- 1Research Division of Infection Biology, Department of Medicine I, Medical University of Vienna, Vienna, Austria
- 2CeMM Research Center for Molecular Medicine of the Austrian Academy of Sciences, Vienna, Austria
Asthma is a chronic respiratory disease of global importance. Mouse models of allergic asthma have been instrumental in advancing research and novel therapeutic strategies for patients. The application of relevant allergens and physiological routes of exposure in such models has led to valuable insights into the complexities of asthma onset and development as well as key disease mechanisms. Furthermore, environmental microbial exposures and infections have been shown to play a fundamental part in asthma pathogenesis and alter disease outcome. In this review, we delve into physiological mouse models of allergic asthma and explore literature reports on most significant interplays between microbial infections and asthma development with relevance to human disease.
Introduction
Asthma is the most prevalent chronic respiratory disease with over 260 million cases worldwide (1). This heterogenous lung disorder leads to airway obstruction that can severely affect basic lung function. Asthma is now recognized as a syndrome encompassing a large array of symptoms, such as shortness of breath, wheezing, coughing, as well as chest tightness and pain with fluctuating severity (2–4).
Classically, asthma has been viewed as the archetype of a type 2 immunity-mediated, allergic disease. Many asthmatic patients show enhanced allergen-specific immunoglobulin E (IgE), eosinophilia and a predominant type 2 T helper (Th2) cell phenotype in the blood and lung lavage fluid (5). However, more recent evidence also suggests the existence of non-Th2-driven asthma (6). For the sake of simplicity, we here use the sole term “asthma” referring to the Th2-driven disease endotype.
Successful translational research requires comprehensive understanding of allergic responses in physiologically relevant contexts. On this basis, murine models have emerged as indispensable tools, unraveling intricate immunopathological and molecular mechanisms while serving as platforms for the evaluation of innovative treatment strategies. In the early 1990s, researchers successfully established mouse models that recreated several key features of allergic asthma, such as IgE production, airway remodeling, eosinophilia and bronchial hyperresponsiveness (7). These tools have greatly facilitated our understanding of key immune mechanisms during acute allergic inflammation in context of the entire organism.
However, most of the early asthma models required unnatural sensitization routes (i.e., intraperitoneal injections) and the use of isolated non-respiratory allergens with low immunogenicity, such as chicken ovalbumin (OVA), alongside powerful chemical adjuvants to achieve a potent type 2 immune response (8, 9).
Over the last decades, more and more physiological asthma mouse models have been developed that use, for instance, extracts of naturally occurring allergen sources and routes of exposure that are relevant for humans. These frameworks could recapitulate the key features of type 2 immune responses and, importantly, some of the complex pathological mechanisms of asthma (3, 10).
In this review, we aimed to provide an overview of adjuvant-free murine asthma models and discuss their relevance to human disease. Special emphasis was placed on the application of naturally occuring allergens (as part of allergen extracts or derived purified components in relation to human disease and sensitization). To integrate the context of physiologic environmental modulators of asthma, we discuss important microbes that can evidently initiate or potentiate allergic immune responses and airway inflammation.
Modeling the key characteristics of allergic asthma
Hallmarks of allergic asthma
Asthma is a multifactorial heterogenous disorder. Its development is thought to be influenced by a variety of predisposing factors, such as genetic background, lifestyle and environmental exposures (11) (Figure 1). Asthma is primarily defined by a narrowing of the airways, which is mediated by two main events: initially, an inflammation within the airway lining manifests, characterized by immune cell activation and infiltration and consequent inflammatory mediator production (3, 10). The chronicity of this inflammatory phase will subsequently allow a complex and prolonged interplay between airway immune and structural cells, leading to tissue remodeling. These changes in pulmonary structure are driven by epithelial cell activation, muscle cell hyperplasia, goblet cell metaplasia and hypervascularity. Pathologic airway remodeling typically associates with excessive collagen deposition, mucus hypersecretion or mucus plug formation and thickening of the airway wall, often resulting in lung dysfunction (10, 12).
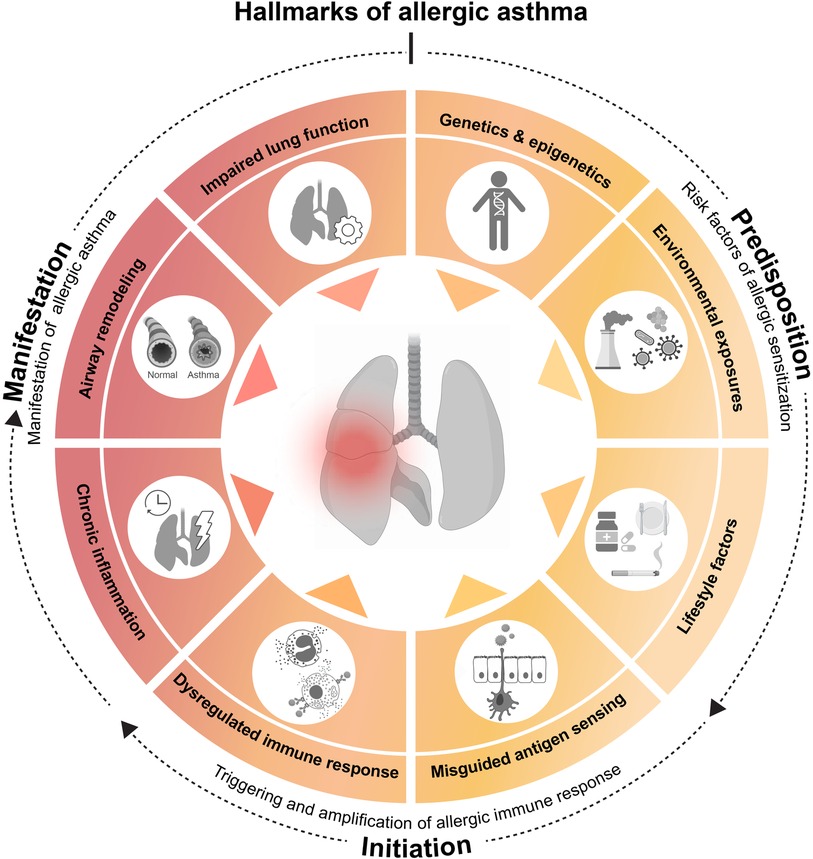
Figure 1. Hallmarks of allergic asthma. Asthma is a multifactorial disorder whose initial development seems influenced by predisposing factors, such as (epi)genetics, environmental exposures and lifestyle. The underlying cascade of immune events is often triggered and amplified by a misdirected immune response against apparently harmless environmental substances. Repeated allergen exposure of the airway then results in the clinical manifestations of the disease. On basis of these observations and in an attempt to simplify a highly complex pathology, we propose the following 8 hallmarks of allergic asthma. Predisposing hallmarks which encompass major risk factors [(1) genetic and epigenetic determinants, (2) environmental influences, such as pollution or microbes and (3) lifestyle factors, such as smoking, medication or dietary habits] that facilitate hallmarks of initiation [based on (4) misguided sensing of environmental substances as noxious, leading to (5) a dysregulated immune response], resulting in allergic sensitization. Finally, recurring airway allergen exposure results in manifestation hallmarks of allergic asthma, consist of major clinical and pathological characteristics [(6) chronic lung inflammation, (7) airway remodeling, and (8) impaired lung function]. This illustration was created with BioRender.com.
The immunological features of asthma have been instrumental in segregating the various endotypes of this disorder (5, 7). The most notable immunological milestones from asthma initiation to manifestation include pulmonary infiltration of immune cells [such as dendritic cells (DCs), eosinophils, neutrophils, mast cells, type 2 innate lymphoid cells (ILC2s) and lymphocytes] as well as secretion of type 2 cytokines [e.g., interleukin (IL)-4, IL-5, IL-13] and chemoattractants (e.g., eotaxins and complement anaphylatoxins). The imbalanced Th cell (Th2 dominant) response often leads to allergen-specific IgE and IgG1 production (5). In this review, we refer to these antibodies collectively as type 2 antibodies due to the dependence of their production on type 2 cytokines (IL-4 and IL-13) (13).
Modeling asthma in mice without adjuvants
A considerable difference to humans is that mice housed in a laboratory environment do not naturally develop allergic airway inflammation and hyperresponsiveness but require an artificial, experimental framework for asthma onset (14). To faithfully and realistically replicate an allergic response in mice, most protocols typically involve a two-phase approach (8). The first phase of initial allergen exposure (referred to as “sensitization”) leads to an induction of antigen-specific type 2 immunity. The second phase (referred to as “challenge”) encompasses secondary allergen exposure(s) of the sensitized individual and results in an allergic reaction and inflammation. The sensitization protocols and experimental approaches can vary greatly between studies. However, one could consider that “physiological” mouse models of asthma rely on three fundamental principles to facilitate clinical and pathological translation (Figure 2): (1) physiologic allergen exposure conditions via barrier organs (skin or airways); (2) use of clinically significant allergens (i.e., with reported sensitization in humans) and allergen extracts and avoidance of clinically irrelevant (i.e., no evidence for driving sensitization in humans) adjuvants; (3) appropriate exposure regimen that consider realistic allergen doses as well as exposure frequency and duration to eventually recreate the main features of human asthma.
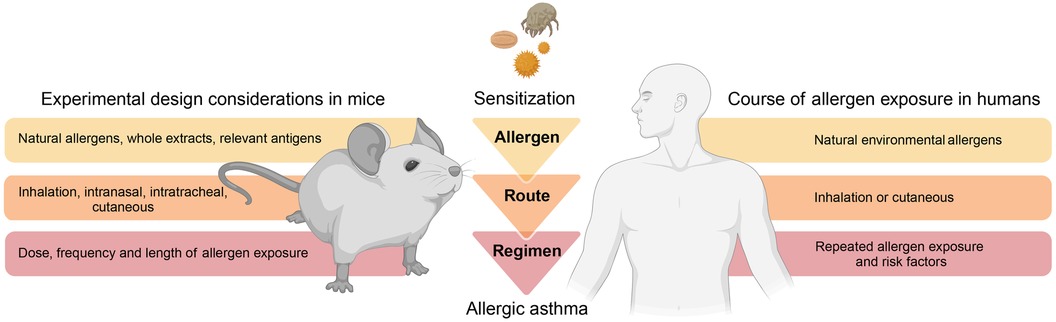
Figure 2. Modelling allergic asthma: mouse vs human. Mouse models of allergic asthma response ideally implement fundamental principles that consider physiologic aspects related to disease onset and progression in humans. As such, mouse models should consider physiologic allergen exposure conditions via barrier organs, utilize relevant allergens and avoid clinically irrelevant adjuvants, as well as incorporate realistic protocols of allergen exposure. This illustration was created with BioRender.com.
Adjuvant-independent mouse models of allergic asthma
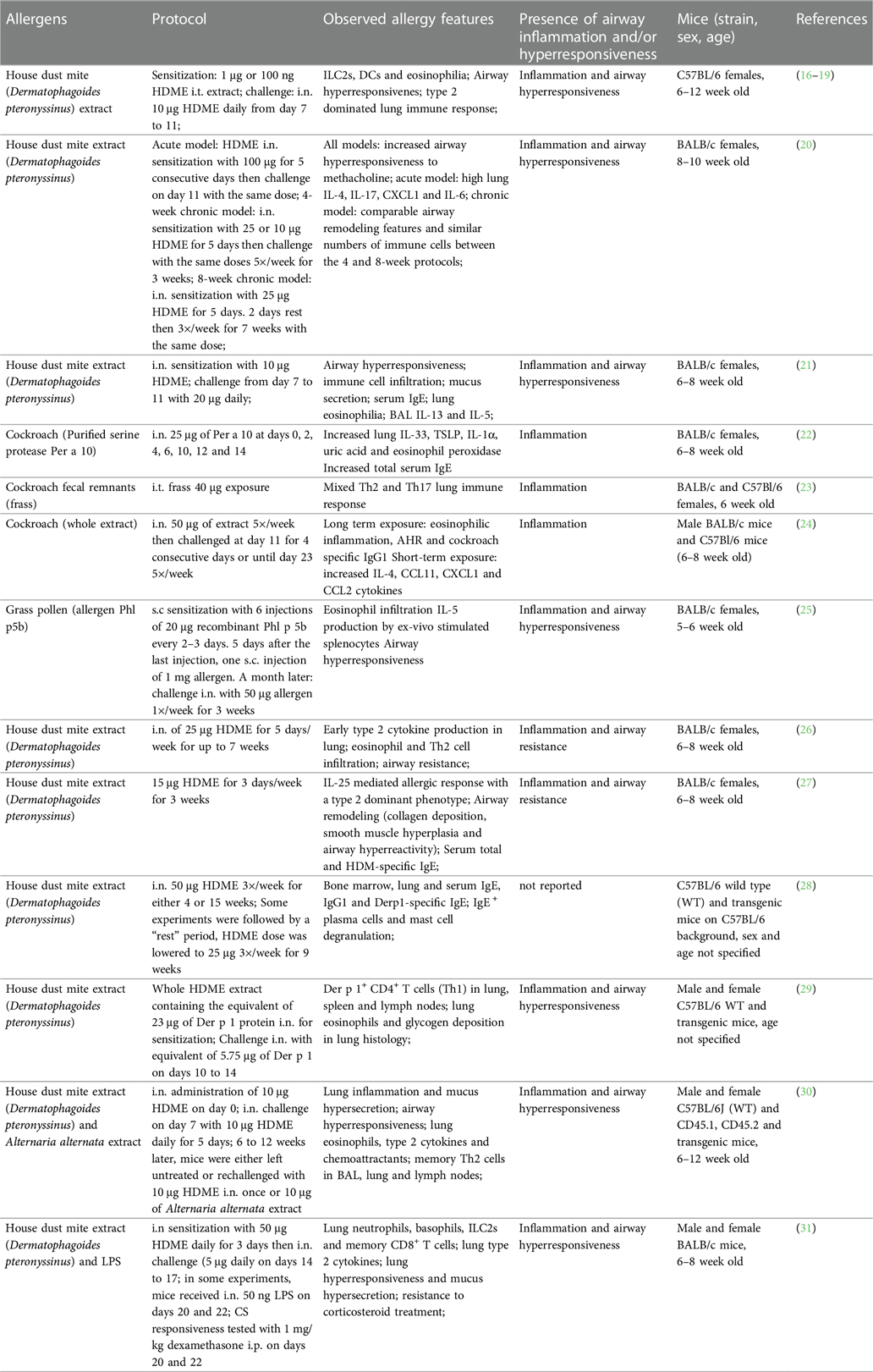
Various mouse models based on natural allergens were developed to investigate the mechanisms of allergen-driven airway immune and functional responses during acute and established inflammation (15). In this section, we review and discuss examples of such protocols that recreate main features of allergic asthma in mice using relevant allergens in absence of chemical adjuvants. All mouse models and treatment protocols are summarized in (Table 1).
Acute allergic airway inflammation models
House dust mite
House dust mites (HDMs; mainly Dermatophagoides pteronyssinus and Dermatophagoides farinae) are among the most prevalent sources of indoor allergens and widely used to replicate allergic asthma in mice (15, 32). HDMs produce diverse allergenic proteins, including the major allergens Der p 1 and Der p 2, most of which are molecularly and functionally characterized (33–35). Additionally, HDMs are known carriers of various bacteria, fungi and pollutants (36, 37). In combination with HDM allergens, these environmental components are potent triggers of the innate and adaptive immune system, thus driving the allergic sensitization to HDMs. Several research groups have demonstrated allergic sensitization against HDMs using a model of intratracheal (i.t.) sensitization with HDM-extract (HDME), followed by intranasal (i.n.) allergen challenges (16–19). Most protocols consisted of one i.t. low dose (1 µg) HDME instillation at day 0, followed by 10-fold higher i.n. allergen doses 6 or 7 days later for 5 consecutive days and endpoint analyses approximately 3 days after the last challenge (16–19) (Table 1). Such treatments induced a robust Th2 immune response, driven by epithelial cell damage and activation of DCs, granulocytes (mast cells, eosinophils, basophils) and ILC2s (16–19). Additionally, most of these studies reported bronchial hyperreactivity, restructured lung architecture and mucus secretion as well as significant HDM-specific IgE production (17, 19). Other protocols rely on i.n. administration of HDME for both, allergen sensitization and challenge (20, 21, 26, 27) with the advantages that i.n. requires less experimental training and equipment than i.t. application while being similarly relevant for respiratory allergen exposure. Compared to models utilizing also i.t. application, these exclusively i.n.-based models use increased allergen doses (10–100 μg) or a higher frequency and longer exposure periods during the sensitization and challenge phases (20, 21, 26, 27) (Table 1). These protocols recreated the main features of asthma, including pulmonary type 2 cytokine secretion and leukocyte infiltration, lung inflammation, impaired lung function and increased IgE and IgG1. In summary, both, i.n. and i.t. HDME instillations reproduce robust type 2 immune-dominated inflammation in the lung with comparable pathology and functional impairment.
Despite standardized production protocols and environments, significant batch-to-batch and lot-to-lot variation in commercial HDME has been observed, reflected by differences in protease activity as well as allergen, lipopolysaccharide (LPS), chitin and ß-glucan content. These variations could be attributed to differential HDM culture conditions. Indeed, several studies have demonstrated that the composition of culture media, growth conditions of HDMs and their diet can alter the production of relevant allergens (38–41).
For instance, two recent studies have compared different HDME lots and their potency to induce allergic immune responses (42, 43). Assessing six HDME lots differing in total protein, antigen, and endotoxin contents, Cyphert-Daly et al. (42) reported that Der p 1 concentration in the HDME correlates with lung IL-5 and eosinophilia, serum IgE as well total protein in bronchoalveolar lavage (BAL) fluid. While only some of the extracts were able to induce airway resistance, all lots induced peribronchial inflammation and increased mucus secretion. Notably, the lots were tested in two different HDM allergy models: an acute asthma model with weekly i.n. HDME instillation for 3 weeks and a setting reflecting chronic sensitization based on one intraperitoneal (i.p.) HDME injection, followed by weekly oropharyngeal allergen administration from week 3 to 7 post sensitization (42). The second study by Pascoe et al. (43) compared two lots of HDME with either low or high LPS levels but similar in Der p 1 abundance. The mice were treated i.n. with 25 μg HDME 5 times a week for two weeks. The authors found that LPS differences did not influence HDME-induced pulmonary immune cell infiltration or IL-4 in the BAL fluid. However, LPS-high HDME resulted in increased transforming growth factor ß (TGFß) and decreased IL-5 (compared to LPS-low HDME) and the exclusive induction (compared to mock treatment) of Eotaxin, IL-17 and RANTES. Remarkably, only LPS-low HDME induced significant vascular endothelial growth factor (VEGF) levels in BAL fluid. In addition, HDME application induced a broad lung transcriptional response that was specifically influenced by LPS levels. Another important observation of the study was that LPS-high HDME appeared to impair lung function more efficiently.
Addressing a related question, Hadebe et al. compared the effects of LPS and ß-glycan on HDME-mediated airway inflammation (44) using a model used was based on two i.t. applications of HDME alone or in combination with LPS and ß-glucan, followed two weeks later by challenges on three consecutive days. Presence of LPS and ß-glycan resulted in significantly higher pulmonary neutrophil infiltration as well as IL-17, CC chemokine ligand (CCL)3 and CCL5. However, HDME alone caused more mucus production, airway resistance and lung tissue cytokines (IL-4, IL-5 and IL-6) (45).
It should be emphasized that most of the referenced studies used female mice due to their enhanced susceptibility for allergy development (46). Additionally, BALB/c mice were mostly the strain of choice. This detail is relevant since genetic mouse strain differences can influence the outcome of type 2 immune responses, for instance against parasites (47, 48). However, a study by HayGlass and colleagues has reported no inherent Th1/Th2 bias in allergen-driven responses in mice immunized with recombinant (r)Der p 1, OVA or human serum albumin with adjuvant in BALB/c vs. C57BL/6J mice (49). More recently, the key role of the MHC II haplotype for allergic sensitization has been reported (50). As such, the H2d MHC II haplotype of BALB/c mice is associated with an inefficient antibody response to Der p 2 while the H2b MHC II haplotype of C57BL/6 mice seems to drive the strong humoral response against the allergen observed in this mouse strain (50). Additionally, these two strains showed differences in the pulmonary cellular response to Der p 2, with a predominantly eosinophilic phenotype in C57BL/6J vs. neutrophilic in BALB/c mice (50). This important finding suggests that the immune response to whole HDM extracts in BALB/c mice is predominantly attributed to Der p 1 (and allergens other than Der p 2) and potentially other environmental extract constituents. Importantly, genetic background has also been tied to inherent structural and physiological differences in the lungs of BALB/c vs. C57BL/6J mice (51–55). For instance, C57BL/6J mice display a greater lung elastance as well as lung collagen content than BALB/c (51–54). Moreover, strain-dependent differences were also reflected by differing eosinophil distribution and localization within the lung (53).
Another important point to consider in context with allergen exposure in mouse models is the dose relevance. Several studies investigated personal indoor mite exposure over time (56–61). The devices used to sample aeroallergens in the environment varied from portable air samplers to intranasal or body-worn samplers (56–61). Based on these investigations, the mean aeroallergen concentration (presented in Der p 1, allergen or protein units) during undisturbed conditions ranged from 0 to 1.7 ng allergen/m3. During human activities (e.g., vacuuming, changing bedsheets, or outdoor transport), it ranged from 0.3 to 190 ng allergen/m3 (56). While most these studies focused on the mean allergen levels as opposed to other statistical measures, such as peak allergen concentrations (56), Swanson and colleagues reported a peak allergen exposure of 736 ng house dust mite allergen/m3 after 5 min of making a bed (61). This is substantially lower than the doses of HDME typically used intranasally in mice which range from 1 to 100 µg (Table 1). Thus, additional mouse models relying on lower HDME doses (but perhaps at a higher frequency) during sensitization and challenge could model human exposure and sensitization in a more relevant way.
Overall, these findings corroborate the relevance of HDME-mediated asthma mouse models to study allergic immune responses and various human disease endotypes (5, 10). A special emphasis must be put on the composition of HDME as it can affect several aspects of the allergic asthma response. In this context, it is advisable to use the same or comparable extract batches throughout a study. In addition, we encourage authors of experimental studies to disclose available details on used extracts, such as batch identifiers as well as allergen or LPS content, in the methods sections of their research papers.
Cockroach
Cockroaches represent another important environmental source of natural allergens with high relevance to human asthma (62). Indeed, cockroach allergens are frequently detected in urban homes in the United States and can trigger positive skin prick tests in the majority of asthmatic inner-city children (62, 63). Several cockroach components have been identified as potent IgE-inducing allergens, mainly in the German (Blatella germanica; Bla g allergen groups) and American cockroaches (Periplaneta americana; Per a allergen groups) (62). Many cockroach-mediated asthma mouse models involve the use of adjuvants (64–67). However, several studies have reported effective adjuvant-free protocols based on i.n. sensitization to cockroach allergen (Per a 10) (22) or whole extract (24), or i.t administration of cockroach fecal remnants (23). These models (Table 1) were associated with increased airway inflammation and resistance, enhanced cytokines [thymic stromal lymphopoietin (TSLP), IL-13, IL-4, IL-5 and Eotaxin] in lung tissue or bronchoalveolar lavage (22–24), and type 2 (IgE (22) and IgG1 (24)) antibody production.
Grass pollen
Pollen allergy is amongst the most important risk factors for asthma onset and exacerbations (68, 69). In patients with asthma and seasonal allergic rhinitis, outdoor pollen exposure can efficiently induce type 2 immune responses, including eosinophilia, type 2 cytokines and IgE antibodies (69, 70).
Most of the currently applied pollen allergy and asthma mouse models rely on i.p. sensitization of allergen extracts mixed with potent adjuvants, such as Alum (71–73). In a recent study, Stiehm and Peters used the major grass pollen allergen Phl p 5b without adjuvant (Table 1). The model was based on six subcutaneous injections every 2–3 days for the sensitization phase, followed by 3 i.n. treatments with 1 week-intervals. The treated mice showed high levels of eosinophils, lymphocytes and IL-5 in the BAL fluid, increased lung inflammation and type 2 antibodies. Allergic animals also exhibited increased airway resistance to methacholine challenge (25).
Chronic allergic airway inflammation and severe asthma models
Asthma is importantly defined by its chronicity and several research groups have strived to recapitulate this aspect in mouse models (9). However, modeling of chronic asthma is notoriously difficult as mice usually develop tolerance to the applied allergen over time and possess considerable disease-relevant physiological and anatomical differences as compared to humans (74, 75). Mouse models were nevertheless used to recapitulate some of the airway remodeling features observed in human asthmatics. Typically, these models require a lengthy and frequent allergen exposure of 7–10 weeks (20, 76, 77). Two of the protocols utilized i.n. HDME treatments with 5 allergen applications per week, resulting a profound lung type 2 cytokine milieu, airway remodeling with eosinophilic inflammation, and increased allergen-specific antibodies. Woo et al., compared a 4-week allergen exposure to the classical 8-week model and found that the lung function and airway remodeling parameters were comparable between the two groups. Sibilano and colleagues used a an 11-week model based on 3 initial i.n. (100 µg) HDME instillations for sensitization in the first week, followed by 10 weekly i.n. applications (20 µg) to maintain the immune response and model chronic exposure (77).
An elegant study by Asrat and colleagues investigated IgE production in short vs. long-term HDME exposure (28). The protocol relied on the i.n. administration of 50 μg HDME 3×/week for 4 or 15 consecutive weeks followed or not by a “resting” period with lowered (25 μg) HDME dose (28). The long-term allergen exposure led to the production of long-lived IgE+ plasma cells that accumulate in the bone marrow and were able to produce anaphylactic IgE. Importantly, these bone marrow IgE+ plasma cells were also found in humans and allergen-specific IgE produced by these cells effectively induced mast cell degranulation following transfer into mice (expressing a partially humanized high affinity IgE receptor FcɛRI) and HDME treatment. In contrast, the short-term HDM exposure led to IgE+ plasma cells that reside in secondary lymphoid organs and recipient mice engrafted with IgE-containing serum did not respond with mast cell degranulation upon allergen challenge (28). These findings highlight the great translational relevance of HDME-driven allergic asthma mouse models in the context of serological memory and IgE responses. In line with memory immune responses to HDM allergens, Hondowicz et al. used a shorter term HDME exposure model to characterize the formation dynamics of Der p 1-specific tissue resident (Trm) and circulating memory (Tcm) Th2 cells and to show their dependence on IL-2 for migration and residence (29). A more recent study by Rahimi et al. applied a long term HDME exposure combined with HDME or Alternaria alternata challenge (Table 1) to highlight the transcriptional and functional differences between Trm and Tcm Th2 cells (30). While Trm Th2 cells drove airway remodeling features including mucus hypersecretion and airway hyperresponsiveness, Tcm Th2 cells induced eosinophil and CD4+ T cell accumulation in the lung (30). These studies provide an important insight into the establishment and function of allergen-specific memory type 2 immune responses and underline the human relevance of these HDME-based models.
Aside from chronicity, severe asthma and asthma exacerbations often exhibit a signature resistance to standard corticosteroid (CS) treatment (78, 79). Resistance to CS is attributable to various mechanisms leading to altered glucocorticoid signaling at different stages (78). CS treatment is known to induce IL-10 production in asthmatics and therefore dampen inflammation. However, this response is dysfunctional or insufficient in patients with severe asthma (80). Additionally, steroid responsiveness seems to be highly dependent on the inflammatory profile present in the lung (80, 81). For instance, many severe asthmatics show an elevated IL-17 signature (82–84) which plays a critical role in Th17/neutrophilic steroid-resistant asthma (85). Numerous mouse models have been used to show features of refractory asthma with most protocols relying on recombinant cytokines, immune cell transfers or allergenic and pathogenic exposures. Key molecules such as LPS, interferon (IFN)γ were linked to the development steroid-resistant asthma (81, 86, 87). Gauthier and colleagues suggested a CXCL10 signature in human and mouse Th1-driven severe asthma (88). Using single cell RNA-sequencing, a more recent study has provided deeper insights into the immune landscape of CS-resistant asthma (31). This work used an HDME-based mouse model followed by LPS challenge (Table 1) and identified basophils, ILC2s and CD8+ memory T cells as primary producers of IL-4 and IL-13 (31). Moreover, inhibition of IL-13 efficiently reduced HDME/LPS-induced airway hyperresponsiveness and inflammation and could be used to bypass CS-resistance (31). Overall, it appears that balanced T cell responses and certain innate immune mediators are crucial for steroid responsiveness.
While murine asthma models still have limitations in reproducing chronic allergen exposure and airway remodeling features, they represent valuable tools to study the immune responses and molecular pathways driving onset and development of allergic asthma.
Environmental pathogens that potentiate allergic asthma
Commensal and pathogenic microorganisms actively interact with our immune system and modulate its specificity, sensitivity and maturity as well as the amplitude of immune responses (89–91). Environmental microbes are therefore prime candidates as modulators of allergy development. As discussed in the previous section, several pathogen constituents, such as bacterial or fungal materials (e.g., LPS and ß-glucan), are potent contributors to asthma induction and exacerbation. Most allergen extracts, for instance those derived from HDM, cockroach and pollen and even OVA preparations, contain variable, but often substantial amounts of LPS which can potentially shape immune responses against allergens (92). Both, the lung and gut microbiota seem to significantly modulate and exacerbate allergic asthma (90, 93–95). In this section, we focus on the current evidence of pathogenic microbes and infections as modulators of asthma development in experimental murine studies. In our search, we considered clinically relevant respiratory pathogens (96–98) and summarize study examples in Tables 2–4.
Viral infections and allergic asthma
Population studies provide evidence that infections with viral pathogens, such as Human Rhinovirus (HRV) can predispose children to onset of asthma (113, 114). Moreover, once asthma is established, subsequent HRV and Respiratory Syncytial Virus (RSV) infections are most likely culprits causing disease exacerbation (96). In childhood, HRV infections are associated with persistent wheezing, a strong indicator for asthma later in life (115, 116). Many studies linked these virus-mediated exacerbations to increased airway hyperactivity and granulocytic infiltration amongst other factors (117).
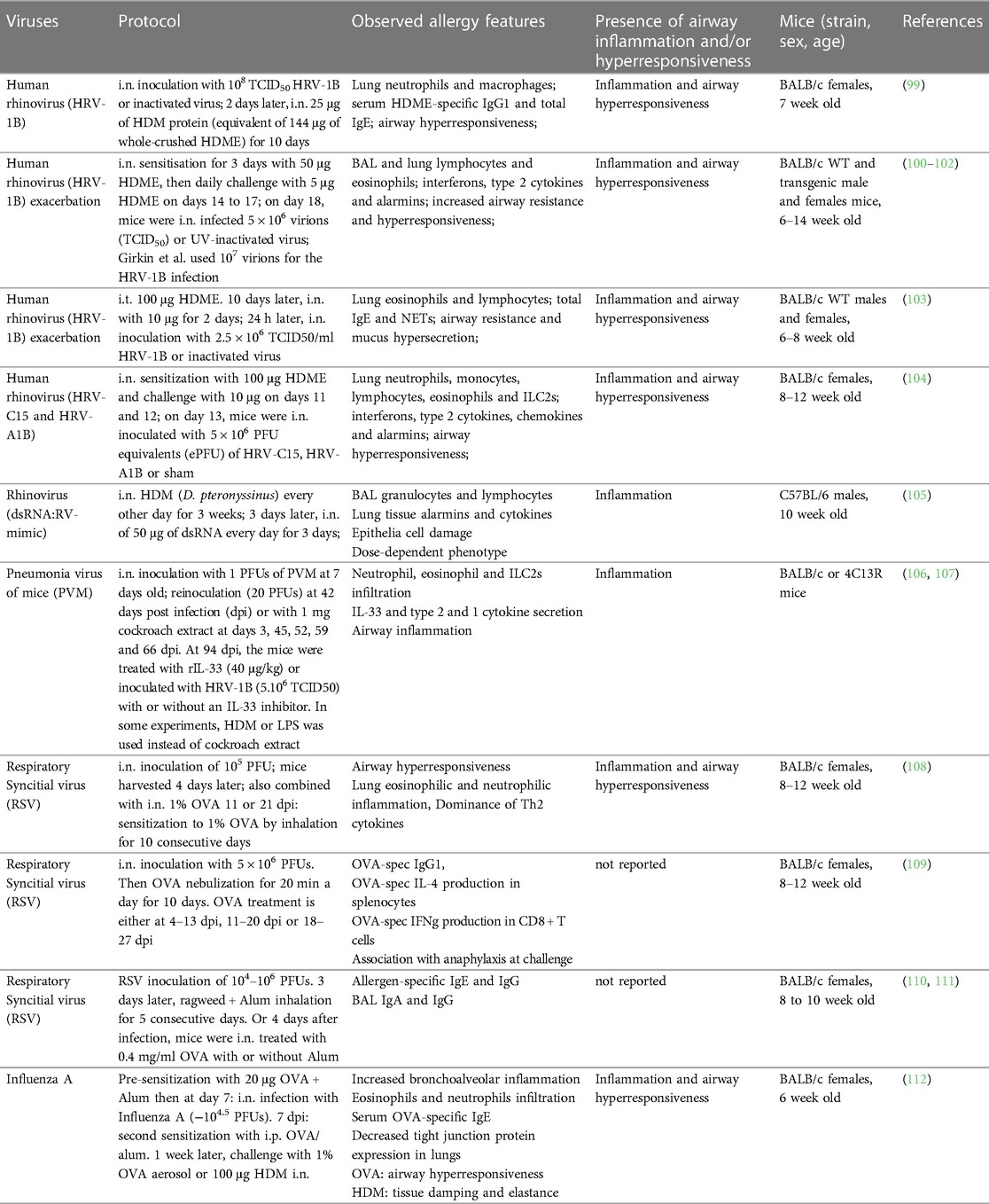
Within the last decade, several mouse models have been established to investigate the importance of viral infections for asthma development and exacerbation. We will discuss examples of relevant protocols combining virus and allergen exposures. All cited models are summarized in Table 2.
Human rhinovirus
Human rhinovirus (HRV) is the most common causative agent of upper airway tract infections and can exacerbate pre-existing chronic pulmonary disorders (118). Rhinoviruses consist of three phylogenetic species: HRV-A, HRV-B and HRV-C (119). HRV-A and -B rhinoviruses are classified into major or minor group viruses based on their receptor binding and tropism (118, 120). Major group viruses utilize intercellular cell adhesion molecule 1 (ICAM-1) and minor group viruses bind to low-density lipoprotein receptor (LDLR) (118). Major group viruses represent 90% of HRV serotypes and do not bind to murine ICAM-1 (118, 121, 122). For a long time, this represented a roadblock for development of mouse models for this group of rhinovirus infections. This led to the development of a chimeric mouse ICAM-1 which contains the terminal immunoglobulin domain of human ICAM-1 to render mouse cells permissive for major group rhinovirus infections (121–123). In contrast, minor group viruses can bind both human and mouse LDLR (118, 124). Phan and colleagues investigated the effects of infection with a minor group rhinovirus (HRV-1B) following a daily 25 μg dose of HDME for 10 consecutive days (Table 2) (99). The combination of HDME and HRV (as compared to HRV alone) resulted in an exacerbation in total lung leukocytes, particularly neutrophils and macrophages, and a marked increase in total IgE and HDM-specific IgG1. Additionally, hyperresponsiveness to methacholine challenge was significantly increased. This corroborates other studies showing minor group HRVs as an exacerbating factor for OVA-induced asthma (125, 126). Other models relied on high doses of HDME early on followed by multiple lower HDME i.n. administrations and viral infection one day after the last allergen challenge (Table 2) (100–102). These protocols showed a virus-mediated increase in type 1 and type 2 immune responses, including elevated pulmonary eosinophil infiltration, lung type 2 cytokines and airway hyperresponsiveness (100–102). Toussaint and colleagues used a similar HDME sensitization procedure to show that minor group HRV infection exacerbated HDME-induced airway inflammation through the release of host double strand (ds) DNA, in association with neutrophil extracellular traps (NETs) (Table 2) (103). Another approach relied on the use of dsRNA (as an RV mimic) in combination with HDME-induced allergic asthma (105). In this setup, C57BL/6 mice were treated with HDME 3 times a week for 3 weeks to establish asthma, followed by either a low or high i.n. dose of dsRNA on 3 consecutive days then sacrificed 24 h later (Table 2). dsRNA treatment increased the granulocyte and lymphocyte numbers in BAL, as well as lung expression of IL-1β, CCL2 and CCL5 as well as of IL-25, IL-33 and TSLP, a specific group of (predominantly) epithelia-derived cytokines referred to as alarmins. While this study did not specifically test lung function, it reported other lung damage measures such as epithelial shedding and protein exudation.
HRV-C, which was first reported on almost 20 years ago (127, 128), uses the cadherin related family member 3 (CDHR3) as entry receptor on human and mouse cells (129). It has since been found that people with a history of asthma are more susceptible to an HRV-C infection (129–131). To date, this HRV serotype has only been investigated in one study by Rajput et al. in relation to its effect on allergic asthma (Table 2) (104). The infection alone induced a strong type 2 immune response with increased lung mRNA and protein expression of type 2 cytokines as well as expansion of ILC2s, eosinophils, monocytes and neutrophils. In mice pre-treated with HDM before infection with HRV-C15, a similar increase in these cell types, type 2 cytokines and alarmins alongside increased airway hyperresponsiveness was observed. This study provided further relevant insight by comparing the immune responses to HRV-A1B and HRV-C15 infections and found that the latter further increased the airway resistance and eosinophilia in an ILC2-dependent manner (104).
Respiratory syncytial virus
In the 1990s, first RSV infection mouse models have shown an exaggerated type 2 immune response after vaccination and natural exposure (132, 133). Through the combination of RSV infection followed by allergen (e.g., ragweed or OVA) sensitization (108–111), these seminal studies increased levels of allergen-specific IgE and IgG as well as BAL IgA and IgG as compared to non-infected mice. Notably, other groups have studied the impact of RSV infection on cockroach extract-mediated sensitization using Incomplete Freund's adjuvant (IFA) during the sensitization phase (134–136). Relying on a similar protocol that includes a systemic (i.p. and s.c.) adjuvant-driven cockroach antigen (CRA) sensitization, followed by RSV infection and CRA challenges, these studies found that RSV indeed exacerbates type 2 immune responses, including eosinophilia and production of cytokines, such as IL-13 and IL-4, as well as airway hyperresponsiveness (134–136). Another recent work used a chronic experimental asthma model combining repeated exposures to pneumonia virus of mice (PVM), a virus closely related to RSV, and cockroach extract for approximately 2 months, followed a month later by the HRV infection [Table 2 (106)]. The infection resulted in a neutrophil-dominated immune response 1 day post infection (d.p.i.), followed by an inflammatory phase between 3 and 7 d.p.i., characterized by increased lung eosinophils and type 2 cytokines as well as mucus hypersecretion. Both phases were abrogated by alarmin depletion and recapitulated by recombinant IL-33 administration. This chronic model, used in other studies, simulates increased airway smooth muscle contraction, a feature of airway remodeling and disease progression observed in humans (107, 137). Combined with the PVM infection, it allowed to identify IL-33 as a main culprit of the observed immune dysregulation. Complex approaches like the one described consider combinations of different stimuli, repeated treatment and resting periods. Therefore, they likely better simulate the exposure processes in humans.
Experimental studies combining RSV and non-adjuvanted allergens such as HDM or pollen extracts could significantly increase our understanding of potential virus-mediated effects on allergic sensitization and asthma.
Influenza A virus
Lower respiratory tract influenza A virus (IAV) infections were shown to be associated with asthma exacerbations (138). However, the interplay between asthma and IAV remains more elusive than for RSV and HRV. Addressing the effects of asthmatic state on infection, studies using OVA-based models have shown that chronic asthma can be detrimental for IAV clearance (139) while viral replication is limited in allergic lungs at the early stages of sensitization (140).
On the other hand, only few studies have experimentally investigated the effect of IAV infection on asthma development (109). A recent work using either OVA or HDM as an allergic asthma model showed that influenza pre-infected mice had an increase in lung neutrophils and eosinophils, with a neutrophil-dominated profile for the HDM-treated mice (112). Only the OVA model showed a bronchial hyperresponsiveness phenotype but both allergens induced a decrease in tight junction proteins, such as claudin-1 and occludin (Table 2).
Overall, there is a significant need for more experimental studies investigating the consequences of respiratory viral infections for (chronic) asthma using relevant infection and allergy protocols. Thus far, it remains unclear, for instance, as to whether viral infections are a key trigger of asthma or simply a marker of an inherent predisposition and prior lung dysfunction (141).
Bacterial infections and allergic asthma
Accumulating evidence suggests that different bacterial species can trigger or exacerbate allergies (142). In the case of asthma, bacteria may aggravate allergic symptoms in conjunction with viruses (HRV or RSV) or alone (96, 143). Since the 1970s, studies documented a correlation between bacterial exposure and allergies. Regarding the common bacterial inhabitants of the human respiratory tract, colonization or infection with Haemophilus influenzae, Streptococcus pneumoniae, Moraxella catharralis and Staphylococcus aureus have been associated with the induction and exacerbation of asthma, chronic obstructive pulmonary diseases and recurrent wheezing early in life (96, 144–150). Moreover, in patients suffering from allergic disorders, such as asthma, atopic dermatitis or nasal polyposis, S. aureus colonization appears to occur more frequently (87%, 90%, 87%, respectively), in contrast to 20%–50% colonization of healthy adults (151–153). In addition, asymptomatic colonization of neonates with S. pneumoniae or M. catarrhalis is associated with later development of recurrent wheezing and asthma (148).
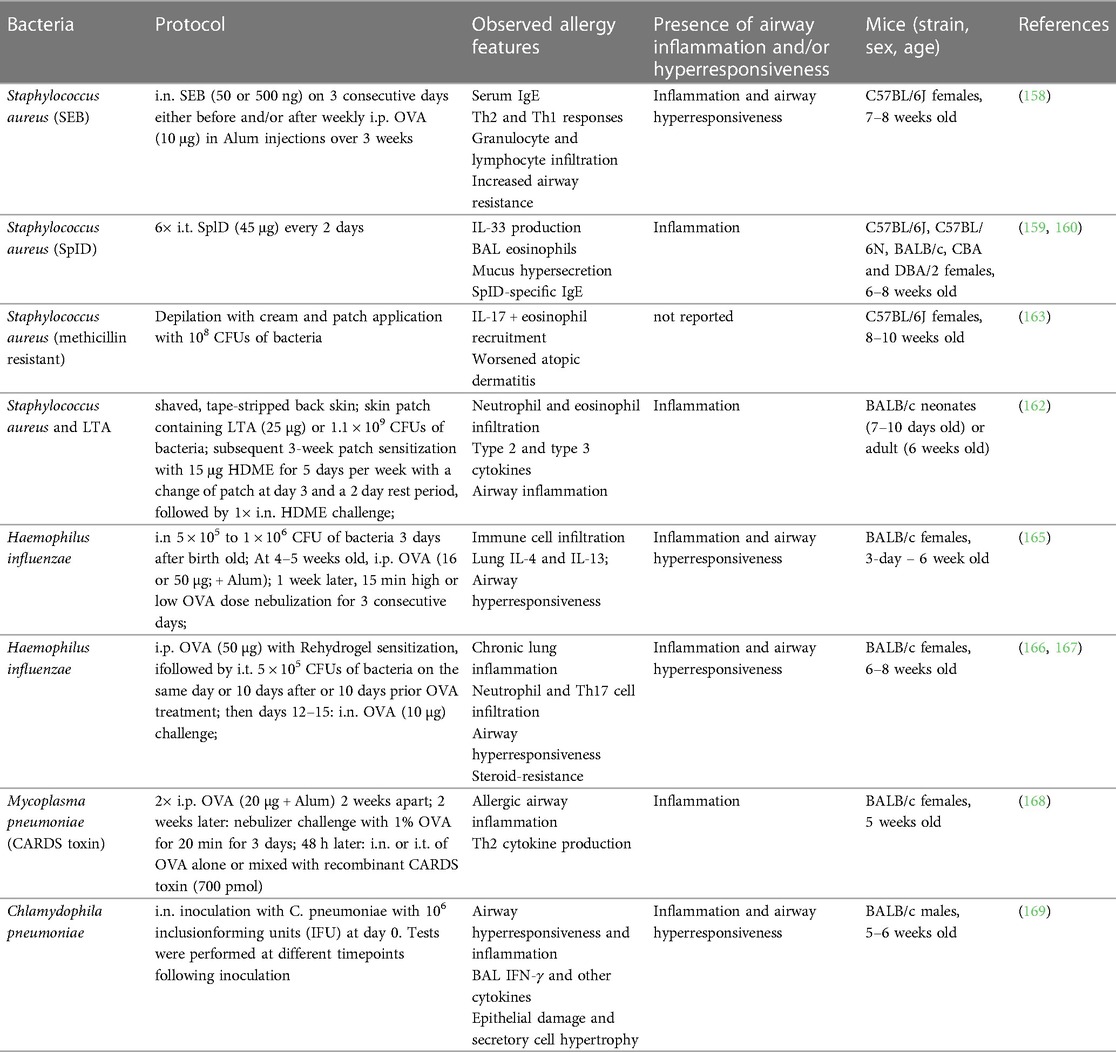
Notably, toxins produced by certain bacteria, such as Staphylococcus aureus, Bordetella pertussis and Streptococcus pneumoniae have the potential to induce and amplify Th2 immune responses and increase IgE levels in both mice and humans (154–156). An excellent resource by Nordengrün et al. gives an extensive overview on the allergenic potential of bacteria and their overall impact on type 2 immune responses (142). In this section, we will dive into mouse models of bacterial infections that elicit or exacerbate allergic immune responses in the context of asthma. All cited models are summarized in Table 3.
Staphylococcus aureus
Several studies have found a correlation between asthma severity and staphylococcal enterotoxin (SE)-specific IgE levels in human patients (110, 144, 157). Experimental work using mouse models has confirmed significant modulation of asthmatic immune responses by skin and airway exposures to S. aureus or its toxins. In a work relying on an OVA-induced allergic airway inflammation, i.n. instillations of staphylococcal enterotoxin B (SEB) were found to have a dual role depending on the dose and the timing of the treatment (before or after OVA sensitization) (158). SEB either aggravated or improved different parameters of allergic sensitization and inflammation (Table 3). Another model, using repeated i.t. treatments of the S. aureus allergen serine protease-like protein D (SplD), leads to robust, IL-33-dependent asthma in C57BL/6 mice. Interestingly, the potency to induce allergic airway inflammation and asthma features was largely absent in BALB/c mice (159, 160).
Other studies investigated effects of S. aureus skin colonization on allergic asthma. The applied models employed either a back skin or ear infection with different strains of the bacterium (161) or exposure to specific bacterial constituents, such as lipoteichoic acid (LTA) (162), serine proteases (163) or toxins (164). S. aureus skin infection of mice induces mast cell degranulation, eosinophil recruitment, IgE and IgG1 specific to certain bacteria-derived products, as well as type 2 cytokines (161, 163, 164).
Ubags and colleagues employed an “atopic march” mouse model that that mimics sequential allergen exposure in different body sites by combining dermal and intranasal HDME treatment with or without prior skin pre-exposure to LTA or live S. aureus in adult mice (162) (Table 3). This study found that dermal HDME application induced a mixed Th2/Th17 response in BALB/c mice, which was exacerbated by S. aureus, with a neutrophil-dominated asthma phenotype. The protocol was also applied in neonates where the S. aureus-mediated effects were partially microbiome dependent (162) (Table 3).
While extensive experimental evidence supports the modulatory effect of S. aureus on allergic immune responses, the molecular mechanisms driving the interplay between S. aureus colonization and asthma exacerbation are still elusive.
Pseudomonas aeruginosa
Pseudomonas aeruginosa is a ubiquitous bacterium and chronic colonizer of the lower respiratory tract and prevails in patients with chronic airway disorders (170, 171). In asthmatic patients, the presence of P. aeruginosa in the sputum is correlated with poorer lung function and increased inflammatory parameters (171). Telford et al. have shown the ability of a P. aeruginosa-derived quorum sensing molecule to induce high levels of IgG1 in vivo and to stimulate IgE production in IL-4-primed human peripheral blood derived mononuclear cells (172).
Haemophilus influenzae
Haemophilus influenzae is one of the most commonly identified bacterial pathogens in the airways of patients with severe asthma (173). Several experimental studies used infection mouse models to explore this connection with allergic airway inflammation primarily using mouse models of OVA-induced allergic asthma (Table 3) (165–167, 174). The results demonstrate the potential of H. influenzae to influence neutrophil infiltration in asthmatic mice through an IL-17 immune axis (166, 167). In juvenile animals, infection induced a bronchial hyperresponsiveness and increased BAL fluid IL-5 and IL-13 (165). However, we were unable to find experimental studies in context of H. influenzae pneumonia using natural allergen (extracts) and omitting adjuvants for sensitization.
Atypical bacteria: Mycoplasma pneumoniae and Chlamydophila pneumoniae
Atypical bacteria, such as Mycoplasma pneumoniae and Chlamydophila pneumoniae, are challenging to detect by standard methodologies (e.g., Gram stain) (175). In addition, they are difficult to isolate and antibiotic treatment is often ineffective due to their intracellular occurrence (175). M. pneumoniae is a common cause of community-acquired pneumonia and can be associated with asthma onset and exacerbations in children and adults (176–179). One experimental study investigated M. pneumoniae lung infection in combination with a allergic asthma (168). In this protocol, the mice were sensitized with an i.p. injection of OVA mixed with Alum and challenged 2 weeks later with nebulized OVA for 3 days. This was followed 48 h later by an i.t. or i.n. treatment with OVA and M. pneumoniae community-acuired respiratory distress syndrome (CARDS) toxin or with OVA only. Presence of CARDS toxin led to enhanced eosinophilic inflammation and type 2 cytokine production (Table 3).
In a study investigating the impact of C. pneumoniae on lung function and inflammation (169), mice were i.n. inoculated with the bacterium, followed by analysis of lung inflammation at different timepoints up to 3 weeks. The infection caused a sustained airway hyperresponsiveness from day 7 to 21, alongside pro-inflammatory cytokine production, expansion of BAL myeloid and lymphoid immune cells and evidence of trachea epithelial cell damage. These results indicate the great potential of C. pneumoniae to prime the airways towards higher susceptibility for allergic sensitization or asthma exacerbation.
In summary, further experimental studies with atypical bacteria in context of allergic asthma would be required to better understand this largely unexplored field.
Fungal infections and allergic asthma
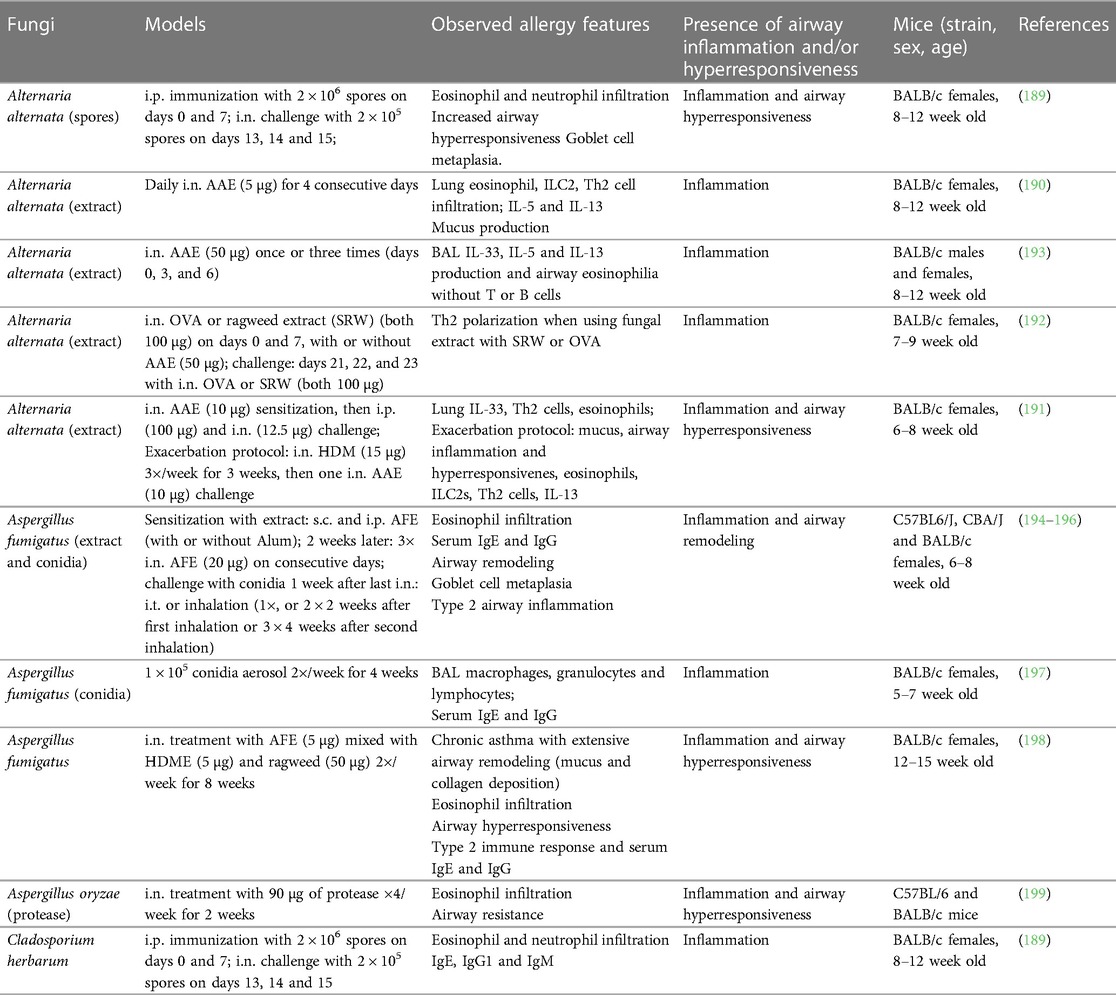
Fungi are ubiquitously present indoors and outdoors and represent one of the largest sources of airborne allergenic substances for humans (180). As such, fungal colonization and exposure to fungal components are considered important factors of asthma development and exacerbations in susceptible individuals (181, 182). Mold allergies are common and can induce a variety of allergic reactions. Several fungal species, such as Alternaria alternata, Aspergiullus fumigatus or Cladosporium herbarum, are potent sensitizing agents in humans (183). Sensitization to some of these fungi is associated with more severe disease, decreased lung function and greater need for corticosteroids for asthma control (184–186). An experimental study investigating the role of chitin, an essential fungal cell wall component (187), has linked this naturally abundant polysaccharide to systemic and pulmonary allergy (188). I.n. chitin instillation of mice led to the lung accumulation of IL-4-producing cells, including eosinophils and basophils. Additionally, this treatment induced alternatively activated macrophages and leukotriene B4 production. This important study illustrates the great potential of fungal elements to mobilize immune components usually implicated in allergic responses. In the following section, we will discuss mouse models using fungal infections alone or in combination with allergens in the context of allergic asthma. All models are summarized in Table 4.
Alternaria alternata
Mouse models of Alternaria alternata infections are relatively well established and generally require either i.p. or i.n. sensitization, followed by repeated i.n. challenges. Asthma features are typically measured one day after the last challenge (Table 4) (189, 190). A study compared the activity of several aeroallergens alone and in combination with Alternaria and found that fungal serine protease activity induced IL-33 production in the airways via protease activated receptor (PAR)-2 and adenosine triphosphate signaling. In combination with HDME, Alternaria potentiated the pulmonary type 2 inflammation after challenge, resulting in decreased lung function (Table 4) (191). Another related work showed the important role of prostaglandin D2 in regulating Alternaria extract (AAE)-mediated eosinophil and ILC2 infiltration, as well as pulmonary type 2 immunity (190). Several studies have also reported Alternaria-mediated signs of airway remodeling with goblet cell metaplasia and increased airway hyperresponsiveness (Table 4) (189, 192, 193).
Aspergillus
Aspergillus fumigatus is a fungal pathogen that can be found in water, soil, food and decomposing vegetation (200). In its conidial (spore) form, A. fumigatus can be readily airborne and inhaled (200). Initially, A. fumigatus infection mouse models used cultured fungus until conidia-based models emerged and were shown to recreate the immunopathological features of fungal asthma (180). The routes of delivery vary between models for sensitization and challenge phases. A study comparing the asthma phenotype between i.t. and inhalation routes of conidia delivery during challenge found that inhalation results in a more enhanced eosinophilic inflammation, serum IgE and airway remodeling (194). Similar models with inhalation challenge showed sustained airway remodeling and type 2 inflammation (195, 196). Importantly, the choice of live vs. irradiated (dead) conidia plays a major role in the character and intensity of the allergic immune response as live conidia elicit more robust inflammation and airway remodeling features in fungal asthma (201).
Another study characterized the lung immune responses to repeated inhalation of conidia aerosolized using an acoustical generator. This approach mimics the exposure to fungal bioaerosols that are typically encountered in the environment (197). This protocol led to increased BAL macrophages, granulocytes, and lymphocytes as well as specific IgG.
Goplen and colleagues established a chronic asthma model utilizing a mixture of one, two or three allergens derived from HDM, ragweed and Aspergillus without adjuvant (198). I.n. exposure to the triple allergen mix twice a week for 8 weeks resulted in chronic airway inflammation accompanied by impaired lung function, a profound type 2 immune response and Aspergillus-specific IgE and IgG production.
Aspergillus oryzae is a fungus of significant economic importance as its α-amylase enzyme is broadly used in the pharmaceutical and food industry. Purified A. oryzae protease is a potent allergen (199) as its i.n. application (4 times a week for 2 weeks) induces robust BAL eosinophilic infiltration and increased airway resistance. This study highlights the great allergenic potential of exogenous proteases upon airway exposure.
Cladosporium herbarum
Cladosporium herbarum is a common fungus of global clinical relevance (202). Alongside Alternaria, Havaux et al. tested the allergenicity of C. herbarum spores (189) and found that two i.p. immunizations with a 1 month interval resulted in significantly increased IgE levels 15 days later. I.n. challenge of sensitized animals led to increased BAL type 2 cytokines (IL-4, IL-5 and IL-13) as well as airway hyperresponsiveness.
Overall, fungi and fungal components are amongst the most potent and important environmental triggers of allergic asthma. However, due to the scarcity of experimental studies, significant questions related to immunopathogenesis and consequences of fungal exposures remain unanswered.
Conclusion and future perspectives
The complexity of pathological immune responses in asthma requires elaborate animal models that consider and reflect this intricacy. In addition, this disease comprises a variety of endotypes and severity grades. The development of protocols that incorporate clinically relevant exposure routes and allergens has brought a significant benefit to this field of research. Furthermore, models of chronic disease are of great importance and have emerged as an indispensable tool to replicate the pathophysiology of asthma and identify the long-term consequences of allergen exposure and repeated exacerbations. However, a significant type of airway allergy that still lacks an adjuvant-free mouse model (independent of chemical adjuvants or systemic immunization) is pollen allergy. Additionally, incorporating a broader range of aeroallergens or utilizing combinations of different allergens would provide a more comprehensive understanding of the complex implications of environmental exposures as observed in humans.
Finally, further research investigating concomitant airway infections and asthma could guide towards new preventative and therapeutic strategies. For instance, potential associations between disease features or clinical biomarkers and pathogen group signatures in mice and humans could address some aspects of treatment resistance and have a great impact on disease management and outcome.
Author contributions
MR: Conceptualization, Visualization, Writing – original draft, Writing – review & editing. PS: Conceptualization, Funding acquisition, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article.
This work was supported by an Austrian Science Fund (FWF) grant to Philipp Starkl (project #P36502).
Acknowledgments
This research was funded in whole, or in part, by the Austrian Science Fund (FWF) [P36502]. For the purpose of open access, the author has applied a CC BY public copyright licence to any Author Accepted Manuscript version arising from this submission. We apologize to colleagues whose valuable work could not be included or highlighted due to space limitations. We thank our colleagues Lisabeth Pimenov, Asma Farhat and Anna Hakobyan for their feedback on the design of the illustration in Figure 1. Illustrations were created in part with BioRender.com and readapted in illustrator.
Conflict of interest
The authors have no commercial affiliation or relationships with any organization or entity with a financial interest to declare and did not receive any financial support for the research, authorship, and publication of this article.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. GBD 2019 Chronic Respiratory Diseases Collaborators. Global burden of chronic respiratory diseases and risk factors, 1990–2019: an update from the global burden of disease study 2019. EClinicalMedicine. (2023) 59:101936. doi: 10.1016/j.eclinm.2023.101936
2. Porsbjerg C, Melén E, Lehtimäki L, Shaw D. Asthma. Lancet. (2023) 401:858–73. doi: 10.1016/S0140-6736(22)02125-0
3. Aegerter H, Lambrecht BN. The pathology of asthma: what is obstructing our view? Annu Rev Pathol Mech Dis. (2023) 18:387–409. doi: 10.1146/annurev-pathol-042220-015902
4. Papi A, Brightling C, Pedersen SE, Reddel HK. Asthma. Lancet. (2018) 391:783–800. doi: 10.1016/S0140-6736(17)33311-1
5. Lambrecht BN, Hammad H, Fahy JV. The cytokines of asthma. Immunity. (2019) 50:975–91. doi: 10.1016/j.immuni.2019.03.018
6. Woodruff PG, Modrek B, Choy DF, Jia G, Abbas AR, Ellwanger A, et al. T-helper type 2-driven inflammation defines major subphenotypes of asthma. Am J Respir Crit Care Med. (2009) 180:388–95. doi: 10.1164/rccm.200903-0392OC
7. Martin RA, Hodgkins SR, Dixon AE, Poynter ME. Aligning mouse models of asthma to human endotypes of disease. Respirology. (2014) 19:823–33. doi: 10.1111/resp.12315
8. Haspeslagh E, Debeuf N, Hammad H, Lambrecht BN. Murine models of allergic asthma. In: Clausen BE, Laman JD, editors. Inflammation. Methods in Molecular Biology. New York, NY: Springer New York (2017). p. 121–36. doi: 10.1007/978-1-4939-6786-5_10
9. Nials AT, Uddin S. Mouse models of allergic asthma: acute and chronic allergen challenge. Dis Model Mech. (2008) 1:213–20. doi: 10.1242/dmm.000323
10. Hammad H, Lambrecht BN. The basic immunology of asthma. Cell. (2021) 184:1469–85. doi: 10.1016/j.cell.2021.02.016
11. Ober C, Yao T. The genetics of asthma and allergic disease: a 21st century perspective. Immunol Rev. (2011) 242:10–30. doi: 10.1111/j.1600-065X.2011.01029.x
12. Dunican EM, Elicker BM, Gierada DS, Nagle SK, Schiebler ML, Newell JD, et al. Mucus plugs in patients with asthma linked to eosinophilia and airflow obstruction. J Clin Invest. (2018) 128:997–1009. doi: 10.1172/JCI95693
13. McKenzie GJ, Fallon PG, Emson CL, Grencis RK, McKenzie AN. Simultaneous disruption of interleukin (IL)-4 and IL-13 defines individual roles in T helper cell type 2-mediated responses. J Exp Med. (1999) 189:1565–72. doi: 10.1084/jem.189.10.1565
14. Fallon PG, Schwartz C. The high and lows of type 2 asthma and mouse models. J Allergy Clin Immunol. (2020) 145:496–8. doi: 10.1016/j.jaci.2019.11.031
15. Gorska MM. Mouse models of asthma. In: Alper S, Janssen WJ, editors. Lung Innate Immunity and Inflammation. Methods in Molecular Biology. New York, NY: Springer New York (2018). p. 351–62. doi: 10.1007/978-1-4939-8570-8_23
16. Schuijs MJ, Willart MA, Vergote K, Gras D, Deswarte K, Ege MJ, et al. Farm dust and endotoxin protect against allergy through A20 induction in lung epithelial cells. Science. (2015) 349:1106–10. doi: 10.1126/science.aac6623
17. Plantinga M, Guilliams M, Vanheerswynghels M, Deswarte K, Branco-Madeira F, Toussaint W, et al. Conventional and monocyte-derived CD11b+ dendritic cells initiate and maintain T helper 2 cell-mediated immunity to house dust Mite allergen. Immunity. (2013) 38:322–35. doi: 10.1016/j.immuni.2012.10.016
18. Paul Chowdhury B, Gorska MM. Modeling asthma in mice using common aeroallergens. In: Gorska MM, editor. Asthma. Methods in Molecular Biology. New York, NY: Springer US (2022). p. 1–18. doi: 10.1007/978-1-0716-2364-0_1
19. Willart MAM, Deswarte K, Pouliot P, Braun H, Beyaert R, Lambrecht BN, et al. Interleukin-1α controls allergic sensitization to inhaled house dust mite via the epithelial release of GM-CSF and IL-33. J Exp Med. (2012) 209:1505–17. doi: 10.1084/jem.20112691
20. Woo LN, Guo WY, Wang X, Young A, Salehi S, Hin A, et al. A 4-week model of house dust mite (HDM) induced allergic airways inflammation with airway remodeling. Sci Rep. (2018) 8:6925. doi: 10.1038/s41598-018-24574-x
21. Tang W, Dong M, Teng F, Cui J, Zhu X, Wang W, et al. Environmental allergens house dust mite-induced asthma is associated with ferroptosis in the lungs. Exp Ther Med. (2021) 22:1483. doi: 10.3892/etm.2021.10918
22. Kale SL, Agrawal K, Gaur SN, Arora N. Cockroach protease allergen induces allergic airway inflammation via epithelial cell activation. Sci Rep. (2017) 7:42341. doi: 10.1038/srep42341
23. Page K, Zhou P, Ledford JR, Day SB, Lutfi R, Dienger K, et al. Early immunological response to German cockroach frass exposure induces a Th2/Th17 environment. J Innate Immun. (2011) 3:167–79. doi: 10.1159/000320718
24. Arizmendi NG, Abel M, Puttagunta L, Asaduzzaman M, Davidson C, Karimi K, et al. Mucosal exposure to cockroach extract induces allergic sensitization and allergic airway inflammation. All Asth Clin Immun. (2011) 7:22. doi: 10.1186/1710-1492-7-22
25. Stiehm M, Peters M. Specific immunotherapy in a murine model of grass pollen (phl p5b)-induced airway inflammation. Front Allergy. (2021) 2:777545. doi: 10.3389/falgy.2021.777545
26. Gregory LG, Causton B, Murdoch JR, Mathie SA, O’Donnell V, Thomas CP, et al. Inhaled house dust mite induces pulmonary T helper 2 cytokine production. Clin Experimental Allergy. (2009) 39:1597–610. doi: 10.1111/j.1365-2222.2009.03302.x
27. Gregory LG, Jones CP, Walker SA, Sawant D, Gowers KHC, Campbell GA, et al. IL-25 drives remodelling in allergic airways disease induced by house dust mite. Thorax. (2013) 68:82–90. doi: 10.1136/thoraxjnl-2012-202003
28. Asrat S, Kaur N, Liu X, Ben L-H, Kajimura D, Murphy AJ, et al. Chronic allergen exposure drives accumulation of long-lived IgE plasma cells in the bone marrow, giving rise to serological memory. Sci Immunol. (2020) 5:eaav8402. doi: 10.1126/sciimmunol.aav8402
29. Hondowicz BD, An D, Schenkel JM, Kim KS, Steach HR, Krishnamurty AT, et al. Interleukin-2-dependent allergen-specific tissue-resident memory cells drive asthma. Immunity. (2016) 44:155–66. doi: 10.1016/j.immuni.2015.11.004
30. Rahimi RA, Nepal K, Cetinbas M, Sadreyev RI, Luster AD. Distinct functions of tissue-resident and circulating memory Th2 cells in allergic airway disease. J Exp Med. (2020) 217:e20190865. doi: 10.1084/jem.20190865
31. Wang L, Netto KG, Zhou L, Liu X, Wang M, Zhang G, et al. Single-cell transcriptomic analysis reveals the immune landscape of lung in steroid-resistant asthma exacerbation. Proc Natl Acad Sci USA. (2021) 118:e2005590118. doi: 10.1073/pnas.2005590118
32. Debeuf N, Haspeslagh E, Van Helden M, Hammad H, Lambrecht BN. Mouse models of asthma. CP Mouse Biology. (2016) 6:169–84. doi: 10.1002/cpmo.4
33. Jacquet A, Robinson C. Proteolytic, lipidergic and polysaccharide molecular recognition shape innate responses to house dust mite allergens. Allergy. (2020) 75:33–53. doi: 10.1111/all.13940
34. Rodinkova VV, Yuriev SD, Kryvopustova MV, Mokin VB, Kryzhanovskyi YM, Kurchenko AI. Molecular profile sensitization to house dust mites as an important aspect for predicting the efficiency of allergen immunotherapy. Front Immunol. (2022) 13:848616. doi: 10.3389/fimmu.2022.848616
35. Huang H-J, Sarzsinszky E, Vrtala S. House dust mite allergy: the importance of house dust mite allergens for diagnosis and immunotherapy. Mol Immunol. (2023) 158:54–67. doi: 10.1016/j.molimm.2023.04.008
36. Barberán A, Dunn RR, Reich BJ, Pacifici K, Laber EB, Menninger HL, et al. The ecology of microscopic life in household dust. Proc R Soc B. (2015) 282:20151139. doi: 10.1098/rspb.2015.1139
37. Wu W, Jin Y, Carlsten C. Inflammatory health effects of indoor and outdoor particulate matter. J Allergy Clin Immunol. (2018) 141:833–44. doi: 10.1016/j.jaci.2017.12.981
38. Rodríguez D, Palacios R, Martínez J, Guisantes JA, Postigo I. Kinetics of dermatophagoides pteronyssinus and dermatophagoides farinae growth and an analysis of the allergen expression in semi-synthetic culture medium. Acarologia. (2021) 61:403–11. doi: 10.24349/acarologia/20214439
39. Batard T, Hrabina A, Bi XZ, Chabre H, Lemoine P, Couret M-N, et al. Production and proteomic characterization of pharmaceutical-grade dermatophagoides pteronyssinus and dermatophagoides farinae extracts for allergy vaccines. Int Arch Allergy Immunol. (2006) 140:295–305. doi: 10.1159/000093707
40. Avula-Poola S, Morgan MS, Arlian LG. Diet influences growth rates and allergen and endotoxin contents of cultured dermatophagoides farinae and dermatophagoides pteronyssinus house dust mites. Int Arch Allergy Immunol. (2012) 159:226–34. doi: 10.1159/000336026
41. Vidal-Quist JC, Ortego F, Rombauts S, Castañera P, Hernández-Crespo P. Dietary shifts have consequences for the repertoire of allergens produced by the European house dust mite. Medical Vet Entomology. (2017) 31:272–80. doi: 10.1111/mve.12234
42. Cyphert-Daly JM, Yang Z, Ingram JL, Tighe RM, Que LG. Physiologic response to chronic house dust mite exposure in mice is dependent on lot characteristics. J Allergy Clin Immunol. (2019) 144:1428–1432.e8. doi: 10.1016/j.jaci.2019.07.019
43. Pascoe CD, Jha A, Basu S, Mahood T, Lee A, Hinshaw S, et al. The importance of reporting house dust mite endotoxin abundance: impact on the lung transcriptome. Am J Physiol Lung Cell Mol Physiol. (2020) 318:L1229–36. doi: 10.1152/ajplung.00103.2020
44. Hadebe S, Kirstein F, Fierens K, Chen K, Drummond RA, Vautier S, et al. Correction: microbial ligand costimulation drives neutrophilic steroid-refractory asthma. PLoS One. (2015) 10:e0137945. doi: 10.1371/journal.pone.0137945
45. Hadebe S, Kirstein F, Fierens K, Chen K, Drummond RA, Vautier S, et al. Microbial ligand costimulation drives neutrophilic steroid-refractory asthma. PLoS One. (2015) 10:e0134219. doi: 10.1371/journal.pone.0134219
46. Melgert BN, Postma DS, Kuipers I, Geerlings M, Luinge MA, Van Der Strate BWA, et al. Female mice are more susceptible to the development of allergic airway inflammation than male mice. Clin Experimental Allergy. (2005) 35:1496–503. doi: 10.1111/j.1365-2222.2005.02362.x
47. Bryan MA, Guyach SE, Norris KA. Specific humoral immunity versus polyclonal B cell activation in trypanosoma cruzi infection of susceptible and resistant mice. PLoS Negl Trop Dis. (2010) 4:e733. doi: 10.1371/journal.pntd.0000733
48. Jutzeler KS, Le Clec’h W, Chevalier FD, Anderson TJC. Contribution of parasite and host genotype to immunopathology of schistosome infections. Parasit Vectors. (2024) 17:203. doi: 10.1186/s13071-024-06286-6
49. HayGlass KT, Nashed B, Haile S, Marshall AJ, Thomas W. C57Bl/6 and BALB/c mice do not represent default Th1 and Th2 strains in allergen-driven immune responses. J Allergy Clin Immunol. (2005) 115:S258. doi: 10.1016/j.jaci.2004.12.1041
50. Lopandić Z, Dragačević L, Kosanović D, Burazer L, Gavrović-Jankulović M, Minić R. Differences in mouse strains determine the outcome of der p 2 allergy induction protocols. J Immunol Methods. (2022) 511:113382. doi: 10.1016/j.jim.2022.113382
51. Rojas-Ruiz A, Boucher M, Gill R, Gélinas L, Tom F-Q, Fereydoonzad L, et al. Lung stiffness of C57BL/6 versus BALB/c mice. Sci Rep. (2023) 13:17481. doi: 10.1038/s41598-023-44797-x
52. Boucher M, Henry C, Khadangi F, Dufour-Mailhot A, Bossé Y. Double-chamber plethysmography versus oscillometry to detect baseline airflow obstruction in a model of asthma in two mouse strains. Exp Lung Res. (2021) 47:390–401. doi: 10.1080/01902148.2021.1979693
53. Takeda K, Haczku A, Lee JJ, Irvin CG, Gelfand EW. Strain dependence of airway hyperresponsiveness reflects differences in eosinophil localization in the lung. Am J Physiol Lung Cell Mol Physiol. (2001) 281:L394–402. doi: 10.1152/ajplung.2001.281.2.L394
54. Evans CM, Raclawska DS, Ttofali F, Liptzin DR, Fletcher AA, Harper DN, et al. The polymeric mucin Muc5ac is required for allergic airway hyperreactivity. Nat Commun. (2015) 6:6281. doi: 10.1038/ncomms7281
55. Duguet A, Biyah K, Minshall E, Gomes R, Wang C-G, Taoudi-Benchekroun M, et al. Bronchial responsiveness among inbred mouse strains: role of airway smooth-muscle shortening velocity. Am J Respir Crit Care Med. (2000) 161:839–48. doi: 10.1164/ajrccm.161.3.9906054
56. Van Boven FE, De Jong NW, Loomans MGLC, Braunstahl GJ, Gerth Van Wijk R, Arends LR. Describing fluctuating indoor aerosol dust measurements with application to house dust mite allergens. Sci Rep. (2020) 10:16897. doi: 10.1038/s41598-020-73839-x
57. Gore RB, Hadi EA, Craven M, Smillie FI, O’Meara TJ, Tovey ER, et al. Personal exposure to house dust mite allergen in bed: nasal air sampling and reservoir allergen levels. Clin Experimental Allergy. (2002) 32:856–9. doi: 10.1046/j.1365-2222.2002.01403.x
58. Tovey ER, Willenborg CM, Crisafulli DA, Rimmer J, Marks GB. Most personal exposure to house dust mite aeroallergen occurs during the day. PLoS One. (2013) 8:e69900. doi: 10.1371/journal.pone.0069900
59. Sakaguchi M, Inouye S, Yasueda H, Irie T, Yoshizawa S, Shida T. Measurement of allergens associated with dust mite allergy. Int Arch Allergy Immunol. (1989) 90:190–3. doi: 10.1159/000235022
60. Tovey ER, Liu-Brennan D, Garden FL, Oliver BG, Perzanowski MS, Marks GB. Time-based measurement of personal Mite allergen bioaerosol exposure over 24 hour periods. PLoS One. (2016) 11:e0153414. doi: 10.1371/journal.pone.0153414
61. Swanson M, Agarwal M, Reed C. An immunochemical approach to indoor aeroallergen quantitation with a new volumetric air sampler: studies with mite, roach, cat, mouse, and Guinea pig antigens. J Allergy Clin Immunol. (1985) 76:724–9. doi: 10.1016/0091-6749(85)90678-5
62. Do DC, Zhao Y, Gao P. Cockroach allergen exposure and risk of asthma. Allergy. (2016) 71:463–74. doi: 10.1111/all.12827
63. Pomés A, Mueller GA, Randall TA, Chapman MD, Arruda LK. New insights into cockroach allergens. Curr Allergy Asthma Rep. (2017) 17:25. doi: 10.1007/s11882-017-0694-1
64. Park YM, Bochner BS. Eosinophil survival and apoptosis in health and disease. Allergy Asthma Immunol Res. (2010) 2:87. doi: 10.4168/aair.2010.2.2.87
65. Prangtaworn P, Chaisri U, Seesuay W, Mahasongkram K, Onlamoon N, Reamtong O, et al. Tregitope-linked refined allergen vaccines for immunotherapy in cockroach allergy. Sci Rep. (2018) 8:15480. doi: 10.1038/s41598-018-33680-9
66. Berlin AA, Lukacs NW. Treatment of cockroach allergen asthma model with imatinib attenuates airway responses. Am J Respir Crit Care Med. (2005) 171:35–9. doi: 10.1164/rccm.200403-385OC
67. Durham CG, Schwiebert LM, Lorenz RG. Use of the cockroach antigen model of acute asthma to determine the immunomodulatory role of early exposure to gastrointestinal infection. In: Allen IC, editor. Mouse Models of Allergic Disease. Methods in Molecular Biology. Totowa, NJ: Humana Press (2013). p. 271–86. doi: 10.1007/978-1-62703-496-8_21
68. Kitinoja MA, Hugg TT, Siddika N, Rodriguez Yanez D, Jaakkola MS, Jaakkola JJK. Short-term exposure to pollen and the risk of allergic and asthmatic manifestations: a systematic review and meta-analysis. BMJ Open. (2020) 10:e029069. doi: 10.1136/bmjopen-2019-029069
69. Idrose NS, Lodge CJ, Erbas B, Douglass JA, Bui DS, Dharmage SC. A review of the respiratory health burden attributable to short-term exposure to pollen. IJERPH. (2022) 19:7541. doi: 10.3390/ijerph19127541
70. Idrose NS, Walters EH, Zhang J, Vicendese D, Newbigin EJ, Douglass JA, et al. Outdoor pollen-related changes in lung function and markers of airway inflammation: a systematic review and meta-analysis. Clin Experimental Allergy. (2021) 51:636–53. doi: 10.1111/cea.13842
71. Xi GP, Zhang Q, Yin J. Establishment and characterization of murine models of asthma and subcutaneous immunotherapy for humulus pollen allergy. Immunity Inflam & Disease. (2021) 9:443–55. doi: 10.1002/iid3.405
72. Zhang Q, Xi G, Yin J. Artemisia sieversiana pollen allergy and immunotherapy in mice. Am J Transl Res. (2021) 13(12):13654–64. PMID: 35035704; PMCID: PMC8748105
73. Yadav UCS, Ramana KV, Aguilera-Aguirre L, Boldogh I, Boulares HA, Srivastava SK. Inhibition of aldose reductase prevents experimental allergic airway inflammation in mice. PLoS One. (2009) 4:e6535. doi: 10.1371/journal.pone.0006535
74. Woodrow JS, Sheats MK, Cooper B, Bayless R. Asthma: the use of animal models and their translational utility. Cells. (2023) 12:1091. doi: 10.3390/cells12071091
75. Wagers S, Lundblad LKA, Ekman M, Irvin CG, Bates JHT. The allergic mouse model of asthma: normal smooth muscle in an abnormal lung? J Appl Physiol. (2004) 96:2019–27. doi: 10.1152/japplphysiol.00924.2003
76. Johnson JR, Wiley RE, Fattouh R, Swirski FK, Gajewska BU, Coyle AJ, et al. Continuous exposure to house dust mite elicits chronic airway inflammation and structural remodeling. Am J Respir Crit Care Med. (2004) 169:378–85. doi: 10.1164/rccm.200308-1094OC
77. Sibilano R, Gaudenzio N, DeGorter MK, Reber LL, Hernandez JD, Starkl PM, et al. A TNFRSF14-FcɛRI-mast cell pathway contributes to development of multiple features of asthma pathology in mice. Nat Commun. (2016) 7:13696. doi: 10.1038/ncomms13696
78. Hew M, Chung KF. Corticosteroid insensitivity in severe asthma: significance, mechanisms and aetiology. Intern Med J. (2010) 40:323–34. doi: 10.1111/j.1445-5994.2010.02192.x
79. Barnes PJ. Corticosteroid resistance in patients with asthma and chronic obstructive pulmonary disease. J Allergy Clin Immunol. (2013) 131:636–45. doi: 10.1016/j.jaci.2012.12.1564
80. Xystrakis E. Reversing the defective induction of IL-10-secreting regulatory T cells in glucocorticoid-resistant asthma patients. J Clin Invest. (2005) 116:146–55. doi: 10.1172/JCI21759
81. Maltby S, Tay HL, Yang M, Foster PS. Mouse models of severe asthma: understanding the mechanisms of steroid resistance, tissue remodelling and disease exacerbation. Respirology. (2017) 22:874–85. doi: 10.1111/resp.13052
82. Nanzer AM, Chambers ES, Ryanna K, Richards DF, Black C, Timms PM, et al. Enhanced production of IL-17A in patients with severe asthma is inhibited by 1α,25-dihydroxyvitamin D3 in a glucocorticoid-independent fashion. J Allergy Clin Immunol. (2013) 132:297–304.e3. doi: 10.1016/j.jaci.2013.03.037
83. Irvin C, Zafar I, Good J, Rollins D, Christianson C, Gorska MM, et al. Increased frequency of dual-positive TH2/TH17 cells in bronchoalveolar lavage fluid characterizes a population of patients with severe asthma. J Allergy Clin Immunol. (2014) 134:1175–1186.e7. doi: 10.1016/j.jaci.2014.05.038
84. Kim HY, Lee HJ, Chang Y-J, Pichavant M, Shore SA, Fitzgerald KA, et al. Interleukin-17–producing innate lymphoid cells and the NLRP3 inflammasome facilitate obesity-associated airway hyperreactivity. Nat Med. (2014) 20:54–61. doi: 10.1038/nm.3423
85. Hong L, Herjan T, Bulek K, Xiao J, Comhair SAA, Erzurum SC, et al. Mechanisms of corticosteroid resistance in type 17 asthma. J Immunol. (2022) 209:1860–9. doi: 10.4049/jimmunol.2200288
86. Yang M, Kumar RK, Foster PS. Pathogenesis of steroid-resistant airway hyperresponsiveness: interaction between IFN-γ and TLR4/MyD88 pathways. J Immunol. (2009) 182:5107–15. doi: 10.4049/jimmunol.0803468
87. Li JJ, Wang W, Baines KJ, Bowden NA, Hansbro PM, Gibson PG, et al. IL-27/IFN-γ induce MyD88-dependent steroid-resistant airway hyperresponsiveness by inhibiting glucocorticoid signaling in macrophages. J Immunol. (2010) 185:4401–9. doi: 10.4049/jimmunol.1001039
88. Gauthier M, Chakraborty K, Oriss TB, Raundhal M, Das S, Chen J, et al. Severe asthma in humans and mouse model suggests a CXCL10 signature underlies corticosteroid-resistant Th1 bias. JCI Insight. (2017) 2:e94580. doi: 10.1172/jci.insight.94580
89. Arleevskaya MI, Aminov R, Brooks WH, Manukyan G, Renaudineau Y. Editorial: shaping of human immune system and metabolic processes by viruses and microorganisms. Front Microbiol. (2019) 10:816. doi: 10.3389/fmicb.2019.00816
90. Perdijk O, Azzoni R, Marsland BJ. The microbiome: an integral player in immune homeostasis and inflammation in the respiratory tract. Physiol Rev. (2024) 104:835–79. doi: 10.1152/physrev.00020.2023
91. Ansaldo E, Farley TK, Belkaid Y. Control of immunity by the microbiota. Annu Rev Immunol. (2021) 39:449–79. doi: 10.1146/annurev-immunol-093019-112348
92. Shin YS, Takeda K, Gelfand EW. Understanding asthma using animal models. Allergy Asthma Immunol Res. (2009) 1:10. doi: 10.4168/aair.2009.1.1.10
93. Campbell CD, Gleeson M, Sulaiman I. The role of the respiratory microbiome in asthma. Front Allergy. (2023) 4:1120999. doi: 10.3389/falgy.2023.1120999
94. Gao Y, Nanan R, Macia L, Tan J, Sominsky L, Quinn TP, et al. The maternal gut microbiome during pregnancy and offspring allergy and asthma. J Allergy Clin Immunol. (2021) 148:669–78. doi: 10.1016/j.jaci.2021.07.011
95. Barcik W, Boutin RCT, Sokolowska M, Finlay BB. The role of lung and gut microbiota in the pathology of asthma. Immunity. (2020) 52:241–55. doi: 10.1016/j.immuni.2020.01.007
96. Darveaux JI, Lemanske RF. Infection-related asthma. The Journal of Allergy and Clinical Immunology: in Practice. (2014) 2:658–63. doi: 10.1016/j.jaip.2014.09.011
97. Pelaia G, Vatrella A, Gallelli L, Renda T, Cazzola M, Maselli R, et al. Respiratory infections and asthma. Respir Med. (2006) 100:775–84. doi: 10.1016/j.rmed.2005.08.025
98. Kloepfer KM, Kennedy JL. Childhood respiratory viral infections and the microbiome. J Allergy Clin Immunol. (2023) 152:827–34. doi: 10.1016/j.jaci.2023.08.008
99. Phan JA, Kicic A, Berry LJ, Fernandes LB, Zosky GR, Sly PD, et al. Rhinovirus exacerbates house-dust-mite induced lung disease in adult mice. PLoS One. (2014) 9:e92163. doi: 10.1371/journal.pone.0092163
100. Hatchwell L, Collison A, Girkin J, Parsons K, Li J, Zhang J, et al. Toll-like receptor 7 governs interferon and inflammatory responses to rhinovirus and is suppressed by IL-5-induced lung eosinophilia. Thorax. (2015) 70:854–61. doi: 10.1136/thoraxjnl-2014-205465
101. Collison A, Hatchwell L, Verrills N, Wark PAB, De Siqueira AP, Tooze M, et al. The E3 ubiquitin ligase midline 1 promotes allergen and rhinovirus-induced asthma by inhibiting protein phosphatase 2A activity. Nat Med. (2013) 19:232–7. doi: 10.1038/nm.3049
102. Girkin JL, Hatchwell LM, Collison AM, Starkey MR, Hansbro PM, Yagita H, et al. TRAIL Signaling is proinflammatory and proviral in a murine model of rhinovirus 1B infection. Am J Physiol Lung Cell Mol Physiol. (2017) 312:L89–99. doi: 10.1152/ajplung.00200.2016
103. Toussaint M, Jackson DJ, Swieboda D, Guedán A, Tsourouktsoglou T-D, Ching YM, et al. Host DNA released by NETosis promotes rhinovirus-induced type-2 allergic asthma exacerbation. Nat Med. (2017) 23:681–91. doi: 10.1038/nm.4332
104. Rajput C, Han M, Ishikawa T, Lei J, Goldsmith AM, Jazaeri S, et al. Rhinovirus C infection induces type 2 innate lymphoid cell expansion and eosinophilic airway inflammation. Front Immunol. (2021) 12:649520. doi: 10.3389/fimmu.2021.649520
105. Mahmutovic Persson I, Akbarshahi H, Menzel M, Brandelius A, Uller L. Increased expression of upstream TH2-cytokines in a mouse model of viral-induced asthma exacerbation. J Transl Med. (2016) 14:52. doi: 10.1186/s12967-016-0808-x
106. Curren B, Ahmed T, Howard DR, Ullah MA, Sebina I, Rashid RB, et al. IL-33-induced neutrophilic inflammation and NETosis underlie rhinovirus-triggered exacerbations of asthma. Mucosal Immunol. (2023) 16:671–84. doi: 10.1016/j.mucimm.2023.07.002
107. Lynch JP, Werder RB, Simpson J, Loh Z, Zhang V, Haque A, et al. Aeroallergen-induced IL-33 predisposes to respiratory virus–induced asthma by dampening antiviral immunity. J Allergy Clin Immunol. (2016) 138:1326–37. doi: 10.1016/j.jaci.2016.02.039
108. Schwarze J, Hamelmann E, Bradley KL, Takeda K, Gelfand EW. Respiratory syncytial virus infection results in airway hyperresponsiveness and enhanced airway sensitization to allergen. J Clin Invest. (1997) 100:226–33. doi: 10.1172/JCI119516
109. O’donnell O. Anaphylactic sensitization to aeroantigen during respiratory virus infection. Clin Experimental Allergy. (1998) 28:1501–8. doi: 10.1046/j.1365-2222.1998.00438.x
110. Leibovitz E, Freihorst J, Piedra PA, Ogra PL. Modulation of systemic and mucosal immune responses to inhaled ragweed antigen in experimentally induced infection with respiratory syncytial virus implication in virally induced allergy. Int Arch Allergy Immunol. (1988) 86:112–6. doi: 10.1159/000234615
111. Freihorst J, Piedra PA, Okamoto Y, Ogra PL. Effect of respiratory syncytial virus infection on the uptake of and immune response to other inhaled antigens. Exp Biol Med. (1988) 188:191–7. doi: 10.3181/00379727-188-42727
112. Looi K, Larcombe AN, Perks KL, Berry LJ, Zosky GR, Rigby P, et al. Previous influenza infection exacerbates allergen specific response and impairs airway barrier integrity in Pre-sensitized mice. IJMS. (2021) 22:8790. doi: 10.3390/ijms22168790
113. Gern JE. Rhinovirus and the initiation of asthma. Curr Opin Allergy Clin Immunol. (2009) 9:73–8. doi: 10.1097/ACI.0b013e32831f8f1b
114. Jartti T, Gern JE. Role of viral infections in the development and exacerbation of asthma in children. J Allergy Clin Immunol. (2017) 140:895–906. doi: 10.1016/j.jaci.2017.08.003
115. Sigurs N, Gustafsson PM, Bjarnason R, Lundberg F, Schmidt S, Sigurbergsson F, et al. Severe respiratory syncytial virus bronchiolitis in infancy and asthma and allergy at age 13. Am J Respir Crit Care Med. (2005) 171:137–41. doi: 10.1164/rccm.200406-730OC
116. Sigurs N, Bjarnason R, Sigurbergsson F, Kjellman B. Respiratory syncytial virus bronchiolitis in infancy is an important risk factor for asthma and allergy at age 7. Am J Respir Crit Care Med. (2000) 161:1501–7. doi: 10.1164/ajrccm.161.5.9906076
117. Adeli M, El-Shareif T, Hendaus M. Asthma exacerbation related to viral infections: an up to date summary. J Family Med Prim Care. (2019) 8:2753. doi: 10.4103/jfmpc.jfmpc_86_19
118. Han M, Rajput C, Ishikawa T, Jarman CR, Lee J, Hershenson MB. Small animal models of respiratory viral infection related to asthma. Viruses. (2018) 10:682. doi: 10.3390/v10120682
119. Palmenberg AC, Gern JE. Classification and evolution of human rhinoviruses. In: Jans DA, Ghildyal R, editors. Rhinoviruses. Methods in Molecular Biology. New York, NY: Springer New York (2015). p. 1–10. doi: 10.1007/978-1-4939-1571-2_1
120. Greve JM, Davis G, Meyer AM, Forte CP, Yost SC, Marlor CW, et al. The major human rhinovirus receptor is ICAM-1. Cell. (1989) 56:839–47. doi: 10.1016/0092-8674(89)90688-0
121. Harris JR, Racaniello VR. Changes in rhinovirus protein 2C allow efficient replication in mouse cells. J Virol. (2003) 77:4773–80. doi: 10.1128/JVI.77.8.4773-4780.2003
122. Tuthill TJ, Papadopoulos NG, Jourdan P, Challinor LJ, Sharp NA, Plumpton C, et al. Mouse respiratory epithelial cells support efficient replication of human rhinovirus. J Gen Virol. (2003) 84:2829–36. doi: 10.1099/vir.0.19109-0
123. Bartlett NW, Walton RP, Edwards MR, Aniscenko J, Caramori G, Zhu J, et al. Mouse models of rhinovirus-induced disease and exacerbation of allergic airway inflammation. Nat Med. (2008) 14:199–204. doi: 10.1038/nm1713
124. Vlasak M, Roivainen M, Reithmayer M, Goesler I, Laine P, Snyers L, et al. The minor receptor group of human rhinovirus (HRV) includes HRV23 and HRV25, but the presence of a lysine in the VP1 HI loop is not sufficient for receptor binding. J Virol. (2005) 79:7389–95. doi: 10.1128/JVI.79.12.7389-7395.2005
125. Chairakaki A-D, Saridaki M-I, Pyrillou K, Mouratis M-A, Koltsida O, Walton RP, et al. Plasmacytoid dendritic cells drive acute asthma exacerbations. J Allergy Clin Immunol. (2018) 142:542–556.e12. doi: 10.1016/j.jaci.2017.08.032
126. Schneider D, Hong JY, Bowman ER, Chung Y, Nagarkar DR, McHenry CL, et al. Macrophage/epithelial cell CCL2 contributes to rhinovirus-induced hyperresponsiveness and inflammation in a mouse model of allergic airways disease. American Journal of Physiology-Lung Cellular and Molecular Physiology. (2013) 304:L162–9. doi: 10.1152/ajplung.00182.2012
127. Lamson D, Renwick N, Kapoor V, Liu Z, Palacios G, Ju J, et al. Masstag polymerase-chain-reaction detection of respiratory pathogens, including a new rhinovirus genotype, that caused influenza-like illness in New York state during 2004–2005. J INFECT DIS. (2006) 194:1398–402. doi: 10.1086/508551
128. Arden KE, McErlean P, Nissen MD, Sloots TP, Mackay IM. Frequent detection of human rhinoviruses, paramyxoviruses, coronaviruses, and bocavirus during acute respiratory tract infections. J Med Virol. (2006) 78:1232–40. doi: 10.1002/jmv.20689
129. Everman JL, Sajuthi S, Saef B, Rios C, Stoner AM, Numata M, et al. Functional genomics of CDHR3 confirms its role in HRV-C infection and childhood asthma exacerbations. J Allergy Clin Immunol. (2019) 144:962–71. doi: 10.1016/j.jaci.2019.01.052
130. Bizzintino J, Lee W-M, Laing IA, Vang F, Pappas T, Zhang G, et al. Association between human rhinovirus C and severity of acute asthma in children. Eur Respir J. (2011) 37:1037–42. doi: 10.1183/09031936.00092410
131. Lambert KA, Prendergast LA, Dharmage SC, Tang M, O’Sullivan M, Tran T, et al. The role of human rhinovirus (HRV) species on asthma exacerbation severity in children and adolescents. Journal of Asthma. (2018) 55:596–602. doi: 10.1080/02770903.2017.1362425
132. Varga SM, Wang X, Welsh RM, Braciale TJ. Immunopathology in RSV infection is mediated by a discrete oligoclonal subset of antigen-specific CD4+ T cells. Immunity. (2001) 15:637–46. doi: 10.1016/S1074-7613(01)00209-6
133. Sparer TE, Matthews S, Hussell T, Rae AJ, Garcia-Barreno B, Melero JA, et al. Eliminating a region of respiratory syncytial virus attachment protein allows induction of protective immunity without vaccine-enhanced lung eosinophilia. J Exp Med. (1998) 187:1921–6. doi: 10.1084/jem.187.11.1921
134. Mukherjee S, Rasky AJ, Lundy PA, Kittan NA, Kunkel SL, Maillard IP, et al. STAT5-Induced Lunatic fringe during Th2 development alters Delta-like 4–mediated Th2 cytokine production in respiratory syncytial virus–exacerbated airway allergic disease. J Immunol. (2014) 192:996–1003. doi: 10.4049/jimmunol.1301991
135. John AE, Berlin AA, Lukacs NW. Respiratory syncytial virus-induced CCL5/RANTES contributes to exacerbation of allergic airway inflammation. Eur J Immunol. (2003) 33:1677–85. doi: 10.1002/eji.200323930
136. Lukacs NW, Tekkanat KK, Berlin A, Hogaboam CM, Miller A, Evanoff H, et al. Respiratory syncytial virus predisposes mice to augmented allergic airway responses via IL-13-mediated mechanisms. J Immunol. (2001) 167:1060–5. doi: 10.4049/jimmunol.167.2.1060
137. O’Reilly R, Ullmann N, Irving S, Bossley CJ, Sonnappa S, Zhu J, et al. Increased airway smooth muscle in preschool wheezers who have asthma at school age. J Allergy Clin Immunol. (2013) 131:1024–1032.e16. doi: 10.1016/j.jaci.2012.08.044
138. Veerapandian R, Snyder JD, Samarasinghe AE. Influenza in asthmatics: for better or for worse? Front Immunol. (2018) 9:1843. doi: 10.3389/fimmu.2018.01843
139. Ahn SY, Lee J, Lee D-H, Ho TL, Le CTT, Ko E-J. Chronic allergic asthma induces T-cell exhaustion and impairs virus clearance in mice. Respir Res. (2023) 24:160. doi: 10.1186/s12931-023-02448-9
140. An S, Jeon YJ, Jo A, Lim HJ, Han YE, Cho SW, et al. Initial influenza virus replication can be limited in allergic asthma through rapid induction of type III interferons in respiratory epithelium. Front Immunol. (2018) 9:986. doi: 10.3389/fimmu.2018.00986
141. Oliver BGG, Robinson P, Peters M, Black J. Viral infections and asthma: an inflammatory interface? Eur Respir J. (2014) 44:1666–81. doi: 10.1183/09031936.00047714
142. Nordengrün M, Michalik S, Völker U, Bröker BM, Gómez-Gascón L. The quest for bacterial allergens. Int J Med Microbiol. (2018) 308:738–50. doi: 10.1016/j.ijmm.2018.04.003
143. Barnes PJ. Intrinsic asthma: not so different from allergic asthma but driven by superantigens? Clin Experimental Allergy. (2009) 39:1145–51. doi: 10.1111/j.1365-2222.2009.03298.x
144. Bachert C, van Steen K, Zhang N, Holtappels G, Cattaert T, Maus B, et al. Specific IgE against Staphylococcus aureus enterotoxins: an independent risk factor for asthma. J Allergy Clin Immunol. (2012) 130:376–381.e8. doi: 10.1016/j.jaci.2012.05.012
145. Bachert C, Gevaert P, Howarth P, Holtappels G, Van Cauwenberge P, Johansson SGO. Ige to Staphylococcus aureus enterotoxins in serum is related to severity of asthma. J Allergy Clin Immunol. (2003) 111:1131–2. doi: 10.1016/S0091-6749(03)70044-X
146. Bisgaard H, Hermansen MN, Bonnelykke K, Stokholm J, Baty F, Skytt NL, et al. Association of bacteria and viruses with wheezy episodes in young children: prospective birth cohort study. Br Med J. (2010) 341:c4978–c4978. doi: 10.1136/bmj.c4978
147. Davis MF, Peng RD, McCormack MC, Matsui EC. Staphylococcus aureus colonization is associated with wheeze and asthma among US children and young adults. J Allergy Clin Immunol. (2015) 135:811–813.e5. doi: 10.1016/j.jaci.2014.10.052
148. Bisgaard H, Hermansen MN, Buchvald F, Loland L, Halkjaer LB, Bønnelykke K, et al. Childhood asthma after bacterial colonization of the airway in neonates. N Engl J Med. (2007) 357:1487–95. doi: 10.1056/NEJMoa052632
149. Hilty M, Burke C, Pedro H, Cardenas P, Bush A, Bossley C, et al. Disordered microbial communities in asthmatic airways. PLoS One. (2010) 5:e8578. doi: 10.1371/journal.pone.0008578
150. Hales BJ, Chai LY, Elliot CE, Pearce LJ, Zhang G, Heinrich TK, et al. Antibacterial antibody responses associated with the development of asthma in house dust mite-sensitised and non-sensitised children. Thorax. (2012) 67:321–7. doi: 10.1136/thoraxjnl-2011-200650
151. Ryu S, Song P, Seo C, Cheong H, Park Y. Colonization and infection of the skin by S. aureus: immune system evasion and the response to cationic antimicrobial peptides. IJMS. (2014) 15:8753–72. doi: 10.3390/ijms15058753
152. Mulcahy ME, McLoughlin RM. Host–bacterial crosstalk determines Staphylococcus aureus nasal colonization. Trends Microbiol. (2016) 24:872–86. doi: 10.1016/j.tim.2016.06.012
153. Krismer B, Weidenmaier C, Zipperer A, Peschel A. The commensal lifestyle of Staphylococcus aureus and its interactions with the nasal microbiota. Nat Rev Microbiol. (2017) 15:675–87. doi: 10.1038/nrmicro.2017.104
154. Qadri F, Asaduzzaman M, Wennerås C, Mohi G, Albert MJ, Abdus Salam M, et al. Enterotoxin-specific immunoglobulin E responses in humans after infection or vaccination with diarrhea-causing enteropathogens. Infect Immun. (2000) 68:6077–81. doi: 10.1128/IAI.68.10.6077-6081.2000
155. Leung DY, Harbeck R, Bina P, Reiser RF, Yang E, Norris DA, et al. Presence of IgE antibodies to staphylococcal exotoxins on the skin of patients with atopic dermatitis. Evidence for a new group of allergens. J Clin Invest. (1993) 92:1374–80. doi: 10.1172/JCI116711
156. Rubin K, Glazer S. The pertussis hypothesis: bordetella pertussis colonization in the etiology of asthma and diseases of allergic sensitization. Med Hypotheses. (2018) 120:101–15. doi: 10.1016/j.mehy.2018.08.006
157. Tomassen P, Jarvis D, Newson R, Van Ree R, Forsberg B, Howarth P, et al. Staphylococcus aureus enterotoxin-specific IgE is associated with asthma in the general population: a GA 2 LEN study. Allergy. (2013) 68:1289–97. doi: 10.1111/all.12230
158. Jorde I, Hildebrand CB, Kershaw O, Lücke E, Stegemann-Koniszewski S, Schreiber J. Modulation of allergic sensitization and allergic inflammation by Staphylococcus aureus enterotoxin B in an ovalbumin mouse model. Front Immunol. (2020) 11:592186. doi: 10.3389/fimmu.2020.592186
159. Teufelberger AR, Nordengrün M, Braun H, Maes T, De Grove K, Holtappels G, et al. The IL-33/ST2 axis is crucial in type 2 airway responses induced by Staphylococcus aureus –derived serine protease–like protein D. J Allergy Clin Immunol. (2018) 141:549–559.e7. doi: 10.1016/j.jaci.2017.05.004
160. Teufelberger AR, Van Nevel S, Hulpiau P, Nordengrün M, Savvides SN, De Graeve S, et al. Mouse strain-dependent difference toward the Staphylococcus aureus allergen serine protease-like protein D reveals a novel regulator of IL-33. Front Immunol. (2020) 11:582044. doi: 10.3389/fimmu.2020.582044
161. Starkl P, Watzenboeck ML, Popov LM, Zahalka S, Hladik A, Lakovits K, et al. Ige effector mechanisms, in concert with mast cells, contribute to acquired host defense against Staphylococcus aureus. Immunity. (2020) 53:793–804.e9. doi: 10.1016/j.immuni.2020.08.002
162. Ubags ND, Trompette A, Pernot J, Nibbering B, Wong NC, Pattaroni C, et al. Microbiome-induced antigen-presenting cell recruitment coordinates skin and lung allergic inflammation. J Allergy Clin Immunol. (2021) 147:1049–1062.e7. doi: 10.1016/j.jaci.2020.06.030
163. Kline SN, Orlando NA, Lee AJ, Wu M-J, Zhang J, Youn C, et al. Staphylococcus aureus proteases trigger eosinophil-mediated skin inflammation. Proc Natl Acad Sci USA. (2024) 121:e2309243121. doi: 10.1073/pnas.2309243121
164. Nakamura Y, Oscherwitz J, Cease KB, Chan SM, Muñoz-Planillo R, Hasegawa M, et al. Staphylococcus δ-toxin induces allergic skin disease by activating mast cells. Nature. (2013) 503:397–401. doi: 10.1038/nature12655
165. McCann JR, Mason SN, Auten RL, St. Geme JW, Seed PC. Early-Life intranasal colonization with nontypeable Haemophilus influenzae exacerbates juvenile airway disease in mice. Infect Immun. (2016) 84:2022–30. doi: 10.1128/IAI.01539-15
166. Essilfie A-T, Simpson JL, Dunkley ML, Morgan LC, Oliver BG, Gibson PG, et al. Combined Haemophilus influenzae respiratory infection and allergic airways disease drives chronic infection and features of neutrophilic asthma. Thorax. (2012) 67:588–99. doi: 10.1136/thoraxjnl-2011-200160
167. Essilfie A-T, Simpson JL, Horvat JC, Preston JA, Dunkley ML, Foster PS, et al. Haemophilus influenzae infection drives IL-17-mediated neutrophilic allergic airways disease. PLoS Pathog. (2011) 7:e1002244. doi: 10.1371/journal.ppat.1002244
168. Medina JL, Coalson JJ, Brooks EG, Le Saux CJ, Winter VT, Chaparro A, et al. Mycoplasma pneumoniae CARDS toxin exacerbates ovalbumin-induced asthma-like inflammation in BALB/c mice. PLoS One. (2014) 9:e102613. doi: 10.1371/journal.pone.0102613
169. Blasi F, Aliberti S, Allegra L, Piatti G, Tarsia P, Ossewaarde JM, et al. Chlamydophila pneumoniae induces a sustained airway hyperresponsiveness and inflammation in mice. Respir Res. (2007) 8:83. doi: 10.1186/1465-9921-8-83
170. Gallego M, Pomares X, Espasa M, Castañer E, Solé M, Suárez D, et al. Pseudomonas aeruginosaisolates in severe chronic obstructive pulmonary disease: characterization and risk factors. BMC Pulm Med. (2014) 14:103. doi: 10.1186/1471-2466-14-103
171. Garcia-Clemente M, De La Rosa D, Máiz L, Girón R, Blanco M, Olveira C, et al. Impact of Pseudomonas aeruginosa infection on patients with chronic inflammatory airway diseases. JCM. (2020) 9:3800. doi: 10.3390/jcm9123800
172. Telford G, Wheeler D, Williams P, Tomkins PT, Appleby P, Sewell H, et al. The Pseudomonas aeruginosa quorum-sensing signal molecule N -(3-oxododecanoyl)- l -homoserine lactone has immunomodulatory activity. Infect Immun. (1998) 66:36–42. doi: 10.1128/IAI.66.1.36-42.1998
173. Brown MA, Jabeen M, Bharj G, Hinks TSC. Non-typeable Haemophilus influenzae airways infection: the next treatable trait in asthma? Eur Respir Rev. (2022) 31:220008. doi: 10.1183/16000617.0008-2022
174. Zhang Y, Wang H, Zhang Y, Zhao P, Li Y. Aerosolization inhalation of non-typeable Haemophilus influenzae outer membrane vesicles contributing to neutrophilic asthma. Front Microbiol. (2023) 14:1226633. doi: 10.3389/fmicb.2023.1226633
175. Blasi F. Atypical pathogens and respiratory tract infections. Eur Respir J. (2004) 24:171–81. doi: 10.1183/09031936.04.00135703
176. Liu X, Wang Y, Chen C, Liu K. Mycoplasma pneumoniae infection and risk of childhood asthma: a systematic review and meta-analysis. Microb Pathog. (2021) 155:104893. doi: 10.1016/j.micpath.2021.104893
177. Yin S-S, Ma F-L, Gao X. Association of Mycoplasma pneumoniae infection with increased risk of asthma in children. Exp Ther Med. (2017) 13:1813–9. doi: 10.3892/etm.2017.4219
178. Cosentini R, Tarsia P, Canetta C, Graziadei G, Brambilla AM, Aliberti S, et al. Severe asthma exacerbation: role of acute Chlamydophila pneumoniae and Mycoplasma pneumoniae infection. Respir Res. (2008) 9:48. doi: 10.1186/1465-9921-9-48
179. Hong S-J. The role of Mycoplasma pneumoniae infection in asthma. Allergy Asthma Immunol Res. (2012) 4:59. doi: 10.4168/aair.2012.4.2.59
180. Tiwary M, Samarasinghe AE. Initiation and pathogenesis of severe asthma with fungal sensitization. Cells. (2021) 10:913. doi: 10.3390/cells10040913
181. Van Tilburg Bernardes E, Gutierrez MW, Arrieta M-C. The fungal microbiome and asthma. Front Cell Infect Microbiol. (2020) 10:583418. doi: 10.3389/fcimb.2020.583418
182. Sharpe RA, Bearman N, Thornton CR, Husk K, Osborne NJ. Indoor fungal diversity and asthma: a meta-analysis and systematic review of risk factors. J Allergy Clin Immunol. (2015) 135:110–22. doi: 10.1016/j.jaci.2014.07.002
183. Bozek A, Pyrkosz K. Immunotherapy of mold allergy: a review. Hum Vaccin Immunother. (2017) 13:2397–401. doi: 10.1080/21645515.2017.1314404
184. Goh KJ, Yii ACA, Lapperre TS, Chan AK, Chew FT, Chotirmall SH, et al. Sensitization to Aspergillus species is associated with frequent exacerbations in severe asthma. J Asthma Allergy. (2017) 10:131–40. doi: 10.2147/JAA.S130459
185. Welsh KG, Holden KA, Wardlaw AJ, Satchwell J, Monteiro W, Pashley CH, et al. Fungal sensitization and positive fungal culture from sputum in children with asthma are associated with reduced lung function and acute asthma attacks respectively. Clin Exp Allergy. (2021) 51:790–800. doi: 10.1111/cea.13799
186. Backman H, Jansson S, Stridsman C, Eriksson B, Hedman L, Eklund B, et al. Severe asthma—a population study perspective. Clin Experimental Allergy. (2019) 49:819–28. doi: 10.1111/cea.13378
187. Lenardon MD, Munro CA, Gow NA. Chitin synthesis and fungal pathogenesis. Curr Opin Microbiol. (2010) 13:416–23. doi: 10.1016/j.mib.2010.05.002
188. Reese TA, Liang H-E, Tager AM, Luster AD, Van Rooijen N, Voehringer D, et al. Chitin induces accumulation in tissue of innate immune cells associated with allergy. Nature. (2007) 447:92–6. doi: 10.1038/nature05746
189. Havaux X, Zeine A, Dits A, Denis O. A new mouse model of lung allergy induced by the spores of alternaria alternata and cladosporium herbarum molds. Clin Exp Immunol. (2004) 139:179–88. doi: 10.1111/j.1365-2249.2004.02679.x
190. Zhou W, Toki S, Zhang J, Goleniewksa K, Newcomb DC, Cephus JY, et al. Prostaglandin I 2 signaling and inhibition of group 2 innate lymphoid cell responses. Am J Respir Crit Care Med. (2016) 193:31–42. doi: 10.1164/rccm.201410-1793OC
191. Snelgrove RJ, Gregory LG, Peiró T, Akthar S, Campbell GA, Walker SA, et al. Alternaria-derived serine protease activity drives IL-33–mediated asthma exacerbations. J Allergy Clin Immunol. (2014) 134:583–592.e6. doi: 10.1016/j.jaci.2014.02.002
192. Kobayashi T, Iijima K, Radhakrishnan S, Mehta V, Vassallo R, Lawrence CB, et al. Asthma-related environmental fungus, alternaria, activates dendritic cells and produces potent Th2 adjuvant activity. J Immunol. (2009) 182:2502–10. doi: 10.4049/jimmunol.0802773
193. Bartemes KR, Iijima K, Kobayashi T, Kephart GM, McKenzie AN, Kita H. IL-33–responsive lineage−CD25+CD44hi lymphoid cells mediate innate type 2 immunity and allergic inflammation in the lungs. J Immunol. (2012) 188:1503–13. doi: 10.4049/jimmunol.1102832
194. Samarasinghe AE, Hoselton SA, Schuh JM. A comparison between intratracheal and inhalation delivery of Aspergillus fumigatus conidia in the development of fungal allergic asthma in C57BL/6 mice. Fungal Biol. (2011) 115:21–9. doi: 10.1016/j.funbio.2010.09.013
195. Hoselton SA, Samarasinghe AE, Seydel JM, Schuh JM. An inhalation model of airway allergic response to inhalation of environmental Aspergillus fumigatus conidia in sensitized BALB/c mice. Med Mycol. (2010) 48:1056–65. doi: 10.3109/13693786.2010.485582
196. Hogaboam CM, Blease K, Mehrad B, Steinhauser ML, Standiford TJ, Kunkel SL, et al. Chronic airway hyperreactivity, goblet cell hyperplasia, and peribronchial fibrosis during allergic airway disease induced by Aspergillus fumigatus. Am J Pathol. (2000) 156:723–32. doi: 10.1016/S0002-9440(10)64775-X
197. Buskirk AD, Green BJ, Lemons AR, Nayak AP, Goldsmith WT, Kashon ML, et al. A murine inhalation model to characterize pulmonary exposure to dry Aspergillus fumigatus conidia. PLoS One. (2014) 9:e109855. doi: 10.1371/journal.pone.0109855
198. Goplen N, Karim MZ, Liang Q, Gorska MM, Rozario S, Guo L, et al. Combined sensitization of mice to extracts of dust mite, ragweed, and Aspergillus species breaks through tolerance and establishes chronic features of asthma. J Allergy Clin Immunol. (2009) 123:925–932.e11. doi: 10.1016/j.jaci.2009.02.009
199. Knight J M, Li E, Tung H-Y, Landers C, Wheeler J, Kheradmand F, et al. A fungal protease model to interrogate allergic lung immunity. In: Reinhardt RL, editor. Type 2 Immunity. Methods in Molecular Biology. New York, NY: Springer New York (2018). p. 1–9. doi: 10.1007/978-1-4939-7896-0_1
200. Latgé J-P, Chamilos G. Aspergillus fumigatus and aspergillosis in 2019. Clin Microbiol Rev. (2019) 33:e00140–18. doi: 10.1128/CMR.00140-18
201. Pandey S, Hoselton SA, Schuh JM. The impact of Aspergillus fumigatus viability and sensitization to its allergens on the murine allergic asthma phenotype. BioMed Res Int. (2013) 2013:1–17. doi: 10.1155/2013/619614
Keywords: allergic asthma, microbial infection, adjuvant-free, natural allergen, mouse model, sensitization
Citation: Radhouani M and Starkl P (2024) Adjuvant-independent airway sensitization and infection mouse models leading to allergic asthma. Front. Allergy 5: 1423938. doi: 10.3389/falgy.2024.1423938
Received: 26 April 2024; Accepted: 5 July 2024;
Published: 2 August 2024.
Edited by:
Friederike Jönsson, Institut Pasteur, FranceReviewed by:
Wayne Robert Thomas, University of Western Australia, AustraliaKapil Sirohi, National Jewish Health (United States), United States
© 2024 Radhouani and Starkl. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Philipp Starkl, cGhpbGlwcC5zdGFya2xAbWVkdW5pd2llbi5hYy5hdA==
 Mariem Radhouani
Mariem Radhouani Philipp Starkl
Philipp Starkl