- 1Molecular Allergology, Paul-Ehrlich-Institut, Langen, Germany
- 2Foundation for Biomedical Research and Innovation, Hospital Universitario Infanta Leonor, Madrid, Spain
- 3Allergy Service, Hospital Universitario Infanta Leonor, Madrid, Spain
Only a small fraction of proteins in plants and animals are classified as allergens. The allergenic properties are frequently attributed to certain functional characteristics of the proteins, such as a role in the plant defense against biotic and abiotic stress, to achieve the systematic acquired resistance. In line with this, eight members out of 17 functional pathogenesis-related (PR) protein families have been characterized as allergens. The present review summarizes the molecular features and allergenic significance of allergens of the PR-1 family. Not many allergens have been identified as belonging to this protein family, with most of them having a pollen origin, like mugwort or Bermuda grass. Molecular and structural features of allergenic PR-1 proteins are discussed and attributed to their IgE-reactive properties, clinical manifestation, and cross-reactivity among different foods and inhalants.
Introduction
Several allergens have been identified as members of the pathogenesis-related (PR) protein families, PR-2 (ß-1-3 Glucanases), PR-3 and PR-4 (Chitinase), PR-5 (Thaumatin-like), PR-8 (Chitinase), PR-10 (Bet v 1-like), PR-14 (nsLTPs) (1, 2), and from the PR-1 family (3). The PR proteins of the same family are known to have highly-homologous sequences and similar functions (4).
The classification of PR-1 proteins is not consistent. On one hand, PR-1 proteins were classified as either cysteine-rich secretory proteins (CRISP) or antigen 5 (Ag5) type proteins (4). These proteins either belong to sperm coating protein (SCP)-like extracellular protein family, PR-1 protein family, or allergen V5/Tpx-1-related protein family (4), as part of the SCP/TAPS (Tpx-1/Ag5/PR-1/Sc7) protein superfamily (5, 6). In another classification, PR-1 proteins are described as members of a CAP protein superfamily, encompassing the Cysteine-rich secretory proteins in vertebrates, Antigen 5 in insects, and PR-1 in plants (7–9). Members of this superfamily are found across the bacterial, fungal, plant, and animal kingdoms, and they have broad physiological functions (10). Combining both classifications, the PR-1 protein family sometimes is termed as CAP/SCP/TAPS superfamily (9).
Pathogenesis-related-1 proteins were initially identified in tobacco plants in response to TMV infection (11). They are ubiquitously abundant, with a role in plant growth and flowering, in cell wall loosening, and with a function in biotic and abiotic stress responses (8, 12). PR-1 expression is induced by salicylic acid signaling, but the molecular mechanism to achieve disease resistance is not known (8). Domain analysis indicated that most PR-1 proteins have a conserved αβα sandwich folded CAP domain of 150 amino acids (4, 9). The CAP domain comprises the whole PR-1 protein and contains four conserved signature motifs (CAP1-4), with six specific cysteine residues encompassing 3 disulfide bonds (Figure 1A) (10) and is attributed to ion-, fatty acid, and sterol-binding capacity (8, 9, 13). Frequently PR-1 proteins are secreted as 14–17 kDa glycoproteins (9), and are classified as acidic or basic PR-1 (8), which are stabilized by hydrophobic interaction, hydrogen bonds, and intramolecular disulfide bonds. In line with this, PR-1 proteins are considered as highly stable proteins (14). The size, stability, and resistance to proteases, along with hydrolytic and membrane permeabilizing ability, have been claimed to make PR-proteins, including PR-1 proteins, excellent candidates to elicit an allergenic response (15). A summary of the PR-1 allergens described in this review is shown in Supplementary Table 1.
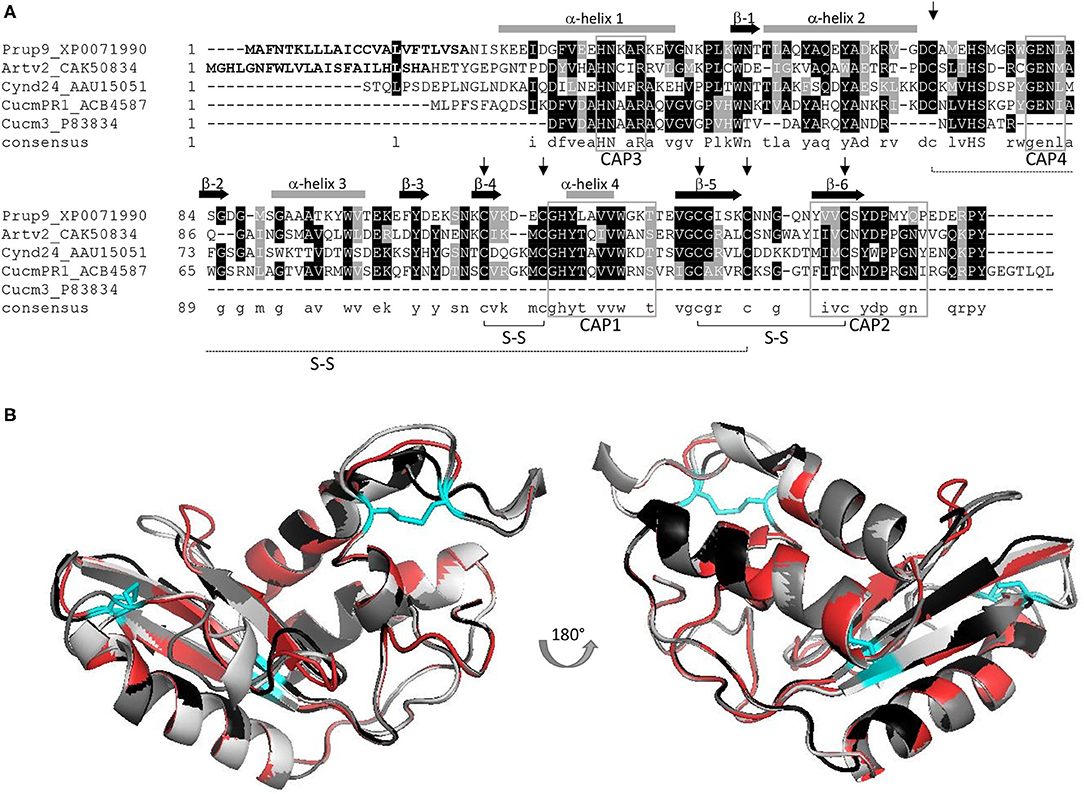
Figure 1. (A) AA alignment of PR-1 allergens Pru p 9, Art v 2, Cyn d 24, and Cuc m 3, including the full-length sequence of a Cuc m PR-1 protein; signal peptides of Pru p 9 and Art v 2 are displayed in bold; six conserved cysteins are indicated by an arrow, and the corresponding S-S bonds are outlined; CAP signature motifs 1–4 are marked (gray box) and α-helix (gray bar) and β-sheets (black arrow) are included; consensus sequence: conserved aa: capital letters, similar aa: small letters, no similarity: no letter; Figure adapted from (7). (B) Superimposition of 3D models including Pru p 9 (red), Art v 2 (black), Cyn d 24 (gray), and Cuc m PR1 (light gray). Disulfide bonds are indicated in cyan. All proteins were modeled using Swiss model and P14a from Lycopersicon esculentum (PDB-1CFE) as reference.
PR-1 Allergens
Cuc m 3, a PR-1 From Melon
Cosensitization to melon (Cucumis melo), belonging to the Cucurbaitaceae family, has been reported in the pollen food allergy syndrome, in particular in ragweed and grass pollen allergic patients (16, 17). In line with this, occupational respiratory allergy to melon flowers and fruit has been described (18).
So far three melon allergens are described: Cuc m 1 (subtilisin-like-protease), Cuc m 2 (profilin), and Cuc m 3. Cuc m 3.0101 (GenBank# P83834,) was the first PR-1 protein characterized as an allergen (19). Initially, a stretch of 41 amino acids (aa) was identified by the N-terminal sequencing of internal peptides of the 17 kDa protein derived from melon juice. A total of 12 out of 17 melon-allergic individuals with positive skin prick test (SPT) to melon extract, melon specific IgE >0.94 kU/L, and double-blind placebo-controlled food challenge (DBPCFC)-confirmed melon allergy showed IgE-binding to purified Cuc m 3. However, the frequency of IgE-sensitization (71%) was not in concordance with a low frequency of positive SPT (14%) to purified Cuc m 3. This discrepancy was attributed to a potential monovalency and/or low IgE-binding affinity. IgE-binding to Cuc m 3 reflected 40 and 70% of the IgE reactivity to extract from pulp and juice, respectively. In 2014, the full cDNA sequence of a muskmelon PR-1 protein derived from the inner layer of the fruit was cloned (GenBank# ACB45874), but never included to the World Health Organization and International Union of Immunological Societies (WHO/IUIS) Allergen Nomenclature database, and therefore in this review named as Cuc m PR-1. The cDNA sequence encodes for a 151 aa polypeptide with a theoretical pI of 9.47 and a predicted molecular mass of 16.97 kDa (20). Cuc m PR-1 expresses one potential glycosylation site, six cysteins, and an SCP-like extracellular protein domain (20). In addition, Cuc m PR-1 shows aa sequence identities with Cyn d 24 (42.6%), and other PR-1 proteins from cucumber (70%) or tomato (61%) (Table 1), which up to now not have been described as allergens, but are discussed as potential cross-reactive allergens (20, 21).
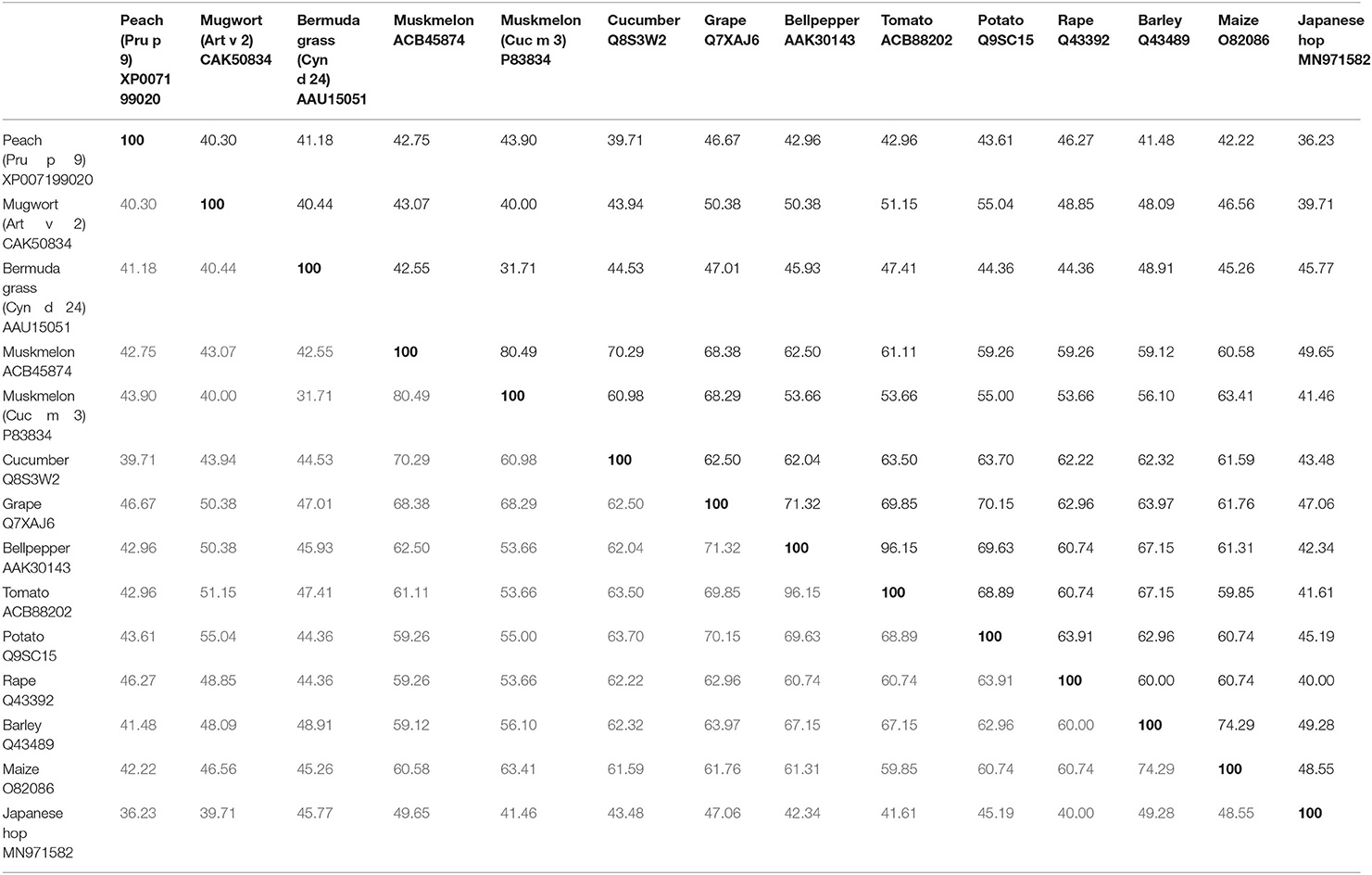
Table 1. AA-identities of Pru p 9, Art v 2, Cyn d 24 and Cuc m 3, including the full length sequence of a Cuc m PR-1 protein, without including the signal peptide.
Cyn d 24, a PR-1 From Bermuda Grass
Pollen from the subtropical Bermuda grass (Cynodon dactylon) is a major source for allergic reactions with a high prevalence of IgE-sensitization in grass pollen allergic patients, ranging from 40% up to 91% in India and Australia, respectively (22). In total, seven allergens were identified (allergen.org): Cyn d 1 (beta-expansin), Cyn d 7 (polcalcin), Cyn d 12 (profilin), Cyn d 15 and Cyn d 23, Cyn d 22 (enolase), and the PR-1 protein Cyn d 24. Shen et al. showed the abundance of an IgE-reactive 21 kDa protein in Bermuda grass pollen, with a frequency of 29% of specific IgE-binding in Bermuda grass allergic patients (23). Using a proteomic approach by 2D LC-MS/MS, N-terminal sequencing, and an anti-PR-1 antibody, the 21 kDa protein (pI = 5.9) was identified as PR-1 protein (24). Four peptides from two spots revealed 39–54% aa similarity with the corresponding region of PR-1 from Hordeum vulgare (P35793). Glycoprotein staining indicated a glycosylation of the PR-1 protein. Finally, natural Cyn d 24 was purified (25) and the cDNA was cloned (GenBank# AAU15051.1). Cyn d 24 consists of 153 aa with an additional signal peptide of 97 aa and it displays an SCP-like extracellular protein domain. Remarkably, Cyn d 24 reveals higher aa identities to SCP proteins than to CRISPs, but low aa-identity to group allergen 5. It contains the conserved region GHYTQVVW, which is responsible for plant defense-related activity (25). Mass spectrometry data of the purified nCyn d 24 resulted in a mass of m/z 18.4 kDa, The pI was estimated to be 5.9 and periodic acid Schiff staining indicated that the protein is glycosylated. Con A binding of nCyn d 24 demonstrated that it likely contains carbohydrate moieties with free α-D-mannopyranoside or α-D-glucopyranoside residues (25). Enzyme-linked immune sorbent assay (ELISA) experiments showed that 65% (23/35) of Bermuda grass pollen allergic patients were sensitized to nCyn d 24. About 5.7% of the mass of Cyn d 24 consists of the oligosaccharide Man3GlcNAc2Fuc, probably a unique feature of PR-1 glycoproteins, which leads to the hypothesis, that these glycan structures could display IgE crossreactive carbohydrates (25). Moreover, Cyn d 24 and other glycoproteins from Bermuda grass pollen carry non-xylosylated glycans, mainly MMF3, which are also found in insect venom (26). Unfortunately, no studies about IgE crossreactivity of Cyn d 24 with other PR-1 allergens from pollen, food, or insect venom have been described until now.
Art v 2 and Artimisia PR-1 Allergens
Artemisia vulgaris (27) belongs to one of the largest families of flowering plants, the Compositae or Asteraceae. It is a weed widely spread in temperate regions and subtropics of Europe, North America, and Asia (28, 29). Among these, Art v 1, Art v 2, Art v 3, and Art v 7 have been shown to be the most relevant allergens (28, 29). However, the function of PR-1 homologous Art v 2 remains unknown (14).
The apparent molecular mass of Art v 2 on gel filtration chromatography is 33 ± 3 kDa, suggesting that it is composed of two mainly identical subunits linked by disulfide bridges (30). Purified nArt v 2 gave a single band with a molecular mass of 19.2 kDa under reduced conditions and 34.2 kDa under non-reduced conditions. It has one potential N-glycosylation site and a calculated pI of 5.23. However, in contrast to Cyn d 24, modification of high-mannose oligosaccharides has been reported to have no significant effect on the binding of IgE. Nevertheless, it could be a stabilizing element of the three-dimensional conformation (31). Art v 2 has six conserved cysteine residues that should be involved in the stability of the 3D structure (27). This protein folds into a sandwich structure with four α-helices and three ß-strands arranged antiparallelly and compacted by three disulfide bridges (Figure 1B) (31, 32).
Art v 2 homologous allergen sequences in Artemisia spp. are highly conserved, indicating a general crossreactivity in all species of this genus (27). In this way, a sequence identity of 40% was found with the allergen Cyn d 24 from Bermuda grass pollen (Table 1). In addition, Art v 2 shares between 47 and 55% aa-identity with PR-1 proteins from tomato (51%), potato (55%), or bell pepper (50%) between others (Table 1) (30), with unknown allergenic properties till now.
Determination of Art v 2-specific IgE-values showed that 63.2% of sera from mugwort-allergic patients reacted with nArt v 2, whereas 10.5% reacted with the recombinant counterpart. The difference in reactivity between the natural and the recombinant allergen is attributed to the lack of the N-glycosylation reported in the nArt v 2 (30). Regarding skin testing, the prevalence of nArt v 2 sensitization was 57.9% at 100 μg/ml and 36.8% at 50 μg/ml, whereas no false positive was detected among the control patients. A positive skin prick test using rArt v 2 was determined only in 21% of the allergic patients (33).
Artemisia pollens have been recognized as a major cause for seasonal allergic respiratory disease worldwide, especially asthma (28, 34). It is relevant along with the Asia–Europe silk-road and in northwestern European countries. Besides, Egger et al. have described the possible role of Art v 2-like protein on pollen food syndromes associated with mugwort pollinosis (32).
However, the information on other PR-1 related proteins in this plant family is limited since studies have been carried out mainly with Artemisia vulgaris. As an example, Artemisia annua (Art an 2) has been recognized as an important source of allergen, and together with Artemisia argyi (Art ar 2) and Artemisia sieversiana (Art si 2), they are recommended for immunotherapy in Chinese allergic patients (29).
Pru p 9, a PR-1 From Peach
Peach tree (Prunus persica) allergens are present in fruit, pollen, leaves, and branches, and can induce systemic, respiratory, gastrointestinal, and cutaneous symptoms. Although classified as entomophilous, like other pollens, recent evidence indicates that in areas with peach tree orchards, pollen grains can be transported by the wind for short distances, facilitated by the presence of pollutants or the disruption of pollen in smaller submicronic particles (35, 36). In addition, in these areas, there is a high prevalence of sensitization and allergy to peach tree pollen (37).
Six peach allergens that are mostly involved in food allergy have been identified and characterized: Pru p 1 (PR-10 protein) (38), Pru p 2 (Thaumatin-like), Pru p 3 (ns-LTP) (39), Pru p 4 (profilin) (40), Pru p 5 (Hev b 5-like) (41), and Pru p 7 (gibberellin-regulated protein) (42). On the other hand, the only aeroallergen described in peach tree pollen is Pru p 9 (GenBank# XP_007199020.1), a PR-1 protein (43).
In a study analyzing 685 children and adolescents between 3 and 19 years and 310 adults from the population, directly or indirectly exposed to peach cultivars, the sensitization to peach tree pollen was estimated to be 20%, being the third most prevalent pollen after olive tree (33%) and grass pollen (26%) (43, 44). According to symptoms, conjunctivitis appeared in 47% of the cases, rhinitis in 42%, and asthma in 14% (43).
Pru p 9 is a 138 aa polypeptide with a theoretical pI of 5.11 and a molecular mass of 18 kDa. This allergen is responsible for the induction of respiratory and ocular symptoms in allergic subjects, independently whether they are directly or indirectly exposed. Reported in another study by Blanca et al., from 62 cases sensitized to peach tree pollen, 41% were positive in SPT to Pru p 9 and presented Pru p 9-specific IgE-antibodies. Besides, the nasal provocation tests showed a decrease in the nasal cavity volume greater than 20% with the new aeroallergen described (42), associated with rhinorrhoea, sneezing, nasal itching, eye redness, and tearing. In addition, Victorio-Puche et al. (45) described the implication of Pru p 9 in the induction of occupational allergic respiratory symptoms, with rhinitis and asthma in 43% of workers in the orchard after handling peach trees during the flowering period. Unfortunately, no studies about IgE crossreactivity of Pru p 9 with other PR-1 allergens from pollen or food (including peach fruit) have been described till now.
Vesp ma 5
Stings of hymenoptera are one of the most frequent triggers for severe IgE-mediated anaphylaxis in adults. Noteworthy, PR-1 proteins share similar features with antigen 5 (Ag5) allergens from the venom of hymenoptera, such as the CAP domain forming a potential CAP cavity for ligand binding. Ag5 showed only moderate aa-ids with Pru p 9: 18% with horsefly Ag5 (Tab y 5), 24% with Savannah tsetse fly Ag5 (Glo m 5), and up to 35% with Vesp ma 5. The Ag5 protein family includes several allergens from hornets, wasps, horseflies, and fire ants: The 25 kDa wasp Vespa magnifica Vesp ma 5 (46) shares 27–87% aa-id with Tab y 5 (Tabanus yao) (47), Vesp m 5 (Vespula maculifrons) (48), Ves v 5 (Vespula vulgaris) (49), Dol m 5 (Dolichovespula maculata) (50), and Vesp c 5 (Vespa crabro) (51), respectively. To date, 26 Vespoidea Ag5 proteins are listed in the official allergen nomenclature database of the WHO/IUIS Allergen Nomenclature database (52). According to the phylogenetic distance between these species, the Ag5 allergens exhibit a varying degree of sequence identity and, therefore, most likely of crossreactivity between them.
Although Ag5 represents one of the most abundant and important major venom allergens in almost all allergy-relevant Vespoidea species (51, 53–56), their function within the venoms remains widely unclear. In blood-feeding ticks, flies, and mosquitoes, Ag5 proteins are salivary proteins that are thought to function either in suppression of the host immune system or in preventing platelet aggregation (57). The presence of Ag5 allergens (as well as hyaluronidases), which exhibit crossreactivity with their homologs of wasp venom, in the saliva of horseflies and mosquitoes (47), may explain the postulated “wasp–mosquito–horsefly-syndrome,” in which wasp venom-allergic patients also experience systemic reactions after bites of mosquitoes or horseflies (58, 59). Due to their outstanding role as major allergens, Ag5 proteins build a key element for the diagnosis of Vespoidea venom allergy (60).
Other Likely Allergenic PR-1 Proteins
The IgE-sensitizing capacity has been shown for additional PR-1 proteins. However, these proteins have not yet been fully characterized and are not included in the IUIS allergen database.
Feverfew (Parthenium hysterophorus), an invasive weed from the Asteraceae family, has been reported as a respiratory allergen. Recently, a pollen-derived PR-1 protein of 20 kDa, which displays IgE-reactivity with sera from feverfew sensitized patients' from India, has been identified by LC-MS/MS and 2D-LC-MS/MS (61). However, the molecular characterization of Par h PR-1 is lacking.
Moreover, screening a cDNA expression library revealed that a PR-1 protein from Japanese hop (Humulus japonicus), an important cause of weed pollinosis in East Asia, is recognized by IgE from 3.4% (1/29) of Japanese hop allergic patients with a positive case history and IgE-sensitization to Japanese hop (62). Hum j PR-1 encodes for a 171 aa peptide (apparent MW of 20 kDa) and shares 40–46% of aa-sequence identity with Art v 2, Cuc m 3, and Cyn d 24 (Table 1). The authors attributed the low IgE-binding frequency of the His-tagged protein with a potential engagement of glycan moieties in IgE-epitopes of the natural counterpart and/or an incorrect protein folding affecting conformational epitopes.
Finally, a 20 kDa IgE-reactive protein derived from tomato pollen has been described to play the role as an occupational allergen for a greenhouse worker (63). Initial experiments showed partial IgE-crossreactivity with purified Pru p 9 (Pedraza et al. unpublished data).
Conclusions
Pathogenesis-related proteins are characterized by certain common biochemical features, such as low molecular weight, stability at low pH, and resistance to proteases. This protein family is widely distributed in mono- and dicotyledonous species (64). Members of the SCP superfamily have been proposed to act in the defense against pathogens as Ca++-chelating serine proteases (3). The Ca++-chelating function would fit with various signaling processes that members of this family, such as the CRISPs, are involved in, supported by sequence and structural evidence of a conserved pocket containing two histidine and glutamate. In plants, PR-1 proteins are among the most abundantly produced proteins during defense responses and have been reported to constitute 2% of the total leaf protein in pathogen-infected tobacco (65). PR-1 proteins are typically 14–17 kDa polypeptides of approximately 135 aa that form disulfide bonds and a compact 3D structure, conferring the protein a high stability (14). In recent years, a variety of PR-1 proteins and their homologs causing allergenicity have been characterized. The size, stability, and resistance to proteases along with hydrolytic and membrane permeabilizing ability make these proteins excellent candidates to elicit allergenic responses (1, 2, 15, 66). However, very little is known about their allergenic properties (4).
The PR-1 proteins from plants and other organisms share structural similarities that may trigger crossreactivity reactions (67). Evident aa-identities within the PR-1 proteins likely could lead to IgE-crossreactivity and even to the presence of further allergenic PR-1 proteins, in e.g., tomato, potato, rape, or maize. Moreover, particularly interesting in the area of allergy is their relationship (approximately 30% of sequence identity) with Ag5 allergens (14), but no studies have been performed yet concerning this.
The effect of posttranslational modification of PR-1 proteins by carbohydrate determinants needs to be further explored. Pru p 9 contains two potential glycosylation sites at aa24 and aa52, whereas Art v 2 (aa91), Cuc m PR-1 (aa33), and Cyn d 24 (aa25) have only one potential glycosylation site, as predicted using the NetNGlyc – 1.0 server (68). Interestingly the N-glycosylation sites of Pru p 9 (aa52), Cyn d 24 and Cuc m PR-1 are conserved between their AA sequence and therefore, could be influencing or be responsible for IgE crossreactivity reactions through crossreactive carbohydrate determinants (25). Besides Cyn d 24 other glycoproteins from Bermuda grass pollen carry non-xylosylated glycans, mainly MMF which are also found in insect venom (26). Similarly, the predicted legume PR-1 protein showed better binding affinity to carbohydrates (glycans) than proteins/lipids, indicating that glycans might act as recognition clues or trigger immune responses in the plant (4).
The clinical relevance of the PR-1 family is partially known. Only few allergens from this widely distributed protein family have been deeply studied, such as Cyn d 24 with 63% of recognition by patients allergic to Bermuda grass pollen and Art v 2 with 63.2% of positive response by patients sensitized to mugwort pollen, or Cuc m 3, a PR-1 food allergen identified in melon allergic patients. In a more complex way, the Ag5 group, encompassing 26 PR-1 allergens, is implicated in hymenoptera allergy, which triggers life-threatening reactions. According to WHO/IUIS Allergen Nomenclature database criteria, all of them could be considered as relevant allergens in their corresponding sources. An important feature of PR-1 proteins is their role as occupational allergens. Of particular interest is the PR-1 protein described in peach tree pollen, Pru p 9, with 43% of IgE binding in patients allergic to this source. In this way, Pru p 9 has been identified as a relevant allergen with critical implications for people working with peach tree cultivars, developing respiratory symptoms (45).
Nevertheless, one of the major difficulties encountered by researchers in studying allergens belonging to the PR-1 protein family lies in the difficulty for identifying cases sensitized, and also for obtaining pollen grains for molecular analysis of their allergenic profile. Probably, this is one reason for the low number of PR-1 allergens identified and registered in databases. However, it is very likely that PR-1 proteins from other sources display allergenic properties. Exploring new PR-1 proteins implicated in allergenicity, and a complete understanding of their structures and IgE-binding epitopes are necessary for improving the diagnoses and treatment in these subjects, reaching precision medicine in allergy.
Author Contributions
LM-P, AW, and SS conceived and supervised the review topics. LM-P, AW, SS, MB, MS, and NB-L wrote the first draft. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We would like to thank the Complutense University of Madrid for its academic support.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/falgy.2022.824717/full#supplementary-material
Abbreviations
PR, pathogenesis-related; CRISP, cysteine-rich secretory proteins; Ag5, antigen 5; SCP, Sperm Coating Protein; TAPS, Tpx-1/Ag5/PR-1/Sc7; CAP, Cysteine-rich secretory proteins; Antigen 5, PR-1; Aa, amino acid; DBPCFC, Double-blind placebo-controlled food challenge.
References
1. Ebner C, Hoffmann-Sommergruber K, Breiteneder H. Plant food allergens homologous to pathogenesis-related proteins. Allergy. (2001) 56:43–4. doi: 10.1034/j.1398-9995.2001.00913.x
2. Midoro-Horiuti T, Brooks EG, Goldblum RM. Pathogenesis-related proteins of plants as allergens. Ann Allergy Asthma Immunol. (2001) 87:261–71. doi: 10.1016/S1081-1206(10)62238-7
3. Sinha M, Singh Rp, Kushwaha Gs, Iqbal N, Singh A, Kaushik S, et al. Current overview of allergens of plant pathogenesis related protein families. Sci World J. (2014) 2014:543195. doi: 10.1155/2014/543195
4. Tellis M, Mathur M, Gurjar G, Kadoo N, Gupta V. Identification and functionality prediction of pathogenesis-related protein 1 from legume family. Proteins. (2017) 85:2066–80. doi: 10.1002/prot.25361
5. Woo HS, Kim TY, Sohn W-M, Yong T-S. A tegument-specific venom allergen-like protein of clonorchis sinensis. Parasitol Res. (2015) 114:329–33. doi: 10.1007/s00436-014-4212-2
6. Chalmers IW, Hoffmann KF. Platyhelminth venom allergen-like (val) proteins: revealing structural diversity, class-specific features and biological associations across the phylum. Parasitology. (2012) 139:1231–45. doi: 10.1017/S0031182012000704
7. Gibbs Gm, Roelants K, O'bryan MK. The cap superfamily: cysteine-rich secretory proteins, antigen 5, and pathogenesis-related 1 proteins–roles in reproduction, cancer, and immune defense. Endocr Rev. (2008) 29:865–97. doi: 10.1210/er.2008-0032
8. Breen S, Williams SJ, Outram M, Kobe B, Solomon PS. Emerging insights into the functions of pathogenesis-related protein 1. Trends Plant Sci. (2017) 22:871–9. doi: 10.1016/j.tplants.2017.06.013
9. Darwiche R, El Atab O, Cottier S, Schneiter R. The function of yeast cap family proteins in lipid export, mating, and pathogen defense. FEBS Lett. (2018) 592:1304–11. doi: 10.1002/1873-3468.12909
10. Gaikwad AS, Hu J, Chapple DG, O'bryan MK. The functions of cap superfamily proteins in mammalian fertility and disease. Hum Reprod Update. (2020) 26:689–723. doi: 10.1093/humupd/dmaa016
11. Van Loon LC, Gerritsen YA, Ritter CE. Identification, purification, and characterization of pathogenesis-related proteins from virus-infected samsun nn tobacco leaves. Plant Mol Biol. (1987) 9:593–609. doi: 10.1007/BF00020536
12. Niderman T, Genetet I, Bruyère T, Gees R, Stintzi A, Legrand M, et al. Pathogenesis-related pr-1 proteins are antifungal. Isolation and characterization of three 14-kilodalton proteins of tomato and of a basic pr-1 of tobacco with inhibitory activity against phytophthora infestans. Plant Physiol. (1995) 108:17–27. doi: 10.1104/pp.108.1.17
13. Gamir J, Darwiche R, Van't Hof P, Choudhary V, Stumpe M, Schneiter R, et al. The sterol-binding activity of pathogenesis-related protein 1 reveals the mode of action of an antimicrobial protein. Plant J. (2017) 89:502–9. doi: 10.1111/tpj.13398
14. Van Loon LC, Van Strien EA. The families of pathogenesis-related proteins, their activities, and comparative analysis of pr-1 type proteins. Physiol Molec Plant Pathol. (1999) 55:85–97. doi: 10.1006/pmpp.1999.0213
15. Hoffmann-Sommergruber K. Plant allergens and pathogenesis-related proteins. What do they have in common? Int Arch Allergy Immunol. (2000) 122:155–66. doi: 10.1159/000024392
16. Anderson L, Dreyfuss EM, Logan J, Johnstone DE, Glaser J. Melon and banana sensitivity coincident with ragweed pollinosis. J Allergy. (1970) 45:310–9. doi: 10.1016/0021-8707(70)90037-7
17. Lipp T, Acar Sahin A, Aggelidis X, Arasi S, Barbalace A, Bourgoin A, et al. Heterogeneity of pollen food allergy syndrome in seven southern european countries: the @it.2020 multicenter study. Allergy. (2021) 76:3041–52. doi: 10.1111/all.14742
18. Gómez Torrijos E, Garcia Rodríguez C, García Rodríguez R, Mendez Díaz Y, Feo Brito FJ. Occupational asthma and rhinoconjunctivitis by melon plant allergy. Ann Allergy Asthma Immunol. (2015) 114:417–8. doi: 10.1016/j.anai.2015.02.006
19. Asensio T, Crespo Jf, Sanchez-Monge R, Lopez-Torrejon G, Somoza ML, Rodriguez J, et al. Novel plant pathogenesis-related protein family involved in food allergy. J Allergy Clin Immunol. (2004) 114:896–9. doi: 10.1016/j.jaci.2004.06.014
20. Sankian M, Hajavi J, Moghadam M, Varasteh A-R. Identification and molecular characterization of the cdna encoding cucumis melo allergen, cuc m 3, a plant pathogenesis-related protein. Rep Biochem Mol Biol. (2014) 2:82–7.
21. Neeharika D, Sunkar S. Computational approach for the identification of putative allergens from cucurbitaceae family members. J Food Sci Technol. (2021) 58:267–80. doi: 10.1007/s13197-020-04539-7
22. Davies JM. Grass pollen allergens globally: the contribution of subtropical grasses to burden of allergic respiratory diseases. Clin Exp Allergy. (2014) 44:790–801. doi: 10.1111/cea.12317
23. Shen HD, Wang SR, Tang RB, Chang ZN, Su SN, Han SH. Identification of allergens and antigens of bermuda grass (cynodon dactylon) pollen by immunoblot analysis. Clin Allergy. (1988) 18:401–9. doi: 10.1111/j.1365-2222.1988.tb02888.x
24. Kao S-H, Su S-N, Huang S-W, Tsai J-J, Chow L-P. Sub-proteome analysis of novel ige-binding proteins from bermuda grass pollen. Proteomics. (2005) 5:3805–13. doi: 10.1002/pmic.200401229
25. Chow L-P, Chiu L-L, Khoo K-H, Peng H-J, Yang S-Y, Huang S-W, et al. Purification and structural analysis of the novel glycoprotein allergen cyn d 24, a pathogenesis-related protein pr-1, from bermuda grass pollen. FEBS J. (2005) 272:6218–27. doi: 10.1111/j.1742-4658.2005.05000.x
26. Altmann F. The role of protein glycosylation in allergy. Int Arch Allergy Immunol. (2007) 142:99–115. doi: 10.1159/000096114
27. Nilsen BM, Grimsøen A, Paulsen BS. Identification and characterization of important allergens from mugwort pollen by ief, sds-page and immunoblotting. Mol Immunol. (1991) 28:733–42. doi: 10.1016/0161-5890(91)90116-2
28. Gao Z-S, Fu W-Y, Zhao L, Gao L, Zhou J-Y, Gao B-Y, et al. Localization of four allergens in artemisia pollen by immunofluorescent antibodies. Int Arch Allergy Immunol. (2019) 179:165–72. doi: 10.1159/000497321
29. Zhao L, Fu W, Gao B, Liu Y, Wu S, Chen Z, et al. Variation in ige binding potencies of seven artemisia species depending on content of major allergens. Clin Transl Allergy. (2020) 10:50. doi: 10.1186/s13601-020-00354-7
30. Arilla MC, Ibarrola I, Puente Y, Daza JC, Martínez A, Asturias JA. Cloning, expression and characterization of mugwort pollen allergen art v 2, a pathogenesis-related protein from family group 1. Mol Immunol. (2007) 44:3653–60. doi: 10.1016/j.molimm.2007.04.022
31. Nilsen BM, Sletten K, Paulsen Bs, O'neill M, Van Halbeek H. Structural analysis of the glycoprotein allergen art v ii from the pollen of mugwort (Artemisia Vulgaris L). J Biol Chem. (1991) 266:2660–8. doi: 10.1016/S0021-9258(18)52295-X
32. Egger M, Mutschlechner S, Wopfner N, Gadermaier G, Briza P, Ferreira F. Pollen-food syndromes associated with weed pollinosis: an update from the molecular point of view. Allergy. (2006) 61:461–76. doi: 10.1111/j.1398-9995.2006.00994.x
33. Gao Z, Fu W-Y, Sun Y, Gao B, Wang H-Y, Liu M, et al. Artemisia pollen allergy in china: component-resolved diagnosis reveals allergic asthma patients have significant multiple allergen sensitization. Allergy. (2019) 74:284–93. doi: 10.1111/all.13597
34. Movérare R, Larsson H, Carlsson R, Holmquist I. Mugwort-sensitized individuals from north europe, south europe and north america show different ige reactivity patterns. Int Arch Allergy Immunol. (2011) 154:164–72. doi: 10.1159/000320231
35. Irañeta SG, Seoane MA, Laucella SA, Apicella C, Alonso A, Duschak VG. Antigenicity and immunocrossreactivity of orange tree pollen and orange fruit allergenic extracts. Int Arch Allergy Immunol. (2005) 137:265–72. doi: 10.1159/000086419
36. Javier F, Emilio F, Montserrat V, Victor S, Purificaci NG. Evolution of the incidence of pollen grains and sensitivity to pollen in the city of Elche (Spain). Asian Pac J Allergy Immunol. (2015) 33:196–202. doi: 10.12932/AP0542.33.3.2015
37. Somoza ML, Pérez-Sánchez N, Victorio-Puche L, Martín-Pedraza L, Esteban Rodríguez A, Blanca-López N, et al. Subjects develop tolerance to pru p 3 but respiratory allergy to pru p 9: a large study group from a peach exposed population. PLoS ONE. (2021) 16:E0255305. doi: 10.1371/journal.pone.0255305
38. Gao Z-S, Zhou X, Yang Z-W, Versteeg SA, Gao L, Fu W-Y, et al. Ige-binding potencies of three peach pru p 1 isoforms. Mol Nutr Food Res. (2016) 60:2457–66. doi: 10.1002/mnfr.201500798
39. García-Casado G, Pacios Lf, Díaz-Perales A, Sánchez-Monge R, Lombardero M, García-Selles FJ, et al. Identification of ige-binding epitopes of the major peach allergen pru P 3. J Allergy Clin Immunol. (2003) 112:599–605. doi: 10.1016/S0091-6749(03)01605-1
40. Rodríguez-Perez R, Fernández-Rivas M, González-Mancebo E, Sánchez-Monge R, Díaz-Perales A, Salcedo G. Peach profilin: cloning, heterologous expression and cross-reactivity with bet V 2. Allergy. (2003) 58:635–40. doi: 10.1034/j.1398-9995.2003.00198.x
41. Giangrieco I, Ricciardi T, Alessandri C, Farina L, Crescenzo R, Tuppo L, et al. Enea, a peach and apricot ige-binding protein cross-reacting with the latex major allergen hev b 5. Mol Immunol. (2019) 112:347–57. doi: 10.1016/j.molimm.2019.05.007
42. Ando Y, Miyamoto M, Kato M, Nakayama M, Fukuda H, Yoshihara S. Pru P 7 predicts severe reactions after ingestion of peach in japanese children and adolescents. Int Arch Allergy Immunol. (2020) 181:183–90. doi: 10.1159/000504367
43. Blanca M, Victorio Puche L, Garrido-Arandia M, Martin-Pedraza L, Romero Sahagún A, López-Sánchez JD, et al. Pru P 9, a new allergen eliciting respiratory symptoms in subjects sensitized to peach tree pollen. PLoS One. (2020) 15:E0230010. doi: 10.1371/journal.pone.0230010
44. Victorio Puche L, Somoza ML, López-Sánchez JD, Garrido-Arandia M, Díaz-Perales A, Blanca M. Peach tree pollen and prunus persica 9 sensitisation and allergy in children and adolescents. Int Arch Allergy Immunol. (2019) 180:212–20. doi: 10.1159/000502352
45. Victorio-Puche L, Somoza ML, Martin-Pedraza L, Fernandez-Caldas E, Abel Fernandez E, Moran M, et al. Prunus persica 9, a new occupational allergen from peach tree pollen involved in rhinitis and asthma. Occup Environ Med. (2021) 78:142–4. doi: 10.1136/oemed-2020-106641
46. An S, Chen L, Wei J-F, Yang X, Ma D, Xu X, et al. Purification and characterization of two new allergens from the venom of vespa magnifica. PLoS ONE. (2012) 7:E31920. doi: 10.1371/journal.pone.0031920
47. Ma D, Li Y, Dong J, An S, Wang Y, Liu C, et al. Purification and characterization of two new allergens from the salivary glands of the horsefly, tabanus yao. Allergy. (2011) 66:101–9. doi: 10.1111/j.1398-9995.2010.02435.x
48. Lu G, Villalba M, Coscia MR, Hoffman DR, King TP. Sequence analysis and antigenic cross-reactivity of a venom allergen, antigen 5, from hornets, wasps, and yellow jackets. J Immunol. (1993) 150:2823–30.
49. Suck R, Weber B, Kahlert H, Hagen S, Cromwell O, Fiebig H. Purification and immunobiochemical characterization of folding variants of the recombinant major wasp allergen ves v 5 (Antigen 5). Int Arch Allergy Immunol. (2000) 121:284–91. doi: 10.1159/000024341
50. King TP, Lu G. Hornet venom allergen antigen 5, dol M 5: its t-cell epitopes in mice and its antigenic cross-reactivity with a mammalian testis protein. J Allergy Clin Immunol. (1997) 99:630–9. doi: 10.1016/S0091-6749(97)70025-3
51. Hoffman DR. Allergens in hymenoptera venom. xxv: the amino acid sequences of antigen 5 molecules and the structural basis of antigenic cross-reactivity. J Allergy Clin Immunol. (1993) 92:707–16. doi: 10.1016/0091-6749(93)90014-7
52. Radauer C, Nandy A, Ferreira F, Goodman Re, Larsen JN, Lidholm J, et al. Update of the who/iuis allergen nomenclature database based on analysis of allergen sequences. Allergy. (2014) 69:413–9. doi: 10.1111/all.12348
53. Hoffman DR. Allergens in hymenoptera venom xv: the immunologic basis of vespid venom cross-reactivity. J Allergy Clin Immunol. (1985) 75:611–3. doi: 10.1016/0091-6749(85)90038-7
54. Hoffman DR. Reactions to less common species of fire ants. J Allergy Clin Immunol. (1997) 100:679–83. doi: 10.1016/S0091-6749(97)70173-8
55. King TP, Kochoumian L, Lam T. Immunochemical observations of antigen 5, a major venom allergen of hornets, yellowjackets and wasps. Mol Immunol. (1987) 24:857–64. doi: 10.1016/0161-5890(87)90188-X
56. King TP, Sobotka AK, Alagon A, Kochoumian L, Lichtenstein LM. Protein allergens of white-faced hornet, yellow hornet, and yellow jacket venoms. Biochemistry. (1978) 17:5165–74. doi: 10.1021/bi00617a016
57. Ribeiro Jm, Francischetti IM. Role of arthropod saliva in blood feeding: sialome and post-sialome perspectives. Annu Rev Entomol. (2003) 48:73–88. doi: 10.1146/annurev.ento.48.060402.102812
58. Quercia O, Emiliani F, Foschi FG, Stefanini GF. The wasp-horsefly syndrome. Eur Ann Allergy Clin Immunol. (2008) 40:61–3.
59. Sabbah A, Hassoun S, Drouet M, Lauret MG, Doucet M. Le syndrome guêpe/moustique: extension de l'allergénicité croisée au taon. Allerg Immunol (Paris). (2000) 32:16–9.
60. Blank S, Bazon ML, Grosch J, Schmidt-Weber CB, Brochetto-Braga MR, Bilò MB, et al. Antigen 5 allergens of hymenoptera venoms and their role in diagnosis and therapy of venom allergy. Curr Allergy Asthma Rep. (2020) 20:58. doi: 10.1007/s11882-020-00954-0
61. Pablos I, Eichhorn S, Briza P, Asam C, Gartner U, Wolf M, et al. Proteomic profiling of the weed feverfew, a neglected pollen allergen source. Sci Rep. (2017) 7:6049. doi: 10.1038/s41598-017-06213-z
62. Jang SW, Jeong KY, Yuk JE, Lee J, Park KH, Park J-W. Allergen homologues, pathogenesis-related 1, polygalacturonase, and pectin methyl esterase from a japanese hop. Protein Pept Lett. (2021) 28:362–71. doi: 10.2174/0929866527666200813201924
63. Vandenplas O, Sohy C, D'alpaos V, Nootens C, Thimpont J, Weigand D, et al. Tomato-induced occupational asthma in a greenhouse worker. J Allergy Clin Immunol. (2008) 122:1229–31. doi: 10.1016/j.jaci.2008.07.035
64. Van Loon LC, Pierpoint WS, Boller T, Conejero V. Recommendations for naming plant pathogenesis-related proteins. Plant Mol Biol Rep. (1994) 12:245–64. doi: 10.1007/BF02668748
65. Alexander D, Goodman RM, Gut-Rella M, Glascock C, Weymann K, Friedrich L, et al. Increased tolerance to two oomycete pathogens in transgenic tobacco expressing pathogenesis-related protein 1a. Proc Natl Acad Sci U S A. (1993) 90:7327–31. doi: 10.1073/pnas.90.15.7327
66. Hoffmann-Sommergruber K. Pathogenesis-related (pr)-proteins identified as allergens. Biochem Soc Trans. (2002) 30:930–5. doi: 10.1042/bst0300930
67. Serrano RL, Kuhn A, Hendricks A, Helms JB, Sinning I, Groves MR. Structural analysis of the human golgi-associated plant pathogenesis related protein gapr-1 implicates dimerization as a regulatory mechanism. J Mol Biol. (2004) 339:173–83. doi: 10.1016/j.jmb.2004.03.015
Keywords: pathogenesis-related protein-1, PR-1, allergen, occupational allergy, allergy diagnosis
Citation: Wangorsch A, Scheurer S, Blanca M, Blanca-Lopez N, Somoza ML and Martín-Pedraza L (2022) Allergenic Properties and Molecular Characteristics of PR-1 Proteins. Front. Allergy 3:824717. doi: 10.3389/falgy.2022.824717
Received: 29 November 2021; Accepted: 03 January 2022;
Published: 08 February 2022.
Edited by:
Maksymilian Chruszcz, University of South Carolina, United StatesReviewed by:
Anna Pomés, Indoor Biotechnologies, United StatesCopyright © 2022 Wangorsch, Scheurer, Blanca, Blanca-Lopez, Somoza and Martín-Pedraza. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Laura Martín-Pedraza, bGF1cmFtcDYyN0BnbWFpbC5jb20=
 Andrea Wangorsch
Andrea Wangorsch Stephan Scheurer
Stephan Scheurer Miguel Blanca
Miguel Blanca Natalia Blanca-Lopez
Natalia Blanca-Lopez María Luisa Somoza
María Luisa Somoza Laura Martín-Pedraza
Laura Martín-Pedraza