- 1King's College London GKT School of Medical Education, London, United Kingdom
- 2Department of Women and Children’s Health (Paediatric Allergy), School of Life Course Sciences, Faculty of Life Sciences and Medicine, King’s College London, London, United Kingdom
- 3Peter Gorer Department of Immunobiology, School of Immunology and Microbial Sciences, King’s College London, London, United Kingdom
- 4Children's Allergy Service, Evelina London Children's Hospital, Guy's and St Thomas' Hospital, London, United Kingdom
Food allergy represents an increasing global health issue, significantly impacting society on a personal and on a systems-wide level. The gold standard for diagnosing food allergy, the oral food challenge, is time-consuming, expensive, and carries risks of allergic reactions, with unpredictable severity. There is, therefore, an urgent need for more accurate, scalable, predictive diagnostic techniques. In this review, we discuss possible future directions in the world of food allergy diagnosis. We start by describing the current clinical approach to food allergy diagnosis, highlighting novel diagnostic methods recommended for use in clinical practice, such as the basophil activation test and molecular allergology, and go on to discuss tests that require more research before they can be applied to routine clinical use, including the mast cell activation test and bead-based epitope assay. Finally, we consider exploratory approaches, such as IgE glycosylation, IgG4, T and B cell assays, microbiome analysis, and plasma cytokines. Artificial intelligence is assessed for potential integrated interpretation of panels of diagnostic tests. Overall, a framework is proposed suggesting how combining established and emerging technologies can effectively enhance the accuracy of food allergy diagnosis in the future.
1 Introduction
The prevalence of food allergy in Europe is increasing, with a recent meta-analysis showing a pooled lifetime prevalence of 19.9%, and a point prevalence of 13.1% for self-reported food allergy (1). US data from 2008 to 2016 show a 3.2-fold increase in emergency department visits connected to anaphylaxis (2). For those diagnosed, there are profound effects on every facet of their lives (3). Effects on mental health and productivity in the workplace are increasingly recognized, with wider implications for a healthy, cohesive society (4, 5). The development of accurate, economical tests for correctly diagnosing food allergy is essential to help combat these issues.
Through the next decade, recent findings and technologies hold immense potential to redefine the paradigm for food allergy diagnosis. Several novel modalities have been proposed to reduce the need for food challenges via non-invasive lab-based tests. These range across a variety of techniques, from conventional serology tests to artificial intelligence (AI) systems. In this narrative review, we will focus on IgE-mediated food allergy. First we will briefly describe the current status of food allergy diagnosis and summarize unmet needs in the field. We shall then discuss each novel approach in turn and explore future directions for food allergy diagnosis, delineating a pathway for the safe and effective integration of novel technologies into clinical practice, in the future. We have also generated Table 1 containing sensitivity and specificity data from selected studies across a range of modalities to provide a contextual background to the review.
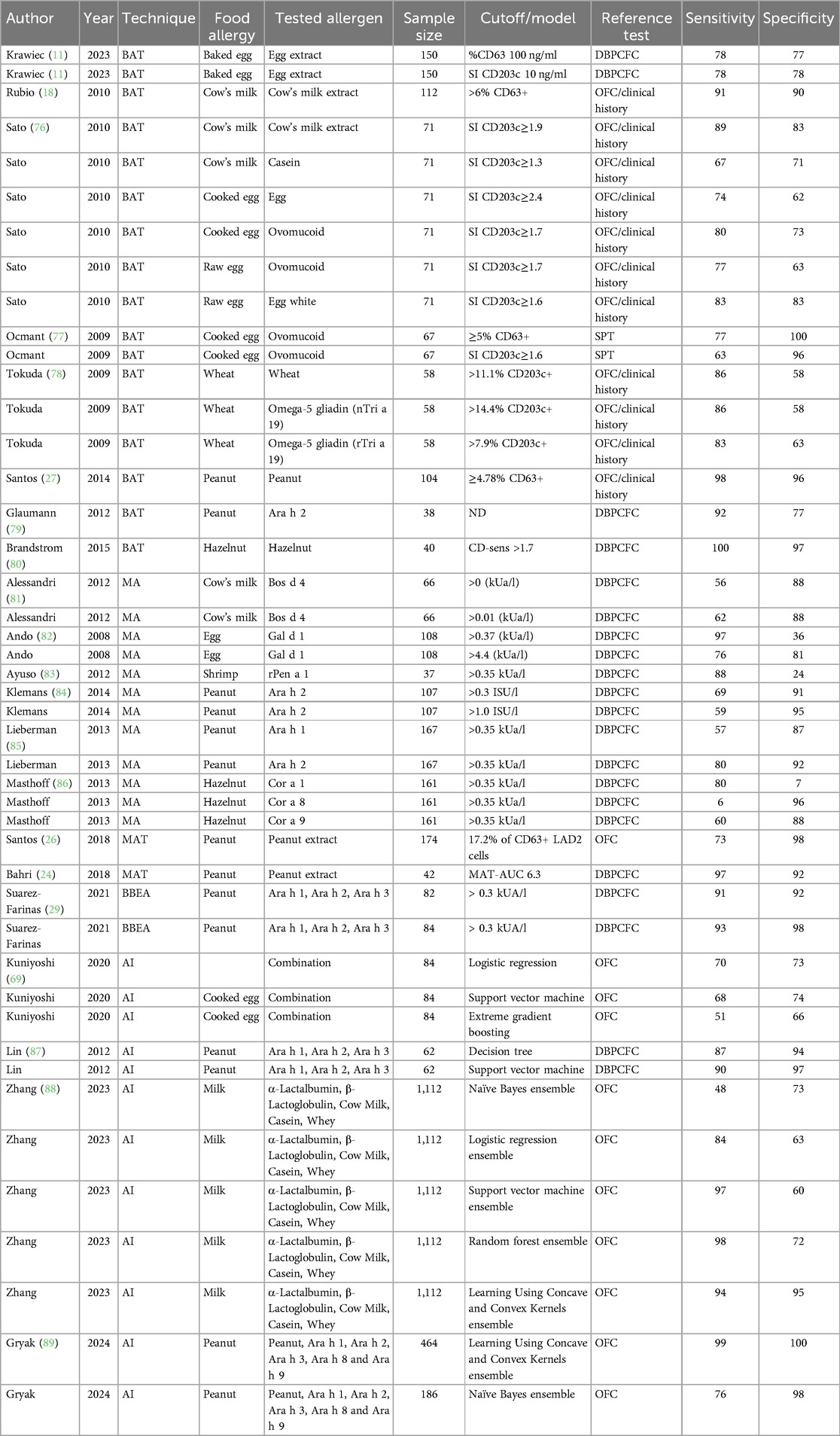
Table 1. Sensitivity and specificity for food allergy diagnostic modalities from selected studies. BAT, basophil activation test; MA, molecular allergology; MAT, mast cell activation test; AI, artificial intelligence; DBPCFC, double-blind placebo-controlled food challenge; OFC, oral food challenge.
2 Current clinical approach to food allergy diagnosis
Typically, food allergy is diagnosed through a combination of a thorough history and physical examination, and evidence of IgE to the suspected allergen, using skin prick testing and/or measurement of food-specific IgE in the serum (6). In patients with a clear history of an immediate reaction to a certain food and evidence of IgE to that food, the diagnosis of food allergy can be confirmed. Conversely, in individuals with no history of reacting to a food and no evidence of IgE sensitization to that food, the diagnosis of food allergy can be excluded. In all intermediate scenarios, additional tests need to be performed.
New guidelines from the European Academy of Allergy and Clinical Immunology (EAACI) (7) recommend initiating the diagnostic process with a detailed allergy-focused clinical history. Following this, skin prick test (SPT) and specific IgE (sIgE) to allergen extracts or the fresh food is strongly recommended. In some cases, these first-line tests do now allow a precise diagnosis, and for foods for which there are “informative components”, molecular allergology (MA) should be used. Examples of such informative components tests are sIgE to Ara h 2, Cor a 14, and Ana o 3, for peanut, hazelnut, and cashew allergies, respectively. For investigation of peanut or sesame seed allergies, basophil activation test (BAT) to peanut or sesame can be used, if available. BAT is a diagnostic technique involving basophil response which will be discussed in more detail in a later section. Of note, this was the first time molecular allergology and the BAT have been included in clinical guidelines to date.
Currently, the reference standard for precise assessment of food allergy is the Oral Food Challenge (OFC). Despite being expensive and carrying risks of anaphylaxis, OFC provides direct confirmation as to whether a patient will react to a given food allergen (8, 9). OFC procedures can also be tailored to answer specific clinical questions as needed. In equivocal cases, at various stages of the diagnostic process, OFC may be indicated and are ultimately the definitive diagnostic test currently. Open OFC are suitable for clinical practice and double-blind placebo-controlled food challenges are reserved for cases in which open OFC are, or have the risk of being inconclusive, and for research purposes.
The systematic review and meta-analysis conducted to inform the new EAACI food allergy clinical guidelines confirmed high sensitivity of SPT for the diagnosis of cow's milk and cooked egg allergy (90% and 94% respectively) (10). Sensitivity of sIgE to whole extract was 81%, 82%, 73/85%, and specificity 83%, 92%, 88/73%, for peanut, cow's milk, and raw/cooked egg respectively. In comparison to whole extracts, molecular allergology for individual components is highly specific for some foods. For instance, Ara h 2-sIgE has specificity of 92%, Cor a 14-sIgE 95%, Ana o 3-sIgE 94%, casein-sIgE 93%, ovomucoid-sIgE 91% for peanut, hazelnut, cashew, cow's milk, and cooked egg allergies, respectively.
The relatively lower sensitivity of components for milk and egg identified (67% and 74% for casein-sIgE and ovomucoid-sIgE respectively) were not superior to extracts, so were not included in the new guidelines. The BAT was both highly specific and highly sensitive for peanut and sesame, the only allergens for which there were sufficient numbers of BAT studies to perform meta-analyses (sensitivity and specificity of 84% and 90% for peanut, 89% and 93% for sesame). Since the review, BAT has also successfully displayed sensitivity and specificity of 78% and 77% to identify reactors to baked egg (11). Summary of the recommended tests in the new EAACI guidelines is shown in Figure 1.
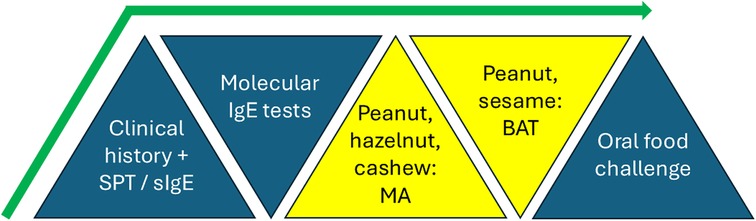
Figure 1. Recommended tests and their sequential use for diagnosis of food allergy in the new clinical guidelines for the diagnosis of IgE-mediated food allergy of the European academy of allergy and clinical immunology (EAACI). SPT, skin prick test; sIgE, specific IgE; MA, molecular allergology; BAT, basophil activation test.
3 Novel approaches in clinical practice
3.1 Molecular allergology
By characterizing molecular components of a given allergen that are recognized by patients' IgE, molecular allergology (MA) can be used to confirm suspected IgE-mediated food allergy (12). One of the main strengths of MA is that it allows for differentiation between co-sensitization and cross-reactivity, improving accuracy (13). The typical example is sensitization to plant foods secondary to pollen allergy—detecting IgE to the cross-reactive component (e.g., Ara h 8, Cor a 1) helps differentiate from primary systemic food allergy, associated with sensitisation to primary food allergens (for example, seed storage proteins Ara h 1/2/3/6 and Cor a 9/14). Further, MA has applicability to risk-stratification, helping to guide clinical management. Despite this, MA has demonstrated comparable specificity, but lower sensitivity, than OFC so far, limiting its value for primary diagnosis, with sensitivity of 67% and specificity 93% for casein, 74% and 91% for hen's egg ovomucoid, 82% and 92% for peanut Ara h 2 respectively (14). Exact values of sIgE titres are of limited clinical value, as severe reactions may still occur with sIgE of low levels and high affinity while conversely sIgE can also be present in the serum of resolved patients (15). Cost of MA is a limitation compared to SPT, though integration of novel multiplex platforms may help to minimize economic barriers, in the cases that require testing to many allergen components.
3.2 Basophil activation test
The BAT assesses degranulation of basophils in vitro following stimulation with the allergen. Basophils drive acute allergic responses via IgE recognition of the allergen and activation via the high-affinity IgE receptor. Following stimulation by allergens, flow cytometry is used to look for upregulation of the key markers CD63 and CD203c as a proxy for basophil degranulation. BAT has shown excellent diagnostic performance in clinical trials, having the potential to reduce the need for OFC (16). BAT has improved specificity compared to SPT and sIgE, while retaining sensitivity- limiting the number of false positives- with optimal cutoffs showing 98% sensitivity and 96% specificity for peanut (17). Further, BAT has potential utility for predicting disease trajectory, with some studies using repeated BAT to monitor acquisition of desensitization and remission of the allergic response (18). The ability of BAT to categorize allergic disease phenotypes through threshold dose reactivities offers opportunities for increased granularity of diagnostic approach, facilitating personalised care (19). Some of the barriers to further integration of BAT into clinical practice include the ca. 10% of individuals with basophils selectively unresponsive to FcεRI-mediated signalling. As an ex-vivo test on fresh (whole) blood, BAT features logistical difficulties in terms of procuring the sample from patients, requiring time-constrained transport to the lab and processing.
3.3 Low-dose and low-risk oral food challenges
Various approaches have been suggested attempting to improve feasibility and deliverability of OFC. In patients with minimal risk of reacting to the allergen, cumulative OFC has been adopted to reduce the time involved and the risk of desensitization during OFC, which has been associated with false-negative OFC. Cumulative OFC consists in a single dose OFC which resembles the age-appropriate portion size. Another approach often adopted for baked milk and baked egg is the low-dose OFC, which offers a way to establish a dose below which patients can continue eating the food at home. A lower dose has been used in a standard OFC, with one study defining the dose as 1%–10% of usual OFC dose (20, 21). The danger of a low dose OFC is the fact that the threshold of reactivity can change over time and allergen-content of foods is not always declared, thus care needs to be taken to avoid the allergen in circumstances known to be associated with a reduction in allergen threshold (e.g., infections, sleep deprivation, intense exercise, menstruation, etc), and to ingest foods with known allergen content that is within the recommended range for that patient.
4 Unmet needs
Primarily, there is a need for a non-invasive test that reduces the need for OFC to when absolutely necessary, due to the potential risk of a life-threatening anaphylactic reaction. Beyond risks to patients, logistically there is also limited capacity for OFCs due to its high resource-burden (22). It is noted that even in allergy centres with the highest capacity a prohibitively large imbalance between supply and demand results in long waiting lists. In the United Kingdom, contemporary data shows only 677 challenges per week over 139 centres (23). Reduced ability to deliver sufficient tests mandates prioritization of the most at-risk patients, and thence delay for much of the patient population. This carries a high cost, as delay in introducing foods back into the diet while immunoplasticity persists in young infants can increase the risk of developing food allergy later, especially in high-risk sensitized infants (9). Equally, late confirmation of food allergy being absent runs the risk of irreversible effects on patients’ food preferences and personal tastes.
There are also issues surrounding geographical equity; robust diagnosis of food allergy is currently limited to developed secondary and tertiary centres. This indicates an accessibility inequality in regions with limited care provision beyond primary care. Additionally, as therapies targeted to specific pathophysiological mechanisms are developed, accurately delineating the course of disease progression, and phenotypically stratifying diagnosed patients into discrete categories, will become increasingly more important.
5 Tests approaching clinical use
5.1 Mast cell activation test
Similarly to the BAT, the mast cell activation test (MAT) uses flow cytometry to observe the in vitro activation of mast cells to ascertain the degree of allergic response to particular allergens. Diagnostic performance of the MAT has been shown to exceed the SPT with an AUC of 0.99 compared to 0.73 for SPT (24). Mast cell lines or primary mast cells derived from peripheral blood hematopoietic stem cells are passively sensitized with the patient's serum or plasma, then incubated with allergen to assess response. The requirement for serum/plasma rather than fresh blood is a key distinguishing factor between BAT and MAT, potentially improving the feasibility of MAT and allowing for testing samples collected prospectively over time in the same experiment. However, growing mast cells can be challenging and primary cells are difficult to isolate, require tissue or large volumes of blood and are highly variable, with passive sensitization limiting sensitivity significantly (25). Requiring transfer of IgE to cells that are not autologous, MAT has lower sensitivity than the BAT with one study showing 75% to peanut compared to over 83% for BAT, so could be more useful in a sequential process following BAT, especially in cases of basophil-non-responders (26, 27).
5.2 Bead-based epitope assays
The bead-based epitope assay (BBEA) represents a scalable, high through-put multiplex assay for IgE and IgG4 to several sequential epitopes, requiring microliter samples only (28). This technology utilises peptides mechanically coupled to beads, which are incubated with serum or plasma samples, followed by quantification of fluorescence by fluorophore-labelled antibodies. Sensitivity and specificity of epitope detection, at 92% and 94% respectively, has been defined for peanut allergy. BBEA can also be used for phenotypic stratification and severity prediction (29, 30). A particular strength of BBEA is its reproducibility, which is key for effective widespread use. Errors pertaining to technical details (well position, reading order), as well as batch effects, are possible limitations.
BBEA requires precise prior knowledge of epitope amino acid sequences corresponding to each allergenic food of interest (29). Further implementation of BBEA is therefore predicated on continued advances in basic research concerning protein sequencing, as well as the sharing and processing of large epitope datasets developed from robustly characterised patient cohorts. Consequently, in the near future, BBEA likely has most applicability to diagnosis of the well-researched peanut allergy. An important limitation is that BBEA is limited to sequential epitopes, whereas in reality conformational epitopes also play a key role in food allergy. BBEA analysis also does not capture the totality of the IgE response, as it does not reflect components such as affinity and cellular response which are better characterized by the cellular assays mentioned earlier, as well as only demonstrating allergen sensitisation rather than overt clinical allergy (31). Utility of BBEA results may therefore be improved with parallel investigation of cellular assays, and correlation with clinical history.
6 Exploratory approaches
6.1 Glycosylation of allergen and glycosylation of IgE
Glycosylation of natural allergens can lead to cross-reactivity mediated by glycan-specific antibodies (14, 32). For instance, sensitization to cross-reactive carbohydrate determinants (CCD) can cause detectable IgE to peanut whilst resulting in negative SPT or BAT to peanut, reflecting the absence of clinical relevance in this CCD sensitisation. In Ghanaian children, CCD was identified as the underlying reason for IgE cross-reactivity between IgE to peanut and IgE to parasites, such as Schistosoma Mansoni (33). While IgE to glycans is often regarded as a barrier to specificity of in vitro diagnostics, it could represent another component of the allergic response to fully investigate in addition to conventional IgE serology, defining distinct patient sensitization profiles (34). For instance, IgE to α-Gal has previously been associated with meat-allergy delayed anaphylaxis (35).
A different prism is that of IgE glycosylation, which has been implicated in the pathogenicity of IgE in its ability to induce mast cell and basophil degranulation in vitro, and anaphylaxis in a mouse model, when compared with non-allergic individuals (36). However, the functional consequences of differences in IgE glycosylation between allergic and tolerant individuals has yet to be assessed between allergen-specific IgE molecules.
6.2 Allergen-specific IgG4 and IgA
Some studies have shown potential significance in testing for specific IgG4 against foods (37, 38). IgG4 is an IgG subclass that is produced under the influence of the same cytokines as IgE and has the unique ability for monovalent bispecific binding via Fab-arm exchange, theorized to be associated with IgE class-switching. However, it has been suggested that IgG4 merely represents a bystander of immunological tolerance rather than a factor driving such immune response (39). In the LEAP study, peanut-specific IgG4 increased over time in the study arm who consumed peanut from the first year of life, who also had less peanut allergy (40). In a recent cross-sectional egg allergy study, sIgG4 to egg, egg white and egg components, ovomucoid and ovalbumin, was higher in allergic compared with tolerant children and in consumers compared with avoiders (41). More research is needed to understand the role of IgG4 in food allergy and, certainly, to understand its utility in food allergy diagnosis.
sIgA is also presumed to play a role in the pathophysiology of food allergy, as sIgA deficiency has been linked to higher rates of allergies including food allergy, and salivary IgA has correlated with oral immunotherapy response for peanut allergy (42, 43). One cohort study indeed revealed higher plasma sIgA counts in children with peanut allergy, though no evidence was found for protection of food-specific IgA against development of food allergy or its potential utility for food allergy diagnosis (44).
6.3 T cell assays
A novel food allergy biomarker involves gauging the frequency of pathogenic T cell subsets and their activation in response to allergen stimulation. T cells are key immune cells in orchestrating food allergic and tolerant responses. In particular, Th2A cells, a subset of terminally differentiated memory CD4 + helper T cells, with characteristic expression of CD molecules and distinct functionality from conventional Th2 cells, have been associated with a specific stable allergic disease-related phenotype (45). One study examining pathophysiological trajectories of peanut allergy in a cohort of patients receiving oral immunotherapy showed clinical utility of subgroup division according to peanut-specific T-cell mediated immune profile (46). Discrete peanut-reactive T cell immunotypes based on relative proportions of Th2A cells were demonstrated, which were significantly related to distinct responses to allergen-specific immunotherapy. Additionally, Tfh13 cells, a subset of T follicular helper cells, may also be implicated in inducing anaphylaxis through induction of high-affinity specific IgE via sequential switching of B cells from IgG1 to IgE (47). In a study elucidating the position of T regulatory cells in spontaneous cow's milk allergy recovery, artificial neural networks were used to identify the markers FoxP3, Nfat-C2, IL-16, and GATA-3 as associated with the persistence of cow's milk allergy, with higher FoxP3 intensity in cow's milk allergic children, linking Th2 and Treg expression profiles with food allergy disease activity (48). An enhanced understanding of further Th subtypes could permit personalised care via assessment of individual T cell phenotypic signatures, designing and delivery of novel immunomodulatory therapies, as well as prediction of treatment response.
6.4 B cell assays
Playing an integral role in the production of allergen-specific antibodies, characterization of B cells is another possible biomarker for food allergy, determining specific attributes of the B cell receptor. IgE + B cells are directly related to the pathogenesis of food allergy, but are low in frequency and therefore difficult to identify. Investigative studies into IgE + B cells have identified the majority of IgE + B cells as immature plasmablasts with a common clonal family (49). IgE + memory B cells have been detected in the circulation, but they can also be present in the tissue, namely in the stomach and duodenum (50, 51). IgE + clonal signatures from gut samples could therefore represent one potential avenue to identify food allergy, though limited by access to relevant tissue sites in the gut. As with T cells, specific B cell subsets may also allow for further insights into food allergy, such as type 2-polarised memory B cells which are implicated in generating sIgE in early sublingual immunotherapy for birch pollen/house dust mite allergy (52). Similarly, a unique population of CD23 + IgG1 + memory B cells has also been described in the long-term persistence of peanut allergy in children, with frequency correlating to serum IgE levels (53, 54).
6.5 Microbiome analysis
In a similar vein to characterization of T cell responses, studying microbiome dynamics could permit discrete categorisation of food allergy trajectories. Disparities in the prevalence of certain bacterial species between peanut-allergic and non-allergic patients were demonstrated in a multidimensional multi-omic study of the oral microbiome, accompanied by significant differences in short-chain fatty acid and IL-4 titres (55). In particular, abundances of Prevotella species were protective against a Th2-mediated response, whereas milieus tending towards Neisseria categorized Th2-mediated allergic response. This corroborates with literature suggesting a role for Neisseria-related mucosal inflammation. In a similar study using faecal samples, a metabolome network analysis from infancy to mid-childhood was conducted, revealing significant associations between reduced microbiome diversity, temporal bacterial variation, and histidine, butyrate, isovalerate pathway metabolites with development of peanut allergy (56).
Iterating on the evidence provided by associative trials, oral and gut microbiomes have been clinically implicated in influencing disease reaction thresholds. A prospective multiscale study revealed characteristic oral and gut microbiome signatures affiliated with peanut allergy, observing significant associations between prevalence of Veillonella and Bacteroides species with high-threshold peanut-allergic patients (57). Consistency of ordinal trends was also noted between these discriminative bacteria and non-allergic participants. These findings suggest integrated analysis of combined oral and gut environments could be worthwhile in exploring crosstalk and categorical summation of host-microbe-immune dynamics. Advances in microbiome analysis may be laterally transferable to other allergenic foods, as pro-allergenic micro-environments are likely to be shared between food allergies without requiring precise knowledge of peptide sequences, as in BBEA. Despite modest predictive performance in preliminary research, the complex and interdependent mechanisms of the gut and oral microbiome create fundamental limitations for the specificity of any proposed diagnostic test involving the microbiome, as confounding effects (diet, ethnicity, comorbidities) may create statistical noise, making microbiome analysis unlikely to be significantly useful outside the context of a broader picture of host metabolites and IgE levels (58, 59).
6.6 Plasma cytokine analysis
As a natural extension from effector immune cell responses, characterization of the cytokine milieu could further define patient-specific immunopathogenetic signatures. Plasma cytokine profiles have been studied as predictors of food allergy diagnosis and resolution among sensitized infants, with specific reference to higher levels of allergy-defining IL-4, IL-13, IL-12p70, and lowered IL-10 (60, 61). Heterogeneity of cytokine profile has also been described between peanut and egg-allergic patients, which has been related to disparities in resolution rate (62). One prospective study explored the clinical utility of these markers, observing significantly raised median TNF- α and IL-6 levels before a therapeutic elimination diet in food-allergic participants, followed by significant decreases post-diet. The existing literature suggests a role for plasma cytokines in quantifying success of immunotherapeutic interventions, with decreases in IL-5 and IL-13 observed after food allergen immunotherapy, though it is worth noting the transience of this immunologic suppression (63, 64). These results are positive toward integration of plasma cytokine levels in initially confirming food allergy and tracking disease trajectory, but must be interpreted with particular consideration to time-course due to their fluctuant nature. More utility may therefore be derived from multiple readings longitudinally and single-cell cytokine studies. The efficacy of single-cell cytokine analysis is highlighted in the work of Chiang et al., who used single-cell profiling to interrogate heterogenous Th2 effector subsets in peanut-allergic patients, revealing a delayed, IL-2-dependent CD154 upregulation on Treg cell marker-expressing cells (65). Similarly, Neeland et al. showed significantly greater Il-6 and TNFα production, and lower IFN-γ production, with single-cell analysis of CD3/CD28 stimulated naïve CD4+ T cells in food allergic adolescents (66).
6.7 Artificial intelligence
AI methods hold a revolutionary potential for precisely identifying food allergy and synthesizing results of various tests for enhanced diagnostic accuracy. In comparison to conventional computerised protocols, artificial intelligence is able to fluidly adapt to provided datasets; learning from prior experience and applying this to novel situations. Specifically, machine learning (ML) frameworks, defined as a subset of AI involving teaching machines to emulate human learning while avoiding explicit programming, have utilized biomarkers including omics, total IgE, sIgE, sIgG4, and SPT data to successfully predict food allergy in infant populations (67). ML has also been efficacious as part of a medical algorithm identifying food allergy from data on methylation of highly discriminating cytosine-phosphate-guanine dinucleotides, reaching 100% accuracy through an ensemble voting scheme (68). Furthermore, prediction of tolerance to oral food challenge has been successfully demonstrated with Random Forest model sensitivity and specificity of 98% and 95% respectively for heated egg (69). ML processes have also been integrated within the novel bead-based epitope profiling mechanism described earlier, with the ability to accurately examine baseline variables influencing response to oral immunotherapy (70).
One of the major benefits of artificial intelligence approaches is the possibility for exploratory, unbiased data-driven analysis such as K-Means Clustering, which is an unsupervised algorithm enabling the grouping of unlabelled datasets into discontinuous clusters. Effective application of this to the MAT technique has been demonstrated, creating distinct clusters of phenotypes within a dataset of patients with similar responses to MAT (24, 71). AI models have also been successfully applied to MA multiplex arrays (72). As described in previous sections, the process of food allergy diagnosis lends itself to categorization of specific phenotypes in continuous unlabelled datasets, indicating high potential to accelerate our understanding of allergic patient presentations across a wide array of allergens and laboratory techniques. As well as this lateral applicability, there is also immense forward capability for analysis of methods that have yet to be discovered or refined, permitting swifter validation and optimization.
7 Conclusion
It is worth noting that no one modality described in this review is likely to be the perfect discriminant of food allergy. Rather, combinations of tests have been postulated to maximize diagnostic accuracy (73, 74). In the short-term, upscaling of existing, proven technologies such as MA and BAT, that have recently begun to be introduced into clinical practice may carry the timeliest return-on-investment in achieving tangible improvements in quality-of-care for patients with suspected food allergy. Laboratory tests made more streamlined, high-throughput, and affordable will be impactful in improving access to food allergy diagnosis in under-resourced communities. This requires a parallel investment in laboratory processing capabilities, and development of rapid, efficient processes on a macroscopic systems level.
Alongside this, ongoing research into the exploratory techniques described in this review will pay dividends for the treatment and management of food allergy on a long-term scale. As advances are made in targeted immunotherapy, increased depth and granularity of food allergy diagnosis will be necessitated to access its full potential. For example, personalized biologic treatments could be selected based on a patient's unique signature of cellular response, cytokine milieu, and microbiome. Underlying this, a data-driven approach guided by high-quality trained ML networks will be essential for enhanced interpretation of intricate omics datasets (75). A suggested pathway for direction of future food allergy diagnostics is shown in Figure 2.
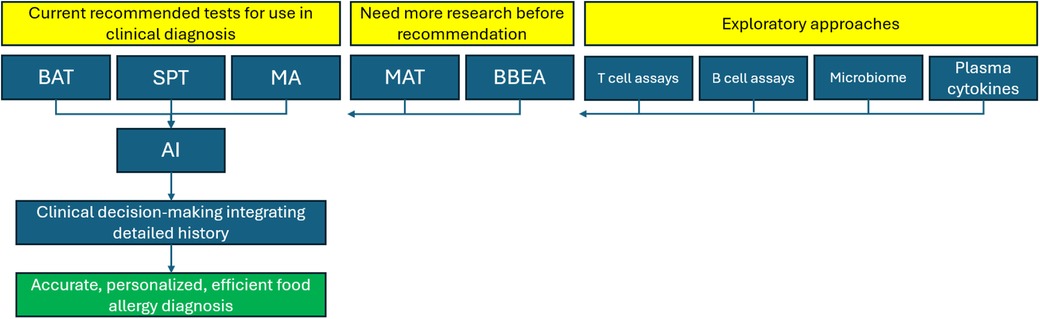
Figure 2. Framework for future directions in the diagnosis of IgE-mediated food allergy. BAT, basophil activation test; SPT, skin prick test; MA, molecular allergology; MAT, mast cell activation test; BBEA, bead-based epitope assay; AI, artificial intelligence.
Overall, this review suggests the rapidly changing face of food allergy diagnosis is well-positioned to address the projected increase in clinical burden in the near future, through an integrated approach combining laboratory technologies, artificial intelligence, and the art of medicine, to make a precise and timely diagnosis for every single food allergy in each individual.
Author contributions
DW: Visualization, Writing – original draft. AS: Conceptualization, Project administration, Resources, Supervision, Visualization, Writing – review & editing.
Funding
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
Conflict of interest
AS reports grants from Medical Research Council (MR/M008517/1; MC/PC/18052; MR/T032081/1), Food Allergy Research and Education (FARE), the Immune Tolerance Network/National Institute of Allergy and Infectious Diseases (NIAID, NIH), Asthma UK (AUK-BC-2015-01), BBSRC, Rosetrees Trust and the NIHR through the Biomedical Research Centre (BRC) award to Guy's and St Thomas' NHS Foundation Trust, during the conduct of the study; personal fees from Thermo Scientific, Nestle, Nutricia, Infomed, Novartis, Allergy Therapeutics, IgGenix, Buhlmann, as well as research support from IgGenix, Buhlmann and Thermo Fisher Scientific through a collaboration agreement with King's College London.
The remaining author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Spolidoro GC, Amera YT, Ali MM, Nyassi S, Lisik D, Ioannidou A, et al. Frequency of food allergy in Europe: an updated systematic review and meta-analysis. Allergy. (2023) 78(2):351–68. doi: 10.1111/all.15560
2. Michelson KA, Dribin TE, Vyles D, Neuman MI. Trends in emergency care for anaphylaxis. J Allergy Clin Immunol Pract. (2020) 8(2):767–8. doi: 10.1016/j.jaip.2019.07.018
3. DunnGalvin A, Dubois AEJ, Flokstra-de Blok BMJ, Hourihane JO. The effects of food allergy on quality of life. Chem Immunol Allergy. (2015) 101:235–52. doi: 10.1159/000375106
4. Ravid NL, Annunziato RA, Ambrose MA, Chuang K, Mullarkey C, Sicherer SH, et al. Mental health and quality-of-life concerns related to the burden of food allergy. Immunol Allergy Clin North Am. (2012) 32(1):83–95. doi: 10.1016/j.iac.2011.11.005
5. Bilaver LA, Chadha AS, Doshi P, O’Dwyer L, Gupta RS. Economic burden of food allergy. Ann Allergy Asthma Immunol. (2019) 122(4):373–80.e1. doi: 10.1016/j.anai.2019.01.014
6. Lopes JP, Sicherer S. Food allergy: epidemiology, pathogenesis, diagnosis, prevention, and treatment. Curr Opin Immunol. (2020) 66:57–64. doi: 10.1016/j.coi.2020.03.014
7. Santos AF, Riggioni C, Halken S, Muraro A, Akdis CA, Roberts G, et al. Executive summary of the EAACI guidelines on diagnosis of IgE-mediated food allergy. Allergy. (2023) 78(12):3053–6. doi: 10.1111/all.15906
8. Grabenhenrich LB, Reich A, Bellach J, Trendelenburg V, Sprikkelman AB, Roberts G, et al. A new framework for the documentation and interpretation of oral food challenges in population-based and clinical research. Allergy. (2016) 72(3):453–61. doi: 10.1111/all.13049
9. Patel N, Shreffler WG, Custovic A, Santos AF. Will oral food challenges still be part of allergy care in 10 years’ time? J Allergy Clin Immunol Pract. (2023) 11(4):988–96. doi: 10.1016/j.jaip.2023.02.010
10. Riggioni C, Ricci C, Moya B, Wong D, van Goor E, Bartha I, et al. Systematic review and meta-analyses on the accuracy of diagnostic tests for IGE-mediated food allergy. Allergy. (2023) 79(2):324–52. doi: 10.1111/all.15939
11. Krawiec M, Radulovic S, Foong RX, Marques-Mejias A, Bartha I, Kwok M, et al. Diagnostic utility of allergy tests to predict baked egg and lightly cooked egg allergies compared to double-blind placebo-controlled food challenges. Allergy. (2023) 78(9):2510–22. doi: 10.1111/all.15797
12. Dramburg S, Hilger C, Santos AF, De L, Aalberse RC, Acevedo N, et al. EAACI molecular allergology user’s guide 2.0. Pediatr Allergy Immunol. (2023) 34(S28):e13854. doi: 10.1111/pai.13854
13. Calamelli E, Liotti L, Beghetti I, Piccinno V, Serra L, Bottau P. Component-resolved diagnosis in food allergies. Medicina (B Aires). (2019) 55(8):498. doi: 10.3390/medicina55080498
14. Kim JF, McCleary N, Nwaru BI, Stoddart A, Sheikh A. Diagnostic accuracy, risk assessment, and cost-effectiveness of component-resolved diagnostics for food allergy: a systematic review. Allergy. (2018) 73(8):1609–21. doi: 10.1111/all.13399
15. Steckelbroeck S, Ballmer-Weber BK, Vieths S. Potential, pitfalls, and prospects of food allergy diagnostics with recombinant allergens or synthetic sequential epitopes. J Allergy Clin Immunol. (2008) 121(6):1323–30. doi: 10.1016/j.jaci.2008.04.008
16. Santos AF, Shreffler WG. Road map for the clinical application of the basophil activation test in food allergy. Clinical & Experimental Allergy. (2017) 47(9):1115–24. doi: 10.1111/cea.12964
17. Santos AF, Bécares N, Wu S-Y, Radulovic S, Stephens A, Turcanu V, et al. Basophil activation test discriminates between allergy and tolerance among peanut sensitized children. J Allergy Clin Immunol. (2013) 131(2):AB130. doi: 10.1016/j.jaci.2012.12.1133
18. Rubio A, Vivinus-Nébot M, Bourrier T, Saggio B, Albertini M, Bernard A. Benefit of the basophil activation test in deciding when to reintroduce cow’s milk in allergic children. Allergy. (2010) 66(1):92–100. doi: 10.1111/j.1398-9995.2010.02432.x
19. Wanich N, Nowak-Wegrzyn A, Sampson HA, Shreffler WG. Allergen-specific basophil suppression associated with clinical tolerance in patients with milk allergy. J Allergy Clin Immunol. (2009) 123(4):789–94. doi: 10.1016/j.jaci.2008.12.1128
20. Yanagida N, Okada Y, Sato S, Ebisawa M. New approach for food allergy management using low-dose oral food challenges and low-dose oral immunotherapies. Allergol Int. (2016) 65(2):135–40. doi: 10.1016/j.alit.2015.10.010
21. Barni S, Mori F, Piccorossi A, Sarti L, Pucci N, Maresca M, et al. Low-dose oral food challenge with hazelnut: efficacy and tolerability in children. Int Arch Allergy Appl Immunol. (2018) 178(1):97–100. doi: 10.1159/000493019
22. Hsu E, Soller L, Abrams EM, Protudjer JLP, Mill C, Chan ES. Oral food challenge implementation: the first mixed-methods study exploring barriers and solutions. J Allergy Clin Immunol Pract. (2020) 8(1):149–156.e1. doi: 10.1016/j.jaip.2019.06.034
23. Wells R, McKay C, Makwana N, Vyas D, Vaughan S, Christopher A, et al. National survey of United Kingdom paediatric allergy services. Clin Exp Allergy. (2022) 52(11):1276–90. doi: 10.1111/cea.14198
24. Bahri R, Custovic A, Korosec P, Tsoumani M, Barron M, Wu J, et al. Mast cell activation test in the diagnosis of allergic disease and anaphylaxis. J Allergy Clin Immunol. (2018) 142(2):485–96.e16. doi: 10.1016/j.jaci.2018.01.043
25. Ebo DG, Bahri R, Tontini C, Van Gasse AL, Mertens C, Hagendorens MM, et al. Mast cell versus basophil activation test in allergy: current status. Clinical & Experimental Allergy. (2024) 54(6):378–87. doi: 10.1111/cea.14487
26. Santos AF, Couto-Francisco N, Bécares N, Kwok M, Bahnson HT, Lack G. A novel human mast cell activation test for peanut allergy. J Allergy Clin Immunol. (2018) 142(2):689–91.e9. doi: 10.1016/j.jaci.2018.03.011
27. Santos AF, Douiri A, Bécares N, Wu S-Y, Stephens A, Radulovic S, et al. Basophil activation test discriminates between allergy and tolerance in peanut-sensitized children. J Allergy Clin Immunol. (2014) 134(3):645–52. doi: 10.1016/j.jaci.2014.04.039
28. Suprun M, Getts R, Raghunathan R, Grishina G, Witmer M, Gimenez G, et al. Novel bead-based epitope assay is a sensitive and reliable tool for profiling epitope-specific antibody repertoire in food allergy. Sci Rep. (2019) 9(1):18425. doi: 10.1038/s41598-019-54868-7
29. Suárez-Fariñas M, Suprun M, Kearney P, Getts R, Grishina G, Hayward C, et al. Accurate and reproducible diagnosis of peanut allergy using epitope mapping. Allergy. (2021) 76(12):3789–97. doi: 10.1111/all.14905
30. Santos AF, Kulis MD, Sampson HA. Bringing the next generation of food allergy diagnostics into the clinic. J Allergy Clin Immunol Pract. (2022) 10(1):1–9. doi: 10.1016/j.jaip.2021.09.009
31. Hemmings O, Niazi U, Kwok M, James LK, Lack G, Santos AF. Peanut diversity and specific activity are the dominant IGE characteristics for effector cell activation in children. J Allergy Clin Immunol. (2021) 148(2):495–505.e14. doi: 10.1016/j.jaci.2021.02.029
32. van Ree R. Carbohydrate epitopes and their relevance for the diagnosis and treatment of allergic diseases. Int Arch Allergy Appl Immunol. (2002) 129(3):189–97. doi: 10.1159/000066770
33. Amoah AS, Obeng BB, Larbi IA, Versteeg SA, Aryeetey Y, Akkerdaas JH, et al. Peanut-specific IGE antibodies in asymptomatic Ghanaian children possibly caused by carbohydrate determinant cross-reactivity. J Allergy Clin Immunol. (2013) 132(3):639–47. doi: 10.1016/j.jaci.2013.04.023
34. Homann A, Schramm G, Jappe U. Glycans and glycan-specific IGE in clinical and molecular allergology: sensitization, diagnostics, and clinical symptoms. J Allergy Clin Immunol. (2017) 140(2):356–68. doi: 10.1016/j.jaci.2017.04.019
35. Commins SP, Satinover SM, Hosen J, Mozena J, Borish L, Lewis BD, et al. Delayed anaphylaxis, angioedema, or urticaria after consumption of red meat in patients with IGE antibodies specific for galactose-α-1,3-galactose. J Allergy Clin Immunol. (2009) 123(2):426–433.e2. doi: 10.1016/j.jaci.2008.10.052
36. Shade KTC, Conroy ME, Washburn N, Kitaoka M, Huynh DJ, Laprise E, et al. Sialylation of immunoglobulin E is a determinant of allergic pathogenicity. Nature. (2020) 582:265–70. doi: 10.1038/s41586-020-2311-z
37. Rowntree S, Plattsmills T, Cogswell J, Mitchell E. A subclass igg4-specific antigen-binding radioimmunoassay (RIA): comparison between IGG and igg4 antibodies to food and inhaled antigens in adult atopic dermatitis after desensitization treatment and during development of antibody responses in children. J Allergy Clin Immunol. (1987) 80(4):622–30. doi: 10.1016/0091-6749(87)90017-0
38. Wilson JM, Workman L, Schuyler AJ, Rifas-Shiman SL, McGowan EC, Oken E, et al. Allergen sensitization in a birth cohort at midchildhood: focus on food component IGE and igg4 responses. J Allergy Clin Immunol. (2018) 141(1):419–23.e5. doi: 10.1016/j.jaci.2017.07.029
39. Stapel SO, Asero R, Ballmer-Weber BK, Knol EF, Strobel S, Vieths S, et al. Testing for igg4 against foods is not recommended as a diagnostic tool: eAACI task force report*. Allergy. (2008) 63(7):793–6. doi: 10.1111/j.1398-9995.2008.01705.x
40. Du Toit G, Roberts G, Sayre PH, Bahnson HT, Radulovic S, Santos AF, et al. Randomized trial of peanut consumption in infants at risk for peanut allergy. N Engl J Med. (2015) 372(9):803–13. doi: 10.1056/NEJMoa1414850
41. Marques-Mejias A, Radulovic S, Foong RX, Bartha I, Krawiec M, Kwok M, et al. Partial egg consumption modifies the diagnostic performance of allergy tests to predict outcome of double-blind placebo-controlled food challenges to egg. J Allergy Clin Immunol Pract. (2024) 12(3):660–9.e5. doi: 10.1016/j.jaip.2023.12.036
42. Janzi M, Kull I, Sjöberg R, Wan J, Melén E, Bayat N, et al. Selective IgA deficiency in early life: association to infections and allergic diseases during childhood. Clin Immunol. (2009) 133(1):78–85. doi: 10.1016/j.clim.2009.05.014
43. Kulis MD, Saba K, Kim E, Bird JA, Kamilaris N, Vickery BP, et al. Increased peanut-specific IgA levels in saliva correlate with food challenge outcomes after peanut sublingual immunotherapy. J Allergy Clin Immunol. (2012) 129(4):1159–62. doi: 10.1016/j.jaci.2011.11.045
44. Liu EG, Zhang B, Martin V, Anthonypillai J, Kraft M, Grishin A, et al. Food-specific immunoglobulin A does not correlate with natural tolerance to peanut or egg allergens. Sci Transl Med. (2022) 14(671):eabq0599. doi: 10.1126/scitranslmed.abq0599
45. Wambre E, Bajzik V, DeLong JH, O’Brien K, Nguyen Q-A, Speake C, et al. A phenotypically and functionally distinct human TH2 cell subpopulation is associated with allergic disorders. Sci Transl Med. (2017) 9(401):eaam9171. doi: 10.1126/scitranslmed.aam9171
46. Calise J, DeBerg H, Garabatos N, Khosa S, Bajzik V, Calderon LB, et al. Distinct trajectories distinguish antigen-specific T cells in peanut-allergic individuals undergoing oral immunotherapy. J Allergy Clin Immunol. (2023) 152(1):155–66.e9. doi: 10.1016/j.jaci.2023.03.020
47. Crespo JF, Cabanillas B. Recent advances in cellular and molecular mechanisms of IGE-mediated food allergy. Food Chem. (2023) 411:135500. doi: 10.1016/j.foodchem.2023.135500
48. Savilahti EM, Karinen S, Salo HM, Klemetti P, Saarinen KM, Klemola T, et al. Combined T regulatory cell and Th2 expression profile identifies children with cow’s milk allergy. Clin Immunol. (2010) 136(1):16–20. doi: 10.1016/j.clim.2010.02.011
49. Croote D, Darmanis S, Nadeau KC, Quake SR. High-affinity allergen-specific human antibodies cloned from single IGE B cell transcriptomes. Science. (2018) 362(6420):1306–9. doi: 10.1126/science.aau2599
50. Jiménez-Saiz R, Ellenbogen Y, Bruton K, Spill P, Sommer DD, Lima H, et al. Human BCR analysis of single-sorted, putative IGE+ memory B cells in food allergy. J Allergy Clin Immunol. (2019) 144(1):336–9.e6. doi: 10.1016/j.jaci.2019.04.001
51. Hoh RA, Joshi SA, Lee J-Y, Martin BA, Varma S, Kwok S, et al. Origins and clonal convergence of gastrointestinal IGE B cells in human peanut allergy. Sci Immunol. (2020) 5(45):eaay4209. doi: 10.1126/sciimmunol.aay4209
52. Koenig JF, Knudsen PH, Phelps A, Bruton K, Hoof I, Lund G, et al. Type 2–polarized memory B cells hold allergen-specific IgE memory. Sci Transl Med. (2024) 16(733):eadi0944. doi: 10.1126/scitranslmed.adi0944
53. von Borstel A, O’Hehir RE, van Zelm MC. Ige in allergy: it takes two. Sci Transl Med. (2024) 16(733):eadl1202. doi: 10.1126/scitranslmed.adl1202
54. Ota M, Hoehn KB, Fernandes-Braga W, Ota T, Aranda CJ, Friedman S, et al. CD23+IgG1+memory B cells are poised to switch to pathogenic IgE production in food allergy. Sci Transl Med. (2024) 16(733):eadi0673. doi: 10.1126/scitranslmed.adi0673
55. Ho H, Chun Y, Jeong S, Jumreornvong O, Sicherer SH, Bunyavanich S. Multidimensional study of the oral microbiome, metabolite, and immunologic environment in peanut allergy. J Allergy Clin Immunol. (2021) 148(2):627–32.e3. doi: 10.1016/j.jaci.2021.03.028
56. Chun Y, Grishin A, Rose R, Zhao W, Arditi Z, Zhang L, et al. Longitudinal dynamics of the gut microbiome and metabolome in peanut allergy development. J Allergy Clin Immunol. (2023) 152(6):1569–80. doi: 10.1016/j.jaci.2023.08.012
57. Zhang L, Chun Y, Ho H, Arditi Z, Lo T, Sajja S, et al. Multiscale study of the oral and gut environments in children with high- and low-threshold peanut allergy. J Allergy Clin Immunol. (2022) 150(3):714–20.e2. doi: 10.1016/j.jaci.2022.04.026
58. Vujkovic-Cvijin I, Sklar J, Jiang L, Natarajan L, Knight R, Belkaid Y. Host variables confound gut microbiota studies of human disease. Nature. (2020) 587(7834):448–54. doi: 10.1038/s41586-020-2881-9
59. Stevenson L. Investigation of the gut microbiome using machine learning as a diagnostic aid for food allergies (Master’s thesis). (2022). Available online at: https://www.proquest.com/docview/2736292775/fulltextPDF/B49FBE919C00454CPQ/1?accountid=11862&sourcetype=Dissertations%20&%20Theses (cited August 24, 2024).
60. Foong RX, Santos A. Biomarkers of diagnosis and resolution of food allergy. Pediatr Allergy Immunol. (2021) 32(2):223–33. doi: 10.1111/pai.13389
61. Radonjic-Hoesli S, Pavlov N, Simon H-U, Simon D. Are blood cytokines reliable biomarkers of allergic disease diagnosis and treatment responses? J Allergy Clin Immunol. (2022) 150(2):251–8. doi: 10.1016/j.jaci.2022.06.008
62. Dang TD, Tang ML, Koplin JJ, Licciardi PV, Eckert JK, Tan T, et al. Characterization of plasma cytokines in an infant population cohort of challenge-proven food allergy. Allergy. (2013) 68(10):1233–40. doi: 10.1111/all.12215
63. Kulis M, Yue X, Guo R, Zhang H, Orgel K, Ye P, et al. High- and low-dose oral immunotherapy similarly suppress pro-allergic cytokines and basophil activation in young children. Clin Exp Allergy. (2018) 49(2):180–9. doi: 10.1111/cea.13256
64. Gorelik M, Narisety SD, Guerrerio AL, Chichester KL, Keet CA, Bieneman AP, et al. Suppression of the immunologic response to peanut during immunotherapy is often transient. J Allergy Clin Immunol. (2015) 135(5):1283–92. doi: 10.1016/j.jaci.2014.11.010
65. Chiang D, Chen X, Jones SM, Wood RA, Sicherer SH, Burks AW, et al. Single-cell profiling of peanut-responsive T cells in patients with peanut allergy reveals heterogeneous effector TH2 subsets. J Allergy Clin Immunol. (2018) 141(6):2107–20. doi: 10.1016/j.jaci.2017.11.060
66. Neeland MR, Andorf S, Dang TD, McWilliam VL, Perrett KP, Koplin JJ, et al. Altered immune cell profiles and impaired CD4T-cell activation in single and multi-food allergic adolescents. Clin Exp Allergy. (2021) 51(5):674–84. doi: 10.1111/cea.13857
67. Metwally AA, Yu PS, Reiman D, Dai Y, Finn PW, Perkins DL. Utilizing longitudinal microbiome taxonomic profiles to predict food allergy via long short-term memory networks. PLoS Comput Biol. (2019) 15(2):e1006693. doi: 10.1371/journal.pcbi.1006693
68. Alag A. Machine learning approach yields epigenetic biomarkers of food allergy: a novel 13-gene signature to diagnose clinical reactivity V1. PLoS One. (2018) 14(6). doi: 10.17504/protocols.io.wa8fahw
69. Kuniyoshi Y, Tokutake H, Takahashi N, Kamura A, Yasuda S, Tashiro M. Machine learning approach and oral food challenge with heated egg. Pediatr Allergy Immunol. (2021) 32(4):776–8. doi: 10.1111/pai.13433
70. Suárez-Fariñas M, Suprun M, Chang HL, Gimenez G, Grishina G, Getts R, et al. Predicting development of sustained unresponsiveness to milk oral immunotherapy using epitope-specific antibody binding profiles. J Allergy Clin Immunol. (2019) 143(3):1038–46. doi: 10.1016/j.jaci.2018.10.028
71. Chong KW, Ruiz-Garcia M, Patel N, Boyle RJ, Turner PJ. Reaction phenotypes in IGE-mediated food allergy and anaphylaxis. Ann Allergy Asthma Immunol. (2020) 124(5):473–8. doi: 10.1016/j.anai.2019.12.023
72. Martinroche G, Goret J, Guemari A, Apoil P-A, Annesi-Maesano I, Fromentin E, et al. Towards an artificial intelligence-assisted allergy diagnosis using semantic attributes and allergen Multiplex technology. J Allergy Clin Immunol. (2024) 153(2):AB373. doi: 10.1016/j.jaci.2023.11.895
73. Martino D, Dang T, Sexton-Oates A, Prescott S, Tang MLK, Dharmage S, et al. Blood DNA methylation biomarkers predict clinical reactivity in food-sensitized infants. J Allergy Clin Immunol. (2015) 135(5):1319–28.e12. doi: 10.1016/j.jaci.2014.12.1933
74. Hemmings O, Niazi U, Kwok M, Radulovic S, Du Toit G, Lack G, et al. Combining allergen components improves the accuracy of peanut allergy diagnosis. J Allergy Clin Immunol Pract. (2022) 10(1):189–99. doi: 10.1016/j.jaip.2021.08.029
75. Bunyavanich S, Schadt EE. Systems biology of asthma and allergic diseases: a multiscale approach. J Allergy Clin Immunol. (2015) 135(1):31–42. doi: 10.1016/j.jaci.2014.10.015
76. Sato S, Tachimoto H, Shukuya A, Kurosaka N, Yanagida N, Utsunomiya T, et al. Basophil activation marker CD203c is useful in the diagnosis of hen’s egg and cow’s milk allergies in children. Int Arch Allergy Appl Immunol. (2010) 152(1):54–61. doi: 10.1159/000312126
77. Ocmant A, Mulier S, Hanssens L, Goldman M, Casimir G, Mascart F, et al. Basophil activation tests for the diagnosis of food allergy in children. Clin Exp Allergy. (2009) 39(8):1234–45. doi: 10.1111/j.1365-2222.2009.03292.x
78. Tokuda R, Nagao M, Hiraguchi Y, Hosoki K, Matsuda T, Kouno K, et al. Antigen-induced expression of CD203c on basophils predicts IgE-mediated wheat allergy. Allergol Int. (2009) 58(2):193–9. doi: 10.2332/allergolint.08-OA-0023
79. Glaumann S, Nopp A, Johansson SGO, Rudengren M, Borres MP, Nilsson C. Basophil allergen threshold sensitivity, CD-sens, IgE-sensitization and DBPCFC in peanut-sensitized children. Allergy. (2011) 67(2):242–7. doi: 10.1111/j.1398-9995.2011.02754.x
80. Brandström J, Nopp A, Johansson SGO, Lilja G, Sundqvist AC, Borres MP, et al. Basophil allergen threshold sensitivity and component-resolved diagnostics improve hazelnut allergy diagnosis. Clin Exp Allergy. (2015) 45(9):1412–8. doi: 10.1111/cea.12515
81. Alessandri C, Sforza S, Palazzo P, Lambertini F, Paolella S, Zennaro D, et al. Tolerability of a fully maturated cheese in cow’s milk allergic children: biochemical, immunochemical, and clinical aspects. PLoS One. (2012) 7(7):e40945. doi: 10.1371/journal.pone.0040945
82. Ando H, Movérare R, Kondo Y, Tsuge I, Tanaka A, Borres MP, et al. Utility of ovomucoid-specific IgE concentrations in predicting symptomatic egg allergy. J Allergy Clin Immunol. (2008) 122(3):583–8. doi: 10.1016/j.jaci.2008.06.016
83. Ayuso R, Sánchez-Garcia S, Pascal M, Lin J, Grishina G, Fu Z, et al. Is epitope recognition of shrimp allergens useful to predict clinical reactivity? Clin Exp Allergy. (2011) 42(2):293–304. doi: 10.1111/j.1365-2222.2011.03920.x
84. Klemans RJB, Liu X, Knulst AC, Knol MJ, Gmelig-Meyling F, Borst E, et al. Ige binding to peanut components by four different techniques: ara h 2 is the most relevant in peanut allergic children and adults. Clin Exp Allergy. (2013) 43(8):967–74. doi: 10.1111/cea.12136
85. Lieberman JA, Glaumann S, Batelson S, Borres MP, Sampson HA, Nilsson C. The utility of peanut components in the diagnosis of IgE-mediated peanut allergy among distinct populations. J Allergy Clin Immunol Pract. (2013) 1(1):75–82. doi: 10.1016/j.jaip.2012.11.002
86. Masthoff LJN, Mattsson L, Zuidmeer-Jongejan L, Lidholm J, Andersson K, Akkerdaas JH, et al. Sensitization to cor a 9 and cor a 14 is highly specific for a hazelnut allergy with objective symptoms in Dutch children and adults. J Allergy Clin Immunol. (2013) 132(2):393–9. doi: 10.1016/j.jaci.2013.02.024
87. Lin J, Bruni FM, Fu Z, Maloney J, Bardina L, Boner AL, et al. A bioinformatics approach to identify patients with symptomatic peanut allergy using peptide microarray immunoassay. J Allergy Clin Immunol. (2012) 129(5):1321–1328.e5. doi: 10.1016/j.jaci.2012.02.012
88. Zhang JZ, Lee DH, Jungles KN, Shaltis D, Najarian K, Ravikumar R, et al. Prediction of oral food challenge outcomes via ensemble learning. Inform Med Unlocked. (2023) 36:101142–2. doi: 10.1016/j.imu.2022.101142
Keywords: food allergy, diagnosis, molecular allergology, basophil activation test, mast cell activation test, bead-based epitope assay, T cell assays, microbiome analysis
Citation: Wong DSH and Santos AF (2024) The future of food allergy diagnosis. Front. Allergy 5:1456585. doi: 10.3389/falgy.2024.1456585
Received: 28 June 2024; Accepted: 7 October 2024;
Published: 7 November 2024.
Edited by:
Takahisa Murata, The University of Tokyo, JapanReviewed by:
Annette Kuehn, Luxembourg Institute of Health, LuxembourgNanae Nagata, The University of Tokyo, Japan
Copyright: © 2024 Wong and Santos. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Alexandra F. Santos, YWxleGFuZHJhLnNhbnRvc0BrY2wuYWMudWs=
 Dominic S. H. Wong1
Dominic S. H. Wong1 Alexandra F. Santos
Alexandra F. Santos