- College of Forestry and Grassland, Jilin Agricultural University, Changchun, China
Populus alba × Populus glandulosa (84K poplar) is model material with excellent genetic engineering resource and ornamental value. In our study, AmRosea1 (Antirrhinum majus) was overexpressed in 84K poplar, and the transgenic 84K (AM) poplar with high content of anthocyanin exhibited red pigmentation leaves. The transcriptome analysis between wild type (WT) and AM showed that 170 differentially expressed genes (DEGs) (86 up-regulated and 84 down-regulated) were found, and some DEGs were involved in flavone and flavonol biosynthesis, flavonoid biosynthesis and anthocyanin biosynthesis. The metabolome analysis showed that 13 anthocyanins-related differentially accumulated metabolites (DAMs) were detected in AM. The correlation analysis between DEGs and DAMs were performed, and the results revealed that 18 DEGs, including 11 MYB genes, two BZ1 genes, one FG2 gene, one ANS gene, and three IF7MAT genes, were negatively or positively correlated with 13 DAMs. The phylogenetic analysis demonstrated that there was high homology between AmRosea1 and PagMYB113, and MYB113 co-expressed with BZ1, ANS and DFR directly. Our results elucidated the molecular mechanism of plant color change mediated by anthocyanin biosynthesis pathway, which laid the foundation for the development and utilization of colorful woody plant.
Introduction
Anthocyanin as natural pigments has abundant physiological and ecological functions, whose content varies greatly depending on the plant variety, season, climate, and plant growth stage (Mobley et al., 2016). Anthocyanin content contributes to the colors such as red, blue and purple of plant vegetative and reproductive organs, thereby determining the application values in landscaping (Quattrocchio et al., 2006; Qiu et al., 2020). Currently, there is growing evidence that anthocyanin has benefits for human health owing to the role as an antioxidant to eliminate free radicals in human body and effectively delaying aging (Chiu et al., 2010; Zhang et al., 2020a). The purple tomato fruit reduces the risks of certain types of cancers and several chronic non-communicable diseases (CNCDs), such as diabetes, hypertension and obesity (Raiola et al., 2014). Anthocyanin biosynthesis is a specific branch of the phenylpropanoid pathway, which is one of the comprehensively studied secondary metabolic pathway (Zong et al., 2019; Yi et al., 2020). The pathway of anthocyanin biosynthesis involves many enzymes, whose coding genes are mainly divided into two classes, one consists of early biosynthetic genes (EBGs, such as CHS, CHI, and F3H), and the other consists of late biosynthetic genes [LBGs, such as DFR, ANS, and BZ1 (encodes UDP glucose flavonoid glucosyl-transferase, UFGT)] (Diao et al., 2004; Zong et al., 2019; Yan et al., 2021).
Studies on numerous species have explored key genes or transcription factors (TFs) that regulate anthocyanin biosynthesis. For example, MYELOBLASTOSIS (MYB) TFs, regulators of plant metabolic network, are used as activators alone or combining with other TFs to form the myeloblastosis-basic helix-loop-helix (bHLH)-tryptophan-aspartate (WD40) (MBW) complex that regulates the expression of anthocyanin biosynthesis structural genes (Xie et al., 2012; Xu et al., 2020). Multiple comparisons of anthocyanin-related genes reveal that the MYB-R3 domain contains a conserved bHLH binding motif (D/E)Lx2 (R/K)x3Lx6Lx3R, and R2R3-MYB usually forming a complex, thereby activating the expression of structural genes and promoting the accumulation of anthocyanin (Albert, et al., 2011; Cao, et al., 2017). In Paeonia suffruticosa, PsMYB12 interacts with bHLH and WD40 to form a protein complex that directly activate the expression of PsCHS. The activation leads to the pigmentation thereby exhibiting blotch, while the loss of the CHS activity contributes to albino flower (Gu, et al., 2019). In Anthurium andraeanum, AaMYB2 is considered to be a potential target that controls corolla color and involves in the joint expression of AaCHS, AaF3H and AaANS (Li, et al., 2016). In other plants, overexpression of MYB118 or MYB119 in poplar may change leaf color (Cho et al., 2016; Wang et al., 2019a). AmRosea1 belonging to MYB family is isolated from snapdragon (A. majus), and the overexpression of this gene is able to affect anthocyanin biosynthesis in snapdragons and tobacco (Schwinn. 2006; Zhang et al., 2013). Meanwhile, Rosea1 can interact with Delia to increase anthocyanin accumulation, which turns orange carrots into purple (Schwinn. 2006; Sharma et al., 2020).
Populus alba x P. glandulosa (84K poplar) is a fast-growing hybrid which derives from a breeding program in South Korea, and it is introduced into China until 1984 (Qiu et al., 2019; Huang et al., 2021). Scientifically, 84K poplar is a model material for studying on growth and development processes of perennial trees due to extensive genetic resources and perfect genetic transformation methods (Feng et al., 2015; Huang et al., 2021). In this study, AmRosa1 was transferred into the 84K male poplar to obtain AmRosea1 overexpression transgenic 84K poplar. The phenotype of AmRosea1-overexpressing plants was charactered, while gene expression profile and metabolites associated with anthocyanin biosynthesis or anthocyanin accumulation were analyzed. The results would enhance the understanding of other significant structural genes in anthocyanin biosynthesis pathway regulated by AmRosa1, which laid the foundation for the development and utilization of colorful woody plants. Furthermore, the preference that 84K male poplar were selected as transgenic receptors for changing plants color could reduce the pollution of poplar catkins in spring, which could optimize the biological safety of poplar as street trees.
Materials and Methods
Plant Materials
Populus alba × P. glandulosa (84K poplar) were obtained from College of Forestry and Grassland, Jilin Agricultural University (Changchun, China). Wild type 84K poplar (named WT) and the highest AmRosea1 overexpression transgenic 84K poplar L-24 (named AM) were cultured from January to June for further analyses (Supplementary Figure S1). The functional leaves (the third to fifth leaves from the main branches) of WT and AM without mechanical damage or disease were mixed respectively for performing transcriptional and metabolic analyses with three biological replicates (named WT-1, WT-2, WT-3 and AM-1, AM-2, AM-3). And storing at −80°C until transcriptional and metabolic analyses were performed. They performed for transcriptional and metabolic analyses.
Vector Construction and Genetic Transformation
The AmRosea1 gene fragment sequence was identified from NCBI database (GenBank: DQ275529, A. majus). The 663 bp gene fragment was synthesized into PUC57 vector (Beijing Genomics Institution, Beijing, China). After confirming sequence, AmRosea1 gene (amplification primers were F: 5′-ATGGAAAAGAATTGTCGTGGAG-3′, R: 5′- TTAATTTCCAATTTGTTGGGCCT-3′) was cloned and inserted into pROKII vector according to Liu et al. (2020). The vector was transferred into 84 K poplar using the Agrobacterium tumefaciens-mediated leaf disc transformation method (Cho et al., 2016). Transgenic shoots were cultured on MS medium with 20 mg/L kanamycin for kanamycin-resistant buds, and wild type line was cultured on MS medium. The genome DNA and total RNA were extracted for PCR using amplification primers and RT-qPCR (the primers were F: 5′-AGAGTATGGTGAAGGGAAATGG-3′, R: 5′-CCGACCTCTTTTGATATTTGGC-3′).
Observation of Stem Cross Section
Cross sections of poplar stems were prepared by hand-cutting, and the anthocyanin pigment accumulation without staining of one-month-old WT and AM was observed (Cho et al., 2016).
The Detection of Total Anthocyanin Content
The contents of total anthocyanin were detected according to Wang et al. (2019a) using functional leaves (the third to fifth leaves from the main branches) of six-month-old WT and AM.
RNA-Seq and RT-qPCR Verification
The total RNA extraction and library construction were carried out according to Li et al. (2021). The libraries were sequenced on the Illumina HiSeq platform with an average reading length of 125 bp/150 bp. As for RT-qPCR verification, the total RNA from each sample was extracted using RNA extraction kit (Tiangen, Beijing, China) according to the manufacturer’s instruction. The cDNA was synthesised using the PrimeScript RT reagent Kit with gDNA Eraser (TaKaRa, Kyoto, Japan). The primers for RT-qPCR were designed using the Integrated DNA Technologies (https://sg.idtdna.com/pages), and the sequences were listed on Supplementary Table S1. PtActin (amplification primers were F: 5′-AATACCCCATTGAGCACGG-3′, R: 5′- ACTCACACCATCACCAGAATC-3′) were selected as an internal reference gene. The amplification system and procedure were carried out according to Li et al. (2021). The relative expression level of genes was calculated using the 2−∆∆CT method (Livak and Schmittgen. 2001).
Transcriptome Analysis
The 84K poplar genome was downloaded and used for the reference genome (Qiu et al., 2019; Huang et al., 2021). Spliced Transcripts Alignments to a Reference (STAR) (Dobin et al., 2014) was used as aligner to map the sequencing reads to the reference genome with parameters recommended by RSEM. The Fragments Per Kilobase of transcript per Million fragments mapped (FPKM) of each gene were calculated based on the gene length. The functions of unigenes were annotated by the NCBI non-redundant protein sequences (Nr), euKaryotic Ortholog Groups (KOG), a manually annotated and curated protein sequence database (Swiss-Prot), Gene Ontology (GO), and Kyoto Encyclopedia of Genes and Genomes (KEGG) databases (Li et al., 2021). The analysis of differential expression genes (DEGs) between two-sample comparison was performed using the DESeq2 package (1.20.0) with p-value < 0.05 and |log2 foldchange| ≥ 1. The resulting p values were adjusted using the Benjamini and Hochberg’s false discovery rate (FDR). The analyses of GO and KEGG were performed according to Li et al. (2021) and Liu et al. (2021). Structural genes related to the anthocyanin biosynthesis pathway were screened from DEGs.
Metabolite Extraction and Metabolite Profiling
The prepared leaf samples were used to extract metabolite according to Zhang et al. (2021). Normalized data of each sample were used to analyzing differentially accumulated metabolites (DAMs). The detailed procedure for metabolite analysis was performed as previously described by Li et al. (2021).
Correlation Analysis of Transcriptome and Metabolome
Pearson correlations were calculated for the integrative analysis between the various DAMs and DEGs related to anthocyanin biosynthesis. The coefficient (r) value >0.9 or < −0.9 represented crucial relationships between metabolome and transcriptome. Correlation Network was constructed using the OmicStudio tools (https://www.omicstudio.cn/tool).
Phylogenetic Tree Analysis and Co-Expression Network Construction
Sequences were obtained from transcriptome for multiple sequence alignment, which was performed by BioEdit. The phylogenetic tree was constructed based on 12 protein sequences using MEGA ver. 7.0. The co-expression network of MYB and other structural genes were constructed by string (https://cn.string-db.org/) using sequences in Arabidopsis thaliana to identify the relationship between MYB and structural genes.
Statistical Analysis of Data
Data analysis were performed using Excel 2021, and SPSS version 26.0 (International Business Machines, NY, United States). Data indicated mean ± SD with three biological replications. Asterisks indicated significant differences between two lines based on student’s t-test (**p < 0.01).
Results
Generation and Characterization of AmRosea1 Transgenic 84K Poplar
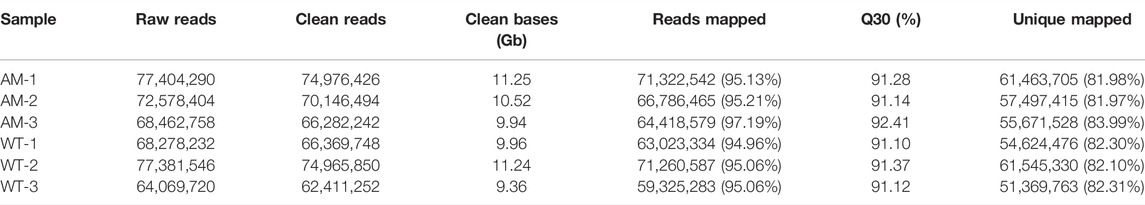
To understand the specific functions of AmRosea1 related to anthocyanin biosynthesis in poplar, AmRosea1 was overexpressed in 84K poplar. The exogenous AmRosea1 gene was detected by PCR and RT-qPCR. Amplified bands about 700 bp were found in positive plasmid (35S: AmRosea1) and AM line, which were not found in negative control and WT (Figure 1A). Meanwhile, the expression of AmRosea1 was extremely significantly up-regulated in AM line (p < 0.01) (Figure 1B). The phenotype of WT and AM was characterized. Compared to WT, red pigmented cells were found in the AM cortex from cross sections of one-month-old plants (Figure 1C). The total anthocyanin content in functional leaves of six-month-old AM was over two times that of WT (Figure 1D). The leaves, stems, and whole plant of six-month-old AM exhibited deeply red pigment, while none pigmentation was found in WT (Figures 1E–G). The total anthocyanin content in functional leaves of six-month-old AM was over two times that of WT (Figure 1F). These results showed that AM was generated and confirmed. The AM line exhibited red pigmentation during the growth stage, which were associated with high anthocyanin content.
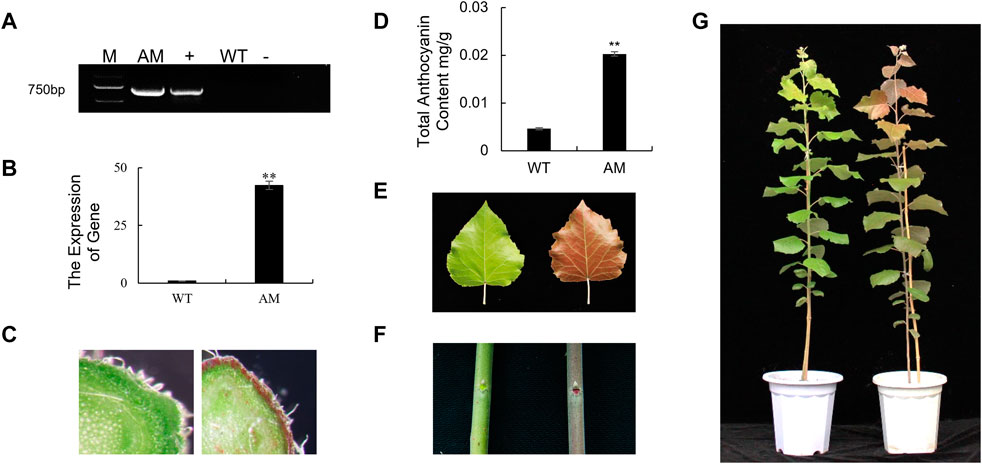
FIGURE 1. Generation and characterization of AmRosea1 transgenic 84K poplar. The whole picture on the left is wild-type (WT) 84K poplar, and AM on the right is AmRosea1 transgenic material. (A). Total DNA was extracted from leaf tissue for PCR verification. M, 2000 bp DNA Marker; AM, AmRosea1 transgenic poplar 84K; +, positive control; WT, wild type poplar 84K; -, negative control. (B). The expression of AmRosea1 in WT and AM. The stem cross cutting of one-month-old WT and AM (C). (D) The total anthocyanin contents of six-month-old WT and AM functional leaves. The phenotypic observation of functional leaves (E), stems (F), and the whole plants (G) of six-month-old WT and AM. Error bars represent the SDs from three biological replicates. Statistical differences were determined by a student’s t-test (**p < 0.01).
Sequencing Quality Evaluation of Transcriptome and RT-qPCR Verification
To analyze the gene expression profiles, RNA-Seq was performed using six-month-old WT and AM leaves. As a result, a total of 428,174,950 raw sequencing reads were generated from six samples, a total of 415,152,012 clean reads were finally obtained after removing low-quality data. The alignment of clean reads in six samples against the reference genome was from 95.06 to 97.19%, the Q30 percentages were all over 91.1% (Table 1). To test the reliability of RNA-Seq, nine genes were selected for verification, using RT-qPCR. The gene expression detected by RT-qPCR showed a similar pattern to that detected by the RNA-Seq (Figure 2). These results proved the accuracy of the transcriptome so that the transcriptome could be used for further analysis.
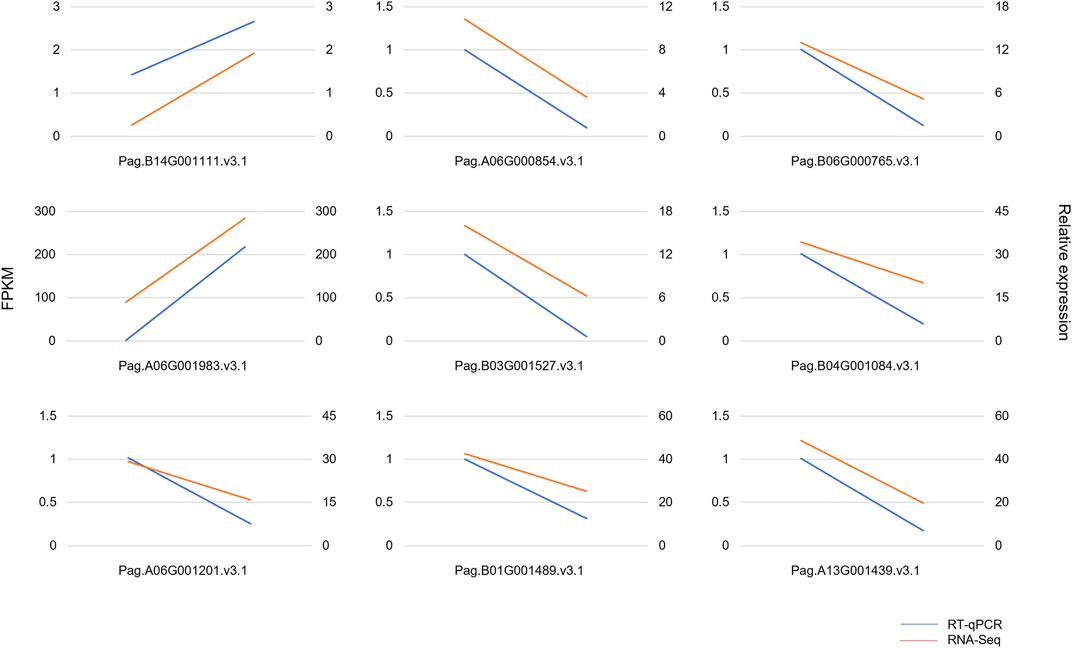
FIGURE 2. Quantitative real-time PCR verification of RNA-Seq. The red lines represent the results of RT-qPCR, and the blue lines represent the results of RNA-Seq. The y-axis on the left represents the FPKM value obtained by RNA-Seq. The y-axis on the right shows the gene relative expression levels analyzed by RT-qPCR. The endpoints on the left represent gene expression in WT, and endpoints on the right represent gene expression in AM.
Transcriptome Analysis of AmRosea1 Overexpression in 84K Poplar
Based on the transcriptome, 170 genes (86 up-regulated and 84 down-regulated) were found to be differentially expressed in AM compared with WT (Figure 3A). KEGG analysis demonstrated that DEGs were mainly enriched in biosynthesis of secondary metabolites, flavonoid biosynthesis and anthocyanin biosynthesis, indicating that AmRosea1 was related to anthocyanin biosynthesis (Figure 3B). DEGs were assigned to the GO categories, according to the biological process, cellular component, and molecular function (Figure 3C; Supplementary Table S2). In the biological process, 20, 38, and eight DEGs were involved in anthocyanin biosynthesis or metabolic pathway, flavonoid biosynthesis or metabolic process, and flavone biosynthesis or metabolic pathway, individually (Supplementary Figure S2). For the molecular function, three DEGs connected with anthocyanidin 3-O-glucosyltransferase activity were found. Additionally, nine DEGs encoding five enzymes, including PagDFR (Pag.A02G002453. v3.1) and PagLAR (Pag.A06G001383. v3.1), PagFG2 (Pag.A06G001188. v3.1), PagIF7MAT (Pag.B01G004171. v3.1, Pag. B01G000997. v3.1, and Pag. B04G000973. v3.1), PagANS (Pag.A16G000305. v3.1), PagBZ1 (Pag.A13G000799. v3.1 and Pag. B03G001191. v3.1), were identified, which participated in flavone and flavonol biosynthesis pathway (ko00944), isoflavonoid biosynthesis pathway (ko00943) and anthocyanin biosynthesis pathway (ko00942) (Supplementary Table S3). Compared to WT, PagDFR were down-regulated in AM, whereas other LBGs, PagANS, PagBZ, PagFG2 and PagIF7MAT were also up-regulated in AM (Figure 3D). These results suggested that the overexpression of AmRosea1 in 84K poplar might affect secondary metabolite biosynthesis and metabolism by regulating gene expression, especially the flavone and flavonol biosynthesis, flavonoid biosynthesis and anthocyanin biosynthesis.
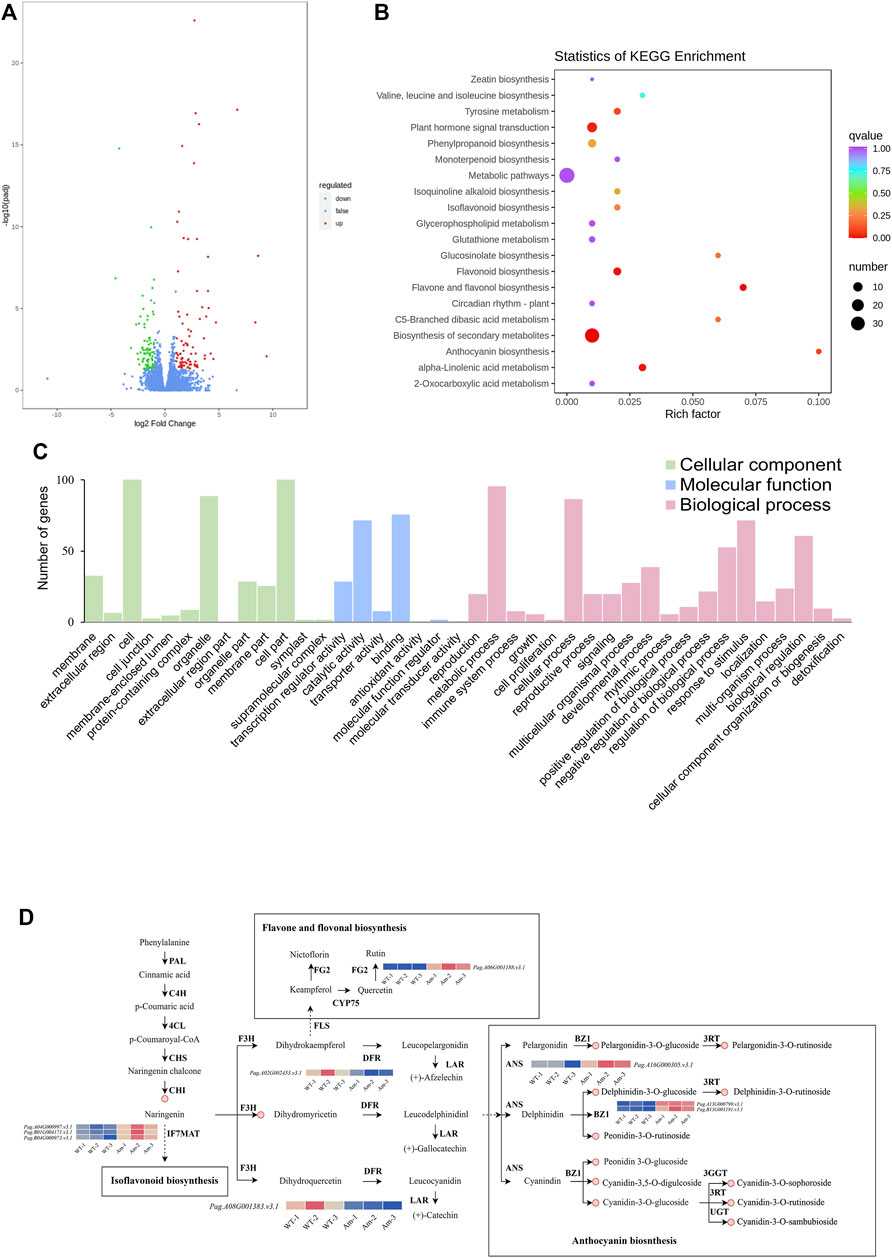
FIGURE 3. Identification of the differentially expressed genes (DEGs) in WT vs. AM. (A). Numbers of DEGs in volcano plots. (B). KEGG enrichment analysis of the DEGs. (C). Gene ontology (GO) classification of DEGs. (D). Differential expression of genes in flavonoid and anthocyanin biosynthesis pathways in poplar 84K leaf (WT vs. AM). PAL, phenylalanine ammonia-lyase; C4H, cinnamic acid 4-hydroxylase; 4CL, 4-coumarate CoA ligase; CHS, chalcone isomerase; CHI, chalcone isomerase; FG2, flavonol-3-O-glucoside l-rhamnosyltransferase; CYP75, flavonoid 3′-monooxygenase; F3H, lavanone 3-hydroxylase; FLS, flavonol synthase; IF7MAT, isoflavone 7-O-glucoside-6″-O-malonyltransferase; DFR, dihydroflavonol 4-reductase; LAR, leucoanthocyanidin reductase; ANS, anthocyanidin synthase; BZ1, anthocyanidin 3-O-glucosyltransferase; 3RT, pelargonidin 3-O-rutinoside + UDP; 3GGT, anthocyanidin 3-O-glucoside 2″-O-glucosyltransferase; UGT, anthocyanidin 3-O-glucoside 5-O-glucosyltransferase.
Metabolome Analysis of AmRosea1 Overexpression Transgenic 84K Poplar
The differentially accumulated metabolites (DAMs) were explored using metabolome. The levels of 13 anthocyanins metabolites increased, including five cyanidin, two delphinidin, two flavonoid, two pelargonidin, and two peonidin. They were cyanidin-3,5-O-diglucoside, cyanidin-3-O-glucoside, cyanidin-3-O-rutinoside, cyanidin-3-O-sambubioside, cyanidin-3-O-sophoroside, delphinidin-3-O-glucoside, delphinidin-3-O-rutinoside, pelargonidin-3-O-glucoside, pelargonidin-3-O-rutinoside, peonidin-3-O-glucoside, peonidin-3-O-rutinoside, dihydromyricetin, and naringenin. Among these metabolites, the levels of cyanidin-3-O-glucoside, delphinidin-3-O-glucoside, and delphinidin-3-O-rutinoside in AM line were significantly higher than WT (Figure 4; Supplementary Table S4). The results proposed that the overexpression of AmRosea1 in 84K poplar increased the level of anthocyanins-related metabolites, especially cyanidin-3-O-glucoside, delphinidin-3-O-glucoside and delphinidin-3-O-rutinoside, thereby controlling anthocyanin biosynthesis.
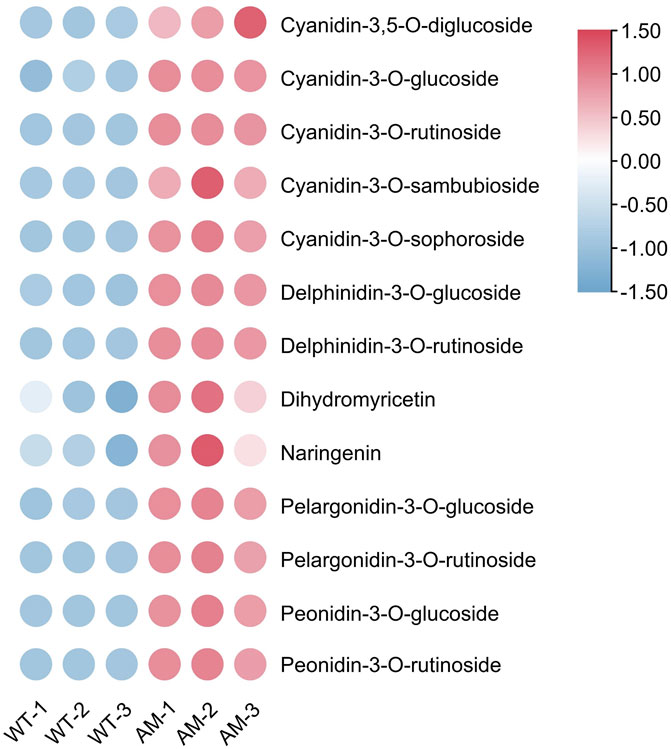
FIGURE 4. The differentially accumulated metabolites (DAMs) of anthocyanin accumulation between WT vs. AM.
Correlation Analysis Between DEGs and DAMs
To understand the relationship between DEGs and DAMs in anthocyanin biosynthesis, the integrative analysis was performed. On the basis of the correlation analysis between DEGs and DAMs with coefficients of r > 0.9 or < -0.9, ANS gene was significantly negatively associated with pelargonidin-3-O-rutinoside and delphinidin-3-O-rutinoside, and IF7MAT genes were positively associated with dihydromyricetin and naringenin. BZ1 genes were significantly positively related to cyanidin-3,5-O-diglucoside, cyanidin-3-O-glucoside, cyanidin-3-O-rutinoside, cyanidin-3-O-sambubioside, cyanidin-3-O-sophoroside, delphinidin-3-O-glucoside, delphinidin-3-O-rutinoside, naringenin, pelargonidin-3-O-glucoside, pelargonidin-3-O-rutinoside, peonidin-3-O-glucoside, and peonidin-3-O-rutinoside. FG2 genes were significantly positively involved in cyanidin-3,5-O-diglucoside, cyanidin-3-O-glucoside, cyanidin-3-O-rutinoside, cyanidin-3-O-sophoroside, cyanidin-3-O-sambubioside, delphinidin-3-O-glucoside, delphinidin-3-O-rutinoside, dihydromyricetin, naringenin, pelargonidin-3-O-glucoside, pelargonidin-3-O-rutinoside, peonidin-3-O-glucoside, and peonidin-3-O-rutinoside. MYB genes were only positively correlated with cyanidin-3-O-sambubioside, naringenin, dihydromyricetin (Supplementary Table S5). The regulatory network where five structural genes regulated 13 mentioned metabolites positively or negatively were constructed (Figure 5). These results showed that the contents of anthocyanin mainly affinitive with MYB genes and other structural genes, FG2 genes, BZI genes, ANS genes and IF7MAT genes as such in AM.
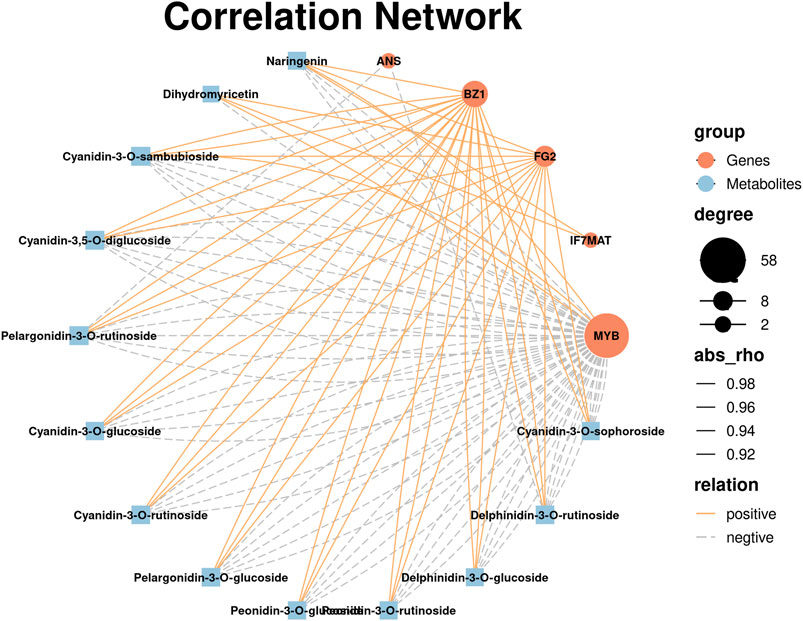
FIGURE 5. The construction of regulatory network between DEGs and DAMs. rho represents the Pearson correlation coefficient (r). Degree represents the gene number. Relation represents the correlations with a coefficient (r) value >0.9 (positive) or < −0.9 (negative).
Co-Expression Network of AmRosea1 in 84K Poplar
In order to explore the mechanism of AmRosea1, we conducted the phylogenetic analysis between AmRosea1 and DEGs belonging to MYB in poplar 84K. The phylogenetic analysis revealed that there was high homology between AmRosea1 and PagMYB113 (Pag.A04G002340. v3.1) (Figure 6A; Supplementary Table S6). Therefore, MYB113 was selected to construct co-expression network in A. thaliana for enhancing the understanding of regulatory relationship, which showed that MYB113 co-expressed with BZ1, ANS and DFR directly (Figure 6B). The results suggested that AmRosea1 might co-express with BZ1, ANS and DFR directly, thereby affecting anthocyanin biosynthesis in AM.
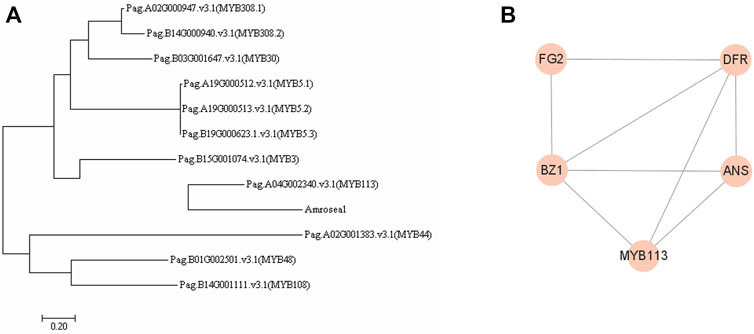
FIGURE 6. Phylogenetic tree analysis and construction of co-expression network. (A). The phylogenetic tree between MYBs and AmRosea1 using MEGA seven software. (B). The co-expression network of MYB113 used String (https://cn.string-db.org/).
Discussion
Populus alba x P. glandulosa originally derived from South Korea, which is one of the major tree species cultivated widely in China recently. 84K poplar is used as a model plant for understanding molecular mechanisms and economic objectives of woody plants. In previous studies, the researches of 84K poplar focused on gene expression patterns under nitrogen deficiency stress (Liu et al., 2021), cavitation and frost fatigue (Feng et al., 2015), molecular mechanisms of wood formation (Xiao et al., 2021), whereas fewer studies that exogenous gene caused the leaves to turn red in 84K poplar were reported. The conjoint analysis of transcriptome and metabolome was a potential method to explore some specific molecular mechanisms based on gene expression patterns and metabolite profiles. This method had been used in many species, for example, the anthocyanin and proanthocyanin accumulation were analyzed by transcriptional and metabolic analysis in red mutant pear (Zhang et al., 2020b), the candidate genes involved in anthocyanin accumulation of green and red leaves in Acer at the growth stage were explored (Zhang et al., 2021), the metabolite accumulation and gene expression patterns in inner and outer seed coats of hard-seeded and soft-seeded pomegranate were detected (Qin et al., 2020). In our study, transcriptome and metabolome of AM exhibiting red pigmentation in leaves, stems and whole plants were integrated to elucidate the molecular mechanism of plant color change.
The Anthocyanin Metabolites Have Effect on Plant Pigment
The various anthocyanins make plants exhibit different colors, six categories of metabolites, including malvidin, pelargonidin, cyanidin, peonidin, delphinidin and leucocyanidin, participate in anthocyanin biosynthesis (Yan et al., 2021). Red phenotype was closely associated with the types and contents of red pigment (Li et al., 2021). The results of Cho et al. (2016) showed that the overexpression of PtrMYB119 affected the accumulation of cyanidin-3-O-glucoside, thereby promoting anthocyanin biosynthesis, which changed leaf color. And the same result was found in the peel of red longan (Yi et al., 2021). Zhang et al. (2021) found that cyanidin 3-O-glucoside, pelargonidin 3-O-glucoside and delphinidin 3-O-glucoside were significantly increased in red-leaf acer. In this study, the AM with high anthocyanin content was generated and confirmed, which exhibited red pigmentation in leaves, stems and whole plant during the growth stage (Figures 1C–E–G). The metabolome analysis showed that 13 anthocyanins-related metabolites increased in transgenic 84K poplar, especially cyanidin-3-O-glucoside improved greatly (Figure 4, Supplementary Table S4), which was consistent with other studies (Cho et al., 2016; Zhang et al., 2021). These results proposed that the levels of anthocyanin-related metabolites, cyanidin-3-O-glucoside as such, affected anthocyanin content, thereby changing plant color.
Integated Analysis Between DEGs and DAMs
The accumulation of anthocyanin is mainly affected by structural genes and TFs (Wang et al., 2019b). Structural genes control the metabolite content in anthocyanin biosynthesis pathway by changing the expression levels of upstream and downstream genes (Li et al., 2019). Transcription factors can influence structural genes to indirectly regulate anthocyanin metabolite accumulation (Zhang et al., 2021) On the basis of correlation network, DFR genes down-regulated in AM were not related to 13 mentioned metabolites (Figure 3D, Figure 5). BZ1, FG2, and IF7MAT genes were positively associated with anthocyanin-related metabolites, whereas ANS genes were negatively associated with anthocyanin-related metabolites. Among these four kinds of genes, FG2 and IF7MAT genes up-regulated in AM were involved in flavone biosynthesis and isoflavonid biosynthesis pathways, which determined the increases of two and 13 anthocyanin-related metabolites, respectively. BZ1 genes up-regulated in AM participated in anthocyanin biosynthesis directly, which also led the accumulation of 13 anthocyanin-related metabolites. Cyanidin-3-O-glucoside with highest level in AM was positively related to FG2 and BZ1 genes (Figure 5; Supplementary Table S5; Figure 3D). These results suggested that the up-regulated IF7MAT genes involved in isoflavonid biosynthesis might promote anthocyanin biosynthesis pathway. Up-regulated BZ1 genes facilitated the synthesis of intermediate products, cyanidin-3-O-glucoside, delphinidin-3-O-glucoside, and pelargonidin-3-O-glucoside as such, followed by improving the levels of end-products, cyanidin-3-O-rutinoside, cyanidin-3-O-sambubioside, cyanidin-3-O-sophoroside, delphinidin-3-O-rutinoside, pelargonidin-3-O-rutinoside, and peonidin-3-O-rutinoside as such. These changes of DEGs and DAMs were major in the increased anthocyanin content.
The Functions of AmRosea1 in Anthocyanin Biosynthesis
Except for mentioned structural genes, MYB TFs used as activators alone or forming ternary-complexes with bHLH and WD40 regulate anthocyanin biosynthesis (Xie et al., 2012; Xu et al., 2020). MrMYB1 (Myricarubra Chinese) activated the promoter of NtDFR depending on the existence of bHLH, and the co-expression of MrMYB1 with the bHLH genes would increase the anthocyanin (Niu, et al., 2010). AmRosea1 as the member of MYB TFs could activate anthocyanin biosynthesis by over-expression in herbaceous plants (Zhang et al., 2013). For example, Rosea1 interacted with Delia to enhance anthocyanin in carrot and Salvia miltiorrhiza, thereby changing plant color (Wang et al., 2013; Sharma et al., 2020). In this study, the anthocyanin content and anthocyanin-related metabolites increased in AM, which demonstrated that the anthocyanin biosynthesis pathway was activated by AM. Most of MYB genes were negatively correlated with anthocyanin-related metabolites except for cyanidin-3-O-sambubioside, dihydromyricetin and naringenin. Additionally, none of genes related to bHLH were identified on the basis of transcriptome, these results proposed that AmRosea1 deriving from snapdragon as an exogenous gene regulated anthocyanin biosynthesis excluding forming classical ternary-complex. In this study, the overexpression of AmRosea1 activated the anthocyanin biosynthesis pathway, and the expression of some structural genes significantly changed, including DFR, ANS, FG2, IF7MAT, BZ1, LAR (Supplementary Figure S3). The MdMYB1, MdDFR and MdUFGT (encoded by BZ1) in apple (Malus domestica) could activate the anthocyanin synthesis pathway (Takos, et al., 2006). The heterologous MYB1 of Muscari botryoides Mill. was inserted in tobacco could independently regulate DFR, and the purple anthocyanin pigment was found in transgenic tobacco leaves, petals, anthers and calyx. (Chen, et al., 2019). FhMYB5 of Freesia hybrida was inserted in tobacco, which specially recognized NtLAR promoter site to promote the anthocyanin accumulation (Li, et al., 2018). These results show that MYB genes activate the anthocyanin pathway by binding to structural genes. The phylogenetic analysis showed that there was high homology between AmRosea1 and PagMYB113 (Pag.A04G002340. v3.1) (Figure 6A, Supplementary Table S6). The co-expression network showed that MYB113 might interact with BZ1, ANS and DFR directly in A. thaliana (Figure 6B). Therefore, we inferred that AmRosea1 might play the similar role with PagMYB113. The overexpression of this gene might activate the expression of ANS and BZ1 genes directly, thereby improving the contents of anthocyanin and anthocyanin-related metabolites, which turned green 84K poplar plants into red. Our results enhanced the understand of anthocyanin biosynthesis pathway to plant color change, which provided references for the development and utilization of colorful woody plant.
Conclusion
In summary, the AmRosea1 overexpression transgenic 84K poplar with high total anthocyanin content exhibited red pigmentation in leaves, stems and whole plant. Genes involved in flavone and flavonol biosynthesis, flavonoid biosynthesis and anthocyanin biosynthesis expressed differentially, while 13 anthocyanins-related metabolites that had affinity with MYB, ANS, FG2, BZ1, and IF7MAT genes increased, especially the levels of cyanidin-3-O-glucoside, delphinidin-3-O-glucoside and delphinidin-3-O-rutinoside improved greatly. PagMYB113 that had high homology with AmRosea1 might be co-expressed with BZ1, ANS and DFR directly in 84K poplar. Overall, all results might suggest that the overexpression of AmRosea1 in 84K poplar increased the contents of anthocyanin and anthocyanins-related metabolites through regulating BZ1, ANS and DFR directly, thereby turning green 84K poplar plants into red.
Data Availability Statement
RNA-Seq raw data from six samples were deposited in the National Center for Biotechnology. Information (NCBI) under the accession number PRJNA822547.
Author Contributions
HY, XL, and XZ was a major contributor in writing the manuscript. PY, XW, HL, and QZ contributed to plant sample collection, DNA/RNA preparation, library construction, and sequencing. HY and XL worked on genome assembly and annotation. RG and XP conducted transcriptome analysis and identified structural genes involved in leaf color formation. XH and RH analyzed the gene family and constructed the evolutionary tree. XZ conceived of the study, participated in its design and data interpretation, and revised the manuscript critically. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by the Scientific research start-up funds of Jilin Agricultural University (No. 2021002).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
Thanks to the members of the College of Forestry and Grassland of Jilin Agricultural University for their assistance during laboratory works and for fruitful discussions. Thanks to the assistance of Wuhan MetWare Biotechnology Co., Ltd. and Frasergen Bioinformatics Co., Ltd. (Wuhan, China).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fbioe.2022.911701/full#supplementary-material
References
Albert, N. W., Lewis, D. H., Zhang, H., Schwinn, K. E., Jameson, P. E., and Davies, K. M. (2011). Members of an R2R3-MYB Transcription Factor Family in Petunia Are Developmentally and Environmentally Regulated to Control Complex Floral and Vegetative Pigmentation Patterning. Plant J. 65, 771–784. doi:10.1111/j.1365-313X.2010.04465.x
Cao, X., Qiu, Z., Wang, X., Van Giang, T., Liu, X., Wang, J., et al. (2017). A Putative R3 MYB Repressor Is the Candidate Gene Underlying Atroviolacium, a Locus for Anthocyanin Pigmentation in Tomato Fruit. J. Exp. Bot. 68, 5745–5758. doi:10.1093/jxb/erx382
Chen, L., Hu, B., Qin, Y., Hu, G., and Zhao, J. (2019). Advance of the Negative Regulation of Anthocyanin Biosynthesis by MYB Transcription Factors. Plant Physiology Biochem. 136 (7), 178–187. doi:10.1016/j.plaphy.2019.01.024
Chiu, L.-W., Zhou, X., Burke, S., Wu, X., Prior, R. L., and Li, L. (2010). The Purple Cauliflower Arises from Activation of a MYB Transcription Factor. Plant Physiol. 154 (3), 1470–1480. doi:10.1104/pp.110.164160
Cho, J.-S., Nguyen, V. P., Jeon, H.-W., Kim, M.-H., Eom, S. H., Lim, Y. J., et al. (2016). Overexpression ofPtrMYB119, a R2R3-MYB Transcription Factor fromPopulus Trichocarpa, Promotes Anthocyanin Production in Hybrid Poplar. Tree Physiol. 36, 1162–1176. doi:10.1093/treephys/tpw046
Diao, Y., Liu, J. Y., Zhou, M. Q., and Li, H. Z. (2004). Chromosomal Localization of Anthocyanin Biosynthetic Genes Bz1, Bz2 in lotus. J. Wuhan Botanical Res. 5, 380–384. doi:10.1007/BF02873086
Dobin, A., Davis, C. A., Schlesinger, F., Drenkow, J., Zaleski, C., Jha, S., et al. (2013). STAR: Ultrafast Universal RNA-Seq Aligner. Bioinformatics 29 (1), 15–21. doi:10.1093/bioinformatics/bts635
Feng, F., Ding, F., and Tyree, M. T. (2015). Investigations Concerning Cavitation and Frost Fatigue in Clonal 84K Poplar Using High-Resolution Cavitron Measurements. Plant Physiol. 168 (1), 144–155. doi:10.1104/pp.114.256271
Gu, Z., Zhu, J., Hao, Q., Yuan, Y.-W., Duan, Y.-W., Men, S., et al. (2019). A Novel R2R3-MYB Transcription Factor Contributes to Petal Blotch Formation by Regulating Organ-specific Expression ofPsCHSin Tree Peony (Paeonia Suffruticosa). Plant & Cell Physiology 60 (3), 599–611. doi:10.1093/pcp/pcy232
Huang, X., Chen, S., Peng, X., Bae, E.-K., Dai, X., Liu, G., et al. (2021). An Improved Draft Genome Sequence of Hybrid Populus Alba × Populus Glandulosa. J. For. Res. 32, 1663–1672. doi:10.1007/s11676-020-01235-2
Li, C., Qiu, J., Yang, G., Huang, S., and Yin, J. (2016). Isolation and Characterization of a R2R3-MYB Transcription Factor Gene Related to Anthocyanin Biosynthesis in the Spathes of Anthurium Andraeanum (Hort.). Plant Cell Rep. 35, 2151–2165. doi:10.1007/s00299-016-2025-8
Li, L., Zhai, Y. H., Luo, X. N., Zhang, Y., and Shi, Q. (2019). Comparative Transcriptome Analyses Reveal Genes Related to Pigmentation in the Petals of Red and White Primula Vulgaris Cultivars. Physiol. Mol. Biol. Plants 25 (4), 1029–1041. doi:10.1007/s12298-019-00664-6
Li, X., Li, Y., Zhao, M., Hu, Y., Meng, F., Song, X., et al. (2021). Molecular and Metabolic Insights into Anthocyanin Biosynthesis for Leaf Color Change in Chokecherry (Padus Virginiana). Ijms 22 (19), 10697. doi:10.3390/ijms221910697
Li, Y., Shan, X., Zhou, L., Gao, R., Yang, S., Wang, S., et al. (2018). The R2R3-MYB Factor FhMYB5 from Freesia Hybrida Contributes to the Regulation of Anthocyanin and Proanthocyanidin Biosynthesis. Front. Plant Sci. 9, 1935. doi:10.3389/fpls.2018.01935
Liu, C., Chen, S., Wang, S., Zhao, X., Li, K., Chen, S., et al. (2021). A Genome Wide Transcriptional Study of Populus Alba X P. Tremula Var. Glandulosa in Response to Nitrogen Deficiency Stress. Physiol. Mol. Biol. Plants 27 (6), 1277–1293. doi:10.1007/s12298-021-01012-3
Liu, Z., Lei, X., Wang, P., Wang, Y., Lv, J., Li, X., et al. (2020). Overexpression of Thsap30bp from Tamarix Hispida Improves Salt Tolerance. Plant Physiology Biochem. 146, 124–132. doi:10.1016/j.plaphy.2019.11.020
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 25, 402–408. doi:10.1006/meth.2001.1262
Mobley, L. R., Scott, L., Rutherford, Y., and Kuo, T.-M. (2017). Using Residential Segregation to Predict Colorectal Cancer Stage at Diagnosis: Two Different Approaches. Ann. Epidemiol. 27 (1), 10–19. doi:10.1016/j.annepidem.2016.11.008
Niu, S.-S., Xu, C.-J., Zhang, W.-S., Zhang, B., Li, X., Lin-Wang, K., et al. (2010). Coordinated Regulation of Anthocyanin Biosynthesis in Chinese Bayberry (Myrica Rubra) Fruit by a R2R3 MYB Transcription Factor. Planta 231, 887–899. doi:10.1007/s00425-009-1095-z
Qin, G., Liu, C., Li, J., Qi, Y., Gao, Z., Zhang, X., et al. (2020). Diversity of Metabolite Accumulation Patterns in Inner and Outer Seed Coats of Pomegranate: Exploring Their Relationship with Genetic Mechanisms of Seed Coat Development. Hortic. Res. 7 (1), 2634–2647. doi:10.1038/s41438-019-0233-4
Qiu, D., Bai, S., Ma, J., Zhang, L., Shao, F., Zhang, K., et al. (2019). The Genome of Populus Alba X Populus Tremula Var. Glandulosa Clone 84K. DNA Res. 26 (5), 423–431. doi:10.1093/dnares/dsz020
Qiu, W., Su, W., Cai, Z., Dong, L., Li, C., Xin, M., et al. (2020). Combined Analysis of Transcriptome and Metabolome Reveals the Potential Mechanism of Coloration and Fruit Quality in Yellow and Purple Passiflora edulis Sims. J. Agric. Food Chem. 68 (43), 12096–12106. doi:10.1021/acs.jafc.0c03619
Quattrocchio, F., Verweij, W., Kroon, A., Spelt, C., Mol, J., and Koes, R. (2006). PH4 of Petunia Is an R2R3 MYB Protein that Activates Vacuolar Acidification through Interactions with Basic-Helix-Loop-Helix Transcription Factors of the Anthocyanin Pathway. Plant Cell 18 (5), 1274–1291. doi:10.1105/tpc.105.034041
Raiola, A., Rigano, M. M., Calafiore, R., Frusciante, L., and Barone, A. (2014). Enhancing the Health-Promoting Effects of Tomato Fruit for Biofortified Food. Mediat. Inflamm. 2014, 139873. doi:10.1155/2014/139873
Schwinn, K., Venail, J., Shang, Y., Mackay, S., Alm, V., Butelli, E., et al. (2006). A Small Family ofMYB-Regulatory Genes Controls Floral Pigmentation Intensity and Patterning in the GenusAntirrhinum. plant cell 18 (4), 831–851. doi:10.1105/tpc.105.039255
Sharma, S., Holme, I. B., Dionisio, G., Kodama, M., Dzhanfezova, T., Joernsgaard, B., et al. (2020). Cyanidin Based Anthocyanin Biosynthesis in Orange Carrot Is Restored by Expression of AmRosea1 and AmDelila, MYB and bHLH Transcription Factors. Plant Mol. Biol. 103 (4-5), 443–456. doi:10.1007/s11103-020-01002-1
Takos, A. M., Jaffé, F. W., Jacob, S. R., Bogs, J., Robinson, S. P., and Walker, A. R. (2006). Light-induced Expression of a MYB Gene Regulates Anthocyanin Biosynthesis in Red Apples. Plant Physiol. 142, 1216–1232. doi:10.1104/pp.106.088104
Wang, D., Song, Y., Chen, Y., Yao, W., Li, Z., Liu, W., et al. (2013). Metabolic Pools of Phenolic Acids in Salvia Miltiorrhiza Are Enhanced by Co-expression of Antirrhinum majus Delila and Rosea1 Transcription Factors. Biochem. Eng. J. 74, 115–120. doi:10.1016/j.bej.2013.02.014
Wang, H., Wang, X., Song, W., Bao, Y., Jin, Y., Jiang, C., et al. (2019a). PdMYB118, Isolated from a Red Leaf Mutant of Populus Deltoids, Is a New Transcription Factor Regulating Anthocyanin Biosynthesis in Poplar. Plant Cell Rep. 38, 927–936. doi:10.1007/s00299-019-02413-1
Wang, L., Lu, W., Ran, L., Dou, L., Yao, S., Hu, J., et al. (2019b). R2R3‐ MYB Transcription Factor MYB 6 Promotes Anthocyanin and Proanthocyanidin Biosynthesis but Inhibits Secondary Cell Wall Formation in Populus Tomentosa. Plant J. 99, 733–751. doi:10.1111/tpj.14364
Xie, X.-B., Li, S., Zhang, R.-F., Zhao, J., Chen, Y.-C., Zhao, Q., et al. (2012). The bHLH Transcription Factor MdbHLH3 Promotes Anthocyanin Accumulation and Fruit Colouration in Response to Low Temperature in Apples. Plant, Cell Environ. 35 (11), 1884–1897. doi:10.1111/j.1365-3040.2012.02523.x
Xu, H., Zou, Q., Yang, G., Jiang, S., Fang, H., Wang, Y., et al. (2020). MdMYB6 Regulates Anthocyanin Formation in Apple Both through Direct Inhibition of the Biosynthesis Pathway and through Substrate Removal. Hortic. Res. 7, 72. doi:10.1038/s41438-020-0294-4
Yan, H., Pei, X., Zhang, H., Li, X., Zhang, X., Zhao, M., et al. (2021). MYB-mediated Regulation of Anthocyanin Biosynthesis. Ijms 22 (6), 3103. doi:10.3390/ijms22063103
Yi, D., Zhang, H., Lai, B., Liu, L., Pan, X., Ma, Z., et al. (2020). Integrative Analysis of the Coloring Mechanism of Red Longan Pericarp through Metabolome and Transcriptome Analyses. J. Agric. Food Chem. 69 (6), 1806–1815. doi:10.1021/acs.jafc.0c05023
Zhang, S., Zhan, W., Sun, A., Xie, Y., Han, Z., Qu, X., et al. (2021). Combined Transcriptome and Metabolome Integrated Analysis of Acer Mandshuricum to Reveal Candidate Genes Involved in Anthocyanin Accumulation. Sci. Rep. 11, 23148. doi:10.1038/s41598-021-02607-2
Zhang, X., Xu, J., Xu, Z., Sun, X., Zhu, J., and Zhang, Y. (2020a). Analysis of Antioxidant Activity and Flavonoids Metabolites in Peel and Flesh of Red-Fleshed Apple Varieties. Molecules 25 (8), 1968. doi:10.3390/molecules25081968
Zhang, Y. Z., Li, Y., Gao, Z. R., Piao, C. L., Song, Q. N., Cui, M. L., et al. (2013). Rose A1-Overexpression Plant Accumulate Anthocyanins and Resist Insect Attack. Amr 726-731, 4418–4422. doi:10.4028/www.scientific.net/amr.726-731.4418
Zhang, Z., Tian, C., Zhang, Y., Li, C., Li, X., Yu, Q., et al. (2020b). Transcriptomic and Metabolomic Analysis Provides Insights into Anthocyanin and Procyanidin Accumulation in Pear. BMC Plant Biol. 20 (1), 129. doi:10.1186/s12870-020-02344-0
Keywords: Populus alba × Populus glandulosa (84K poplar), anthocyanin, AmRosea1, metabolome, transcriptome
Citation: Yan H, Zhang X, Li X, Wang X, Li H, Zhao Q, Yin P, Guo R, Pei X, Hu X, Han R and Zhao X (2022) Integrated Transcriptome and Metabolome Analyses Reveal the Anthocyanin Biosynthesis Pathway in AmRosea1 Overexpression 84K Poplar. Front. Bioeng. Biotechnol. 10:911701. doi: 10.3389/fbioe.2022.911701
Received: 03 April 2022; Accepted: 05 May 2022;
Published: 06 June 2022.
Edited by:
Mulualem Tigabu, Swedish University of Agricultural Sciences, SwedenReviewed by:
Qingzhang Du, Beijing Forestry University, ChinaChangjun Ding, Chinese Academy of Forestry, China
Copyright © 2022 Yan, Zhang, Li, Wang, Li, Zhao, Yin, Guo, Pei, Hu, Han and Zhao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rui Han, aGFucnVpNDU3OUBqbGF1LmVkdS5jbg==; Xiyang Zhao, emhhb3h5cGhkQDE2My5jb20=
†These authors have contributed equally to this work and share first authorship
 Huiling Yan
Huiling Yan Xinxin Zhang
Xinxin Zhang Xiang Li
Xiang Li Xuelai Wang
Xuelai Wang Hanxi Li
Hanxi Li Qiushuang Zhao
Qiushuang Zhao Peng Yin
Peng Yin Ruixue Guo
Ruixue Guo Xiaona Pei
Xiaona Pei Xiaoqing Hu
Xiaoqing Hu Rui Han
Rui Han Xiyang Zhao
Xiyang Zhao