- 1European Food Safety Authority (EFSA), Parma, Italy
- 2Experimental Allergy Laboratory, Department of Dermatology, Medical University of Vienna, Vienna, Austria
- 3Instituto de Investigación en Ciencias de la Alimentación (CIAL), CSIC-UAM, CEI (UAM+CSIC), Madrid, Spain
The allergenicity and protein risk assessments in food safety are facing new challenges. Demands for healthier and more sustainable food systems have led to significant advances in biotechnology, the development of more complex foods, and the search for alternative protein sources. All this has increased the pressure on the safety assessment prediction approaches anchored into requirements defined in the late 90's. In 2022, the EFSA's Panel on Genetically Modified Organisms published a scientific opinion focusing on the developments needed for allergenicity and protein safety assessments of new products derived from biotechnology. Here, we further elaborate on the main elements described in this scientific opinion and prioritize those development needs requiring critical attention. The starting point of any new recommendation would require a focus on clinical relevance and the development of a fit-for-purpose database targeted for specific risk assessment goals. Furthermore, it is imperative to review and clarify the main purpose of the allergenicity risk assessment. An internationally agreed consensus on the overall purpose of allergenicity risk assessment will accelerate the development of fit-for-purpose methodologies, where the role of exposure should be better clarified. Considering the experience gained over the last 25 years and recent scientific developments in the fields of biotechnology, allergy, and risk assessment, it is time to revise and improve the allergenicity safety assessment to ensure the reliability of allergenicity assessments for food of the future.
1 Introduction
More than 400 genetically modified organisms (GMOs) have been approved worldwide (1) (Supplementary Material). Since the early 2000s, over 100 GMOs have been approved in the European Union (EU) (2, 3). To date, EFSA's allergenicity risk assessment for approved GMOs has not identified any hazards. However, the scientific community is facing new challenges, starting with the population's demands for healthier and more sustainable systems (4–7), leading to significant advances in biotechnology and the development of more complex foods, like products with multiple events containing a high number of new proteins, that, in some cases, are also difficult to test, e.g., membrane-bound proteins, transcription factors; and in a broader context, the assessment of proteins in a new whole food, such as insects. Consequently, the prediction of potential adverse allergic reactions to novel proteins (allergenicity) becomes more difficult.
The current strategies for the allergenicity and safety assessments of new/novel proteins are based on principles adapted from the chemical risk assessment and guidelines of Codex Alimentarius for the safety assessment of foods derived from “modern” biotechnology from 2003 (Figure 1). The assessment is performed for newly expressed proteins in GMOs as well as for whole novel foods. The weight-of-evidence approach is the most robust strategy used for all products, as no single piece of information or experimental method provides sufficient evidence for assessing allergenicity.
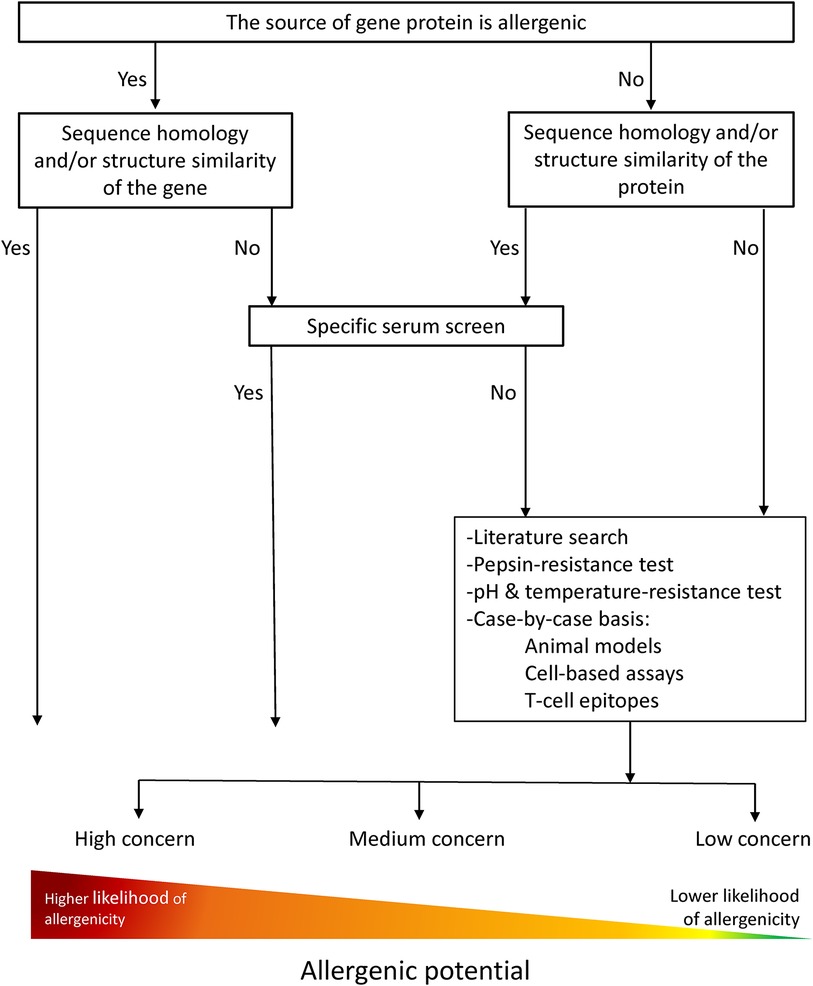
Figure 1. Allergenicity risk assessment current flow chart modified from FAO/WHO (2001) and Davies H. (2005). A weight-of-evidence approach is followed where information of different nature, e.g. in silico, in vitro, in vivo, is considered in the overall assessment to conclude on the allergenic potential of novel proteins.
In 2022, EFSA published a scientific opinion focusing on the development needs for the allergenicity and protein safety assessment of food and feed products derived from biotechnology (8). A series of short-term and long-term recommendations were provided. These would include the need to: (i) update in silico tools that are linked to more targeted databases, (ii) better integrate and standardise test materials and in vitro/in vivo assays, (iii) better clarity on the use of the weight-of-evidence approach for protein safety and the role of expert judgment, and (iv) (re)define the allergenicity safety objectives.
Here, we follow-up the EFSA scientific opinion and prioritise the main research gaps and future needs for in silico, in vitro and in vivo allergenicity assessment tools, and other elements, such as dietary exposure, that needs urgent development. It is timely and necessary to revise and improve the allergenicity safety assessment.
2 Allergenicity prediction in risk assessment—current state and development needs
2.1 In silico analysis
Primary amino acid sequence similarity searches against an allergen database are still the current practice for the in silico assessment of a novel protein and allergenicity prediction (Figure 1). A threshold value of >35% amino acid identity over at least 80 amino acids was established by a joint FAO/WHO expert consultation in 2001 (9) and embedded in Codex Alimentarius (10). This strategy is considered highly conservative and demanding when hits above the threshold are identified. Furthermore, these in silico tools used in the allergenicity assessment inform about the potential capacity of a protein to cross-react with a known allergen (e.g., cross-react and elicit a response in a previously sensitized individual), but they do not provide information on the capacity of proteins for de novo sensitization.
Advanced bioinformatic tools different from those defined by Codex (10), the classical FASTA algorithm, are for example similarity searches of 3D protein structure (11, 12), machine learning based on mapping of IgE epitope and motif search (13) or new approaches considering allergen-IgE interaction (14). It is highly likely that these advanced bioinformatic tools will provide higher sensitivity, specificity, accuracy, and improve allergenicity prediction. Furthermore, bioinformatic screening should also consider additional characteristics of proteins beyond its potential for cross-reactivity. These tools can also be used to provide information on the relatedness of a novel protein with commonly consumed proteins and the evolutionary distance between proteins relevant for allergenicity (15). However, advanced bioinformatic tools are not routinely used in the risk assessment process.
Exceptions exist and progressive bioinformatic tools have been developed for predicting the risk of proteins triggering celiac disease (16). The main elements which improved the bioinformatics tools used for celiac disease, for example, are: (i) a definition of clear inclusion criteria for database formation (17); (ii) a ranking strategy of immunodominant T-cell epitopes according to their clinical relevance and related features (18); and (iii) the development of a software tool for peptide binding prediction to HLA proteins (19). However, for allergenicity, current in silico approaches heavily rely on expert judgement to interpret a posteriori the outcome of the bioinformatic analysis. Because similarity search outcomes may change depending on the database used, (e.g., Allergenonline1, CompareDatabase2, Allergome3, WHO/IUIS4), it can lead to a lack of harmonisation, reproducibility, and transparency in the risk assessment process (8). It is imperative to refine databases so that they are fit-for-purpose for the allergenicity assessment (8, 20–24). To this end, the clinical relevance of known allergens in a given database should be defined a priori where allergens are ranked in terms of their clinical relevance, and are associated with specific risk assessment follow-up actions depending on the clinical relevance of the findings (20, 25). It will also be necessary to validate new bioinformatic tools using a comprehensive set of positive and negative control allergens.
2.2 In vitro tests
In vitro methods for the allergenicity assessment include protein stability measurements, e.g., classical pepsin resistance test and denaturation under differing pH and temperature conditions, and immunological assays, e.g., ELISAs and immunoblotting with human sera (Figure 1) (10, 26, 27). The most commonly used is the classical pepsin resistance test, which provides information on the stability of the proteins under acidic conditions and is useful in the weight-of-evidence approach. However, the test is poorly predictive of allergy, possibly because there is not a single intrinsic characteristic of proteins leading to allergenicity, and it does not mimic the physiologic conditions of gastric digestion (16, 28, 29). It is likely that understanding the influence of intestinal digestion on the fate of the proteins in the gastrointestinal tract and how they interact with relevant cells may improve predictability (29, 30), which could be achieved by improving the characterization of digestion products, e.g., molecular size, persistence, abundance, etc (16, 31). For instance, one new interesting approach is in vitro protein degradation studies, which simulate sequential gastric digestion followed by an intestinal digestion phase (32, 33). Because one of the most prominent traits attributed to known food allergens is protein stability (34–37), it will be crucial to optimize in vitro testing taking into account the following aspects: protein stability during heating and other processing procedures, pH changes and proteolysis, and physical stability, including aggregation. Consideration of industrial processing is critical and is emphasised in the guidelines on the effects of industrial processing of milk protein allergens, e.g., denaturation, the generation of new antigenic epitopes (38).
In GMO risk assessment, testing of the newly expressed proteins with human sera must be performed for the assessment if the source of the introduced gene is allergenic or if there is sequence homology similarity >35% with a known allergen (Figure 1) (10, 26, 27). However, it remains unclear i) how the testing should be specifically carried out; ii) why it is necessary to test human sera on all these cases; and iii) how additional elements such as the quality of the sequence homology similarity and the clinical relevance of the known allergen can be used to wave such requirements. Moreover, the difficulties identified in the assessment of newly expressed proteins become more complex when applied to whole foods.
There are an assortment of additional human cell and tissue models that might potentially be relevant for an allergenicity risk assessment such as biopsy-based models, coculture systems with epithelial and immune cells, precision cut organ slices, organoids, e.g., mini-gut cultures and organ-on-a chip models (gut-on-chip) (39–41). Moreover, there are in vitro models evaluating the potential sensitising capacity of food proteins such as antigen uptake via the intestinal mucosal barrier (42, 43). However, some of these models might need considerable work to ensure predictability and cost-effectiveness.
While in vitro assays are potentially invaluable, they require optimisation. For instance, test items and conditions will need to be standardized, information on interactions between proteins/fragments and the gastrointestinal tract/immune system need to be provided for the risk assessment process to ensure predictability.
2.3 In vivo studies
Mouse models of food allergy have been developed to understand further and elucidate underlying disease mechanisms (44). To date, it is not clear whether any of the models fully replicate human disease or whether they are able to predict protein allergenicity or adjuvanticity despite being used to assess the allergenicity and adjuvanticity risks of GMOs (45, 46). Nevertheless, where these models might be most useful is for further understanding the sensitizing potential of proteins, their cross-reactivity with other food proteins (46, 47), and the potential of novel proteins to act as adjuvants (46, 48–53). However, attention to experimental design, e.g., mouse strains, allergens, administration methods, and environmental factors, is crucial. Additionally, the model choice should be fit-for-purpose, multiple models might be needed, and combining data from in silico, in vitro, and in vivo models will likely improve predictability.
2.4 Other elements
Additional information from other sources may also improve the current risk assessment approach. For example, dietary exposure and eliciting dose data could be useful in the risk assessment process which are not clearly defined at the moment. Current regulatory guidelines focus on the hazard identification step of risk assessment. In future, we should explore possibilities to define more clearly what the role of exposure is in the overall risk assessment (8–10). Another possibility is the building of a framework with threshold levels of the most common and potent allergens, which could provide protection for people with food allergies (54). Indeed, a joint FAO/WHO expert Committee recently established recommended reference doses, based on the ED05 (max. 5% of the affected persons showing allergic reactions), for a series of major allergenic foods that meet the criterion of “exposure without appreciable health risk” (55). However, there are challenges that need be addressed, such as the lack of information for individual allergens and for food sources not considered common allergenic foods, as well as issues with inter-individual variability and with quality of clinical data. Nevertheless, it has been proposed that the use of information on the most common and potent allergens, as a worst-case scenario, should be able to cover other foods for which there is less available data (54).
Although pre-market monitoring has been successful, a post-market monitoring strategy could potentially prevent allergic reactions in subgroups of vulnerable individuals in the general population (56) and could address specific uncertainties arising from the pre-market assessment phase. However, it is crucial to consider the feasibility and practicality of including post-market monitoring requirements in the risk assessment process.
Specific risk assessment requirements might differ depending on the product under assessment and the regulatory frame under which it is evaluated. For example, the assessment of a simple protein or simple protein mixture vs. a complex protein mixtures or whole food leads to different challenges to the risk assessment process. Furthermore, the exposure scenario might differ depending on the product assessed. For instance, the assessment of a novel staple food is the most difficult allergenicity risk assessment scenario because staple foods are widely consumed and/or processed in different manners. Thus, a novel, widely consumed staple food is challenging to do a hazard, exposure or risk-based assessment.
3 Future needs for improving the allergenicity assessment
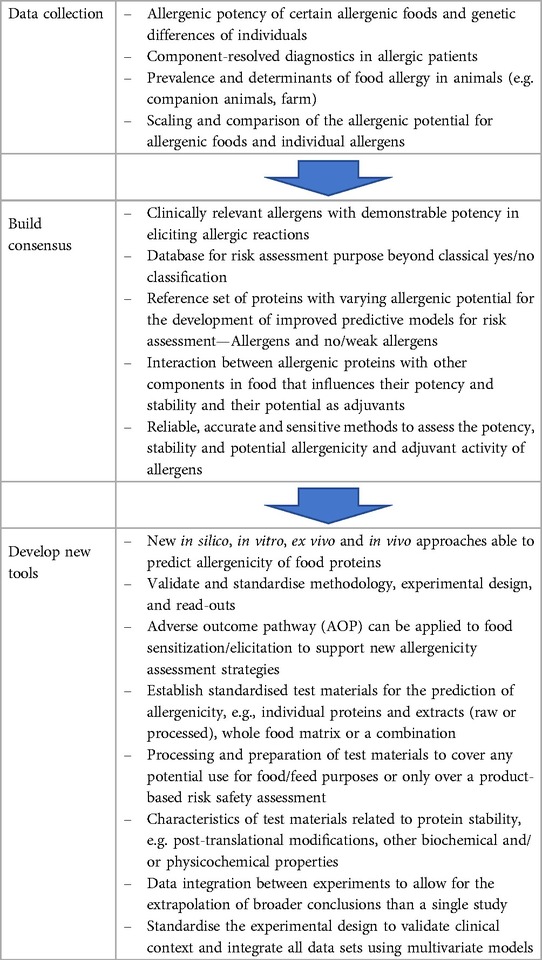
Continuous scientific advances over the last two decades have led to a functional asynchrony between the availability of safety standards and available scientific knowledge. As the numbers and complexity of new GMOs and new novel foods grow, there is a need for an overall revision of the allergenicity assessment objectives, still anchored on requirements and methodology established in late 90's. In 2022, EFSA published a scientific opinion on development needs for the allergenicity and protein safety assessment of food and feed products derived from biotechnology that provided short-and long-term recommendations. Therefore, it is necessary to revise and improve the allergenicity safety assessment. Here, we expand and prioritize advanced developmental stages, ready for implementation approaches to improve the current risk assessment including alternative/complementary methods to those already in place (e.g., in silico tools for cross-reactivity), and others that will need more development, research and consensus (e.g., in vitro tools for de novo sensitisation). Table 1 illustrates the priorities of these developments of which the top three are as follows:
(i) The development of a fit-for-purpose database, based on reliable and consensual inclusion criteria ensuring that only well-defined and characterised allergens are included. Ideally, the database should contain specific follow-up actions when similarities above thresholds with known allergens are identified depending on the clinical relevance and the quality of the similarity matches. Data curation and maintenance should also be specified;
(ii) The definition of a set of positive and negative control allergens together with the development of a specific validation testing process for in silico, in vitro and in vivo models. This process will need the development of a clear hypothesis relevant for allergenicity assessment and standardised experimental design ensuring appropriate statistical power under precise conditions and proper controls; and
(iii) Consensus on the purpose of an allergenicity risk assessment. A new frame of the purpose of the allergenicity assessment should be identified and internationally agreed where the role of exposure should be clarified, and consideration of the desired risk management outcome (e.g., preventing allergen sensitisation, accepting rare, potentially fatal reactions).
New tools developed for allergenicity prediction should consider models for cross-reactivity (e.g., elicitation), sensitization and adjuvanticity, providing more precise information and clarity on how the weight-of-evidence approach is used, and the role of expert judgment in the overall safety assessment. Most importantly, any new tool/approach developed for its use in risk assessment should be proven to have better sensitivity, specificity and accuracy than current methods as well as being reproducible and cost-effective.
Author contributions
AF: Writing – original draft, Writing – review & editing. ED: Writing – original draft, Writing – review & editing. MT: Writing – original draft, Writing – review & editing. SK: Writing – original draft, Writing – review & editing. FM: Writing – original draft, Writing – review & editing. ME: Writing – original draft, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article.
The authors acknowledge funding from the European Union's Horizon 2020 research and innovation program under grant agreement no. 965173 (Imptox) and European Union's Horizon Europe research and innovation programme under the Marie Skłodowska-Curie grant agreement No 101072377.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Author disclaimer
AF is employed by the European Food Safety Authority (EFSA). The positions and opinions presented in this article are those of the authors alone and do not necessarily represent the views or scientific works of the EFSA.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/falgy.2024.1297547/full#supplementary-material
Footnotes
1http://www.allergenonline.org/
References
1. ISAAA. Available online at: https://www.isaaa.org/gmapprovaldatabase/ (accessed July 15, 2023).
2. European Food Safety Authority—EFSA. Available online at: https://www.efsa.europa.eu/en/publications (accessed July 15, 2023).
3. European Commission—EC. Available online at: https://webgate.ec.europa.eu/dyna2/gm-register/ (accessed July 15, 2023).
4. European Commission—EC. Available online at: https://commission.europa.eu/strategy-and-policy/priorities-2019-2024/european-green-deal_en (accessed July 15, 2023).
5. Devos Y, Arena M, Ashe S, Blanck M, Bray E, Broglis A, et al. Addressing the need for safe, nutritious and sustainable food: outcomes of the “ONE—health, environment & society—conference 2022”. Trends Food Sci Technol. (2022) 129:164–78. doi: 10.1016/j.tifs.2022.09.014
6. European Food Safety Authority, Bronzwaer S, Kass G, Robinson T, Tarazona J, Verhagen H, et al. Editorial on food safety regulatory research needs 2030. EFSA J. (2019) 17:e170622. doi: 10.2903/j.efsa.2019.e170622
7. van Zanten HHE, Simon W, van Selm B, Wacker J, Maindl TI, Frehner A, et al. Circularity in Europe strengthens the sustainability of the global food system. Nat Food. (2023) 4:320–30. doi: 10.1038/s43016-023-00734-9
8. EFSA Panel on Genetically Modified Organisms (GMO), Mullins E, Bresson J-L, Dalmay T, Dewhurst IC, Epstein MM, et al. Scientific opinion on development needs for the allergenicity and protein safety assessment of food and feed products derived from biotechnology. EFSA J. (2022) 20:7044. doi: 10.2903/j.efsa.2022.7044
9. FAO/WHO. Evaluation of Allergenicity of Genetically Modified Foods. Report of a Joint FAO/WHO Expert Consultation on Allergenicity of Foods Derived from Biotechnology. Food and Agricultural Organization/World Health Organization. Rome, Italy (2001).
10. Codex Alimentarius. Foods Derived from Modern Biotechnology. Rome: Codex Alimentarius Commission, Joint FAO/WHO Food Standards Programme (2003–2009).
11. Nguyen MN, Krutz NL, Limviphuvadh V, Lopata AL, Gerberick GF, Maurer-Stroh S. Allercatpro 2.0: a web server for predicting protein allergenicity potential. Nucleic Acids Res. (2022) 50(W1):W36–43. doi: 10.1093/nar/gkac446
12. Maurer-Stroh S, Krutz NL, Kern PS, Gunalan V, Nguyen MN, Limviphuvadh V, et al. AllerCatPro-prediction of protein allergenicity potential from the protein sequence. Bioinformatics. (2019) 35(17):3020–7. doi: 10.1093/bioinformatics/btz029
13. Sharma N, Patiyal S, Dhall A, Pande A, Arora C, Raghava GPS. Algpred 2.0: an improved method forpredicting allergenic proteins and mapping of IgE epitopes. Brief Bioinformatics. (2021) 22:bbaa294. doi: 10.1093/bib/bbaa294
14. Pomés A, Smith SA, Chruszcz M, Mueller GA, Brackett NF, Chapman MD. Precision engineering for localization, validation, and modification of allergenic epitopes. J Allergy Clin Immunol. (2024):S0091–6749(24)00001-0. doi: 10.1016/j.jaci.2023.12.017 [Epub ahead of print].
15. Jenkins JA, Breiteneder H, Mills EN. Evolutionary distance from human homologs reflects allergenicity of animal food proteins. J Allergy Clin Immunol. (2007) 120(6):1399–405. doi: 10.1016/j.jaci.2007.08.019
16. EFSA Panel on Genetically Modified Organisms (GMO), Naegeli H, Birch AN, Casacuberta J, De Schrijver A, Gralak MA, et al. Guidance on allergenicity assessment of genetically modified plants. EFSA J. (2017) 15:4862. doi: 10.2903/j.efsa.2017.4862
17. Sollid LM, Tye-Din JA, Qiao SW, Anderson RP, Gianfrani C, Koning F. Update 2020: nomenclature and listing of celiac disease-relevant gluten epitopes recognized by CD4(+) T cells. Immunogenetics. (2020) 72:85–8. doi: 10.1007/s00251-019-01141-w
18. Vriz R, Moreno FJ, Koning F, Fernandez A. Ranking of immunodominant epitopes in celiac disease: identification of reliable parameters for the safety assessment of innovative food proteins. Food Chem Toxicol. (2021) 157:112584. doi: 10.1016/j.fct.2021.112584
19. Doytchinova I, Dimitrov I, Atanasova M. PreDQ—a software tool for peptide binding prediction to HLA-DQ2 and HLA-DQ8. EFSA Support Publ. (2023) 20:EN-8108. doi: 10.2903/sp.efsa.2023.EN-8108
20. Fernandez A, Mills ENC, Koning F, Moreno FJ. Allergenicity assessment of novel food proteins: what should be improved? Trends Biotechnol. (2021) 39:4–8. doi: 10.1016/j.tibtech.2020.05.011
21. Dimitrov I, Bangov I, Flower DR, Doytchinova I. AllerTOP vol 2—a server for in silico prediction of allergens. J Mol Model. (2014) 20:2278. doi: 10.1007/s00894-014-2278-5
22. Dimitrov I, Naneva L, Bangov I, Doytchinova I. AllergenFP: allergenicity prediction by descriptor fingerprints. Bioinformatics. (2014) 30:846–51. doi: 10.1093/bioinformatics/btt619
23. Westerhout J, Krone T, Snippe A, Babé L, McClain S, Ladics GS, et al. Allergenicity prediction of novel and modified proteins: not a mission impossible! Development of a random forest allergenicity prediction model. Regul Toxicol Pharm. (2019) 107:104422. doi: 10.1016/j.yrtph.2019.104422
24. Krutz NL, Kimber I, Winget J, Nguyen MN, Limviphuvadh V, Maurer-Stroh S, et al. Application of AllerCatPro 2.0 for protein safety assessments of consumer products. Front Allergy. (2023) 11:1209495. doi: 10.3389/falgy.2023.1209495
25. Dribin TE, Schnadower D, Spergel JM, Campbell RL, Shaker M, Neuman MI, et al. Severity grading system for acute allergic reactions: a multidisciplinary Delphi study. J Allergy Clin Immunol. (2021) 148(1):173–81. doi: 10.1016/j.jaci.2021.01.003
26. EFSA Panel on Genetically Modified Organisms (GMO). Scientific opinion on the assessment of allergenicity of GM plants and microorganisms and derived food and feed. EFSA J. (2010) 8:1700. doi: 10.2903/j.efsa.2010.1700
27. EFSA Panel on Genetically Modified Organisms (GMO). Scientific opinion on guidance for risk assessment of food and feed from genetically modified plants. EFSA J. (2011) 9:2150. doi: 10.2903/j.efsa.2011.2150
28. Verhoeckx K, Bøgh KL, Dupont D, Egger L, Gadermaier G, Larré C, et al. The relevance of a digestibility evaluation in the allergenicity risk assessment of novel proteins. Opinion of a joint initiative of COST action ImpARAS and COST action INFOGEST. Food Chem Toxicol. (2019) 129:405–23. doi: 10.1016/j.fct.2019.04.052
29. EFSA Panel on Genetically Modified Organisms (GMO), Naegeli H, Bresson JL, Dalmay T, Dewhurst IC, Epstein MM. Statement on in vitro protein digestibility tests in allergenicity and protein safety assessment of genetically modified plants. EFSA J. (2021) 19:e06350. doi: 10.2903/j.efsa.2021.6350
30. Wang K, Crevel RWR, Mills ENC. Assessing protein digestibility in allergenicity risk assessment: a comparison of in silico and high throughput in vitro gastric digestion assays. Food Chem Toxicol. (2022) 167:113273. doi: 10.1016/j.fct.2022.113273
31. Fernandez A, Mills ENC, Koning F, Moreno FJ. Safety assessment of immune-mediated adverse reactions to novel food proteins. Trends in Biotech. (2019) 37:796–800. doi: 10.1016/j.tibtech.2019.03.010
32. EFSA Panel on Genetically Modified Organisms (GMO), Mullins E, Bresson JL, Dalmay T, Dewhurst IC, Epstein MM, et al. Assessment of genetically modified Maize MON 87429 for food and feed uses, under regulation (EC) No 1829/2003 (application EFSA-GMO-NL-2019-161). EFSA J. (2022) 20:e07589. doi: 10.2903/j.efsa.2022.7589
33. EFSA Panel on Genetically Modified Organisms (GMO), Mullins E, Bresson J-L, Dalmay T, Dewhurst IC, Epstein MM. Scientific opinion on the assessment of genetically modified maize MON 95379 for food and feed uses, under regulation (EC) No 1829/2003 (application EFSA-GMO-NL-2020-170). EFSA J. (2022) 20:7588. doi: 10.2903/j.efsa.2022.7588
34. Helm RM. Topic 5: Stability of Known Allergens (Digestive and Heat Stability). Report of a Joint FAO, WHO expert consultation on allergenicity of food derived from biotechnology, 22–25 (2001).
35. Breiteneder H, Mills EN. Molecular properties of food allergens. J Allergy Clin Immunol. (2005) 115:14–23. doi: 10.1016/j.jaci.2004.10.022
36. Foo ACY, Mueller GA. Abundance and stability as common properties of allergens. Front Allergy. (2021) 2:769728. doi: 10.3389/falgy.2021.769728
37. Costa J, Bavaro SL, Benede S, Diaz-Perales A, Bueno-Diaz C. Are physicochemical properties shaping the allergenic potency of plant allergens? Clin Rev Allergy Immunol. (2022) 62:37–63. doi: 10.1007/s12016-020-08810-9
38. Fiocchi A, Bognanni A, Brożek J, Ebisawa M, Schünemann H; WAO DRACMA guideline group. World allergy organization (WAO) diagnosis and rationale for action against cow’s milk allergy (DRACMA) guidelines update—i—plan and definitions. World Allergy Organ J. (2022) 15(1):100609. doi: 10.1016/j.waojou.2021.100609
39. Hung L, Obernolte H, Sewald K, Eiwegger T, et al. Humanex vivo and in vitro disease models to study food allergy. Asia Pac Allergy. (2019) 9:pe4. doi: 10.5415/apallergy.2019.9.e4
40. Suber J, Zhang Y, Ye P, Guo R, Burks AW, Kulis MD, et al. Novel peanut-specific human IgE monoclonal antibodies enable screens for inhibitors of the effector phase in food allergy. Front Immunol. (2022) 13:974374. doi: 10.3389/fimmu.2022.974374
41. Zuurveld M, Díaz CB, Redegeld F, Folkerts G, Garssen J, Van't Land B, et al. An advanced in vitro human mucosal immune model to predict food sensitizing allergenicity risk: a proof of concept using ovalbumin as model allergen. Front Immunol. (2023) 13:1073034. doi: 10.3389/fimmu.2022.1073034
42. Lozano-Ojalvo D, Benedé S, Antunes CM, Bavaro SL, Bouchaud G, Costa A, et al. Applying the adverse outcome pathway (AOP) for food sensitization to support in vitro testing strategies. Trends Food Sci Technol. (2019) 85:307–19. doi: 10.1016/j.tifs.2019.01.014
43. Dijk W, Villa C, Benedé S, Vassilopoulou E, Mafra I, Garrido-Arandia M, et al. Critical features of an in vitro intestinal absorption model to study the first key aspects underlying food allergen sensitization. Compr Rev Food Sci Food Saf. (2023) 22:971–1005. doi: 10.1111/1541-4337.13097
44. Kazemi S, Danisman E, Epstein MM. Animal models for the study of food allergies. Curr Protoc. (2023) 3:e685. doi: 10.1002/cpz1.685
45. Marsteller N, Bøgh KL, Goodman RE, Epstein MM. A review of animal models used to evaluate potential allergenicity of genetically modified organisms (GMOs). Drug Discov Today Dis Models. (2015) 17–18:81–8. doi: 10.1016/j.ddmod.2016.11.001
46. Lee RY, Reiner D, Dekan G, Moore AE, Higgins TJ, Epstein MM. Genetically modified alpha-amylase inhibitor peasare not specifically allergenic in mice. PlosOne. (2013) 8:e52972. doi: 10.1371/journal.pone.0052972
47. Smaldini P, Curciarello R, Candreva A, Rey MA, Fossati CA, Petruccelli S, et al. In vivo evidence of cross-reactivity between cow’s milk and soybeanproteins in a mouse model of food allergy. Int Arch Allergy Immunol. (2012) 158:335–46. doi: 10.1159/000333562
48. Vazquez-Padron RI, Moreno-Fierros L, Neri-Bazan L, de la Riva GA, Lopez-Revilla R. Intragastric and intraperitoneal administration of Cry1Ac protoxin from Bacillus thuringiensis induces systemic and mucosal antibody responses in mice. LifeSciences. (1999) 64:1897–912. doi: 10.1016/s0024-3205(99)00136-8
49. Prescott VE, Campbell PM, Moore A, Mattes J, Rothenberg ME, Foster PS, et al. Transgenic expression of bean alpha-amylase inhibitor in peas results in altered structure and immunogenicity. J Agric Food Chem. (2005) 53:9023–30. doi: 10.1021/jf050594v
50. Guimaraes V, Drumare MF, Ah-Leung S, Lereclus D, Bernard H, Créminon C, et al. Comparative study of the adjuvanticity of Bacillus thuringiensis Cry1Ab protein and cholera toxin on allergic sensitisation and elicitation to peanut. Food Agric Immunol Food Agric Immunol. (2008) 19:325–37. doi: 10.1080/09540100802495651
51. Reiner D, Lee RY, Dekan G, Epstein MM. No adjuvant effect of bacillus thuringiensis-maize on allergic responses inmice. Plos One. (2014) 9:e103979. doi: 10.1371/journal.pone.0103979
52. Andreassen M, Bohn T, Wikmark OG, Bodin J, Traavik T, Løvik M, et al. Investigations of immunogenic, allergenic and adjuvant properties of Cry1Ab protein after intragastric exposure in a food allergy model in mice. BMC Immunol. (2016) 17(10). doi: 10.1186/s12865-016-0148-x
53. Tulinska J, Adel-Patient K, Bernard H, Liskova A, Kuricova M, Ilavská S, et al. Humoral and cellular immune response in Wistar Han RCC rats fed two genetically modified maize MON810 varieties for 90 days (EU 7th framework programme project GRACE). Arch Toxicol. (2018) 92:2385–99. doi: 10.1007/s00204-018-2230-z
54. Madsen CB, van den Dungen MW, Cochrane S, Houben GF, Knibb RC, Knulst AC, et al. Can we define a level of protection for allergic consumers that everyone can accept? Regul Toxicol Pharmacol. (2020) 117:104751. doi: 10.1016/j.yrtph.2020.104751
55. FAO/WHO. Risk Assessment of Food Allergens—Part 2: Review and Establish Threshold Levels in Foods for the Priority Allergens. Meeting Report. Food Safety and Quality Series No. 15. Rome. Available online at: https://doi.org/10.4060/cc2946en (2022).
56. FAO. Available online at: https://www.fao.org/food-safety/news/news-details/en/c/1607274/ (accessed September 12, 2023).
Keywords: sensitisation, elicitation, bioinformatics, predictive, risk assessment, allergy, protein safety, food allergy
Citation: Fernandez A, Danisman E, Taheri Boroujerdi M, Kazemi S, Moreno FJ and Epstein MM (2024) Research gaps and future needs for allergen prediction in food safety. Front. Allergy 5:1297547. doi: 10.3389/falgy.2024.1297547
Received: 20 September 2023; Accepted: 5 February 2024;
Published: 19 February 2024.
Edited by:
Geoffrey Mueller, National Institute of Environmental Health Sciences (NIH), United StatesReviewed by:
Stefano Passanisi, University of Messina, Italy© 2024 Fernandez, Danisman, Taheri Boroujerdi, Kazemi, Moreno and Epstein. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: A. Fernandez YW50b25pby5mZXJuYW5kZXpkdW1vbnRAZWZzYS5ldXJvcGEuZXU=
 A. Fernandez
A. Fernandez E. Danisman
E. Danisman M. Taheri Boroujerdi
M. Taheri Boroujerdi S. Kazemi
S. Kazemi F. J. Moreno
F. J. Moreno M. M. Epstein
M. M. Epstein