 (1), 171–181. doi:10.1038/nprot.2014.006
Pillarisetty, L. S., and Mahdy, H. (2022). Recurrent pregnancy loss, in StatPearls. Treasure Island (FL): StatPearls Publishing.
Pique-Regi, R., Romero, R., Tarca, A. L., Sendler, E. D., Xu, Y., Garcia-Flores, V., et al. (2019). Single cell transcriptional signatures of the human placenta in term and preterm parturition. Elife 8, e52004. doi:10.7554/eLife.52004
Pollheimer, J., Vondra, S., Baltayeva, J., Beristain, A. G., and Knofler, M. (2018). Regulation of placental extravillous trophoblasts by the maternal uterine environment. Front. Immunol. 9, 2597. doi:10.3389/fimmu.2018.02597
Potter, S. S. (2018). Single-cell RNA sequencing for the study of development, physiology and disease. Nat. Rev. Nephrol. 14 (8), 479–492. doi:10.1038/s41581-018-0021-7
PrabhuDas, M., Bonney, E., Caron, K., Dey, S., Erlebacher, A., Fazleabas, A., et al. (2015). Immune mechanisms at the maternal-fetal interface: Perspectives and challenges. Nat. Immunol. 16 (4), 328–334. doi:10.1038/ni.3131
Practice Committee of the American Society for Reproductive Medicine. Electronic address (2020). Definitions of infertility and recurrent pregnancy loss: A committee opinion. Fertil. Steril. 113 (3), 533–535. doi:10.1016/j.fertnstert.2019.11.025
Raghupathy, R., Makhseed, M., Azizieh, F., Omu, A., Gupta, M., and Farhat, R. (2000). Cytokine production by maternal lymphocytes during normal human pregnancy and in unexplained recurrent spontaneous abortion. Hum. Reprod. 15 (3), 713–718. doi:10.1093/humrep/15.3.713
Ramskold, D., Luo, S., Wang, Y. C., Li, R., Deng, Q., Faridani, O. R., et al. (2012). Full-length mRNA-Seq from single-cell levels of RNA and individual circulating tumor cells. Nat. Biotechnol. 30 (8), 777–782. doi:10.1038/nbt.2282
Redman, C. W., and Sargent, I. L. (2010). Immunology of pre-eclampsia. Am. J. Reprod. Immunol. 63 (6), 534–543. doi:10.1111/j.1600-0897.2010.00831.x
Saito, S., and Nakashima, A. (2013). Review: The role of autophagy in extravillous trophoblast function under hypoxia. Placenta 34, S79–S84. doi:10.1016/j.placenta.2012.11.026
Sarno, L., Maruotti, G. M., Saccone, G., Sirico, A., Mazzarelli, L. L., and Martinelli, P. (2015). Pregnancy outcome in proteinuria-onset and hypertension-onset preeclampsia. Hypertens. Pregnancy 34 (3), 284–290. doi:10.3109/10641955.2015.1015731
Setty, M., Kiseliovas, V., Levine, J., Gayoso, A., Mazutis, L., and Pe'er, D. (2019). Characterization of cell fate probabilities in single-cell data with Palantir. Nat. Biotechnol. 37 (4), 451–460. doi:10.1038/s41587-019-0068-4
Shulse, C. N., Cole, B. J., Ciobanu, D., Lin, J., Yoshinaga, Y., Gouran, M., et al. (2019). High-throughput single-cell transcriptome profiling of plant cell types. Cell Rep. 27 (7), 2241–2247. doi:10.1016/j.celrep.2019.04.054
Smith, S. D., Dunk, C. E., Aplin, J. D., Harris, L. K., and Jones, R. L. (2009). Evidence for immune cell involvement in decidual spiral arteriole remodeling in early human pregnancy. Am. J. Pathol. 174 (5), 1959–1971. doi:10.2353/ajpath.2009.080995
Sun, F., Wang, S., and Du, M. (2021). Functional regulation of decidual macrophages during pregnancy. J. Reprod. Immunol. 143, 103264. doi:10.1016/j.jri.2020.103264
Suryawanshi, H., Morozov, P., Straus, A., Sahasrabudhe, N., Max, K. E. A., Garzia, A., et al. (2018). A single-cell survey of the human first-trimester placenta and decidua. Sci. Adv. 4 (10), eaau4788. doi:10.1126/sciadv.aau4788
Tamblyn, J. A., Pilarski, N. S. P., Markland, A. D., Marson, E. J., Devall, A., Hewison, M., et al. (2022). Vitamin D and miscarriage: A systematic review and meta-analysis. Fertil. Steril. 118 (1), 111–122. doi:10.1016/j.fertnstert.2022.04.017
Than, N. G., Romero, R., Tarca, A. L., Kekesi, K. A., Xu, Y., Xu, Z., et al. (2018). Integrated systems biology approach identifies novel maternal and placental pathways of preeclampsia. Front. Immunol. 9, 1661. doi:10.3389/fimmu.2018.01661
Tilburgs, T., Crespo, A. C., van der Zwan, A., Rybalov, B., Raj, T., Stranger, B., et al. (2015). Human HLA-G+ extravillous trophoblasts: Immune-activating cells that interact with decidual leukocytes. Proc. Natl. Acad. Sci. U. S. A. 112 (23), 7219–7224. doi:10.1073/pnas.1507977112
Toothaker, J. M., Olaloye, O., McCourt, B. T., McCourt, C. C., Silva, T. N., Case, R. M., et al. (2022). Immune landscape of human placental villi using single-cell analysis. Development 149 (8), dev200013. doi:10.1242/dev.200013
Triggianese, P., Perricone, C., Chimenti, M. S., De Carolis, C., and Perricone, R. (2016). Innate immune system at the maternal-fetal interface: Mechanisms of disease and targets of therapy in pregnancy syndromes. Am. J. Reprod. Immunol. 76 (4), 245–257. doi:10.1111/aji.12509
Turco, M. Y., and Moffett, A. (2019). Development of the human placenta. Development 146 (22), dev163428. doi:10.1242/dev.163428
van 't Hof, L. J., Schotvanger, N., Haasnoot, G. W., van der Keur, C., Roelen, D. L., Lashley, L., et al. (2021). Maternal-fetal HLA compatibility in uncomplicated and preeclamptic naturally conceived pregnancies. Front. Immunol. 12, 673131. doi:10.3389/fimmu.2021.673131
Vazquez, J., Chasman, D. A., Lopez, G. E., Tyler, C. T., Ong, I. M., and Stanic, A. K. (2019). Transcriptional and functional programming of decidual innate lymphoid cells. Front. Immunol. 10, 3065. doi:10.3389/fimmu.2019.03065
Vento-Tormo, R., Efremova, M., Botting, R. A., Turco, M. Y., Vento-Tormo, M., Meyer, K. B., et al. (2018). Single-cell reconstruction of the early maternal-fetal interface in humans. Nature 563 (7731), 347–353. doi:10.1038/s41586-018-0698-6
Vince, G. S., Starkey, P. M., Jackson, M. C., Sargent, I. L., and Redman, C. W. (1990). Flow cytometric characterisation of cell populations in human pregnancy decidua and isolation of decidual macrophages. J. Immunol. Methods 132 (2), 181–189. doi:10.1016/0022-1759(90)90028-t
Wang, F., Jia, W., Fan, M., Shao, X., Li, Z., Liu, Y., et al. (2021a). Single-cell immune landscape of human recurrent miscarriage. Genomics Proteomics Bioinforma. 19 (2), 208–222. doi:10.1016/j.gpb.2020.11.002
Wang, S., Li, M., Sun, F., Chen, C., Ye, J., Li, D., et al. (2021b). Th17/Treg-cell balance in the peripheral blood of pregnant females with a history of recurrent spontaneous abortion receiving progesterone or cyclosporine A. Exp. Ther. Med. 21 (1), 37. doi:10.3892/etm.2020.9469
Williams, P. J., Searle, R. F., Robson, S. C., Innes, B. A., and Bulmer, J. N. (2009). Decidual leucocyte populations in early to late gestation normal human pregnancy. J. Reprod. Immunol. 82 (1), 24–31. doi:10.1016/j.jri.2009.08.001
Yang, F., Zheng, Q., and Jin, L. (2019). Dynamic function and composition changes of immune cells during normal and pathological pregnancy at the maternal-fetal interface. Front. Immunol. 10, 2317. doi:10.3389/fimmu.2019.02317
Yang, Y., Guo, F., Peng, Y., Chen, R., Zhou, W., Wang, H., et al. (2021). Transcriptomic profiling of human placenta in gestational diabetes mellitus at the single-cell level. Front. Endocrinol. (Lausanne) 12, 679582. doi:10.3389/fendo.2021.679582
Yasen, A., Aini, A., Wang, H., Li, W., Zhang, C., Ran, B., et al. (2020). Progress and applications of single-cell sequencing techniques. Infect. Genet. Evol. 80, 104198. doi:10.1016/j.meegid.2020.104198
Yu-Ju Wu, C., Chen, C. H., Lin, C. Y., Feng, L. Y., Lin, Y. C., Wei, K. C., et al. (2020). CCL5 of glioma-associated microglia/macrophages regulates glioma migration and invasion via calcium-dependent matrix metalloproteinase 2. Neuro Oncol. 22 (2), 253–266. doi:10.1093/neuonc/noz189
Yuan, J. Q., Wang, Y. C., Wang, Z. M., Li, L., and Gao, L. (2020). The immune microenvironment in maternal-fetal interface contributes to the parturition and preterm labor. Sheng Li Xue Bao 72 (1), 1–10.
Zeng, J., Ning, Z., Wang, Y., and Xiong, H. (2020). Implications of CD39 in immune-related diseases. Int. Immunopharmacol. 89, 107055. doi:10.1016/j.intimp.2020.107055
Zhang, T., Bian, Q., Chen, Y., Wang, X., Yu, S., Liu, S., et al. (2021). Dissecting human trophoblast cell transcriptional heterogeneity in preeclampsia using single-cell RNA sequencing. Mol. Genet. Genomic Med. 9 (8), e1730. doi:10.1002/mgg3.1730
Zhang, Y. H., He, M., Wang, Y., and Liao, A. H. (2017). Modulators of the balance between M1 and M2 macrophages during pregnancy. Front. Immunol. 8, 120. doi:10.3389/fimmu.2017.00120
Zhao, T., Fu, Y., Zhu, J., Liu, Y., Zhang, Q., Yi, Z., et al. (2018a). Single-cell RNA-seq reveals dynamic early embryonic-like programs during chemical reprogramming. Cell Stem Cell 23 (1), 31–45. e37. doi:10.1016/j.stem.2018.05.025
Zhao, X., Jiang, Y., Wang, L., Li, Z., Li, Q., and Feng, X. (2018b). Advances in understanding the immune imbalance between T-lymphocyte subsets and NK cells in recurrent spontaneous abortion. Geburtshilfe Frauenheilkd 78 (7), 677–683. doi:10.1055/a-0634-1813
Zheng, G. X., Terry, J. M., Belgrader, P., Ryvkin, P., Bent, Z. W., Wilson, R., et al. (2017). Massively parallel digital transcriptional profiling of single cells. Nat. Commun. 8, 14049. doi:10.1038/ncomms14049
Zheng, Z., Chen, E., Lu, W., Mouradian, G., Hodges, M., Liang, M., et al. (2020). Single-cell transcriptomic analysis. Compr. Physiol. 10 (2), 767–783. doi:10.1002/cphy.c190037
Zhou, W., Wang, H., Yang, Y., Guo, F., Yu, B., and Su, Z. (2022a). Trophoblast cell subtypes and dysfunction in the placenta of individuals with preeclampsia revealed by SingleCell RNA sequencing. Mol. Cells 45, 317–328. doi:10.14348/molcells.2021.0211
Keywords: maternal-fetal interface, single-cell RNA sequencing, trophoblast cell, decidual stromal cells, NK cells, macrophages, T cells
Citation: Chen Q, Shan D, Xie Y, Luo X, Wu Y, Chen Q, Dong R and Hu Y (2023) Single cell RNA sequencing research in maternal fetal interface. Front. Cell Dev. Biol. 10:1079961. doi: 10.3389/fcell.2022.1079961
Received: 25 October 2022; Accepted: 27 December 2022;
Published: 10 January 2023.
Edited by:
Xianwen Ren, Peking University, ChinaReviewed by:
Wangsheng Wang, Shanghai Jiao Tong University, ChinaRen-Wei Su, South China Agricultural University, China
Copyright © 2023 Chen, Shan, Xie, Luo, Wu, Chen, Dong and Hu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qian Chen, Y3FpYW4yMDE3QDE2My5jb20=; Yayi Hu, eWF5aS5odUAxNjMuY29t
 Qian Chen
Qian Chen Dan Shan1,2
Dan Shan1,2 Yayi Hu
Yayi Hu