- Ministry of Education Key Laboratory of Cell Activities and Stress Adaptations, School of Life Sciences, Lanzhou University, Lanzhou, China
Low temperature is a major adverse environment that affects normal plant growth. Previous reports showed that the actin cytoskeleton plays an important role in the plant response to low-temperature stress, but the regulatory mechanism of the actin cytoskeleton in this process is not clear. C-repeat binding factors (CBFs) are the key molecular switches for plants to adapt to cold stress. However, whether CBFs are involved in the regulation of the actin cytoskeleton has not been reported. We found that Arabidopsis actin depolymerizing factor 5 (ADF5), an ADF that evolved F-actin bundling function, was up-regulated at low temperatures. We also demonstrated that CBFs bound to the ADF5 promoter directly in vivo and in vitro. The cold-induced expression of ADF5 was significantly inhibited in the cbfs triple mutant. The freezing resistance of the adf5 knockout mutant was weaker than that of wild type (WT) with or without cold acclimation. After low-temperature treatment, the actin cytoskeleton of WT was relatively stable, but the actin cytoskeletons of adf5, cbfs, and adf5 cbfs were disturbed to varying degrees. Compared to WT, the endocytosis rate of the amphiphilic styryl dye FM4-64 in adf5, cbfs, and adf5 cbfs at low temperature was significantly reduced. In conclusion, CBFs directly combine with the CRT/DRE DNA regulatory element of the ADF5 promoter after low-temperature stress to transcriptionally activate the expression of ADF5; ADF5 further regulates the actin cytoskeleton dynamics to participate in the regulation of plant adaptation to a low-temperature environment.
Introduction
Low temperature is a major plant stress factor that causes plant growth delay, stagnation, and retrogression and reduces grain yield (Dolferus, 2014; Zhang et al., 2019). Over long evolutionary process, plants have created a series of complex mechanisms to adapt to low-temperature stress. Cold acclimation is one of these mechanisms. When plants are exposed to non-freezing low temperatures for a period of time, their tolerance to lower temperatures improves (Thomashow, 1999; Shi et al., 2018). C-repeat binding factors (CBFs) are the key molecular switches in this process (Liu J. et al., 2018). CBF1, CBF2, and CBF3, also known as DREB1B, DREB1C, and DREB1A, belong to the AP2/ERF transcription factor family, which recognizes the C-repeat (CRT)/dehydration responsive element (DRE) DNA regulatory element (CCGAC) (Stockinger et al., 1997; Gilmour et al., 1998; Liu et al., 1998; Medina et al., 1999; Thomashow, 1999). Low temperature rapidly induces the expression of CBFs. CBFs further activate the expression of a set of COLD-REGULATED (COR) genes (Chinnusamy et al., 2007; Ding et al., 2019). These genes generally encode some cryoprotective proteins, reactive oxygen species scavenging proteins, enzymes for osmolyte biosynthesis and photosynthetic membrane protective proteins, which enhance the freezing resistance of plants (Lin and Thomashow, 1992; Jaglo-Ottosen et al., 1998; Liu et al., 1998; Janmohammadi et al., 2015; Bremer et al., 2017).
The maintenance and establishment of specific structures and highly dynamic changes in the plant actin cytoskeleton are necessary for plant cellular processes, such as cell division, cytoplasmic streaming, and vesicular transport (Staiger, 2000). The rapid remodeling of actin cytoskeleton is directly regulated by diverse actin-binding proteins (ABPs), such as profilin, ADF/cofilin, capping protein, villin/gelsolin, formin, and actin-related protein2/3 (Arp2/3) complex (Staiger and Blanchoin, 2006; Qian and Xiang, 2019). Actin depolymerizing factors (ADFs) are conserved actin binding proteins in eukaryotes that regulate the actin cytoskeleton by forming more pointed ends and monomer actins via their severing/depolymerizing activity (Staiger et al., 1997; Hussey et al., 2002; Andrianantoandro and Pollard, 2006). The Arabidopsis genome encodes 11 ADF genes, which are divided into 4 subfamilies (Ruzicka et al., 2007). Our laboratory reported that the third subfamily of Arabidopsis thaliana (ADF5, ADF9) have developed new functionalization during the process of evolution. They lost the conservative severing/depolymerizing activity of the family and evolved F-actin bundling function (Nan et al., 2017). The other three subfamilies retain the conserved function of severing/depolymerizing activities (Tholl et al., 2011; Nan et al., 2017). The physiological functions of plant ADFs were reported in recent years, such as pollen germination, pollen tube polar growth (Chen et al., 2002, 2003; Bou Daher et al., 2011; Zheng et al., 2013; Jiang et al., 2017; Zhu et al., 2017), hypocotyl elongation (Dong et al., 2001; Henty et al., 2011), stomatal movement (Zhao S. et al., 2016; Qian et al., 2019), innate immunity (Tian et al., 2009; Porter et al., 2012; Fu et al., 2014; Inada et al., 2016), and nematode and aphid infection (Clément et al., 2009; Mondal et al., 2018). The new function of ADF5 plays an important role in regulating the actin bundling process during certain plant-specific physiological activities, such as pollen germination, pollen tube polar growth and ABA-induced stomatal closure (Zhu et al., 2017; Qian et al., 2019). ADF9 is primarily involved in plant growth and development. Under long-day light cycles, the adf9 mutant showed an early flowering phenotype. Compared to the WT, adf9 had delayed growth, a reduced number of lateral branches and a weakened callus formation ability (Burgos-Rivera et al., 2008). Previous studies showed that wheat ADF (TaADF) was rapidly and strongly up-regulated under low temperature (Danyluk et al., 1996). Bioinformatics analysis and previous reports showed that the expression of ADFs in Arabidopsis thaliana changed under low temperature stress (Fan et al., 2015), which suggests that members of this family also participate in the plant response and adaptation to low-temperature stress. However, the physiological function and molecular mechanisms of ADF members after low-temperature stress are not clear.
We found that CBFs directly bound to the CRT/DRE DNA regulatory element of the ADF5 promoter to induce the up-regulation of ADF5 expression and finely regulated actin cytoskeleton dynamics at low temperature and affected the endocytosis rate of the FM4-64-labeled plasma membrane. The present study identified the molecular module of the CBF pathway regulating the actin cytoskeleton at low temperature, which enriched the physiological function of ADF5 and revealed the potential mechanism of the actin cytoskeleton response to cold stress.
Results
ADF5 Promotes Basic and Acquired Freezing Resistance in Arabidopsis thaliana
The bioinformatics data showed that low temperature treatment up-regulated the expression of ADF5 and inhibited ADF9 expression (Nan et al., 2017), which suggests that this subfamily is involved in the plant response to low-temperature stress. To reveal the physiological function of Arabidopsis ADF5 under low-temperature stress, we used three adf5 knockout/gene editing mutants: adf5-1, adf5-2, and adf5-3. Our laboratory previously reported adf5-1 (Zhu et al., 2017). The adf5-2 is a T-DNA insertion mutant (SALK_030145) from ABRC. PCR and sequencing showed that two T-DNAs were inserted after the first G of the second intron (Figure 1A). RT-PCR showed that the full-length ADF5 gene in adf5-2 was not expressed (Figure 1B). The adf5-3 was generated by CRISPR/Cas9 technology in which the 236 bp of ADF5 genome was deleted (221 to 455 bp after ATG) and only 21 amino acids were correctly translated (Figure 1A).
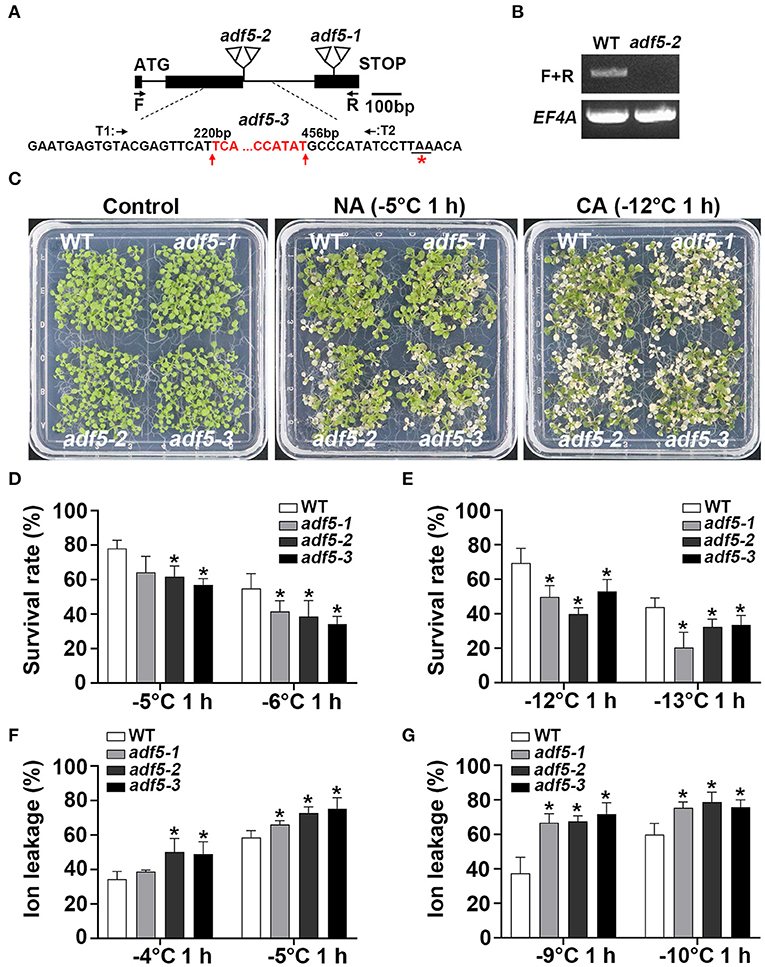
Figure 1. Mutation of ADF5 results in increased freezing sensitivity. (A) Sketch map of adf5 mutants. The black square represents the exon, and the black line represents the intron. The positions of adf5-1 and adf5-2 T-DNA are shown in the triangle. F and R are amplification sites of ADF5 for RT-PCR or identification of adf5-3. T1 and T2 are specific Cas9-splicing sites. The red base is deleted in adf5-3, and the red asterisk is the edited termination site. (B) Expression of full length ADF5 in WT and adf5-2. EF4A is used as an internal control gene. (C) Freezing phenotypes of WT and adf5s, each material contains ~40 seedlings, which were directly used for freezing treatments (non-cold-acclimated, NA), or treated at 4°C for 3 days (cold-acclimated, CA) before freezing treatment. (D,E) Survival rate of WT and adf5s. After 3–5 days of recovery under normal growth conditions, the survival rate was calculated. The data are the means of three biological replicates ± SD (n = 40 for each replicate). Asterisks indicate statistically significant differences (P < 0.05, one-way ANOVA with a Dunnett's multiple comparisons test). (F,G) Ion leakage of WT and adf5s. After 3 weeks of normal growth in soil, the fully developed rosettes of seedlings were used for freezing treatment to obtain ion leakage under NA and CA conditions. The data are the means of three biological replicates ± SD (n = 5 for each replicate). Asterisks indicate statistically significant differences (P < 0.05, one-way ANOVA with a Dunnett's multiple comparisons test).
To verify the phenotype of these mutants under low temperature stress, we first used 12 to 15-day-old seedlings grown on 1/2 MS plates for freezing analyses. The results showed that the three adf5 knockout mutants had lower freezing resistance and lower survival rate than WT with or without cold acclimation, and the difference was statistically significant (Figures 1C–E). Ion leakage generally represents damage to the cell membrane under stress, and it negatively correlates with the survival rate of plants after freezing (Ye et al., 2019). Second, we used seedlings that grew in soil for 21 days to perform independent freezing treatment experiments and obtained the ion leakage of WT and adf5 with or without cold acclimation. The ion leakage of adf5 was higher than WT under both treatment conditions, and the difference was statistically significant (Figures 1F,G). In conclusion, the results of freezing experiments and physiological data indicate that ADF5 promotes basic and acquired freezing resistance in Arabidopsis thaliana.
Low Temperature Can Induce the Expression of ADF5 Through a Partial CBFs Dependent Pathway
To verify whether the bioinformatics analysis is correct, we used qRT-PCR to detect the expression of ADF5 in the WT at low temperature. ADF5 was up-regulated nearly 10-fold after 12 h at 4°C, which indicates that ADF5 is a cold-induced gene (Figure 2). To examine the transcriptional regulatory factors upstream of ADF5, we analyzed its promoter sequence and found a CCGAC element at −231 to −227 bp and −472 to −468 bp sites, which could be recognized by CBFs (Figure 3A). To demonstrate that CBFs are the upstream transcription activator of ADF5, we used the cbf3 single mutant, cbf1 cbf3 double mutant and cbf1 cbf2 cbf3 triple mutant (cbfs-1) to detect the induction of ADF5 at 4°C for different durations. The induction amount of ADF5 in the single mutant was lower than that in WT, and the double mutant was further reduced. ADF5 induction was significantly inhibited in the triple mutant (Figure 2). Together, these results indicate that ADF5 may participate in the CBFs signaling pathway.
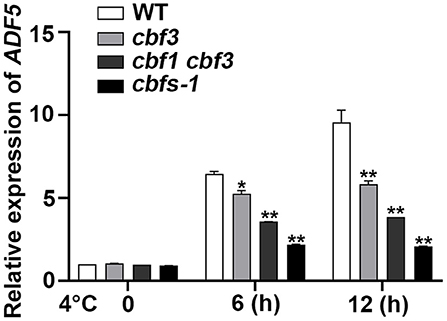
Figure 2. ADF5 expression is induced by cold in WT, cbf3, cbf1/3, and cbfs. Ten-day-old Arabidopsis seedlings were treated at 4°C for different durations, and the leaves were detached for RNA extraction. UBQ10 was used as an internal standard. Data presented are the means ± SD of three independent biological replicates (*P < 0.05; **P < 0.01; Student's two-tailed t-test).
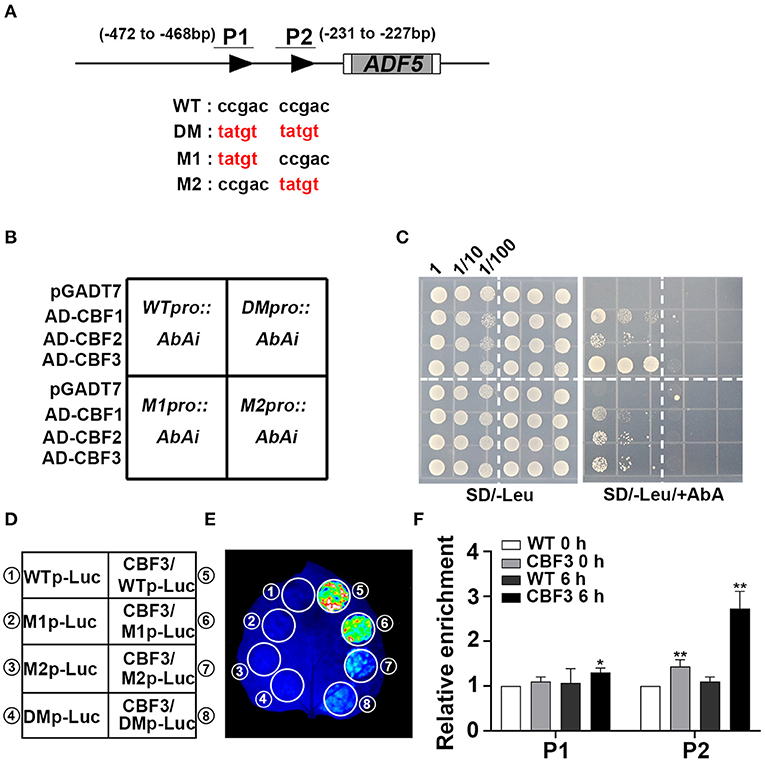
Figure 3. CBFs activate ADF5 expression via direct binding to the ADF5 promoter. (A) Schematic diagram of the CRT/DRE DNA regulatory element of the ADF5 promoter. The black triangle represents the CRT/DRE DNA regulatory element. M1 is a mutation at the −472 to −468 bp sites. M2 is a mutation at the −231 to −227 bp sites. DM is a simultaneous mutation of M1 and M2. P1 and P2 are specific primers for ChIP assay. (B) Diagram of Y1H colony. (C) Y1H assay of the interaction between CBFs and the ADF5 promoter showing the growth of yeast cells on SD/-Leu medium containing 850 ng/mL Aureobasidin A (AbA). The numbers above the images indicate the dilutions. (D) Schematic diagram of the transcriptional activation experiment. (E) CBF3 activates the expression of ADF5 in tobacco leaves via transient expression. (F) ChIP analysis of the interaction between CBFs and the ADF5 promoter under normal conditions or 4°C for 6 h. P1 contains −472 to −468 bp sites of the CRT/DRE DNA regulatory element. P2 contains −231 to −227 bp sites of the CRT/DRE DNA regulatory element. Data presented are the means ± SD of three independent biological replicates (*P < 0.05; **P < 0.01; Student's two-tailed t-test).
ADF5 Is a Downstream Target Gene of CBFs
To further confirm that CBFs bind to the corresponding region of the ADF5 promoter, we performed yeast one-hybrid (Y1H) experiment. The results show that CBF1, CBF2, and CBF3 bind to the ADF5 promoter (Figures 3B,C). We further mutated the CBF recognition sites on the ADF5 promoter to determine the key binding sites (Stockinger et al., 1997) (Figure 3A). We found the mutation of the two binding sites recognized by CBFs or the binding sites at −231 to −227 bp, CBFs no longer bound to the ADF5 promoter in yeast, but the sites at −472 to −468 bp remained functional (Figures 3B,C). These results indicate that CBFs bind to the ADF5 promoter via the CRT/DRE DNA regulatory element at −231 to −227 bp sites.
To determine the effect of CBFs on ADF5 activation, we performed transient transcriptional activation experiments in tobacco leaves. Because Y1H analysis found that CBF1, CBF2, and CBF3 bound to the ADF5 promoter, and their function had a certain redundancy (Park et al., 2015), we selected the CBF3 with the strongest binding ability in the Y1H experiments for follow-up experiments (Figures 3B,C). Transcriptional activation analysis showed that CBF3 directly transcriptionally activated the expression of ADF5 in tobacco mesophyll cells (Figures 3D,E). Subsequent mutation analysis showed that the recognition sites at −472 to −468 bp did not affect the transcriptional activation efficiency of CBF3 to ADF5 and the mutation at −231 to −227 bp sites or both significantly reduced the transcriptional activation efficiency (Figure 3E). The results indicate that CBF3 activates the expression of ADF5 via binding to the CBF recognition sites of the ADF5 promoter at −231 to −227 bp.
To further confirm that CBFs bound to the ADF5 promoter in vivo, ChIP experiments were performed. Col/pSuper::CBF3-Myc was treated at 4°C for different times. Chromatin was precipitated using anti-c-myc antibody magnetic beads. P1 primers containing −472 to −468 bp sites and P2 primers containing −231 to −227 bp sites were used to analyze the enrichment by qRT-PCR (Figure 3A). The results showed no significant difference between the CBF3 overexpression line and the WT at the −472 to −468 bp sites at 0 h, but the CBF3 overexpression line was 1.3 times more enriched than the WT at the −231 to −227 bp sites. After 6 h of low-temperature treatment, the enrichment of WT at the two recognition sites did not change significantly, and CBF3 overexpression line showed a significant increase that was nearly three times the enrichment at the −231 to −227 bp sites (Figure 3F). These results suggest that CBF3 binds to the CBF recognition element of the ADF5 promoter in vivo, and this binding activity is regulated by low temperature. In conclusion, CBFs primarily bind to the CBF recognition sites at the −231 to −227 bp of the ADF5 promoter in vivo, and ADF5 is a downstream target gene of CBFs.
The Effect of ADF5 on Freezing Resistance in Arabidopsis thaliana Partially Depends on the CBF Pathway
To further investigate the genetic relationship between ADF5 and CBFs, adf5 cbfs quadruple mutant was obtained by crossing adf5-1 with the triple mutant cbfs-1. Freezing experiments showed that adf5-1, cbfs-1, and adf5 cbfs were more sensitive than WT under non-cold-acclimated (NA) conditions, and the survival rate was significantly different. The freezing resistance of adf5 cbfs was closer to adf5-1 with decreasing temperature and there was no significant difference between the two groups (Figures 4A,B). Under cold-acclimated (CA) conditions, adf5-1, cbfs-1, and adf5 cbfs were more sensitive to freezing than WT, and their survival rate was significantly different. The freezing resistance of the adf5 cbfs showed the freezing resistance of cbfs-1 and there was no significant difference between the two groups (Figures 4A,C). Seedlings grown in soil for 21 days were used for freezing treatment to obtain ion leakage. Under NA conditions, the ion leakage of WT and cbfs-1 treated at −4°C was similar, and there was no significant difference. adf5-1 and adf5 cbfs had higher ion leakage than WT and were significantly different. At −5°C, the ion leakage of cbfs-1 increased significantly relative to WT. The adf5-1 and adf5 cbfs had significantly higher ion leakage than WT, and the adf5 cbfs had significantly higher ion leakage than adf5-1 (Figure 4D). Under CA conditions, the ion leakage of adf5-1, cbfs-1, and adf5 cbfs was significantly higher than WT. The ion leakage of adf5 cbfs resembled cbfs-1, and there was no significant difference (Figure 4E). In conclusion, ADF5 positively regulates the freezing tolerance ability of Arabidopsis thaliana by partially relying on the CBF pathway.
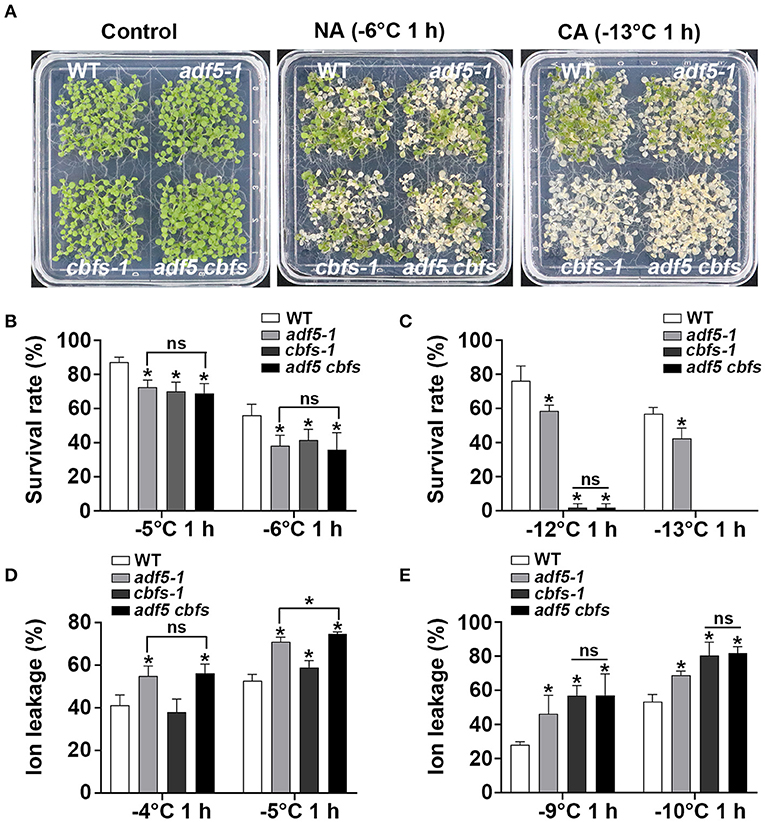
Figure 4. ADF5 is the genetic downstream of CBFs. (A) Freezing phenotypes of WT, adf5-1, cbfs-1, and adf5 cbfs under NA or CA conditions. (B,C) Survival rate of WT, adf5-1, cbfs-1, and adf5 cbfs. After 3–5 days of recovery under normal growth conditions, the survival rate was calculated. The data are the means of three biological replicates ± SD (n = 40 for each replicate). Asterisks indicate statistically significant differences and ns is not significant (P < 0.05, one-way ANOVA with a Tukey's multiple comparisons test). (D,E) Ion leakage of WT, adf5-1, cbfs-1, and adf5 cbfs. After 3 weeks of normal growth in soil, the fully developed rosettes of seedlings were used for freezing treatment to obtain ion leakage under NA or CA conditions. The data are the means of three biological replicates ±SD (n = 5 for each replicate). Asterisks indicate statistically significant differences and ns is not significant (P < 0.05, one-way ANOVA with a Tukey's multiple comparisons test).
The Actin Cytoskeleton of the Mutant Was Disordered During Cold Acclimation
To determine whether the phenotypes of the adf5 and cbfs mutants are directly related to the actin cytoskeleton, we observed the actin cytoskeleton morphology in epidermal cells from root transition and elongation zone of WT and mutants under normal growth and different durations of 4°C treatment (Figure 5A). ABD2-GFP was expressed in all mutants after crossing Col/pUBQ10::ABD2-GFP. The average fluorescence intensity was measured directly. Similar to previous reports, the fluorescence intensity of adf5-1 under normal growth conditions was significantly lower than the WT (Zhu et al., 2017; Qian et al., 2019). The fluorescence intensity of cbfs-1 was slightly higher than WT, but adf5 cbfs had the highest fluorescence intensity compared to WT, with a significant difference. After 6 h of low temperature treatment, the fluorescence intensity of WT and cbfs-1 decreased, and adf5 cbfs obviously decreased, but the fluorescence intensity of adf5-1 was not changed. There was no significant difference among the materials at this time. After 12 h of low-temperature treatment, the fluorescence values of all materials recovered, and there was no significant difference among WT, adf5-1, and cbfs-1. The fluorescence value of the adf5 cbfs quadruple mutant increased less, and there was a significant difference between WT and adf5 cbfs (Figure 5B).
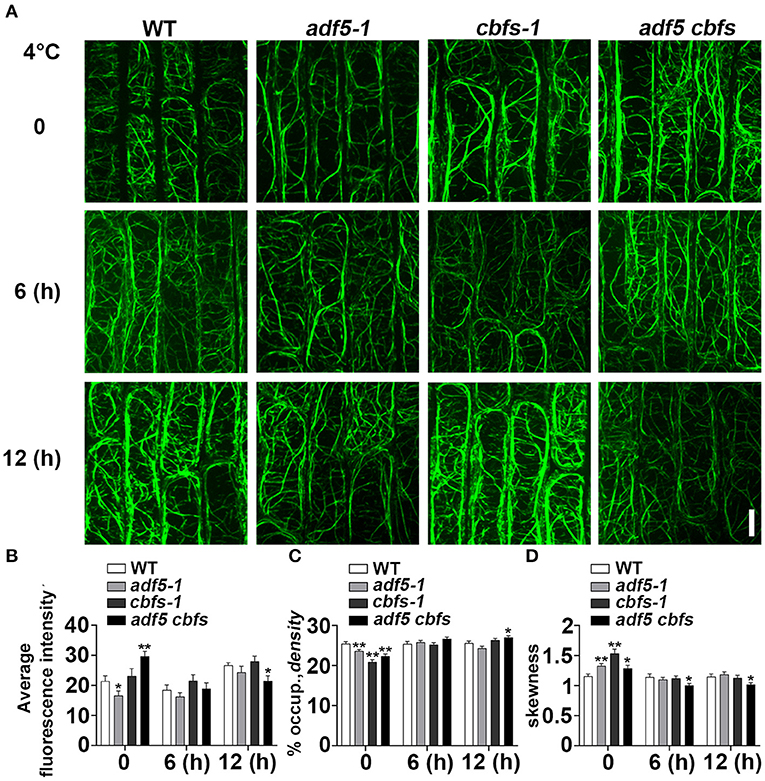
Figure 5. Actin cytoskeleton array rearranges in response to cold treatment. (A) Five-day-old seedlings expressing ABD2-GFP were used for fluorescence collection. Images of the cortical actin cytoskeleton array in epidermal cells from the root elongation transition zone of WT, adf5-1, cbfs-1, and adf5 cbfs treated at 4°C for different durations. Scale bar = 10 μm. (B) The average intensity of fluorescence of the GFP signal in WT, adf5-1, cbfs-1, and adf5 cbfs root elongation transition zone cells. (C) Average actin cytoskeleton density of WT, adf5-1, cbfs-1, and adf5 cbfs root elongation transition zone cells. (D) The extent of actin cytoskeleton bundling (skewness) of WT, adf5-1, cbfs-1, and adf5 cbfs root elongation transition zone cells. The values represent the means ± SEM (n = 60 cells per genotype. *P < 0.05; **P < 0.01; Student's two-tailed t-test).
We analyzed the actin cytoskeleton density and skewness after different time points of low-temperature treatment (Figures 5C,D). Consistent with a previous report, the structure of the actin cytoskeleton in WT was relatively stable (Shibasaki et al., 2009), and its density and degree of filament bundling (skewness) were not changed. Compared to WT, the mutants had lower actin cytoskeleton density and a higher degree of bundled actin filament at 0 h, which corresponded to the function of ADF5 in forming a more stable actin cytoskeleton network (Zhu et al., 2017; Qian et al., 2019). After 6 hours of low-temperature treatment, the actin cytoskeleton density in the adf5-1, cbfs-1, and adf5 cbfs mutants was relatively changed. However, there was no significant difference between WT and mutants. The degree of bundled actin filament was reduced, there was no significant differences among the adf5-1, cbfs-1, and WT, but significant differences between adf5 cbfs and WT. After 12 h of treatment, the density of adf5-1 decreased slightly and cbfs-1 increased, but there was no significant difference compared to WT. The quadruple mutant also increased and was significantly different than WT. The skewness value of adf5-1 increased, and the values of cbfs-1 and adf5 cbfs decreased. Only the quadruple mutant was significantly different from WT (Figures 5C,D). In conclusion, the actin cytoskeleton structure of WT is relatively stable, but the mutants change significantly compared to WT under low temperature treatment, and actin cytoskeleton dynamic process in mutants is affected.
The Endocytosis Rate of the Mutants Decreased During Cold Acclimation
The dynamics of the plasma membrane are very important for the plant response to stress, and the actin cytoskeleton is important for plant endocytosis (Baluška et al., 2002; Wang et al., 2020). Considering the effect of low temperature on actin cytoskeleton dynamics, then to track the endocytosis process, we used amphiphilic styryl dye FM4-64 to label epidermal cells from root transition zone and quantified the cytosol/PM FM4-64 signal intensity ratio to represent the endocytosis rate (Figures 6A,B). The results showed that the endocytosis rate of adf5-1, cbfs-1, and adf5 cbfs mutants was faster than WT, and there was significant differences between adf5-1, adf5 cbfs, and WT, respectively, but there was no significant difference in cbfs-1. The endocytosis rate of WT, adf5-1, cbfs-1, and adf5 cbfs decreased after 6 h of low-temperature treatment, and the difference between WT and mutants was reduced. There was a significant difference between adf5 cbfs and WT. After 12 h of low-temperature treatment, the endocytosis rate of WT and adf5-1 increased slightly, and there was no significant difference between adf5 and WT. The endocytosis rate of cbfs-1 and adf5 cbfs was further reduced, and there was a significant difference between cbfs-1 and WT, but not in the quadruple mutant (Figure 6B). The results indicate that the abnormal endocytosis rate of adf5-1, cbfs-1, and adf5 cbfs under normal growth conditions may be the reason for their freezing sensitivity. During the cold training process, the endocytosis rate of non-synchronous change in mutants may cause disorders of cold signals and cryoprotective substance transport, which may affect the freezing resistance of the mutants.
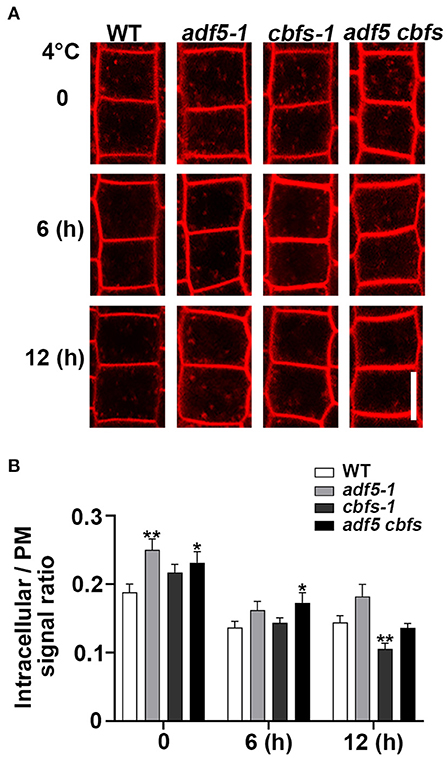
Figure 6. Low temperature reduced the rate of endocytosis in mutants compared to WT. (A) Five-day-old seedlings were used for amphiphilic styryl dye FM4-64 signal intensity collection. Seedling roots of WT, adf5-1, cbfs-1, and adf5 cbfs treated at 4°C for different durations were labeled with FM4-64 to measure internalization. Scale bar = 10 μm. (B) The rate of cytosol average fluorescence intensity to plasma membrane average fluorescence intensity. The values represent the means ± SEM (n = 60 cells per genotype. *P < 0.05; **P < 0.01; Student's two-tailed t-test).
Discussion
Previous reports showed that low temperature disturbed the actin cytoskeleton of plant cells and reduced all cell processes (Das et al., 1966; Pokorná et al., 2004; Fan et al., 2015). The present study provided molecular genetic and physiological evidences for ADF5, which is an ABP that evolved F-actin-bundling function, regulation of actin cytoskeleton dynamics at low temperature partially via a CBF-dependent pathway. The molecular components of fine regulation of the actin cytoskeleton at low temperature were demonstrated.
Cold acclimation reshapes the physiological and biochemical status of cells (Janmohammadi et al., 2015; Ding et al., 2019). CBFs are the core regulatory factor in this process and activate many downstream COR genes during cold acclimation (Shi et al., 2018). Published transcriptome data revealed that CBFs regulated a number of potential actin cytoskeleton regulatory genes including ADF5 (Jia et al., 2016). Previous evidence also showed that TaADF accumulated more in freezing-resistant seedlings (Ouellet et al., 2001). Our qRT-PCR data also directly indicated that ADF5 was partially up-regulated by CBFs at low temperature treatment (Figure 2). Genetic analyses also showed that the freezing sensitivity of adf5 cbfs was closer to adf5-1 under NA conditions (Figure 4A). Under this condition, the ion leakage of adf5 cbfs was higher than adf5-1, and the difference was statistically significant (Figure 4D). These results suggested that ADF5 promotes the freezing resistance of Arabidopsis thaliana via a non-CBF pathway. Under CA conditions, CBFs are the core transcription factors of the cold response pathway, and there are many downstream induced genes. The adf5 cbfs primarily showed the freezing sensitivity and physiological characteristics of cbfs-1 (Figures 4A,E). In summary, ADF5 promotes freezing resistance in Arabidopsis thaliana partially via the CBF signaling pathway. Moreover, transcriptome data clearly showed that some non-CBF-dependent ABPs were cold regulated (Jia et al., 2016; Zhao C. et al., 2016). For example, AtFH16 was downregulated, and FIM4 and CROLIN were up-regulated (Jia et al., 2013; Wang J. et al., 2013; Ding et al., 2018). Therefore, coordination of actin cytoskeleton dynamics is an important plant cell process under low-temperature stress.
Membrane vesicle transport is a conserved basic cell process in eukaryotes, and it plays an important role in the plant response to stress. Under stress, it is responsible for the correct sorting and positioning of specific proteins and the stress response (Rosquete and Drakakaki, 2018; Wang et al., 2020). For example, low temperature reduces the intracellular transport of auxin transporters PIN2 and PIN3, which affects auxin transport and changes the gravitropism of roots (Shibasaki et al., 2009). Recent studies also showed that stable GNOM ARF-GEF-mediated endosomal trafficking helped Arabidopsis adapt to low temperature (Ashraf and Rahman, 2019). The cryoprotective protein RCl2A also depends on clathrin-mediated endocytosis (Wang C. et al., 2013), and the transport of its homologous protein RCI2B at low temperature is selectively regulated to maintain a normal transport rate (Shibasaki et al., 2009). The actin cytoskeleton plays an important role in endocytosis and intracellular transport (Šamaj et al., 2004; Kim et al., 2005). Previous studies showed that BRI1 and CRPK1 were essential for the regulation of plant freezing resistance (Liu et al., 2017; Unterholzner et al., 2017). BRI1 is internalized by clathrin and sorted by the SYP61/VHA-a1 endosomal compartment, which also sorts the auxin transporters PIN2 and AUX1(Robert et al., 2008). This process requires coordination of the actin cytoskeleton (Lanza et al., 2012; Arieti and Staiger, 2020). The number of actin cytoskeletons decreased in the rice vln2 mutant, and the polarity distribution and cycle of PIN2 changed (Wu et al., 2015). FH5-regulated actin cytoskeleton polymerization and elongation processes are necessary for the movement of FH5-labeled vesicles in Arabidopsis pollen tubes (Liu C. et al., 2018). Therefore, the actin cytoskeleton is very important for the integrity of the endocytosis and sorting processes, which ultimately affects the correct positioning and distribution of endocytic cargos and the plant response to stress. However, the processes of vesicle transport and sorting controlled by the actin cytoskeleton under low temperature are rarely reported.
The present study showed that the morphology of the actin cytoskeleton in WT was relatively stable consistent with previous reports (Shibasaki et al., 2009; Figure 5), and its endocytosis rate was reduced during low-temperature treatment (Figure 6), which may be related to the slowing of molecular thermal movement at low temperature. However, the endocytosis rate in adf5-1 had a greater reduction than WT at 6 h, which may be related to the disorder of the adf5-1 actin cytoskeleton (Figures 5, 6). The endocytosis rate of cbfs-1 and adf5 cbfs continuously decreased significantly compared to WT at low temperature (Figure 6), which may be due to the influence of their disturbed actin cytoskeleton and additional regulatory pathways regulated downstream of CBFs. In summary, the damaged actin cytoskeleton of mutants changes the endocytosis rate during cold acclimation and the change in the endocytosis rate is not consistent with WT, in which the distribution, recovery and sorting of receptors and cryoprotective substances on the plasma membrane are directly changed. Therefore, the cold acclimation process of mutants is affected, and the freezing resistance of mutants is reduced.
Methods
Plant Materials and Growth Conditions
Arabidopsis seeds were vernalized for 3 days at 4°C with 2% PPM-Preservative (Plant Cell Technology) and grown on half-strength Murashige and Skoog medium (MS, PhytoTech M519) containing 0.8% agar and 1.5% sucrose for 12–15 days at 22°C with an optical density of 80–100 μmol m−2 s−1 under a 16 h light/8 h dark LD photoperiod. Arabidopsis thaliana Col-0 and Nicotiana benthamiana were used in this experiment. Mutants adf5-1 (Salk_018325) and adf5-2 (SALK_030145) were obtained from the Arabidopsis Biological Resource Center. adf5-3 was obtained using two specific Cas9-cleaved targets of ADF5 to mutate Col-0, and this method was obtained from the report (Xing et al., 2014). The cbf3, cbf1 cbf3, cbfs-1, and Col/pSuper::CBF3-Myc were obtained from the Prof. Shuhua Yang Laboratory (China Agricultural University) (Jia et al., 2016; Liu et al., 2017). The adf5 cbfs was obtained by crossing adf5-1 with cbfs-1. The primers used to identify homozygous lines and for CRISPR are listed in Supplementary Table 1. The materials for ion leakage measurements were grown on MS plates containing 1% agar for 5–7 days, transferred to soil, then grown at 23°C, 16 h light/8 h dark LD photoperiod to an optical density of ~100 μmol m−2 s−1 and relative humidity of 60% for 3 weeks. Nicotiana benthamiana had similar growth conditions.
Freezing Tolerance and Ion Leakage Assays
This experiment was performed according to the protocol described previously (Shi et al., 2012). Briefly, 12 to 15-day-old seedlings were used to obtain phenotypes by freezing treatment. For cold acclimation (CA) treatment, the normally growing seedlings were grown at 4°C for an additional 3 days at a light density of ~25 μmol m−2 s−1, 16 h light/ 8 h dark LD photoperiod and transferred to the freezing incubator (PERCIVAL, LT-36VLC8) for treatment according to the following procedure. The seedlings were kept at 0°C for 1 h, then decreased by 1°C per hour until the temperature shown in the figure was maintained for the corresponding times. Non-cold-acclimation (NA) processing was directly performed according to a program. After freezing treatment, the seedlings were shifted to 4°C, kept in darkness for 12 h, and transferred to a normal growth environment for growth recovery for 3–5 days. The survival rate was obtained by calculating the proportion of seedlings that grew new leaves.
Ion leakage assays was performed as described (Guo et al., 2002). Briefly, seedlings grown for 3 weeks in soil were treated for ion leakage analyses with or without cold acclimation (4°C for 3 d). A fully and well-developed rosette leaf was washed with deionized water, and placed in the bottom of a 15-mL sterile centrifuge tube containing 100 μL deionized water, then placed in a low-temperature circulator (SCIENTZ, DC-2030). After holding at 0°C for 30 min, a small amount of tiny and pure ice crystal was added to the centrifuge tube, and the temperature was reduced by 1°C per 30 min until the temperature shown in the figure and maintained for 1 h. The centrifuge tube was removed and kept in darkness at 4°C for 12 h. Then, 10 mL of deionized water was added to the sterile tube, and the tubes were shaken at normal temperature for 2 h. The electrical conductivity S1 was measured. After sterilization at 121°C for 15 min, S2 conductivity was measured after shaking for 2 h. The S0 conductivity of deionized water used in the experiment, and the conductivity S0′ after sterilization was measured. The ratio of S1–S0 to S2–S0′ was used as the ion leakage.
RNA Extraction and Real-Time Quantitative PCR Analysis
Seedlings grown in a normal environment for 10 days on MS plates containing 1% agar were used for gene expression analyses with or without treatment at 4°C. Total RNA was extracted using a MiniBEST Plant RNA Extraction kit (TaKaRa), and total RNA was reverse transcribed into cDNA using M-MLV reverse transcriptase (TaKaRa). qRT-PCR was performed using SYBR Premix Ex Taq (TaKaRa), and UBQ10 was used as an internal reference gene. The primers used for qRT-PCR are listed in Supplementary Table 1.
Yeast One-Hybrid Assays
Yeast one-hybrid (Y1H) assays were performed as described previously (Qian et al., 2019). Briefly, the ADF5 promoter containing the CRT/DRE DNA regulatory element was amplified using PCR and cloned into the pAbAi vector. The CRT/DRE mutation sequence was obtained using specific primers and cloned into the pAbAi vector. The vector was linearized and transformed into the Y1H GOLD (Clontech) strain. The full-length CBF coding sequence was amplified using PCR, cloned into the pGADT7 vector, and transformed into the constructed Y1H strain. Aureobasidin A (AbA, 850 ng/mL, Clontech) was used for the reporter gene activation test. The primers used to clone are listed in Supplementary Table 1.
Transcriptional Activation Assays
This experiment was performed as described previously (Qian et al., 2019). PCR amplified and cloned the ADF5 promoter sequence into pGWB235-LUC to construct the reporter vector. The full-length CBF3 coding sequence was amplified using PCR, and it was cloned into the pBIB-35s-GWR-flag vector as the effect vector. The reporter and effector vectors were co-transformed into N. benthamiana leaves for transcriptional activation analyses. A reporting vector alone was used as a negative control. Fluorescence of the luciferase and luciferin (Promega) reaction was obtained using a Lumazone CA1300B camera (Photometrics). The primers used to clone are listed in Supplementary Table 1.
Chromatin Immunoprecipitation Assays
Under normal growth conditions, seedlings grown on 1% agar MS plates for 12 days were used in this experiment. The seedlings of Col/pSuper::CBF3-Myc and Col-0 (without Myc-tag, as negative control) were treated at 4°C or not, and a ChIP experiment was performed according to a previously described method (Ni et al., 2009). The relative enrichment of the ADF5 promoter region was detected by qRT-PCR using specific primers. The specific primers are listed in Supplementary Table 1.
Visualization of Actin Filaments by Confocal Laser Scanning Microscopy and Quantitative Analyses of Actin Filament
To visualize the actin cytoskeleton, we crossed Col-0/pUBQ10::ABD2-GFP (Tian et al., 2015) with adf5-1, cbfs-1, and adf5 cbfs and expressed ABD2-GFP in mutants for GFP fluorescence collection. Under normal growth conditions, seedlings grown on 1% agar MS plates for 5 days were treated with or without 4°C for the duration shown in the figure for fluorescence acquisition. Images of epidermal cells from root transition and elongation zone were acquired using a spinning disk microscope (Andor) equipped with a 63 × NA oil immersion lens, and the Z-series images were captured with the step size set at 0.5 μm (Zhou et al., 2020). GFP was excited with a 488-nm laser and observed using a 514-nm emission filter. Images were processed and analyzed using ImageJ. To measure the extra actin cytoskeleton structure, the percentage of occupancy (density) and skewness were measured using ImageJ software as previously described (Higaki et al., 2010; Henty et al., 2011).
FM4-64 Labeling and Endocytosis Measurements
Under normal growth conditions, the seedlings growing on 1% agar MS plates for 5 days were treated with or without 4°C for different times then used in this experiment. Seedlings were held on ice for 2 min with 2 μM FM4-64 and stained at 22 or 4°C for an additional 5 min. After FM4-64 washout, dye internalization was imaged using a Zeiss LSM880 confocal microscope equipped with 40 × 1.3 NA oil immersion lens. FM4-64 signals were excited with a 488-nm laser and observed using a 600 to 630-nm emission filter. The endocytosis rate was measured and calculated in ImageJ software as previously described (Zhang et al., 2020).
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author Contributions
YuN conceived the study and designed the research. PZ, DQ, CL, YiN, TL, and CL performed the research. PZ, DQ, CL, YX, XW, and YiN analyzed the data. PZ, YuN, YX, and DQ wrote the manuscript.
Funding
This work was supported by grants from the National Natural Science Foundation of China (31722005 and 31670180).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Professor Qijun Chen (China Agricultural University) for sharing CRISPR/Cas9 vector. We thank Professor Shuhua Yang (China Agricultural University) for sharing the cbf related mutants. We thank the Core Facility of the School of Life Sciences, Lanzhou University for technical assistance.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2021.635533/full#supplementary-material
References
Andrianantoandro, E., and Pollard, T. D. (2006). Mechanism of actin filament turnover by severing and nucleation at different concentrations of ADF/Cofilin. Mol. Cell 24, 13–23. doi: 10.1016/j.molcel.2006.08.006
Arieti, R. S., and Staiger, C. J. (2020). Auxin-induced actin cytoskeleton rearrangements require AUX1. New Phytol. 226, 441–459. doi: 10.1111/nph.16382
Ashraf, M. A., and Rahman, A. (2019). Cold stress response in Arabidopsis thaliana is mediated by GNOM ARF-GEF. Plant J. 97, 500–516. doi: 10.1111/tpj.14137
Baluška, F., Hlavacka, A., Šamaj, J., Palme, K., Robinson, D. G., Matoh, T., et al. (2002). F-actin-dependent endocytosis of cell wall pectins in meristematic root cells. Insights from brefeldin A-induced compartments. Plant Physiol. 130, 422–431. doi: 10.1104/pp.007526
Bou Daher, F., Van Oostende, C., and Geitmann, A. (2011). Spatial and temporal expression of actin depolymerizing factors ADF7 and ADF10 during male gametophyte development in Arabidopsis thaliana. Plant Cell Physiol. 52, 1177–1192. doi: 10.1093/pcp/pcr068
Bremer, A., Wolff, M., Thalhammer, A., and Hincha, D. K. (2017). Folding of intrinsically disordered plant LEA proteins is driven by glycerol-induced crowding and the presence of membranes. FEBS J. 284, 919–936. doi: 10.1111/febs.14023
Burgos-Rivera, B., Ruzicka, D. R., Deal, R. B., McKinney, E. C., King-Reid, L., and Meagher, R. B. (2008). Actin depolymerizing factor9 controls development and gene expression in Arabidopsis. Plant Mol. Biol. 68, 619–632. doi: 10.1007/s11103-008-9398-1
Chen, C. Y., Wong, E. I., Vidali, L., Estavillo, A., Hepler, P. K., Wu, H. M., et al. (2002). The regulation of actin organization by actin-depolymerizing factor in elongating pollen tubes. Plant Cell 14, 2175–2190. doi: 10.1105/tpc.003038
Chen, C. Y. H., Cheung, A. Y., and Wu, H. M. (2003). Actin-depolymerizing factor mediates Rac/Rop GTPase-regulated pollen tube growth. Plant Cell 15, 237–249. doi: 10.1105/tpc.007153
Chinnusamy, V., Zhu, J., and Zhu, J. K. (2007). Cold stress regulation of gene expression in plants. Trends Plant Sci. 12, 444–451. doi: 10.1016/j.tplants.2007.07.002
Clément, M., Ketelaar, T., Rodiuc, N., Banora, M. Y., Smertenko, A., Engler, G., et al. (2009). Actin-depolymerizing factor2-mediated actin dynamics are essential for root-knot nematode infection of Arabidopsis. Plant Cell 21, 2963–2979. doi: 10.1105/tpc.109.069104
Danyluk, J., Carpentier, E., and Sarhan, F. (1996). Identification and characterization of a low temperature regulated gene encoding an actin-binding protein from wheat. FEBS Lett. 389, 324–327. doi: 10.1016/0014-5793(96)00599-6
Das, T. M., Hildebrandt, A. C., and Riker, A. J. (1966). Cine-photomicrography of low temperature effects on cytoplasmic streaming, nucleolar activity and mitosis in single tobacco cells in microculture. Am. J. Bot. 53, 253–259. doi: 10.1002/j.1537-2197.1966.tb07331.x
Ding, X., Zhang, S., Liu, J., Liu, S., and Su, H. (2018). Arabidopsis FIM4 and FIM5 regulates the growth of root hairs in an auxin-insensitive way. Plant Signal. Behav. 13:e1473667. doi: 10.1080/15592324.2018.1473667
Ding, Y., Shi, Y., and Yang, S. (2019). Advances and challenges in uncovering cold tolerance regulatory mechanisms in plants. New Phytol. 222, 1690–1704. doi: 10.1111/nph.15696
Dolferus, R. (2014). To grow or not to grow: a stressful decision for plants. Plant Sci. 229, 247–261. doi: 10.1016/j.plantsci.2014.10.002
Dong, C. H., Xia, G. X., Hong, Y., Ramachandran, S., Kost, B., and Chua, N. H. (2001). ADF proteins are involved in the control of flowering and regulate F-actin organization, cell expansion, and organ growth in Arabidopsis. Plant Cell 13, 1333–1346. doi: 10.1105/TPC.010051
Fan, T. T., Ni, J. J., Dong, W. C., An, L. Z., Xiang, Y., and Cao, S. Q. (2015). Effect of low temperature on profilins and ADFs transcription and actin cytoskeleton reorganization in Arabidopsis. Biol. Plant 59, 793–796. doi: 10.1007/s10535-015-0546-6
Fu, Y., Duan, X., Tang, C., Li, X., Voegele, R. T., Wang, X., et al. (2014). TaADF7, an actin-depolymerizing factor, contributes to wheat resistance against Puccinia striiformis f. sp. tritici. Plant J. 78, 16–30. doi: 10.1111/tpj.12457
Gilmour, S. J., Zarka, D. G., Stockinger, E. J., Salazar, M. P., Houghton, J. M., and Thomashow, M. F. (1998). Low temperature regulation of the Arabidopsis CBF family of AP2 transcriptional activators as an early step in cold-induced COR gene expression. Plant J. 16, 433–442. doi: 10.1046/j.1365-313x.1998.00310.x
Guo, Y., Xiong, L., Ishitani, M., and Zhu, J. K. (2002). An Arabidopsis mutation in translation elongation factor 2 causes superinduction of CBF/DREB1 transcription factor genes but blocks the induction of their downstream targets under low temperatures. Proc. Natl. Acad. Sci. U.S.A. 99, 7786–7791. doi: 10.1073/pnas.112040099
Henty, J. L., Bledsoe, S. W., Khurana, P., Meagher, R. B., Day, B., Blanchoin, L., et al. (2011). Arabidopsis actin depolymerizing factor4 modulates the stochastic dynamic behavior of actin filaments in the cortical array of epidermal cells. Plant Cell 23, 3711–3726. doi: 10.1105/tpc.111.090670
Higaki, T., Kutsuna, N., Sano, T., Kondo, N., and Hasezawa, S. (2010). Quantification and cluster analysis of actin cytoskeletal structures in plant cells: role of actin bundling in stomatal movement during diurnal cycles in Arabidopsis guard cells. Plant J. 61, 156–165. doi: 10.1111/j.1365-313X.2009.04032.x
Hussey, P. J., Allwood, E. G., and Smertenko, A. P. (2002). Actin-binding proteins in the Arabidopsis genome database: properties of functionally distinct plant actin-depolymerizing factors/cofilins. Philos. Trans. R. Soc. B Biol. Sci. 357, 791–798. doi: 10.1098/rstb.2002.1086
Inada, N., Higaki, T., and Hasezawa, S. (2016). Nuclear function of subclass I actin-depolymerizing factor contributes to susceptibility in Arabidopsis to an adapted powdery mildew fungus. Plant Physiol. 170, 1420–1434. doi: 10.1104/pp.15.01265
Jaglo-Ottosen, K. R., Gilmour, S. J., Zarka, D. G., Schabenberger, O., and Thomashow, M. F. (1998). Arabidopsis CBF1 overexpression induces COR genes and enhances freezing tolerance. Science 280, 104–106. doi: 10.1126/science.280.5360.104
Janmohammadi, M., Zolla, L., and Rinalducci, S. (2015). Low temperature tolerance in plants: changes at the protein level. Phytochemistry 117, 76–89. doi: 10.1016/j.phytochem.2015.06.003
Jia, H., Li, J., Zhu, J., Fan, T., Qian, D., Zhou, Y., et al. (2013). Arabidopsis CROLIN1, a novel plant actin-binding protein, functions in cross-linking and stabilizing actin filaments. J. Biol. Chem. 288, 32277–32288. doi: 10.1074/jbc.M113.483594
Jia, Y., Ding, Y., Shi, Y., Zhang, X., Gong, Z., and Yang, S. (2016). The cbfs triple mutants reveal the essential functions of CBFs in cold acclimation and allow the definition of CBF regulons in Arabidopsis. New Phytol. 212, 345–353. doi: 10.1111/nph.14088
Jiang, Y., Wang, J., Xie, Y., Chen, N., and Huang, S. (2017). ADF10 shapes the overall organization of apical actin filaments by promoting their turnover and ordering in pollen tubes. J. Cell Sci. 130, 3988–4001. doi: 10.1242/jcs.207738
Kim, H., Park, M., Kim, S. J., and Hwang, I. (2005). Actin filaments play a critical role in vacuolar trafficking at the Golgi complex in plant cells. Plant Cell 17, 888–902. doi: 10.1105/tpc.104.028829
Lanza, M., Garcia-Ponce, B., Castrillo, G., Catarecha, P., Sauer, M., Rodriguez-Serrano, M., et al. (2012). Role of actin cytoskeleton in brassinosteroid signaling and in its integration with the auxin response in plants. Dev. Cell 22, 1275–1285. doi: 10.1016/j.devcel.2012.04.008
Lin, C., and Thomashow, M. F. (1992). DNA sequence analysis of a complementary DNA for cold-regulated Arabidopsis Gene cor15 and characterization of the COR15 polypeptide. Plant Physiol. 99, 519–525. doi: 10.1104/pp.99.2.519
Liu, C., Zhang, Y., and Ren, H. (2018). Actin polymerization mediated by AtFH5 directs the polarity establishment and vesicle trafficking for pollen germination in Arabidopsis. Mol. Plant 11, 1389–1399. doi: 10.1016/j.molp.2018.09.004
Liu, J., Shi, Y., and Yang, S. (2018). Insights into the regulation of C-repeat binding factors in plant cold signaling. J. Integr. Plant Biol. 60, 780–795. doi: 10.1111/jipb.12657
Liu, Q., Kasuga, M., Sakuma, Y., Abe, H., Miura, S., Yamaguchi-Shinozaki, K., et al. (1998). Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell 10, 1391–1406. doi: 10.1105/tpc.10.8.1391
Liu, Z., Jia, Y., Ding, Y., Shi, Y., Li, Z., Guo, Y., et al. (2017). Plasma membrane CRPK1-mediated phosphorylation of 14-3-3 proteins induces their nuclear import to fine-tune CBF signaling during cold response. Mol. Cell 66, 117–128.e5. doi: 10.1016/j.molcel.2017.02.016
Medina, J., Bargues, M., Terol, J., Pérez-Alonso, M., and Salinas, J. (1999). The Arabidopsis CBF gene family is composed of three genes encoding AP2 domain-containing proteins whose expression is regulated by low temperature but not by abscisic acid or dehydration. Plant Physiol. 119, 463–469. doi: 10.1104/pp.119.2.463
Mondal, H. A., Louis, J., Archer, L., Patel, M., Nalam, V. J., Sarowar, S., et al. (2018). Arabidopsis ACTIN-DEPOLYMERIZING FACTOR3 is required for controlling aphid feeding from the phloem. Plant Physiol. 176, 879–890. doi: 10.1104/pp.17.01438
Nan, Q., Qian, D., Niu, Y., He, Y., Tong, S., Niu, Z., et al. (2017). Plant actin-depolymerizing factors possess opposing biochemical properties arising from key amino acid changes throughout evolution. Plant Cell 29, 395–408. doi: 10.1105/tpc.16.00690
Ni, Z., Kim, E.-D., Ha, M., Lackey, E., Liu, J., Zhang, Y., et al. (2009). Altered circadian rhythms regulate growth vigour in hybrids and allopolyploids. Nature 457, 327–331. doi: 10.1038/nature07523
Ouellet, F., Carpentier, E., Cope, M. J. T. V., Monroy, A. F., and Sarhan, F. (2001). Regulation of a wheat actin-depolymerizing factor during cold acclimation. Plant Physiol. 125, 360–368. doi: 10.1104/pp.125.1.360
Park, S., Lee, C. M., Doherty, C. J., Gilmour, S. J., Kim, Y., and Thomashow, M. F. (2015). Regulation of the Arabidopsis CBF regulon by a complex low-temperature regulatory network. Plant J. 82, 193–207. doi: 10.1111/tpj.12796
Pokorná, J., Schwarzerová, K., Zelenková, S., Petrášek, J., Janotová, I., Capková, V., et al. (2004). Sites of actin filament initiation and reorganization in cold-treated tobacco cells. Plant Cell Environ. 27, 641–653. doi: 10.1111/j.1365-3040.2004.01186.x
Porter, K., Shimono, M., Tian, M., and Day, B. (2012). Arabidopsis actin-depolymerizing factor-4 links pathogen perception, defense activation and transcription to cytoskeletal dynamics. PLoS Pathog. 8:e1003006. doi: 10.1371/journal.ppat.1003006
Qian, D., and Xiang, Y. (2019). Actin cytoskeleton as actor in upstream and downstream of calcium signaling in plant cells. Int. J. Mol. Sci. 20:1403. doi: 10.3390/ijms20061403
Qian, D., Zhang, Z., He, J., Zhang, P., Ou, X., Li, T., et al. (2019). Arabidopsis ADF5 promotes stomatal closure by regulating actin cytoskeleton remodeling in response to ABA and drought stress. J. Exp. Bot. 70, 435–446. doi: 10.1093/jxb/ery385
Robert, S., Chary, S. N., Drakakaki, G., Li, S., Yang, Z., Raikhel, N. V., et al. (2008). Endosidin1 defines a compartment involved in endocytosis of the brassinosteroid receptor BRI1 and the auxin transporters PIN2 and AUX1. Proc. Natl. Acad. Sci. U.S.A. 105, 8464–8469. doi: 10.1073/pnas.0711650105
Rosquete, M. R., and Drakakaki, G. (2018). Plant TGN in the stress response: a compartmentalized overview. Curr. Opin. Plant Biol. 46, 122–129. doi: 10.1016/j.pbi.2018.09.003
Ruzicka, D. R., Kandasamy, M. K., McKinney, E. C., Burgos-Rivera, B., and Meagher, R. B. (2007). The ancient subclasses of Arabidopsis ACTIN DEPOLYMERIZING FACTOR genes exhibit novel and differential expression. Plant J. 52, 460–472. doi: 10.1111/j.1365-313X.2007.03257.x
Šamaj, J., Baluška, F., Voigt, B., Schlicht, M., Volkmann, D., and Menzel, D. (2004). Endocytosis, actin cytoskeleton, and signaling. Plant Physiol. 135, 1150–1161. doi: 10.1104/pp.104.040683
Shi, Y., Ding, Y., and Yang, S. (2018). Molecular regulation of CBF signaling in cold acclimation. Trends Plant Sci. 23, 623–637. doi: 10.1016/j.tplants.2018.04.002
Shi, Y., Tian, S., Hou, L., Huang, X., Zhang, X., Guo, H., et al. (2012). Ethylene signaling negatively regulates freezing tolerance by repressing expression of CBF and type-A ARR genes in Arabidopsis. Plant Cell 24, 2578–2595. doi: 10.1105/tpc.112.098640
Shibasaki, K., Uemura, M., Tsurumi, S., and Rahman, A. (2009). Auxin response in Arabidopsis under cold stress: underlying molecular mechanisms. Plant Cell 21, 3823–3838. doi: 10.1105/tpc.109.069906
Staiger, C. J. (2000). Signaling to the actin cytoskeleton in plants. Annu. Rev. Plant Biol. 51, 257–288. doi: 10.1146/annurev.arplant.51.1.257
Staiger, C. J., and Blanchoin, L. (2006). Actin dynamics: old friends with new stories. Curr. Opin. Plant Biol. 9, 554–562. doi: 10.1016/j.pbi.2006.09.013
Staiger, C. J., Gibbon, B. C., Kovar, D. R., and Zonia, L. E. (1997). Profilin and actin-depolymerizing factor: modulators of actin organization in plants. Trends Plant Sci. 2, 275–281. doi: 10.1016/S1360-1385(97)86350-9
Stockinger, E. J., Gilmour, S. J., and Thomashow, M. F. (1997). Arabidopsis thaliana CBF1 encodes an AP2 domain-containing transcriptional activator that binds to the C-repeat/DRE, a cis-acting DNA regulatory element that stimulates transcription in response to low temperature and water deficit. Proc. Natl. Acad. Sci. U.S.A. 94, 1035–1040. doi: 10.1073/pnas.94.3.1035
Tholl, S., Moreau, F., Hoffmann, C., Arumugam, K., Dieterle, M., Moes, D., et al. (2011). Arabidopsis actin-depolymerizing factors (ADFs) 1 and 9 display antagonist activities. FEBS Lett. 585, 1821–1827. doi: 10.1016/j.febslet.2011.05.019
Thomashow, M. F. (1999). Plant cold acclimation: freezing tolerance genes and regulatory mechanisms. Annu. Rev. Plant Biol. 50, 571–599. doi: 10.1146/annurev.arplant.50.1.571
Tian, J., Han, L., Feng, Z., Wang, G., Liu, W., Ma, Y., et al. (2015). Orchestration of microtubules and the actin cytoskeleton in trichome cell shape determination by a plant-unique kinesin. Elife 4:e09351. doi: 10.7554/eLife.09351
Tian, M., Chaudhry, F., Ruzicka, D. R., Meagher, R. B., Staiger, C. J., and Day, B. (2009). Arabidopsis actin-depolymerizing factor AtADF4 mediates defense signal transduction triggered by the Pseudomonas syringae effector AvrPphB1. Plant Physiol. 150, 815–824. doi: 10.1104/pp.109.137604
Unterholzner, S. J., Rathnayake, A. I., Khan, M., Kugler, K. G., May, S. T., Mayer, K. F. X., et al. (2017). Brassinosteroids participate in the control of basal and acquired freezing tolerance of plants. Proc. Natl. Acad. Sci. U.S.A. 114, E1038–E1089. doi: 10.1073/pnas.1700593114
Wang, C., Yan, X., Chen, Q., Jiang, N., Fu, W., Ma, B., et al. (2013). Clathrin light chains regulate clathrin-mediated trafficking, auxin signaling, and development in Arabidopsis. Plant Cell 25, 499–516. doi: 10.1105/tpc.112.108373
Wang, J., Zhang, Y., Wu, J., Meng, L., and Ren, H. (2013). At FH16, an Arabidopsis type II formin, binds and bundles both microfilaments and microtubules, and preferentially binds to microtubules. J. Integr. Plant Biol. 55, 1002–1015. doi: 10.1111/jipb.12089
Wang, X., Xu, M., Gao, C., Zeng, Y., Cui, Y., Shen, W., et al. (2020). The roles of endomembrane trafficking in plant abiotic stress responses. J. Integr. Plant Biol. 62, 55–69. doi: 10.1111/jipb.12895
Wu, S., Xie, Y., Zhang, J., Ren, Y., Zhang, X., Wang, J., et al. (2015). VLN2 regulates plant architecture by affecting microfilament dynamics and polar auxin transport in rice. Plant Cell. 27, 2829–2845. doi: 10.1105/tpc.15.00581
Xing, H. L., Dong, L., Wang, Z. P., Zhang, H. Y., Han, C. Y., Liu, B., et al. (2014). A CRISPR/Cas9 toolkit for multiplex genome editing in plants. BMC Plant Biol. 14:327. doi: 10.1186/s12870-014-0327-y
Ye, K., Li, H., Ding, Y., Shi, Y., Song, C., Gong, Z., et al. (2019). BRASSINOSTEROID-INSENSITIVE2 negatively regulates the stability of transcription factor ICE1 in response to cold stress in Arabidopsis. Plant Cell 31, 2682–2696. doi: 10.1105/tpc.19.00058
Zhang, J., Li, X. M., Lin, H. X., and Chong, K. (2019). Crop improvement through temperature resilience. Annu. Rev. Plant Biol. 70, 753–780. doi: 10.1146/annurev-arplant-050718-100016
Zhang, X., Adamowski, M., Marhava, P., Tan, S., Zhang, Y., Rodriguez, L., et al. (2020). Arabidopsis flippases cooperate with ARF GTPase exchange factors to regulate the trafficking and polarity of PIN auxin transporters. Plant Cell 32, 1644–1664. doi: 10.1105/tpc.19.00869
Zhao, C., Zhang, Z., Xie, S., Si, T., Li, Y., and Zhu, J. K. (2016). Mutational evidence for the critical role of CBF transcription factors in cold acclimation in Arabidopsis. Plant Physiol. 171, 2744–2759. doi: 10.1104/pp.16.00533
Zhao, S., Jiang, Y., Zhao, Y., Huang, S., Yuan, M., Zhao, Y., et al. (2016). CASEIN KINASE1-LIKE PROTEIN2 regulates actin filament stability and stomatal closure via phosphorylation of actin depolymerizing factor. Plant Cell 28, 1422–1439. doi: 10.1105/tpc.16.00078
Zheng, Y., Xie, Y., Jiang, Y., Qu, X., and Huang, S. (2013). Arabidopsis ACTIN-DEPOLYMERIZING FACTOR7 severs actin filaments and regulates actin cable turnover to promote normal pollen tube growth. Plant Cell 25, 3405–3423. doi: 10.1105/tpc.113.117820
Zhou, Y., Yang, Y., Niu, Y., Fan, T., Qian, D., Luo, C., et al. (2020). The tip-localized phosphatidylserine established by Arabidopsis ALA3 is crucial for Rab GTPase-mediated vesicle trafficking and pollen tube growth. Plant Cell 32, 3170–3187. doi: 10.1105/tpc.19.00844
Keywords: actin depolymerizing factor, cold stress, C-repeat binding factors genes, actin cytoskeleton, regulatory mechanism
Citation: Zhang P, Qian D, Luo C, Niu Y, Li T, Li C, Xiang Y, Wang X and Niu Y (2021) Arabidopsis ADF5 Acts as a Downstream Target Gene of CBFs in Response to Low-Temperature Stress. Front. Cell Dev. Biol. 9:635533. doi: 10.3389/fcell.2021.635533
Received: 30 November 2020; Accepted: 08 January 2021;
Published: 28 January 2021.
Edited by:
Yi Zhang, Beijing Normal University, ChinaReviewed by:
Xiangfeng Wang, China Agricultural University, ChinaZhaosheng Kong, Chinese Academy of Sciences, China
Copyright © 2021 Zhang, Qian, Luo, Niu, Li, Li, Xiang, Wang and Niu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yue Niu, bml1eUBsenUuZWR1LmNu
 Pan Zhang
Pan Zhang Dong Qian
Dong Qian Changxin Luo
Changxin Luo Yun Xiang
Yun Xiang Yue Niu
Yue Niu