
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell Dev. Biol., 03 November 2022
Sec. Developmental Epigenetics
Volume 10 - 2022 | https://doi.org/10.3389/fcell.2022.1010601
This article is part of the Research TopicEpigenetic and Molecular Control of Development and Germ Cell Fate DeterminationView all 7 articles
 Yibo Wang1
Yibo Wang1 Wenji Wang2
Wenji Wang2 Kaixin Cheng1
Kaixin Cheng1 Kaiying Geng1
Kaiying Geng1 Jing Liang1
Jing Liang1 Peike Wang1,3
Peike Wang1,3 Jiawei Zhang1
Jiawei Zhang1 Shudong Niu1
Shudong Niu1 Longzhong Jia1
Longzhong Jia1 Shuo Zhang1
Shuo Zhang1 Lingyu Li1
Lingyu Li1 Xiean Feng1
Xiean Feng1 Chao Wang1
Chao Wang1 Haibin Wang3
Haibin Wang3 Hua Zhang1
Hua Zhang1 Yan Zhang1*
Yan Zhang1*Ovarian follicles are the fundamental structure to support oocyte development, which provides mature oocytes for offspring. This process requires granulosa cells (GCs) to respond to the midcycle surge of hormones, leading to GC proliferation and differentiation by a series of genes’ transcriptional expression changes. Epigenetic mediator, Polycomb Repressive Complex 1 (PRC1) has been reported to function in fetal ovarian development. However, its functional relevance to folliculogenesis and ovulation remains unknown. In this study, we demonstrated that GC-selective depletion of PCGF2, a key component of PRC1, led to the loss of follicles, ovulation defects, and a lengthened estrus cycle, resulting in subfertility in female mice. The expression of PCGF2 is in the GCs of growing follicles and increases after human chorionic gonadotropin (hCG) stimulation. PCGF2 bound to the promoter of the key ovulation gene progesterone receptor (Pgr) and upregulated the expression of Pgr by targeting the epigenetic modification of H2AK119ub1 after hCG surge. Consistently, the expression of downstream genes of Pgr also sharply decreased, which resulted in the follicular rupture failed and oocyte entrapped in corpus luteum in GC-specific Pcgf2 knockout mice. Together, our study identified that PCGF2 is essential for folliculogenesis and ovulation via modulating hormone receptor expression.
In mammalian ovaries, the ovarian follicle is the structural basis of female reproduction. It is composed of an oocyte and surrounded by granulosa cells (GCs) and theca cells (Eppig, 1991). The functions of GCs are crucial for folliculogenesis, including primordial follicles growing to preovulatory follicles, ovulation, and the luteal process under the effects of hormones (Zhang and Liu, 2015; Liu W. et al., 2017; Richards and Ascoli, 2018). GCs are also responsible for synthesizing and secreting hormones (Tureck and Strauss, 1982). The accurate control of specific gene transcriptional states is the basis for establishing a GC identity for folliculogenesis, ovulation, and corpus luteum formation (Park et al., 2004; Fan et al., 2009). The abnormal expression of genes of GC proliferation and differentiation results in serious ovarian dysfunction diseases such as polycystic ovary syndrome (PCOS) and premature ovarian insufficiency (POI) (Murdoch and Cavender, 1989; Wu et al., 2007; Schmidt et al., 2014; Duffy et al., 2019). However, the physiological significance of epigenetic regulators in GCs during follicular development has remained largely unexplored.
Polycomb group (PcG) proteins are chromatin-modifying proteins that act as critical epigenetic modulators (Entrevan et al., 2016; Piunti and Shilatifard, 2016). PcG proteins are classified into two multiprotein repressive complexes, namely polycomb repressive complexes 1 and 2 (PRC1 and PRC2) (Scheuermann et al., 2010; Aranda et al., 2015). In mammalian cells, PcG proteins mediate gene silencing through at least two distinct enzymatic activities directed to histone tails: PRC1 contains E3 ubiquitin ligase RING1A/B, which specifically targets histone H2A mono-ubiquitination at K119 (H2AK119ub1) (Cao et al., 2005; Taherbhoy et al., 2015), while PRC2 mediates histone H3 tri-methylation at K27 (H3K27me3) (Shen et al., 2008; Jiao and Liu, 2015).
The polycomb protein PCGF2 (polycomb group ring finger 2), also known as Mel-18, is a component of PRC1 and regulates stem cell differentiation and cancerous cell development (Morey et al., 2015). PCGF2 in luminal breast cancer cells positively regulates the promoter activities of ER-α (estrogen receptor-α) and PGR (progesterone receptor) by suppressing SUMOylation (Lee et al., 2015). Several recent studies reported that the PRC1 complex plays various roles in the female reproductive system (Baumann and De La Fuente, 2011; Yokobayashi et al., 2013; Tang et al., 2016). PCGF4 (polycomb group ring finger 4, also known as Bmi-1), a homolog of PCGF2, has redundant functional properties with PCGF2 (Liu X. et al., 2017). PCGF4 deficiency results in female infertility by activating p16/p19 signaling and increasing oxidative stress in mice (Wang et al., 2019). In female mice, PCGF4 interacts with PGR and E3 ligase, which is essential for normal embryo implantation independent of PRC1 repression function (Xin et al., 2018). Moreover, PRC1 regulates germ cell viability, meiosis onset, homologous chromosome synapsis, and sexual differentiation in different species (Baumann and De La Fuente, 2011; Yokobayashi et al., 2013; Tang et al., 2016; Maezawa et al., 2017). The findings suggest that PRC1 is a crucial epigenetic mediator for regulating hormone receptors during reproduction system development in female mice. However, it remains unclear whether the PRC1 complex is involved in regulating follicular development in the ovary.
In this study, we observed that Pcgf2 is mainly expressed in ovarian GCs and then we established a GC-specific-Pcgf2 knockout mouse model to investigate its physiological functions in the ovary in vivo. Our results showed that Pcgf2 deficiency in granulosa cells results in subfertility, lengthened estrus cycle, and ovulation defects. In addition, we identified that Pcgf2 regulates Pgr transcription in GCs under hCG surge, which is a key regulator in follicular rupture. Our study highlights that the mutation of Pcgf2 may be a pathogenic factor of secondary premature ovarian insufficiency (POI) and anovulation.
The C57BL/6J mice were acquired from the Laboratory Animal Center of the Institute of Genetics (Beijing, China). The Foxl2-Cre knock-in mice were obtained from the laboratory animal center, institute of zoology, Chinese Academy of Sciences. Detailed information of the generation of the Foxl2-Cre knock-in mouse was provided in Cen et al. (2020). The Pcgf2fl/fl mice were provided by Dr. Rongwen Xi (National Institute of Biological Sciences, Beijing). We crossed Foxl2-Cre with Pcgf2fl/fl to get Foxl2-Cre;Pcgf2fl/fl GC-specific Pcgf2-knockout mouse model. In Pcgf2fl/fl mice, loxP-flanked upstream the exon 5 expression in cells. When bred to Foxl2-Cre mice, the exon 5 of Pcgf2 is deleted in the CRE-positive granulosa cells of all follicles. Mice were housed in mouse facilities that conformed to the standards and requirements of the Institutional Animal Care and Use Committee of China Agricultural University, No. AW92702202-3-1.
Mating trials were started at age of 6 months for female mice. Healthy males at 8–10 weeks of age were mated with females (1:2) for 7 months. The size and number of litter from each female were recorded to analyze the reproductive capacity.
The estrus cycle stage was evaluated every day by vaginal smear (Cora et al., 2015) for continuously 22 days. Using Wright’s stain, the stage was classified into 4 phases: Pro, proestrus; Est, estrus; Met, metestrus; Di, diestrus. The result of the estrus cycle was analyzed by using Graphpad prism 6.0 software.
The female mice were superovulated by intraperitoneal injection of 5 IU pregnant mare serum gonadotropin (PMSG, Sansheng Biological Technology) at postnatal day 21 (PD21), and 46 h later the mice were additionally injected with 5 IU human chorionic gonadotropin (hCG, Sansheng Biological Technology). The oocytes were then collected from the oviducts, counted, and captured under the stereoscope. The number of oocytes was statistical analysis by using Graphpad prism 6.0 software. The ovaries were collected and analyzed the ovarian histology at PMSG 46 h, hCG 8 h, 12 h, 16 h, 24 h and 48 h.
Ovaries were collected at specific ages and then fixed in 4% Paraformaldehyde solution in PBS (PFA, sc-281692, Santa Cruz Biotechnology, United States). The embedded ovaries were dehydrated and then sectioned to obtain 8-µm thickness serial paraffin sections. The ovarian sections were processed to deparaffinize and rehydrate, the ovarian sections were stained with hematoxylin (sc-24973A, Santa Cruz Biotechnology, United States) to analyze the histology of the ovaries. The images were all analyzed under the microscope (DM500, Leica). Every five slices were counted for the number of primordial and primary follicles under 100 μm. Then, the counting numbers were multiplied by five to calculate the number of all primordial or primary follicles. Each slice was counted for the number of secondary follicles, antral follicles and the structure of the corpus luteum. The structures of the preovulatory follicle and oocyte-trapped CL were also counted in each slice. The number of follicles was statistically analyzed by using Graphpad prism 6.0 software.
The ovaries were collected at 8 h after human chorionic gonadotropin (hCG, Sansheng Biological Technology) injection when the meiotic resumption and cumulus expansion happened in most preovulatory follicles (Fan et al., 2009). The ovarian slices were stained with hematoxylin (sc-24973A, Santa Cruz Biotechnology, United States). To check the capacity of meiotic resumption, the number of germinal vesicle breakdown (GVBD) was counted in preovulatory follicles. The percent of GVBD was statistically analyzed by using Graphpad prism 6.0 software. In preovulatory follicles, the state of cumulus cell expansion was also checked at this time under the microscope (DM500, Leica).
For immunofluorescence staining, the ovarian sections were subjected to deparaffinize and rehydrate. Using high temperature (95–98°C) in 0.01% sodium citrate buffer (pH 6.0) to retrieve the antigen of ovarian sections. Then, the ovarian sections were blocked by 10% donkey serum (Jackson ImmunoResearch) for 1 h and incubated with different primary antibodies at 4°C overnight. The primary antibodies used in this experiment were as follows: CX43 antibody (ab11370, rabbit, 1:300, Abcam, United Kingdom) and COLLAGEN IV antibody (ab6586, rabbit, 1:300, Abcam, United Kingdom), CYP11A1 antibody (Ab272494, rabbit, 1:300, Abcam, United Kingdom). Then the sections were incubated with Alexa Fluor 555- or 488-conjugated donkey secondary antibody (1:200, Life Technologies, United States) at 37°C for 60 min and then were counterstained with Hoechst 33342 (1:200, Beyotime, China). The images were photographed by using the Nikon Eclipse Ti digital fluorescence microscope or Andor Dragonfly spinning-disc confocal microscope and analyzed by software of Image J or Imaris.
For immunohistochemistry staining, we used the Rabbit two-step assay kit (PV-9001, ORIGENE, China). The ovarian sections were subjected to deparaffinize and rehydrate, and then retrieved the antigen. Then, the ovarian sections were incubated with endogenous peroxidase blocker for 10 min to block endogenous peroxidase. After washing with PBS for 9 min, the ovarian sections were then incubated with primary antibody H2AK119ub1 (D27C4, rabbit, 1:100, Cell signaling technology, United States) at 4°C overnight. Then the ovarian sections were incubated with reaction enhancing solution for 20 min at 37°C. After incubating goat anti-rabbit IgG polymer for 20 min at 37°C, we used the freshly prepared DAB color rendering solution to detect the H2AK119ub1 positive cells. The histological analysis was then checked after hematoxylin staining under the microscope (DM500, Leica).
In situ hybridization was performed as described in Wang et al. (2006). For in situ hybridization, using Primer 5 software to design gene-specific primers using the following criteria if possible: the optimal fragment length is 200-300bp, the best annealing temperature is 60°C, and the GC content is 55%. The designed primers should preferably span exons, which can ensure that the mRNA is mature RNA and improve its specificity. We added T7 RNA polymerase promoter sequence 5′-AATTAACCCTCACTAAAGGG-3′ to sense primer and added T3 RNA polymerase promoter sequence 5′-TAATACGACTCACTATAGGG-3′ to antisense primer (See Supplementary Table S4) and order oligos. Using ovary cDNA sample to do standard PCR with primers for getting more than 400 ng product. Running PCR reaction out on a 1% agarose gel and cut out band corresponding to expected size amplicon. And then, we purified DNA from the gel using a gel extraction kit (TIANgel Purification Kit, Lot#W9812) and used nanodrop to quantify DNA concentration. The concentration of DNA should be higher than 20 ng/mL for doing the next step. Mouse-specific Pcgf2 cRNA probe was labeled with digoxin by using DIG Northern Starter Kit (12039672910, Roche, Switzerland). To generate the sense probe of Pcgf2, we used T7 RNA polymerase in one reaction and used T3 RNA polymerase in a separate reaction to generate the antisense probe of Pcgf2. 10-μm ovarian cryosections hybridized with Pcgf2 antisense probe to detect the Pcgf2 mRNA localization and the sense probe served as negative control. After hybridization with cRNA probes for 24 h at 65°C, we used anti-digoxigenin-AP (75 mU/mL) to bind the DIG-label at 4°C overnight. After washing slices with MABT (maleic acid with 10%v/v Tween-20) solution for 1 h, we then incubated the slices with NBT/BCIP solution to react with the AP for color rendering, the positive signals were existed in purple in cytoplasm of the cells. To terminate the color rendering at a proper time, the slices were washed with PBS and then stained with Nuclear Fast Red solution for 10 min to display the nuclear of cells. And eventually the slices were dehydrated and transparent. The images were all analyzed under the microscope (DM500, Leica).
Granulosa cells were isolated by a follicular puncture from PMSG (5 IU, 46 h) and hCG (5 IU, 6 h and 16 h)-primed PD21 immature mice. Briefly, GCs at different times were released from antral follicles or corpus luteum by puncturing with a needle and the GCs were then stored at −80°C until being used.
For quantitative real-time PCR (QRT-PCR), total RNA was isolated from GCs of at least 3 additional GC-Pcgf2−/− and GC-Pcgf2+/+ female mice. Reverse transcription was done using the PrimeScript™RT reagent Kit with gDNA Eraser (TAKARA, PR047Q, Japan). The PCR reaction included 5 µL of SYBR Green Mix (Roche, 04913914001, Switzerland), cDNA product (1:10 dilution), and 0.5 µM of primers. Relative levels of mRNAs were calculated and normalized to the levels of endogenous β-actin in the same samples. The relative transcript levels of other samples were compared to the control, and the relative expression levels were shown in the graphs. For each experiment, QRT-PCR reactions were done in triplicate. The primers used in this paper were listed in Supplementary Table S4.
The GCs in different groups were collected and then lysed in WIP lysis solution (Cell signaling technology, United States). Separated the prepared protein samples by electrophoresis with 10% SDS-PAGE and transferred the protein to polyvinylidene fluoride (PVDF) membranes (IPVH00010, Millipore, United States). Then, blocked the membranes by using 5% nonfat-dry milk for 1 h at 37°C and incubated the membranes at 4°C overnight with the primary antibodies to be tested. The used primary antibody in the western blot was H2AK119ub1 (D27C4, rabbit, 1:1000, Cell signaling technology, United States). After overnight incubating, the membrane was washed with tris-buffered saline with 0.05% tween (TBST) for 30 min and then processed to incubate with the secondary antibody at room temperature for 1 h (1:5000, Beyotime, China). The level of α-Tubulin (A0208, rabbit, 1:1000, Beyotime, China) was used as an internal control. The membrane was washed with TBST for 30 min. The protein on membrane was captured by the Super Signal detection system (Prod 34080, ThermoFisher Scientific, United States). The images were then quantified by Image J software.
Mice were anesthetized and blood was collected by orbital blood. The level of progesterone (P4) in serum was analyzed by the Beijing Northern Institute of Biotechnology.
ChIP-Sequence data were downloaded from Scelfo et al. (2019) (GEO: GSE122715) (Scelfo et al., 2019). Reads were aligned to the mouse reference genome mm9 for ChIP-Sequence of PCGF2 and H2AK119ub1 samples, using Bowtie2 v2.4.5 with default parameters without allowing for multi-mapping (–m 1). The PCR duplicates were removed by using PICARD (http://broadinstitute.github.io/picard/). The peaks were called by using MACS2 v2.2.7.1 with default parameters. A list containing the final PCGF2 and H2AK119ub1 peaks used in the analyses could be found in Supplementary Tables S1, S3. Genomic peak annotation was performed with the R package ChIPseeker v1.30.3, considering the region ± 2.0 kb around the TSS as the promoter. Gene ontology (Lydon et al.) analysis of PCGF2 targets was performed using the R package clusterProfiler setting as threshold an adjusted p value and q-value of 0.05. For visualization of PCGF2 enrichment of ChIP-seq, the BigWig file with input signal subtracted was generated using the function bamCompare from deepTools 3.5.1 with default parameters. Heatmap was performed using the functions computeMatrix followed by plotHeatmap from deepTools excluding blacklisted regions by ENCODE. Known motif discovery was performed by using HOMER v4.11 with default parameters.
To address the functional role of Pcgf2 in folliculogenesis, we first analyzed the Pcgf2 expression pattern in ovaries by in situ hybridization. Pcgf2 was mainly detected in the GCs of growing follicles (Figure 1A and Supplementary Figure S1, arrowheads), suggesting as a potential role in folliculogenesis and fertility in females.
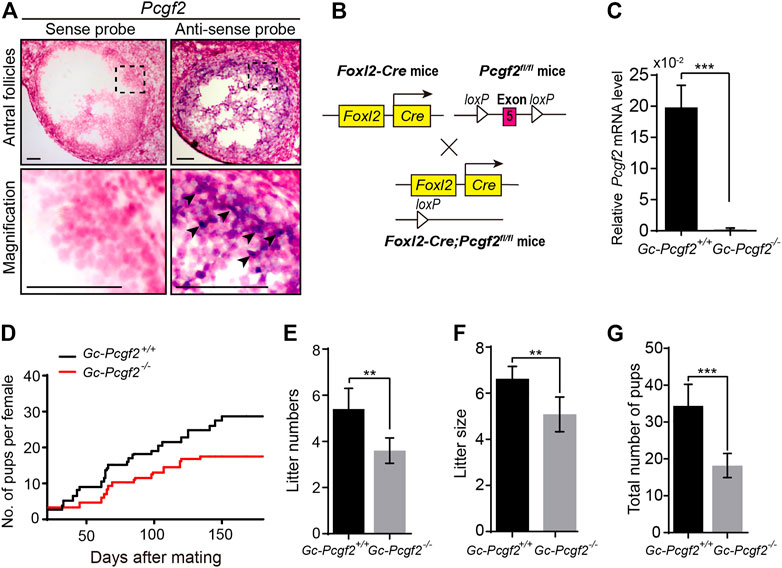
FIGURE 1. Deleting Pcgf2 from ovarian follicle granulosa cells decreases fertility. (A) In situ hybridization showing Pcgf2 mainly expressed in granulosa cells (arrowheads) of growing follicles (n = 3 per group). Scale bars: 50 µm. (B) Schematic diagram of deletion of Pcgf2 exon 5 by Foxl2-Cre-mediated recombination in granulosa cells in GC-Pcgf2−/− ovaries. (C) Quantitative real-time PCR (QRT-PCR) result showing the mRNA level of Pcgf2 sharply reduced in granulosa cells of GC-Pcgf2−/− ovaries (n = 3 per group). (D–G) Fertility checking showing a subfertility phenotype in the GC-Pcgf2−/− female mice after mating with healthy wild-type males at 6 months. (D) Fertility checking result showing a subfertility phenotype in the GC-Pcgf2−/− female mice (n = 5) with significantly decreased number and size of litters, compared with in GC-Pcgf2+/+ mice (n = 5), during 7 months of mating. (E) Quantitative analysis result showing a significantly decreased number of litters in GC-Pcgf2−/− female mice (4 ± 0) compared with the controls (5 ± 1; n = 5 per group). (F) Quantitative analysis results showing reduced litter size in GC-Pcgf2−/− female mice (5 ± 1) compared with the controls (7 ± 0; n = 5 per group). (G) Quantitative analysis results showing a significantly decreased total number of pups in GC-Pcgf2−/− female mice (18 ± 3) compared with the controls (34 ± 5; n = 5 per group). **p < 0.01 and ***p < 0.001 by two-tailed unpaired Student’s t test.
Next, we generated a GC-specific Pcgf2 knockout mouse model, to explore the role of Pcgf2 in ovaries. By flanking the exon 5 of Pcgf2 with loxP sequences (Pcgf2 flox/flox) crossed with Foxl2-Cre mice, as illustrated in Figure 1B, we attained the resulting Foxl2-Cre;Pcgf2fl/fl mice, referred to as GC-Pcgf2−/−. The littermates of No-Cre;Pcgf2fl/fl mice, referred to as GC-Pcgf2+/+, were used as controls. Quantitative real-time PCR (QRT-PCR) was performed to detect the mRNA level of Pcgf2 in GCs to validate the efficiency of deletion in GC-Pcgf2−/− female mice. QRT-PCR analysis showed that Pcgf2 was effectively deleted in the GCs of ovarian follicles in GC-Pcgf2−/− mice (Figure 1C). These data suggested that the GC-Pcgf2−/− mouse model could be used to detect the functional role of Pcgf2 in ovarian GCs. We tested female fertility by mating GC-Pcgf2−/− and GC-Pcgf2+/+ female mice with healthy wild-type males. GC-Pcgf2−/− female mice showed a subfertility phenotype from the age of 6 months (Figure 1D). Compared with GC-Pcgf2+/+ female mice, we found lower litter numbers (4 ± 0 in GC-Pcgf2−/− vs. 5 ± 1 in GC-Pcgf2+/+; Figure 1E) and reduced litter sizes (5 ± 1 in GC-Pcgf2−/− vs. 7 ± 0 in GC-Pcgf2+/+; Figure 1F) produced by GC-Pcgf2−/− female mice. These resulted in a decreased total number of pups in GC-Pcgf2−/− female mice (18 ± 3) compared with GC-Pcgf2+/+ female mice (34 ± 5) after 7 months of mating trial tracing (Figure 1G). These results indicate that the Pcgf2 function in GCs is involved in regulating female reproductive capacity.
To find out the cause for subfertility in GC-Pcgf2−/− female mice, we detected the development of follicles in GC-Pcgf2−/− ovaries at different ages. A similar morphology of ovaries (Supplementary Figure S2A, PD6) and an identical total number of follicles were found in GC-Pcgf2−/− (3925 ± 812) and GC-Pcgf2+/+(3748 ± 471) mice at postnatal day 6 (PD6) (Supplementary Figure S2B), suggesting that deletion of Pcgf2 in (pre)GCs did not affect reproductive reserves formation. In PD35 and 2-month female mice, both ovarian size (Supplementary Figure S2A, PD35 and 2 months) and total numbers of follicles in GC-Pcgf2−/− were similar to those in the controls (Supplementary Figure S2B). However, follicle counting showed that the number of primordial follicles (PFs) was significantly reduced in GC-Pcgf2−/− ovaries (751 ± 126) compared with those in the controls (1181 ± 271) at 6 months (Supplementary Figure S2C). In 10-month female mice, fewer PFs and GFs distribution were found in GC-Pcgf2−/− ovaries (Figure 2A and Supplementary Figures S2B,C; PFs: 460 ± 115 in GC-Pcgf2−/− vs. 719 ± 171 in GC-Pcgf2+/+; GFs: 418 ± 84 in GC-Pcgf2−/− vs. 591 ± 130 in GC-Pcgf2+/+), which resulted in sharply decreased total follicle numbers in GC-Pcgf2−/− mice (878 ± 156 in GC-Pcgf2−/− vs. 1310 ± 270 in GC-Pcgf2+/+) at 10 months (Figure 2B). These results indicated that Pcgf2 deletion in GCs lead to accelerate follicle loss in female mice. However, histological analysis showed identical numbers of corpus luteum were formed in the ovaries of GC-Pcgf2−/− and GC-Pcgf2+/+ female mice at 6 and 10 months (Figures 2A,C; 6 months: 9 ± 2 in GC-Pcgf2−/− vs. 9 ± 1 in GC-Pcgf2+/+; 10 months: 9 ± 2 in GC-Pcgf2−/− vs. 7 ± 2 in GC-Pcgf2+/+). These results suggest that the decline of GC-Pcgf2−/− fertility is endowed with an undefined mechanism.
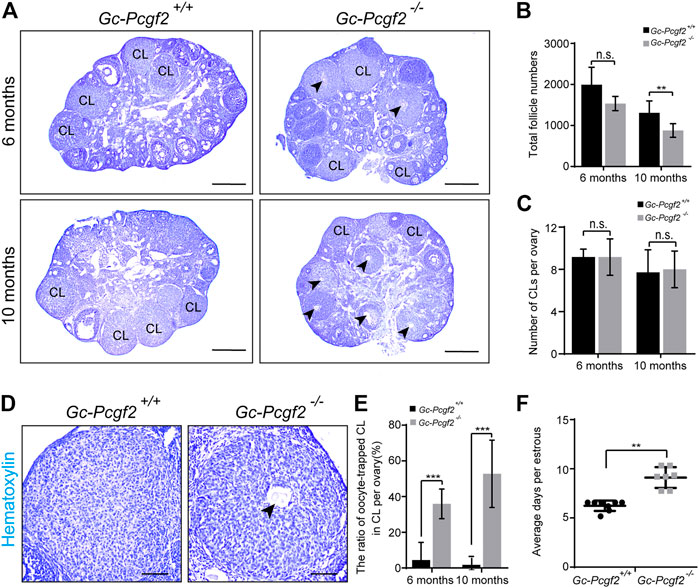
FIGURE 2. Deleting Pcgf2 from ovarian follicle granulosa cells accelerates follicle loss and disrupts the estrus cycle. (A) Histological analysis showing the corpus luteum contained entrapped oocytes (arrowheads) were found in GC-Pcgf2−/− ovaries at 6 months and 10 months. CL, corpus luteum. Arrowheads, oocytes entrapped in CLs. Scale bars: 500 µm. (B) Follicle counting result showing identical numbers of the total follicle in female mice at 6 months (1585 ± 182 in GC-Pcgf2−/− vs. 1994 ± 379 in GC-Pcgf2+/+; n = 5 per group) and a significantly decreased total follicle number in GC-Pcgf2−/− mice at 10 months (878 ± 156 in GC-Pcgf2−/− vs. 1310 ± 270 in GC-Pcgf2+/+; n = 8 per group). (C–E) Histological analysis of corpus luteum showing significantly higher ratio of oocytes entrapped in CLs of GC-Pcgf2−/− ovaries, compared with those in the controls at 6 months and 10 months. (C) CL counting results showing an identical number of CLs in ovaries at 6 months (9 ± 2 in GC-Pcgf2−/− vs. 9 ± 1 in GC-Pcgf2+/+; n = 6 per group) and 10 months (9 ± 2 in GC-Pcgf2−/− vs. 7 ± 2 in GC-Pcgf2+/+; n = 8 per group). (D) Detailed histological analysis of CLs showing the structures of CL entrapped with oocyte (arrowheads) of GC-Pcgf2−/− ovaries at 6 months. Scale bars: 100 µm. (E) Quantitative result showing significantly higher ratio of oocyte-trapped-CL in GC-Pcgf2−/− (6 months: 36% ± 7%; 10 months: 53% ± 17%) compared with in the controls (6 months: 4% ± 9%; 10 months: 2% ± 4%; 6 months: n = 6 per group, 10 months: n = 8 per group). (F) Quantitative result showing a longer estrus cycle in GC-Pcgf2−/− (9 ± 1 day; n = 7) compared with in the controls (6 ± 0 days; n = 8). The experiments were repeated at least three times. The data in (B,C,E,F) represent the results (mean ± SD) of the biological triplicate experiments. n.s. p > 0.05, **p < 0.01, and ***p < 0.001 by two-tailed unpaired Student’s t test.
We checked the histology of CLs and found the abnormal entrapped oocytes were present inside CLs in GC-Pcgf2−/− ovaries at 6 months (36% ± 7% in GC-Pcgf2−/− vs. 4% ± 9% in GC-Pcgf2+/+) (Figures 2A,D,E, arrowheads). In 10-month GC-Pcgf2−/− ovaries, CL structures entrapped with oocyte were found, compared with those in the controls, at 10 months (53% ± 17% in GC-Pcgf2−/− vs. 2% ± 4% in GC-Pcgf2+/+) (Figures 2A,E, arrowheads). These results revealed ovulation disorders in GC-Pcgf2−/− female mice. Moreover, the relative mRNA level of Pcgf2 was significantly increased at the estrus stage compared with that in other cycling stages (Supplementary Figure S2E). We further monitored the estrus cycle of GC-Pcgf2−/− and control female mice for 22 days. We found that GC-Pcgf2−/− mice exhibited a lengthened estrus cycle, compared with the control mice (9 ± 1 day in GC-Pcgf2−/− vs. 6 ± 0 days in GC-Pcgf2+/+) (Figure 2F), which lead to a lower number of litters. These results suggest that Pcgf2 knockout in GCs results in defects of the ovulation and estrus cycle, thus leading to subfertility in GC-Pcgf2−/− female mice.
To examine the ovulatory capacity in GC-Pcgf2−/−mice, we established the superovulation mouse model by injecting pregnant mare serum gonadotropin (PMSG) and human chorionic gonadotropin (hCG) at PD21. Statistical analysis of ovulated oocytes from oviducts showed that GC-Pcgf2−/− females only ovulated 16 ± 8 oocytes, which was significantly lower than that in GC-Pcgf2+/+ mice (55 ± 14 oocytes) at 16 h after hCG treatment (hCG 16 h) (Figures 3A,B).
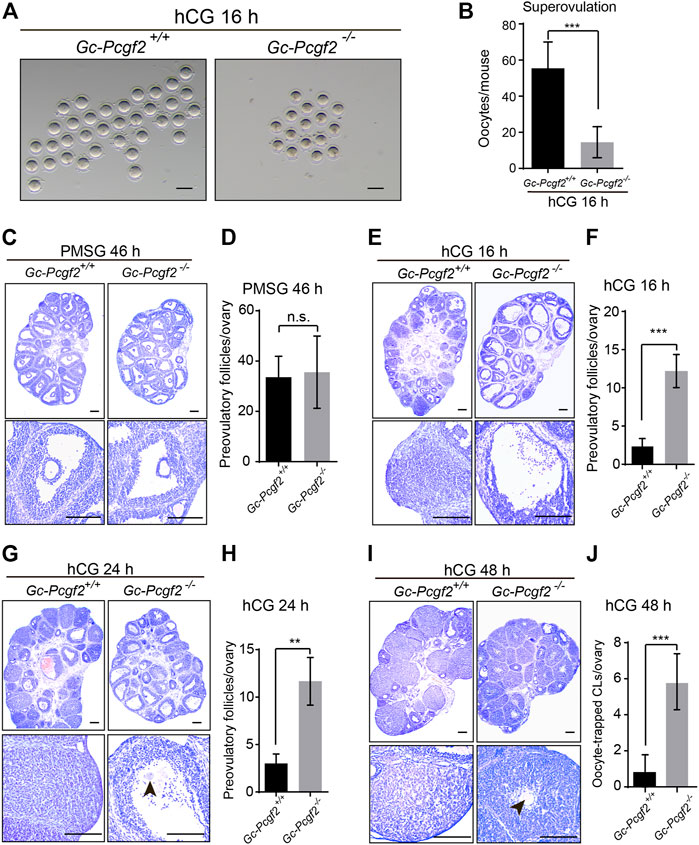
FIGURE 3. Abnormal response to hCG in granulosa cells of GC-Pcgf2−/− female mice. (A) Images showing ovulated oocytes from GC-Pcgf2+/+ and GC-Pcgf2−/− female mice after superovulation. Scale bars: 100 µm. (B) Quantitative analysis results showing a decreased number of oocytes ovulated in GC-Pcgf2−/− female mice (16 ± 8) compared with in the controls (55 ± 14; n = 9 per group). (C) Histological analysis showing normal preovulatory follicles distributed in the ovaries of GC-Pcgf2−/− female mice at 46 h after PMSG (PMSG 46 h) treatment (n = 5 per group). Scale bars: 200 µm. (D) Follicle counting result showing an identical number of preovulatory follicles in GC-Pcgf2−/− (36 ± 13) and GC-Pcgf2+/+ (34 ± 7) female mice at PMSG 46 h (n = 5 per group). (E) Histological analysis showing more preovulatory follicles in GC-Pcgf2−/− ovaries at hCG 16 h compared with those in the controls (n = 8 per group). Scale bars: 200 µm. (F) Statistical analysis result showing a significantly increased number of preovulatory follicles in ovaries in GC-Pcgf2−/− (12 ± 2) female mice, compared with those in the controls (2 ± 1) at hCG 16 h (n = 8 per group). (G) Histological analysis showing more preovulatory follicles in GC-Pcgf2−/− at hCG 24 h compared to those in the controls (n = 3 per group). Scale bars: 200 µm. (H) Quantitative analysis results showing a significantly increased number of preovulatory follicles in GC-Pcgf2−/− (12 ± 2) female mice, compared with those in the controls (3 ± 1) at hCG 24 h (n = 3 per group). (I) Histological analysis showing oocyte-trapped-CL (arrow) in ovaries in GC-Pcgf2−/− female mice at hCG 48 h compared with those in the controls (n = 7 per group). Scale bars: 200 µm. (J) Quantitative analysis results showing a significantly increased number of oocyte-trapped CL in GC-Pcgf2−/− (6 ± 1) ovaries at hCG 48 h compared with those in the controls (1 ± 1; n = 7 per group). The experiments were repeated at least three times. The data in (B,D,F,H,J) represent the results (mean ± SD) of the biological triplicate experiments. n.s. p > 0.05, **p < 0.01, ***p < 0.001 by two-tailed unpaired Student’s t test.
The ovarian histological observation showed that the number of preovulatory follicles in GC-Pcgf2−/− ovaries (36 ± 13) were no significance with the controls (34 ± 7) at 46 h after PMSG treatment (PMSG 46 h) at PD21 (Figures 3C,D). Next, hCG induced effective meiotic resumption (Supplementary Figures S3A,B, arrowheads) and cumulus expansion in GC-Pcgf2−/− ovaries (Supplementary Figure S3A) at 8 h after hCG treatment (hCG 8 h). Nonetheless, most of the preovulatory follicles failed to rupture in GC-Pcgf2−/− ovaries at 16 h after hCG treatment, compared with CL formation in GC-Pcgf2+/+ ovaries (Figures 3E,F). We traced the fate of preovulatory follicles in GC-Pcgf2−/− ovaries and found that these follicles of GC-Pcgf2−/− did not release oocytes complex at 24 h after hCG injection (Figures 3G,H, arrowhead). Moreover, we counted the number of ovulated oocytes from oviducts at 24 h after hCG injection. Statistical analysis showed an identical number of ovulated oocytes at hCG 24 h compared to that at hCG 16 h in GC-Pcgf2−/− mice (Supplementary Figure S3C), indicating no more oocytes released after 16 h of hCG injection. Subsequently, we found oocyte-entrapped in the CLs (arrowheads) of GC-Pcgf2−/− ovaries at 48 h after hCG injection (Figures 3I,J; 6 ± 1 in GC-Pcgf2−/− vs. 1 ± 1 in GC-Pcgf2+/+), which were similar to the structures of oocyte-trapped CLs in GC-Pcgf2−/− ovaries at 6 and 10 months. These data demonstrated that most of the preovulatory follicle fails to rupture in the GC-Pcgf2−/− ovaries of superovulation mouse models, even though the processes of meiotic resumption and cumulus expansion were normal.
To investigate the mechanism of follicular rupture defects, we compared the morphology of GCs at the ovulated side (Ovu-side) of preovulatory follicles in GC-Pcgf2−/− and GC-Pcgf2+/+ ovaries at 12 h after hCG stimulation, when they began to ovulate oocytes and form corpus haemorrhagicum (Sekulovski et al., 2020). We found that only 2 ± 0 layers of GCs remained at the Ovu-side of preovulatory follicles in GC-Pcgf2+/+ovaries (Figures 4A,B, left). In contrast, 5 ± 1 layers of GCs existed at the Ovu-side of preovulatory follicles in GC-Pcgf2−/− ovaries (Figures 4A,B, right). This result showed that the loosening process of mural granulosa cells (mGCs) at the Ovu-side defected in GC-Pcgf2−/− female mice. We detected the distribution of gap junction protein connexin 43 (CX43) at the Ovu-side of preovulatory follicles at 12 h after hCG stimulation. In the preovulatory follicles of the controls, we observed almost no distribution of CX43 in the GCs of the Ovu-side (Figure 4C, left), indicating that the connection of GCs repressed before follicle ruptured. However, CX43 was highly expressed in the plasma membrane of GCs at the Ovu-side in GC-Pcgf2−/− mice compared with those in the controls (Figures 4C,D). These data suggested that GCs connect tightly with each other by gap junctions in GC-Pcgf2−/− ovaries, which results in the failure of follicular rupture.
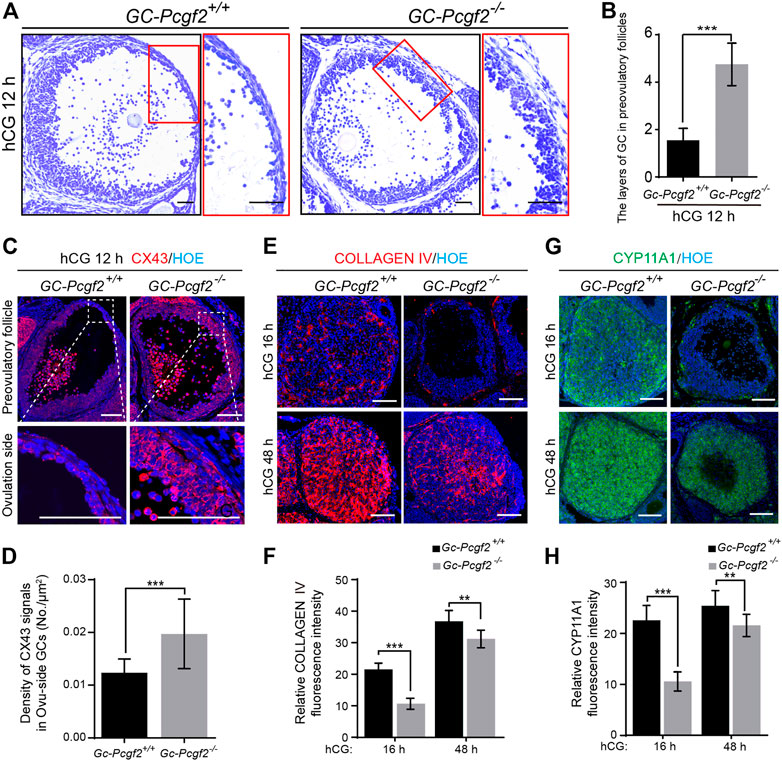
FIGURE 4. Deletion of Pcgf2 in granulosa cells leading to follicular rupture fail and corpus luteum formation delay. (A) Histological analysis showing a thicker GC layer in ovulated-side (Ovu-side) of GC-Pcgf2−/− preovulatory follicles at hCG 12 h. Scale bars: 50 µm. (B) Statistical analysis result showing more layers of GC in Ovu-side of GC-Pcgf2−/− (5 ± 1) compared with that of the controls (2 ± 0; n = 40 per group). (C) Immunostaining showing decreased density of CX43 signals in Ovu-side GCs of GC-Pcgf2−/− compared with that of the controls. Red: CX43; Blue: Hoechst. Scale bars: 100 µm. (D) Quantitative analysis result showing the decreased density of CX43 signals in Ovu-side GCs of GC-Pcgf2−/− compared with that of the controls (n = 27 per group). (E) Immunostaining analysis result showing decreased expression of COLLAGEN IV in CLs of GC-Pcgf2−/− compared with that of the controls. Red: COLLAGEN IV; Blue: Hoechst. Scale bars: 100 µm. (F) Quantitative analysis result showing decreased fluorescence intensity of COLLAGEN IV in CLs of GC-Pcgf2−/− compared with that of the controls (hCG 16 h: n = 9 per group; hCG 48 h: n = 7 per group). (G) Immunostaining analysis result showing decreased expression of CYP11A1 in CLs of GC-Pcgf2−/− compared with that of the controls. Green: CYP11A1; Blue: Hoechst. Scale bars: 100 µm. (H) Quantitative analysis result showing decreased fluorescence intensity of CYP11A1 in CLs of GC-Pcgf2−/− compared with that of the controls (hCG 16 h: n = 10 per group; hCG 48 h: n = 12 per group). The data in (B,D,F,H) represent the results (mean ± SD) of the biological triplicate experiments. **p < 0.01, and ***p < 0.001 by two-tailed unpaired Student’s t test.
Furthermore, we detected the subsequent luteinization process in GC-Pcgf2−/− and GC-Pcgf2+/+ovaries. The immunofluorescent staining of CL markers, COLLAGEN IV (Figures 4E,F), and Cytochrome P450 family 11 subfamily A member 1 (CYP11A1, Figures 4G,H) showed a significant decline of fluorescence intensity in GC-Pcgf2−/− ovaries, compared with GC-Pcgf2+/+ovaries at 16 h and 48 h after hCG treatment. These data showed that the luteinization process was delayed in GC-Pcgf2−/− ovaries. CYP11A1 is a member of the cytochrome P450 superfamily of enzymes that regulates the production of progesterone (P4), which is crucial for female fertility. Hormone detection showed significantly decreased concentrations of progesterone (P4) in GC-Pcgf2−/− female mice than in the controls both at 16 h and 48 h after hCG treatment (Supplementary Figure S4). These results indicate that Pcgf2 in GCs is essential for follicular rupture and luteinization.
To uncover the molecular mechanism of Pcgf2 mediating ovulation, we performed QRT-PCR to detect the mRNA expression dynamics of Pcgf2 in GCs at different times after hCG injection. The highest mRNA level of Pcgf2 in GCs was at 6 h after hCG injection (hCG 6 h) compared with hCG 0 h, 12 h, and 16 h (Figure 5A). This result implies Pcgf2 mainly functions at 6 h after hCG stimulation during the ovulation process.
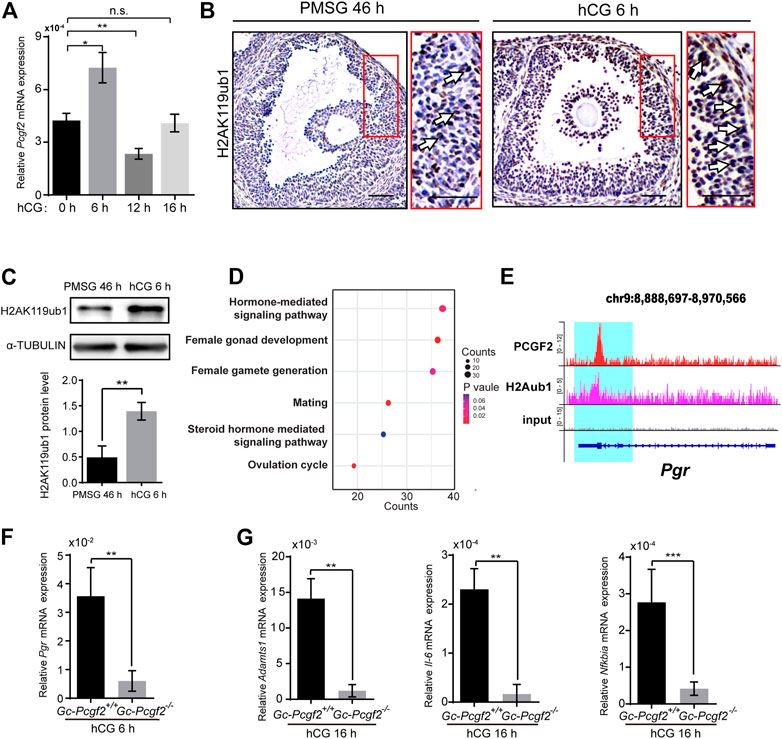
FIGURE 5. PCGF2 binding to the promoter of progesterone receptor and mediating the process of ovulation in mice. (A) QRT-PCR result showing a Pcgf2 mRNA expression peak at hCG 6 h. (B) The immunohistochemical histological analysis of H2AK119ub1 showing an increasing signal of H2AK119ub1 (arrows) in the GCs of preovulatory follicles at hCG 6 h compared with at PMSG 46 h. Scale bars: 50 µm. (C) Quantitative analysis of western blot showing a significantly increasing accumulation level of H2AK119ub1 in GCs at hCG 6 h compared with at PMSG 46 h. (D) The GO analysis of PCGF2 target genes showing that the target genes were associated with female fertility, including ovulation and hormone related pathways. (E) Genomic snapshot of the indicated ChIP-seq profiles of PCGF2 and H2Aub1 (H2AK119ub1) at gene loci of Pgr. (F) The statistical analysis of the QRT-PCR result showing a sharply decreasing of Pgr in GCs of GC-Pcgf2−/− at hCG 6 h compared with that in controls (n = 3 per group). (G) The statistical analysis of QRT-PCR results showing significantly decreasing the mRNA levels of Adamts1, Il-6, and Nfκbia in GCs of GC-Pcgf2−/− at hCG 16 h. The experiments were repeated at least three times. The data in (A,C,F,G) represent the results (mean ± SD) of the biological triplicate experiments. n.s. p > 0.05, *p < 0.05, **p < 0.01, and ***p < 0.001 by two-tailed unpaired Student’s t test.
Pcgf2 is required for stability of the PRC1 complex core which targeting at H2AK119ubl (Cao et al., 2005; Taherbhoy et al., 2015). Immunohistochemical staining showed significantly increased accumulation level of H2AK119ub1 in the GCs of preovulatory follicles at 6 h after hCG stimulation (hCG 6 h), compared with at 46 h after PMSG stimulation (PMSG 46 h) (Figure 5B, arrows). Western blot confirmed the sharply increased the level of H2AK119ub1 in GCs at hCG 6 h compared with PMSG 46 h (Figure 5C). These results imply that Pcgf2 may function in ovulation by targeting H2AK119ub1 epigenetic modification at 6 h after hCG stimulation.
To find potential target genes associated with the ovulation pathways of PCGF2, we analyzed the publicly available ChIP-seq data of PCGF2 using the GEO database (GEO: GSE122715) (Scelfo et al., 2019). Visualization of PCGF2 enrichment by ChIP-seq analysis showed that PCGF2 had unique target loci around ± 2.0 kb of the TSS of common (Supplementary Figures S5A,B). PCGF2 had an extensive binding repertoire with 4341 target genes (Supplementary Table S1). We next enriched these PCGF2 target genes in a gene ontology analysis and selected female reproduction biology pathways to screen ovulation-related genes (Figure 5D and Supplementary Table S2). PCGF2 target genes were enriched with GO terms including “reproductive structure development”, “positive regulation of hormone secretion”, and “ovulation cycle” (Figure 5D). Among the enriched PCGF2 target genes of the ovulation cycle biology pathway, we found one of the key ovulation genes that were reported to be the main mediator of follicular rupture, progesterone receptor (Pgr), which expression was induced by luteinizing hormone (LH) (Park et al., 2020) (Supplementary Table S2). Park et al. (2020) reported the phenotypes of ovulation failed and the oocyte remains within CL in GC-specific Pgr knockout mice, which was similar to the phenotype of GC-Pcgf2−/− mice. The call peak result of PCGF2 detected a binding peak (Figure 5E) and a specific binding motif (Supplementary Figure S5C) at the promoter of Pgr (+457) (Supplementary Table S1). These results imply that PCGF2 regulates the expression of Pgr to affect ovulation.
We then collected GCs of preovulatory follicles and detected the mRNA level of Pgr at 6 h after hCG stimulation. The QRT-PCR result showed the mRNA level of Pgr in GCs of GC-Pcgf2−/− unexpectedly sharply decreased compared with that in control mice (Figure 5F), which was inconsistent with PRC1 mediated gene silencing as previous studies reported. This result suggested that PCGF2 upregulated the expression of Pgr during the ovulation process through nonclassical PRC1 functions. Furthermore, we found that the mRNA level of Pgr downstream genes, including Adamts1 (Robker et al., 2000), Il-6 and Nfκbia (Park et al., 2020) significantly reduced at 16 h after hCG treatment (Figure 5G). These data uncover that PCGF2 mediates ovulation by transcriptional regulation of key ovulation gene Pgr in mice.
Ovulation is a complex process with multiple cell lineage responding to luteinizing hormone (LH) surge, including the resumption of oocyte meiosis, the expansion of cumulus oocyte complex, preovulatory follicle rupture, and release of COCs. Many studies focus on the molecular mechanism of meiotic resumption and COC expansion, but little is known about the regulation mechanism of follicle rupture (Richards and Ascoli, 2018). Our study demonstrated that PCGF2 is highly expressed in GCs and affects follicle rupture through loosening GCs at the Ovu-side of preovulatory follicles. Ovulated cell behaviors are dependent on tissue remodeling hormone receptor interactions and specifically timed gene expression in distinct type cells. We provide physiological evidence that PCGF2 affects follicular rupture under LH surge via modulating Pgr transcriptional expression level.
The LH surge induces the high expression of key ovulation gene Pgr in the mGCs of preovulatory follicles (Park et al., 2020). As a nuclear receptor transcription factor, PGR regulates ovulation, the development of mammary glands, and embryo implantation through tissue-specific transcriptionally regulating downstream genes (Lydon et al., 1995; Lee et al., 2015; Xin et al., 2018). Pgr deletion in GCs does not affect the expansion of COC and the fertilization ability of oocytes. PGR regulates follicle rupture through induction of ovulation target genes, such as Adamts1 (Doyle et al., 2004), Edn2 (Ko et al., 2006), IL-6 (Park et al., 2020), and Pparg (Kim et al., 2008). These are consistent with the phenotype of GC-Pcgf2−/− female mice. However, the transcriptional regulation mechanisms of Pgr expression in GCs induced by the LH surge are still not clear. We found that PCGF2 transcriptionally upregulates the expression of Pgr after hCG stimulation, rather than the classic function of PRC1 complex repressed gene expression. Moreover, the activation of Pgr promoter needs the interactions of the obligatory Sp1/Sp3 loci between the proximal promoter and the distal promoter by LH surge induction (Sriraman et al., 2003). Our ChIP analysis showed that PCGF2 and H2AK119ub1 enriched at the promoter region of PGR (+361/+437) between the proximal promoter and the distal promoter (Figure 5E, Supplementary Table S3). These data imply that the PCGF2 serve as a coregulator with LH mediated the transcription of Pgr in GCs by promoting Sp1/Sp3 interactions. However, Pgr gene expression was not completely suppressed in GC-Pcgf2−/− ovaries, perhaps because of the redundant functional properties of PCGF4 with PCGF2.
The ChIP-seq data showed that PCGF2 extensively binds to genes with accumulation of H2AK119ub1, suggesting that PCGF2 is involved in multiple functions of follicle development. We found that the reproductive capacity and follicle reserve of GC-Pcgf2−/− female mice decreased after 6 months compared to the controls. These results suggested that GC-Pcgf2−/− ovaries exhibited some symptoms of secondary POI after the age of 6 months. Previous studies discovered that the expression of aging-associated of genes such as Egfr, Egr2, Tmed2, and Ptgs2 in GCs of aged primate ovaries were increased compared with young GCs (Wang et al., 2020). Our ChIP-seq data supports that these genes to be target of PCGF2 (Supplementary Table S1). This result implies that PCGF2 represses silencing of these aging-associated genes’ expression in GCs for maintaining the reproductive capacity of ovaries.
In summary, our study provides systematic in vivo evidence that polycomb subunit Pcgf2 mediates ovulation and fertility through transcriptionally regulation progesterone receptor expression in female mice. Deletion of Pcgf2 in GCs affects the follicular rupture and leads to a series of defects in female fertility. These results remind us that the mutation of Pcgf2 may be a pathogenic factor of anovulation, such as luteinized unruptured follicle (LUF) syndrome. This study provides a potential target for finding and curing patients with anovulation in clinical treatment.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
The animal study was reviewed and approved by the Institutional Animal Care and Use Committee of China Agricultural University.
YW, HZ, and YZ designed the research; YW, WW, KC, KG, JL, PW, YZ, JZ, SN, LL, LJ, SZ, and XF performed the experiments; YW, WW, YZ, and HZ analyzed the data; YW, WW, CW, HW, HZ, and YZ wrote the paper. All authors have seen and approved the final version.
This study was supported by the National Key Research and Development Program of China to HZ and YZ (2022YFC2703800; 2018YFC1003700; 2018YFC1003800), the National Natural Science Foundation of China (81873815). This study was also supported by the Innovative Project of State Key Laboratory of Agrobiotechnology (2022SKLAB1-2 to HZ) and the 2115 Talent Development Program of China Agricultural University (1021-00109022 to YZ).
The authors are grateful to Fei Gao (Institute of Zoology, Chinese Academy of Sciences, Beijing, China) for kindly sharing the Foxl2-Cre mice. The authors also appreciate Rongwen Xi (Investigator, National Institute of Biological Sciences, Beijing, China) for providing the Pcgf2fl/fl mice.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2022.1010601/full#supplementary-material
Aranda, S., Mas, G., and Di Croce, L. (2015). Regulation of gene transcription by Polycomb proteins. Sci. Adv. 1 (11), e1500737. doi:10.1126/sciadv.1500737
Baumann, C., and De La Fuente, R. (2011). Role of polycomb group protein cbx2/m33 in meiosis onset and maintenance of chromosome stability in the Mammalian germline. Genes (Basel) 2 (1), 59–80. doi:10.3390/genes2010059
Cao, R., Tsukada, Y., and Zhang, Y. (2005). Role of bmi-1 and Ring1A in H2A ubiquitylation and hox gene silencing. Mol. Cell 20 (6), 845–854. doi:10.1016/j.molcel.2005.12.002
Cen, C., Chen, M., Zhou, J., Zhang, L., Duo, S., Jiang, L., et al. (2020). Inactivation of Wt1 causes pre-granulosa cell to steroidogenic cell transformation and defect of ovary development. Biol. Reprod. 103 (1), 60–69. doi:10.1093/biolre/ioaa042
Cora, M. C., Kooistra, L., and Travlos, G. (2015). Vaginal cytology of the laboratory rat and mouse: Review and criteria for the staging of the estrous cycle using stained vaginal smears. Toxicol. Pathol. 43 (6), 776–793. doi:10.1177/0192623315570339
Doyle, K. M., Russell, D. L., Sriraman, V., and Richards, J. S. (2004). Coordinate transcription of the ADAMTS-1 gene by luteinizing hormone and progesterone receptor. Mol. Endocrinol. 18 (10), 2463–2478. doi:10.1210/me.2003-0380
Duffy, D. M., Ko, C., Jo, M., Brannstrom, M., and Curry, T. E. (2019). Ovulation: Parallels with inflammatory processes. Endocr. Rev. 40 (2), 369–416. doi:10.1210/er.2018-00075
Entrevan, M., Schuettengruber, B., and Cavalli, G. (2016). Regulation of genome architecture and function by polycomb proteins. Trends Cell Biol. 26 (7), 511–525. doi:10.1016/j.tcb.2016.04.009
Eppig, J. J. (1991). Intercommunication between mammalian oocytes and companion somatic cells. Bioessays 13 (11), 569–574. doi:10.1002/bies.950131105
Fan, H. Y., Liu, Z., Shimada, M., Sterneck, E., Johnson, P. F., Hedrick, S. M., et al. (2009). MAPK3/1 (ERK1/2) in ovarian granulosa cells are essential for female fertility. Science 324 (5929), 938–941. doi:10.1126/science.1171396
Jiao, L., and Liu, X. (2015). Structural basis of histone H3K27 trimethylation by an active polycomb repressive complex 2. Science 350 (6258), aac4383. doi:10.1126/science.aac4383
Kim, J., Sato, M., Li, Q., Lydon, J. P., Demayo, F. J., Bagchi, I. C., et al. (2008). Peroxisome proliferator-activated receptor gamma is a target of progesterone regulation in the preovulatory follicles and controls ovulation in mice. Mol. Cell. Biol. 28 (5), 1770–1782. doi:10.1128/mcb.01556-07
Ko, C., Gieske, M. C., Al-Alem, L., Hahn, Y., Su, W., Gong, M. C., et al. (2006). Endothelin-2 in ovarian follicle rupture. Endocrinology 147 (4), 1770–1779. doi:10.1210/en.2005-1228
Lee, J. Y., Won, H. Y., Park, J. H., Kim, H. Y., Choi, H. J., Shin, D. H., et al. (2015). MEL-18 loss mediates estrogen receptor-α downregulation and hormone independence. J. Clin. . 125 (5), 1801–1814. doi:10.1172/jci73743
Liu, W., Xin, Q., Wang, X., Wang, S., Wang, H., Zhang, W., et al. (2017). Estrogen receptors in granulosa cells govern meiotic resumption of pre-ovulatory oocytes in mammals. Cell Death Dis. 8 (3), e2662. doi:10.1038/cddis.2017.82
Liu, X., Wei, W., Li, X., Shen, P., Ju, D., Wang, Z., et al. (2017). BMI1 and MEL18 promote colitis-associated cancer in mice via REG3B and STAT3. Gastroenterology 153 (6), 1607–1620. doi:10.1053/j.gastro.2017.07.044
Lydon, J. P., DeMayo, F. J., Funk, C. R., Mani, S. K., Hughes, A. R., Montgomery, C. A., et al. (1995). Mice lacking progesterone receptor exhibit pleiotropic reproductive abnormalities. Genes Dev. 9 (18), 2266–2278. doi:10.1101/gad.9.18.2266
Maezawa, S., Hasegawa, K., Yukawa, M., Sakashita, A., Alavattam, K. G., Andreassen, P. R., et al. (2017). Polycomb directs timely activation of germline genes in spermatogenesis. Genes Dev. 31 (16), 1693–1703. doi:10.1101/gad.302000.117
Morey, L., Santanach, A., Blanco, E., Aloia, L., Nora, E. P., Bruneau, B. G., et al. (2015). Polycomb regulates mesoderm cell fate-specification in embryonic stem cells through activation and repression mechanisms. Cell Stem Cell 17 (3), 300–315. doi:10.1016/j.stem.2015.08.009
Murdoch, W. J., and Cavender, J. L. (1989). Effect of indomethacin on the vascular architecture of preovulatory ovine follicles: Possible implication in the luteinized unruptured follicle syndrome. Fertil. Steril. 51 (1), 153–155. doi:10.1016/s0015-0282(16)60445-4
Park, C. J., Lin, P. C., Zhou, S., Barakat, R., Bashir, S. T., Choi, J. M., et al. (2020). Progesterone receptor serves the ovary as a trigger of ovulation and a terminator of inflammation. Cell Rep. 31 (2), 107496. doi:10.1016/j.celrep.2020.03.060
Park, J. Y., Su, Y. Q., Ariga, M., Law, E., Jin, S. L., and Conti, M. (2004). EGF-like growth factors as mediators of LH action in the ovulatory follicle. Science 303 (5658), 682–684. doi:10.1126/science.1092463
Piunti, A., and Shilatifard, A. (2016). Epigenetic balance of gene expression by Polycomb and COMPASS families. Science 352 (6290), aad9780. doi:10.1126/science.aad9780
Richards, J. S., and Ascoli, M. (2018). Endocrine, paracrine, and autocrine signaling pathways that regulate ovulation. Trends Endocrinol. Metab. 29 (5), 313–325. doi:10.1016/j.tem.2018.02.012
Robker, R. L., Russell, D. L., Espey, L. L., Lydon, J. P., O'Malley, B. W., and Richards, J. S. (2000). Progesterone-regulated genes in the ovulation process: ADAMTS-1 and cathepsin L proteases. Proc. Natl. Acad. Sci. U. S. A. 97 (9), 4689–4694. doi:10.1073/pnas.080073497
Scelfo, A., Fernández-Pérez, D., Tamburri, S., Zanotti, M., Lavarone, E., Soldi, M., et al. (2019). Functional landscape of PCGF proteins reveals both RING1A/B-Dependent-and RING1A/B-Independent-Specific activities. Mol. Cell 74 (5), 1037e1037–1052. doi:10.1016/j.molcel.2019.04.002
Scheuermann, J. C., de Ayala Alonso, A. G., Oktaba, K., Ly-Hartig, N., McGinty, R. K., Fraterman, S., et al. (2010). Histone H2A deubiquitinase activity of the Polycomb repressive complex PR-DUB. Nature 465 (7295), 243–247. doi:10.1038/nature08966
Schmidt, J., Weijdegård, B., Mikkelsen, A. L., Lindenberg, S., Nilsson, L., and Brännström, M. (2014). Differential expression of inflammation-related genes in the ovarian stroma and granulosa cells of PCOS women. Mol. Hum. Reprod. 20 (1), 49–58. doi:10.1093/molehr/gat051
Sekulovski, N., Whorton, A. E., Shi, M., Hayashi, K., and MacLean, J. A. (2020). Periovulatory insulin signaling is essential for ovulation, granulosa cell differentiation, and female fertility. FASEB J. 34 (2), 2376–2391. doi:10.1096/fj.201901791R
Shen, X., Liu, Y., Hsu, Y. J., Fujiwara, Y., Kim, J., Mao, X., et al. (2008). EZH1 mediates methylation on histone H3 lysine 27 and complements EZH2 in maintaining stem cell identity and executing pluripotency. Mol. Cell 32 (4), 491–502. doi:10.1016/j.molcel.2008.10.016
Sriraman, V., Sharma, S. C., and Richards, J. S. (2003). Transactivation of the progesterone receptor gene in granulosa cells: Evidence that sp1/sp3 binding sites in the proximal promoter play a key role in luteinizing hormone inducibility. Mol. Endocrinol. 17 (3), 436–449. doi:10.1210/me.2002-0252
Taherbhoy, A. M., Huang, O. W., and Cochran, A. G. (2015). BMI1-RING1B is an autoinhibited RING E3 ubiquitin ligase. Nat. Commun. 6, 7621. doi:10.1038/ncomms8621
Tang, X., Xu, S., Li, R., Zhang, H., Chen, Q., Wu, W., et al. (2016). Polycomb repressive complex 1 (PRC1) regulates meiotic initiation of ovarian germ cells in chick embryos. Mol. Cell. Endocrinol. 437 (5), 171–182. doi:10.1016/j.mce.2016.08.024
Tureck, R. W., and Strauss, J. F. (1982). Progesterone synthesis by luteinized human granulosa cells in culture: The role of de novo sterol synthesis and lipoprotein-carried sterol. J. Clin. Endocrinol. Metab. 54 (2), 367–373. doi:10.1210/jcem-54-2-367
Wang, H., Xie, H., Guo, Y., Zhang, H., Takahashi, T., Kingsley, P. J., et al. (2006). Fatty acid amide hydrolase deficiency limits early pregnancy events. J. Clin. . 116 (8), 2122–2131. doi:10.1172/jci28621
Wang, R., Xue, X., Wang, Y., Zhao, H., Zhang, Y., Wang, H., et al. (2019). BMI1 deficiency results in female infertility by activating p16/p19 signaling and increasing oxidative stress. Int. J. Biol. Sci. 15 (4), 870–881. doi:10.7150/ijbs.30488
Wang, S., Zheng, Y., Li, J., Yu, Y., Zhang, W., Song, M., et al. (2020). Single-cell transcriptomic atlas of primate ovarian aging. Cell 180 (3), 585–600. e519. doi:10.1016/j.cell.2020.01.009
Wu, R., Fujii, S., Ryan, N. K., Van der Hoek, K. H., Jasper, M. J., Sini, I., et al. (2007). Ovarian leukocyte distribution and cytokine/chemokine mRNA expression in follicular fluid cells in women with polycystic ovary syndrome. Hum. Reprod. 22 (2), 527–535. doi:10.1093/humrep/del371
Xin, Q., Kong, S., Yan, J., Qiu, J., He, B., Zhou, C., et al. (2018). Polycomb subunit BMI1 determines uterine progesterone responsiveness essential for normal embryo implantation. J. Clin. 128 (1), 175–189. doi:10.1172/jci92862
Yokobayashi, S., Liang, C. Y., Kohler, H., Nestorov, P., Liu, Z., Vidal, M., et al. (2013). PRC1 coordinates timing of sexual differentiation of female primordial germ cells. Nature 495 (7440), 236–240. doi:10.1038/nature11918
Keywords: PCGF2, granulosa cell, progesteron receptor, ovulation defect, subfertility
Citation: Wang Y, Wang W, Cheng K, Geng K, Liang J, Wang P, Zhang J, Niu S, Jia L, Zhang S, Li L, Feng X, Wang C, Wang H, Zhang H and Zhang Y (2022) Polycomb subunit Pcgf2 mediates ovulation and fertility through transcriptional regulation progesterone receptor. Front. Cell Dev. Biol. 10:1010601. doi: 10.3389/fcell.2022.1010601
Received: 03 August 2022; Accepted: 24 October 2022;
Published: 03 November 2022.
Edited by:
Kui Liu, The University of Hong Kong, ChinaReviewed by:
Zhenmin Lei, University of Louisville, United StatesCopyright © 2022 Wang, Wang, Cheng, Geng, Liang, Wang, Zhang, Niu, Jia, Zhang, Li, Feng, Wang, Wang, Zhang and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yan Zhang, eWFuemhhbmcxMDExQGNhdS5lZHUuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.