- 1Allergy-Immunology Division, Northwestern University Feinberg School of Medicine, Chicago, IL, United States
- 2Herman B Wells Center for Pediatric Research, Department of Pediatrics, Indiana University School of Medicine, Indianapolis, IN, United States
- 3Department of Microbiology and Immunology, Indiana University School of Medicine, Indianapolis, IN, United States
- 4University of Minnesota Genomics Center, Minneapolis, MN, United States
- 5Department of Medicine, University of California, San Francisco, San Francisco, CA, United States
- 6Department of Pediatrics, Section of Pulmonology, Baylor College of Medicine, Texas Children’s Hospital, Houston, TX, United States
- 7Veterans Caribbean Health Care System, San Juan, PR, United States
- 8Centro de Neumologia Pediatrica, CSP, San Juan, PR, United States
- 9Pediatric Pulmonary Division, Jacobi Medical Center, Bronx, NY, United States
- 10Department of Pediatrics, University of California, San Francisco, San Francisco, CA, United States
- 11Center for Genes, Environment, and Health and the Department of Pediatrics, National Jewish Health, Denver, CO, United States
- 12Division of Pulmonary Sciences and Critical Care Medicine, Department of Medicine, University of Colorado, Denver, CO, United States
- 13Division of Allergy and Clinical Immunology, Ann and Robert H Lurie Children’s Hospital of Chicago, Chicago, IL, United States
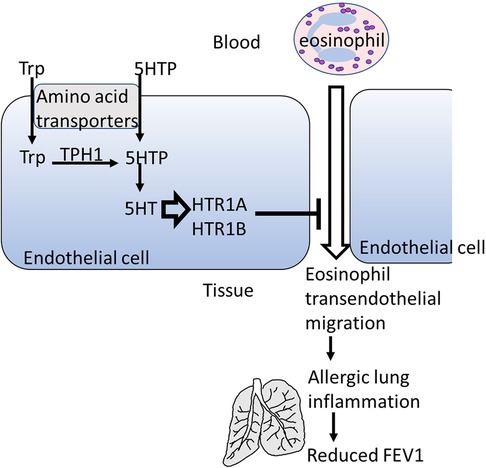
Background: Previous research showed that 5-hydroxytryptophan (5HTP), a metabolic precursor of serotonin, reduces allergic lung inflammation by inhibiting eosinophil migration across endothelial monolayers.
Objective: It is unknown if serotonin receptors are involved in mediating this 5HTP function or if serotonin receptor (HTR) single nucleotide polymorphisms (SNPs) associate with lung function in humans.
Methods: Serotonin receptor subtypes were assessed by qPCR, western blot, confocal microscopy, pharmacological inhibitors and siRNA knockdown. HTR SNPs were assessed in two cohorts.
Results: Pharmacological inhibition or siRNA knockdown of the serotonin receptors HTR1A or HTR1B in endothelial cells abrogated the inhibitory effects of 5HTP on eosinophil transendothelial migration. In contrast, eosinophil transendothelial migration was not inhibited by siRNA knockdown of HTR1A or HTR1B in eosinophils. Surprisingly, these HTRs were intracellular in endothelial cells and an extracellular supplementation with serotonin did not inhibit eosinophil transendothelial migration. This is consistent with the inability of serotonin to cross membranes, the lack of selective serotonin reuptake receptors on endothelial cells, and the studies showing minimal impact of selective serotonin reuptake inhibitors on asthma. To extend our HTR studies to humans with asthma, we examined the CHIRAH and GALA cohorts for HTR SNPs that affect HTR function or are associated with behavior disorders. A polygenic index of SNPs in HTRs was associated with lower lung function in asthmatics.
Conclusions: Serotonin receptors mediate 5HTP inhibition of transendothelial migration and HTR SNPs associate with lower lung function. These results may serve to aid in design of novel interventions for allergic inflammation.
3 Key messages
• Serotonin receptors in endothelial cells are intracellular.
• Intracellular serotonin receptors in endothelial cells mediate 5-hydroxytryptophan inhibition of eosinophil transendothelial migration.
• In asthmatics, serotonin receptor SNPs associate with lower lung function.
Introduction
Asthma affects up to 8% of adults and 7% of children (1). The most common subtype of asthma is allergic asthma, affecting up to 80% of children with asthma (2–5). A challenging aspect of asthma is that mental health disorders like anxiety and depression are more prevalent in people with asthma (6–11). A link between asthma and anxiety may lie within the serotonin pathway (12–19). Decreased serotonin synthesis or polymorphisms of the multiple classes of serotonin receptors (HTR's) are associated with anxiety and depression (20–29). Furthermore, serotonin receptors are found on the surface of immune cells, endothelial cells, and immunoregulatory functions have been attributed to serotonin (15–19, 30, 31).
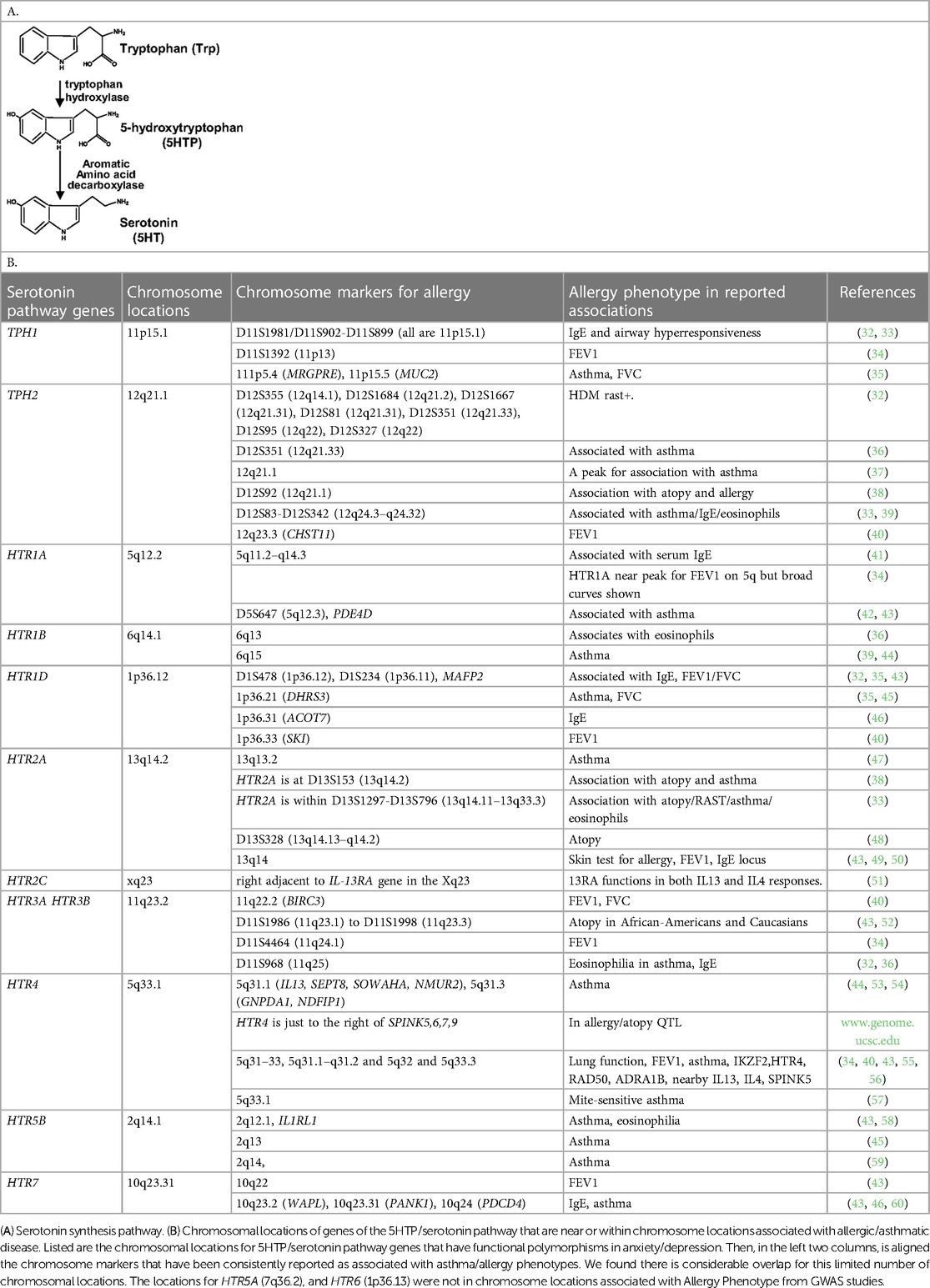
For serotonin synthesis, the rate limiting step is tryptophan hydroxylase (TPH) of metabolism of L-tryptophan to 5-hydroxytryptophan (5HTP). 5HTP is then rapidly metabolized to serotonin (Table 1A). Decreased TPH function reduces generation of 5HTP and serotonin, which is associated with depression (20–24). An imbalance in 5-HTP and serotonin levels can result in changes in activation of 14 subtypes of inhibitory and stimulatory HTR's in peripheral tissues (22, 61–64). Furthermore, single nucleotide polymorphisms (SNPs) in the HTRs alter HTR function or are associated with behavior disorders (21–23, 26, 27, 29, 65–71).
HTRs and serotonin synthesis also exist in peripheral tissues. Interestingly, systemic administration of serotonin and 5HTP have opposing effects in the vascular and immune systems. Systemic administration of serotonin induces smooth muscle constriction in the airway (72–74). Also, vasoconstriction of the pulmonary arteries and aorta (75–77) can occur during treatment of anxiety and depression with selective serotonin reuptake inhibitors (SSRI's), which block the neuronal SERT transporter and elevate systemic serotonin levels (78). On the other hand, 5HTP is associated with vasodilation in animal studies (79–82). The opposing functions of 5HTP and serotonin on vasoconstriction may occur as an outcome of changes in local microenvironment concentrations of 5HTP and serotonin that would generate a change in the balance of activation of the multiple inhibitory and stimulatory HTRs that are differentially expressed by cells. Pharmacological inhibitors of the individual serotonin receptors HTR1A and HTR2A had only marginal benefit for asthma as summarized in a review by Cazzola et al. (83). It is anticipated that because there are multiple HTR receptors, a balance of multiple HTRs functioning in concert may be needed to regulate asthma.
It has been shown that 5HTP is acquired by endothelial cells and is metabolized to serotonin within the endothelial cells and that 5HTP reduces leukocyte transendothelial migration and reduces eosinophilic inflammation in four preclinical models and reduces airway hyper-responsiveness (18). Although 5HTP supplementation blocked leukocyte recruitment in the preclinical models, it did not alter leukocyte and eosinophil adhesion molecule expression, chemokine receptor expression, chemotaxis, or chemokinesis (18). However, whether HTRs have a role in 5HTP regulation of endothelial cells and leukocytes in allergic inflammation is not known. Moreover, these preclinical studies used the natural product 5HTP, derived from the plant Griffonia simplifonia, suggesting that it may act as an innovative novel therapy to address the current treatment gaps for allergic asthma and mood disorder.
We report subtypes of HTRs for mouse endothelium and eosinophils. Furthermore, selective HTR1A and HTR1B pharmacological inhibitors or siRNA knock down of these receptors blocked 5HTP inhibition of migration without altering eosinophil adhesion. In contrast, inhibition or siRNA knock down of these receptors in eosinophils did not block the 5HTP inhibition of transendothelial migration or eosinophil binding. Thus, endothelial serotonin receptors rather than eosinophil serotonin receptors, mediated 5HTP inhibition of transendothelial migration. Furthermore, 5HTP was catabolized to serotonin in endothelial cells and, intriguingly, HTR expression was intracellular in the endothelial cells. Consistent with intracellular HTR expression in endothelial cells and the inability of serotonin to cross cell membranes, exogenous addition of serotonin did not inhibit eosinophil transendothelial migration. In the CHIRAH and GALA cohorts, indexes of SNPs in HTRs and TPH associated with decreased lung function. These results will help design clinical studies addressing mechanisms for 5HTP inhibition of allergic asthmatic responses and the common comorbidities of anxiety or depression.
Methods
Animals
C57BL/6J mice (female, 6–8 weeks old) and NJ1638 mice were from Jackson Laboratory, Bar Harbor, Maine. The studies are approved by the Indiana University Institutional Review Committee for animals (protocol number: 21109).
CHIRAH and GALA cohorts
CHIRAH
This analysis is based on the cohort established by the Chicago Initiative to Raise Asthma Health Equity (CHIRAH) study (84–86). The institutional review board of Northwestern University approved the CHIRAH protocol (IRB Project Number: STU00042117) (84–86). Written informed consent was obtained. The CHIRAH cohort is a community-based longitudinal cohort study of urban children and adults with persistent asthma enrolled from February 2004 to July 2005. The cohort was established by a broad community-based screening for households with persons with asthma using a school-based sampling technique. Adults were analyzed in this study. The adults were 18–40 years of age and self-reported race/ethnicity as non-Hispanic/African American, Hispanic/Latino, and non-Hispanic white/other. Our analyses examined the adults that had lung function analyses and DNA samples and were self-identified as African Americans (n = 103) or Latinx (n = 73) (85, 87, 88). Key SNPs in the HTR1A/1B/2A/2C/3B and TPH1 and TPH2 were chosen by effects on gene function or associations with behavior disorders (21–23, 26, 27, 29, 65–71). SNP analyses were carried out using the sequenom iPLEX platform.
GALA II
The Genes-environments and Admixture in Latino Americans (GALA II) is a clinic-based multicenter asthma case-control study designed to examine the genetic and environmental risk factors for asthma and asthma-related phenotypes in Latino children (Institutional Review Board of University of California San Francisco, IRB project number 10-00889; Ann and Robert H Lurie Children's Hospital of Chicago, IRB project number 2008-13531) (89–91). Written informed consent was obtained. Asthma cases were defined as participants aged 8–21 years with a history of physician-diagnosed asthma and the presence of 2 or more symptoms of coughing, wheezing, or shortness of breath in the 2 years preceding enrollment. Healthy control subjects were recruited from the community and clinics with the same catchment area as cases. Subjects were recruited from five centers (Chicago, Illinois; Bronx, New York; Houston, Texas; the San Francisco Bay Area, California; and Puerto Rico) from July 2008 through November 2011. Control subjects were frequency matched on age (within 1 year), sex, and study center. 4,477 subject participants were considered for inclusion in the study sample. After excluding those with missing values for key variables, the final sample included 2,126 participants (90). For the GALA 2 analysis, genotyping had been carried out by the Affymetrix Axiom World Array 4™ (92). GALA protocols were approved by institutional review boards for the centers as previously described (89–91).
Endothelial cells and eosinophils
Human microvascular endothelial cells from the lung (HMVEC-Ls) (CC-Lonza, Walkersville, MD) were grown in EGM-MV endothelial growth medium plus 5% FCS (catalog #CC-3125, Lonza) and were used at passage 2–6. To prepare human eosinophils, human peripheral blood was diluted 1/2.5 with PBS containing 2 mM EDTA and then separated on 1.090 g/ml Percoll with centrifugation at 1,200 rpm for 20 min at room temperature. The layer containing eosinophils and neutrophils was collected and red blood cells removed by hypotonic lysis. The cells were further isolated by depletion of neutrophils using human CD16 negative selection using Miltenyi microbeads (catalog #130-045-701) to yield >95% eosinophils as determined by morphology and eosin staining of cytospins. These cells were used for western blot analyses of serotonin receptor expression.
The murine endothelial cell line mHEVa was cultured as previously described (93, 94). The murine endothelial cells were maintained in RPMI 1,640 (Corning 15-040-CV) supplemented with 20% fetal calf serum, 2 mM L-glutamine (Corning), 1 mM HEPES (Corning), 10 mM Sodium bicarbonate (Sigma), 50 µg/ml gentamycin (Lonza), 1X Penicillin/Streptomycin (Corning). Spleen eosinophils from NJ1638 mice were used for transendothelial migration assays. To generate murine eosinophils for knockdown of eosinophil HTRs, bone marrow cells were flushed from tibia and femur of C57Bl/6 mice and cultured in RPMI 1,640 supplemented with 20% fetal calf serum, 2 mM L-glutamine, 2.5 mM HEPES, 50 µg/ml gentamycin, 1X Penicillin/Streptomycin (Corning), 0.1 mM Sodium Pyruvate (Corning), 5 µM beta-mercaptoethanol (Sigma), 100 ng/ml SCF (250-03-100UG), and 100 ng/ml FLT3-L (Peprotech 250-31l-100UG). After five days of bone marrow in culture the cells were treated with siRNAs (1 µM) or untreated, bone marrow cells are centrifuged and replaced with fresh media containing 10 ng/ml IL5 (Peprotech 215-15-50UG). On day 8, the cells were >85% eosinophils as determined by morphology and eosin staining of cytospins.
Immunolabeling HTRs in cells for confocal microscopy
Cultured mouse endothelial cell monolayers were fixed in −20°C methanol for 15 min and rehydrated with PBS for 1 h, which removes free serotonin. The samples were blocked with goat serum in PBS-0.3% BSA, incubated with rabbit anti-mouse HTR1A (catalog #MBS2528551, MyBiosource), rabbit anti-mouse HTR1B (catalog #NB100-56350, Novus Biologicals), or isotype control antibodies for 1 h at room temperature, washed, labeled with FITC-conjugated goat anti-rabbit IgG antibodies (catalog #554020, BD Biosciences), washed and then labeled with Alexa Fluor® 532-conjugated rat anti-mouse VCAM-1 (catalog #NBP2-50620AF53, Novus Biologicals), washed and cover slipped with ProLong™ Gold Antifade Mountant with DAPI (catalog #P36935, ThermoFisher). Images were analyzed by fluorescence microscopy.
Transfection with siRNA
The cell/RNA mixture was added to an electroporation cuvette (catalog #12358-346, Bulldog-bio) and pulsed twice at 125 V for 2.5 ms at a 50.0 ms interval followed by five pulses at 20 V for 50 ms at a 50.0 ms interval in a Nepa Gene electroporator. Cells were immediately placed in fresh media in a parallel plate flow chamber for transendothelial migration assays or transferred to cell culture flask for growth prior to western blot analysis. After 72 h in culture, cells were assayed for transendothelial cell migration or for HTR expression by western blot analysis.
Western blots
For western blot analysis of siRNA knockdown of HTRs at 72 h after siRNA transfection, the cells were suspended with 0.03% EDTA, centrifuged and lysed in 1X RIPA buffer (Thermo 89900), on ice for 30 min. Protein concentration for each sample was determined via BCA assay (ThermoFisher 23227) and normalized. Loading buffer was added and 10 µg total protein was loaded onto 4%–20% Tris-Glycine gels (ThermoFisher XP04200) for western blot analysis of HTR1A expression or 40 µg total protein was loaded for western blot analysis of HTR1B expression. Blots were transferred to PVDF and blocked with 5% bovine serum albumin (Sigma) or 5% milk (BioRad) in TBS (Fisher) with 0.1% Tween20 (Fisher). Primary antibodies included rabbit anti-HTR1A (catalog #MBS2528551, MyBiosource) at 1:1,000, rabbit anti-HTR1B (catalog #NB100-56350, Novus Biologics) at 1:1,000, and rabbit anti-beta actin (catalog #12620, Cell Signaling) at 1:10,000. The secondary antibody was alpaca anti-rabbit conjugated to HRP (catalog #611-035-251, Jackson ImmunoResearch) at 1:10,000. Chemiluminescence was performed using WesternBright ECL substrate (Advansta R-03027-C50).
To examine endothelial cell surface HTR1A and HTR1B, serum-containing medium was removed from the endothelial cell monolayers by washing 3 times with phosphate buffered saline supplemented with 0.2 mM CaCl2 and 0.1 mM MgCl2 (PBS-Ca-Mg) because cations are required for cell adhesion. Then the surface of endothelial cells was biotinylated with 0.5 mg/ml sulfo-NHS-biotin (catalog #A39256, Fisher Scientific) in PBS-Ca-Mg for 30 min (95). Then, the cells were washed 3 times with 4°C 10 mM glycine in PBS-Ca-Mg to quench unbound sulfo-NHS-biotin and then wash once with 4°C PBS-Ca-Mg. The cells were collected by gentle scraping, centrifuged 1,200 rpm for 8 min, and lysed with a lysis buffer (150 mM NaCl, 20 mM Tris-HCl, 0.5% NP40, 10% glycerol, pH7.5 with protease phosphatase inhibitors) as described (95). The samples were vortexed briefly and incubated 10 min; this was repeated for a total of 3 times. The samples were centrifuged 13,000 rpm for 30 min at 4°C. The supernatant was collected and placed in a pre-chilled tube and then 100 µl of Pierce High-Capacity Streptavidin Agarose (catalog #20357, Thermo Scientific) was added and end-over-end rotation was applied for 2 h at 4°C. The samples were centrifuged 10,000 × g for 2 min at 4°C, supernatants were removed, and agarose beads were washed twice with lysis buffer, twice with 1.5 M guanidine HCl, and then twice with lysis buffer. SDS loading buffer was added to the samples, boiled for 5 min and centrifuged 5 min 10,500 × g for 2 min at 4°C. Samples were examined by western blot for HTR1A, HTR1B, VCAM-1 and βactin as described above.
In vitro eosinophil migration assays in transwells or under laminar flow
For Transwell migration assays, confluent monolayers of mouse endothelial cells grown in slide flasks were treated with siRNA (HTR1A, HTR1B or scrambled) and grown for 3 days to confluence in slide flasks or confluent monolayers of endothelial were treated overnight with or without small molecule pharmacological inhibitors of HTR1A (6 nM NAD-299 and 150 nM WAY-100135), HTR1B (500 nM NAS-181 and 88 nM SB224289) or HTR3 (75 nM Dolasetron Mesylate and 62 nM Ondansetron Hydrochloride) (Tocris Bioscience, MN). These inhibitors are lipid soluble (datasheets, Tocris Bioscience, MN), and have been described to cross tissue membranes (96, 97). The treatments had no effect on cell viability as determined by trypan blue exclusion. At 18 h after treatment, eosinophil binding to endothelial cells and eosinophil transendothelial migration were assessed as we previously describe in detail (98, 99). For eosinophil binding, mouse eosinophils (>85% eosinophils) were added to a endothelial cell monolayers in a parallel plate flow changed and incubated at 37°C for 5 min; then nonbound eosinophils were removed by washing 3 times with PBS with 200 µM MgCl2 and 150 µM CaCl2. The bound cells were fixed with methanol, coverslipped and counted by microscopy. Eosinophil transendothelial migration was examined using a parallel plate flow chamber under conditions of laminar flow of 2 dynes/cm2, as previously described (98, 99). This migration assay is dependent on endothelial cell expression of VCAM-1 and the chemokine MCP-1 (93, 100). Eosinophils were added to the endothelial cell monolayers, laminar flow applied for 15 min, cells fixed with methanol, slides coverslipped and then cells that had migrated under the endothelial monolayer examined by phase contrast microscopy as we previously described (98, 99).
Statistics
For the preclinical studies, data were analyzed by a one-way ANOVA followed by Tukey's or Dunnett's multiple comparisons test (SigmaStat, Jandel Scientific, San Ramon, CA). p < 0.05 is considered significant. Presented are the means ± the standard errors.
For analyses of the GALA2 and CHIRAH cohorts, the minor variants of the identified SNPs were evaluated for direction of association with percent predicted FEV1 as a positive or inverse association. To evaluate significance of the association of individual SNPs with lung function, linear regression was carried out in each cohort with FEV1 as the outcome. Covariates for the CHIRAH analyses included duration of asthma, household income, private insurance, household smoke exposure, and use of an inhaled corticosteroid. Covariates included for the GALA2 study are as follows: ancestry (African and European), whether the subject ever smoked tobacco, site, annual household income, and maternal education. The outcome for these analyses in both cohorts was percent predicted FEV1 using Hankinson NHANES equations (101). For the GALA2 and CHIRAH cohorts, the polygenic index is the sum of SNPs whereby coding for each SNP was 0, 1, or 2 for number of alleles with the SNP. SNPs included in the index in CHIRAH had a nominal association with FEV1 (p < 0.2). This p-value was chosen to allow for inclusion of SNPs which would not have met significance at the 0.2 level, to allow for inclusion on the basis of putative biological functional because these SNPs affect protein function or are associated with behavior disorders (21–23, 26, 27, 29, 65–71). For the larger cohort GALA, we included all SNPs with p-values <0.2. Then, using the allele coding for these SNPs in HTRs and TPH, a polygenic risk score was calculated for subjects in CHIRAH and GALA. In both cohorts, we carried out linear regression analyses of the polygenic risk scores on the outcome of percent predicted FEV1.
Results
HTRs are in chromosome locations associated with asthma/allergic disease in humans
5HTP blocks recruitment of eosinophils in four models of allergic inflammation and blocks increased airway hyperresponsiveness in models of allergic airway inflammation (18). Endothelial cells express enzymes for synthesis and degradation of serotonin (18, 102–106). There are two TPH genes for the metabolism of tryptophan to 5HTP which is then metabolized to serotonin and there are 14 receptors (HTRs) for serotonin (61, 62). To focus our studies on HTRs that may be involved in eosinophil recruitment during allergic diseases and regulation of airway function, we determined if there are HTRs or TPHs in human chromosomal locations that are also chromosomal locations associated with asthma/allergic disease in genome-wide association studies (GWAS). Interestingly, genes in the serotonin pathway (63, 107–109) are located within chromosome locations that are also associated with allergy/asthma (32–34, 36–38, 41, 42, 48, 49, 52, 55–57, 65, 66, 68–71, 110–132) (Table 1B), suggesting that this pathway may be an important pathway in regulation of asthma and allergic disease.
HTR expression by endothelial cells and eosinophils
We determined expression of serotonin receptors (HTRs) in endothelial cells and eosinophils by qPCR. Mouse brain tissue was a positive control for detection of HTRs. Mouse endothelial cells expressed HTR1A, HTR1B and HTR3A but not the other HTRs. Mouse eosinophils expressed HTR1A, HTR1B, HTR2A and HTR3A but not the other HTRs (Figure 1A). Also mouse endothelial cells were treated with 5HTP (125 µm) overnight as we previously described (18); this did not alter mRNA (Figure 1A) or protein expression of HTR1A, HTR1B or HTR3A (Figures 1B–D). These receptors are also expressed by human endothelial cells and human eosinophils in Figures 1E,F and as previously described (16, 31, 133–139).
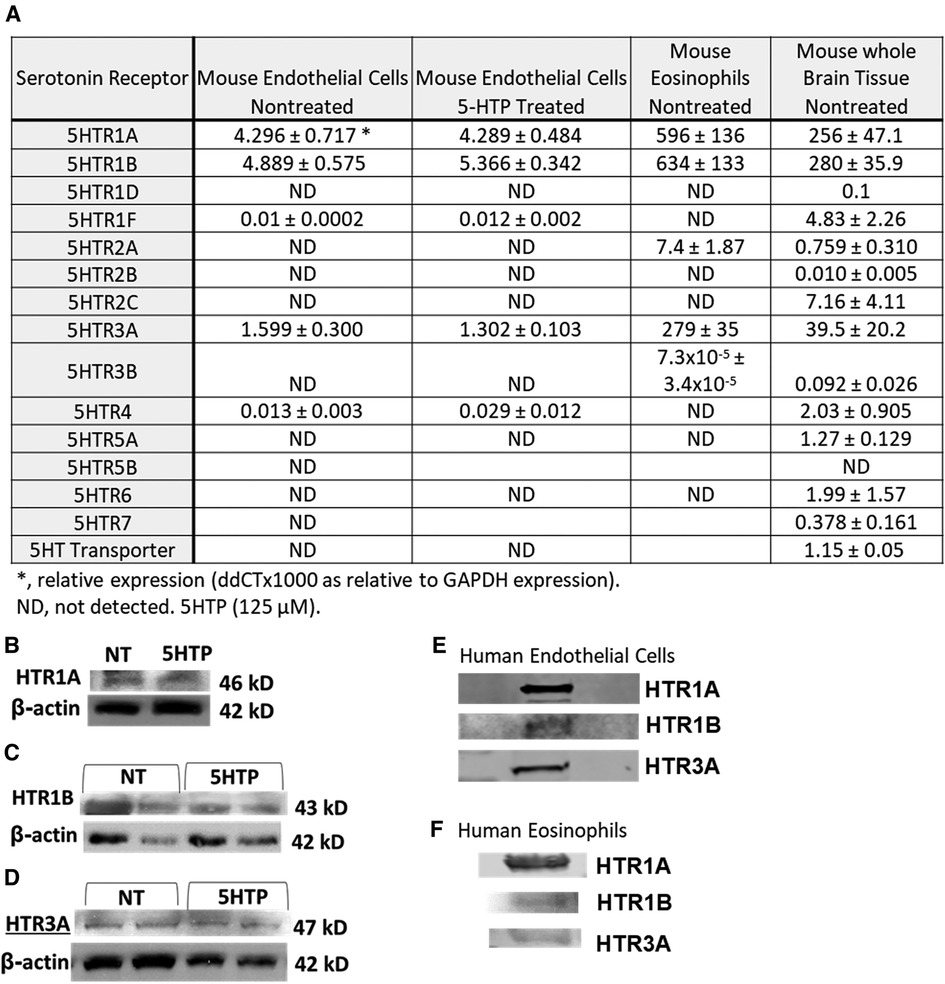
Figure 1. Expression of serotonin receptors in endothelial and eosinophils (mouse and brain control). (A) Mouse endothelial cell line, NJ1638 mouse eosinophils and C57BL/6 mouse brain tissue were analyzed for HTRs by qPCR. (B–D) Mouse endothelial cells with and without 5HTP (125 µM) were examined for HTR1A, HTR1B and HTR3A protein expression was analyzed by western blot. (E) Human microvascular endothelial cell and F) Human peripheral blood eosinophil protein expression of HTRs was analyzed by western blot. Panel (E,F) were not normalized to GAPDH because regulation of expression was not assessed.
Pharmacologic inhibition of HTR1A and HTR1B blocked the 5HTP inhibition of eosinophil transendothelial migration
We have demonstrated that 5HTP is metabolized in endothelial cells to serotonin and that 5HTP overnight pretreatment of endothelial cells in vitro inhibits eosinophil transendothelial migration (18). In contrast, we demonstrated that 5HTP does not alter interaction of eosinophils with the endothelial cell apical surface in vitro (18) and does not alter expression of the adhesion molecules, including VCAM-1, or chemokines that promote eosinophil transendothelial migration (18), suggesting that 5HTP may regulate endothelial function during eosinophil transendothelial migration. In Figure 2, endothelial cells expressed HTRs. Therefore, we determined whether HTR1A, HTR1B and HTR3A regulate 5HTP inhibition of eosinophil transendothelial migration in a parallel plate flow chamber at flow rates in postcapillary venules, major sites of leukocyte recruitment during allergic inflammation. The endothelial cells were treated with highly selective pharmacological inhibitors of these HTRs at doses 10-fold the IC50 to sufficiently block receptors. The endothelial cells were pretreated with the HTR inhibitors for 15 min before addition of 5HTP and during the migration assay because the inhibitor binding to HTRs is reversible. Pharmacological inhibitors included the HTR1A inhibitors NAD-299 (NAD) and WAY-100135 (WAY), the HTR1B inhibitors NAS-181 (NAS) and SB224289 (SB), and the HTR3A inhibitors Dolasetron Mesylate (Dola) and Ondansetron Hydrochloride (Ond). The pharmacologic inhibition of HTR1A, HTR1B but not HTR3A blocked 5HTP inhibition of eosinophil transendothelial migration (Figure 2), indicating that HTR1A and HTR1B in endothelial cells or eosinophils mediated inhibition of eosinophil transendothelial migration.
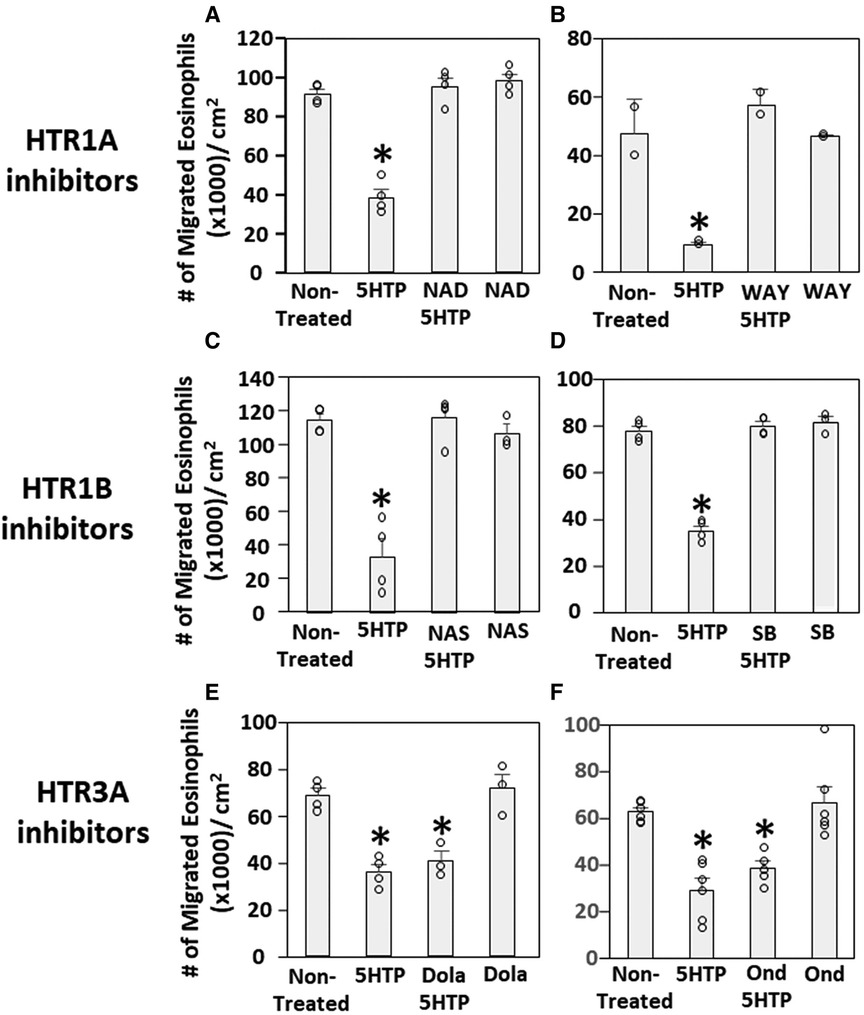
Figure 2. Pharmacologic inhibition of HTR1A and HTR1B blocked the 5HTP inhibition of eosinophil transendothelial migration. The pharmacological inhibitors were added to endothelial cells overnight and were present during the eosinophil transendothelial migration assay because these inhibitors are reversible. (A,B) HTR1A (6 nM NAD-299 and 150 nM WAY-100135) and (C,D) HTR1B (500 nM NAS-181 and 88 nM SB224289) blocked 5HTP (125 µM) inhibition of eosinophil transendothelial migration. In contrast, pharmacological inhibitors of (E,F) HTR3A (75 nM Dolasetron Mesylate and 62 nM Ondansetron Hydrochloride) did not block 5HTP (125 µM) inhibition of eosinophil transendothelial migration.
siRNA knock down of HTR1A and HTR1B expression in endothelial cells but not eosinophils blocked 5HTP inhibition of eosinophil transendothelial migration
Pharmacologic inhibition of HTR1A and HTR1B blocked 5HTP inhibition of eosinophil transendothelial migration in Figure 2. However, because the reversible inhibitors were present during eosinophil transendothelial migration, it is not known whether the HTRs in eosinophils or endothelial cells mediated the 5HTP inhibition of migration. To address HTR1A and HTR1B function in endothelial cells, the expression of these HTRs was knocked down in endothelial cells or in eosinophils by siRNAs. Also, because HTR1A dimerizes and HTR1B dimerizes (140), we determined expression of HTR1A and HTR1B for each siRNA knockdown. The siRNA for HTR1A only reduced the expression of the HTR1A (Figure 3A), but the siRNA for HTR1B reduced expression of both HTR1A and HTR1B (Figure 3B). Importantly, the knockdown of HTR1A or HTR1B in endothelial cells blocked 5HTP inhibition of eosinophil transendothelial migration (Figures 3C,D), suggesting that these endothelial cell HTRs functioned in 5HTP inhibition of transendothelial migration.
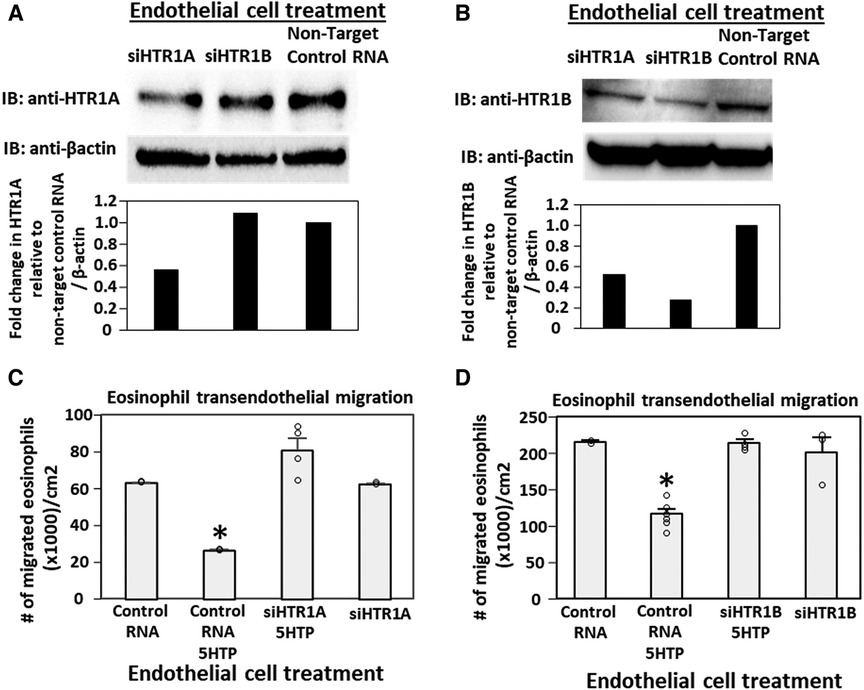
Figure 3. Endothelial cell HTR1A and HTR1B mediate 5HTP inhibition of eosinophil transendothelial migration. (A,B) HTR1A siRNA or HTR1B siRNA in endothelial cells knocked down HTR1A expression in the endothelial cells by western blot (HTR1A, 46kD; HTR1B, 43kD; βactin, 42kD) as compared to non-target control siRNA. Shown are representative blots of 2 experiments. (C,D) siRNA of HTR1A or HTR1B in endothelial cells blocked the 5HTP (125 µM) inhibition of eosinophil transendothelial migration. N = 3 for representative experiment of 2 experiments. *p < 0.05 compared to control siRNA.
To address the function of HTR1A and HTR1B in eosinophils, HTR1A siRNA or HTR1B siRNA was added to 5-day cultures of bone marrow generation of eosinophils because this is the stage of pre-eosinophils when IL5 was added to the culture. On day 8 of the culture, the cells were >85% eosinophils as determined by morphology and eosin staining of cytospins (data not shown). The treatment with siRNA for HTR1A and HTR1B during generation of eosinophils reduced expression of these HTRs in the eosinophils as determined by western blot (Figures 4A,B). Interestingly, siRNA of HTR1A or HTR1B in eosinophils did not block the 5HTP inhibition of eosinophil transendothelial migration (Figures 4C,D), indicating that HTR1A and HTR1B in eosinophils were not required for the 5HTP inhibition of eosinophil transendothelial migration.
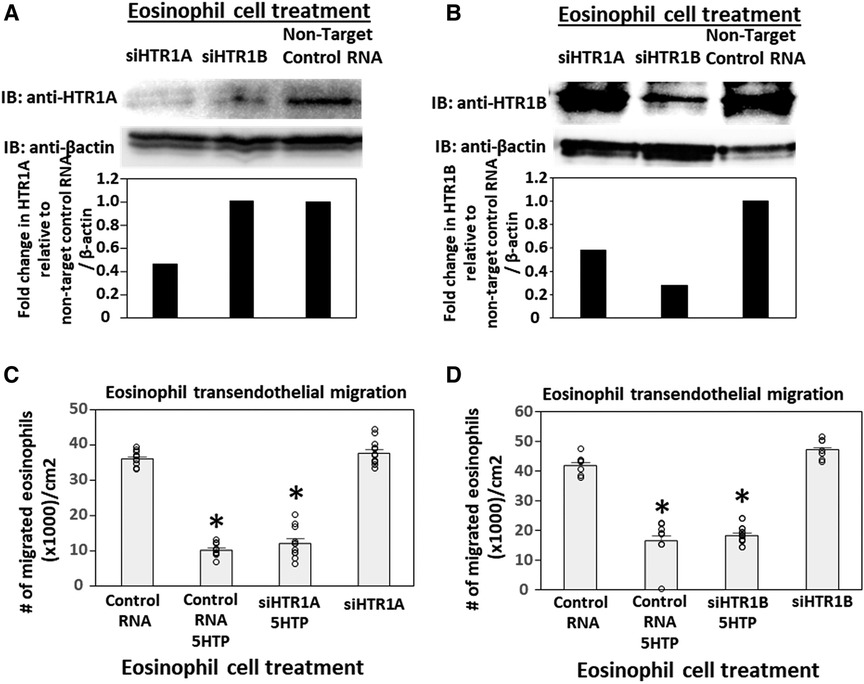
Figure 4. Eosinophil HTR1A and HTR1B do not mediate the inhibitory function of 5HTP on eosinophil transendothelial migration. (A,B) HTR1A siRNA or HTR1B siRNA in eosinophils knocked down HTR expression in the eosinophils by western blot as compared to non-target control siRNA. (C,D) siRNA of HTR1A or HTR1B in eosinophils did not block the 5HTP (125 µM) inhibition of eosinophil transendothelial migration.
HTRs are expressed intracellularly in endothelial cells
HTR1A and HTR1B protein expression by mouse endothelial cells and eosinophils was determined by immunolabeling and confocal microscopy. Unexpectedly, these receptors, which have 7 membrane spanning domains, were not expressed on the cell surface but were expressed intracellularly with punctate localization in endothelial cells consistent with intracellular membranes (Figure 5A). To further demonstrate that the receptors were not expressed on the cell surface or expressed at low undetectable levels on the surface of endothelial cells, we biotinylated the endothelial cell surface, precipitated with streptavidin beads and examined expression of HTRs by western blot. The streptavidin-isolated biotinylated-proteins from the cell surface did not contain HTR1A and HTR1B by western blot, but the proteins from the cell lysate that were not pulled down with the streptavidin beads did contain these HTRs (Figure 5B), suggesting that the HTRs are not on the cell surface.
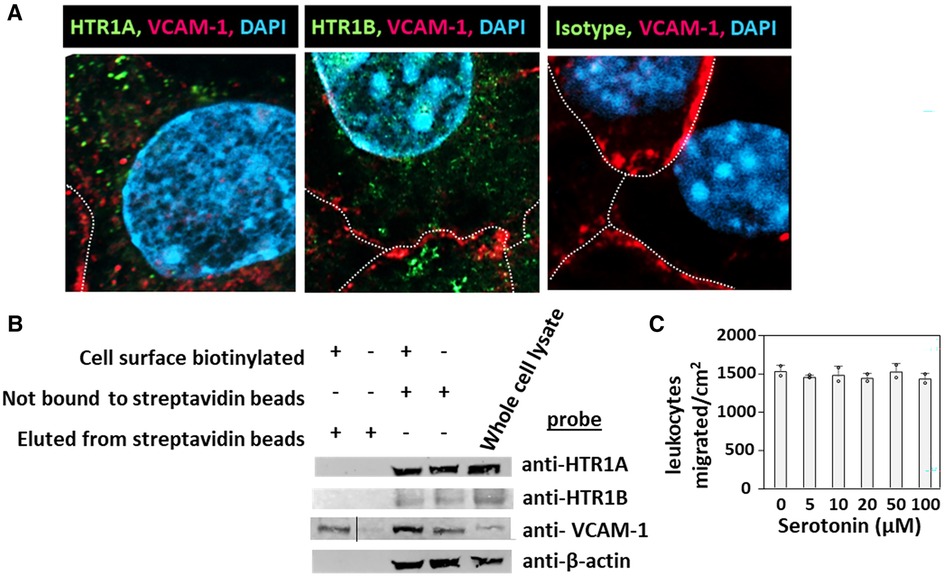
Figure 5. Exogenous serotonin did not block eosinophil transendothelial migration, consistent with intracellular HTR expression in endothelial cells. (A) Optical slices through the center of immunolabeled endothelial cell monolayers. Cells were fixed and immunolabeled with rabbit anti-mouse HTR1A, rabbit anti-mouse HTR1B or isotype control antibodies. Then the cells were washed and immunolabeled with FITC-conjugated goat anti-rabbit IgG. Then the cells were washed and immunolabeled with Alexa 647-conjugated rat anti-mouse VCAM-1 to mark the cell surface. The cells were cover-slipped with DAPI Prolong Gold to label the nuclei and coverslip the slides. The dotted white lines indicate location of cell membrane with VCAM-1 surface expression. (B) Biotinylation of endothelial surface proteins and western blot for HTR1A and HTR1B. (HTR1A, 46kD; HTR1B, 43kD; VCAM-1, 110kD; βactin, 42kD). The vertical line in the anti-VCAM-1 blot indicates removal of an empty lane where no sample was loaded on the gel. (C) Exogenous serotonin treatment overnight and during the transendothelial migration assay did not block leukocyte transendothelial migration.
Because serotonin is transported across membranes by SERT but peripheral venule endothelium, which mediates leukocyte recruitment into inflammatory sites and synthesizes serotonin (102–106), does not express functional SERT (141) and because Figures 5A,B demonstrate intracellular localization of HTR1A and HTR1B, we determined whether exogenous serotonin regulated leukocyte transendothelial migration. We used a dose curve for exogenous addition of serotonin to the leukocyte transendothelial migration assay, because 5HTP, which is metabolized within these endothelial cells to serotonin (18), blocks both eosinophil (Figure 2) (18) and lymphocyte transendothelial migration (18) at 5HTP doses (75–125 µM). These doses are reported to regulate other functions of endothelial cells and other cell types in vitro (142, 143). Also, serotonin levels are increased up to µM levels by platelet activation during inflammation (31) and µM levels induce bronchoconstriction (83, 133, 144). To examine exogenous serotonin regulation of endothelial function during leukocyte migration, endothelial cells were treated overnight and during the transendothelial migration assay with serotonin (5–100 µM). Interestingly, serotonin, which does not cross peripheral venule endothelial cell membranes (141), did not block eosinophil transendothelial migration (Figure 5C), suggesting that metabolism of 5HTP to serotonin within endothelial cells functions within endothelial cells to block eosinophil recruitment.
SNPs in HTRs and TPH associate with lower lung function in asthmatics
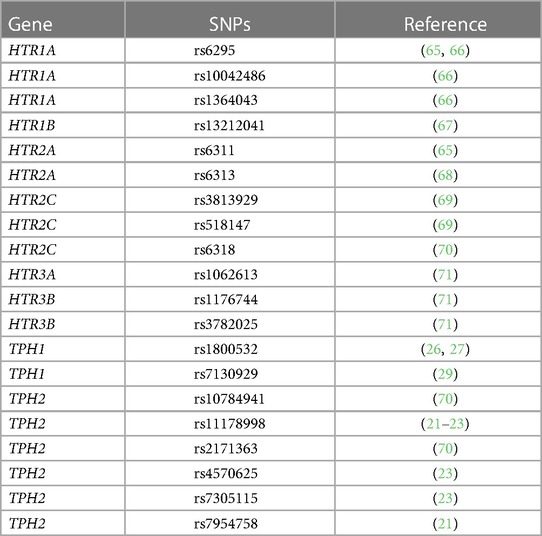
5HTP is metabolized to serotonin in endothelial cells (18), 5HTP functioned through HTRs to regulate eosinophil transendothelial migration in Figures 3–5, 5HTP regulates lung function and allergic inflammation in the lungs in preclinical models (18), and in Table 1B, HTRs and TPHs are located in chromosome locations that are also found in GWAS studies of asthmatics. Moreover, some of these HTRs are adjacent to genes that regulate immune responses (Table 1B). Therefore, to determine whether the findings in the mechanistic preclinical studies on HTRs and lung function may extend to HTR function in asthmatics, we determined whether SNPs in HTRs and TPHs associate with lung function in asthmatics. We chose only SNPs in the HTRs that have functional consequences for HTR signaling or associate with behavior disorders (21–23, 26, 27, 29, 65–71) (Table 2). We chose a test asthma cohort CHIRAH from Chicago (85, 87) and a larger nationwide Latinx cohort, GALA (90), for proof-of-concept of the HTR and TPH genes with SNPs and associations with percent predicted FEV1. A polygenic risk score for subjects was generated using the SNPs in Figures 6C,F for CHIRAH and GALA. In CHIRAH, the polygenic risk score for HTRs and TPHs was significantly associated with lower lung function for adult black subjects (n = 103, 88 males, 15 females, age 31.8 ± 5.8) and adult Latinx subjects (n = 73, 54 male, 19 female, age 31.8 ± 5.4) (Figures 6A,B). These initial analyses in CHIRAH adjusted for duration of asthma in the Latinx subjects, and did not include adjustments other than ancestry in the African American analyses (due to the fact that smoking, inhaled steroid use, insurance status, severity classification, maternal education and household income were not confounders). In the larger cohort of Latinx asthmatic children GALA (n = 2,126, 1,128 female, 998 male, age 13.1 ± 3.5), the polygenic risk scores for HTRs and TPH were also significantly associated with lower lung function (Figures 6D,E). The GALA analyses were corrected for ancestry, smoking, inhaled steroid use, household income, and maternal education as potential confounders of the association. Although in the GALA Latinx dataset only one SNP overlapped with the CHIRAH SNPs and the index only consisted of 3 SNPs in the GALA sample compared to the 7 SNPs in the CHIRAH sample, the SNPs in the GALA cohort are within the same genes of the serotonin pathway as in the CHIRAH cohort. Given that the polygenic risk scores are not identical due to differences in SNPs genotyped due to different genotyping platforms, ethnic specific differences, and age in the cohorts in these associations, this represents a proof of concept and not a true replication of association.
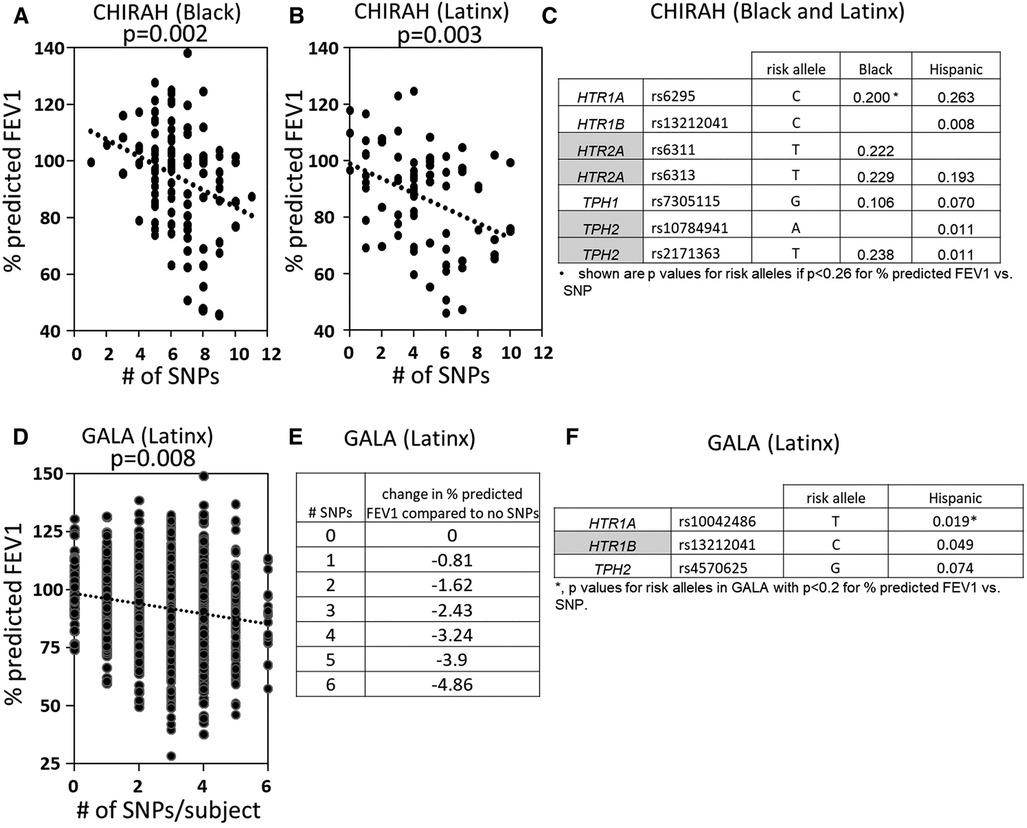
Figure 6. SNPs in HTRs and TPH significantly associated with lower lung function in humans. A polygenic index was calculated as the sum of SNPs with coding of 0, 1, or 2 for alleles with the SNP. SNPs included in the index had a nominal association with FEV1 (p < 0.2). (A) SNP index vs. % predicted FEV1 for CHIRAH black subjects. (B) SNP index vs. % predicted FEV1 for CHIRAH latinx subjects. (C) SNPs in HTRs and TPHs that had p < 0.26 for association with FEV1 for CHIRAH black and latinx subjects. (D) SNP index vs. % predicted FEV1 for GALA latinx subjects. (E) SNP index vs. change in % predicted FEV compared to no SNPs). (F) SNPs in HTRs and TPHs that had p < 0.2 for association of FEV1 for GALA latinx subjects.
Discussion
Endothelial cells have an active function in eosinophil recruitment during allergic inflammation (145, 146). This recruitment of eosinophils in preclinical models is blocked by 5HTP supplementation (1). 5HTP is metabolized to serotonin within endothelial cells (18) by TPH1 (147). Furthermore, we demonstrate that the serotonin receptors HTR1A, HTR1B and HTR3A are expressed by endothelial cells and eosinophils. Inhibitors of HTR3A did not affect eosinophil transendothelial migration. However, the knock-down of expression of HTR1A or HTR1B in endothelial cells blocked 5HTP inhibition of eosinophil transendothelial migration. Also, the pharmacological inhibitors of the HTR1A and HTR1B receptors blocked 5HTP inhibition of eosinophil transendothelial migration. In contrast, knockdown of HTR1A and HTR1B in eosinophils did not modify 5HTP inhibition of eosinophil transendothelial migration, indicating that 5HTP inhibition of eosinophil transendothelial migration is mediated through HTRs in endothelial cells but not eosinophils. We also demonstrated that expression of HTR1A and HTR1B was within the cytoplasmic compartments of endothelial cells. Consistent with intracellular HTR localization in endothelial cells, 5HTP, which is metabolized to serotonin within endothelial cells, blocked eosinophil transendothelial migration but extracellular addition of serotonin, which is not taken up by endothelial cells, did not alter eosinophil transendothelial migration. Together these results indicate that to regulate leukocyte recruitment, 5HTP is metabolized to serontonin within the cytoplasm of endothelial cells and that this stimulates HTRs located within cytoplasmic compartments of endothelial cells, whereas HTR1A and HTR1B in eosinophils do not mediate 5HTP inhibition of eosinophil transendothelial migration. Furthermore, TPHs and HTRs are in chromosome locations that are near GWAS locations for allergic inflammation and asthma. In studies to extend HTR analyses to humans with asthma, we generated a polygenic index with SNPs that regulate TPH or HTR function or have been reported to associate with behavior disorders that associated with lower lung function. In humans with asthma, lung function is lower in asthmatics when the subjects are polygenic for SNPs in HTRs that alter HTR function or are associated with behavior disorders. This finding would suggest that TPHs and HTRs may serve as one possible link between asthma phenotypes and behavior disorders.
Allergic asthma occurs in about 80% of children with asthma and about 60% of asthma overall (3–5) and those with asthma have an increased prevalence of anxiety or depression (6–11). GWAS analyses of allergy and asthma have identified multiple chromosomal locations that are consistently associated with allergy/asthma (32–34, 36–38, 41, 42, 48, 49, 52, 55–57, 65, 66, 68–71, 110–132). These loci in Table 1B are associated with lung function and mediators of allergic inflammation, including IgE, eosinophils, IL4 and IL13. Interestingly, 10 of the genes for HTRs and TPHs (63, 107–109) are located near the dozen chromosome locations consistently associated with allergy/asthma (32–34, 36–38, 41, 42, 48, 49, 52, 55–57, 65, 66, 68–71, 110–132). This raises the question of whether the 5HTP/serotonin pathway is an important pathway in allergic asthma, and whether the perturbations of this pathway in subjects with allergic asthma may be a potential link explaining the increased rates of anxiety and depression observed in subjects with allergic asthma.
Anxiety/depression is often treated, with selective serotonin reuptake inhibitors (SSRIs) which are pharmacological inhibitors to block neuronal SERT transporter-mediated reuptake/removal of serotonin. It is reported that SSRIs inhibit eotaxin-induced eosinophil transendothelial migration through human dermal or lung microvascular endothelial cells in vitro (148), but the mechanisms are not known, especially since microvascular endothelial cells do not express a functional SERT (141). In a study of adult participants with both asthma and major depressive disorder, an SSRI had minimal effect on asthma control questionnaire scores and, for a subset of severe asthmatics, significantly lowered percent predicted FEV1 (149). Whether this is related to SSRIs having vasoconstriction side effects (75, 150) is not known. Also, in a mouse model of allergic asthma, serotonin derived from mast cells increases airway hyperresponsiveness (151). In contrast, 5HTP reduces airway hyperresponsiveness in several mouse models of allergic asthma (18), is not vasoconstricting (79–82, 152, 153), and still clinically effective for behavior disorders (154–164), but whether 5HTP can affect asthma is not known. Clinical studies with inhibitors of individual HTRs such as HTR1A or HTR2A have had only a small benefit for asthma (83), but this may reflect involvement of multiple HTRs during asthma. Because endothelial HTRs regulate inflammation which impacts lung function and because HTRs on smooth muscle cells regulate airway constriction and lung function, SNPs in HTRs and TPHs that are expressed in multiple cell types in the lung may impact lung function in asthmatics. We demonstrated that increased numbers of SNPs in HTRs and TPHs, which affect protein function or associate with behavior disorders, associated with lower FEV1 in asthmatic children and adults.
It is also reported that dietary supplementation with 5HTP blocks allergic inflammation in four mouse models, whereby the 5HTP was administered before or after sensitization for 2 different allergens or blocks eosinophil migration into the lungs after intratracheal IL4 administration (18). In vitro pretreatment of endothelial cells with 5HTP blocks eosinophil transendothelial migration (18); also 5HTP is taken up by endothelial cells and metabolized to serotonin within endothelial cells (18), but it was not reported whether endothelial cell generation of serotonin stimulates endothelial cell HTRs. We demonstrate that eosinophils and endothelial cells express intracellular HTR1A and HTR1B and that pharmacological inhibitors of these receptors block eosinophil transendothelial migration in vitro. We have previously reported that this eosinophil migration on the endothelial cells used in our studies is dependent on binding to VCAM-1, which reflects the VCAM-1 dependent recruitment of eosinophils into the lung in vivo (18, 145). The 5HTP inhibition of eosinophil transendothelial migration was through HTRs in endothelial cells but not eosinophils, because knockdown of HTR1A or HTR1B in endothelial cells blocked 5HTP inhibition of eosinophil transendothelial migration but knockdown of HTR1A or HTR1B in eosinophils did not block 5HTP inhibition of eosinophil transendothelial migration. Knockdown of either HTR1A or HTR1B in endothelial cells blocked the effects of 5HTP, which likely reflect that these HTRs can dimerize (140).
Others have reported that human eosinophils express HTR1A, HTR1B and HTR2A by RT-PCR (139). It is reported that treatment of human or mouse eosinophils in vitro with exogenous serotonin or an HTR2A agonist increases eosinophil chemotaxis and increases eosinophil rolling on recombinant VCAM-1-coated plastic surfaces (16, 139). Another report indicates that in vivo infusion of serotonin in mice, enhances eosinophil interaction with endothelial surfaces with increases in numbers of eosinophils rolling on the endothelial surface, decreases in eosinophil rolling velocity and increases in number of eosinophils adhered to the endothelium (16, 139), but transendothelial migration was not reported. We showed that exogenous serotonin did not alter eosinophil transendothelial migration, which is consistent with lack of functional SERT by venule endothelium (141) and our demonstration in this current report that endothelial HTRs are intracellular. It is reported that pharmacological inhibition of HTR2A in mice reduces eosinophil recruitment to lungs of mice challenged with the allergen chicken egg ovalbumin (OVA), but it was not shown whether plasma or tissue serotonin levels were increased in the mice. It another report, mice challenged with inhalation of OVA or the allergen house dust mite did not change serotonin levels in the mouse plasma, lungs, intestine or brain (18). In studies with 5HTP, supplementation of 5HTP in vivo does not increase plasma serotonin concentrations but decreases eosinophil recruitment into the lung during allergic inflammation (18); in vitro 5HTP is metabolized within endothelial cells to serotonin and decreases eosinophil transendothelial migration but does not alter chemokine-induced leukocyte chemotaxis or chemokinesis (18). We demonstrated that 5HTP inhibition of eosinophil transendothelial migration requires intracellular HTRs in endothelial cells and that in humans, SNPs in these receptors associate with lower FEV1 in asthmatic adults and children.
In conclusion, we have demonstrated on a cellular and molecular level that HTRs are expressed intracellularly in endothelial cells and that these receptors mediate 5HTP inhibition of eosinophil transendothelial migration (Graphical Abstract). Moreover, in asthmatic adults and children, a polygenic index of SNPs in TPHs and HTRs correlated with lower FEV1. A limitation is that the analyses in the Latinx subjects in CHIRAH was not corrected for ancestry due to limited genotyping being carried out in these subjects. Notably, similar polygenic associations were noted in the analyses of African American subjects in CHIRAH and the analyses in GALA II which were corrected for ancestry. Furthermore, the two human cohorts are not replication cohorts for the polygenic index because of differences in the population demographics and historical differences in genotyping platforms used. Nevertheless, both cohorts indicate that increasing the number of SNPs in the serotonin pathway associate with lower lung function within subjects with asthma. These studies form a framework for future studies to assess effectiveness of 5HTP for asthmatic subjects, especially since 5HTP has minimal side effects in other human studies (162), is not vasoconstricting (79–82, 152, 153), and is clinically effective in studies of anxiety and depression (154–160).
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/gap/, phs001274.v2.p1.
Ethics statement
The studies involving humans were approved by Institutional review board of Northwestern University and the Institutional Review Board of University of California San Francisco. The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent for participation in this study was provided by the participants’ legal guardians/next of kin. The animal study was approved by Indiana University Institutional Review Committee. The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
MW: Data curation, Investigation, Methodology, Writing – original draft, Writing – review & editing. JB: Data curation, Investigation, Writing – original draft, Writing – review & editing. TK: Data curation, Investigation, Methodology, Writing – review & editing. SM: Data curation, Investigation, Writing – review & editing. JG: Data curation, Investigation, Writing – review & editing. RF: Data curation, Investigation, Writing – review & editing. JC: Data curation, Investigation, Writing – review & editing. SA: Data curation, Investigation, Writing – review & editing. KB: Writing – review & editing. LG: Data curation, Writing – review & editing. CE: Writing – review & editing. PA: Writing – review & editing. HF: Writing – review & editing. WR-C: Writing – review & editing. JR-S: Writing – review & editing. DS: Writing – review & editing. ST: Writing – review & editing. MS: Writing – review & editing. EB: Writing – review & editing. RK: Data curation, Formal Analysis, Methodology, Writing – original draft, Writing – review & editing. JC-M: Conceptualization, Investigation, Methodology, Supervision, Writing – original draft, Writing – review & editing.
Funding
The authors declare financial support was received for the research, authorship, and/or publication of this article.
These studies were supported by National Institutes of Health Grant R01 AT004837 (J.M.C-M) and by American Heart Association 0855583G (J.M.C-M). CHIRAH was supported by National Institutes of Health Grant 1UO1 HL072496 and The Ernest S Bazley Grant to Northwestern Memorial Hospital and Northwestern University. GALA was supported in part by the National Institutes of Health grants (ES015794, AI077439, HL088133, HL078885, CA113710, AI079139, HL004464, HL104608, AI061774, HL079055, DK064695, and M01-RR00188; HHSN26120080001E; and MD006902); the Flight Attendant Medical Research Institute (FAMRI); an RWJF Amos Medical Faculty Development Award (to E.G.B.); the Sandler Foundation; the American Asthma Foundation (to E.G.B); In addition, the research reported in this publication was also supported by the National Institute on Minority Health and Health Disparities of the National Institutes of Health under award no. P60MD006902.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abbreviations
5HTP, 5-hydroxytryptophan; CHIRAH, Chicago initiative to raise asthma health equity study; Dola, HTR3A inhibitor dolasetron mesylate; GALA, genes-environments and admixture in latino Americans cohort; GWAS, genome-wide association studies; HMVEC-Ls, human microvascular endothelial cells from the lung; HTR, 5-hydroxytryptomine receptor or serotonin receptor; NAD, HTR1A inhibitor NAD-299; NAS, HTR1B inhibitor NAS-181; Ond, HTR3A inhibitor ondansetron hydrochloride; SB, HTR1B inhibitors SB224289; siRNA, short interfering RNA; SNPs, single nucleotide polymorphism; TG2, transglutaminase 2; TPH, tryptophan hydroxylase; WAY, HTR1A inhibitor WAY-100135.
References
1. Prevention CfDCa. 2019 national health interview survey data. US department of health & human services (2020). Available online at: https://www.aafa.org/asthma-facts/ https://www.cdc.gov/asthma/nhis/2019/data.htm
2. Wenzel SE. Asthma phenotypes: the evolution from clinical to molecular approaches. Nat Med. (2012) 18(5):716–25. doi: 10.1038/nm.2678
3. Ross MK, Romero T, Sim MS, Szilagyi PG. Obese- and allergic-related asthma phenotypes among children across the United States. J Asthma. (2019) 56(5):512–21. doi: 10.1080/02770903.2018.1466317
4. Pakkasela J, Ilmarinen P, Honkamäki J, Tuomisto LE, Andersén H, Piirilä P, et al. Age-specific incidence of allergic and non-allergic asthma. BMC Pulm Med. (2020) 20(1):9. doi: 10.1186/s12890-019-1040-2
5. Akar-Ghibril N, Casale T, Custovic A, Phipatanakul W. Allergic endotypes and phenotypes of asthma. J Allergy Clin Immunol Pract. (2020) 8(2):429–40. doi: 10.1016/j.jaip.2019.11.008
6. Vila G, Nollet-Clemencon C, de Blic J, Mouren-Simeoni MC, Scheinmann P. Asthma severity and psychopathology in a tertiary care department for children and adolescent. Eur Child Adolesc Psychiatry. (1998) 7(3):137–44. doi: 10.1007/s007870050059
7. Strine TW, Mokdad AH, Balluz LS, Gonzalez O, Crider R, Berry JT, et al. Depression and anxiety in the United States: findings from the 2006 behavioral risk factor surveillance system. Psychiatr Serv. (2008) 59(12):1383–90. doi: 10.1176/ps.2008.59.12.1383
8. Cheung TK, Lam B, Lam KF, Ip M, Ng C, Kung R, et al. Gastroesophageal reflux disease is associated with poor asthma control, quality of life, and psychological status in Chinese asthma patients. Chest. (2009) 135(5):1181–5. doi: 10.1378/chest.08-1702
9. Sansone RA, Sansone LA. Asthma: wheezing, woes, and worries. Psychiatry. (2008) 5(10):48–52. 2695759.19724724
10. Di Marco F, Verga M, Santus P, Giovannelli F, Busatto P, Neri M, et al. Close correlation between anxiety, depression, and asthma control. Respir Med. (2010) 104(1):22–8. doi: 10.1016/j.rmed.2009.08.005
11. Cordina M, Fenech AG, Vassallo J, Cacciottolo JM. Anxiety and the management of asthma in an adult outpatient population. Ther Adv Respir Dis. (2009) 3(5):227–33. doi: 10.1177/1753465809347038
12. Felger JC, Lotrich FE. Inflammatory cytokines in depression: neurobiological mechanisms and therapeutic implications. Neuroscience. (2013) 246:199–229. doi: 10.1016/j.neuroscience.2013.04.060
13. Rawdin BJ, Mellon SH, Dhabhar FS, Epel ES, Puterman E, Su Y, et al. Dysregulated relationship of inflammation and oxidative stress in major depression. Brain Behav Immun. (2013) 31:143–52. doi: 10.1016/j.bbi.2012.11.011
14. Miller GE, Rohleder N, Stetler C, Kirschbaum C. Clinical depression and regulation of the inflammatory response during acute stress. Psychosom Med. (2005) 67(5):679–87. doi: 10.1097/01.psy.0000174172.82428.ce
15. De Bie JJ, Henricks PA, Cruikshank WW, Hofman G, Jonker EH, Nijkamp FP, et al. Modulation of airway hyperresponsiveness and eosinophilia by selective histamine and 5-HT receptor antagonists in a mouse model of allergic asthma. Br J Pharmacol. (1998) 124(5):857–64. doi: 10.1038/sj.bjp.0701901
16. Boehme SA, Lio FM, Sikora L, Pandit TS, Lavrador K, Rao SP, et al. Cutting edge: serotonin is a chemotactic factor for eosinophils and functions additively with eotaxin. J Immunol. (2004) 173(6):3599–603. doi: 10.4049/jimmunol.173.6.3599
17. Das AM, Flower RJ, Perretti M. Eotaxin-induced eosinophil migration in the peritoneal cavity of ovalbumin-sensitized mice: mechanism of action. J Immunol. (1997) 159:1466–73. doi: 10.4049/jimmunol.159.3.1466
18. Abdala-Valencia H, Berdnikovs S, McCary CA, Urick D, Mahadevia R, Marchese ME, et al. Inhibition of allergic inflammation by supplementation with 5-hydroxytryptophan. Am J Physiol Lung Cell Mol Physiol. (2012) 303:L642–60. doi: 10.1152/ajplung.00406.2011
19. León-Ponte M, Ahern GP, O’Connell PJ. Serotonin provides an accessory signal to enhance T-cell activation by signaling through the 5-HT7 receptor. Blood. (2007) 109(8):3139–46. doi: 10.1182/blood-2006-10-052787
20. Tzvetkov MV, Brockmoller J, Roots I, Kirchheiner J. Common genetic variations in human brain-specific tryptophan hydroxylase-2 and response to antidepressant treatment. Pharmacogenet Genomics. (2008) 18(6):495–506. doi: 10.1097/FPC.0b013e3282fb02cb
21. Grigoroiu-Serbanescu M, Diaconu CC, Herms S, Bleotu C, Vollmer J, Muhleisen TW, et al. Investigation of the tryptophan hydroxylase 2 gene in bipolar I disorder in the Romanian population. Psychiatr Genet. (2008) 18(5):240–7. doi: 10.1097/YPG.0b013e3283053045
22. Chen GL, Vallender EJ, Miller GM. Functional characterization of the human TPH2 5′ regulatory region: untranslated region and polymorphisms modulate gene expression in vitro. Hum Genet. (2008) 122(6):645–57. doi: 10.1007/s00439-007-0443-y
23. Lin YM, Chao SC, Chen TM, Lai TJ, Chen JS, Sun HS. Association of functional polymorphisms of the human tryptophan hydroxylase 2 gene with risk for bipolar disorder in Han Chinese. Arch Gen Psychiatry. (2007) 64(9):1015–24. doi: 10.1001/archpsyc.64.9.1015
24. Hennig J, Reuter M, Netter P, Burk C, Landt O. Two types of aggression are differentially related to serotonergic activity and the A779C TPH polymorphism. Behav Neurosci. (2005) 119(1):16–25. doi: 10.1037/0735-7044.119.1.16
25. Manuck SB, Flory JD, Ferrell RE, Dent KM, Mann JJ, Muldoon MF. Aggression and anger-related traits associated with a polymorphism of the tryptophan hydroxylase gene. Biol Psychiatry. (1999) 45(5):603–14. doi: 10.1016/S0006-3223(98)00375-8
26. Jernej B, Stefulj J, Hranilovic D, Balija M, Skavic J, Kubat M. Intronic polymorphism of tryptophan hydroxylase and serotonin transporter: indication for combined effect in predisposition to suicide. J Neural Transm. (2004) 111(6):733–8. doi: 10.1007/s00702-003-0114-7
27. Rujescu D, Giegling I, Bondy B, Gietl A, Zill P, Moller HJ. Association of anger-related traits with SNPs in the TPH gene. Mol Psychiatry. (2002) 7(9):1023–9. doi: 10.1038/sj.mp.4001128
28. Serretti A, Artioli P. From molecular biology to pharmacogenetics: a review of the literature on antidepressant treatment and suggestions of possible candidate genes. Psychopharmacology. (2004) 174(4):490–503. doi: 10.1007/s00213-004-1822-x
29. Rotondo A, Schuebel K, Bergen A, Aragon R, Virkkunen M, Linnoila M, et al. Identification of four variants in the tryptophan hydroxylase promoter and association to behavior. Mol Psychiatry. (1999) 4(4):360–8. doi: 10.1038/sj.mp.4000578
30. Melnikov M, Sviridova A, Rogovskii V, Oleskin A, Boziki M, Bakirtzis C, et al. Serotoninergic system targeting in multiple sclerosis: the prospective for pathogenetic therapy. Mult Scler Relat Disord. (2021) 51:102888. doi: 10.1016/j.msard.2021.102888
31. Herr N, Bode C, Duerschmied D. The effects of serotonin in immune cells. Front Cardiovasc Med. (2017) 4:48. doi: 10.3389/fcvm.2017.00048
32. Altmuller J, Seidel C, Lee YA, Loesgen S, Bulle D, Friedrichs F, et al. Phenotypic and genetic heterogeneity in a genome-wide linkage study of asthma families. BMC Pulm Med. (2005) 5:1. doi: 10.1186/1471-2466-5-1
33. Ober C, Moffatt MF. Contributing factors to the pathobiology: the genetics of asthma. Clin Chest Med. (2000) 21(2):245–61. doi: 10.1016/S0272-5231(05)70264-1
34. Postma DS, Meyers DA, Jongepier H, Howard TD, Koppelman GH, Bleecker ER. Genomewide screen for pulmonary function in 200 families ascertained for asthma. Am J Respir Crit Care Med. (2005) 172(4):446–52. doi: 10.1164/rccm.200407-864OC
35. Herrera-Luis E, Li A, Mak ACY, Perez-Garcia J, Elhawary JR, Oh SS, et al. Epigenome-wide association study of lung function in latino children and youth with asthma. Clin Epigenetics. (2022) 14(1):9. doi: 10.1186/s13148-022-01227-5
36. Immervoll T, Loesgen S, Dutsch G, Gohlke H, Herbon N, Klugbauer S, et al. Fine mapping and single nucleotide polymorphism association results of candidate genes for asthma and related phenotypes. Hum Mutat. (2001) 18(4):327–36. doi: 10.1002/humu.1194
37. Malerba G, Lauciello MC, Scherpbier T, Trabetti E, Galavotti R, Cusin V, et al. Linkage analysis of chromosome 12 markers in Italian families with atopic asthmatic children. Am J Respir Crit Care Med. (2000) 162(4 Pt 1):1587–90. doi: 10.1164/ajrccm.162.4.9909031
38. Lonjou C, Collins A, Ennis S, Tapper W, Morton NE. Meta-analysis and retrospective collaboration: two methods to map oligogenes for atopy and asthma. Clin Exp Allergy. (1999) 29(Suppl 4):57–9. 10641569.10641569
39. Schoettler N, Rodríguez E, Weidinger S, Ober C. Advances in asthma and allergic disease genetics: is bigger always better? J Allergy Clin Immunol. (2019) 144(6):1495–506. doi: 10.1016/j.jaci.2019.10.023
40. Lee M, Huan T, McCartney DL, Chittoor G, de Vries M, Lahousse L, et al. Pulmonary function and blood DNA methylation: a multiancestry epigenome-wide association meta-analysis. Am J Respir Crit Care Med. (2022) 206(3):321–36. doi: 10.1164/rccm.202108-1907OC
41. Denham S, Koppelman GH, Blakey J, Wjst M, Ferreira MA, Hall IP, et al. Meta-analysis of genome-wide linkage studies of asthma and related traits. Respir Res. (2008) 9:38. doi: 10.1186/1465-9921-9-38
42. Bouzigon E, Ulgen A, Dizier MH, Siroux V, Lathrop M, Kauffmann F, et al. Evidence for a pleiotropic QTL on chromosome 5q13 influencing both time to asthma onset and asthma score in French EGEA families. Hum Genet. (2007) 121(6):711–9. doi: 10.1007/s00439-007-0363-x
43. García-Sánchez A, Isidoro-García M, García-Solaesa V, Sanz C, Hernández-Hernández L, Padrón-Morales J, et al. Genome-wide association studies (GWAS) and their importance in asthma. Allergol Immunopathol (Madr). (2015) 43(6):601–8. doi: 10.1016/j.aller.2014.07.004
44. Sordillo JE, Lutz SM, Jorgenson E, Iribarren C, McGeachie M, Dahlin A, et al. A polygenic risk score for asthma in a large racially diverse population. Clin Exp Allergy. (2021) 51(11):1410–20. doi: 10.1111/cea.14007
45. Zayed H. Novel comprehensive bioinformatics approaches to determine the molecular genetic susceptibility profile of moderate and severe asthma. Int J Mol Sci. (2020) 21(11):4022. doi: 10.3390/ijms21114022
46. Chen W, Wang T, Pino-Yanes M, Forno E, Liang L, Yan Q, et al. An epigenome-wide association study of total serum IgE in hispanic children. J Allergy Clin Immunol. (2017) 140(2):571–7. doi: 10.1016/j.jaci.2016.11.030
47. Herrera-Luis E, Mak ACY, Perez-Garcia J, Martin-Gonzalez E, Eng C, Beckman KB, et al. Admixture mapping of severe asthma exacerbations in hispanic/latino children and youth. Thorax. (2023) 78(3):233–41. doi: 10.1136/thorax-2022-218755
48. Bhattacharyya S, Leaves NI, Wiltshire S, Cox R, Cookson WO. A high-density genetic map of the chromosome 13q14 atopy locus. Genomics. (2000) 70(3):286–91. doi: 10.1006/geno.2000.6398
49. Koppelman GH, Stine OC, Xu J, Howard TD, Zheng SL, Kauffman HF, et al. Genome-wide search for atopy susceptibility genes in Dutch families with asthma. J Allergy Clin Immunol. (2002) 109(3):498–506. doi: 10.1067/mai.2002.122235
50. Holt RJ, Vandiedonck C, Willis-Owen SA, Knight JC, Cookson WO, Moffatt MF, et al. A functional AT/G polymorphism in the 5′-untranslated region of SETDB2 in the IgE locus on human chromosome 13q14. Genes Immun. (2017) 18(1):57. doi: 10.1038/gene.2016.22
51. González-Castro TB, Hernandez-Diaz Y, Juárez-Rojop IE, López-Narváez L, Tovilla-Zárate CA, Rodriguez-Perez JM, et al. The role of the Cys23Ser (rs6318) polymorphism of the HTR2C gene in suicidal behavior: systematic review and meta-analysis. Psychiatr Genet. (2017) 27(6):199–209. doi: 10.1097/YPG.0000000000000184
52. Blumenthal MN, Langefeld CD, Beaty TH, Bleecker ER, Ober C, Lester L, et al. A genome-wide search for allergic response (atopy) genes in three ethnic groups: collaborative study on the genetics of asthma. Hum Genet. (2004) 114(2):157–64. doi: 10.1007/s00439-003-1030-5
53. Do AR, An J, Jo J, Kim WJ, Kang HY, Lee S, et al. A genome-wide association study implicates the pleiotropic effect of NMUR2 on asthma and COPD. Sci Rep. (2022) 12(1):22073. doi: 10.1038/s41598-022-24766-6
54. Wjst M. Exome variants associated with asthma and allergy. Sci Rep. (2022) 12(1):21028. doi: 10.1038/s41598-022-24960-6
55. Ryu HJ, Jung HY, Park JS, Ryu GM, Heo JY, Kim JJ, et al. Gene-based single nucleotide polymorphisms and linkage disequilibrium patterns of 29 asthma candidate genes in the chromosome 5q31–33 region in Koreans. Int Arch Allergy Immunol. (2006) 139(3):209–16. doi: 10.1159/000091166
56. Zervou MI, Tzortzaki EG, Makris D, Gaga M, Zervas E, Economidou E, et al. Differences in microsatellite DNA level between asthma and chronic obstructive pulmonary disease. Eur Respir J. (2006) 28(3):472–8. doi: 10.1183/09031936.06.00127305
57. Noguchi E, Yokouchi Y, Zhang J, Shibuya K, Shibuya A, Bannai M, et al. Positional identification of an asthma susceptibility gene on human chromosome 5q33. Am J Respir Crit Care Med. (2005) 172(2):183–8. doi: 10.1164/rccm.200409-1223OC
58. Ntontsi P, Photiades A, Zervas E, Xanthou G, Samitas K. Genetics and epigenetics in asthma. Int J Mol Sci. (2021) 22(5):2412. doi: 10.3390/ijms22052412
59. Murk W, DeWan AT. Genome-wide search identifies a gene-gene interaction between 20p13 and 2q14 in asthma. BMC Genet. (2016) 17(1):102. doi: 10.1186/s12863-016-0376-3
60. Herrera-Luis E, Ortega VE, Ampleford EJ, Sio YY, Granell R, de Roos E, et al. Multi-ancestry genome-wide association study of asthma exacerbations. Pediatr Allergy Immunol. (2022) 33(6):e13802. doi: 10.1111/pai.13802
61. Adayev T, Ranasinghe B, Banerjee P. Transmembrane signaling in the brain by serotonin, a key regulator of physiology and emotion. Biosci Rep. (2005) 25(5-6):363–85. doi: 10.1007/s10540-005-2896-3
62. Ozdemir E. The pathophysiological role of serotonin receptor systems in opioid analgesia and tolerance. Int J Basic Clin Pharmacol. (2017) 6(2):217–28. doi: 10.18203/2319-2003.ijbcp20170312
63. Brandt EB, Strait RT, Hershko D, Wang Q, Muntel EE, Scribner TA, et al. Mast cells are required for experimental oral allergen-induced diarrhea. J Clin Invest. (2003) 112(11):1666–77. doi: 10.1172/JCI19785
64. Sanger GJ, Banner SE, Smith MI, Wardle KA. SB-207266: 5-HT4 receptor antagonism in human isolated gut and prevention of 5-HT-evoked sensitization of peristalsis and increased defaecation in animal models. Neurogastroenterol Motil. (1998) 10(4):271–9. doi: 10.1046/j.1365-2982.1998.00106.x
65. Lenze EJ, Shardell M, Ferrell RE, Orwig D, Yu-Yahiro J, Hawkes W, et al. Association of serotonin-1A and 2A receptor promoter polymorphisms with depressive symptoms and functional recovery in elderly persons after hip fracture. J Affect Disord. (2008) 111(1):61–6. doi: 10.1016/j.jad.2008.02.005
66. Kato M, Fukuda T, Wakeno M, Okugawa G, Takekita Y, Watanabe S, et al. Effect of 5-HT1A gene polymorphisms on antidepressant response in major depressive disorder. Am J Med Genet B Neuropsychiatr Genet. (2009) 150B(1):115–23. doi: 10.1002/ajmg.b.30783
67. Jensen KP, Covault J, Conner TS, Tennen H, Kranzler HR, Furneaux HM. A common polymorphism in serotonin receptor 1B mRNA moderates regulation by miR-96 and associates with aggressive human behaviors. Mol Psychiatry. (2009) 14(4):381–9. doi: 10.1038/mp.2008.15
68. Levitan RD, Masellis M, Basile VS, Lam RW, Jain U, Kaplan AS, et al. Polymorphism of the serotonin-2A receptor gene (HTR2A) associated with childhood attention deficit hyperactivity disorder (ADHD) in adult women with seasonal affective disorder. J Affect Disord. (2002) 71(1-3):229–33. doi: 10.1016/S0165-0327(01)00372-X
69. Li J, Wang Y, Zhou R, Zhang H, Yang L, Wang B, et al. Association between polymorphisms in serotonin 2C receptor gene and attention-deficit/hyperactivity disorder in han Chinese subjects. Neurosci Lett. (2006) 407(2):107–11. doi: 10.1016/j.neulet.2006.08.022
70. Ni X, Chan D, Chan K, McMain S, Kennedy JL. Serotonin genes and gene-gene interactions in borderline personality disorder in a matched case-control study. Prog Neuropsychopharmacol Biol Psychiatry. (2009) 33(1):128–33. doi: 10.1016/j.pnpbp.2008.10.022
71. Yamada K, Hattori E, Iwayama Y, Ohnishi T, Ohba H, Toyota T, et al. Distinguishable haplotype blocks in the HTR3A and HTR3B region in the Japanese reveal evidence of association of HTR3B with female major depression. Biol Psychiatry. (2006) 60(2):192–201. doi: 10.1016/j.biopsych.2005.11.008
72. Pype JL, Verleden GM, Demedts MG. 5-HT modulates noncholinergic contraction in Guinea pig airways in vitro by prejunctional 5-HT1-like receptor. J Appl Physiol. (1994) 77(3):1135–41. doi: 10.1152/jappl.1994.77.3.1135
73. Nagase T, Fukuchi Y, Dallaire MJ, Martin JG, Ludwig MS. In vitro airway and tissue response to antigen in sensitized rats. Role of serotonin and leukotriene D4. Am J Respir Crit Care Med. (1995) 152(1):81–6. doi: 10.1164/ajrccm.152.1.7599867
74. Wohlsen A, Uhlig S, Martin C. Immediate allergic response in small airways. Am J Respir Crit Care Med. (2001) 163(6):1462–9. doi: 10.1164/ajrccm.163.6.2007138
75. Delaney C, Gien J, Grover TR, Roe G, Abman SH. Pulmonary vascular effects of serotonin and selective serotonin reuptake inhibitors in the late-gestation ovine fetus. Am J Physiol Lung Cell Mol Physiol. (2011) 301(6):L937–44. doi: 10.1152/ajplung.00198.2011
76. Wanstall JC, Fiore SA, Gambino A, Chess-Williams R. Potentiation of 5-hydroxytryptamine (5-HT) responses by a 5-HT uptake inhibitor in pulmonary and systemic vessels: effects of exposing rats to hypoxia. Naunyn Schmiedebergs Arch Pharmacol. (2003) 368(6):520–7. doi: 10.1007/s00210-003-0823-3
77. Gruetter CA, Lemke SM, Anestis DK, Szarek JL, Valentovic MA. Potentiation of 5-hydroxytryptamine-induced contraction in rat aorta by chlorpheniramine, citalopram and fluoxetine. Eur J Pharmacol. (1992) 217(2-3):109–18. doi: 10.1016/0014-2999(92)90827-Q
78. Veenstra-VanderWeele J, Anderson GM, Cook EH Jr. Pharmacogenetics and the serotonin system: initial studies and future directions. Eur J Pharmacol. (2000) 410(2-3):165–81. doi: 10.1016/S0014-2999(00)00814-1
79. Itskovitz HD, Werber JL, Sheridan AM, Brewer TF, Stier CT Jr. 5-hydroxytryptophan and carbidopa in spontaneously hypertensive rats. J Hypertens. (1989) 7(4):311–5. doi: 10.1097/00004872-198904000-00011
80. Diaz J, Ni W, Thompson J, King A, Fink GD, Watts SW. 5-hydroxytryptamine lowers blood pressure in normotensive and hypertensive rats. J Pharmacol Exp Ther. (2008) 325(3):1031–8. doi: 10.1124/jpet.108.136226
81. Baron A, Riesselmann A, Fregly MJ. Reduction in the elevated blood pressure of dahl salt-sensitive rats treated chronically with L-5-hydroxytryptophan. Pharmacology. (1991) 42(1):15–22. doi: 10.1159/000138763
82. Ding XR, Stier CT Jr, Itskovitz HD. Serotonin and 5-hydroxytryptophan on blood pressure and renal blood flow in anesthetized rats. Am J Med Sci. (1989) 297(5):290–3. doi: 10.1097/00000441-198905000-00004
83. Cazzola I, Matera MG. 5-HT modifiers as a potential treatment of asthma. Trends Pharmacol Sci. (2000) 21(1):13–6. doi: 10.1016/S0165-6147(99)01408-X
84. Evans AT, Sadowski LS, VanderWeele TJ, Curtis LM, Sharp LK, Kee RA, et al. Ethnic disparities in asthma morbidity in Chicago. J Asthma. (2009) 46(5):448–54. doi: 10.1080/02770900802492061
85. Weiss KB, Shannon JJ, Sadowski LS, Sharp LK, Curtis L, Lyttle CS, et al. The burden of asthma in the Chicago community fifteen years after the availability of national asthma guidelines: the design and initial results from the CHIRAH study. Contemp Clin Trials. (2009) 30(3):246–55. doi: 10.1016/j.cct.2009.01.006
86. Shalowitz MU, Sadowski LM, Kumar R, Weiss KB, Shannon JJ. Asthma burden in a citywide, diverse sample of elementary schoolchildren in Chicago. Ambul Pediatr. (2007) 7(4):271–7. doi: 10.1016/j.ambp.2007.03.004
87. Grammer LC, Weiss KB, Pedicano JB, Kimmel LG, Curtis LS, Catrambone CD, et al. Obesity and asthma morbidity in a community-based adult cohort in a large urban area: the Chicago initiative to raise asthma health equity (CHIRAH). J Asthma. (2010) 47(5):491–5. doi: 10.3109/02770901003801980
88. Washington DM, Curtis LM, Waite K, Wolf MS, Paasche-Orlow MK. Sociodemographic factors mediate race and ethnicity-associated childhood asthma health disparities: a longitudinal analysis. J Racial Ethn Health Disparities. (2018) 5(5):928–38. doi: 10.1007/s40615-017-0441-2
89. Celebi Sözener Z, Cevhertas L, Nadeau K, Akdis M, Akdis CA. Environmental factors in epithelial barrier dysfunction. J Allergy Clin Immunol. (2020) 145(6):1517–28. doi: 10.1016/j.jaci.2020.04.024
90. Thakur N, Borrell LN, Ye M, Oh SS, Eng C, Meade K, et al. Acculturation is associated with asthma burden and pulmonary function in latino youth: the GALA II study. J Allergy Clin Immunol. (2019) 143(5):1914–22. doi: 10.1016/j.jaci.2018.12.1015
91. Torgerson DG, Gignoux CR, Galanter JM, Drake KA, Roth LA, Eng C, et al. Case-control admixture mapping in latino populations enriches for known asthma-associated genes. J Allergy Clin Immunol. (2012) 130(1):76–82. doi: 10.1016/j.jaci.2012.02.040
92. Gignoux CR, Torgerson DG, Pino-Yanes M, Uricchio LH, Galanter J, Roth LA, et al. An admixture mapping meta-analysis implicates genetic variation at 18q21 with asthma susceptibility in latinos. J Allergy Clin Immunol. (2019) 143(3):957–69. doi: 10.1016/j.jaci.2016.08.057
93. Matheny HE, Deem TL, Cook-Mills JM. Lymphocyte migration through monolayers of endothelial cell lines involves VCAM-1 signaling via endothelial cell NADPH oxidase. J Immunol. (2000) 164:6550–9. doi: 10.4049/jimmunol.164.12.6550
94. Abdala-Valencia H, Cook-Mills JM. VCAM-1 signals activate endothelial cell protein kinase ca via oxidation. J Immunol. (2006) 177:6379–87. doi: 10.4049/jimmunol.177.9.6379
95. Xiao J, Bergson C. Detection of cell surface dopamine receptors. Methods Mol Biol. (2013) 964:3–13. doi: 10.1007/978-1-62703-251-3_1
96. Elkomy MH, Sultan P, Carvalho B, Peltz G, Wu M, Clavijo C, et al. Ondansetron pharmacokinetics in pregnant women and neonates: towards a new treatment for neonatal abstinence syndrome. Clin Pharmacol Ther. (2015) 97(2):167–76. doi: 10.1002/cpt.5
97. Gan LS, Hsyu PH, Pritchard JF, Thakker D. Mechanism of intestinal absorption of ranitidine and ondansetron: transport across caco-2 cell monolayers. Pharm Res. (1993) 10(12):1722–5. doi: 10.1023/A:1018965929419
98. Abdala-Valencia H, Earwood J, Bansal S, Jansen M, Babcock G, Garvy B, et al. Nonhematopoietic NADPH oxidase regulation of lung eosinophilia and airway hyperresponsiveness in experimentally induced asthma. Am J Physiol Lung Cell Mol Physiol. (2007) 292(5):L1111–25. doi: 10.1152/ajplung.00208.2006
99. Deem TL, Abdala-Valencia H, Cook-Mills JM. VCAM-1 activation of PTP1B in endothelial cells. J Immunol. (2007) 178:3865–73. doi: 10.4049/jimmunol.178.6.3865
100. Qureshi MH, Cook-Mills J, Doherty DE, Garvy BA. TNF-alpha-dependent ICAM-1- and VCAM-1-mediated inflammatory responses are delayed in neonatal mice infected with {I}pneumocystis carinii. J Immunol. (2003) 171:4700–7. doi: 10.4049/jimmunol.171.9.4700
101. Hankinson JL, Odencrantz JR, Fedan KB. Spirometric reference values from a sample of the general U.S. Population. Am J Respir Crit Care Med. (1999) 159(1):179–87. doi: 10.1164/ajrccm.159.1.9712108
102. MacLean MR, Dempsie Y. Serotonin and pulmonary hypertension–from bench to bedside? Curr Opin Pharmacol. (2009) 9(3):281–6. doi: 10.1016/j.coph.2009.02.005
103. MaassenVanDenBrink A, Centurion D, Villalon CM. Crosstalk of vascular 5-HT1 receptors with other receptors: clinical implications. Neuropharmacology. (2008) 55(6):986–93. doi: 10.1016/j.neuropharm.2008.06.051
104. Kawahara K, Hashiguchi T, Kikuchi K, Tancharoen S, Miura N, Ito T, et al. Induction of high mobility group box 1 release from serotonin-stimulated human umbilical vein endothelial cells. Int J Mol Med. (2008) 22(5):639–44. doi: 10.3892/ijmm_00000066
105. Dempsie Y, MacLean MR. Pulmonary hypertension: therapeutic targets within the serotonin system. Br J Pharmacol. (2008) 155(4):455–62. doi: 10.1038/bjp.2008.241
106. MacLean MR. Pulmonary hypertension and the serotonin hypothesis: where are we now? Int J Clin Pract Suppl. (2007) 156:27–31. doi: 10.1111/j.1742-1241.2007.01497.x
107. Kojim S, Ikeda M, Shibukawa A, Kamikawa Y. Modification of 5-hydroxytryptophan-evoked 5-hydroxytryptamine formation of Guinea pig colonic mucosa by reactive oxygen species. Jpn J Pharmacol. (2002) 88(1):114–8. doi: 10.1254/jjp.88.114
108. Kojima S, Ikeda M, Kamikawa Y. Investigation into the 5-hydroxytryptophan-evoked luminal 5-hydroxytryptamine release from the Guinea pig colon. Jpn J Pharmacol. (2000) 84(2):174–8. doi: 10.1254/jjp.84.174
109. Jonnakuty C, Gragnoli C. What do we know about serotonin? J Cell Physiol. (2008) 217(2):301–6. doi: 10.1002/jcp.21533
110. Bouzigon E, Siroux V, Dizier MH, Lemainque A, Pison C, Lathrop M, et al. Scores of asthma and asthma severity reveal new regions of linkage in EGEA study families. Eur Respir J. (2007) 30(2):253–9. doi: 10.1183/09031936.00162206
111. Zhang Y, Leaves NI, Anderson GG, Ponting CP, Broxholme J, Holt R, et al. Positional cloning of a quantitative trait locus on chromosome 13q14 that influences immunoglobulin E levels and asthma. Nat Genet. (2003) 34(2):181–6. doi: 10.1038/ng1166
112. Anderson GG, Leaves NI, Bhattacharyya S, Zhang Y, Walshe V, Broxholme J, et al. Positive association to IgE levels and a physical map of the 13q14 atopy locus. Eur J Hum Genet. (2002) 10(4):266–70. doi: 10.1038/sj.ejhg.5200801
113. Blumenthal MN, Ober C, Beaty TH, Bleecker ER, Langefeld CD, King RA, et al. Genome scan for loci linked to mite sensitivity: the collaborative study on the genetics of asthma (CSGA). Genes Immun. (2004) 5(3):226–31. doi: 10.1038/sj.gene.6364063
114. Bellenguez C, Ober C, Bourgain C. A multiple splitting approach to linkage analysis in large pedigrees identifies a linkage to asthma on chromosome 12. Genet Epidemiol. (2009) 33(3):207–16. doi: 10.1002/gepi.20371
115. Shao C, Suzuki Y, Kamada F, Kanno K, Tamari M, Hasegawa K, et al. Linkage and association of childhood asthma with the chromosome 12 genes. J Hum Genet. (2004) 49(3):115–22. doi: 10.1007/s10038-003-0118-z
116. Chi PB, Duggal P, Kao WH, Mathias RA, Grant AV, Stockton ML, et al. Comparison of SNP tagging methods using empirical data: association study of 713 SNPs on chromosome 12q14.3–12q24.21 for asthma and total serum IgE in an African Caribbean population. Genet Epidemiol. (2006) 30(7):609–19. doi: 10.1002/gepi.20172
117. Burt SA, Mikolajewski AJ. Preliminary evidence that specific candidate genes are associated with adolescent-onset antisocial behavior. Aggress Behav. (2008) 34(4):437–45. doi: 10.1002/ab.20251
118. Yoon HK, Yang JC, Lee HJ, Kim YK. The association between serotonin-related gene polymorphisms and panic disorder. J Anxiety Disord. (2008) 22(8):1529–34. doi: 10.1016/j.janxdis.2008.03.006
119. Beretta L, Cossu M, Marchini M, Cappiello F, Artoni A, Motta G, et al. A polymorphism in the human serotonin 5-HT2A receptor gene may protect against systemic sclerosis by reducing platelet aggregation. Arthritis Res Ther. (2008) 10(5):R103. doi: 10.1186/ar2495
120. Kling A, Seddighzadeh M, Arlestig L, Alfredsson L, Rantapaa-Dahlqvist S, Padyukov L. Genetic variations in the serotonin 5-HT2A receptor gene (HTR2A) are associated with rheumatoid arthritis. Ann Rheum Dis. (2008) 67(8):1111–5. doi: 10.1136/ard.2007.074948
121. Benedetti F, Barbini B, Bernasconi A, Fulgosi MC, Colombo C, Dallaspezia S, et al. Serotonin 5-HT2A receptor gene variants influence antidepressant response to repeated total sleep deprivation in bipolar depression. Prog Neuropsychopharmacol Biol Psychiatry. (2008) 32(8):1863–6. doi: 10.1016/j.pnpbp.2008.08.017
122. Krzywkowski K, Davies PA, Irving AJ, Brauner-Osborne H, Jensen AA. Characterization of the effects of four HTR3B polymorphisms on human 5-HT3AB receptor expression and signalling. Pharmacogenet Genomics. (2008) 18(12):1027–40. doi: 10.1097/FPC.0b013e328310f950
123. Fakra E, Hyde LW, Gorka A, Fisher PM, Munoz KE, Kimak M, et al. Effects of HTR1A C(-1019)G on amygdala reactivity and trait anxiety. Arch Gen Psychiatry. (2009) 66(1):33–40. doi: 10.1001/archpsyc.66.1.33
124. Gunes A, Melkersson KI, Scordo MG, Dahl ML. Association between HTR2C and HTR2A polymorphisms and metabolic abnormalities in patients treated with olanzapine or clozapine. J Clin Psychopharmacol. (2009) 29(1):65–8. doi: 10.1097/JCP.0b013e31819302c3
125. Ducci F, Enoch MA, Yuan Q, Shen PH, White KV, Hodgkinson C, et al. HTR3B is associated with alcoholism with antisocial behavior and alpha EEG power–an intermediate phenotype for alcoholism and co-morbid behaviors. Alcohol. (2009) 43(1):73–84. doi: 10.1016/j.alcohol.2008.09.005
126. Savitz J, Lucki I, Drevets WC. 5-HT(1A) receptor function in major depressive disorder. Prog Neurobiol. (2009) 88(1):17–31. doi: 10.1016/j.pneurobio.2009.01.009
127. Jacobsen KX, Vanderluit JL, Slack RS, Albert PR. HES1 regulates 5-HT1A receptor gene transcription at a functional polymorphism: essential role in developmental expression. Mol Cell Neurosci. (2008) 38(3):349–58. doi: 10.1016/j.mcn.2008.03.007
128. Kato M, Zanardi R, Rossini D, De Ronchi D, Okugawa G, Kinoshita T, et al. 5-HT2A gene variants influence specific and different aspects of antidepressant response in Japanese and Italian mood disorder patients. Psychiatry Res. (2009) 167(1-2):97–105. doi: 10.1016/j.psychres.2007.12.012
129. Ohtsuki T, Ishiguro H, Detera-Wadleigh SD, Toyota T, Shimizu H, Yamada K, et al. Association between serotonin 4 receptor gene polymorphisms and bipolar disorder in Japanese case-control samples and the NIMH genetics initiative bipolar pedigrees. Mol Psychiatry. (2002) 7(9):954–61. doi: 10.1038/sj.mp.4001133
130. Li J, Wang Y, Zhou R, Wang B, Zhang H, Yang L, et al. Association of attention-deficit/hyperactivity disorder with serotonin 4 receptor gene polymorphisms in han Chinese subjects. Neurosci Lett. (2006) 401(1-2):6–9. doi: 10.1016/j.neulet.2006.02.053
131. Suzuki T, Iwata N, Kitamura Y, Kitajima T, Yamanouchi Y, Ikeda M, et al. Association of a haplotype in the serotonin 5-HT4 receptor gene (HTR4) with Japanese schizophrenia. Am J Med Genet B Neuropsychiatr Genet. (2003) 121B(1):7–13. doi: 10.1002/ajmg.b.20060
132. Niesler B, Flohr T, Nothen MM, Fischer C, Rietschel M, Franzek E, et al. Association between the 5′ UTR variant C178T of the serotonin receptor gene HTR3A and bipolar affective disorder. Pharmacogenetics. (2001) 11(6):471–5. doi: 10.1097/00008571-200108000-00002
133. Nikolić J, Vukojević K, Šoljić V, Mišković J, Orlović Vlaho M, Saraga-Babić M, et al. Expression patterns of serotonin receptors 5-HT1A, 5-HT2A, and 5-HT3A during human fetal lung development. Int J Mol Sci. (2023) 24(3):2965. doi: 10.3390/ijms24032965
134. Ayme-Dietrich E, Lawson R, Côté F, de Tapia C, Da Silva S, Ebel C, et al. The role of 5-HT(2B) receptors in mitral valvulopathy: bone marrow mobilization of endothelial progenitors. Br J Pharmacol. (2017) 174(22):4123–39. doi: 10.1111/bph.13981
135. Iwabayashi M, Taniyama Y, Sanada F, Azuma J, Iekushi K, Kusunoki H, et al. Role of serotonin in angiogenesis: induction of angiogenesis by sarpogrelate via endothelial 5-HT1B/Akt/eNOS pathway in diabetic mice. Atherosclerosis. (2012) 220(2):337–42. doi: 10.1016/j.atherosclerosis.2011.10.042
136. Rybakova EY, Avdonin PP, Trufanov SK, Goncharov NV, Avdonin PV. Synergistic interaction of 5-HT(1B) and 5-HT(2B) receptors in cytoplasmic Ca2+ regulation in human umbilical vein endothelial cells: possible involvement in pathologies. Int J Mol Sci. (2023) 24(18):13833. doi: 10.3390/ijms241813833
137. Morecroft I, White K, Caruso P, Nilsen M, Loughlin L, Alba R, et al. Gene therapy by targeted adenovirus-mediated knockdown of pulmonary endothelial Tph1 attenuates hypoxia-induced pulmonary hypertension. Mol Ther. (2012) 20(8):1516–28. doi: 10.1038/mt.2012.70
138. Lazarus HM, Denning J, Wring S, Palacios M, Hoffman S, Crizer K, et al. A trial design to maximize knowledge of the effects of rodatristat ethyl in the treatment of pulmonary arterial hypertension (ELEVATE 2). Pulm Circ. (2022) 12(2):e12088. doi: 10.1002/pul2.12088
139. Kang BN, Ha SG, Bahaie NS, Hosseinkhani MR, Ge XN, Blumenthal MN, et al. Regulation of serotonin-induced trafficking and migration of eosinophils. PLoS ONE. (2013) 8(1):e54840. doi: 10.1371/journal.pone.0054840
140. Salim K, Fenton T, Bacha J, Urien-Rodriguez H, Bonnert T, Skynner HA, et al. Oligomerization of G-protein-coupled receptors shown by selective co-immunoprecipitation. J Biol Chem. (2002) 277(18):15482–5. doi: 10.1074/jbc.M201539200
141. Linder AE, Ni W, Szasz T, Burnett R, Diaz J, Geddes TJ, et al. A serotonergic system in veins: serotonin transporter-independent uptake. J Pharmacol Exp Ther. (2008) 325(3):714–22. doi: 10.1124/jpet.107.135699
142. Gomes P, Soares-da-Silva P. L-DOPA transport properties in an immortalised cell line of rat capillary cerebral endothelial cells, RBE 4. Brain Res. (1999) 829(1-2):143–50. doi: 10.1016/S0006-8993(99)01387-6
143. Zabel M. Studies on in vitro effect of serotonin on calcitonin secretion by rat thyroid C cells. Histochemistry. (1985) 83(1):71–5. doi: 10.1007/BF00495303
144. Keshavarz M, Schwarz H, Hartmann P, Wiegand S, Skill M, Althaus M, et al. Caveolin-1: functional insights into its role in muscarine- and serotonin-induced smooth muscle constriction in murine airways. Front Physiol. (2017) 8:295. doi: 10.3389/fphys.2017.00295
145. Cook-Mills JM, Marchese ME, Abdala-Valencia H. Vascular cell adhesion molecule-1 expression and signaling during disease: regulation by reactive oxygen species and antioxidants. Antioxid Redox Signal. (2011) 15(6):1607–38. doi: 10.1089/ars.2010.3522
146. Abdala-Valencia H, Kountz TS, Marchese ME, Cook-Mills JM. VCAM-1 induces signals that stimulate ZO-1 serine phosphorylation and reduces ZO-1 localization at lung endothelial cell junctions. J Leukoc Biol. (2018) 104(1):215–28. doi: 10.1002/JLB.2MA1117-427RR
147. Maclean MR, Dempsie Y. The serotonin hypothesis of pulmonary hypertension revisited. Adv Exp Med Biol. (2010) 661:309–22. doi: 10.1007/978-1-60761-500-2_20
148. Thomson L, Blaylock MG, Sexton DW, Campbell A, Walsh GM. Cetirizine and levocetirizine inhibit eotaxin-induced eosinophil transendothelial migration through human dermal or lung microvascular endothelial cells. Clin Exp Allergy. (2002) 32(8):1187–92. doi: 10.1046/j.1365-2745.2002.01444.x
149. Brown ES, Sayed N, Van Enkevort E, Kulikova A, Nakamura A, Khan DA, et al. A randomized, double-blind, placebo-controlled trial of escitalopram in patients with asthma and Major depressive disorder. J Allergy Clin Immunol Pract. (2018) 6(5):1604–12. doi: 10.1016/j.jaip.2018.01.010
150. Brenner B, Harney JT, Ahmed BA, Jeffus BC, Unal R, Mehta JL, et al. Plasma serotonin levels and the platelet serotonin transporter. J Neurochem. (2007) 102(1):206–15. doi: 10.1111/j.1471-4159.2007.04542.x
151. Mendez-Enriquez E, Alvarado-Vazquez PA, Abma W, Simonson OE, Rodin S, Feyerabend TB, et al. Mast cell-derived serotonin enhances methacholine-induced airway hyperresponsiveness in house dust mite-induced experimental asthma. Allergy. (2021) 76(7):2057–69. doi: 10.1111/all.14748
152. Fregly MJ, Lockley OE, Sumners C. Chronic treatment with L-5-hydroxytryptophan prevents the development of DOCA-salt-induced hypertension in rats. J Hypertens. (1987) 5(5):621–8. doi: 10.1097/00004872-198710000-00018
153. Echizen H, Freed CR. Long-term infusion of L-5-hydroxytryptophan increases brain serotonin turnover and decreases blood pressure in normotensive rats. J Pharmacol Exp Ther. (1982) 220(3):579–84. 6977639.6977639
154. Birdsall TC. 5-hydroxytryptophan: a clinically-effective serotonin precursor. Altern Med Rev. (1998) 3(4):271–80. 9727088.9727088
155. Schruers K, van Diest R, Overbeek T, Griez E. Acute L-5-hydroxytryptophan administration inhibits carbon dioxide-induced panic in panic disorder patients. Psychiatry Res. (2002) 113(3):237–43. doi: 10.1016/S0165-1781(02)00262-7
156. Gijsman HJ, van Gerven JM, de Kam ML, Schoemaker RC, Pieters MS, Weemaes M, et al. Placebo-controlled comparison of three dose-regimens of 5-hydroxytryptophan challenge test in healthy volunteers. J Clin Psychopharmacol. (2002) 22(2):183–9. doi: 10.1097/00004714-200204000-00012
157. Lowe SL, Yeo KP, Teng L, Soon DK, Pan A, Wise SD, et al. L-5-hydroxytryptophan augments the neuroendocrine response to a SSRI. Psychoneuroendocrinology. (2006) 31(4):473–84. doi: 10.1016/j.psyneuen.2005.11.005
158. Jacobs GE, Kamerling IM, de Kam ML, Derijk RH, van Pelt J, Zitman FG, et al. Enhanced tolerability of the 5-hydroxytryptophane challenge test combined with granisetron. J Psychopharmacol. (2010) 24(1):65–72. doi: 10.1177/0269881108094299
159. Smarius LJ, Jacobs GE, Hoeberechts-Lefrandt DH, de Kam ML, van der Post JP, de Rijk R, et al. Pharmacology of rising oral doses of 5-hydroxytryptophan with carbidopa. J Psychopharmacol. (2008) 22(4):426–33. doi: 10.1177/0269881107082025
160. Freedman RR. Treatment of menopausal hot flashes with 5-hydroxytryptophan. Maturitas. (2010) 65(4):383–5. doi: 10.1016/j.maturitas.2009.11.025
161. Turner EH, Loftis JM, Blackwell AD. Serotonin a la carte: supplementation with the serotonin precursor 5-hydroxytryptophan. Pharmacol Ther. (2006) 109(3):325–38. doi: 10.1016/j.pharmthera.2005.06.004
162. Shaw K, Turner J, Del Mar C. Tryptophan and 5-hydroxytryptophan for depression. Cochrane Database Syst Rev. (2002) 1:Cd003198. doi: 10.1002/14651858.CD003198
163. Shaw K, Turner J, Del Mar C. Tryptophan and 5-hydroxytryptophan for depression. Cochrane Database Syst Rev. (2001) 3:Cd003198.
Keywords: 5-hydroxytryptophan, eosinophil, endothelial cell, serotonin receptors, FEV1
Citation: Walker MT, Bloodworth JC, Kountz TS, McCarty SL, Green JE, Ferrie RP, Campbell JA, Averill SH, Beckman KB, Grammer LC, Eng C, Avila PC, Farber HJ, Rodriguez-Cintron W, Rodriguez-Santana JR, Serebrisky D, Thyne SM, Seibold MA, Burchard EG, Kumar R and Cook-Mills JM (2024) 5-HTP inhibits eosinophilia via intracellular endothelial 5-HTRs; SNPs in 5-HTRs associate with asthmatic lung function. Front. Allergy 5:1385168. doi: 10.3389/falgy.2024.1385168
Received: 12 February 2024; Accepted: 8 May 2024;
Published: 23 May 2024.
Edited by:
Luan D. Vu, Louisiana State University, United StatesReviewed by:
Daniel P. Potaczek, University of Marburg, GermanyDeepak A. Deshpande, Thomas Jefferson University, United States
© 2024 Walker, Bloodworth, Kountz, McCarty, Green, Ferrie, Campbell, Averill, Beckman, Grammer, Eng, Avila, Farber, Rodriguez-Cintron, Rodriguez-Santana, Serebrisky, Thyne, Seibold, Burchard, Kumar and Cook-Mills. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Joan M. Cook-Mills, am9hbmNvb2tAaXUuZWR1
 Matthew T. Walker
Matthew T. Walker Jeffrey C. Bloodworth2,3
Jeffrey C. Bloodworth2,3 Kenneth B. Beckman
Kenneth B. Beckman Celeste Eng
Celeste Eng Shannon M. Thyne
Shannon M. Thyne Max A. Seibold
Max A. Seibold Rajesh Kumar
Rajesh Kumar Joan M. Cook-Mills
Joan M. Cook-Mills