
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Biomater. Sci., 12 July 2024
Sec. Biomaterials Manufacturing and Technology
Volume 3 - 2024 | https://doi.org/10.3389/fbiom.2024.1380153
This article is part of the Research TopicRising Stars in Biomaterials ScienceView all 3 articles
Periodontitis is a chronic inflammatory disease associated with dysbiosis in subgingival plaque biofilm, characterised by damage to the periodontal tissues, eventually leading to tooth loss. Hence, the pathophysiology of periodontitis and interaction between subgingival plaque and host tissue under various environmental cues are central to the pathogenesis of periodontitis. Therefore, engineering biofilm models that mimic in vivo pathophysiology is crucial to obtaining a clear insight into the pathology and developing targeted therapeutic methods. In this review, we provide a comprehensive overview of the engineering strategies employed of modelling oral biofilms focusing on surface attachment, fluid microenvironment, gas environment, shear force, microbial-host interaction and offer insights into the ongoing challenges and future perspectives, which will enable the development of novel physiological relevant models for oral biofilms.
Periodontitis is an inflammatory disease associated with dysbiosis within the plaque that colonises the subgingival area, leading to the destruction of both soft (i.e., gum) and hard tissues (i.e., bone and cementum) (Darveau, 2010). Beyond causing tissue damage in the oral cavity, periodontitis has been linked to several diseases, including cardiovascular, cerebrovascular and respiratory conditions (Wu et al., 2000; Scannapieco and Ho, 2001; Ohyama et al., 2009; Li et al., 2017). These correlations imply a potential systemic connection between the dental plaque and the host’s organs. The ability to simulate microbiome dysbiosis within the oral cavity could provide vital mechanistic insights into the progression of periodontitis and systemic association across the host body, thereby aiding in the discovery of novel treatments. However, existing models for periodontitis are constrained by the inherent complexity of the biology and pathophysiology associated with the progression of the disease.
A key engineering challenge in modelling microbial dysbiosis within the oral cavity is the multitude of players involved in the pathophysiology, including the oral biofilm formed by hundreds of oral microorganism species and host cells such as immune cells, bone cells, and epithelial cells. Unlike traditional theories attributing diseases to specific pathogenic bacteria, the development of periodontitis is linked to shifts in the oral microbial community (Berezow and Darveau, 2011). The subsequent immune responses triggered by this microbial dysbiosis lead to severe destruction of periodontal tissues (Berezow and Darveau, 2011). Consequently, there is a significant need for periodontitis models that can effectively capture the intricate microbial-microbial and host-microbial interactions. Such models are essential not only for unravelling the molecular mechanisms underpinning the disease but also for serving as a foundation for the development of new treatments.
In vivo models have long been the gold standard for periodontitis research. Animal models, including dogs, non-human primates and small rodents such as rats and hamsters, are often used for in vivo studies (Tariq et al., 2012). However, these models come with inherent limitations, such as high costs, challenges in standardising individual differences and the complexity of the system. These drawbacks render animal models not suitable for every scenario, particularly in early-stage periodontitis studies. Therefore, researcher s’ attention has shifted towards the utilisation of in vitro models.
In comparison to animal models, in vitro models serve as simplified and more focused platforms that are ideal for isolating and examining specific factors or pathways in a controlled manner. While in vitro models may not offer the same comprehensiveness as animal models, they perform better at dissecting underlying mechanisms at a molecular or cellular level, allowing for a precise and clear understanding of specific disease components. However, constructing in vitro models faces significant challenges when dealing with complex scenarios.
To date, various in vitro models for oral biofilms have been developed to facilitate pathology studies and drug screenings. While existing reviews by Luo and Pan extensively cover in vitro biofilm models (Luo et al., 2022) and the pathophysiology of periodontitis (Pan et al., 2019), there is limited coverage of in vitro models that encompass the diverse aspects of periodontitis pathophysiology. This review aims to provide a landscape perspective on current engineering approaches that enable the modelling of various aspects of periodontitis. Additionally, we present additional strategies that could potentially be integrated into the model for biofilm development in the context of periodontitis.
To thoroughly understand the pathophysiology of periodontitis, it is crucial to clarify the oral microbial dysbiosis and the subsequent response of the host immune system. This clarification includes all factors involved in the development of periodontitis (Figure 1) including microbiome interspecies interaction (Jakubovics, 2015; Marsh and Zaura, 2017), biofilm surface adhesion (Hao et al., 2018; Sterzenbach et al., 2020), local liquid and dissolved gas environment (Pöllänen et al., 2013; Stacy et al., 2016; Mark Welch et al., 2020), and microbiome-host cell interactions (Pan et al., 2019; Lamont et al., 2023). These aspects have been extensively discussed in multiple reviews; hence, they will only be briefly covered in this review.
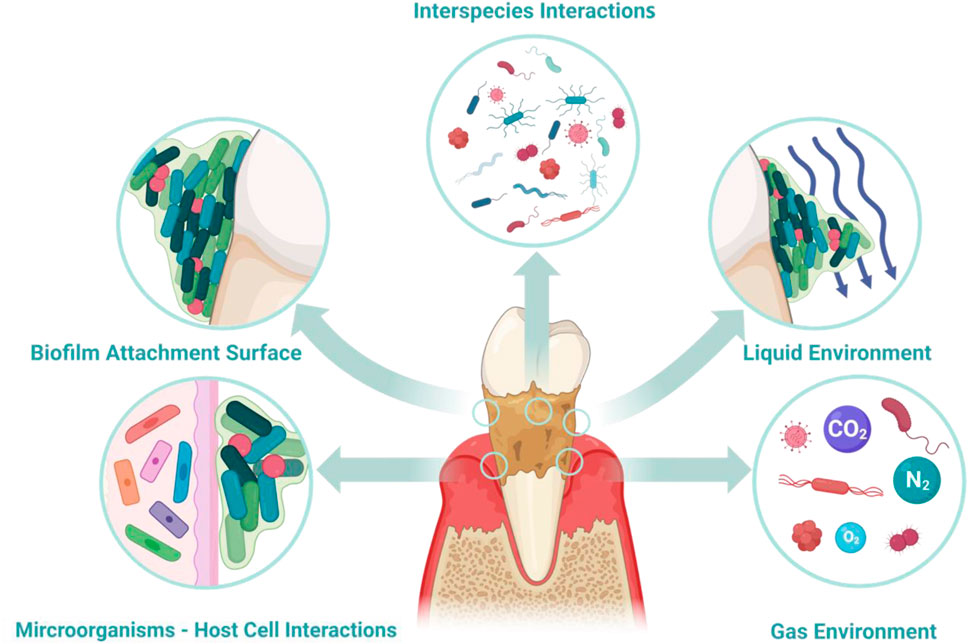
Figure 1. Environmental factors contributing to periodontitis. Key factors involved in the pathological mechanisms include inter-bacterial interactions, microbial adhesion to the host, the fluid environment (both liquid and gas) within periodontal tissues, and the interaction between host cells and microorganisms. When developing in vitro models for periodontal diseases, selecting suitable methods to simulate these factors becomes essential.
The oral microbiome, comprising roughly 700 species, plays a crucial role in both health and disease (Zhao et al., 2017). Among them subgingival plaque was found most relevant to periodontitis (Zijnge et al., 2012). Initial research identified six microbial complexes within the plaque, correlating their presence with health or disease states, among which red complex species [Porphyromonas gingivalis (P. gingivalis), Treponema denticola (T. denticola), Tannerella forsythia (T. forsythia)] are considered most relevant to periodontitis (Socransky S. et al., 1998; Hajishengallis et al., 2012). However, with the detection of red complex species in healthy individuals, it is evident that attributing disease solely to these species is not a comprehensive theory (Lamont and Jenkinson, 1998; Carrouel et al., 2016). The subsequent Keystone theory suggests that pathogens like P. gingivalis do not directly cause inflammation in the body. Instead, they induce a noticeable shift from Gram-positive facultative anaerobic bacteria to Gram-negative anaerobic bacteria in the subgingival microbiome (Balan et al., 2023), causing a microbiome dysbiosis and then triggers complement-dependent inflammation (Hajishengallis et al., 2012). The polymicrobial synergy and dysbiosis (PSD) model, on the other hand, points out that the pathogenicity of red complex species is only manifested within a synergistic microbial community (Hajishengallis and Lamont, 2012). Therefore, in modern theories, the concept of pathogens and commensals is gradually becoming blurred. Researchers tend to focus more on analysing the species abundance, microbial dynamics, and complex interspecies interactions including nutritional symbiosis and competitive antagonism within the subgingival plaque under different disease states (Christensen et al., 2002; Coppenhagen-Glazer et al., 2015; Marsh and Zaura, 2017). These findings shift the focus from individual pathogens to the dynamics of microbial communities, underscores the significance of microbial interactions within the oral ecosystem, highlighting periodontitis as a condition driven by the collective pathogenic potential of microbial communities rather than the presence of a single pathogen. Figure 2 shows the shift in subgingival plaque.
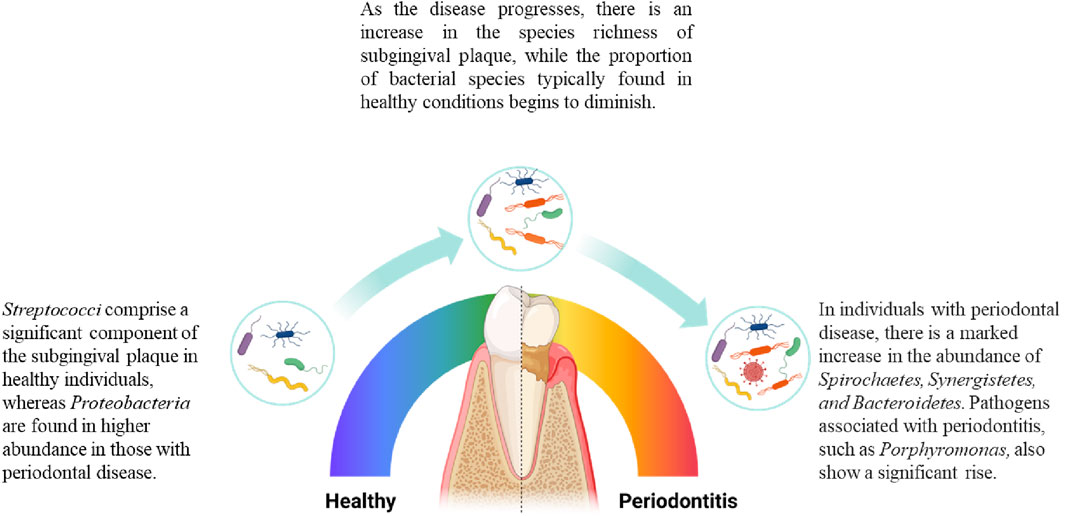
Figure 2. Changes in the composition of subgingival microbiome during periodontitis. As periodontitis progresses, there are changes in the microbial species composition of subgingival plaque. Microbial species associated with health decrease in proportion as the disease develops, while the number of disease-associated species gradually increases. In terms of species richness, the diversity of species in subgingival plaque increases as the disease worsens (Griffen et al., 2012).
Oral microorganisms predominantly form biofilms in the oral cavity, creating complex, three-dimensional structures that enhance their survival. These biofilms begin with the adhesion of bacteria to oral surfaces such as teeth and epithelial cells, facilitated by salivary glycoproteins that attract microbes through electrochemical forces (Weerkamp et al., 1988; Ren et al., 2018), primarily involving Gram-positive facultative anaerobic cocci and rods (Kriebel et al., 2018). Biofilm formation includes not only passive adhesion but also active surface attachment mechanisms, such as the involvement of flagella that allow bacteria to sense and move toward surfaces (Yang et al., 2016). Through cell proliferation and a phenotypic shift towards increased extracellular polymeric substance (EPS) production, biofilm starts to mature, enhancing the structural stability and enabling further microbial aggregation (Flemming et al., 2007; Bowen and Koo, 2011; Laventie et al., 2019). This process is facilitated by high-affinity surface adhesins, which help transition from reversible to irreversible attachment (Bowen and Koo, 2011; Groeger et al., 2022), and by dispersion mechanisms that allow divided cells to detach and form new biofilms, significantly increasing the biofilms’ resistance to external stresses (Stoodley et al., 1999; Tolker-Nielsen et al., 2000).
Periodontitis takes place in the gingival pockets, an area characterised by a mix of host tissues, oral biofilms, saliva, and various gases, creating a complex environment crucial for microbial survival and interaction. Saliva, acting as the primary liquid medium, facilitates material exchange, while gingival crevicular fluid (GCF), comprising serum, leukocytes, and other cellular components, maintains an alkaline pH that aids in microbial adhesion (Uitto, 2003; Barros et al., 2016; Carpenter, 2020). Liquid shear forces within this niche play a crucial role in microbial adsorption and the dispersal of bacteria, enabling the formation of new biofilms (Weerkamp et al., 1988; Stoodley et al., 1999; Ren et al., 2018).
The oxygen gradient within gingival pockets significantly influences the progression of periodontitits (Celik and Kantarci, 2021). Most pathogenic bacteria in subgingival plaque are anaerobes thriving in the low-oxygen conditions exacerbated by inflammation (Socransky S. S. et al., 1998; Celik and Kantarci, 2021). This environment promotes the proliferation of anaerobic pathogens, further fuelling the inflammatory process (Celik and Kantarci, 2021).
Furthermore, cross-species communication within biofilms alters the local environment, supporting biofilm growth (Joshi et al., 2021). For example, a notable “hedgehog” structure observed within oral biofilms illustrates the complex spatial organisation, with Corynebacterium at the core surrounded by various bacteria, creating gradients that cater to both aerobic and anaerobic species (Mark Welch et al., 2016). This intricate interaction within biofilms and the surrounding microenvironment underscores the complexity of periodontal inflammation and the challenges in managing periodontal disease (Mark Welch et al., 2016).
Subgingival plaque dysbiosis, triggered by factors such as bad dietary habits, alcohol, and smoking, prompts a host immune response to oral biofilms harbouring pathogenic bacteria like P. gingivalis, T. forsythia, and T. denticola (Sedghi et al., 2000). These pathogens activate toll-like receptors (TLR-2 and TLR-4), inducing inflammatory responses and disrupting epithelial barrier proteins through proteases, leading to periodontal tissue damage (Kawai and Akira, 2005; Yilmaz et al., 2006; Yoshioka et al., 2008; Zheng et al., 2021). The immune response includes neutrophil accumulation and altered functions due to bacterial interference, affecting phagocytosis and promoting inflammation through cytokine secretion and macrophage activation (Kudrin and Ray, 2008; Bostanci et al., 2013; Li et al., 2013; Maekawa et al., 2014; Olsen and Hajishengallis, 2016; Abe-Yutori et al., 2017; Papadopoulos et al., 2017; Pan et al., 2019; Takeuchi et al., 2021a). The ongoing inflammation disrupts alveolar bone homeostasis by tipping the balance between bone-forming osteoblasts and bone-resorbing osteoclasts, mediated by imbalances in RANKL and osteoprotegerin (OPG), resulting in alveolar bone loss (Vernal et al., 2005). The complex interactions between various periodontal bacteria and the host immune system underline the challenges in understanding and treating periodontal disease. Developing models that mimic the periodontal microenvironment could enhance our understanding and lead to better prevention and treatment strategies.
Animal models are a good option for periodontitis study as they may effectively simulate the full spectrum of periodontitis progression, offering valuable insights into the condition (Tariq et al., 2012). For instance, ligature-induced periodontitis models are widely applied in mice as they presented high availability for genetically engineered strains and high-quality immunochemical and cellular reagents (Marchesan et al., 2018). Monkeys have been proven to possess gingival immunological and histological characteristics identical to humans (Weinberg and Bral, 1999). They also exhibit microbiological characteristics of subgingival plaque in various states—healthy, gingivitis, or periodontitis—mirroring those in humans (Weinberg and Bral, 1999). However, animal models have inherent drawbacks. Firstly, there are xenotypical and physiological differences between animal models and human (Schou et al., 1993). Furthermore, the complex nature of animal models introduces challenges in isolating and analysing individual factors to provide a systematic approach to identify the mechanism of the disease. Beyond these limitations, animal experiments are known to involve high costs, lengthy turnaround times, and ethical controversies, prompting researchers to develop in vitro models suitable for various scenarios.
As the subgingival plaque plays a crucial role in the development of periodontitis, extensive research has been dedicated to constructing in vitro models of pathogenic microbes. These models aim to replicate the morphology of microorganisms in the oral cavity or the interaction between oral microorganisms and host cells. In contrast to animal models, in vitro periodontitis models offer a versatile platform that enables better control over the microenvironment, including parameters such as liquid shear stress, surface stiffness and the availability of essential biomolecules crucial for the development and maintenance of the biofilm.
Being simpler, in vitro periodontitis models are more focused, stable, and provide strong observability and ease of operation. However, current in vitro models cannot comprehensively mimic all the microenvironment factors contributing to the disease progression compared to in vivo models. Therefore, microbiologists and tissue engineers need to consider the research question and experiment needs when choosing the appropriate in vitro models.
Till today, there are various models have been developed to recapitulate different essential aspects of microenvironments in periodontitis. McBain conducted a comprehensive review of biofilm models developed across various domains (McBain, 2009), including simple agar plate models (Verhamme et al., 2009; Chai et al., 2011), well plate-based models (Friedman and Kolter, 2004; Sánchez et al., 2011; Sánchez et al., 2014), flow cell models (Aspiras et al., 2000), constant-depth film fermenter models (McBain et al., 2005), and drip-flow biofilm reactor models (McBain et al., 2005), among others. However, not all models are equally suitable for modelling periodontitis-associated biofilms. A concise overview of common models for in vitro modelling of oral biofilms, their operating principles and their respective advantages and disadvantages, is presented in Table 1.
As periodontitis entails dynamic interactions among multiple bacterial species and host cells in a heterogeneous environment, in vitro models capable of mimicking the native physiology of periodontitis will be ideal to facilitate a systematic understanding of the disease pathophysiology. This review primarily explores the methodologies used to simulate various aspects of periodontal diseases. Figure 3 provides a concise overview of some typical methodologies. The integration of new techniques has expanded the possibilities for creating biofilm models and addressing pertinent factors.
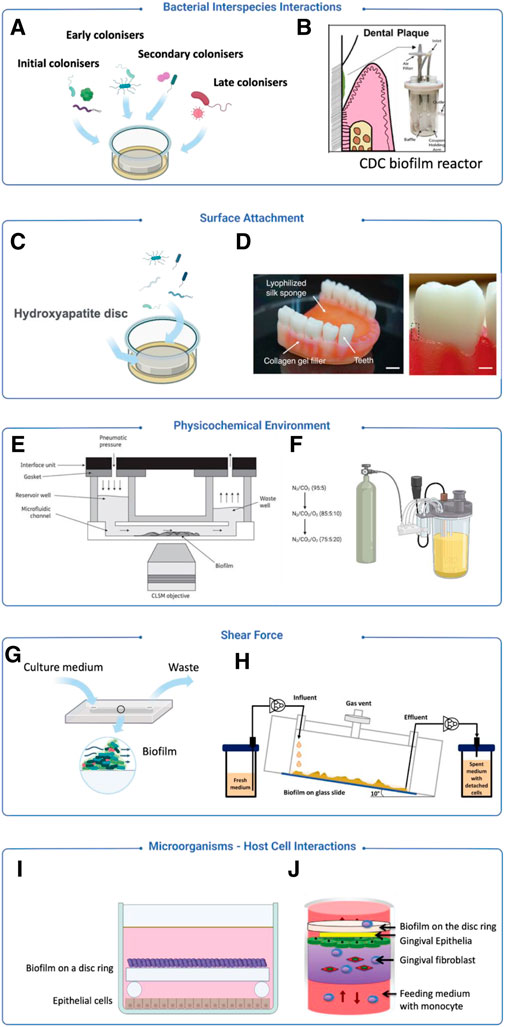
Figure 3. Examples of modelling strategies for periodontitis focusing on different aspects. (A) A model introduced bacteria from initial, early, secondary and late stage of biofilm colonisation (Sánchez et al., 2011). (B) The CDC model introduced dental plaque samples for oral biofilm culture (Ramachandra et al., 2023a). (C) Hydroxyapatite discs are used for simulating human teeth (Sánchez et al., 2011). (D) A 3D-printed silk sponge and collagen, incorporating teeth to mimic soft and hard tissues for biofilm attachment (Adelfio et al., 2023). (E) A model using sterile human saliva as the sole culture medium to mimic an in vivo liquid environment (Nance et al., 2013). (F) A bioreactor equipped with a gaseous control system for the regulation of gas components (Diaz et al., 2002). (G) A flow cell system is used to generate a low shear force environment (Ali Mohammed et al., 2013). (H) A drip-flow reactor creating a low shear force environment through inclined channels (Ghesquière et al., 2023). (I) Zurich model applies biofilm on a disc ring to co-culture with host cells to observe microbial-host interactions (Guggenheim et al., 2009) (J) A co-culturing system integrating mammalian cell and biofilm culture to observe microbial-host interactions (Bao et al., 2015).
When modelling the dynamics within subgingival plaque, it is essential to first identify the key bacterial species and understand their potential interactions within the microbial community. In the context of microbiome and biofilm modelling, it is important to consider the role of early colonisers, which establish the initial biofilm matrix, and the subsequent cross-feeding of metabolites. These interactions are critical for the development and maturation of biofilms. For instance, Mishra et al. (2010) co-cultured fimA-, fimB- and srtC2-deficient Actinomyces oris (A. oris) with streptococci demonstrating the role of A. oris in synthesising fimbriae, essential for the adhesion of the streptococci during biofilm formation. Apart from the synergistic relationship of the bacteria, the potential antagonistic effect between the bacterial species is important to help researchers identify strategies to treat and halt periodontitis. Duran-Pinedo et al. (2014) co-cultured Streptococcus mitis (S. mitis), a member of the yellow complex commonly found in healthy gingival biofilms, along with members of the red complex, P. gingivalis and T. forsythia. At high populations, P. gingivalis and T. forsythia induce an inflammatory response in host cells that leads to the death of S. mitis. This facilitates the colonisation of P. gingivalis and T. forsythia and contributes to the development of periodontitis. These studies underscore the importance of modelling interactions between bacterial species to advance our understanding of disease progression and to identify potential treatment strategies.
The simplest strategy to model the interaction between bacterial species is to mix a defined ratio of microbial species into a culture broth. Due to their consistency, ease of analysis, and the scalability of microplates, this platform has been routinely applied to screen for drug treatments that require multiple dose-response studies. For instance, Li and Kumbar observed the inhibitory effect of curcumin on the biofilm formation of S. mutans and P. gingivalis (Li et al., 2018; Kumbar et al., 2021) while Izui et al. (2016) constructed a P. gingivalis- S. gordonii (Streptococcus gordonii) interbacterial biofilm model in microtiter plates using a similar approach. This verification demonstrated that curcumin also inhibits the ability of P. gingivalis to form biofilms based on early colonisers such as S. gordonii in complex biofilms (Izui et al., 2016). These models are often used to observe specific interactions between a limited number of species, typically involving only two or three microbial species. To better capture all interactions of microbials species in periodontitis, it is suggested that the criteria for selection can be based on the prevalence of these species in periodontal disease sites and their ability to be cultivated and quantified. Guggenheim and colleagues developed the Zurich model, selecting five bacterial species and Candida albicans to represent supragingival plaque (Shapiro et al., 2002). The Zurich model’s key principle involves selecting a broad range of microbial species in a controlled and quantifiable way to create a biofilm that is complex yet precisely composed. The species selected to represent the subgingival microbiome were Campylobacter rectus (C. rectus), Fusobacterium nucleatum (F. nucleatum), P. gingivalis, Prevotella intermedia (P. intermedia), T. forsythia, Veillonella dispar (V. dispar), Actinomyces naeslundii (A. naeslundii), Staphylococcus intermedius (S. intermedius), and Streptococcus oralis (S. oralis) (Guggenheim et al., 2009). Sanchez and team adopted a similar approach but expanded the oral biofilm species to include different stages of colonisation: initial (S. oralis and A. naeslundii), early (V. parvula), secondary (F. nucleatum), and late (P. gingivalis and A. actinomycetemcomitans) stages (Sánchez et al., 2011). The aim was to encompass the diversity of the subgingival plaque and the stages of the biofilm formation and development over time, enabling researchers to replicate and study this process in a temporal context.
The Zurich model is commonly set up by dipping hydroxyapatite discs into the mixture of bacterial suspension containing a defined ratio of each species of bacteria. To facilitate bacterial adhesion on the surface, an incubation period of 16 h–24 h is typically required before their subsequent culture and use for experimentation. However, these models might have limitations in accurately simulating the intricate nature of an oral biofilm in a laboratory setting. To address this, researchers have experimented with using patient samples to create more representative in vitro models of undefined multispecies biofilms. For instance, Ramachandra et al. (2023a) introduced subgingival plaque from patient sources into a CDC bioreactor, a widely recognised device for biofilm research. This approach more effectively mirrors the complexity of natural biofilms compared to traditional defined models. One challenge with this approach is adjusting the culture broth to support all included bacterial species (Ammann et al., 2012), supplements like human saliva and blood are commonly used to facilitate co-culture (Love, 2010). However, while viable, this strategy introduces batch-to-batch variation due to the variability in human samples. Additionally, this co-culture strategy does not readily allow for the spatial-temporal tracking of bacterial dynamics or individual metabolite release because all species share the same liquid environment. These limitations can pose significant challenges in studying the mechanisms underlying interbacterial relationships in periodontitis.
It is important to note that using this strategy while viable introduces batch-to-batch variation that results from the human samples during the culture. Additionally, this co-culture strategy of the bacterial mixture does not easily allow researchers to spatio-temporally track the dynamics of all bacterial species during the culture as well as the metabolite release of individual species due to sharing a single liquid environment. This can pose challenges to studying the mechanism of the interbacterial relationship in periodontitis.
One engineering approach to enable the study of interbacterial cross-feeding relationships is through the use of a flow cell system. In the flow cell system, differently cultured biofilms are linked together by a fluid conduit. The culture broth is then flowed through from 1 cell to another. In Zainal-Abidin’s study, T. denticola, P. gingivalis and T. forsythia, classified as red complexes, were inoculated in a single-channel flow cells system that allowed continuous cultivation for up to 90 h, enabling visual observation of the biofilm (Zainal-Abidin et al., 2012). By pairing up the P. gingivalis in series with T. denticola, and T. forsythia, it was observed that the upregulation of glycine catabolism in P. gingivalis resulted in a change in the structure of flagella in T. denticola. This work showcased a critical benefit in providing mechanical insights between interacting bacterial species that are otherwise challenging to be implemented in microplate platforms.
Surface adhesion plays an essential role in biofilm development, marking a key difference from planktonic microbes (Ammann et al., 2012). In periodontitis, the adhesion of early colonisers plays a significant role in the formation and maturation of biofilm by altering the immediate microenvironment and generating additional adhesion sites for other bacterial species (Love, 2010). There’s extensive research on how microbes attach at the molecular biology level which was covered by existing reviews (Kuboniwa and Lamont, 2000). For the oral microbial and periodontitis model, a common substrate that enables biofilm adhesion is hydroxyapatite (Pratten and WillsBarnettWilson, 1998; Shapiro et al., 2002; Guggenheim et al., 2009) for its similarity in chemical structure to tooth enamel. In periodontitis, P. gingivalis is known to adhere to hydroxyapatite through fimbriae (Lee et al., 1992) which can be altered by the hag genes (Connolly et al., 2017). Jaffar et al. (2016) created dense and porous HA discs to replicate enamel and dentin at the microscopic structural level respectively, and cultured biofilms of Aggregatibacter actinomycetemcomitans (A. actinomycetemcomitans) and P. gingivalis on the discs. The advantage of using commercial HA products was easier standardisation, making them ideal for studying the effects of surface roughness on biofilm adhesion. Additionally, HA can be constructed in various form factors, allowing biofilm adhesion and culture in many culture configurations including microplates (Kumbar et al., 2021), flow cells (Zainal-Abidin et al., 2012) and CDC bioreactors (Ramachandra et al., 2023a). Beyond HA, biofilm adhesion on other material surfaces such as titanium and zirconium are important in studying biofilm development on dental implants. Although massive literature suggests titanium and zirconium provide antimicrobial properties (Siddiqi et al., 2016), exposed rough surfaces of titanium and zirconium are found to enable P. gingivalis to form biofilm (Kniha et al., 2021).
Apart from hydroxyapatite, common laboratory materials like glass and polypropylene are frequently used for their availability and simplicity (Hamada and Torii, 1978; Saito et al., 2008). However, they do not closely mimic the hard tissues of the periodontium. Hägi et al. (2015) processed human teeth into dentin slices and exposed them to a bacterial culture with various strains (Pratten and WillsBarnettWilson, 1998). Using human dentin slices, this material provides an excellent model for dentists to investigate the impact of biofilm removal techniques including hand curettes, ultrasonication, and subgingival air-polishing, on the dentin structure (Hägi et al., 2015). However, the limitations of relying on human dentin including limited and variable sources would impart significant issues for standardised assay development and scalability.
While hydroxyapatite and dentin have demonstrated good utilities for researchers to model biofilm on tooth surfaces, they do not reflect the development of biofilm along the gingival margin which can result in the formation of periodontal pockets (Haffajee et al., 2009). In vitro models to capture this phenomenon would require researchers to use softer substrate that can mimic the gum tissue stiffness. Recent progress in 3D printing and bioprinting has proven to be an effective tool to fabricate adhesive surfaces with tuneable surface stiffness. Ramachandra et al. (2023b) demonstrated this feasibility by constructing 3D printed polycaprolactone (PCL) scaffolds, enabling saliva biofilm development. In addition to selecting the 3D printing material, the surface properties of the scaffold can also be tuned by mixing biological-derived material such as collagen, gelatine or alginate. A notable study used silk proteins and type I collagen, combined with 3D printing techniques, to replicate the gingival tissue stiffness in vitro (Adelfio et al., 2023).
The oral cavity is a complex ecosystem, where elements like gingival crevices, tooth surfaces, oral epithelium, saliva flow, diet, and individual habits result in a unique and intricate microenvironment for each individual. This environment is crucial for the attachment and survival of oral biofilms. In periodontitis, disruptions to this microenvironment, often caused by unhealthy lifestyle choices such as smoking and poor oral hygiene, play a key role in disturbing the balance of the subgingival microbiome (Genco, 1996; Janakiram and Dye, 2020). These disturbances manifest as alterations in fluid composition, shifts in the partial pressures of different gases, and changes in the shear forces acting on the oral biofilm. Accordingly, accurately replicating these chemical and physical aspects of the oral microenvironment is vital when developing in vitro models for studying oral biofilms.
Saliva is a key component in the oral fluid environment, providing essential nutrients, unique fluid dynamics, and a specific chemical and electrochemical milieu for oral biofilms (Kolenbrander, 2011). One early study used saliva as the sole nutritional source for culturing randomly isolated oral bacteria (De Jong and Van der Hoeven, 1987). In this setup, subgingival microbial strains were enriched and then cultured in sterile saliva, requiring daily replenishment of 25 mL for static growth (De Jong and Van der Hoeven, 1987). The study highlighted the critical interactions between the oral microbiome and saliva, particularly in the binding and degradation of salivary glycoproteins, underscoring saliva’s indispensable role in the survival of oral microbiome (De Jong and Van der Hoeven, 1987). A more common technique involves using sterile saliva to mimic the oral environment for bacterial adhesion on surfaces like HA discs, before moving them to culture media for growth (Guggenheim et al., 2001; Guggenheim et al., 2009; Sánchez et al., 2011). However, natural saliva’s variability and unknown components limit experiment reproducibility. To overcome this, artificial saliva has been developed, replicating natural saliva’s viscosity and buffering properties using ingredients like inorganic salts (e.g., potassium chloride, ammonium chloride, calcium chloride, and magnesium chloride), cellular metabolites (e.g., creatinine and choline), amino acids, microorganisms, and significant amount of glycoproteins as the source of saliva viscosity and electrochemical properties and primary nutritional source for oral bacteria (Shellis, 1978). This has subsequently been used in the cultivation of many in vitro oral biofilm models and has been continuously improved to meet various requirements (Weyell et al., 2019). For instance, Blanc used hog gastric mucin to mimic salivary glycoproteins due to their similar oligosaccharide structures (Blanc et al., 2014). In Wong’s study, two artificial saliva bacterial culture media, defined medium mucin (DMM) and designed basal medium mucin (BMM), were compared in subgingival biofilm cultivation (Wong and Sissions, 2001). BMM, which has long been widely used for oral biofilm cultivation, is composed of yeast extract, proteose (meat hydrolysate), and trypticase, containing much of unknown components (Wong and Sissions, 2001). In the two liquid environments, the inoculated subgingival plaque showed similar growth rates, but there were subtle differences in specific metabolism. The biofilm grown in BMM, as opposed to DMM, had four more enzymes detected, which may be attributed to the abundant unknown peptides from yeast extract and protease in BMM (Wong and Sissions, 2001).
Microfluidic platforms partially resolve the limitation of human saliva, leveraging on the microlitre scale culture chambers requiring significantly low volume of saliva for biofilm cultures. For instance, Nance et al. used the BioFlux microfluidic system, inoculating human saliva and then culturing biofilms in sterilised saliva, to assess antibacterial effects (Nance et al., 2013). This system’s small dimensions (48 wells, each 70 μm deep and 370 μm wide) reduce the saliva volume needed, though it introduces challenges like high equipment costs and complexity (Nance et al., 2013).
Biochemical contents introduced through external factors such as diet, smoking and oral hygiene habits are known to influence microbial behaviour contributing to periodontitis. Huang et al. (2014) explored the connection between smoking and oral biofilms by cultivating S. gordonii, an early coloniser of dental biofilms (commonly found in subgingival and supragingival biofilms), in a nicotine-infused culture medium. This study revealed that S. gordonii enhanced biofilm growth and formation in the presence of nicotine, suggesting a direct link between smoking and increased oral biofilm accumulation (Huang et al., 2014). Zilm and Rogers (2007) used a more sophisticated chemostat-based biofilm culture system with a pH controller to study F. nucleatum, a significant pathogen in oral biofilms mimicking dietary induced pH changes on the biofilm. By maintaining the culture medium’s pH between 7.8 and 8.2 through the gradual addition of potassium hydroxide (KOH), the biofilm production of F. nucleatum in different pH conditions could be observed (Zilm and Rogers, 2007). The study found that an increase in pH led to changes in cell morphology and intracellular polyglucose levels, with bacterial flocculation and biofilm growth peaking at pH 8.2. This aligns with the alkaline environment reported in the gingival sulcus (Marsh and Martin, 1992), thus demonstrating the influence of pH on F. nucleatum biofilm production (Zilm and Rogers, 2007). Dynamic cultivation systems that continuously control the liquid environment during biofilm growth offer a more realistic simulation. Bradshaw et al. (1996a) developed an oral biofilm model using a chemostat, where a community of up to ten oral microorganisms formed biofilms on HA discs. In this system, the medium, supplied continuously by a 75 mL chemostat for several days, included glucose pulsing and pH control to study their effects on the biofilm (Bradshaw et al., 1996a). The use of a chemostat not only introduced liquid shear forces, mimicking saliva flow, but also provided a stable and adjustable liquid composition for more precise control over the biofilm growth environment.
A diverse range of anaerobic bacterial species exists in the subgingival plaque. In conditions where inflammatory responses intensify and periodontal pockets deepen, a low-oxygen environment emerges, ideal for the growth of anaerobic bacteria linked to periodontal diseases like P. gingivalis, T. forsythia, and T. denticola (Celik and Kantarci, 2021). This necessitates the creation of suitable anaerobic conditions for in vitro simulation of biofilms associated with periodontitis. Commonly, subgingival microbiome studies use anaerobic cultivation settings, placing culture plates or flow cells in an anaerobic chamber with a typical gas mix (10% H2, 10% CO2, and balanced N2) (Sánchez et al., 2011; Blanc et al., 2014; Sánchez et al., 2019). However, these chambers offer a uniform gas environment, which may not always suffice for multi-species bacteria co-culture and host-bacteria co-culture when differential oxygen environments are necessary to support the co-cultures.
The culture of obligate anaerobes will necessitate the use of anaerobic chambers which are not always accessible to all research groups. To resolve this limitation, many groups have employed the use of physically purging oxygen from the culture broth with nitrogen (N2). This setup depends on constant gas and liquid flow to ensure purging of the oxygen from the broth effectively before delivery to the cultivation setup. In Bradshaw’s study, the team linked two chemostats with the first chamber exposed to anaerobic media (aerated with CO2 and N2) and the second chamber received the oxygen-rich air supplementation to investigate the impact of oxygen on oral microbes like Neisseria subflava (N. subflava) and streptococci (Bradshaw et al., 1996b). Even after transferring the culture of ten mixed microorganisms from the first to the second chemostat, anaerobic species continued to grow, suggesting some degree of oxygen tolerance among these bacteria (Bradshaw et al., 1996b). In another study, chemostat was used for the co-culture of P. gingivalis and F. nucleatum (Diaz et al., 2002). The chemostat provided a controlled medium and gaseous environment over an extended period. This experiment exposed P. gingivalis and F. nucleatum to increasing oxygen levels (from 0% to 10%–20%), observing the oxygen tolerance of P. gingivalis in the presence of F. nucleatum (Diaz et al., 2002). It was noted that while P. gingivalis has lower oxygen tolerance in a single-species culture, its survival is supported by F. nucleatum in oxygen-rich environments (Diaz et al., 2002). The chemostat serves as the basis for this dynamic culture model, providing longer sustained biofilm growth as well as stable and controllable gas environmental control.
Microfluidic platforms have demonstrated the potential to facilitate the spatial distribution of oxygen through their microchannel networks (Whitesides, 2006). Many microfluidic devices also demonstrate their high-throughput bacteria culture with multiple microscale chambers while enabling sophisticated control of the bacterial microenvironment. An artificial device containing up to 128 incubation chambers based on a microfluidic device has been developed for biofilm culture and enables the control of various microenvironmental parameters such as culture solution composition and growth factors (Lam et al., 2016). Equipped with liquid flow channels and gas flow channels, 128 individual chambers with a diameter of 1 mm and a height of 210 μm were set up to achieve independent nutrient and gas supply to each chamber. The dissolved oxygen concentration in the chambers was regulated through the microvalves (Lam et al., 2016). This setup has been used to study both single-species and multi-species oral biofilms, including F. nucleatum, Streptococci, and samples of undefined oral plaque, under dynamically controlled oxygen conditions (Lam et al., 2016). This system replicates the impact of daily fluctuations in oxygen levels within the oral environment (aerobic during the day and microaerobic to anaerobic during the night) on oral biofilm growth and composition (Lam et al., 2016). It is important to note however, that due to the small footprint of the microfluidic device, the assembly of microfluidic devices with functioning valves and tubing assembly is technically demanding, making their adoption among microbiologists challenging.
Due to dietary activity, the movement of saliva within the oral cavity imparts liquid shear forces that can influence the biofilm development, including spatial structuring, nutrient uptake, and surface area expansion. Evidence of the importance of mimicking liquid shear stress in oral biofilm has been demonstrated by Maezono’s team where it was observed that under liquid shear stress, P. gingivalis biofilms showed resistance to erythromycin compared to the non-shear stress counterpart (Maezono et al., 2011). Given that modelling liquid shear stress requires the setup to integrate external pumps, well plate cultures are usually not suitable (Christensen et al., 1985). Maezono’s team achieved a controlled shear stress model using a Modified Robbin’s Device (MRD) connected to an external peristaltic pump (Maezono et al., 2011).
There are also existing platforms where an impeller is integrated within the bioreactor to generate liquid flow and their shear stress (Song et al., 2017; Ramachandra et al., 2023a). Zilm’s group has showcased the use of a chemostat-based model for F. nucleatum biofilms, with a precise flow rate of 27.5 mL/h−1, tailored to the growth and generation rate of dental plaque (Zilm and Rogers, 2007). Song et al. (2017), demonstrated the use of CDC bioreactors to control the liquid shear stress exposed to biofilms of P. gingivalis and F. nucleatum grown on HA discs. In these models, the shear stress of the liquid is tuned by controlling the rotation rate of the impeller rotating within the bioreactors. While these models provide good liquid shear stress, the position of the impellers and the size of the bioreactor should be taken into consideration as improper setup tend to generate local dead volumes with low shear stress.
Apart from integrating impellers to generate flow, other platforms rely on external pumps or flow system to enable liquid flow on the biofilm culture. Flow cell bioreactors which offer a distinct advantage over larger bioreactors by facilitating controlled liquid flow through small-volume chambers, more stable and precise fluid dynamics. Zainal-Abidin utilised a flow cell setup in their study on the synergistic effects of the red complex bacteria, employing cylindrical growth tubes with a flow rate of 3 mL/h. This design was intended to more accurately mimic saliva flow and allow for clear observation of biofilm development on the walls of glass tubes (Zainal-Abidin et al., 2012). Unlike an impeller-based bioreactors, flow cells enable sequential feeding of the culture broth from one unit to another. This setup provides a clear-cut method for researchers to investigate the dynamics of biofilm development. Foster’s team linked two flow cells with different inoculated bacterial biofilms [S. gordonii, A. naeslundii, Veillonella atypica (V. atypica), and F. nucleatum] to identify the primary and late colonisers by tracking the population changes at the biofilm downstream of the first bioreactors (Foster and Kolenbrander, 2004). In this study, the flow rate was set at 200 μL per minute (Foster and Kolenbrander, 2004), determined based on previous calculations of saliva flow rates in various parts of the oral cavity (Dawes et al., 1989). Similarly, Ali Mohammed et al. (2013) constructed flow chambers measuring 1 × 4 × 40 mm and cultivated biofilms of F. nucleatum and P. gingivalis within. A peristaltic pump was employed to deliver a stable cultivation medium at a rate of 3.3 mL/h through the system (Ali Mohammed et al., 2013). Modified flow cell reactors such as the drip-flow reactors offer a low-shear setting more representative of the oral environment (Ghesquière et al., 2023). Here, biofilms of key periodontitis-asociated bacterial species are formed under a gentle, continuous flow in inclined channels (Ghesquière et al., 2023) enabling the spatial-temporal formation of biofilms along the direction of liquid flow.
The small and uniform structure of the flow cell ensures precise control over the liquid medium’s flow over the biofilm surface, minimising flow disturbances. This level of control is a significant advantage, often difficult to attain with other models like MRD or chemostat devices.
Using the same strategy of mounting bioreactors to fluid pumps, microfluidics have also been recently used to investigate in vitro biofilm models. Compared to the flow cells, microfluidic platforms typically consist of microscale chamber arrays (<500 µm in size), enabling the simultaneous cultivation of large numbers of biofilms under precise control (Eun and Weibel, 2009; Janakiraman et al., 2009). Leveraging on the flexible assembly and customisation of these platforms, microfluidics devices can be modified to mimic multiple aspects of biofilm microenvironments. Makkar et al. (2023) leveraged microfluidic techniques to accurately mimic the fluid dynamics of the human gingival crevice, including specific parameters like liquid pressure and flow velocity. In their experiment, gingival fibroblasts encapsulated in a human fibrin-based 3D matrix, along with S. oralis biofilms, were placed in a microfluidic channel designed to simulate the gingival sulcus. These channels were 400 µm wide and 100 µm high, facilitating the study of dental plaque adherence to gingival tissues (Makkar et al., 2023). The flow of simulated gingival crevicular fluid (s-GCF) through these channels was then meticulously adjusted. Under conditions of high flow rates and shear forces, the researchers observed that biofilm clearance was facilitated, growth was constrained, and a stable co-culture of host tissue and symbiotic microorganisms was sustained (Makkar et al., 2023). This study highlights the utility of microfluidics in replicating complex oral environments for the study of the dynamics between oral tissues and microbial communities.
Beyond modelling liquid shear stress, the ability of modelling physical shear stress can be important to allow researchers to study biofilm retention and removal from various physical activities. The constant depth film fermentor (CDFF) incorporates a scraping mechanism to maintain a consistent biofilm thickness, simulating the mechanical disturbances experienced in the oral cavity from actions like brushing or tongue movement (Pratten and WillsBarnettWilson, 1998).
The abundance of microorganisms colonising the oral environment poses a long-term risk of microbial invasion to host oral tissues, underscoring the importance of maintining host-microbe homeostasis for overall host health (Cai et al., 2023). Disruption of the homeostasis between the host and microorganisms is a key factor in the development of periodontitis. Therefore, the interplay between subgingival biofilms and host cells, the two most important participants in periodontitis, is the key to the pathological study of periodontal disease. The most convenient and common practice is to simultaneously culture target bacteria in cell culture media. One study conducted co-culturing wild-type P. gingivalis with its fimbriae-deficient mutant and a monolayer of gingival epithelial cells (GECs) to obtain P. gingivalis-infected cells (Yilmaz et al., 2003). GECs were cultured as a monolayer, and the P. gingivalis strain was added to culture media to observe P. gingivalis’ invasion capability on GECs (Yilmaz et al., 2003). This simple and easy-to-operate model has therefore widely adopted (Takeuchi et al., 2021b; Zhang et al., 2022).
However, this experimental setup inaeduqately capture the in vivo survival patterns at both the microbial and host cell levels. Firstly, host cells do not grow as monolayers physiologically but exist within the complex 3D structure of periodontal tissues. To address this, a series of experiments have attempted to improve this method. For example, the introduction of 3D culture with materials like collagen allows cells to be in a spatial environment more similar to physiological conditions. One such study layered multiple strata of oral epithelial cells over rat-tail type I collagen-embedded fibroblasts to create a 3D soft tissue model (Pinnock et al., 2014). This was compared with a traditional 2D monolayer cell culture. In both models, P. gingivalis was introduced into the same culture media and incubated for specific durations to observe bacterial invasion into mammalian cells (Pinnock et al., 2014). The outcomes, including staining and chemokine array analysis, revealed notable differences between the 3D and 2D cultures. The 3D model more closely resembled in vivo conditions in aspects like bacterial invasion, intracellular viability, bacterial release, and cytokine production (Pinnock et al., 2014). Similar methods involving 3D culturing with collagen or collagen-like materials are widely used to simulate environments such as oral mucosa or periodontal soft tissues, providing cells with a 3D spatial structure similar to these tissues (Bao et al., 2015; Adelfio et al., 2023).
Microorganisms do not simply float in liquid; instead, they attach to the surfaces of periodontal tissues in the form of biofilms, a characteristic not accurately depicted in previous models. To tackle this issue, a common approach involves introducing biofilms into the co-culture system by first cultivating mature biofilms on surfaces such as HA discs or coverslips, before co-culturing them with cells. Guggenheim et al. (2009) developed such a coculture model using a ten-strain biofilm cultured on HA discs. Mature biofilms on HA discs were introduced into a well plate containing human gingival epithelial cells, positioned closely but not in direct contact, to simulate subgingival conditions. This setup enabled the study of biofilm-induced apoptosis and the enhanced expression of virulence factors by the microorganisms in a biofilm state compared to planktonic forms (Guggenheim et al., 2009). This result also confirms the limitations of previous models, highlighting that planktonic bacterium, due to their phenotypic differences, cannot effectively replicate the virulence characteristics of biofilms in vitro. Therefore, this method has subsequently been adopted in various pathological investigations (Thurnheer et al., 2014) and expanded to diverse fields, including drug screening research (Millhouse et al., 2014). In a similar manner, Lang et al. (2022) developed a periodontal pocket model by dipping and culturing 12-strain bacterial biofilm on dentin discs and epithelial cells on glass slides in pockets with cell culture media, assessing the therapeutic efficacy of ultrasonics and hand instruments on root surface treatment. In addition to more accurately reproducing bacterial survival patterns and phenotypes, another advantage of introducing mature biofilms into the culture system is the ability to better quantify and visualise the microorganisms before and after the experiment. For instance, Millhouse et al. set up a co-culture model with a composite biofilm including P. gingivalis, F. nucleatum, A. actinomycetemcomitans, and S. mitis on coverslips, alongside immortalised human oral keratinocyte cell lines, to test the antimicrobial and anti-inflammatory effects of naturally derived polyphenol resveratrol and chlorhexidine (Millhouse et al., 2014). The untreated and drug-treated biofilms, as well as host cells, were quantified not only for biomass and cell viability using the AlamarBlue assay and absorbance measurements, but also visualised through SEM imaging, which revealed morphological differences and aggregation patterns between different bacterial species at the microscopic level (Millhouse et al., 2014).
It can be envisioned that in attempting to simulate the spatial distribution of host cells and the microbial colonisation in biofilm, these efforts are being integrated to obtain a faithful reflection of the microbial-host cell crosstalk. Integrating collagen-based 3D cell culture with oral biofilms adhered to hard surfaces in the same culture system is a straightforward approach. Bao et al. (2015) further enhanced this model by incorporating 3D cell culture techniques, combining biofilm-host cell coculture with a 3D periodontal pocket tissue model to better mimic the spatial organisation of cells. A bioreactor system, composed of gingival fibroblast cells supported by 3D collagen sponge scaffolds, along with gingival epithelial cells on the scaffold surface and monocytes flowing through, simulated the tissue structure of the periodontal pocket. Subsequently, biofilms composed of 11 oral microbial species grown on HA discs were introduced to mimic conditions of periodontal inflammation (Bao et al., 2015). In addition to biofilm, this periodontal pocket model also incorporates the 3D structure of periodontal soft tissues and immune cells interacting with the external environment through GCF flow. At the level of both microorganism and host tissue, it further accurately reproduces the participants of periodontitis and their spatial distribution within the periodontal pocket.
One could argue that the current approach to constructing co-culture models entails the continual refinement of basic host cell-microbe models in standard well plates. This effort aims to align the survival patterns of both host cells and microorganisms more closely with those observed in the physiological environment. However, some challenges persist in such models. For example, these co-cultures can only provide a uniform oxygen environment for both host cells and microorganisms, while in reality, pathogenic microorganisms thrive in gingival pockets where oxygen concentration gradually decreases due to the unique anatomical structure. Previous studies have reported that oxygen-sensitive microorganisms such as P. gingivalis and T. forsythia struggle to maintain stable colonisation in normoxic environments (Guggenheim et al., 2009). The emergence of 3D printing technology enables not only the replication of cell spatial distribution, but also the construction of macroscopic anatomical structures of the periodontium for in vitro models (Adelfio et al., 2023). Adelfio et al. (2023) utilised 3D printing technology and silk biopolymer material to create a replica mold of the adult human mandibular gingiva, providing a spatial environment consistent with physiological conditions for host cells and microorganisms cultured within it. This allows for precise replication of gas diffusion and nutrient distribution in the oral environment. Built on high-fidelity anatomical molds, this model addresses challenges encountered in previous studies, such as reproducing oxygen gradients generated with increased depth of gingival pockets.
Until today, simulating periodontal biofilm formation and its interactions with host tissues perfectly from various aspects in vitro still faces many unresolved challenges.
The ability of in vitro models to mimic a microenvironment similar to human physiology is important for accurately understanding the progression of periodontitis, posing an ongoing engineering challenge. Apart from mimicking the microenvironment of the oral cavity, the engineering of in vitro biofilm models should also consider the potential scale-up of the platforms to enable parallel experiments and enhance repeatability. This wishlist has driven various engineering developments of in vitro models to enable periodontal biofilm cultures (Table 2).
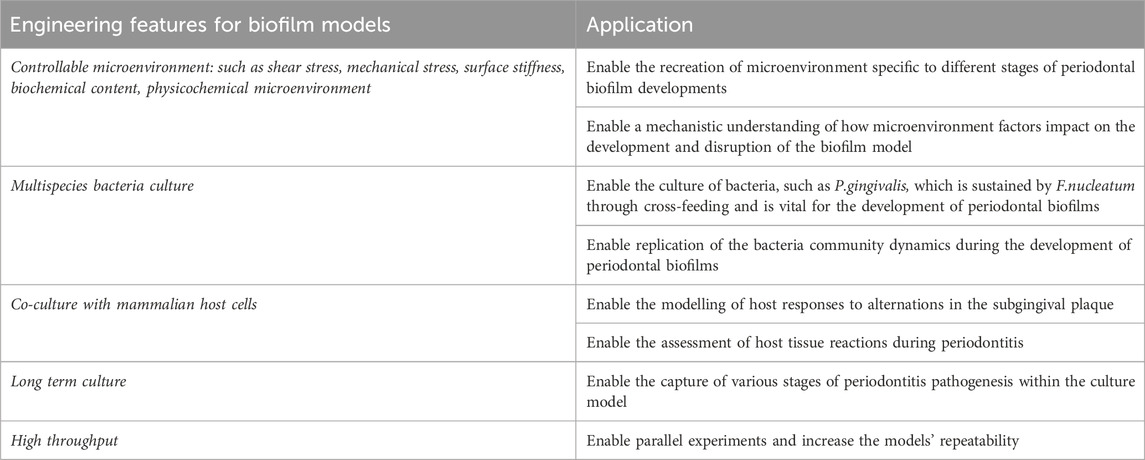
Table 2. Characteristics of an ideal in vitro model for periodontitis. It is widely acknowledged that achieving the “perfect” in vitro biofilm model is challenging, as it is fundamentally an emulation of the physiological environment rather than a replica.
Modelling of the inter-bacterial and bacterial-host tissue interactions in many cases requires a culture setup where the spatial distribution of culture nutrients is required. This is particular the case in bacteria-host tissue co-cultures where the culture media composition can vary significantly (Mountcastle et al., 2020). While mixing culture media at various ratio has been the common strategies implemented by many groups, this approach can be particularly challenging for co-culture of more than 2 types of bacteria and/or cells. Furthermore, co-cultures that leverage on mixing of culture media limit mechanistically investigation that involves cross-feeding of bacteria due to the metabolite generation diffused to the bulk liquid.
As the periodontitis-associated biofilm develops, there is a significant change to the local pH and oxygen concentration which enables further maturation of the biofilm. While hypoxic chambers are effective platforms for obligate anaerobes in vitro, the uniform low oxygen level throughout the culture volume can be limited to model for early stages of periodontitis requiring aerobic colonisers, as well as the modelling for host tissue interaction due to the lack of oxygen supporting cell growth and metabolism.
The growth rate of the bacteria cultures is significantly higher than mammalian culture resulting in the short-term co-cultures of bacteria and mammalian cells due to the accumulation of endotoxins released by the bacteria. The swift increase in microbial biomass in vitro can induce a level of cell toxicity that surpasses what is typically observed from oral microbiomes in conditions of chronic inflammation, such as periodontitis. To manage bacterial population levels, manual interventions such as physical removal or the application of fluid flow are often used. However, these methods considerably heighten the technical complexity involved in managing the experimental setups.
In the context of complex periodontal disease models, different microbial species and mammalian cells require distinct mimicking strategies and culture modalities. The integration of these diverse culture modalities is essential to meet the demands of multi-species co-cultures in periodontal disease models. While microfluidic platforms can be a suitable alternative platform to emulate multispecies culture interactions, the reliance of auxiliary equipment, tubing connections and sensitivity to microscale bubbles are among the well-known technical barriers limiting their adoption among the microbiologist to adopt this platform.
For the cultivation of multispecies bacteria, accurately replicating complex and diverse microenvironments is vital to cater to the survival needs of various microorganisms. Microbial patterning emerges as a key technique in establishing distinct microenvironments within a model, with 3D bioprinting presenting substantial promise for such applications. At present, there are only a few examples conducted the bioprinting of multiple periodontitis-related bacterial colonies that effectively illustrate cross-feeding interactions and biofilm development.
Challenges in regulating biochemical and physicochemical microenvironments is leading researchers to the use of new materials and technologies for improved microenvironment control, including managing biomacromolecule distribution, regulating liquids and gases gradients and even shear force control. These advancements will not only solve existing problems but also expand the models’ capabilities, such as supporting diverse culture modes for different microbial or mammalian cells.
As research progresses, periodontitis biofilm models will surely become more complex and integrated, incorporating and innovating technologies to simulate a broader range of physiological conditions and variables. This includes introducing diverse nutritional environments and using 3D printing for precise spatial arrangements, enhancing the understanding of periodontitis from multiple angles. Such comprehensive systems are increasingly crucial in periodontitis research, highlighting the significance of such technologies in enhancing our grasp of the disease and its underlying mechanisms.
Meanwhile, the applications of periodontal disease models can also be expanded. There have been reports regarding the potential of treating periodontitis through the transplantation of oral microbiome (Nath et al., 2021). It can be seen that the use of in vitro microbial models in new areas such as pathological examinations and even disease treatments hold great promise in the near future.
CW: Conceptualization, Visualization, Writing–original draft, Writing–review and editing. TX: Writing–original draft. JS: Funding acquisition, Supervision, Writing–review and editing. LO: Conceptualization, Data curation, Formal Analysis, Methodology, Project administration, Software, Supervision, Visualization, Writing–review and editing. YZ: Supervision, Writing–review and editing.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was supported by the International Team for Implantology (ITI) Foundation Research Grant (1699-2022) and Osteology Foundation Advanced Researcher Grant (22-007). LO was supported via the QUT DVC ECR (Early Career Researchers) grant (323100-0235) and The Max Planck Queensland Centre (MPQC) (324912-0007/07).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abe-Yutori, M., Chikazawa, T., Shibasaki, K., and Murakami, S. (2017). Decreased expression of E-cadherin by Porphyromonas gingivalis-lipopolysaccharide attenuates epithelial barrier function. J. Periodontal Res. 52 (1), 42–50. doi:10.1111/jre.12367
Adelfio, M., Martin-Moldes, Z., Erndt-Marino, J., Tozzi, L., Duncan, M. J., Hasturk, H., et al. (2023). Three-dimensional humanized model of the periodontal gingival pocket to study oral microbiome. Adv. Sci. 10 (12), 2205473. doi:10.1002/advs.202205473
Ali Mohammed, M. M., Nerland, A. H., Al-Haroni, M., and Bakken, V. (2013). Characterization of extracellular polymeric matrix, and treatment of Fusobacterium nucleatum and Porphyromonas gingivalis biofilms with DNase I and proteinase K. J. Oral Microbiol. 5, 20015. doi:10.3402/jom.v5i0.20015
Ammann, T. W., Gmür, R., and Thurnheer, T. (2012). Advancement of the 10-species subgingival Zurich Biofilm model by examining different nutritional conditions and defining the structure of the in vitrobiofilms. BMC Microbiol. 12 (1), 227. doi:10.1186/1471-2180-12-227
Aspiras, M. B., Kazmerzak, K. M., Kolenbrander, P. E., McNab, R., Hardegen, N., and Jenkinson, H. F. (2000). Expression of green fluorescent protein in Streptococcus gordonii DL1 and its use as a species-specific marker in coadhesion with Streptococcus oralis 34 in saliva-conditioned biofilms in vitro. Appl. Environ. Microbiol. 66 (9), 4074–4083. doi:10.1128/aem.66.9.4074-4083.2000
Balan, P., Belibasakis, G., Ivanovski, S., Bostanci, N., and Seneviratne, C. J. (2023). Community dynamics of subgingival microbiome in periodontitis and targets for microbiome modulation therapy. Crit. Rev. Microbiol. 49, 726–738. doi:10.1080/1040841x.2022.2133594
Bao, K., Papadimitropoulos, A., Akgül, B., Belibasakis, G. N., and Bostanci, N. (2015). Establishment of an oral infection model resembling the periodontal pocket in a perfusion bioreactor system. Virulence 6 (3), 265–273. doi:10.4161/21505594.2014.978721
Barros, S. P., Williams, R., Offenbacher, S., and Morelli, T. (2016). Gingival crevicular fluid as a source of biomarkers for periodontitis. Periodontology 70 (1), 53–64. doi:10.1111/prd.12107
Berezow, A. B., and Darveau, R. P. (2011). Microbial shift and periodontitis. Periodontol 55 (1), 36–47. doi:10.1111/j.1600-0757.2010.00350.x
Blanc, V., Isabal, S., Sánchez, M. C., Llama-Palacios, A., Herrera, D., Sanz, M., et al. (2014). Characterization and application of a flow system for in vitro multispecies oral biofilm formation. J. Periodontal Res. 49 (3), 323–332. doi:10.1111/jre.12110
Bostanci, N., Thurnheer, T., Aduse-Opoku, J., Curtis, M. A., Zinkernagel, A. S., and Belibasakis, G. N. (2013). Porphyromonas gingivalis regulates TREM-1 in human polymorphonuclear neutrophils via its gingipains. PLoS One 8 (10), e75784. doi:10.1371/journal.pone.0075784
Bowen, W., and Koo, H. (2011). Biology of Streptococcus mutans-derived glucosyltransferases: role in extracellular matrix formation of cariogenic biofilms. Caries Res. 45 (1), 69–86. doi:10.1159/000324598
Bradshaw, D. J., Marsh, P., Schilling, K., and Cummins, D. (1996a). A modified chemostat system to study the ecology of oral biofilms. J. Appl. Bacteriol. 80 (2), 124–130. doi:10.1111/j.1365-2672.1996.tb03199.x
Bradshaw, D. J., Marsh, P. D., Allison, C., and Schilling, K. M. (1996b). Effect of oxygen, inoculum composition and flow rate on development of mixed-culture oral biof ilms. Microbiology 142 (3), 623–629. doi:10.1099/13500872-142-3-623
Cai, J.-N., and Kim, D. (2023). “Chapter Two - biofilm ecology associated with dental caries: understanding of microbial interactions in oral communities leads to development of therapeutic strategies targeting cariogenic biofilms,” in Advances in applied microbiology. Editors G. M. Gadd, and S. Sariaslani (Academic Press), 27–75.
Carpenter, G. (2020). Salivary factors that maintain the normal oral commensal microflora. J. Dent. Res. 99 (6), 644–649. doi:10.1177/0022034520915486
Carrouel, F., Viennot, S., Santamaria, J., Veber, P., and Bourgeois, D. (2016). Quantitative molecular detection of 19 major pathogens in the interdental biofilm of periodontally healthy young adults. Front. Microbiol. 7, 840. doi:10.3389/fmicb.2016.00840
Celik, D., and Kantarci, A. (2021). Vascular changes and hypoxia in periodontal disease as a link to systemic complications. Pathogens 10 (10), 1280. doi:10.3390/pathogens10101280
Chai, L., Vlamakis, H., and Kolter, R. (2011). Extracellular signal regulation of cell differentiation in biofilms. MRS Bull. 36 (5), 374–379. doi:10.1557/mrs.2011.68
Christensen, B. B., Haagensen, J. A. J., Heydorn, A., and Molin, S. (2002). Metabolic commensalism and competition in a two-species microbial consortium. Appl. Environ. Microbiol. 68 (5), 2495–2502. doi:10.1128/aem.68.5.2495-2502.2002
Christensen, G. D., Simpson, W. A., Younger, J. J., Baddour, L. M., Barrett, F. F., Melton, D. M., et al. (1985). Adherence of coagulase-negative staphylococci to plastic tissue culture plates: a quantitative model for the adherence of staphylococci to medical devices. J. Clin. Microbiol. 22 (6), 996–1006. doi:10.1128/jcm.22.6.996-1006.1985
Connolly, E., Millhouse, E., Doyle, R., Culshaw, S., Ramage, G., and Moran, G. (2017). The Porphyromonas gingivalis hemagglutinins HagB and HagC are major mediators of adhesion and biofilm formation. Mol. Oral Microbiol. 32 (1), 35–47. doi:10.1111/omi.12151
Coppenhagen-Glazer, S., Sol, A., Abed, J., Naor, R., Zhang, X., Han, Y. W., et al. (2015). Fap2 of Fusobacterium nucleatum is a galactose-inhibitable adhesin involved in coaggregation, cell adhesion, and preterm birth. Infect. Immun. 83 (3), 1104–1113. doi:10.1128/iai.02838-14
Darveau, R. P. (2010). Periodontitis: a polymicrobial disruption of host homeostasis. Nat. Rev. Microbiol. 8 (7), 481–490. doi:10.1038/nrmicro2337
Dawes, C., Watanabe, S., Biglow-Lecomte, P., and Dibdin, G. (1989). Estimation of the velocity of the salivary film at some different locations in the mouth. J. Dent. Res. 68 (11), 1479–1482. doi:10.1177/00220345890680110201
De Jong, M. H., and Van der Hoeven, J. S. (1987). The growth of oral bacteria on saliva. J. Dent. Res. 66 (2), 498–505. doi:10.1177/00220345870660021901
Diaz, P. I., Zilm, P. S., and Rogers, A. H. (2002). Fusobacterium nucleatum supports the growth of Porphyromonas gingivalis in oxygenated and carbon-dioxide-depleted environments. Microbiology 148 (2), 467–472. doi:10.1099/00221287-148-2-467
Duran-Pinedo, A. E., Baker, V. D., and Frias-Lopez, J. (2014). The periodontal pathogen Porphyromonas gingivalis induces expression of transposases and cell death of Streptococcus mitis in a biofilm model. Infect. Immun. 82 (8), 3374–3382. doi:10.1128/iai.01976-14
Eun, Y.-J., and Weibel, D. B. (2009). Fabrication of microbial biofilm arrays by geometric control of cell adhesion. Langmuir 25 (8), 4643–4654. doi:10.1021/la803985a
Flemming, H. C., Neu, T. R., and Wozniak, D. J. (2007). The EPS matrix: the "house of biofilm cells. J. Bacteriol. 189 (22), 7945–7947. doi:10.1128/jb.00858-07
Foster, J. S., and Kolenbrander, P. E. (2004). Development of a multispecies oral bacterial community in a saliva-conditioned flow cell. Appl. Environ. Microbiol. 70 (7), 4340–4348. doi:10.1128/aem.70.7.4340-4348.2004
Friedman, L., and Kolter, R. (2004). Two genetic loci produce distinct carbohydrate-rich structural components of the Pseudomonas aeruginosa biofilm matrix. Am. Soc. Microbiol. 186, 4457–4465. doi:10.1128/jb.186.14.4457-4465.2004
Genco, R. J. (1996). Current view of risk factors for periodontal diseases. J. periodontology 67, 1041–1049. doi:10.1902/jop.1996.67.10.1041
Ghesquière, J., Simoens, K., Koos, E., Boon, N., Teughels, W., and Bernaerts, K. (2023). Spatiotemporal monitoring of a periodontal multispecies biofilm model: demonstration of prebiotic treatment responses. Appl. Environ. Microbiol. 89 (10), 010811–e1123. doi:10.1128/aem.01081-23
Griffen, A. L., Beall, C. J., Campbell, J. H., Firestone, N. D., Kumar, P. S., Yang, Z. K., et al. (2012). Distinct and complex bacterial profiles in human periodontitis and health revealed by 16S pyrosequencing. ISME J. 6 (6), 1176–1185. doi:10.1038/ismej.2011.191
Groeger, S., Zhou, Y., Ruf, S., and Meyle, J. (2022). Pathogenic mechanisms of Fusobacterium nucleatum on oral epithelial cells. Front. Oral Health 3, 831607. doi:10.3389/froh.2022.831607
Guggenheim, B., Giertsen, E., Schüpbach, P., and Shapiro, S. (2001). Validation of an in vitro biofilm model of supragingival plaque. J. Dent. Res. 80 (1), 363–370. doi:10.1177/00220345010800011201
Guggenheim, B., Gmür, R., Galicia, J. C., Stathopoulou, P. G., Benakanakere, M. R., Meier, A., et al. (2009). In vitromodeling of host-parasite interactions: the 'subgingival' biofilm challenge of primary human epithelial cells. BMC Microbiol. 9 (1), 280. doi:10.1186/1471-2180-9-280
Haffajee, A. D., Teles, R. P., Patel, M. R., Song, X., Veiga, N., and Socransky, S. S. (2009). Factors affecting human supragingival biofilm composition. I. Plaque mass. J. Periodontal Res. 44 (4), 511–519. doi:10.1111/j.1600-0765.2008.01154.x
Hägi, T. T., Klemensberger, S., Bereiter, R., Nietzsche, S., Cosgarea, R., Flury, S., et al. (2015). A biofilm pocket model to evaluate different non-surgical periodontal treatment modalities in terms of biofilm removal and reformation, surface alterations and attachment of periodontal ligament fibroblasts. PLOS ONE 10 (6), e0131056. doi:10.1371/journal.pone.0131056
Hajishengallis, G., Darveau, R. P., and Curtis, M. A. (2012). The keystone-pathogen hypothesis. Nat. Rev. Microbiol. 10 (10), 717–725. doi:10.1038/nrmicro2873
Hajishengallis, G., and Lamont, R. J. (2012). Beyond the red complex and into more complexity: the polymicrobial synergy and dysbiosis (PSD) model of periodontal disease etiology. Mol. Oral Microbiol. 27 (6), 409–419. doi:10.1111/j.2041-1014.2012.00663.x
Hamada, S., and Torii, M. (1978). Effect of sucrose in culture media on the location of glucosyltransferase of Streptococcus mutans and cell adherence to glass surfaces. Infect. Immun. 20 (3), 592–599. doi:10.1128/iai.20.3.592-599.1978
Hao, Y., Huang, X., Zhou, X., Li, M., Ren, B., Peng, X., et al. (2018). Influence of dental prosthesis and restorative materials interface on oral biofilms. Int. J. Mol. Sci. 19 (10), 3157. doi:10.3390/ijms19103157
Huang, R., Li, M., Ye, M., Yang, K., Xu, X., and Gregory, R. L. (2014). Effects of nicotine on Streptococcus gordonii growth, biofilm formation, and cell aggregation. Appl. Environ. Microbiol. 80 (23), 7212–7218. doi:10.1128/aem.02395-14
Izui, S., Sekine, S., Maeda, K., Kuboniwa, M., Takada, A., Amano, A., et al. (2016). Antibacterial activity of curcumin against periodontopathic bacteria. J. Periodontology 87 (1), 83–90. doi:10.1902/jop.2015.150260
Jaffar, N., Miyazaki, T., and Maeda, T. (2016). Biofilm formation of periodontal pathogens on hydroxyapatite surfaces: implications for periodontium damage. J. Biomed. Mater. Res. Part A 104 (11), 2873–2880. doi:10.1002/jbm.a.35827
Jakubovics, N. S. (2015). Intermicrobial interactions as a driver for community composition and stratification of oral biofilms. J. Mol. Biol. 427 (23), 3662–3675. doi:10.1016/j.jmb.2015.09.022
Janakiram, C., and Dye, B. A. (2020). A public health approach for prevention of periodontal disease. Periodontology 84 (1), 202–214. doi:10.1111/prd.12337
Janakiraman, V., Englert, D., Jayaraman, A., and Baskaran, H. (2009). Modeling growth and quorum sensing in biofilms grown in microfluidic chambers. Ann. Biomed. Eng. 37 (6), 1206–1216. doi:10.1007/s10439-009-9671-8
Joshi, R. V., Gunawan, C., and Mann, R. (2021). We are one: multispecies metabolism of a biofilm consortium and their treatment strategies. Front. Microbiol. 12, 635432. doi:10.3389/fmicb.2021.635432
Kawai, T., and Akira, S. (2005). Pathogen recognition with Toll-like receptors. Curr. Opin. Immunol. 17 (4), 338–344. doi:10.1016/j.coi.2005.02.007
Kniha, K., Heussen, N., Modabber, A., Hölzle, F., and Möhlhenrich, S. (2021). The effect of zirconia and titanium surfaces on biofilm formation and on host-derived immunological parameters. Int. J. Oral Maxillofac. Surg. 50 (10), 1361–1374. doi:10.1016/j.ijom.2021.01.021
Kolenbrander, P. E. (2011). Multispecies communities: interspecies interactions influence growth on saliva as sole nutritional source. Int. J. Oral Sci. 3 (2), 49–54. doi:10.4248/ijos11025
Kriebel, K., Hieke, C., Müller-Hilke, B., Nakata, M., and Kreikemeyer, B. (2018). Oral biofilms from symbiotic to pathogenic interactions and associated disease–connection of periodontitis and rheumatic arthritis by peptidylarginine deiminase. Front. Microbiol. 9, 53. doi:10.3389/fmicb.2018.00053
Kuboniwa, M., and Lamont, R. J. (2000). Subgingival biofilm formation. Periodontol 52 (1), 38–52. doi:10.1111/j.1600-0757.2009.00311.x
Kudrin, A., and Ray, D. (2008). Cunning factor: macrophage migration inhibitory factor as a redox-regulated target. Immunol. Cell Biol. 86 (3), 232–238. doi:10.1038/sj.icb.7100133
Kumbar, V. M., Peram, M. R., Kugaji, M. S., Shah, T., Patil, S. P., Muddapur, U. M., et al. (2021). Effect of curcumin on growth, biofilm formation and virulence factor gene expression of Porphyromonas gingivalis. Odontology 109 (1), 18–28. doi:10.1007/s10266-020-00514-y
Lam, R. H., Cui, X., Guo, W., and Thorsen, T. (2016). High-throughput dental biofilm growth analysis for multiparametric microenvironmental biochemical conditions using microfluidics. Lab. Chip 16 (9), 1652–1662. doi:10.1039/c6lc00072j
Lamont, R. J., Hajishengallis, G., and Koo, H. (2023). Social networking at the microbiome-host interface. Infect. Immun. 91 (9), e0012423–23. doi:10.1128/iai.00124-23
Lamont, R. J., and Jenkinson, H. F. (1998). Life below the gum line: pathogenic mechanisms of Porphyromonas gingivalis. Microbiol. Mol. Biol. Rev. 62 (4), 1244–1263. doi:10.1128/mmbr.62.4.1244-1263.1998
Lang, K. N., Sculean, A., Eick, S., and Stähli, A. (2022). A novel in vitro periodontal pocket model to evaluate the effect of root surface instrumentation on biofilm-epithelial cell interactions. Clin. Oral Investig. 26 (5), 4021–4029. doi:10.1007/s00784-022-04371-7
Laventie, B.-J., Sangermani, M., Estermann, F., Manfredi, P., Planes, R., Hug, I., et al. (2019). A surface-induced asymmetric program promotes tissue colonization by Pseudomonas aeruginosa. Cell Host Microbe 25 (1), 140–152.e6. doi:10.1016/j.chom.2018.11.008
Lee, J. Y., Sojar, H. T., Bedi, G. S., and Genco, R. J. (1992). Synthetic peptides analogous to the fimbrillin sequence inhibit adherence of Porphyromonas gingivalis. Infect. Immun. 60 (4), 1662–1670. doi:10.1128/iai.60.4.1662-1670.1992
Li, B., Li, X., Lin, H., and Zhou, Y. (2018). Curcumin as a promising antibacterial agent: effects on metabolism and biofilm formation in S. mutans. BioMed Res. Int. 2018, 1–11. doi:10.1155/2018/4508709
Li, C., Lv, Z., Shi, Z., Zhu, Y., Wu, Y., Li, L., et al. (2017). Periodontal therapy for the management of cardiovascular disease in patients with chronic periodontitis. Cochrane Database Syst. Rev. 11 (11), Cd009197. doi:10.1002/14651858.cd009197.pub3
Li, X., Lan, H. Y., Huang, X. R., Zhang, C., and Jin, L. J. (2013). Expression profile of macrophage migration-inhibitory factor in human gingiva and reconstituted human gingival epithelia stimulated by Porphyromonas gingivalis lipopolysaccharide. J. Periodontal Res. 48 (4), 527–532. doi:10.1111/jre.12035
Love, R. M. (2010). Biofilm–substrate interaction: from initial adhesion to complex interactions and biofilm maturity. Endod. Top. 22 (1), 50–57. doi:10.1111/j.1601-1546.2012.00280.x
Luo, T. L., Vanek, M. E., Gonzalez-Cabezas, C., Marrs, C. F., Foxman, B., and Rickard, A. H. (2022). In vitro model systems for exploring oral biofilms: from single-species populations to complex multi-species communities. J. Appl. Microbiol. 132 (2), 855–871. doi:10.1111/jam.15200
Maekawa, T., Krauss, J., Abe, T., Jotwani, R., Triantafilou, M., Triantafilou, K., et al. (2014). Porphyromonas gingivalis manipulates complement and TLR signaling to uncouple bacterial clearance from inflammation and promote dysbiosis. Cell Host Microbe 15 (6), 768–778. doi:10.1016/j.chom.2014.05.012
Maezono, H., Noiri, Y., Asahi, Y., Yamaguchi, M., Yamamoto, R., Izutani, N., et al. (2011). Antibiofilm effects of azithromycin and erythromycin on Porphyromonas gingivalis. Antimicrob. Agents Chemother. 55 (12), 5887–5892. doi:10.1128/aac.05169-11
Makkar, H., Zhou, Y., Tan, K. S., Lim, C. T., and Sriram, G. (2023). Modeling crevicular fluid flow and host-oral microbiome interactions in a gingival crevice-on-chip. Adv. Healthc. Mater. 12 (6), 2202376. doi:10.1002/adhm.202202376
Marchesan, J., Girnary, M. S., Jing, L., Miao, M. Z., Zhang, S., Sun, L., et al. (2018). An experimental murine model to study periodontitis. Nat. Protoc. 13 (10), 2247–2267. doi:10.1038/s41596-018-0035-4
Mark Welch, J. L., Ramírez-Puebla, S. T., and Borisy, G. G. (2020). Oral microbiome geography: micron-scale habitat and niche. Cell Host Microbe 28 (2), 160–168. doi:10.1016/j.chom.2020.07.009
Mark Welch, J. L., Rossetti, B. J., Rieken, C. W., Dewhirst, F. E., and Borisy, G. G. (2016). Biogeography of a human oral microbiome at the micron scale. Proc. Natl. Acad. Sci. 113 (6), E791–E800. doi:10.1073/pnas.1522149113
Marsh, P., and Martin, M. (1992). The mouth as a microbial habitat. Oral Microbiol., 6–26. doi:10.1007/978-1-4615-7556-6_2
Marsh, P. D., and Zaura, E. (2017). Dental biofilm: ecological interactions in health and disease. J. Clin. Periodontology 44 (S18), S12–S22. doi:10.1111/jcpe.12679
McBain, A., Sissons, C., Ledder, R., Sreenivasan, P., De Vizio, W., and Gilbert, P. (2005). Development and characterization of a simple perfused oral microcosm. J. Appl. Microbiol. 98 (3), 624–634. doi:10.1111/j.1365-2672.2004.02483.x
McBain, A. J. (2009). Chapter 4: in vitro biofilm models: an overview. Adv. Appl. Microbiol. 69, 99–132. doi:10.1016/S0065-2164(09)69004-3
Millhouse, E., Jose, A., Sherry, L., Lappin, D. F., Patel, N., Middleton, A. M., et al. (2014). Development of an in vitroperiodontal biofilm model for assessing antimicrobial and host modulatory effects of bioactive molecules. BMC Oral Health 14 (1), 80. doi:10.1186/1472-6831-14-80
Mishra, A., Wu, C., Yang, J., Cisar, J. O., Das, A., and Ton-That, H. (2010). The Actinomyces oris type 2 fimbrial shaft FimA mediates co-aggregation with oral streptococci, adherence to red blood cells and biofilm development. Mol. Microbiol. 77 (4), 841–854. doi:10.1111/j.1365-2958.2010.07252.x
Mountcastle, S. E., Cox, S. C., Sammons, R. L., Jabbari, S., Shelton, R. M., and Kuehne, S. A. (2020). A review of co-culture models to study the oral microenvironment and disease. J. Oral Microbiol. 12 (1), 1773122. doi:10.1080/20002297.2020.1773122
Nance, W. C., Dowd, S. E., Samarian, D., Chludzinski, J., Delli, J., Battista, J., et al. (2013). A high-throughput microfluidic dental plaque biofilm system to visualize and quantify the effect of antimicrobials. J. Antimicrob. Chemother. 68 (11), 2550–2560. doi:10.1093/jac/dkt211
Nath, S., Zilm, P., Jamieson, L., Kapellas, K., Goswami, N., Ketagoda, K., et al. (2021). Development and characterization of an oral microbiome transplant among Australians for the treatment of dental caries and periodontal disease: a study protocol. PLOS ONE 16 (11), e0260433. doi:10.1371/journal.pone.0260433
Ohyama, H., Nakasho, K., Yamanegi, K., Noiri, Y., Kuhara, A., Kato-Kogoe, N., et al. (2009). An unusual autopsy case of pyogenic liver abscess caused by periodontal bacteria. Jpn. J. Infect. Dis. 62 (5), 381–383. doi:10.7883/yoken.jjid.2009.381
Olsen, I., and Hajishengallis, G. (2016). Major neutrophil functions subverted by Porphyromonas gingivalis. J. Oral Microbiol. 8 (1), 30936. doi:10.3402/jom.v8.30936
Pan, W., Wang, Q., and Chen, Q. (2019). The cytokine network involved in the host immune response to periodontitis. Int. J. oral Sci. 11 (3), 30. doi:10.1038/s41368-019-0064-z
Papadopoulos, G., Shaik-Dasthagirisaheb, Y., Huang, N., Viglianti, G., Henderson, A., Kantarci, A., et al. (2017). Immunologic environment influences macrophage response to Porphyromonas gingivalis. Mol. oral Microbiol. 32 (3), 250–261. doi:10.1111/omi.12168
Pinnock, A., Murdoch, C., Moharamzadeh, K., Whawell, S., and Douglas, C. I. (2014). Characterisation and optimisation of organotypic oral mucosal models to study Porphyromonas gingivalis invasion. Microbes Infect. 16 (4), 310–319. doi:10.1016/j.micinf.2014.01.004
Pöllänen, M. T., Paino, A., and Ihalin, R. (2013). Environmental stimuli shape biofilm formation and the virulence of periodontal pathogens. Int. J. Mol. Sci. 14 (8), 17221–17237. doi:10.3390/ijms140817221
Pratten, J., Wills, K., Barnett, P., and Wilson, M. (1998). In vitro studies of the effect of antiseptic-containing mouthwashes on the formation and viability of Streptococcus sanguis biofilms. J. Appl. Microbiol. 84 (6), 1149–1155. doi:10.1046/j.1365-2672.1998.00462.x
Ramachandra, S. S., Abdal-hay, A., Han, P., Lee, R. S., and Ivanovski, S. (2023b). Fabrication and characterization of a 3D polymicrobial microcosm biofilm model using melt electrowritten scaffolds. Biomater. Adv. 145, 213251. doi:10.1016/j.bioadv.2022.213251
Ramachandra, S. S., Sime, F. B., Naicker, S., Han, P., Lee, R. S., C. Wallis, S., et al. (2023a). An in vitro dynamic bioreactor model for evaluating antimicrobial effectiveness on periodontal polymicrobial biofilms: a proof-of-concept study. J. Periodontol. 95, 384–396. doi:10.1002/jper.23-0086
Ren, Y., Wang, C., Chen, Z., Allan, E., van der Mei, H. C., and Busscher, H. J. (2018). Emergent heterogeneous microenvironments in biofilms: substratum surface heterogeneity and bacterial adhesion force-sensing. FEMS Microbiol. Rev. 42 (3), 259–272. doi:10.1093/femsre/fuy001
Saito, Y., Fujii, R., Nakagawa, K., Kuramitsu, H. K., Okuda, K., and Ishihara, K. (2008). Stimulation of Fusobacterium nucleatum biofilm formation by Porphyromonas gingivalis. Oral Microbiol. Immunol. 23 (1), 1–6. doi:10.1111/j.1399-302x.2007.00380.x
Sánchez, M. C., Llama-Palacios, A., Blanc, V., León, R., Herrera, D., and Sanz, M. (2011). Structure, viability and bacterial kinetics of an in vitro biofilm model using six bacteria from the subgingival microbiota. J. Periodontal Res. 46 (2), 252–260. doi:10.1111/j.1600-0765.2010.01341.x
Sánchez, M. C., Llama-Palacios, A., Fernández, E., Figuero, E., Marín, M., León, R., et al. (2014). An in vitro biofilm model associated to dental implants: structural and quantitative analysis of in vitro biofilm formation on different dental implant surfaces. Dent. Mater. 30 (10), 1161–1171. doi:10.1016/j.dental.2014.07.008
Sánchez, M. C., Toledano-Osorio, M., Bueno, J., Figuero, E., Toledano, M., Medina-Castillo, A., et al. (2019). Antibacterial effects of polymeric PolymP-n Active nanoparticles. An in vitro biofilm study. Dent. Mater. 35 (1), 156–168. doi:10.1016/j.dental.2018.11.015
Scannapieco, F. A., and Ho, A. W. (2001). Potential associations between chronic respiratory disease and periodontal disease: analysis of National Health and Nutrition Examination Survey III. J. Periodontol. 72 (1), 50–56. doi:10.1902/jop.2001.72.1.50
Schou, S., Holmstrup, P., and Kornman, K. S. (1993). Non-human primates used in studies of periodontal disease pathogenesis: a review of the literature. J. Periodontology 64 (6), 497–508. doi:10.1902/jop.1993.64.6.497
Sedghi, L., DiMassa, V., Harrington, A., Lynch, S. V., and Kapila, Y. L. (2000). The oral microbiome: role of key organisms and complex networks in oral health and disease. Periodontol 87 (1), 107–131. doi:10.1111/prd.12393
Shapiro, S., Giertsen, E., and Guggenheim, B. P. D. (2002). An in vitro oral biofilm model for comparing the efficacy of antimicrobial mouthrinses. Caries Res. 36, 93–100. doi:10.1159/000057866
Shellis, R. P. (1978). A synthetic saliva for cultural studies of dental plaque. Archives Oral Biol. 23 (6), 485–489. doi:10.1016/0003-9969(78)90081-x
Siddiqi, A., Milne, T., Cullinan, M. P., and Seymour, G. J. (2016). Analysis of P. gingivalis, T. forsythia and S. aureus levels in edentulous mouths prior to and 6 months after placement of one-piece zirconia and titanium implants. Clin. Oral Implants Res. 27 (3), 288–294. doi:10.1111/clr.12536
Socransky, S., Haffajee, A., Cugini, M., Smith, C., and Kent, R. L. (1998a). Microbial complexes in subgingival plaque. J. Clin. periodontology 25 (2), 134–144. doi:10.1111/j.1600-051x.1998.tb02419.x
Socransky, S. S., Haffajee, A., Cugini, M., Smith, C., and Kent, R. L. (1998b). Microbial complexes in subgingival plaque. J. Clin. Periodontol. 25 (2), 134–144. doi:10.1111/j.1600-051x.1998.tb02419.x
Song, W., Lee, J. K., Park, S. H., Um, H. S., Lee, S. Y., and Chang, B. S. (2017). Comparison of periodontitis-associated oral biofilm formation under dynamic and static conditions. jpis 47 (4), 219–230. doi:10.5051/jpis.2017.47.4.219
Stacy, A., McNally, L., Darch, S. E., Brown, S. P., and Whiteley, M. (2016). The biogeography of polymicrobial infection. Nat. Rev. Microbiol. 14 (2), 93–105. doi:10.1038/nrmicro.2015.8
Sterzenbach, T., Helbig, R., Hannig, C., and Hannig, M. (2020). Bioadhesion in the oral cavity and approaches for biofilm management by surface modifications. Clin. oral Investig. 24, 4237–4260. doi:10.1007/s00784-020-03646-1
Stoodley, P., Lewandowski, Z., Boyle, J. D., and Lappin-Scott, H. M. (1999). The formation of migratory ripples in a mixed species bacterial biofilm growing in turbulent flow. Environ. Microbiol. 1 (5), 447–455. doi:10.1046/j.1462-2920.1999.00055.x
Takeuchi, H., and Amano, A. (2021b). “Invasion of gingival epithelial cells by porphyromonas gingivalis,” in Periodontal pathogens: methods and protocols. Editors K. Nagano, and Y. Hasegawa (New York, NY: Springer US), 215–224.
Takeuchi, H., Yamaga, S., Sasaki, N., Kuboniwa, M., Matsusaki, M., and Amano, A. (2021a). Porphyromonas gingivalis induces penetration of lipopolysaccharide and peptidoglycan through the gingival epithelium via degradation of coxsackievirus and adenovirus receptor. Cell Microbiol. 23 (11), e13388. doi:10.1111/cmi.13388
Tariq, M., Talegaonkar, S., Ahmad, Z., Ali, J., Baboota, S., Iqbal, Z., et al. (2012). Treatment modalities and evaluation models for periodontitis. Int. J. Pharm. investigation 2 (3), 106. doi:10.4103/2230-973x.104394
Thurnheer, T., Belibasakis, G. N., and Bostanci, N. (2014). Colonisation of gingival epithelia by subgingival biofilms in vitro: role of “red complex” bacteria. Archives Oral Biol. 59 (9), 977–986. doi:10.1016/j.archoralbio.2014.05.023
Tolker-Nielsen, T., Brinch, U. C., Ragas, P. C., Andersen, J. B., Jacobsen, C. S., and Molin, S. (2000). Development and dynamics of Pseudomonas sp. biofilms. J. Bacteriol. 182 (22), 6482–6489. doi:10.1128/jb.182.22.6482-6489.2000
Uitto, V.-J. (2003). Gingival crevice fluid – an introduction. Periodontology 31 (1), 9–11. doi:10.1034/j.1600-0757.2003.03101.x
Verhamme, D. T., Murray, E. J., and Stanley-Wall, N. R. (2009). DegU and Spo0A jointly control transcription of two loci required for complex colony development by Bacillus subtilis. J. Bacteriol. 191 (1), 100–108. doi:10.1128/jb.01236-08
Vernal, R., Dutzan, N., Chaparro, A., Puente, J., Antonieta Valenzuela, M., and Gamonal, J. (2005). Levels of interleukin-17 in gingival crevicular fluid and in supernatants of cellular cultures of gingival tissue from patients with chronic periodontitis. J. Clin. periodontology 32 (4), 383–389. doi:10.1111/j.1600-051x.2005.00684.x
Weerkamp, A., Uyen, H., and Busscher, H. (1988). Effect of zeta potential and surface energy on bacterial adhesion to uncoated and saliva-coated human enamel and dentin. J. Dent. Res. 67 (12), 1483–1487. doi:10.1177/00220345880670120801
Weinberg, M. A., and Bral, M. (1999). Laboratory animal models in periodontology. J. Clin. periodontology 26 (6), 335–340. doi:10.1034/j.1600-051x.1999.260601.x
Weyell, P., Beekmann, U., Küpper, C., Dederichs, M., Thamm, J., Fischer, D., et al. (2019). Tailor-made material characteristics of bacterial cellulose for drug delivery applications in dentistry. Carbohydr. Polym. 207, 1–10. doi:10.1016/j.carbpol.2018.11.061
Whitesides, G. M. (2006). The origins and the future of microfluidics. Nature 442 (7101), 368–373. doi:10.1038/nature05058
Wong, L., and Sissions, C. H. (2001). A comparison of human dental plaque microcosm biofilms grown in an undefined medium and a chemically defined artificial saliva. Archives Oral Biol. 46 (6), 477–486. doi:10.1016/s0003-9969(01)00016-4
Wu, T., Trevisan, M., Genco, R. J., Dorn, J. P., Falkner, K. L., and Sempos, C. T. (2000). Periodontal disease and risk of cerebrovascular disease: the first national health and nutrition examination survey and its follow-up study. Archives Intern. Med. 160 (18), 2749–2755. doi:10.1001/archinte.160.18.2749
Yang, D. C., Blair, K. M., and Salama, N. R. (2016). Staying in shape: the impact of cell shape on bacterial survival in diverse environments. Microbiol. Mol. Biol. Rev. 80 (1), 187–203. doi:10.1128/mmbr.00031-15
Yilmaz, Ö., Verbeke, P., Lamont, R. J., and Ojcius, D. M. (2006). Intercellular spreading of porphyromonas gingivalis infection in primary gingival epithelial cells. Infect. Immun. 74 (1), 703–710. doi:10.1128/iai.74.1.703-710.2006
Yilmaz, Ö., Young, P. A., Lamont, R. J., and Kenny, G. E. (2003). Gingival epithelial cell signalling and cytoskeletal responses to Porphyromonas gingivalis invasion. Microbiology 149 (9), 2417–2426. doi:10.1099/mic.0.26483-0
Yoshioka, H., Yoshimura, A., Kaneko, T., Golenbock, D. T., and Hara, Y. (2008). Analysis of the activity to induce Toll-like receptor (TLR) 2-and TLR4-mediated stimulation of supragingival plaque. J. periodontology 79 (5), 920–928. doi:10.1902/jop.2008.070516
Zainal-Abidin, Z., Veith, P. D., Dashper, S. G., Zhu, Y., Catmull, D. V., Chen, Y. Y., et al. (2012). Differential proteomic analysis of a polymicrobial biofilm. J. proteome Res. 11 (9), 4449–4464. doi:10.1021/pr300201c
Zhang, Z., Liu, S., Zhang, S., Li, Y., Shi, X., Liu, D., et al. (2022). Porphyromonas gingivalis outer membrane vesicles inhibit the invasion of Fusobacterium nucleatum into oral epithelial cells by downregulating FadA and FomA. J. Periodontol. 93 (4), 515–525. doi:10.1002/jper.21-0144
Zhao, H., Chu, M., Huang, Z., Yang, X., Ran, S., Hu, B., et al. (2017). Variations in oral microbiota associated with oral cancer. Sci. Rep. 7 (1), 11773. doi:10.1038/s41598-017-11779-9
Zheng, S., Yu, S., Fan, X., Zhang, Y., Sun, Y., Lin, L., et al. (2021). Porphyromonas gingivalis survival skills: immune evasion. J. periodontal Res. 56 (6), 1007–1018. doi:10.1111/jre.12915
Zijnge, V., Ammann, T., Thurnheer, T., and Gmür, R. (2012). Subgingival biofilm structure. Front. Oral Biol. 15, 1–16. doi:10.1159/000329667
Keywords: periodontitis, dental plaque biofilms, pathophysiology, in vitro models, microbiome
Citation: Wang C, Xu T, Seneviratne CJ, Ong LJY and Zhou Y (2024) Modelling periodontitis in vitro: engineering strategies and biofilm model development. Front. Biomater. Sci. 3:1380153. doi: 10.3389/fbiom.2024.1380153
Received: 01 February 2024; Accepted: 21 June 2024;
Published: 12 July 2024.
Edited by:
Keiji Nagano, Health Sciences University of Hokkaido, JapanReviewed by:
Anilei Hoare, University of Chile, ChileCopyright © 2024 Wang, Xu, Seneviratne, Ong and Zhou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Louis Jun Ye Ong, bG91aXMub25nanVueWVAcXV0LmVkdS5hdSYjeDAyMDBhOw==; Yinghong Zhou, eWluZ2hvbmcuemhvdUB1cS5lZHUuYXU=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.