- 1State Key Laboratory of Biobased Material and Green Papermaking (LBMP), Qilu University of Technology, Jinan, Shandong, China
- 2Department of Biological Engineering, Qilu University of Technology, Jinan, Shandong, China
- 3Lanling Meijiu Co., Ltd., Lanling, Shandong, China
Daqu is a spontaneous, solid-state cereal fermentation product used for saccharification and as a starter culture for Chinese Baijiu production. Bacillus and Acinetobacter, two dominant microbial genera in Daqu, produce enzymes and organic acids that influence the Daqu quality. However, there are no rapid analytical methods for detecting Bacillus and Acinetobacter. We designed primers specific to the genera Bacillus and Acinetobacter to perform genetic comparisons using the 16 S rRNA. After amplification of polymerase chain reaction using specific primers, high-throughput sequencing was performed to detect strains of Bacillus and Acinetobacter. The results showed that the effective amplification rates for Bacillus and Acinetobacter in Daqu were 86.92% and 79.75%, respectively. Thus, we have devised and assessed a method to accurately identify the species associated with Bacillus and Acinetobacter in Daqu, which can also hold significance for bacterial typing and identification.
1 Introduction
Baijiu is known as an ancient and distinct Chinese distilled spirit worldwide (Tu et al., 2022). It comprises one or more grains, typically sorghum, rice, wheat, barley, or maize. White wine fermentation is a complex process involving a variety of microorganisms in an open environment (Mao et al., 2022). During fermentation, barley is used as a microbial fermenting agent (Zhang et al., 2021) and is an integral part of the fermentation process. In fermentation, Daqu, which is used as a microbial fermenting agent (Zhang et al., 2021), is critical in the formation of Baijiu and is essential to develop the particular aroma associated with Baijiu (Xia et al., 2023). Baijiu and Daqu are highly associated with each other; since ancient times, good quality Daqu has been indispensable for producing good quality Baijiu, and the flavor of Baijiu depends on the quality of Daqu. Various enzymes and fungi are abundant in Daqu (Gao et al., 2022). The quality of Daqu greatly affects its yield and rate of product formation, and Bacillus and Acinetobacter play important roles. Bacillus can hydrolyze proteins and starch (Li et al., 2014), and few Bacillus species can metabolize the aromatic components in Baijiu, such as diacetyl (Shibamoto, 2014). Acinetobacter is the dominant bacteria in the early stage of liquor brewing and can secrete esterases, lipases (Doolittle and Peterfy, 2010), and pectinases (Hu et al., 2010), in addition to other enzymes. Organic acids (Bangar et al., 2022), fatty acids (Yang et al., 2022), amino acids, higher alcohols (Cordente et al., 2021), oligosaccharides (Rastall, 2010), and other small-molecule precursors are conducive to the production of flavored substances in Baijiu (Xiao et al., 2009).
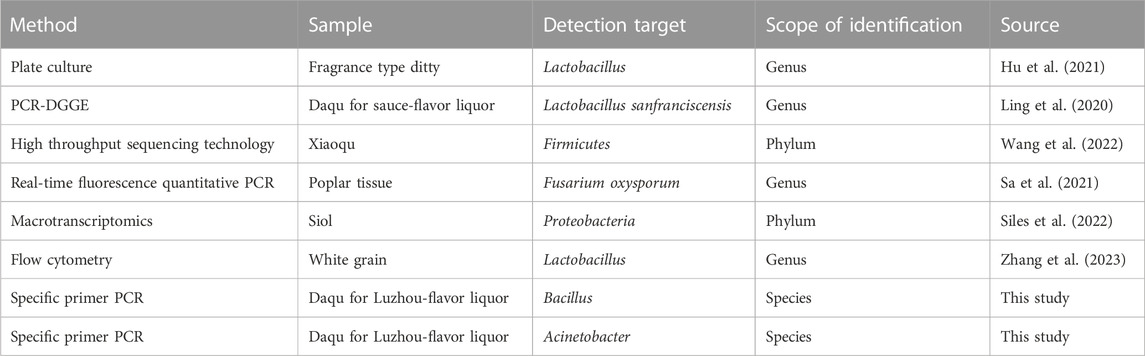
Microbiological testing is a crucial component of biological and technological research. The most common detection methods for microorganisms in the Baijiu fermentation include agar plate culture counting (Hu et al., 2021) and microscopy methods (Maceda and Terrazas, 2022). These two methods can be used to detect the number of microbial colonies and the individual morphology of Baijiu microorganisms. To overcome the limitations of traditional microbial detection technologies, high-throughput sequencing technologies (Jones, 2010), such as polymerase chain reaction (PCR) (Zhu et al., 2020), real-time fluorescence quantitative PCR (Hancock et al., 2010), flow cytometry (McKinnon, 2018), and other techniques are used to further detect and evaluate the number and size of microorganisms. PCR implemented in this study used a specific primer design resulting in a shorter cycle time, lower cost, and faster detection results than high-throughput sequencing and flow cytometry. Specific primer PCR involves designing the upstream and downstream primers for the target microbial DNA, enabling the analysis of trace amounts of DNA to be substantially amplified. The DNA of microorganisms that was amplified by PCR using specific primers has been reported for C. perfringens. Specific downstream primers for Clostridium perfringens (Weisburg et al., 1991) were designed for selective amplification of a bacterial strain from the myriad microflora in the sample.
Currently, there are two strategies to determine the presence of Bacillus and Acinetobacter species in a sample. The first involves plate coating using Bacillus or Acinetobacter screening media. However, this method is time-consuming and does not enable quantitative analysis. In the second strategy, all microbial communities in the sample can be detected using high-throughput sequencing (Xu et al., 2022), but the protocol is expensive and time-consuming, and the species and genus of Bacillus and Acinetobacter cannot be accurately determined. Therefore, it is necessary to develop highly specific and rapid analytical methods for Bacillus and Acinetobacter detection.
In this study, by designing specific primers for amplification and high-throughput sequencing, we obtained information about different species, performed accurate species-level analysis, devised a rapid detection method, and developed tools for designing specific primers for other species.
2 Materials and methods
2.1 Samples and reagents
Daqu samples were obtained from mature Daqu at the Shandong Lanling Fine Wine Co., Ltd. production plant. Five Daqu samples from five different points in each room were taken, crushed, mixed as parallel samples (Figure 1), placed in airtight bags, and frozen at −20°C. FastDNA SPIN Kit for Soil and FastPrep were purchased from MP Biomedical lnc. DL2000 Plus DNA Marker, DL 15000 DNA Marker, Phanta Max Super-Fidelity DNA Polymerase and Vazyme Gel Extraction Kit were purchased from Vozymes Biotech Co., Ltd. The sequence alignment software used was Clustalw (https://www.genome.jp/tools-bin/clustalw). Primer synthesis was completed by Sangon Biotech (Shanghai) Co., Ltd.
2.2 Method for extracting the Daqu genome
DNA was extracted using the FastDNA SPIN Kit for Soil. First, the solid distillate was lysed, and 500 mg of soil was added to the Lysing Matrix E tube (Lysed DNA) and mixed with the reagents. The mixture was mixed in the FastPrep apparatus for 40 s at a speed setting of 6.0. The supernatant was separated by centrifugation, and 250 μL of PPS was added. The supernatant was separated by centrifugation and the DNA was eluted by adding l mL of the Binding Matrix Supension, while the DNA binding matrix was left in place. Subsequently, we removed the supernatant and transferred 500 μL the mixture to the SPINTM Filter, which was centrifuged and air-dried. Finally, 50 μL of DES was added to elute the DNA (the reagents mentioned in this section are included in the FastDNA SPIN Kit for Soil).
2.3 High-throughput macro-genome taxonomic sequencing of Daqu currants
A total of 500 mg of Daqu was placed in a sterilized 2 mL tube, 1× PBS solution was added, and the mixture was shaken and mixed. The sample was centrifuged at 13,000 RCF for 3 min at 25°C, and the top layer was discarded. The 2 mL centrifuge tube was inverted onto blotting paper for 1 min until no liquid was further removed. The first round of PCR amplification was performed after genome extraction using the FastDNA SPIN Kit for Soil Gene Extraction. We used 16 S V3 to V4 zone (Caporaso et al., 2011) universal primers for amplification (338F: 5′-ACT CCT ACG GGA GGC AGC AG-3′ and 806R: 5′-GGA CTA CHV GGG TWT CTA AT-3′). The PCR products were purified and sequenced on the Illumina MiSeq high-throughput sequencing platform.
2.4 Primer design and PCR amplification specific to Bacillus and Acinetobacter
Bacillus subtilis 16 S rRNA was used as the template. 16 S rRNA is relatively conserved in structure and function and has changed relatively little during evolution. Although the 16 S rRNA gene sequence is relatively conserved, there are still differences in the 16 S rRNA gene sequence in different microorganisms. In total, 100 16 S rRNA genes of Bacillus were searched using NCBI GenBank, and multiple sequence matching was performed using Clustalw to identify gene regions that are highly conserved in the 16 S rRNA of Bacillus. Using the identified gene region as a template, Clustalw was used to design primer pairs specific for Bacillus. The sequence of this primer pair was: F: 5′-GTC TGT AAC TGA CGC TGA GGC-3′, R: 5′-GCG ATT ACT AGC GAT TCC A-3′. The primers were used to amplify the Daqu DNA.
Acinetobacter 16 S rRNA was used as the template. NCBI GenBank was used to search for 100 16 S rRNA genes of Acinetobacter, and multiple sequence comparisons were conducted using Clustalw to identify highly conserved gene regions in Acinetobacter encoding 16 S rRNA. Using the identified gene region as a template, specific Acinetobacter primer pairs were designed using Clustalw. The primer pair sequences used were: F2: (5′-ATG TGA AAT CCC CGA GCT T-3′) and R2: (5′-AGT TTG TCA CTG GCA GTA TCC T-3′). Two specific primers with high homology in Bacillus and Acinetobacter, but with little homology in non-Bacillus and non-Acinetobacter with similar sequences, were used for amplification. (Supplementary Figures S1, S2).
The amplification procedure was as follows: pre-denaturation at 94°C for 5 min; 20 cycles of denaturation at 94°C for 1 min, annealing at 55°C for 1 min, and extension at 72°C for 150 s; and extension at 72°C for 10 min using the Phanta Max Super-Fidelity DNA Polymerase (25 µL reactions).
2.5 PCR product recovery and sequencing using specific primers
The PCR amplified sample (10 µL) was analyzed on a 2% agarose gel along with the DL 2000 Plus DNA Marker. The Vazyme Gel Extraction Kit was used to recover the PCR amplification products. The Bacillus primers used were F: 5′-GTC TGT AAC TGA CGC TGA GGC-3′, R: 5′-GCG ATT ACT AGC GAT TCC A-3′, and Acinetobacter primer pairs included F2: 5′-ATG TGA AAT CCC CGA GCT T-3′, R2: 5′-AGT TTG TCA CTG GCA GTA TCC T-3′, which were used to process and analyze Illumina MiSeq high-throughput sequencing data from Bacillus and Acinetobacter, respectively.
3 Results
3.1 High-throughput sequencing of bacterial community structure and diversity analysis of bacterial communities in macrophytes
The community structure of the Daqu samples at the phylum level was determined based on taxonomic information of the species obtained from the different microbes within the sample (Figure 2A). At the phylum level, a total of 10 community structures relating to bacteria were detected in the two types of Daqu, with a total of four dominant phyla (≥1% relative abundance), namely, Firmicutes (82.67%), Proteobacteria (13.22%), Actinobacteria (7.06%), and Bacteroidetes (1.65%). Firmicutes were the dominant group of bacteria in the macrophyte samples (Chen et al., 2020).
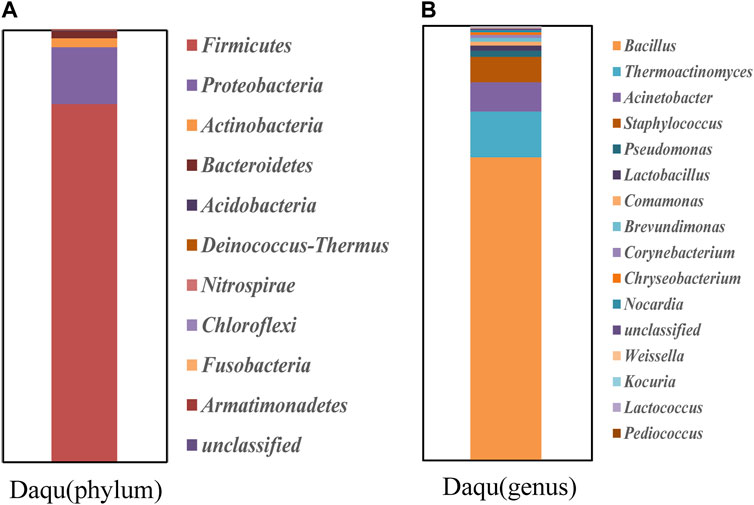
FIGURE 2. High-throughput sequencing of bacterial community structure and analysis of bacterial community diversity. (A) Classification of Daqu samples at phylum level. (B) Classification of Daqu samples at the generic level.
The community structure of the Daqu samples at the genus level was analyzed from the taxonomic information of the species in the samples (Figure 2B). In the Daqu samples, the top five prokaryotic communities in terms of relative abundance were Bacillus (69.61%), Weissella (34.33%), Lactobacillus (22.07%), Thermoactinomyces (19.9%), and Acinetobacter (11.56%). Bacillus was the dominant genera in the Daqu samples. The protein and starch in Daqu can be decomposed by Bacillus, and the aromatic substances in Daqu mainly originate from its action.
3.2 PCR analysis of Bacillus and Acinetobacter from Daqu
DNA was extracted from Daqu samples. The PCR amplification products of Bacillus and Acinetobacter were 626 and 536 bp, respectively, as shown in Figure 3. Meanwhile PCR experiments at 50, 52, 55, 57 and 60°C were also performed in this paper, and the annealing temperature was chosen at 55°C according to the experimental results (Supplementary Figure S3). The results also showed no spurious bands; thus, the primer pairs for Bacillus and Acinetobacter were specific.
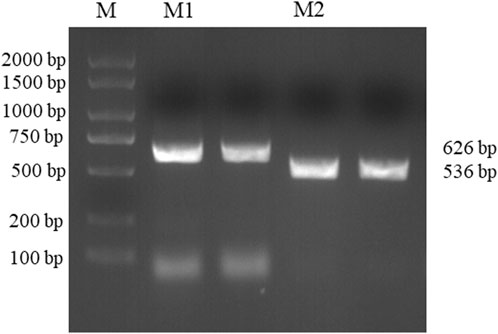
FIGURE 3. Bacillus and Acinetobacter agarose gel electrophoresis results. (Lane M: KB Ladder, Lane M1: Bacillus, Lane M2: Acinetobacter).
The amplified bands were purified using a Vazyme Gel Extraction Kit, and the purified products were sent to Sangon Biotech (Shanghai) for sequencing using the Illumina MiSeq PE300 platform and for high-throughput data processing and analysis.
3.3 High throughput sequencing for detecting Bacillus in Daqu
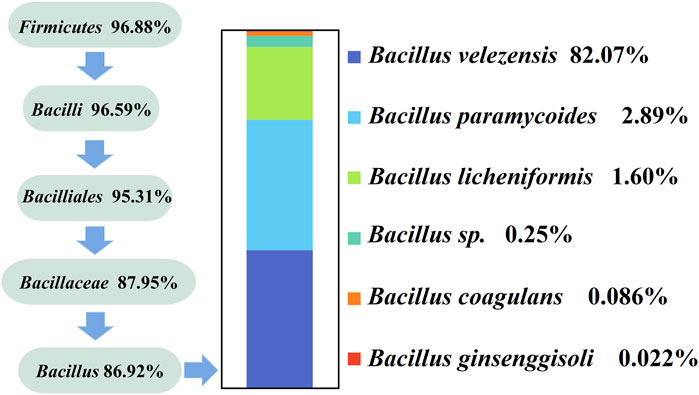
The results in Figure 2A show that Bacillus mostly contains macromolecules and produces proteases, amylases, and cellulases. We designed specific primers for Bacillus, performed specific primer PCR to amplify 16 S rRNA from only one microorganism, and verified the specificity of these primers using high-throughput sequencing (Figure 4). We identified six Bacillus species using high-throughput sequencing, and the resulting sequences were uploaded to the NCBI database (PRJNA985236). All six Bacillus species, namely, B. velezensis (82.07%), Bacillus paramycoides (2.89%), Bacillus licheniformis (1.6%), Bacillus sp. (0.25%), Bacillus coagulans (0.086%), and Bacillus ginsenggisoli (0.022%), belonged to the phylum Firmicutes, with Bacillus velezensis being the dominant strain.
3.4 High-throughput sequencing for detecting Acinetobacter in Daqu
As shown in Figure 2B, Acinetobacter was the dominant bacterial group in Daqu. We designed specific primers for Acinetobacter, performed PCR to selectively amplify the 16 S rRNA, verified the specificity of these primers after amplification, and conducted high-throughput sequencing, as shown in Figure 5. We identified five species of Acinetobacter by high-throughput sequencing and the resulting sequences were uploaded to NCBI database (PRJNA985236). These five species were classified as Proteobacteria at the phylum level and Acinetobacter at the genus level. The five species were A. baumannii (74.34%), Acinetobacter calcoaceticus (3.17%), Acinetobacter septicus (1.01%), Acinetobacter sp. GeSchum1Abdr4 (0.94%), and Acinetobacter bereziniae (0.29%), with Acinetobacter baumannii being the dominant strain.
4 Discussion
Microbial detection in modern liquor mainly includes high-throughput sequencing technology, PCR, real-time fluorescence quantitative PCR, flow cytometry, and other techniques that can detect the number and biological classification of microorganisms (Table 1). This study used specific primers and high-throughput sequencing analysis to identify Daqu microbial species. At the genus level, two dominant genera (Bacillus and Acinetobacter) were selected for further high-throughput sequencing. We found that Bacillus detected in Daqu included six species: B. velezensis (82.07%), B. paramycoides (2.89%), B. licheniformis (1.6%), Bacillus sp. (0.25%), B. coagulans (0.086%), and B. ginsenggisoli (0.022%). Bacillus species are protein hydrolase and amylase producers (Yi et al., 2019) that can enhance the production of organic acids in barley. Additionally, Bacillus species can produce protein hydrolases and amylases (He et al., 2019). Of these species, B. velezensis (Nam et al., 2009) is an important functional microorganism in winemaking and is the dominant bacterium found in Daqu at high temperature. It has high protease and amylase production capacity and can change the native flora, enzyme activity, and flavor composition of Daqu by regulating the metabolic activity. As an emerging functional strain, it is widely used for food fermentation (Ye et al., 2018). B. licheniformis can increase the amount of flavor substances in fermented liquor grains and improve sensory scores (Zhang et al., 2013). High-throughput sequencing showed that the primers F1 (5′-GTC TGT AAC TGA CGC TGA GGC-3′) and R1 (5′-GCG ATT ACT AGC GAT TCC A-3′) were suitable for performing PCR to specifically detect the Bacillus species.
In this study, we identified five species of Acinetobacter, namely, A. baumannii (74.34%), A. calcoaceticus (3.17%), A. septicus (1.01%), Acinetobacter sp. GeSchum1Abdr4 (0.94%), and A. bereziniae (0.29%) via high-throughput sequencing. Acinetobacter, the dominant bacterial genus in white wine barley, oxidizes glucose to produce acetic acid, which is one of the main flavor components in Baijiu. Acinetobacter, which is widely distributed in nature, is an aerobic, power-negative, gram-negative Coccobacillus that has been the focus of several research studies in the medical field (Wong et al., 2017). Actinobacter is mostly aerobic and, to a lesser extent, anaerobic, having the capacity to produce antibiotics (Lin and Lan, 2014). Actinobacter is also widely present in Daqu, fermented grains, and pit mud and drives the production of metabolites during fermentation. The specific primers F2 (5′-ATG TGA AAT CCC CGA GCT T-3′) and R2 (5′-AGT TTG TCA CTG GCA GTA TCC T-3′) for Acinetobacter are useful for high-throughput second-generation sequencing analysis. This method can be used to detect more types of Acinetobacter and better understand their role as dominant strains in Daqu.
Weisburg et al. (1991) amplified Clostridium 16 S rRNA using specific primers and determined the genus specificity of the bacteria, enabling distinction between strains. By ensuring genus specificity, data can be improved, especially for 16 S rRNA-based typing of bacteria (Sanschagrin and Yergeau, 2014). Liu et al. (2022) screened the 16 S rDNA fragments of Lactobacillus obtained using Lactobacillus-specific primer PCR with a reporter system to assess the activation of natural target sequences by different crRNAs and detected the production of fluorescent signals. In the study, two specific primer pairs were designed to target the highly conserved region of the prokaryotic gene 16 S rRNA: Bacillus F (5′-GTC TGT AAC TGA CGC TGA GGC-3′), R (5′-GCG ATT ACT AGC GAT TCC A-3′) and Actinobacter F2 (5′-ATG TGA AAT CCC CGA GCT T-3′), R2 (5′-AGT TTG TCA CTG GCA GTA TCC T-3′). The results showed that the prevalence of Bacillus and Acinetobacter in Daqu was 86.92% and 79.75%, respectively.
In summary, we performed PCR using specific primers to identify the species of various microorganisms and obtain nucleotide sequences of the bacteria, demonstrating an important method for bacterial typing. Compared with traditional 16 S rRNA taxonomic sequencing and high-throughput sequencing with specific primers, the method used in this study can reveal more phylum and genus classes. Therefore, for specific microbial genera such as Bacillus, additional information for different species can be obtained, along with a more accurate analysis of the species level. Our present study demonstrates that this method can be used as a rapid detection method when designing specific primers for other species. Relative to high-cost flow cytometry and long-cycle bacterial sequencing, specific primer PCR can amplify the required strains in a short time at a low cost and achieve 79.75% accuracy, making it a widely applicable method.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/genbank/, PRJNA985236.
Author contributions
YW: Writing–original draft. SZ: Writing–review and editing. GG: Writing–review and editing. ZW: Writing–review and editing. RW: Writing–review and editing. PL: Writing–review and editing. YL: Writing–review and editing. JW: Writing–review and editing. GJ: Writing–review and editing. HW: Writing–review and editing. CS: Writing–review and editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the Focus on Research and Development Plan in Shandong Province (2021ZDSYS10, 2022CXGC020206), Taishan Scholar Foundation of Shandong Province (tscx202306067), Innovation Fund for Small and Medium-sized Technology Innovation Capacity Enhancement Project of Shandong Province (2023TS1047), Key innovation Project of Qilu University of Technology (Shandong Academy of Sciences) (2022JBZ01-06), and National Natural Science Foundation of China (31801527).
Acknowledgments
We would like to thank the State Key Laboratory of Bio-based Materials and Green Papermaking, Qilu University of Technology for its help and support, and the Taishan industry leading talent funding.
Conflict of interest
GG, ZW, and YL are employed by the Lanling Meijiu Co., Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fbioe.2023.1261563/full#supplementary-material
References
Bangar, S. P., Suri, S., Trif, M., and Ozogul, F. (2022). Organic acids production from lactic acid bacteria: a preservation approach. Food Biosci. 46, 2212–4249. doi:10.1016/j.fbio.2022.101615
Caporaso, J. G., Lauber, C. L., Walters, W. A., Berg-Lyons, D., Lozupone, C. A., Turnbaugh, P. J., et al. (2011). Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci. U. S. A. 108, 4516–4522. doi:10.1073/pnas.1000080107
Chen, L., Li, Y. Z., Jin, L., He, L., Ao, X. L., Liu, S. L., et al. (2020). Analyzing bacterial community in pit mud of Yibin Baijiu in China using high throughput sequencing. Peer J. 8 (1), e9122. doi:10.7717/peerj.9122
Cordente, A. G., Nandorfy, D. E., Solomon, M., Schulkin, A., Kolouchova, R., Francis, I. L., et al. (2021). Aromatic higher alcohols in wine: implication on aroma and palate attributes during chardonnay aging. Molecules 26 (16), 4979. doi:10.3390/molecules26164979
Doolittle, M. H., and Peterfy, M. (2010). Mechanisms of lipase maturation. Clin. Lipidol. 5 (1), 117–130. doi:10.2217/clp.09.84
Gao, L., Zhou, J., and He, G. (2022). Effect of microbial interaction on flavor quality in Chinese baijiu fermentation. Front. Nutr. 9, 960712. doi:10.3389/fnut.2022.960712
Hancock, D., Funnell, A., Jack, B., and Johnston, J. (2010). Introducing undergraduate students to real-time PCR. Biochem. Mol. Biol. Educ. 38 (5), 309–316. doi:10.1002/bmb.20414
He, G. Q., Dong, Y., Huang, J., Wang, X. J., Zhang, S. Y., Wu, C. D., et al. (2019). Alteration of microbial community for improving flavor character of Daqu by inoculation with Bacillus velezensis and Bacillus subtilis. Lwt-Food Sci. Technol. 111, 1–8. doi:10.1016/j.lwt.2019.04.098
Hu, T. L. (2010). Study on enzymatic properties of pectinase. 12th International Wool Research Conference (IWRC 2010), Shanghai, China.
Hu, Y. L., Wang, L. Y., Zhang, Z. J., Yang, Q., Chen, S. X., Zhang, L., et al. (2021). Microbial community changes during the mechanized production of light aroma Xiaoqu Baijiu. Biotechnol. Biotechnol. Equip. 35 (1), 487–495. doi:10.1080/13102818.2021.1892525
Jones, W. J. (2010). High-throughput sequencing and metagenomics. Estuaries Coasts 33 (4), 944–952. doi:10.1007/s12237-009-9182-8
Li, Z., Bai, Z., Wang, D., Zhang, W., Zhang, M., Lin, F., et al. (2014). Cultivable bacterial diversity and amylase production in three typical D aqus of C hinese spirits. Int. J. Food Sci. Technol. 49 (3), 776–786. doi:10.1111/ijfs.12365
Lin, M. F., and Lan, C. Y. (2014). Antimicrobial resistance in Acinetobacter baumannii: from bench to bedside. World J. Clin. Cases 2 (12), 787–814. doi:10.12998/wjcc.v2.i12.787
Ling, Y. X., Li, W. Y., Tong, T., Li, Z. M., Li, Q., Bai, Z. H., et al. (2020). Assessing the microbial communities in four different Daqus by using PCR-DGGE, PLFA, and biolog analyses. Pol. J. Microbiol. 69 (1), 27–37. doi:10.33073/pjm-2020-004
Liu, Y. F., Wang, M. C., Yang, F., Zhang, X. L., Li, J. H., Du, G. C., et al. (2022). A CRISPR-cas12a-based assay for efficient quantification of Lactobacillus panis in Chinese Baijiu brewing microbiome. Fermentation-Basel 8 (2), 88. doi:10.3390/fermentation8020088
Maceda, A., and Terrazas, T. (2022). Fluorescence microscopy methods for the analysis and characterization of lignin. Polym. -Basel 14 (5), 961. doi:10.3390/polym14050961
Mao, F., Huang, J., Zhou, R., Qin, H., Zhang, S., Cai, X., et al. (2022). Effects of different Daqu on microbial community domestication and metabolites in Nongxiang Baijiu brewing microecosystem. Front. Microbiol. 13, 939904–904. doi:10.3389/fmicb.2022.939904
McKinnon, K. M. (2018). Flow cytometry: an overview. Curr. Opin. Immunol. 120, 5.1.1–5. doi:10.1002/cpim.40
Nam, M. H., Park, M. S., Kim, H. G., and Yoo, S. J. (2009). Biological control of strawberry Fusarium wilt caused by Fusarium oxysporum f. sp. fragariae using Bacillus velezensis BS87 and RK1 formulation. J. Microbiol. Biotechnol. 19 (5), 520–524. doi:10.4014/jmb.0805.333
Rastall, R. A. (2010). Functional oligosaccharides: application and manufacture. Annu. Rev. Food Sci. Technol. 1, 305–339. doi:10.1146/annurev.food.080708.100746
Sa, R. B., Zhang, J. L., Sun, J. Z., and Gao, Y. X. (2021). Colonization characteristics of poplar fungal disease biocontrol bacteria N6-34 and the inhibitory effect on pathogenic fungi by real-time fluorescence quantitative PCR detection. Curr. Microbiol. 78 (8), 2916–2925. doi:10.1007/s00284-021-02529-2
Sanschagrin, S., and Yergeau, E. (2014). Next-generation sequencing of 16S ribosomal RNA gene amplicons. J. Vis. Exp. (90), 51709. doi:10.3791/51709
Shibamoto, T. (2014). Diacetyl: occurrence, analysis, and toxicity. J. Agric. Food Chem. 62 (18), 4048–4053. doi:10.1021/jf500615u
Siles, J. A., Starke, R., Martinovic, T., Fernandes, M. L. P., Orgiazzi, A., and Bastida, F. (2022). Distribution of phosphorus cycling genes across land uses and microbial taxonomic groups based on metagenome and genome mining. Soil Biol. Biochem. 174, 108826. doi:10.1016/j.soilbio.2022.108826
Tu, W., Cao, X., Cheng, J., Li, L., Zhang, T., Wu, Q., et al. (2022). Chinese baijiu: the perfect works of microorganisms. Front. Microbiol. 13, 919044. doi:10.3389/fmicb.2022.919044
Wang, Q., Wang, C. Y., Xiang, X. Q., Xu, H. L., and Han, G. Q. (2022). Analysis of microbial diversity and succession during Xiaoqu Baijiu fermentation using high-throughput sequencing technology. Eng. Life Sci. 22 (7), 495–504. doi:10.1002/elsc.202200015
Weisburg, W. G., Barns, S. M., Pelletier, D. A., and Lane, D. J. (1991). 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 173 (2), 697–703. doi:10.1128/jb.173.2.697-703.1991
Wong, D., Nielsen, T. B., Bonomo, R. A., Pantapalangkoor, P., Luna, B., and Spellberg, B. (2017). Clinical and pathophysiological overview of acinetobacter infections: a century of challenges. Clin. Microbiol. Rev. 30 (1), 409–447. doi:10.1128/cmr.00058-16
Xia, Y., Luo, H., Wu, Z., and Zhang, W. (2023). Microbial diversity in jiuqu and its fermentation features: saccharification, alcohol fermentation and flavors generation. Appl. Microbiol. Biotechnol. 107 (1), 25–41. doi:10.1007/s00253-022-12291-5
Xiao, Z., Ma, C., Xu, P., and Lu, J. R. (2009). Acetoin catabolism and acetylbutanediol formation by Bacillus pumilus in a chemically defined medium. Public Libr. Sci. One 4 (5), e5627. doi:10.1371/journal.pone.0005627
Xu, B. Y., Xu, S. S., Cai, J., Sun, W., Mu, D. D., Wu, X. F., et al. (2022). Analysis of the microbial community and the metabolic profile in medium-temperature Daqu after inoculation with Bacillus licheniformis and Bacillus velezensis. Lwt-Food Sci. Technol. 160, 113214. doi:10.1016/j.lwt.2022.113214
Yang, K., Yu, M. L., Zhu, X. L., Xia, Y., Li, F. H., Li, Y. Z., et al. (2022). Genetic Incorporation of fluorescent amino acid into fatty acid binding protein for fatty acid detection. J. Mol. Biol. 434 (8), 167498. doi:10.1016/j.jmb.2022.167498
Ye, M., Tang, X., Yang, R., Zhang, H., Li, F., Tao, F., et al. (2018). Characteristics and application of a novel species of Bacillus: bacillus velezensis. ACS Chem. Biol. 13 (3), 500–505. doi:10.1021/acschembio.7b00874
Yi, Z. L., Jin, Y. L., Xiao, Y., Chen, L. C., Tan, L., Du, A. P., et al. (2019). Unraveling the contribution of high temperature stage to Jiang-flavor Daqu, a liquor starter for production of Chinese Jiang-flavor Baijiu, with special reference to metatranscriptomics. Front. Microbiol. 10, 472. doi:10.3389/fmicb.2019.00472
Zhang, Q., Chen, X., Ding, Y., Ke, Z., Zhou, X., and Zhang, J. (2021). Diversity and succession of the microbial community and its correlation with lipid oxidation in dry-cured black carp (Mylopharyngodon piceus) during storage. Food Microbiol. 98, 103686. doi:10.1016/j.fm.2020.103686
Zhang, R., Wu, Q., and Xu, Y. (2013). Aroma characteristics of Moutai-flavour liquor produced with Bacillus licheniformis by solid-state fermentation. Lett. Appl. Microbiol. 57 (1), 11–18. doi:10.1111/lam.12087
Zhang, Z. Y., Wei, Y. W., Peng, Z. H., Du, P., Du, X. Y., Zuo, G. Y., et al. (2023). Exploration of microbiome diversity of stacked fermented grains by flow cytometry and cell sorting. Front. Microbiol. 14, 1160552. doi:10.3389/fmicb.2023.1160552
Keywords: specific primer PCR, high-throughput sequencing, Daqu, Baijiu production, Bacillus, Acinetobacter
Citation: Wei Y, Zhang S, Guan G, Wan Z, Wang R, Li P, Liu Y, Wang J, Jiao G, Wang H and Sun C (2023) A specific and rapid method for detecting Bacillus and Acinetobacter species in Daqu. Front. Bioeng. Biotechnol. 11:1261563. doi: 10.3389/fbioe.2023.1261563
Received: 19 July 2023; Accepted: 11 September 2023;
Published: 25 September 2023.
Edited by:
Selin Şahin Sevgili, Istanbul University, TürkiyeReviewed by:
İrem Toprakçı Yüksel, Istanbul University-Cerrahpasa, TürkiyeEbru Kurtulbaş, Istanbul University-Cerrahpasa, Türkiye
Copyright © 2023 Wei, Zhang, Guan, Wan, Wang, Li, Liu, Wang, Jiao, Wang and Sun. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Junqing Wang, d2pxdHQuNjA4MkAxNjMuY29t
 Yanwei Wei1,2
Yanwei Wei1,2 Ruiming Wang
Ruiming Wang Piwu Li
Piwu Li Junqing Wang
Junqing Wang Hao Wang
Hao Wang