- 1APC Microbiome Ireland, University College Cork, Cork, Ireland
- 2Division of Dermatology, University of Cape Town, Cape Town, South Africa
- 3Department of Peadiatrics, University of Cape Town, Cape Town, South Africa
- 4Division of Otorhinolaryngology, University of Witwatersrand, Johannesburg, South Africa
- 5Department of Medicine, University College Cork, Cork, Ireland
- 6School of Microbiology, University College Cork, Cork, Ireland
Atopic dermatitis is a complex inflammatory condition characterized by synergist interactions between epidermal and immune related genotypes, skin barrier defects and immune dysregulation as well as microbial dysbiosis. Ethnicity-specific variations in clinical presentation, immune endotypes and genetic susceptibility have been described in diverse populations. We summarize available data with specific consideration of AD in populations of African ancestry. Some highlights include the observation of AD lesions on extensor surfaces, lichen planus-like AD, prurigo type AD and follicular AD in African populations. In addition, a consistent absence of dominant filaggrin gene defects has been reported. The detection of normal filaggrin protein content in AD skin implicates the contribution of alternative mechanisms in the pathogenesis of AD in African patients. Markedly high IgE has been described in paediatric and adult African AD. While Th2, Th22 and Th17 activation in African AD skin shares the same direction as with other populations, it has been noted that the magnitude of activation is dissimilar. Reduced Th17 cytokines have been observed in the circulation of moderate to severe paediatric AD.
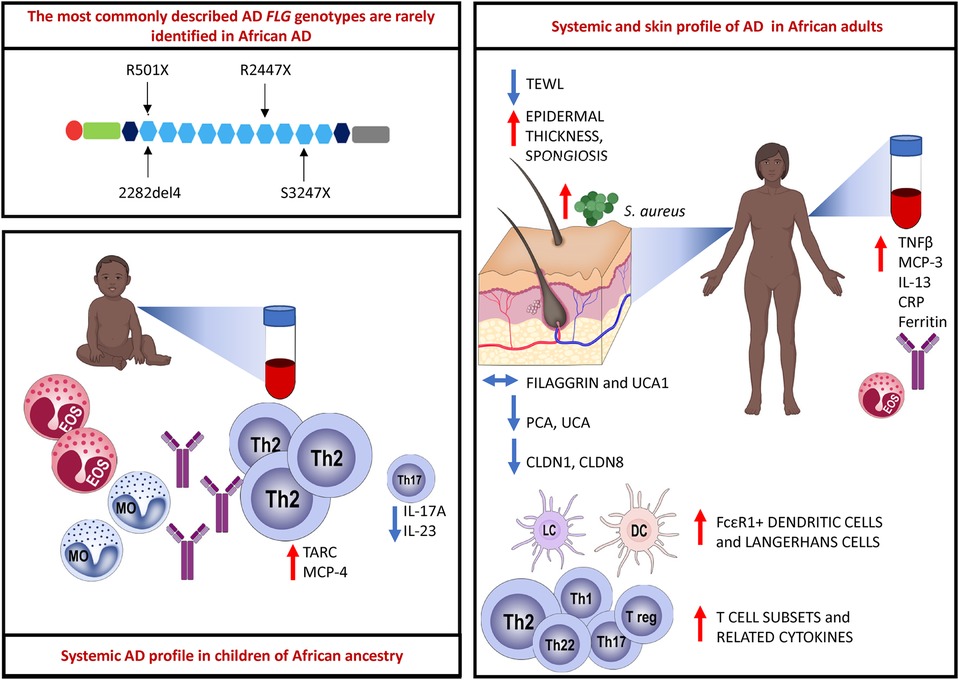
Graphical Abstract. The most commonly described FLG loss-of-function (LoF) mutations located in the epidermal differentiation complex (EDC) on chromosome 1q21 are rarely identified in African AD. Paediatric AD is characterized by an abundant Th2-related cytokine milieu, and reduced Th17-related cytokines have been observed. Eosinophils, monocytes and total and specific IgE are elevated in the circulation of children with AD. There is a multipolar cytokine profile in adult AD with increased eosinophils and total IgE also noted. Adult AD skin biopsy samples show increased epidermal thickness and spongiosis. The content of filaggrin protein and filaggrin breakdown products such as urocanic acid-1 (UCA1) are unchanged in African AD skin. Filaggrin breakdown products such as pyrrolidone-5-carboxylic acid (PCA) and urocanic acid (UCA) are reduced. Tight junction proteins such as claudin-1 (CLDN1) and claudin-8 (CLDN8) expression is reduced in AD in adult Africans. There is increased influx of FcεR1 bearing dendritic cells and Langerhans cells as well as increased expression of diverse T cell subsets and related cytokines. Created with BioRender.com.
Introduction
Atopic dermatitis (AD) is a highly complex multifactorial inflammatory skin disease that follows a relapsing-remitting chronic course (1). AD disproportionately affects children globally (2) and its incidence has increased rapidly during recent years in African countries (3, 4). Current data from South Africa as an example suggests that the changing diversity of environmental exposures, socioeconomic factors, diet, and lifestyles across African societies might provide unique disease promoting effects in early life, in populations where a low risk of developing AD and allergic disease was previously documented (4).
Features of atopic dermatitis
The hallmark features of AD are well established. It is characterized by intense pruritus (2). Notable skin lesions at predilection sites such as the flexural surfaces include xerosis, erythema, oedema, excoriations, oozing, crusting and lichenification. A wide spectrum of clinical AD phenotypes that vary with age and ethnicity are also key clinical features (5). Atopic dermatitis is thought to arise from gene-gene interactions and gene-environment interactions that impact the epidermal barrier and host immunity (6, 7). The increasing prevalence of the disease supports the role of environmental factors interacting with host immune responses, microbial composition, and epidermal barrier factors in the pathogenesis of AD (6, 7).
Pruritus responses may be more intense in patients of African descent (8). Larger mast cell granules and differences in the subgranular structure, and localization of the proteases, tryptase and cathepsin G in the mast cells of African individuals compared to Europeans, may account for this (9). Other mechanisms of itch may account for differences in itch perception among different ethnicities however these have not been adequately tested (8, 9).
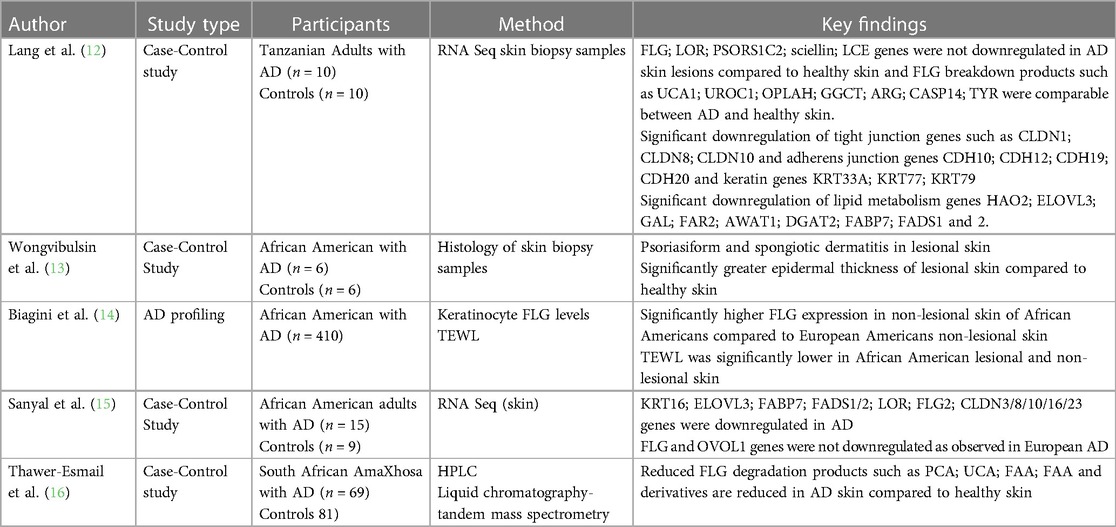
Morphological variations in AD phenotype have been noted with increased frequency in individuals of African and Asian ancestry (5, 7). AD patients of Asian ancestry may present with parakeratosis which is the incomplete maturation of keratinocytes and the retention of nuclei in the stratum corneum (10). It is an atypical morphological feature of AD and is more consistent with psoriasis (10). It has been observed that African individuals may have a pronounced treatment-resistant lichenified type and often present with severe forms of AD (Figure 1) (11). The follicular type variant characterised by densely aggregated follicular papules, has been described in darkly pigmented skin (Figure 3B) (5, 7, 9). Long-standing disease may present with prurigo AD type (Figure 3C) (8). Perifollicular accentuation, papulation, scaling, lichenification, and pigmentary changes may also be more prominent in patients with darker skin (8). Transepidermal water loss (TEWL) is notably higher in individuals of African descent compared to European descent due to the lower ceramide content in the skin in addition to several other more granular phenotypic characteristics (Table 1) (8, 12–16). Clinical variations in AD phenotypes are also age-dependent (Figures 1–3) (5, 11).
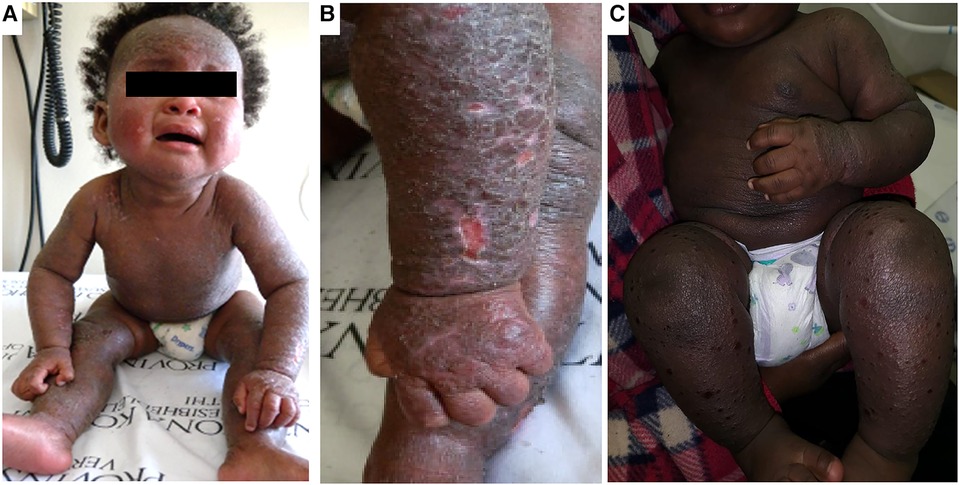
Figure 1. Clinical aspects of atopic dermatitis in infancy. Panels (A,B) shows an infant with marked lichenification of the skin with erythroderma and hyper accentuation of skin markings. There are also areas of hyperpigmentation. Panel (C) shows another infant with extensive erythroderma and well demarcated shallow ulcerative lesions of eczema coxsackium.
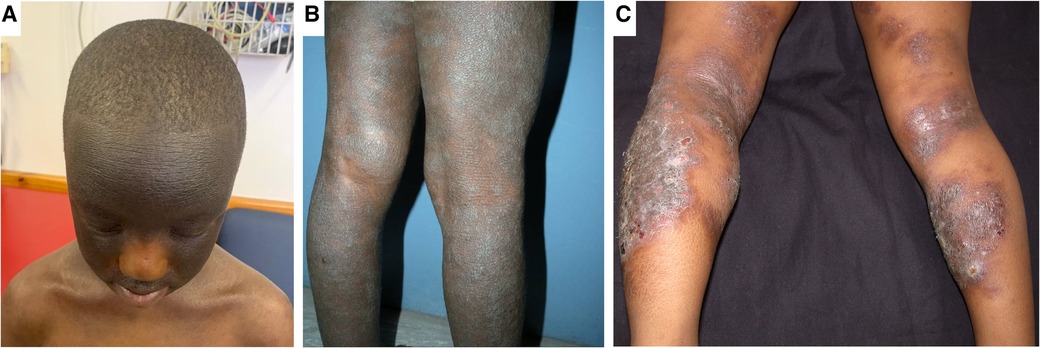
Figure 2. Clinical aspects of atopic dermatitis in childhood. Panel (A) shows marked hyperpigmentation of the face and hyper accentuation of skin markings with sparing of the central face. This patient does not fully typify the central facial sparing as the sparing is limited to the nose. Panel (B) shows lichenification, in addition to hyperpigmentation and dryness of the legs. Panel (C) shows eczematous plaques of eczema in both extensors and flexors with superimposed crusting in an older child.
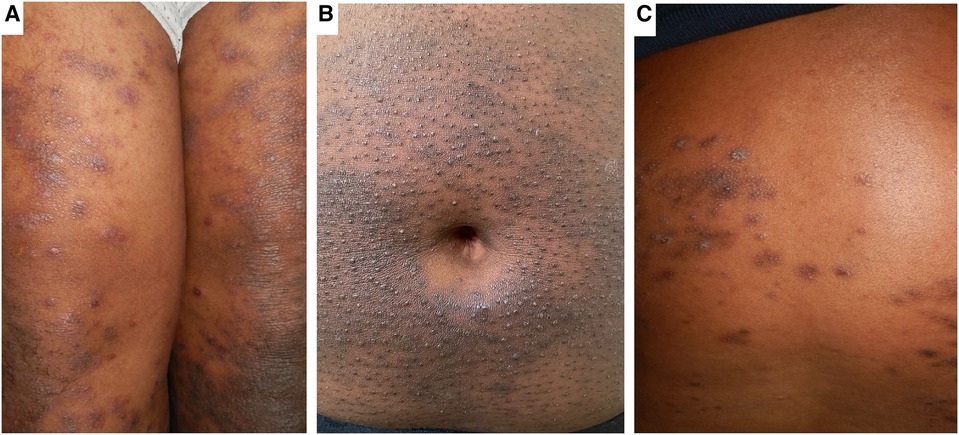
Figure 3. Clinical phenotypes of AD in African populations. Panel (A) shows Lichen-Planus like AD with hyperpigmented flat topped papules with shiny surface distributed over the thighs. Panel (B) shows hyperpigmentation and densely aggregated follicular papules (folliculocentric) characteristic of the follicular AD phenotype on the abdominal wall. Panel (C) shows the prurigo-like AD phenotype characterised by dome shaped papules with hyperpigmented areas.
Diagnostic criteria and disease severity scoring of AD
Clinical AD diagnostic criteria have not been adequately evaluated in African populations in the developed and developing settings (17). The Hanifin and Rajka criteria is the earliest formal diagnostic criteria (18). It has been distilled into the UK Working Party diagnostic criteria which is widely used especially in epidemiology research (17). However, the UK Working Party was a poor predictor of AD in a community-based validation study in South Africa suggesting that it may not accurately identify cases of African AD (19). There is a general consensus amongst key stakeholders in AD care that AD severity scoring indices are important tools in AD management (20). However, these indices or outcomes measures also have limited application in darkly pigmented skin as they are reliant on the presence of erythema (21). Erythema is not readily appreciable in darkly pigmented skin, and this leads to gross underestimation of disease activity and severity (21, 22). This situation is not mitigated by assessments conducted by well experienced dermatologists who predominantly treat patients with darkly pigmented skin types (23). Recently the Patient-Oriented SCORing Atopic Dermatitis (PO-SCORAD) has been validated for evaluating AD severity in a multicenter study conducted in seven African countries (24). This subjective tool is useful for monitoring disease activity between physician consultations and guiding treatment strategies (24). Nevertheless, objective biomarkers of disease and disease severity are desirable to accurately diagnose, guide clinical management and monitor response to novel treatments in clinical trials.
Prevalence of atopic dermatitis
This debilitating dermatosis affects up to 30% of children and up to 10% of adults throughout the world (2). 1%–3% of the elderly population in industrialised countries has AD with a 2:1 male predominance observed (25). Globally the prevalence of AD has been on the rise parallel to industrialization with 2–3-fold increase over the past few decades (1). The increasing trend continues in low-middle income countries (1). Onset of disease is commonly between 3 and 6 months of age (2). The International Study of Asthma and Allergies in Childhood (ISAAC) produced the most robust global epidemiology data for children and adolescents (3). ISAAC revealed that the prevalence of AD varies greatly throughout the world with the highest prevalence (15%) of AD in both 6–7-year-olds and 13–14-year-olds reported for urban Africa, the Baltics, Australasia, and Northern and Western Europe (3). An overall female: male ratio of 1.3:1 was also reported, consistent with findings elsewhere (3).
The high urban prevalence is consistent with observations made in South Africa showing an urban-rural gradient characterized by high allergic sensitization and allergic disease in urban settings compared to rural communities of the same ethno-linguistic background (4). A growing number of reports even dating back to the mid-90s already identified a higher prevalence of AD in populations of African ancestry in the UK (26). More recent data, from the US and UK also report an increasing prevalence and a significantly higher risk of severe forms of AD and asthma in individuals of African ancestry compared with those of European descent (22, 26–30). Based on ISAAC phase I and III data AD prevalence in Latin America ranges from 4% to 25% (3). A positive association between African genetic admixture (independent of socioeconomic factors) and atopy and asthma has been noted in some Latin American countries where African ancestry make up a significant composition of the population (31).
Environmental risk factors
The increased prevalence of AD in developed or urbanised settings highlights the role of the environmental exposures in altering disease risk (1). A wealth of literature has drawn parallels between the increase in respiratory allergy and environmental factors specifically ambient air pollutants associated with urbanization (32). However, fewer studies examined the role of environmental pollutants on allergic disease in the skin. Some of these show that air pollution, characteristic of urbanization, positively associates with AD prevalence (33, 34). Prenatal exposure to certain air pollutants may link to allergy development through dysregulated immune responses mediated by microRNA and DNA methylation (34). Birth cohort studies suggest that indoor and outdoor air pollution associates with AD development (33, 35). Longitudinal studies also suggest that various air pollutants associate with AD exacerbations and disease persistence (34). Environmental exposures such as polluted air and maternal cigarette smoking have been shown to exert their effects on the epigenome to influence AD risk (34, 35). The role of environmental pollutants in AD pathogenesis is supported by the higher prevalence of disease in urban settings. Children living in urban centres of Latin America as an example, have been shown to have a much higher prevalence of AD than their rural counterparts (36). Traffic related air pollutants (TRAP), volatile organic compounds (VOCs) and second-hand smoke (SHS) exposure increases the risk of atopic dermatitis and other atopic diseases in children including asthma (32–38). These pollutants induce oxidative stress to protein and lipids resulting in epidermal barrier disruption and triggering an inflammatory cascade (32, 34, 35, 39). Itching and scratching is also elicited following oxidative stress in animal models (40). Environmental oxidants such as air pollutants and solar radiation were shown to cause oxidative protein damage in the stratum corneum (41). It is anticipated that this scenario can be much worse in regions that experience more sunlight and high temperatures which transform air pollutants and particulate matter. Increased IL-4 producing T cells were observed in children following VOC exposure (42). Exposure to tobacco smoke was associated with hypomethylation of thymic stromal lymphopoietin (TSLP) 5′ CpG island leading to increased TSLP expression and AD development (43). The impact of these environmental exposures and others on allergy development is complex and variable, likely reflecting differences in timing and duration of exposure, dose of exposure and host genetic and immune-mediated susceptibility factors.
Research within South Africa demonstrates a lower overall prevalence of atopy in rural areas compared with urban areas over the decades (4), which highlights the importance of environmental factors on allergy risk or protection, but this rural-urban gradient is rapidly diminishing over time (4). While allergy rates have been shown to be different between South African rural and urban settings, allergic diseases still do arise in both these diverse environments. The farming environment has been shown to have generally protective effects in other parts of the world, potentially due to exposure to a more microbially diverse environment (44–46). In addition, a recent intervention study in an urban daycare centre modified its outdoor environment to include sod, forest floor segments and planters. They observed modification in skin and gut microbial composition which coincided with an increased tolerogenic immune profile (47).
Genetic risk factors
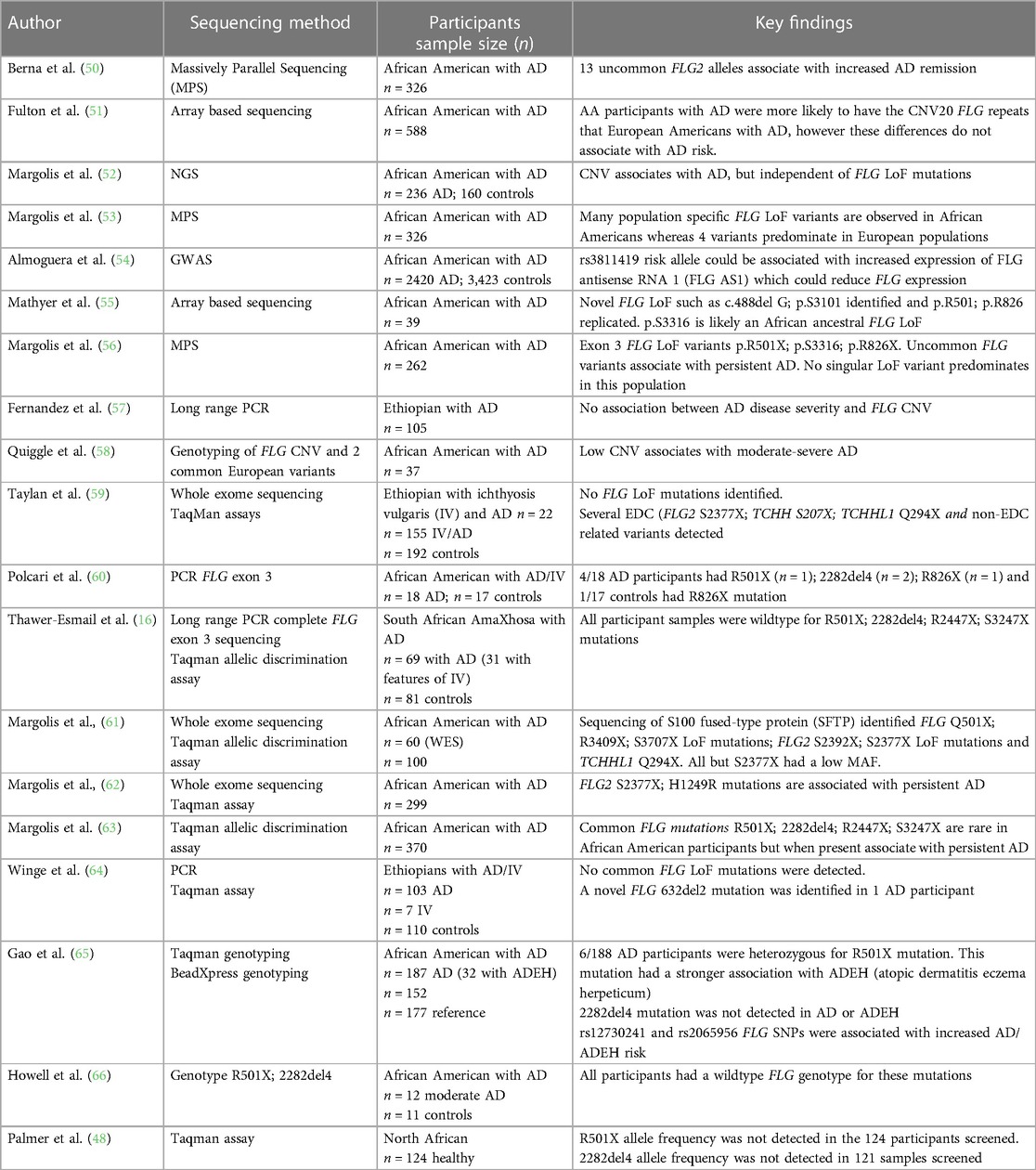
Discovery of the association between filaggrin gene mutations with epidermal disruption in AD has improved our understanding of the disease (48). However, this genotype plays a role only in a minority of patients. The important role of loss-of-function (LoF) mutations and copy number variations in the FLG gene located within the epidermal differentiation complex (EDC) on chromosome 1q21 and AD risk was first documented in 2006 (48). However, FLG mutations are not uniformly distributed globally. p.R501X and c.2282del4 are commonly described variants in European populations (49) and have not been shown to predominate in African populations (Table 2) (16, 48, 50–66).
Recent studies have suggested that polymorphisms in other barrier associated genes, such as Filaggrin-2 gene variants (62) and uncommon FLG exon 3 loss-of function (LoF) variants have been described in a minority of individuals of African ancestry and are thought to collectively confer a more persistent disease phenotype (56). Ethno-specific mutations have been reported in Japanese, Chinese, Korean and Bangladeshi populations that associate with specific phenotypes such as dryness (67–69). In particular the c.3321delA is specific to Asian population and is the most common variant in the Chinese Han population (69). A few studies have been conducted but were unable to reproduce a strong AD link to these specific variants in individuals of African ancestry (Table 2) (16, 48, 50–66).
FLG intragenic copy number variation (CNV) impact gene dosage and consequently the amount of filaggrin produced in the epidermis (70). CNV has been shown to associate with AD risk in a large study of Irish paediatric AD (70). whereas CNV in Ethiopian AD was not associated with AD disease severity (57). CNV vary with ethnicity however this difference did not associate with AD risk in a study including African American and European American participants (58). Thus far, to our knowledge, CNV have not been sufficiently studied in African ancestry and have not shown an association with AD in the few studies reported on African populations (57, 58).
Genome-wide association studies (GWAS) analysis identified over 30 further genetic risk loci including FLG (71). Notably, IL-13 also demonstrated the most significant association with AD among the susceptible gene loci identified. In addition, some of the susceptibility loci overlap with known asthma risk loci, highlighting shared “atopy” risk. Novel risk loci identified were related to immune regulation (71). Other AD associated polymorphisms occur in genes encoding elements of innate immune signaling e.g., TLR-2 and TLR-4 as well as T-cell immunity (72). Variants of Th2 cytokine genes (IL-4; IL-5; IL-9; IL-13 including RAD50) found on chromosome 5q31-33 associating with AD have been described but need confirmation in large studies (73–75). These observations underscore the important role of immune mediated pathogenesis of AD. This accumulating evidence supports the robust contribution of factors other than epidermal barrier defects in the pathogenesis of AD, such as a Th2 predominant multipolar immune activation as this can deplete epidermal barrier proteins and tight junction protein expression (75–79).
Immune mechanisms in AD
Innate and adaptive immune responses contribute to AD pathogenesis. Polymorphisms in Pattern Recognition Receptors (PRRs) such as Toll-Like Receptor 2 (TLR2) - the main receptor responsible for recognizing staphylococcal ligands, Nucleotide - binding oligomerization domains 1 (NOD1) and NOD2 are associated with AD in a way that enables S. aureus overgrowth (80). Low levels of CD14, a multifunctional receptor that induces innate immune cell activation via TLR dependent pathway have been reported in atopic children (81). Keratinocytes, sebocytes, eccrine glands and mast cells contribute to innate immune function by producing antimicrobial peptides (AMPs) such as cathelicidin (LL-37), human beta defensins (hBD2; hBD3) and dermcidin to protect the host from infection (82, 83). Levels of multiple AMPs are reduced in AD patients and have defective antimicrobial function. This likely enables S. aureus colonization as it has been observed that over 90% of AD patients exhibit S. aureus colonization in lesional and non-lesional skin (44).
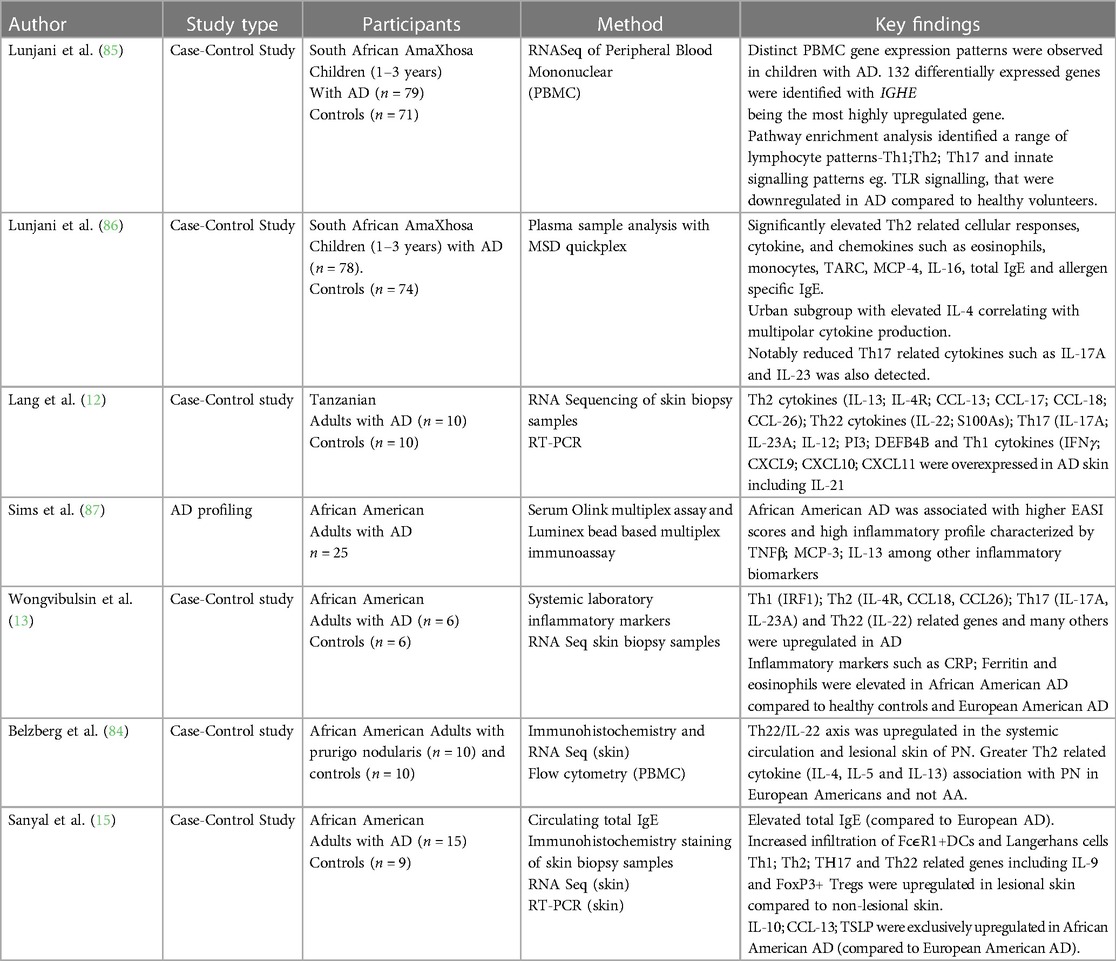
T cell infiltration is well documented in AD and accompanies the histological features of spongiosis (7). In recent years there has been a growing appreciation that in addition to dominant Th2 immune responses, there is multipolar immune activation that include Th1 (IFNγ), Th17 (IL-17A; IL-23) and Th22 (IL-22) activation (7). These immune pathways correspond with particular features (5, 10, 11). Th1 activation has been described in chronic AD lesions, Th17 activation has been described as feature of Asian AD that corresponds with psoriasiform features in AD, Th22 activation has been demonstrated in epidermal hyperplasia of lichenification (5, 10, 11). Interestingly, prurigo is often a co-occurring phenotype in AD especially in African AD, however the immune activation of prurigo nodularis in African skin feature Th22 immune responses with no Th2 activation-a departure from prurigo observed in European patients (84). There is a dearth of literature providing detailed data on immune mechanisms of AD in populations of African ancestry globally, nevertheless a handful of studies in African adults and children provide some evidence of differential host immune responses in this disease phenotype (Table 3) (12, 13, 15, 84–87). It is important to describe immune pathways activated in AD patients of African ancestry to determine the potential efficacy of currently available biologics targeting specific pathway in this demographic.
Atopic dermatitis and the Th2 axis
AD is often associated with elevated serum immunoglobulin (IgE) and considered a foreshadow of the atopic diathesis - IgE-mediated food allergy, asthma and allergic rhinitis (6). Moderate to severe AD carries a higher risk of developing atopic co-morbidities. About 20%–30% of infants follow this disease course. Additionally, AD susceptibility loci increase the risk of the atopic march (88). Notably this group of children did not outgrow AD. It has recently been reported that children with AD who progress along the atopic march have a 2.7-fold increased risk of neurodevelopmental disorders (89). African populations are thought to represent a high IgE producer genotype (90–93). There is a strong association between IgE mediated sensitization to food allergens, aeroallergens and AD but the functional mechanisms of IgE in AD needs further clarification (2, 94). South African children for example have similar IgE-mediated food allergy rates as described in industrialized countries (4) however it has been noted that African children display population specific IgE sensitization characterized by large positive food allergen skin prick test (SPT) wheals and high ImmunoCAP IgE levels with no clinically relevant food allergy (i.e., Allergen tolerance) and distinct atopic march trajectories have been observed in children of African ancestry (95–98). These population-specific IgE-related observations have been confirmed in children with low detection of cross-reactive carbohydrate determinants (CCD) (85, 86) in direct contrast to the common argument that high total and specific IgE in the tropics/subtropics reflect endemic helminth infection. Furthermore African-specific allele frequencies that amplify Th2 immune responses have been identified (91–93). This may underpin the increased allergy risk. IL-10 and IL-4 polymorphisms that associate with allergic disease in African children have also been observed (92).
Recently, we described an AD immune endotype in African children with moderate to severe AD that was characterized by high circulating Th2 related cytokines and chemokines TARC, MCP-4, IL-16 and low Th17 related cytokines IL-17A and IL-23 (86). Sanyal et al. described high amplitude of total IgE in adult African Americans compared to European Americans (15). Furthermore, they noted that Th17 related cytokines were elevated in the skin of African American AD compared to African American healthy counterparts but not to the same magnitude as European AD. Differences in paediatric and adult AD have also been described, mainly characterized by similar polarization of Th2 cells (5). IL22/Th22 expression and Th17 subsets, which contribute to disease chronicity, exhibited lower expression in children compared to adults. Absence of Th1 activation in paediatric AD compared to adult AD has also been documented (5, 11).
The atopic march
The atopic march refers to the sequential progression to allergic rhinitis, food allergy or asthma with AD as the entry point. It is thought to arise from shared atopic genetic predisposition, immunological responses, and environmental risk factors (6). In an American study of 18,596 AD participants where 51% were African American, it was observed that African American children had an atopic trajectory that was characterized by progression from AD to asthma whereas European American children progressed from AD to allergic rhinitis and the AD -IgE mediated food allergy trajectory was predominant amongst Asian children (98). A smaller study (n = 601) reproduced the “AD to asthma” atopic trajectory observed in African American children (14). Furthermore, they determined that the observed asthma risk in African American children was six times higher than European American children. This was associated with higher reports of parental AD, parental asthma and increased exposure to environmental pollutants such as secondhand smoke and traffic related air pollution (14, 98). Region-specific variations in allergic disease presentation have been reported. Colombian researchers have observed that the clinical presentation of allergy in their region is not consistent with the putative “atopic march”, but the natural history of allergy is skewed towards a debut with respiratory symptoms (99). This is possibly influenced by the tropical climate, which supports high mold, cockroach, and house dust mite allergen load in humid conditions. It is important to note that the Colombian population has a significant population of African descent, and this genetic background may have distinct clinical presentation in this regional environment (25). It has also been noted that urbanization in developing countries often occurs in the context of economic deprivation and this promotes exposure to noxious substances that might influence immune function and modify allergy susceptibility (25, 99).
Microbiome disruption in AD
Diverse communities of commensal microbes on internal and external body surfaces play essential roles in regulation of host metabolic responses, epithelial barrier function, immune education, and immune regulation. Within the gut, individual microbes, microbial components and individual metabolites (e.g., short-chain fatty acids, histamine) are continuously being newly described that influence host immune responses, including those that impact disease processes within the skin (100–102). Associations between AD risk and gut microbiota composition and metabolism are evident across multiple studies (103), but it is still unknown if gut microbiota alterations are a cause or consequence of AD. Importantly, different microbial strains can dominate in different populations across the world, with some microbial populations now absent from industrialised populations (104). Indeed, there is evidence that immunologically relevant gut microbes such as Prevotella copri, Eubacterium rectale, and Bifidobacterium longum codiversified with humans, but it is not known how these variations in functional capacity might impact AD risk or severity in different populations (105). Population specific dietary habits can indirectly influence immune responses via the gut microbiota, as microbial fermentation generates metabolites with immune modulatory effects (106). Consumption of a higher diversity of fruits, vegetables and fermented foods were associated with a reduced risk of atopic disorders and asthma in children, potentially mediated in part by microbial-derived butyrate and propionate (107). However, the specific plant-based substrates (e.g., fiber type, fatty acids, polyphenols) that are responsible for these positive associations are unknown. It is hypothesized that the transition to reduced diversity, low-quality, highly processed, high-energy diets typical of urban or industralised populations have altered the metacommunity, its processes that underpin assembly and activity of the human microbiome and, consequently, increased risk of inappropriate and uncontrolled immune responses that damage host tissues and function (108). Population specific dietary differences may underpin many of the microbial changes in AD, but these links remain to be proven.
The skin microbiome is also an important component in AD pathogenesis (44). Pathogenic organisms such as Staphylococcus aureus and certain strains of Malassezzia species have been associated with AD and disease severity. Lifestyle factors, UV exposure, climate, age, gender, ethnicity and skin site influence the composition of the skin microbiome (109). Differences in skin microbiome composition in AD as it relates to different ethnicities and different regions that may have distinct microbial exposures have not been well documented. Due to the close relationship and bidirectional interaction between host immunity and skin microbiome, it is plausible that differences may exist. Single nucleotide polymorphisms in human beta defensins (hβDs) have been described in different ethnicities including African ethnicity (110). This may account for differences in colonization with pathogenic organisms as hβDs are important host antimicrobial peptides that are first line innate immune responses (80, 82). S. aureus genotyping showed differential distribution in different American ethnicities. S. aureus strains lacking the virulence factor gene encoding Toxic Shock Syndrome Toxin 1 (TSST-1) were detected in African Americans with AD compared to European Americans and Mexican Americans with AD (86). This may be a function of host immunity. Significant temporal shifts in S. aureus superantigen prevalence were also notable possibly in keeping with S. aureus strains cycling through the population approximately every ten years (111). The S. aureus genes lukE, lukD, splA, splB, ssl8, and sasG were more frequent in isolates from AD patients compared to controls in one study, but it's unknown if these molecular signatures are also present in isolates from African populations (112). Given these considerations it would be valuable to monitor S. aureus profiles in AD patients in different regions.
Health disparities
Several studies have shown the association of structural racism and disproportionally high allergy prevalence in marginalized communities. In a Detroit based study in a diverse demographic of children, African American children had significantly higher allergic outcomes such as IgE sensitization to at least one food/aeroallergen skin prick test; elevated serum IgE levels; presence of atopic dermatitis and reported symptoms of wheeze compared to European Americans (113). In a study of African American paediatric AD, residential segregation as a marker of structural racism was associated with AD severity (114), which is not dissimilar to observation in South Africa, where high asthma rates were documented in marginalized communities living in heavily industrialized settings in keeping with apartheid spatial planning (115). Notably, in this study, estimated particulate matter concentrations were more than 6 times above the USA National Ambient air quality standard. Although AD was not assessed as an outcome, given the shared pathogenic environmental risk factors it is plausible that the skin could be similarly affected.
Future perspectives
In addition to systemic immunosuppressive drugs such as methotrexate, azathioprine, cyclosporin used in moderate to severe disease, novel therapies may be useful in severe disease forms and where there are multiple comorbid allergic conditions. Recent advances in biologics for AD have ushered in the era of precision medicine owing to a more granular understanding of immune mechanisms of disease. Modulation of appropriately targeted immune mechanisms relevant to specific patients holds the most promise for AD therapies for all AD sufferers. Indeed, African populations may stand to benefit the most from precision medicine particularly in clinically heterogenous conditions like AD as Africans are the most genetically diverse population in the world (116).
In addition, within the context of pharmacogenetics biomedical variation in drug response in African populations have also been documented (116). Under the current state-of-the-art, precision medicine efforts, particularly the burgeoning use of biologics may be less well-targeted in individuals of African ancestry, as they are not well represented in the experimental data and clinical studies underlying these initiatives. There is a growing appreciation of population/ethnicity-specific disease phenotypes and corresponding immune endotypes that may additionally be unique to certain geographies and early life exposures.
Collectively this data suggests that the role of genetic background generally needs careful consideration in terms of clinical presentation of disease, diagnostic criteria, disease mechanisms and indeed treatment responses and this consideration is relevant in AD and allergic disease.
Unprecedented urbanization and urban migration are ongoing throughout the African continent and will represent the fastest urban growth rate in the world. It is estimated that the population of Africa will double by the year 2050 with two thirds of this growth concentrated in urban areas. It has been noted that 70 per cent of Africans live in urban informal settlements (117). Therefore, it will be important to study AD and allergy development as well as healthy immune development in the African urban and rural settings; to document the influence of local and specific environmental exposures, identify interventions that may mitigate against adverse health outcomes related to living environment and anticipate future healthcare needs. In addition, clarifying the contribution of genetic and epigenetic susceptibility, immune responses, socio-economic status, environmental factors, and their collective interactions to account for the disparate observations in African populations is needed.
Ethics statement
Written informed consent was obtained from the minor's legal guardian for the publication of any potentially identifiable images or data included in this article.
Author contributions
NL contributed to the conceptualization, writing and revision of the manuscript. TK contributed to the writing and revision of the manuscript. FM contributed to the writing and revision of the manuscript. CH contributed to the conceptualization, writing and revision of the manuscript as well as providing educational photographs. LO’M contributed to the conceptualization, writing and revisions of the manuscript. All authors contributed to the article and approved the submitted version.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Nutten S. Atopic dermatitis: global epidemiology and risk factors. Ann Nutr Metab. (2015) 66(Suppl 1):8–16. doi: 10.1159/000370220
3. Williams H, Robertson C, Stewart A, Aït-Khaled N, Anabwani G, Anderson R, et al. Worldwide variations in the prevalence of symptoms of atopic eczema in the international study of asthma and allergies in childhood. J Allergy Clin Immunol. (1999) 103(1):125–38. doi: 10.1016/S0091-6749(99)70536-1
4. Botha M, Basera W, Facey-Thomas HE, Gaunt B, Gray CL, Ramjith J, et al. Rural and urban food allergy prevalence from the South African food allergy (SAFFA) study. J Allergy Clin Immunol. (2019) 143(2):662–8.e2. doi: 10.1016/j.jaci.2018.07.023
5. Leung D. Atopic dermatitis: age and race do matter!. J Allergy Clin Immunol. (2015) 136(5):1265–7. doi: 10.1016/j.jaci.2015.09.011
6. Weidinger S, Novak N. Atopic dermatitis. Lancet. (2016) 387(10023):1109–22. doi: 10.1016/S0140-6736(15)00149-X
7. Langan S, Irvine A, Weidinger S. Atopic dermatitis. Lancet. (2020) 396(10247):345–60. doi: 10.1016/S0140-6736(20)31286-1
8. Vachiramon V, Tey HL, Thompson AE, Yosipovitch G. Atopic dermatitis in African American children: addressing unmet needs of a common disease. Pediatr Dermatol. (2012) 29(4):395–402. doi: 10.1111/j.1525-1470.2012.01740.x
9. Sueki H, Whitaker-Menezes D, Kligman AM. Structural diversity of mast cell granules in black and white skin. Br J Dermatol. (2001) 144(1):85–93. doi: 10.1046/j.1365-2133.2001.03957.x
10. Noda S, Suárez-Fariñas M, Ungar B, Kim SJ, de Guzman Strong C, Xu H, et al. The Asian atopic dermatitis phenotype combines features of atopic dermatitis and psoriasis with increased TH17 polarization. J Allergy Clin Immunol. (2015) 136(5):1254–64. doi: 10.1016/j.jaci.2015.08.015
11. Czarnowicki T, He H, Krueger JG, Guttman-Yassky E. Atopic dermatitis endotypes and implications for targeted therapeutics. J Allergy Clin Immunol. (2019) 143(1):1–11. doi: 10.1016/j.jaci.2018.10.032
12. Lang CCV, Renert-Yuval Y, Del Duca E, Pavel AB, Wu J, Zhang N, et al. Immune and barrier characterization of atopic dermatitis skin phenotype in Tanzanian patients. Ann Allergy Asthma Immunol. (2021) 127(3):334–41. doi: 10.1016/j.anai.2021.04.023
13. Wongvibulsin S, Sutaria N, Kannan S, Alphonse MP, Belzberg M, Williams KA, et al. Transcriptomic analysis of atopic dermatitis in African Americans is characterized by Th2/Th17-centered cutaneous immune activation. Sci Rep. (2021) 11(1):11175. doi: 10.1038/s41598-021-90105-w
14. Biagini JM, Kroner JW, Baatyrbek Kyzy A, Gonzales A, He H, Stevens M, et al. Longitudinal atopic dermatitis endotypes: an atopic march paradigm that includes black children. J Allergy Clin Immunol. (2022) 149(5):1702–10.e4. doi: 10.1016/j.jaci.2021.09.036
15. Sanyal RD, Pavel AB, Glickman J, Chan TC, Zheng X, Zhang N, et al. Atopic dermatitis in African American patients is TH2/TH22-skewed with TH1/TH17 attenuation. Ann Allergy Asthma Immunol. (2019) 122(1):99–110.e6. doi: 10.1016/j.anai.2018.08.024
16. Thawer-Esmail F, Jakasa I, Todd G, Wen Y, Brown SJ, Kroboth K, et al. South African Amaxhosa patients with atopic dermatitis have decreased levels of filaggrin breakdown products but no loss-of-function mutations in filaggrin. J Allergy Clin Immunol. (2014) 133(1):280–2.e22. doi: 10.1016/j.jaci.2013.09.053
17. Vakharia P, Chopra R, Silverberg J. Systematic review of diagnostic criteria used in atopic dermatitis randomized controlled trials. Am J Clin Dermatol. (2017) 19(1):15–22. doi: 10.1007/s40257-017-0299-4
18. Bieber T. How to define atopic dermatitis? Dermatol Clin. (2017) 35(3):275–81. doi: 10.1016/j.det.2017.02.001
19. Chalmers DA, Todd G, Saxe N, Milne JT, Tolosana S, Ngcelwane PN, et al. Validation of the U.K. working party diagnostic criteria for atopic eczema in a Xhosa-speaking African population [published correction appears in Br J Dermatol. 2007 Mar;156(3):612]. Br J Dermatol. (2007) 156(1):111–6. doi: 10.1111/j.1365-2133.2006.07606.x
20. Schmitt J, Langan S, Stamm T, Williams HC. Harmonizing outcome measurements in eczema (HOME) delphi panel. Core outcome domains for controlled trials and clinical recordkeeping in eczema: international multiperspective delphi consensus process. J Invest Dermatol. (2011) 131(3):623–30. doi: 10.1038/jid.2010.303
21. Oranje AP, Glazenburg EJ, Wolkerstorfer A, de Waard-van der Spek FB. Practical issues on interpretation of scoring atopic dermatitis: the SCORAD index, objective SCORAD and the three-item severity score. Br J Dermatol. (2007) 157(4):645–8. doi: 10.1111/j.1365-2133.2007.08112.x
22. Ben-Gashir MA, Hay RJ. Reliance on erythema scores may mask severe atopic dermatitis in black children compared with their white counterparts. Br J Dermatol. (2002) 147(5):920–5. doi: 10.1046/j.1365-2133.2002.04965.x
23. Zhao CY, Wijayanti A, Doria MC, Harris AG, Jain SV, Legaspi KN, et al. The reliability and validity of outcome measures for atopic dermatitis in patients with pigmented skin: a grey area. Int J Womens Dermatol. (2015) 1(3):150–4. doi: 10.1016/j.ijwd.2015.05.002
24. Faye O, Meledie N'Djong AP, Diadie S, Coniquet S, Niamba PA, Atadokpede F, et al. Validation of the patient-oriented SCORing for atopic dermatitis tool for black skin. J Eur Acad Dermatol Venereol. (2020) 34(4):795–9. doi: 10.1111/jdv.15999
25. Tanei R, Hasegawa Y. Atopic dermatitis in older adults: a viewpoint from geriatric dermatology. Geriatr Gerontol Int. (2016) 16(Suppl 1):75–86. doi: 10.1111/ggi.12771
26. Williams H, Pembroke A, Forsdyke H, Boodoo G, Hay R, Burney P. London-born black Caribbean children are at increased risk of atopic dermatitis. J Am Acad Dermatol. (1995) 32(2):212–7. doi: 10.1016/0190-9622(95)90128-0
27. Shaw TE, Currie GP, Koudelka CW, Simpson EL. Eczema prevalence in the United States: data from the 2003 national survey of children’s health. J Invest Dermatol. (2011) 131(1):67–73. doi: 10.1038/jid.2010.251
28. Wegienka G, Johnson CC, Zoratti E, Havstad S. Racial differences in allergic sensitization: recent findings and future directions. Curr Allergy Asthma Rep. (2013) 13(3):255–61. doi: 10.1007/s11882-013-0343-2
29. Levin AM, Wang Y, Wells KE, Padhukasahasram B, Yang JJ, Burchard EG, et al. Nocturnal asthma and the importance of race/ethnicity and genetic ancestry. Am J Respir Crit Care Med. (2014) 190(3):266–73. doi: 10.1164/rccm.201402-0204OC
30. Ardura-Garcia C, Stolbrink M, Zaidi S, Cooper PJ, Blakey JD. Predictors of repeated acute hospital attendance for asthma in children: a systematic review and meta-analysis. Pediatr Pulmonol. (2018) 53(9):1179–92. doi: 10.1002/ppul.24068
31. Acevedo N, Sánchez J, Zakzuk J, Bornacelly A, Quiróz C, Alvarez Á, et al. Particular characteristics of allergic symptoms in tropical environments: follow up to 24 months in the FRAAT birth cohort study. BMC Pulm Med. (2012) 12:13. doi: 10.1186/1471-2466-12-13
32. Murrison LB, Brandt EB, Myers JB, Hershey GKK. Environmental exposures and mechanisms in allergy and asthma development. J Clin Invest. (2019) 129(4):1504–15. doi: 10.1172/JCI124612
33. Kramer U, Sugiri D, Ranft U, Krutmann J, von Berg A, Berdel D, et al. Eczema, respiratory allergies, and traffic-related air pollution in birth cohorts from small-town areas. J Dermatol Sci. (2009) 56(2):99–105. doi: 10.1016/j.jdermsci.2009.07.014
34. Ahn K. The role of air pollutants in atopic dermatitis. J Allergy Clin Immunol. (2014) 134(5):993–1000. doi: 10.1016/j.jaci.2014.09.023
35. Herberth G, Gubelt R, Röder S, Krämer U, Schins RP, Diez U, et al. Increase of inflammatory markers after indoor renovation activities: the LISA birth cohort study. Pediatr Allergy Immunol. (2009) 20(6):563–70. doi: 10.1111/j.1399-3038.2008.00819.x
36. Sanchez J, Cherrez-Ojeda I, Galvan C, Garcia E, Hernández-Mantilla N, Londoño Garcia A, et al. The unmet needs in atopic dermatitis control in Latin America: a multidisciplinary expert perspective. Dermatol Ther. (2021) 11(5):1521–40. doi: 10.1007/s13555-021-00595-9
37. Sood A. Indoor fuel exposure and the lung in both developing and developed countries. Clin Chest Med. (2012) 33(4):649–65. doi: 10.1016/j.ccm.2012.08.003
38. Lam N, Smith K, Gauthier A, Bates M. Kerosene: a review of household uses and their hazards in low- and middle-income countries. J Toxicol Environ Health B Crit Rev. (2012) 15(6):396–432. doi: 10.1080/10937404.2012.710134
39. Pastore S, Korkina L. Redox imbalance in T cell-mediated skin diseases. Mediators Inflamm. (2010) 2010:861949. doi: 10.1155/2010/861949
40. Liu T, Ji RR. Oxidative stress induces itch via activation of transient receptor potential subtype ankyrin 1 in mice. Neurosci Bull. (2012) 28(2):145–54. doi: 10.1007/s12264-012-1207-9
41. Niwa Y, Sumi H, Kawahira K, Terashima T, Nakamura T, Akamatsu H. Protein oxidative damage in the stratum corneum: evidence for a link between environmental oxidants and the changing prevalence and nature of atopic dermatitis in Japan. Br J Dermatol. (2003) 149(2):248–54. doi: 10.1046/j.1365-2133.2003.05417.x
42. Lehmann I, Thoelke A, Rehwagen M, Rolle-Kampczyk U, Schlink U, Schulz R, et al. The influence of maternal exposure to volatile organic compounds on the cytokine secretion profile of neonatal T cells. Environ Toxicol. (2002) 17(3):203–10. doi: 10.1002/tox.10055
43. Wang IJ, Chen SL, Lu TP, Chuang EY, Chen PC. Prenatal smoke exposure, DNA methylation, and childhood atopic dermatitis. Clin Exp Allergy. (2013) 43(5):535–43. doi: 10.1111/cea.12108
44. Lunjani N, Satitsuksanoa P, Lukasik Z, Sokolowska M, Eiwegger T, O'Mahony L. Recent developments and highlights in mechanisms of allergic diseases: microbiome. Allergy. (2018) 73(12):2314–27. doi: 10.1111/all.13634
45. Walter J, O'Mahony L. The importance of social networks—an ecological and evolutionary framework to explain the role of microbes in the aetiology of allergy and asthma. Allergy. (2019) 74(11):2248–51. doi: 10.1111/all.13845
46. Frei R, Ferstl R, Roduit C, Ziegler M, Schiavi E, Barcik W, et al. Exposure to nonmicrobial N-glycolylneuraminic acid protects farmers’ children against airway inflammation and colitis. J Allergy Clin Immunol. (2018) 141(1):382–90.e7. doi: 10.1016/j.jaci.2017.04.051
47. Roslund MI, Puhakka R, Grönroos M, Nurminen N, Oikarinen S, Gazali AM, et al. Biodiversity intervention enhances immune regulation and health-associated commensal microbiota among daycare children. Sci Adv. (2020) 6(42):eaba2578. doi: 10.1126/sciadv.aba2578
48. Palmer CN, Irvine AD, Terron-Kwiatkowski A, Zhao Y, Liao H, Lee SP, et al. Common loss-of-function variants of the epidermal barrier protein filaggrin are a major predisposing factor for atopic dermatitis. Nat Genet. (2006) 38(4):441–6. doi: 10.1038/ng1767
49. Drislane C, Irvine AD. The role of filaggrin in atopic dermatitis and allergic disease. Ann Allergy Asthma Immunol. (2020) 124(1):36–43. doi: 10.1016/j.anai.2019.10.008
50. Berna R, Mitra N, Hoffstad O, Wubbenhorst B, Nathanson KL, Margolis DJ. Uncommon variants in FLG2 and TCHHL1 are associated with remission of atopic dermatitis in a large longitudinal US cohort. Arch Dermatol Res. (2022) 314(10):953–9. doi: 10.1007/s00403-021-02319-7
51. Fulton RL, Margolis DJ, Sockler PG, Mitra N, Wong XFCC, Common JE. No association of filaggrin copy number variation and atopic dermatitis risk in white and black Americans. Exp Dermatol. (2022) 31(2):233–6. doi: 10.1111/exd.14449
52. Margolis DJ, Mitra N, Berna R, Hoffstad O, Kim BS, Yan A, et al. Associating filaggrin copy number variation and atopic dermatitis in African-Americans: challenges and opportunities. J Dermatol Sci. (2020) 98(1):58–60. doi: 10.1016/j.jdermsci.2020.01.014
53. Margolis DJ, Mitra N, Wubbenhorst B, D'Andrea K, Kraya AA, Hoffstad O, et al. Association of filaggrin loss-of-function variants with race in children with atopic dermatitis. JAMA Dermatol. (2019) 155(11):1269–76. doi: 10.1001/jamadermatol.2019.1946
54. Almoguera B, Vazquez L, Mentch F, March ME, Connolly JJ, Peissig PL, et al. Novel locus for atopic dermatitis in African Americans and replication in European Americans. J Allergy Clin Immunol. (2019) 143(3):1229–31. doi: 10.1016/j.jaci.2018.10.038
55. Mathyer ME, Quiggle AM, Wong XFCC, Denil SLIJ, Kumar MG, Ciliberto HM, et al. Tiled array-based sequencing identifies enrichment of loss-of-function variants in the highly homologous filaggrin gene in African-American children with severe atopic dermatitis. Exp Dermatol. (2018) 27(9):989–92. doi: 10.1111/exd.13691
56. Margolis DJ, Mitra N, Gochnauer H, Wubbenhorst B, D'Andrea K, Kraya A, et al. Uncommon filaggrin variants are associated with persistent atopic dermatitis in African Americans [published correction appears in J Invest Dermatol. 2018 Sep;138(9):2084–2085]. J Invest Dermatol. (2018) 138(7):1501–6. doi: 10.1016/j.jid.2018.01.029
57. Fernandez K, Asad S, Taylan F, Wahlgren CF, Bilcha KD, Nordenskjöld M, et al. Intragenic copy number variation in the filaggrin gene in Ethiopian patients with atopic dermatitis. Pediatr Dermatol. (2017) 34(3):e140–1. doi: 10.1111/pde.13095
58. Quiggle AM, Goodwin ZA, Marfatia TR, Kumar MG, Ciliberto H, Bayliss SJ, et al. Low filaggrin monomer repeats in African American pediatric patients with moderate to severe atopic dermatitis. JAMA Dermatol. (2015) 151(5):557–9. doi: 10.1001/jamadermatol.2014.4916
59. Taylan F, Nilsson D, Asad S, Lieden A, Wahlgren CF, Winge MC, et al. Whole-exome sequencing of Ethiopian patients with ichthyosis vulgaris and atopic dermatitis. J Allergy Clin Immunol. (2015) 136(2):507–9.e19. doi: 10.1016/j.jaci.2015.02.010
60. Polcari I, Becker L, Stein SL, Smith MS, Paller AS. Filaggrin gene mutations in African Americans with both ichthyosis vulgaris and atopic dermatitis. Pediatr Dermatol. (2014) 31(4):489–92. doi: 10.1111/pde.12355
61. Margolis DJ, Gupta J, Apter AJ, Hoffstad O, Papadopoulos M, Rebbeck TR, et al. Exome sequencing of filaggrin and related genes in African-American children with atopic dermatitis. J Invest Dermatol. (2014) 134(8):2272–4. doi: 10.1038/jid.2014.126
62. Margolis DJ, Gupta J, Apter AJ, Ganguly T, Hoffstad O, Papadopoulos M, et al. Filaggrin-2 variation is associated with more persistent atopic dermatitis in African American subjects. J Allergy Clin Immunol. (2014) 133(3):784–9. doi: 10.1016/j.jaci.2013.09.015
63. Margolis DJ, Apter AJ, Gupta J, Hoffstad O, Papadopoulos M, Campbell LE, et al. The persistence of atopic dermatitis and filaggrin (FLG) mutations in a US longitudinal cohort. J Allergy Clin Immunol. (2012) 130(4):912–7. doi: 10.1016/j.jaci.2012.07.008
64. Winge MC, Bilcha KD, Liedén A, Shibeshi D, Sandilands A, Wahlgren CF, et al. Novel filaggrin mutation but no other loss-of-function variants found in Ethiopian patients with atopic dermatitis. Br J Dermatol. (2011) 165(5):1074–80. doi: 10.1111/j.1365-2133.2011.10475.x
65. Gao PS, Rafaels NM, Hand T, Murray T, Boguniewicz M, Hata T, et al. Filaggrin mutations that confer risk of atopic dermatitis confer greater risk for eczema herpeticum. J Allergy Clin Immunol. (2009) 124(3):507–13.e5137. doi: 10.1016/j.jaci.2009.07.034
66. Howell MD, Kim BE, Gao P, Grant AV, Boguniewicz M, Debenedetto A, et al. Cytokine modulation of atopic dermatitis filaggrin skin expression. J Allergy Clin Immunol. (2007) 120(1):150–5. doi: 10.1016/j.jaci.2007.04.031
67. Park KY, Li K, Seok J, Seo SJ. An analysis of the filaggrin gene polymorphism in Korean atopic dermatitis patients. J Korean Med Sci. (2016) 31(7):1136–42. doi: 10.3346/jkms.2016.31.7.1136
68. Pigors M, Common JEA, Wong XFCC, Malik S, Scott CA, Tabarra N, et al. Exome sequencing and rare variant analysis reveals multiple filaggrin mutations in Bangladeshi families with atopic eczema and additional risk genes. J Invest Dermatol. (2018) 138(12):2674–7. doi: 10.1016/j.jid.2018.05.013
69. Zhong WL, Wu X, Yu B, Zhang J, Zhang W, Xu N, et al. Filaggrin gene mutation c.3321delA is associated with dry phenotypes of atopic dermatitis in the Chinese han population. Chin Med J. (2016) 129(12):1498–500. doi: 10.4103/0366-6999.183424
70. Brown SJ, Kroboth K, Sandilands A, Campbell LE, Pohler E, Kezic S, et al. Intragenic copy number variation within filaggrin contributes to the risk of atopic dermatitis with a dose-dependent effect. J Invest Dermatol. (2012) 132(1):98–104. doi: 10.1038/jid.2011.342
71. Paternoster L, Standl M, Waage J, Baurecht H, Hotze M, Strachan DP, et al. Multi-ancestry genome-wide association study of 21,000 cases and 95,000 controls identifies new risk loci for atopic dermatitis. Nat Genet. (2015) 47(12):1449–56. doi: 10.1038/ng.3424
72. Zhang Y, Wang HC, Feng C, Yan M. Analysis of the association of polymorphisms rs5743708 in TLR2 and rs4986790 in TLR4 with atopic dermatitis risk. Immunol Invest. (2019) 48(2):169–80. doi: 10.1080/08820139.2018.1508228
73. He JQ, Chan-Yeung M, Becker AB, Dimich-Ward H, Ferguson AC, Manfreda J, et al. Genetic variants of the IL13 and IL4 genes and atopic diseases in at-risk children. Genes Immun. (2003) 4(5):385–9. doi: 10.1038/sj.gene.6363985
74. Tsunemi Y, Saeki H, Nakamura K, Sekiya T, Hirai K, Kakinuma T, et al. Interleukin-13 gene polymorphism G4257A is associated with atopic dermatitis in Japanese patients. J Dermatol Sci. (2002) 30(2):100–7. doi: 10.1016/s0923-1811(02)00065-8
75. Liu X, Nickel R, Beyer K, Wahn U, Ehrlich E, Freidhoff LR, et al. An IL13 coding region variant is associated with a high total serum IgE level and atopic dermatitis in the German multicenter atopy study (MAS-90). J Allergy Clin Immunol. (2000) 106(1 Pt 1):167–70. doi: 10.1067/mai.2000.107935
76. Belgrave D, Granell R, Simpson A, Guiver J, Bishop C, Buchan I, et al. Developmental profiles of eczema, wheeze, and rhinitis: two population-based birth cohort studies. PLoS Med. (2014) 11(10):e1001748. doi: 10.1371/journal.pmed.1001748
77. Brown SJ, Irvine AD. Atopic eczema and the filaggrin story. Semin Cutan Med Surg. (2008) 27(2):128–37. doi: 10.1016/j.sder.2008.04.001
78. Løset M, Brown SJ, Saunes M, Hveem K. Genetics of atopic dermatitis: from DNA sequence to clinical relevance. Dermatology. (2019) 235(5):355–64. doi: 10.1159/000500402
79. Brown SJ. Molecular mechanisms in atopic eczema: insights gained from genetic studies. J Pathol. (2017) 241(2):140–5. doi: 10.1002/path.4810
80. Bitschar K, Wolz C, Krismer B, Peschel A, Schittek B. Keratinocytes as sensors and central players in the immune defense against Staphylococcus aureus in the skin. J Dermatol Sci. (2017) 87(3):215–20. doi: 10.1016/j.jdermsci.2017.06.003
81. Zdolsek HA, Jenmalm MC. Reduced levels of soluble CD14 in atopic children. Clin Exp Allergy. (2004) 34(4):532–9. doi: 10.1111/j.1365-2222.2004.1921.x
82. Elias PM. The skin barrier as an innate immune element. Semin Immunopathol. (2007) 29(1):3–14. doi: 10.1007/s00281-007-0060-9
83. Kim BE, Leung DYM. Significance of skin barrier dysfunction in atopic dermatitis. Allergy Asthma Immunol Res. (2018) 10(3):207–15. doi: 10.4168/aair.2018.10.3.20
84. Belzberg M, Alphonse MP, Brown I, Williams KA, Khanna R, Ho B, et al. Prurigo nodularis is characterized by systemic and cutaneous T helper 22 immune polarization. J Invest Dermatol. (2021) 141(9):2208–18.e14. doi: 10.1016/j.jid.2021.02.749
85. Lunjani N, Ambikan AT, Hlela C, Levin M, Mankahla A, Heldstab-Kast JI, et al. Rural and urban exposures shape early life immune development in South African children with atopic dermatitis and nonallergic children. Allergy. (2024) 79(1):65–79. doi: 10.1111/all.15832
86. Lunjani N, Tan G, Dreher A, Sokolowska M, Groeger D, Warwyzniak M, et al. Environment-dependent alterations of immune mediators in urban and rural South African children with atopic dermatitis. Allergy. (2022) 77(2):569–81. doi: 10.1111/all.14974
87. Sims JT, Chang CY, Higgs RE, Engle SM, Liu Y, Sissons SE, et al. Insights into adult atopic dermatitis heterogeneity derived from circulating biomarker profiling in patients with moderate-to-severe disease. Exp Dermatol. (2021) 30(11):1650–61. doi: 10.1111/exd.14389
88. Marenholz I, Esparza-Gordillo J, Rüschendorf F, Bauerfeind A, Strachan DP, Spycher BD, et al. Meta-analysis identifies seven susceptibility loci involved in the atopic march. Nat Commun. (2015) 6:8804. doi: 10.1038/ncomms9804
89. Jackson-Cowan L, Cole EF, Silverberg JI, Lawley LP. Childhood atopic dermatitis is associated with cognitive dysfunction: a national health interview survey study from 2008 to 2018. Ann Allergy Asthma Immunol. (2021) 126(6):661–5. doi: 10.1016/j.anai.2020.11.008
90. Haus M, Heese HD, Weinberg EG, Potter PC, Hall JM, Malherbe D. The influence of ethnicity, an atopic family history, and maternal ascariasis on cord blood serum IgE concentrations. J Allergy Clin Immunol. (1988) 82(2):179–89. doi: 10.1016/0091-6749(88)90997-9
91. Wenzel SE, Balzar S, Ampleford E, Hawkins GA, Busse WW, Calhoun WJ, et al. IL4R alpha mutations are associated with asthma exacerbations and mast cell/IgE expression. Am J Respir Crit Care Med. (2007) 175(6):570–6. doi: 10.1164/rccm.200607-909OC
92. Laurence C, van der Merwe L, Zhang G, Le Souef P, Levin M. Association between pro-inflammatory alleles and allergic phenotypes in Xhosa adolescents. Pediatr Allergy Immunol. (2018) 29(3):311–7. doi: 10.1111/pai.12859
93. Russell SB, Smith JC, Huang M, Trupin JS, Williams SM. Pleiotropic effects of immune responses explain variation in the prevalence of fibroproliferative diseases. PLoS Genet. (2015) 11(11):e1005568. doi: 10.1371/journal.pgen.1005568
94. Werfel T, Allam JP, Biedermann T, Eyerich K, Gilles S, Guttman-Yassky E, et al. Cellular and molecular immunologic mechanisms in patients with atopic dermatitis. J Allergy Clin Immunol. (2016) 138(2):336–49. doi: 10.1016/j.jaci.2016.06.010
95. Gray C, Levin M, Zar H, Potter P, Khumalo N, Volkwyn L, et al. Food allergy in South African children with atopic dermatitis. Pediatr Allergy Immunol. (2014) 25:572–9. doi: 10.1111/pai.12270
96. Gray CL, Levin ME, du Toit G. Egg sensitization, allergy and component patterns in African children with atopic dermatitis. Pediatr Allergy Immunol. (2016) 27(7):709–15. doi: 10.1111/pai.12615
97. Gray CL, Levin ME, Du Toit G. Which test is best for diagnosing peanut allergy in South African children with atopic dermatitis? S Afr Med J. (2016) 106(2):214–20. doi: 10.7196/SAMJ.2016.v106i2.10125
98. Gabryszewski SJ, Chang X, Dudley JW, Mentch F, March M, Holmes JH, et al. Unsupervised modeling and genome-wide association identify novel features of allergic march trajectories. J Allergy Clin Immunol. (2021) 147(2):677–85.e10. doi: 10.1016/j.jaci.2020.06.026
99. Caraballo L, Zakzuk J, Lee BW, Acevedo N, Soh JY, Sánchez-Borges M, et al. Particularities of allergy in the tropics. World Allergy Organ J. (2016) 9:20. doi: 10.1186/s40413-016-0110-7
100. Lunjani N, Ahearn-Ford S, Dube FS, Hlela C, O'Mahony L. Mechanisms of microbe-immune system dialogue within the skin. Genes Immun. (2021) 22(5-6):276–88. doi: 10.1038/s41435-021-00133-9
101. Barcik W, Wawrzyniak M, Akdis CA, O'Mahony L. Immune regulation by histamine and histamine-secreting bacteria. Curr Opin Immunol. (2017) 48:108–13. doi: 10.1016/j.coi.2017.08.011
102. Forde B, Yao L, Shaha R, Murphy S, Lunjani N, O'Mahony L. Immunomodulation by foods and microbes: unravelling the molecular tango. Allergy. (2022) 77(12):3513–26. doi: 10.1111/all.15455
103. Kim JE, Kim HS. Microbiome of the skin and gut in atopic dermatitis (AD): understanding the pathophysiology and finding novel management strategies. J Clin Med. (2019) 8(4):444. doi: 10.3390/jcm8040444
104. Carter MM, Olm MR, Merrill BD, Dahan D, Tripathi S, Spencer SP, et al. Ultra-deep sequencing of hadza hunter-gatherers recovers vanishing gut microbes. Cell. (2023) 186(14):3111–24.e13. doi: 10.1016/j.cell.2023.05.046
105. Suzuki TA, Fitzstevens JL, Schmidt VT, Enav H, Huus KE, Mbong Ngwese M, et al. Codiversification of gut microbiota with humans. Science. (2022) 377(6612):1328–32. doi: 10.1126/science.abm7759
106. Venter C, Meyer RW, Greenhawt M, Pali-Schöll I, Nwaru B, Roduit C, et al. Role of dietary fiber in promoting immune health-an EAACI position paper. Allergy. (2022) 77(11):3185–98. doi: 10.1111/all.15430
107. Roduit C, Frei R, Ferstl R, Loeliger S, Westermann P, Rhyner C, et al. High levels of butyrate and propionate in early life are associated with protection against atopy. Allergy. (2019) 74(4):799–809. doi: 10.1111/all.13660
108. Armet AM, Deehan EC, O'Sullivan AF, Mota JF, Field CJ, Prado CM, et al. Rethinking healthy eating in light of the gut microbiome. Cell Host Microbe. (2022) 30(6):764–85. doi: 10.1016/j.chom.2022.04.016
109. Lunjani N, Hlela C, O'Mahony L. Microbiome and skin biology. Curr Opin Allergy Clin Immunol. (2019) 19(4):328–33. doi: 10.1097/ACI.0000000000000542
110. Jurevic RJ, Chrisman P, Mancl L, Livingston R, Dale BA. Single-nucleotide polymorphisms and haplotype analysis in beta-defensin genes in different ethnic populations. Genet Test. (2002) 6(4):261–9. doi: 10.1089/10906570260471787
111. Merriman JA, Mueller EA, Cahill MP, Beck LA, Paller AS, Hanifin JM, et al. Temporal and racial differences associated with atopic dermatitis Staphylococcus aureus and encoded virulence factors. mSphere. (2016) 1(6):e00295–16. doi: 10.1128/mSphere.00295-16
112. Rojo A, Aguinaga A, Monecke S, Yuste JR, Gastaminza G, España A. Staphylococcus aureus genomic pattern and atopic dermatitis: may factors other than superantigens be involved? Eur J Clin Microbiol Infect Dis. (2014) 33(4):651–8. doi: 10.1007/s10096-013-2000-z
113. Wegienka G, Havstad S, Joseph CL, Zoratti E, Ownby D, Woodcroft K, et al. Racial disparities in allergic outcomes in African Americans emerge as early as age 2 years. Clin Exp Allergy. (2012) 42(6):909–17. doi: 10.1111/j.1365-2222.2011.03946.x
114. Tackett KJ, Jenkins F, Morrell DS, McShane DB, Burkhart CN. Structural racism and its influence on the severity of atopic dermatitis in African American children. Pediatr Dermatol. (2020) 37(1):142–6. doi: 10.1111/pde.14058
115. Nriagu J, Robins T, Gary L, Liggans G, Davila R, Supuwood K, et al. Prevalence of asthma and respiratory symptoms in South-central Durban, South Africa. Eur J Epidemiol. (1999) 15(8):747–55. doi: 10.1023/a:1007653709188
116. Baker JL, Shriner D, Bentley AR, Rotimi CN. Pharmacogenomic implications of the evolutionary history of infectious diseases in Africa. Pharmacogenomics J. (2017) 17(2):112–20. doi: 10.1038/tpj.2016.78
117. Kariuki R, Bakalian M, Lall AE, White S, Parby R, Huang JI, et al. Harnessing urbanization to end poverty and boost prosperity in Africa: an action agenda for transformation (English). Africa region sustainable development series Washington, D.C.: World Bank Group. Available at: http://documents.worldbank.org/curated/en/710431468191672231/Harnessing-urbanization-to-end-poverty-and-boost-prosperity-in-Africa-an-action-agenda-for-transformation
Keywords: atopic dermatitis, microbiome, immune mechanism, skin barrier disruption, atopic dermatitis in African populations
Citation: Lunjani N, Kerbelker T, Mdletshe FB, Hlela C and O’Mahony L (2024) Phenotypes, endotypes and genotypes of atopic dermatitis and allergy in populations of African ancestry on the continent and diaspora. Front. Allergy 4:1203304. doi: 10.3389/falgy.2023.1203304
Received: 10 April 2023; Accepted: 11 December 2023;
Published: 24 January 2024.
Edited by:
Andrew Glory Mtewa, Malawi University of Science and Technology, MalawiReviewed by:
Kavita Reginald, Sunway University, MalaysiaMarie-Charlotte Brüggen, University Hospital Zürich, Switzerland
© 2024 Lunjani, Kerbelker, Mdletshe, Hlela and O'Mahony. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: N. Lunjani bHVuamFuaUBnbWFpbC5jb20=
 N. Lunjani
N. Lunjani T. Kerbelker3
T. Kerbelker3 L. O’Mahony
L. O’Mahony