
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Cell Dev. Biol., 25 May 2021
Sec. Epigenomics and Epigenetics
Volume 9 - 2021 | https://doi.org/10.3389/fcell.2021.679662
This article is part of the Research TopicEpigenomic and Epitranscriptomic Basis of Development and Human DiseaseView all 14 articles
N6-methyladenosine (m6A) modification, as the most prevalent internal modification on mRNA, has been implicated in many biological processes through regulating mRNA metabolism. Given that m6A modification is highly enriched in the mammalian brain, this dynamic modification provides a crucial new layer of epitranscriptomic regulation of the nervous system. Here, in this review, we summarize the recent progress on studies of m6A modification in the mammalian nervous system ranging from neuronal development to basic and advanced brain functions. We also highlight the detailed underlying mechanisms in each process mediated by m6A writers, erasers, and readers. Besides, the involvement of dysregulated m6A modification in neurological disorders and injuries is discussed as well.
Messenger RNAs (mRNAs) play crucial roles in biological processes, which not only serve as messengers that pass genetic information from DNA to protein but also bear various post-transcriptional regulation mechanisms. Modifications on mRNA have been studied for several decades (Boccaletto et al., 2018). Other than 5′ cap and 3′ polyadenylation, numerous modified nucleotides such as N6-methyladenosine (m6A), N1-methyladenosine (m1A), N6,2′-O-dimethyladenosine (m6Am), 5-methylcytosine (m5C), and 5-hydroxymethylcytosine (hm5C) have been identified (Roundtree et al., 2017a). Modifications on mRNAs can change the structural properties of modified mRNAs, which affects the accessibility and affinity to specific RNA binding proteins (RBPs). Similar to chemical modifications on DNA and histone proteins, mRNA modifications have profound significance to biological processes.
m6A modification, as the most prevalent internal chemical modification on mRNA, was found more than four decades ago (Desrosiers et al., 1974; Adams and Cory, 1975; Furuichi et al., 1975; Wei et al., 1975). However, due to the lack of detection methods, functional studies on m6A were greatly hindered. The discovery of the first m6A demethylase in 2011 led to a resurgence in exploring m6A modification (Cao et al., 2016). Moreover, with the advances in biochemistry and sequencing technology in recent years, much progress has been achieved on m6A modification.
The abundance of m6A was estimated in a ratio of 0.1–0.4% of adenosine in mammals (about 3∼5 m6A modification per mRNA) (Rottman et al., 1974; Wei et al., 1975; Fu et al., 2014). It occurs on the consensus motif DRACH (D means a non-cytosine base, R refers to G/A, A is the m6A modified site, and H represents a non-guanine base) (Fu et al., 2014; Livneh et al., 2020). m6A modification is preferentially distributed in long coding exons, 3′ untranslated regions (UTR), and near the stop codon of mRNAs (Dominissini et al., 2012; Meyer et al., 2012). m6A has been found to be dynamically regulated and involved in many biological processes by affecting the fate of modified mRNA. In this review, we will summarize the recent findings of m6A modification in the nervous system from development to higher functions and from neurological disorders to injuries.
The deposition of m6A modification on mRNA is mediated by a multi-component methyltransferase complex. The methyltransferases are also called m6A writers, including methyltransferase-like 3 (METTL3), methyltransferase-like 14 (METTL14), and Wilms tumor 1-associated protein (WTAP) (Bokar et al., 1997; Liu et al., 2014; Ping et al., 2014; Schwartz et al., 2014). During the methylation process, METTL3 and METTL14 form a stable heterodimer complex and work synergistically to regulate adenosine methylation. METTL3 is the catalytically active enzymatic component, while METTL14 is an allosteric activator (Sledz and Jinek, 2016; Wang P. et al., 2016; Wang X. et al., 2016). This METTL3-METTL14 complex catalyzes the vast majority of m6A modification on mRNA, as ablation of METTL3 or inactivation of METTL14 in mouse embryonic stem cells leads to the loss of more than 99% of total m6A in mRNA (Geula et al., 2015). The remaining modified m6A residues in mRNA could be catalyzed by METTL16 or other potential methyltransferases (Zaccara et al., 2019). WTAP is a critical adaptor that translocates the METTL3-METTL14 complex into nuclear speckles, thus facilitating the methylation efficiency (Ping et al., 2014; Schwartz et al., 2014).
The discovery of m6A erasers (demethylases) proves that m6A is a dynamic and reversible modification. The first m6A eraser, fat mass and obesity-associated (FTO), was discovered in 2011 (Jia et al., 2011). FTO belongs to the Fe (II) and α-ketoglutarate-dependent AlkB family (Gerken et al., 2007), which was initially found to be associated with body weight and food intake in mice (Fischer et al., 2009; Church et al., 2010). It can effectively demethylate m6A in both RNA and DNA in vitro (Jia et al., 2011). In vivo, FTO also demethylates specific mRNAs that affect neuronal signaling in the mouse brain (Hess et al., 2013). However, FTO was further found to preferentially demethylate m6Am in the 5′ cap of mRNA (Mauer et al., 2017). Thus, more studies from the third parties would be required to solve this scientific dispute.
The second eraser of m6A, alkB homolog 5 (ALKBH5), was related to fertility in mice (Zheng et al., 2013). It also belongs to the Fe (II) and α-ketoglutarate-dependent AlkB family. ALKBH5 can catalyze the demethylation of m6A modification on mRNA both in vitro and in vivo, which influences the nuclear RNA export and metabolism (Zheng et al., 2013). Unlike FTO, ALKBH5 cannot demethylate m6Am (Mauer et al., 2017).
N6-methyladenosine modification exerts its function by recruiting m6A-binding proteins, which are also called m6A readers. There are two ways of reader proteins to bind to m6A modification: direct binding and indirect binding. Direct binding relies on a specialized domain within the readers, which can directly recognize and bind to m6A. The first direct reader proteins identified were the YTH (YT521-B homology) domain-containing proteins (Dominissini et al., 2012). The YTH domain is a highly conserved RNA binding domain identified in a wide range of eukaryotes (Stoilov et al., 2002). There are three classes of YTH domain-containing proteins in mammals, including the YTH domain-containing family protein (YTHDF) family, YTH domain-containing protein 1 (YTHDC1), and YTH domain-containing protein 2 (YTHDC2) (Patil et al., 2018). The indirect reader proteins include HNRNPC, HNRNPG, HNRNPA2B1, and IGF2bp proteins, which can bind m6A through the mechanism of m6A-dependent mRNA structural change (Zaccara et al., 2019).
Transcriptome-wide binding sequencing studies of endogenous (Patil et al., 2016) or overexpressed YTH proteins (Wang et al., 2014, 2015) using crosslinking and immunoprecipitation (CLIP) experiments showed that most YTH proteins bind to the m6A consensus motif in mRNA. The distribution of the YTHDF family proteins’ binding sites is similar to the distribution pattern of m6A on mRNA (Patil et al., 2016). YTHDF proteins and YTHDC1 can recognize and selectively bind m6A through an aromatic cage (hydrophobic pocket) formed by three tryptophans in the YTH domain (Li et al., 2014; Luo and Tong, 2014; Theler et al., 2014; Xu et al., 2014).
YTHDF family proteins contain three members: YTHDF1, YTHDF2, and YTHDF3. YTHDF proteins have the same binding specificity toward m6A-modified mRNA (Xu et al., 2015). These three proteins share high similarity in amino acid sequence over their entire length and are expressed mainly in the cytoplasm (Patil et al., 2018). YTHDF proteins have almost identical YTH domains at C-terminal. Apart from the YTH domain, YTHDF family proteins contain a low-complexity region with no recognizable modular protein domain and include several P/Q/N-rich domains (Patil et al., 2018). The function of this low-complexity region is to lead mRNA-YTHDF complexes to undergo liquid-liquid phase separation to different endogenous compartments, like processing bodies (P-bodies), neuronal RNA granules, or stress granules (Ries et al., 2019).
The role of YTHDF1 was found to promote the translation efficiency of m6A-modified mRNA (Wang et al., 2015). It was shown that YTHDF1 plays a dual role in this process by delivering m6A-modified mRNA to translation machinery and enhancing translation initiation (Wang et al., 2015). This could be possibly caused by the loop structure mediated by eIF4G and the interaction between YTHDF1 and eIF3 (Wang et al., 2015).
YTH domain-containing protein 2 was found to be implicated in enhancing the degradation of m6A-modified mRNA (Wang et al., 2014). In this process, YTHDF2 binds to m6A-modified mRNA and translocates those mRNA from the translatable pool into P-bodies, which are mRNA decay sites (Wang et al., 2014). However, other studies did not find the existence of YTHDF2 in P-bodies (Hubstenberger et al., 2017). The possible explanation is that the association between YTHDF2 and P-bodies is transient, which results in the difficulty to detect YTHDF2 in P-bodies. Another study found that YTHDF2 regulates mRNA stability by mediating mRNA deadenylation first and then translocating to P-bodies (Du et al., 2016). The N-terminal region of YTHDF2 is capable of recruiting the CCR4-NOT deadenylase complex, causing the deadenylation of mRNA (Du et al., 2016), which finally degrades mRNA. YTHDF2 also regulate endoribonucleolytic cleavage of m6A-modified mRNA through interaction with RNase P/MRP, which is bridged by HRSP12 (Park et al., 2019).
The role of YTHDF3 was characterized as working together with YTHDF1 and YTHDF2 to regulate the metabolism of m6A-modified mRNA (Shi et al., 2017). It has a combined effect of YTHDF1 and YTHDF2, promoting both translation and decay of m6A-modified mRNA (Shi et al., 2017). Knockdown of YTHDF3 reduces the RNA-binding specificity of both YTHDF1 and YTHDF2 (Shi et al., 2017). Compared with YTHDF1 and YTHDF2, YTHDF3 exerts its function on the early life cycle of RNA in the cytosol (Shi et al., 2017). When m6A-modified mRNA is transported to the cytoplasm, it might be initially recognized by YTHDF3. The binding of YTHDF3 could then facilitate YTHDF1 binding to the mRNA and together promote translation. Subsequently, the mRNA might be bound and partitioned among YTHDF proteins and eventually recognized by YTHDF2 for degradation.
However, a recent study has argued that all the YTHDF proteins function redundantly to mediate mRNA degradation (Zaccara and Jaffrey, 2020). Thus, more studies are needed to explore their functions in detail.
YTH domain-containing protein 1 is predominantly expressed in nuclear speckles (Hartmann et al., 1999). It has been shown that YTHDC1 mediates splicing (Xiao et al., 2016), and nuclear export of mRNA (Roundtree et al., 2017b). As active transcription occurs in nuclear speckles, YTHDC1 may bind m6A-modified mRNAs and affect their splicing. By recruiting pre-mRNA splicing factor SRSF3 and inhibiting SRSF10, YTHDC1 facilitates exon inclusion in target m6A-modified mRNA (Xiao et al., 2016). YTHDC1 can also selectively promote the transport of m6A-modified mRNA from nuclear to cytoplasm through interacting with nuclear mRNA receptors NXF1 and SRSF3 (Roundtree et al., 2017b). Besides, transcriptome-wide UV crosslinking immunoprecipitation (CLIP) study showed that YTHDC1 preferentially binds m6A residues in long non-coding RNAs (Patil et al., 2016), whereas the YTHDF family readers prefer to bind m6A sites on mRNA. The proper function of the long non-coding RNA, X-inactive specific transcript (XIST), which regulates X chromosome inactivation and transcriptional silencing of genes on the X chromosome, needs YTHDC1 to bind its m6A sites (Patil et al., 2016). In addition, YTHDC1 can interact with the H3K9me2 demethylase KDM3B to promote H3K9me2 demethylation and gene expression (Li et al., 2020b).
YTH domain-containing protein 2 is expressed both in the nucleus and cytoplasm. Unlike the other m6A readers that are universally expressed, YTHDC2 is enriched in testes (Bailey et al., 2017; Hsu et al., 2017; Wojtas et al., 2017; Jain et al., 2018). YTHDC2 can promote the translation efficiency of its target mRNAs and also decrease mRNA levels (Hsu et al., 2017; Kretschmer et al., 2018). YTHDC2 promotes germ cell fate transition from mitosis to meiosis (Bailey et al., 2017). Ythdc2 knockout mice show defects in spermatogenesis (Bailey et al., 2017; Hsu et al., 2017).
N6-methyladenosine is widely distributed in many mouse tissues, with the highest expression in the brain (Meyer et al., 2012; Chang et al., 2017). Immunostaining with a specific m6A antibody revealed wide-spread and strong m6A signals in the developing mouse brain, spinal cord, and dorsal root ganglion (DRG) (Figure 1). Transcriptome-wide m6A sequencing showed that distinct m6A methylation patterns occur in different brain regions or at different stages in the same region, suggesting the critical dynamic involvement of m6A modification in neuronal development (Chang et al., 2017). In the mouse cerebral cortex and cerebellum, neurons have relatively higher m6A levels than glia cells (Chang et al., 2017). The highly methylated mRNAs are associated with processes such as synapse assembly and axon guidance, suggesting that m6A modification plays an essential role in neuronal development and brain functions (Chang et al., 2017).
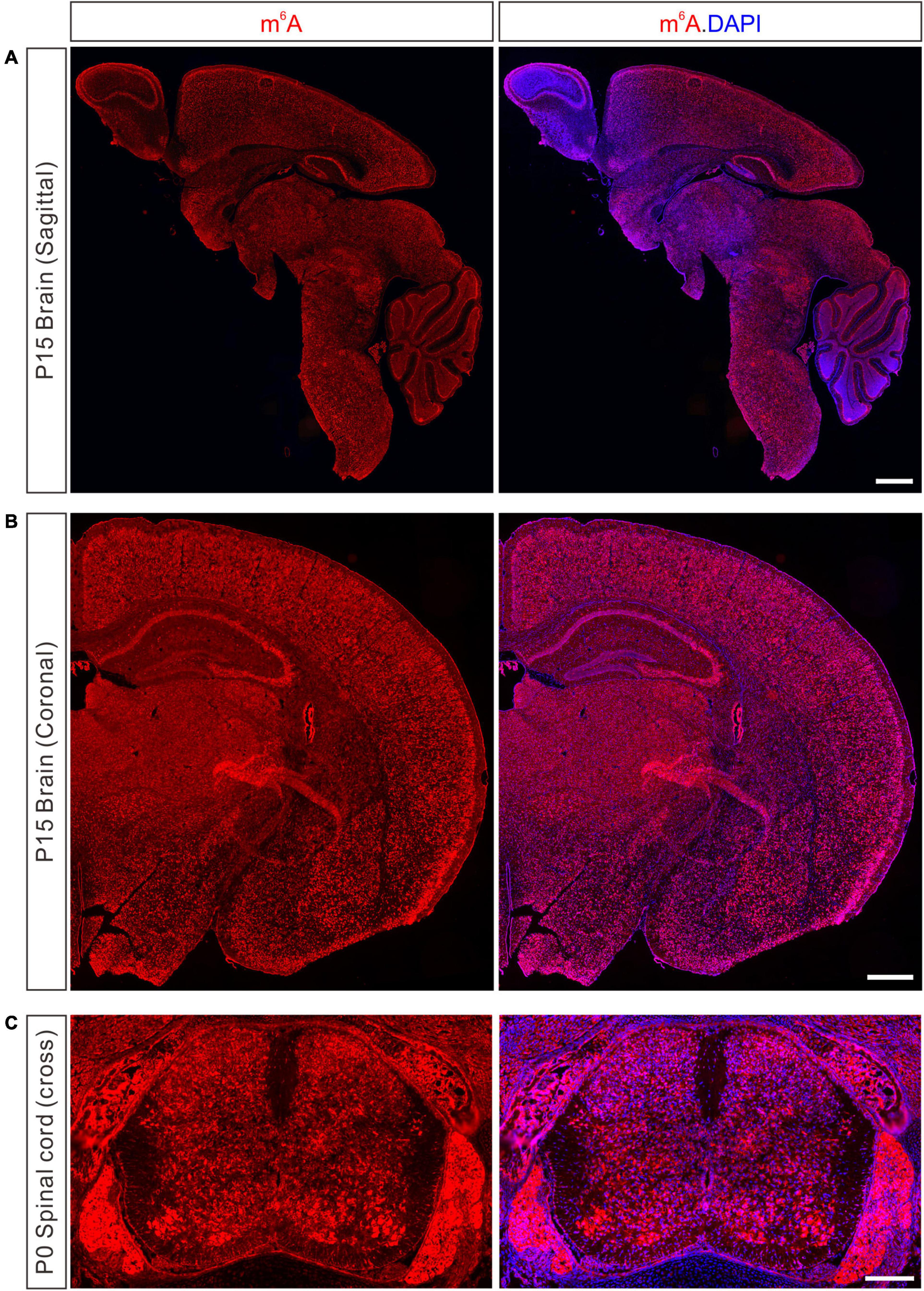
Figure 1. Detection of m6A modification in the developing nervous system of mouse. (A) Immunostaining of m6A in sagittal section of P15 mouse brain. Scale bar, 1 mm. (B) Immunostaining of m6A in coronal section of P15 mouse brain. Scale bar, 500 μm. (C) Immunostaining of m6A in cross-section of P0 mouse spinal cord and DRG. Scale bar, 200 μm.
For the past 5 years, tremendous progress has been made, showing that m6A modification can regulate multiple neuronal developmental processes and functions in mammals. We summarize these findings in Table 1 and discuss the details in the following parts.
During neuronal development, neurogenesis is a precisely orchestrated process (Kohwi and Doe, 2013). In the cerebral cortex, radial glia cells (RGCs), also known as neural stem cells (NSCs), are the principal progenitor cells that generate consecutive different types of neurons which further migrate to different layers. m6A modification has been shown to regulate this exquisitely timed process (Yoon et al., 2017; Wang Y. et al., 2018). When m6A modification was ablated in the nervous system using Nestin-Cre;Mettl14f/f conditional knockout (cKO) mice, the cell cycle of RGCs was prolonged and cortical neurogenesis extended into postnatal stages, causing brutal postnatal death (Yoon et al., 2017). These were due to the failure of m6A-dependent decay of mRNAs encoding proteins related to stem cell, cell cycle and neurogenesis in neural stem cells (Yoon et al., 2017). Another study also found cortical neurogenesis defects in Nestin-Cre;Mettl14f/f cKO mice which were characterized with decreased cortical thickness and postnatal death (Wang Y. et al., 2018). In this study, m6A modification was found to mediate NSCs self-renewal, as deletion of Mettl14 in NSCs led to reduced proliferation and premature differentiation, causing NSC pool loss and insufficient late-born neurons (Wang Y. et al., 2018). The underlying mechanism was that m6A modification could regulate histone modification, inhibiting proliferation-related genes and promoting differentiation-related genes (Wang Y. et al., 2018). These studies provided direct proof that m6A modification can regulate mouse embryonic cortical neurogenesis. However, the seemingly opposite mechanisms described in these two studies after using the same Nestin-cre to conditionally knock out Mettl14 (“prolonged cell cycle and maintenance of radial glia cells” vs. “decreased proliferation and premature differentiation”) suggest that further exploration and clarification are needed. In addition, as Nestin-cre is generally expressed in the nervous system, how m6A modification specifically affects mouse cortical neurogenesis without affecting other areas in the brain needs further elucidation.
In addition, disrupting the recognition and reading of m6A modification can also phenocopy the effect on cortex neurogenesis (Li et al., 2018; Edens et al., 2019). Self-renewal and spatiotemporal neurogenesis of NSCs were severely affected in the cortex of Ythdf2 knockout mice, causing retarded cortical development and lethality at late embryonic stages (Li et al., 2018). Both proliferation and differentiation abilities were decreased in Ythdf2–/– NSCs, which were indeed caused by the impaired clearance of m6A-modified genes (Li et al., 2018). Another reader protein involved in regulating cortical neurogenesis is fragile X mental retardation protein (FMRP) (Edens et al., 2019). The role of FMRP was to preferentially bind m6A-modified mRNAs and facilitate them to export from nuclear (Edens et al., 2019). Deletion of Fmr1 cause nuclear retention of m6A-modified mRNAs associated with neural differentiation (Edens et al., 2019). Thus, Fmr1 KO mice exhibited extended maintenance of NSCs into postnatal stages with delayed NSC cell cycle progression and differentiation.
Unlike cortical neurogenesis that occurs in embryonic stages, the development of the cerebellum mainly begins postnatally. The cerebellum has generally higher m6A levels than the cerebral cortex (Ma et al., 2018). The expression of m6A modifiers (writers, erasers, and readers) is developmentally regulated and differentially expressed in different cell types and regions in the cerebellum (Ma et al., 2018). Similarly, the mRNAs in the cerebellum show dynamic methylation levels throughout the developmental stages (Chang et al., 2017; Ma et al., 2018). These findings demonstrate that m6A might be required for the development and function of the cerebellum.
Specific deletion of Mettl3 in the mouse nervous system leads to cerebellar hypoplasia caused by increased apoptosis of newly generated cerebellar granule cells (CGCs) in the external granular layer (Wang C.X. et al., 2018). Due to the loss of m6A, the half-lives of mRNA associated with cerebellar development and apoptosis are extended. In addition, synapse-associated mRNAs show abnormal splicing after Mettl3 depletion. Those events finally contribute to incorrect cell differentiation and cell death (Wang C.X. et al., 2018). Knockdown of METTL3 results in disorganized structures of Purkinje cells and glial cells in cerebellum (Ma et al., 2018). In addition, deletion of Alkbh5 in mice exposed to hypobaric hypoxia leads to aberrant proliferation and differentiation due to the dysregulated mRNA nuclear export (Ma et al., 2018). Those findings together prove that m6A acts as a crucial regulator during cerebellar development.
Fat mass and obesity-associated was unexpectedly found expressed in the axons of mouse DRG neurons, which can dynamically regulate m6A modification on axonal mRNA (Yu et al., 2018). Despite the previous finding that demethylation occurs in nuclear speckles, m6A modification can be dynamically regulated in the highly compartmentalized subcellular structures such as axons. Axonally derived FTO regulates the level of m6A modification on GAP-43 mRNA and further affects the local translation of GAP-43 mRNA in axons, eventually controlling axon growth (Yu et al., 2018). This study is the first example of mRNA modification regulating local translation in axons.
Axon guidance cues provided by the floor plate enable the right pathfinding of commissural axons in the developing spinal cord (Colamarino and Tessier-Lavigne, 1995). Robo3.1 is one of the axon guidance receptors from Roundabout (Robo) family that facilitate midline crossing of commissural axons (Chen et al., 2008). The precise spatiotemporal expression of Robo3.1 has been found to be regulated by m6A modification (Zhuang et al., 2019). YTHDF1 can bind to Robo3.1 mRNA in an m6A dependent manner and promote its translation (Zhuang et al., 2019). Specific deletion of YTHDF1 in commissural neurons using Atoh1-cre;Ythdf1f/f cKO mice led to axon guidance defects (Zhuang et al., 2019).
Axon regeneration of mouse DRG neurons in the peripheral nervous system (PNS) requires de novo gene transcription and translation of regeneration-associated genes (RAGs) (Costigan et al., 2002; Mahar and Cavalli, 2018). Similar to other epigenetic mechanisms, such as DNA methylation (Weng et al., 2017) and histone modification (Gaub et al., 2011; Puttagunta et al., 2014), m6A modification has also been shown to participate in the activation of RAGs (Weng et al., 2018). Sciatic nerve lesion (SNL) substantially increases levels of m6A-modified transcripts in vivo. Those transcripts can be divided into three categories: transcripts encoding RAGs, injury-induced retrograde signaling molecules, and translation machinery components (Weng et al., 2018). Either loss of METTL14 or YTHDF1 can delay the injury-induced protein translation of RAGs, such as Atf3, and cause defective axon regeneration and function recovery (Weng et al., 2018). These findings indicate that m6A modification may affect response to pathological stimulus in the adult nervous system.
Low input m6A sequencing of mouse forebrain synaptosomes has revealed a synaptic m6A epitranscriptome (SME) in which 2921 synaptosomal transcripts are m6A-modified (Merkurjev et al., 2018). Transcripts in SME are most significantly enriched in central nervous system development. More than half of the genes in the SME overlapped with the synaptic transcriptome. Surprisingly, those genes are functionally annotated to synapse-associated functions, such as “synapse assembly,” “postsynaptic membrane,” “long-term synaptic potentiation.” In contrast, those hypomethylated transcripts in the synaptic transcriptome were mainly related to metabolic pathways. These findings suggest that m6A modification possibly regulates synapse formation and synaptic function. Dendrite localization of m6A writers, erasers, and readers was detected in mouse cortical pyramidal neurons in brain slices, which further indicates that m6A modification could be dynamically and locally regulated in synapses (Merkurjev et al., 2018). Either knockdown of YTHDF1 or YTHDF3 in cultured hippocampal neurons leads to abnormal dendrite spine morphology, reduced PSD95 clustering, decreased expression of GluA1, thus compromising synaptic transmission (Merkurjev et al., 2018).
Adult neurogenesis occurs (yet still in debate) limitedly, and has been shown to be related to neurological and psychiatric disorders (Apple et al., 2017; Kempermann et al., 2018). m6A has also been reported to function in adult neurogenesis. FTO is expressed in adult neural stem cells (aNSCs), and deletion of Fto reduces the proliferation and neuronal differentiation of aNSCs (Li et al., 2017; Cao et al., 2020). This is due to the altered expression of several key components that are modified by m6A in the brain-derived neurotrophic factor (BDNF) pathway (Li et al., 2017) and the Pdgfra/Socs5-Stat3 pathway (Cao et al., 2020). On the other hand, depletion of METTL3 also inhibits the proliferation of aNSCs (Chen J. et al., 2019). The mRNA of histone methyltransferase Ezh2 is modified by m6A (Chen J. et al., 2019). Upon deletion of Mettl3, the protein level of Ezh2 deceased, further causing reduced H3K27me3 levels (Chen J. et al., 2019). These studies showed that m6A modification is involved in adult neurogenesis. However, how writers and erasers work together under normal conditions to regulate adult neurogenesis still needs more investigation.
Glia cells account for more than 50% of cells in the human brain (Nave, 2010; Rowitch and Kriegstein, 2010). Oligodendrocytes and astrocytes are the two major macroglial cells derived from the neuroepithelium (Rowitch and Kriegstein, 2010). Oligodendrocytes are responsible for the myelination of axons. m6A modification has been shown to control the oligodendrocyte lineage progression. Specific deletion of Mettl14 in developing oligodendrocyte lineage cells or in postmitotic oligodendrocytes leads to loss of mature oligodendrocytes and hypomyelination (Xu H. et al., 2020). This is because the loss of METTL14 results in abnormal splicing of many mRNAs which encode proteins associated with the myelinating process, such as protein tyrosine phosphate receptor type Z1 (Ptprz1) and neurofascin 155 (NF155) (Xu H. et al., 2020). Another study discovered a novel m6A reader, Proline-rich coiled-coil 2A (PRRC2A), which regulates oligodendrocyte specification and myelination (Wu et al., 2019). Deletion of Prrc2a in oligodendrocyte progenitor cells (OPCs) leads to hypomyelination, locomotive and cognitive defects in mice. Interestingly, PRRC2A directly stabilizes the Olig2 mRNA in an m6A-dependent manner. Olig2 is known to regulate OPC specification, differentiation and myelination (Lu et al., 2002; Zhou and Anderson, 2002). However, the mechanism of how PRRC2A stabilizes m6A-modified mRNA remains unclear.
In addition to oligodendrocytes, m6A modification also functions in the gliogenesis of astrocytes, which provide structural support, modulate synaptic transmission, and maintain the blood-brain barrier (Rowitch and Kriegstein, 2010). Global deletion of Mettl14 in the mouse nervous system significantly reduces astrocytes in the cortex (Yoon et al., 2017). The scaffold organization pattern provided by Bergmann glia (a highly diversified type of astrocytes) in the mouse cerebellum is abolished after deleting Mettl3 in the nervous system (Wang C.X. et al., 2018). As for the m6A readers, in vitro differentiation assay of neurospheres showed that deletion of Ythdf2 in neural stem/progenitor cell (NSPC) caused dramatic reduction of GFAP+ astrocytes (Li et al., 2018). These studies indicate that m6A modification also controls the production of astrocytes. However, the underlying mechanism needs further investigation.
Learning and memory require coordinated regulation of gene expression and protein translation. The substantial increase of m6A level from the embryonic brain to the adult brain (Meyer et al., 2012) suggests that the dynamic m6A epitranscriptome could be involved in the regulation of the advanced brain functions.
Fat mass and obesity-associated is highly expressed in the dendrites and synapses of mouse CA1 hippocampal neurons (Walters et al., 2017). Interestingly, the expression of FTO protein decreased in the synaptic fraction, not the nuclear fraction of hippocampus 0.5 h after contextual fear conditioning, indicating that behavioral training-induced memory preferentially decreases FTO levels near synapses (Walters et al., 2017). As expected, with the decrease of FTO, the m6A level on mRNA is significantly increased. Knockdown of FTO in hippocampus facilitated contextual fear memory, suggesting that synaptic FTO could normally restrict memory formation and experience-induced increase of m6A modification could enhance memory formation (Walters et al., 2017). Another study also shows that cue fear conditioning increases m6A level in mouse medial prefrontal cortex (mPFC) (Widagdo et al., 2016). Knockdown of FTO in mPFC results in increased fear memory consolidation (Widagdo et al., 2016). These studies demonstrate that experience or behavior-induced upregulation of m6A modification might participate in the regulation of memory. However, as FTO was also reported to preferentially demethylate m6Am (Mauer et al., 2017) and m6Am participates in fear memory (Engel et al., 2018), the possibility that m6Am may contribute to some of the phenotypes cannot be ruled out.
The study of m6A writer METTL3 provides direct evidence that m6A modification regulates memory formation. Using CaMKIIa-cre;Mettl3f/f cKO mice, specific deletion of METTL3 in the forebrain excitatory neurons decreases the formation of hippocampus-dependent long-term memory without changing short-term memory and learning ability when adequate training is provided (Zhang et al., 2018). The hippocampus-dependent memory consolidation ability exquisitely relies on the function of METTL3, as the expression of METTL3 in wild-type (WT) mice positively associates with the learning efficacy and overexpression of METTL3 facilitates long-term memory consolidation (Zhang et al., 2018). The formation of long-term memory requires de novo protein synthesis of immediate early genes (IEGs), such as Arc, Egr1, c-Fos, Npas4, and Nr4a1. By affecting the m6A levels on those IEGs, METTL3 eventually promotes their translation, thus enhancing memory (Zhang et al., 2018).
Regarding the roles of m6A reader protein, YTHDF1 was reported to be required in the process of m6A enhanced learning and memory in the hippocampus (Shi et al., 2018). Ythdf1 mRNA is preferentially located in the mouse hippocampus (Lein et al., 2007), suggesting that it might be involved in learning and memory. When YTHDF1 is ablated entirely from the hippocampus, hippocampus histology, neurogenesis, motor ability, and emotional state are not altered in Ythdf1 KO mice (Shi et al., 2018). However, by compromising basal synaptic transmission and long-term potentiation (LTP), the learning and memory abilities of Ythdf1 KO mice in Morris water maze (MWM) and contextual fear conditioning tests are impaired (Shi et al., 2018). Restoring the expression of YTHDF1 in the hippocampus of Ythdf1 KO mice can successfully rescue the synaptic and behavioral defects (Shi et al., 2018). Further analysis of the underlying molecular mechanism showed that YTHDF1 preferentially recognizes m6A modified mRNAs and facilitates their translation in a neuronal-stimulus-dependent manner. More interestingly, YTHDF1 could translocate into the postsynaptic density (PSD) fraction to facilitate protein synthesis locally in synapses of the hippocampus in response to fear conditioning, thus promote synaptic plasticity and memory formation (Shi et al., 2018).
Learning and memory-related synaptic plasticity requires local translation at synapses (Wang et al., 2009). Due to the dynamic SME (Merkurjev et al., 2018) and the localization of m6A writers, erasers, and readers in synapses, it’s highly likely that m6A-dependent local translation of synaptic mRNAs is the central event that occurs in synapses in response to stimuli.
Besides the forebrain, m6A modification also affects synaptic transmission in the midbrain and striatum. It has been shown that FTO can regulate the activity of the dopaminergic (DA) signaling in the mouse midbrain, which controls complex behaviors (Hess et al., 2013). Deletion of Fto attenuates neuronal activity controlled by dopamine D2-like receptor and behavioral responses (Hess et al., 2013). Compared with WT mice, Fto-deficient mice showed impaired cocaine-induced behavioral activity and synaptic dopamine release (Hess et al., 2013). Transcriptome-wide m6A sequencing showed that the m6A level of many genes involved in the DA signal pathway is increased in Fto-deficient mice. In the adult mouse striatum, specific deletion of Mettl14 in two distinct but related types of neurons, striatonigral and striatopallidal neurons, leads to deficiency in striatum-mediated learning and dopamine signaling without affecting cell numbers and morphology (Koranda et al., 2018). Interestingly, neuronal and synaptic mRNAs are downregulated in either type of neurons after deletion of Mettl14, while upregulated mRNAs are mainly associated with translational regulation and metabolism (Koranda et al., 2018). These m6A-dependent gene regulation increases neuronal excitability and decreases spike frequency adaptation, which finally attenuates striatum-mediated learning and behavioral performance (Koranda et al., 2018). Considering activity-dependent synaptic protein synthesis is vital to synaptic plasticity and learning, it is important to decipher how m6A readers are involved in this process to spatial temporally regulate protein synthesis in response to neuronal activities.
Transcriptome-wide sequencing of human and mouse brains showed that m6A modification is spatiotemporally regulated during neurodevelopment and aging (Shafik et al., 2021). Increased m6A sites are observed as age increases. The dynamically regulated m6A sites are enriched in alternatively untranslated regions of genes involved in aging-related pathways (Shafik et al., 2021). Alzheimer’s disease (AD) is the most common form of dementia among elderly people (Bateman et al., 2012). The m6A levels of many transcripts involved in AD-associated pathways are decreased in the brain of a familial AD mouse model (5XFAD) (Shafik et al., 2021). In contrast, m6A levels are elevated in the cortex and hippocampus of APP/PS1 transgenic mice, another AD mouse model, compared with WT mice (Han et al., 2020). Interestingly, the expression of METTL3 increased, and FTO is decreased in the APP/PS1 mice (Han et al., 2020). These studies show that m6A modification is involved in AD. However, the mechanism by which m6A regulates the progression of AD remains almost unknown.
Parkinson’s disease (PD) is a common neurodegenerative disorder characterized by the early prominent death of dopaminergic neurons (Lees, 2017). The global m6A levels of mRNAs are decreased in the striatum of the PD rat brain and a cellular PD model (6-OHDA-induced PC12 cells), which is mainly due to the increase of FTO protein (Chen X. et al., 2019). The decrease of m6A level could induce the expression of N-methyl-D-aspartate (NMDA) receptor 1, and increase oxidative stress and Ca2+ influx, which finally leads to dopaminergic neuron apoptosis (Chen X. et al., 2019). Conversely, knockdown of FTO in PC12 cells decreases NMDAR1 expression and exhibits anti-apoptosis effect (Chen X. et al., 2019). This study suggests that m6A modification via FTO may play a crucial role in the pathogenesis of PD.
Ischemic stroke is a severe neurological disease, which is a leading cause of disability in humans (Wang et al., 2017). Cerebral ischemia/reperfusion (I/R) injury rapidly triggers different types of programmed cell death in neurons. Several studies have shown that m6A modification was involved in I/R injury (Si et al., 2020). The expression of METTL3 is significantly decreased at the reperfusion injury period. Decreased m6A level leads to the reduction of miR-335 and stress granule formation (Si et al., 2020). Therefore, by upregulating the expression of miR-335, METTL3-mediated m6A modification can normally promote stress granule formation and improve cell survival of neurons. Contradictorily, another study reported increased m6A levels after I/R injury (Xu K. et al., 2020). The expression of m6A erasers, ALKBH5 and FTO, are decreased but not writers. Overexpression of m6A erasers can alleviate neuronal damage induced by I/R injury (Xu S. et al., 2020). A third study found that oxygen-glucose deprivation/re-oxygenation (OGD/R) increased METTL3-dependent m6A modification of long non-coding RNA D63785 (lnc-D63785), thus causing reduced expression of lnc-D63785 (Xu S. et al., 2020). Downregulation of lnc-D63785 further induces accumulation of miR-422a, which results in neuronal cell apoptosis (Xu S. et al., 2020).
Hypothermia is an effective therapeutic method to alleviate I/R injury (Callaway et al., 2015; Donnino et al., 2015). Hypoxia/reoxygenation (H/R) caused m6A-dependent increase of PTEN mRNA stability and consequently upregulation of its protein level, which could be reversed by hypothermia (Diao et al., 2020). Thus hypothermia could activate PI3K/Akt signaling to protect neurons from I/R-induced pyroptosis (Diao et al., 2020). Another study reported that the m6A reader YTHDC1 protects ischemic stroke through mediating PTEN mRNA degradation, which further promotes Akt phosphorylation and facilitates neuronal survival (Zhang Z. et al., 2020). These two studies demonstrate that m6A modification could modulate PTEN expression to regulate PI3K/Akt signaling in I/R injury.
Taken together, all these studies demonstrated that m6A modification is involved in the process of I/R injury, which could provide potential therapeutic targets for I/R injury.
Traumatic brain injury (TBI), one of the most severe types of injury, is a major public health threat (Majdan et al., 2017). After TBI, the mRNA and protein levels of METTL3 were significantly decreased in the hippocampus of mice, but not other writers and erasers (Wang et al., 2019). Correspondingly, the m6A level of RNA was downregulated in the hippocampus after TBI. Genome-wide m6A profiling identified that altered peaks of m6A-modified transcripts after TBI were mainly related to the regulation of the metabolic process (Wang et al., 2019). Metabolism alteration induced by brain injury could lead to long-term cognitive and neurological disabilities (McKenna et al., 2015). Therefore, this study indicates that m6A modification-induced metabolic alteration might be the underlying mechanism of TBI. Thus rectifying altered m6A level might be a potential therapeutic strategy for TBI.
N6-methyladenosine modification has been shown to participate in both inflammatory and neuropathic pain (Li et al., 2020a; Zhang C. et al., 2020). The m6A levels of spinal mRNAs are significantly increased in Complete Freund’s Adjuvant (CFA)-induced chronic inflammatory pain mouse model, which shows strong thermal and mechanical hyperalgesia (Zhang C. et al., 2020). The upregulated m6A level is due to the increase of METTL3 in CFA-injected mice, which can modulate the pain sensitization by regulating m6A-dependent pri-miRNA processing. Meanwhile, another study reported that FTO contributes to nerve injury-induced neuropathic pain in the primary sensory neurons in DRG (Li et al., 2020a). Nerve injury activates the transcription factor Runx1, which can bind to Fto gene promoter and activate its expression but not m6A readers. Upregulated FTO then demethylates m6A modification on the Ehmt2 mRNA encoding euchromatic histone lysine methyltransferase 2 and elevates its protein level, thus resulting in neuropathic pain symptoms. Conversely, knockdown of FTO could alleviate nerve injury-associated pain hypersensitivities (Li et al., 2020a). These two studies indicate that m6A modification regulates different pain responses through different mechanisms.
N6-methyladenosine modification has been implicated in various types of cancer including brain tumor (Deng et al., 2018). Glioblastoma is the most common and severe brain tumor. The proliferation and tumorigenesis of glioblastoma stem cells (GSCs) require high expression of the m6A eraser ALKBH5 (Zhang et al., 2017). ALKBH5 demethylates FOXM1 nascent transcript, maintaining expression of FOXM1 that preserves GSC properties (Zhang et al., 2017). Knockdown of ALKBH5 reduces proliferation of patient-derived GSCs (Zhang et al., 2017). In addition, knockdown of the m6A writers METTL3 and METTL14 significantly increases GSC-initiated tumor progression in vivo (Cui et al., 2017). Interestingly, treatment with MA2, an FTO inhibitor, inhibits GSC-initiated tumorigenesis and extends the lifespan of GSC-engrafted mice (Cui et al., 2017). Controversially, another two studies found that clinical aggressiveness of glioblastoma is related to increased expression of METTL3 (Visvanathan et al., 2018; Li et al., 2019). METTL3 promotes GSC stemness by enhancing SOX2 stability in glioblastoma, and METTL3 silencing inhibits tumor growth (Visvanathan et al., 2018). Knockdown of METTL3 suppresses aggressive and tumorigenic properties of GSCs by causing YTHDC1-dependent nonsense-mediated mRNA decay of SRSF, and subsequent decrease of SRSF protein level (Li et al., 2019). m6A modification also regulates neuroblastoma, another common malignant brain tumor (Cheng et al., 2020). MYCN is a genetic biomarker of high risk and poor outcome in neuroblastoma. m6A modification in the 3′UTR of MYCN promotes its interaction with miR-98, decreasing MYCN expression and inhibiting neuroblastoma progression (Cheng et al., 2020). These studies indicate that m6A modification could be a promising target for anti-brain tumor therapy.
The nervous system is the most complex and diverse system, with exceptional capabilities that control higher cognitive and emotional functions. The development of the nervous system is a highly coordinated process in which epigenetic mechanisms exert significant effects by spatiotemporally regulating gene expression. Apart from DNA methylation and histone modifications, dynamic mRNA m6A modification provides an additional regulatory layer to regulate gene expression. As described above, m6A modification regulates the development and functions of the nervous system.
The higher function of the nervous system relies on synaptic plasticity. In response to stimuli, the nervous system undergoes extremely swift reactions to adapt its proteome. Neurons are highly compartmentalized cells, and local translation plays a central role in rapidly changing subcellular proteomes in response to extrinsic cues and stimuli. Accumulating evidence has suggested that m6A modification modulates the local translation of mRNAs in axons and synapses. This m6A-dependent local translation could be the principal mechanism that regulates the plasticity of the nervous system. This highlights the requirement of comprehensive studies of m6A modification and local translation of the nervous system. How m6A writers, erasers, and readers function together to spatiotemporally regulate local proteome needs more investigation.
Up to now, there are controversial findings regarding the functions of m6A reader proteins. As YTH proteins share very high similarity in the YTH domains, the mechanism of how these reader proteins select their target mRNA remains unknown. Therefore, it is crucial to deeply decipher the roles and mechanisms of reader proteins on how they divide jobs and coordinate to mediate m6A signaling.
Dysregulation of m6A modification causes neurological diseases. The involvement of m6A in neurological diseases and injuries provides new potential therapeutic targets for treatment. However, the roles of m6A in injury-induced neuronal diseases, psychiatric disorders, and aging-related neurodegenerative disorders are still far beyond understanding. In-depth studies of how m6A signaling modulates neuronal physiology and pathology in the adult brain are in great demand.
JY and S-JJ drafted and revised the manuscript. S-JJ conceived and designed the review. YS helped to edit and revise the manuscript. All authors read and approved the final manuscript.
This work was supported by the National Natural Science Foundation of China (31871038 to S-JJ), Shenzhen-Hong Kong Institute of Brain Science-Shenzhen Fundamental Research Institutions (2019SHIBS0002 and 2021SHIBS0002), High-Level University Construction Fund for Department of Biology (Internal grant no. G02226301), and Science and Technology Innovation Commission of Shenzhen Municipal Government (ZDSYS2020081114400 2008).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank Prof. Chi Wai Lee at The University of Hong Kong (HKU) for co-mentoring the Ph.D. student in the SUSTech-HKU Joint Ph.D. Program.
Adams, J. M., and Cory, S. (1975). Modified nucleosides and bizarre 5’-termini in mouse myeloma mRNA. Nature 255, 28–33. doi: 10.1038/255028a0
Apple, D. M., Fonseca, R. S., and Kokovay, E. (2017). The role of adult neurogenesis in psychiatric and cognitive disorders. Brain Res. 1655, 270–276. doi: 10.1016/j.brainres.2016.01.023
Bailey, A. S., Batista, P. J., Gold, R. S., Chen, Y. G., De Rooij, D. G., Chang, H. Y., et al. (2017). The conserved RNA helicase YTHDC2 regulates the transition from proliferation to differentiation in the germline. Elife 6:e26116.
Bateman, R. J., Xiong, C., Benzinger, T. L., Fagan, A. M., Goate, A., Fox, N. C., et al. (2012). Clinical and biomarker changes in dominantly inherited Alzheimer’s disease. N. Engl. J. Med. 367, 795–804.
Boccaletto, P., Machnicka, M. A., Purta, E., Piatkowski, P., Baginski, B., Wirecki, T. K., et al. (2018). MODOMICS: a database of RNA modification pathways. 2017 update. Nucleic Acids Res. 46, D303–D307.
Bokar, J. A., Shambaugh, M. E., Polayes, D., Matera, A. G., and Rottman, F. M. (1997). Purification and cDNA cloning of the AdoMet-binding subunit of the human mRNA (N6-adenosine)-methyltransferase. RNA 3, 1233–1247.
Callaway, C. W., Donnino, M. W., Fink, E. L., Geocadin, R. G., Golan, E., Kern, K. B., et al. (2015). Part 8: post-cardiac arrest care: 2015 American heart association guidelines update for cardiopulmonary resuscitation and emergency cardiovascular care. Circulation 132, S465–S482.
Cao, G., Li, H. B., Yin, Z., and Flavell, R. A. (2016). Recent advances in dynamic m6A RNA modification. Open Biol. 6:160003. doi: 10.1098/rsob.160003
Cao, Y., Zhuang, Y., Chen, J., Xu, W., Shou, Y., Huang, X., et al. (2020). Dynamic effects of Fto in regulating the proliferation and differentiation of adult neural stem cells of mice. Hum. Mol. Genet. 29, 727–735. doi: 10.1093/hmg/ddz274
Chang, M., Lv, H., Zhang, W., Ma, C., He, X., Zhao, S., et al. (2017). Region-specific RNA m(6)A methylation represents a new layer of control in the gene regulatory network in the mouse brain. Open Biol. 7:170166. doi: 10.1098/rsob.170166
Chen, J., Zhang, Y. C., Huang, C., Shen, H., Sun, B., Cheng, X., et al. (2019). m(6)A regulates neurogenesis and neuronal development by modulating histone methyltransferase Ezh2. Genomics Proteomics Bioinformatics 17, 154–168. doi: 10.1016/j.gpb.2018.12.007
Chen, X., Yu, C., Guo, M., Zheng, X., Ali, S., Huang, H., et al. (2019). Down-regulation of m6A mRNA methylation is involved in dopaminergic neuronal death. ACS Chem. Neurosci. 10, 2355–2363. doi: 10.1021/acschemneuro.8b00657
Chen, Z., Gore, B. B., Long, H., Ma, L., and Tessier-Lavigne, M. (2008). Alternative splicing of the Robo3 axon guidance receptor governs the midline switch from attraction to repulsion. Neuron 58, 325–332. doi: 10.1016/j.neuron.2008.02.016
Cheng, J., Xu, L., Deng, L., Xue, L., Meng, Q., Wei, F., et al. (2020). RNA N(6)-methyladenosine modification is required for miR-98/MYCN axis-mediated inhibition of neuroblastoma progression. Sci. Rep. 10:13624.
Church, C., Moir, L., Mcmurray, F., Girard, C., Banks, G. T., Teboul, L., et al. (2010). Overexpression of Fto leads to increased food intake and results in obesity. Nat. Genet. 42, 1086–1092. doi: 10.1038/ng.713
Colamarino, S. A., and Tessier-Lavigne, M. (1995). The role of the floor plate in axon guidance. Annu. Rev. Neurosci. 18, 497–529. doi: 10.1146/annurev.ne.18.030195.002433
Costigan, M., Befort, K., Karchewski, L., Griffin, R. S., D’urso, D., Allchorne, A., et al. (2002). Replicate high-density rat genome oligonucleotide microarrays reveal hundreds of regulated genes in the dorsal root ganglion after peripheral nerve injury. BMC Neurosci. 3:16. doi: 10.1186/1471-2202-3-16
Cui, Q., Shi, H., Ye, P., Li, L., Qu, Q., Sun, G., et al. (2017). m(6)A RNA methylation regulates the self-renewal and tumorigenesis of glioblastoma stem cells. Cell Rep. 18, 2622–2634. doi: 10.1016/j.celrep.2017.02.059
Deng, X., Su, R., Feng, X., Wei, M., and Chen, J. (2018). Role of N(6)-methyladenosine modification in cancer. Curr. Opin. Genet. Dev. 48, 1–7. doi: 10.1016/j.gde.2017.10.005
Desrosiers, R., Friderici, K., and Rottman, F. (1974). Identification of methylated nucleosides in messenger RNA from Novikoff hepatoma cells. Proc. Natl. Acad. Sci. U.S.A. 71, 3971–3975. doi: 10.1073/pnas.71.10.3971
Diao, M. Y., Zhu, Y., Yang, J., Xi, S. S., Wen, X., Gu, Q., et al. (2020). Hypothermia protects neurons against ischemia/reperfusion-induced pyroptosis via m6A-mediated activation of PTEN and the PI3K/Akt/GSK-3beta signaling pathway. Brain Res. Bull. 159, 25–31. doi: 10.1016/j.brainresbull.2020.03.011
Dominissini, D., Moshitch-Moshkovitz, S., Schwartz, S., Salmon-Divon, M., Ungar, L., Osenberg, S., et al. (2012). Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 485, 201–206. doi: 10.1038/nature11112
Donnino, M. W., Andersen, L. W., Berg, K. M., Reynolds, J. C., Nolan, J. P., Morley, P. T., et al. (2015). Temperature management after cardiac arrest: an advisory statement by the advanced life support task force of the International Liaison Committee on resuscitation and the American heart association emergency cardiovascular care committee and the council on cardiopulmonary, critical care, perioperative and resuscitation. Circulation 132, 2448–2456. doi: 10.1161/cir.0000000000000313
Du, H., Zhao, Y., He, J., Zhang, Y., Xi, H., Liu, M., et al. (2016). YTHDF2 destabilizes m(6)A-containing RNA through direct recruitment of the CCR4-NOT deadenylase complex. Nat. Commun. 7:12626.
Edens, B. M., Vissers, C., Su, J., Arumugam, S., Xu, Z., Shi, H., et al. (2019). FMRP modulates neural differentiation through m(6)A-dependent mRNA nuclear export. Cell Rep. 28, 845–854.e5.
Engel, M., Eggert, C., Kaplick, P. M., Eder, M., Roh, S., Tietze, L., et al. (2018). The role of m(6)A/m-RNA methylation in stress response regulation. Neuron 99, 389–403.e9.
Fischer, J., Koch, L., Emmerling, C., Vierkotten, J., Peters, T., Bruning, J. C., et al. (2009). Inactivation of the Fto gene protects from obesity. Nature 458, 894–898.
Fu, Y., Dominissini, D., Rechavi, G., and He, C. (2014). Gene expression regulation mediated through reversible m(6)A RNA methylation. Nat. Rev. Genet. 15, 293–306. doi: 10.1038/nrg3724
Furuichi, Y., Morgan, M., Shatkin, A. J., Jelinek, W., Salditt-Georgieff, M., and Darnell, J. E. (1975). Methylated, blocked 5 termini in HeLa cell mRNA. Proc. Natl. Acad. Sci. U.S.A. 72, 1904–1908. doi: 10.1073/pnas.72.5.1904
Gaub, P., Joshi, Y., Wuttke, A., Naumann, U., Schnichels, S., Heiduschka, P., et al. (2011). The histone acetyltransferase p300 promotes intrinsic axonal regeneration. Brain 134, 2134–2148. doi: 10.1093/brain/awr142
Gerken, T., Girard, C. A., Tung, Y. C., Webby, C. J., Saudek, V., Hewitson, K. S., et al. (2007). The obesity-associated FTO gene encodes a 2-oxoglutarate-dependent nucleic acid demethylase. Science 318, 1469–1472. doi: 10.1126/science.1151710
Geula, S., Moshitch-Moshkovitz, S., Dominissini, D., Mansour, A. A., Kol, N., Salmon-Divon, M., et al. (2015). Stem cells. m6A mRNA methylation facilitates resolution of naive pluripotency toward differentiation. Science 347, 1002–1006. doi: 10.1126/science.1261417
Han, M., Liu, Z., Xu, Y., Liu, X., Wang, D., Li, F., et al. (2020). Abnormality of m6A mRNA methylation is involved in Alzheimer’s disease. Front. Neurosci. 14:98. doi: 10.3389/fnins.2020.00098
Hartmann, A. M., Nayler, O., Schwaiger, F. W., Obermeier, A., and Stamm, S. (1999). The interaction and colocalization of Sam68 with the splicing-associated factor YT521-B in nuclear dots is regulated by the Src family kinase p59(fyn). Mol. Biol. Cell 10, 3909–3926. doi: 10.1091/mbc.10.11.3909
Hess, M. E., Hess, S., Meyer, K. D., Verhagen, L. A., Koch, L., Bronneke, H. S., et al. (2013). The fat mass and obesity associated gene (Fto) regulates activity of the dopaminergic midbrain circuitry. Nat. Neurosci. 16, 1042–1048. doi: 10.1038/nn.3449
Hsu, P. J., Zhu, Y., Ma, H., Guo, Y., Shi, X., Liu, Y., et al. (2017). Ythdc2 is an N(6)-methyladenosine binding protein that regulates mammalian spermatogenesis. Cell Res. 27, 1115–1127. doi: 10.1038/cr.2017.99
Hubstenberger, A., Courel, M., Benard, M., Souquere, S., Ernoult-Lange, M., Chouaib, R., et al. (2017). P-body purification reveals the condensation of repressed mRNA regulons. Mol. Cell 68, 144–157.e5.
Jain, D., Puno, M. R., Meydan, C., Lailler, N., Mason, C. E., Lima, C. D., et al. (2018). Ketu mutant mice uncover an essential meiotic function for the ancient RNA helicase YTHDC2. Elife 7:e30919.
Jia, G., Fu, Y., Zhao, X., Dai, Q., Zheng, G., Yang, Y., et al. (2011). N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol. 7, 885–887. doi: 10.1038/nchembio.687
Kempermann, G., Gage, F. H., Aigner, L., Song, H., Curtis, M. A., Thuret, S., et al. (2018). Human adult neurogenesis: evidence and remaining questions. Cell Stem Cell 23, 25–30. doi: 10.1016/j.stem.2018.04.004
Kohwi, M., and Doe, C. Q. (2013). Temporal fate specification and neural progenitor competence during development. Nat. Rev. Neurosci. 14, 823–838. doi: 10.1038/nrn3618
Koranda, J. L., Dore, L., Shi, H., Patel, M. J., Vaasjo, L. O., Rao, M. N., et al. (2018). Mettl14 is essential for epitranscriptomic regulation of striatal function and learning. Neuron 99, 283–292.e5.
Kretschmer, J., Rao, H., Hackert, P., Sloan, K. E., Hobartner, C., and Bohnsack, M. T. (2018). The m(6)A reader protein YTHDC2 interacts with the small ribosomal subunit and the 5’-3’ exoribonuclease XRN1. RNA 24, 1339–1350. doi: 10.1261/rna.064238.117
Lein, E. S., Hawrylycz, M. J., Ao, N., Ayres, M., Bensinger, A., Bernard, A., et al. (2007). Genome-wide atlas of gene expression in the adult mouse brain. Nature 445, 168–176.
Li, F., Yi, Y., Miao, Y., Long, W., Long, T., Chen, S., et al. (2019). N(6)-methyladenosine modulates nonsense-mediated mRNA decay in human glioblastoma. Cancer Res. 79, 5785–5798. doi: 10.1158/0008-5472.can-18-2868
Li, F., Zhao, D., Wu, J., and Shi, Y. (2014). Structure of the YTH domain of human YTHDF2 in complex with an m(6)A mononucleotide reveals an aromatic cage for m(6)A recognition. Cell Res. 24, 1490–1492. doi: 10.1038/cr.2014.153
Li, L., Zang, L., Zhang, F., Chen, J., Shen, H., Shu, L., et al. (2017). Fat mass and obesity-associated (FTO) protein regulates adult neurogenesis. Hum. Mol. Genet. 26, 2398–2411. doi: 10.1093/hmg/ddx128
Li, M., Zhao, X., Wang, W., Shi, H., Pan, Q., Lu, Z., et al. (2018). Ythdf2-mediated m(6)A mRNA clearance modulates neural development in mice. Genome Biol. 19:69.
Li, Y., Guo, X., Sun, L., Xiao, J., Su, S., Du, S., et al. (2020a). N(6)-methyladenosine demethylase FTO contributes to neuropathic pain by stabilizing G9a expression in primary sensory neurons. Adv. Sci. (Weinh) 7, 1902402. doi: 10.1002/advs.201902402
Li, Y., Xia, L., Tan, K., Ye, X., Zuo, Z., Li, M., et al. (2020b). N(6)-methyladenosine co-transcriptionally directs the demethylation of histone H3K9me2. Nat. Genet. 52, 870–877. doi: 10.1038/s41588-020-0677-3
Liu, J., Yue, Y., Han, D., Wang, X., Fu, Y., Zhang, L., et al. (2014). A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat. Chem. Biol. 10, 93–95. doi: 10.1038/nchembio.1432
Livneh, I., Moshitch-Moshkovitz, S., Amariglio, N., Rechavi, G., and Dominissini, D. (2020). The m(6)A epitranscriptome: transcriptome plasticity in brain development and function. Nat. Rev. Neurosci. 21, 36–51. doi: 10.1038/s41583-019-0244-z
Lu, Q. R., Sun, T., Zhu, Z., Ma, N., Garcia, M., Stiles, C. D., et al. (2002). Common developmental requirement for Olig function indicates a motor neuron/oligodendrocyte connection. Cell 109, 75–86. doi: 10.1016/s0092-8674(02)00678-5
Luo, S., and Tong, L. (2014). Molecular basis for the recognition of methylated adenines in RNA by the eukaryotic YTH domain. Proc. Natl. Acad. Sci. U.S.A. 111, 13834–13839. doi: 10.1073/pnas.1412742111
Ma, C., Chang, M., Lv, H., Zhang, Z. W., Zhang, W., He, X., et al. (2018). RNA m(6)A methylation participates in regulation of postnatal development of the mouse cerebellum. Genome Biol. 19;68.
Mahar, M., and Cavalli, V. (2018). Intrinsic mechanisms of neuronal axon regeneration. Nat. Rev. Neurosci. 19, 323–337. doi: 10.1038/s41583-018-0001-8
Majdan, M., Plancikova, D., Maas, A., Polinder, S., Feigin, V., Theadom, A., et al. (2017). Years of life lost due to traumatic brain injury in Europe: a cross-sectional analysis of 16 countries. PLoS Med. 14:e1002331. doi: 10.1371/journal.pmed.1002331
Mauer, J., Luo, X., Blanjoie, A., Jiao, X., Grozhik, A. V., Patil, D. P., et al. (2017). Reversible methylation of m(6)Am in the 5’ cap controls mRNA stability. Nature 541, 371–375. doi: 10.1038/nature21022
McKenna, M. C., Scafidi, S., and Robertson, C. L. (2015). Metabolic alterations in developing brain after injury: knowns and unknowns. Neurochem. Res. 40, 2527–2543. doi: 10.1007/s11064-015-1600-7
Merkurjev, D., Hong, W. T., Iida, K., Oomoto, I., Goldie, B. J., Yamaguti, H., et al. (2018). Synaptic N(6)-methyladenosine (m(6)A) epitranscriptome reveals functional partitioning of localized transcripts. Nat. Neurosci. 21, 1004–1014. doi: 10.1038/s41593-018-0173-6
Meyer, K. D., Saletore, Y., Zumbo, P., Elemento, O., Mason, C. E., and Jaffrey, S. R. (2012). Comprehensive analysis of mRNA methylation reveals enrichment in 3’ UTRs and near stop codons. Cell 149, 1635–1646. doi: 10.1016/j.cell.2012.05.003
Nave, K. A. (2010). Myelination and support of axonal integrity by glia. Nature 468, 244–252. doi: 10.1038/nature09614
Park, O. H., Ha, H., Lee, Y., Boo, S. H., Kwon, D. H., Song, H. K., et al. (2019). Endoribonucleolytic cleavage of m(6)A-containing RNAs by RNase P/MRP complex. Mol. Cell 74, 494–507.e8.
Patil, D. P., Chen, C. K., Pickering, B. F., Chow, A., Jackson, C., Guttman, M., et al. (2016). m(6)A RNA methylation promotes XIST-mediated transcriptional repression. Nature 537, 369–373. doi: 10.1038/nature19342
Patil, D. P., Pickering, B. F., and Jaffrey, S. R. (2018). Reading m(6)A in the transcriptome: m(6)A-binding proteins. Trends Cell Biol. 28, 113–127. doi: 10.1016/j.tcb.2017.10.001
Ping, X. L., Sun, B. F., Wang, L., Xiao, W., Yang, X., Wang, W. J., et al. (2014). Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res. 24, 177–189. doi: 10.1038/cr.2014.3
Puttagunta, R., Tedeschi, A., Soria, M. G., Hervera, A., Lindner, R., Rathore, K. I., et al. (2014). PCAF-dependent epigenetic changes promote axonal regeneration in the central nervous system. Nat. Commun. 5:3527.
Ries, R. J., Zaccara, S., Klein, P., Olarerin-George, A., Namkoong, S., Pickering, B. F., et al. (2019). m(6)A enhances the phase separation potential of mRNA. Nature 571, 424–428. doi: 10.1038/s41586-019-1374-1
Rottman, F., Shatkin, A. J., and Perry, R. P. (1974). Sequences containing methylated nucleotides at the 5’ termini of messenger RNAs: possible implications for processing. Cell 3, 197–199. doi: 10.1016/0092-8674(74)90131-7
Roundtree, I. A., Evans, M. E., Pan, T., and He, C. (2017a). Dynamic RNA modifications in gene expression regulation. Cell 169, 1187–1200. doi: 10.1016/j.cell.2017.05.045
Roundtree, I. A., Luo, G. Z., Zhang, Z., Wang, X., Zhou, T., Cui, Y., et al. (2017b). YTHDC1 mediates nuclear export of N(6)-methyladenosine methylated mRNAs. Elife 6:e31311.
Rowitch, D. H., and Kriegstein, A. R. (2010). Developmental genetics of vertebrate glial-cell specification. Nature 468, 214–222. doi: 10.1038/nature09611
Schwartz, S., Mumbach, M. R., Jovanovic, M., Wang, T., Maciag, K., Bushkin, G. G., et al. (2014). Perturbation of m6A writers reveals two distinct classes of mRNA methylation at internal and 5’ sites. Cell Rep. 8, 284–296. doi: 10.1016/j.celrep.2014.05.048
Shafik, A. M., Zhang, F., Guo, Z., Dai, Q., Pajdzik, K., Li, Y., et al. (2021). N6-methyladenosine dynamics in neurodevelopment and aging, and its potential role in Alzheimer’s disease. Genome Biol. 22:17.
Shi, H., Wang, X., Lu, Z., Zhao, B. S., Ma, H., Hsu, P. J., et al. (2017). YTHDF3 facilitates translation and decay of N(6)-methyladenosine-modified RNA. Cell Res. 27, 315–328. doi: 10.1038/cr.2017.15
Shi, H., Zhang, X., Weng, Y. L., Lu, Z., Liu, Y., Lu, Z., et al. (2018). m(6)A facilitates hippocampus-dependent learning and memory through YTHDF1. Nature 563, 249–253. doi: 10.1038/s41586-018-0666-1
Si, W., Li, Y., Ye, S., Li, Z., Liu, Y., Kuang, W., et al. (2020). Methyltransferase 3 mediated miRNA m6A methylation promotes stress granule formation in the early stage of acute ischemic stroke. Front. Mol. Neurosci. 13:103. doi: 10.3389/fnmol.2020.00103
Sledz, P., and Jinek, M. (2016). Structural insights into the molecular mechanism of the m(6)A writer complex. Elife 5:e18434.
Stoilov, P., Rafalska, I., and Stamm, S. (2002). YTH: a new domain in nuclear proteins. Trends Biochem. Sci. 27, 495–497. doi: 10.1016/s0968-0004(02)02189-8
Theler, D., Dominguez, C., Blatter, M., Boudet, J., and Allain, F. H. (2014). Solution structure of the YTH domain in complex with N6-methyladenosine RNA: a reader of methylated RNA. Nucleic Acids Res. 42, 13911–13919. doi: 10.1093/nar/gku1116
Visvanathan, A., Patil, V., Arora, A., Hegde, A. S., Arivazhagan, A., Santosh, V., et al. (2018). Essential role of METTL3-mediated m(6)A modification in glioma stem-like cells maintenance and radioresistance. Oncogene 37, 522–533. doi: 10.1038/onc.2017.351
Walters, B. J., Mercaldo, V., Gillon, C. J., Yip, M., Neve, R. L., Boyce, F. M., et al. (2017). The role of the RNA demethylase FTO (Fat Mass and Obesity-Associated) and mRNA methylation in hippocampal memory formation. Neuropsychopharmacology 42, 1502–1510. doi: 10.1038/npp.2017.31
Wang, C. X., Cui, G. S., Liu, X., Xu, K., Wang, M., Zhang, X. X., et al. (2018). METTL3-mediated m6A modification is required for cerebellar development. PLoS Biol. 16:e2004880. doi: 10.1371/journal.pbio.2004880
Wang, D. O., Kim, S. M., Zhao, Y., Hwang, H., Miura, S. K., Sossin, W. S., et al. (2009). Synapse- and stimulus-specific local translation during long-term neuronal plasticity. Science 324, 1536–1540. doi: 10.1126/science.1173205
Wang, P., Doxtader, K. A., and Nam, Y. (2016). Structural basis for cooperative function of Mettl3 and Mettl14 methyltransferases. Mol. Cell 63, 306–317. doi: 10.1016/j.molcel.2016.05.041
Wang, W., Jiang, B., Sun, H., Ru, X., Sun, D., Wang, L., et al. (2017). Prevalence, incidence, and mortality of stroke in China: results from a nationwide population-based survey of 480 687 adults. Circulation 135, 759–771. doi: 10.1161/circulationaha.116.025250
Wang, X., Feng, J., Xue, Y., Guan, Z., Zhang, D., Liu, Z., et al. (2016). Structural basis of N(6)-adenosine methylation by the METTL3-METTL14 complex. Nature 534, 575–578. doi: 10.1038/nature18298
Wang, X., Lu, Z., Gomez, A., Hon, G. C., Yue, Y., Han, D., et al. (2014). N6-methyladenosine-dependent regulation of messenger RNA stability. Nature 505, 117–120. doi: 10.1038/nature12730
Wang, X., Zhao, B. S., Roundtree, I. A., Lu, Z., Han, D., Ma, H., et al. (2015). N(6)-methyladenosine Modulates Messenger RNA Translation Efficiency. Cell 161, 1388–1399. doi: 10.1016/j.cell.2015.05.014
Wang, Y., Li, Y., Yue, M., Wang, J., Kumar, S., Wechsler-Reya, R. J., et al. (2018). N(6)-methyladenosine RNA modification regulates embryonic neural stem cell self-renewal through histone modifications. Nat. Neurosci. 21, 195–206. doi: 10.1038/s41593-017-0057-1
Wang, Y., Mao, J., Wang, X., Lin, Y., Hou, G., Zhu, J., et al. (2019). Genome-wide screening of altered m6A-tagged transcript profiles in the hippocampus after traumatic brain injury in mice. Epigenomics 11, 805–819. doi: 10.2217/epi-2019-0002
Wei, C. M., Gershowitz, A., and Moss, B. (1975). Methylated nucleotides block 5’ terminus of HeLa cell messenger RNA. Cell 4, 379–386. doi: 10.1016/0092-8674(75)90158-0
Weng, Y. L., An, R., Cassin, J., Joseph, J., Mi, R., Wang, C., et al. (2017). An intrinsic epigenetic barrier for functional axon regeneration. Neuron 94, 337–346.e6.
Weng, Y. L., Wang, X., An, R., Cassin, J., Vissers, C., Liu, Y., et al. (2018). Epitranscriptomic m(6)A regulation of axon regeneration in the adult mammalian nervous system. Neuron 97, 313–325.e6.
Widagdo, J., Zhao, Q. Y., Kempen, M. J., Tan, M. C., Ratnu, V. S., Wei, W., et al. (2016). Experience-dependent accumulation of N6-methyladenosine in the prefrontal cortex is associated with memory processes in mice. J. Neurosci. 36, 6771–6777. doi: 10.1523/jneurosci.4053-15.2016
Wojtas, M. N., Pandey, R. R., Mendel, M., Homolka, D., Sachidanandam, R., and Pillai, R. S. (2017). Regulation of m(6)A transcripts by the 3’–>5’ RNA helicase YTHDC2 is essential for a successful meiotic program in the mammalian germline. Mol. Cell 68, 374–387.e12.
Wu, R., Li, A., Sun, B., Sun, J. G., Zhang, J., Zhang, T., et al. (2019). A novel m(6)A reader Prrc2a controls oligodendroglial specification and myelination. Cell Res. 29, 23–41. doi: 10.1038/s41422-018-0113-8
Xiao, W., Adhikari, S., Dahal, U., Chen, Y. S., Hao, Y. J., Sun, B. F., et al. (2016). Nuclear m(6)A reader YTHDC1 regulates mRNA splicing. Mol. Cell 61, 507–519. doi: 10.1016/j.molcel.2016.01.012
Xu, C., Liu, K., Ahmed, H., Loppnau, P., Schapira, M., and Min, J. (2015). Structural basis for the discriminative recognition of N6-methyladenosine RNA by the human YT521-B homology domain family of proteins. J. Biol. Chem. 290, 24902–24913. doi: 10.1074/jbc.m115.680389
Xu, C., Wang, X., Liu, K., Roundtree, I. A., Tempel, W., Li, Y., et al. (2014). Structural basis for selective binding of m6A RNA by the YTHDC1 YTH domain. Nat. Chem. Biol. 10, 927–929. doi: 10.1038/nchembio.1654
Xu, H., Dzhashiashvili, Y., Shah, A., Kunjamma, R. B., Weng, Y. L., Elbaz, B., et al. (2020). m(6)A mRNA methylation is essential for oligodendrocyte maturation and CNS myelination. Neuron 105, 293–309.e5.
Xu, K., Mo, Y., Li, D., Yu, Q., Wang, L., Lin, F., et al. (2020). N(6)-methyladenosine demethylases Alkbh5/Fto regulate cerebral ischemia-reperfusion injury. Ther. Adv. Chronic. Dis. 11:2040622320916024.
Xu, S., Li, Y., Chen, J. P., Li, D. Z., Jiang, Q., Wu, T., et al. (2020). Oxygen glucose deprivation/re-oxygenation-induced neuronal cell death is associated with Lnc-D63785 m6A methylation and miR-422a accumulation. Cell Death Dis. 11:816.
Yoon, K. J., Ringeling, F. R., Vissers, C., Jacob, F., Pokrass, M., Jimenez-Cyrus, D., et al. (2017). Temporal control of mammalian cortical neurogenesis by m(6)A methylation. Cell 171, 877–889.e17.
Yu, J., Chen, M., Huang, H., Zhu, J., Song, H., Zhu, J., et al. (2018). Dynamic m6A modification regulates local translation of mRNA in axons. Nucleic Acids Res. 46, 1412–1423. doi: 10.1093/nar/gkx1182
Zaccara, S., and Jaffrey, S. R. (2020). A unified model for the function of YTHDF proteins in regulating m(6)A-modified mRNA. Cell 181, 1582– 1595.e18.
Zaccara, S., Ries, R. J., and Jaffrey, S. R. (2019). Reading, writing and erasing mRNA methylation. Nat. Rev. Mol. Cell. Biol. 20, 608–624. doi: 10.1038/s41580-019-0168-5
Zhang, C., Wang, Y., Peng, Y., Xu, H., and Zhou, X. (2020). METTL3 regulates inflammatory pain by modulating m(6)A-dependent pri-miR-365-3p processing. FASEB J. 34, 122–132. doi: 10.1096/fj.201901555r
Zhang, S., Zhao, B. S., Zhou, A., Lin, K., Zheng, S., Lu, Z., et al. (2017). m(6)A Demethylase ALKBH5 maintains tumorigenicity of glioblastoma stem-like cells by sustaining FOXM1 expression and cell proliferation program. Cancer Cell 31, 591–606.e6.
Zhang, Z., Wang, M., Xie, D., Huang, Z., Zhang, L., Yang, Y., et al. (2018). METTL3-mediated N(6)-methyladenosine mRNA modification enhances long-term memory consolidation. Cell Res. 28, 1050–1061. doi: 10.1038/s41422-018-0092-9
Zhang, Z., Wang, Q., Zhao, X., Shao, L., Liu, G., Zheng, X., et al. (2020). YTHDC1 mitigates ischemic stroke by promoting Akt phosphorylation through destabilizing PTEN mRNA. Cell Death Dis. 11:977.
Zheng, G., Dahl, J. A., Niu, Y., Fedorcsak, P., Huang, C. M., Li, C. J., et al. (2013). ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol. Cell 49, 18–29. doi: 10.1016/j.molcel.2012.10.015
Zhou, Q., and Anderson, D. J. (2002). The bHLH transcription factors OLIG2 and OLIG1 couple neuronal and glial subtype specification. Cell 109, 61–73. doi: 10.1016/s0092-8674(02)00677-3
Keywords: m6A modification, nervous system, development, neurological disorders, learning and memory
Citation: Yu J, She Y and Ji S-J (2021) m6A Modification in Mammalian Nervous System Development, Functions, Disorders, and Injuries. Front. Cell Dev. Biol. 9:679662. doi: 10.3389/fcell.2021.679662
Received: 12 March 2021; Accepted: 03 May 2021;
Published: 25 May 2021.
Edited by:
Xuekun Li, Zhejiang University, ChinaReviewed by:
Ki-Jun Yoon, Korea Advanced Institute of Science and Technology, South KoreaCopyright © 2021 Yu, She and Ji. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sheng-Jian Ji, amlzakBzdXN0ZWNoLmVkdS5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.