
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
PROTOCOLS article
Front. Plant Sci. , 26 February 2019
Sec. Technical Advances in Plant Science
Volume 10 - 2019 | https://doi.org/10.3389/fpls.2019.00219
An improved cowpea transformation method utilizing Agrobacterium-mediated gene delivery to explants derived from the cotyledonary nodes of imbibed cowpea seed is described. The explants were regenerated following a sonication procedure and a stringent selection comprising alternating regimes of kanamycin and geneticin. The method was reproducible and led to the recovery of independent fertile transgenic plants in the greenhouse at a level of about one per cent of starting explants. A transgene encoding an insecticidal protein from Bacillus thuringiensis was used to demonstrate the efficacy of the system.
Cowpea is a drought-tolerant grain legume that plays an important role in the diets of many people in Africa and in other parts of the developing world (Murdock et al., 2008). The grain has a protein content of 25% and its leaves and crop residues are used as a vegetable and animal feed, respectively (Nielsen et al., 1997). Although cowpea is important in tropical regions, its low productivity is attributed to biotic and abiotic stresses (Boukar et al., 2016). The majority of cowpea losses are due to insect pests (Murdock et al., 2008). Insect pests have been managed by a variety of approaches including spraying with synthetic and natural products as well as other cultural practices (Ofuya, 1997; Balachandra et al., 2012; Tiroesele et al., 2015). Additionally, breeding for host plant resistance to control thrips, aphids and beetles has been successful, as resistance genes to these exist in the primary gene pool of cowpea (Singh et al., 2002; Hall et al., 2003). However, this is not feasible for a pest such as the Maruca pod borer (MPB) as genes conferring resistance to MPB are not present in cowpea although they have been detected in the genome of a wild Vigna species (Vigna vexillata), which is sexually incompatible with cowpea. In such circumstances modern biotechnology-based approaches, such as genetic engineering, complement the existing methods to control insect pests for cowpea as proposed by Machuka et al. (2000).
Incorporating useful genes into cowpea germplasm using genetic engineering depends on the availability of a reproducible genetic transformation system to generate transgenic plants (Popelka et al., 2006; Aragão and Campos, 2007; Citadin et al., 2013). In vitro plant regeneration techniques have been applied to grain legumes via direct shoot organogenesis (Le et al., 2002; Raveendar et al., 2009; Mekala et al., 2016) and indirect organogenesis comprising the establishment of callus cultures, somatic embryogenesis or embryogenic cell suspension cultures (Anand et al., 2000; Ramakrishnan et al., 2005; Aasim et al., 2010). In these studies, different organs or tissues have been used as the starting material including mature seeds (Raveendar et al., 2009; Das et al., 2016), cotyledonary nodal cuttings (Le et al., 2002; Popelka et al., 2006; Chaudhury et al., 2007) mature embryos (Penza et al., 1991; Kaur et al., 2016), plumules (Aasim et al., 2010) and shoot apices or tips (Mao et al., 2006; Mekala et al., 2016). Despite several reports over nearly three decades, the genetic engineering of cowpea is still challenging, consistent with the generally recalcitrant nature of legumes to in vitro manipulation (Somers et al., 2003; Bakshi and Sahoo, 2013). This is well illustrated by the many methods that have been trialed using cowpea regeneration via somatic embryogenesis and direct multiple shoot organogenesis (Garcia et al., 1987; Kononowicz et al., 1997; Brar et al., 1999; Popelka et al., 2006; Chaudhury et al., 2007; Raveendar et al., 2009; Behura et al., 2015). Garcia et al. (1987) were among the first to attempt transformation experiments in cowpeas using Agrobacterium. Although transgene expression in callus cultures was reported, no regenerated plants were obtained. More recently, Agrobacterium-mediated and biolistic transformation have been used successfully to introduce genes conferring traits of potential agronomic importance into cowpea. These traits include insect resistance, herbicide resistance and virus resistance (Popelka et al., 2006; Adesoye et al., 2008; Solleti et al., 2008; Higgins et al., 2012; Citadin et al., 2013; Cruz and Aragao, 2014; Bett et al., 2017). In some of these cowpea transformation protocols, genes encoding neomycin phosphotransferase II (NPT II) coupled with a gene of interest, were used to select cells and tissues that had taken up the transgenes. In these instances, an antibiotic (such as kanamycin or geneticin) resistance gene was used and putatively transformed tissues cultured in selective medium (Garcia et al., 1986; Solleti et al., 2008).
Here, we describe an improved step by step transformation and regeneration system for cowpea. Imbibed mature cowpea seeds were sonicated prior to incubation with Agrobacterium tumefaciens carrying a binary T-DNA vector. The explants were then regenerated under a selection regime involving the selectable marker nptII combined with alternating antibiotics on plant culture media. The procedures represent a major improvement on the previous protocol (Popelka et al., 2006).
(1) A. tumefaciens strain AGL1 in glycerol stock
(2) Ten sterile 250 mL flasks
(3) Orbital shaker
(4) Mannitol Glutamate Luria (MGL) liquid medium, pH 7 (Table 1)
(5) Spectinomycin (50 mg/L) (Sigma-Aldrich, St. Louis, MO, United States)
(6) Floor centrifuge
(7) Cowpea seed (IT86D-1010)
(8) Cowpea co-cultivation medium (CCM), pH 5.4 (Tables 2, 3)
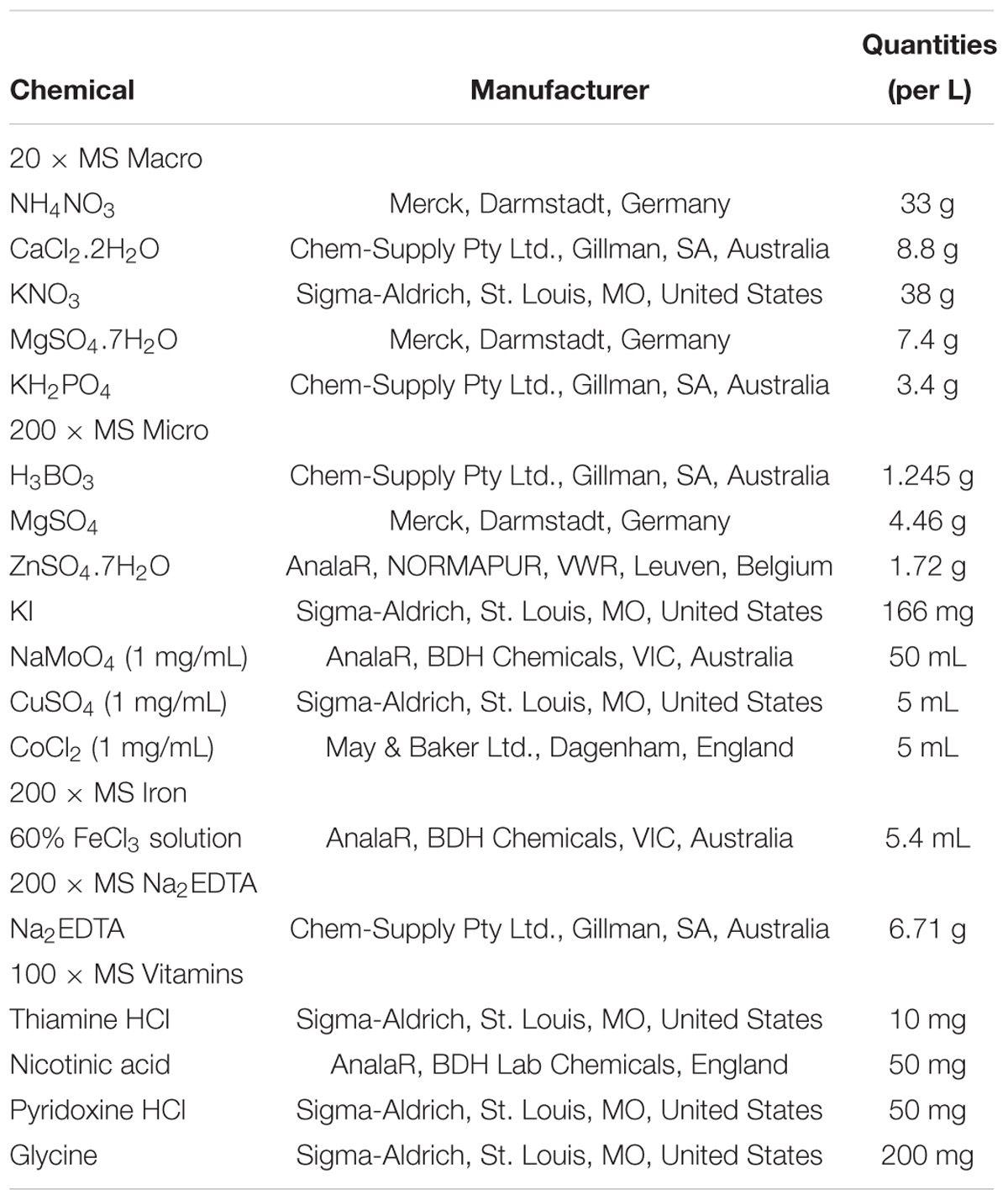
Table 2. Components of Murashige and Skoog (1962) Macro stock solutions.
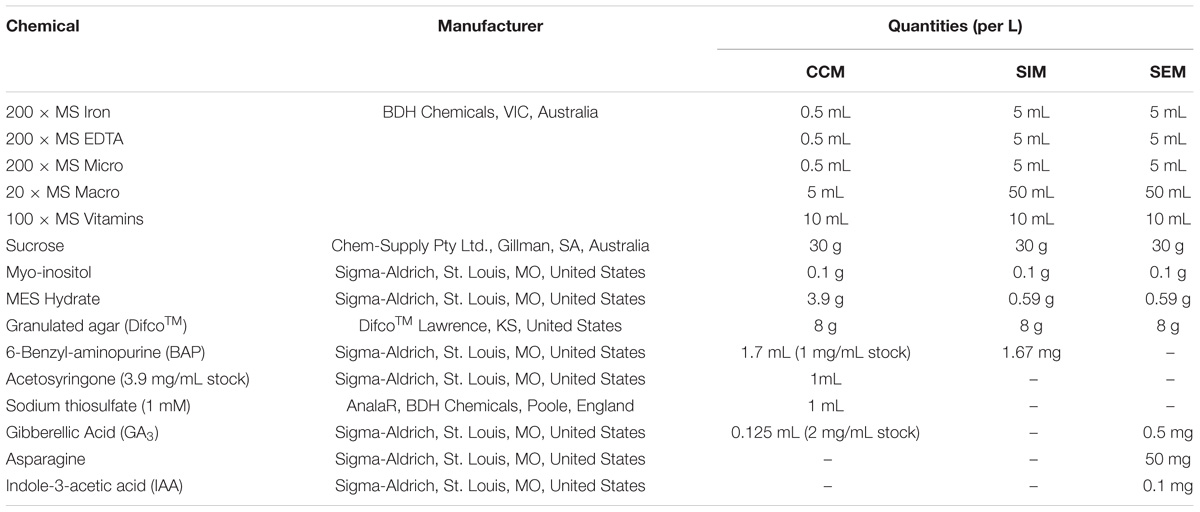
Table 3. Composition of cowpea co-cultivation medium (CCM), shoot induction medium (SIM) and shoot elongation and rooting medium (SEM).
(9) Weighing balance
(10) Absolute ethanol
(11) Commercial bleach
(12) Reverse-osmosis (RO) purified water
(13) Petri plates
(14) Aluminum foil
(15) Laminar flow chamber/hood, with sterilizing unit and beads
(16) Growth room chamber with controlled temperatures of 24°C and controlled cool lighting (16 h photoperiod) of 1000–1200 lux provided by white fluorescent bulbs.
(17) Scalpel blades, spatula and scalpel holder
(18) Sonicator (Bransonic Ultrasonic Cleaner, Model 1510E-DTH; output 70 W)
(19) Medical tape
(20) Permanent marker (for labeling cultures)
(21) Personal protective equipment (Latex hand gloves; face masks, safety goggles and labcoat)
(22) Measuring cylinders (500-, 1000- and 2000 mL) (Pyrex)
(23) Cylindrical glass beakers (500-, 1000- and 2000 mL) (Pyrex)
(24) Pipettes and pipette tips (10-, 50-, 100-, 500- and 1000 μL)
(25) Shoot induction medium (SIM), pH 5.6 (Table 3)
(26) Shoot elongation and rooting medium (SERM), pH 5.6 (Table 3)
(27) Kanamycin (Sigma-Aldrich, St. Louis, MO, United States)
(28) Spectinomycin (Sigma-Aldrich, St. Louis, MO, United States)
(29) Merrem (Ranbaxy Australia Pty, Sydney, NSW, Australia)
(30) Geneticin (G-418) (Sigma-Aldrich, St. Louis, MO, United States)
(31) Light sandy soil mix (25% perlite, 25% vermiculite, 30% coarse sand and 20% commercial potting mix (Bunnings, Canberra, ACT, Australia)
(32) Soil (70% commercial potting mix, 10% perlite, 10% river sand, 10% peat, 1.5 g/L Osmocote, 0.5 g/L Nitram and 3 g/L Carb lime)
(33) Pots (7 cm diameter × 9 cm height and 20 cm diameter × 20 cm height)
(34) Nucleic acid isolation kit (Puregene, MN, United States)
(35) Phusion® Hot Start Flex 2X master mix (Cat # M0536S, New England Biolabs)
(36) Vip3Ba forward and reverse primers: 5′GAACGCTCAGCTCAACTCCA3′ and 5′GGTGGAGTTGATGAGCACGT3′
(37) nptII forward and reverse primers: 5′GCTTGGGTGGAGAGGCTATT3′ and 5′TCATTTCGAACCCCAGAGTC3′
(38) PCR thermocycler (BioRad)
(39) SDS Gels (Thermo Fisher Scientific)
(40) Gel tanks and appropriate combs
(41) Gel electrophoresis equipment
(42) Agarose gel
(43) Trans-illuminator gel documentation system (GelDoc 2000 software, BioRad)
(44) Pestle and mortar
(45) Protein extraction buffer (0.1 M N-[Tris(hydroxymethyl) methyl]-2-aminoethanesulfonic acid (TES) (AnalaR, NORMAPUR, VWR, Leuven, Belgium) pH 7.6, 0.2 M NaCl, 1 mM phenylmethanesulfonyl fluoride (PMSF) (Sigma-Aldrich, St. Louis, MO, United States), 1 mM EDTA) in a 1.5 mL Eppendorf tube
(46) Escherichia coli-expressed Vip3Ba protein
(47) Vip3Ba-specific monoclonal antibody (AbMart Inc., Shanghai, China)
(1) An aliquot (400 μL) of a glycerol stock of the A. tumefaciens strain AGL1 (Lazo et al., 1991) containing a Vip3Ba gene construct (Bett et al., 2017) was added to 100 mL of MGL liquid medium, pH 7 (Table 1), in a sterile 250 mL flask, in the laminar hood.
(2) Spectinomycin (to 50 mg/L) was added to the Agrobacterium culture as the selective agent.
(3) Aluminum foil was used as a lid to cover the flasks containing the cultures.
(4) The culture was allowed to grow overnight in an orbital shaker at 28°C at 200 rpm and then centrifuged for 15 min at 7500 g at room temperature.
(5) The pellet was re-suspended in 100 mL of cowpea co-cultivation medium (CCM) (Tables 2, 3), pH 5.4 using an orbital shaker at 200 rpm for a minimum of 1 h at 28°C.
(6) Prior to inoculation of explants, an additional 100 mL of CCM was added to the Agrobacterium suspension to make a total of 200 mL.
The transformation method was a modification of the protocol described by Popelka et al. (2006) (Table 4). The sterilization procedures commenced 18 h prior to the co-cultivation step, to give sufficient time for the seeds to imbibe and soften for explant preparation.
(1) Dry cowpea seed was weighed (30 g) into a 250 mL Schott bottle.
(2) 50 mL of 70% ethanol was added for 1 min.
(3) The mixture was shaken vigorously for 30 s.
(4) The ethanol was poured off, replaced with 50 mL of 20% commercial bleach and incubated for 30 min at room temperature.
(5) The seeds were rinsed 5 times in sterile RO-purified water.
(6) Seeds were allowed to imbibe in 50 mL of sterile RO water overnight.
(7) Imbibed seeds were drained, and seed coats aseptically removed.
(8) Each seed was split in two by separating the cotyledons.
(9) Using the cotyledon with the attached embryonic axis (hereafter referred to as the explant), the lower 2/3 of the radicle was excised (Figure 1A).
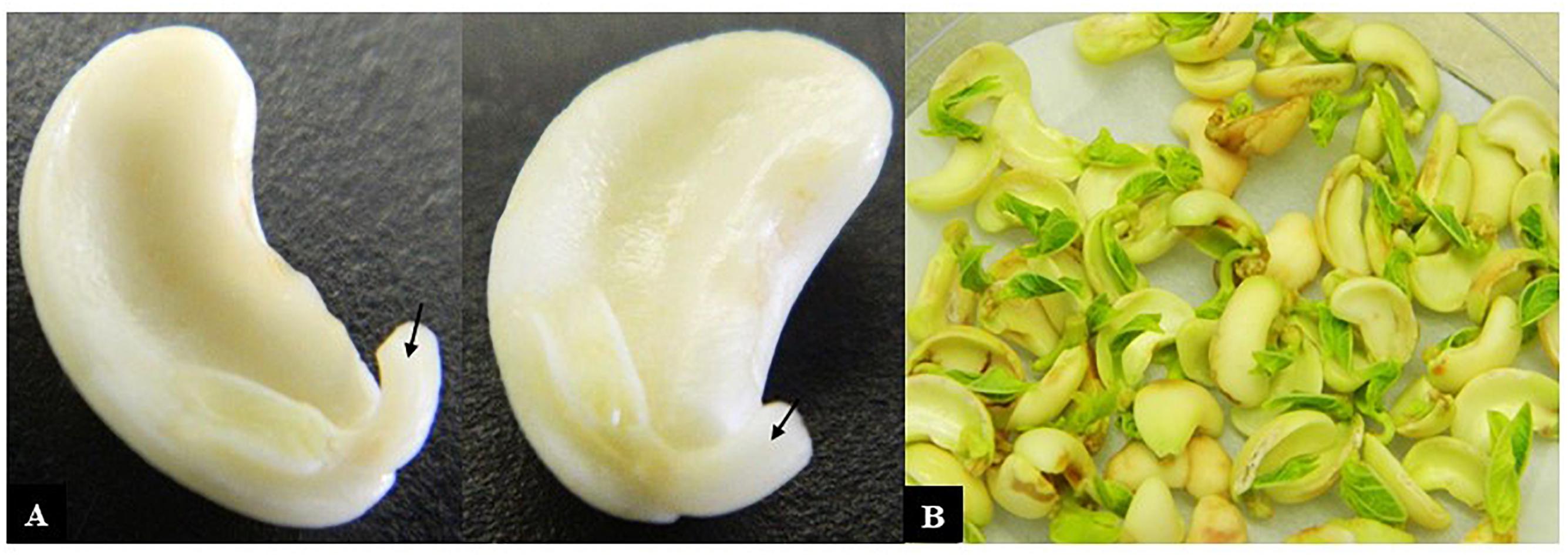
Figure 1. Cowpea explants used for transformation. (A) Cotyledons with attached axes that had their radicle tips removed (arrows) ready for incubation with Agrobacterium and (B) after co-cultivation with Agrobacterium for 3 days.
(10) Approximately 50 explants were placed in sterile 250 ml flasks containing 10 mL of CCM, to keep the explants hydrated until all 400 explants were prepared.
(11) To transform the explants, CCM was replaced with approximately 25 mL of the Agrobacterium suspension to submerge 50 explants.
(12) The explants were sonicated for 30 s using a Bransonic Ultrasonic Cleaner, Model 1510E-DTH; output 70 W.
(13) The sonicated explants were incubated for 1 h on a rotary shaker at 28°C and 200 rpm.
(14) The explants were placed onto sterile filter papers to blot the excess medium.
(15) Co-cultivation plates were prepared by pouring 30 mL of CCM media solidified with 8 g/L granulated agar and placing two new sterile filter papers on the media.
(16) 30 to 40 explants were transferred onto each filter paper plates and co-cultivated for 3 days at 28°C with a 16 h photoperiod.
(1) The explants were placed with the growing shoot facing down onto SIM, pH 5.6 (Table 3) containing 100 mg/L kanamycin for selection and 25 mg/L Merrem to prevent the growth of Agrobacterium.
(2) Explants were incubated in a growth chamber at 28°C with a 16 h photoperiod for 12 to 14 days.
(3) At the next transfer (day 17), the cotyledon, primary shoots and any regrown radicle and was removed and transferred the remaining portion of the explants to SIM with increased kanamycin (150 mg/L) and Merrem (25 mg/L).
(4) Explants were incubated in a growth chamber at 28°C with a 16 h photoperiod for 12 to 14 days.
(5) At the third transfer (day 29), any brown callus was removed. The remaining tissue supporting multiple small shoots was transferred to SIM with kanamycin (150 mg/L) and Merrem (25 mg/L).
(6) Explants were incubated in a growth chamber at 28°C with a 16 h photoperiod for 12 to 14 days.
(7) At the fourth transfer (day 45), multiple shoots were transferred to SIM with Merrem (25 mg/L) and more stringent selection using geneticin (30 mg/L).
(8) Explants were incubated in a growth chamber at 28°C with a 16 h photoperiod for 12 to 14 days.
(9) Two further transfers (each of 12–14 days) were made onto SIM with 30 mg/L geneticin. Large clumps consisting of many shoots were separated into smaller clumps or selected as single shoots depending on their vigor. Dead shoots were removed and discarded in these transfers.
(10) In the seventh transfer, multiple shoots were separated and incubated on SIM with 30 mg/L geneticin at 28°C with a 16 h photoperiod for 12 to 14 days.
(11) For the eighth transfer, single shoots were placed onto shoot elongation and rooting medium (SEM), pH 5.6 (Table 3) with geneticin (30 mg/L) and 25 mg/L Merrem for 14 days to allow elongation and rooting. Depending on their height, shoots were placed on either Petri plates or in 250 mL containers with lids and incubated in a growth chamber at 28°C with a 16 h photoperiod.
(12) Shoots developing healthy roots were selected, and solid SEM rinsed off their roots with lukewarm water. These were transferred into small pots (7 cm diameter × 9 cm height) containing a light sandy soil mix. Subsequently, these were maintained in the culture room for up to 4 weeks for acclimatization after which they were transferred to the glasshouse in larger pots (20 cm diameter × 20 cm height) containing soil.
(1) Genomic DNA was extracted from putatively transgenic cowpea plants using a nucleic acid isolation kit (Puregene, MN, 55447, United States).
(2) PCR mixes were prepared using 5 μL of Phusion® Hot Start Flex 2X master mix (Cat # M0536S, New England Biolabs), forward and reverse Vip3Ba or nptII primers (2 μL each) and 10 μL dH2O.
(3) Approximately 100 ng genomic DNA was added, and samples subjected to the following PCR cycling regime: 98°C for 30 s, 98°C for 10 s, 60°C for 20 s and 72°C for 90 s (30 cycles), followed by 72°C for 10 min and 25°C for 1 min.
(4) Amplicons were analyzed by electrophoresis through a 1.8% agarose gel at 60 mA and photographed using a trans-illuminator gel documentation system.
(1) T1 seeds were harvested from the T0 transgenic lines to obtain T1 and T2 plants for protein isolation.
(2) Non-transgenic line (IT86D-1010) was propagated as a negative control.
(3) Young fully expanded unifoliate leaves (7-days old) were harvested from the transgenic and non-transgenic lines.
(4) Total soluble protein (TSP) was extracted by macerating 80–110 mg of cowpea leaf (from individual plants) in 500 μL of protein extraction buffer.
(5) A 40 μg aliquot of TSP was subjected to SDS-PAGE, together with known levels (1, 3, 10, and 30 ng) of E. coli-expressed Vip3Ba protein.
(6) The SDS-PAGE gel was blotted to nitrocellulose membranes and assessed for the presence of Vip3Ba by western blot using a Vip3Ba-specific monoclonal antibody.
(7) The amount of Vip3Ba protein was estimated by visual comparison with the known concentrations of E. coli-expressed Vip3Ba protein previously included on the gels.
(8) Data was expressed as ng Vip3Ba per mg of TSP.
The following unforeseen circumstances may occur while carrying out transformation and regeneration experiments:
(i) Contamination of growth media on Petri plates and explant contamination. Apply sterilization techniques during media dispensation and explant sterilization procedures to overcome these. In addition, hormones and antibiotics should be filter-sterilized and added to the plant culture media after autoclaving and cooling to 50°C.
(ii) The Agrobacterium culture may not grow to an optimum Optical Density (OD) prior to inoculation of explants. To avoid this, inoculate the MGL medium with Agrobacterium no less than 24 h prior to the co-cultivation step and grow overnight at 28°C.
(iii) Cowpea seeds may be too hard to prepare explants, therefore commence sterilization procedures 18 h prior the co-cultivation step. This will give sufficient time for the seeds to imbibe for explant preparation.
(iv) The explants may dry up during preparation. Keep the explants hydrated in co-cultivation medium prior to the Agro-inoculation procedure.
The transformation procedures described above were repeated fourteen times. A total of 6696 explants were co-cultivated with Agrobacterium containing the vip3Ba transgene. After the 3-day co-cultivation period, primary shoots were observed developing from the embryonic axis (Figure 1B). The explants were then transferred to SIM with kanamycin for a maximum of 14 days. By this time, primary shoots had elongated (Figure 2A) and potentially transformed small shoot buds appeared at the cotyledonary node (Figure 2A), and callus had formed at the radicle end (Figure 2A). The cotyledons, primary shoots and radicle-associated callus were removed leaving a clump with the small shoot buds (Figure 2B) on a callus base. The remaining part of the explants was transferred to SIM with kanamycin for two transfers (on Days 17 and 29). All subsequent transfers were onto medium containing geneticin. The clumps of multiple shoot buds (Figure 2C) were divided and any dead tissue and surplus callus was removed (Figure 2D). By transfer 6 (Day 70), single shoots were separated onto fresh SEM (Figure 2E). Roots developed during two cycles on shoot elongation media (Figure 2F). The rooted plantlets were acclimatized in a tissue culture room prior to transfer to the glasshouse (Figure 2G). Following selection, 77 independent putative transgenic lines were obtained. All plants that regenerated from one explant were considered to be clones (siblings) belonging to one transgenic event. Using this transformation protocol, whole plantlets (approximately 7 to 10 cm in height) belonging to independent transgenic lines were obtained in approximately 4 months.

Figure 2. In vitro regeneration of cowpea explants following co-cultivation with Agrobacterium. (A) Cowpea explant with cotyledon and primary shoots (arrow 1) on shoot induction medium (SIM) with selection at 2 weeks after co-cultivation, regenerated small buds (arrow 2) and callus (arrow 3) are visible (B) explant with cotyledon and primary shoot removed at 4 weeks leaving a clump with shoot buds (arrows) (C) multiple shoots formed on callus (D) multiple shoots separated onto SIM with 30 mg/L geneticin at 8 weeks (E) individual shoots grown with 30 mg/L geneticin at 10 weeks, (F) individual shoots rooting on elongation and rooting medium at 14 weeks and (G) rooted plantlets in soil at 16 weeks.
The successful transformation of many plant species, including cowpea, is dependent on the genotype, the type of explant used, the antibiotic or herbicide selection regime and plant growth media composition (Manman et al., 2013). In this study, significant modifications were made to the previous cowpea transformation protocol (Popelka et al., 2006) to improve its efficiency. These technical changes resulted in a cowpea transformation system with an efficiency of just over 1%, with 73 independent transgenic lines obtained. This rate is similar to, or better than some other cowpea transformation studies in which efficiencies ranged from 0.15 to 3.9% (Popelka et al., 2006; Chaudhury et al., 2007; Ivo et al., 2008; Solleti et al., 2008; Adesoye et al., 2010; Raveendar and Ignacimuthu, 2010; Behura et al., 2015). The sonication of legume cotyledonary nodes has been shown to increase the transformation efficiency. Using sonication-assisted Agrobacterium-mediated transformation in soybean, Trick and Finer (1998) generated hygromycin-resistant clones and reported enhanced transformation compared to very low transient expression of GUS gene when sonication procedures were not employed. Further, a 100% increase in GUS expression frequency was also achieved following sonication treatment from 5 to 25 s in the transformation of winter cherry (Sivanandhan et al., 2015). In the current study the co-cultivated cowpea explants were sonicated and also subjected to 100 mg/L kanamycin for early stage selection, and then kanamycin at 150 mg/L and geneticin at 30 mg/L for the remaining transfers thereby obtaining transgenic plants in soil within approximately 4 months.
Genomic DNA was extracted from the 77 putative transgenic cowpeas and used as a template in PCR with primers designed to amplify a 187 bp fragment of the vip3Ba gene and a 970 bp region of the nptII gene (Figure 3). Of the 77 plants, 73 were positive for both the vip3Ba and nptII genes (Table 5), resulting in an average transformation frequency of 1.1%. All 73 T0 plants were fertile and seed from 42 lines was selected for further analysis.
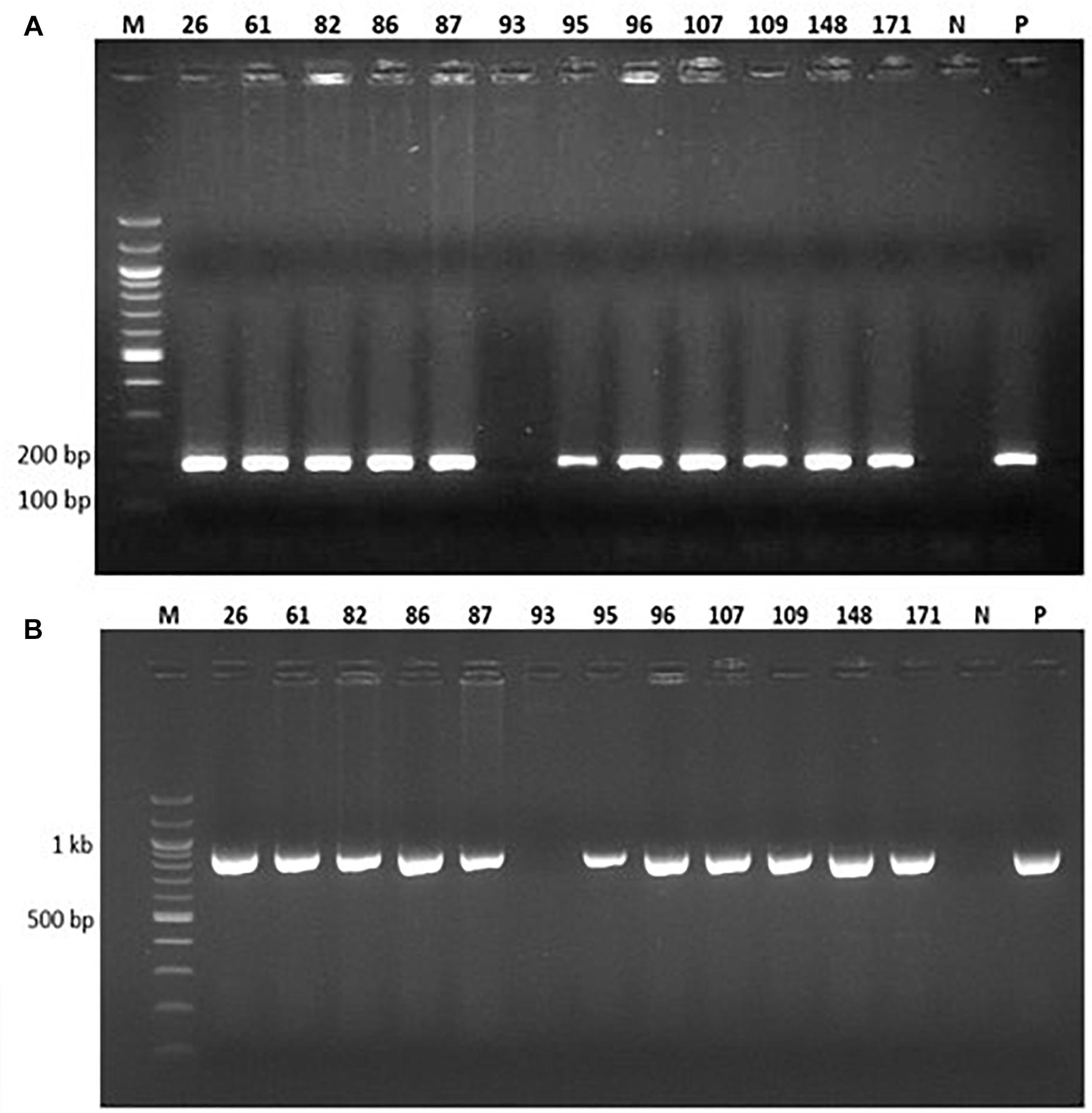
Figure 3. PCR analysis of 12 representative putatively transgenic cowpea lines using (A) vip3Ba-specific primers designed to amplify a 187 bp product and (B) nptII-specific primers designed to amplify a 970 bp product. Lane M: DNA Molecular weight marker; Lane N: No template (negative control); Lane P: Plasmid DNA containing both vip3Ba and nptII genes (positive control); numbers above lanes represent the line number of randomly selected, independent transgenic cowpea plants.
Of 42 independent T0 lines analyzed by western blotting, a band of the expected size for Vip3Ba (∼75 kDa) was detected in extracts from 9 lines (Table 5) namely V9, V24, V25, V43, V56, V87, V107, V176 and V191. Based on comparison to the standards included in the gels, the amount of Vip3Ba present in seven (V9, V24, V25, V43, V56, V87, V107) of these lines ranged between 0.25 and 5.0 μg/mg TSP (Bett et al., 2017). A representative blot of total soluble leaf protein extracts from seven T1 progeny derived from one line (V43) is presented in Figure 4. Subsequently, four lines (V24-9, V25-8, V43-3, V87-2) of the T1 progeny produced T2 seed and further produced T2 plants. Following a Western blot analysis of at least five T2 plants from each of the four T1 lines, a band of the expected size for Vip3Ba (∼75 kDa) was detected in extracts from these progenies indicating expression in the segregating progeny of the four lines (data not shown).
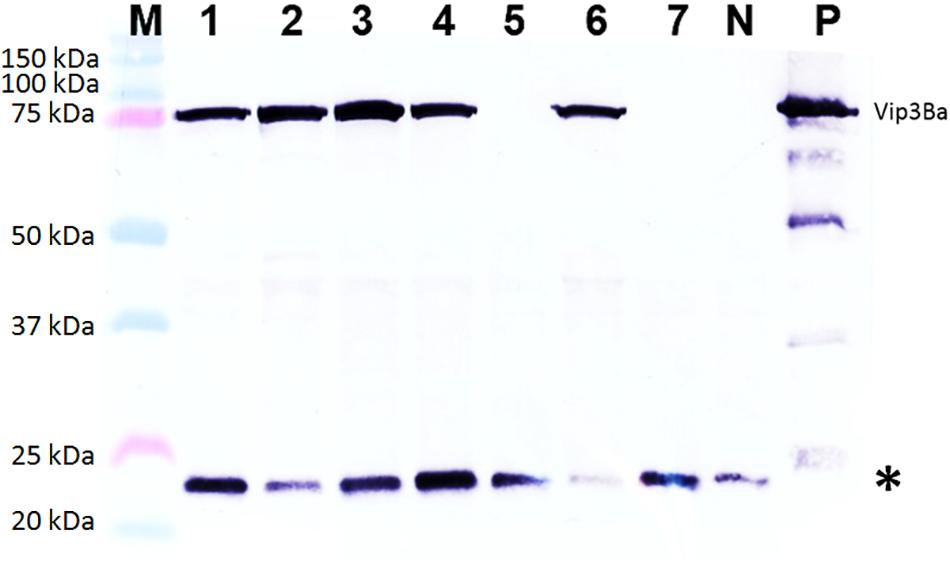
Figure 4. Western blot analysis of Vip3Ba expression in seven T1 progeny derived from transgenic cowpea line 43. Lane M: Protein precision markers; Lanes 1 to 7: Protein (40 μg) extracts from leaves of seven T1 plants of line V43; Lane N: protein from line IT86D-1010 (negative control); Lane P: 100 ng of Vip3Ba protein from Escherichia coli expressing Vip3Ba (positive control). Vip3Ba band migrates at ∼75 kDa. The 22 kDa band (∗) in all lanes including the negative control represents an unknown protein that cross-reacts with the monoclonal antibody.
There were varying levels of Vip3Ba protein amongst the independent transgenic cowpea lines (data not shown). Variable transgene expression is a common phenomenon in genetically modified plants and levels can vary widely between lines (Matzke and Matzke, 1998; Schuler et al., 1998). This variation is generally considered to be the result of (i) the position effect i.e., where in the genome the transgene cassette has integrated, and (ii) complex integration events which can trigger post-transcriptional gene silencing (PTGS) and/or transcriptional gene silencing (TGS) (Stam et al., 1997; Matzke and Matzke, 1998; Schuler et al., 1998). The latter of these phenomena are of particular importance as both PTGS and TGS can result in the complete silencing of transgene expression.
The levels of Vip3Ba measured in this study were within the range of those observed in cowpea lines expressing Cry 1Ab, but higher than those reported in chickpea expressing Cry 1Ac (Kar et al., 1997; Sanyal et al., 2005; Indurker et al., 2010; Mehrotra et al., 2011; Ganguly et al., 2014).
This improved protocol for cowpea transformation yielded a higher transformation efficiency than that described in Popelka et al. (2006), Chaudhury et al. (2007) and Ivo et al. (2008). Chaudhury et al. (2007) employed kanamycin (85 mg/L) for shoot regeneration and subsequently obtained shoots rooted on media containing kanamycin at a reduced concentration of 10 mg/L, resulting in inheritance of transgenes to progeny in Mendelian fashion at a rate of 0.76%. The system presented here is a modification of existing protocols (Popelka et al., 2006; Citadin et al., 2011), which included using a growth medium in the absence of certain reducing agents (L-cysteine and sodium thiosulfate at co-cultivation and shoot induction stages, respectively), a different antibiotic selection regime, and employing sonication to permeate the plant cell wall thereby facilitating Agrobacterium-mediated T-DNA transfer into the host plant cell. Higgins et al. (2012) employed a kanamycin/geneticin regime at 150 and 25–50 mg/L, respectively, for shoot initiation in cowpea. The surviving green shoots were further subjected to 50 mg/L geneticin or 150 mg/L kanamycin for shoot elongation and 1–3 transgenic plants per 1000 explants were obtained, which is equivalent to 0.1–0.3% transformation efficiency (Higgins et al., 2012). In contrast, Bakshi et al. (2011) used sonication and vacuum infiltration assisted Agrobacterium-mediated transformation for cowpea, which substantially increased the transformation efficiency.
With this protocol, rooted plants were recovered within 4 months of explant preparation. This duration is within the range of other researchers, who reported varied timelines in recovery of rooted plants between 1 and 8 months (Chaudhury et al., 2007; Raveendar and Ignacimuthu, 2010; Bakshi et al., 2011; Higgins et al., 2012).
The improved result in this study is attributed to wounding of the cotyledonary explants by sonication, using alternating antibiotics for selection and the use of growth media without certain reducing agents previously incorporated by Popelka et al. (2006).
TH and RH conceived and designed the study. BB and SG performed the transformation experiments. BB and AM performed the molecular characterization experiments. BB, RH, and TH analyzed and interpreted the results. BB, SG, RH, and TH prepared the manuscript. All authors read and approved the manuscript.
This research was supported by Commonwealth Scientific and Industrial Research Organisation (CSIRO) and the Australian Government (AusAID) through a scholarship to Queensland University of Technology (QUT).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors thank Lisa Molvig and Wendy Welfare for their technical advice and support. The authors acknowledge the original source of the work cited in the Bett (2016) doctoral thesis submitted to QUT for examination in 2016.
Aasim, M., Khawar, K. M., and Ozcan, S. (2010). Efficient in vitro propagation from preconditioned embryonic axes of Turkish cowpea (Vigna unguiculata L.) cultivar Akkiz. Arch. Biol. Sci. 62, 1047–1052. doi: 10.2298/ABS1004047A
Adesoye, A., Machuka, J., and Togun, A. (2008). CRY 1AB transgenic cowpea obtained by nodal electroporation. Afr. J. Biotechnol. 7, 3200–3210.
Adesoye, A. I., Togun, A. O., and Machuka, J. (2010). Transformation of cowpea (Vigna unguiculata L. Walp.) by Agrobacterium infiltration. J. Appl. Biosci. 30, 1845–1860.
Anand, R. P., Ganapathi, A., Anbazhagan, V. R., Vengadesan, G., and Selvaraj, N. (2000). High frequency plant regeneration via somatic embryogenesis in cell suspension cultures of cowpea, Vigna unguiculata (L.) Walp. In Vitro Cell. Dev. Biol. Plant 36, 475–480. doi: 10.1007/s11627-000-0085-4
Aragão, F. J. L., and Campos, F. A. P. (2007). “Common bean and cowpea,” in Transgenic Crops IV. Biotechnology in Agriculture and Forestry, Vol. 59, eds E. C. Pua and M. R. Davey (Berlin: Springer).
Bakshi, S., Sadhukhan, A., Mishra, S., and Sahoo, L. (2011). Improved Agrobacterium-mediated transformation of cowpea via sonication and vacuum infiltration. Plant Cell Rep. 30, 2281–2292. doi: 10.1007/s00299-011-1133-8
Bakshi, S., and Sahoo, L. (2013). How relevant is recalcitrance for the recovery of transgenic cowpea: implications of selection strategies. J. Plant Growth Regul. 32, 148–158. doi: 10.1007/s00344-012-9284-6
Balachandra, B. A. H. E., Pathiratha, P. U., and Paranagama, P. A. (2012). Control of stored grain pest, Callosobruchus maculatus (F.) (Coleoptera: Bruchidae) using the essential oil isolated from Plectranthus zeylanicus. Nat. Prod. Res. 26, 2219–2222. doi: 10.1080/14786419.2011.643883
Behura, R., Kumar, S., Saha, B., Panda, M. K., Dey, M., Sadhukhan, A., et al. (2015). Cowpea [Vigna unguiculata (L.) Walp.]. Methods Mol. Biol. 1223, 255–264. doi: 10.1007/978-1-4939-1695-5_20
Bett, B., Gollasch, S., Moore, A., James, W., Armstrong, J., Walsh, T., et al. (2017). Transgenic cowpeas (Vigna unguiculata L. Walp) expressing Bacillus thuringiensis Vip3Ba protein are protected against the Maruca pod borer (Maruca vitrata). Plant Cell Tissue Organ Cult. 131, 335–345. doi: 10.1007/s11240-017-1287-3
Bett, B. B. (2016). Enhancing the Resilience of Bt Cowpea [Vigna unguiculata (l.) Walp] for Insect Resistance Management. Doctoral dissertation, Queensland University of Technology, Brisbane.
Boukar, O., Fatokun, C. A., Huynh, B.-L., Roberts, P. A., and Close, T. J. (2016). Genomic tools in cowpea breeding programs: status and perspectives. Front. Plant Sci. 7:757. doi: 10.3389/fpls.2016.00757
Brar, M. S., Al-Khayri, J. M., Morelock, T. E., and Anderson, E. J. (1999). Genotypic response of cowpea Vigna unguiculata (L.) to in vitro regeneration from cotyledon explants. In Vitro Cell. Dev. Biol. Plant 35, 8–12. doi: 10.1007/s11627-999-0002-4
Chaudhury, D., Madanpotra, S., Jaiwal, R., Saini, R., Kumar, P. A., and Jaiwal, P. K. (2007). Agrobacterium tumefaciens-mediated high frequency genetic transformation of an Indian cowpea (Vigna unguiculata L. Walp) cultivar and transmission of transgenes into progeny. Plant Sci. 172, 692–700. doi: 10.1016/j.plantsci.2006.11.009
Citadin, C. T., Cruz, A. R. R., and Aragao, F. J. L. (2013). Development of transgenic imazapyr-tolerant cowpea (Vigna unguiculata). Plant Cell Rep. 32, 537–543. doi: 10.1007/s00299-013-1385-6
Citadin, C. T., Ibrahim, A. B., and Aragao, F. J. L. (2011). Genetic engineering in cowpea (Vigna unguiculata): history, status and prospects. GM Crops 2, 144–149. doi: 10.4161/gmcr.2.3.18069
Cruz, A. R. R., and Aragao, F. J. L. (2014). RNAi-based enhanced resistance to cowpea severe mosaic virus and cowpea aphid-borne mosaic virus in transgenic cowpea. Plant Pathol. 63, 831–837. doi: 10.1111/ppa.12178
Das, A., Datta, S., Sujayanand, G. K., Kumar, M., Singh, A. K., Arpan, et al. (2016). Expression of chimeric Bt gene, Cry1Aabc in transgenic pigeonpea (cv. Asha) confers resistance to gram pod borer (Helicoverpa armigera Hubner.). Plant Cell Tissue Organ Cult. 127, 705–715. doi: 10.1007/s11240-016-1131-1
Ganguly, M., Molla, K. A., Karmakar, S., Datta, K., and Datta, S. K. (2014). Development of pod borer-resistant transgenic chickpea using a pod-specific and a constitutive promoter-driven fused cry1Ab/Ac gene. Theor. Appl. Genet. 127, 2555–2565. doi: 10.1007/s00122-014-2397-5
Garcia, J. A., Hille, J., and Goldbach, R. (1986). Transformation of cowpea Vigna unguiculata cells with an antibiotic resistance gene using a Ti-plasmid-derived vector. Plant Sci. 44, 37–46. doi: 10.1016/0168-9452(86)90166-4
Garcia, J. A., Hille, J., Vos, P., and Goldback, R. (1987). Transformation of cowpea Vigna unguiculata with a full length DNA copy of cowpea mosaic virus m-RNA. Plant Sci. 48, 89–98. doi: 10.1016/0168-9452(87)90135-X
Hall, A. E., Cisse, N., Thiaw, S., Elawad, H. O. A., Ehlers, J. D., Ismail, A. M., et al. (2003). Development of cowpea cultivars and germplasm by the Bean/Cowpea CRSP. Field Crop Res. 82, 103–134. doi: 10.1016/S0378-4290(03)00033-9
Higgins, T. J. V., Gollasch, S., Molvig, L., Moore, A., Popelka, C., Watkins, P., et al. (2012). “Insect-protected cowpeas using gene technology,” in Innovative Research Along the Cowpea Value Chain. Proceedings of the Fifth World Cowpea Conference on Improving Livelihoods in the Cowpea Value Chain Through Advancement in Science, eds O. Boukar, O. Coulibaly, C. A. Fatokun, K. Lopez, and M. Tamò (Ibadan: International Institute of Tropical Agriculture), 131–137.
Indurker, S., Misra, H. S., and Eapen, S. (2010). Agrobacterium-mediated transformation in chickpea (Cicer arietinum L.) with an insecticidal protein gene: optimisation of different factors. Physiol. Mol. Biol. Plants 16, 273–284. doi: 10.1007/s12298-010-0030-x
Ivo, N. L., Naschimento, C. P., Vieira, L. S., Campos, F. A. P., and Aragao, F. J. L. (2008). Biolistic-mediated genetic transformation of cowpea (Vigna unguiculata) and stable mendelian inheritance of transgenes. Plant Cell Rep. 27, 1475–1483. doi: 10.1007/s00299-008-0573-2
Kar, S., Basu, D., Das, S., Ramkrishnan, N. A., Mukherjee, P., Nayak, P., et al. (1997). Expression of cry1Ac gene of Bacillus thuringiensis in transgenic chickpea plants inhibits development of pod-borer (Heliothis armigera) larvae. Transgenic Res. 6, 177–185. doi: 10.3389/fpls.2017.01423
Kaur, A., Sharma, M., Sharma, C., Kaur, H., Kaur, N., Sharma, S., et al. (2016). Pod borer resistant transgenic pigeon pea (Cajanus cajan L.) expressing cry1Ac transgene generated through simplified Agrobacterium transformation of pricked embryo axes. Plant Cell Tissue Organ Cult. 127, 717–727. doi: 10.1007/s11240-016-1055-9
Kononowicz, A. K., Cheah, K. T., Narasimhan, M. L., Murdock, L. L., Shade, R. E., Chrispeels, M. J., et al. (1997). “Developing a transformation system for cowpea (Vigna unguiculata [L.] Walp),” in Advances in Cowpea Research, eds B. B. Singh, D. R. Mohan Raj, K. E. Dashiell, and L. E. N. Jackai (Ibadan: IITA), 361–371.
Lazo, G. R., Stein, P. A., and Ludwig, R. A. (1991). A DNA transformation-competent Arabidopsis genomic library in Agrobacterium. Biotechnol. 9, 963–967. doi: 10.1038/nbt1091-963
Le, B.-V., Cruz de Carvalho, M. H., Zuily-Fodil, Y., Thi, A. T. P., and Van, K. T. T. (2002). Direct whole plant regeneration of cowpea [Vigna unguiculata (L.) Walp] from cotyledonary node thin cell layer explants. J. Plant Physiol. 159, 1255–1258. doi: 10.1078/0176-1617-00789
Machuka, J., Adesoye, A., and Obembe, O. O. (2000). “Regeneration and genetic transformation in cowpea,” in Challenges and Opportunities for Enhancing Sustainable Cowpea Production. Proceedings of the World Cowpea Conference III, eds C. A. Fatokun, S. A. Tarawali, B. B. Singh, P. M. Kormawa, and M. Tamo (Ibadan: IITA), 185–196.
Manman, T., Qian, L., Huaqiang, T., Yongpeng, Z., Jia, L., and Huanxiu, L. (2013). A review of regeneration and genetic transformation in cowpea (Vigna unguiculata L. Walp). Afr. J. Agric. Res. 8, 1115–1122. doi: 10.5897/AJAR12.2059
Mao, J. Q., Zaidi, M. A., Arnason, J. T., and Altosaar, I. (2006). In vitro regeneration of Vigna unguiculata (L.) Walp. Cv. Blackeye cowpea via shoot organogenesis. Plant Cell Tissue Organ Cult. 87, 121–125. doi: 10.1007/s11240-006-9145-8
Matzke, A. J. M., and Matzke, M. A. (1998). Position effects and epigenetic silencing of plant transgenes. Curr. Opin. Plant Biol. 1, 142–148. doi: 10.1016/S1369-5266(98)80016-2
Mehrotra, M., Singh, A. K., Sanyal, I., Altosaar, I., and Amla, D. V. (2011). Pyramiding of modified cry1Ab and cry1Ac genes of Bacillus thuringiensis in transgenic chickpea (Cicer arietinum L.) for improved resistance to pod borer insect Helicoverpa armigera. Euphytica 182, 87–102. doi: 10.1007/s10681-011-0501-3
Mekala, G. K., Juturu, V. N., and Mallikarjuna, G. (2016). Optimization of Agrobacterium-mediated genetic transformation of shoot tip explants of green gram (Vigna radiata (L.) Wilczek). Plant Cell Tissue Organ Cult. 127, 651–663. doi: 10.1007/s11240-016-1085-3
Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol. Plant. 15, 473–497. doi: 10.1111/j.1399-3054.1962.tb08052.x
Murdock, L. L., Coulibaly, O., Higgins, T. J. V., Huesing, J. E., Ishiyaku, M., and Sithole-Niang, I. (2008). “Cowpea,” in Compendium of Transgenic Crop Plants: Transgenic Legume Grains and Forages, eds C. Kole and T. C. Hall (Oxford: Blackwell Publishing), 23–56. doi: 10.1002/9781405181099.k0302
Nielsen, S. S., Ohler, T. A., and Mitchell, C. A. (1997). “Cowpea leaves for human consumption: production, utilization, and nutrient composition,” in Advances in Cowpea Research, eds B. B. Singh, R. D. R. Mohan, K. E. Dashiell, and L. E. N. Jackai (Ibadan: IITA), 326–332.
Ofuya, T. I. (1997). Control of the cowpea aphid, Aphis craccivora Koch (Homoptera:Aphididae), in cowpea, Vigna unguiculata (L.) Walp. Integrated Pest Manag. Rev. 2, 199–207. doi: 10.1093/jee/toy135
Penza, R., Lurquin, P. F., and Filippone, E. (1991). Gene transfer by co-cultivation of mature embryos with Agrobacterium tumefaciens: application to cowpea (Vigna unguiculata L. Walp). J. Plant Physiol. 138, 39–43. doi: 10.1016/S0176-1617(11)80727-3
Popelka, J. C., Gollasch, S., Moore, A., Molvig, L., and Higgins, T. J. V. (2006). Genetic transformation of cowpea (Vigna unguiculata L.) and stable transmission of the transgenes to progeny. Plant Cell Rep. 25, 304–312. doi: 10.1007/s00299-005-0053-x
Ramakrishnan, K., Gnanam, R., Sivakumar, P., and Manickam, A. (2005). In vitro somatic embryogenesis from cell suspension cultures of cowpea [Vigna unguiculata (L.) Walp]. Plant Cell Rep. 24, 449–461. doi: 10.1007/s00299-005-0965-5
Raveendar, S., and Ignacimuthu, S. (2010). Improved Agrobacterium mediated transformation in Cowpea Vigna unguiculata L. Walp. Asian J. Plant Sci. 9, 256–263. doi: 10.3923/ajps.2010.256.263
Raveendar, S., Premkumar, A., Sasikumar, S., Ignacimuthu, S., and Agastian, P. (2009). Development of a rapid, highly efficient system of organogenesis in cowpea Vigna unguiculata (L.) Walp. S. Afr. J. Bot. 75, 17–21. doi: 10.1016/j.sajb.2008.05.009
Sanyal, I., Singh, A. K., Kaushik, M., and Amla, D. V. (2005). Agrobacterium-mediated transformation of chickpea (Cicer arietinum L.) with Bacillus thuringiensis cry1Ac gene for resistance against pod borer insect Helicoverpa armigera. Plant Sci. 168, 1135–1146. doi: 10.1016/j.plantsci.2004.12.015
Schuler, T. H., Poppy, G. M., Kerry, B. R., and Denholm, I. (1998). Insect-resistant transgenic plants. Trends Biotechnol. 16, 168–175. doi: 10.1016/S0167-7799(97)01171-2
Singh, B. B., Ehlers, J. D., Sharma, B., and Freire Filho, F. R. (2002). “Recent progress in cowpea breeding,” in Challenges and Opportunities for Enhancing Sustainable Cowpea Production. Proceedings of the World Cowpea Conference III held at the International Institute of Tropical Agriculture (IITA), eds C. A. Fatokun, S. A. Tarawali, B. B. Singh, P. M. Kormawa, and M. Tamo (Ibadan: IITA), 22–51.
Sivanandhan, G., Dev, G. K., Thoral, J., Selvaraj, N., Ganapathi, A., and Manickavasagam, M. (2015). Sonication, vacuum infiltration and thiol compounds enhance the Agrobacterium-mediated transformation frequency of Withania somnifera (L.) Dunal. PLoS One 10:e0124693. doi: 10.1371/journal.pone.0124693
Solleti, S. K., Bakshi, S., Purkayastha, J., Panda, S. K., and Sahoo, L. (2008). Transgenic cowpea (Vigna unguiculata) seeds expressing a bean alpha-amylase inhibitor 1 confer resistance to storage pests, bruchid beetles. Plant Cell Rep. 27, 1841–1850. doi: 10.1007/s00299-008-0606-x
Somers, D. A., Samac, D. A., and Olhoft, P. M. (2003). Recent advances in legume transformation. Plant Physiol. 131, 892–899. doi: 10.1104/pp.102.017681
Stam, M., Mol, J. N. M., and Kooter, J. M. (1997). The silence of genes in transgenic plants. Ann. Bot. 79, 3–12. doi: 10.1006/anbo.1996.0295
Tiroesele, B., Thomas, K., and Seketeme, S. (2015). Control of cowpea weevil, Callosobruchus maculatus (F.) (Coleoptera: Bruchidae), using natural plant products. Insects 6, 77–84. doi: 10.3390/insects6010077
Keywords: cowpea transformation, Agrobacterium, sonication, kanamycin-based selection, geneticin-based selection, cowpeas
Citation: Bett B, Gollasch S, Moore A, Harding R and Higgins TJV (2019) An Improved Transformation System for Cowpea (Vigna unguiculata L. Walp) via Sonication and a Kanamycin-Geneticin Selection Regime. Front. Plant Sci. 10:219. doi: 10.3389/fpls.2019.00219
Received: 06 August 2018; Accepted: 08 February 2019;
Published: 26 February 2019.
Edited by:
Roger Deal, Emory University, United StatesReviewed by:
Lingaraj Sahoo, Indian Institute of Technology Guwahati, IndiaCopyright © 2019 Bett, Gollasch, Moore, Harding and Higgins. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bosibori Bett, Ym9zaWJvcmliZXR0QGdtYWlsLmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.