- 1Division of Rhinology, Department of Otolaryngology, Mayo Clinic in Arizona, Phoenix, AZ, United States
- 2Division of Education, Department of Library Services, Mayo Clinic, Phoenix, AZ, United States
Background: Epigenetics facilitates insights on the impact of host environment on the genesis of chronic rhinosinusitis (CRS) through modulations of host gene expression and activity. Epigenetic mechanisms such as DNA methylation cause reversible but heritable changes in gene expression over generations of progeny, without altering the DNA base-pair sequences. These studies offer a critical understanding of the environment-induced changes that result in host predisposition to disease and may help in developing novel biomarkers and therapeutics. The goal of this systematic review is to summarize the current evidence on epigenetics of CRS with a focus on chronic rhinosinusitis with nasal polyps (CRSwNP) and highlight gaps that merit further research.
Methods: A systematic review of the English language literature was performed to identify investigations related to epigenetic studies in subjects with CRS.
Results: The review identified 65 studies. These have focused on DNA methylation and non-coding RNAs, with only a few on histone deacetylation, alternative polyadenylation, and chromatin accessibility. Studies include those investigating in vivo and in vitro changes or both. Studies also include animal models of CRS. Almost all have been conducted in Asia. The genome-wide studies of DNA methylation found differences in global methylation between CRSwNP and controls, while others specifically found significant differences in methylation of the CpG sites of the thymic stromal lymphopoietin (TSLP), IL-8, and PLAT. In addition, DNA methyltransferase inhibitors and histone deacetylase inhibitors were studied as potential therapeutic agents. Majority of the studies investigating non-coding RNAs focused on micro-RNAs (miRNA) and found differences in global expression of miRNA levels. These studies also revealed some previously known as well as novel targets and pathways such as tumor necrosis factor alpha, TGF beta-1, IL-10, EGR2, aryl hydrocarbon receptor, PI3K/AKT pathway, mucin secretion, and vascular permeability. Overall, the studies have found a dysregulation in pathways/genes involving inflammation, immune regulation, tissue remodeling, structural proteins, mucin secretion, arachidonic acid metabolism, and transcription.
Conclusions: Epigenetic studies in CRS subjects suggest that there is likely a major impact of the environment. However, these are association studies and do not directly imply pathogenesis. Longitudinal studies in geographically and racially diverse population cohorts are necessary to quantify genetic vs. environmental risks for CRSwNP and CRS without nasal polyps and assess heritability risk, as well as develop novel biomarkers and therapeutic agents.
Introduction
Epigenetics is the study of reversible molecular mechanisms that modify gene activity and expression without altering the DNA base-pair nucleotide structure. These epigenetic changes are a consequence of the impact of the host environment. The commonly involved mechanisms through which epigenetic modulations are deployed include DNA methylation, histone modifications, non-coding RNAs, and alternative polyadenylation (APA). DNA methylation decreases gene expression by blocking specific sites on DNA, which prevents the binding of transcription factors on the DNA (1). Thus, hypermethylation may repress gene transcription, and hypomethylation potentially increases gene transcription. Histone modifications also control transcription by regulating the winding and unwinding of DNA. Tightly coiled DNA around histones is unable to undergo transcription. Chemical modifications (acetylation and deacetylation) in histones can alter the amount of DNA available for transcription (2). In general, histone acetylation causes transcriptional activation, and deacetylation causes transcriptional repression (3). Non-coding RNAs such as micro-RNAs (miRNAs) and long non-coding RNA (lncRNA) may control post-transcription protein synthesis, in addition to transcription. miRNAs can bind to target mRNAs leading to their degradation. Epi-miRNAs can repress key enzymes that drive epigenetic remodeling and also bind to complementary sequences in gene promoters, recruiting specific protein complexes that modulate chromatin structure and gene expression. miRNAs themselves can also be epigenetically regulated by DNA methylation and/or specific histone modifications (4). In turn, non-coding RNAs also have regulatory epigenetic roles. Non-coding RNAs such as lncRNAs and circular RNAs (circRNAs) can affect the methylation pattern of downstream genes (5), and short interfering RNA (siRNAs) and miRNAs can lead to transcriptional gene silencing through DNA methylation and histone modifications (6). APA at the 3′ untranslated region (UTR) affects mRNA stability and protein translation (7). Thus, the environment (microbes, allergens, pollutants, smoke, etc.) can induce changes in the “epigenome” through various mechanisms, ultimately affecting gene expression without disturbing the “genome”. Figure 1 illustrates some of the common epigenetic mechanisms that have been investigated in chronic rhinosinusitis (CRS).
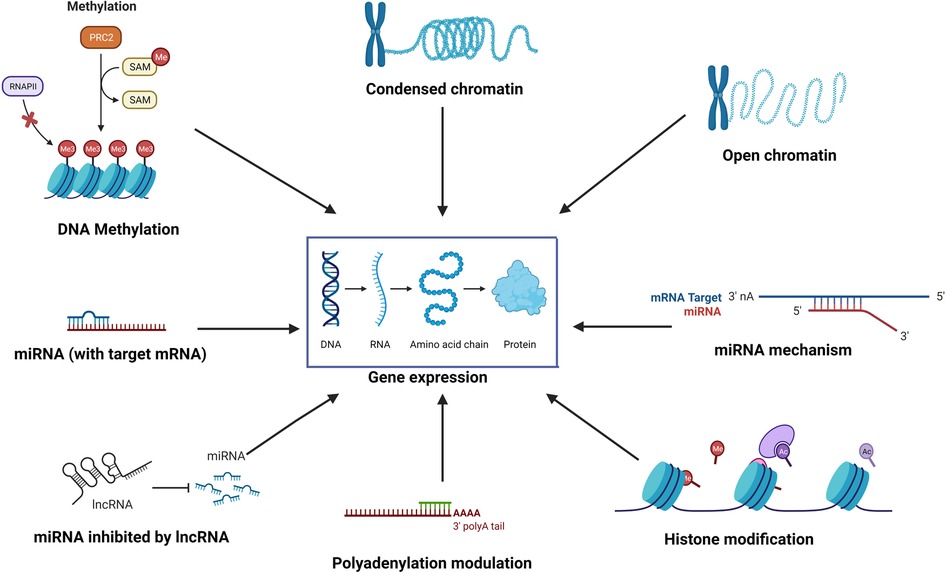
Figure 1. The environment affects the “epigenome” which in turn affects gene expression through various epigenetic mechanisms such as DNA methylation, histone modification, role of non-coding RNAs such as miRNA and lncRNA, alternative polyadenylation, and chromatin accessibility.
While epigenetic changes occur as a result of the host–environment interaction, but they can be passed on to progeny for a few generations (8). However, because these changes are pharmacogenetically reversible (9), there exists a huge potential in developing biomarkers and novel treatments and optimizing personalized therapeutics. Currently, seven epigenetic agents are FDA-approved for treatment of certain cancers, while research on several other drugs is underway for oncologic and non-oncologic indications (10).
CRS is a common condition requiring surgical management in the United States and worldwide, with many subtypes being particularly recalcitrant to standard of care treatment modalities (11, 12). The disease is heterogenous in both clinical features as well as underlying etiopathogenic factors. Factors that may attribute to disease pathophysiology include altered host responses at epithelial surfaces, which may lead to changes in epithelial barrier integrity and permeability and altered innate and adaptive immune responses (13). The role of the epithelium cannot be emphasized enough, as it is also the interface for the host–environment interaction.
CRS with nasal polyps (CRSwNP) is a subtype that has been shown to be associated with familial clustering (14, 15). Whether this clustering reflects genetic susceptibility (16) or a shared and common environment is yet to be fully deciphered as familial clustering also affects spouses (14). Thus, understanding the role of both genetic susceptibility and the environment is important in CRS. The study of epigenetics is thus valuable in CRS as it can help elucidate genetic factors that are driven through environmental changes rather than changes in the actual gene structure (17). Recent reviews on the role of epigenetics in CRS have emphasized the immense potential of these studies in laying the foundation to the development of novel therapeutics in CRS (17, 18). Thus, the identification of these reversible mechanisms that can alter gene expression can lead to a paradigm shift in treatment approaches in the future.
The goal of our systematic review was to search the literature for all studies that have investigated the role of epigenetic mechanisms in CRS and CRSwNP, since a better understanding of these may assist with quantifying the disease risk and prognostication, as well as developing biomarkers and personalized treatment for the management of this condition.
Methods
A review of the English language literature was performed on PubMed to identify all original studies that investigated epigenetic mechanisms in CRS from inception to September 2022. References of these articles were also further reviewed to include additional original reports on epigenetics in CRS. For this project, a medical librarian searched the following databases: Ovid MEDLINE, Ovid EMBASE, Scopus, and Web of Science. All searches were conducted in January 2023, with no date limitations but limited to English language. The search was a combination of Medical Subject Headings (MeSH) and keywords. The MeSH terms used included the following: Epigenesis, Genetic; DNA Methylation; MicroRNAs; Histones; RNA, Untranslated; Polyadenylation; Sinusitis; and Nasal Polyps. The keywords used included the following: epigenetic*; methylation; micro-RNA*; MiR; histone; non coding rna; non-coding rna; noncoding rna; alternative polyadenylation; lncrna; sinusitis; nasal polyp*, and chronic rhinosinusitis. The epigenetic terms were combined using the Boolean operator “OR”, and the sinusitis terms were also combined using the Boolean operator of “OR”. The epigenetic terms and the sinusitis terms were then combined using the Boolean operator “AND” resulting in 790 references. Duplicate references were then removed leaving a total of 420 references for review to meet the inclusion or exclusion criteria. (* indicates truncation of word or phrase).
After screening the titles and abstracts, we retrieved published full-text articles and extracted data. Two independent reviewers reviewed the articles for inclusion, and disagreements were resolved by consensus.
Results
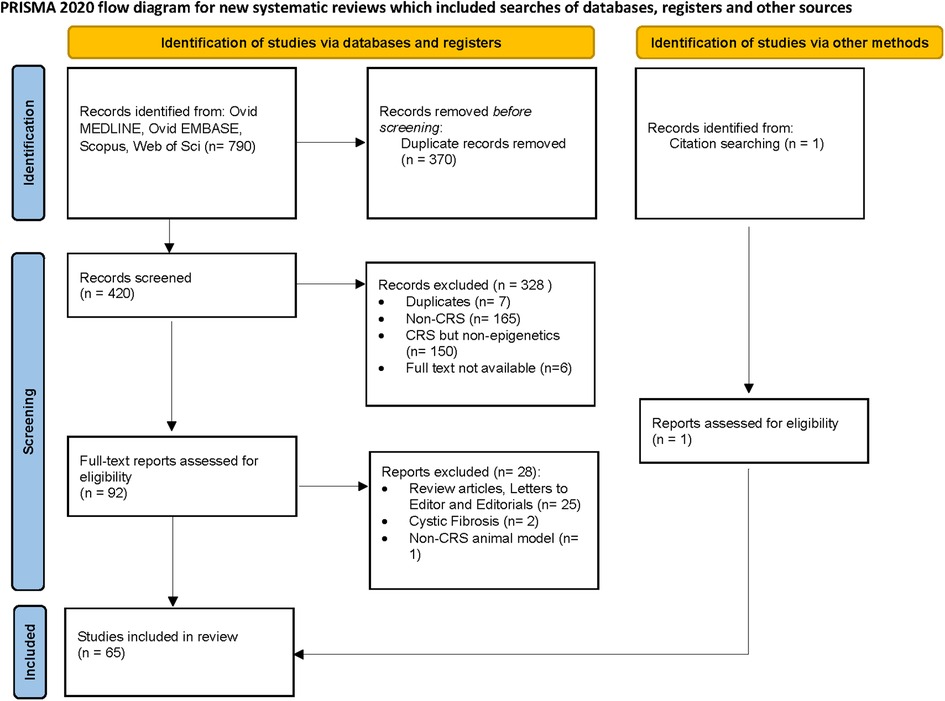
A total of 420 studies were reviewed. After excluding non-CRS studies, non-epigenetic studies, studies without full texts, duplicates, review articles, editorials, and cystic fibrosis, we identified 65 studies that investigated the role of epigenetics in CRS. Please refer to PRISMA diagram illustrated in Figure 2. A majority of studies investigated the role of non-coding RNAs (45 studies) followed by DNA methylation (13 studies). Four studies investigated histone modifications, two studies were on APA, and one study on chromatin accessibility. Most studies focused on CRSwNP, while very few included CRS agnostic of polyp status or CRS without nasal polyps (CRSsNP). Most studies were reported on populations from China (44) followed by Korea (12), Japan (1), Spain (2), Belgium (1), Brazil (2), Poland (1), and the United States (2). Details of the studies are presented below, categorized by the epigenetic mechanism investigated and listed in Tables 1–4 and Supplementary Table S1. Figure 3 illustrates the epigenetic mechanism investigated in CRS and summarizes the genes, proteins, or pathways that were identified or studied.
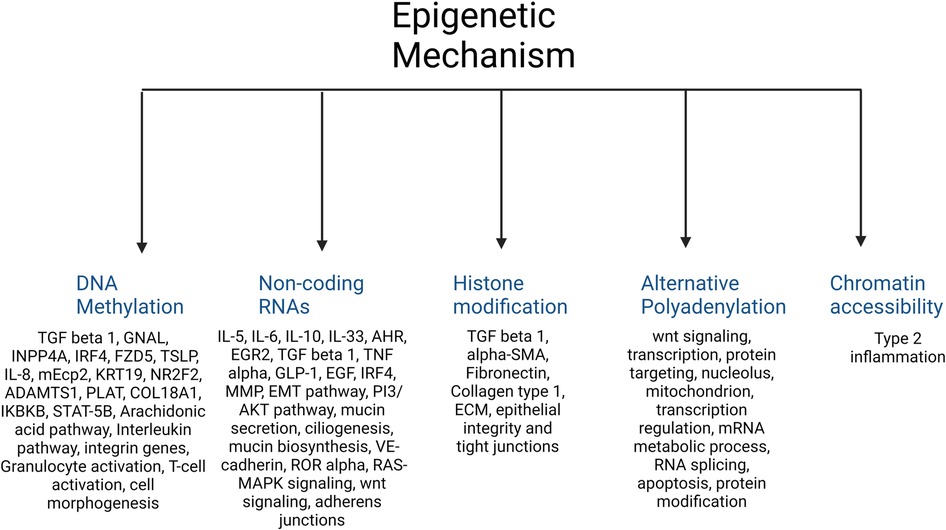
Figure 3. Illustration of the major epigenetic mechanisms investigated in CRS and a summary of the main genes, proteins, or pathways that were identified in each.
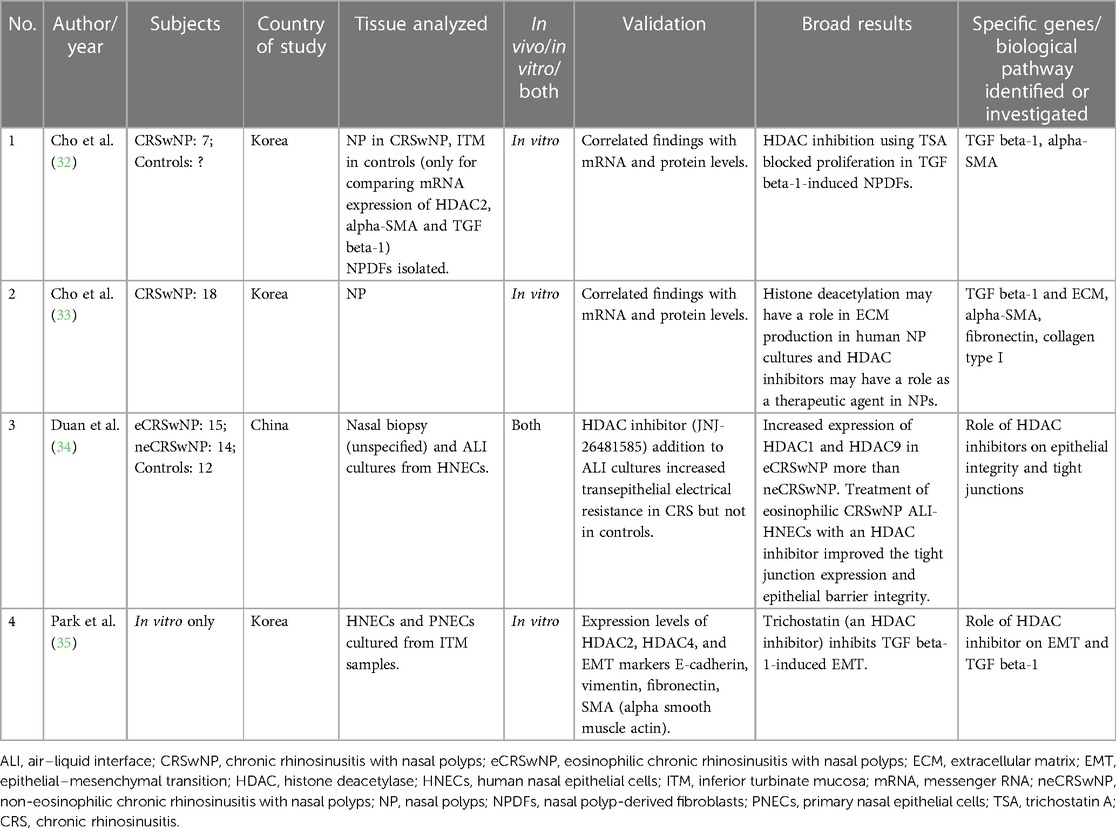
Table 2. Studies evaluating the role of histone modifications (deacetylation) in chronic rhinosinusitis.
DNA methylation studies
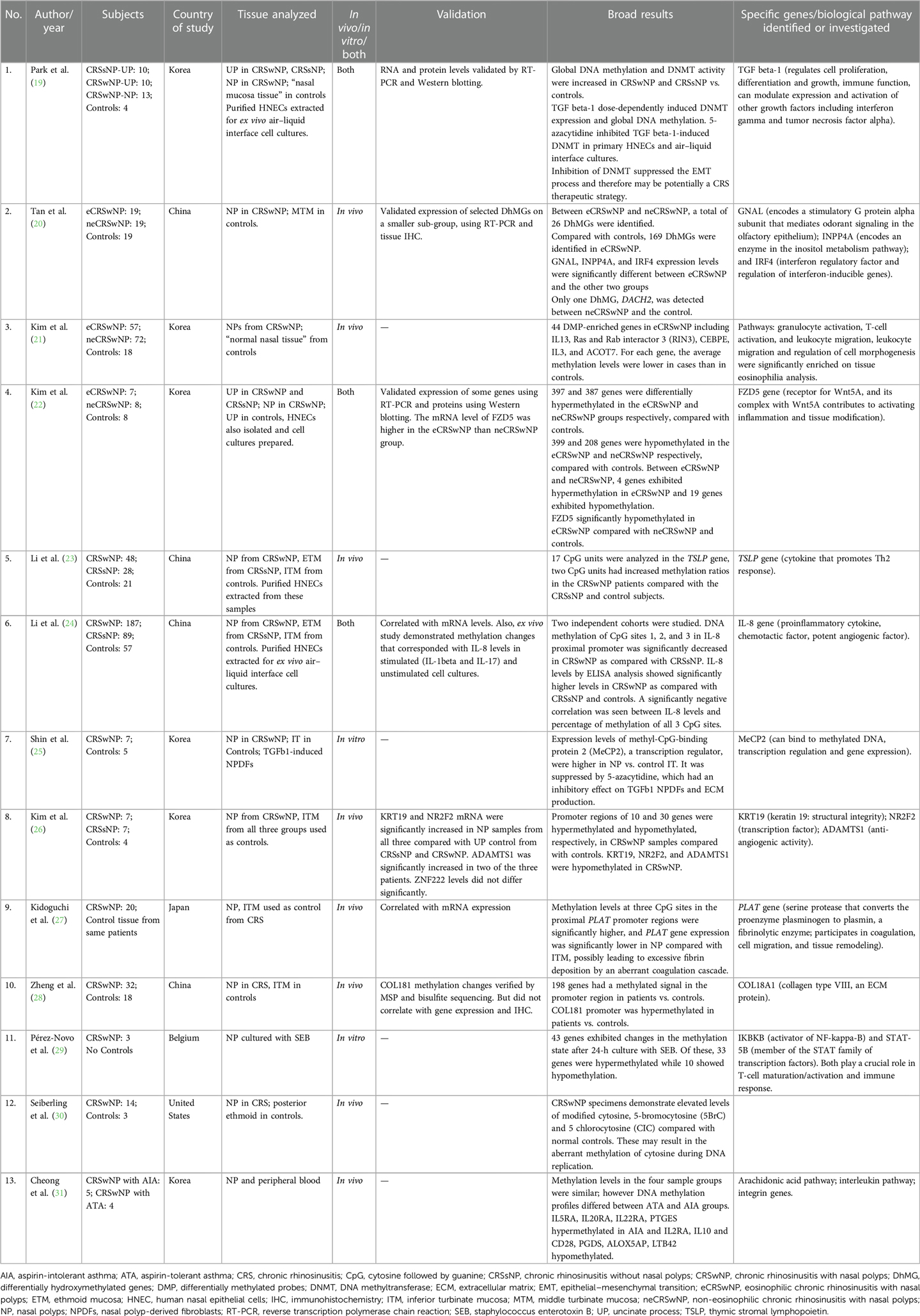
The number of CRS subjects in the studies ranged from 3 to 187. Eleven of 13 studies used a control arm with subject numbers ranging from 3 to 57. The majority (including the genome-wide studies) used inferior turbinate mucosa (ITM) as a control tissue; however, one study used posterior ethmoid (30), one used middle turbinate mucosa (20), one used uncinate process, and one used “normal nasal tissue” (19) as controls. Eight of the 13 studies looked at in vivo changes in nasal polyps (NPs), two looked at in vitro changes, and three studies investigated both in vivo and in vitro. The in vitro studies extracted human nasal epithelial cells (HNECs) from the NP tissue to prepare air–liquid interface cell cultures. Geographically, most of these studies were from the Asian populations (China, Korea, and Japan), one from the United States, and one from Europe (Belgium). For additional details, please refer to Table 1.
Six genome-wide DNA methylation studies found differences in global methylation between CRSwNP and controls (19–22, 26, 28, 31), while others that specifically investigated TSLP (23) (a cytokine involved in dendritic cell maturation and allergic inflammation), IL-8 (24) (proinflammatory cytokine and a potent chemotactic factor, rapidly induced by bacterial and viral products), and PLAT (27) (encodes for a protein that converts the proenzyme plasminogen to plasmin, a fibrinolytic enzyme) found significant differences in methylation of CpG sites of these genes.
The first genome-wide DNA methylation study was published by Cheong et al. in a Korean population and compared CRSwNP with aspirin-intolerant asthma (AIA) and CRSwNP with aspirin-tolerant asthma (ATA) (31). Significant differences were found in methylation of interleukin (IL) genes, IL receptor genes, and genes involved in the arachidonic acid metabolism. IL5RA, IL20RA, IL22RA, and PTGES were hypermethylated in CRSwNP with AIA, and IL2RA, IL10 and CD28, PGDS, ALOX5AP, and LTB42 hypomethylated, as compared with those with CRSwNP with ATA (31). Zheng et al. identified that the COLI8A1 gene was hypermethylated in CRSwNP vs. controls (28). However, this did not correlate with gene expression levels and immunohistochemistry. Kim et al. found hypermethylation in promoter regions of 10 genes and hypomethylation in 30 genes in CRSwNP compared with controls. They selected four genes for validation (KRT19, NR2F2, ADAMTS1, and ZNF222) and found that only some correlated with mRNA expression levels (26). Another genome-wide study investigated differential methylation in eosinophilic CRSwNP (eCRSwNP) vs. non-eosinophilic CRSwNP (neCRSwNP), and also compared them with controls. On comparing with controls, each group showed more than 500 differentially methylated genes, but on comparing with each other, only four were hypermethylated and 19 hypomethylated in eCRSwNP vs. neCRSwNP. FZD5 (a protein that is a receptor for the Wnt5A ligand) was significantly hypomethylated in the eCRSwNP, and the mRNA level of FZD5 was higher in the eCRSwNP than in the neCRSwNP group (22). Another group of investigators that studied eCRSwNP, neCRSwNP, and controls found differences in hydroxymethylated DNA levels. They found 26 differentially hydroxymethylated genes (DhMGs) between eCRSwNP and neCRSwNP, 169 DhMGs in eCRSwNP and controls, and only one DhMG between neCRSwNP and controls. They also found that the GNAL (encodes a stimulatory G protein alpha subunit that mediates odorant signaling in the olfactory epithelium), INPP4A (encodes an enzyme in the inositol metabolism pathway), and IRF4 [interferon (IFN) regulatory factor and regulation of interferon-inducible genes] expression levels were significantly different between eCRSwNP and the other two groups. The middle turbinate mucosa was the control tissue used, as opposed to ITM that was used by others (20). Kim et al. also found 44 differentially methylated probes (DMPs) in eCRSwNP including IL13 (21). Some studies investigated the role of DNA methyltransferase (DNMT) and DNMT inhibitors. Global DNA methylation and DNMT activity were significantly increased in CRSwNP vs. controls and in CRSsNP vs. controls, along with an increased tissue expression level of DNMTs. In addition, nasal epithelial cells were cultured, and TGF beta-1 was shown to induce DNMT activity, global DNA methylation, and mRNA and protein levels of DNMT1, DNMT3A, and DNMT3B, after these had been knocked down using siRNA transfection. The authors also demonstrated that 5-azacytidine, a DNMT inhibitor, inhibited TGF beta-1- induced DNMT activity and epithelial–mesenchymal transition (EMT) in primary HNECs and air–liquid interface cultures (19). Shin et al. also demonstrated the use of DNMT inhibitors as potential therapeutic agents. They demonstrated that expression levels of MeCP2, a transcription regulator, were higher in NP vs. control ITM and was suppressed by 5-azacytidine, which had an inhibitory effect on TGF beta-1 nasal polyp-derived fibroblasts (NPDFs) and extracellular matrix (ECM) production (25).
PLAT was found to be hypermethylated in NP tissue of CRSwNP compared with control ITM from the same group of patients. Gene expression levels were also decreased in NP tissue. The authors suggested that this could possibly lead to excessive fibrin deposition and an aberrant coagulation cascade (27). Li et al. found that methylation levels at CpG sites in the proximal promoter region of IL-8 were significantly decreased in NP of CRSwNP vs. ethmoid tissue of CRSsNP and ITM of control subjects, and this also correlated with IL-8 levels on ELISA, which were higher in CRSwNP vs. CRSsNP and controls (24). It has been found that despite the geographic diversity in the TH cytokine profiles across the globe, IL-8 levels were significantly upregulated in all population groups (39). TSLP showed hypermethylation in CRSwNP in two of 17 CpG units, as compared with CRSsNP and controls. However, gene expression or protein levels were not analyzed (23).
Only two DNA methylation studies originated outside of Asia (29, 30), of which one was an in vitro study (29). Seiberling et al. from the United States investigated and found elevated levels of modified forms of cytosine (5BrC and CIC), which are by-products of activated neutrophils and eosinophils and can result in an aberrant methylation of cytosine. The authors used posterior ethmoid tissue in controls and NP in CRSwNP (30). Pérez-Novo et al. from Belgium sought to investigate pathogen-induced methylation changes in NPs cultured with Staphylococcus enterotoxin B (SEB). They found that after 24 h of incubation, 43 genes exhibited changes in the methylation state, with 33 being hypermethylated and 10 hypomethylated. The two genes highlighted in their data analysis were IKBKB (activator of NF-kappa-B) and STAT-5B (member of the STAT family of transcription factors). Both play a crucial role in T-cell maturation/activation and immune response (29). Microbes and microbial dysbiosis have long been implicated in CRS etiopathogenesis, and further studies are needed to investigate pathogen-induced epigenetic changes. Similarly, smoking can lead to epigenetic changes as has been demonstrated in published literatures, but such studies are lacking in CRS.
To validate results, mRNA levels of some differentially methylated genes were investigated in 6 out of 13 studies and found to be generally consistent. A few studies also correlated protein expression using Western blotting or immunohistochemistry.
Non-coding RNA studies
Out of the 44 studies investigating the role of non-coding RNAs, most studies investigated miRNAs, and only a few investigated other RNA types such as lncRNAs (40–44), siRNAs (45), and circRNAs (46, 47). Six studies used pre-existing GEO data sets to identify transcriptome-wide miRNA or lncRNA profiles of CRSwNP vs. controls, either exclusively or in conjunction with in vivo studies on CRS subjects (40–42, 44, 46, 48). Most studies had control subjects. The number of CRSwNP subjects in studies ranged from 4 to 164, and number of controls ranged from 3 to 50 subjects. A few studies investigated peripheral blood dendritic cells (DCs) (49–51), but most investigated sinonasal tissue (majority used NP in CRSwNP), and one study (the only pediatric epigenetic study) used NPs, serum, and eosinophils (52). For control sinonasal tissue, most used ITM. Only five studies originated outside of Asia. Refer to Supplementary Table S1 for additional details.
Studies either compared transcriptome-wide miRNA profiles (some of which additionally looked at specific miRNAs and their target genes), or only looked at specific miRNA levels [such as miR-19a (50), miR-150-5p (51), miR-124 (53), miR-663 (52), mir-142-3p (54), miR-21 (42), and miR-21-5p (55)] and their potential target genes. A total of 18 out of 44 studies looked at in vivo changes, 4 were only in vitro, 19 were both, and 3 exclusively performed a bioinformatic analysis on pre-existing GEO data sets. One study was exclusively done in a murine model of CRS (56), whereas five studies looked at murine models in addition to human subjects (42, 57–60). Overall, differences in miRNA and lncRNA profiles were found between CRSwNP and controls (40–44, 46, 47, 49, 61, 62, 63–66). The specific miRNAs that were investigated found some of the target genes and pathways as follows: IL-10 (67, 68), IL-5 (64), IL-6 (69), aryl hydrocarbon receptor (AHR) (transcription factor that also modulates immune response) (53), EGR2 (51), TGF beta-1 (52, 62, 70–73) and tumor necrosis factor (TNF) alpha (54, 60), glucagon-like peptide-1 and IL-33 (55), EGF (74), IRF4 (42), PI3K/AKT pathway (40, 56, 58, 65, 75), mucin secretion (76), MMP (77), EMT, VE-cadherin (78), and ROR alpha (61).
Zhang et al. compared global miRNA expression levels between eCRSwNP, neCRSwNP, CRSsNP, and controls. Distinct miRNA profiles were seen in eCRSwNP and CRSsNP. MiR-125b was upregulated in CRSwNP, and the authors showed that miR-125b inhibitor decreased interferon-alpha in vitro and IFN-beta in vivo. Further, IFN-β expression was increased in eosinophilic CRSwNP, and IFN-β mRNA levels positively correlated with IL-5 mRNA levels and eosinophil infiltration in sinonasal mucosa. They concluded that upregulated miR-125b can enhance type I interferon expression in airway epithelial cells (63). Though in another study that compared eCRSwNP with neCRSwNP, CRSsNP, and controls, no differences in miRNA expression were found (79). Luo et al. studied dendritic cells from peripheral blood in CRSwNP vs. controls and found lower IL-10 expression and higher miR-19a expression in subjects and concluded that miR-19a plays a role in the suppression of IL-10 in peripheral dendritic cells. They also found that knocking down the miR-19a gene could abolish the effect of recombinant IL-4-induced suppression of IL-10 expression (50). Ma et al. also studied dendritic cells from peripheral blood and found upregulated expression of miR-150-5p in CRS. They also found that its target gene was EGR2 (early growth response 2) in DCs (80). These authors had previously compared and found distinct miRNA expression patterns in dendritic cells from peripheral blood in atopic CRSwNP, non-atopic CRSwNP, and CRSsNP as compared with controls, with 31 commonly changed miRNAs in all CRS subjects (49).
Liu et al. found that AHR, which is essential for modulating the immune response, was significantly higher in NPs and had an inverse correlation with the expression changes of miR124 (53). Qing et al. in 2021 suggested the role of miR-142-3p in the body's inflammatory response through the TNF alpha signaling pathway in CRSwNP. They studied ITM in subjects as well as controls (54). Liu et al. found miR-21 higher in NPs of CRSwNP vs. controls but no difference between CRSsNP and controls. MiR-21 also positively correlated with IL-10 and negatively with IL-1 beta, IL-6, IL-8, IL-33, TSLP, and Lund Mackay and Lund Kennedy scores. They concluded that miR-21 could be a prominent negative feedback regulator in the inflammatory process (67). Li et al. identified six differentially expressed miRNAs between CRSwNP and CRSsNP, one (miR-4492) between CRSwNP and controls, and none between CRSsNP and controls. They demonstrated that miR-4492 is downregulated, and IL-10 is upregulated in NPs. Using a bioinformatic analysis, they predicted its targets genes as IL-10 and TNF alpha. They found elevated levels of IL-10 in CRSwNP and a negative correlation of miR-4492 and IL-10 but no correlation with TNF alpha (68). Luo et al. had found lower IL-10 expression in peripheral blood dendritic cells of CRSwNP (50).
Xuan et al. found 25 differentially expressed miRNAs between CRSwNP and controls. On pathway analysis, they found that upregulated miRNAs were involved in mucin type O-glycan biosynthesis, and downregulated miRNAs were involved in the TGF beta-1 signaling pathway (62). Another study found the miRNA transcriptome related to ciliogenesis and ciliary function impaired in during differentiation of CRSwNP nasal epithelium as compared with controls (81). Other non-coding RNA studies have also implicated dysregulation in ciliogenesis (81), mucin secretion (76), or mucin type-O-glycan biosynthesis (44, 65, 66).
Yu et al. performed the only pediatric epigenetic study in CRSwNP and compared 35 CRSwNP with 46 controls. In addition to NP (in CRSwNP) and ITM (in controls), they also studied serum and peripheral eosinophils in both groups. They found TGF beta-1 mRNA and protein to be higher in all types of samples from subjects and decreased miR-663 expression in NPs and eosinophils. They concluded that miR-663 may be a negative regulator of TGF beta-1 in children with NPs (52). Several other miRNA studies have implicated or investigated the role of EMT and/or TGF beta-1 (48, 65, 70–73, 75, 82).
Other pathways investigated or identified in these studies include those related to T cells and cytokines (40, 43, 48, 55, 60, 66), eosinophil trafficking (59), PI3K/AKT signaling (40, 56, 58, 65, 75), Ras–MAPK signaling (47, 66, 69, 75), Wnt signaling (47, 57), ECM (83, 84), adherens junctions (65, 83, 84), focal adhesions (47, 65, 84), matrix metalloproteinases (77), vascular permeability (45, 78), cell cycle and apoptosis (64, 85), EGF (74), ROR alpha (on exposure to pollution) (61), and IRF4 (42).
Histone modification studies
There are only four studies on the role of histone modifications, and all have looked at histone deacetylation. All performed in vitro experiments using NPDFs or HNEC cultures. Kindly refer to Table 2 for additional details.
Both studies by Cho et al. on NPDFs illuminate the potential role of trichostatin A (TSA), a histone deacetylase (HDAC) inhibitor, for reversing TGF beta-1-induced ECM accumulation. In their first study, they demonstrated that TSA treatment reversed the morphological changes, ECM accumulation, and myofibroblast differentiation in NPDFs that were stimulated with TGF beta-1. They correlated their findings with mRNA expression and found that TSA reduced alpha-SMA and collagen at mRNA and protein level. They also demonstrated elevated HDAC2 levels in NP vs. ITM tissue and that TSA inhibited HDAC levels. Using histone levels, they also demonstrated that TSA leads to hyperacetylation (32). In their subsequent study, the authors also demonstrated that TSA inhibited TGF beta-1-induced changes in NPDFs, and this was done through the suppression of protein expression of HDAC2 and HDAC4 (33). The other studies by Duan et al. (34) and Park et al. (35) also investigated the use of HDAC inhibitors. Duan et al. similarly found increased expression of HDAC1 and HDAC9 in nasal biopsy specimens of eCRSwNP as compared with neCRSwNP (34). All these studies have found a potential role of these drugs in treating CRSwNP. With the availability of HDAC inhibitors as therapeutic agents in other diseases, new possibilities for treatment of CRS using HDAC inhibitors may be tapped in the future.
APA studies
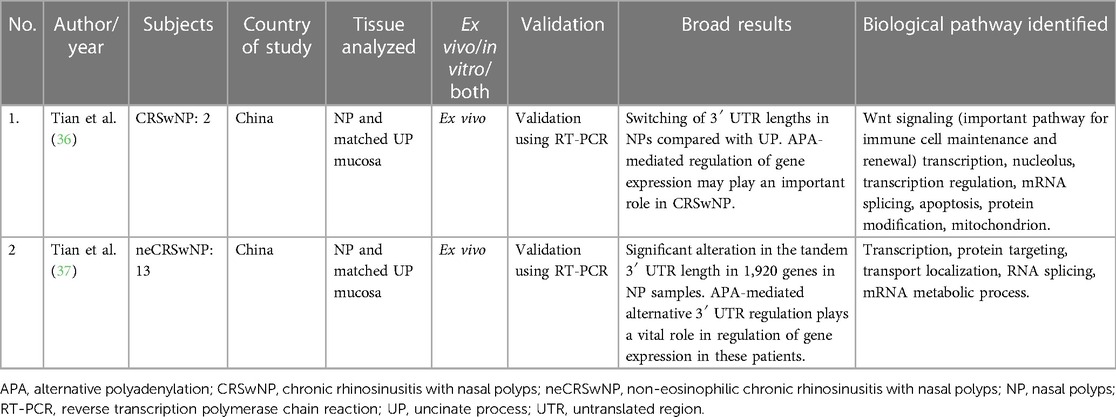
Only two studies on the role of APA in CRS were found, both by the same authors from China. Both looked at in vivo changes and used matched control tissue from the uncinate process mucosa (UPM). Sample sizes were small, with two CRSwNP subjects in the first study and 13 non-eosinophilic CRSwNP patients in their second study. They have shown significant differences in 3′ UTR lengths in NPs compared with control tissue. The pathways that were enriched in these genes were those related to regulation of transcription, macromolecule catabolic localization, and mRNA processing (36, 37). Refer to Table 3 for additional details.
Chromatin accessibility
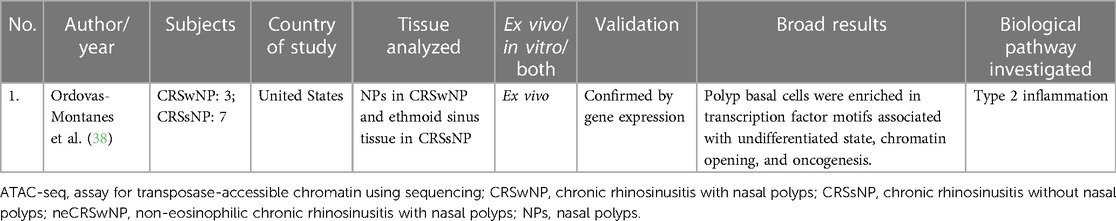
Ordovas-Montanes studied chromatin accessibility in their transcriptomic study in CRS. They found that polyp basal cells were enriched in transcription factor motifs associated with undifferentiated state, chromatin opening, and oncogenesis. This study was primarily an RNA-sequencing study but additionally performed an epigenetic profiling using the assay for transposase-accessible chromatin using sequencing (ATAC-seq) profiling (38). Refer to Table 4 for details.
Discussion
The studies identified in the systematic review were heterogeneous in sample sizes, type of tissue analyzed, control tissue used, and epigenetic mechanism studied. Interestingly, a common theme of principal biological pathways involved in inflammation, cytokine signaling, type 2 response, tissue remodeling, structural integrity, arachidonic acid metabolism, ciliary function, coagulation, and transcription were noted on these studies. In a recent comprehensive review on the role of genetics and epigenetics in CRS, Lal et al. also identified several genes and pathways in CRS that were common in both genetic and epigenetic studies (17). Therefore, the familial predisposition to CRS that has been noted (14, 86) may be a result of genetic factors, epigenetic factors, or both. Epigenetics in CRS has been investigated through different epigenetic mechanisms, but the most commonly investigated is the role of miRNAs followed by DNA methylation, as was also noted by Li et al. in a recent review (18).
The role of TGF beta-1 was identified or investigated through almost all epigenetic mechanisms. It plays an important role in tissue remodeling (potent stimulator of osteoblastic bone formation) and can also induce EMT. In addition, it can regulate cell proliferation, differentiation, and growth; immune function; and expression and activation of other growth factors including interferon gamma and tumor necrosis factor alpha. It promotes either T-helper 17 cells (Th17) or regulatory T cells (Treg) lineage differentiation in a concentration-dependent manner (87). The role of TGF beta-1 has been extensively investigated in asthma (87–90). In addition, its role as a biomarker in diagnosing severe asthma has also been investigated (91). TGF beta ligands have found to be upregulated in several lung conditions including asthma, pulmonary fibrosis, emphysema, and lung cancer (92). Park et al. demonstrated that TGF beta-1 induced DNMT expression and DNA methylation. However, 5-azacytidine (a DNMT inhibitor) was able to inhibit TGF beta-1-induced DNMT and EMT, thus having a potential therapeutic role in CRS (19, 25). In addition, miRNA studies were able to identify TGF beta-1 as a target of miR-663 in pediatric NPs (52). Several other adult miRNA studies have implicated or investigated the role of EMT and/or TGF beta-1 (48, 65, 70–73, 75, 82). Histone deacetylation studies revealed that HDAC inhibitors like trichostatin could block proliferation in TGF beta-1-induced NPDFs (32, 33, 35). In addition to epigenetic studies, genetic studies that have been replicated have also found polymorphisms in the TGF beta-1 gene to be associated with CRS (93, 94) even after excluding asthma (94). Its role in pathophysiology of NPs has been studied in animal models as well (95). Both DNMT inhibitors and HDAC inhibitors have a promising role in targeting TGF beta-1-induced EMT in NPs.
Several interleukins and members of the cytokine family have also been investigated and identified in epigenetic studies. These include IL-10, IL-8, IL-1 beta, IL-5, IL-6, IL-33, IL2RA, IL5RA, IL20RA, IL22RA1, CD28, PIK3CG, TSLP, CCL18, IRF4, and type I interferons (23, 24, 31, 50). IL-10 (a cytokine with pleiotropic effects in immunoregulation and inflammation that downregulates the expression of Th1 cytokines, enhances B-cell survival and proliferation, and antibody production) has been shown to be epigenetically regulated through DNA methylation (31), as well as micro-RNAs (50). It was shown to be hypomethylated in AIA, implying higher levels of IL-10 (31). In other studies related to miRNAs, its expression was reported as higher in NPs in two studies, and it was regulated bymiR-4492 (68) and miR-21 (67) and lower in one study and regulated by miR-19a (50). The promoter region of IL-8 gene, a proinflammatory cytokine, chemotactic factor, and potent angiogenic factor, was significantly hypomethylated and was associated with significantly higher IL-8 expression levels (24). Other proinflammatory cytokines mentioned above include those with a predominant role against microbes (type I IFN, IL-6) and Th2 response (IL5RA, CCL18, TSLP, CD28, and IL22RA). TNF alpha, a potent proinflammatory cytokine, was also found to be higher in CRSwNP and also associated with higher miR-142-3p levels (54). Polymorphisms in TNF alpha gene have also previously been identified in geographically diverse populations in CRS (96–98). TNF alpha inhibitors are currently used to treat multiple inflammatory conditions such as rheumatoid arthritis, psoriasis, inflammatory bowel disease, and ankylosing spondylitis. They have been investigated in vivo for the treatment of asthma and chronic obstructive pulmonary disease (COPD) (99). Given the availability of drugs that target some of the key molecules implied in CRS pathophysiology, there is a great potential to develop novel therapeutics in CRS targeting these proteins. The pathways identified in non-coding RNA studies also relate to immune function, T cells and cytokines (40, 43, 48, 55, 60, 66), and eosinophil trafficking (59).
Dysregulations in ciliogenesis and ciliary function have also been identified. These include involvement of the mucin type-O-glycan biosynthesis pathway (44, 62, 65, 66, 81), mucin secretion (76), and identification of GALNT7 through a lncRNA study (44). Impairment of pathways and implication of genes involved in structural integrity include COL18A1 (28), integrin genes (ITGB2, ITGAL, and ITGAM) (31), KRT19 (26), alpha-SMA, fibronectin, and collagen type 1 (32, 33), and miRNA studies have found pathways involving adherens junctions (65, 83, 84) and focal adhesions (47, 65, 84). An impaired epithelial barrier is a commonly accepted pathophysiological mechanism in CRS, and whether the epigenetic modifications in genes involved in the nasal epithelial structure is a cause or consequence of the inflammatory process is yet to be discerned. Other pathways identified include those involved in coagulation. PLAT gene encodes for a serine protease that converts the proenzyme plasminogen to plasmin, a fibrinolytic enzyme. It participates in coagulation, cell migration, and tissue remodeling. Methylation levels in its promoter were significantly higher and its expression levels lower, possibly leading to the deposition of fibrin (27). Alterations in vascular permeability (45, 78) have also been associated with CRS through non-coding RNA studies. In addition, ADAMST1 (anti-angiogenic activity and active metalloprotease) was hypomethylated, implying increase in angiogenic activity (26). An miRNA study also found that miR-29b-3p expression was positively correlated with the expression of MMP-2 and MMP-9 in CRSwNP (77).
One of the first epigenetic studies in CRS that compared AIA with ATA found aberrations in the arachidonic acid metabolism pathway. Hypomethylation in PGDS (a potent inhibitor of platelet aggregation), ALOX5AP (a protein that converts arachidonic acid to ALOX5), LTB4R (leukotriene B4 receptor involved in inflammatory response and the neuropeptide signaling pathway), and hypermethylation in PTGES [terminal enzyme of the cyclooxygenase (COX)-2-mediated prostaglandin E2 biosynthetic pathway] were identified (31).
Genes encoding for transcription factors, which can in turn regulate expression levels of other proteins, have also been identified. These include MeCP2 (can bind to methylated DNA, transcription regulation, and gene expression) (25), NR2F2 (ligand-inducible transcription factor that is involved in the regulation of many different genes) (26), AHR (transcription factor; among its related pathways are beta-2 adrenergic-dependent CFTR expression and adipogenesis) (53), EGR2 (transcription factor, plays a role in peripheral nerve myelination, may play a role in adipogenesis) (51), and FZD5 (receptor for Wnt5A, and its complex with Wnt5A contributes to activating inflammation and tissue modification) (22). Both APA studies found differences in pathways related to Wnt signaling (important pathway for immune cell maintenance and renewal) transcription, nucleolus, transcription regulation, mRNA splicing, apoptosis, and protein modification (36, 37). Non-coding RNA studies have also found differences in Wnt signaling (47, 57) and cell cycle and apoptosis (64, 85). Other pathways like PI3K/AKT signaling (40, 56, 58, 65, 75) and Ras–MAPK signaling (47, 66, 69, 75), which are important in cell cycle regulation, have also been identified in non-coding RNA studies.
Limitations
Our study has limitations. Due to the heterogeneity of sample sizes, methodology, and control tissue utilized, comparing studies with each other or drawing common conclusions is challenging. In addition, epigenetics may vary based on race, ethnicity, and geographical location, and since most of these studies have been performed in Asia, it may be difficult to generalize the findings across other populations. Just as genetic differences between races can affect the interpretation of genetic studies in CRS from different parts of the world, similarly geographical and environmental differences can lead to different epigenetic changes. Type 2 endotype is more common in patients with NPs in the western world vs. Asians, where mixed endotype remains dominant (39). In addition, most studies classify CRS based on phenotype (CRSwNP and CRSsNP); however, there is emerging evidence, including transcriptomic data, to show that polyp status may not reveal true pathogenetic mechanisms at play (100). This makes it imperative to study genetic and epigenetic mechanisms across racially and geographically diverse populations. The presence of co-morbid conditions like asthma and allergic rhinitis may influence results of studies, and this may limit our understanding of CRSwNP-specific epigenetic changes. Bioinformatic analyses on pre-existing GEO data sets need to be consistent in including studies that have used the same disease and control tissue, and this has not been done uniformly across these studies (40–42). In a recent review on epigenetics in CRS, authors have elaborated that several miRNA studies have shown controversial results in whether a specific miRNA was upregulated or downregulated in CRS (18). Because of the conflicting data, the interpretation of epigenetic studies in CRS remains a challenge.
Areas of further research
Epigenetic studies on racially and geographically distinct populations are the need of the hour. In addition, studying the epigenetic effects of different environmental stimuli, including different microbes, smoking, and the effects of air pollution (performed in vitro by one study) (61) may lead to further insights on disease etiopathogenesis, as may longitudinal studies. Further research also needs to account for confounding factors such as asthma and allergic rhinitis, the presence of which may make interpretation of results more complicated. Studies on pediatric age groups have also not been undertaken, barring one miRNA study from China (52).
Since epigenetic modifications are reversible, and greater research on developing targeted therapeutics using HDAC inhibitors and DNMT inhibitors may lead to a paradigm shift in the treatment of the subsets of CRS.
In addition, existing data sets of transcriptomic studies in CRS from different regions of the world can be used to identify and compare non-coding RNA profiles across diverse populations; however, caution in matching disease and control tissue should be exerted.
Conclusions
Differences in methylation patterns, non-coding RNAs, histone deacetylation, APA, and chromatin accessibility studies suggest that in CRS subjects, there is likely a major impact of the environment. Overall, the main genes and biological pathways implicated were those involved in inflammation, cytokine signaling, type 2 response, tissue remodeling, arachidonic acid metabolism, ciliogenesis and mucin secretion, coagulation, and regulation of transcription. Since epigenetic changes are pharmaceutically reversible, it is possible that future studies can be leveraged to develop targeted therapeutics in CRS.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author contributions
TB: conceptualization, methodology, data curation, investigation, and writing—original draft preparation. LM: data curation, software, and resources. DL: conceptualization, methodology, writing—review and editing, and supervision. All authors contributed to the article and approved the submitted version.
Conflict of interest
DL has received consulting fee from GSK.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/falgy.2023.1165271/full#supplementary-material.
Abbreviations
APA, alternative polyadenylation; CRS, chronic rhinosinusitis; eCRSwNP, eosinophilic chronic rhinosinusitis with nasal polyps; neCRSwNP, non-eosinophilic chronic rhinosinusitis with nasal polyps; CRSwNP, chronic rhinosinusitis with nasal polyps; DNMT, DNA methyltransferase; ECM, extracellular matrix; EMT, epithelial–mesenchymal transition; HDAC, histone deacetylase; HNEC, human nasal epithelial cells; IFN, interferon; ITM, inferior turbinate mucosa; lncRNA, long non-coding RNA; miRNA, micro-RNA; NP, nasal polyps; TSA, trichostatin A; UTR, untranslated region.
References
1. Moore LD, Le T, Fan G. DNA methylation and its basic function. Neuropsychopharmacology. (2013) 38(1):23–38. doi: 10.1038/npp.2012.112
2. Annunziato AT, Hansen JC. Role of histone acetylation in the assembly and modulation of chromatin structures. Gene Expr. (2000) 9(1–2):37–61. doi: 10.3727/000000001783992687
3. Struhl K. Histone acetylation and transcriptional regulatory mechanisms. Genes Dev. (1998) 12(5):599–606. doi: 10.1101/gad.12.5.5996/
4. Yao Q, Chen Y, Zhou X. The roles of microRNAs in epigenetic regulation. Curr Opin Chem Biol. (2019) 51:11–7. doi: 10.1016/j.cbpa.2019.01.024
5. Peng Y, Zi XX, Tian TF, Lee B, Lum J, Tang SA, et al. Whole-transcriptome sequencing reveals heightened inflammation and defective host defence responses in chronic rhinosinusitis with nasal polyps. Eur Respir J. (2019) 54(5). doi: 10.1183/13993003.00732-2019/
6. Wei JW, Huang K, Yang C, Kang CS. Non-coding RNAs as regulators in epigenetics (Review). Oncol Rep. (2017) 37(1):3–9. doi: 10.3892/or.2016.5236
7. Yeh HS, Yong J. Alternative polyadenylation of mRNAs: 3′-untranslated region matters in gene expression. Mol Cells. (2016) 39(4):281–5. doi: 10.14348/molcells.2016.0035
8. Skinner MK. Environmental epigenetic transgenerational inheritance and somatic epigenetic mitotic stability. Epigenetics. (2011) 6(7):838–42. doi: 10.4161/epi.6.7.16537
9. Zhang L, Lu Q, Chang C. Epigenetics in health and disease. Adv Exp Med Biol. (2020) 1253:3–55. doi: 10.1007/978-981-15-3449-2_1
10. Nepali K, Liou JP. Recent developments in epigenetic cancer therapeutics: clinical advancement and emerging trends. J Biomed Sci. (2021) 28(1):27. doi: 10.1186/s12929-021-00721-x
11. Orlandi RR, Kingdom TT, Smith TL, Bleier B, DeConde A, Luong AU, et al. International consensus statement on allergy and rhinology: rhinosinusitis 2021. Int Forum Allergy Rhinol. (2021) 11(3):213–739. doi: 10.1002/alr.22741
12. Fokkens WJ, Lund VJ, Hopkins C, Hellings PW, Kern R, Reitsma S, et al. Executive summary of EPOS 2020 including integrated care pathways. Rhinology. (2020) 58(2):82–111. doi: 10.4193/Rhin20.601
13. Tan BK, Schleimer RP, Kern RC. Perspectives on the etiology of chronic rhinosinusitis. Curr Opin Otolaryngol Head Neck Surg. (2010) 18(1):21–6. doi: 10.1097/MOO.0b013e3283350053
14. Oakley G, Curtin K, Orb Q, Schaefer C, Orlandi R, Alt J. Familial risk of chronic rhinosinusitis with and without nasal polyposis: genetics or environment. Int Forum Allergy Rhinol. (2015) 5(4):276–82. doi: 10.1002/alr.21469
15. Bohman A, Oscarsson M, Holmberg K, Johansson L, Millqvist E, Nasic S, et al. Heredity of nasal polyps. Rhinology. (2015) 53(1):25–8. doi: 10.4193/Rhino14.032
16. Brar T, Marino MJ, Lal D. Unified airway disease: genetics and epigenetics. Otolaryngol Clin North Am. (2023) 56(1):23–38. doi: 10.1016/j.otc.2022.09.002
17. Lal D, Brar T, Ramkumar SP, Li J, Kato A, Zhang L. Genetics and epigenetics of chronic rhinosinusitis. J Allergy Clin Immunol. (2023) 151(4):848–68. doi: 10.1016/j.jaci.2023.01.004
18. Li J, Qiu CY, Tao YJ, Cheng L. Epigenetic modifications in chronic rhinosinusitis with and without nasal polyps. Front Genet. (2023) 13:1089647. doi: 10.3389/fgene.2022.1089647
19. Park JH, Shin JM, Yang HW, Park IH. DNMTs are involved in TGF-β1-induced epithelial-mesenchymal transitions in airway epithelial cells. Int J Mol Sci. (2022) 23(6):3003. doi: 10.3390/ijms23063003
20. Tan H, Tong X, Gao Z, Xu Y, Tan L, Zhang W, et al. The hMeDIP-seq identified INPP4A as a novel biomarker for eosinophilic chronic rhinosinusitis with nasal polyps. Epigenomics. (2022) 14(12):757–75. doi: 10.2217/epi-2022-005
21. Kim KW, Park SC, Cho HJ, Jang H, Park J, Shim HS, et al. Integrated genetic and epigenetic analyses uncover MSI2 association with allergic inflammation. J Allergy Clin Immunol. (2021) 147(4):1453–63. doi: 10.1016/j.jaci.2020.06.040
22. Kim JY, Cha MJ, Park YS, Kang J, Choi JJ, In SM, et al. Upregulation of FZD5 in eosinophilic chronic rhinosinusitis with nasal polyps by epigenetic modification. Mol Cells. (2019) 42(4):345–55. doi: 10.14348/molcells.2019.2418
23. Li J, Jiao J, Gao Y, Zhang Y, Zhang L. Association between methylation in nasal epithelial TSLP gene and chronic rhinosinusitis with nasal polyps. Allergy Asthma Clin Immunol. (2019) 15(1):1–8. doi: 10.1186/s13223-019-0389-3
24. Li J, Jiao J, Wang M, Gao Y, Li Y, Wang Y, et al. Hypomethylation of the IL8 promoter in nasal epithelial cells of patients with chronic rhinosinusitis with nasal polyps. J Allergy Clin Immunol. (2019) 144(4):993–1003.e12. doi: 10.1016/j.jaci.2019.06.042
25. Shin JM, Um JY, Lee SA, Park IH, Lee SH, Lee HM. Effect of MeCP2 on TGF-β1-induced extracellular matrix production in nasal polyp-derived fibroblasts. Am J Rhinol Allergy. (2018) 32(4):228–35. doi: 10.1177/1945892418770291
26. Kim JY, Kim DK, Yu MS, Cha MJ, Yu SL, Kang J. Role of epigenetics in the pathogenesis of chronic rhinosinusitis with nasal polyps. Mol Med Rep. (2018) 17(1):1219–27. doi: 10.3892/mmr.2017.8001
27. Kidoguchi M, Noguchi E, Nakamura T, Ninomiya T, Morii W, Yoshida K, et al. DNA methylation of proximal PLAT promoter in chronic rhinosinusitis with nasal polyps. Am J Rhinol Allergy. (2018) 32(5):374–9. doi: 10.1177/1945892418782236
28. Zheng YB, Zhao Y, Yue LY, Lin P, Liu YF, Xian JM, et al. Pilot study of DNA methylation in the pathogenesis of chronic rhinosinusitis with nasal polyps. Rhinology. (2015) 53(4):345–52. doi: 10.4193/Rhino14.086
29. Pérez-Novo CA, Zhang Y, Denil S, Trooskens G, De Meyer T, Van Criekinge W, et al. Staphylococcal enterotoxin B influences the DNA methylation pattern in nasal polyp tissue: a preliminary study. Allergy Asthma Clin Immunol. (2013) 9(1):48. doi: 10.1186/1710-1492-9-48
30. Seiberling KA, Church CA, Herring JL, Sowers LC. Epigenetics of chronic rhinosinusitis and the role of the eosinophil. Int Forum Allergy Rhinol. (2012) 2(1):80–4. doi: 10.1002/alr.20090
31. Cheong HS, Park SM, Kim MO, Park JS, Lee JY, Byun JY, et al. Genome-wide methylation profile of nasal polyps: relation to aspirin hypersensitivity in asthmatics. Allergy Eur J Allergy Clin Immunol. (2011) 66(5):637–44. doi: 10.1111/j.1398-9995.2010.02514.x
32. Cho JS, Moon YM, Park IH, Um JY, Moon JH, Park SJ, et al. Epigenetic regulation of myofibroblast differentiation and extracellular matrix production in nasal polyp-derived fibroblasts. Clin Exp Allergy. (2012) 42(6):872–82. doi: 10.1111/j.1365-2222.2011.03931.x
33. Cho JS, Moon YM, Park IH, Um JY, Kang JH, Kim TH, et al. Effects of histone deacetylase inhibitor on extracellular matrix production in human nasal polyp organ cultures. Am J Rhinol Allergy. (2013) 27(1):18–23. doi: 10.2500/ajra.2013.27.3827
34. Duan S, Han X, Jiao J, Wang M, Li Y, Wang Y, et al. Histone deacetylase activity is a novel target for epithelial barrier defects in patients with eosinophilic chronic rhinosinusitis with nasal polyps. Clin Exp Allergy. (2023) 53(4):443–54. doi: 10.1111/cea.14258
35. Park IH, Kang JH, Shin JM, Lee HM. Trichostatin A inhibits epithelial mesenchymal transition induced by TGF-β1 in airway epithelium. PLoS One. (2016) 11(8):e0162058. doi: 10.1371/journal.pone.0162058
36. Tian P, Sun Y, Li Y, Liu X, Wan L, Li J, et al. A global analysis of tandem 3′UTRs in eosinophilic chronic rhinosinusitis with nasal polyps. PLoS One. (2012) 7(11):1–12. doi: 10.1371/journal.pone.0048997
37. Tian P, Li J, Liu X, Li Y, Chen M, Ma Y, et al. Tandem alternative polyadenylation events of genes in non-eosinophilic nasal polyp tissue identified by high-throughput sequencing analysis. Int J Mol Med. (2014) 33(6):1423–30. doi: 10.3892/ijmm.2014.1734
38. Ordovas-Montanes J, Dwyer DF, Nyquist SK, Buchheit KM, Vukovic M, Deb C, et al. Allergic inflammatory memory in human respiratory epithelial progenitor cells. Nature. (2018) 560(7720):649–54. doi: 10.1038/s41586-018-0449-8
39. Wang X, Zhang N, Bo M, Holtappels G, Zheng M, Lou H, et al. Diversity of TH cytokine profiles in patients with chronic rhinosinusitis: a multicenter study in Europe, Asia, and Oceania. J Allergy Clin Immunol. (2016) 138(5):1344–53. doi: 10.1016/j.jaci.2016.05.041
40. Wang X, Zhu X, Peng L, Zhao Y. Identification of lncRNA biomarkers and LINC01198 promotes progression of chronic rhinosinusitis with nasal polyps through sponge miR-6776-5p. Biomed Res Int. (2022) 2022:9469207. doi: 10.1155/2022/9469207
41. Li K, Liu FF. Analysis of competing endogenous RNA (ceRNA) crosstalk in eosinophilic chronic rhinosinusitis with nasal polyps. Int Forum Allergy Rhinol. (2022) 12(12):1468–79. doi: 10.1002/alr.23008
42. Chen JC, Xing QL, Yang HW, Yang F, Luo Y, Kong WJ, et al. Construction and analysis of a ceRNA network and patterns of immune infiltration in chronic rhinosinusitis with nasal polyps: based on data mining and experimental verification. Sci Rep. (2022) 12(1):9735. doi: 10.1038/s41598-022-13818-6
43. Wang M, Bu X, Luan G, Lin L, Wang Y, Jin J, et al. Distinct type 2-high inflammation associated molecular signatures of chronic rhinosinusitis with nasal polyps with comorbid asthma. Clin Transl Allergy. (2020) 10:26. doi: 10.1186/s13601-020-00332-z
44. Liu M, Guo P, An J, Guo C, Lu F, Lei Y. Genome-wide profiling of lncRNA and mRNA expression in CRSwNP. Mol Med Rep. (2019) 49(5):3855–63. doi: 10.3892/mmr.2019.10005
45. Cao C, Yan C, Hu Z, Zhou S. Potential application of injectable chitosan hydrogel treated with siRNA in chronic rhinosinusitis therapy. Mol Med Rep. (2015) 12(5):6688–94. doi: 10.3892/mmr.2015.4237
46. Sun Q, Liu Z, Xu X, Yang Y, Han X, Wang C, et al. Identification of a circRNA/miRNA/mRNA ceRNA network as a cell cycle-related regulator for chronic sinusitis with nasal polyps. J Inflamm Res. (2022) 15:2601–15. doi: 10.2147/JIR.S358387
47. Yu J, Kang X, Xiong Y, Luo Q, Dai D, Ye J. Gene expression profiles of circular RNAs and microRNAs in chronic rhinosinusitis with nasal polyps. Front Mol Biosci. (2021) 8:643504. doi: 10.3389/fmolb.2021.643504
48. Hao W, Zhu Y, Guo Y, Wang H. miR-1287-5p upregulation inhibits the EMT and pro-inflammatory cytokines in LPS-induced human nasal epithelial cells (HNECs). Transpl Immunol. (2021) 68:101429. doi: 10.1016/j.trim.2021.101429
49. Ma ZX, Tan X, Shen Y, Ke X, Yang YC, He XB, et al. MicroRNA expression profile of mature dendritic cell in chronic rhinosinusitis. Inflamm Res. (2015) 64(11):885–93. doi: 10.1007/s00011-015-0870-5
50. Luo XQ, Shao JB, Di XR, Zeng L, Li XX, Qiu SQ, et al. Micro RNA-19a interferes with IL-10 expression in peripheral dendritic cells of patients with nasal polyposis. Oncotarget. (2017) 8(30):48915–21. doi: 10.18632/oncotarget.16555
51. Ma Z, Shen Y, Zeng Q, Liu J, Yang L, Fu R, et al. MiR-150-5p regulates EGR2 to promote the development of chronic rhinosinusitis via the DC-Th axis. Int Immunopharmacol. (2018) 1(54):188–97. doi: 10.1016/j.intimp.2017.11.011
52. Yu H, Ju J, Liu J, Li D. Aberrant expression of miR-663 and transforming growth factor-β1 in nasal polyposis in children. Exp Ther Med. (2018) 15(5):4550–6. doi: 10.3892/etm.2018.5927
53. Liu CC, Xia M, Zhang YJ, Jin P, Zhao L, Zhang J, et al. Micro124-mediated AHR expression regulates the inflammatory response of chronic rhinosinusitis (CRS) with nasal polyps. Biochem Biophys Res Commun. (2018) 500(2):145–51. doi: 10.1016/j.bbrc.2018.03.204
54. Qing X, Zhang Y, Peng Y, He G, Liu A, Liu H. Mir-142-3p regulates inflammatory response by contributing to increased TNF-α in chronic rhinosinusitis with nasal polyposis. Ear Nose Throat J. (2021) 100(1):NP50–6. doi: 10.1177/0145561319847972
55. Luan G, Wang M, Yuan J, Bu X, Wang Y, Ying S, et al. MicroRNA-21-5p promotes mucosal type 2 inflammation via regulating GLP1R/IL-33 signaling in chronic rhinosinusitis with nasal polyps. J Allergy Clin Immunol. (2022) 150(6):1460–75. doi: 10.1016/j.jaci.2022.05.030
56. Gu X, Yao X, Liu D. Up-regulation of microRNA-335-5p reduces inflammation via negative regulation of the TPX2-mediated AKT/GSK3β signaling pathway in a chronic rhinosinusitis mouse model. Cell Signal. (2020) 70:109596. doi: 10.1016/j.cellsig.2020.109596
57. Song L, Wang X, Qu X, Lv C. Transcription factor specificity protein 1 regulates inflammation and fibrin deposition in nasal polyps via the regulation of microRNA-125b and the Wnt/β-catenin signaling pathway. Inflammation. (2022) 45(3):1118–32. doi: 10.1007/s10753-021-01605-w
58. Jian B, Yin P. STAT1 mediates the PI3K/AKT pathway through promoting microRNA-18a in nasal polyps. Immunopharmacol Immunotoxicol. (2022) 44(2):194–205. doi: 10.1080/08923973.2021.2025388
59. Korde A, Ahangari F, Haslip M, Zhang X, Liu Q, Cohn L, et al. An endothelial microRNA-1-regulated network controls eosinophil trafficking in asthma and chronic rhinosinusitis. J Allergy Clin Immunol. (2020) 145(2):550–62. doi: 10.1016/j.jaci.2019.10.031
60. Du J, Lv H, Dou X, Cao Z. Nuclear factor κB/microRNA-155 upregulates the expression pattern of cytokines in regulating the relapse of chronic sinusitis with nasal polyps and the underlying mechanism of glucocorticoid. Med Sci Monit. (2020) 26:e923618. doi: 10.12659/MSM.923618
61. Shin C-H, Byun J, Lee K, Kim B, Noh YK, Tran NL, et al. Exosomal miRNA-19a and miRNA-614 induced by air pollutants promote proinflammatory M1 macrophage polarization via regulation of RORα expression in human respiratory mucosal microenvironment. J Immunol. (2020) 205(11):3179–90. doi: 10.4049/jimmunol.2000456
62. Xuan L, Luan G, Wang Y, Lan F, Zhang X, Hao Y, et al. MicroRNAs regulating mucin type O-glycan biosynthesis and transforming growth factor β signaling pathways in nasal mucosa of patients with chronic rhinosinusitis with nasal polyps in northern China. Int Forum Allergy Rhinol. (2019) 9(1):106–13. doi: 10.1002/alr.22230
63. Zhang XH, Zhang YN, Bin LH, Hu CY, Wang N, Cao PP, et al. Overexpression of miR-125b, a novel regulator of innate immunity, in eosinophilic chronic rhinosinusitis with nasal polyps. Am J Respir Crit Care Med. (2012) 185(2):140–51. doi: 10.1164/rccm.201103-0456OC
64. Silveira MLC, Tamashiro E, Santos ARD, Martins RB, Faria FM, Silva LECM, et al. miRNA-205-5p can be related to T2-polarity in chronic rhinosinusitis with nasal polyps. Rhinology. (2021) 59(6):567–76. doi: 10.4193/Rhin21.109
65. Cha S, Seo EH, Lee SH, Kim KS, Oh CS, Moon JS, et al. MicroRNA expression in extracellular vesicles from nasal lavage fluid in chronic rhinosinusitis. Biomedicines. (2021) 9(5):471. doi: 10.3390/biomedicines9050471
66. Bu X, Wang M, Luan G, Wang Y, Wang C, Zhang L. Integrated miRNA and mRNA expression profiling reveals dysregulated miRNA-mRNA regulatory networks in eosinophilic and non-eosinophilic chronic rhinosinusitis with nasal polyps. Int Forum Allergy Rhinol. (2021) 11(8):1207–19. doi: 10.1002/alr.22781
67. Liu R, Du J, Zhou J, Zhong B, Ba L, Zhang J, et al. Elevated microRNA-21 is a brake of inflammation involved in the development of nasal polyps. Front Immunol. (2021) 12:530488. doi: 10.3389/fimmu.2021.530488
68. Li L, Feng J, Zhang D, Yong J, Wang Y, Yao J, et al. Differential expression of miR-4492 and IL-10 is involved in chronic rhinosinusitis with nasal polyps. Exp Ther Med. (2019) 18(5):3968–76. doi: 10.3892/etm.2019.8022
69. Zhang J, Han L, Chen F. Let-7a-5p regulates the inflammatory response in chronic rhinosinusitis with nasal polyps. Diagn Pathol. (2021) 16(1):27. doi: 10.1186/s13000-021-01089-0
70. Li X, Li C, Zhu G, Yuan W, Xiao ZA. TGF-β1 induces epithelial-mesenchymal transition of chronic sinusitis with nasal polyps through microRNA-21. Int Arch Allergy Immunol. (2019) 179(4):304–19. doi: 10.1159/000497829
71. Yang N, Cheng H, Mo Q, Zhou X, Xie M. Mir-155-5p downregulation inhibits epithelial-to-mesenchymal transition by targeting SIRT1 in human nasal epithelial cells. Mol Med Rep. (2020) 22(5):3695–704. doi: 10.3892/mmr.2020.11468
72. Shin JM, Park JH, Yang HW, Moon JW, Lee HM, Park IH. miR-29b regulates TGF-β1-induced epithelial-mesenchymal transition by inhibiting heat shock protein 47 expression in airway epithelial cells. Int J Mol Sci. (2021) 22(21):11535. doi: 10.3390/ijms222111535
73. Jiang W, Zhou C, Ma C, Cao Y, Hu G, Li H. TGF-β1 induces epithelial-to-mesenchymal transition in chronic rhinosinusitis with nasal polyps through microRNA-182. Asian Pac J Allergy Immunol. (2021). doi: 10.12932/AP-040921-1224
74. Chen J, Liu D, Yang J, Jin C, Zhao C, Cheng J. Epidermal growth factor activates a hypoxia-inducible factor 1α-microRNA-21 axis to inhibit aquaporin 4 in chronic rhinosinusitis. Ann N Y Acad Sci. (2022) 1518(1):299–314. doi: 10.1111/nyas.14914
75. Xia G, Bao L, Gao W, Liu S, Ji K, Li J. Differentially expressed miRNA in inflammatory mucosa of chronic rhinosinusitis. J Nanosci Nanotechnol. (2015) 15(3):2132–9. doi: 10.1166/jnn.2015.9161
76. Yan D, Ye Y, Zhang J, Zhao J, Yu J, Luo Q. Human neutrophil elastase induces MUC5AC overexpression in chronic rhinosinusitis through miR-146a. Am J Rhinol Allergy. (2020) 34(1):59–69. doi: 10.1177/1945892419871798
77. Liu Z, Liu H, Yu D, Gao J, Ruan B, Long R. Downregulation of miR-29b-3p promotes α-tubulin deacetylation by targeting the interaction of matrix metalloproteinase-9 with integrin β1 in nasal polyps. Int J Mol Med. (2021) 48(1):126. doi: 10.3892/ijmm.2021.4959
78. Zhang W, Zhang T, Yan Y, Zhang J, Zhou Y, Pei Y, et al. Exosomal miR-22-3p derived from chronic rhinosinusitis with nasal polyps regulates vascular permeability by targeting VE-cadherin. Biomed Res Int. (2020) 2020:1237678. doi: 10.1155/2020/1237678
79. Zhang YN, Cao PP, Zhang XH, Lu X, Liu Z. Expression of microRNA machinery proteins in different types of chronic rhinosinusitis. Laryngoscope. (2012) 122(12):2621–7. doi: 10.1002/lary.23517
80. Ma Z, Shen Y, Zeng Q, Liu J, Yang L, Fu R, et al. MiR-150-5p regulates EGR2 to promote the development of chronic rhinosinusitis via the DC-Th axis. Int Immunopharmacol. (2018) 54:188–97. doi: 10.1016/j.intimp.2017.11.011
81. Callejas-Díaz B, Fernandez G, Fuentes M, Martínez-Antón A, Alobid I, Roca-Ferrer J, et al. Integrated mRNA and microRNA transcriptome profiling during differentiation of human nasal polyp epithelium reveals an altered ciliogenesis. Allergy Eur J Allergy Clin Immunol. (2020) 75(10):2548–61. doi: 10.1111/all.14307
82. Zhang T, Zhou Y, You B, You Y, Yan Y, Zhang J, et al. miR-30a-5p inhibits epithelial-to-mesenchymal transition by targeting CDK6 in nasal polyps. Am J Rhinol Allergy. (2021) 35(2):152–63. doi: 10.1177/1945892420939814
83. Mimmi S, Lombardo N, Maisano D, Piazzetta G, Pelaia C, Pelaia G, et al. Spotlight on a short-time treatment with the IL-4/IL-13 receptor blocker in patients with CRSwNP: microRNAs modulations and preliminary clinical evidence. Genes (Basel). (2022) 13(12):2366. doi: 10.3390/genes13122366
84. He S, Wu J, Han D, Li Y, Wang T, Wei H, et al. Differential expression profile of plasma exosomal microRNAs in chronic rhinosinusitis with nasal polyps. Exp Biol Med (Maywood). (2022) 247(12):1039–46. doi: 10.1177/15353702221090184
85. Morawska-Kochman M, Śmieszek A, Marcinkowska K, Marycz KM, Nelke K, Zub K, et al. Expression of apoptosis-related biomarkers in inflamed nasal Sinus epithelium of patients with chronic rhinosinusitis with nasal polyps (CRSwNP)-evaluation at mRNA and miRNA levels. Biomedicines. (2022) 10(6):1400. doi: 10.3390/biomedicines10061400
86. Bohman A, Juodakis J, Oscarsson M, Bacelis J, Bende M, Torinsson Naluai Å. A family-based genome-wide association study of chronic rhinosinusitis with nasal polyps implicates several genes in the disease pathogenesis. PLoS One. (2017) 12(12):e0185244. doi: 10.1371/journal.pone.0185244
87. Musiol S, Alessandrini F, Jakwerth CA, Chaker AM, Schneider E, Guerth F, et al. TGF-β1 drives inflammatory Th cell but not Treg cell compartment upon allergen exposure. Front Immunol. (2022) 7(12):763243. doi: 10.3389/fimmu.2021.763243
88. Yuan J, Zhang W. Expression and significance of TGF-β1 in infant asthma model. Cell Mol Biol (Noisy-le-grand). (2022) 68(7):51–5. doi: 10.14715/cmb/2022.68.7.9
89. Panek MG, Karbownik MS, Górski KM, Koćwin M, Kardas G, Marynowski M, et al. New insights into the regulation of TGF-β/Smad and MPK signaling pathway gene expressions by nasal allergen and methacholine challenge test in asthma. Clin Transl Allergy. (2022) 12(7):e12172. doi: 10.1002/clt2.12172
90. Riemma MA, Cerqua I, Romano B, Irollo E, Bertolino A, Camerlingo R, et al. Sphingosine-1-phosphate/TGF-β axis drives epithelial mesenchymal transition in asthma-like disease. Br J Pharmacol. (2022) 179(8):1753–68. doi: 10.1111/bph.15754
91. Prabha A, Lokesh KS, Chaya SK, Jayaraj BS, Malamardi S, Subbarao MVSST, et al. Pilot study investigating diagnostic utility of serum MMP-1 and TGF-β1 in asthma in “real world” clinical practice in India. J Clin Pathol. (2022) 75(4):222–5. doi: 10.1136/jclinpath-2020-206821
92. Saito A, Horie M, Nagase T. TGF-β signaling in lung health and disease. Int J Mol Sci. (2018) 19(8):2460. doi: 10.3390/ijms19082460
93. Kim SH, Park HS, Holloway JW, Shin HD, Park CS. Association between a TGFbeta1 promoter polymorphism and rhinosinusitis in aspirin-intolerant asthmatic patients. Respir Med. (2007) 101(3):490–5. doi: 10.1016/j.rmed.2006.07.002
94. Henmyr V, Vandeplas G, Halldén C, Säll T, Olze H, Bachert C, et al. Replication study of genetic variants associated with chronic rhinosinusitis and nasal polyposis. J Allergy Clin Immunol. (2014) 133(1):273–5. doi: 10.1016/j.jaci.2013.08.011
95. Qin D, Liu P, Zhou H, Jin J, Gong W, Liu K, et al. TIM-4 in macrophages contributes to nasal polyp formation through the TGF-β1-mediated epithelial to mesenchymal transition in nasal epithelial cells. Front Immunol. (2022) 5(13):941608. doi: 10.3389/fimmu.2022.941608
96. Erbek SS, Yurtcu E, Erbek S, Atac FB, Sahin FI, Cakmak O. Proinflammatory cytokine single nucleotide polymorphisms in nasal polyposis. Arch Otolaryngol Head Neck Surg. (2007) 133(7):705–9. doi: 10.1001/archotol.133.7.705
97. Bernstein JM, Anon JB, Rontal M, Conroy J, Wang C, Sucheston L. Genetic polymorphisms in chronic hyperplastic sinusitis with nasal polyposis. Laryngoscope. (2009) 119(7):1258–64. doi: 10.1002/lary.20239
98. Batikhan H, Gokcan MK, Beder E, Akar N, Ozturk A, Gerceker M. Association of the tumor necrosis factor-alpha-308 G/A polymorphism with nasal polyposis. Eur Arch Otorhinolaryngol. (2010) 267(6):903–8. doi: 10.1007/s00405-009-1167-5
99. Matera MG, Calzetta L, Cazzola M. TNF-alpha inhibitors in asthma and COPD: we must not throw the baby out with the bath water. Pulm Pharmacol Ther. (2010) 23(2):121–8. doi: 10.1016/j.pupt.2009.10.007
Keywords: epigenetics, CRSwNP, sinusitis, nasal polyps, chronic rhinosinusitis, DNA methylation, miRNA, non-coding RNA
Citation: Brar T, Marks L and Lal D (2023) Insights into the epigenetics of chronic rhinosinusitis with and without nasal polyps: a systematic review. Front. Allergy 4:1165271. doi: 10.3389/falgy.2023.1165271
Received: 13 February 2023; Accepted: 3 May 2023;
Published: 22 May 2023.
Edited by:
Asif H. Khan, Sanofi, FranceReviewed by:
Lei Cheng, Nanjing Medical University, ChinaYu Xu, Renmin Hospital of Wuhan University, China
© 2023 Brar, Marks and Lal. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Devyani Lal bGFsLmRldnlhbmlAbWF5by5lZHU=
 Tripti Brar
Tripti Brar Lisa Marks
Lisa Marks Devyani Lal
Devyani Lal