
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Bioeng. Biotechnol. , 04 December 2023
Sec. Synthetic Biology
Volume 11 - 2023 | https://doi.org/10.3389/fbioe.2023.1261832
L-tryptophan and its derivatives are widely used in the chemical, pharmaceutical, food, and feed industries. Microbial fermentation is the most commonly used method to produce L-tryptophan, which calls for an effective cell factory. The mechanism of L-tryptophan biosynthesis in Escherichia coli, the widely used producer of L-tryptophan, is well understood. Saccharomyces cerevisiae also plays a significant role in the industrial production of biochemicals. Because of its robustness and safety, S. cerevisiae is favored for producing pharmaceuticals and food-grade biochemicals. However, the biosynthesis of L-tryptophan in S. cerevisiae has been rarely summarized. The synthetic pathways and engineering strategies of L-tryptophan in E. coli and S. cerevisiae have been reviewed and compared in this review. Furthermore, the information presented in this review pertains to the existing understanding of how L-tryptophan affects S. cerevisiae’s stress fitness, which could aid in developing a novel plan to produce more resilient industrial yeast and E. coli cell factories.
L-tryptophan (L-Trp) is an essential aromatic amino acid required for protein synthesis in humans and animals (Modoux et al., 2021). In recent years, the application of L-Trp in food additives and feed and in medical fields has attracted much attention (Xiao et al., 2023). L-Trp is typically produced through chemical synthesis, direct fermentation, and enzymatic conversion (Liu et al., 2012). However, with the advent and expansion of the green business, chemical synthesis has become less desired. Enzymatic conversion is an efficient method for manufacturing L-Trp, but its high cost is its main downside. Microbial fermentation is often favored over chemical synthesis and enzymatic conversion processes because it allows for the environmentally benign synthesis of L-Trp from inexpensive and renewable carbon sources (Niu et al., 2019). Then, a pressing issue for the growth of this industry is how to enhance the yield of L-Trp through the microbial fermentation process. Although the production of L-Trp has significantly grown in recent years due to the ongoing development of superior genome sequencing and synthetic biology technologies, methods for maximizing L-Trp yield still need to be refined (Liu et al., 2022).
The metabolic engineering alteration of core strains is one of the essential components of microbial fermentation. Escherichia coli (E. coli) and Corynebacterium glutamicum (C. glutamicum) are well-studied chassis cell factories for producing L-Trp (Wendisch et al., 2006; Guo et al., 2022). Saccharomyces cerevisiae (S. cerevisiae) is a prominent representative among eukaryotic microbial hosts, and aromatic amino acid metabolism by yeast is of interest for some industrial applications (Luttik et al., 2008b). S. cerevisiae has also been utilized to produce L-Trp and its derivativesin bioprocessing. This choice is attributed to the organism’s resilience and enhanced ability to withstand challenging fermentation conditions. Additionally, S. cerevisiae can express P450 oxidase in eukaryotic secondary metabolic pathways, further supporting its suitability (Zhang et al., 2016). The metabolic engineering of the L-Trp biosynthetic pathway has emerged as a research hotspot to generate the L-Trp overproducing microbial strains for industrial production of L-Trp (Niu et al., 2019). However, additional investigation is required in order to elucidate the metabolic pathways and ascertain the specific targets for alteration.
Another way to enhance L-Trp production is to make fermentation strains more resilient and adapted to the environment. During fermentation, E. coli and S. cerevisiae are exposed to various environmental stresses, such as osmotic, ethanol, oxidation, and heat stress, which can seriously affect the growth and metabolism of microbes (Shen et al., 2022). Studies have shown increasing interest in studying amino acid metabolism related to signaling and metabolic adaptation to stress in plants and microbes in recent years (Zeng et al., 2022). To improve the robustness of E. coli, strategies such as adaptive laboratory evolution and genome-wide evolutionary engineering are commonly used (Yao et al., 2022). Whether E. coli tolerance to stressful settings can be improved by regulating L-Trp to expand L-Trp production capacity further has received less attention, with most studies focused on regulating L-Trp for stress adaption in S. cerevisiae. Exogenous L-Trp addition and modulation of overexpression or downregulation of L-Trp-related genes were discovered to successfully improve S. cerevisiae adaptability to environmental stress and boost L-Trp synthesis (Kavšček et al., 2015).
The biosynthetic pathway and the main regulatory mechanisms involved in L-Trp production in E. coli and S. cerevisiae have been reviewed and compared in this review. In addition, the latest strategies used to enhance the production of L-Trp in E. coli and S. cerevisiae have been summarized. Lastly, the relationship between L-Trp and the stress tolerance of S. cerevisiae has been highlighted, along with a discussion of the current strategies for regulating the metabolism of amino acids, especially L-Trp, to improve the robustness of S. cerevisiae and E. coli.
Both S. cerevisiae and E. coli are jointly involved in three synthetic pathways of L-Trp synthesis: the central metabolic pathway, the shikimic acid (SK) pathway, and the chorismate (CHO) pathway. Under oxygen-rich conditions, the Embden-Meyerhof-Parnas pathway transforms a portion of glucose into phosphoenolpyruvate (PEP), while the tricarboxylic acid cycle (TCA) consumes the other portion of glucose (Carvalho et al., 2019). Hexose monophosphate and erythrose-4-phosphate (E4P) are eradicated to generate 3-deoxy-D-arabinogenose-7-phosphate (DAHP). SK pathway starts with DAHP, and the resulting 3-dehydrogenate, 3-dehydrooxalate (3-DHS) and 3-phosphate are converted to 5-enol propyl oxalate 3-phosphate (Gottardi et al., 2017). In the CHO pathway, anthranilate (ANTH) and phenylpyruvate (PPA) are first generated, leading to the production of L-Glutamic acid (L-Glu). ANTH is converted into indoglycerophosphate and anthranilate-5-phosphoribosyl pyrophosphate (PRA), while indole and serine together synthesize L-Trp. Three aromatic amino acids are produced in the CHO pathway: L-Phenylalanine (L-Phe), L-Tyrosine (L-Tyr), and L-Trp. The other two aromatic amino acids, i.e., L-Phe and L-Tyr, negatively affect L-Trp (Tröndle et al., 2020). This study undertakes a comparative analysis of L-Trp biosynthesis and regulation in E. coli and S. cerevisiae, focusing on three distinct synthetic pathways (Figure 1).
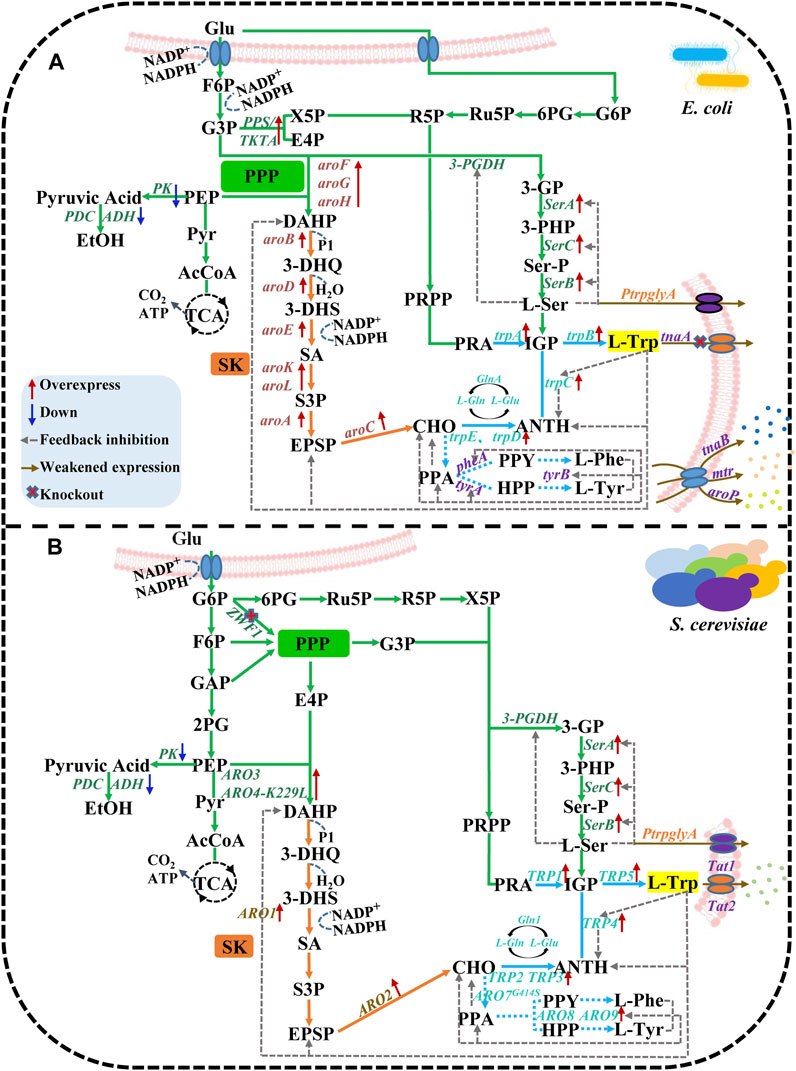
FIGURE 1. The biosynthetic pathway of L-Trp in E. coli and S. cerevisiae. (A) E. coli and (B) S. cerevisiae are jointly involved in three synthetic pathways: central metabolic, shikimic acid, and chorismate. The red solid line arrow indicates overexpress, the blue solid line arrow indicates down, the gray dotted line arrow indicates feedback inhibition, the brown solid line arrow indicates weakened expression, and × indicates knockout.
In E. coli, 70% of glucose entering the cell is consumed to produce fructose-6-phosphate (F6P) and glyceraldehyde-3-phosphate (G3P). The remaining glucose enters into the Pentose Phosphate Pathway (PPP) pathway. PEP and E4P are essential precursors of L-Trp synthesis. The L-Trp production can be enhanced by directly upregulating the concentration of these precursors or reducing the competing reactions (Ikeda, 2006). When E. coli is grown in a restricted medium with glucose as a carbon source, about 50% of PEP is consumed by the phosphotransferase system (PTS) to carry out glucose transport and phosphorylation (Gosset, 2005). Only 3% of PEP synthesizes DAHP (Gosset, 2005). Low intracellular PEP concentration in E. coli leads to decreased carbon fusion in the TCA cycle and PPP pathway, resulting in poor cell growth (Li et al., 2020a). The introduction of the glf and glk genes encoding glucose transporter and glucokinase, respectively, in exogenous efficient transport systems like Pseudomonas is less efficient than the modification of the PTS system, which results in glucose no longer consuming PEP and an increase in intracellular PEP (Escalante et al., 2012). Therefore, PTS mutant strains (ptsHIcrr−, glf-glk+, and ptsG−) were constructed in E. coli for batch fermentation (Wu et al., 2017). The growth of ptsHIcrr− and glf-glk mutant bacteria was up to an OD600 value of 125, which was 47% higher than the original strain. The glucose conversion rate was also increased by 26.5%. On the other hand, ptsG− mutant bacteria grew up to an OD600 value of 100, which was 17.6% higher than the original strain. The glucose conversion rate was also increased by 17.4% (Wu et al., 2017).
In another study, Glf in Z. mobilis and Glk in E. coli for glucose utilization were used to replace the PTS system (Tang et al., 2013; Noda et al., 2016; Noda et al., 2017). According to reports, intracellular glucose can undergo phosphorylation by using ATP as a phosphoric acid donor. This process facilitates the integration of Glf into the ptsG site of SX2 strains, thereby exerting control over the production of glf genes. These genes are regulated by promoters exhibiting varying degrees of expression intensity (Lu et al., 2012). Among these promotors, the weak promoters PM1-12 and Ptrc-pycP458S overexpressed glf and glk corresponding to strain SX11, fermented in a 5 L bioreactor for 40 h. SX11 strain produced L-Trp titers as high as 41.7 g/L, yielding 34.1 times than the L-Trp yield in the original strain SX2 (Xiong et al., 2021). Phosphoenolpyruvate synthase (PPS) and transketolase (tktA) are the main enzymes required to form PEP and E4P in E. coli directly. By overexpressing pps and/or tktA in E. coli, the expressions of PEP and E4P have been upregulated, resulting in 2 times higher production of DAHP (Shen et al., 2012). Similarly, the upregulation of the citric acid pathway and TCA cycle resulted in the upregulation of citT, which transports citric acid into cells. After 24 h of culture, the accumulated L-Trp was 7 g/L (Du et al., 2019). Knocking out the gene encoding PEP to degrade pyruvate kinases pykA and pykF, as well as blocking the metabolism of PEP, also facilitated the carbon metabolic flow and led to higher aromatics and DAHP yields (Gosset et al., 1996; Meza et al., 2012).
After balancing the effect of PEP on TCA, 5 g/L L-Trp was obtained from E. coli strain TRP07 with a glucose yield of 0.186 g/g after 36 h of fermentation in a 5 L bioreactor (Chen et al., 2018b; Du et al., 2019). L-Serine (L-Ser), a precursor in L-Trp synthesis, is often inhibited by D-3-phosphoglycerate dehydrogenase (3-PGDH) during synthesis process (Grant, 2018). L-Ser is mostly undersupplied, and thus, L-Trp cannot be overproduced (Chen and Zeng, 2017). However, removal of the SerA gene encoding 3-PGDH regulatory domain can stop the inhibition of L-Ser (Bell et al., 2002).
In S. cerevisiae, glucose enters the cell to synthesize glucose-6-phosphate (G6P), F6P, G3P, glyceraldehyde-3-phosphate (GAP), 2-phosphate glycerate (2PG), and finally PEP. Like E. coli, S. cerevisiae needs to transform the non-oxidized part of PPP to optimize the synthesis of E4P, overexpression of tkl1 (encoding transketolase isoenzyme 1), and deletion of Zwf1 (Curran et al., 2013). This resulted in increased carbon flux into PPP at the non-oxidizing node, which further enhanced the flux of glycolysis intermediates F6P and G3P to E4P and X5P by 7 times and increased the 3-dehydroshikimic acid (3-DHS) titer by 7.8 times (Deaner and Alper, 2017). In addition, the Crabtree effect also exists in S. cerevisiae. Pyruvate kinase converts the majority of the precursor PEP into pyruvate. Pyruvate is then converted to ethanol by pyruvate decarboxylase and alcohol dehydrogenase (Dai et al., 2018). Previous studies have reported that only 1% of PEP is consumed via the entry into the AAP in S. cerevisiae (Blank et al., 2005). However, 13C labeled metabolic flux analysis combined with quantitative PCR revealed that expressing the ARO4-K229L (ARO4 allele) could increase the availability of PEP and E4P and reduce the activity of pyruvate kinase or pyruvate decarboxylase, resulting in the accumulation of 17.9 mg/g glucose and 358 mg/L DAHP (Suástegui et al., 2016).
Significant rate-limiting steps in the central metabolic pathway are overcome using various genetic techniques, such as the overexpression and downregulation of crucial enzymes. It is possible to control the carbon flux into L-Trp biosynthesis in the central metabolic pathway (Báez-Viveros et al., 2007) or to increase the availability of the L-Trp biosynthesis precursors PEP and E4P to increase the yield of L-Trp synthesis from glucose (Bongaerts et al., 2001). However, the PPP pathway, where tktA is the primary isozyme and overexpression of tktA dramatically boosted the yield of aromatic compounds, and consequently, the availability of E4P accounts for a portion of the difference between the pathways for DAHP synthesis in E. coli and S. cerevisiae. Differences in tktA transcript levels between strains may impact L-Trp synthesis (Báez-Viveros et al., 2007).
The SK pathway starts with DAHP, and 3-Deoxy-D-Arabino-Heptulosonate 7-Phosphate Synthase (DAHPS) is the first rate-limiting enzyme in this pathway. DAHPS in E. coli comprises three isoenzymes encoded by genes aroG, aroF, and aroH, which catalyze 79%, 20%, and 1% of DAHPS activity, respectively. Activities of these three isoenzymes are inhibited by L-Phe, L-Tyr, and L-Trp (Helmstaedt et al., 2005) (Figure 2). The construction of aroGfbr and aroFfbr mutants in the WF1234 strain lacking aroB, attenuated the feedback inhibition of isoenzymes by L-Phe and L-Tyr. The constructed mutant strain also overexpressed tktA and increased the net carbon flux from CCM to SK twice (Lin et al., 2014; Tyagi et al., 2017). An aroG variant resistant to L-Phe can be constructed by mutating the SerA at position 180. Eliminating the feedback inhibition of L-Phe could ultimately increase L-Trp production by 38.5% (Doroshenko et al., 2010; Chen et al., 2019). By reducing the copy number of L-Trp hydroxylated plasmids and replacing the promoter of the aroHfbr gene encoding DAHPS, the L-Trp conversion to 5-Hydroxytryptophan (5-HTP) in the recombinant strain TRPmut/pSCHTP-LMT was increased by 24.8%, compared to the original strain (Xu et al., 2020). Deregulating the natively regulated wild-type trp operon resulted in a maximum L-Trp yield of 40 g/L in batch fermentation (Chen and Zeng, 2017).
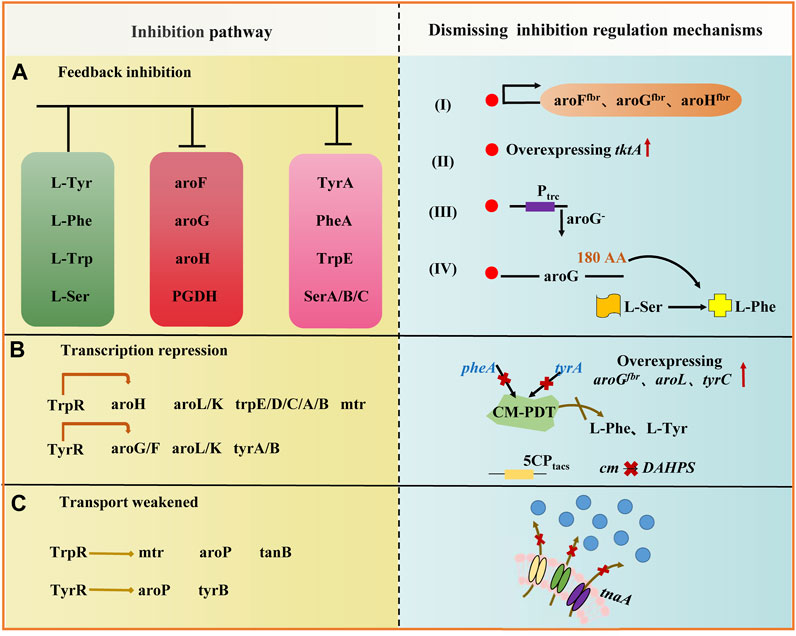
FIGURE 2. Inhibition pathway and dismissing inhibition regulation mechanism of L-Trp biosynthesis pathway in E. coli. Inhibition pathways include feedback inhibition (A), transcriptional inhibition (B), and transport weakened (C). Activities of three isoenzymes (aroG, aroF, and aroH) are inhibited by L-Phe, L-Tyr, and L-Trp. TyrR and TrpR suppress the expression of several crucial genes in the l-tryptophan biosynthesis pathway and l-tryptophan transport system. The right side shows derepression regulation mechanisms. The red solid line upward arrow indicates over-expression, the black solid line downward arrow indicates down-regulation, and × indicates knockout.
There are two isoenzymes of DAHPS in S. cerevisiae encoded by ARO3 and ARO4 genes. However, both isoenzymes face feedback inhibition by L-Phe and L-Tyr (Chrzanowski, 2020). The promoters of the genes involved in amino acid biosynthesis are usually regulated by Gcn4 (Lou et al., 2022). However, the transcriptional effects of Gcn4 on ARO3 and ARO4 genes are different under amino acid starvation. While Gcn4 reduces the expression levels of ARO3, there are no inhibitory effects on the expression of ARO4. The feedback-resistant mutant ARO4-K229L causes the DAHPS to be insensitive to L-Phe and L-Tyr. Furthermore, reduced expression of ARO3 gene in ARO4-K229L mutant can increase the SK flux by 4–5 times (Luttik et al., 2008a). In E. coli, several reactions in the SK pathway are catalyzed by individual enzymes.
On the contrary, in S. cerevisiae, conversion reactions are mainly carried out by the action of a five-function enzyme encoded by ARO1 (Kuplińska and Rząd, 2021). The final step of the SK is catalyzed by ARO2-encoded chorionic synthase. After overexpression of ARO2 in S. cerevisiae, the extraction of aromatic compounds from tyrosine resulted in a 2-folds increase in the yield of coumaric acid (Mao et al., 2017). Thus, overexpressing ARO2 is advantageous to raise CHO levels. L-Tyr and L-Phe will gradually accumulate more due to the CHO pathway’s increased carbon flow, which will rise with the increased carbon flow. On the other hand, DAHPS feedback inhibition is an inevitable consequence of an excessive build-up of L-Trp. Therefore, it is essential to remove the feedback inhibition of DAHPS by L-Phe, L-Tyr, and L-Trp to facilitate the higher productivity of L-Trp in the SK pathway.
A study of the key genes involved in the production of DAHP in E. coli Trp01 indicated that the aroF, aroG, and aroH were considerably in the logarithmic and steady phase, indicating that separate enzymes catalyzed PEP and E4P. The new strain Trp07 produced 3.94 g/L of DAHP in shake flasks when the S180F mutation was added to the aroG gene to eliminate feedback inhibition of DAHP by L-Trp (Tang et al., 2023) and through the introduction of ARO3 with ARO4-K229L, which is insensitive to DAHPS, in S. cerevisiae, overexpression of the ARO4-K229L allele increased the total flux of branched aromatic amino acid metabolism 3 to 4 times in comparison to the control bacterium, CEN. PK (Luttik et al., 2008b; Yang et al., 2021).
In E. coli, a part of the CHO pathway synthesizes L-Trp. The remaining reactions of the pathway are catalyzed by the Chorismate mutase (CM) to generate prebenzic acid (PPA). Furthermore, PPA is subsequently transformed into L-Phe and L-Tyr by prephenate dehydrogenase (encoded by tyrA) and prephenate dehydratase (encoded by pheA). By knocking out the pheA gene involved in the competing pathway encoding the Chorismate mutase-prephenate dehydrogenase (CM-PDT), the carbon flow to L-Tyr and L-Phe is blocked. In contrast, the carbon flow to the L-Trp producing reactions indirectly increases (Patnaik et al., 2008). Anthranilic acid (ANTA) synthase catalyzes the formation of ANTA from CHO, which enters the CHO pathway for L-Trp biosynthesis. ANTA synthase is encoded by the gene trpED, and L-Trp inhibits the enzyme activity of ANTA synthase. Gene transcription in the trpEDCBA operon is inhibited by TrpR. When the co-inhibitor L-Trp is present in large quantities, trpR and tyrR are active. TyrR and TrpR suppress the expression of several crucial genes in the L-tryptophan biosynthesis pathway and L-tryptophan transport system. Thus, these two repressors are typically detected concurrently or separately to improve L-tryptophan production (Jing et al., 2018) (Figure 2). Knocking out trpR and tnaA can remove the inhibitory effects of repressor protein and the degradation of tryptophan. Both trpR knockout strains and double-gene (trpR and tnaA) knockout strains produced a higher quantity of tryptophan (10- and 20-folds, respectively) (Yu et al., 2008). The inactivation of tryptophan attenuators and modification of the tryptophan operon promoter significantly improved L-Trp biosynthesis, and the engineered E. coli strain GPT1002 produced 15.48 g/L of L-Trp in 10 h (Gu et al., 2012). Constitutive plasmids containing tryptophan operons (trpEfbr, trpDfbr) were transformed into JB102 strains lacking trpR and tnaA genes. After 27 h of fermentation, 6.2 g/L of L-Trp was produced by the JB102 strain (Dodge and Gerstner, 2002). After multiple mutagenesis breeding and adding surfactant L61, the L-Trp yield reached 54.5 g/L.
The mechanism of the CHO pathway in S. cerevisiae and E. coli is similar, and the generation of L-Phe and L-Tyr inhibits CHO in both microbes. To obtain the target product L-Trp, ARO7G141S must be overexpressed, increasing the anti-feedback inhibition of DAHPS (Cao et al., 2020). Genes involved in amino acid biosynthesis are widely distributed in yeast genomes, and both Trp2 and Trp3 contain binding sites for Gcn4 (Mittal et al., 2017). Three Gcn4 binding sites have been found in Trp4; one can also bind to the transcriptional regulator Pho2 (also known as Bas2) to participate in phosphate Pi metabolism (Cao et al., 2020). In phosphate Pi metabolism, PRPP is used as a Trp4 substrate. When Pi is limited in the case of amino acid starvation, Trp4 expression is determined by the Pho2, which occupies the binding site. Consequently, Trp4 cannot bind with Gcn4 to fully activate TRP4 expression. Thus, Pho2 prevents the production of excess anthranilate phosphoribosyl transferase (Perveen et al., 2017). ANTH is the first intermediate in the tryptophan biosynthesis pathway, produced by Trp2/Trp3 complexes in an amide group reaction (Blank et al., 2005).
The feedback-resistant DAHP synthase encoded by aroF was produced by eliminating its residue Ile11. For the production of a feedback-resistant ANTA synthase encoded by trpED, the substitution of the Ser40 residue with Phe was performed. The inhibition of the transcriptional regulation of the trp repressor and the degradation process of L-Trp were achieved by suppressing the trpR and tnaA-producing genes, respectively. By eliminating their respective essential genes, pheA and tyrA, two competing pathways that produce L-Phe and L-Tyr, were also inhibited (Zhao et al., 2011a). Contrarily, TyrR and the tryptophan manipulator are absent in S. cerevisiae. As a result, the carbon flux from the chorismate to ANTA can only be boosted by overexpressing Trp2 and Trp3 to increase the production of L-Trp (Kuivanen et al., 2021).
Research has indicated that there is an ongoing rise in intracellular tryptophan synthesis, which may be attributed to the reuptake and accumulation of specific tryptophan molecules by tryptophan isozyme or the retrieval of excreted tryptophan from the surrounding environment (Li et al., 2020b). During the fermentation, intracellular L-Trp is secreted extracellularly. Therefore, the L-Trp transport system must be modified to improve tryptophan production. Both TnaB and Mtr are specific tryptophan osmozymes. While TnaB is a low-affinity transporter encoding the TnaA operon, Mtr is a high-affinity tryptophan isozyme (Figures 1, 2) (Koyanagi et al., 2004). In the E. coli S015 strain, the expression of tnaA in the degradation pathway was blocked, and tryptophan-specific transporters mtr and tnaB were knocked out (van der Stel et al., 2021). Afterward, two feedback resistance genes, aroGS180F, and SerAH344A/N364A were generated by site-directed mutagenesis. Subsequently, aroGS180F and SerAH344A/N364A were linked to a strong ribosomal binding site (rbs, AAGGAG) and controlled by the strong promoter tac. An enhanced operon (PJ23119-rpsL-tac-aroGS180F-SerAH344A/N364A) was formed in the plasmid strp015A, and the yield of L-Trp was increased to 40.3 g/L (Chen and Zeng, 2017). Furthermore, the robust promoter trc was employed to substitute the regulatory region of the aforementioned operons for the purpose of regulating the L-Trp branching pathway, enhancing inhibition and attenuation, and attaining consistent overexpression of L-Trp operons (Du et al., 2019). It was found that the naturally regulated wild-type L-Trp operon in E. coli S019 was released, and the L-Trp yield increased to 2.14 ± 0.03 g/L. In another study, Ptrc-trpE and Ptrc-aroG were inserted into the trpE and tyrR locus. Consequently, feedback inhibition of ANTHS by tyrR was alleviated and 49 g/L L-Trp was accumulated after 36 h of fermentation in a 5 L bioreactor (Du et al., 2019).
AroP in E. coli TRP1 strain is a universal aromatic amino acid osmotic enzyme encoded by the aroP gene. aroP can also transport L-Phe and L-Tyr with high affinity. By knocking out the aroP gene and overexpressing the yddG gene in E. coli TRTH ΔaroP mutant, the L-Trp yield increased by 13.3% compared to the original strain (Liu et al., 2012). Increasing the efflux of L-Trp is an effective strategy for reducing the intracellular concentration of L-Trp and enhancing the glucose consumption rate. Nevertheless, these procedures cannot completely mitigate the feedback inhibitory effect of elevated levels of intracellular L-Trp. Hence, additional investigation is warranted to regulate the feedback-inhibitory impacts of L-Trp to enhance its production (Table 1).
L-Trp is degraded by the serotonin pathway and kynurenine pathway. The serotonin pathway is initiated by tryptophan hydroxylase, and the 5′hydrogen atom on the tryptophan benzene ring is replaced by a hydroxyl group to produce 5-HTP. 5-HTP is further converted into melatonin, the final product of the serotonin pathway (Barik, 2020). On the other hand, the kynurenine pathway is initiated by one of two heme-containing oxidoreductases, indole 2,3-dioxygenase (IDO) or the highly related tryptophan 2,3-dioxygenase, producing kynurenine and niacin as primary metabolites (Munn and Mellor, 2013). IDO serves as the initial enzyme that restricts the pace of the metabolic process known as the kynurenine pathway. IDO was previously hypothesized to function as a modulator of the inflammatory response. The subsequent research demonstrated that immune cells that express IDO1, such as macrophages and dendritic cells, exerted an inhibitory effect on the proliferation of T cells (Hwu et al., 2000; Frumento et al., 2002; Munn et al., 2002). It has also been shown that exogenously added kynurenine binds to transforming growth factors and acts as an immunosuppressive metabolite (Orabona and Grohmann, 2011). L-Trp regulates gluconeogenesis to maintain blood sugar levels and prevent hypoglycemia (Badawy, 2017).
In the industrial setting, microbial fermentation is widely employed to create L-Trp and synthesize several derivatives. Researchers are interested in increasing the industrial yield of L-Trp by examining potential metabolic regulatory networks of L-Trp biosynthesis and altering them. Because of their high output and easier regulation of the L-tryptophan biosynthesis pathway, E. coli and C. glutamicum are frequently utilized as hosts for L-Trp production (Liu et al., 2016). The industrial production of L-Trp in E. coli, C. glutamicum, and S. cerevisiae is summarized (Table 2).
After 48 h of fermentation in shake flasks, the industrial strain of E. coli TS-1 produced 1.710 g/L L-Trp due to the overexpression of icd and gdhA, the addition of prs, the re-expression of mutant serA and thrA to increase the supply of serine precursors, and eventually the overexpression of syhA and pntAB (Li et al., 2020b). In E. coli TRTH0709/pMEL03, overexpression of yddG and suppression of phosphate acetyltransferase-high affinity tryptophan transporter (pta-mtr) led to L-Trp titers of up to 48.68 g/L when batch fermentation was added after 48 h in 30 L fermenter (Wang et al., 2013). It was discovered that PHB could upregulate the transcription of the tryptophan manipulator by producing L-Trp-producing E. coli GPT1002, into which the PHB biosynthesis pathway containing the phaCAB manipulator gene was introduced. Xylose was added as a co-substrate to the medium to increase the supply of precursors for PHB biosynthesis. By replenishing batch culture, 14.4 g/L of L-Trp was produced in GPT2000 (Gu et al., 2013) by overpressing feedback-resistant genes for AroF and TrpED, deleting the tryptophan repressor, and preventing the manufacture of L-Phe and L-Tyr. The finished modified E. coli FB-04/pSV03 can generate 13.3 g/L of L-Trp (Zhao et al., 2011b). After 55 h of batch fermentation with supplementation, the final L-Trp titer was 44.0 g/L due to the phosphate acetyltransferase knockdown in E. coli FB-04 (Liu et al., 2016; van der Stel et al., 2021). By co-expressing the coding genes acnBA and icd under the control of PacnB, the TRP07 collected 49 g/L L-Trp after 36 h in a 5 L bioreactor (Du et al., 2019). The L-Trp concentration was increased up to 40.3 g/L by augmenting the initial inoculum of the S028 strain. The expression of anthranilate-activated TrpC from A. niger in the S092 resulted in a significant increase in L-Trp concentration, elevating it from 19 to 29 g/L within 42 h (Chen et al., 2018a). After fermentation in a 5 L bioreactor for 40 h, the high-yield strain TRP9 achieved an L-Trp yield of 36.08 g/L by improving the expression of the L-Trp operon and optimizing the essential genes serA and prs for the synthesis of serine and PRPP in the chorismate to L-Trp module (Ding et al., 2023).
The gene for 3-phosphoglycerate dehydrogenase (PGD), the first enzyme in the serine pathway, was cloned from wild-type C. glutamicum ATCC 31833 and linked onto pKW99 to generate pKW9901. This increased the amount of carbon flow into serine. The best recombinant strain thus created can produce L-Trp at a titer of 50 g/L after 80 h of growth in jar fermentors without antibiotics (Ikeda et al., 1994). The final L-Trp titer for pIK9960 was 58 g/L, 15% higher than for carriers pKW9901 and pSW9911 (Ikeda and Katsumata, 1999). UV radiation of the ACGEB1 and ACGEB2 C. glutamicum. L-Trp levels before mutation in C. glutamicum ACGEB1 were 19.6 g/mL and 20.48 g/mL in ACGEB2, respectively. Particularly in C.G. ACGEB2b (278.4 g/mL), followed by C.G. ACGEB2c (81.6 g/mL), the L-Trp output is noticeably boosted (Mohamed et al., 2018).
L-tryptophan synthesis in S. cerevisiae for using in industry has been documented less frequently and with lower yields. It was discovered that engineering S. cerevisiae for the de novo production of Halogenated Tryptophan and Tryptamine Derivatives (Gong et al., 2022). Integration of S. cerevisiae ST7574 (CEN.PK113-7D) with SrPyrH, SttH, and LaRebH. The highest titers obtained for halogenated tryptophan and tryptamine were 3.56 ± 0.24 mg/L 7-bromotryptophan and 2.42 ± 0.31 mg/L 7-chlorotryptamine (Milne et al., 2023).
Metabolic engineering strategy has significantly increased the production of L-Trp by introducing new pathways and rewiring native metabolism. However, a number of unfavorable factors significantly restrict microbial growth and metabolic activity during the fermentation process (Mohedano et al., 2022). Therefore, improving the robustness and tolerance of microbial cell factory is essential to fulfill the requirements of industrial applications and gain maximum economic benefits (Gong et al., 2017). Numerous strategies, such as the most used adaptive laboratory evolution, have been developed and employed to study stress tolerance mechanisms and develop robust industrial E. coli and S. cerevisiae (Pereira et al., 2019). Recent academic research have suggested that the metabolism of amino acids may improve the resilience of microbes to stress (Horinouchi et al., 2010; Ohashi and Chaleckis, 2021). Therefore, regulation of amino acids may provide a novel strategy to improve the microbial ability to ferment in challenging fermentaion conditions.
S. cerevisiae is widely used to produce biochemical products, biofuels, high-value natural products, and fermented foods. During the fermentation process, S. cerevisiae often faces various environmental stresses, including extreme temperature, ethanol, oxidation, and replication stress (Deparis et al., 2017; Shen et al., 2022) (Figure 3). In recent years, academic studies have noted a notable focus on investigating the relationship between amino acid metabolism and stress fitness. In the following subsections, the effects of L-Trp on the stress tolerance of yeast have been summarized (Figure 3; Table 3).
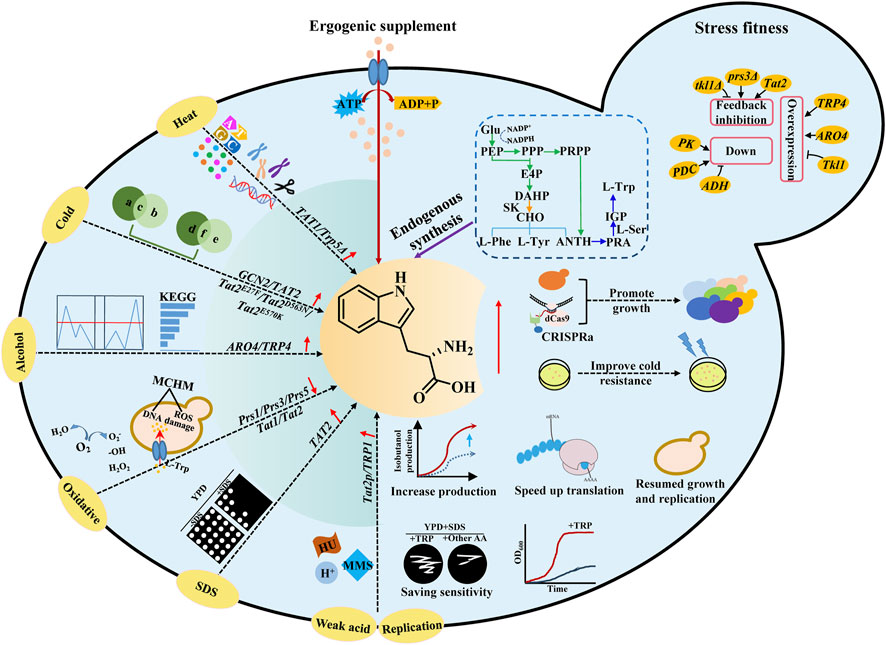
FIGURE 3. Effects of L-Trp regulation on stress fitness of S. cerevisiae. S. cerevisiae often faces various environmental stresses, including extreme temperature, ethanol, oxidation, and replication stress. Stress fitness of S. cerevisiae could be improved by exogenous supplement and endogenous synthesis of L-Trp. TAT1 and TAT2 encode tryptophan amino acid transporters. TRP1, TRP2, TRP4, and TRP5 are key genes for L-Trp biosynthesis. ARO4 encodes DAHP synthase, catalyzes the first step in aromatic amino acid biosynthesis, and is feedback-inhibited by high concentrations of L-Trp. PRS1, PRS3, and PRS5 encode ribose phosphate diphosphokinase subunits. GCN2 encodes serine/threonine-protein kinase.
The suitable temperature for the growth and metabolism of S. cerevisiae is 30°C–32°C. S. cerevisiae uses complex regulatory mechanisms to respond to temperature stress. One such mechanism involves the regulation of tryptophan metabolism (Pan et al., 2019; Shen et al., 2020; Choo et al., 2021). S. cerevisiae commonly encounters high-temperature stress during the process of fermentation. According to reports, the heat-shock response is activated in the trp5Δ strain, which is deficient in tryptophan synthesis due to starvation. Additionally, it has been observed that mutants with impaired L-Trp metabolism exhibit resistance to heat stress during both growth stages (Jarolim et al., 2013). As compared to standard Synthetic Complete (SC) media (composed of 20 mg/L tryptophan and 100 mg/mL leucine), high tryptophan and leucine supplementation during heat shock treatment resulted in >50% reduction in Tribromoethanol-mediated thermotolerance (Luethy et al., 2020).
Yeast cells frequently experience low-temperature stress during the accumulation of volatile compounds. However, low-temperature stress may result in a reduced growth rate and increased energy consumption (Xu et al., 2022). For tryptophan auxotrophic yeast, tryptophan appears to be the primary limiting factor for low-temperature resistance (Lopez-Malo et al., 2014). Eliminating the TRP1 gene, which encodes a phospho-ribosyl anthranilate isomerase involved in tryptophan biosynthesis, increases susceptibility to low temperatures (Ballester-Tomás et al., 2017). However, overexpression of the tryptophan permeases TAT1 and TAT2, also known as SCM2, can enhance the cold tolerance of yeast (Vicent et al., 2015; Kuroda et al., 2019). The tryptophan auxotrophic strain YPH499 could transport tryptophan-containing oligopeptides via its peptide transporter to compensate for the inability to grow at low temperatures (Kitagawa et al., 2013). High levels of Tat2 protein (a high-affinity tryptophan permease) expression resulted in cells having the capacity to grow at low temperatures of 10°C–15°C (Abe and Horikoshi, 2000). In addition, the N- and C-terminal mutagenesis of HPG2 (a TAT2 allele) improved Tat2 stability during low-temperature incubation, leading to cell growth under these demanding circumstances (Nagayama et al., 2004).
The most famous application of S. cerevisiae is to produce bioethanol using biomass resources such as cellulose and starch. However, high levels of ethanol affect the growth and productivity of yeast cells. Ethanol concentrations above 9% (v/v) have been reported to influence the development of S. cerevisiae cells (Li et al., 2019). Excessive concentrations of alcohols mainly affect cell membrane integrity, vacuole structure, and function (Stanley et al., 2010; IngoLFSSON Helgi and Andersen Olaf., 2011). Ethanol stress tolerance-related gene in S. cerevisiae was studied based on DNA microarray, and yeast cells showed tolerance to 5% ethanol after overexpression of TRP1, TRP2, and TRP3 (Hirasawa et al., 2007). Upregulation of differentially expressed genes (DEGs) and differentially expressed proteins (DEPs) associated with aromatic amino acids, such as ARO2, ARO4, ARO9, and TRP4, is conducive to the upregulation of TRP4 genes, especially under ethanol stress. Consequently, upregulating DEGs and DEPs facilitates the catalysis of tryptophan synthesis and serves as a protective mechanism against ethanol-induced stress in S. cerevisiae cells. This protective mechanism is achieved by creating hydrophobic areas, which ultimately contribute to the survival of the cells, as mentioned above (Li et al., 2019).
Isobutanol is also considered a potential biofuel, and S. cerevisiae is one of the most suitable hosts to produce isobutanol (Wess et al., 2019). However, branched-chain alcohols may cause severe cytotoxic effects by disrupting the cell membranes, thus becoming a prominent barrier during isobutanol production (Kuroda et al., 2019). Kuroda et al. (2019) produced S. cerevisiae strains by eliminating the TRP genes, which encode tryptophan production enzymes. Isobutanol sensitivity is raised in the altered strains. Moreover, the regulation of tryptophan metabolism improved the isobutanol tolerance, thus increasing the yield of isobutanol in engineered S. cerevisiae strains (Kuroda et al., 2019). Liu et al. (2021) restored the isobutanol tolerance in the damaged strains by adding exogenous tryptophan to the growth medium, demonstrating the significance of tryptophan in improving the isobutanol tolerance in yeast (Liu et al., 2021).
Moreover, transcriptome studies demonstrated that in conditions similar to nitrogen deficiency, the genes involved in tryptophan synthesis and transportation were increased (Liu et al., 2021). In summary, tryptophan and associated synthetic regulatory networks are critical in yeast’s ability to withstand alcohol stress.
Reactive oxygen species (ROS), the partially reduced forms of molecular oxygen, are the primary cause of oxidative stress in all aerobically growing organisms. High levels of oxidative stress may damage cellular constituents, such as DNA, lipid, and protein lesions (Alves et al., 2023). To cope with ROS, yeast cells quickly adjust and synthesize self-healing substanceslike tryptophan. Other than malnutrition, L-Trp has been suggested to be connected to stress tolerance (Ayers et al., 2020). The expressions of genes involved in amino acid synthesis and transportation, such as Prs1, Prs3, Tat1, and Tat2, must be interrupted to weaken the metabolism of tryptophan and other amino acids. The proliferation of yeast cells experiences a state of stagnation due to inadequate nutrient availability. However, this growth inhibition can be alleviated by introducing supplementary tryptophan to the growth media. This supplementation facilitates reinstating a balanced distribution of nitrogen sources within the medium (Hernando et al., 1999).
When exposed to different external pressures, yeast cells rely on cell walls and membranes as the primary protective barrier against these shocks. The activation of the multifactorial stress response in S. cerevisiae occurs when any disruption hinders the normal functioning of the cell wall or cell membrane (Levin, 2005; Levin, 2011). Sodium dodecyl sulfate (SDS) is a detergent that destroys the cell membranes (Cócera et al., 1999; Cócera et al., 2004), activates the cell wall integrity (CWI) signaling, and restricts the growth of S. cerevisiae cells (Igual et al., 1996; Bickle et al., 1998). Cell wall and plasma membrane integrity stress in yeast involving the MCK1 effector is sensitive to SDS. This stress depends on tryptophan biosynthesis and levels of L-Trp and L-Tyr in media and cells (Schroeder and Ikui, 2019). In S. cerevisiae, ΔMCK1 was used in the presence of SDS to screen inhibitory genes. The results showed that the overexpression of TAT2 tryptophan isoenzyme resulted in higher L-Trp synthesis in cells, thereby restoring the growth of SDS-treated ΔMCK1 cells.
S. cerevisiae can grow with weak organic acid preservatives, such as sorbate and benzoate, which also cause food and beverage deterioration (Piper et al., 2001). In the context of industrial utilization of S. cerevisiae, exposure to weak acid stress predominantly has deleterious consequences on cellular function by compromising the integrity of the cellular membrane. Additionally, this stress condition hinders yeast cells’ absorption of aromatic amino acids. The issue of weak acid hypersensitivity in the tryptophan biosynthesis pathway is resolved by introducing elevated concentrations of L-Trp into the growth medium. The overexpression of Tat2p, a high-affinity tryptophan permease, is employed to mitigate the sensitivity associated with sorbate (Bauer et al., 2003).
L-Trp is a precursor for the production of serotonin as well as NAD+. In S. cerevisiae, Tat2 is sub-morphic at colder temperatures. After disrupting tryptophan biosynthetic genes, growth at room temperature induces tryptophan depletion (Abe and Horikoshi, 2000). To determine whether the L-Trp biosynthetic gene is involved in the resistance of yeast cells to replication stress, the sensitivity of trp1-1 strain to DNA damaging agents, such as methyl mesylate (MMS) and hydroxyurea (HU), was detected (Godin et al., 2016). In contrast, the sensitivity of trp1-1 strain to replication stress was inhibited by overexpressing TRP1.
Moreover, the yeast cells grew more slowly in the tryptophan-deficient medium at 23°C than at 28°C and 30°C. Therefore, the DNA damage sensitivity of trp1-1 cells was specific for tryptophan biosynthesis. This proved that both low temperature and tryptophan consumption led to the sensitivity of cells to replication stress (Polleys and Bertuch, 2015). The study further suggested that the DNA damage sensitivity of L-Trp depletion might be due to impaired protein synthesis. However, DNA damage repair and L-Trp consumption are still being explored while applying L-Trp as a nutritional marker.
With the continuous advancements in metabolic engineering and synthetic biology, green biosynthesis has gradually replaced L-Trp chemical synthesis. With the improvements in the anabolism of L-Trp, the systematic metabolic network of L-Trp in E. coli is more clearly understood. However, the mechanism of L-Trp biosynthesis and metabolic regulation in S. cerevisiae remains unclear and needs further exploration. In the CHO pathway, the knock out of branched acid isomerase inhibits the production of L-Phe and L-Tyr, and the efflux mechanism of L-Trp is critical for Trp production. Secondly, the associated citric acid pathway and TCA cycle play essential roles in the supply of precursors.
Further investigation is required to examine the accumulation of L-Trp in meticulously controlled settings, such as compromised L-Trp metabolic pathways. Furthermore, the proliferation of S. cerevisiae cells is influenced by various environmental stressors. Additionally, L-Trp is crucial for enhancing stress resistance in S. cerevisiae. On one hand, it can promote L-Trp synthesis or add exogenous L-Trp to play a “detoxifying” role. On the other hand, it can encourage yeast cell growth through L-Trp derivatives such as 5-HTP.
The L-Trp biosynthetic system still needs further expansion and improvement, especially in terms of metabolic regulation and engineering of S. cerevisiae to regulate some key enzymes and genes and inhibit the decomposition of L-Trp. The involvement of L-Trp in S. cerevisiae’s response to other stressful environments also needs to be explored further.
Regulating tryptophan metabolism has been proven to be an efficient method to enhance the stress fitness of S. cerevisiae, especially the defective mutants of tryptophan synthesis. Thus, the regulation of tryptophan metabolism should be explored to develop novel strategies for producing stress-tolerant yeasts. Notably, the dosage of tryptophan in the media seems to be a crucial factor affecting the stress tolerance of yeasts. In recent research, exogenous supplementation of tryptophan has been commonly used to improve yeast stress tolerance. Furthermore, the development of dCas9-assisted enzyme perturbation provides a powerful tool for the graded expression of pathway enzymes. Compared to exogenous supplementation of tryptophan, regulating endogenous genes to synthesize appropriate quantity of tryptophan would be more cost-effective and stable. Thus, in further studies, the graded regulation of the tryptophan metabolism pathway needs to be explored to develop robust industrial cell factories. Previous results have signified that the external supply of critical amino acids may decrease cellular stress and improve process productivity in E. coli (Kumar et al., 2020). However, there are rare investigations to investigate the relationship between L-Trp metabolism and stress fitness in E. coli. In the future, the mechanism of tryptophan-mediated stress fitness should be clarified, which may provide novel insights to enhance the stress tolerance of yeast and E. coli.
XR: Writing–original draft. YW: Writing–original draft. HZ: Writing–original draft. JS: Writing–original draft. FZ: Writing–review and editing. ZW: Writing–review and editing, Funding acquisition. LL: Writing–review and editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the National Natural Science Foundation of China (32200067), Hebei Natural Science Foundation (C2020204013), the Key Research and Development Program Projects of Hebei Province (22322905D), and the Specialized Research Fund for the Doctoral Program of Hebei Agricultural University (YJ201950).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abe, F., and Horikoshi, K. (2000). Tryptophan permease gene TAT2 confers high-pressure growth in Saccharomyces cerevisiae. Mol. Cell Biol. 20, 8093–8102. doi:10.1128/.20.21.8093-8102.2000
Alves, L. F., Bortolucci, J., Reginato, V., Guazzaroni, M. E., and Mussatto, S. I. (2023). Improving Saccharomyces cerevisiae acid and oxidative stress resistance using a prokaryotic gene identified by functional metagenomics. Heliyon 9, e14838. doi:10.1016/j.heliyon.2023.e14838
Ayers, M. C., Sherman, Z. N., and Gallagher, J. E. G. (2020). Oxidative stress responses and nutrient starvation in MCHM treated Saccharomyces cerevisiae. G3 (Bethesda) 10, 4665–4678. doi:10.1534/g3.120.401661
Badawy, A. A. (2017). Kynurenine pathway of tryptophan metabolism: regulatory and functional aspects. Int. J. Tryptophan Res. 10, 117864691769193. doi:10.1177/1178646917691938
BáEZ-Viveros, J. L., Flores, N., JuáREZ, K., Castillo-España, P., Bolivar, F., and Gosset, G. (2007). Metabolic transcription analysis of engineered Escherichia coli strains that overproduce L-phenylalanine. Microb. Cell Fact. 6, 30. doi:10.1186/1475-2859-6-30
Ballester-TomáS, L., Prieto, J. A., Alepuz, P., González, A., Garre, E., and Randez-Gil, F. (2017). Inappropriate translation inhibition and P-body formation cause cold-sensitivity in tryptophan-auxotroph yeast mutants. Biochim. Biophys. Acta Mol. Cell Res. 1864, 314–323. doi:10.1016/j.bbamcr.2016.11.012
Barik, S. (2020). The uniqueness of tryptophan in biology: properties, metabolism, interactions and localization in proteins. Int. J. Mol. Sci. 21, 8776. doi:10.3390/ijms21228776
Bauer, B. E., Rossington, D., Mollapour, M., Mamnun, Y., Kuchler, K., and Piper, P. W. (2003). Weak organic acid stress inhibits aromatic amino acid uptake by yeast, causing a strong influence of amino acid auxotrophies on the phenotypes of membrane transporter mutants. Eur. J. Biochem. 270, 3189–3195. doi:10.1046/j.1432-1033.2003.03701.x
Bell, J. K., Pease, P. J., Bell, J. E., Grant, G. A., and Banaszak, L. J. (2002). De-regulation of D-3-phosphoglycerate dehydrogenase by domain removal. Eur. J. Biochem. 269, 4176–4184. doi:10.1046/j.1432-1033.2002.03075.x
Berry, A. (1996). Improving production of aromatic compounds in Escherichia coli by metabolic engineering. Trends Biotechnol. 14, 250–256. doi:10.1016/0167-7799(96)10033-0
Bickle, M., Delley, P. A., Schmidt, A., Hall, M. N., et al. (1998). Cell wall integrity modulates RHO1 activity via the exchange factor ROM2. Embo J. 17, 2235–2245. doi:10.1093/emboj/17.8.2235
Blank, L. M., Kuepfer, L., and Sauer, U. (2005). Large-scale 13C-flux analysis reveals mechanistic principles of metabolic network robustness to null mutations in yeast. Genome Biol. 6, R49. doi:10.1186/gb-2005-6-6-r49
Bongaerts, J., KräMER, M., MüLLER, U., Raeven, L., and Wubbolts, M. (2001). Metabolic engineering for microbial production of aromatic amino acids and derived compounds. Metab. Eng. 3, 289–300. doi:10.1006/mben.2001.0196
Cao, M., Gao, M., SuáSTEGUI, M., Mei, Y., and Shao, Z. (2020). Building microbial factories for the production of aromatic amino acid pathway derivatives: from commodity chemicals to plant-sourced natural products. Metab. Eng. 58, 94–132. doi:10.1016/j.ymben.2019.08.008
Carvalho, S. M., Marques, J., RomãO, C. C., and Saraiva, L. M. (2019). Metabolomics of Escherichia coli treated with the antimicrobial carbon monoxide-releasing molecule CORM-3 reveals tricarboxylic acid cycle as major target. Antimicrob. Agents Chemother. 63, 63. doi:10.1128/aac.00643-19
Chen, L., Chen, M., Ma, C., and Zeng, A. P. (2018a). Discovery of feed-forward regulation in L-tryptophan biosynthesis and its use in metabolic engineering of E. coli for efficient tryptophan bioproduction. Metab. Eng. 47, 434–444. doi:10.1016/j.ymben.2018.05.001
Chen, L., and Zeng, A. P. (2017). Rational design and metabolic analysis of Escherichia coli for effective production of L-tryptophan at high concentration. Appl. Microbiol. Biotechnol. 101, 559–568. doi:10.1007/s00253-016-7772-5
Chen, M., Chen, L., and Zeng, A. P. (2019). CRISPR/Cas9-facilitated engineering with growth-coupled and sensor-guided in vivo screening of enzyme variants for a more efficient chorismate pathway in E. coli. Metab. Eng. Commun. 9, e00094. doi:10.1016/j.mec.2019.e00094
Chen, Y., Liu, Y., Ding, D., Cong, L., and Zhang, D. (2018b). Rational design and analysis of an Escherichia coli strain for high-efficiency tryptophan production. J. Ind. Microbiol. Biotechnol. 45, 357–367. doi:10.1007/s10295-018-2020-x
Choo, J. H., Lee, S. B., Moon, H. Y., Lee, K. H., Yoo, S. J., Kim, K. P., et al. (2021). Molecular characterization of Hsf1 as a master regulator of heat shock response in the thermotolerant methylotrophic yeast Ogataea parapolymorpha. J. Microbiol. 59, 151–163. doi:10.1007/s12275-021-0646-2
Chrzanowski, G. (2020). Saccharomyces cerevisiae-an interesting producer of bioactive plant polyphenolic metabolites. Int. J. Mol. Sci. 21, 7343. doi:10.3390/ijms21197343
CóCERA, M., Lopez, O., Coderch, L., Parra, J., and de la Maza, A. (1999). Influence of the level of ceramides on the permeability of stratum corneum lipid liposomes caused by a C12-betaine/sodium dodecyl sulfate mixture. Int. J. Pharm. 183, 165–173. doi:10.1016/s0378-5173(99)00084-8
CóCERA, M., LóPEZ, O., Pons, R., Amenitsch, H., and de la Maza, A. (2004). Effect of the electrostatic charge on the mechanism inducing liposome solubilization: a kinetic study by synchrotron radiation SAXS. Langmuir 20, 3074–3079. doi:10.1021/la035972+
Curran, K. A., Leavitt, J. M., Karim, A. S., and Alper, H. S. (2013). Metabolic engineering of muconic acid production in Saccharomyces cerevisiae. Metab. Eng. 15, 55–66. doi:10.1016/j.ymben.2012.10.003
Dai, Z., Huang, M., Chen, Y., Siewers, V., and Nielsen, J. (2018). Global rewiring of cellular metabolism renders Saccharomyces cerevisiae Crabtree negative. Nat. Commun. 9, 3059. doi:10.1038/s41467-018-05409-9
Deaner, M., and Alper, H. S. (2017). Systematic testing of enzyme perturbation sensitivities via graded dCas9 modulation in Saccharomyces cerevisiae. Metab. Eng. 40, 14–22. doi:10.1016/j.ymben.2017.01.012
Deparis, Q., Claes, A., Foulquié-Moreno, M. R., and Thevelein, J. M. (2017). Engineering tolerance to industrially relevant stress factors in yeast cell factories. FEMS Yeast Res, 17. doi:10.1093/femsyr/fox036
Ding, S., Chen, X., Gao, C., Song, W., Wu, J., Wei, W., et al. (2023). Modular engineering of Escherichia coli for high-level production of l-tryptophan. Sheng Wu Gong Cheng Xue Bao 39, 2359–2374. doi:10.13345/j.cjb.221005
Dodge, T. C., and Gerstner, J. M. (2002). Optimization of the glucose feed rate profile for the production of tryptophan from recombinant E coli. J. Chem. Technol. Biotechnol. 77, 1238–1245. doi:10.1002/jctb.698
Doroshenko, V. G., Tsyrenzhapova, I. S., Krylov, A. A., Kiseleva, E. M., Ermishev, V. Y., Kazakova, S. M., et al. (2010). Pho regulon promoter-mediated transcription of the key pathway gene aroGFbr improves the performance of an L-phenylalanine-producing Escherichia coli strain. Appl. Microbiol. Biotechnol. 88, 1287–1295. doi:10.1007/s00253-010-2794-x
Du, L., Zhang, Z., Xu, Q., and Chen, N. (2019). Central metabolic pathway modification to improve L-tryptophan production in Escherichia coli. Bioengineered 10, 59–70. doi:10.1080/21655979.2019.1592417
Escalante, A., Salinas Cervantes, A., Gosset, G., and Bolívar, F. (2012). Current knowledge of the Escherichia coli phosphoenolpyruvate-carbohydrate phosphotransferase system: peculiarities of regulation and impact on growth and product formation. Appl. Microbiol. Biotechnol. 94, 1483–1494. doi:10.1007/s00253-012-4101-5
Frumento, G., Rotondo, R., Tonetti, M., Damonte, G., Benatti, U., and Ferrara, G. B. (2002). Tryptophan-derived catabolites are responsible for inhibition of T and natural killer cell proliferation induced by indoleamine 2,3-dioxygenase. J. Exp. Med. 196, 459–468. doi:10.1084/jem.20020121
Godin, S. K., Lee, A. G., Baird, J. M., Herken, B. W., and Bernstein, K. A. (2016). Tryptophan biosynthesis is important for resistance to replicative stress in Saccharomyces cerevisiae. Yeast 33, 183–189. doi:10.1002/yea.3150
Gong, X., Luo, H., Hong, L., Wu, J., Wu, H., Song, C., et al. (2022). Effects of tryptophan and phenylalanine on tryptophol production in Saccharomyces cerevisiae revealed by transcriptomic and metabolomic analyses. J. Microbiol. 60, 832–842. doi:10.1007/s12275-022-2059-2
Gong, Z., Nielsen, J., and Zhou, Y. J. (2017). Engineering robustness of microbial cell factories. Biotechnol J, 12. doi:10.1002/biot.201700014
Gosset, G. (2005). Improvement of Escherichia coli production strains by modification of the phosphoenolpyruvate:sugar phosphotransferase system. Microb. Cell Fact. 4, 14. doi:10.1186/1475-2859-4-14
Gosset, G., Yong-Xiao, J., and Berry, A. (1996). A direct comparison of approaches for increasing carbon flow to aromatic biosynthesis in Escherichia coli. J. Ind. Microbiol. 17, 47–52. doi:10.1007/bf01570148
Gottardi, M., Reifenrath, M., Boles, E., and Tripp, J. (2017). Pathway engineering for the production of heterologous aromatic chemicals and their derivatives in Saccharomyces cerevisiae: bioconversion from glucose. FEMS Yeast Res. 17, 17. doi:10.1093/femsyr/fox035
Grant, G. A. (2018). D-3-Phosphoglycerate dehydrogenase. Front. Mol. Biosci. 5, 110. doi:10.3389/fmolb.2018.00110
Guo, L., Ding, S., Liu, Y., Gao, C., Hu, G., Song, W., et al. (2022). Enhancing tryptophan production by balancing precursors in Escherichia coli. Biotechnol. Bioeng. 119, 983–993. doi:10.1002/bit.28019
Gu, P., Kang, J., Yang, F., Wang, Q., Liang, Q., and Qi, Q. (2013). The improved L-tryptophan production in recombinant Escherichia coli by expressing the polyhydroxybutyrate synthesis pathway. Appl. Microbiol. Biotechnol. 97, 4121–4127. doi:10.1007/s00253-012-4665-0
Gu, P., Yang, F., Kang, J., Wang, Q., and Qi, Q. (2012). One-step of tryptophan attenuator inactivation and promoter swapping to improve the production of L-tryptophan in Escherichia coli. Microb. Cell Fact. 11, 30. doi:10.1186/1475-2859-11-30
Helmstaedt, K., Strittmatter, A., Lipscomb, W. N., and Braus, G. H. (2005). Evolution of 3-deoxy-D-arabino-heptulosonate-7-phosphate synthase-encoding genes in the yeast Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U. S. A. 102, 9784–9789. doi:10.1073/pnas.0504238102
Hernando, Y., Carter, A. T., Parr, A., Hove-Jensen, B., and Schweizer, M. (1999). Genetic analysis and enzyme activity suggest the existence of more than one minimal functional unit capable of synthesizing phosphoribosyl pyrophosphate in Saccharomyces cerevisiae. J. Biol. Chem. 274, 12480–12487. doi:10.1074/jbc.274.18.12480
Hirasawa, T., Yoshikawa, K., Nakakura, Y., Nagahisa, K., Furusawa, C., Katakura, Y., et al. (2007). Identification of target genes conferring ethanol stress tolerance to Saccharomyces cerevisiae based on DNA microarray data analysis. J. Biotechnol. 131, 34–44. doi:10.1016/j.jbiotec.2007.05.010
Horinouchi, T., Tamaoka, K., Furusawa, C., Ono, N., Suzuki, S., Hirasawa, T., et al. (2010). Transcriptome analysis of parallel-evolved Escherichia coli strains under ethanol stress. BMC Genomics 11, 579. doi:10.1186/1471-2164-11-579
Hwu, P., Du, M. X., Lapointe, R., Do, M., Taylor, M. W., and Young, H. A. (2000). Indoleamine 2,3-dioxygenase production by human dendritic cells results in the inhibition of T cell proliferation. J. Immunol. 164, 3596–3599. doi:10.4049/jimmunol.164.7.3596
Igual, J. C., Johnson, A. L., and Johnston, L. H. (1996). Coordinated regulation of gene expression by the cell cycle transcription factor Swi4 and the protein kinase C MAP kinase pathway for yeast cell integrity. Embo J. 15, 5001–5013. doi:10.1002/j.1460-2075.1996.tb00880.x
Ikeda, M. (2006). Towards bacterial strains overproducing L-tryptophan and other aromatics by metabolic engineering. Appl. Microbiol. Biotechnol. 69, 615–626. doi:10.1007/s00253-005-0252-y
Ikeda, M., and Katsumata, R. (1999). Hyperproduction of tryptophan by Corynebacterium glutamicum with the modified pentose phosphate pathway. Appl. Environ. Microbiol. 65, 2497–2502. doi:10.1128/aem.65.6.2497-2502.1999
Ikeda, M., Nakanishi, K., Kino, K., and Katsumata, R. (1994). Fermentative production of tryptophan by a stable recombinant strain of Corynebacterium glutamicum with a modified serine-biosynthetic pathway. Biosci. Biotechnol. Biochem. 58, 674–678. doi:10.1271/bbb.58.674
IngoLFSSON Helgi, I., and Andersen Olaf, S. (2011). Alcohol's effects on lipid bilayer properties. Biophysical J. 101, 847–855. doi:10.1016/j.bpj.2011.07.013
Jarolim, S., Ayer, A., Pillay, B., Gee, A. C., Phrakaysone, A., Perrone, G. G., et al. (2013). Saccharomyces cerevisiae genes involved in survival of heat shock. G3 (Bethesda) 3, 2321–2333. doi:10.1534/g3.113.007971
Jing, K., Tang, Y., Yao, C., del Rio-Chanona, E. A., Ling, X., and Zhang, D. (2018). Overproduction of L-tryptophan via simultaneous feed of glucose and anthranilic acid from recombinant Escherichia coli W3110: kinetic modeling and process scale-up. Biotechnol. Bioeng. 115, 371–381. doi:10.1002/bit.26398
Kavšček, M., Stražar, M., Curk, T., Natter, K., and Petrovič, U. (2015). Yeast as a cell factory: current state and perspectives. Microb. Cell Fact. 14, 94. doi:10.1186/s12934-015-0281-x
Kitagawa, S., Sugiyama, M., Motoyama, T., and Abe, F. (2013). Soy peptides enhance yeast cell growth at low temperatures. Biotechnol. Lett. 35, 375–382. doi:10.1007/s10529-012-1088-z
Koyanagi, T., Katayama, T., Suzuki, H., and Kumagai, H. (2004). Identification of the LIV-I/LS system as the third phenylalanine transporter in Escherichia coli K-12. J. Bacteriol. 186, 343–350. doi:10.1128/jb.186.2.343-350.2004
Kuivanen, J., Kannisto, M., Mojzita, D., Rischer, H., Toivari, M., and Jäntti, J. (2021). Engineering of Saccharomyces cerevisiae for anthranilate and methyl anthranilate production. Microb. Cell Fact. 20, 34. doi:10.1186/s12934-021-01532-3
Kumar, J., Chauhan, A. S., Shah, R. L., Gupta, J. A., and Rathore, A. S. (2020). Amino acid supplementation for enhancing recombinant protein production in E. coli. Biotechnol. Bioeng. 117, 2420–2433. doi:10.1002/bit.27371
Kuplińska, A., and Rząd, K. (2021). Molecular targets for antifungals in amino acid and protein biosynthetic pathways. Amino Acids 53, 961–991. doi:10.1007/s00726-021-03007-6
Kuroda, K., Hammer, S. K., Watanabe, Y., Montaño López, J., Fink, G. R., Stephanopoulos, G., et al. (2019). Critical roles of the pentose phosphate pathway and GLN3 in isobutanol-specific tolerance in yeast. Cell Syst. 9, 534–547.e5. doi:10.1016/j.cels.2019.10.006
Levin, D. E. (2005). Cell wall integrity signaling in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 69, 262–291. doi:10.1128/mmbr.69.2.262-291.2005
Levin, D. E. (2011). Regulation of cell wall biogenesis in Saccharomyces cerevisiae: the cell wall integrity signaling pathway. Genetics 189, 1145–1175. doi:10.1534/genetics.111.128264
Lin, S., Liang, R., Meng, X., OuYang, H., Yan, H., Wang, Y., et al. (2014). Construction and expression of mutagenesis strain of aroG gene from Escherichia coli K-12. Int. J. Biol. Macromol. 68, 173–177. doi:10.1016/j.ijbiomac.2014.04.034
Li, R., Miao, Y., Yuan, S., Li, Y., Wu, Z., and Weng, P. (2019). Integrated transcriptomic and proteomic analysis of the ethanol stress response in Saccharomyces cerevisiae Sc131. J. Proteomics 203, 103377. doi:10.1016/j.jprot.2019.103377
Liu, H. L., Wang, C. H., Chiang, E. I., Huang, C. C., and Li, W. H. (2021). Tryptophan plays an important role in yeast's tolerance to isobutanol. Biotechnol. Biofuels 14, 200. doi:10.1186/s13068-021-02048-z
Liu, L., Duan, X., and Wu, J. (2016). L-tryptophan production in Escherichia coli improved by weakening the pta-AckA pathway. PLoS One 11, e0158200. doi:10.1371/journal.pone.0158200
Liu, Q., Cheng, Y., Xu, Q., et al. (2012). Effects of aroP gene disruption on L-tryptophan fermentation. Frontiers of Chemical Science & Engineering. doi:10.1007/s11705-012-1275-4
Liu, S., Xu, J. Z., and Zhang, W. G. (2022). Advances and prospects in metabolic engineering of Escherichia coli for L-tryptophan production. World J. Microbiol. Biotechnol. 38, 22. doi:10.1007/s11274-021-03212-1
Li, Z., Ding, D., Wang, H., Liu, L., Fang, H., Chen, T., et al. (2020a). Engineering Escherichia coli to improve tryptophan production via genetic manipulation of precursor and cofactor pathways. Synth. Syst. Biotechnol. 5, 200–205. doi:10.1016/j.synbio.2020.06.009
Li, Z., Ding, D., Wang, H., Liu, L., Fang, H., Chen, T., et al. (2020b). Engineering Escherichia coli to improve tryptophan production via genetic manipulation of precursor and cofactor pathways. Synthetic Syst. Biotechnol. 5, 200–205. doi:10.1016/j.synbio.2020.06.009
Lopez-Malo, M., Garcia-Rios, E., Chiva, R., Guillamon, J. M., and Martí-Raga, M. (2014). Effect of deletion and overexpression of tryptophan metabolism genes on growth and fermentation capacity at low temperature in wine yeast. Biotechnol. Prog. 30, 776–783. doi:10.1002/btpr.1915
Lou, H., Yang, Y., Zheng, S., Ma, Z., Chen, W., Yu, C., et al. (2022). Identification of key genes contributing to amino acid biosynthesis in Torreya grandis using transcriptome and metabolome analysis. Food Chem. 379, 132078. doi:10.1016/j.foodchem.2022.132078
Luethy, A., Kindler, C. H., and Cotten, J. F. (2020). Anesthetic pretreatment confers thermotolerance on Saccharomyces cerevisiae yeast. Biochem. Biophys. Res. Commun. 522, 479–484. doi:10.1016/j.bbrc.2019.11.083
Lu, J., Tang, J., Liu, Y., Zhu, X., Zhang, T., and Zhang, X. (2012). Combinatorial modulation of galP and glk gene expression for improved alternative glucose utilization. Appl. Microbiol. Biotechnol. 93, 2455–2462. doi:10.1007/s00253-011-3752-y
Luttik, M. A., Vuralhan, Z., Suir, E., Braus, G., Pronk, J., and Daran, J. (2008a). Alleviation of feedback inhibition in Saccharomyces cerevisiae aromatic amino acid biosynthesis: quantification of metabolic impact. Metab. Eng. 10, 141–153. doi:10.1016/j.ymben.2008.02.002
Luttik, M. A. H., Vuralhan, Z., Suir, E., Braus, G., Pronk, J., and Daran, J. (2008b). Alleviation of feedback inhibition in Saccharomyces cerevisiae aromatic amino acid biosynthesis: quantification of metabolic impact. Metab. Eng. 10, 141–153. doi:10.1016/j.ymben.2008.02.002
Mao, J., Liu, Q., Song, X., Wang, H., Feng, H., Xu, H., et al. (2017). Combinatorial analysis of enzymatic bottlenecks of L-tyrosine pathway by p-coumaric acid production in Saccharomyces cerevisiae. Biotechnol. Lett. 39, 977–982. doi:10.1007/s10529-017-2322-5
Meza, E., Becker, J., Bolivar, F., Gosset, G., and Wittmann, C. (2012). Consequences of phosphoenolpyruvate:sugar phosphotranferase system and pyruvate kinase isozymes inactivation in central carbon metabolism flux distribution in Escherichia coli. Microb. Cell Fact. 11, 127. doi:10.1186/1475-2859-11-127
Milne, N., SáEZ-SáEZ, J., Nielsen, A. M., Dyekjær, J. D., Rago, D., Kristensen, M., et al. (2023). Engineering Saccharomyces cerevisiae for the de novo production of halogenated tryptophan and tryptamine derivatives. ChemistryOpen 12, e202200266. doi:10.1002/open.202200266
Mittal, N., Guimaraes, J. C., Gross, T., Schmidt, A., Vina-Vilaseca, A., Nedialkova, D. D., et al. (2017). The Gcn4 transcription factor reduces protein synthesis capacity and extends yeast lifespan. Nat. Commun. 8, 457. doi:10.1038/s41467-017-00539-y
Modoux, M., Rolhion, N., Mani, S., and Sokol, H. (2021). Tryptophan metabolism as a pharmacological target. Trends Pharmacol. Sci. 42, 60–73. doi:10.1016/j.tips.2020.11.006
Mohamed, H. B., Nady, G. H., Ali, A., Abdel-Razik, A. B., and Ibrahim, S. A. (2018). Production of L-tryptophan by mutants of corynebacterium glutamicum [89]. Arab Univ. J. Agric. Sci. 26, 1187–1201. doi:10.21608/ajs.2018.28383
Mohedano, M. T., Konzock, O., and Chen, Y. (2022). Strategies to increase tolerance and robustness of industrial microorganisms. Synth. Syst. Biotechnol. 7, 533–540. doi:10.1016/j.synbio.2021.12.009
Munn, D. H., and Mellor, A. L. (2013). Indoleamine 2,3 dioxygenase and metabolic control of immune responses. Trends Immunol. 34, 137–143. doi:10.1016/j.it.2012.10.001
Munn, D. H., Sharma, M. D., Lee, J. R., Jhaver, K. G., Johnson, T. S., Keskin, D. B., et al. (2002). Potential regulatory function of human dendritic cells expressing indoleamine 2,3-dioxygenase. Science 297, 1867–1870. doi:10.1126/science.1073514
Nagayama, A., Kato, C., and Abe, F. (2004). The N- and C-terminal mutations in tryptophan permease Tat2 confer cell growth in Saccharomyces cerevisiae under high-pressure and low-temperature conditions. Extremophiles 8, 143–149. doi:10.1007/s00792-003-0373-0
Niu, H., Li, R., Liang, Q., Qi, Q., Li, Q., and Gu, P. (2019). Metabolic engineering for improving L-tryptophan production in Escherichia coli. J. Ind. Microbiol. Biotechnol. 46, 55–65. doi:10.1007/s10295-018-2106-5
Noda, S., Shirai, T., Mori, Y., Oyama, S., and Kondo, A. (2017). Engineering a synthetic pathway for maleate in Escherichia coli. Nat. Commun. 8, 1153. doi:10.1038/s41467-017-01233-9
Noda, S., Shirai, T., Oyama, S., and Kondo, A. (2016). Metabolic design of a platform Escherichia coli strain producing various chorismate derivatives. Metab. Eng. 33, 119–129. doi:10.1016/j.ymben.2015.11.007
Ohashi, K., and Chaleckis, R. (2021). High levels of Tryptophan reduce cell wall or membrane stress tolerance in Saccharomyces cerevisiae. Biosci. Biotechnol. Biochem. 85, 2131–2136. doi:10.1093/bbb/zbab142
Orabona, C., and Grohmann, U. (2011). Indoleamine 2,3-dioxygenase and regulatory function: tryptophan starvation and beyond. Methods Mol. Biol. 677, 269–280. doi:10.1007/978-1-60761-869-0_19
Palmer, L. K., Wolfe, D., Keeley, J. L., and Keil, R. L. (2002). Volatile anesthetics affect nutrient availability in yeast. Genetics 161, 563–574. doi:10.1093/genetics/161.2.563
Pan, D., Wiedemann, N., and Kammerer, B. (2019). Heat stress-induced metabolic remodeling in Saccharomyces cerevisiae. Metabolites 9, 266. doi:10.3390/metabo9110266
Patnaik, R., and Liao, J. C. (1994). Engineering of Escherichia coli central metabolism for aromatic metabolite production with near theoretical yield. Appl. Environ. Microbiol. 60, 3903–3908. doi:10.1128/aem.60.11.3903-3908.1994
Patnaik, R., Zolandz, R. R., Green, D. A., and Kraynie, D. F. (2008). L-tyrosine production by recombinant Escherichia coli: fermentation optimization and recovery. Biotechnol. Bioeng. 99, 741–752. doi:10.1002/bit.21765
Pereira, R., Wei, Y., Mohamed, E., Radi, M., Malina, C., Herrgård, M. J., et al. (2019). Adaptive laboratory evolution of tolerance to dicarboxylic acids in Saccharomyces cerevisiae. Metab. Eng. 56, 130–141. doi:10.1016/j.ymben.2019.09.008
Perveen, S., Rashid, N., Tang, X. F., Imanaka, T., and Papageorgiou, A. C. (2017). Anthranilate phosphoribosyltransferase from the hyperthermophilic archaeon Thermococcus kodakarensis shows maximum activity with zinc and forms a unique dimeric structure. FEBS Open Bio 7, 1217–1230. doi:10.1002/2211-5463.12264
Piper, P., Calderon, C. O., Hatzixanthis, K., and Mollapour, M. (2001). Weak acid adaptation: the stress response that confers yeasts with resistance to organic acid food preservatives. Microbiol. Read. 147, 2635–2642. doi:10.1099/00221287-147-10-2635
Polleys, E. J., and Bertuch, A. A. (2015). Tryptophan-dependent control of colony formation after DNA damage via sea3-regulated TORC1 signaling in Saccharomyces cerevisiae. G3 (Bethesda) 5, 1379–1389. doi:10.1534/g3.115.018721
Schroeder, L., and Ikui, A. E. (2019). Tryptophan confers resistance to SDS-associated cell membrane stress in Saccharomyces cerevisiae. PLoS One 14, e0199484. doi:10.1371/journal.pone.0199484
Shen, D., He, X., Weng, P., Liu, Y., and Wu, Z. (2022). A review of yeast: high cell-density culture, molecular mechanisms of stress response and tolerance during fermentation. FEMS Yeast Res, 22. doi:10.1093/femsyr/foac050
Shen, L., Wang, Y. T., Tang, X. X., Zhang, K., Wang, P. M., Sui, Y., et al. (2020). Heat shock drives genomic instability and phenotypic variations in yeast. Amb. Express 10, 146. doi:10.1186/s13568-020-01091-7
Shen, T., Liu, Q., Xie, X., Xu, Q., and Chen, N. (2012). Improved production of tryptophan in genetically engineered Escherichia coli with TktA and PpsA overexpression. J. Biomed. Biotechnol. 2012, 1–8. doi:10.1155/2012/605219
Stanley, D., Bandara, A., Fraser, S., Chambers, P., and Stanley, G. (2010). The ethanol stress response and ethanol tolerance of Saccharomyces cerevisiae. J. Appl. Microbiol. 109, 13–24. doi:10.1111/j.1365-2672.2009.04657.x
SuáSTEGUI, M., Guo, W., Feng, X., and Shao, Z. (2016). Investigating strain dependency in the production of aromatic compounds in Saccharomyces cerevisiae. Biotechnol. Bioeng. 113, 2676–2685. doi:10.1002/bit.26037
Tang, J., Zhu, X., Lu, J., Liu, P., Xu, H., Tan, Z., et al. (2013). Recruiting alternative glucose utilization pathways for improving succinate production. Appl. Microbiol. Biotechnol. 97, 2513–2520. doi:10.1007/s00253-012-4344-1
Tang, M., Pan, X., Yang, T., You, J., Zhu, R., Yang, T., et al. (2023). Multidimensional engineering of Escherichia coli for efficient synthesis of L-tryptophan. Bioresour. Technol. 386, 129475. doi:10.1016/j.biortech.2023.129475
TröNDLE, J., Schoppel, K., Bleidt, A., Trachtmann, N., Sprenger, G. A., and Weuster-Botz, D. (2020). Metabolic control analysis of L-tryptophan production with Escherichia coli based on data from short-term perturbation experiments. J. Biotechnol. 307, 15–28. doi:10.1016/j.jbiotec.2019.10.009
Tyagi, N., Saini, D., Guleria, R., and Mukherjee, K. J. (2017). Designing an Escherichia coli strain for phenylalanine overproduction by metabolic engineering. Mol. Biotechnol. 59, 168–178. doi:10.1007/s12033-017-9999-5
Van Der Stel, A. X., Gordon, E. R., Sengupta, A., Martínez, A. K., Klepacki, D., Perry, T. N., et al. (2021). Structural basis for the tryptophan sensitivity of TnaC-mediated ribosome stalling. Nat. Commun. 12, 5340. doi:10.1038/s41467-021-25663-8
Vicent, I., Navarro, A., Mulet, J. M., Sharma, S., and Serrano, R. (2015). Uptake of inorganic phosphate is a limiting factor for Saccharomyces cerevisiae during growth at low temperatures. FEMS Yeast Res. 15, fov008. doi:10.1093/femsyr/fov008
Wang, J., Cheng, L. K., Wang, J., Liu, Q., and Shen, T. (2013). Genetic engineering of Escherichia coli to enhance production of L-tryptophan. Appl. Microbiol. Biotechnol. 97, 7587–7596. doi:10.1007/s00253-013-5026-3
Wendisch, V. F., Bott, M., Kalinowski, J., Oldiges, M., and Wiechert, W. (2006). Emerging Corynebacterium glutamicum systems biology. J. Biotechnol. 124, 74–92. doi:10.1016/j.jbiotec.2005.12.002
Wess, J., Brinek, M., and Boles, E. (2019). Improving isobutanol production with the yeast Saccharomyces cerevisiae by successively blocking competing metabolic pathways as well as ethanol and glycerol formation. Biotechnol. Biofuels 12, 173. doi:10.1186/s13068-019-1486-8
Wu, T., Zhao, J., and Mao, X. (2017). Effect of PTS modifications on L-tryptophan production in Escherichia coli. Sheng Wu Gong Cheng Xue Bao 33, 1877–1882. doi:10.13345/j.cjb.160460
Xiao, S., Wang, Z., Wang, B., Hou, B., Cheng, J., Bai, T., et al. (2023). Expanding the application of tryptophan: industrial biomanufacturing of tryptophan derivatives. Front. Microbiol. 14, 1099098. doi:10.3389/fmicb.2023.1099098
Xiong, B., Zhu, Y., Tian, D., Jiang, S., Fan, X., Ma, Q., et al. (2021). Flux redistribution of central carbon metabolism for efficient production of l-tryptophan in Escherichia coli. Biotechnol. Bioeng. 118, 1393–1404. doi:10.1002/bit.27665
Xu, D., Fang, M., Wang, H., Huang, L., Xu, Q., and Xu, Z. (2020). Enhanced production of 5-hydroxytryptophan through the regulation of L-tryptophan biosynthetic pathway. Appl. Microbiol. Biotechnol. 104, 2481–2488. doi:10.1007/s00253-020-10371-y
Xu, K., Zhang, Y. F., Guo, D. Y., Qin, L., Ashraf, M., and Ahmad, N. (2022). Recent advances in yeast genome evolution with stress tolerance for green biological manufacturing. Biotechnol. Bioeng. 119, 2689–2697. doi:10.1002/bit.28183
Yang, J., Tian, Y., Liu, H., Kan, Y., Zhou, Y., Wang, Y., et al. (2021). Harnessing the endogenous 2μ plasmid of Saccharomyces cerevisiae for pathway construction. Front. Microbiol. 12, 679665. doi:10.3389/fmicb.2021.679665
Yao, X., Liu, P., Chen, B., Wang, X., Tao, F., Lin, Z., et al. (2022). Synthetic acid stress-tolerance modules improve growth robustness and lysine productivity of industrial Escherichia coli in fermentation at low pH. Microb. Cell Fact. 21, 68. doi:10.1186/s12934-022-01795-4
Yu, J., Wang, J., Li, J., Guo, C., Huang, Y., and Xu, Q. (2008). Regulation of key enzymes in tryptophan biosynthesis pathway in Escherichia coli]. Sheng Wu Gong Cheng Xue Bao 24, 844–850. doi:10.1016/S1004-9541(08)60026-9
Zeng, L., Huang, J., Feng, P., Zhao, X., Si, Z., Long, X., et al. (2022). Transcriptomic analysis of formic acid stress response in Saccharomyces cerevisiae. World J. Microbiol. Biotechnol. 38, 34. doi:10.1007/s11274-021-03222-z
Zhang, J., Wu, C., Sheng, J., and Feng, X. (2016). Molecular basis of 5-hydroxytryptophan synthesis in Saccharomyces cerevisiae. Mol. Biosyst. 12, 1432–1435. doi:10.1039/c5mb00888c
Zhao, Z.-J., Zou, C., Zhu, Y.-X., Dai, J., Chen, S., Wu, D., et al. (2011a). Development of l-tryptophan production strains by defined genetic modification in Escherichia coli. J. Industrial Microbiol. Biotechnol. 38, 1921–1929. doi:10.1007/s10295-011-0978-8
Keywords: L-tryptophan, Saccharomyces cerevisiae, Escherichia coli, biosynthesis, stress fitness
Citation: Ren X, Wei Y, Zhao H, Shao J, Zeng F, Wang Z and Li L (2023) A comprehensive review and comparison of L-tryptophan biosynthesis in Saccharomyces cerevisiae and Escherichia coli. Front. Bioeng. Biotechnol. 11:1261832. doi: 10.3389/fbioe.2023.1261832
Received: 20 July 2023; Accepted: 22 November 2023;
Published: 04 December 2023.
Edited by:
Si-Yu Li, National Chung Hsing University, TaiwanReviewed by:
Dongsoo Yang, Korea University, Republic of KoreaCopyright © 2023 Ren, Wei, Zhao, Shao, Zeng, Wang and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhen Wang, wangzhen@hebau.edu.cn; Li Li, lgxlili@hebau.edu.cn
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.