- 1Division of Allergy and Immunology, Children's Hospital of Philadelphia, Philadelphia, PA, United States
- 2Department of Pediatrics and Institute for Immunology, Perelman School of Medicine at the University of Pennsylvania, Philadelphia, PA, United States
- 3Division of Gastroenterology, Children's Hospital of Philadelphia, Philadelphia, PA, United States
- 4Department of Pediatrics, Perelman School of Medicine at the University of Pennsylvania, Philadelphia, PA, United States
Two sides of the same coin
“Eosinophilic esophagitis (EoE) is an immune-mediated disease of the esophagus”… is how hundreds of articles, reviews, and clinical guidelines have introduced EoE over the past several decades—and with good reason! There is unequivocal evidence of the immune system's role in EoE well beyond its sine qua non of esophageal eosinophilia (1). Clinically, murine and human studies support that a large proportion of EoE is the result of transepithelial antigen exposure as mice will develop EoE-like inflammation after epidermal sensitization and children with atopic dermatitis are at increased risk of EoE development (2–5). There is also robust molecular evidence of immune involvement in EoE including transcriptomic (6–8) and mechanistic (9, 10) studies of type 2 (T2) inflammatory pathways such as those mediated by IL-5 (11) and IL-13 (12). Most recently, in-depth interrogations of patient T cells have established the role of the adaptive immune system in EoE (13–15). Taken together, this body of work has formed the basis of our understanding of EoE immunopathology and led to the utilization of steroids, T2-targeting biologics, and other immune-modulatory medications as the backbone of EoE therapy (16).
However, key leaders in the field have focused attention on EoE as a disease of the epithelium (17). This perspective is justified by recent data that has identified epithelial cells as critical to EoE pathogenesis. For example, many of the most high-risk disease loci for EoE encode epithelial proteins including calpain 14, thymic stromal lymphopoietin (TSLP), Desmoglein 1, Filaggrin, and STAT6 (12, 17–20—all) of which have been shown to be relevant to epithelial barrier integrity and/or T2 inflammation (2, 21). Furthermore, genetic and functional data establish a primary role for impaired epithelial barrier function in disease susceptibility and pathoetiology (21–24). Additionally, the EoE transcriptome (a set of genes dysregulated in the esophagi of patients with EoE) is enriched in genes that encode for proteins involved in esophageal epithelial cell differentiation (8, 25). Taken together, these studies suggest that the epithelium is more than just a passive respondent to the inflammation of EoE, but rather an active participant.
The Mucosa as a barrier
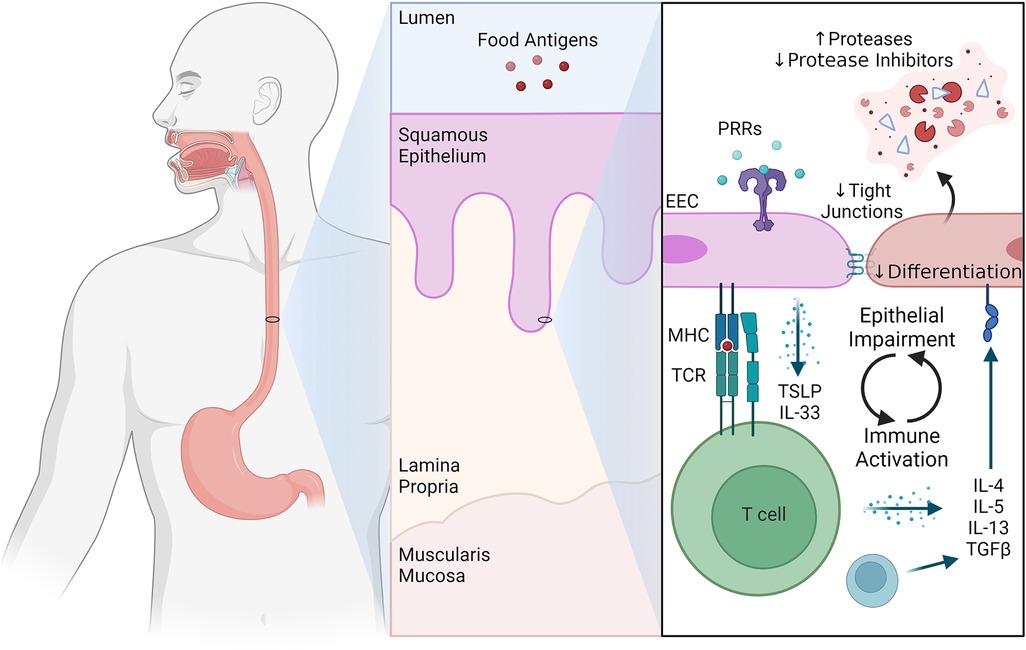
Integrity of the mucosal barrier throughout the gastrointestinal tract is critical as it provides protection against invading microbes and exposure to harmless food proteins. In the esophagus, barrier function is maintained by a stratified epithelium in which differentiation from basal to squamous cells is exquisitely regulated. The epithelium is made up of basal cells (which are positioned atop the lamina propria), a proliferative layer of transit amplifying cells, and more apical layers of increasingly differentiated cells that become anucleate and eventually slough off into the lumen. Molecular evidence in the form of single cell analysis of the active EoE esophageal epithelium has demonstrated a halted differentiation process that persists in remission despite decreased inflammation (8). Further, clinical studies demonstrate that there are disruptions in normal differentiation in EoE with basal cell hyperplasia (BCH), as well as decreased expression of tight junctions, leading to dilated intercellular spaces (26). These changes are attenuated as the inflammation decreases in the setting of disease remission, however, differentiation does not fully normalize (8, 27). Even in the setting of remission, there remains persistent basal cell hyperplasia in 28% of patients with EoE, and patients with persistent basal cell hyperplasia have increased symptomatology compared to those who regain normal differentiation (8, 27). Together, these observations demonstrate that there are primary defects in the EoE epithelium that are independent of the degree of inflammation.
Epithelial cells as immune sentinels
Like epithelial cells elsewhere in the gastrointestinal tract (28), esophageal epithelial cells (EECs) themselves can act as detectors of inflammatory stimuli and directors of inflammatory responses. EECs are ideally located for immunological surveillance as they have the potential to sample food components, commensal and pathogenic microorganisms, and toxins from luminal contents. EECs express several innate pathogen recognition receptors (PRRs) including Toll-like receptors (TLR) (29, 30), NOD-like receptors (31, 32), and G-protein-coupled receptors (33). We know that these pathways are both active and relevant to EEC biology as stimulation of EECs with TLR ligands augments esophageal barrier integrity (30).
There is also emerging evidence that under pathological conditions, EECs (like other gastrointestinal epithelial cells) can act as non-professional antigen presenting cells and modulate adaptive immune responses. For example, EECs from patients with active EoE express major histocompatibility complex (MHC) class II, CD80, and CD86 (34). Notably, both elevated interferon-γ (IFNγ) levels and elevated IFNγ response gene signatures have been detected in active EoE (34, 35). This is relevant as IFNγ is known to induce MHCII expression by non-professional APCs in other settings (36). Indeed, IFNγ stimulation of the human esophageal epithelial line HET-1A increases expression of MHCII, as well as the processing and presentation of ovalbumin and causes T helper cell activation (34).
Finally, EECs can both sense and release cytokines and chemokines to augment their own and the immune system's response to food antigens. For example, loss of tonic regulatory signals, such as TGFβ, can lead to hyperproliferation, failure of differentiation, and overexpression of innate proinflammatory mediators by EECs (37). Further, IL-13 signaling on EECs leads to induction of an EoE-like transcriptional program (25), increased epithelial protease activity, and impaired barrier function (38, 39). EECs are also a critical source of early, innate inflammatory cytokines such as TSLP and IL-33 (2, 40–42), which direct esophageal inflammatory responses (24, 43). Together, these data highlight the central role for EECs, and the epithelium in general, as central modifiers of the mucosal immune response.
The conversation
Understandably, immunologists and epithelial cell biologists have historically focused on their respective areas of expertise when arguing the relative importance of the immune system or the epithelium to EoE pathogenesis. However, the truth is likely somewhere in the middle: that EoE represents the culmination of a complex and dynamic conversation between epithelial cells and the immune system (Figure 1). As an extension, investigations of the immune-epithelial interface in EoE will provide new discoveries that can be exploited therapeutically to enhance beneficial and abrogate pathogenic communication between immune and epithelial cells.
Future research efforts in this space would be best served by collaborative teams of immunologists and epithelial cell biologists that can complement each other to drive new and innovative science at the immune-epithelial interface. Specific areas of focus could include: (1) identification of novel molecules and pathways that mediate the bidirectional crosstalk between epithelial and immune cells; (2) more studies of the role that EECs play in antigen sampling, presentation, and modulation of adaptive immune responses; and (3) longitudinal studies of the immune-epithelial interface to understand how it changes during the transition from acute to chronic disease. In doing so, researchers should consider limitations and biases that can accompany investigations of the esophageal epithelium including that biopsies are taken at random, are limited to the epithelium providing inadequate sampling of lamina propria and muscularis (44), and as a result mostly contain epithelial cells and less fibroblast or nerves which may skew our understanding of the disease etiology. Hopefully, functional evaluations of esophageal distensibility and motility (e.g., functional lumen imaging probe and manometry) will improve our understanding of esophageal dysfunction below the epithelial surface (45). These research efforts will be accelerated by requests for collaborative research proposals by the NIH focused on the immune-epithelial interface of EoE. Ultimately, this line of research has the potential to introduce a new class of EoE-specific therapeutics to the field that can complement immune or epithelial-targeted medications to treat refractory endotypes and improve clinical outcomes.
Author Contributions
DH: Conceptualization, Writing – original – draft, Writing – review & editing; AM: Writing – original – draft, Writing – review & editing.
Funding
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
Acknowledgments
Figure made in Biorender.
Conflict of interest
AM is on the medical advisory board for Regeneron, Bristol Meyers Squib, and Nexstone Immunology and her lab receives research funding from Allakos and Morphic.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abbreviations
EoE, eosinophilic esophagitis; T2, type 2; BCH, basal cell hyperplasia, EECs, esophageal epithelial cells; TSLP, thymic stromal lymphopoietin; MHC, major histocompatibility complex; IFN
References
1. Hill DA, Spergel JM. The immunologic mechanisms of eosinophilic esophagitis. Curr Allergy Asthma Rep. (2016) 16(2):9. doi: 10.1007/s11882-015-0592-3
2. Noti M, Wojno EDT, Kim BS, Siracusa MC, Giacomin PR, Nair MG, et al. Thymic stromal lymphopoietin-elicited basophil responses promote eosinophilic esophagitis. Nat Med. (2013) 19(8):1005–13. doi: 10.1038/nm.3281
3. Akei HS, Mishra A, Blanchard C, Rothenberg ME. Epicutaneous antigen exposure primes for experimental eosinophilic esophagitis in mice. Gastroenterology. (2005) 129(3):985–94. doi: 10.1053/j.gastro.2005.06.027
4. Venturelli N, Lexmond WS, Ohsaki A, Nurko S, Karasuyama H, Fiebiger E, et al. Allergic skin sensitization promotes eosinophilic esophagitis through the IL-33-basophil axis in mice. J Allergy Clin Immunol. (2016) 138(5):1367–80. doi: 10.1016/j.jaci.2016.02.034
5. Hill DA, Grundmeier RW, Ramos M, Spergel JM. Eosinophilic esophagitis is a late manifestation of the allergic march. J Allergy Clin Immunol Pract. (2018) 6(5):1528–33. doi: 10.1016/j.jaip.2018.05.010
6. Blanchard C, Wang N, Stringer KF, Mishra A, Fulkerson PC, Abonia JP, et al. Eotaxin-3 and a uniquely conserved gene-expression profile in eosinophilic esophagitis. J Clin Invest. (2006) 116(2):536–47. doi: 10.1172/JCI26679
7. Sherrill JD, Kiran KC, Blanchard C, Stucke EM, Kemme KA, Collins MH, et al. Analysis and expansion of the eosinophilic esophagitis transcriptome by RNA sequencing. Genes Immun England. (2014) 15(6):361–9. doi: 10.1038/gene.2014.27
8. Rochman M, Wen T, Kotliar M, Dexheimer PJ, Ben-Baruch Morgenstern N, Caldwell JM, et al. Single-cell RNA-Seq of human esophageal epithelium in homeostasis and allergic inflammation. JCI Insight. (2022) 7(11):e159093. doi: 10.1172/jci.insight.159093
9. Yamazaki K, Murray JA, Arora AS, Alexander JA, Smyrk TC, Butterfield JH, et al. Allergen-specific in vitro cytokine production in adult patients with eosinophilic esophagitis. Dig Dis Sci. (2006) 51(11):1934–41. doi: 10.1007/s10620-005-9048-2
10. Blanchard C, Stucke EM, Rodriguez-Jimenez B, Burwinkel K, Collins MH, Ahrens A, et al. A striking local esophageal cytokine expression profile in eosinophilic esophagitis. J Allergy Clin Immunol. (2011) 127(1):208–7. doi: 10.1016/j.jaci.2010.10.039
11. Mishra A, Hogan SP, Brandt EB, Rothenberg ME. IL-5 promotes eosinophil trafficking to the esophagus. J Immunol Baltim Md 1950. (2002) 168(5):2464–9. doi: 10.4049/jimmunol.168.5.2464
12. Blanchard C, Mingler MK, Vicario M, Abonia JP, Wu YY, Lu TX, et al. IL-13 involvement in eosinophilic esophagitis: transcriptome analysis and reversibility with glucocorticoids. J Allergy Clin Immunol. (2007) 120(6):1292–300. doi: 10.1016/j.jaci.2007.10.024
13. Wen T, Aronow BJ, Rochman Y, Rochman M, Kc K, Dexheimer PJ, et al. Single-cell RNA sequencing identifies inflammatory tissue T cells in eosinophilic esophagitis. J Clin Invest. (2019) 129(5):2014–28. doi: 10.1172/JCI125917
14. Dilollo J, Rodríguez-López EM, Wilkey L, Martin EK, Spergel JM, Hill DA. Peripheral markers of allergen-specific immune activation predict clinical allergy in eosinophilic esophagitis. Allergy. (2021) 76(11):3470–78. doi: 10.1111/all.14854
15. Morgan DM, Ruiter B, Smith NP, Tu AA, Monian B, Stone BE, et al. Clonally expanded, GPR15-expressing pathogenic effector TH2 cells are associated with eosinophilic esophagitis. Sci Immunol. (2021) 6(62):eabi5586. doi: 10.1126/sciimmunol.abi5586
16. Dellon ES, Rothenberg ME, Collins MH, Hirano I, Chehade M, Bredenoord AJ, et al. Dupilumab in adults and adolescents with eosinophilic esophagitis. N Engl J Med. (2022) 387(25):2317–30. doi: 10.1056/NEJMoa2205982
17. Rochman M, Azouz NP, Rothenberg ME. Epithelial origin of eosinophilic esophagitis. J Allergy Clin Immunol. (2018) 142(1):10–23. doi: 10.1016/j.jaci.2018.05.008
18. Kottyan LC, Davis BP, Sherrill JD, Liu K, Rochman M, Kaufman K, et al. Genome-wide association analysis of eosinophilic esophagitis provides insight into the tissue specificity of this allergic disease. Nat Genet. (2014) 46(8):895–900. doi: 10.1038/ng.3033
19. Blanchard C, Stucke EM, Burwinkel K, Caldwell JM, Collins MH, Ahrens A, et al. Coordinate interaction between IL-13 and epithelial differentiation cluster genes in eosinophilic esophagitis. J Immunol Baltim Md 1950. (2010) 184(7):4033–41. doi: 10.4049/jimmunol.0903069
20. Kottyan LC, Parameswaran S, Weirauch MT, Rothenberg ME, Martin LJ. The genetic etiology of eosinophilic esophagitis. J Allergy Clin Immunol. (2020) 145(1):9–15. doi: 10.1016/j.jaci.2019.11.013
21. Davis BP, Stucke EM, Khorki ME, Litosh VA, Rymer JK, Rochman M, et al. Eosinophilic esophagitis-linked calpain 14 is an IL-13-induced protease that mediates esophageal epithelial barrier impairment. JCI Insight. (2016) 1(4):e86355. doi: 10.1172/jci.insight.86355
22. Sherrill JD, Kc K, Wu D, Djukic Z, Caldwell JM, Stucke EM, et al. Desmoglein-1 regulates esophageal epithelial barrier function and immune responses in eosinophilic esophagitis. Mucosal Immunol. (2014) 7(3):718–29. doi: 10.1038/mi.2013.90
23. Fahey LM, Chandramouleeswaran PM, Guan S, Benitez AJ, Furuta GT, Aceves SS, et al. Food allergen triggers are increased in children with the TSLP risk allele and eosinophilic esophagitis. Clin Transl Gastroenterol. (2018) 9(3):139. doi: 10.1038/s41424-018-0003-x
24. Doyle AD, Masuda MY, Pyon GC, Luo H, Putikova A, LeSuer WE, et al. Detergent exposure induces epithelial barrier dysfunction and eosinophilic inflammation in the esophagus. Allergy. (2023) 78(1):192–201. doi: 10.1111/all.15457
25. Rochman M, Travers J, Miracle CE, Bedard MC, Wen T, Azouz NP, et al. Profound loss of esophageal tissue differentiation in patients with eosinophilic esophagitis. J Allergy Clin Immunol. (2017) 140(3):738–49. doi: 10.1016/j.jaci.2016.11.042
26. Collins MH, Martin LJ, Alexander ES, Boyd JT, Sheridan R, He H, et al. Newly developed and validated eosinophilic esophagitis histology scoring system and evidence that it outperforms peak eosinophil count for disease diagnosis and monitoring. Dis Esophagus. (2017) 30(3):1–8. doi: 10.1111/dote.12470
27. Whelan KA, Godwin BC, Wilkins B, Elci OU, Benitez A, DeMarshall M, et al. Persistent basal cell hyperplasia is associated with clinical and endoscopic findings in patients with histologically inactive eosinophilic esophagitis. Clin Gastroenterol Hepatol. (2020) 18(7):1475–82. doi: 10.1016/j.cgh.2019.08.055
28. Hill DA, Artis D. Intestinal bacteria and the regulation of immune cell homeostasis. Annu Rev Immunol. (2010) 28:623–67. doi: 10.1146/annurev-immunol-030409-101330
29. Lim DM, Narasimhan S, Michaylira CZ, Wang ML. TLR3-mediated NF-{kappa}B signaling in human esophageal epithelial cells. Am J Physiol Gastrointest Liver Physiol. (2009) 297(6):G1172-1180. doi: 10.1152/ajpgi.00065.2009
30. Ruffner MA, Song L, Maurer K, Shi L, Carroll MC, Wang JX, et al. Toll-like receptor 2 stimulation augments esophageal barrier integrity. Allergy. (2019) 74(12):2449–60. doi: 10.1111/all.13968
31. Uehara A, Fujimoto Y, Fukase K, Takada H. Various human epithelial cells express functional toll-like receptors, NOD1 and NOD2 to produce anti-microbial peptides, but not proinflammatory cytokines. Mol Immunol. (2007) 44(12):3100–11. doi: 10.1016/j.molimm.2007.02.007
32. Nadatani Y, Huo X, Zhang X, Yu C, Cheng E, Zhang Q, et al. NOD-Like Receptor protein 3 inflammasome priming and activation in Barrett's Epithelial cells. Cell Mol Gastroenterol Hepatol. (2016) 2(4):439–53. doi: 10.1016/j.jcmgh.2016.03.006
33. Kleuskens MTA, Haasnoot ML, Herpers BM, van Ampting MTJ, Bredenoord AJ, Garssen J, et al. Butyrate and propionate restore interleukin 13-compromised esophageal epithelial barrier function. Allergy. (2022) 77(5):1510–21. doi: 10.1111/all.15069
34. Mulder DJ, Pooni A, Mak N, Hurlbut DJ, Basta S, Justinich CJ. Antigen presentation and MHC class II expression by human esophageal epithelial cells: role in eosinophilic esophagitis. Am J Pathol. (2011) 178(2):744–53. doi: 10.1016/j.ajpath.2010.10.027
35. Ruffner MA, Hu A, Dilollo J, Benocek K, Shows D, Gluck M, et al. Conserved IFN signature between adult and pediatric eosinophilic esophagitis. J Immunol Baltim Md 1950. (2021) 206(6):1361–71. doi: 10.4049/jimmunol.2000973
36. Stanifer ML, Guo C, Doldan P, Boulant S. Importance of type I and III interferons at respiratory and intestinal barrier surfaces. Front Immunol. (2020) 11:608645. doi: 10.3389/fimmu.2020.608645
37. Laky K, Kinard JL, Li JM, Moore IN, Lack J, Fischer ER, et al. Epithelial-intrinsic defects in TGFβR signaling drive local allergic inflammation manifesting as eosinophilic esophagitis. Sci Immunol. (2023) 8(79):eabp9940. doi: 10.1126/sciimmunol.abp9940
38. D’Mello RJ, Caldwell JM, Azouz NP, Wen T, Sherrill JD, Hogan SP, et al. LRRC31 Is induced by IL-13 and regulates kallikrein expression and barrier function in the esophageal epithelium. Mucosal Immunol. (2016) 9(3):744–56. doi: 10.1038/mi.2015.98
39. Kc K, Rothenberg ME, Sherrill JD. In vitro model for studying esophageal epithelial differentiation and allergic inflammatory responses identifies keratin involvement in eosinophilic esophagitis. PloS One. (2015) 10(6):e0127755. doi: 10.1371/journal.pone.0127755
40. Judd L.M., Heine R.G., Menheniott T.R., Buzzelli J., O'Brien-Simpson N., Pavlic D., O'Connor L., Al Gazali K., Hamilton O., Scurr M. and Collison A.M., et al. Elevated IL-33 expression is associated with pediatric eosinophilic esophagitis, and exogenous IL-33 promotes eosinophilic esophagitis development in mice. Am J Physiol Gastrointest Liver Physiol. (2016) 310(1):G13–25. doi: 10.1152/ajpgi.00290.2015 (Cited 30 July, 2023).26514775
41. Travers J, Rochman M, Caldwell JM, Besse JA, Miracle CE, Rothenberg ME. IL-33 is induced in undifferentiated, non-dividing esophageal epithelial cells in eosinophilic esophagitis. Sci Rep. (2017) 7(1):17563. doi: 10.1038/s41598-017-17541-5
42. Azouz NP, Ynga-Durand MA, Caldwell JM, Jain A, Rochman M, Fischesser DM, et al. The antiprotease SPINK7 serves as an inhibitory checkpoint for esophageal epithelial inflammatory responses. Sci Transl Med. (2018) 10(444):eaap9736. doi: 10.1126/scitranslmed.aap9736
43. Doherty TA, Baum R, Newbury RO, Yang T, Dohil R, Aquino M, et al. Group 2 innate lymphocytes (ILC2) are enriched in active eosinophilic esophagitis. J Allergy Clin Immunol. (2015) 136(3):792–4. doi: 10.1016/j.jaci.2015.05.048
44. Wang J, Park JY, Huang R, Souza RF, Spechler SJ, Cheng E. Obtaining adequate lamina propria for subepithelial fibrosis evaluation in pediatric eosinophilic esophagitis. Gastrointest Endosc. (2018) 87(5):1207–14. doi: 10.1016/j.gie.2017.12.020
Keywords: eosinophilic esophagitis, esophageal epithelial cells, epithelium, food allergy, type 2 inflammation
Citation: Hill DA and Muir AB (2023) The immune-epithelial interface in eosinophilic esophagitis: a conversation. Front. Allergy 4:1270581. doi: 10.3389/falgy.2023.1270581
Received: 1 August 2023; Accepted: 19 September 2023;
Published: 3 October 2023.
Edited by:
Quan M. Nhu, The Scripps Research Institute, United StatesReviewed by:
Nurit P. Azouz, Cincinnati Children's Hospital Medical Center, United StatesMargaret H. Collins, Cincinnati Children's Hospital Medical Center, United States
© 2023 Hill and Muir. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: David A. Hill aGlsbGQzQGNob3AuZWR1 Amanda B. Muir bXVpcmFAY2hvcC5lZHU=
 David A. Hill
David A. Hill Amanda B. Muir3,4*
Amanda B. Muir3,4*