
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
PERSPECTIVE article
Front. Allergy, 22 March 2022
Sec. Allergens
Volume 3 - 2022 | https://doi.org/10.3389/falgy.2022.863172
This article is part of the Research TopicWhat makes an Allergen an Allergen?View all 7 articles
The introduction of plant extracts to mitigate the symptoms of “hay fever”, about a century ago, led to discoveries beginning sixty years ago on determining the sequences and eventually structures of allergenic proteins. As more proteins were cloned, there was a need to rapidly identify and categorize those with significant similarity to known allergens. The Structural Database of Allergenic Proteins (SDAP) was created at the beginning of the 21st century as the first cross-referenced website to allow rapid overview of the structures and sequences of allergenic proteins. SDAP provides a way to identify sequence and functional similarities between these proteins, despite the complex nomenclature system based on the Latin names of their different sources. A rapid FASTA search simplifies grouping allergens from the same structural or functional family. SDAP also provides an overview of the rapidly expanding literature on the sequence, structure and epitopes of allergenic proteins and a way to estimate the potential allergenicity of novel proteins based on rules provided by the IUIS. Twenty years and a pandemic later, the list of allergenic proteins and their attributes continues to grow. SDAP is expanding and improving to allow rapid access to all this information.
This report marks 20 years since the online version of the Structural Database of Allergenic Proteins (SDAP) was first established. SDAP's original purpose was to provide a cross referenced website to classify allergens according to their names, structure and function, a need that had been building for over 100 years. Although allergic reactions are described in print as early as the 16th century, or even ancient times (1, 2), the word allergy, and the beginning of treatments for allergy with plant extracts, began in the first years of the 20th century (3). Extracts of ragweed helped many individuals with “hay fever,” but injecting patients with whole pollen extracts could also induce dangerous anaphylaxis. Thus researchers, aided by modern protein chemistry, set out to make safer, simpler, “component resolved” extracts. Molecular studies of the protein components in the extracts that could contribute to reactivity began about 60 years ago. During the standardization of these extracts, it was found that there were several components that bound IgE in sera from hypersensitized individuals, leading to modern tests for categorizing the types of allergens a patient might react to (4). The first isolated allergenic proteins, Amb a 1 and Amb a 2 of ragweed (5, 6) were soon followed by many others from many different sources.
A standardized nomenclature based on the Latin names of their plant, animal, insect or venom source was first published in 1984 (7). The nomenclature they agreed upon was to designate highly purified allergenic proteins by the first three letters of the genus followed by a space, the first letter of the species name, followed by a space, both italicized, followed by a Roman numeral to indicate the order of importance (or isolation) of the protein. For example, the first perennial rye grass allergen, from the plant with the Latin name Lolium perenne was called Lol p I.
The list of proteins in the paper, mostly aeroallergens from dander and pollen, covered less than a journal page. The only “ingested” allergens were parvalbumin from cod (Gad c 1), three egg white proteins (Gal d 1-3) and a surface protein of round worm (Asc s 1).
The allergen field expanded rapidly (some called it a “data explosion”), thanks to innovations in immunology, protein sequencing and the recognition that IgE in serum bound specifically to allergens (Figure 1). Multiple allergens were isolated from peanuts (8) and other sources (9, 10).
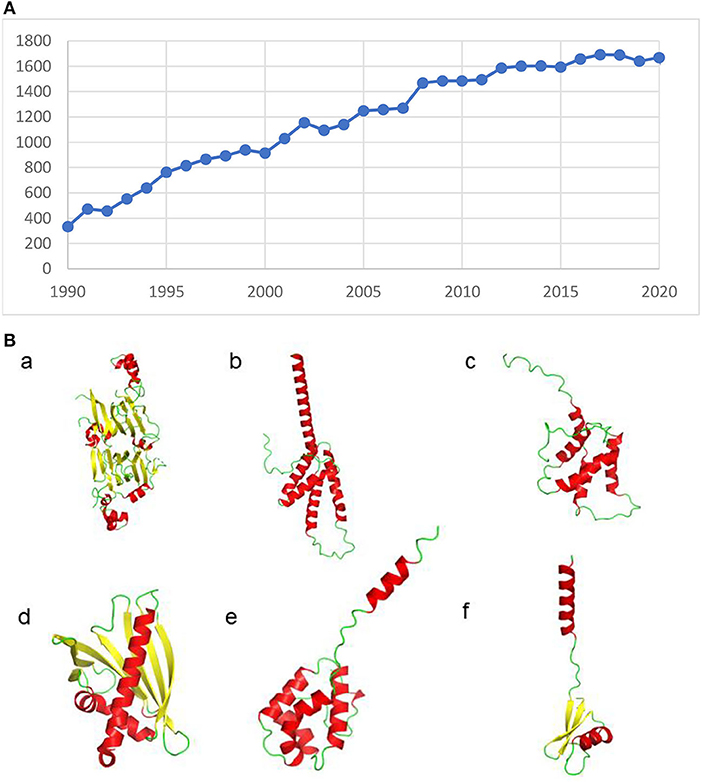
Figure 1. (A) Publications per year with the words Allergenic Proteins in Pubmed are steadily increasing. (B) Allergens have diverse structures, as ribbon structures of some of the allergens identified in peanuts show. a) Ara h 1 (PDB id: 2SMH), b) Ara h 2 (PDB id: 2OB4), c) Ara h 6 (PDB id: 1W2Q), d) Ara h 8 (PDB id: 4M9B) e) Ara h 9 and f) Ara h 12. The loop region in Ara h 2 and structures of Ara h 9 and Ara h 12 were modeled using Alphafold.
The nomenclature was simplified a few years later (11), in that names were no longer italicized, and Arabic, not Roman numbers were used (e.g., Ara h 8 from peanuts instead of VIII). The list of allergens in the 1994 paper stretched over more than four journal pages, with additional recommendations, not just for the sequences of the whole protein, but for peptides from the sequence that were epitopes for IgE from patient sera. But numbering remained a problem. The small table in the 1984 publication shows that Api m I, Ves g I, Pol a I all clearly referred to phospholipases of honey bee, yellow jacket and wasps, respectively, while Api m II, Ves g II, and Pol a II refer to hyaluronidases. But new allergens of different families, such as pectate lyases and from various pathogen related (PR) groupings (12, 13) were named as they were isolated, so the numbers no longer reliably predicted similarity in function or structure. For example, the vicilin allergen of peanut, Ara h 1, corresponded best in structure to Jug r 2 of walnut. Later identified allergens, Ara h 6 and Ara h 7, had similarity to Ara h 2 and other 2S albumins.
The first question in assembling SDAP was which proteins should be included. For clinicians, the word allergen refers simply to a food or pollen the patient reacts to, such as milk, shrimp, peanut or ryegrass. SDAP's goal is to aid researchers or regulators who need a more molecular definition by cataloging all the proteins or protein fragments that contribute to the allergenicity of the plant or animal source (which is specified for all entries). For inclusion, SDAP relies primarily on the WHO/IUIS list provided at their website (http://allergen.org/), as these proteins have been reviewed by a committee of experts in the field. Due to potential anaphylactic reactions in direct assays, such as patch testing or oral food challenge, most proteins are classified as allergenic if they bind IgE from sera of a sufficient number of patients with clinically diagnosed reactivity to the source. However, to quote from the WHO/IUIS website:
“The primary goal of a systematic nomenclature is to define a common language for scientists. As such, assessment of new allergen candidates for inclusion into this database does not involve a judgement on their clinical significance (my italics). A minimal criterion of demonstrated IgE binding to the suggested allergen using sera from patients allergic to the specific source is required.”
In addition to the IUIS list, other proteins have been included if they were listed in one of the existing data bases containing allergens [see Table 1 in (14) and for a more recent discussion of databases, see (15)]. These “non-IUIS” entries (clearly marked as such) are kept in SDAP as a service for researchers who are exploring and studying proteins that might have a potential allergenic response. Some proteins of wheat that cause non-IgE mediated symptoms in sensitive individuals have also been included, again to help those seeking to define the potential relationship between these proteins and known allergenic ones. The files in SDAP contain more information and literature references for highly studied component proteins, such as those of peanut or shrimp. Additional literature searching may be needed for the less studied and especially “non-IUIS” proteins.
The first job of SDAP was thus to provide a cross-referenced list of the sequences and associated information of all the proteins acknowledged to be allergenic by the IUIS (16). This was done by a series of cross referenced MySQL lists, most of which were assembled tediously by human effort (17). Later versions of the database could use automatic identification (14), but many proteins were found to be allergenic or IgE_binding only after their isolation and naming.
The need for bioinformatic tools to identify such potential allergens was brought to the forefront by an attempt to enhance the methionine content of grains, by inserting a newly identified gene from a brazil nut (18, 19). These projects were terminated when allergic responses were found to the “genetically modified” foods. Thus, a sequence FASTA search in SDAP was implemented to rapidly show whether a test protein had significant identity to any known allergen, using a set of rules established by the IUIS (20). The user could then decide whether to proceed with using the protein or drop projects before problems arose.
Many allergens known to cross react, such as those from peanut and walnut, have very low sequence identity. The next SDAP innovation, the peptide similarity scale, found similar sequences using a physicochemical property (PCP) scale of the amino acids determined by the Braun group (21, 22). The scale was first used to identify common motifs (23), similar regions within allergens, that could be used to identify potentially cross reacting epitopes (24) even in allergens with very different structures (25–29). Other webserver or downloadable tools can be used to further analyze SDAP results. Episearch maps peptide mimotopes from phage libraries to allergenic proteins (30) and DGraph allows one to view the “property distances (PD)” between, for example, IgE reactive sequences or whole related protein sequences, as a 2D-map, without an initial sequence alignment (27, 31, 32).
Allergens can have many diverse structures (Figure 1B) (33–37) with functional (38) or even “disordered” (39) regions that contain epitopes. One of the most important and distinguishing features of SDAP is the incorporation of structural data, through direct links to files in the Protein Database (PDB) or model structures made from suitable templates (34, 36).
The information on allergens' structures and epitopes continues to grow at a rapid rate. SDAP was created to help understand the similarities and differences in these proteins. Twenty years after its start, there is now a major push to update its software and list of allergenic proteins and their isomers, to be a tool for researchers, regulatory agencies and patients.
CHS wrote and edited the manuscript with help from WB. SN prepared Figure 1B. All authors contributed to the article and approved the submitted version.
SDAP was originally funded by a Research Development Grant (#2535-01) from the John Sealy Memorial Endowment Fund for Biomedical Research, grants from the US-Food and Drug Administration (FD-U-002249), and the Texas Higher Education Coordinating Board (ATP grant 004952-0036-2003) to WB. Our work with allergenic proteins and SDAP upkeep has been supported by grants from the U.S. Environmental Protection Agency under STAR Research Assistance Agreements (RD-833137, RD-834823-01 to WB, RE-83406601 to CHS), the US Department of Agriculture (ARS 58-6435-9-40 to CHS), the National Institute of Health (R01 AI 064913 to WB, 1R01AI165866-01 to Stephen Dreskin; subcontract to CHS), and funding for this article was provided by the Margaret Maccallum Gage and Tracy Davis Gage Professorship in Biochemistry and Allergies to WB.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
SDAP was originally designed and compiled by Drs. Ovidiu Ivanciuc and WB and is presently maintained by SN. Three dimensional models were originally generated by Numan S. Oezguen and Trevor D. Power and are currently being updated by SN. The PD search began with the Ph.D. thesis work of Venkatarajan A. Mathura. The D-graph program was written by Benjamin A. Braun, Ph.D., with physics advice from WB and validated for use with protein sequences of allergens and viruses by CHS. We thank all our collaborators over the 20 years of SDAP's existence as well as those using its tools in research and regulatory matters.
1. Ring J. History of allergy in antiquity. Chem Immunol Allergy. (2014) 100:2-14. doi: 10.1159/000358422
2. Ring J. History of allergy: clinical descriptions, pathophysiology, and treatment. Handb Exp Pharmacol. (2022) 268:3-19. doi: 10.1007/164_2021_509
3. Ring J, Grosber M, Brockow K, Bergmann K C. Anaphylaxis. Chem Immunol Allergy. (2014) 100:54-61. doi: 10.1159/000358503
4. Johansson SGO. The discovery of IgE. J Allergy Clin Immunol. (2016) 137:1671-3. doi: 10.1016/j.jaci.2016.04.004
5. Lowenstein H. Characterization and standardization of allergen extracts. Chem Immunol Allergy. (2014) 100:323-32. doi: 10.1159/000359989
6. King TP, Norman PS, Lichtenstein LM. Studies on ragweed pollen allergens. Ann Allergy. (1967) 25:541-53.
7. Marsh DG, Goodfriend L, King TP, Lowenstein H, Platts-Mills TA. Allergen nomenclature. Bull World Health Organ. (1986) 64:767-74.
8. Burks AW, Williams LW, Connaughton C, Cockrell G, O'Brien TJ, Helm RM. Identification and characterization of a second major peanut allergen, Ara h II, with use of the sera of patients with atopic dermatitis and positive peanut challenge. J Allergy Clin Immunol. (1992) 90:962-9. doi: 10.1016/0091-6749(92)90469-I
9. Andersen KE, Lowenstein H. An investigation of the possible immunological relationship between allergen extracts from birch pollen, hazelnut, potato and apple. Contact Dermatitis. (1978) 4:73-9. doi: 10.1111/j.1600-0536.1978.tb03739.x
10. Valenta R, Breiteneder H, Petternburger K, Breitenbach M, Rumpold H, Kraft D, et al. Homology of the major birch-pollen allergen, Bet v I, with the major pollen allergens of alder, hazel, and hornbeam at the nucleic acid level as determined by cross-hybridization. J Allergy Clin Immunol. (1991) 87:677-82. doi: 10.1016/0091-6749(91)90388-5
11. King TP, Hoffman D, Lowenstein H, Marsh DG, Platts-Mills TA, Thomas W. Allergen nomenclature. Allergy. (1995) 50:765-74. doi: 10.1111/j.1398-9995.1995.tb01222.x
12. Soman KV, Midoro-Horiuti T, Ferreon JC, Goldblum RM, Brooks EG, Kurosky A, et al. Homology modeling and characterization of IgE binding epitopes of mountain cedar allergen Jun a 3. Biophys J. (2000) 79:1601-9. doi: 10.1016/S0006-3495(00)76410-1
13. Midoro-Horiuti T, Goldblum RM, Kurosky A, Wood TG, Schein CH, Brooks EG. Molecular cloning of the mountain cedar (Juniperus ashei) pollen major allergen, Jun a 1. J Allergy Clin Immunol. (1999) 104:613-7. doi: 10.1016/S0091-6749(99)70332-5
14. Ivanciuc O, Gendel SM, Power TD, Schein CH, Braun W. AllerML: markup language for allergens. Regul Toxicol Pharmacol. (2011) 60:151-60. doi: 10.1016/j.yrtph.2011.03.006
15. van Ree R, Sapiter Ballerda D, Berin MC, Beuf L, Chang A, Gadermaier G, et al. The COMPARE database: A public resource for allergen identification, adapted for continuous improvement. Front Allergy. (2021) 2:700533. doi: 10.3389/falgy.2021.700533
16. Ivanciuc O, Schein CH, Braun W. Data mining of sequences and 3D structures of allergenic proteins. Bioinformatics. (2002) 18:1358-64. doi: 10.1093/bioinformatics/18.10.1358
17. Ivanciuc O, Schein CH, Braun W. SDAP: database and computational tools for allergenic proteins. Nucleic Acids Res. (2003) 31:359-62. doi: 10.1093/nar/gkg010
18. Saalbach I, Pickardt T, Machemehl F, Saalbach G, Schieder O, Muntz K. A chimeric gene encoding the methionine-rich 2S albumin of the Brazil nut (Bertholletia excelsa H.B.K.) is stably expressed and inherited in transgenic grain legumes. Mol Gen Genet. (1994) 242:226-36. doi: 10.1007/BF00391017
19. Tu HM, Godfrey LW, Sun SS. Expression of the Brazil nut methionine-rich protein and mutants with increased methionine in transgenic potato. Plant Mol Biol. (1998) 37:829-38. doi: 10.1023/A:1006098524887
20. Ivanciuc O, Mathura V, Midoro-Horiuti T, Braun W, Goldblum RM, Schein CH. Detecting potential IgE-reactive sites on food proteins using a sequence and structure database, SDAP-food. J Agric Food Chem. (2003) 51:4830-7. doi: 10.1021/jf034218r
21. Mathura VS, Schein CH, Braun W. Identifying property based sequence motifs in protein families and superfamilies: application to DNase-1 related endonucleases. Bioinformatics. (2003) 19:1381-90. doi: 10.1093/bioinformatics/btg164
22. Venkatarajan MS, Braun W. New quantitative descriptors of amino acids based on multidimensional scaling of a large number of physical-chemical properties. Mol Model. (2001) 7:445–53. doi: 10.1007/s00894-001-0058-5
23. Ivanciuc O, Oezguen N, Mathura VS, Schein CH, Xu Y, Braun W. Using property based sequence motifs and 3D modeling to determine structure and functional regions of proteins. Curr Med Chem. (2004) 11:583-93. doi: 10.2174/0929867043455819
24. Midoro-Horiuti T, Schein CH, Mathura V, Braun W, Czerwinski EW, Togawa A, et al. Structural basis for epitope sharing between group 1 allergens of cedar pollen. Mol Immunol. (2006) 43:509-18. doi: 10.1016/j.molimm.2005.05.006
25. Ivanciuc O, Garcia T, Torres M, Schein CH, Braun W. Characteristic motifs for families of allergenic proteins. Mol Immunol. (2009) 46:559-68. doi: 10.1016/j.molimm.2008.07.034
26. Ivanciuc O, Midoro-Horiuti T, Schein CH, Xie L, Hillman GR, Goldblum RM, et al. The property distance index PD predicts peptides that cross-react with IgE antibodies. Mol Immunol. (2009) 46:873-83. doi: 10.1016/j.molimm.2008.09.004
27. Nesbit JB, Schein CH, Braun BA, Gipson SAY, Cheng H, Hurlburt BK, et al. Epitopes with similar physicochemical properties contribute to cross reactivity between peanut and tree nuts. Mol Immunol. (2020) 122:223-31. doi: 10.1016/j.molimm.2020.03.017
28. Lu W, Negi SS, Schein CH, Maleki SJ, Hurlburt BK, Braun W. Distinguishing allergens from non-allergenic homologues using Physical-Chemical Property (PCP) motifs. Mol Immunol. (2018) 99:1-8. doi: 10.1016/j.molimm.2018.03.022
29. Maleki SJ, Teuber SS, Cheng H, Chen D, Comstock SS, Ruan S, et al. Computationally predicted IgE epitopes of walnut allergens contribute to cross-reactivity with peanuts. Allergy. (2011) 66:1522-9. doi: 10.1111/j.1398-9995.2011.02692.x
30. Tiwari R, Negi SS, Braun B, Braun W, Pomes A, Chapman MD, et al. Validation of a phage display and computational algorithm by mapping a conformational epitope of Bla g 2. Int Arch Allergy Immunol. (2012) 157:323-30. doi: 10.1159/000330108
31. Nesbit JB, Hurlburt BK, Schein CH, Cheng H, Wei H, Maleki SJ. Ara h 1 structure is retained after roasting and is important for enhanced binding to IgE. Mol Nutr Food Res. (2012) 56:1739-47. doi: 10.1002/mnfr.201100815
32. Braun BA, Schein CH, Braun W. D-Graph clusters flaviviruses and beta-coronaviruses according to their hosts, disease type, and human cell receptors. Bioinform Biol Insights. (2021) 15:11779322211020316. doi: 10.1177/11779322211020316
33. Schein CH, Ivanciuc O, Midoro-Horiuti T, Goldblum RM, Braun W. An allergen portrait gallery: representative structures and an overview of IgE binding surfaces. Bioinform Biol Insights. (2010) 4:113-25. doi: 10.4137/BBI.S5737
34. Power TD, Ivanciuc O, Schein CH, Braun W. Assessment of 3D models for allergen research. Proteins. (2013) 81:545-54. doi: 10.1002/prot.24239
35. Ivanciuc O, Schein CH, Garcia T, Oezguen N, Negi SS, Braun W. Structural analysis of linear and conformational epitopes of allergens. Regul Toxicol Pharmacol. (2009) 54(3 Suppl):S11-19. doi: 10.1016/j.yrtph.2008.11.007
36. Oezguen N, Zhou B, Negi SS, Ivanciuc O, Schein CH, Labesse G, et al. Comprehensive 3D-modeling of allergenic proteins and amino acid composition of potential conformational IgE epitopes. Mol Immunol. (2008) 45:3740-7. doi: 10.1016/j.molimm.2008.05.026
37. Foo ACY, Nesbit JB, Gipson SAY, Cheng H, Bushel P, CeRose EF, et al. Structure, immunogenicity, and IgE cross-reactivity among walnut and peanut vicilin-buried peptides. J Agri Food Chem. (2022) 70:2389–400. doi: 10.1021/acs.jafc.1c07225
38. Midoro-Horiuti T, Mathura V, Schein CH, Braun W, Yu S, Watanabe M, et al. Major linear IgE epitopes of mountain cedar pollen allergen Jun a 1 map to the pectate lyase catalytic site. Mol Immunol. (2003) 40:555-62. doi: 10.1016/S0161-5890(03)00168-8
Keywords: allergenic protein nomenclature, sequence and structure, physicochemical property scale, IgE epitopes, history of allergen studies, peanut and nut allergens, property distance scale, component resolved extracts
Citation: Schein CH, Negi SS and Braun W (2022) Still SDAPing Along: 20 Years of the Structural Database of Allergenic Proteins. Front. Allergy 3:863172. doi: 10.3389/falgy.2022.863172
Received: 26 January 2022; Accepted: 28 February 2022;
Published: 22 March 2022.
Edited by:
Soheila June Maleki, Agricultural Research Service (USDA), United StatesReviewed by:
Philip Johnson, University of Nebraska-Lincoln, United StatesCopyright © 2022 Schein, Negi and Braun. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Catherine H. Schein, Y2hzY2hlaW5AdXRtYi5lZHU=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.