- 1School of Materials Science and Engineering, Gwangju Institute of Science and Technology (GIST), Gwangju, South Korea
- 2Research Center for Innovative Energy and Carbon Optimized Synthesis for Chemicals (Inn-ECOSysChem), Gwangju Institute of Science and Technology (GIST), Gwangju, South Korea
Hydrogen gas obtained from cheap or sustainable sources has been investigated as an alternative to fossil fuels. By using hydrogenase (H2ase) and formate dehydrogenase (FDH), H2 and CO2 gases can be converted to formate, which can be conveniently stored and transported. However, developing an enzymatic process that converts H2 and CO2 obtained from cheap sources into formate is challenging because even a very small amount of O2 included in the cheap sources damages most H2ases and FDHs. In order to overcome this limitation, we investigated a pair of oxygen-tolerant H2ase and FDH. We achieved the cascade reaction between H2ase from Ralstonia eutropha H16 (ReSH) and FDH from Rhodobacter capsulatus (RcFDH) to convert H2 and CO2 to formate using in situ regeneration of NAD+/NADH in the presence of O2.
1 Introduction
The development of renewable energy technologies to replace fossil fuels is essential for the sustainable growth of the economy and society. Hydrogen (H2), obtained from various sources such as solar (Song et al., 2022), algae (Wang and Yin, 2018), biomass (Lepage et al., 2021), and by-product gas (Lee and Elgowainy, 2018), is expected to be an alternative fuel with high gravimetric energy density and net-zero carbon dioxide (CO2) production (Eppinger and Huang, 2017). However, owing to the low volumetric energy capacity of H2, its transportation and storage as a fuel are limited (Eppinger and Huang, 2017). Therefore, converting H2 into a chemical with a high volumetric energy capacity while maintaining the molar energy capacity is advantageous for the commercialization of alternative energy.
Suitable materials for converting H2 energy should satisfy the following conditions: 1) high energy/volume capacity, 2) low energy loss during the conversion process, 3) liquid material at ambient pressure and temperature, and 4) non-flammable chemicals for safety. Thus, the conversion of H2 and CO2 into formate is an appropriate approach (Ping et al., 2013; Eppinger and Huang, 2017; Mihet et al., 2020). Because formate is a non-flammable liquid at ambient temperature and pressure, it is convenient to transport and store. However, the reaction requires a catalyst, and synthetic catalysts cannot be applied to various H2 sources because of their low selectivity, low efficiency, and requirement for precious metals (Loges et al., 2008; Kuehnel et al., 2015; Sordakis et al., 2018). Thus, the substrate specificity and high reaction rate of the enzymatic process reveal the potential for H2 and CO2 as alternative synthetic catalysts. The oxidation of H2 and reduction of CO2 occur in hydrogenase (H2ase) (Lubitz et al., 2014) and formate dehydrogenases (FDH) (Appel et al., 2013; Amao, 2018; Moon et al., 2020), respectively, among oxidoreductases.
Formate hydrogenlyase (FHL) is a natural FDH and H2ase-linked enzyme complex that catalyzes formate/H2 interconversion (McDowall et al., 2014). The catalytic bias of the FHL is H2 production from the oxidation of formate (Pinske and Sargent, 2016; Schwarz et al., 2018). The semi-artificial coupling of H2ase and FDH from Desulfovibrio vulgaris Hildenborough successfully demonstrated the interconversion of H2 and CO2 into formate (Sokol et al., 2019). Cheap and sustainable H2 sources, such as coke oven gas generated from steel industries, contain a small portion of O2 (0.4–1.7%) (Li et al., 2019; García García et al., 2020). Because of the transition metal active sites and low potential electrons, most H2ases and FDHs are inhibited or irreversibly damaged by a trace amount of O2 (Fontecilla-Camps et al., 2007; Niks and Hille, 2018), limiting the application of H2 conversion obtained from various renewable sources.
H2ases and FDHs from aerobic organisms maintain their catalytic activities under aerobic conditions. H2ase can be classified according to the metal ion composition of their active sites in [NiFe], [FeFe], and [Fe] H2ases (Lubitz et al., 2014). [NiFe] H2ase inactivation under aerobic condition was shown to form an inactive state by O2 bridging to Ni-Fe through X-Ray crystallography, electron paramagnetic resonance (EPR) studies, and the density function theory calculations (Volbeda et al., 2005; Shafaat et al., 2013; Qiu et al., 2018). Well-studied O2-tolerant [NiFe] H2ases from Escherichia coli (Sargent, 2016), Aquifex aeolicus (Pandelia et al., 2010), and Ralstonia eutropha (Burgdorf et al., 2005) have potential biotechnological applications such as biofuel cells and H2 production. Their O2 tolerance was attributed to the reduction of O2 bound to NiFe active site into either hydrogen peroxide or water (Lauterbach and Lenz, 2013; Wulff et al., 2014; Horch et al., 2015). The soluble H2ase from the R. eutropha (ReSH) complex contains heterodimeric [NiFe] hydrogenase (HoxHY) subunits and diaphorase (HoxFU) subunits, which reduce NAD(P)+ while oxidizing H2 (Lauterbach and Lenz, 2013). FDH can be classified according to the metal ion composition of their active sites in molybdenum (Mo) containing and tungsten (W) containing formate dehydrogenase (Moon et al., 2020). Under aerobic conditions, the inactivation of FDH occurs by substitution of oxo ligand for sulfide ligand at the active site by O2 (Duffus et al., 2020). It was proposed that the O2 tolerance of W-containing FDH2 from Desulfovibrio vulgaris Hildenborough results from reduction of O2 to hydrogen peroxide by formate oxidase activity (Graham et al., 2022). The FDHs from Clostridium carboxidivorans strain P7T (Alissandratos et al., 2013), Methylobacterium extorquens AM1 (Laukel et al., 2003; Baccour et al., 2020), and Rhodobacter capsulatus (Hartmann and Leimkühler, 2013) maintain high CO2-reducing activity under aerobic conditions. FDH from R. capsulatus (RcFDH) consists of FdsA subunit containing the bis(molybdopterin guanine dinucleotide) cofactor and FdsGB diaphorase subunit for oxidizing NADH while reducing CO2. We hypothesized that H2 and CO2 are converted to formate through a cascade reaction of O2-tolerant H2ases and FDHs under oxic conditions. Here, we demonstrated a cascade reaction of ReSH and RcFDH with NAD+ regeneration (Figure 1). Formate production was observed under anaerobic and O2 concentration-controlled conditions.
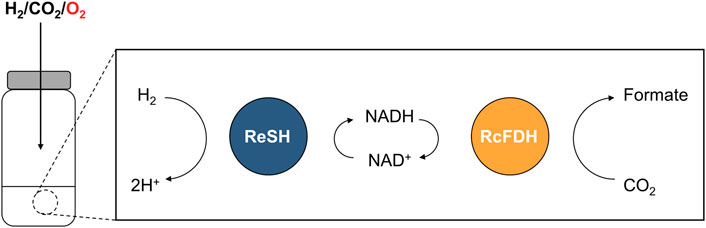
FIGURE 1. Schematic showing the NAD+-dependent cascade reaction of ReSH and RcFDH in the presence of O2.
2 Materials and methods
2.1 Materials
The 5X In-Fusion® HD Enzyme Premix was purchased from Takara Bio (Kusatsu, Japan). Strep-Tactin XT 4 Flow high-capacity resin was obtained from IBA Life Sciences (Göttingen, Germany). Disposable PD-10 desalting columns were purchased from Cytiva (Marlborough, MA, United States). Vivaspin 6 centrifugal concentrators with a molecular weight cutoff (MWCO) of 100 kDa were purchased from Sartorius (Göttingen, Germany). A polypropylene column (1 ml) was purchased from Qiagen (Hilden, Germany). The Ziptip C18 resin was purchased from Millipore (Burlington, MA, United States). All other chemical reagents were purchased from Sigma-Aldrich (St. Louis, MO, United States) unless otherwise stated.
2.2 Construction of plasmids and strains
To construct the strep-tag II-fused RcFDH expression plasmid, pTrcHis-RcFDH (Choi et al., 2018) was used as a template. Infusion cloning was performed to substitute the hexahistidine-tag for strep tag II. pTrcHis-RcFDH was amplified by PCR with the in-fusion primer (FW: 5′-GCCACCCGCAGTTCGAAAAAGGTATGGCTAGCATGACGGATACC-3′, RV: 5′-CGAACTGCGGGTGGCTCCAAGAACCCCCCATGGTTTATTCCTCC-3′). The PCR product was mixed with 5X In-Fusion HD Enzyme Premix to generate pTrcHis-strep-RcFDH. The E. coli MC1061 strain was transformed with pTrcHis-Strep-RcFDH, and the R. eutropha HF210 [pGE771] strain (Lauterbach and Lenz, 2013) was used as the ReSH-expressing strain.
2.3 Expression of ReSH and RcFDH
For the expression of ReSH and RcFDH, a 7 L scale fermenter was used. Previously, Lenz described the heterotrophic cultivation of R. eutropha derivatives (Lenz et al., 2018). A 10X H16 buffer (pH 7.0) consisting of 250 mM Na2HPO4 and 110 mM KH2PO4 was used as the medium. For a 1 L of fructose-ammonium (FN) medium, 100 ml of 10X H16 buffer was mixed with 850 ml of sterilized water (additional 13% (w/v) of Bacto agar in case of solid agar plates) and autoclaved. Next, 10 ml of 20% (w/v) NH4Cl, 1 ml each of 20% (w/v) NH4Cl, 20% (w/v) MgSO4*7H2O, 1% (w/v) CaCl2*H2O, 0.5% (w/v) FeCl3*6H2O (in 0.1 N HCl), 1 mM NiCl2, and 1.25 ml of 40% (w/v) D-fructose were mixed and filled up to 1000 ml with sterile H2O. A single colony of R. eutropha was pre-cultured in 50 ml of FN medium containing 10 μg ml−1 tetracyclin until the OD436nm reached 1. For the main culture, 5 L of modified fructose-glycerol-ammonium (FGNmod) with 0.05% (w/v) glycerol, 5 ml of SL6 trace element solution (Lenz et al., 2018), and 5 ml of 1 mM ZnCl2 (added to the FN medium containing 10 μg/ml tetracycline) were prepared in the fermenter. The pre-culture was inoculated into the FGNmod medium and subjected to 300 rpm shaking and 1 VVM aeration at 30°C. The pH range was maintained between 6.9 to 7.0 through automatic injection of 1 N NaOH. After 24 h, 5 ml of 1 mM NiCl2 was added. When the OD at 436 nm reached 9–11, the cells were harvested by centrifugation at 6,000 × g for 10 min before storage at −80°C.
For RcFDH expression, a single-cell colony was pre-cultured in Luria-Bertani (LB) medium containing 150 μg ml−1 ampicillin for 12 h at 37°C. For the main culture, 5 L of LB medium containing 150 μg ml−1 ampicillin, 1 mM sodium molybdate, and 20 μM isopropyl β-D-1-thiogalactopyranoside was prepared in the fermenter. The pre-culture was inoculated into the LB medium and subjected to 100 rpm shaking and 0.1 VVM aeration at 30°C. After 24 h, the cells were harvested by centrifugation at 6,000 × g for 10 min before storage at -80°C.
2.4 Purification of ReSH and RcFDH
To purify ReSH and RcFDH, cell pellets were resuspended in 50 mM potassium phosphate buffer (pH 7.0) (Kpi buffer) containing 1 mg/ml lysozyme to a concentration of 1 g/10 ml. The resuspended cells were lysed by sonication (amplitude 28%, on/off 2 s/4 s) for 1 h. Insoluble cell debris was removed by centrifugation at 13,000 × g for 30 min. Strep-Tactin XT 4Flow high-capacity resin (2 ml) was mixed with the clear supernatants and incubated at 4°C for 30 min. The resin was washed with Kpi buffer containing 300 mM potassium chloride on a gravity-flow polypropylene column to remove any impurities. The proteins were eluted with 3 ml of Kpi buffer containing 50 mM biotin and buffer-exchanged with Kpi buffer containing 10 mM potassium nitrate using a PD-10 column. Protein purity was verified by SDS-PAGE (Figure 2). The concentrations of purified proteins were determined by measuring their absorbance at 280 nm using a microplate reader (Synergy, BioTek, Winooski, VT, United States), as previously reported for other proteins (Kim et al., 2019; 2021; Bak et al., 2020). The extinction coefficients of ReSH and RcFDH were calculated to be 165,710 and 350,000 M−1⋅cm−1, respectively, based on their amino acid sequences.
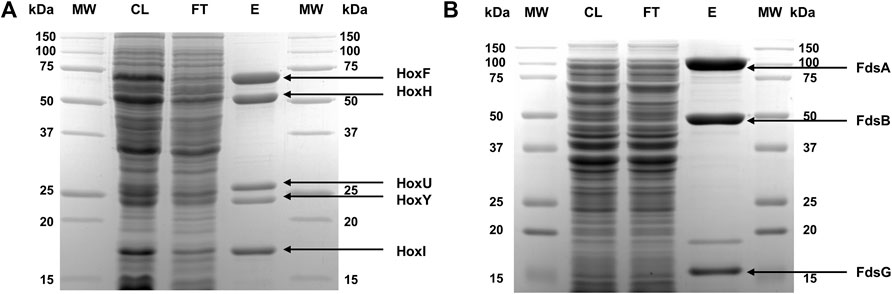
FIGURE 2. SDS-PAGE of purified proteins. (A) ReSH and (B) RcFDH stained with Coomassie blue. The lanes are molecular weight markers (MW), cell lysate after sonication (CL), flow-through streptavidin resin (FT), and eluted protein (E).
2.5 Matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) mass spectrometry
Proteins in buffer were desalted using Ziptip C18 according to the manufacturer’s protocol. Purified ReSH and RcFDH were mixed in a 1:1 (v/v) ratio with a sinapinic acid-saturated matrix solution consisting of 30% acetonitrile, 0.1% trifluoroacetic acid (TFA), and 70% water (v/v). The mixtures were subjected to mass characterization by Autoflex speed (Bruker Corporation, Billerica, United States).
2.6 Enzyme kinetics
The enzyme reaction kinetics of ReSH were measured for the NAD+-dependent oxidation of H2 to H+ in the presence or absence of O2. The sealing cuvette was filled with 900 μL of Kpi buffer containing NAD+ and sealed; then, 100% H2 and a mixed gas consisting of 10% O2 and 90% N2 (or 100% N2 for anaerobic conditions) were injected simultaneously for 30 min at 10 ml/min. ReSH (2 ml, 80 nM) was purged with 10 ml/min N2 gas bubbling in a 10 ml sealing vial for 30 min to remove O2 from the air. The reaction was initiated by mixing 100 μL of 80 nM ReSH with a gas-saturated solution in a sealed cuvette. The final concentration of NAD+ was varied from 0 to 2 mM.
The enzyme reaction kinetics of RcFDH were measured for NADH-dependent reduction of CO2 to formate in the presence or absence of O2. The sealing cuvette was filled with 900 μL of Kpi buffer containing NADH and sealed; then, 100% CO2 and a mixed gas consisting of 4% O2 and 96% N2 (or 100% N2 for anaerobic conditions) were injected simultaneously for 30 min at 10 ml/min, respectively. RcFDH (2 ml, 2 μM) was purged with 10 ml/min N2 gas bubbling in a 10 ml sealing vial for 30 min to remove O2 from the air. The reaction was initiated by mixing 100 μL of 2 μM RcFDH with a gas-saturated solution in a sealing cuvette. The final concentration of NADH was varied from 0 to 1 mM.
All measurements were performed in triplicate based on the change in the absorbance at 365 nm in the cuvette, measured using a T60 UV-Vis spectrophotometer (PG Instruments Ltd., Lutterworth, UK). The change in absorbance over 1 min was plotted using the Michaelis-Menten equation to calculate the kinetic parameters.
2.7 Formate production and quantification
For the cascade reaction in the presence or absence of O2, the gas content was controlled in a 20 ml polytetrafluoroethylene (PTFE) septa sealing vial. The vials were filled with 500 μL of reaction solution containing 3.2 U/mL ReSH, 0.16 U/mL RcFDH, 1 mM NAD+, and 0.5 M Kpi buffer and sealed. A needle was inserted into the septa for gas evacuation. Then, 10 ml/min CO2 and 20 ml/min N2/O2 mixed gas were injected for 30 min (the needle did not enter the reaction solution). The O2 ratios of the mixed gas varied from 0%–2%–4%; therefore, the final concentrations of O2 were 0, 1, and 2%. The reaction was initiated by a 10 ml/min H2 gas injection. Formate production was sampled every 20 min during incubation for 1 h, and 10 μL of 6 N H2SO4 was added to the 100 μL sample to inactivate the enzymes immediately. Additionally, 240 μL of distilled water was mixed with the sample, and the aggregate enzymes were removed by centrifugation at 13,000 × g. Formate production was quantified by HPLC (1260, Agilent, CA, United States) equipped with a diode-array detector and an Aminex HPX-87H column (BIO-RAD, CA, United States) with a mobile phase of 5 μM H2SO4 at a flow rate of 0.6 ml/min. The retention time of formate was 13.010 min. The formate concentration was calculated using a formate calibration curve (Supplementary Figure S1).
3 Results and discussion
3.1 Preparation of ReSH and RcFDH
ReSH and RcFDH are expressed in R. eutropha and E. coli, respectively. They were purified using affinity resins, as described in the Materials and methods. Five bands of purified ReSH subunits were observed, which matched the expected molecular weights (HoxF, 68,110 Da; HoxH, 54,863 Da; HoxU, 26,173 Da; HoxY, 22,881 Da; HoxI, 18,567 Da) (Figure 2A). Similarly, three bands of purified RcFDH subunits were observed, which were consistent with the expected molecular weights (FdsA, 104,466 Da; FdsB, 52,699 Da; FdsG, 17,304 Da) (Figure 2B). Both enzymes showed high purity. The identity of the purified enzymes was confirmed by MALDI-TOF mass spectrometry. The experimentally determined masses of ReSH subunits were 67,542, 54,492, 26,038, 22,836, and 18,545 m/z, which matched well with the expected masses (68,111, 54,864, 26,174, 22,882, and 18,568 m/z, respectively) with less than 1% deviation (Supplementary Figures S2A–C). The experimentally determined masses of RcFDH subunits were 104,259, 52,385, and 17,136 m/z, which matched well with the expected masses (104,467, 52,700, and 17,305 m/z, respectively) with less than 1% deviation (Supplementary Figures S2D, E). These results showed that the purified ReSH and RcFDH were successfully prepared.
3.2 Enzyme kinetics in the presence or absence of O2
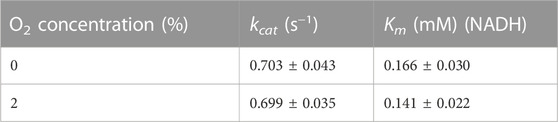
We investigated the enzymatic activities of ReSH and RcFDH in the presence or absence of O2. The NAD+-dependent H2 oxidation reaction rate by ReSH was measured, and the Michaelis-Menten curve was fitted to calculate the kinetic parameters using Origin 2022 program (Figure 3A). Both kcat and Km values of ReSH showed an insignificant difference under the 0% and 5% O2 conditions (Table 1). Similarly, The NADH-dependent CO2 reduction reaction rate by RcFDH was measured, and the Michaelis-Menten curve was fitted to calculate the kinetic parameters (Figure 3B). Likewise, kcat and Km values of RcFDH showed an insignificant difference between the 0% and 2% O2 conditions (Table 2). These results show that purified ReSH and RcFDH retained the enzymatic activity at least under less than 2% O2.
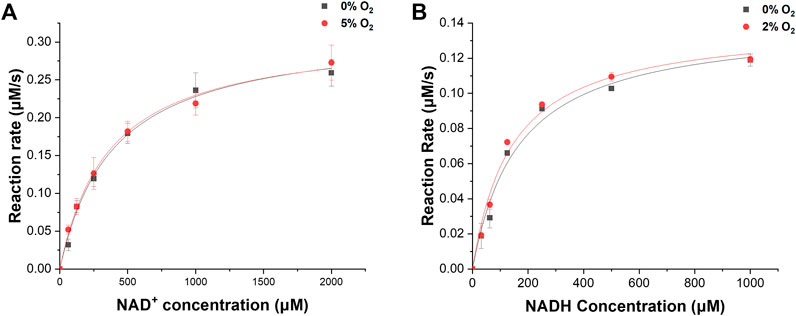
FIGURE 3. Kinetic analysis. (A) NAD+-dependent H2 oxidation of ReSH in the presence of O2, or not. (B) NADH-dependent CO2 reduction of RcFDH in the presence of O2, or not.
3.3 Cascade reaction condition control
We determined the NAD+, ReSH, and RcFDH contents for the cascade reaction of ReSH and RcFDH. Owing to the relatively low kcat value (Tables 1, 2), the rate-determining step was the CO2 reduction by RcFDH. Because the reaction rate of RcFDH was saturated at NADH concentrations above 1 mM (Figure 3B), the NAD+ concentration was determined to be 1 mM. For the continuous CO2 reduction by RcFDH, the concentration of ReSH was determined to maintain a state in which all NAD+ was reduced to NADH. The concentration of RcFDH was fixed at 0.08 U/mL and the amount of ReSH was adjusted to 0, 0.08, 0.8, and 1.6 U/mL (U/mL ratio of ReSH:RcFDH = 0:1, 1:1, 5:1, 10:1, 20:1). Reaction solutions were placed in a 20 ml sealing vial, and 10 ml/min CO2 and 10 ml/min H2 were injected for 1 h simultaneously, after which formate was measured (Supplementary Figure S3). Formate production was not observed in the reaction solution without ReSH. In contrast, substantial formate production was observed in the reaction solution with the three components (ReSH, RcFDH, and NAD+). Formate production was saturated above a 5:1 ratio. At higher ReSH concentrations, NAD+ was immediately converted to NADH through H2 oxidation. This result set the cascade reaction content to 1 mM NAD+, and the U/mL ratio of ReSH:RcFDH = 20:1.
3.4 Formate production under O2 conditions
We demonstrated H2 and CO2 conversion into formate under 0%–2% O2 conditions. ReSH, RcFDH, and 1 mM NAD+ were mixed and placed in a 20 ml sealing vial. Changes in the concentrations of NADH and formate over time were investigated when O2 (at a controlled concentration), H2, and CO2 were simultaneously and continuously injected into the vial. During the injection of the gases, under all O2 conditions from 0% to 2%, NAD+ was reduced to NADH and maintained at 1 mM by H2 oxidation of ReSH (Figure 4A). Furthermore, the formate concentration increased continuously (Figure 4B) owing to the CO2 reduction of RcFDH. Approximately 230 μM of formate was produced after 1 h, which showed a statistically insignificant difference at 0, 1, or 2% O2 conditions (p > 0.05). In order to investigate the O2-tolerant limit of the system, we tested the formate production in a higher concentration of O2 (Supplementary Figure S4). We observed a substantial reduction in formate production at 5% O2 compared to 0%. Therefore, in the specific enzyme systems we chose, the O2-tolerance limit was between 2% and 5%. The O2-tolerance of both H2ase and FDH is attributed to the reduction of O2 bound to the active site of enzymes, leading to the reactivation of active site. Therefore, we speculated that the substantial loss of enzymatic activities at 5% O2 results from that O2 binding to the active site is more favorable than O2 reduction at the active site. These results demonstrate, as hypothesized, the plausibility of a cascade reaction using ReSH and RcFDH, even in the presence of O2. Of course, greater O2-tolerance limit would be beneficial in developing practical processes. We speculate that there are ways to increase the O2-tolerance limit of enzymes. First, the enzyme concentration can be adjusted to increase O2-tolerance limit. O2-tolerance is likely attributed to the reduction mechanism of O2 to either H2O or H2O2. In this case, O2 is a co-substrate of these enzymes. Therefore, if the concentrations of enzymes were sufficiently high, the enzymes would quickly reduce O2, leading to the increased O2-tolerance limit. Another possible approach to increase O2-tolerance is engineering enzyme. Recently it was reported that the simple point mutations in the gas tunnel region of O2-sensitive CO dehydrogenase greatly increased the O2-tolerance limit (Kim et al., 2022). We speculate that such enzyme engineering strategy can be applied to ReSH and RcFDH to increase O2-tolerance limit.
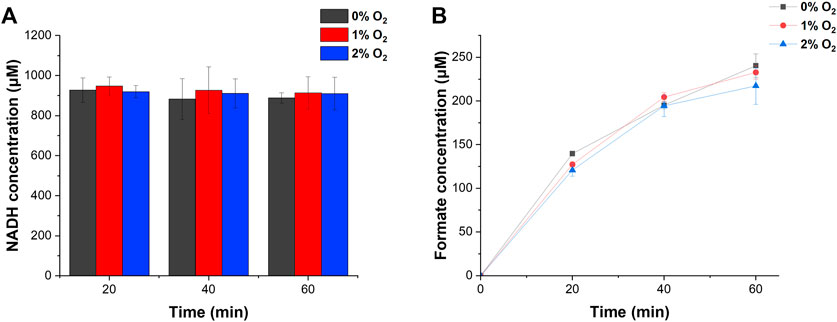
FIGURE 4. H2 and CO2 conversion into formate by NAD+-dependent ReSH and RcFDH cascade reactions. (A) NADH concentration. (B) Formate concentration. All measurements were performed in triplicate.
4 Conclusion
We demonstrated the conversion of H2 and CO2 into formate using an NAD+-dependent cascade reaction of O2-tolerant H2ase and FDH in the presence of O2. However, in order to produce formate using H2 and CO2 obtained from cheap sources, such as by-product gas from steel industries, we may need to tackle other obstacles. For instances, it was reported that H2ases are often damaged by CO (Bagley et al., 1994; Vincent et al., 2007), one of components in by-product gas. We plan to investigate the enzymatic process which is tolerant to both O2 and CO in future. Furthermore, we could not obtain the kinetic parameters for both CO2 and H2 due to difficulty in determining the actual concentration of the gases in the aqueous reaction solution. We plan to determine the kinetic parameters for CO2 and H2 once suitable gas control facilities are in place.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author contributions
JC participated in the design of this study, performed analysis, and drafted the manuscript. HB participated in the analysis. IK was involved in the design and supervision of this study and preparation of the manuscript draft.
Funding
This work was supported by the National Research Foundation of Korea (NRF), the Ministry of Science, and ICT [grant numbers 2021R1A5A1028138].
Acknowledgments
The authors thank Oliver Lenz (Technische Universität Berlin) for providing R.eutropha HF210 [pGE771] cell and Kyoungseon Min (Korea Institute of Energy Research) for providing MC1061 E.coli cell and pTrcHis-RcFDH plasmid.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fbioe.2022.1078164/full#supplementary-material
References
Alissandratos, A., Kim, H.-K., Matthews, H., Hennessy, J. E., Philbrook, A., and Easton, C. J. (2013). Clostridium carboxidivorans strain P7T recombinant formate dehydrogenase catalyzes reduction of CO(2) to formate. Appl. Environ. Microbiol. 79, 741–744. doi:10.1128/AEM.02886-12
Amao, Y. (2018). Formate dehydrogenase for CO2 utilization and its application. J. CO2 Util. 26, 623–641. doi:10.1016/j.jcou.2018.06.022
Appel, A. M., Bercaw, J. E., Bocarsly, A. B., Dobbek, H., DuBois, D. L., Dupuis, M., et al. (2013). Frontiers, opportunities, and challenges in biochemical and chemical catalysis of CO2 fixation. Chem. Rev. 113, 6621–6658. doi:10.1021/cr300463y
Baccour, M., Lamotte, A., Sakai, K., Dubreucq, E., Mehdi, A., Kano, K., et al. (2020). Production of formate from CO2 gas under ambient conditions: Towards flow-through enzyme reactors. Green Chem. 22, 3727–3733. doi:10.1039/D0GC00952K
Bagley, K. A., Van Garderen, C. J., Chen, M., Duin, E. C., Albracht, S. P., and Woodruff, W. H. (1994). Infrared studies on the interaction of carbon monoxide with divalent nickel in hydrogenase from Chromatium vinosum. Biochemistry 33, 9229–9236. doi:10.1021/bi00197a026
Bak, M., Park, J., Min, K., Cho, J., Seong, J., Hahn, Y. S., et al. (2020). Recombinant peptide production platform coupled with site-specific albumin conjugation enables a convenient production of long-acting therapeutic peptide. Pharmaceutics 12, 364. doi:10.3390/pharmaceutics12040364
Burgdorf, T., Lenz, O., Buhrke, T., van der Linden, E., Jones, A. K., Albracht, S. P. J., et al. (2005). [NiFe]-hydrogenases of Ralstonia eutropha H16: Modular enzymes for oxygen-tolerant biological hydrogen oxidation. J. Mol. Microbiol. Biotechnol. 10, 181–196. doi:10.1159/000091564
Choi, E., Electrochem, J., Soc, H., Choi, E., Yeon, Y. J., Min, K., et al. (2018). Communication—CO2 reduction to formate: An electro-enzymatic approach using a formate dehydrogenase from Rhodobacter capsulatus. J. Electrochem. Soc. 165, 446–H448. doi:10.1149/2.0531809jes
Duffus, B. R., Schrapers, P., Schuth, N., Mebs, S., Dau, H., Leimkühler, S., et al. (2020). Anion binding and oxidative modification at the molybdenum cofactor of formate dehydrogenase from Rhodobacter capsulatus studied by X-ray absorption spectroscopy. Inorg. Chem. 59, 214–225. doi:10.1021/acs.inorgchem.9b01613
Eppinger, J., and Huang, K.-W. (2017). Formic acid as a hydrogen energy carrier. ACS Energy Lett. 2, 188–195. doi:10.1021/acsenergylett.6b00574
Fontecilla-Camps, J. C., Volbeda, A., Cavazza, C., and Nicolet, Y. (2007). Structure/function relationships of [NiFe]- and [FeFe]-Hydrogenases. Chem. Rev. 107, 4273–4303. doi:10.1021/cr050195z
García García, S., Rodríguez Montequín, V., Morán Palacios, H., and Mones Bayo, A. (2020). A mixed integer linear programming model for the optimization of steel waste gases in cogeneration: A combined coke oven and converter gas case study. Energies 13, 3781. doi:10.3390/en13153781
Graham, J. E., Niks, D., Zane, G. M., Gui, Q., Hom, K., Hille, R., et al. (2022). How a formate dehydrogenase responds to oxygen: Unexpected O2 insensitivity of an enzyme harboring tungstopterin, selenocysteine, and [4Fe–4S] clusters. ACS Catal. 12, 10449–10471. doi:10.1021/acscatal.2c00316
Hartmann, T., and Leimkühler, S. (2013). The oxygen-tolerant and NAD+-dependent formate dehydrogenase from Rhodobacter capsulatus is able to catalyze the reduction of CO2 to formate. FEBS J. 280, 6083–6096. doi:10.1111/febs.12528
Horch, M., Lauterbach, L., Mroginski, M. A., Hildebrandt, P., Lenz, O., and Zebger, I. (2015). Reversible active site sulfoxygenation can explain the oxygen tolerance of a NAD+-Reducing [NiFe] hydrogenase and its unusual infrared spectroscopic properties. J. Am. Chem. Soc. 137, 2555–2564. doi:10.1021/ja511154y
Kim, S., Kim, M., Jung, S., Kwon, K., Park, J., Kim, S., et al. (2019). Co-delivery of therapeutic protein and catalase-mimic nanoparticle using a biocompatible nanocarrier for enhanced therapeutic effect. J. Control. release Off. J. Control. Release Soc. 309, 181–189. doi:10.1016/j.jconrel.2019.07.038
Kim, S., Kwon, K., Tae, G., and Kwon, I. (2021). Nano-entrapping multiple oxidoreductases and cofactor for all-in-one nanoreactors. ACS Sustain. Chem. Eng. 9, 6741–6747. doi:10.1021/acssuschemeng.1c00843
Kim, S. M., Lee, J., Kang, S. H., Heo, Y., Yoon, H.-J., Hahn, J.-S., et al. (2022). O2-tolerant CO dehydrogenase via tunnel redesign for the removal of CO from industrial flue gas. Nat. Catal. 5, 807–817. doi:10.1038/s41929-022-00834-y
Kuehnel, M. F., Wakerley, D. W., Orchard, K. L., and Reisner, E. (2015). Photocatalytic formic acid conversion on CdS nanocrystals with controllable selectivity for H2 or CO. Angew. Chem. Int. Ed. 54, 9627–9631. doi:10.1002/anie.201502773
Laukel, M., Chistoserdova, L., Lidstrom, M. E., and Vorholt, J. A. (2003). The tungsten-containing formate dehydrogenase from Methylobacterium extorquens AM1: Purification and properties. Eur. J. Biochem. 270, 325–333. doi:10.1046/j.1432-1033.2003.03391.x
Lauterbach, L., and Lenz, O. (2013). Catalytic production of hydrogen peroxide and water by oxygen-tolerant [NiFe]-Hydrogenase during H2 cycling in the presence of O2. J. Am. Chem. Soc. 135, 17897–17905. doi:10.1021/ja408420d
Lee, D.-Y., and Elgowainy, A. (2018). By-product hydrogen from steam cracking of natural gas liquids (NGLs): Potential for large-scale hydrogen fuel production, life-cycle air emissions reduction, and economic benefit. Int. J. Hydrogen Energy 43, 20143–20160. doi:10.1016/j.ijhydene.2018.09.039
Lenz, O., Lauterbach, L., and Frielingsdorf, S. (2018). “Chapter Five - O2-tolerant [NiFe]-hydrogenases of Ralstonia eutropha H16: Physiology, molecular biology, purification, and biochemical analysis,” in Enzymes of energy Technology. Editors F. B. T. -M., and E. Armstrong (Academic Press), 117–151. doi:10.1016/bs.mie.2018.10.008
Lepage, T., Kammoun, M., Schmetz, Q., and Richel, A. (2021). Biomass-to-hydrogen: A review of main routes production, processes evaluation and techno-economical assessment. Biomass Bioenergy 144, 105920. doi:10.1016/j.biombioe.2020.105920
Li, W., He, S., and Li, S. (2019). Experimental study and thermodynamic analysis of hydrogen production through a two-step chemical regenerative coal gasification. Appl. Sci. 9, 3035. doi:10.3390/app9153035
Loges, B., Boddien, A., Junge, H., and Beller, M. (2008). Controlled generation of hydrogen from formic acid amine adducts at room temperature and application in H 2/O 2 fuel cells. Angew. Chem. Int. Ed. 4, 3962–3965. doi:10.1002/anie.200705972
Lubitz, W., Ogata, H., Rüdiger, O., and Reijerse, E. (2014). Hydrogenases. Chem. Rev. 114, 4081–4148. doi:10.1021/cr4005814
McDowall, J. S., Murphy, B. J., Haumann, M., Palmer, T., Armstrong, F. A., and Sargent, F. (2014). Bacterial formate hydrogenlyase complex. Proc. Natl. Acad. Sci. U. S. A. 111, E3948–E3956. doi:10.1073/pnas.1407927111
Mihet, M., Dan, M., Barbu-Tudoran, L., Lazar, M. D., and Blanita, G. (2020). Controllable H2 generation by formic acid decomposition on a novel Pd/templated carbon catalyst. Hydrogen 1, 22–37. doi:10.3390/hydrogen1010003
Moon, M., Park, G. W., Lee, J., Lee, J.-S., and Min, K. (2020). Recent progress in formate dehydrogenase (FDH) as a non-photosynthetic CO2 utilizing enzyme: A short review. J. CO2 Util. 42, 101353. doi:10.1016/j.jcou.2020.101353
Niks, D., and Hille, R. (2018). “Chapter Eleven - reductive activation of CO2 by formate dehydrogenases,” in Enzymes of energy Technology methods in enzymology. Editor F. Armstrong (Academic Press), 277–295. doi:10.1016/bs.mie.2018.10.013
Pandelia, M.-E., Infossi, P., Giudici-Orticoni, M. T., and Lubitz, W. (2010). The oxygen-tolerant hydrogenase I from Aquifex aeolicus weakly interacts with carbon monoxide: An electrochemical and time-resolved FTIR study. Biochemistry 49, 8873–8881. doi:10.1021/bi1006546
Ping, Y., Yan, J.-M., Wang, Z.-L., Wang, H.-L., and Jiang, Q. (2013). Ag0.1-Pd0.9/rGO: An efficient catalyst for hydrogen generation from formic acid/sodium formate. J. Mat. Chem. A 1, 12188–12191. doi:10.1039/C3TA12724A
Pinske, C., and Sargent, F. (2016). Exploring the directionality of Escherichia coli formate hydrogenlyase: A membrane-bound enzyme capable of fixing carbon dioxide to organic acid. Microbiologyopen 5, 721–737. doi:10.1002/mbo3.365
Qiu, S., Olsen, S., MacFarlane, D. R., and Sun, C. (2018). The oxygen reduction reaction on [NiFe] hydrogenases. Phys. Chem. Chem. Phys. 20, 23528–23534. doi:10.1039/C8CP04160A
Sargent, F. (2016). “Chapter eight - the model [NiFe]-Hydrogenases of Escherichia coli,” in Advances in bacterial electron transport systems and their regulation. Editors M. P. Poole (Academic Press), 433–507. doi:10.1016/bs.ampbs.2016.02.008
Schwarz, F. M., Schuchmann, K., and Müller, V. (2018). Hydrogenation of CO2 at ambient pressure catalyzed by a highly active thermostable biocatalyst. Biotechnol. Biofuels 11, 237. doi:10.1186/s13068-018-1236-3
Shafaat, H. S., Rüdiger, O., Ogata, H., and Lubitz, W. (2013). [NiFe] hydrogenases: A common active site for hydrogen metabolism under diverse conditions. Biochim. Biophys. Acta - Bioenerg. 1827, 986–1002. doi:10.1016/j.bbabio.2013.01.015
Sokol, K. P., Robinson, W. E., Oliveira, A. R., Zacarias, S., Lee, C.-Y., Madden, C., et al. (2019). Reversible and selective interconversion of hydrogen and carbon dioxide into formate by a semiartificial formate hydrogenlyase mimic. J. Am. Chem. Soc. 141, 17498–17502. doi:10.1021/jacs.9b09575
Song, H., Luo, S., Huang, H., Deng, B., and Ye, J. (2022). Solar-driven hydrogen production: Recent advances, challenges, and future perspectives. ACS Energy Lett. 7, 1043–1065. doi:10.1021/acsenergylett.1c02591
Sordakis, K., Tang, C., Vogt, L. K., Junge, H., Dyson, P. J., Beller, M., et al. (2018). Homogeneous catalysis for sustainable hydrogen storage in formic acid and alcohols. Chem. Rev. 118, 372–433. doi:10.1021/acs.chemrev.7b00182
Vincent, K. A., Parkin, A., and Armstrong, F. A. (2007). Investigating and exploiting the electrocatalytic properties of hydrogenases. Chem. Rev. 107, 4366–4413. doi:10.1021/cr050191u
Volbeda, A., Martin, L., Cavazza, C., Matho, M., Faber, B. W., Roseboom, W., et al. (2005). Structural differences between the ready and unready oxidized states of [NiFe] hydrogenases. J. Biol. Inorg. Chem. JBIC a Publ. Soc. Biol. Inorg. Chem. 10, 239–249. doi:10.1007/s00775-005-0632-x
Wang, J., and Yin, Y. (2018). Fermentative hydrogen production using pretreated microalgal biomass as feedstock. Microb. Cell. Fact. 17, 22. doi:10.1186/s12934-018-0871-5
Keywords: hydrogen, carbon dioxide, formate, oxygen-tolerant, hydrogenase, formate dehydrogenase
Citation: Cha J, Bak H and Kwon I (2023) Hydrogen-fueled CO2 reduction using oxygen-tolerant oxidoreductases. Front. Bioeng. Biotechnol. 10:1078164. doi: 10.3389/fbioe.2022.1078164
Received: 24 October 2022; Accepted: 22 December 2022;
Published: 05 January 2023.
Edited by:
Ning Li, South China University of Technology, ChinaReviewed by:
Hongling Shi, Nanyang Normal University, ChinaFeng Cheng, Zhejiang University of Technology, China
Copyright © 2023 Cha, Bak and Kwon. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Inchan Kwon, aW5jaGFuQGdpc3QuYWMua3I=
 Jaehyun Cha
Jaehyun Cha Hyeonseon Bak1
Hyeonseon Bak1 Inchan Kwon
Inchan Kwon