- 1Helmholtz International Lab for Anti-infectives, Shandong University-Helmholtz Institute of Biotechnology, State Key Laboratory of Microbial Technology, Shandong University, Qingdao, China
- 2State Key Laboratory of Genetic Engineering, School of Life Sciences, Fudan University, Shanghai, China
Antimicrobial resistance is one of the most serious public health issues in the worldwide and only a few new antimicrobial drugs have been discovered in recent decades. To overcome the ever-increasing emergence of multidrug-resistant (MDR) pathogens, discovery of new natural products (NPs) against MDR pathogens with new technologies is in great demands. Lanthipeptides which are ribosomally synthesized and post-translationally modified peptides (RiPPs) display high diversity in their chemical structures and mechanisms of action. Genome mining and biosynthetic engineering have also yielded new lanthipeptides, which are a valuable source of drug candidates. In this review we cover the recent advances in the field of microbial derived lanthipeptide discovery and development.
Introduction
With the emergence of multidrug-resistant (MDR) pathogens, antimicrobial resistance for many clinically-used antibiotics, even the most famous daptomycin, has been reported (Heidary et al., 2018). An increasing and urgent demand for new antibiotics arises clinically. Nevertheless, the frequent rediscovery of known compounds triggered a drastic decrease in new antibiotic detection using traditional drug discovery pipelines and only around 30 antibiotics were approved as new drugs during the last two decades (Weber, 2016; Butler et al., 2017).
Serving as the last line of defense against MDR pathogens, most of peptide antibiotics target highly stable cell walls or cell membranes of bacteria and it is widely considered not easy to develop antimicrobial resistance for peptide antibiotics. Several new important antibiotics reported in recent years, such as cadasides, malacidins and teixobactin, are all peptides with defense capability against a variety of MDR bacteria and without antimicrobial resistance by far (Ling et al., 2015; Guo et al., 2018; Hover et al., 2018; Wu et al., 2019).
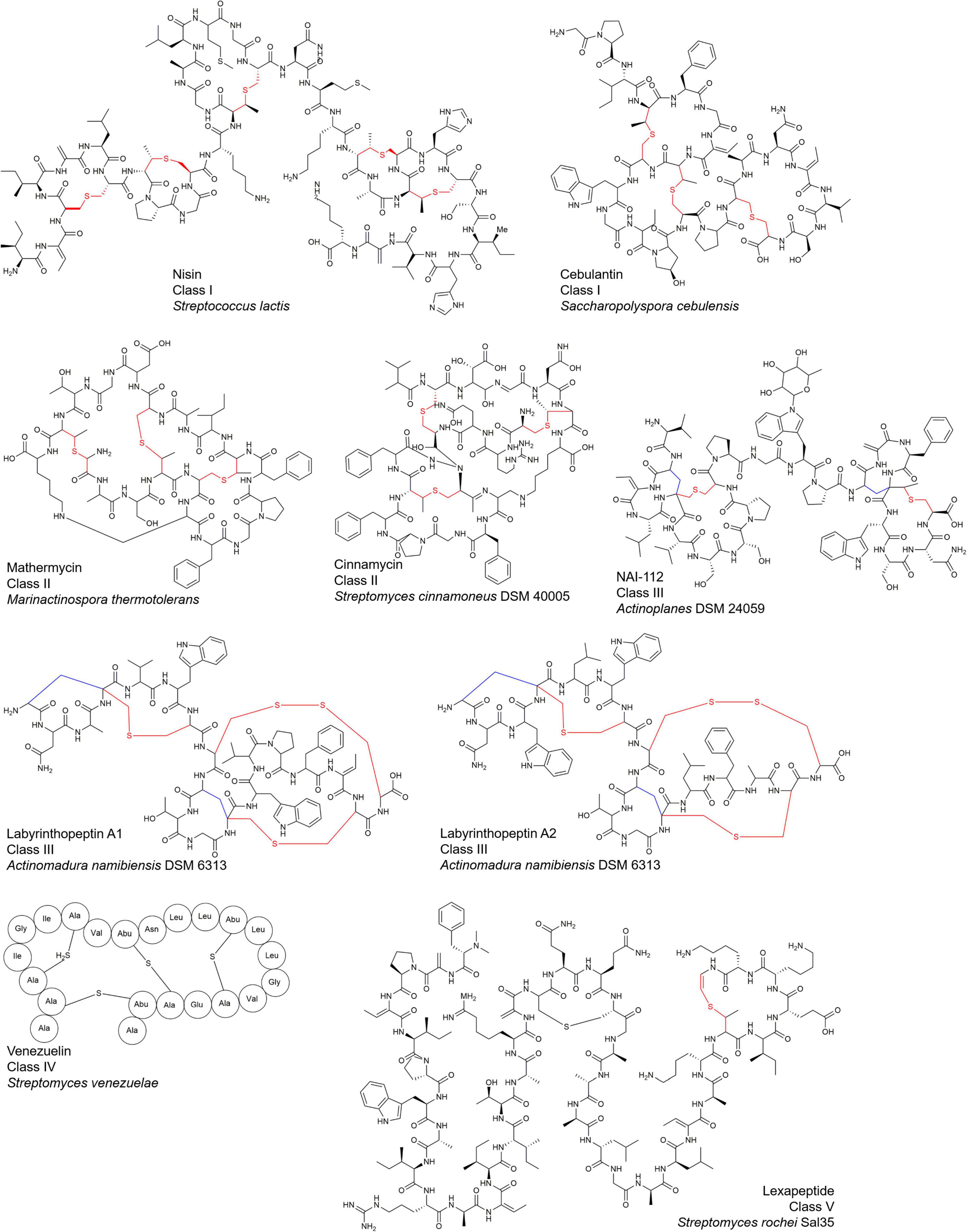
Lanthipeptides, the largest sub-family of ribosomally synthesized and post-translational modification modified peptides (RiPPs) (Arnison et al., 2013), are characterized by the presence of multiple lanthionine (Lan) or (methyl-) lanthionine rings (MeLan) to form thioether bonds (Figure 1). Most of them possess multiple antibacterial mechanisms, endowing them with a high potential in the development of anti-infective drugs (Mo et al., 2016). Nisin (Figure 1), as the first-identified lanthipeptide containing 34 amino acid residues, has been used as a preservative food additive for over 60 years in more than 80 countries, but so far, few nisin-resistant bacteria have been found (Mo et al., 2016; van Staden et al., 2021).
Lanthipeptide biosynthetic gene clusters (BGCs) are widespread within the genomes of microorganisms, providing a substantial repository for novel bioactive peptides. In order to utilize lanthipeptide BGCs, especially, those silent or not accessible in native producers (Zhang et al., 2015), genome-mining (genome-based predication and functional identification of BGCs) has become a commonly used technique in recent years to explore lanthipeptide-derived drugs (Huo and van der Donk, 2016; Chu et al., 2020; Montalbán-López et al., 2021).
Structures, Microorganism Producers and Bioactivities of Lanthipeptides
Since the discovery of nisin in 1928, about a little more than one hundred of lanthipeptides have been reported from microorganisms, and they display complex and highly diverse structures and bioactivities (Figure 1; van Staden et al., 2021).
Structural Diversity of Lanthipeptides
Significant difference in peptide chain lengths, amino acid composition of primary sequences, location of thioether bonds, types of unnatural amino acids, and other various structural modification generated a high degree of structural diversity of lanthipeptides:
Lanthipeptides contain varying numbers of characteristic lanthionine (Lan) or methyllanthionine (MeLan) residues (Figure 1), both of which contain a characteristic thioether bond (thioether ring or thioether bridge) formed by linking dehydrated serine or threonine residues (Ser/Thr) to the thiol group of cysteine residues (Cys). The topology of some lanthipeptides containing multiple thioether rings exhibits either linear forms where thioether rings are non-interlaced spatially or complex interlaced ring patterns. Thioether bonds create more stable structures with improved pharmacodynamic properties by protecting peptides from proteolytic degradation (Mo et al., 2016). Besides Lan and MeLan, most lanthipeptides also contain two unnatural amino acid residues including dehydrobutyrine (Dhb) and dehydroalanine (Dha) from dehydrated serine or threonine residues (Ser/Thr).
Furthermore, many lanthipeptides contain special AviCys (S-[(Z)-aminovinyl]-D- cysteine) structures at the C-terminus (Mo et al., 2016, 2019; Lagedroste et al., 2020; Figures 1, 2). The tailoring structural modification further increase structural diversity of lanthipeptides, such as glycosylation, hydroxylation, halogenation, decarboxylation, acylation, disulfide bond formation, and α-C configuration transformation on some specific amino acid residues (Mo et al., 2016; Figure 2A).
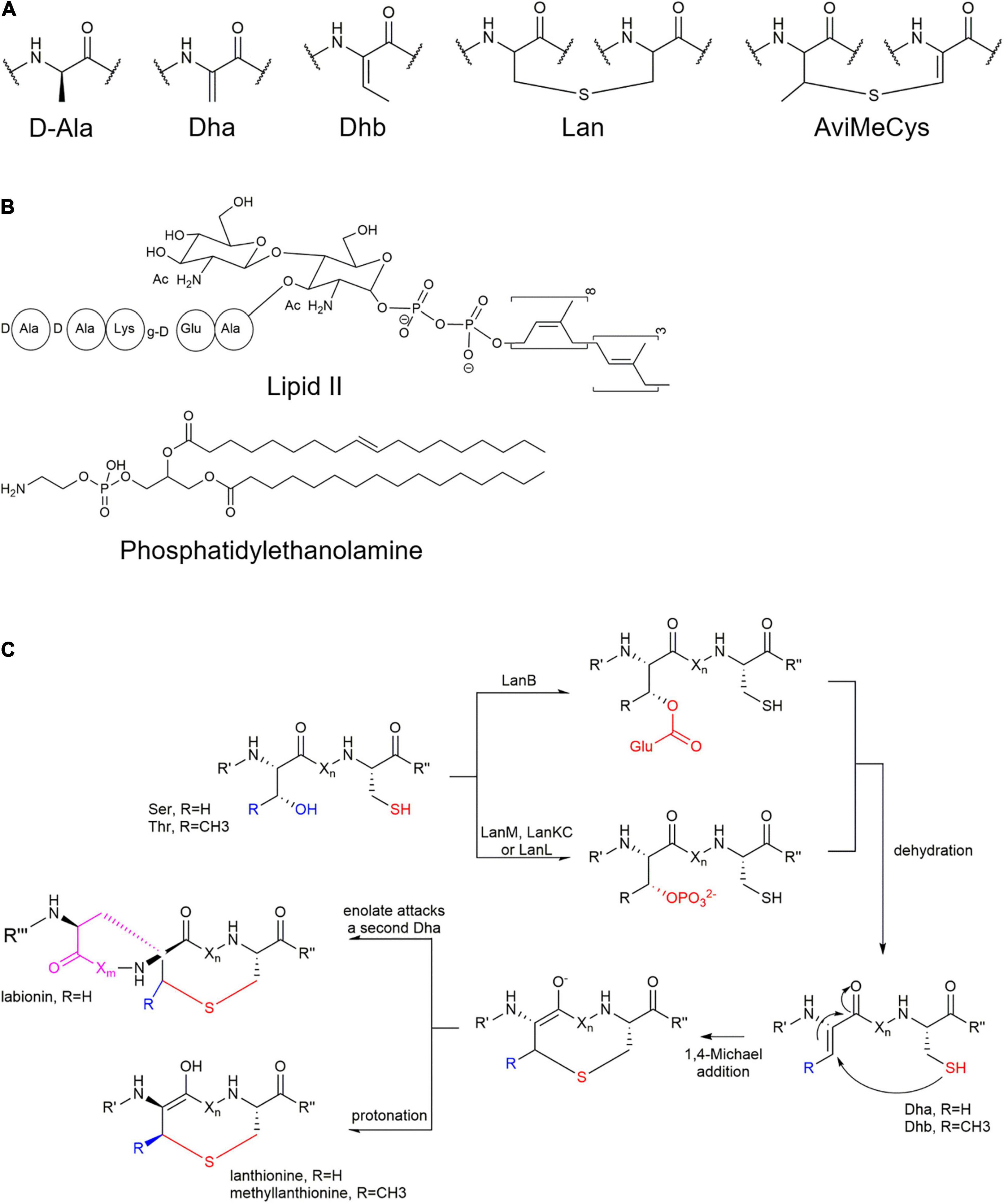
Figure 2. Formation of unusual amino acid residues and action targets of lanthipeptides. (A) Unusual amino acid residues in the lanthipeptides. (B) Two main action targets of lanthipeptides. (C) Formation of characteristic lanthionine (Lan) or methyllanthionine (MetLan). Firstly, Ser/Thr-OH is dehydrated and followed by Cys-SH nucleophilic attack and cyclization. In class I lanthipeptide, Ser/Thr-OH is activated by glutamination at the presence of tRNAGlu, followed by deglutamination and dehydration while in class II-IV, Ser/Thr-OH is phosphorylated by LanM/LanKC/LanL and then undergoes dephosphorylation and dehydration. In addition, the labionin in the class III structure is formed by the nucleophilic attack of another Dha enol anion (Ren et al., 2020). It is speculated that class V is also dehydrated through phosphorylation and dephosphorylation mechanisms.
These structural features determine the structural diversity and biological activity of lanthipeptides (Biswas et al., 2017; van Staden et al., 2021; Figures 1, 2).
Microorganism Producers of Lanthipeptides
Lanthipeptides have been found to accumulate in a variety of microorganisms, and bacteria remain to be the major producers for lanthipeptides (Figure 1 and Table 1).
Actinobacteria: Lanthipeptides were identified with the majority in Actinobacteria (Gomes et al., 2017) and genome sequencing revealed that lanthipeptide BGCs are extremely abundant in the genomes of Actinobacteria (Vikeli et al., 2020; Walker et al., 2020). Several examples reported in recent years include lexapeptide isolated from Streptomyces rochei Sal35 (Xu et al., 2020), roseocin isolated from Streptomyces roseosporus (Singh et al., 2020), mathermycin isolated from Marinactinospora thermotolerans (Chen et al., 2017), cebulantin derived from Saccharopolyspora cebuensis (Moon et al., 2019), microbisporicin obtained from Microbisporus corallina (Fernández-Martínez et al., 2015) and so on.
Firmicutes: Firmicutes are also a large source for lanthipeptides, among which, Bacillus species were identified as notably producers. As an example, bicereucin was produced by Bacillus cereus (Huo and van der Donk, 2016). Nisin was firstly isolated from Streptococcus lactis, and its analogs have been found in many kinds of bacteria. Recently, it has been discovered that nisin analogs could be derived from Staphylococcus capitis residing in human skins (O’Sullivan et al., 2020).
Bacteroides: wide distribution of lanthipeptides was also observed in Bacteroides (Walker et al., 2020). For instance, abundant lanthipeptide BGCs have been found in the genomes of Chryseobacterium, Pedobacter and Flavobacterium. Multiple lanthipeptides including pinensins have been found in Chitinophaga pinensis (Caetano et al., 2020).
Besides general bacteria, cyanobacteria and even archaea also are attracting more attentions in recent years as potential producers of lanthipeptides. Exemplified are prochlorosins, a group of lanthipeptides, which were produced by certain strains of the ubiquitous marine picocyanobacteria, like species in Prochlorococcus and Synechococcus. And picocyanobacteria are found to produce remarkably thousands of different cyclic peptides, few of which would display similar ring topologies (Cubillos-Ruiz et al., 2017). Landornamide A synthesized by a silent by BGC from a cyanobacterium Kamptonema sp. PCC 6506 was identified via pathway reconstruction in E. coli (Bösch et al., 2020). Archaea might be producers for lanthipeptides because a high number of lanthipeptides gene clusters were detected (Walker et al., 2020).
High Diversity of Bioactivities of Lanthipeptides
Most of known lanthipeptides have antibacterial activity, especially against MDR strains, while some have antiviral, antitumor, antifungal, and immunomodulatory properties and some display the potential to alleviate cystic fibrosis symptoms. Other activities, such as morphogenetic and antinociceptive actions, have also been reported (van Staden et al., 2021; Table 1).
For instance, microbisporicin, NAI-107, mersacidin and penisin exhibit inhibition against antibiotic-resistant pathogens, such as methicillin-resistant Staphylococcus aureus (Baindara et al., 2018). Cinnamycin could kill Bacillus and also has antifungal ability (Chen et al., 2017; Vestergaard et al., 2019). Cebulantin is effective against the growth of various Vibrio pathogens (Moon et al., 2019). Deoxyactagardine B derivatives have entered clinical trials as antibacterial infection drugs. Nisin, as the first food additive, has a strong inhibitory effect on many G+ bacteria including Streptococcus pneumoniae, and also has certain inhibitory activity on some G– bacteria (Ahmadi et al., 2017; Li et al., 2018). However, compared to the effects against G+ bacteria, most of lanthipeptides show relatively weak inhibitory activity against G– bacteria (Liu et al., 2020).
In addition to antibacterial activity, nisin has inhibitory effects on head-neck squamous cell carcinoma (Ahmadi et al., 2017). Mathermycin shows inhibitory activity on non-small cell lung cancer and other tumors (Cheng et al., 2021). Bicereucin possesses hemolytic activity (Huo and van der Donk, 2016). Labyrinthopeptin is effective against a variety of viral infections caused by HIV, hepatitis C virus, herpes simplex virus, and respiratory syncytial virus (Prochnow et al., 2020). NAI-112 has been proved to reduce inflammation and relieve pain (Iorio et al., 2014). Duramycin is particularly effective in the treatment of cystic fibrosis, and some lanthipeptides have other properties such as lowering blood pressure or regulating immunity (Ahmadi et al., 2017).
Therapeutic Action Mechanisms of Lanthipeptides
It has been proposed that the target affinity of lanthipeptides is of great importance and crucial for their therapeutic potential (van Staden et al., 2021).
Most lanthipeptides exhibit antibacterial activity by binding to lipid II, an essential intermediate in peptidoglycan biosynthesis (Figure 2B) to result in blocking of cell wall biosynthesis and disruption of the cell membrane integrity. Taking nisin as an example, compared to vancomycin, nisin has a different binding site with lipid II. The two thioether rings of nisin can bind to the pyrophosphate group of lipid II to form a cage-like complex. In addition, its binding with lipid II leads to the formation of a polymerized complex, which causes cell membrane perforation and electrolyte outflow (Dickman et al., 2019). The essential role of cyclic structures of nisin for lipid II-binding proposed that such constrained structures caused by thioether rings imposed on lanthipeptides is suitable or required for target binding (Bosma et al., 2019). As one of the peptidoglycan precursors, the target lipid II is not prone to mutate to reduce resistance to lanthipeptides.
Many lanthipeptides exert bioactivity by recognizing and combining phosphatidylethanolamine which is an important component of membrane structures in for instance cell membranes and viral envelopes (Figure 2B). The antiviral labyrinthopeptin A1 was proved to bind to the specific motif of phosphatidylethanolamines in the virus envelope, leading to the lysis of the virus and interfering with the structural integrity of the virus envelope (Blockus et al., 2020; Prochnow et al., 2020). Cinnamycin with bactericidal activity can target the primary amino group of phosphatidylethanolamine on the bacterial cell membrane to form a hydrogen-bonding complex. This complex could also specifically bind to the phosphate group of phosphatidylethanolamines (Vestergaard et al., 2019). Since the expression of phosphatidylethanolamines on the inner and outer surfaces of tumor cell membranes is abnormally abundant, mathermycin is able to interfere with the metabolic activity of tumor cells and induce tumor cell necrosis by targeting phosphatidylethanolamines (Cheng et al., 2021; van Staden et al., 2021).
The two-component synergistic mechanism is a special mode for certain lanthipeptides. Unlike most lanthipeptides exerting their biological activity through a single peptide chain component, the bioactivity of a small but increasing number of lanthipeptides relies on synergistic action of two components in a 1:1 ratio while the single component has no or only very weak activity. Taking haloduracin α and haloduracin β from alkalophilic Bacillus as an example, two peptide chains and lipid II are combined in a 2:2:1 ratio to form a complex, which inhibits cell wall biosynthesis and mediates cell membrane perforation (McClerren et al., 2006; Oman et al., 2011). Bicereucin, roseocin and lacticin 3147 also showed synergistic antibacterial activity against G+ bacteria at a ratio of 1:1 in a two-component mode, probably by targeting the formation of the cell wall (Martin et al., 2004; Huo and van der Donk, 2016; Singh et al., 2020). It has been revealed recently that combination between labyrinthopeptin A1/A2 could significantly improve its antiviral activity (Figure 1; Blockus et al., 2020).
It is noteworthy that some lanthipeptides could target other targets. The sugar transport system (sugar PTS proteins) on the cell membranes is also the target for some lanthipeptides, which display cell wall lysis activity by activating N-acetyl-1-alanine amidase and N-acetylglucosaminidase. Furthermore, some lanthipeptide antibiotics are capable of binding to cell mitochondrial membranes, leading to cell autophagy (Zhang and Liu, 2013; Sandiford, 2019), and some lanthipeptides, such as nisin and subtilin, were found to covalently modify the targets on the spore wall of Bacillus to inhibit spore germination. In recent years, it has also been reported that nisin may interfere with DNA replication, recombination and repair in E. coli, though the mechanism has yet to be elucidated (Galván Márquez et al., 2020).
Biosynthesis Mechanism and Classification of Lanthipeptides
The biosynthesis of lanthipeptides were generally divided into four stages (Figures 2, 3): (i) synthesis of small molecule precursor peptides directed by ribosomes; (ii) formation of thioether rings (dehydration and ring formation); (iii) other post-translational modifications, and (iv) removal of the N-terminal leader peptides. The formation of the thioether ring requires the presence of the leader peptides (Figure 2C and Figure 3A). The common enzymes encoded by lanthipeptide BGCs include the precursor peptide synthetase (LanA), key enzymes (such as LanB, LanC and so on) for dehydration and cyclization to catalyze the formation of the thioether rings (Figure 2C), the protease (LanP) for cleaving the leader peptide and LanT for peptide transport (Figure 2C). In addition, some lanthipeptide BGCs also encode some special enzymes for tailoring modification, such as glycosylase, oxidase, methyl transferase and decarboxylase. According to the different key enzymes that catalyze dehydration and cyclization required for the synthesis of thioether rings, lanthipeptides can be divided into five classes at present (Figures 1–3).
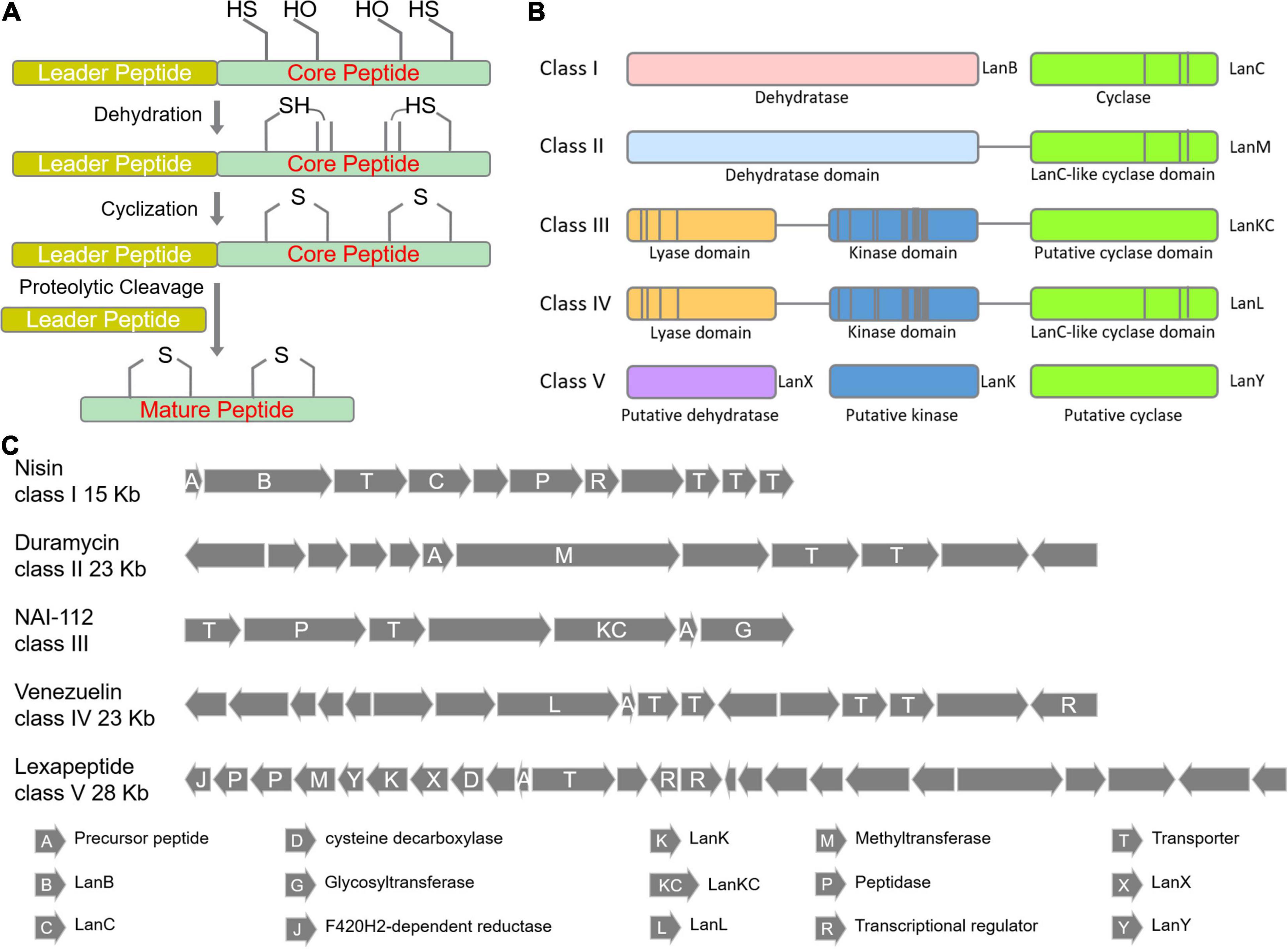
Figure 3. Biosynthetic pathways of lanthipeptides. (A) The general steps of lanthipeptide biosynthesis. (B) Five classes of key enzymes for the formation of thioether ring. (C) Representative BGCs for lanthipeptide biosynthesis.
As shown in Figure 3B and Figure 3C, lanthipeptides in Class I and Class II are more frequently identified, most of which show antibacterial properties. Both dehydratase LanB and cyclase LanC are single-functional separate enzymes for class I while LanM has dual functions for dehydration and cyclization for class II. LanB for class I is characterized by its role to activate serine and threonine residues by glutamylation in a tRNA-dependent manner (Zhang and Liu, 2013; Figure 2C). LanKC for class III and LanL for class IV have different key motif regions, though both of them are multifunctional enzymes and catalyze dehydration using dedicated kinase and lyase domains to perform similar functions (cleavage, phosphorylation and cyclization) (Figure 2C; Müller et al., 2013). Unlike lanthipeptides in class I and II, class III and IV lanthipeptides are quite rare, most of which possess other actions than antibacterial activity (Goto et al., 2010). Lexapeptide A as first member in class V by far was identified in 2020 (Figure 1). Its BGC encodes three single-functional key enzymes responsible for phosphorylation, dehydration and cyclization respectively, remarkably different from class I-IV (Xu et al., 2020; Figure 2C). Deciphering of NCBI Datasets for these key enzymes encoded by class V BGCs demonstrated a high degree of diversity in their structures and catalytic mechanisms. Their classification might be far more complex than those in class I-IV, implying the rich structural and biosynthetic diversity of class V lanthipeptides in nature (Walker et al., 2020; Xu et al., 2020).
A structural view on the maturation of lanthipeptides concerning the mechanisms of these key enzymes (class I-IV) was well reviewed (Lagedroste et al., 2020).
Compared to relatively clear biosynthetic mechanisms of key enzymes in class I-II, those in class III-V possess some unusual features yet to be elucidated in the terms of structure and biosynthesis.
The following are three typical biosynthetic enzymatic systems newly reported for insight into comparatively rare class III-V lanthipeptides:
(i) Class III: NAI-112 contains two labionin macrocyclic structures. Labionin is a characteristic feature for class III lanthipeptides and its formation is mediated by three amino acid residues (Ser-Ser/Thr-Cys) which undergo dehydration and cyclization by two steps of Michael addition reactions to form a thioether ring and a carbocyclic ring (Figure 2C; Müller et al., 2013). The formation of such labionin macrocycles in NAI-112 is catalyzed by multifunctional AplKC. A rare N-deoxyhexose glycosylation was identified on Try residue of NAI-112 and completed by N-glycosyltransferase AplG. AplP, as a broad-substrate-spectrum protease with dual functions, is responsible for the cleavage of the leader peptide of NAI-112 (Iorio et al., 2014; Chen et al., 2019; Sheng et al., 2020; Figure 3C).
(ii) Class IV: Modified-SflA isolated from Streptomyces contains two non-interlaced thioether rings. Although its BGC contains no aminopeptidase gene, it was found that an unknown aminopeptidase gene outside of its BGC can assist the excision of the leader peptide. The cyclization process was revealed to occur simultaneously at multiple sites of its core peptide (Ren et al., 2020).
(iii) Class V: Lexapeptide has a thioether ring formed by three independent modification enzymes, LxmK, LxmX and LxmY. It also contains several rare amino acid residues, such as N-terminal (N, N)-dimethyl-Phe and C-terminal (2-aminovinyl)-3-methyl-Cys and D-Ala. The formation of its rare D-Ala was catalyzed by a novel coenzyme F420-dependent oxidoreductase LxmJ, which is responsible for the reduction of dehydrated alanine to form this unnatural amino acid (Xu et al., 2020; Figure 3C).
Mining and Biosynthetic Reconstruction of New Lanthipeptides
Though the number of lanthipeptides uncovered is still small, they are attracting more attentions in solving MDR problem. Analysis of expanding bacterial genome sequencing data revealed that a great wealth of lanthipeptide BGCs present in microorganisms have become important resources for exploration of new lanthipeptides, despite of that a majority of these BGCs are silent, expressed inefficiently in native strains, or present in unculturable strains or in metagenomic samples (Blin et al., 2014).
Genome-mining has become a promising solution for NP discovery and broadened the concept of NP producers. It involves (i) predication in silico, (ii-a) cloning and heterologous expression or (ii-b) activation in situ of previously uncharacterized BGCs, followed by experimental identification of the products of the gene clusters and further (iii) biosynthetic pathway engineering to produce derivative products with higher productivity or improved therapeutic actions (Figure 3; Abbasi et al., 2020).
In silico Analysis of Lanthipeptides BGCs
Various bioinformatics methods such as NCBI BLAST, antiSMASH, RiPPMiner, PRISM, RiPP-PRISM, RODEO, BAGEL, Natural product peptidogenomics (NPP) and RiPPquest, have been developed for lanthipeptide discovery (An and van der Donk, 2020).
Among them, RODEO (Rapid ORF Description and Evaluation Online) and antiSMASH (antibiotics and secondary metabolite analysis shell) are most commonly used tools to identify BGCs via large-scale mining of RiPP data (Tietz et al., 2017).
An expanded RODEO was used to search the RefSeq database, 8405 precursors peptide synthase (LanA) were predicted in silico successfully, though lanA identification has been a challenge due to their variability and small size often not annotated as genes. By searching non-redundant RefSeq database (release 93) with LanC domain shared in all known classes of lanthipeptides and BGC categorization, plenty of lanthipeptide BGCs were identified, including 2753 putative class I lanthipeptide BGCs, 3708 class II BGCs, 2377 class III BGCs, 815 class IV BGCs. Application of this approach revealed that lanthipeptide BGCs are distributed widely across bacterial phyla (Tietz et al., 2017).
antiSMASH is a use-friendly tool for mining of lanthipeptide BGCs by simply submitting genome sequences into this NP BGC analytic platform.1 Many lanthipeptide BGCs have been identified using antiSMASH (Arnison et al., 2013; Blin et al., 2014). The current version of the antiSMASH database contains annotations for 6200 full bacterial genomes and 18,576 bacterial draft genomes (Blin et al., 2019a,b), although antiSMASH does not distinguish between class III and class IV BGCs.
Functional Identification of Lanthipeptide BGCs
Generally, cultural condition optimization still seems quite useful for a supportive and first- trial method to accelerate accumulation of lanthipeptide of interest (Li et al., 2010; Cubillos-Ruiz et al., 2017). Otherwise, BGC direct capture or pathway reconstruction and heterologous expression or in situ activation are required in most of cases (Table 2).
To capture the whole lanthipeptide BGCs, TAR (transformation-assisted recombination) and Recombineering (Red/ET-mediated recombination engineering) have been developed into most commonly-used cloning techniques. Based on the indigenous yeast recombinases, TAR is capable of one-step acquisiting NP BGCs larger than 50 kb in size from microorganism genomes, or used for reconstruction of BGCs from the environmental DNA libraries. Recombineering based on phage recombinases Red/ET displays advantageous potential in mining of NP BGCs in size up to 106 kb (Wang et al., 2018; Abbasi et al., 2020). Recently, combining CRISPR/Cas9-derived systems with TAR or Recombineering has proved to improve significantly the efficiency in direct cloning, refactoring and heterologous expression of NP BGCs (Wang et al., 2018; Figure 4).
Using these strategies in recent years, some new lanthipeptides have been identified by cloning or reconstructing their biosynthetic pathways and expressing them in heterologous hosts, while some lanthipeptides were identified by inserting strong promoters into the genome in the original bacteria to activate the silent BGCs of these compounds (van Heel et al., 2016; Bobeica and van der Donk, 2018; Hegemann and van der Donk, 2018; Huo et al., 2019; Bösch et al., 2020; Sandiford, 2020; Table 2).
In addition, some high-throughput genome-mining strategies have been established for discovery of lanthipeptides, as demonstrated, for instance, by LEXAS (library expression analysis system). LEXAS was used to successfully identify Streptomyces-derived lexapeptide as the first example in class V lanthipeptide, highlighted with high-throughput mobilization of BAC clones into Streptomyces hosts and high-throughput bioassay for antimicrobial activity (Xu et al., 2016, 2020). Many lantipeptide display systems were ever established to allow preparation of different lanthionine-containing peptides, including in vitro display systems using RNA/phage and in vivo display systems using bacterial/yeast (Hofmann et al., 2012; Bosma et al., 2019). Currently, some in vitro cell free biosynthesis platforms were well-designed for mining of lanthipeptides with the superiority of independent of cell growth, especially combined with high-throughput bioactivity screening systems. For instance, by use of CFPS (cell-free protein synthesis) set up based on E. coli cell extract, searching all nisin-related gene sequences in NCBI database resulted in biosynthesis of four new lanthipeptides with antibacterial activity in a single day (Zhang et al., 2018; Liu et al., 2020).
Biosynthetic Pathway Engineering of Lanthipeptides
Compared with other types of antibiotic BGCs, it is easier to carry out genetic manipulation on lanthipeptide BGCs due to their smaller size (<30 kb), which facilitates pathway engineering of lanthipeptide BGCs to yield new bioactive products (Table 2).
Precursor peptide synthase (LanA) was a common target for pathway engineering. For example, Schmitt S group divided these enzymes from biosynthetic pathways of 12 natural lanthipeptides into 33 modules which differs in sites of thioether rings and target binding, and performed “combinatorial shuffling” of these modules at the DNA level. Finally, they obtained the synthesis of 6,000 derived precursor peptides. Then heterologous expression by combining with the post-translational modification system for nisin and a high-throughput micro-screening system led to identification of 11 new lanthipeptides with enhanced antibacterial activity (Schmitt et al., 2019).
Some studies also focus on the engineering of key enzymes for thioether ring formation. For instance, in 2019, Kuipers OP group fused the Ser/Thr-dehydratase NisB and the cyclase NisC for nisin production to the biosynthetic pathway of vasopressin, and obtained a vasopressin analog with replacement of an original disulfide bridge with a thioether bridge, providing the proof for applying the post-translational modification systems of lanthipeptides into generation of new bioactive products (Li et al., 2019).
In addition, in order to enhance the outer membrane-traversing efficiency of nisin, by fusing the C-terminal of nisin with several relatively short peptides with activity against G– bacteria reported in references, a derivative compound was generated with increased activity by 4- to 12-fold compared to that of nisin, when inhibiting against several important G– pathogens, including Escherichia coli, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa and Enterobacter aerogenes (Li et al., 2018).
Structural Determination of Lanthipeptides
Generally, similar to other type NPs, most commonly used approaches for structures elucidation of newly discovered lanthipeptides include nuclear magnetic resonance (NMR) spectroscopy, mass spectrometry (MS) and some new approaches (Leenders et al., 2015; Wang et al., 2016).
To determine the thioether ring topology of new lanthipeptides, NMR spectroscopy is the most reliable approach, if regardless of its longer operation and requirements for considerable quantities of target products. To distinguish multiple overlapped rings in lanthipeptides, some new strategies based on mass spectrometry, such as Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF-MS) combined with LIFT tandem MS sequencing or LC-ESI-MS/MS have been established to fragment modified LanAs (Zhang et al., 2014; Tang et al., 2015; Schrimpe-Rutledge et al., 2018).
To confirm the formation and stereochemistry of Lan or MeLan, modified peptides (LanAs) are typically hydrolyzed under acidic conditions into the individual amino acids and reduced to yield disulfide bonds. Then the hydrolyzed mixtures containing free Cys thiols are derivatized into more volatile components with mass change, then analyzed by gas chromatography mass spectrometry (GC-MS) (An and van der Donk, 2020).
It is notable that Global Natural Product Social Molecular Networking (GNPS) as a generic metabolomics portal to analyze the MS/MS data would likely be used for structural analysis of lanthipeptides (Wang et al., 2016).
Conclusion and Prospects
Over the past years, tremendous advances have been reported in the understanding of lanthipeptides at the levels of genetics, bioactivity, structure and enzymology.
Though the number of known lantibiotics is still limited, it is still greatly advantageous to explore anti-infective drugs from lanthipeptides, due to the rarely observed antimicrobial resistance and abundance of their microbial-derived BGCs in nature. Using genome-mining, it is expected to isolate and identify new bioactive lanthipeptides from microorganisms.
By far, most of lanthipeptides with antibacterial activity have displayed very potent activity against G+ bacteria. But their activity against G– bacteria is relatively lower. Precise engineering lanthipeptide available at DNA levels could be an applicable solution to get new lanthipeptides against G– bacteria.
Compared to class I-II, biosynthesis of most of class III-V lanthipeptides still remains unclear and their activity other than antibacterial actions highlighted the importance of their potential value as new antiviral or antitumor agents. More insights into the novel mechanisms of their post-translational modification systems will be considerably required, especially in the molecular coordination and timing of the maturation enzymes and their interplay with the exporter proteins. As well, it is of importance to precisely engineer their biosynthetic pathways to enhance their activity and production yields.
Author Contributions
CL and AL: draft writing. KA, YiZ, and JH: figure drawing. YoZ and RL: draft planning and organization. AL: draft organization and manuscript writing. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by the National Key R&D Program of China (No. 2019YFA0905700, 2018YFA0900400, and 2019YFA0904000) and the Shandong Province Natural Science Foundation (ZR2017MC031 and ZR2020MC015).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We are thankful to National Natural Science Foundation of China (31170050), the Open Project Program of the State Key Laboratory of Bio-based Material and Green Papermaking (KF201825), the 111 Project (B16030), and The Fundamental Research Funds of Shandong University (2018GN021) for financial support.
Footnotes
References
Abbasi, M. N., Fu, J., Bian, X., Wang, H., Zhang, Y. M., and Li, A. Y. (2020). Recombineering for genetic engineering of natural product biosynthetic pathways. Trends Biotechnol. 38, 715–728. doi: 10.1016/j.tibtech.2019.12.018
Ahmadi, S., Ghollasi, M., and Hosseini, H. M. (2017). The apoptotic impact of nisin as a potent bacteriocin on the colon cancer cells. Microb. Pathog. 111, 193–197. doi: 10.1016/j.micpath.2017.08.037
An, L., and van der Donk, W. A. (2020). Recent progress in lanthipeptide biosynthesis. Dis. Eng. Chem. Biol. 2, 119–165. doi: 10.1016/B978-0-12-409547-2.14625-6
Arnison, P. G., Bibb, M. J., Bierbaum, G., Bowers, A. A., Bugni, T. S., Bulaj, G., et al. (2013). Ribosomally synthesized and post-translationally modified peptide natural products: overview and recommendations for a universal nomenclature. Nat. Prod. Rep. 30, 108–160. doi: 10.1039/c2np20085f
Baindara, P., Chaudhry, V., Mittal, G., Liao, L. M., Matos, C. O., Khatri, N., et al. (2015). Characterization of the antimicrobial peptide penisin, a class ia novel lantibiotic from paenibacillus sp. strain A3. Antimicrob. Agents Chemother. 60, 580–591. doi: 10.1128/AAC.01813-15
Baindara, P., Korpole, S., and Grover, V. (2018). Bacteriocins: perspective for the development of novel anticancer drugs. Appl. Microbiol. Biotechnol. 102, 10393–10408. doi: 10.1007/s00253-018-9420-8
Biswas, S., Garcia De Gonzalo, C. V., Repka, L. M., and van der Donk, W. A. (2017). Structure-activity relationships of the S-linked glycocin sublancin. ACS Chem. Biol. 12, 2965–2969. doi: 10.1021/acschembio.7b00819
Blin, K., Kazempour, D., Wohlleben, W., and Weber, T. (2014). Improved lanthipeptide detection and prediction for antiSMASH. PLoS One. 9:e89420. doi: 10.1371/journal.pone.0089420
Blin, K., Pascal Andreu, V., de Los Santos, E., Del Carratore, F., Lee, S. Y., Medema, M. H., et al. (2019a). The antiSMASH database version 2: a comprehensive resource on secondary metabolite biosynthetic gene clusters. Nucleic Acids Res. 47, D625–D630. doi: 10.1093/nar/gky1060
Blin, K., Shaw, S., Steinke, K., Villebro, R., Ziemert, N., Lee, S. Y., et al. (2019b). antiSMASH 5.0: updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res. 47, W81–W87. doi: 10.1093/nar/gkz310
Blockus, S., Sake, S. M., Wetzke, M., Grethe, C., Graalmann, T., Pils, M., et al. (2020). Labyrinthopeptins as virolytic inhibitors of respiratory syncytial virus cell entry. Antiviral. Res. 177:104774. doi: 10.1016/j.antiviral.2020.104774
Boakes, S., Appleyard, A. N., Cortés, J., and Dawson, M. J. (2010). Organization of the biosynthetic genes encoding deoxyactagardine B (DAB), a new lantibiotic produced by Actinoplanes liguriae NCIMB41362. J. Antibiot. 63, 351–358. doi: 10.1038/ja.2010.48
Bobeica, S. C., and van der Donk, W. A. (2018). The enzymology of prochlorosin biosynthesis. Methods Enzymol. 604, 165–203. doi: 10.1016/bs.mie.2018.01.038
Bösch, N. M., Borsa, M., Greczmiel, U., Morinaka, B. I., Gugger, M., Oxenius, A., et al. (2020). Landornamides: antiviral ornithine-containing ribosomal peptides discovered through genome mining. Angew. Chem. Int. Ed. Engl. 59, 11763–11768. doi: 10.1002/anie.201916321
Bosma, T., Rink, R., Moosmeier, M. A., and Moll, G. N. (2019). Genetically encoded libraries of constrained peptides. Chembiochem 20, 1754–1758. doi: 10.1002/cbic.201900031
Butler, M. S., Blaskovich, M. A. T., and Cooper, M. A. (2017). Antibiotics in the clinical pipeline at the end of 2015. J. Antibiot. (Tokyo). 70, 3–24. doi: 10.1038/ja.2016.72
Caetano, T., van der Donk, W. A., and Mendo, S. (2020). Bacteroidetes can be a rich source of novel lanthipeptides: the case study of Pedobacter lusitanus. Microbiol. Res. 235:126441. doi: 10.1016/j.micres.2020.126441
Chen, E., Chen, Q., Chen, S., Xu, B., Ju, J., and Wang, H. (2017). Mathermycin, a lantibiotic from the marine actinomycete Marinactinospora thermotolerans SCSIO 00652. Appl. Environ. Microbiol. 83:e926-17. doi: 10.1128/AEM.00926-17
Chen, S., Xu, B., Chen, E., Wang, J., Lu, J., Donadio, S., et al. (2019). Zn-dependent bifunctional proteases are responsible for leader peptide processing of class III lanthipeptides. Proc. Natl. Acad. Sci. U.S.A. 116, 2533–2538. doi: 10.1073/pnas.1815594116
Cheng, C., Chen, H., Tong, L., Li, Z., Yang, Y., Wu, S., et al. (2021). Mathermycin, an anti-cancer molecule that targets cell surface phospholipids. Toxicol. Appl. Pharmacol. 413:115410. doi: 10.1016/j.taap.2021.115410
Chu, L., Huang, J., Muhammad, M., Deng, Z., and Gao, J. (2020). Genome mining as a biotechnological tool for the discovery of novel marine natural products. Crit. Rev. Biotechnol. 40, 571–589. doi: 10.1080/07388551.2020.1751056
Cubillos-Ruiz, A., Berta-Thompson, J. W., Becker, J. W., van der Donk, W. A., and Chisholm, S. W. (2017). Evolutionary radiation of lanthipeptides in marine cyanobacteria. Proc. Natl. Acad. Sci. U.S.A. 114, E5424–E5433. doi: 10.1073/pnas.1700990114
Dickman, R., Mitchell, S. A., Figueiredo, A. M., Hansen, D. F., and Tabor, A. B. (2019). Molecular recognition of lipid II by lantibiotics: synthesis and conformational studies of analogues of nisin and mutacin rings A and B. J. Org. Chem. 84, 11493–11512. doi: 10.1021/acs.joc.9b01253
Fernández-Martínez, L. T., Gomez-Escribano, J. P., and Bibb, M. J. (2015). A relA-dependent regulatory cascade for auto-induction of microbisporicin production in Microbispora corallina. Mol. Microbiol. 97, 502–514. doi: 10.1111/mmi.13046
Foulston, L. C., and Bibb, M. J. (2010). Microbisporicin gene cluster reveals unusual features of lantibiotic biosynthesis in actinomycetes. Proc. Natl. Acad. Sci. U.S.A. 107, 13461–13466. doi: 10.1073/pnas.1008285107
Galván Márquez, I. J., McKay, B., Wong, A., Cheetham, J. J., Bean, C., Golshani, A., et al. (2020). Mode of action of nisin on Escherichia coli. Can. J. Microbiol. 66, 161–168. doi: 10.1139/cjm-2019-0315
Gomes, K. M., Duarte, R. S., and de Freire Bastos, M. (2017). Lantibiotics produced by Actinobacteria and their potential applications (a review). Microbiology 163, 109–121. doi: 10.1099/mic.0.000397
Goto, Y., Li, B., Claesen, J., Shi, Y., Bibb, M. J., and van der Donk, W. A. (2010). Discovery of unique lanthionine synthetases reveals new mechanistic and evolutionary insights. PLoS Biol. 8:e1000339. doi: 10.1371/journal.pbio.1000339
Guo, C., Mandalapu, D., Ji, X., Gao, J., and Zhang, Q. (2018). Chemistry and biology of teixobactin. Chemistry 24, 5406–5422. doi: 10.1002/chem.201704167
Hegemann, J. D., and van der Donk, W. A. (2018). Investigation of substrate recognition and biosynthesis in class IV lanthipeptide systems. J. Am. Chem. Soc. 140, 5743–5754. doi: 10.1021/jacs.8b01323
Heidary, M., Khosravi, A. D., Khoshnood, S., Nasiri, M. J., Soleimani, S., and Goudarzi, M. (2018). Daptomycin. J. Antimicrob. Chemother. 73, 1–11. doi: 10.1093/jac/dkx349
Hofmann, F. T., Szostak, J. W., and Seebeck, F. P. (2012). In vitro selection of functional lantipeptides. J. Am. Chem. Soc. 134, 8038–8041. doi: 10.1021/ja302082d
Hover, B. M., Kim, S. H., Katz, M., Charlop-Powers, Z., Owen, J. G., Ternei, M. A., et al. (2018). Culture-independent discovery of the malacidins as calcium-dependent antibiotics with activity against multidrug-resistant Gram-positive pathogens. Nat. Microbiol. 3, 415–422. doi: 10.1038/s41564-018-0110-1
Huo, L., Hug, J. J., Fu, C., Bian, X., Zhang, Y., and Müller, R. (2019). Heterologous expression of bacterial natural product biosynthetic pathways. Nat. Prod. Rep. 36, 1412–1436. doi: 10.1039/c8np00091c
Huo, L., and van der Donk, W. A. (2016). Discovery and characterization of bicereucin, an unusual d-Amino scid-containing mixed two-component lantibiotic. J. Am. Chem. Soc. 138, 5254–5257. doi: 10.1021/jacs.6b02513
Iorio, M. I., Sasso, O., and Maffioli, S. I. (2014). A glycosylated, labionin-containing lanthipeptide with marked antinociceptive activity. ACS Chem. Biol. 9, 398–404. doi: 10.1021/cb400692w
Lagedroste, M., Reiners, J., Knospe, C. V., Smits, S., and Schmitt, L. (2020). A structural view on the maturation of lanthipeptides. Front. Microbiol. 11:1183. doi: 10.3389/fmicb.2020.01183
Lawton, E. M., Cotter, P. D., Hill, C., and Ross, R. P. (2007). Identification of a novel two-peptide lantibiotic, Haloduracin, produced by the alkaliphile Bacillus halodurans C-125. FEMS Microbiol. Lett. 267, 64–71. doi: 10.1111/j.1574-6968.2006.00539.x
Leenders, J., Frédérich, M., and de Tullio, P. (2015). Nuclear magnetic resonance: a key metabolomics platform in the drug discovery process. Drug. Discov. Today 13, 39–46. doi: 10.1016/j.ddtec.2015.06.005
Li, B., Sher, D., Kelly, L., Shi, Y., Huang, K., Knerr, P. J., et al. (2010). Catalytic promiscuity in the biosynthesis of cyclic peptide secondary metabolites in planktonic marine cyanobacteria. Proc. Natl. Acad. Sci. U.S.A. 107, 10430–10435. doi: 10.1073/pnas.0913677107
Li, Q., Montalban-Lopez, M., and Kuipers, O. P. (2018). Increasing the antimicrobial activity of nisin-based lantibiotics against gram-negative pathogens. Appl. Environ. Microbiol. 84:e52-18. doi: 10.1128/AEM.00052-18
Li, Q., Montalban-Lopez, M., and Kuipers, O. P. (2019). Feasability of introducing a thioether ring in vasopressin by nisBTC co-expression in Lactococcus lactis. Front. Microbiol. 10:1508. doi: 10.3389/fmicb.2019.01508
Ling, L. L., Schneider, T., Peoples, A. J., Spoering, A. L., Engels, I., Conlon, B. P., et al. (2015). A new antibiotic kills pathogens without detectable resistance. Nature 517, 455–459. doi: 10.1038/nature14098
Liu, R., Zhang, Y., Zhai, G., Fu, S., Xia, Y., Hu, B., et al. (2020). A cell-free platform based on nisin biosynthesis for discovering novel lanthipeptides and guiding their overproduction in vivo. Adv. Sci. 7:2001616. doi: 10.1002/advs.202001616
Martin, N. I., Sprules, T., Carpenter, M. R., Cotter, P. D., Hill, C., Ross, R. P., et al. (2004). Structural characterization of lacticin 3147, a two-peptide lantibiotic with synergistic activity. Biochemistry 43, 3049–3056. doi: 10.1021/bi0362065
McClerren, A. L., Cooper, L. E., Quan, C., Thomas, P. M., Kelleher, N. L., and van der Donk, W. A. (2006). Discovery and in vitro biosynthesis of haloduracin, a two-component lantibiotic. Proc. Natl. Acad. Sci. U.S.A. 103, 17243–17248. doi: 10.1073/pnas.0606088103
Merwin, N. J., Mousa, W. K., Dejong, C. A., Skinnider, M. A., Cannon, M. J., Li, H. X., et al. (2020). DeepRiPP integrates multiomics data to automate discovery of novel ribosomally synthesized atural products. Proc. Natl. Acad. Sci. U.S.A. 117, 371–380. doi: 10.1073/pnas.1901493116
Mo, T., Xue, L., and Zhang, Q. (2016). New progress in the biosynthesis of lanthipeptide compounds. Acta Microbii Sinica 56, 373–382. doi: 10.13343/j.cnki.wsxb.20150427
Mo, T., Yuan, H., Wang, F., Ma, S., Wang, J., Li, T., et al. (2019). Convergent evolution of the Cys decarboxylases involved in aminovinyl-cysteine (AviCys) biosynthesis. FEBS Lett. 593, 573–580. doi: 10.1002/1873-3468.13341
Monciardini, P., Iorio, M., Maffioli, S., Sosio, M., and Donadio, S. (2014). Discovering new bioactive molecules from microbial sources. Microb. Biotechnol. 7, 209–220. doi: 10.1111/1751-7915.12123
Montalbán-López, M., Scott, T. A., Ramesh, S., Rahman, I. R., van Heel, A. J., Viel, J. H., et al. (2021). New developments in RiPP discovery, enzymology and engineering. Nat. Prod. Rep. 38, 130–139. doi: 10.1039/d0np00027b
Moon, K., Xu, F., and Seyedsayamdost, M. R. (2019). Cebulantin, a cryptic lanthipeptide antibiotic uncovered using bioactivity-coupled HiTES. Angew. Chem. Int. Ed. 58, 5973–5977. doi: 10.1002/anie.201901342
Müller, W. M., Schmiederer, T., Ensle, P., and Süssmuth, R. D. (2013). In vitro biosynthesis of the prepeptide of type-III lantibiotic labyrinthopeptin A2 including formation of a C-C bond as a post-translational modification. Angew. Chem. Int. Ed. 49, 2436–2440. doi: 10.1002/anie.200905909
Ökesli, A., Cooper, L. E., Fogle, E. J., and van der Donk, W. A. (2011). Nine post-translational modifications during the biosynthesis of cinnamycin. J. Am. Chem. Soc. 133, 13753–13760. doi: 10.1021/ja205783f
Oman, T. J., Boettcher, J. M., Wang, H., Okalibe, X. N., and van der Donk, W. A. (2011). Sublancin is not a lantibiotic but an S-linked glycopeptide. Nat. Chem. Biol. 7, 78–80. doi: 10.1038/nchembio.509
Ortega, M. A., Hao, Y., Walker, M. C., Donadio, S., Sosio, M., Nair, S. K., et al. (2016). Structure and tRNA specificity of MibB, a lantibiotic dehydratase from Actinobacteria involved in NAI-107 biosynthesis. Cell Chem. Biol. 23, 370–380. doi: 10.1016/j.chembiol.2015.11.017
O’Sullivan, J. N., O’Connor, P. M., Rea, M. C., O’Sullivan, O., Walsh, C. J., Healy, B., et al. (2020). Nisin J, a novel natural nisin variant, is produced by Staphylococcus capitis sourced from the human skin microbiota. J. Bacteriol. 202:e639-19. doi: 10.1128/JB.00639-19
Prochnow, H., Rox, K., Birudukota, N., Weichert, L., Hotop, S. K., Klahn, P., et al. (2020). Labyrinthopeptins exert broad-spectrum antiviral activity through lipid-binding-mediated virolysis. J. Virol. 94:e1471-19. doi: 10.1128/JVI.01471-19 ∗e1471-19
Ren, H., Shi, C., Bothwell, I. R., van der Donk, W. A., and Zhao, H. (2020). Discovery and characterization of a class IV lanthipeptide with a nonoverlapping ring pattern. ACS Chem. Biol. 15, 1642–1649. doi: 10.1021/acschembio.0c00267
Repka, L. M., Chekan, J. R., Nair, S. K., and van der Donk, W. A. (2017). Mechanistic understanding of lanthipeptide biosynthetic enzymes. Chem. Rev. 117, 5457–5520. doi: 10.1021/acs.chemrev.6b00591
Rupcic, Z., Hüttel, S., Bernecker, S., Kanaki, S., and Stadler, M. (2018). Large scale production and downstream processing of labyrinthopeptins from the Actinobacterium Actinomadura namibiensis. Bioengineering. 5:42. doi: 10.3390/bioengineering5020042
Sandiford, S. (2020). An overview of lantibiotic biosynthetic machinery promiscuity and its impact on antimicrobial discovery. Expert. Opin. Drug. Dis. 15, 373–382. doi: 10.1080/17460441.2020.1699530
Sandiford, S. K. (2019). Current developments in lantibiotic discovery for treating Clostridium difficile infection. Expert. Opin. Drug. Dis. 14, 71–79. doi: 10.1080/17460441.2019.1549032
Schmitt, S., Montalbán-López, M., Peterhoff, D., Deng, J., Wagner, R., Held, M., et al. (2019). Analysis of modular bioengineered antimicrobial lanthipeptides at nanoliter scale. Nat. Chem. Biol. 15, 437–443. doi: 10.1038/s41589-019-0250-5
Schmitz, S., Hoffmann, A., Szekat, C., Rudd, B., and Bierbaum, G. (2006). The lantibiotic mersacidin is an autoinducing peptide. Appl. Environ. Microbiol. 72, 7270–7277. doi: 10.1128/AEM.00723-06
Schrimpe-Rutledge, A. C., Sherrod, S. D., and McLean, J. A. (2018). Improving the discovery of secondary metabolite natural products using ion mobility-mass spectrometry. Curr. Opin. Chem. Biol. 42, 160–166. doi: 10.1016/j.cbpa.2017.12.004
Sheng, W., Xu, B., Chen, S., Li, Y., Liu, B., and Wang, H. (2020). Substrate tolerance of the biosynthetic enzymes of glycosylated lanthipeptide NAI-112. Org. Biomol. Chem. 18, 6095–6099. doi: 10.1039/D0OB01215G
Shin, J. M., Gwak, J. W., Kamarajan, P., Fenno, J. C., Rickard, A. H., and Kapila, Y. L. (2016). Biomedical applications of nisin. J. Appl. Microbiol. 120, 1449–1465. doi: 10.1111/jam.13033
Singh, M., Chaudhary, S., and Sareen, D. (2020). Roseocin, a novel two-component lantibiotic from an actinomycete. Mol. Microbiol. 113, 326–337. doi: 10.1111/mmi.14419
Spieß, T., Korn, S. M., Kötter, P., and Entian, K. D. (2015). Autoinduction specificities of the lantibiotics subtilin and nisin. Appl. Environ. Microbiol. 81, 7914–7923. doi: 10.1128/AEM.02392-15
Tang, W., Jiménez-Osés, G., Houk, K. N., and van der Donk, W. A. (2015). Substrate control in stereoselective lanthionine biosynthesis. Nat. Chem. 7, 57–64. doi: 10.1038/nchem.2113
Tietz, J. I., Schwalen, C. J., Patel, P. S., Maxson, T., Blair, P. M., Tai, H. C., et al. (2017). A new genome-mining tool redefines the lasso peptide biosynthetic landscape. Nat. Chem. Biol. 13, 470–478. doi: 10.1038/nchembio.2319
van Heel, A. J., Kloosterman, T. G., Montalban-Lopez, M., Deng, J., Plat, A., Baudu, B., et al. (2016). Discovery, production and modification of five novel lantibiotics using the promiscuous nisin modification machinery. ACS Synth. Biol. 5, 1146–1154. doi: 10.1021/acssynbio.6b00033
van Staden, A. D. P., van Zyl, W. F., Trindade, M., Dicks, L. M. T., and Smith, C. (2021). Therapeutic application of lantibiotics and other lanthipeptides: old and new findings. Appl. Environ. Microbiol. 87:e0018621. doi: 10.1128/AEM.00186-21
Vestergaard, M., Berglund, N. A., Hsu, P. C., Song, C., Koldsø, H., Schiøtt, B., et al. (2019). Structure and dynamics of cinnamycin-lipid complexes: mechanisms of selectivity for phosphatidylethanolamine lipids. ACS Omega. 4, 18889–18899. doi: 10.1021/acsomega.9b02949
Vikeli, E., Widdick, D. A., Batey, S. F. D., Heine, D., Holmes, N. A., Bibb, M. J., et al. (2020). In situ activation and heterologous production of a cryptic lantibiotic from an african plant ant-derived Saccharopolyspora species. Appl. Environ. Microbiol. 86:e1876-19. doi: 10.1128/AEM.01876-19
Walker, M. C., Eslami, S. M., Hetrick, K. J., Ackenhusen, S. E., Mitchell, D. A., and van der Donk, W. A. (2020). Precursor peptide-targeted mining of more than one hundred thousand genomes expands the lanthipeptide natural product family. BMC Genom. 21:387. doi: 10.1186/s12864-020-06785-7
Wang, H., Li, Z., Jia, R., Yin, J., Li, A., Xia, L., et al. (2018). ExoCET: exonuclease in vitro assembly combined with RecET recombination for highly efficient direct DNA cloning from complex genomes. Nucleic Acids Res. 46:e28. doi: 10.1093/nar/gkx1296
Wang, M., Carver, J. J., Phelan, V. V., Sanchez, L. M., Garg, N., Peng, Y., et al. (2016). Sharing and community curation of mass spectrometry data with global natural products social molecular networking. Nat. Biotechnol. 34, 828–837. doi: 10.1038/nbt.3597
Weber, T. (2016). Introduction to the special issue “bioinformatic tools and approaches for synthetic biology of natural products”. Synth. Syst. Biotechnol. 1, 67–68. doi: 10.1016/j.synbio.2016.04.001
Wu, C., Shang, Z., Lemetre, C., Ternei, M. A., and Brady, S. F. (2019). Cadasides, calcium-dependent acidic lipopeptides from the soil metagenome that are active against multidrug-resistant bacteria. J. Am. Chem. Soc. 141, 3910–3919. doi: 10.1021/jacs.8b12087
Xu, M., Wang, Y., Zhao, Z., Gao, G., Huang, S. X., Kang, Q., et al. (2016). Functional genome mining for metabolites encoded by large gene vlusters through heterologous expression of a whole-genome bacterial srtificial chromosome library in streptomyces spp. Appl. Environ. Microbiol. 82, 5795–5805. doi: 10.1128/AEM.01383-16
Xu, M., Zhang, F., Cheng, Z., Bashiri, G., Wang, J., Hong, J., et al. (2020). Functional genome mining reveals a class V lanthipeptide containing a d-amino acid introduced by an F420 H2 -dependent reductase. Angew. Chem. Int. Ed. Eng. 59, 18029–18035. doi: 10.1002/ange.202008035
Zhang, Q., Doroghazi, J. R., Zhao, X., Walker, M. C., and van der Donk, W. A. (2015). Expanded natural product diversity revealed by analysis of lanthipeptide-like gene clusters in actinobacteria. Appl. Environ. Microbiol. 81, 4339–4350. doi: 10.1128/AEM.00635-15
Zhang, Q., and Liu, W. (2013). Biosynthesis of thiopeptide antibiotics and their pathway engineering. Nat. Prod. Rep. 30, 218–226. doi: 10.1039/C2NP20107K
Zhang, Q., Ortega, M., Shi, Y., Wang, H., Melby, J. O., Tang, W., et al. (2014). Structural investigation of ribosomally synthesized natural products by hypothetical structure enumeration and evaluation using tandem MS. Proc. Natl. Acad. Sci. U.S.A. 111, 12031–12036. doi: 10.1073/pnas.1406418111
Keywords: natural product, lanthipeptide, biosynthesis, genome mining, pathway engineering
Citation: Li C, Alam K, Zhao Y, Hao J, Yang Q, Zhang Y, Li R and Li A (2021) Mining and Biosynthesis of Bioactive Lanthipeptides From Microorganisms. Front. Bioeng. Biotechnol. 9:692466. doi: 10.3389/fbioe.2021.692466
Received: 08 April 2021; Accepted: 29 June 2021;
Published: 29 July 2021.
Edited by:
Jose Ruben Morones-Ramirez, Autonomous University of Nuevo León, MexicoReviewed by:
Xinqing Zhao, Shanghai Jiao Tong University, ChinaZheng-Jun Li, Beijing University of Chemical Technology, China
Copyright © 2021 Li, Alam, Zhao, Hao, Yang, Zhang, Li and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Aiying Li, YXlsaUBzZHUuZWR1LmNu; Youming Zhang, emhhbmd5b3VtaW5nQHNkdS5lZHUuY24=; Ruijuan Li, bGlydWlqdWFuQHNkdS5lZHUuY24=
 Caiyun Li1
Caiyun Li1 Jinfang Hao
Jinfang Hao Youming Zhang
Youming Zhang Aiying Li
Aiying Li