
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell Dev. Biol. , 14 October 2021
Sec. Developmental Epigenetics
Volume 9 - 2021 | https://doi.org/10.3389/fcell.2021.751154
This article is part of the Research Topic Monoallelic Expression in Development and Diseases View all 10 articles
Non-coding Xist RNA plays an essential role in X chromosome inactivation (XCI) in female mammals. It coats the X chromosome in cis and mediates the recruitment of many proteins involved in gene silencing and heterochromatinization. The molecular basis of how Xist RNA initiates chromosomal silencing and what proteins participate in this process has been extensively studied and elucidated. Its involvement in the establishment and maintenance of the X-inactivated state is, however, less understood. The XistIVS allele we previously reported is peculiar in that it can initiate XCI but fails to establish the inactive state that is stably maintained and, therefore, may provide an opportunity to explore how Xist RNA contributes to establish a robust heterochromatin state. Here we demonstrate that ectopic splicing taking place to produce XistIVS RNA disturbs its function to properly establish stable XCI state. This finding warrants the potential of XistIVS RNA to provide further insight into our understanding of how Xist RNA contributes to establish sustainable heterochromatin.
A subset of long non-coding RNA (lncRNA) associates with chromatin and regulates chromatin state. Xist RNA is one of the most extensively studied lncRNAs (Borsani et al., 1991; Brockdorff et al., 1991), which becomes upregulated from one of the two X chromosomes during the early development of female mammals (Kay et al., 1993) and induces X chromosome inactivation (XCI) to compensate for the dosage difference of X-linked genes between the sexes by associating with the X chromosome it originates from (Lyon, 1961; Clemson et al., 1996; Penny et al., 1996; Marahrens et al., 1997). In the mouse, XCI is imprinted in the extraembryonic lineages (Takagi and Sasaki, 1975), which contributes to the placenta and a part of the extraembryonic membranes, whereas it takes place in a random fashion to either X in the embryonic lineage, giving rise to whole tissues of the fetus (Monk and Harper, 1979). Xist RNA plays a pivotal role in both types of XCI by serving as a scaffold for recruiting many proteins involved in heterochromatin formation and maintenance.
Female mouse embryonic stem cells (ESCs) have been extensively used for the study of XCI as they carry two active X chromosomes, one of which undergoes chromosome-wide silencing with monoallelic upregulation of Xist and its association with the chromosome in cis upon induction of differentiation (Panning and Jaenisch, 1996). Functional domains of Xist RNA have been identified by studying ESCs introduced with a transgene expressing mutant Xist RNA with a deletion of various conserved domains among species. Deletion of the most proximal conserved repeat called the A-repeat not only totally compromises the silencing function of Xist RNA in the context of differentiating ESCs and developing embryos (Wutz et al., 2002; Sakata et al., 2017) but also abolishes an interaction of SPEN with Xist RNA (Chu et al., 2015; Moindrot et al., 2015; Monfort et al., 2015). A more recent study showed that the B-repeat interacts with hnRNPK to recruit PGF3/5-PRC1 to establish Polycomb-mediated chromosomal silencing (Pintacuda et al., 2017). Genetic screening and a more comprehensive proteomic analysis using ESCs have revealed many proteins that interact with Xist RNA (Chu et al., 2015; Moindrot et al., 2015; Pandya-Jones et al., 2020).
We previously reported a partial loss-of-function allele of Xist, XistIVS, which initiates XCI in the extraembryonic tissues upon paternal transmission, but the XCI state initiated by XistIVS is not stable, resulting in derepression of X-inactivated genes in the extraembryonic tissues (Hoki et al., 2011). The embryos carrying the paternally derived XistIVS eventually die at the midgestation stage, at least, partly due to malfunction of the placenta. XistIVS was generated by introducing the 0.9-kb second intron of human γ-globin gene (IVS2) 0.9 kb downstream of the major transcription start site of Xist as a control allele for the analysis of antisense regulation by Tsix (Ohhata et al., 2008). When this allele is transcribed, the introduced IVS2 sequence is spliced out to produce essentially the same transcript as that transcribed from the wild-type allele except for the presence of an additional 16-base insertion derived from the targeting vector at the splicing junction. Although it was unknown whether or not the presence of such small insertion affects the function of Xist RNA, it was no doubt that the XistIVS allele is not fully functional. Unlike other mutant alleles of Xist, XistIVS is peculiar in that its RNA product can initiate appreciable levels of XCI in the extraembryonic tissues, which supports early postimplantation development, but fail to establish the stable XCI state. The XistIVS allele would, therefore, provide a unique opportunity to understand the molecular basis of how Xist RNA contributes to establishing the heritable robust heterochromatin, which allows stable maintenance of the X-inactivated state essential for embryonic development and healthy life after birth.
In this study, we addressed how XistIVS behaves in the embryonic tissue, in which random XCI takes place, and the impact of the presence of the 16-base insertion in Xist RNA on its function. The results demonstrate that the XistIVS allele is not upregulated in the embryonic tissue in contrast to that in the extraembryonic tissues, suggesting some difference in the mechanisms for upregulation of Xist between the embryonic and extraembryonic tissues. In addition, forced expression of XistIVS RNA from the newly generated XistCAGIVS allele and another allele, XistCAG16in, which produces the transcript containing the same 16-base insertion at the same position as XistIVS RNA without splicing, in differentiating ESCs demonstrates that the 16-base insertion per se does not affect the function of Xist RNA, indicating that splicing in the former transcript to remove the IVS2 sequence brings the qualitative difference between these transcripts. We discuss the potential of the XistIVS allele, which could facilitate our further understanding of how Xist RNA contributes to establish robust heterochromatin state of the inactive X chromosome.
XistIVS and Xist1lox mice were described elsewhere (Sado et al., 2005; Hoki et al., 2011). The JF1 strain was maintained in-house and C57BL/6J strain purchased from Japan SLC, Inc. (Shizuoka, Japan). All mice were maintained and used in accordance with the Guidelines for the Care and Use of Laboratory Animals of Kindai University (KDAS-26-0006).
Embryonic stem cells used in this study were established from E3.5 blastocysts according to Ying et al. (2008). Blastocysts were cultured on four-well plates containing feeder cells with N2B27 medium supplemented with 1 μM of PD0325901 (Cayman Chemical, Ann Arbor, MI, United States) and 3 μM of CHIR99021 (Cayman) as well as 1,000 U/ml of LIF (Nacalai USA, San Diego, CA, United States). Outgrowths of undifferentiated cells were dissociated with TrypLE (Invitrogen, Carlsbad, CA, United States) and seeded on a four-well plate containing feeder cells. Many ESC colonies that appeared in several days were dissociated by TrypLE and seeded on a 35-mm dish containing feeder cells as passage 1 (P1). Blastocysts used for the establishment of ESCs homozygous for XistIVS were prepared from a cross between females heterozygous for XistIVS and males hemizygous for XistIVS. Those used for the establishment of F1 hybrid female ESCs were prepared from a cross between JF1 (Mus m. molossinus) females and C57BL/6J (B6) males. JB4, one of the established ESCs, was adapted to grow on a gelatin-coated dish with conventional ESC medium containing 2i (DMEM containing 15% FBS, 1× non-essential amino acids, 1× penicillin/streptomycin, 0.1 mM β-mercaptoethanol, 1,000 U/ml of LIF, 1 μM of PD0325901, 3 μM of CHIR99021) for some passages. For induction of differentiation, ESCs were cultured with N2B27 medium on gelatin-coated dishes for up to 7 days.
For construction of a targeting vector for Eif2s3x, a 938-bp fragment present downstream of a termination codon of Eif2s3x was amplified by PCR using primers, Eif2s3x-F (5′-actctgtaga caaggctggc-3′) and Eif2s3x-R (5′-TTCTGTAGGGAGAATTGG CC-3′), on B6 genomic DNA and cloned into pBluescriptII-SK(+), in which the SpeI site present in MCS had been destroyed. This plasmid was linearized at the SpeI site present in the Eif2s3x fragment, and an IRES neo cassette was subsequently cloned in an appropriate orientation to generate p3′EifIRESneo. For construction of a targeting vector for Hprt, a 763-bp fragment containing an ATG start codon of Hprt was amplified by PCR using primers, gHprt-F1 (5′-agacgacagagggcctgggggctgc-3′) and gHprt-R1 (5′-ttgtagagctgggcctctcccagga-3′), on JF1 genomic DNA. This plasmid was used for inverse PCR using primers Hprt-invR(Spe) (5′-gggaaacttactagtCGGCAAAAAGC GGTCTGAGGAGGAAGC-3′) and Hprt-invF(Pst) (5′-gggaaact tctgcagCGACCCGCAGTCCCAGCGTCGTGgtga-3′), and the amplified fragment was circulized by self-ligation. The resultant plasmid was digested by PstI and SpeI and used to clone a CAG-Zeo-pA cassette to produce pHprt_invF/R-Zeo.
For construction of a targeting vector to generate the XistCAGIVS2lox allele, a 0.9-kb IVS fragment amplified on genomic DNA carrying the XistIVS allele by PCR according to Ohhata et al. (2008) was cloned into a targeting vector, pCAG-CΔM20, which was used for replacing the endogenous Xist promoter with the CAG promoter (Amakawa et al., 2015), to generate pCAG-CΔM20-IVS#5. For construction of a targeting vector to generate the XistCAG16in2lox allele, an extra 16-base double-stranded DNA XistIVS RNA was introduced in pCAG-CΔM20 at the unique XhoI site to generate pCAG16in#3.
A 20-bp double-stranded DNA fragment corresponding to the respective specific guide RNA sequence was cloned into pX330 (Add genes) linearized by BbsI to generate pX330-EifgRNA1, pHprt-gRNA, and pX330-Xist(-20). sgRNA sequences are as follows: Eif2s3x (5′-ATTTATAGCTGCTACTAGTA-3′), Hprt (5′-TGACGGGACCGGTCGGCTCG-3′), and Xist (5′-GATCAGTTAAAGGCGTGCAA-3′).
The Eif2s3x locus, which is known to escape XCI, and the Hprt locus were selected for the site to introduce an IRESneo cassette and a CAGzeo-pA cassette, respectively. JB4 ESCs were used at P8 to introduce an IRESneo cassette at the Eif2s3x locus, and 5 × 105 cells were transfected with 1 μg of pX330-EifgRNA1 and 1 μg of p3′EifIRESneo using FuGENE HD Transfection Reagent (Promega, Madison, WI, United States) and seeded on two gelatin-coated 60-mm dishes. Twenty-four hours later, selection was started with conventional ES medium containing 2i/LIF and 200 μg/ml of G418 for 8 days. Of the 40 colonies isolated, 12 turned out to harbor expected homologous recombination at one of the two Eif2s3x alleles. One of the 12 clones, termed JB4/EI7, which were confirmed to have an IRESneo cassette integrated on the X chromosome derived from B6, was used for targeting a CAGzeo-pA cassette at the Hprt locus on the X chromosome derived from JF1. Five hundred thousand cells were transfected with 1 μg of pHprt-gRNA and 1 μg of pHprt_invF/R-Zeo, as above. Selection was carried out using conventional ES medium containing 2i/LIF and 25 μg/ml of Zeocin for 10 days. Of the 40 colonies screened by PCR, one was found to harbor a correct homologous recombination at the Hprt locus on the X chromosome derived from JF1 and termed JB4/EI7HZ2. JB4/EI7HZ2 retained two X chromosomes in 99% of the population after 70 days of culture in the presence of G418 and Zeocin.
Five hundred thousand JB4/EI7HZ2 cells were transfected with 1 μg of pX330-Xist(-20) and 1 μg of either pCAG-CΔM20-IVS#5 or pCAG16in#3, as above. Selection was carried out using conventional ES medium containing 2i/LIF and 2 μg/ml of puromycin for 7–10 days. Twelve of 48 and 24 of 96 colonies isolated turned out to harbor XistCAGIVS2lox and XistCAG16in2lox, respectively. To identify which of the B6 or JF1 alleles was targeted, a relevant region containing a single-nucleotide polymorphism (SNP) was amplified by PCR and analyzed by restriction digestion and sequencing. A floxed PacECFP-pA cassette was removed by transient expression of Cre recombinase to convert XistCAGIVS2lox and XistCAG16in2lox into XistCAGIVS and XistCAG16in, respectively.
RNA-FISH was performed using cells fixed with either 4% paraformaldehyde (PFA) or Carnoy’s fixative as previously described (Sado et al., 2001). For PFA fixation, cells grown on a coverslip were fixed with 4% PFA and subsequently permeabilized with 0.5 % Triton X-100/0.5% BSA/PBS for 30 min and dehydrated through 70 and 100% ethanol. Xist and Atrx probes were prepared using pXist_SS12.9 and a BAC clone P23-260I15, respectively, as previously described (Sakata et al., 2017).
Whole-mount RNA-FISH was carried out according to Shiura et al. (2018). Briefly, embryos were first permeabilized in 0.1% Triton X-100/PBS for 10 s and fixed in 4% paraformaldehyde/PBS containing 0.1% Triton X-100 for 10 min. Following incubation in 2xSCC/0.05% Tween 20, 2xSSC/25% formamide/0.05% Tween 20, and 2xSSC/50% formamide/0.05% Tween 20 for 10 min each, the embryos were hybridized with Xist and Atrx probes overnight at 37°C, followed by washes for 5 min twice at 37°C in each of 2xSSC/50% formamide and 2xSSC/0.05% Tween 20 and subsequent counterstaining with Hoechst 33258.
For allele-specific RNA-FISH for Xist, probes were prepared according to Harris et al. (2019). Briefly, five B6- and JF1-specific oligonucleotides, the sequences of which were exactly the same as those designed by Harris et al. (2019) and contained an SNP between B6 and JF1, were labeled with Cy5 and Cy3, respectively, at their 3′-end. Five mask oligonucleotides complementary to the common 3′ part of the labeled oligonucleotides were also prepared. These Cy5-labeled B6-specific oligonucleotides, Cy3-labeled JF1-specific oligonucleotides, and mask oligonucleotides were included in hybridization buffer (2xSSC/10% dextran sulfate/2 mg/ml BSA/25% formamide) at a concentration of 5 μM for labeled and 10 μM for mask oligonucleotides. In the hybridization reaction, a Green-dUTP-labeled strand-specific probe for Xist prepared according to Shiura et al. (2018) was also included to validate the hybridization signal produced by Cy5- and Cy3-labeled oligonucleotides. Washing was carried out in 2xSSC/10% formamide for 30 min at 42°C twice and in 2xSSC for 5 min at room temperature.
cDNA synthesis was carried out using random hexamer on 1 μg of total RNA treated with DNaseI in the presence or absence of SuperScript III (Invitrogen). One-fiftieth of the reaction was used as a template for PCR using a primer set, R700P2, and F1063AS (Sado et al., 2005). The PCR product was subsequently digested with XhoI, whose restriction site is present in the fragment derived from wild-type Xist RNA but destroyed in that derived from XistIVS and Xist16in (Hoki et al., 2011).
To investigate the effect of XistIVS on XCI in the embryonic lineage, we established wild-type and XistIVS/XistIVS ESCs from blastocysts obtained from a cross between XistIVS/+ females and XistIVS/Y males. They were induced to differentiate for 5 days and examined for Xist expression by RNA-FISH. While Xist was monoallelically upregulated in about 40% of wild-type cells, the majority of XistIVS/XistIVS cells exhibited two pinpoint signals and none contained the Xist cloud (Supplementary Figure 1). Extension of differentiation did not change the situation. This raised a possibility that although the XistIVS allele induced XCI by coating the X chromosome in the extraembryonic lineages of the embryo, it was not upregulated in the embryonic lineage. To make a more direct assessment of this issue, we set out to examine whether or not the XistIVS allele became upregulated in the epiblast of the postimplantation embryo. We crossed females heterozygous for Xist1lox with males hemizygous for XistIVS, and embryos were recovered at embryonic day (E) 6.5 for whole-mount RNA-FISH. Since the Xist1lox allele is functionally null and does not form the Xist cloud (Sado et al., 2005), RNA-FISH allowed us to evaluate the ability of the XistIVS allele to be upregulated in Xist1lox/XistIVS embryos. These embryos were morphologically reminiscent of previously reported XistIVS/XistIVS embryos (Hoki et al., 2011), in which the epiblast is diminished, and therefore, they were differentiated from morphologically normal +/XistIVS female embryos, in which wild-type Xist is uniformly upregulated, and male embryos (Xist1lox/Y or +/Y) by visual inspection. Figure 1 shows the distal part of +/XistIVS and Xist1lox/XistIVS embryos examined at E6.5 for the expression of Xist and another X-linked Atrx gene by whole-mount RNA-FISH. In the +/XistIVS embryo, the Xist cloud was detected in the epiblast as well as the visceral endoderm layer surrounding the epiblast. Atrx was monoallelically expressed in these tissues, suggesting that one Atrx allele was silenced by XCI. In contrast, the Xist cloud was detected in the visceral endoderm but not in the epiblast of Xist1lox/XistIVS embryos. A pinpoint Xist signal detected was, however, often juxtaposed to a pinpoint signal of Atrx. Since even if RNA was transcribed from the Xist1lox allele, it was truncated due to the insertion of an IRESEGFP-pA cassette in exon1 and would not be detected by the Xist probe used, and the pinpoint signals detected by the Xist probe should represent RNA transcribed from the XistIVS allele, suggesting a defect of the XistIVS allele to undergo upregulation. It was, therefore, likely that XCI was not induced in the epiblast of Xist1lox/XistIVS embryos, resulting in developmental failure of the epiblast. Taken together with the result in female ESCs homozygous for XistIVS, we concluded that the XistIVS allele was defective in upregulating its transcription in the epiblast lineage. This contrasts with the fact that the XistIVS allele is upregulated and capable of inducing XCI in the extraembryonic lineages and suggests some differences in the mechanism of Xist upregulation between the embryonic and extraembryonic lineages.
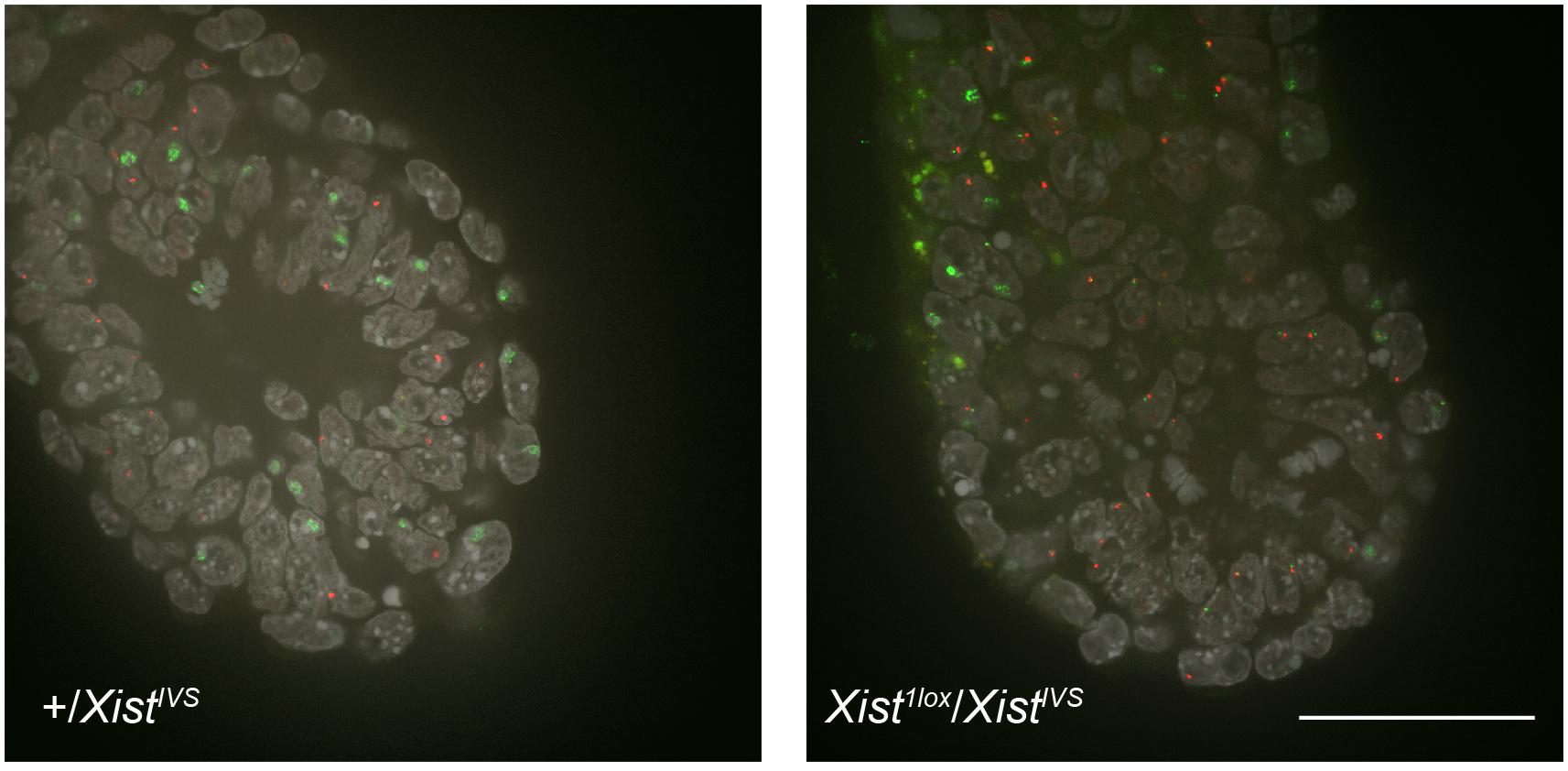
Figure 1. Whole-mount RNA-FISH in the distal part of E6.5 embryos. Expression of Xist (green) and Atrx (red) was examined in +/XistIVS and Xist1lox/XistIVS embryos recovered on E6.5. Six confocal sections of a 0.2-μm interval obtained by confocal microscopy were projected into a single image. epi, epiblast; ve, vesceral endoderm. Scale bar = 100 um.
To circumvent the defect of the XistIVS allele to be upregulated, we attempted to replace the endogenous Xist promoter with the CAG promoter (Niwa et al., 1991) in F1 hybrid female ESCs between JF1 and B6 by homologous recombination with the aid of CRIPSR/Cas9 genome editing (Supplementary Figure 2). ESCs used here carried the neomycin resistance gene on the X chromosome derived from B6 (XB6) and the Zeocin resistance gene on the X derived from JF1 (XJF1) and, therefore, stably maintained both X chromosomes in the presence of G418 and Zeocin in culture medium (see Materials and methods). We isolated several lines harboring the XistCAGIVS2lox allele on either X chromosome and chose two lines, one targeted on the XB6 (IVS-2L-B47) and the other on the XJF1 (IVS-2L-J19), for further analyses. A floxed PacECFPpA cassette was removed by transient expression of Cre recombinase in IVS-2L-B47 and IVS-2L-J19 lines to obtain IVS-B47#24 and IVS-J19#3, respectively, in which the XistCAGIVS2lox allele was converted into the XistCAGIVS allele.
These ESC lines thus generated carrying either XistCAGIVS2lox (IVS-2L-B47 and IVS-2L-J19) or XistCAGIVS (IVS-B47#24 and IVS-J19#3) were allowed to differentiate for up to 7 days in N2B27 medium without G418 and Zeocin, and expression of Xist and X-linked Atrx was examined by RNA-FISH. Given our previous studies of the targeted Xist alleles generated in a similar scheme, XistCAG2L and XistCAGΔ5′–2L (Amakawa et al., 2015; Sakata et al., 2017), it was reasonable to expect that the XistCAGIVS2L allele would behave as a functionally null allele and the other wild-type allele would be upregulated upon differentiation. In IVS-2L-B47 and IVS-2L-J19 cells, the proportion of cells containing the Xist cloud increased over time to reach 65–70% of the population at day 7 (d7) (Figures 2A,B). Expression of Atrx was detected as a single pinpoint signal, which did not overlap with the Xist cloud, suggesting that Atrx on the Xist RNA-coated X was silenced. In IVS-B47#24 and IVS-J19#3 cells, although the XistCAGIVS allele was driven by the CAG promoter, it was not upregulated prior to differentiation as previously reported (Amakawa et al., 2015). Following differentiation, although gradual upregulation of Xist was similarly observed in IVS-B47#24 and IVS-J19#3 cells over time, a large fraction of cells with a single Xist cloud expressed Atrx biallelically with expression of one allele overlapping with the Xist cloud at d5 and d7. In the remaining fraction of cells with a single Xist cloud, Atrx was monoallelically expressed, and its signal did not overlap with the Xist cloud. In addition, there was a gradual increase of cells that contained two Xist clouds, one of which overlapped with an Atrx signal. Such cells represented ∼30% of the population by d7. These results suggested that one of the two Xist clouds detected in a fraction of differentiating IVS-B47#24 and IVS-J19#3 cells allowed misexpression of Atrx and, therefore, was defective in silencing the X chromosome.
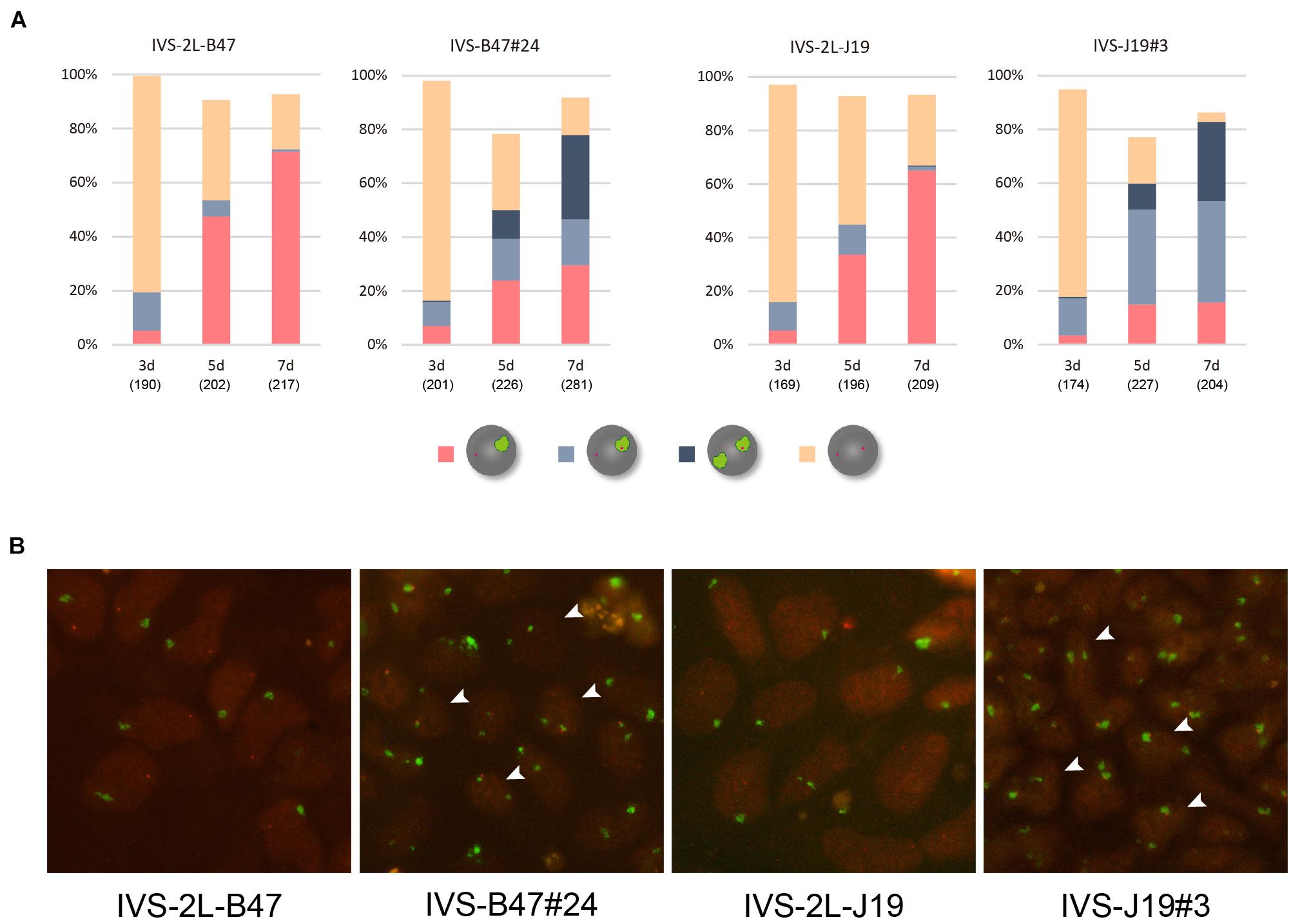
Figure 2. RNA-FISH in differentiating ESCs heterozygous for either XistCAGIVS2lox or XistCAGIVS. (A) Expression of Xist and Atrx was examined in respective differentiating ESCs for day 3 (d3) up to day 7 (d7). IVS-2L-B47 and IVS-B47#24 carry the targeted Xist allele on the XB6; IVS-2L-J19 and IVS-J19#3 on the XJF1. The number of nuclei examined is indicated in parentheses under each day point. (B) Representative images of RNA-FISH in each ESC line on d7. Xist in green and Atrx in red. Arrowhead indicates some of the nuclei containing two Xist clouds, one of which juxtaposes an Atrx signal.
To identify the allelic origin of Xist RNA forming the Xist cloud in differentiating female ESCs, we employed allele-specific RNA-FISH using labeled B6- and JF1-specific oligo probes that differentially hybridized to Xist RNA derived from the B6 and JF allele, respectively (Harris et al., 2019). This confirmed that only the wild-type Xist allele expressed from the XJF1 and XB6 formed the cloud in IVS-2L-B47 and IVS-2L-J19 cells, respectively, at day 7 of differentiation.
Subsequently performed allele-specific Xist RNA-FISH revealed that Xist RNA forming the cloud in IVS-B47#24 cells was unexpectedly biased toward the RNA occurring from the wild-type allele on the XJF1 rather than the one from the XistCAGIVS allele on the XB6 on d3 (Figures 3A,B). Although this bias was more pronounced on d5, two Xist clouds detected in a subset of cells were indeed originated from the wild-type and XistCAGIVS alleles on the XB6 and XJF1, respectively. The population of cells containing two Xist clouds increased from d5 to d7 with a decrease in the proportion of cells containing the single cloud of wild-type Xist on the XJF1. This suggested that the XistCAGIVS allele became upregulated in a subset of cells, which had undergone differentiation and initiated XCI via upregulation of wild-type Xist on the XJF1, in IVS-B47#24 cells.
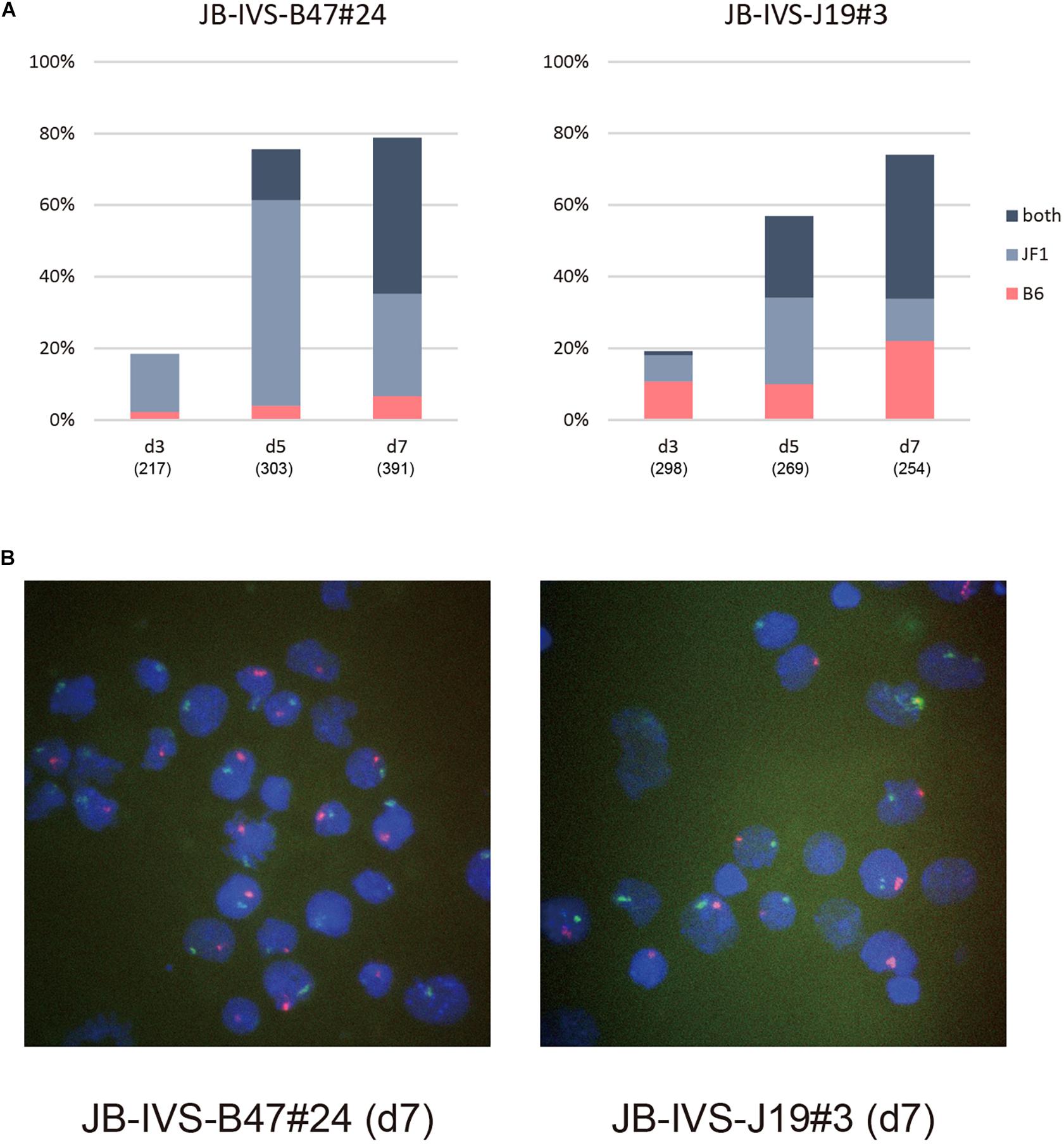
Figure 3. Allele-specific RNA-FISH for Xist in IVS-B47#24 and IVS-J19#3. (A) Proportion of nuclei harboring the Xist cloud of the XB6 or XJF1 in origin or both in respective differentiating ESCs for d3 up to d7. IVS-B47#24 carries XistCAGIVS on the XB6 and IVS-J19#3 on the XJF1. (B) Representative images of allele-specific RNA-FISH for Xist in each ESC line on d7. Xist RNA originated from the XB6 and XJF1 is shown in red and green, respectively.
The kinetics of allelic expression of Xist in IVS-J19#3 cells was different. Cells that had initiated XCI by d3 seemed to have selected either the wild-type allele on the XB6 or the XistCAGIVS allele on the XJF1 for upregulation in a random fashion (Figures 3A,B). Those that upregulated the XistCAGIVS allele, however, had dominated in the population carrying the single Xist cloud by d5. As was the case in IVS-B47#24 cells, the proportion of these cells subsequently decreased from d5 to d7 with an increase of those containing two Xist clouds, which were derived from the wild-type and XistCAGIVS allele on the XB6 and XJF1, respectively. This suggested that the wild-type Xist became upregulated in a subset of cells, which had upregulated the XistCAGIVS first during differentiation.
Although we expected that the XistCAGIVS allele driven by the CAG promoter was preferentially upregulated upon differentiation, allele-specific RNA-FISH suggested that it was not necessarily the case and either allele could be chosen for upregulation when XCI initiated. Although two Xist clouds were subsequently formed in both IVS-B47#24 and IVS-J19#3 cells, the kinetics to become the two cloud states appeared different. Nonetheless, allele-specific RNA-FISH confirmed that the two Xist clouds were indeed formed by wild-type Xist RNA and XistIVS RNA derived from the XistCAGIVS allele. It was most likely that XistIVS RNA was the one forming the cloud defective in Atrx silencing, and therefore, it was functionally compromised.
XistIVS RNA inevitably contains an additional 16-base insertion derived from the targeting vector after splicing of the IVS2 sequence (Ohhata et al., 2008). To address the impact of the presence of the 16-base insertion on the function of Xist RNA, we generated another female ESCs harboring the XistCAG16in allele, which produced Xist16in RNA containing exactly the same 16-base insertion at the same position without splicing under the control of the CAG promoter (Supplementary Figure 3). The ESC lines thus generated, 16in-B11#2 and 16in-B38#2, which carried the targeted allele on the XB6 (those carrying the targeted allele on the XJF1 were not recovered), and their parental line containing a floxed PacECFPpA cassette, 16in-2L-B11 and 16in-2L-B38, respectively, were allowed to differentiate in N2B27 medium and examined for Xist and Atrx expression by RNA-FISH. In all cases, the proportion of cells containing the Xist cloud gradually increased over time (Figure 4) and Xist RNA forming a single cloud essentially silenced Atrx as few overlaps were observed. This suggested that Xist16in RNA was capable of inducing and establishing the stably silenced state of the X chromosome. In contrast to IVS-B47#24 and IVS-J19#3 cells, only a minor population of the cells showed two Xist clouds in 16in-B11#2 and 16in-B38#2.
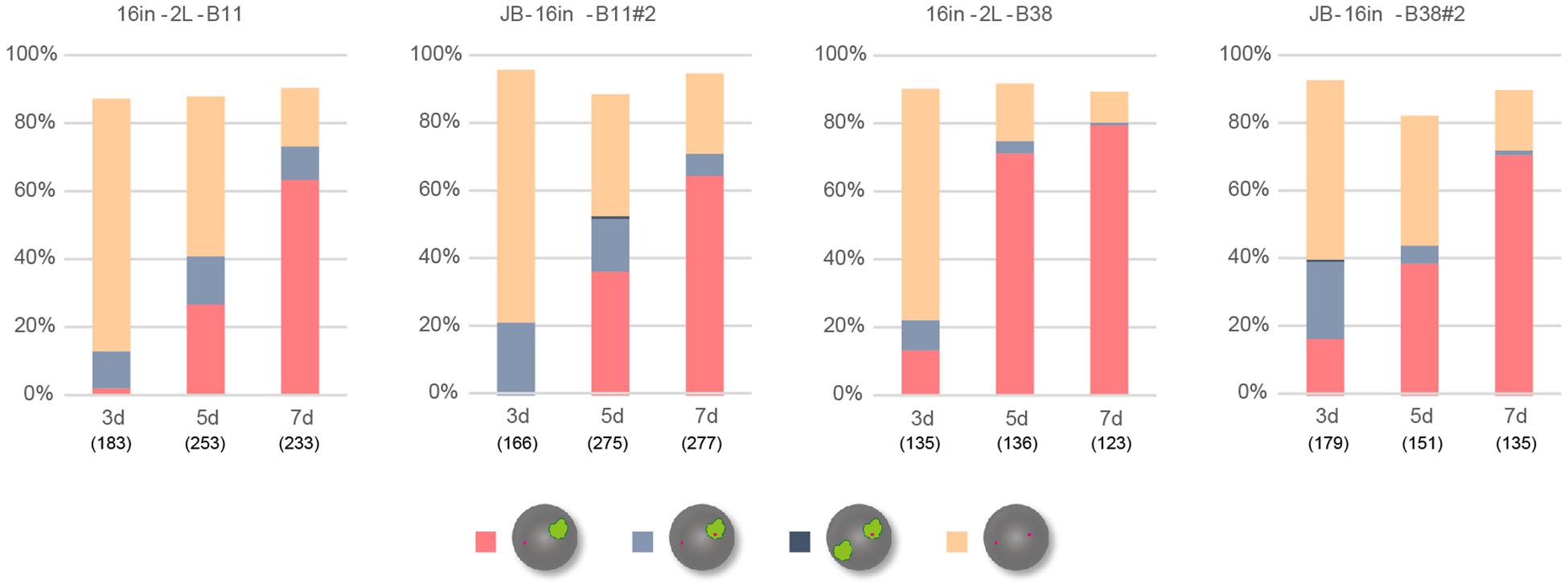
Figure 4. RNA-FISH in differentiating ESCs heterozygous for either XistCAG16in2lox or XistCAG16in. Expression of Xist and Atrx was examined in respective differentiating ESCs for day 3 (d3) up to day 7 (d7). Each cell line examined (16in-2L-B11, 16in-B11#2, 16in-2L-B38, and 16in-B38#2) carried the targeted Xist allele on the XB6. The number of nuclei examined is indicated in parentheses under each day point.
Allele-specific RNA-FISH for Xist revealed that the RNA forming the majority of the cloud derived from XB6, on which the Xist16in allele had been introduced, in 16in-B38#2 on d7 (Figures 5A,B). RT-PCR and subsequent restriction digestion of the amplified fragment further confirmed the expression of Xist16in RNA in 16in-B11#2 and 16in-B38#2 (Figure 5C). The same analysis also demonstrated the production of not only the expected spliced product from the XistCAGIVS allele but also wild-type Xist RNA in IVS-B47#24 and IVS-J19#3 (Figure 5C), consistent with allele-specific RNA-FISH for Xist, shown in Figure 3. These results indicated that Xist16in RNA was the one that formed the cloud stably silencing Atrx in 16in-B38#2 cells. Xist16in RNA was therefore competent to induce stable XCI even though it contains exactly the same 16-base insertion at the same position as XistIVS RNA. It is, therefore, reasonable to conclude that the presence of the 16-base insertion per se does not compromise the function of Xist RNA, and it is splicing that deteriorates the function of the RNA expressed from the XistCAGIVS allele.
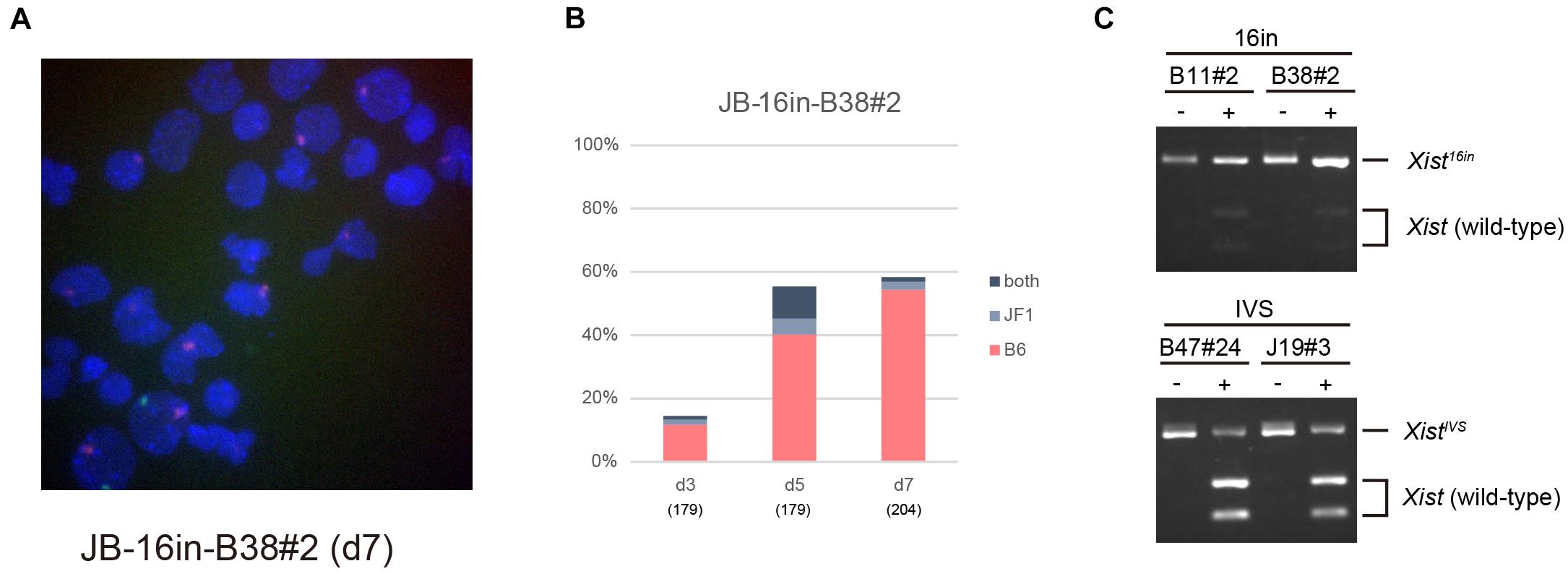
Figure 5. Xist16in RNA induces stable XCI. (A) Representative images of allele-specific RNA-FISH for Xist in 16in-B38#2 differentiated for 7 days. Xist RNA originated from the XB6 and XJF1 is shown in red and green, respectively. (B) Proportion of nuclei harboring the Xist cloud of the XB6 or XJF1 in origin or both in differentiating 16in-B38#2 ESCs for d3 up to d7. (C) Expression of Xist16in and XistIVS RNA in differentiating ESCs heterozygous for the Xist16in and XistCAGIVS allele, respectively. The unique XhoI site present in the amplified fragment derived from wild-type Xist RNA was destroyed by the presence of the 16-base insertion in the amplified fragment derived from Xist16in and XistIVS RNA. The presence of an undigested band by XhoI indicates the expression of Xist16in and XistIVS RNA. (–) and (+) indicate undigested and digested, respectively.
We previously reported that although XistIVS RNA expressed in the extraembryonic tissues could not establish a stable XCI state, it could support the early postimplantation development by inducing appreciable levels of XCI (Hoki et al., 2011). We also described that the Xist cloud was detected in 70% of the nuclei but absent in the remaining 30% in the distal part of E7.5 XistIVS/XistIVS embryos dissociated by treatment with lactic acid. This led us to speculate that although the XistIVS allele was initially monoallelically upregulated, the transcripts coating one of the two Xs was lost over time in the embryonic tissue. However, the relative abundance of the epiblast cells in the distal part of an E7.5 embryo, which consists of the epiblast and visceral endoderm, would be much lower in XistIVS/XistIVS embryos characterized by the small epiblast that fails to expand than in the morphologically normal wild-type embryos. Inevitable contamination of a relatively large proportion of the visceral endoderm in the mutant embryos might have led us to misinterpretation. In this study, however, whole-mount RNA-FISH using E6.5 compound heterozygotes, Xist1lox/XistIVS, unequivocally demonstrated that the upregulation of the XistIVS allele was confined only to the visceral endoderm and no Xist cloud was formed in the epiblast in the distal part of the embryo. This finding together with RNA-FISH analysis of ESCs homozygous for XistIVS indicated that XistIVS was not upregulated in the embryonic tissue. The differential behavior of the XistIVS allele between the embryonic and extraembryonic tissues is most probably related to the mode of XCI, that is, imprinted or random. While in the tissues that undergo imprinted XCI, Tsix, an antisense RNA of Xist, is expressed on the maternal X but not on the paternal X, it is biallelically expressed prior to upregulation of Xist in undifferentiated epiblast cells and ESCs, which undergo random XCI as cells differentiate (Lee, 2000; Sado et al., 2001). Given that Tsix negatively regulates Xist through modification of chromatin structure (Navarro et al., 2005; Sado et al., 2005), it is possible that the presence or absence of Tsix RNA or its transcription affects the potential of the XistIVS allele to be upregulated in response to cellular differentiation. Tsix expression is downregulated on one of the two Xs in the epiblast or ESCs during differentiation, and Xist becomes upregulated on that X to induce XCI (Lee et al., 1999). A series of these events may be compromised on the X chromosome carrying the XistIVS allele in the epiblast cells or ESCs. In the extraembryonic tissues, on the other hand, since Tsix is imprinted not to be expressed on the paternal X, there would not be any influence of Tsix on the paternal XistIVS allele, allowing its upregulation.
We expected that when female ESCs heterozygous for XistCAGIVS were allowed to differentiate, the XistCAGIVS allele would become preferentially upregulated and its transcript, XistIVS RNA, would coat the X chromosome. Allele-specific RNA-FISH, however, revealed that this was not the case and either the wild-type or the XistCAGIVS allele could be monoallelically upregulated at the onset of XCI. Although we had to admit that the XistCAGIVS allele behaved a little different way from the one that we expected during the early phase of differentiation, still we could express the XistIVS RNA in differentiating ESCs as a result of the formation of two Xist clouds in both cell lines, IVS-B47#24 and IVS-J19#3. We speculate that in IVS-B47#24, although the wild-type Xist allele on the XJF1 was initially preferentially upregulated upon initiation of XCI, the subsequent differentiated state of the cells allowed them to activate the CAG promoter, which barely drives transcription at the Xist locus in undifferentiated state, to express XistIVS RNA from the XistCAGIVS allele. In IVS-J19#3, on the other hand, one of either allele of Xist was selected for upregulation at the onset of XCI; the wild-type Xist allele appeared secondarily activated in those that upregulated the XistCAGIVS allele first to compensate for the insufficient XCI initiated by XistIVS RNA. It is likely that those that upregulated the wild-type Xist first in the population of IVS-J19#3 formed two Xist clouds in the same manner as IVS-B47#24. Whatever the reason for the formation of two Xist clouds is, upregulation of the XistCAGIVS allele allowed us to evaluate the function of XistIVS RNA in the embryonic lineage. Since one of the two Xist clouds seemed to be defective in silencing Atrx, it was most likely that XistIVS RNA produced from the XistCAGIVS allele was dysfunctional and failed to establish the proper XCI state. In contrast, Xist16in RNA containing the same 16-base insertion at the same position as XistIVS RNA was indistinguishable from wild-type Xist RNA in terms of the kinetics of Atrx silencing. This strongly suggests that it is not the 16-base insertion per se that compromises the function of XistIVS RNA. The difference between XistIVS RNA and Xist16in RNA is whether or not the RNA undergoes splicing to remove the IVS2 sequence introduced 0.9 kb downstream of the major transcription start site of Xist. When an intron is removed and two exons are connected by splicing, many splicing-related proteins such as an exon junction complex (EJC) bind in the vicinity of the exon-exon junction. It is, therefore, reasonable to expect that such proteins would bind to the processed XistIVS RNA but not to Xist16in RNA. The IVS2 sequence was located about 0.2 kb downstream of the A-repeat, which is essential for the silencing function of Xist RNA (Wutz et al., 2002) and mediates binding of some important proteins required for XCI such as SPEN (Chu et al., 2015; McHugh et al., 2015; Moindrot et al., 2015; Monfort et al., 2015) and RBM15 (Patil et al., 2016). We are tempted to speculate that binding of these factors to Xist RNA is disturbed by the presence of EJC and other factors brought on XistIVS RNA as a result of splicing to remove the IVS2 sequence. It would be therefore particularly interesting to compare the proteins assembled on XistIVS RNA with those on the wild-type Xist RNA. We cannot, however, exclude the possibilities that the inefficient silencing associated with XistIVS RNA could result from the efficiency of the expected splicing event on the premature XistIVS RNA or the difference of the overall levels of wild-type Xist, XistIVS, and Xist16in RNA. The fact that differentiating ESCs carrying the XistCAGIVS allele give rise to the populations with biallelic expression of Xist (wild-type and XistIVS) as well as monoallelic expression of either allele makes it difficult to compare the quantity of the RNA produced from the respective wild-type and XistCAGIVS allele. To circumvent this problem, we are currently attempting to derive a unique cell population with monoallelic expression of either allele or that with biallelic expression of both by inducing differentiation of ESCs into neural stem cells and subsequent cloning.
Since most of the Xist mutant allele so far generated compromise the initiation process of XCI, XistIVS RNA is peculiar in that it can initiate XCI but fails to maintain the XCI state. This is most probably due to the failure of the establishment of robust heterochromatin. It is likely that proteins recruited by Xist RNA contribute to not only the initiation of XCI but also the establishment of a sustainable heterochromatin state. XistIVS RNA would provide a unique opportunity to explore such factors involved in the latter process and how Xist participates in the establishment of the chromatin state required for the stable maintenance of the X-inactivated state.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
The animal study was reviewed and approved by the committee for the Care and Use of Laboratory Animals of Kindai University.
TS and TN designed the study. RM, TN, SI, and TS performed the experiments and analyzed the data. TS wrote the manuscript. All authors contributed to the article and approved the submitted version.
This work was supported partly by Grants-in-Aid for Scientific Research (A) from the Japan Society for the Promotion of Science (JSPS) (17H01588 and 20H00550 to TS) and Takeda Science Foundation to TS.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2021.751154/full#supplementary-material
Amakawa, Y., Sakata, Y., Hoki, Y., Arata, S., Shioda, S., Fukagawa, T., et al. (2015). A new Xist allele driven by a constitutively active promoter is dominated by Xist locus environment and exhibits the parent-of-origin effects. Development 142, 4299–4308. doi: 10.1242/dev.128819
Borsani, G., Tonlorenzi, R., Simmler, M. C., Dandolo, L., Arnaud, D., Capra, V., et al. (1991). Characterization of a murine gene expressed from the inactive X chromosome. Nature 351, 325–329. doi: 10.1038/351325a0
Brockdorff, N., Ashworth, A., Kay, G. F., Cooper, P., Smith, S., McCabe, V. M., et al. (1991). Conservation of position and exclusive expression of mouse Xist from the inactive X chromosome. Nature 351:351329a0. doi: 10.1038/351329a0
Chu, C., Zhang, Q. C., da Rocha, S. T., Flynn, R. A., Bharadwaj, M., Calabrese, J. M., et al. (2015). Systematic discovery of Xist RNA binding proteins. Cell 161, 404–416. doi: 10.1016/j.cell.2015.03.025
Clemson, C. M., McNeil, J. A., Willard, H. F., and Lawrence, J. B. (1996). XIST RNA paints the inactive X chromosome at interphase: evidence for a novel RNA involved in nuclear/chromosome structure. J. Cell. Biol. 132, 259–275. doi: 10.1083/jcb.132.3.259
Harris, C., Cloutier, M., Trotter, M., Hinten, M., Gayen, S., Du, Z., et al. (2019). Conversion of random X-inactivation to imprinted X-inactivation by maternal PRC2. Elife 8:e44258. doi: 10.7554/eLife.44258
Hoki, Y., Ikeda, R., Mise, N., Sakata, Y., Ohhata, T., Sasaki, H., et al. (2011). Incomplete X-inactivation initiated by a hypomorphic Xist allele in the mouse. Development 138, 2649–2659. doi: 10.1242/dev.061226
Kay, G. F., Penny, G. D., Patel, D., Ashworth, A., Brockdorff, N., and Rastan, S. (1993). Expression of Xist during mouse development suggests a role in the initiation of X chromosome inactivation. Cell 72, 171–182. doi: 10.1016/0092-8674(93)90658-D
Lee, J., Davidow, L. S., and Warshawsky, D. (1999). Tsix, a gene antisense to Xist at the X-inactivation centre. Nat. Genet. 21, 400–404. doi: 10.1038/7734
Lee, J. T. (2000). Disruption of imprinted X inactivation by parent-of-origin effects at tsix. Cell 103, 17–27. doi: 10.1016/S0092-8674(00)00101-X
Lyon, M. F. (1961). Gene action in the X-chromosome of the mouse (Mus musculus L.). Nature 190, 372–373. doi: 10.1038/190372a0
Marahrens, Y., Panning, B., Dausman, J., Strauss, W., and Jaenisch, R. (1997). Xist-deficient mice are defective in dosage compensation but not spermatogenesis. Genes Dev. 11, 156–166. doi: 10.1101/gad.11.2.156
McHugh, C. A., Chen, C.-K., Chow, A., Surka, C. F., Tran, C., McDonel, P., et al. (2015). The Xist lncRNA interacts directly with SHARP to silence transcription through HDAC3. Nature 521:232. doi: 10.1038/nature14443
Moindrot, B., Cerase, A., Coker, H., Masui, O., Grijzenhout, A., Pintacuda, G., et al. (2015). A pooled shRNA screen identifies Rbm15, Spen, and Wtap as factors required for Xist RNA-mediated silencing. Cell Rep. 12, 562–572. doi: 10.1016/j.celrep.2015.06.053
Monfort, A., Di Minin, G., Postlmayr, A., Freimann, R., Arieti, F., Thore, S., et al. (2015). Identification of spen as a crucial factor for xist function through forward genetic screening in haploid embryonic stem cells. Cell Rep. 12, 554–561. doi: 10.1016/j.celrep.2015.06.067
Monk, M., and Harper, M. I. (1979). Sequential X chromosome inactivation coupled with cellular differentiation in early mouse embryos. Nature 281, 311–313. doi: 10.1038/281311a0
Navarro, P., Pichard, S., Ciaudo, C., Avner, P., and Rougeulle, C. (2005). Tsix transcription across the Xist gene alters chromatin conformation without affecting Xist transcription: implications for X-chromosome inactivation. Gene Dev. 19, 1474–1484. doi: 10.1101/gad.341105
Niwa, H., Yamamura, K., and Miyazaki, J. (1991). Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene 108, 193–199. doi: 10.1016/0378-1119(91)90434-D
Ohhata, T., Hoki, Y., Sasaki, H., and Sado, T. (2008). Crucial role of antisense transcription across the Xist promoter in Tsix-mediated Xist chromatin modification. Development 135, 227–235. doi: 10.1242/dev.008490
Pandya-Jones, A., Markaki, Y., Serizay, J., Chitiashvili, T., Leon, W. R. M., Damianov, A., et al. (2020). A protein assembly mediates Xist localization and gene silencing. Nature 587, 145–151. doi: 10.1038/s41586-020-2703-0
Panning, B., and Jaenisch, R. (1996). DNA hypomethylation can activate Xist expression and silence X-linked genes. Genes Dev. 10, 1991–2002. doi: 10.1101/gad.10.16.1991
Patil, D. P., Chen, C.-K., Pickering, B. F., Chow, A., Jackson, C., Guttman, M., et al. (2016). m6A RNA methylation promotes XIST-mediated transcriptional repression. Nature 537, 369–373. doi: 10.1038/nature19342
Penny, G. D., Kay, G. F., Sheardown, S. A., Rastan, S., and Brockdorff, N. (1996). Requirement for Xist in X chromosome inactivation. Nature 379:379131a0. doi: 10.1038/379131a0
Pintacuda, G., Wei, G., Roustan, C., Kirmizitas, B. A., Solcan, N., Cerase, A., et al. (2017). hnRNPK recruits PCGF3/5-PRC1 to the Xist RNA B-repeat to establish polycomb-mediated chromosomal silencing. Mol. Cell 68, 955–969.e10. doi: 10.1016/j.molcel.2017.11.013
Sado, T., Hoki, Y., and Sasaki, H. (2005). Tsix silences Xist through modification of chromatin structure. Dev. Cell 9, 159–165. doi: 10.1016/j.devcel.2005.05.015
Sado, T., Wang, Z., Sasaki, H., and Li, E. (2001). Regulation of imprinted X-chromosome inactivation in mice by Tsix. Development 128, 1275–1286. doi: 10.1242/dev.128.8.1275
Sakata, Y., Nagao, K., Hoki, Y., Sasaki, H., Obuse, C., and Sado, T. (2017). Defects in dosage compensation impact global gene regulation in the mouse trophoblast. Development 144, 2784–2797. doi: 10.1242/dev.149138
Shiura, H., Sakata, Y., Abe, K., and Sado, T. (2018). RNA-FISH and immunofluorescence of mouse preimplantation and postimplantation embryos. Methods Mol. Biol. 1861, 161–176. doi: 10.1007/978-1-4939-8766-5_13
Takagi, N., and Sasaki, M. (1975). Preferential inactivation of the paternally derived X chromosome in the extraembryonic membranes of the mouse. Nature 256, 640–642. doi: 10.1038/256640a0
Wutz, A., Rasmussen, T. P., and Jaenisch, R. (2002). Chromosomal silencing and localization are mediated by different domains of Xist RNA. Nat. Genet. 30, 167–174. doi: 10.1038/ng820
Keywords: Xist RNA, embryonic stem cells (ESCs), X chromosome inactivation, heterochromatin, gene silencing, RNA-FISH
Citation: Matsuura R, Nakajima T, Ichihara S and Sado T (2021) Ectopic Splicing Disturbs the Function of Xist RNA to Establish the Stable Heterochromatin State. Front. Cell Dev. Biol. 9:751154. doi: 10.3389/fcell.2021.751154
Received: 31 July 2021; Accepted: 20 September 2021;
Published: 14 October 2021.
Edited by:
Louis Lefebvre, University of British Columbia, CanadaReviewed by:
Joseph Mauro Calabrese, University of North Carolina at Chapel Hill, United StatesCopyright © 2021 Matsuura, Nakajima, Ichihara and Sado. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Takashi Sado, dHNhZG9AbmFyYS5raW5kYWkuYWMuanA=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.