- 1Department of Neurobiology, Basic Medical College, Qingdao University, Qingdao, China
- 2Department of Laboratory, Women and Children’s Hospital of Qingdao, Qingdao, China
- 3Department of Biochemistry and Molecular Biology, Basic Medical College, Qingdao University, Qingdao, China
- 4Department of Breast Surgery, The Affiliated Hospital of Qingdao University, Qingdao University, Qingdao, China
A regulator of ribosome synthesis 1 (RRS1) was discovered in yeast and is mainly localized in the nucleolus and endoplasmic reticulum. It regulates ribosomal protein, RNA biosynthesis, and protein secretion and is closely involved in cellular senescence, cell cycle regulation, transcription, translation, oncogenic transformation etc., Mutations in the RRS1 gene are associated with the occurrence and development of Huntington’s disease and cancer, and overexpression of RRS1 promotes tumor growth and metastasis. In this review, the structure, function, and mechanisms of RRS1 in various diseases are discussed.
Introduction
Ribosomes are the sites of protein synthesis and consist of ribosomal RNA (rRNA) and ribosomal proteins (RPs). In mammalian ribosomes, rRNA and RPs are arranged into large (60S) and small (40S) subunits. The RPs also have functions independent of ribosomes, such as the regulation of DNA repair, cell proliferation, apoptosis, and differentiation (Warner and Mcintosh, 2009). Furthermore, RPs participate in the coordination of protein synthesis in the cytoplasm and mitochondria (Surguchov, 1987). Warner and McIntosh suggested that the extra-ribosomal functions of RPs can be classified as follows: (1) regulation and maintenance of the ribosome assembly, (2) identification of the nucleolar stress responses to mediate G phase cell cycle arrest, and (3) apoptosis and other specific or non-specific functions. Tsuno et al. discovered the regulator of the ribosome synthesis 1 (RRS1) protein in yeast cells. It is located in the nucleolus and endoplasmic reticulum, and is involved in ribosome biogenesis, 25S rRNA maturation, and the assembly of the 60S subunit (Surguchov, 1987). Aberrant expression levels of RRS1 affect ribosome biogenesis, which in turn has pathological consequences.
The Structure of RRS1 Gene and Its Coding Protein
The human RRS1 gene is 1,714 bp long and is located in chromosome 8q13.1. It contains only one exon (NCBI accession number: NC_000008.11) that is transcribed into a single mRNA transcript without any alternatively spliced isoforms. The 1,079 bp open reading frame (ORF) of RRS1 encodes for the RRS1 protein containing 365 amino acid residues with a relative molecular mass of 41,193 KDa. The RRS1 protein is mainly localized to the nucleolus and endoplasmic reticulum. The C-terminal residues 302–365 are rich in arginine, glycine and lysine (Arg/Gly/Lys). The first methionine residue at the N-terminus can be modified by acetylation, and Arg344 at the C-terminus is the phosphorylation site.
The Biological Function of RRS1
RRS1 Is Involved in Ribosome Biosynthesis
Cellular proliferation requires high rates of protein synthesis, which in turn is dependent on ribosome biogenesis (Warner and Mcintosh, 2009). RRS1 regulates 25S rRNA maturation and 60S subunit assembly (Tsuno et al., 2000). Tsujii et al. (2000) found that RRS1 formed a complex with the nucleolus protein EBP2 in yeast cells, which bound to and stabilized the pre-ribosomal subunit and promoted ribosome biosynthesis. RRS1 also binds to the ribosomal protein RPL11 on the 60S subunit, indicating its important role in ribosome biogenesis (Miyoshi et al., 2002). In Saccharomyces cerevisiae, RRS1 interacts with RPF2 (ribosome production factor 2) to recruit RPL5 and RPL11, to synthesize the 60S subunit (Fatica and Tollervey, 2002; Fromont-Racine et al., 2003; Granneman and Baserga, 2004). In the absence of RRS1 and RPF2 binding, the immature pre-RNA is exported from the nucleolus to the nuclear mass but not to the cytoplasm, thereby affecting 60S subunit biosynthesis (Zhang et al., 2007). Missense mutations in the C-terminal of RRS1 have been detected in secretion-deficient yeast cells, indicating that it plays an important role in protein synthesis and secretion. Morita et al. showed that Rpf2 binds to both RRS1 and RPL11 in yeast cells and regulates the 27-SB pre-rRNA transformation during 25S and 60S ribosomal subunit biosynthesis (Morita et al., 2002). Miyoshi et al. found that RRS1 interacts with the 60S subunit during biogenesis, maturation, assembly, and export from the nucleus to the cytoplasm (Miyoshi et al., 2004; Nariai et al., 2005). Zhang et al. found that RRS1 and Rpf2 recruit 5S rRNA, RPL5, and RPL11 to the newly produced ribosomes (Zhang et al., 2007). Therefore, the main role of RRS1 in ribosomal biosynthesis is to recruit the necessary factors and to form a stable structure, especially for the assembly and nucleation of 60S subunit.
RRS1 Regulates Chromosomal and Telomeric Arrangement During the Cell Cycle
The process of mitosis is divided into distinct stages namely interphase, prophase, metaphase, anaphase, and telophase. During the prophase, the chromosomes start to condense and gradually converge to the equatorial plate by a metaphase (Kuriyama and Borisy, 1982). Gambe et al. (2009) found that RRS1 silencing in HeLa cell lines increased the proportion of cells in the early metaphase and disrupted chromosomal arrangement on the equatorial plate, resulting in a delayed anaphase. The accumulation of chromosomes in the equatorial plate depends on the cohesion of sister chromatids, and their attachment to the spindle microtubules. RRS1-knockdown HeLa cells treated with colchicine and MG132 showed premature separation of the sister chromatids and lower levels of the centromere protein Shugoshin 1, resulting in abnormally arranged chromosomes on the equatorial plate (Losada, 2008). Thus, RRS1 controls chromosomal accumulation on the equatorial plate during cell cycle (Gambe et al., 2009).
The telomeres are located at the termini of linear chromosomes, and consist of TTAGGG repeat sequences and related proteins that are protected by a cap structure (Chan and Blackburn, 2002; Palm and De, 2008). SIR4 and yKu are partially bound to the N-terminus of the membrane protein s3 (MPs3) during the S phase of the cell cycle to create a niche for anchoring telomeres (Hediger et al., 2002; Taddei et al., 2004; Bupp et al., 2007; Schober et al., 2009). This process is stabilized by the EBNA1 binding protein 2 (EBP2) and RRS1, which interact with both MPs3 and the C-terminus of Sir4. Yeast cells with mutated EBP2 and RRS1 show aberrant telomere aggregation (Horigome et al., 2011). Therefore, RRS1 and EBP2 also affect telomere aggregation.
RRS1 Weakens Cell Senescence
Due to a limitation of the number of proliferating cycles, the inactivation of oncogenes, activation of tumor suppressor genes, drugs that induce DNA damage, and cell senescence affection, most mammalian cells cannot proliferate indefinitely (Campisi and d’Adda di Fagagna, 2007; Kuilman et al., 2010; Pérezmancera et al., 2014). Although induction of senescence in cancer cells can inhibit tumor growth or elicit an immune response to clear precancerous cells, recent studies show that senescent cells lead to age-related dysfunction through inflammatory reactions (Campisi and d’Adda di Fagagna, 2007; Xue et al., 2007; Baker et al., 2011; Kang et al., 2015). Telomerase dysfunction, DNA damage response, and oxidative stress induce senescence via the p53-p14/p19Arf and p16INK4A-RB pathways (Schramek et al., 2011). The tumor suppressor p53 is normally expressed at low levels due to its interaction with the E3 ubiquitin ligase murine double minute 2 (MDM2) (Kuilman et al., 2010; Lee and Gu, 2010). It is activated in response to cellular stresses with the detachment of MDM2, and transcriptionally activates downstream targets effectors of cell cycle arrest, apoptosis, DNA repair, and cell senescence (Levine and Oren, 2009). Recent studies show that the nucleoli can sense various stresses and activate the p53 pathway (Lee and Gu, 2010). Following nuclear stress, several ribosomal proteins including nucleolar phosphate protein, nucleolar protein, nuclear factor, RL11, RPL5, RPL23, and RPS7 are released. These proteins directly bind to MDM2 to prevent ubiquitin-mediated degradation of p53, thereby maintaining normal cell proliferation (Boulon et al., 2010). Bhat’s study showed that RPL11 is involved in the p53 nuclear stress response pathway (Manfredi, 2010). Increased RPL11 translocation is observed in cells with aberrant 40S ribosomal subunit biogenesis, and RPL11 translation promotes p53 activation (Bhat et al., 2004). Some studies show that RPL11 regulates MDM2 as part of the 5S ribonucleoprotein (RNP) complex consisting of RPL11, RPL5, and 5S rRNA (Wilson et al., 1996; Sugawara et al., 1999; Fumagalli et al., 2012; Donati et al., 2013). Miyoshi et al. found that RRS1 binds to RPL11, and enhances the binding of p53 to MDM2, which downregulates p53 activity (Miyoshi et al., 2002). Thus, overexpression of RRS1 diminishes p53-mediated cellular senescence.
The Relationship Between RRS1 and Diseases
The Relationship Between RRS1 and Huntington Disease
Huntington disease (HD) is a neurodegenerative disease characterized by involuntary limb movement and progressive dementia. Endoplasmic reticulum stress (ERS) has recently been identified as the molecular basis of HD pathogenesis (Duennwald and Lindquist, 2008; Reijonen et al., 2008). The ER is extremely sensitive to cellular stresses such as low energy levels, abnormal redox state, and high calcium influx, and can trigger apoptosis. Therefore, ERS is closely related to the genesis and progression of various diseases (Harding and Ron, 2002; Mouw et al., 2003). RRS1 mRNA levels were significantly increased in the brain tissues of a HD (Hdh+/– and Hdh –/–) mouse model, as well as in the brain tissues of HD patients relative to age-matched healthy controls (Fossale et al., 2002). Furthermore, RRS1 and metadherin are co-localized on the ER in HD mice. RRS1 is activated during ERS and relays the signals to metadherin. Therefore, RRS1 plays an important role in the pathogenesis of HD by promoting ERS (Carnemolla et al., 2009).
The Role of RRS1 and Cancer
Abnormal ribosome biosynthesis can trigger the ribosomal stress response, which activates the oncogenic p53-HDM2 feedback pathway (Macias et al., 2010; Montanaro et al., 2012). Aberrant expression of RPL5, RPL11, RPL23, and RPS7 can also promote the ribosomal stress response and p53-HDM2 feedback pathway (Lohrum et al., 2003; Dai and Lu, 2004; Jin et al., 2004; Chen et al., 2007). Several RPs are aberrantly expressed in human tumors. For instance, RPL29 is significantly up-regulated in colorectal cancer (Wang et al., 1999), and RPS8, RPL12, RPL23A, RPL27, and RPL30 show high expression levels in diverse cancers. In addition, overexpression of RPS3A promotes tumor formation in nude mice (Naora et al., 1998).
RRS1 is distributed in the periphery of the nucleus in HeLa cells unlike the nuclear localization seen in normal cells. RRS1 silencing delays cell cycle progression in HeLa cells (Gambe et al., 2009). In addition, RRS1 regulates the balance between cytoplasmic membrane and endoplasmic reticulum, and its dysregulation in tumor cells can significantly alter their physiological functions.
The Relationship Between RRS1 and Cervical Cancer
Cervical cancer is one of the leading causes of death among women worldwide (Jemal et al., 2011), and its incidence and fatality rates has increased significantly in developing countries. It ranks second after breast cancer as the most common cancer in females (Li et al., 2013). Although the exact pathological mechanism is unknown, virus infection and genetic mutations have been implicated in the development of cervical cancer. Gambe et al. (2009) detected high levels of RRS1 in the nuclear periphery of cervical cancer Hela cells, which bound to other nucleolar proteins during mitosis and interphase. The expression level of RRS1 was particularly increased at prophase during nucleolus breakage. RRS1 silencing significantly increased the number of tetraploid cells and significantly prolonged cell division, indicating a role in chromosome segregation (Gambe et al., 2009). Finally, down-regulation of RRS1 by miRN-148a inhibited the proliferation, invasion, and migration of cervical cancer cells (Zhang et al., 2019). Therefore, RRS1 promotes cervical cancer development and progression.
The Relationship Between RRS1 and HCC
Hepatocellular carcinoma is a lethal malignancy and ranks third in terms of mortality rates after gastric cancer and esophageal cancer (Zimber and GesPaeh, 2008). The incidence of liver cancer has increased significantly in recent years, and patient prognosis is dismal due to a < 10% 5 year survival rate. RRS1 is overexpressed in liver cancer tissues relative to the para-carcinoma tissues, and the overall and disease-free survival rates of patients with high RRS1 expression are significantly lower than that in RRS1low patients. Wang et al. found that RRS1 gene silencing in the liver cancer cell line SMMC7721 induced G1 phase arrest, which effectively blocked cell division and proliferation (Wang et al., 2017). In addition, RRS1 promoted HCC cell proliferation and colony formation, and inhibited apoptosis (Wang et al., 2017). Therefore, RRS1 functions as a pro-tumorigenic factor in liver cancer.
The Relationship Between RRS1 and Breast Cancer
Breast cancer is a common malignancy among females, and its incidence rate is increasing every year. It exhibits high genetic heterogeneity (Miller et al., 2016; Turashvili and Brogi, 2017), and targeted therapies against specific gene expression patterns have significantly improved the survival rate of breast cancer patients (Staaf et al., 2010). We found that the RRS1 gene is highly expressed in breast cancer cells, and its knockdown significantly reduced the proliferation rate and increased apoptosis (Hua et al., 2019). RRS1 may promote proliferation of breast cancer through p53 activation via RPL11/MDM2 (Figure 1) (Song et al., 2018). Taken together, RRS1 is a novel oncogene of breast cancer and a promising therapeutic target.
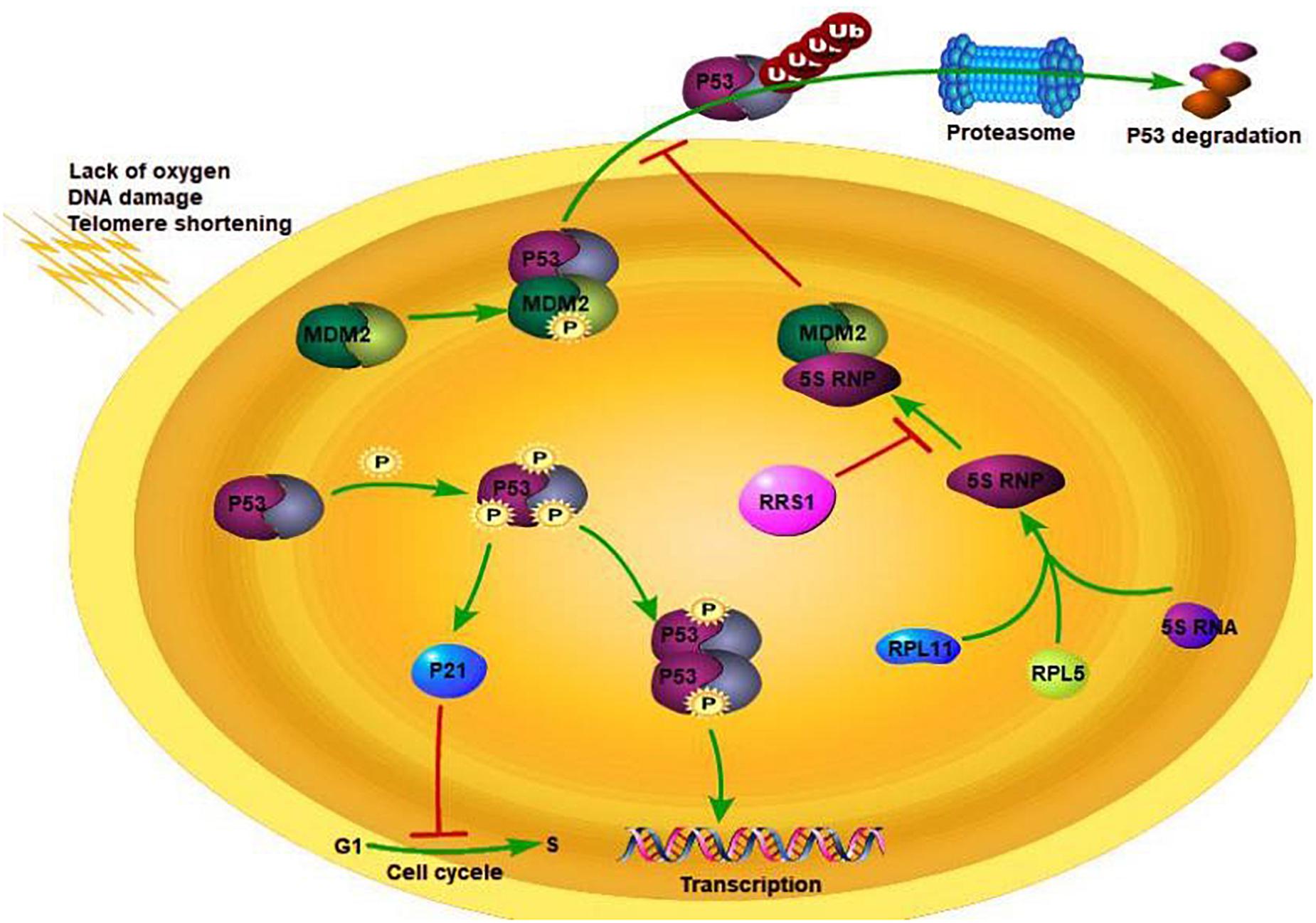
Figure 1. RSS1 regulation mechanism. Adapted from Cao (2019).
The Relationship Between RRS1 and PTC
Papillary thyroid carcinoma (PTC) is one of the most common malignancies of the endocrine system, though the mechanisms underlying the pathogenesis, progression, and prognosis are still largely unclear (Burman and Wartofsky, 2015). Chen et al. recently found that RRS1 silencing in PTC cells inhibited their proliferation by inducing G2/M phase arrest and promoting apoptosis. In addition, RRS1 knockdown promoted metabolism and inhibited vascular development in the in vivo PTC model (Chen et al., 2018). Thus, RRS1 is a potential oncogene in PTC and should be further explored.
The Relationship Between RRS1 and Colorectal Cancer
Colorectal cancer (CRC) is the third most common malignancy and the fourth leading cause of cancer-related deaths globally (Alwan, 2007; Siegel et al., 2012). Several mutations have been identified as risk factors for the occurrence and progression of CRC (Fearon, 2011; Vermeulen et al., 2013), and their biological relevance have been validated in functional in vitro and in vivo studies (Hussain et al., 2000; von Karstedt et al., 2015). Therapies targeting proto-oncogenes or tumor suppressors have been developed but their efficacy is limited (Amado et al., 2008; Karapetis et al., 2008; Grothey et al., 2013). Therefore, there is an urgent need to explore new drug targets to treat CRC. Wu et al. found that RRS1 is overexpressed in CRC tissues compared to that in normal tissues and knocking down RRS1 in CRC cells inhibited their proliferation and tumorigenesis in nude mice (Wu et al., 2017). Thus, RRS1 is a potential therapeutic target for CRC.
The Relationship Between RRS1 and Gastric Cancer
Gastric cancer (GC) is one of the most common malignant tumors in the digestive system (Song et al., 2017), and the second leading cause of cancer-related deaths worldwide (Karimi et al., 2014). Due to the metastatic potential, recurrence, and chemoresistance of gastric cancer cells, the prognosis of patients with advanced gastric cancer is poor and the overall survival rate is dismal (Fujita, 2009). Studies show that cancer stem cells (CSCs) are the major determinant of gastric tumor metastasis and invasion (Yu et al., 2012). Ma et al. (2019) recently found that miRNA-598 regulates the growth of gastric CSCs by regulating the expression of RRS1.
Conclusion
RRS1 plays an important role in ribosome biosynthesis, chromosome aggregation in the equatorial plate, and telomere aggregation during cell cycle. In addition, it regulates the cell cycle, ERS, and ribosomal stress response, which affects the p53 signaling pathway. The aberrant expression of RRS1 is associated with Huntington’s disease and cancer development. RRS1 is a potential oncogene in various cancers, and therefore a promising therapeutic target.
Author Contributions
YH: conceptualization, writing of original draft, project administration, and editing. JS and CP: data curation, formal analysis, funding acquisition, and editing. RW, ZM, JZ, ZZ, and NL: data curation, validation, and editing. LH: conceptualization, supervision, and editing. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by the National Natural Science Foundation of China (Grant No. 81472542), the Focus on Research and Development Plan in Shandong (Grant No. 2019GSF107025), the Qingdao Startup and Innovation Leader Talent Plan (Grant No. 13-CX-3, 201409-201709), the Qingdao Applied Basic Research Program Youth Project (Grant No. 19-6-2-59-cg), and the Qingdao Science and Technology Benefiting Project (Grant No. 19-6-1-47-nsh).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Amado, R. G., Wolf, M., Peeters, M., Van, C. E., Siena, S., Freeman, D. J., et al. (2008). Wild-type KRAS is required for panitumumab efficacy in patients with metastatic colorectal cancer. J. Clin. Oncol. 26, 1626–1634.
Baker, D. J., Wijshake, T., Tchkonia, T., Brasseur, N. K. L., Childs, B. G., Sluis, B. V. D., et al. (2011). Clearance of p16Ink4a-positive senescent cells delays ageing-associated disorders. J. Agric. Biotechnol. 479, 232–236. doi: 10.1038/nature10600
Bhat, K. P., Itahana, K., Jin, A., and Zhang, Y. P. (2004). Essential role of ribosomal protein L11 in mediating growth inhibition-induced p53 activation. Embo J. 23, 2402–2412. doi: 10.1038/sj.emboj.7600247
Boulon, S., Westman, B. J., Hutten, S., Boisvert, F. M., and Lamond, A. I. (2010). The nucleolus under stress. Mol. Cell 40, 216–227. doi: 10.1016/j.molcel.2010.09.024
Bupp, J. M., Martin, A. E., Stensrud, E. S., and Jaspersen, S. L. (2007). Telomere anchoring at the nuclear periphery requires the budding yeast Sad1-UNC-84 domain protein Mps3. J. Cell Biol. 179, 845–854. doi: 10.1083/jcb.200706040
Burman, K. D., and Wartofsky, L. (2015). Clinical practice. Thyroid nodules. N. Engl. J. Med. 373, 2347–2356.
Campisi, J., and d’Adda di Fagagna, F. (2007). Cellular senescence: when bad things happen to good cells. Nat. Rev. Mol. Cell Biol. 8, 729–740. doi: 10.1038/nrm2233
Cao, Y. (2009). The function and mechanism of ribosomal regulatory protein 1 in neuroblastoma. doi: 10.27262/d.cnki.gqdau.2019.002235
Carnemolla, A., Fossale, E., Agostoni, E., Michelazzi, S., Calligaris, R., Maso, L. D., et al. (2009). Rrs1 is involved in endoplasmic reticulum stress response in huntington disease. J. Biol. Chem. 284, 18167–18173. doi: 10.1074/jbc.m109.018325
Chan, S. R. W. L., and Blackburn, E. H. (2002). Telomeres and Telomerase. Hoboke, NJ: John Wiley & Sons, Inc.
Chen, D., Zhang, Z., Li, M., Wang, W., Li, Y., Rayburn, E. R., et al. (2007). Ribosomal protein S7 as a novel modulator of p53-MDM2 interaction: binding to MDM2, stabilization of p53 protein, and activation of p53 function. Oncogene 26, 5029–5037. doi: 10.1038/sj.onc.1210327
Chen, F., Jin, Y., Feng, L., Zhang, J., Tai, J., Shi, J., et al. (2018). RRS1 gene expression involved in the progression of papillary thyroid carcinoma. Cancer Cell Int. 18:20.
Dai, M. S., and Lu, H. (2004). Inhibition of MDM2-mediated p53 ubiquitination and degradation by ribosomal protein L5. J. Biol. Chem. 279, 44475–44482. doi: 10.1074/jbc.m403722200
Donati, G., Peddigari, S., Mercer, C. A., and Thomas, G. (2013). 5S Ribosomal RNA is an essential component of a nascent ribosomalprecursor complex that regulates the Hdm2-p53 checkpoint. Cell Rep. 4, 87–98. doi: 10.1016/j.celrep.2013.05.045
Duennwald, M. L., and Lindquist, S. (2008). Impaired ERAD and ER stress are early and specific events in polyglutamine toxicity. Genes Dev. 22, 3308–3319. doi: 10.1101/gad.1673408
Fatica, A., and Tollervey, D. (2002). Making ribosomes. Curr. Opin. Cell Biol. 14, 313–318. doi: 10.1016/s0955-0674(02)00336-8
Fossale, E., Wheeler, V. C., Vrbanac, V., Lebel, L. A., Teed, A., Mysore, J. S., et al. (2002). Identification of a presymptomatic molecular phenotype in Hdh CAG knock-in mice. Hum. Mol. Genet. 11, 2233–2241. doi: 10.1093/hmg/11.19.2233
Fromont-Racine, M., Senger, B., Saveanu, C., and Fasiolo, F. (2003). Ribosome assembly in eukaryotes. Gene 313, 17–42. doi: 10.1016/s0378-1119(03)00629-2
Fujita, T. (2009). Gastric cancer. Lancet 374, 1593–1594. author reply 1594–1595,Google Scholar
Fumagalli, S., Ivanenkov, V. V., Teng, T., and Thomas, G. (2012). Suprainduction of p53 by disruption of 40S and 60S ribosome biogenesis leads to the activation of a novel G2/M checkpoint. Genes Dev. 26, 1028–1040. doi: 10.1101/gad.189951.112
Gambe, A. E., Matsunaga, S., Takata, H., Maniwa, R. O., Baba, A., Uchiyama, S., et al. (2009). A nucleolar protein. R. R.S1 contributes to chromosome congression. Febs Lett. 583, 1951–1956. doi: 10.1016/j.febslet.2009.05.033
Granneman, S., and Baserga, S. J. (2004). Ribosome biogenesis: of knobs and. R. N.A processing. Exp. Cell Res. 296, 43–50. doi: 10.1016/j.yexcr.2004.03.016
Grothey, A., Van Cutsem, E., Sobrero, A., Siena, S., Falcone, A., Ychou, M., et al. (2013). Regorafenib monotherapy for previously treated metastatic colorectal cancer (CORRECT), an international, multicentre, randomised, placebo-controlled, phase 3 trial. Lancet. 381, 303–312.
Harding, H. P., and Ron, D. (2002). Endoplasmic reticulum stress and the development of diabetes: a review. Diabetes 3(Suppl. 3), S455–S461. doi: 10.2337/diabetes.51.2007.S455
Hediger, F., Neumann, F. R., Houwe, G. V., Dubrana, K., and Gasser, S. M. (2002). Live imaging of telomeres: yKu and Sir proteins define redundant telomere-anchoring pathways in yeast. Curr. Biol. 12, 2076–2089.
Horigome, C., Okada, T., Shimazu, K., Gasser, S. M., and Mizuta, K. (2011). Ribosome biogenesis factors bind a nuclear envelope SUN domain protein to cluster yeast telomeres. Embo J. 30, 3799–3811. doi: 10.1038/emboj.2011.267
Hua, Y. N., Song, J. L., Ma, Z. L., Wu, L., Zhang, Z., Zhang, L., et al. (2019). Effect of RRS1 gene knockdown on BT549 cell line proliferation and apoptosis in breast cancer. Neoplasma 66, 28–32.
Hussain, S. P., Amstad, P., Raja, K., Ambs, S., Nagashima, M., Bennett, W. P., et al. (2000). Increased p53 mutation load in noncancerous colon tissue from ulcerative colitis: a cancerprone chronic inflammatory disease. Cancer Res. 60, 3333–3337.
Jemal, A., Bray, F., Center, M. M., Ferlay, J., Ward, E., and Forman, D. (2011). Global cancer statistics. Ca A Cancer J. Clin. 61, 69–90.
Jin, A., Itahana, K., O’Keefe, K., and Zhang, Y. P. (2004). Inhibition of HDM2 and activation of p53 by ribosomal protein L23. Mol. Cell Biol. 24, 7669–7680. doi: 10.1128/MCB.24.17.7669-7680.2004
Kang, T. W., Yevsa, T., Woller, N., Hoenicke, L., Wuestefeld, T., Dauch, D., et al. (2015). Senescence surveillance of pre-malignant hepatocytes limits liver cancer development. Nature 4, 547–551.
Karapetis, C. S., Khambata, F. S., Jonker, D. J., O’Callaghan, C. J., Tu, D., Tebbutt, N. C., et al. (2008). K-ras mutations and benefit from cetuximab in advanced colorectal cancer. N. Engl. J. M. 359, 1757–1765.
Karimi, P., Islami, F., Anandasabapathy, S., Freedman, N. D., and Kamangar, F. (2014). Gastric cancer: descriptive epidemiology, risk factors, screening, and prevention. Cancer Epidemiol. Biomarkers Prev. 23, 700–713. doi: 10.1158/1055-9965.EPI-13-1057
Kuilman, T., Michaloglou, C., Mooi, W. J., and Peeper, D. S. (2010). The essence of senescence. Genes Dev. 24, 2463–2479. doi: 10.1101/gad.1971610
Kuriyama, R., and Borisy, G. G. (1982). Microtubule-nucleating activity of centrosomes in Chinese hamster ovary cells is independent of the centriole cycle but coupled to the mitotic cycle. J. Cell Biol. 91(3 Pt 1), 822–826. doi: 10.1083/jcb.91.3.822
Lee, J. T., and Gu, W. (2010). The multiple levels of regulation by p53 ubiquitination. Cell Death Differ. 17, 86–92.
Levine, A. J., and Oren, M. (2009). The first 30 years of p53: growing ever more complex. Nat. Rev. Cancer 9, 749–758. doi: 10.1038/nrc2723
Li, S., Hu, T., Lv, W., Zhou, H., Li, X., Yang, R., et al. (2013). Changes in prevalence and clinical characteristics of cervical cancer in the People’s Republic of China: a study of 10,012 cases from a nationwide working group. Oncologist 18, 1101–1107.
Lohrum, M. A., Ludwig, R. L., Kubbutat, M. H., Hanlon, M., and Vousden, K. H. (2003). Regulation of HDM2 activity by the ribosomal protein L11. CancerCell 3, 577–587. doi: 10.1016/S1535-6108(03)00134-X
Losada, A. (2008). The regulation of sister chromatid cohesion. BBA Rev. Cancer 1786, 41–48. doi: 10.1016/j.bbcan.2008.04.003
Ma, Y., Yan, F., Wei, W., Deng, J., Li, L., Liu, L., et al. (2019). Affiliations expand. MicroRNA-598 inhibits the growth and maintenance of gastric cancer stem-like cells by down-regulating RRS1. Cell Cycle 18, 2757–2769.
Macias, E., Jin, A., Deisenroth, C., Bhat, K., Mao, H., Lindström, M. S., et al. (2010). An ARF-independent c-MYC-activated tumor suppression pathway mediated by ribosomal protein-Mdm2 interaction. Cancer Cell 18, 231–243.
Manfredi, J. J. (2010). The Mdm2-p53 relationship evolves: Mdm2 swings both ways as an oncogene and a tumor suppressor. Genes Dev. 24, 1580–1589. doi: 10.1101/gad.1941710
Miller, K. D., Siegel, R. L., Lin, C. C., Mariotto, A. B., Kramer, J. L., Rowland, J. L. H., et al. (2016). Affiliations expand. Cancer treatment and survivorship statistics. CA Cancer J. Clin. 66, 271–289.
Miyoshi, K., Shirai, C., Horigome, C., Takenami, K., Kawasaki, J., and Mizuta, K. (2004). Rrs1p, a ribosomal protein L11-binding protein, is required for nuclear export of the 60S pre-ribosomal subunit in Saccharomyces cerevisiae. Febs Lett. 565, 106–110. doi: 10.1016/j.febslet.2004.03.087
Miyoshi, K., Tsujii, R., Yoshida, H., Maki, Y., Wada, A., Matsui, Y., et al. (2002). Normal assembly of 60 S ribosomal subunits is required for the signaling in response to a secretory defect in Saccharomyces cerevisiae. J. Biol. Chem. 277, 18334–18339.
Montanaro, L., Trere, D., and Derenzini, M. (2012). Changes in ribosome biogenesis may induce cancer by down-regu lating the cell tumor suppressor potential. Biochim. Biophys. Acta 1825, 10l–110. doi: 10.1016/j.bbcan.2011.10.006
Morita, D., Miyoshi, K., Matsui, Y., Toh-E, A., Shinkawa, H., Miyakawa, T., et al. (2002). Rpf2p, an evolutionarily conserved protein, interacts with ribosomal protein L11 and is essential for the processing of 27 SB Pre?rRNA to 25 S rRNA and the 60 S ribosomal subunit assembly in Saccharomyces cerevisiae. J. Biol. Chem. 277, 28780–28786. doi: 10.1074/jbc.M203399200
Mouw, G., Zechel, J. L., Gamboa, J., Lust, W. D., Selman, W. R., and Ratcheson, R. A. (2003). Activation of caspase-12, an endoplasmic reticulum resident caspase, after permanent focal ischemia in rat. Neuroreport 14, 183–186. doi: 10.1097/00001756-200302100-00004
Naora, H., Takai, I., Adachi, M., and Naora, H. (1998). Altered cellular responses by varying expression of a ribosomal protein gene:sequential co-ordination of enhancement and suppression of ribosomal protein S3a gene expression induces apoptosis. J. Cell Biol. 141, 741–753. doi: 10.1083/jcb.141.3.741
Nariai, M., Tanaka, T., Okada, T., Shirai, C., Horigome, C., and Mizuta, K. (2005). Synergistic defect in 60S ribosomal subunit assembly caused by a mutation of Rrs1p, a ribosomal protein L11-binding protein, and 3′-extension of 5S rRNA in Saccharomyces cerevisiae. Nucleic Acids Res. 33, 4553–4562. doi: 10.1093/nar/gki772
Palm, W., and De, L. T. (2008). How shelterin protects mammalian telomeres. Annu. Rev. Genet. 42, 301–334. doi: 10.1146/annurev.genet.41.110306.130350
Pérezmancera, P. A., Young, A. R., and Narita, M. (2014). Inside and out: the activities of senescence in cancer. Nat. Revi. Cancer 14, 547–558. doi: 10.1038/nrc3773
Reijonen, S., Putkonen, N., Nørremølle, A., Lindholm, D., and Korhonen, L. (2008). Inhibition of endoplasmic reticulum stress counteracts neuronal cell death and protein aggregation caused by N?terminal mutant huntingtin proteins. Exp. Cell Res. 314, 950–960. doi: 10.1016/j.yexcr.2007.12.025
Schober, H., Ferreira, H., Kalck, V., Gehlen, L. R., and Gasser, S. M. (2009). Yeast telomerase and the SUN domain protein Mps3 anchor telomeres and repress subtelomeric recombination. Genes Dev. 23, 928–938. doi: 10.1101/gad.1787509
Schramek, D., Kotsinas, A., Meixner, A., Wada, T., Elling, U., Pospisilik, J. A., et al. (2011). The stress kinase MKK7 couples oncogenic stress to p53 stability and tumor suppression. Nat. Genet. 43, 212–219.
Siegel, R., Naishadham, D., and Jemal, A. (2012). Cancer statistics,. CA Cancer J. Clin. 62, 10–29. doi: 10.3322/caac.20138
Song, J., Ma, Z., Hua, Y., Xu, J. T., Li, N., Ju, C. X., et al. (2018). Functional role of RRS1 in breast cancer cell proliferation. J. Cell Mol. Med. 22, 6304–6313.
Song, Z., Wu, Y., Yang, J., Yang, D. Q., and Fang, X. D. (2017). Progress in the treatment of advanced gastric cancer. Tumour Biol. 39:1010428317714626.
Staaf, J., Ringnér, M., Vallon-Christersson, J., Jönsson, G., Bendahl, P., Holm, K., et al. (2010). Identification of subtypes in human epidermal growth factor receptor 2–positive breast cancer reveals a gene signature prognostic of outcome. J. Clin. Oncol. 28, 1813–1820.
Sugawara, Y., Makuuchi, M., Kato, N., Shimotohno, K., and Takada, K. (1999). Enhancement of hepatitis C virus replication by Epstein| [ndash]| Barr virus-encoded nuclear antigen 1. Embo J. 18, 5755–5760. doi: 10.1093/emboj/18.20.5755
Surguchov, A. P. (1987). Common genes for mitochondrial and cytoplasmic proteins. Biochem. Sci. 12, 335–338. doi: 10.1016/0968-0004(87)90157-5
Taddei, A., Hediger, F., Neumann, F. R., Bauer, C., and Gasser, S. M. (2004). Separation of silencing from perinuclear anchoring functions in yeast Ku80, Sir4 and Esc1 proteins. Embo J. 23, 1301–1312. doi: 10.1038/sj.emboj.7600144
Tsujii, R., Miyoshi, K., Tsuno, A., Matsui, Y., Toh-e, A., Miyakawa, T., et al. (2000). Ebp2p, yeast homologue of a human protein that interacts with Epstein-Barr virus nuclear antigen 1, is required for pre-rRNA processing and ribosomal subunit assembly. Genes to Cells 5, 543–553. doi: 10.1046/j.1365-2443.2000.00346.x
Tsuno, A., Miyoshi, K., Tsujii, R., Miyakawa, T., and Mizuta, K. (2000). RRS1, a conserved essential gene, encodes a novel regulatory protein required for ribosome biogenesis inSaccharomyces cerevisiae. Mol. Cell. Biol. 20, 2066–2074. doi: 10.1128/MCB.20.6.2066-2074.2000
Vermeulen, L., Morrissey, E., van der Heijden, M., Nicholson, A. M., Sottoriva, A., Buczacki, S., et al. (2013). Defining stem cell dynamics in models of intestinal tumor initiation. Science 342, 995–998.
von Karstedt, S., Conti, A., Nobis, M., Montinaro, A., Hartwig, T., Lemke, J., et al. (2015). Cancer cell-autono mous, T. R.AIL-R signaling promotes KRAS-driven cancer progression, invasion, and metastasis. Cancer Cell 27, 561–573.
Wang, J., Li, Z., Zuo, C., Xie, Q. F., Li, H., Jia, J. H., et al. (2017). Knockdown of RRS1 by lentiviral-mediated RNAi promotes apoptosis and suppresses proliferation of human hepatocellular carcinoma cells. Oncol. Rep. 38, 2166–2172.
Wang, Y., Cheong, D., Chan, S., and Hooi, S. C. (1999). Heparin/heparan sulfate interacting protein gene expression is up-regulated in human colorecta1 carcinoma and correlated with differentiation status and metastasis. Cancer Res. 59, 2989–2994.
Warner, J. R., and Mcintosh, K. B. (2009). How common are extraribosomal functions of ribosomal proteins? Mol. Cell 34, 3–11. doi: 10.1016/j.molcel.2009.03.006
Wilson, J. B., Bell, J. L., and Levine, A. J. (1996). Expression of epstein-barr virus nuclear antigen-1 induces B cell neoplasia in transgenic mice. Embo J. 15, 3117–3126. doi: 10.1002/j.1460-2075.1996.tb00674.x
Wu, X. L., Yang, Z. W., He, L., Dong, P. D., Hou, M. X., Meng, X. K., et al. (2017). RRS1 silencing suppresses colorectal cancer cell proliferation and tumorigenesis by inhibiting G2/M progression and angiogenesis. Oncotarget 8, 82968–82980.
Xue, W., Zender, L., Miething, C., Dickins, R. A., Hernando, E., Krizhanovsky, V., et al. (2007). Senescence and tumour clearance is triggered by p53 restoration in murine liver carcinomas. Nature 445, 656–660.
Yu, Z., Pestell, T. G., Lisanti, M. P., and Pestell, R. G. (2012). Cancer stem cells. Int. J. Biochem. Cell Biol. 44, 2144–2151.
Zhang, J., Harnpicharnchai, P., Jakovljevic, J., Tang, L., Guo, Y. R., Oeffinger, M., et al. (2007). Assembly factors Rpf2 and Rrs1 recruit 5S rRNA and ribosomal proteins rpL5 and rpL11 into nascent ribosomes. Genes Dev. 21, 2580–2592. doi: 10.1101/gad.1569307
Zhang, Y., Sun, B., Zhao, L., Liu, Z. L., Xu, Z. L., Tian, Y. H., et al. (2019). Up-regulation of miRNA-148a inhibits proliferation, invasion, and migration while promoting apoptosis of cervical cancer cells by down-regulating RRS1. Biosci. Rep. 39:BSR20181815.
Keywords: RRS1, ribosome, disease, cancer, molecular mechanism
Citation: Hua Y, Song J, Peng C, Wang R, Ma Z, Zhang J, Zhang Z, Li N and Hou L (2021) Advances in the Relationship Between Regulator of Ribosome Synthesis 1 (RRS1) and Diseases. Front. Cell Dev. Biol. 9:620925. doi: 10.3389/fcell.2021.620925
Received: 14 November 2020; Accepted: 20 January 2021;
Published: 25 February 2021.
Edited by:
Andrei Surguchov, University of Kansas Medical Center, United StatesReviewed by:
Irina G. Sourgoutcheva, University of Kansas Medical Center, United StatesSunny Sharma, Rutgers, The State University of New Jersey, United States
Copyright © 2021 Hua, Song, Peng, Wang, Ma, Zhang, Zhang, Li and Hou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lin Hou, cWluZ3lpMDAxQDEyNi5jb20=
 Yanan Hua
Yanan Hua Jinlian Song2
Jinlian Song2