
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
BRIEF RESEARCH REPORT article
Front. Allergy , 16 August 2021
Sec. Rhinology
Volume 2 - 2021 | https://doi.org/10.3389/falgy.2021.724328
This article is part of the Research Topic Rising Stars in Asthma and Rhinology Research View all 5 articles
 Simranjit K. Samra1,2,3
Simranjit K. Samra1,2,3 Ashwini Rajasekaran2,3
Ashwini Rajasekaran2,3 Andrew J. Sandford1,2,4
Andrew J. Sandford1,2,4 Anne K. Ellis5,6
Anne K. Ellis5,6 Scott J. Tebbutt1,2,3,4*
Scott J. Tebbutt1,2,3,4*Allergic rhinitis (AR) is characterized by an early-phase response (EPR), and in a subgroup of individuals, a late-phase response (LPR). We sought to investigate polymorphisms in cholinergic synapse pathway genes, previously associated with late-asthmatic responses, in the LPR. Twenty healthy participants and 74 participants with AR underwent allergen exposure using the Environmental Exposure Unit. Allergic participants were sub-phenotyped using self-reported nasal congestion scores; congestion is the predominant symptom experienced during the LPR. Acute congestion (AC, n = 36) participants developed only an EPR, while persistent congestion (PC, n = 38) participants developed both allergic responses. We interrogated blood samples collected before allergen exposure with genotyping and gene expression assays. Twenty-five SNPs located in ADCY3, AKT3, CACNA1S, CHRM3, CHRNB2, GNG4, and KCNQ4 had significantly different allele frequencies (P < 0.10) between PC and AC participants. PC participants had increased minor allele content (P = 0.009) in the 25 SNPs compared to AC participants. Two SNPs in AKT3 were associated with gene expression differences (FDR < 0.01) in PC participants. This study identified an association between the LPR and polymorphisms in the cholinergic synapse pathway genes, and developed a novel method to sub-phenotype AR using self-reported nasal congestion scores.
Allergic rhinitis (AR) is the most prevalent clinical manifestation of allergy, affecting up to 40% of the global population (1). Significant economic and quality of life impacts are associated with AR including decreased productivity, cognitive function, and sleep (2, 3). AR is a heterogeneous disorder defined by IgE-mediated inflammation of the nasal mucosa. Allergic responses are initiated by environmental allergen exposure in genetically predisposed individuals, resulting in symptoms of rhinorrhea, sneezing, nasal congestion, and exacerbation of comorbid asthma (4). Children and adults with AR have an increased risk of developing allergic asthma (5, 6).
AR is characterized by an immediate early-phase response (EPR) and, in some individuals, a subsequent late-phase response (LPR). Allergen exposure in IgE-sensitized individuals causes degranulation of mast cells and basophils, inducing the EPR. The LPR occurs hours after allergen exposure and is caused by infiltration of inflammatory cells, including Th2 T lymphocytes, eosinophils, and basophils, into the nasal mucosa. Unlike the EPR, nasal congestion is the predominant symptom experienced during the LPR (7). Distinct immunological changes have been identified in the EPR (8, 9). However, there is limited understanding about the LPR, and how certain individuals may be protected from developing this additional allergic response. We have previously shown that polymorphisms in the cholinergic synapse pathway are associated with late asthmatic responses (10). Given the pathobiological overlap between allergic asthma and rhinitis, we hypothesized that this pathway might also be associated with AR sub-phenotypes.
In this investigation, we used the Environmental Exposure Unit (EEU) (11), a controlled allergen challenge facility, to study the development of the LPR. The EEU is a precise, replicable model of AR that offers the ability to study the development of allergic responses and symptoms, while removing confounding variables present in the natural environment. Using self-reported nasal congestion scores collected during EEU studies, allergic participants were sub-phenotyped either as acute congestion (AC) participants if they developed only an EPR, or as persistent congestion (PC) participants if they developed both early and late allergic responses. Genotyping and statistical analysis revealed an association between single nucleotide polymorphisms (SNP) in genes comprising the cholinergic synapse pathway and the LPR. This study also identified SNPs in AKT3, a cholinergic synapse pathway gene, which were associated with gene expression differences in PC participants.
The institutional review boards of the University of British Columbia and Queen's University approved this study, and all participants provided written informed consent. In this investigation, 74 allergic and 20 healthy, non-allergic participants underwent allergen exposure. Allergic participants had a 2-year documented history of allergic rhinoconjunctivitis symptoms. Additionally, allergic participants had a positive skin prick test to either birch (Betula pendula), or rye grass (Lolium perenne), or house dust mite (Dermatophagoides pteronyssinus and Dermatophagoides farinae) at screening (defined as a wheal diameter of 3 mm or greater than that produced by the negative control) (12). Healthy participants had a negative skin prick test to common environmental allergens. Participants were of Caucasian ethnicity and between the ages of 18–65 years. Participants were excluded if they had an upper respiratory tract infection within 1 week of allergen exposure, asthma requiring the use of a short-acting beta-agonist greater than twice a week, and if they were unable to adhere to medication washout periods before screening and allergen exposure. Participants were also excluded if they had a known history of positive test results for Hepatitis B, Hepatitis C, HIV or tuberculosis, a significant history of alcohol or drug abuse, or clinically relevant abnormalities on physical exam.
Using the EEU, participants were exposed to either birch pollen (3,500 grains/m3 for 4 h), or rye grass pollen (2,500–3,500 grains/m3 for 3 h), or house dust mite (2.07–6.66 ng/m3 of D. pteronyssinus and 2.67–3.80 ng/m3 of D. farina for 3 h). The air in the EEU was continuously circulated using fans, and allergen levels were measured every 30 min using impact-type particle samplers. The allergen emission rate was modified based on these counts to maintain target allergen concentrations. Participants self-reported nasal congestion scores (along with other AR symptoms) at baseline, every 30 min in the EEU, and every hour after leaving the EEU until 12 h had passed since allergen onset. AR symptoms were scored using a four-point scale (0–3). The scores were defined as the following: 0 = absence of symptoms, 1 = mild symptoms, 2 = moderate symptoms, and 3 = severe and intolerable symptoms.
Participants measured their Peak Nasal Inspiratory Flow (PNIF), an objective measure of nasal patency, at the same time intervals as AR symptoms using the In-Check meter (Clement Clark International Ltd, Essex, UK). Variation in raw PNIF scores was minimized by calculating the percent change in PNIF from baseline.
Whole peripheral blood samples were collected from participants before allergen exposure using PAXgene Blood RNA tubes (PreAnalytiX, Hombrechtikon, Switzerland). First, intracellular RNA was extracted from 5 mL of each PAXgene tube using the PAXgene Blood miRNA kit (PreAnalytiX). Then, from the remaining PAXgene tube solution (5 mL), DNA was extracted using a modified protocol. The solution was centrifuged, the resulting nucleic acid pellets were washed, resuspended, and incubated in buffers and proteinase K from the PAXgene Blood miRNA kit (PreAnalytiX). After protein digestion, the cell lysate was homogenized, and DNA was extracted using the PAXgene Blood DNA kit (PreAnalytiX).
DNA (500 ng) from allergic and healthy participants were interrogated using Axiom SNP arrays (Affymetrix, Santa Clara, USA). We used the Axiom Analysis Suite Software to perform quality control checks and export genotypes into PLINK format. Using PLINK, SNPs that were not in Hardy-Weinberg equilibrium and had minor allele frequency <10% were excluded from the dataset. Allele frequencies of 218 SNPs that were located <50,000 bp upstream of the transcription start site and downstream of the 3′ untranslated region of ADCY3, AKT3, CACNA1S, CHRM3, CHRNB2, GNB1, GNG4, and KCNQ4 were analyzed using logistic regression. Sex was controlled for in this analysis and we considered a nominal P value of <0.10 to be significant. Next, minor allele content (MAC) of PC, AC, and healthy participants was calculated (total number of minor alleles divided by the total number of SNPs analyzed), and the mean MAC values of each subgroup were compared using Mann–Whitney U-tests. Linkage disequilibrium was calculated using the LDheatmap R-library.
Peripheral blood RNA samples (100 ng) from allergic and healthy participants were interrogated using a custom NanoString nCounter Elements assay (NanoString, Seattle, USA). The assay measures the relative expression of 166 genes and has previously been used to develop blood-based biomarker panels to predict late-asthmatic responses. Reporter and Capture probes of the 166 target genes were mixed with each RNA sample for hybridization at 67°C for 18 h. The hybridized samples were processed using the nCounter Prep Station with the High Sensitivity protocol and then quantified using the nCounter Digital Analyzer. Raw data was normalized using positive controls and housekeeping genes. A cis-expression quantitative trait loci (cis-eQTL) analysis was performed using the MatrixEQTL R-library, and we considered an FDR < 0.01 to be significant.
Our goal was to test if polymorphisms in the cholinergic synapse pathway were associated with underlying nasal congestion development. Participants recorded their nasal congestion and PNIF scores over a 12-h period following allergen onset. A strong correlation was observed between nasal congestion and percent PNIF change from baseline (Pearson correlation, R2 = 0.96).
Allergic participants were sub-phenotyped using nasal congestion scores (Figure 1A). AC participants developed only an EPR; they experienced a 50% decrease in congestion scores between hours 3–6 and returned to baseline by hour 12. PC participants developed both allergic responses; they did not experience a 50% decrease in nasal congestion scores by hour 6 compared to hour 3 and did not return to baseline by hour 12. Of the 74 allergic participants as part of this investigation, 38 participants had PC and 36 participants had AC. During the first 5 h of allergen exposure, there were no significant differences among the sub-phenotypes (Wilcoxon Signed Rank test with Bonferroni corrections). Significant differences were seen at 6 h (P < 0.1), 7 and 8 h (P < 0.01), 9 h (P < 0.001), and from 10 to 12 h (P < 0.0001). Factors such as age, sex, height, weight, and complete blood cell counts did not differ between PC, AC, and healthy participants (Table 1).
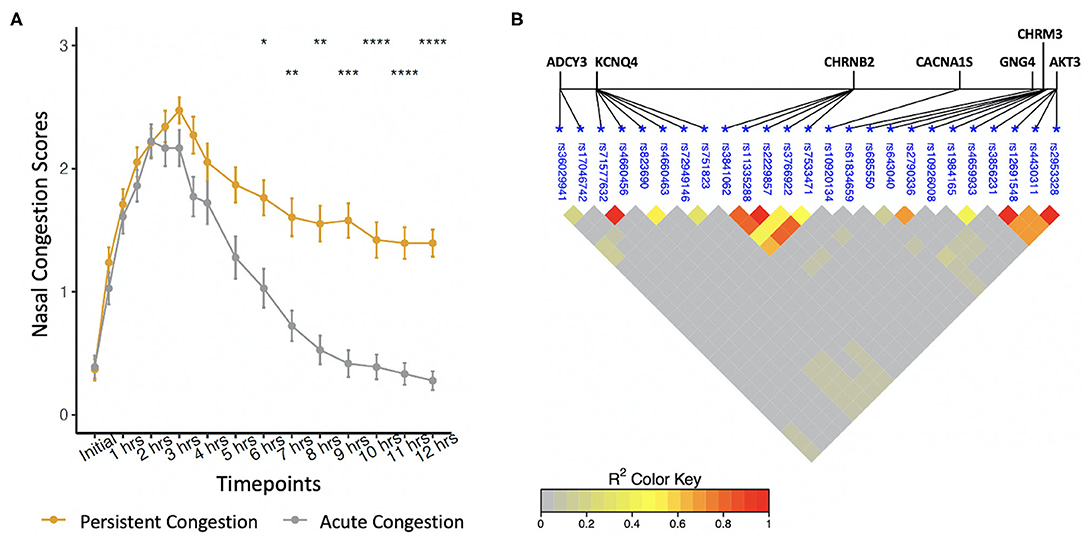
Figure 1. (A) Phenotyping of self-reported nasal congestion scores over a 12-h period from the start of allergen exposure. Significant differences were found between the sub-phenotypes at hours 6–12. *P < 0.1, **P < 0.01, ***P < 0.001, ****P < 0.0001, Wilcoxon Signed Rank test with Bonferroni corrections. (B) Linkage disequilibrium between polymorphisms associated with the late-phase response.
Allele frequencies of 25 SNPs were significantly different between PC and AC sub-phenotypes (Table 2). Identified SNPs were in seven genes comprising the cholinergic synapse pathway: ADCY3, AKT3, CACNA1S, CHRM3, CHRNB2, GNG4, and KCNQ4. Several of the SNPs were in linkage disequilibrium (Figure 1B). The average MAC of the PC sub-phenotype (MAC = 0.71) was significantly higher than that of the AC sub-phenotype (MAC = 0.61, P = 0.009). The 25 SNPs were not significantly different between healthy participants and either PC or AC participants.
We were able to measure the relative RNA abundance (gene expression) for one of the seven cholinergic synapse pathway genes, AKT3. Thus, we performed a cis-eQTL analysis to identify genetic variants in AKT3 associated with gene expression differences in PC, AC, and healthy participants. Using an additive and dominant genetic model, we identified two cis-eQTLs associated with the PC sub-phenotype (Figure 2A). The cis-eQTLs were in moderate linkage disequilibrium (r = 0.6). PC participants who were heterozygous for rs10927033 and rs320320 had significantly increased AKT3 expression (Figure 2B).
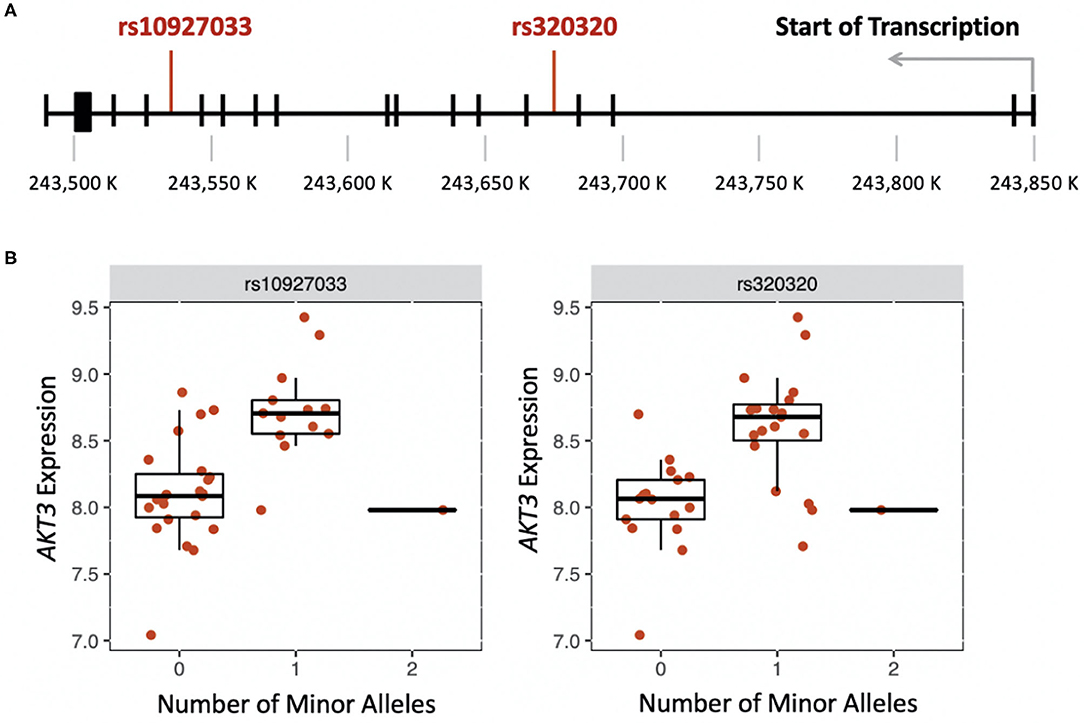
Figure 2. (A) Location of genetic variants that influence expression of AKT3 in the late-phase response (FDR < 0.01). (B) Boxplots showing the effect of the number of minor alleles on AKT3 expression for each genetic variant.
Stimulation of parasympathetic cholinergic nerves results in tissue remodeling, reduced airflow, and hypersecretion. In this investigation, we were interested in investigating the cholinergic synapse pathway in AR. We studied nasal congestion during and after allergen exposure in a controlled setting, using the Environmental Exposure Unit (EEU), and developed a protocol to sub-phenotype AR. We identified an association between the late-phase response (LPR) and polymorphisms in cholinergic synapse pathway genes (ADCY3, AKT3, CACNA1S, CHRM3, CHRNB2, GNG4, and KCNQ4). Persistent congestion (PC) participants who developed the LPR had a significantly increased minor allele content (MAC) than acute congestion (AC) participants who only developed the early-phase response (EPR).
Acetylcholine mediates cholinergic synapse transmission and is the predominant parasympathetic neurotransmitter in the airways. In the nose, acetylcholine causes nasal discharge and congestion (13). Increased activity of choline acetyltransferase, an enzyme that catalyzes acetylcholine biosynthesis, is associated with AR (14). This increased activity suggests that elevated neurotransmitter levels are present in individuals with AR, contributing to the development of rhinorrhea and nasal congestion symptoms after allergen exposure. CHRM3 encodes a muscarinic receptor (M3) and CHRNB2 encodes a subunit of nicotinic receptors, and both receptors bind acetylcholine. Muscarinic receptors are a class of G-protein-coupled receptor subtypes; M3 is the dominant muscarinic receptor in the nose and mediates secretion and vasodilation (14). Nicotinic receptors mediate the influx of cations using voltage-gated calcium (CACNA1S), potassium (KCNQ4), and sodium channels.
ADCY3, AKT3, and GNG4 encode genes involved in G-protein coupled receptor signaling and mediate multiple signaling pathways. The AKT3 gene is also involved in platelet activation (15), essential for leukocyte recruitment during allergen-induced inflammatory responses (16, 17). In addition to cytokines and chemokines, platelets sustain nasal inflammation by inducing an influx of inflammatory cells toward the nasal mucosa, a characteristic of the LPR (18). In AR, eosinophils adhere to cholinergic nerves, leading to their activation and degranulation. This interaction induces the expression of several cholinergic genes, such as choline acetyltransferase (19), resulting in elevated levels of acetylcholine and increased nasal symptoms. We also identified genetic variants in AKT3 associated with gene expression differences in the blood of PC participants. This association was not observed in AC and healthy participants. To our knowledge, these cis-eQTLs have previously not been associated with AR or other conditions.
This investigation has several limitations. Although participants' scoring of nasal congestion strongly correlated with percent PNIF change from baseline, nasal congestion scores are a subjective measure. Despite this limitation, allergic participants were sub-phenotyped using self-reported nasal congestion scores, in accordance with the FDA guidance on clinical outcomes for AR studies (20). Additionally, previous studies have also used self-reported symptom scores to stratify AR participants (21, 22). Another limitation was combining allergic and healthy participants from several different EEU studies, which used either seasonal or perennial allergens. However, for each study, allergen concentrations and exposure times were previously determined and proven to generate a mean peak of six for total nasal symptom scores (TNSS; a composite score of sneezing, rhinorrhea, and nasal congestion and itching) (23–25). Participants achieving a TNSS of 6 is a standard for controlled allergen challenge facility studies (26). Future studies should investigate the cholinergic synapse pathway separately in seasonal and perennial AR. This study also had a relatively small sample size which limited our statistical power. We investigated the cholinergic synapse pathway in AR sub-phenotypes because it has been associated with the LPR in allergic asthma (10). Our results are consistent with the findings in asthma, and the sample sizes of these two studies are similar. A strength of this investigation was the inclusion of healthy individuals, which helped to ensure that the identified polymorphisms were specific to AR and the LPR.
We have shown for the first time that polymorphisms in cholinergic synapse pathway genes are associated with the LPR in AR. We have also demonstrated that nasal congestion scores are a useful measurement to sub-phenotype the LPR. Anticholinergic therapy in AR reduces the duration of rhinorrhea (27, 28), and our results suggest that similar treatments may also reduce the duration of nasal congestion.
The data analyzed in this study is subject to the following licenses/restrictions: Datasets relevant to the results presented in this paper will be made available through direct request to the corresponding author. Requests to access these datasets should be directed to Scott Tebbutt, c2NvdHQudGViYnV0dEBobGkudWJjLmNh.
The studies involving human participants were reviewed and approved by the University of British Columbia and Queen's University. The patients/participants provided their written informed consent to participate in this study.
SS, AR, AE, and ST designed the study. SS and AE performed experiments. SS, AS, and ST participated in the statistical analysis. SS wrote the first draft of the manuscript. All authors contributed to the final version of the manuscript.
This research was supported by funding from AllerGen NCE Inc. (Allery, Genes and Environment Network), British Columbia Lung Association, Mitacs and PROOF Centre of Excellence.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Brozek JL, Bousquet J, Baena-Cagnani CE, Bonini S, Canonica GW, Casale TB, et al. Allergic Rhinitis and its Impact on Asthma (ARIA) guidelines-−2016 revision. J Allergy Clin Immunol. (2017) 40:950–8. doi: 10.1016/j.jaci.2017.03.050
2. Thompson AK, Juniper E, Meltzer EO. Quality of life in patients with allergic rhinitis. Ann Allergy Asthma Immunol. (2000) 1:338–48. doi: 10.1016/S1081-1206(10)62543-4
3. Bachert C, Bousquet J, Canonica GW, Durham SR, Klimek L, Mullol J, et al. Levocetirizine improves quality of life and reduces costs in long-term management of persistent allergic rhinitis. J Allergy Clin Immunol. (2004) 114:838–44. doi: 10.1016/j.jaci.2004.05.070
4. Broide DH. Allergic rhinitis: pathophysiology. Allergy Asthma Proc. (2010) 31:370–4. doi: 10.2500/aap.2010.31.3388
5. Rochat MK, Illi S, Ege MJ, Lau S, Keil T, Wahn U, et al. Allergic rhinitis as a predictor for wheezing onset in school-aged children. J Allergy Clin Immunol. (2010) 126:1170–5. doi: 10.1016/j.jaci.2010.09.008
6. Burgess JA, Walters EH, Byrnes GB, Matheson MC, Jenkins MA, Wharton CL, et al. Childhood allergic rhinitis predicts asthma incidence and persistence to middle age: a longitudinal study. J Allergy Clin Immunol. (2007) 120:863–9. doi: 10.1016/j.jaci.2007.07.020
7. Wallace DV, Dykewicz MS, Bernstein DI, Blessing-Moore J, Cox L, Khan DA, et al. The diagnosis and management of rhinitis: an updated practice parameter. J Allergy Clin Immunol. (2008) 122:S1–84. doi: 10.1016/j.jaci.2008.06.003
8. Kim YW, Singh A, Shannon CP, Thiele J, Steacy LM, Ellis AK, et al. Investigating immune gene signatures in peripheral blood from subjects with allergic rhinitis undergoing nasal allergen challenge. J Immunol. (2017) 199:3395–405. doi: 10.4049/jimmunol.1700378
9. Kim YW, Tonti E, Hickey P, Ellis AK, Neighbour H, Larché M, et al. Immunological changes in peripheral blood following nasal allergen challenge in subjects with allergic rhinitis pre- and post-peptide immunotherapy: an open-label clinical study. Allergy. (2021) 76:1907–11. doi: 10.1111/all.14710
10. Rajasekaran A, He D, Yue A, Singh A, Shannon CP, FitzGerald JM, et al. Cholinergic synapse pathway gene polymorphisms associated with allergen-induced late asthmatic responses. ERJ Open Res. (2019) 5:107. doi: 10.1183/23120541.00107-2019
11. Ellis AK, North ML, Walker T, Steacy LM. Environmental exposure unit: a sensitive, specific, and reproducible methodology for allergen challenge. Ann Allergy Asthma Immunol. (2013) 111:323–8. doi: 10.1016/j.anai.2013.07.019
12. Bousquet J, Heinzerling L, Bachert C, Papadopoulos NG, Bousquet PJ, Burney PG, et al. Practical guide to skin prick tests in allergy to aeroallergens. Allergy. (2012) 67:18–24. doi: 10.1111/j.1398-9995.2012.02772.x
13. White MV. Muscarinic receptors in human airways. J Allergy Clin Immunol. (1995) 95:1065–8. doi: 10.1016/S0091-6749(95)70209-1
14. Kubo N, Minami T, Hori Y, Yamashita T, Kumazawa T. Enhanced parasympathetic nerve activities in experimentally-induced nasal hypersensitivity. Acta Oto-laryngologica. (1989) 108:14–20. doi: 10.3109/00016488909138628
15. O'Brien KA, Stojanovic-Terpo A, Hay N, Du X. An important role for Akt3 in platelet activation and thrombosis. Blood. (2011) 18:4215–23. doi: 10.1182/blood-2010-12-323204
16. Pitchford SC, Yano H, Lever R, Riffo-Vasquez Y, Ciferri S, Rose MJ, et al. Platelets are essential for leukocyte recruitment in allergic inflammation. J Allergy Clin Immunol. (2003) 112:109–18. doi: 10.1067/mai.2003.1514
17. Pan D, Amison RT, Riffo-Vasquez Y, Spina D, Cleary SJ, Wakelam MJ, et al. P-Rex and Vav Rac-GEFs in platelets control leukocyte recruitment to sites of inflammation. Blood. (2015) 125:1146–58. doi: 10.1182/blood-2014-07-591040
18. Galli SJ, Tsai M, Piliponsky AM. The development of allergic inflammation. Nature. (2008) 45:445–54. doi: 10.1038/nature07204
19. Durcan N, Costello RW, McLean WG, Blusztajn J, Madziar B, Fenech AG, et al. Eosinophil-mediated cholinergic nerve remodeling. Am J Respir Cell Mol Biol. (2006) 34:775–86. doi: 10.1165/rcmb.2005-0196OC
20. Food and Drug Administration (FDA). Allergic Rhinitis, Developing Drug Products for Treatment: Guidance for Industry. Silver Spring, MD: Office of Communications, Division of Drug Information, Center for Drug Evaluation and Research (2016).
21. Soliman M, Ellis AK. Phenotyping allergic rhinitis as early- or dual-phase responses using the environmental exposure unit. Ann Allergy Asthma Immunol. (2015) 114:344–5. doi: 10.1016/j.anai.2014.12.020
22. Jacobs RL, Ramirez DA, Rather CG, Andrews CP, Jupiter DC, Trujillo F, et al. Redness response phenotypes of allergic conjunctivitis in an allergen challenge chamber. Ann Allergy Asthma Immunol. (2017) 118:86–93. doi: 10.1016/j.anai.2016.10.023
23. Ellis AK, Soliman M, Steacy LM, Adams DE, Hobsbawn B, Walker TJB. Clinical validation of controlled exposure to birch pollen in the Environmental Exposure Unit (EEU). Allergy Asthma Clin Immunol. (2016) 12:53–61. doi: 10.1186/s13223-016-0156-7
24. Ellis AK, Steacy LM, Hobsbawn B, Conway CE, Walker TJ. Clinical validation of controlled grass pollen challenge in the Environmental Exposure Unit (EEU). Allergy Asthma Clin Immunol. (2015) 11:5–12. doi: 10.1186/s13223-015-0071-3
25. Hossenbaccus L, Linton S, Thiele J, Steacy L, Walker T, Malone C, et al. Clinical validation of controlled exposure to house dust mite in the environmental exposure unit (EEU). Allergy Asthma Clin Immunol. (2021) 17:34–48. doi: 10.1186/s13223-021-00536-3
26. Hohlfeld JM, Holland-Letz T, Larbig M, Lavae-Mokhtari M, Wierenga E, Kapsenberg M, et al. Diagnostic value of outcome measures following allergen exposure in an environmental challenge chamber compared with natural conditions. Clin Exp Allergy. (2010) 40:998–1006. doi: 10.1111/j.1365-2222.2010.03498.x
27. Finn AF Jr., Aaronson D, Korenblat P, Lumry W, Settipane G, Spector S, et al. Ipratropium bromide nasal spray 0.03% provides additional relief from rhinorrhea when combined with terfenadine in perennial rhinitis patients; a randomized, double-blind, active-controlled trial. Am J Rhinol. (1998) 12:441–9. doi: 10.2500/105065898780707919
Keywords: environmental exposure unit, genetics, nasal congestion, allergic rhinitis, inflammation, late-phase response
Citation: Samra SK, Rajasekaran A, Sandford AJ, Ellis AK and Tebbutt SJ (2021) Cholinergic Synapse Pathway Gene Polymorphisms Associated With Late-Phase Responses in Allergic Rhinitis. Front. Allergy 2:724328. doi: 10.3389/falgy.2021.724328
Received: 12 June 2021; Accepted: 26 July 2021;
Published: 16 August 2021.
Edited by:
Peter Valentin Tomazic, Medical University of Graz, AustriaReviewed by:
Aleksandar Peric, Military Medical Academy, SerbiaCopyright © 2021 Samra, Rajasekaran, Sandford, Ellis and Tebbutt. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Scott J. Tebbutt, c2NvdHQudGViYnV0dEBobGkudWJjLmNh
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.