
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Allergy , 12 September 2022
Sec. Asthma
Volume 3 - 2022 | https://doi.org/10.3389/falgy.2022.993937
This article is part of the Research Topic Non-coding RNAs in Allergy and Asthma View all 4 articles
 Constanze A. Jakwerth
Constanze A. Jakwerth Hannah Kitzberger
Hannah Kitzberger Dimitrii Pogorelov
Dimitrii Pogorelov Annika Müller
Annika Müller Simon Blank
Simon Blank Carsten B. Schmidt-Weber
Carsten B. Schmidt-Weber Ulrich M. Zissler*
Ulrich M. Zissler*
MicroRNAs (miRs) have gained scientific attention due to their importance in the pathophysiology of allergic diseases as well as their potential as biomarkers in allergen-specific treatment options. Their function as post-transcriptional regulators, controlling various cellular processes, is of high importance since any single miR can target multiple mRNAs, often within the same signalling pathway. MiRs can alter dysregulated expression of certain cellular responses and contribute to or cause, but in some cases prevent or repress, the development of various diseases. In this review article, we describe current research on the role of specific miRs in regulating immune responses in epithelial cells and specialized immune cells in response to various stimuli, in allergic diseases, and regulation in the therapeutic approach of allergen-specific immunotherapy (AIT). Despite the fact that AIT has been used successfully as a causative treatment option since more than a century, very little is known about the mechanisms of regulation and its connections with microRNAs. In order to fill this gap, this review aims to provide an overview of the current knowledge.
As upper and lower airways are considered a morphologic and functional unit, not only do allergic rhinitis and asthma share mechanisms of allergic inflammation (1, 2). Also transition of allergic rhinitis into asthma can represent a steady continuum depending on endotype and is therefore of relevance with respect to allergic pathomechanisms (3, 4). Allergic diseases are characterized by an uncontrolled immune reaction towards harmless environmental antigens to which the body is exposed either via airways, as seen in allergic rhinitis and allergic asthma (5). Type 2-driven diseases are characterized by the infiltration of inflammatory cells into the lung, elevated IgE serum levels, mucus hypersecretion, bronchial hyperresponsiveness (BHR), airway obstruction, and chronic inflammation (6, 7). Type 2 cytokines including interleukin-4 (IL-4), IL-5, IL-13, IL-24, and IL-33 mediate allergic inflammation and airway eosinophil infiltration, which affect the function of airway wall epithelial and smooth muscle cells (1, 4, 8). These mediators can be detected in different biomatrices, which can be assessed more or less invasively (8). A functional connection between miR expression and the pathogenesis of asthma was reported for a high number of genes (9). Importantly, mRNA transcripts of multiple genes can be targeted by a single miR (9), which enables miRs to act broadly on controlling cell function.
98% of the human genome consists of non-coding DNA sequences (10). Non-coding RNAs (ncRNAs) have various functions such as gene regulation and can be divided into different groups. Long non-coding RNAs (lncRNA) are defined by being composed of more than 200 nucleotides and not being translated into protein. Their function is often not clear, because it is a big and heterogenous group. One process lncRNAs are involved in is gene regulation on the epigenetic level via chromatin remodelling (11–13). Small nucleolar RNAs (snoRNAs) are responsible for the fine-tuning of the ribosome and spliceosome function. They do this mostly by post-transcriptional regulation of ribosomal RNA (rRNA) and small nuclear RNA (snRNA) (14, 15). Another group are the circular RNAs (ciRNAs), which are single-stranded, covalently closed RNA molecules. They were first discovered in 1976 and thought to be viroids, pathogens of certain plants (16). The first detection in humans was in the human cancer cell line HeLa by electron microscopy in 1979 (17). The most prominent and best-investigated group are miRs, which are about 22 nucleotides long and function as post-transcriptional suppressors of gene expression by binding to a complementary mRNA sequence (18). miRs are transcribed mostly by RNA polymerase II (19), only a few by RNA polymerase III (20). They carry a 5′-cap and are polyadenylated at the 3′ tail (21). miRs are processed by RNAase II enzymes DROSHA and DICER to result in their mature form (22, 23). Its precursors are located most frequently in intergenic regions and introns of protein-coding genes (24). It is also known that one miR can have multiple mRNAs as its target (25).
miRs have been associated with different disease phenotypes. The role of miRs in human diseases has primarily been studied in cancer, showing that defects in their processing can lead to huge damage. This highlights the role of the let-7 miR family, which targets oncogenes and is reduced in cancer cells (26). This reduced post-transcriptional suppression leads to increased expression of oncogenes and thus supports tumour growth (27–29). However, the role of non-coding RNAs in allergic diseases and in central pathways of allergy pathophysiology remains largely unknown.
Allergic diseases are caused by an excessive response to pathogens, allergens, or toxins. Allergic rhinitis, allergic asthma, and atopic dermatitis are the most common allergic diseases (30). Asthma is a very heterogenous chronic airway disease characterized by airway hyperreactivity, smooth muscle hyperresponsiveness and inflammation (31). It is characterized by a type 2 immune response that includes prototypic type 2 cytokines and elevated levels of eosinophils and IgE. miRs can contribute to the development of asthma by regulating inflammation, cell migration, and proliferation, which was shown in animal models and isolated human cells (8, 32–34). In allergic diseases characterized by a cytokine milieu dominated by type 2 mediators such as IL-4 and IL-13, epithelial cells may express miRs (Table 1), which may also be associated with a decline in lung function parameters (4). Recently, it was shown that small extracellular vesicles (sEV)-associated epithelial miRNAs are involved in regulating the immune responses by underlying dendritic cells (DCs) (35). Decreased levels of miR-92b, miR-210, and miR-34a in epithelial-derived sEVs upon asthma development, would allow DCs to polarize Th2 cells, perpetuating the asthmatic phenotype in the lung microenvironment (35). Furthermore, an association of certain miRs with airway obstruction measurements as well as with the gene expression amplitude of their predicted target genes was found. The latter are relevant to type 2 airway polarization in children and therefore suggest a role for miRs in the development of asthma. In addition, airway remodeling and loss of epithelial integrity in asthma affect airway obstruction and hyperreactivity, which may be regulated by epithelial small extracellular vesicles containing specific miRs.
Since asthma is a disease that occurs at a young age, some studies have focused on circulating miRs correlated to lung function parameters: such as miR-15b, -126, -139, -142, -186, -191, -342, -409, -660, -942, -1290 (36). The identification of these miRs, their sex-dependent association with lung function parameters as well as their interactions could be useful in finding a treatment option for type 2-related diseases. A functional associaten between miR expression and the pathogenesis of asthma has been reported for miR-1248, miR-1291, miR-570-3p, and the previously mentioned miR let-7a, which were specifically expressed in affected lungs of asthmatic patients when compared to healthy subjects (37). The let-7 family is known as a modulator in Th2 inflammation and is believed to have a pro-inflammatory effect as it is abundantly expressed in the airway tissues of asthmatic patients and is involved in regulating the expression of IL-33 (38, 39). In a mouse model, miR-21 was found to be increased and to regulate IL-12, which is known to propagate Th1 polarization (28). By analyzing the serum levels of miR-21 and IL-4 in asthmatic patients and healthy controls, a strong positive correlation was detected, as both were significantly higher expressed in asthmatic patients (40). miR-21 is expressed at high levels in different cell types and can be increasingly be induced via STAT3 and NF-κB (41).
miR-19a contributes to the production of Th2 cytokines and was found to be upregulated in airway infiltrating T cells in vitro differentiation experiments. It interacts with mRNAs encoding PTEN, SOCS1, and A20 (TNFAIP3) by deregulating and derepressing their signalling pathways (42). Another mechanism involved in maintaining Th1/Th2 balance is the impact of miR-155 on macrophages. Its targets have been shown to include IL-13, SOCS1, and SHIP1. When miR-155 is reduced in macrophages, STAT6 becomes more active, resulting in an alternative M2 phenotype (43). In a miR-155 knockout mouse model, mice tended to develop asthmatic inflammation and increased levels of IL-4 and IL-5 in T cells (44). In addition, an association between miR-3935 and its predicted target gene, the prostaglandin E3 receptor, was revealed, which could mediate allergen-specific immunotherapy (AIT) effects through suppression of the PGE2-PTGER3 axis (8). Furthermore, miR-221 was found to be reduced in epithelia and sputum of asthma patients and associated with eosinophilic airway inflammation. Therefore, miR-221 might serve a potential biomarker in allergic inflammation. In house dust mite-challenged mice, airway overexpression of miR-221 suppressed CXCL17 expression and thereby enhanced expression of CCL24, CCL26 and POSTN (45). This could potentially affect the epithelial cytokine-mediated priming of epithelial cells towards an E1 or E2 epithelial response, a phenomenon named after the causative Th1 and Th2-derived cytokines (1, 3, 4, 46).
A major factor in the pathogenesis of asthma are changes in gene expression and secreted protein patterns in the airway epithelium (1, 46). Several studies have examined the expression pattern of miRs in airway epithelial cells of type 2-related diseases targeting genes such as TNF-α, IL-8, or IL-6 (34). MiR-19, a well-studied microRNA is able to promote type 2 cytokine production of IL-33, IL-5, and IL-13 through direct targeting of the signaling inhibitors PTEN, SOCS1, and TNFAIP3 (A20) (47). These signaling inhibitors are not only regulators of Th2 cells but also of ILC2 cells (47). Further, miR-449 is able to repress the expression of IL-13 in asthma (48). Interestingly, also some miRs are known to correlate with changes in lung function parameters in a sex-specific manner, such as miR-485, miR-660-5p, or miR-942 (36). miR-1248, which can increase the expression of IL-5, was also described to negatively correlate with lung function parameters in asthma (49). However, also miRs like miR-34 (38) or miR-299 (50) downregulated in asthma are known. Downregulation of miR-133a contributed to upregulation of RhoA in bronchial smooth muscle cells of asthmatic patients (51) and alleviates airway remodeling in asthma by targeting IGF1R (52). miR-192 expression was reduced in the peripheral blood of asthmatic patients undergoing allergen inhalation challenge (53). It is further able to attenuated airway remodeling and autophagy in asthma by targeting MMP-16 and ATG7 (54). miR-21 has been shown to be upregulated in allergic airway inflammation and to regulate IL-12p35 expression, which appears to promote Th2 and attenuates Th1 responses by targeting IL-12 expression (28). An important regulator of Th17 responses was identified with miR-182, which was shown to be upregulated upon Th17 differentiation, targeting FoxO family members 1 and 3 (55). miR-375 is associated with an increase in the expression of TSLP in primary bronchial epithelial cells, a key cytokine in asthmatic airway inflammation (56). Moreover, miR-221 was described to correlate with airway eosinophilia in asthma and further increased CCL-24, CCL-26, and POSTN in asthmatic airways (45). Counterregulatory mechanisms could be induced by upregulation of miR-146a, which could potentially enhance the T regulatory (Treg) cell-mediated suppression of Th1 responses and result in unhindered Th2 activation (57). The let-7 family members appear to target IL-13 expression, as downregulation of let-7 could enhance Th2 responses by upregulating IL-13 expression (39, 58). However, also regulators of epithelial genes are known. A prominent example is miR-1303, which regulates the epithelial-derived ADAM33 known to be involved in asthma and related bronchial hyperreactivity (59). Taken together, miRNAs are involved in a number of regulatory processes in airway epithelial cells during inflammation.
In addition to airway inflammation, airway remodeling is also a pathological hallmark in asthma, driven by eosinophils, neutrophils, and other inflammatory cells (60–62). Increasing evidence suggests that airway remodeling can occur early in childhood, concomitantly with, but not necessarily subsequent to, airway inflammation. Features of airway remodeling include increased smooth muscle cell mass, thickening of airway walls, and epithelial barrier dysfunction (63). In the remodeling process, matrix metalloproteinases (MMPs) were implicated in the degradation of the extracellular matrix in the process of tissue remodeling. MMP-2 and MMP-9 activities were increased around inflamed airways, which were the main site of tissue remodeling (64). In addition, miR-192-5p can alleviate airway inflammation and airway remodeling in asthma by targeting MMP-16 (54). Furthermore, it has also been reported that airway inflammation and airway remodeling are exacerbated by activation of JNK1/2-MMP-9 pathway associated with ORMDL3 knockdown in asthmatic mice, suggesting that miR-192-5p mediates airway remodeling and autophagy by other signaling pathways (65). Abnormal expression has been reported for miR-451a in pulmonary diseases associated with remodeling processes, as elevated levels of expression have been demonstrated in patients with chronic obstructive pulmonary disease (COPD) (66). Several miRs, such as miR-19a, miR-142, and miR-221, have been found to be differentially expressed in asthma and associated with airway remodeling (67–69). miR-142-3p in bronchial biopsies from patients with early- or late-onset severe asthma was consistent with a differential WNT signature, suggesting that miR-142 is involved in regulating the balance between proliferation and differentiation of airway smooth muscle cells in asthma, possibly via control of WNT signaling (68). Also involved in the WNT pathway is β-catenin, which has been shown to be elevated in Aspergillus fumigatus-associated asthma, as the 3′-UTR of the β-catenin transcript is a genuine miR-3162-3p binding site (70). Endogenous miR-3162-3p could aggravate the severity of allergic asthma caused by miR-3162-3p antagomir by alleviating the reduction in β-catenin expression in asthmatic mice and suppressing airway hyperresponsiveness and airway inflammation (70). Furthermore, miR-221 is hypothesized to play a crucial role in driving the differentiation of the Th17/Treg ratio via RORγt/Foxp3 by targeting SOCS1 (69). In addition, mir-221 reduced the Th17 cell function by directly inhibiting RORγt/SOCS1 and promoted the function of Treg cells via induction of Foxp3/SOCS1 in asthma (69). Asthma-related airway remodeling was reduced by miR-451a overexpression, which was been shown to target ETS1, while miR-451a downregulation promotes differentiation of CD4+ T Cells towards Th2 cells through ETS1 upregulation in childhood asthma (71). Among these phases, the airway remodeling phase is irreversible, and directly related to the prognosis of affected children (61).
Additionally, the extracellular matrix plays an important role in asthma-induced airway remodeling, including type I collagen (COL-I) and fibronectin (FN), since secretion of COL-I and FN results in thickening of the basal membrane and subcutaneous fibrosis of the airway, eventually leading to airway remodeling (61). MiR-146a and miR-146b are negative regulators of inflammatory gene expression in lung fibroblasts, epithelial cells, monocytes, and endothelial cells. They negatively regulate the expression of cyclooxygenase-2 (COX-2) and IL-1β (72). These findings suggest that miR-146a and/or miR-146b are able to modulate the expression of inflammatory mediators in airway smooth muscle cells, thereby contributing to the pathogenesis of asthma (72). Elevated levels of miR-378 in the serum of asthmatic children have been shown to promote cell proliferation and resistance to apoptosis. Upregulation of miR-378 promotes smooth muscle cell proliferation and resistance to apoptosis, and increases the expression of COL-I and FN proteins (61).
Thymic stromal lymphopoietin (TSLP) acts as a key epithelial-derived cytokine involved in remodeling of the asthmatic airway. Decreased TSLP expression reduced the production of inflammatory cytokines and thereby inhibited STAT3 expression and phosphorylation. STAT3 upregulation and activation in the airways are closely associated with the onset and development of asthma, while STAT3 activation facilitates TSLP in asthma-associated airway remodeling (73). TSLP can be negatively regulated by miR-19b by binding to the upstream non-coding region of TSLP (74). Therefore, miR-19b is able to reduce airway remodeling, airway inflammation, and levels of oxidative stress by inhibiting TSLP signaling through STAT3. Since TSLP as well as the other epithelium-derived alarmins IL-25 and IL-33 also affect the expression and secretion of IL-5 and IL-13 from innate lymphoid cells type-2 (ILC2), it directly influences the miR-mediated regulation of immune cells.
The function of ILC2s by targeting TSLP can be regulated by miR-375, which has been reported to be downregulated in patients with Th2-associated diseases such as atopic dermatitis, ulcerative colitis and allergic rhinitis (75). Systemic miR-15b levels were shown to be associated with lung function, while miR-16-5p levels correlated with bronchial hyperresponsiveness (BHR). Local T cells from asthma patients expressed diminished levels of miR-19a-3p, the upregulated levels of which promote the production of Th2 cytokines (42). miR-126 increases GATA-3 expression in T cells in an indirect manner that might promote a Th2 response and is believed to increase IL-13 levels. This suggests that it is associated with an excessive activation of Th2 cells in asthmatic children (76). In contrast, miR-21-3p detected in exhaled breath condensate of asthmatics showed reduced levels compared to healthy controls (77). In addition, a pediatric study observed elevated levels of miR-191-5p targeting CEBPB, which correlated with the lung function parameter FEV1/FVC (Tiffeneau) (36). Furthermore, miR-223-3p was elevated in asthmatics compared to healthy controls and correlated with lung function parameters (36). MiR-210-3p has been shown to be involved in mast cell activation (78), while miR-223-3p is involved in neutrophil maturation and function by inhibiting IL-18 expression in macrophages, suggesting a fine-tuned mechanism involving inflammatory IL-18-induced neutrophil extracellular traps (NETs) (79).
Since both the innate and adaptive immune responses show altered IL-13 levels, the expression of the interlinked microRNA could also be altered. IL-13, produced by Th2 cells but also by group 2 ILC2s, has been found to increase neutrophil-derived PGE2 levels by upregulating COX-2 gene expression (8). The suppressive effects of AIT on type-2 immune responses not only affect Th2 cells but also induce a shift from ILC2 to ILC1. For example, miR-155 can affect the Th1/Th2 balance by targeting IL-13RA, SOCS1, and SHIP1 of macrophages, since specific modulation of miR-155 expression may be able to reduce exaggerated inflammation (80). In addition, IL-10 induces miR-187, which is able to negatively regulate the expression of the pro-inflammatory mediators TNF-α, IL-6 and IL-12p40, as it directly affects the stability and translation of TNF-α mRNA targets and indirectly decreases IL-6 and IL-12p40 expression via down-modulation of IκBζ, a major regulator of the transcription of these two cytokines (81). Furthermore, miR-21 expression increased remarkably in an OVA-induced asthma model and suppressed the expression of IL-12/STAT4 proteins (82). Therefore, miR-21 expression increased significantly and both IL-12p70 and IL-12p35 were down-regulated in mice with OVA-induced asthma (82). In addition, suppression of miR-21 by intranasal administration of the antagomir improved asthma symptoms including airway inflammation and BHR, inhibited Th2 polarization of CD4+/CD8− cells, and altered cytokine levels in BAL fluid (83). These immune cell-related regulatory changes are key targets of treatment options such as AIT, which also initiate changes in miR expression and regulation.
miRs have been intensively studied in recent years in the context of allergic and respiratory diseases (9, 84). There is a wealth of research focusing on the role of miRs in the pathogenesis of asthma and allergies, and thus to our understanding and delineation of disease endotypes (85) in preclinical (28) and clinical (86) settings.
No less important was the work aimed at elucidating the differences in specific miR levels in health and disease (40), revealing the association between specific miRs and disease risk, severity (71, 87) and exacerbation (88). Further, also characterizing of miR profiles of patients presenting with one or two allergic comorbidities (89), and lastly predicting the clinical remission of childhood disease in early adulthood (71). Further, the effect of prophylactic sublingual immunotherapy revealed changes of miRs in asymptomatic subjects, who were sensitized to cedar pollen (90). As cedar pollinosis is the predominant seasonal allergic rhinitis in Japan, it is of interest that miR-223 was significantly up-regulated in pollen season and let-7b was down-regulated in sensitized subjects and may possibly mirror the exposure to antigen during pollen season (90). Another important line of research lies in the investigation of miRs as potential biomarkers for monitoring allergy treatments and therapeutic outcomes, particularly in relation to resource-intensive therapies, such as AIT (Figure 1; Table 2).
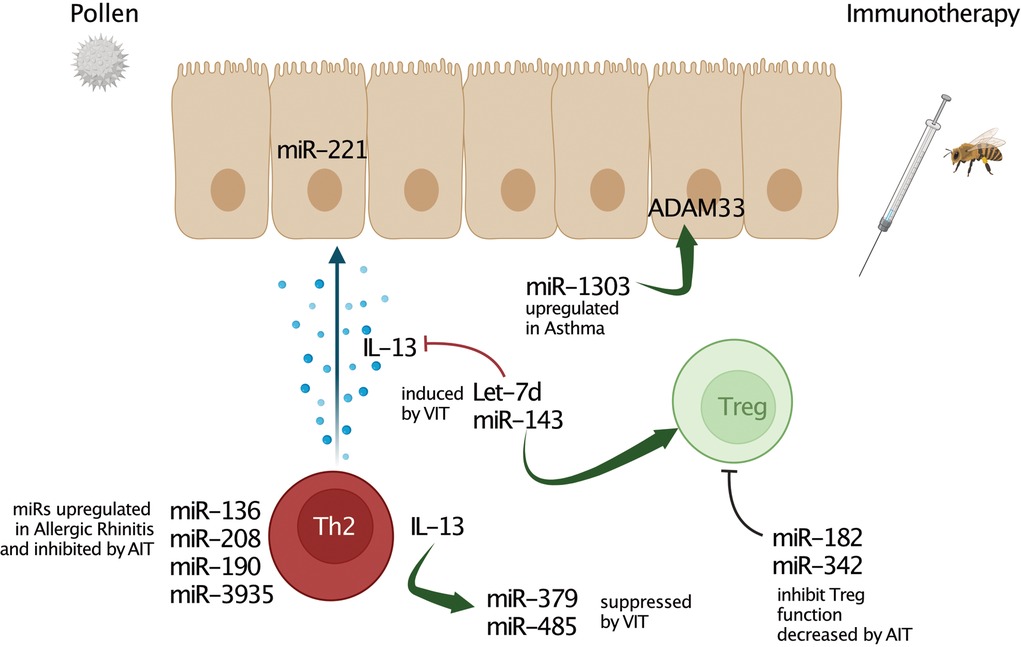
Figure 1. MicroRNA interactions in allergic inflammation and allergen-specific immunotherapy (AIT). Several miRs play a crucial role in regulating multiple processes and characteristics of allergy-associated disease pathology. Deregulation of multiples miRs affects to inflammatory processes, Th1/Th2 response balance, cytokine, chemokine, lipid mediator production, and remodelling processes. However, miRs up-regulated by immunotherapy such as Let-7d or miR-143 can also alleviate airway inflammation and decrease pro-allergic cytokine production by promoting regulatory T cells (Tregs). These miRs directly inhibit type 2 key mediators such as IL-13. Further, negative regulatory association of miR-3935 and its predicted target gene, the PGE2 receptor EP3, revealed this miR as a potential AIT-mediated mechanism in the airways of AA patients. Moreover, there is also evidence for miRs targeting regulatory factors of asthma such as ADAM33.
Allergen-specific immunotherapy is an effective disease-modifying therapy used for the treatment of allergic sensitizations, however of limited use in severe or uncontrolled asthma. Targeting specialized treatment regimens, individual genetics, environment, and lifestyle must be incorporated in therapeutic decision-making (91). These include comprehensive and accurate allergy diagnostic testing tools, a better understanding of the mechanisms, the identification of biomarkers to predict and monitor response, and the development of safe, effective, affordable and convenient treatments (91). In addition to changes in decreased allergen-specific IgE and increased allergen-specific IgG production, AIT has been associated with several other immunological events, including alterations in allergen-specific T- and B-cell population and associated cytokine responses, production of antibodies capable of blocking allergen presentation, thought to be of the IgG4 subset, reduction in tissue eosinophils and mast cells, as well as decreased basophil activation (91–93). Some published works observed these immunological events take place at different time points in the AIT course (93, 94). As mast cell and basophil desensitization are early events in AIT treatment, they are followed by the induction of IL-10 producing T and B regulatory cells that induce a B cell isotype switch that shifts the immunoglobulin production from IgE to IgG. There is also a shift in cytokine production which results in suppression of effector type 1 and type 2 cells reflecting the antagonistic cytokine pattern (1, 93, 95). These changes in the cytokine milieu are in turn affecting also e.g. the airway epithelium, which was shown to express an antagonistic response following a cytokine-dependent polarization (1, 2, 93). AIT has been used in clinical practice for more than 100 years, and although it has been used very successfully to treat severe insect sting reactions with a response rate of approximately 90%, it has variable responses in patients with respiratory allergies and is considered moderately effective (96). However, considering this fact, it must be taken into account that the efficacy of therapy for insect venom allergy and allergies to inhaled allergens are assessed in different ways. Insect venom allergy is a disease with infrequent or even absent allergen exposure for years and the efficacy of VIT is assessed based on the lack of systemic sting reaction. In contrast, allergies to inhalant allergens are characterized by seasonal or perennial allergen exposure, and treatment efficacy is assessed based on a symptom and/or medication score. Due to these differences, the efficacy of VIT and AIT with inhalant allergens is difficult to compare in a purposeful manner. With the advent and rapid development of miR technologies, the question of AIT efficacy in airway diseases and its biomarkers thas been revisited in several studies. Specjalski et al. determined the expression of 48 miRs in whole blood samples from 16 allergic rhinitis (AR) patients receiving grass pollen AIT and seven healthy controls to establish the possible correlation between miR upregulation and clinical outcomes (97). AR patients showed upregulation of miR-136, miR-208, and miR-190 compared to healthy controls, but no differences between good and poor responders were observed after six months of treatment, despite the overall reduction in pro-inflammatory miRs.
Recently, Jakwerth et al. sought to decipher the effects of grass pollen AIT on miR expression in the sputum of AR patients with or without asthma (98). While over two thousand miRs were upregulated in patients compared to controls, this number was only four in AIT-treated individuals. The prostaglandin EP3 receptor, which is the target of one of those upregulated miRs, was downregulated in AIT-treated compared to untreated patients. Of note, PGE2 levels were elevated even in the AR group, decreased after AIT, and correlated with type 2 markers in sputum and symptom severity.
Although miR-based approaches have enabled considerable progress in improving our understanding of the role of finely tuned mechanisms regulating gene expression in health and in a variety of allergic diseases, there remains an unmet need for large-cohort double-blind placebo-controlled AIT studies to determine which miR signatures are the most promising candidates for identifying therapy responders and predicting treatment outcomes in the clinical practice.
Like AIT with inhalant allergens, venom-specific immunotherapy (VIT) is a highly effective causative treatment for allergy that can restore allergen tolerance and protect the patients against future potentially fatal allergic sting (99). In contrast to allergy e.g. to airborne allergen sources, venom allergy is not considered an atopic disease. To date, very few studies addressed changes in the expression of miRs over the course of VIT (Table 2).
A 2016 study examined the changes in 740 miRs 24 h after completion of the up-dosing phase of VIT using an ultra-rush protocol in seven patients with yellow jacket venom allergy (100). Out of the 740 miRs, 440 could be detected in the peripheral blood of the patients. Five miRs had significantly altered expression 24 h after completion of the up-dosing phase compared to baseline. A significant increase was observed for miR-299 and miR-29c and significant decreases for miR-370, miR-539, and miR-502-3p, none of which were previously mentioned in the context of allergen-specific immunotherapy. Interestingly, the miR-29 family is expressed in T and B cells and thus might be involved in a variety of immunological responses (101, 102), while miR-502-3p is induced by IL-4 (103). In addition, the same study found another 62 miRs that changed 2-fold in some patients, although these changes were not significant. Nevertheless, these miRs could be interesting candidates in connection with the protective effect of VIT, since they are associated with e.g. stimulation of FoxP3 expression and CD4 + CD25+ lymphocyte proliferation (up-regulation of miR-143) or inhibition of IL-13 and IL-13Rα1 expression (up-regulation of let-7d and miR-143) (58, 73).
In a follow-up study, the same authors addressed changes in miR expression after three months of the maintenance phase in 5 patients with yellow jacket venom allergy (104). At this time point, six miRs showed significantly lower and 11 higher expressions compared to the baseline before the start of VIT. Again, it was found that miR-143 and let-7d are up-regulated by VIT. Additionally, among the most upregulated miRs was miR-34b, which has been shown to be downregulated in asthma (38) and suppressed by IL-13 (48). Other upregulated miRs included amongst others miR-1303, which regulates ADAM33 (59), miR-204, which regulates bronchial smooth muscle cell proliferation (105), miR-299, which is down-regulated in asthma (49), IL-13-induced miR-379 (106) and miR-485 up-regulated in asthma (102). Significantly down-regulated miRs included miR-182, which promotes clonal expansion of activated Th cells and regulates Treg function (12, 107), miR-342, which regulates Treg function (108), and miR-375 up-regulated in bronchial epithelial cells during pollutant-induced asthma exacerbations (56).
Limitations of the studies described above in elucidating potential miR-associated mechanisms contributing to the high therapeutic efficacy of VIT and long-lasting allergen tolerance include the small patient numbers and early sampling time points. Karpinski et al. compared the expression pattern of 2549 miRs from 13 patients with venom allergy one year after the start of VIT with the baseline values before VIT (109). Here, both correlation and principal component analysis indicated a limited effect of VIT on the overall miR expression pattern. After 12 months of VIT, the expression pattern in whole blood was broadly similar to that observed before VIT. Taking into account the differences between the studies, the authors conclude that the short- and long-term effects of VIT on the miR pattern appear to differ. Therefore, larger controlled studies are needed to examine the contribution of miRs to the tolerogenic effects of VIT in both immediate and long-lasting effects.
New techniques have led to major advances in the field of miR research, resulting in a deluge of information and long lists of potentially interesting miRs. However, mechanistic studies on miR targeting and function are often lacking. Furthermore, miRs alone may not or only rarely be the key to explaining the pathology of asthma or allergic diseases, so that complex interaction networks are required to elucidate the pathogenesis of diseases and their heterogeneity. The expression of miR in certain cell populations, in defined disease states, in therapy models and in certain phenotypes must also be researched intensively to decode the pathophysiological consequences of altered miR expression, for example in allergic diseases. However, this review article summarizes the critical role of miR in the pathogenesis of asthma and allergic diseases and their associated comorbidities, making them interesting targets for therapeutic interventions such as allergen-specific immunotherapy. Since asthma and allergies are very heterogeneous diseases and current treatments are still ineffective in controlling severe forms of these diseases, there is a great need for precise and more effective therapies based on a deep understanding of the disease-underlying mechanisms. Interest in using miR profiles as biomarkers in lung diseases continues to grow. However, in order to implement the use of miRs as biomarkers, they should be very specific for certain signaling pathways or sub-cell types that, through their association with the disease or therapy underlying mechanisms, allow for a more detailed characterization of particular disease phenotypes. The position that the individual miRs can have in the hierarchy as biomarkers for predicting the course of a disease or the response to therapeutic interventions must be clarified in the coming years.
UMZ formulated the outline of the review, CAJ, HK, DP, AM, SB, CSW, and UMZ wrote sections of the manuscript, and edited the manuscript. All authors contributed to the article and approved the submitted version.
UMZ and CSW are supported by German Center for Lung Research (DZL) and German Research Foundation (DFG; 398577603).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Zissler UM, Chaker AM, Effner R, Ulrich M, Guerth F, Piontek G, et al. Interleukin-4 and interferon-gamma orchestrate an epithelial polarization in the airways. Mucosal Immunol. (2016) 9(4):917–26. doi: 10.1038/mi.2015.110
2. Zissler UM, Jakwerth CA, Guerth F, Lewitan L, Rothkirch S, Davidovic M, et al. Allergen-specific immunotherapy induces the suppressive secretoglobin 1A1 in cells of the lower airways. Allergy. (2021) 76(8):2461–74. doi: 10.1111/all.14756
3. Zissler UM, Ulrich M, Jakwerth CA, Rothkirch S, Guerth F, Weckmann M, et al. Biomatrix for upper and lower airway biomarkers in patients with allergic asthma. J Allergy Clin Immunol. (2018) 142(6):1980–3. doi: 10.1016/j.jaci.2018.07.027
4. Jakwerth CA, Ordovas-Montanes J, Blank S, Schmidt-Weber CB, Zissler UM. Role of respiratory epithelial cells in allergic diseases. Cells. (2022) 11(9):1387. doi: 10.3390/cells11091387
5. Musiol S, Alessandrini F, Jakwerth CA, Chaker AM, Schneider E, Guerth F, et al. TGF-beta1 Drives inflammatory th cell but not treg cell compartment upon allergen exposure. Front Immunol. (2021) 12:763243. doi: 10.3389/fimmu.2021.763243
6. Rondon C, Eguiluz-Gracia I, Shamji MH, Layhadi JA, Salas M, Torres MJ, et al. Ige test in secretions of patients with respiratory allergy. Curr Allergy Asthma Rep. (2018) 18(12):67. doi: 10.1007/s11882-018-0821-7
7. Trejo Bittar HE, Yousem SA, Wenzel SE. Pathobiology of severe asthma. Annu Rev Pathol. (2015) 10:511–45. doi: 10.1146/annurev-pathol-012414-040343
8. Baumann R, Untersmayr E, Zissler UM, Eyerich S, Adcock IM, Brockow K, et al. Noninvasive and minimally invasive techniques for the diagnosis and management of allergic diseases. Allergy. (2021) 76(4):1010–23. doi: 10.1111/all.14645
9. Weidner J, Bartel S, Kilic A, Zissler UM, Renz H, Schwarze J, et al. Spotlight on microRNAs in allergy and asthma. Allergy. (2021) 76(6):1661–78. doi: 10.1111/all.14646
10. Mattick JS. Non-coding RNAs: the architects of eukaryotic complexity. EMBO Rep. (2001) 2(11):986–91. doi: 10.1093/embo-reports/kve230
11. Lee JT. Epigenetic regulation by long noncoding RNAs. Science. (2012) 338(6113):1435–9. doi: 10.1126/science.1231776
12. Bothur E, Raifer H, Haftmann C, Stittrich AB, Brustle A, Brenner D, et al. Antigen receptor-mediated depletion of FOXP3 in induced regulatory T-lymphocytes via PTPN2 and FOXO1. Nat Commun. (2015) 6:8576. doi: 10.1038/ncomms9576
13. Li P, Lang X, Xia S. Elevated expression of microRNA-378 in children with asthma aggravates airway remodeling by promoting the proliferation and apoptosis resistance of airway smooth muscle cells. Exp Ther Med. (2019) 17(3):1529–36. doi: 10.3892/etm.2018.7141
14. Busch H, Reddy R, Rothblum L, Choi YC. SnRNAs, SnRNPs, and RNA processing. Annu Rev Biochem. (1982) 51:617–54. doi: 10.1146/annurev.bi.51.070182.003153
15. Jack K, Bellodi C, Landry DM, Niederer RO, Meskauskas A, Musalgaonkar S, et al. rRNA pseudouridylation defects affect ribosomal ligand binding and translational fidelity from yeast to human cells. Mol Cell. (2011) 44(4):660–6. doi: 10.1016/j.molcel.2011.09.017
16. Sanger HL, Klotz G, Riesner D, Gross HJ, Kleinschmidt AK. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc Natl Acad Sci U S A. (1976) 73(11):3852–6. doi: 10.1073/pnas.73.11.3852
17. Hsu MT, Coca-Prados M. Electron microscopic evidence for the circular form of RNA in the cytoplasm of eukaryotic cells. Nature. (1979) 280(5720):339–40. doi: 10.1038/280339a0
18. Ambros V. The functions of animal microRNAs. Nature. (2004) 431(7006):350–5. doi: 10.1038/nature02871
19. Saini HK, Griffiths-Jones S, Enright AJ. Genomic analysis of human microRNA transcripts. Proc Natl Acad Sci U S A. (2007) 104(45):17719–24. doi: 10.1073/pnas.0703890104
20. Borchert GM, Lanier W, Davidson BL. RNA Polymerase III transcribes human microRNAs. Nat Struct Mol Biol. (2006) 13(12):1097–101. doi: 10.1038/nsmb1167
21. Cai X, Hagedorn CH, Cullen BR. Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA. (2004) 10(12):1957–66. doi: 10.1261/rna.7135204
22. Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, et al. The nuclear RNase III drosha initiates microRNA processing. Nature. (2003) 425(6956):415–9. doi: 10.1038/nature01957
23. Basyuk E, Suavet F, Doglio A, Bordonne R, Bertrand E. Human let-7 stem-loop precursors harbor features of RNase III cleavage products. Nucleic Acids Res. (2003) 31(22):6593–7. doi: 10.1093/nar/gkg855
24. Lagos-Quintana M, Rauhut R, Meyer J, Borkhardt A, Tuschl T. New microRNAs from mouse and human. RNA. (2003) 9(2):175–9. doi: 10.1261/rna.2146903
25. Shalgi R, Lieber D, Oren M, Pilpel Y. Global and local architecture of the mammalian microRNA-transcription factor regulatory network. PLoS Comput Biol. (2007) 3(7):e131. doi: 10.1371/journal.pcbi.0030131
26. Sun X, Liu J, Xu C, Tang SC, Ren H. The insights of let-7 miRNAs in oncogenesis and stem cell potency. J Cell Mol Med. (2016) 20(9):1779–88. doi: 10.1111/jcmm.12861
27. Hammond SM. MicroRNAs as tumor suppressors. Nat Genet. (2007) 39(5):582–3. doi: 10.1038/ng0507-582
28. Croce CM. Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet. (2009) 10(10):704–14. doi: 10.1038/nrg2634
29. Kumar MS, Lu J, Mercer KL, Golub TR, Jacks T. Impaired microRNA processing enhances cellular transformation and tumorigenesis. Nat Genet. (2007) 39(5):673–7. doi: 10.1038/ng2003
30. Holgate ST, Polosa R. Treatment strategies for allergy and asthma. Nat Rev Immunol. (2008) 8(3):218–30. doi: 10.1038/nri2262
32. Garbacki N, Di Valentin E, Huynh-Thu VA, Geurts P, Irrthum A, Crahay C, et al. MicroRNAs profiling in murine models of acute and chronic asthma: a relationship with mRNAs targets. PLoS One. (2011) 6(1):e16509. doi: 10.1371/journal.pone.0016509
33. Collison A, Siegle JS, Hansbro NG, Kwok CT, Herbert C, Mattes J, et al. Epigenetic changes associated with disease progression in a mouse model of childhood allergic asthma. Dis Model Mech. (2013) 6(4):993–1000. doi: 10.1242/dmm.011247
34. Jardim MJ, Dailey L, Silbajoris R, Diaz-Sanchez D. Distinct microRNA expression in human airway cells of asthmatic donors identifies a novel asthma-associated gene. Am J Respir Cell Mol Biol. (2012) 47(4):536–42. doi: 10.1165/rcmb.2011-0160OC
35. Bartel S, La Grutta S, Cilluffo G, Perconti G, Bongiovanni A, Giallongo A, et al. Human airway epithelial extracellular vesicle miRNA signature is altered upon asthma development. Allergy. (2020) 75(2):346–56. doi: 10.1111/all.14008
36. Kho AT, Sharma S, Davis JS, Spina J, Howard D, McEnroy K, et al. Circulating MicroRNAs: association with lung function in asthma. PLoS One. (2016) 11(6):e0157998. doi: 10.1371/journal.pone.0157998
37. Roff AN, Craig TJ, August A, Stellato C, Ishmael FT. MicroRNA-570-3p regulates HuR and cytokine expression in airway epithelial cells. Am J Clin Exp Immunol. (2014) 3(2):68–83. ISSN: 2164-7712/AJCEI000055025143867
38. Levanen B, Bhakta NR, Torregrosa Paredes P, Barbeau R, Hiltbrunner S, Pollack JL, et al. Altered microRNA profiles in bronchoalveolar lavage fluid exosomes in asthmatic patients. J Allergy Clin Immunol. (2013) 131(3):894–903. doi: 10.1016/j.jaci.2012.11.039
39. Polikepahad S, Knight JM, Naghavi AO, Oplt T, Creighton CJ, Shaw C, et al. Proinflammatory role for let-7 microRNAS in experimental asthma. J Biol Chem. (2010) 285(39):30139–49. doi: 10.1074/jbc.M110.145698
40. ElKashef S, Ahmad SE, Soliman YMA, Mostafa MS. Role of microRNA-21 and microRNA-155 as biomarkers for bronchial asthma. Innate Immun. (2021) 27(1):61–9. doi: 10.1177/1753425920901563
41. Iliopoulos D, Jaeger SA, Hirsch HA, Bulyk ML, Struhl K. STAT3 Activation of miR-21 and miR-181b-1 via PTEN and CYLD are part of the epigenetic switch linking inflammation to cancer. Mol Cell. (2010) 39(4):493–506. doi: 10.1016/j.molcel.2010.07.023
42. Simpson LJ, Patel S, Bhakta NR, Choy DF, Brightbill HD, Ren X, et al. A microRNA upregulated in asthma airway T cells promotes TH2 cytokine production. Nat Immunol. (2014) 15(12):1162–70. doi: 10.1038/ni.3026
43. Martinez-Nunez RT, Louafi F, Sanchez-Elsner T. The interleukin 13 (IL-13) pathway in human macrophages is modulated by microRNA-155 via direct targeting of interleukin 13 receptor alpha1 (IL13Ralpha1). J Biol Chem. (2011) 286(3):1786–94. doi: 10.1074/jbc.M110.169367
44. Kim HJ, Park SO, Byeon HW, Eo JC, Choi JY, Tanveer M, et al. T cell-intrinsic miR-155 is required for Th2 and Th17-biased responses in acute and chronic airway inflammation by targeting several different transcription factors. Immunology. (2022) 166(3):357–79. doi: 10.1111/imm.13477
45. Zhang K, Liang Y, Feng Y, Wu W, Zhang H, He J, et al. Decreased epithelial and sputum miR-221-3p associates with airway eosinophilic inflammation and CXCL17 expression in asthma. Am J Physiol Lung Cell Mol Physiol. (2018) 315(2):L253–L64. doi: 10.1152/ajplung.00567.2017
46. Frey A, Lunding LP, Ehlers JC, Weckmann M, Zissler UM, Wegmann M. More than just a barrier: the immune functions of the airway epithelium in asthma pathogenesis. Front Immunol. (2020) 11:761. doi: 10.3389/fimmu.2020.00761
47. Singh PB, Pua HH, Happ HC, Schneider C, von Moltke J, Locksley RM, et al. MicroRNA regulation of type 2 innate lymphoid cell homeostasis and function in allergic inflammation. J Exp Med. (2017) 214(12):3627–43. doi: 10.1084/jem.20170545
48. Solberg OD, Ostrin EJ, Love MI, Peng JC, Bhakta NR, Hou L, et al. Airway epithelial miRNA expression is altered in asthma. Am J Respir Crit Care Med. (2012) 186(10):965–74. doi: 10.1164/rccm.201201-0027OC
49. Panganiban RP, Pinkerton MH, Maru SY, Jefferson SJ, Roff AN, Ishmael FT. Differential microRNA epression in asthma and the role of miR-1248 in regulation of IL-5. Am J Clin Exp Immunol. (2012) 1(2):154–65.23885321
50. Panganiban RP, Wang Y, Howrylak J, Chinchilli VM, Craig TJ, August A, et al. Circulating microRNAs as biomarkers in patients with allergic rhinitis and asthma. J Allergy Clin Immunol. (2016) 137(5):1423–32. doi: 10.1016/j.jaci.2016.01.029
51. Chiba Y, Tanabe M, Goto K, Sakai H, Misawa M. Down-regulation of miR-133a contributes to up-regulation of rhoa in bronchial smooth muscle cells. Am J Respir Crit Care Med. (2009) 180(8):713–9. doi: 10.1164/rccm.200903-0325OC
52. Shao Y, Chong L, Lin P, Li H, Zhu L, Wu Q, et al. MicroRNA-133a alleviates airway remodeling in asthtama through PI3K/AKT/mTOR signaling pathway by targeting IGF1R. J Cell Physiol. (2019) 234(4):4068–80. doi: 10.1002/jcp.27201
53. Yamamoto M, Singh A, Ruan J, Gauvreau GM, O'Byrne PM, Carlsten CR, et al. Decreased miR-192 expression in peripheral blood of asthmatic individuals undergoing an allergen inhalation challenge. BMC Genomics. (2012) 13:655. doi: 10.1186/1471-2164-13-655
54. Lou L, Tian M, Chang J, Li F, Zhang G. MiRNA-192-5p attenuates airway remodeling and autophagy in asthma by targeting MMP-16 and ATG7. Biomed Pharmacother. (2020) 122:109692. doi: 10.1016/j.biopha.2019.109692
55. Kastle M, Bartel S, Geillinger-Kastle K, Irmler M, Beckers J, Ryffel B, et al. microRNA cluster 106a∼363 is involved in T helper 17 cell differentiation. Immunology. (2017) 152(3):402–13. doi: 10.1111/imm.12775
56. Bleck B, Grunig G, Chiu A, Liu M, Gordon T, Kazeros A, et al. MicroRNA-375 regulation of thymic stromal lymphopoietin by diesel exhaust particles and ambient particulate matter in human bronchial epithelial cells. J Immunol. (2013) 190(7):3757–63. doi: 10.4049/jimmunol.1201165
57. Lu LF, Boldin MP, Chaudhry A, Lin LL, Taganov KD, Hanada T, et al. Function of miR-146a in controlling treg cell-mediated regulation of Th1 responses. Cell. (2010) 142(6):914–29. doi: 10.1016/j.cell.2010.08.012
58. Kumar M, Ahmad T, Sharma A, Mabalirajan U, Kulshreshtha A, Agrawal A, et al. Let-7 microRNA-mediated regulation of IL-13 and allergic airway inflammation. J Allergy Clin Immunol. (2011) 128(5):1077–85.e1-10. doi: 10.1016/j.jaci.2011.04.034
59. Vishweswaraiah S, Veerappa AM, Mahesh PA, Jayaraju BS, Krishnarao CS, Ramachandra NB. Molecular interaction network and pathway studies of ADAM33 potentially relevant to asthma. Ann Allergy Asthma Immunol. (2014) 113(4):418–24.e1. doi: 10.1016/j.anai.2014.07.009
60. Pelaia G, Vatrella A, Busceti MT, Gallelli L, Calabrese C, Terracciano R, et al. Cellular mechanisms underlying eosinophilic and neutrophilic airway inflammation in asthma. Mediators Inflamm. (2015) 2015:879783. doi: 10.1155/2015/879783
61. Haddad A, Gaudet M, Plesa M, Allakhverdi Z, Mogas AK, Audusseau S, et al. Neutrophils from severe asthmatic patients induce epithelial to mesenchymal transition in healthy bronchial epithelial cells. Respir Res. (2019) 20(1):234. doi: 10.1186/s12931-019-1186-8
62. Khalfaoui L, Symon FA, Couillard S, Hargadon B, Chaudhuri R, Bicknell S, et al. Airway remodelling rather than cellular infiltration characterizes both type2 cytokine biomarker-high and -low severe asthma. Allergy. (2022). doi: 10.1111/all.15376. [Online ahead of print]35579040
63. Heijink IH, Kuchibhotla VNS, Roffel MP, Maes T, Knight DA, Sayers I, et al. Epithelial cell dysfunction, a major driver of asthma development. Allergy. (2020) 75(8):1902–17. doi: 10.1111/all.14421
64. Chakrabarti S, Patel KD. Matrix metalloproteinase-2 (MMP-2) and MMP-9 in pulmonary pathology. Exp Lung Res. (2005) 31(6):599–621. doi: 10.1080/019021490944232
65. Wang H, Liu Y, Shi J, Cheng Z. ORMDL3 Knockdown in the lungs alleviates airway inflammation and airway remodeling in asthmatic mice via JNK1/2-MMP-9 pathway. Biochem Biophys Res Commun. (2019) 516(3):739–46. doi: 10.1016/j.bbrc.2019.06.122
66. O'Farrell HE, Bowman RV, Fong KM, Yang IA. Plasma extracellular vesicle miRNAs can identify lung cancer, current smoking Status, and stable COPD. Int J Mol Sci. (2021) 22(11):5803. doi: 10.3390/ijms22115803
67. Haj-Salem I, Fakhfakh R, Berube JC, Jacques E, Plante S, Simard MJ, et al. MicroRNA-19a enhances proliferation of bronchial epithelial cells by targeting TGFbetaR2 gene in severe asthma. Allergy. (2015) 70(2):212–9. doi: 10.1111/all.12551
68. Bartel S, Carraro G, Alessandrini F, Krauss-Etschmann S, Ricciardolo FLM, Bellusci S. miR-142-3p is associated with aberrant WNT signaling during airway remodeling in asthma. Am J Physiol Lung Cell Mol Physiol. (2018) 315(2):L328–L33. doi: 10.1152/ajplung.00113.2018
69. Guan Y, Ma Y, Tang Y, Liu X, Zhao Y, An L. MiRNA-221-5p suppressed the Th17/treg ratio in asthma via RORgammat/Foxp3 by targeting SOCS1. Allergy Asthma Clin Immunol. (2021) 17(1):123. doi: 10.1186/s13223-021-00620-8
70. Fang C, Lu W, Li C, Peng X, Wang Y, Huang X, et al. MiR-3162-3p is a novel MicroRNA that exacerbates asthma by regulating beta-catenin. PLoS One. (2016) 11(3):e0149257. doi: 10.1371/journal.pone.0149257
71. Wang AL, Li J, Kho AT, McGeachie MJ, Tantisira KG. Enhancing the prediction of childhood asthma remission: integrating clinical factors with microRNAs. J Allergy Clin Immunol. (2021) 147(3):1093–5.e1. doi: 10.1016/j.jaci.2020.08.019
72. Comer BS, Camoretti-Mercado B, Kogut PC, Halayko AJ, Solway J, Gerthoffer WT. MicroRNA-146a and microRNA-146b expression and anti-inflammatory function in human airway smooth muscle. Am J Physiol Lung Cell Mol Physiol. (2014) 307(9):L727–34. doi: 10.1152/ajplung.00174.2014
73. Wu J, Liu F, Zhao J, Wei Y, Lv J, Dong F, et al. Thymic stromal lymphopoietin promotes asthmatic airway remodelling in human lung fibroblast cells through STAT3 signalling pathway. Cell Biochem Funct. (2013) 31(6):496–503. doi: 10.1002/cbf.2926
74. Ye L, Mou Y, Wang J, Jin ML. Effects of microRNA-19b on airway remodeling, airway inflammation and degree of oxidative stress by targeting TSLP through the Stat3 signaling pathway in a mouse model of asthma. Oncotarget. (2017) 8(29):47533–46. doi: 10.18632/oncotarget.17258
75. Luo X, Zeng Q, Yan S, Liu W, Luo R. MicroRNA-375-mediated regulation of ILC2 cells through TSLP in allergic rhinitis. World Allergy Organ J. (2020) 13(8):100451. doi: 10.1016/j.waojou.2020.100451
76. Mendes FC, Paciencia I, Ferreira AC, Martins C, Rufo JC, Silva D, et al. Development and validation of exhaled breath condensate microRNAs to identify and endotype asthma in children. PLoS One. (2019) 14(11):e0224983. doi: 10.1371/journal.pone.0224983
77. Pinkerton M, Chinchilli V, Banta E, Craig T, August A, Bascom R, et al. Differential expression of microRNAs in exhaled breath condensates of patients with asthma, patients with chronic obstructive pulmonary disease, and healthy adults. J Allergy Clin Immunol. (2013) 132(1):217–9. doi: 10.1016/j.jaci.2013.03.006
78. Just J, Munk Ipsen P, Kruhoffer M, Lykkemark S, Skjold T, Schiotz PO, et al. Human mast cell sensitization with IgE increases miRNA-210 expression. Int Arch Allergy Immunol. (2019) 179(2):102–7. doi: 10.1159/000496513
79. Liao TL, Chen YM, Tang KT, Chen PK, Liu HJ, Chen DY. MicroRNA-223 inhibits neutrophil extracellular traps formation through regulating calcium influx and small extracellular vesicles transmission. Sci Rep. (2021) 11(1):15676. doi: 10.1038/s41598-021-95028-0
80. Billeter AT, Hellmann J, Roberts H, Druen D, Gardner SA, Sarojini H, et al. MicroRNA-155 potentiates the inflammatory response in hypothermia by suppressing IL-10 production. FASEB J. (2014) 28(12):5322–36. doi: 10.1096/fj.14-258335
81. Rossato M, Curtale G, Tamassia N, Castellucci M, Mori L, Gasperini S, et al. IL-10-induced microRNA-187 negatively regulates TNF-alpha, IL-6, and IL-12p40 production in TLR4-stimulated monocytes. Proc Natl Acad Sci U S A. (2012) 109(45):E3101–10. doi: 10.1073/pnas.1209100109
82. Wu SQ, Wang GL, Li LY, Ji J. Effects of microRNA-21 on the interleukin 12/signal transducer and activator of transcription 4 signaling pathway in asthmatic mice. Cent Eur J Immunol. (2014) 39(1):40–5. doi: 10.5114/ceji.2014.42121
83. Lee HY, Lee HY, Choi JY, Hur J, Kim IK, Kim YK, et al. Inhibition of MicroRNA-21 by an antagomir ameliorates allergic inflammation in a mouse model of asthma. Exp Lung Res. (2017) 43(3):109–19. doi: 10.1080/01902148.2017.1304465
84. Ogulur I, Pat Y, Ardicli O, Barletta E, Cevhertas L, Fernandez-Santamaria R, et al. Advances and highlights in biomarkers of allergic diseases. Allergy. (2021) 76(12):3659–86. doi: 10.1111/all.15089
85. Martinez-Anton A, Mullol J. MicroRNA: endotyping united airways? Int Arch Allergy Immunol. (2014) 164(1):10–2. doi: 10.1159/000362333
86. Rebane A. microRNA and allergy. Adv Exp Med Biol. (2015) 888:331–52. doi: 10.1007/978-3-319-22671-2_17
87. Wu X, Zhao S, Huang W, Huang L, Huang M, Luo X, et al. Aberrant expressions of circulating lncRNA NEAT1 and microRNA-125a are linked with Th2 cells and symptom severity in pediatric allergic rhinitis. J Clin Lab Anal. (2022) 36(3):e24235. doi: 10.1002/jcla.24235
88. Kho AT, McGeachie MJ, Moore KG, Sylvia JM, Weiss ST, Tantisira KG. Circulating microRNAs and prediction of asthma exacerbation in childhood asthma. Respir Res. (2018) 19(1):128. doi: 10.1186/s12931-018-0828-6
89. Suojalehto H, Lindstrom I, Majuri ML, Mitts C, Karjalainen J, Wolff H, et al. Altered microRNA expression of nasal mucosa in long-term asthma and allergic rhinitis. Int Arch Allergy Immunol. (2014) 163(3):168–78. doi: 10.1159/000358486
90. Hou B, Murata M, Said AS, Sakaida H, Masuda S, Takahashi T, et al. Changes of micro-RNAs in asymptomatic subjects sensitized to Japanese cedar pollen after prophylactic sublingual immunotherapy. Allergy Rhinol (Providence). (2015) 6(1):33–8. doi: 10.2500/ar.2015.6.0107
91. Cox L. Grand challenges in allergen immunotherapy. Front Allergy. (2021) 2:710345. doi: 10.3389/falgy.2021.710345
92. van de Veen W, Stanic B, Yaman G, Wawrzyniak M, Sollner S, Akdis DG, et al. Igg4 production is confined to human IL-10-producing regulatory B cells that suppress antigen-specific immune responses. J Allergy Clin Immunol. (2013) 131(4):1204–12. doi: 10.1016/j.jaci.2013.01.014
93. Zissler UM, Jakwerth CA, Guerth FM, Pechtold L, Aguilar-Pimentel JA, Dietz K, et al. Early IL-10 producing B-cells and coinciding th/Tr17 shifts during three year grass-pollen AIT. EBioMedicine. (2018) 36:475–88. doi: 10.1016/j.ebiom.2018.09.016
94. Akdis M, Akdis CA. Mechanisms of allergen-specific immunotherapy: multiple suppressor factors at work in immune tolerance to allergens. J Allergy Clin Immunol. (2014) 133(3):621–31. doi: 10.1016/j.jaci.2013.12.1088
95. Shamji MH, Layhadi JA, Sharif H, Penagos M, Durham SR. Immunological responses and biomarkers for allergen-specific immunotherapy against inhaled allergens. J Allergy Clin Immunol Pract. (2021) 9(5):1769–78. doi: 10.1016/j.jaip.2021.03.029
96. Incorvaia C, Ridolo E, Mauro M, Pucciarini F, Heffler E, Canonica GW. Venom immunotherapy and aeroallergen immunotherapy: how do their outcomes differ? Front Allergy. (2022) 3:854080. doi: 10.3389/falgy.2022.854080
97. Specjalski K, Maciejewska A, Romantowski J, Pawlowski R, Jassem E, Niedoszytko M. miRNA profiles change during grass pollen immunotherapy irrespective of clinical outcome. Immunotherapy. (2022) 14(6):433–44. doi: 10.2217/imt-2021-0217
98. Jakwerth CA, Chaker AM, Guerth F, Oelsner M, Pechtold L, Zur Bonsen LS, et al. Sputum microRNA-screening reveals prostaglandin EP3 receptor as selective target in allergen-specific immunotherapy. Clin Exp Allergy. (2021) 51(12):1577–91. doi: 10.1111/cea.14013
99. Blank S, Grosch J, Ollert M, Bilo MB. Precision medicine in hymenoptera venom allergy: diagnostics, biomarkers, and therapy of different endotypes and phenotypes. Front Immunol. (2020) 11:579409. doi: 10.3389/fimmu.2020.579409
100. Specjalski K, Maciejewska A, Pawlowski R, Chelminska M, Jassem E. Changes in the expression of MicroRNA in the buildup phase of wasp venom immunotherapy: a pilot study. Int Arch Allergy Immunol. (2016) 170(2):97–100. doi: 10.1159/000447637
101. Horita M, Farquharson C, Stephen LA. The role of miR-29 family in disease. J Cell Biochem. (2021) 122(7):696–715. doi: 10.1002/jcb.29896
102. Liston A, Papadopoulou AS, Danso-Abeam D, Dooley J. MicroRNA-29 in the adaptive immune system: setting the threshold. Cell Mol Life Sci. (2012) 69(21):3533–41. doi: 10.1007/s00018-012-1124-0
103. Ruiz-Lafuente N, Alcaraz-Garcia MJ, Sebastian-Ruiz S, Garcia-Serna AM, Gomez-Espuch J, Moraleda JM, et al. IL-4 Up-Regulates MiR-21 and the MiRNAs hosted in the CLCN5 gene in chronic lymphocytic leukemia. PLoS One. (2015) 10(4):e0124936. doi: 10.1371/journal.pone.0124936
104. Specjalski K, Maciejewska A, Pawlowski R, Zielinski M, Trzonkowski P, Pikula M, et al. Changing microRNA expression during three-month wasp venom immunotherapy. Immunol Invest. (2019) 48(8):835–43. doi: 10.1080/08820139.2019.1617303
105. Courboulin A, Paulin R, Giguere NJ, Saksouk N, Perreault T, Meloche J, et al. Role for miR-204 in human pulmonary arterial hypertension. J Exp Med. (2011) 208(3):535–48. doi: 10.1084/jem.20101812
106. Ooi AT, Ram S, Kuo A, Gilbert JL, Yan W, Pellegrini M, et al. Identification of an interleukin 13-induced epigenetic signature in allergic airway inflammation. Am J Transl Res. (2012) 4(2):219–28.22611474
107. Stittrich AB, Haftmann C, Sgouroudis E, Kuhl AA, Hegazy AN, Panse I, et al. The microRNA miR-182 is induced by IL-2 and promotes clonal expansion of activated helper T lymphocytes. Nat Immunol. (2010) 11(11):1057–62. doi: 10.1038/ni.1945
108. Hezova R, Slaby O, Faltejskova P, Mikulkova Z, Buresova I, Raja KR, et al. microRNA-342, microRNA-191 and microRNA-510 are differentially expressed in T regulatory cells of type 1 diabetic patients. Cell Immunol. (2010) 260(2):70–4. doi: 10.1016/j.cellimm.2009.10.012
109. Karpinski P, Kahraman M, Ludwig N, Skiba P, Kosinska M, Rosiek-Biegus M, et al. Limited long-term impact of insect venom immunotherapy on the micro-RNA landscape in whole blood. J Investig Allergol Clin Immunol. (2019) 29(3):206–12. doi: 10.18176/jiaci.0303
110. Lu TX, Munitz A, Rothenberg ME. MicroRNA-21 is up-regulated in allergic airway inflammation and regulates IL-12p35 expression. J Immunol. (2009) 182(8):4994–5002. doi: 10.4049/jimmunol.0803560
111. Wang R, Xue S, Liu Y, Peng M, Guo B. The correlation of long non-coding RNA NEAT1 and its targets microRNA (miR)-21, miR-124, and miR-125a with disease risk, severity, and inflammation of allergic rhinitis. Medicine (Baltimore). (2021) 100(4):e22946. doi: 10.1097/MD.0000000000022946
112. Bao L, Chau C, Bao J, Tsoukas MM, Chan LS. IL-4 dysregulates microRNAs involved in inflammation, angiogenesis and apoptosis in epidermal keratinocytes. Microbiol Immunol. (2018) 62(11):732–6. doi: 10.1111/1348-0421.12650
113. Tian M, Ji Y, Wang T, Zhang W, Zhou Y, Cui Y. Changes in circulating microRNA-126 levels are associated with immune imbalance in children with acute asthma. Int J Immunopathol Pharmacol. (2018) 32:2058738418779243. doi: 10.1177/2058738418779243
114. Zhu MX, Huang LH, Zhu YK, Cai XJ. LncRNA NEAT1 promotes airway smooth muscle cell inflammation by activating the JAK3/STAT5 pathway through targeting of miR-139. Exp Lung Res. (2021) 47(4):161–72. doi: 10.1080/01902148.2021.1876792
115. Siddiqui S, Johansson K, Joo A, Bonser LR, Koh KD, Le Tonqueze O, et al. Epithelial miR-141 regulates IL-13-induced airway mucus production. JCI Insight. (2021) 6(5):e139019. doi: 10.1172/jci.insight.139019
116. Yu S, Zhang R, Zhu C, Cheng J, Wang H, Wu J. MicroRNA-143 downregulates interleukin-13 receptor alpha1 in human mast cells. Int J Mol Sci. (2013) 14(8):16958–69. doi: 10.3390/ijms140816958
117. Collison A, Mattes J, Plank M, Foster PS. Inhibition of house dust mite-induced allergic airways disease by antagonism of microRNA-145 is comparable to glucocorticoid treatment. J Allergy Clin Immunol. (2011) 128(1):160–7.e4. doi: 10.1016/j.jaci.2011.04.005
118. Arroyo M, Salka K, Chorvinsky E, Xuchen X, Abutaleb K, Perez GF, et al. Airway mir-155 responses are associated with TH1 cytokine polarization in young children with viral respiratory infections. PLoS One. (2020) 15(5):e0233352. doi: 10.1371/journal.pone.0233352
119. Alexandrova E, Miglino N, Hashim A, Nassa G, Stellato C, Tamm M, et al. Small RNA profiling reveals deregulated phosphatase and tensin homolog (PTEN)/phosphoinositide 3-kinase (PI3K)/akt pathway in bronchial smooth muscle cells from asthmatic patients. J Allergy Clin Immunol. (2016) 137(1):58–67. doi: 10.1016/j.jaci.2015.05.031
120. Shaoqing Y, Ruxin Z, Guojun L, Zhiqiang Y, Hua H, Shudong Y, et al. Microarray analysis of differentially expressed microRNAs in allergic rhinitis. Am J Rhinol Allergy. (2011) 25(6):e242–6. doi: 10.2500/ajra.2011.25.3682
121. Wu G, Yang G, Zhang R, Xu G, Zhang L, Wen W, et al. Altered microRNA expression profiles of extracellular vesicles in nasal mucus from patients with allergic rhinitis. Allergy Asthma Immunol Res. (2015) 7(5):449–57. doi: 10.4168/aair.2015.7.5.449
122. Rutledge H, Baran-Gale J, de Villena FP, Chesler EJ, Churchill GA, Sethupathy P, et al. Identification of microRNAs associated with allergic airway disease using a genetically diverse mouse population. BMC Genomics. (2015) 16:633. doi: 10.1186/s12864-015-1732-9
123. Wang L, Xia JW, Ke ZP, Zhang BH. Blockade of NEAT1 represses inflammation response and lipid uptake via modulating miR-342-3p in human macrophages THP-1 cells. J Cell Physiol. (2019) 234(4):5319–26. doi: 10.1002/jcp.27340
124. Li J, Tiwari A, Mirzakhani H, Wang AL, Kho AT, McGeachie MJ, et al. Circulating MicroRNA: incident asthma prediction and vitamin D effect modification. J Pers Med. (2021) 11(4):307. doi: 10.3390/jpm11040307
125. Rupani H, Martinez-Nunez RT, Dennison P, Lau LC, Jayasekera N, Havelock T, et al. Toll-like receptor 7 is reduced in severe asthma and linked to an altered MicroRNA profile. Am J Respir Crit Care Med. (2016) 194(1):26–37. doi: 10.1164/rccm.201502-0280OC
126. Liu F, Qin HB, Xu B, Zhou H, Zhao DY. Profiling of miRNAs in pediatric asthma: upregulation of miRNA-221 and miRNA-485-3p. Mol Med Rep. (2012) 6(5):1178–82. doi: 10.3892/mmr.2012.1030
127. Ma ZX, Tan X, Shen Y, Ke X, Yang YC, He XB, et al. MicroRNA expression profile of mature dendritic cell in chronic rhinosinusitis. Inflamm Res. (2015) 64(11):885–93. doi: 10.1007/s00011-015-0870-5
128. Mai J, Virtue A, Maley E, Tran T, Yin Y, Meng S, et al. MicroRNAs and other mechanisms regulate interleukin-17 cytokines and receptors. Front Biosci (Elite Ed). (2012) 4(4):1478–95. doi: 10.2741/e474
129. Zhu D, Gao W, Zhang Z. MicroRNA-1180 is associated with growth and apoptosis in prostate cancer via TNF receptor associated factor 1 expression regulation and nuclear factor-kappaB signaling pathway activation. Oncol Lett. (2018) 15(4):4775–80. doi: 10.3892/ol.2018.7914
130. Bozack AK, Colicino E, Rodosthenous RS, Bloomquist TR, Baccarelli AA, Wright RO, et al. Breast milk-derived extracellular vesicle miRNAs are associated with maternal asthma and atopy. Epigenomics. (2022) 14(12):727–39. doi: 10.2217/epi-2022-0090
131. Jun H, Ying H, Daiwen C, Bing Y, Xiangbing M, Ping Z, et al. miR-628, a microRNA that is induced by toll-like receptor stimulation, regulates porcine innate immune responses. Sci Rep. (2015) 5:12226. doi: 10.1038/srep12226
132. Sheu CC, Tsai MJ, Chen FW, Chang KF, Chang WA, Chong IW, et al. Identification of novel genetic regulations associated with airway epithelial homeostasis using next-generation sequencing data and bioinformatics approaches. Oncotarget. (2017) 8(47):82674–88. doi: 10.18632/oncotarget.19752
133. Liu C, Lai Y, Ying S, Zhan J, Shen Y. Plasma exosome-derived microRNAs expression profiling and bioinformatics analysis under cross-talk between increased low-density lipoprotein cholesterol level and ATP-sensitive potassium channels variant rs1799858. J Transl Med. (2020) 18(1):459. doi: 10.1186/s12967-020-02639-8
Keywords: microRNA, airway epithelial cells (AECs), type 2 inflammation, immune crosstalk, allergen specific immunotherapy (ASIT), venom immunotherapy (VIT), induced sputum, nasal secretions
Citation: Jakwerth CA, Kitzberger H, Pogorelov D, Müller A, Blank S, Schmidt-Weber CB and Zissler UM (2022) Role of microRNAs in type 2 diseases and allergen-specific immunotherapy. Front. Allergy 3:993937. doi: 10.3389/falgy.2022.993937
Received: 14 July 2022; Accepted: 11 August 2022;
Published: 12 September 2022.
Edited by:
Kristina Johansson, University of Gothenburg, SwedenReviewed by:
Marek Jutel, Wroclaw Medical University, Poland© 2022 Jakwerth, Kitzberger, Pogorelov, Müller, Blank, Schmidt-Weber and Zissler. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ulrich M. Zissler dWxyaWNoLnppc3NsZXJAdHVtLmRl
Specialty Section: This article was submitted to Asthma, a section of the journal Frontiers in Allergy
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.