- 1Department of Pathogenic Biology and Immunology, College of Integrated Chinese and Western Medicine, Anhui University of Chinese Medicine, Hefei, China
- 2Key Laboratory of Chinese Herbal Compound Formula in Anhui Province, Anhui University of Chinese Medicine, Hefei, China
- 3Division of Molecular and Cell Biophysics, Hefei National Science Center for Physical Sciences, University of Science and Technology of China, Hefei, China
As a major opportunistic pathogen, Pseudomonas aeruginosa can produce various virulence factors and form biofilms. These processes are controlled by the quorum sensing (QS) system. Sodium new houttuyfonate (SNH) is an adduct of houttuyfonate, the main component of the common Chinese medicine plant Houttuynia cordata, which has antibacterial and anti-inflammatory effects. We evaluated the effect of SNH on P. aeruginosa biofilms, virulence factors, and transcription. Transcriptome analysis showed that the key rhlI and pqsA genes of the P. aeruginosa QS system were down-regulated after SNH treatment. SNH reduces proteases and pyocyanin production and inhibits biofilm formation by regulating the P. aeruginosa QS system. SNH also changes the expression of genes related to virulence factors and biofilms (lasA, lasB, lecA, phzM, pqsA, and pilG). These results suggested that the mechanism of SNH against P. aeruginosa by affecting the expression of biofilm and virulence factors controlled by quorum sensing.
Introduction
Pseudomonas aeruginosa is a common clinical opportunistic pathogen (Denning et al., 2003; Xiaoming et al., 2012; Gurunathan et al., 2014). Patients with severe burns (Ramsey and Wozniak, 2005; Bentzmann and Patrick, 2011) or metabolic diseases are sensitive to infection when the body resistance decreases. P. aeruginosa infections has a high morbidity and mortality in the clinic because of its secretion of a large number of virulence factors and increasing antibiotic resistance (Breidenstein et al., 2011). Therefore, there is an urgent need to develop new drugs against P. aeruginosa infection.
Bacterial quorum sensing (QS) is a cellular communication mechanism that depends on the concentration of signal molecules (Ma et al., 2012; Asfahl and Schuster, 2017). There are main three QS regulation systems in P. aeruginosa (Kim et al., 2015), the Las, Rhl, and PQS systems (Schuster et al., 2013; Kalia et al., 2015; Rajkumari et al., 2018), whose transcription regulators are LasR, RhlR, and PqsR respectively. The P. aeruginosa QS system is involved in regulating the expression of virulence factors and biofilms (Van Delden and Iglewski, 1998; Williams and Camara, 2009). At an appropriate concentration, these receptor proteins bind to their respective signal molecules and induce transcription encoding different virulence products such as elastase, rhamnolipid, and pyocyanin. The LasI/Rhl system contains the synthase LasI/RhlI and the transcription activator LasR/RhlR. The Las system can synthesize and recognize N-3-oxo-C12-HSL through LasI synthetase and transcription regulator LasR. The second AHL system is the Rhl system, which can synthesize C4-HSL. The combination of RhlR and C4-HSL activates the expression of antibiotics, rhamnolipids, alkaline proteases, elastase, lectin LecA, cyanide, and genes produced by mass movement. Therefore, it is important to understand the relationship between antibacterial drugs and the P. aeruginosa QS system.
Houttuynia cordata is a traditional Chinese medicine, which is derived from Saururaceae. H. cordata has the functions of antibacterial, antiviral, improving immunity, diuresis and so on. And it is one of the varieties officially identified medicine and food by the Ministry of Health (Figure 1). Sodium new houttuyfonate (SNH) is a sodium bisulfite adduct of the active ingredient of the Chinese herbal medicine H. cordata (Shao et al., 2012) and dodecanoylacetaldehyde. SNH has spectrum wide antibacterial activity, few adverse reactions, and improves immunity (Juanjuan, 2009; Yumei et al., 2012; Yang et al., 2016). Previously, we confirmed that sodium houttuyfonate (SH) has a certain inhibitory effect on P. aeruginosa (Shao et al., 2013; Wu et al., 2014; Wu et al., 2015). SNH and SH structures are highly similar, but SNH is more chemically stable than does SH (Yumei et al., 2012). Therefore, it is important to understand the inhibitory effects and mechanisms of SNH on P. aeruginosa. We speculate that SNH may also inhibit the activity of P. aeruginosa by regulating the QS system.
Figure 1 Houttuynia cordata Thunb and related compounds. (A) Houttuynia cordata Thunb (www.cqcqfn.com). (B) Houttuynin. (C) Sodium houttuyfonate and (D) Sodium new houttuyfonate.
Materials and Methods
Chemicals, Bacteria, and Culture Medium
SNH was purchased from Shanghai Yuanye Biological Technology Co., Ltd. (Shanghai, China), with the content ≥ 98%. Azithromycin (AZM) was acquired from the Northeast Pharmaceutical Group Shenyang No. 1 Pharmaceutical Co., Ltd. (Shenyang, China). P. aeruginosa strain ATCC 27853 was purchased from the China National Institute for the Control of Pharmaceutical and Biological Products (Beijing, China). PDP medium was acquired from Qingdao Hi-Tech Park Haibo Biotechnology Co., Ltd. (Qingdao, China). A single ATCC 27853 colony was selected from Luria-Bertani (LB) solid medium (Hangzhou, China) and cultured in 5 ml LB liquid media (Hangzhou, China) at 37°C, with shaking at 220 r/min for 24 h, then by centrifugation at 10,800 g for 1 min. The supernatant was discarded and the pellet was resuspended in sterile saline solution for optic density detection at 600 nm(OD600) in a UV spectrophotometer. The absorbance of cell suspensions was adjusted to 0.08–0.1, and diluted to 10^7 times for later use for further experiments.
Measurement of Minimum Inhibitory Concentration and Minimum Bactericidal Concentration
The minimum inhibitory concentration (MIC) (Wiegand et al., 2008) and minimum bactericidal concentration (MBC) of SNH were determined using the microdilution method. Different concentrations of SNH solution (100 μl; 2,048, 1,024, 512, 256, 128, 64, 32, 16, 8, and 4 μg/ml) were added with an equal volume of diluted bacterial solution to a 96-well plate. AZM treatment was used as a positive control and culture medium and bacterial solution were used as a blank control group. Plates were incubated at 37°C for 24 h, and the minimum drug concentration with aseptic growth was taken as the MIC. An aliquot (100 μl) was transferred from the sterile growth well to the nutrient agar plate, and cultured at 37°C. After 24 h, MBC was determined as less than 5 colonies on the plate. The experiment was repeated three times.
Growth Curve
SNH solutions (100 μl) were prepared to final concentrations of 2,048, 1,024, and 512 μg/ml. Equal volumes of SNH bad bacterial solutions were added to 96-well plates, and the control groups established. Plates were incubated at 37°C, the OD600 (Nalca et al., 2006) measured at different time points, and the growth curves were drawn.
Drug Treatment and Sample Collection
SNH (2 ml) and an equal volume of bacterial solution was added to the test tube to obtain a concentration of 1 x MIC SNH and the bacteria was cultured at 37°C for 24 h. The blank control group was the SNH treatment control group and no drugs were added. Centrifuge to remove supernatant, and rinsed three times with sterile water. The collected bacterial sample was placed in a centrifuge tube, sealed with sealing film, and placed directly onto dry ice to perform RNA-seq (Poluri et al., 2019). For each group, four samples were prepared under the same experimental conditions.
Transcriptome Sequencing
SNH-induced sequencing of P. aeruginosa transcription was performed using the BGISEQ-500 platform (Huada Gene Technology). First, total RNA was processed and purified, and reverse transcription performed to synthesize cDNA which was then used to produce double-stranded DNA. The end of the synthesized double-stranded DNA was flattened and phosphorescence added to the 5′ end. The 3′ end then forms a sticky end protruding from an “a”, and a bubbly joint with a protruding “t” at the 3′ end is connected. PCR amplification is then performed using specific primers, PCR products thermally denatured into single strands, and the single-stranded DNA is circularized to obtain a single-stranded circular DNA library.
Data are obtained by transcription sequencing was counted using the SOAPnuke (Yuxin et al., 2018) filtering software and filtered using trimmomatic (Bolger et al., 2014). First, reads containing the linker, with unknown base N content greater than 5%, and those of low-quality were removed. NC_002516.2 was used as reference genome. Bowtie2 software was used to align clean reads to the reference gene sequence, and RSEM (Dewey and Bo, 2011; Langmead and Salzberg, 2012) was used to calculate expression levels.
Differential Gene Expression Analysis
Differentially expressed genes (DEGs) were detected based in the method described by Wang et al. (2010). For comparison, the P-values calibrated for multiple tests were performed according to the Benjamini and Hochberg method. False discovery rate (FDR) threshold ≤ 0.05 and | log2fold change (FC)| >1 were defined as significant DEGs (Sun et al., 2015).
Gene Ontology and Kyoto Encyclopedia of Genes and Genomes Enrichment Analysis
DEGs were subjected to Gene Ontology (GO) enrichment analysis and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis (Sun et al., 2017). In general, DEGs are annotated into GO entries, and the number of genes under each entry is counted to analyze GO and KEGG pathway enrichment. GO entries and KEGG pathways are corrected for P-value by FDR, and the Q-value ≤ 0.05 is considered significantly enriched. The major biological functions that DEGs perform can be evaluated by analysis of significant GO and KEGG pathway enrichment.
Determination of Total Protease, LasA Staphylococcal Protease, LasB Elastase, LecA Lectin, Pyocyanin, and Floating Swimming Ability
Bacterial supernatants (500 μl) extracted following treatment with different SNH concentrations were incubated with 1 ml of skim milk (1.25%) at 37°C for 20 min, and their OD600 measured (El-Mowafy et al., 2014). The overnight Staphylococcus aureus culture was boiled for 10 min, centrifuged for 10 min at 10,000 g, and the precipitate was resuspended in 10 mM with Na2HPO4 (pH4.5) to an OD600 of 0.8. The supernatants with different SNH concentrations and S. aureus suspensions were mixed at a ratio of 1:9, and OD600 was measured at different time points to calculate the activity of LasA Staphylococcus protease (Kessler et al., 1993). Bacterial supernatants with different SNH concentrations were used to measure the amounts of secreted elastase LasB and lectin LecA using ELISA kits (Shanghai, China). Different concentrations of pyocyanin assay medium, PAO1, cultured at 37°C for 24 h at 220 r/min were removed from the shaker and centrifuged for 1 min at 10,800 g to remove the supernatant, diluted with sterile water to 0.5 M colorimetric tube, and diluted to 104 times for later use, inoculated on the slant of the medium, and cultured at 37°C for 24 h. Chloroform (3 ml) was added to the test tube, mixed thoroughly and allowed to stand for 15 min. When the chloroform layer turned green they were transferred to another test tube. Then 1 ml of 1 mol/L HCL was added, mixed, and left to stand for 5 min. The OD520 was measured when the solution turned pink. The pyocyanin content was determined by: OD520 * 17.072 (Kong et al., 2005). In a word, the supernatant samples were tested for virulence factors 24 h after coincubation with bacteria and drugs, the expression was standardized by growth OD600. Floatation exercise plates with different drug concentrations (10 g/L tryptone, 10 g/L NaCl, 0.3% agar) were prepared. The strain cultured for 24 h at 37°C and 220 r/min was taken from the shaker and the supernatant was centrifuged at 12,000 r/min, diluted to 0.5 M, and serially diluted 100 times. Using a pipette gun, 2 μl of bacterial liquid was inoculated onto the surface of the floating motion medium, and was placed at 37°C for 24 h to measure colony diameter (Rashid and Kornberg, 2000).
Biofilm Formation
SNH solution (2 ml) with final concentrations of 2,048, 1,024, and 512 μg/ml and an equal volume of diluted bacterial solution were added to a 6-well plate and placed a sterile cover glass. The positive control AZM and blank control group were set up simultaneously. Plates were incubated at 37°C for 24 h, then the planktonic bacteria were washed 3 times on the slide with PBS. The samples were then placed in pre-cooled 2.5% glutaraldehyde solution at least 2 hours in the dark at 4°C, and placed in 30%, 50%, 70%, 90% and 100% ethanol for gradient dehydration for 10 minutes. After drying at room temperature, the biofilm samples were sprayed gold after vacuum drying, and observed and imaged (×5000) with Scanning Electron Microscope (GeminiSEM 500, Germany). Crystal violet (0.4%) was used for staining, 30% acetic acid was used for dissolution, and absorbance was measured at 540 nm as previously described (Corral-Lugo et al., 2016).
RT-PCR and qRT-PCR
Expression of rhlI, pqsA, lasA, lasB, lecA, phzM, and pilG genes in the QS system was evaluated using PCR. Preparations of different concentrations of bacteria and drugs were used for coculture. In the control group, Mueller-Hinton Broth (MH (B)) medium was added instead of SNH. After culturing at 37°C for 24 h, the supernatant was removed and total RNA extracted from the bacteria using TIANGEN′s RNAprep Pure Cell/Bacteria Kit and following the manufacturer′s instructions. cDNA was reverse transcribed using TIANGEN′s FastQuant RT Kit. RT-PCR was performed using 2 x EasyTaq PCR SuperMix (TransGen Biotech, Beijing, China) and a GENETEST Series Gene Amplifier (Hangzhou, China). The reaction involved 35 amplification cycles of: 94°C 5 min, 94°C 30 s, 56°C 30 s, 72°C 30 s, and 72°C 10 min. The synthesized DNA was electrophoresed on a 1.5% agarose gel, and then imaged on a gel imager. SYBR dye was used for qRT-PCR experiments and the 25 μl reaction system included 12.5 μl SYBR dye, 1 μl upstream primer and 1 μl downstream primer (10 μM), 0.5 μl cDNA, and 11 μl DEPC water. The reaction was performed on an Applied Biosystems 7,500 real-time PCR system (USA) and the reaction steps were: 95°C 15 s, 56°C 30 s, 72°C 30 s, for 40 amplification cycles. Reactions were gradient cooled at 95°C–60°C and the dissolution curve detected. rpoD was used as an internal reference, and the gene expression determined by 2-Δ Δ CT. Primer sequences are shown in Table 1.
In Vivo Evaluation of Efficacy of SNH Against P. aeruginosa
Galleria mellonella larvae were obtained from Tianjin Huiyude Biological Technology Co., Ltd., with size of 2-3 cm and weight of 0.2-0.3 g. The larvae were stored at 4°C for later use. SNH and AZM were dissolved in PBS without any cosolvent to avoid the possible toxic effects of cosolvent against larvae. The treatment groups of larvae were set as follows: PBS group (without any drugs and infection, infection (P. aeruginosa 10^4 CFU per one larvae, the rest of infection groups were treated with same concentration of P. aeruginosa) + PBS group, infection + SNH 2048 μg/ml group, infection + SNH 1024 µg/ml group, infection + SNH 512 µg/ml group, infection + AZM 64 µg/ml group. The vivo assays were carried as previously described (Wu et al., 2020).
Statistical Analysis
SPSS 23.0 statistical software was used for analysis, and the results are reported as mean ± standard deviation. All procedures were performed in triplicate.
Results
Effect of SNH on P. aeruginosa Growth
We evaluated the antibacterial potential of SNH against P. aeruginosa and determined its MIC and MBC. At a concentration of 2048 μg/ml, aseptic growth is visible to the naked eye, indicating that the MIC of SNH to P. aeruginosa is 2048 μg/ml. These results show that SNH has certain antibacterial potential against P. aeruginosa. MBC was evaluated based on MIC results, and the bactericidal activity of SNH under MIC was observed. By counting observing plated ATCC 27853 bacteria, it was found that all long bacteria were long and that the number of colonies was > 5. The MBC results suggested that SNH had a bacteriostatic effect on ATCC 27853 and not a bactericidal effect. Therefore, the antibacterial effect of SNH is suitable for use against infections caused by P. aeruginosa.
SNH had no effect on the growth of P. aeruginosa below the MIC (Figure 2). Compared with the untreated blank control, the OD value decreased in the presence of SNH before 24 h, but there was no statistical difference (Data not shown), and OD600 decreased slightly after 30 h, but compared with AZM group, the data showed significant difference, and AZM group decreased significantly from 36 h to 42 h indicating that SNH did not affect the growth of P. aeruginosa, and the change of biofilm formation was not caused by the change of growth.
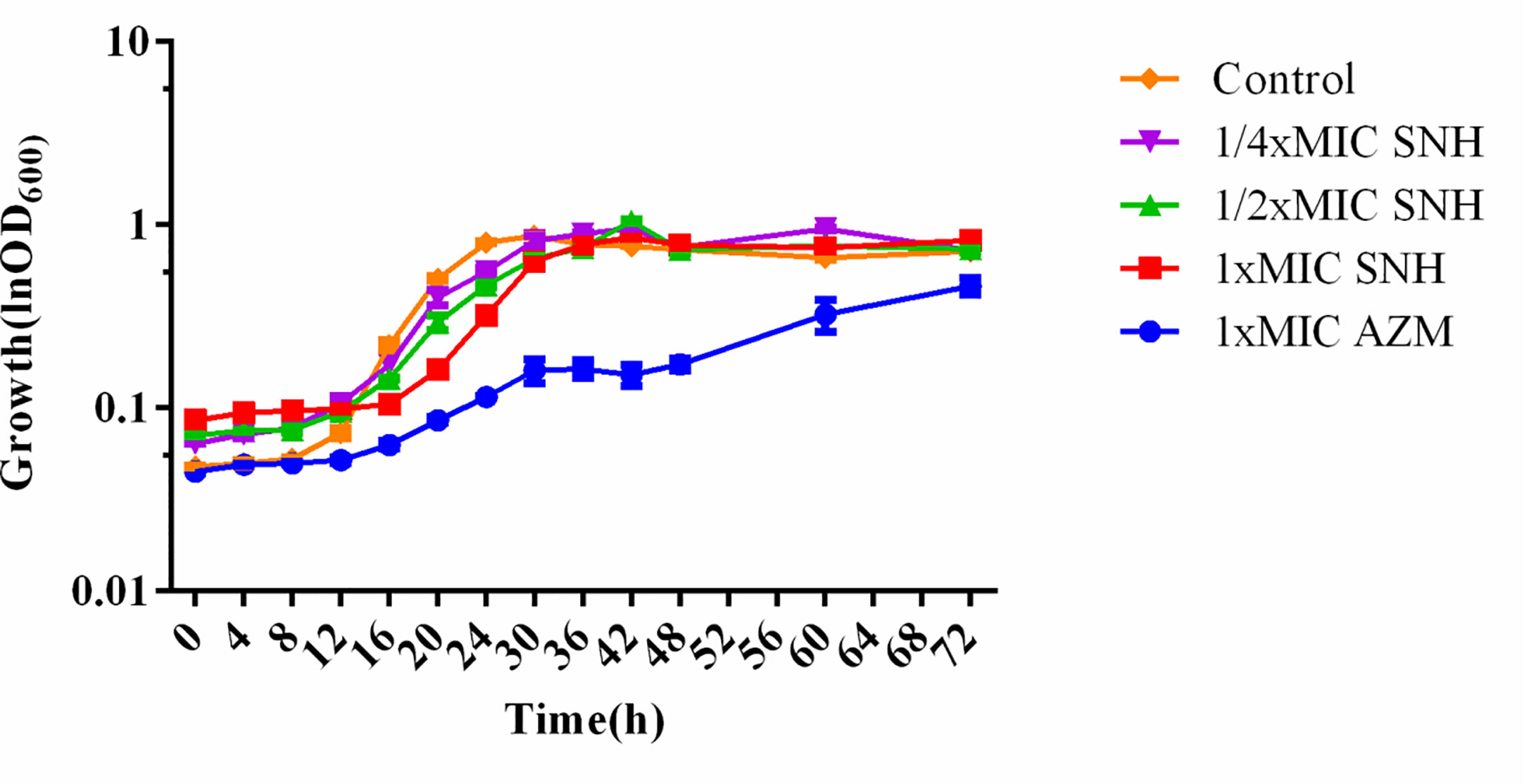
Figure 2 Growth curves of P. aeruginosa treated with different concentrations SNH.The drug concentration of treatments was as follows: Control (without any drugs), 512 μg/ml (1/4×MIC) SNH, 1,024 μg/ml (1/2×MIC) SNH, 2,048 μg/ml (1×MIC) sodium new houttuyfonate (SNH), and 64 μg/ml (1×MIC) azithromycin (AZM).
Transcriptome Sequencing and Clustering DEGs
To further understand the antibacterial mechanism of SNH, we performed transcriptome sequencing. Totally 170.44 MB of transcriptome sequencing data was obtained after RNA-seq applying BGISEQ-500 platform. The original sequencing sequence data has been deposited into the NCBI SRA database (SRA accession: PRJNA542020) (http://identifiers.org/ncbi/insdc.sra:SRP197195.). In total, 5940 reference genes (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE133428.) were assembled from sequencing data, with 5,937 (99.95%) known genes in the control group and 5,934 (99.9%) in the SNH treated group. Principal component analysis was based on FDR threshold ≤ 0.05 and | log2 fold change (FC) | > 1, and 1,674 significant DEGs were identified. Of these DEGs, 1,454 were down-regulated and the remaining 220 were up-regulated (Figure 3).
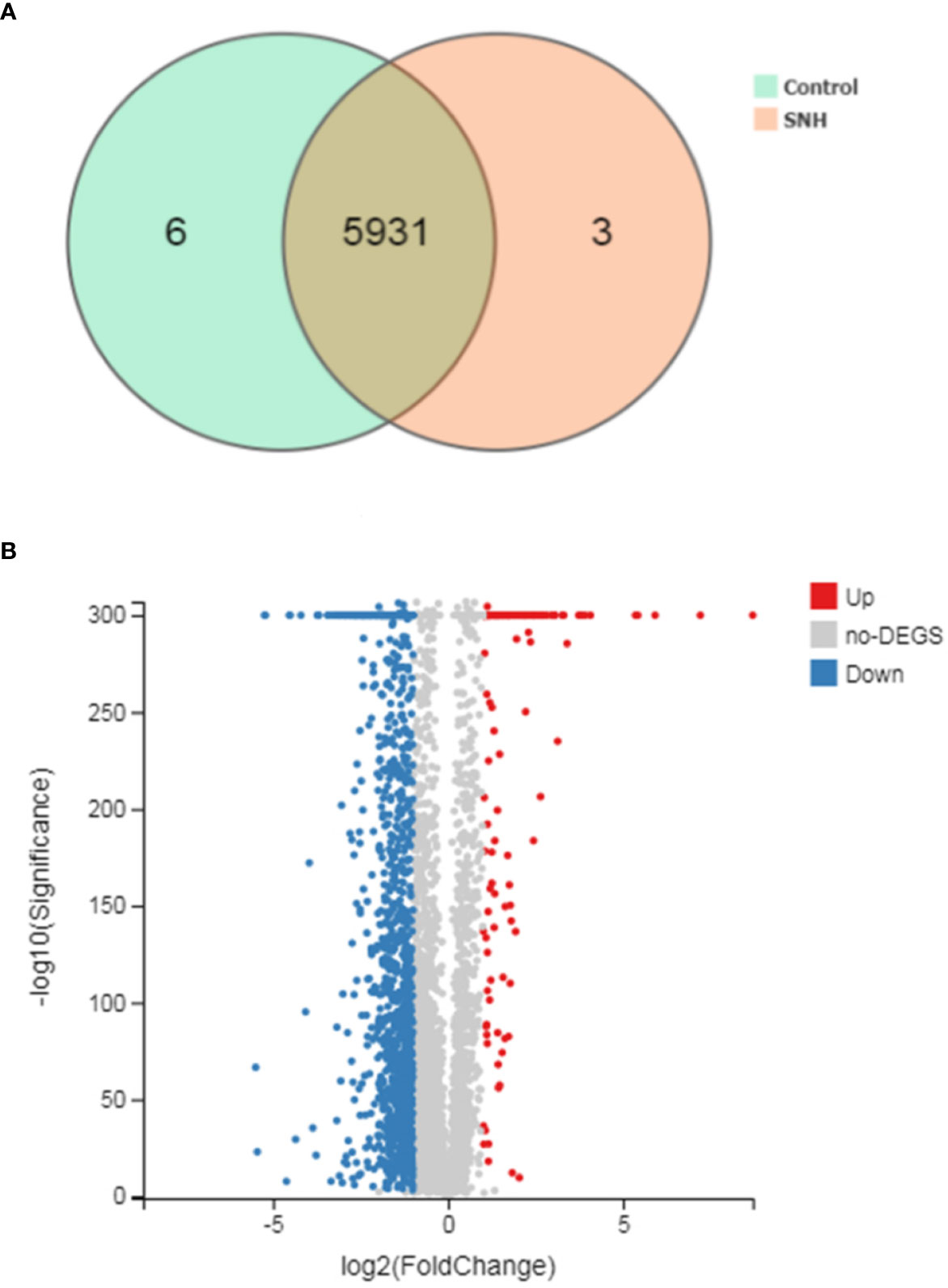
Figure 3 (A) Venn picture of differentially expressed genes (DEGs) and (B) Volcano graph between sodium new houttuyfonate (SNH) treated group and control group.
Analysis of GO and KEGG Pathway of DEGs
GO analysis provides reliable gene product descriptions from various databases and offers a set of dynamic, controlled, and structured terminologies to describe gene functions and products in organism. According to GO functions, all DEGs were classified into: biological process, cellular component, and molecular function. The most annotated functions were “catalytic activity” (GO: 0003824), “membrane” (GO: 0016020), “binding” (GO: 0005488), “cellular process” (GO: 0009987), “membrane part” (GO: 0044425), “metabolic process” (GO: 0008152), and “cell” (GO: 0005623). There were 664 genes in “catalytic activity”, 445 genes in “membrane”, 442 in “binding”, 396 in “membrane part”, 415 in “cell process”, and 294 in “metabolic process” (Figure 4A). DEGs were also enriched in KEGG pathways, which provide data on biological systems and their relationships at molecular, cellular, and organism levels. The KEGG pathways were annotated using the assembled P. aeruginosa transcriptome, and the results were mapped with GO terms. The first three functions that were most significantly enriched in KEGG pathway analysis were the QS system, biofilm formation, and bacterial secretion system Type I, Type II, Type III, and Type VI was changed under SNH treatment (Supplementary Material Figure 2). Among them, 39 genes were enriched in the QS system, 37 genes were enriched in biofilm formation, and 34 genes were enriched in the secretion system. In addition, degradation of fluorobenzoic acid, degradation of chlorocyclohexane and chlorobenzene, degradation of benzoic acid, and biosynthesis of non-ribosomal peptides containing iron carriers are the most significantly abundant signal pathways (Figure 4B). These results show that P. aeruginosa gene expression was up-regulated or down-regulated after SNH treatment compared with the control group. These results indicate that there are certain interactions between SNH and P. aeruginosa, and suggest that SNH may further inhibit the expression of virulence factors and the biofilm formation by influencing the QS system and bacterial metabolic pathways.
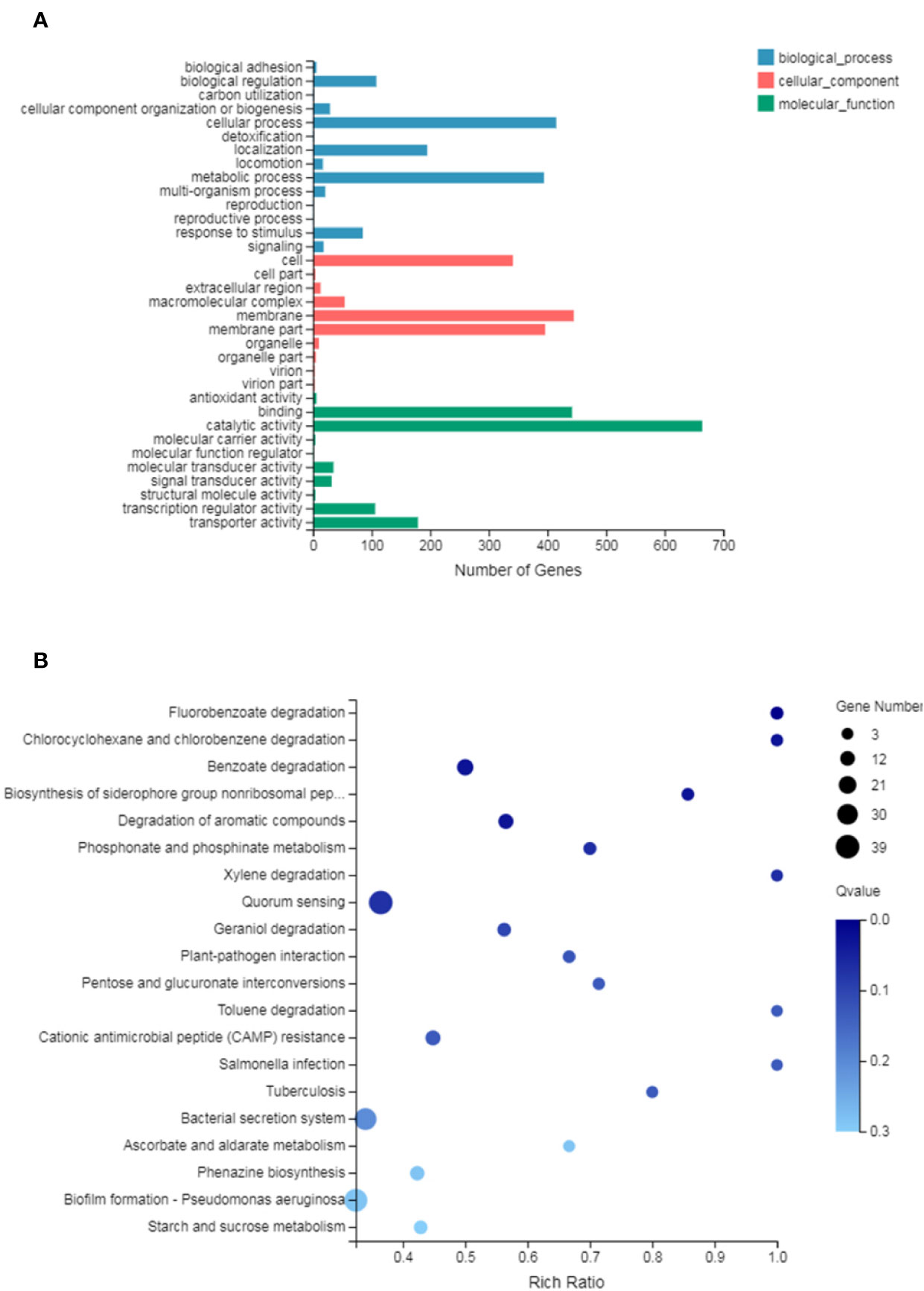
Figure 4 (A) Differentially expressed genes (DEGs) of biological processes, cellular component and molecular function Gene Ontology (GO) terms. (B) Statistical enrichment of DEGs in Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways.
Effect of SNH on P. aeruginosa DEGs
To better understand P. aeruginosa transcription changes under SNH treatment conditions, genes with expression differences were screened using GO and KEGG analyses. Expression genes involved in biofilm formation, the QS system, and phenazine biosynthesis were selected and examined further to reveal the effect of SNH on P. aeruginosa, and to classify their relative expression changes. The results show that among the 32 genes related to biofilm formation, QS system, and phenazine synthesis, 10 were up-regulated and 22 were down-regulated (Table 2), suggesting that SNH may have an inhibitory effect on the production of related virulence factors.
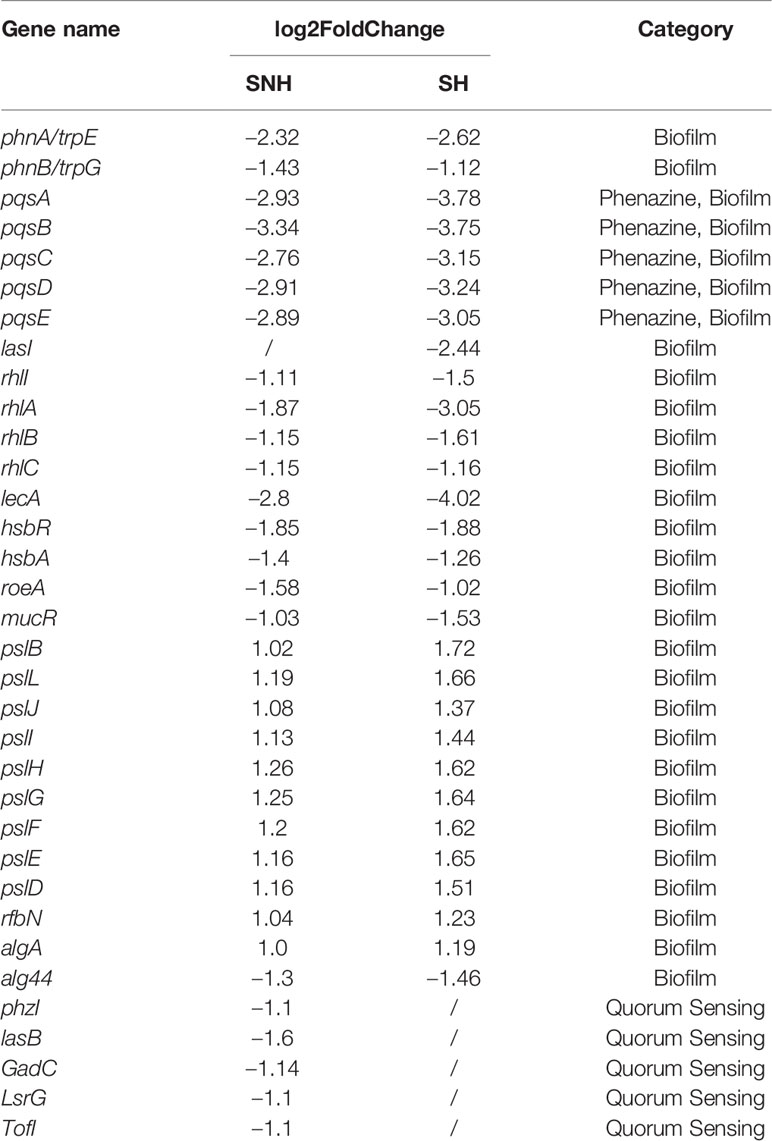
Table 2 Genes differentially expressed under sodium new houttuyfonate (SNH) and sodium houttuyfonate (SH) treatment.
Effect of SNH on P. aeruginosa Virulence Factor
The total protease, LasA staphylococcus protease, LasB elastase, LecA lectin, and pyocyanin levels and swimming ability were assessed (Figure 5). Total protease, LasA staphylococcal protease, LasB elastase, LecA lectin, and pyocyanin production, along with swimming ability, were all inhibited. This inhibition was dose-dependent, and was significant for all measured values when compared with the blank control group (p < 0.01). These results indicate that SNH can inhibit the synthesis of total protease, LasA staphylococcal protease, LasB elastase, LecA lectin, and pyocyanin, and inhibit and swimming ability in P. aeruginosa. Taken together, these results suggest that SNH may inhibit the activity of P. aeruginosa by inhibiting the expression of P. aeruginosa virulence factors.
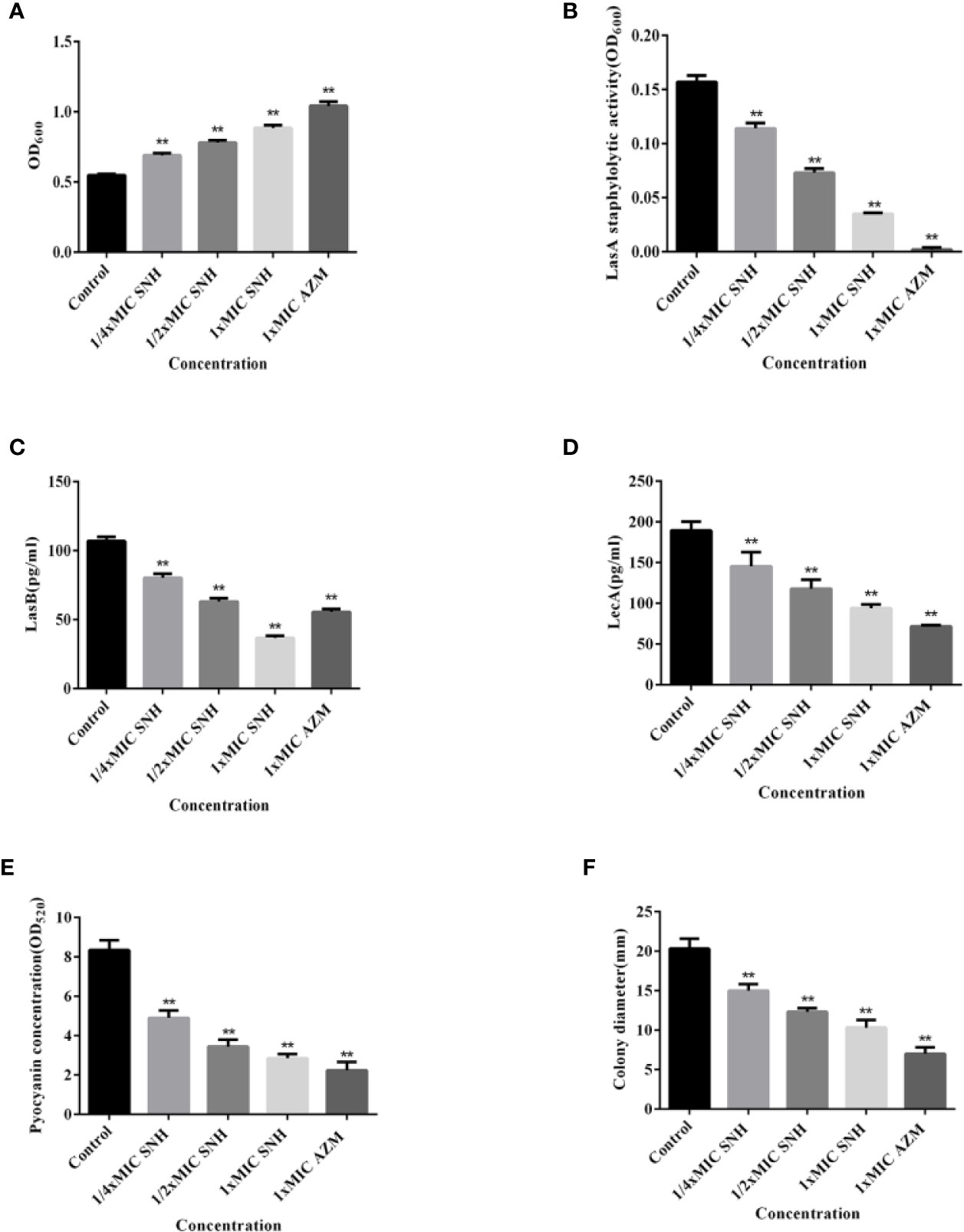
Figure 5 Effect of sodium new houttuyfonate (SNH) on virulence factor of P. aeruginosa. (A) Total protease. (B) LasA staphylococcal protease. (C) LasB elastase. (D) LecA lectin. (E) Pyocyanin. (F) Float swimming. The drug concentration of treatments was as follows: Control, 512 μg/ml (1/4×MIC) SNH, 1024 μg/ml (1/2×MIC) SNH, 2,048 μg/ml (1×MIC) SNH, and 64 μg/ml (1×MIC) azithromycin (AZM). Data represent the means ± SD (n=3, **p < 0.01 vs. control group).
Effect of SNH on P. aeruginosa Biofilm Formation
P. aeruginosa causes chronic infection because it can form a biofilm. Therefore, inhibiting P. aeruginosa biofilm formation may be an effective way to inhibit chronic infection of P. aeruginosa. To explain the effect of SNH on P. aeruginosa biofilm, we used an inverted SEM and crystal violet staining. At 24 h, P. aeruginosa had begun the initial adhesion state. Control group bacteria gathered together in large numbers and had a mucus layer that surrounded a large number of bacteria. The biofilm treated by SNH gradually dispersed with increasing SNH concentration and the number of bacteria decreased (Figure 6A). The amount of biofilm formed in the control group was the largest. The amount of biofilm formed gradually decreased as the SNH concentration increased, showing a dose-dependent characteristic, and a significant difference was observed when compared with the blank control group (p < 0.01) (Figure 6B). These results confirm that SNH can inhibit the formation of P. aeruginosa biofilm.
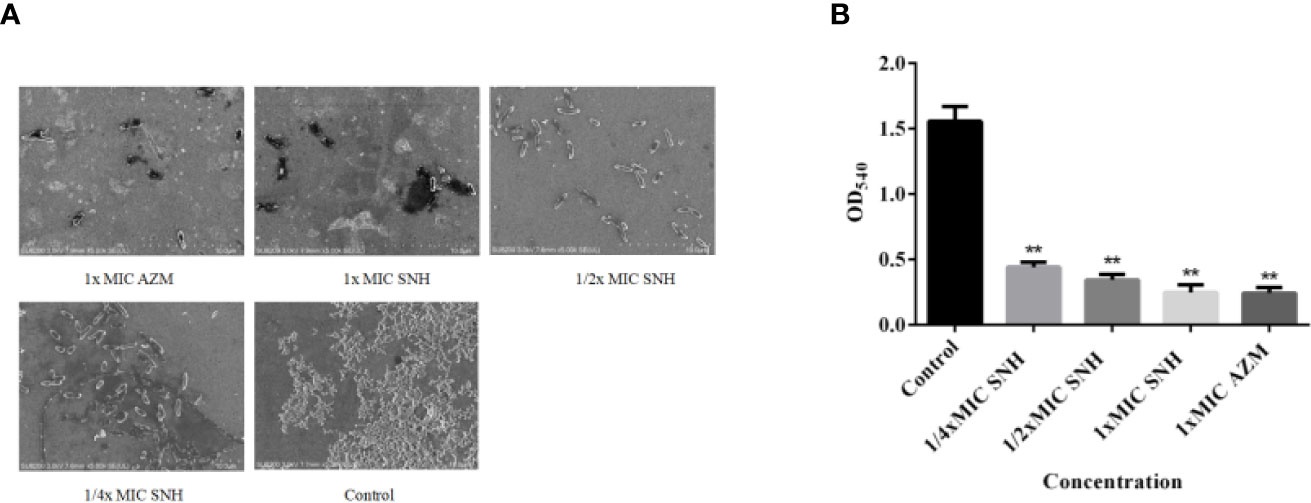
Figure 6 Sodium new houttuyfonate (SNH) effect on P. aeruginosa biofilm. (A) Morphology of ATCC 27853 biofilm under SEM (×5,000). (B) Effect of SNH on ATCC 27853 biofilm production. The drug concentration of treatments was as follows: Control, 512 μg/ml (1/4×MIC) SNH, 1,024 μg/ml (1/2×MIC) SNH, 2,048 μg/ml (1×MIC) SNH, and 64 μg/ml (1×MIC) azithromycin (AZM). Data represent the means ± SD (n=3, **p < 0.01 vs. control group).
Effect of SNH on P. aeruginosa Gene Expression
RT-PCR and qRT-PCR analysis were performed to verify gene expression in bacteria. The virulence genes, biofilms, and related genes controlled by the QS system were selected as the RT-PCR and qRT-PCR analysis targets. RT-PCR results showed that lasA, lasB, lecA, rhlI, phzM, pqsA, and pilG gene expression was down-regulated to different degrees after SNH treatment, and that the down regulation was dose dependent (Figure 7A). qRT-PCR results showed that increasing SNH concentration caused dose-dependent down regulation of the lasA, lasB, lecA, rhlI, phzM, pqsA, and pilG genes (Figure 7B). Although there are some differences in the RT-PCR, qRT-PCR, and RNA-seq results, the expression trends are basically the same. These results indicate that SNH can inhibit the expression of some virulence genes, and biofilm related genes regulated by the QS system. Therefore, SNH may inhibit P. aeruginosa pathogenicity by inhibiting the expression of virulence genes, biofilm genes, and related genes regulated by the QS system.
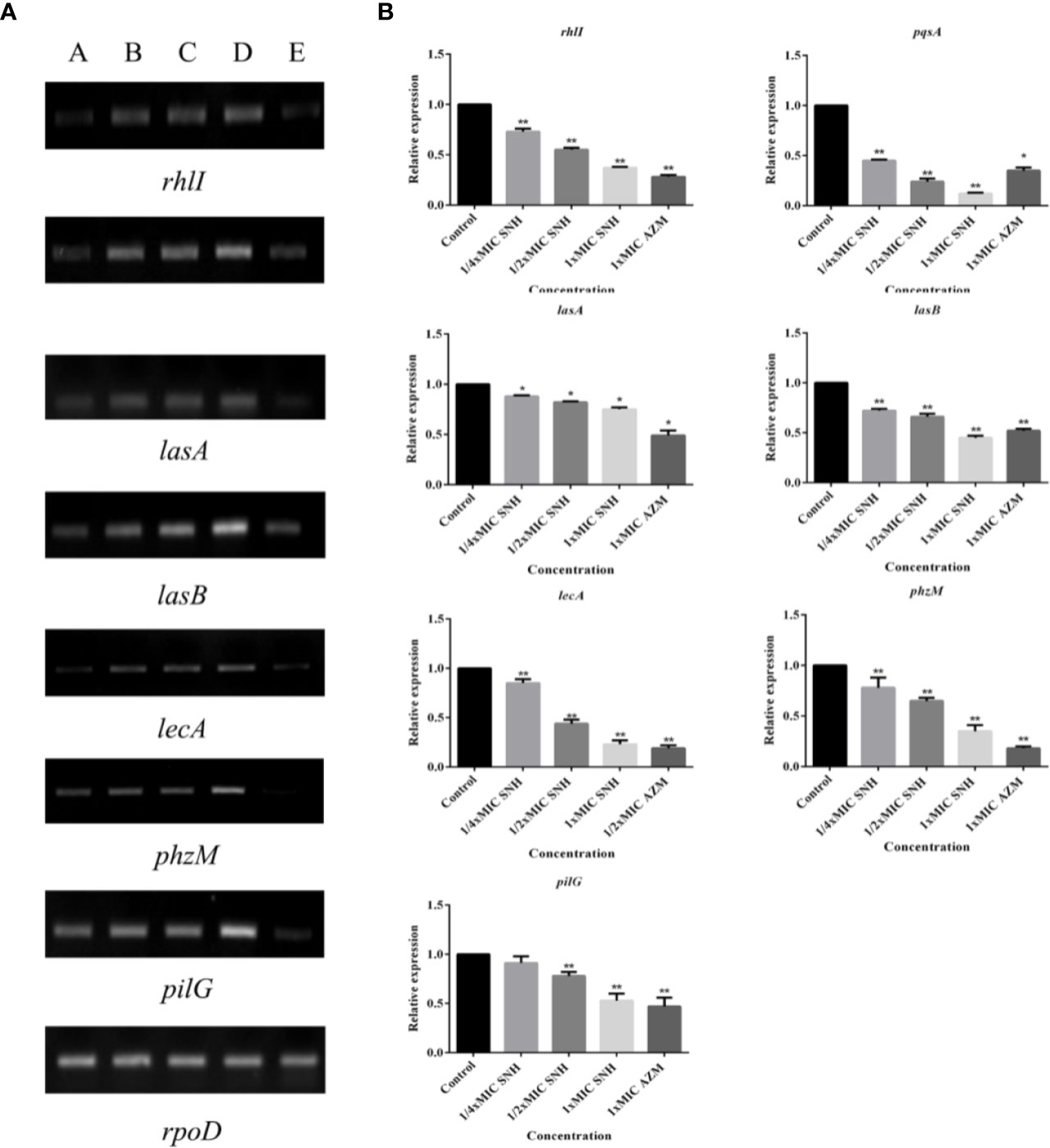
Figure 7 (A) RT-PCR results of sodium new houttuyfonate (SNH) inhibiting biofilm and related genes and virulence factors. The lanes from left to right were as follows: A: 2,048 μg/ml (1×MIC) SNH, B: 1,024 μg/ml (1/2×MIC) SNH, C: 512 μg/ml (1/4×MIC) SNH, D: Control, E: 1xMIC AZM. (B) qRT-PCR results of SNH inhibiting biofilm and related genes and virulence factors. Expression of the house-keeping gene, rpoD, was used as the internal control for each sample. The drug concentration of treatments was as follows: Control, 512 μg/ml (1/4×MIC) SNH, 1024 μg/ml (1/2×MIC) SNH, 2,048μg/ml (1×MIC) SNH, and 64 μg/ml (1×MIC) azithromycin (AZM). Data represent the means ± SD (n=3, *p < 0.05, **p < 0.01 vs. control group).
In Vivo Evaluation of Efficacy of SNH Against P. aeruginosa
After pre-experiment study, we determined 10^4 CFU per one larvae as the applied concentration LD50 of G. mellonella larvae. As shown in Figure 8, we investigated the effects of SNH treatment at 24 h post-infection of P. aeruginosa against G. mellonella. The results show that the survival rate of the infection + PBS group was 0%, and the infection + SNH 512 μg/mL group was 50%, and the infection + SNH 1024 μg/mL group was 60%, and the infection + SNH 2048 μg/mL group was 70%, and the infection + AZM 64 μg/mL group was 70%, and the PBS group was 100% at 24 h. These results also indicate that compared with untreated larvae, infected larvae showed significantly higher survival rate in drug treatment groups, and the difference was statistically significant (p < 0.01). Hence, these results suggest that SNH can improve the survival rate of larvae infected by P. aeruginosa, thereby effectively inhibit P. aeruginosa infection in vivo.
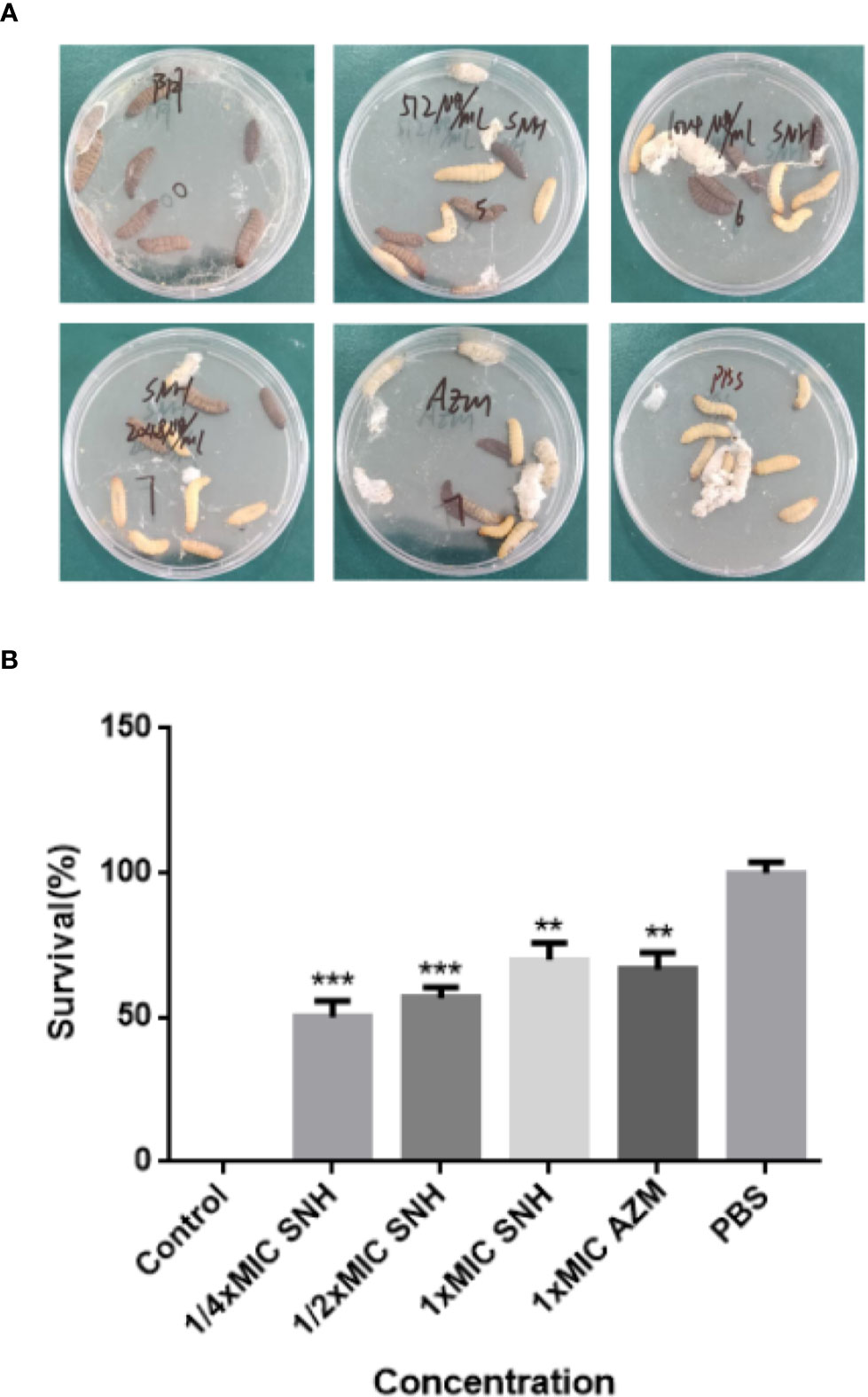
Figure 8 Survival rate of larvae infected by P. aeruginosa. (A) Survival status of larvae under different concentrations of SNH. (B) Survival rate of larvae at different concentrations of SNH. Data represent the means ± SD (n=3, **p < 0.01, ***p < 0.001 vs. control group).
Discussion
SH may play an antibacterial role by regulating the P. aeruginosa QS system to inhibit the synthesis of the virulence factors LasA protease and pyocyanin. SNH is a new generation product of Houttuynia cordata. Compared with SH, SNH has higher safety and effectiveness, is less toxic, has fewer side effects, and has better clinical promotion value (Yefei and Meixian, 2016). It has been recently reported that some fatty acids as antibiofilm and antivirulence agents (Kumar et al., 2020) as Houttuyfonate controlled QS-dependent phenotypes. The structures of Houttuyfonate and SNH are similar to long-chain fatty acids. Therefore, we speculate that SNH may also play an anti-P. aeruginosa effect based on QS-controlled signal transduction.
In order to determine the effect of SNH on the P. aeruginosa QS system and biofilm formation, we performed transcriptional analysis. Principle component analysis and correlation charts were used to evaluate the correlation between the blank control and SNH treatment groups (Supplementary Material Figure 1). Paired Pearson correlation coefficients for each group were higher than 0.7, proving that the samples were well correlated. Transcriptome analysis indicated that SNH may also play an antibacterial role by regulating the QS system of P. aeruginosa. Therefore, based on transcription analysis and experiments to examine the relationship between biofilm formation, the QS system, and pathogenicity, it may be possible to discover the mechanism of SNH action.
The virulence factors regulated by P. aeruginosa QS include protease, elastase, pyocyanin, lectin, rhamnolipid, toxin, and biofilm formation. These virulence factors can affect the formation and maintenance of biofilms and cluster movement. The mutual adjustment is complex, involving many internal and external environmental factors. Total protease can destroy the immunoglobulin that protects the mucous membrane, and destroy the tight junctions between host epithelial cells, which leads to invasion and injury of organism (Heras et al., 2014). Elastase is encoded by the lasB gene and plays an important role in P. aeruginosa infection, which can help bacteria damage host tissues and degrade immune proteins (Szamosvári et al., 2016). LecA is a galactose tetramer lectin produced by P. aeruginosa. It has affinity for extracellular polysaccharides, mediates the adhesion to host cells and infection and biofilm formation in P. aeruginosa (Huang et al., 2018). Pyocyanin is a blue-green redox secondary metabolite produced by P. aeruginosa, which can induce host neutrophils to accelerate apoptosis and reduce local inflammatory reaction, thus providing advantages for the survival of bacteria in the host. In the presence of biofilms, pyocyanin can induce the deposition of extracellular DNA (eDNA), which is the main component of biofilm extracellular polysaccharides and is essential for the formation and stability of biofilms. We further analyzed the effect of SNH on its virulence factors and biofilm formation. In P. aeruginosa biofilm, SNH affects the expression of a large number of genes. SNH reduced the synthesis of total protease, LasA staphylococcal protease, LasB elastase, LecA lectin, pyocyanin, and biofilm in P. aeruginosa. RT-PCR and qRT-PCR revealed that expression of QS regulated genes rhlI, pqsA, lecA, lasA, lasB, phzM, and pilG were down-regulated after SNH treatment.
In P.aeruginosa, SNH can inhibit the expression of some virulence factors and the formation of biofilms. Experimental results show that expression of these virulence genes, biofilms, and related genes is dose-dependent at concentrations of 1 x MIC SNH, ½ x MIC SNH and, ¼ x MIC SNH. Additionally, analysis of virulence factor, biofilm, and related gene expression was consistent with transcriptome sequencing and PCR analysis, further supporting that SNH has an inhibitory effect on P. aeruginosa.
Larval model is an important tool to study the pathogenesis of P. aeruginosa infection in vivo. We simulated the larval model, which proved that SNH significantly reduced the mortality of larvae. The difference of virulence between the drug groups and the blank control group proved by previous growth and virulence testing experiments can be confirmed in the larval model, which proves that the larval model is a suitable model for P. aeruginosa infection. The larval model can be used to study the pathogenic mechanism of P. aeruginosa. Once the genome sequence of larvae is available, the model of larvae will become more valuable in the future.
The MIC of SNH to P. aeruginosa was 2,048 μg/ml, which was four times higher than SH. This possible because our research group has been doing drug resistance tests on P. aeruginosa in recent years, which may gradually adapt to drugs and produce resistance. Previous studies found that SH could down-regulate lasI gene and up-regulate rhlI gene after 72 h of treatment by our research group. However, through transcriptome sequencing data, it was found that lasI, rhlI, and pqsA were down-regulated by 2.44, 1.5, and 3.78 times after SH treatment for 24 h in this study, while no change in lasI gene expression was detected after SNH treatment, and rhlI gene was down-regulated by 1.11 times and pqsA was down-regulated by 2.93 times, which was inconsistent with the previous research results, possibly due to the different action time selected in the experiment (72 h was selected in the previous experiment, while the action time selected in this experiment was 24 h). And it can be seen that when the action time is 24 h, the ways of SH and SNH on inhibiting P. aeruginosa are not completely the same. SH works together by regulating the Las/Rhl and PQS system, in which Las system plays a major role, while SNH plays a regulatory role by regulating Rhl and PQS system. However, due to the limitations of transcriptome sequencing, we also suspect that the relevant data may not be detected because of the small change multiple of lasI.
Importantly, we found that the expression of virulence genes at the transcription level is regulated in different ways among SNH and SH. Although LasR is considered as the main regulator in P. aeruginosa, the cross-regulation between the Las and Rhl system was observed in this experiment. For example, low but detectable C4-HSL is produced without LasR (Wang et al., 2018). The specific reason the expression of lasR is not detected in SNH is still unclear, which may be caused by other highly interactive QS regulatory systems, such as Vfr, VqsR, MvaT, GacA, RpoN, RpoS, and RsmA (Balasubramanian et al., 2013). The existence of Rhl signal without Las signal in clinical isolates suggests that las-independent QS activity may play a role in maintaining P. aeruginosa infection (Feltner et al., 2016). There are researchers reported that P. aeruginosa may enhance some virulence genes and reduce the expression of other genes as part of its survival strategy (Wang et al., 2018). We observed that the yield of total protease and lasB decreased in this experiment, which may be related to the high level of Psl. Therefore, exploring whether the presence of Psl levels in SNH is related to P. aeruginosa infection would be an interesting research direction in the future.
Our research shows that further study on QS system of P. aeruginosa may be helpful to understand its pathogenic mechanism. Nevertheless, there was no change of lasR gene in ATCC 27853 under the action of SNH, which indicated that Las system was not absolutely necessary for the establishment of P. aeruginosa infection. In addition, our study also questioned the previously recognized level understanding of the QS system, and the mechanism should be understood more comprehensively in many aspects and combination with specific situations i.e., different strains and hosts.
Combined with the above experiments, we infer that these virulence factors and biofilms are inhibited by SNH inhibiting the production of C4-HSL. The difference of action mechanism between SH and SNH on P. aeruginosa may lie in the number of C atoms in its structure, which is more than SH 2 C atoms in SNH, which may cause SNH not combining with 3-oxo-C12-HSL, combining with RhlR and C4-HSL complex to play an antibacterial role. SNH has replaced houttuynin, and to a large extent SH due to its improved chemical and pharmacological properties, which has been used as an effective therapeutic agent for respiratory infections and inflammatory diseases such as acute or chronic bronchitis and pneumonia (Lu et al., 2013). Through the experimental study, it was found that SNH had a certain inhibitory effect on P. aeruginosa, but its MIC was high, which provided a certain basis for its structural modification and transformation in the future.
Conclusion
These results show that SNH may inhibit the P. aeruginosa QS system, reduce the synthesis of virulence factors and biofilms, and down-regulate the expression of key genes (Figure 9) to inhibit P. aeruginosa infection. Additionally, this study can be used as a basis for designing new QS-targeted drug candidates using SNH, while minimizing the chance of generating resistance (Rodolfo et al., 2013). These results are currently being expanded upon through research on animal infection models in vivo in our research group.
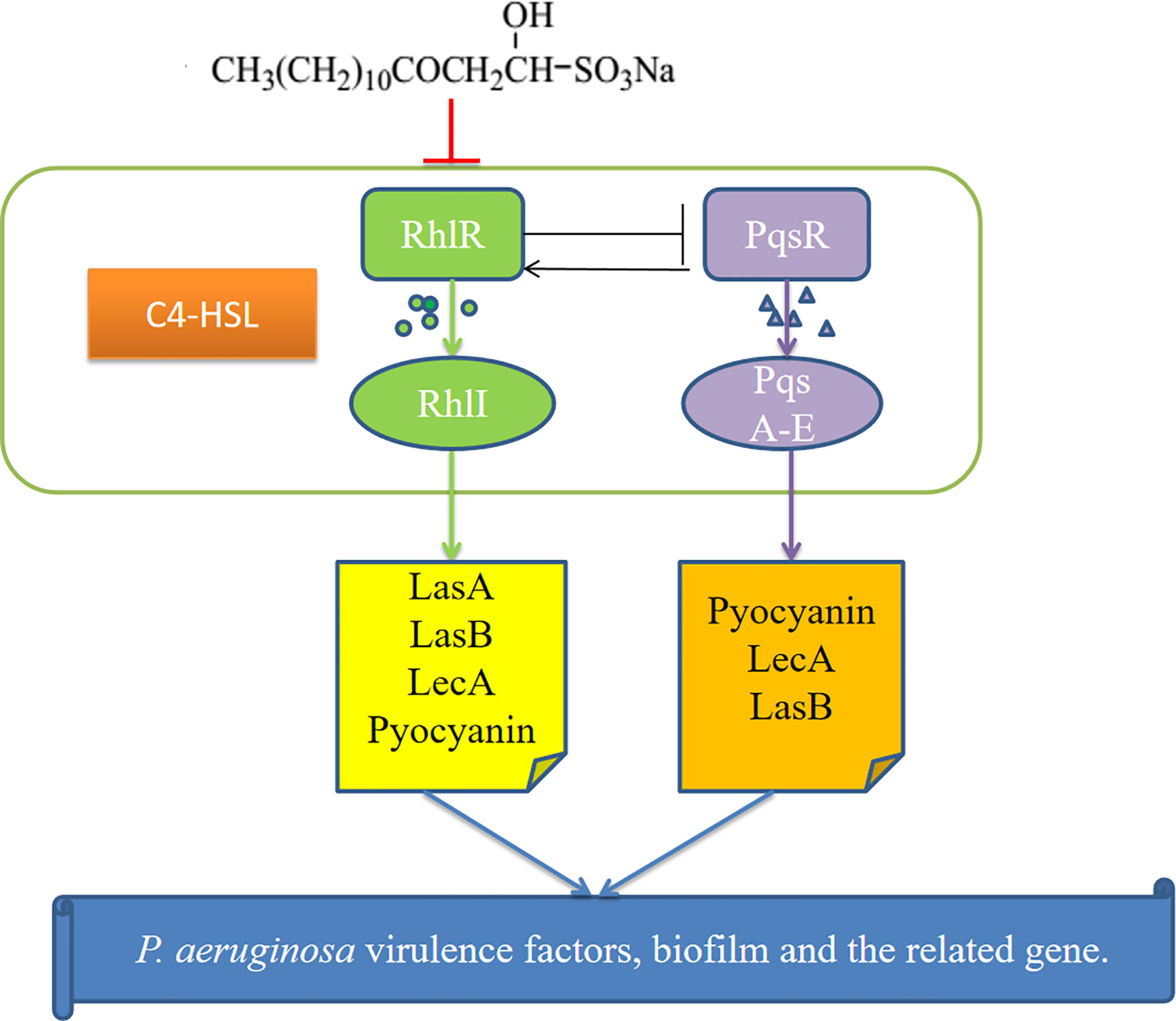
Figure 9 Possible mechanism for the inhibition of P.aeruginosa quorum sensing (QS) system by sodium new houttuyfonate (SNH). The figure shows the Rhl and PQS systems and the virulence genes regulated by them. Our results show that SNH inhibits the expression of rhlI, and pqsA. The scheme shows quorum sensing(QS)-controlled gene regulation, Rhl can produce virulence factors LasA, elastase B, lectin LecA, pyocyanin, etc. Alone or in cooperation with PQS. These results confirm that SNH interferes with the production of QS and QS-related virulence factors of P.aeruginosa. Therefore, the pathogenicity of P.aeruginosa may be weakened by inhibiting the formation of virulence factors and biofilm.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material; further inquiries can be directed to the corresponding author.
Author Contributions
DW, GY, and CW conceived and designed the study. YZ and DW wrote the manuscript. JS and TW critically reviewed the manuscript and provided general advice. YZ, LM, YS, and JW performed the experiments and analyzed data. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the project funded by the China Postdoctoral Science Foundation under Grant No. 2019M662185, the key discipline of Anhui University of Chinese Medicine (DC18100042), the Outstanding Talent Support Program in University (Key project) of Anhui Province under Grant No. gxyqZD2020024, and the Natural Science Foundation (Key project) of Anhui University of Chinese Medicine under Grant No.2020zrzd07. The Funding is used to cover laboratory expenses, sample preparation and sequencing. These funding agencies have no role in research design, data collection, analysis of results, or manuscript writing.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank the sequencing facilities of Huada Gene Cloud Platform for the preparation and sequencing of the library.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphar.2020.572375/full#supplementary-material
Supplementary Figure 1 | (A) PCA and (B) Correlation coefficient diagram between SNH treated group and control group.
Supplementary Figure 2 | The bacterial secretion system Type I, Type II, Type III, and Type VI was changed under SNH treatment.
References
Asfahl, K. L., Schuster, M. (2017). Additive effects of quorum sensing anti-activators on Pseudomonas aeruginosa virulence traits and transcriptome. Front. Microbiol. 8, 2654. doi: 10.3389/fmicb.2017.02654
Balasubramanian, D., Schneper, L., Kumari, H., Mathee, K. (2013). A dynamic and intricate regulatory network determines Pseudomonas aeruginosa virulence. Nucleic Acids Res. 41, 1–20. doi: 10.1093/nar/gks1039
Bentzmann, S. D., Patrick, P. (2011). The Pseudomonas aeruginosa opportunistic pathogen and human infections. Environ. Microbiol. 13, 1655–1665. doi: 10.1111/j.1462-2920.2011.02469.x
Bolger, A. M.,., Lohse, M., Usadel, B. (2014). Trimmomatic: a flexible trimmer for illumina sequence data. Bioinf. (Oxford England) 30 (15), 2114–2120. doi: 10.1093/bioinformatics/btu170
Breidenstein, E. B., De La Fuente-Nunez, C., Hancock, R. E. (2011). Pseudomonas aeruginosa: all roads lead to resistance. Trends Microbiol. 19, 419–426. doi: 10.1016/j.tim.2011.04.005
Corral-Lugo, A., Daddaoua, A., Ortega, A., Espinosa-Urgel, M., Krell, T. (2016). Rosmarinic acid is a homoserine lactone mimic produced by plants that activates a bacterial quorum-sensing regulator. Sci. Signal. 9, ra1–ra1. doi: 10.1126/scisignal.aaa8271
Denning, G. M., Iyer, S. S., Reszka, K. J., O′Malley, Y., Britigan, B. E. (2003). Phenazine-1-carboxylic acid, a secondary metabolite of Pseudomonas aeruginosa, alters expression of immunomodulatory proteins by human airway epithelial cells. Am. J. Physiol. Lung Cell Mol. Physiol. 285, 584–592. doi: 10.1152/ajplung.00086.2003
Dewey, C. N., Bo, L. (2011). RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 12:323. doi: 10.1186/1471-2105-12-323
El-Mowafy, S. A., Abd, E. G. K. H., El-Messery, S. M., Shaaban, M., II (2014). Aspirin is an efficient inhibitor of quorum sensing, virulence and toxins in Pseudomonas aeruginosa. Microb Pathog. 74, 25–32. doi: 10.1016/j.micpath.2014.07.008
Feltner, J. B., Wolter, D. J., Pope, C. E.,., Groleau, M. C., Smalley, N. E., Greenberg, E. P., et al. (2016). LasR variant cystic fibrosis isolates reveal an adaptable quorum-sensing hierarchy in Pseudomonas aeruginosa. mBio 7, 7 (5), e01513–e01516. doi: 10.1128/mBio.01513-16
Gurunathan, S., Han, J. W., Kwon, D. N., Kim, J. H. (2014). Enhanced antibacterial and anti-biofilm activities of silver nanoparticles against Gram-negative and Gram-positive bacteria. Nanoscale Res. Lett. 9, 1–17. doi: 10.1186/1556-276X-9-373
Harding, C. R., Schroeder, G. N., Collins, J. W., Frankel, G. (2013). Use of Galleria mellonella as a model organism to study Legionella pneumophila infection. J. Vis. Exp. 22 (81), e50964. doi: 10.3791/50964
Heras, B., Scanlon, M. J., Martin, J. L. (2014). Targeting virulence not viability in the search for future antibacterials. Br. J. Clin. Pharmacol. 79 (2), 208–215. doi: 10.1111/bcp.12356
Hickey, C., Schaible, B., Nguyen, S., Hurley, D., Srikumar, S., Fanning, S., et al. (2018). Increased virulence of bloodstream over peripheral isolates of P.aeruginosa identified through post-transcriptional regulation of virulence factors. Front. Cell Infect. Microbiol. 8, 357. doi: 10.3389/fcimb.2018.00357
Huang, S. F.,., Lin, C. H., Yu, L. T., Tsai, C. L.,., Cheng, T. J.,. R., Wang, S. K.,. (2018). Development of Pseudomonas aeruginosa lectin LecA inhibitor by using bivalent galactosides supported on polyproline peptide scaffolds. Chem. Asian J. 13 (6), 686–700. doi: 10.1002/asia.201701724
Juanjuan, H. (2009). Effect of sodium new houttuyfonate injection combined with Interferon on children′s autumn diarrhea. Modern Chin. Drug Application 1, 138–139. doi: 10.3969/j.issn.1673-9523.2009.01.114
Kalia, M., Yadav, V. K., Singh, P. K., Sharma, D., Pandey, H., Narvi, S. S., et al. (2015). Effect of cinnamon oil on quorum sensing-controlled virulence factors and biofilm formation in Pseudomonas aeruginosa. PloS One 10, e0135495. doi: 10.1371/journal.pone.0135495
Kessler, E., Safrin, M., Olson, J. C., Ohman, D. E. (1993). Secreted LasA of Pseudomonas aeruginosa is a staphylolytic protease. J. Biol. Chem. 268, 7503–7508. doi: 10.1016/0014-5793(93)80087-B
Kim, H. S., Lee, S. H., Byun, Y., Park, H. D. (2015). 6-Gingerol reduces Pseudomonas aeruginosa biofilm formation and virulence via quorum sensing inhibition. Sci. Rep. 5, 8656. doi: 10.1038/srep08656
Kong, K. F., Jayawardena, S. R., Indulkar, S. D., Del Puerto, A., Koh, C. L., Høiby, N., et al. (2005). Pseudomonas aeruginosa AmpR is a global transcriptional factor that regulates expression of AmpC and PoxB beta-lactamases, proteases, quorum sensing, and other virulence factors. Antimicrob. Agents Chemother. 49, 4567–4575. doi: 10.1128/AAC.49.11.4567-4575.2005
Kumar, P., Lee, J. H., Beyenal, H., Lee, J. (2020). Fatty Acids as Antibiofilm and Antivirulence Agents. Trends Microbiol. 28 (9), 753–768. doi: 10.1016/j.tim.2020.03.014
Langmead, B., Salzberg, S. L. (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359. doi: 10.1038/nmeth.1923
Lu, X., Yang, X., Li, X., Lu, Y., Ren, Z., Zhao, L., et al. (2013). In vitro activity of sodium new houttuyfonate alone and in combination with oxacillin or netilmicin against methicillin-resistant Staphylococcus aureus. PloS One 8, e68053. doi: 10.1371/journal.pone.0068053
Ma, L., Liu, X., Liang, H., Che, Y., Chen, C., Dai, H., et al. (2012). Effects of 14-alpha-lipoyl andrographolide on quorum sensing in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 56, 6088–6094. doi: 10.1128/AAC.01119-12
Nalca, Y., Jansch, L., Bredenbruch, F., Geffers, R., Buer, J., Haussler, S. (2006). Quorum-sensing antagonistic activities of azithromycin in Pseudomonas aeruginosa PAO1:a global approach. Antimicrob. Agents Chemother. 50, 1680–1688. doi: 10.1128/AAC.50.5.1680-1688.2006
Poluri, R. T. K., Beauparlant, C. J., Droit, A., Etienne, A.-W. (2019). RNA sequencing data of human prostate cancer cells treated with androgens. Data Brief. 25:104372. doi: 10.1016/j.dib.2019.104372
Rajkumari, J., Borkotoky, S., Murali, A., Suchiang, K., Mohanty, S. K., Busi, S. (2018). Cinnamic acid attenuates quorum sensing associated virulence factors and biofilm formation in Pseudomonas aeruginosa PAO1. Biotechnol. Lett. 40, 1087–1100. doi: 10.1007/s10529-018-2557-9
Ramsey, D. M., Wozniak, D. J. (2005). Understanding the control of Pseudomonas aeruginosa alginate synthesis and the prospects for management of chronic infections in cystic fbrosis. Mol. Microbiol. 56, 309–322. doi: 10.1111/j.1365-2958.2005.04552.x
Rashid, M. H., Kornberg, A. (2000). Inorganic polyphosphate is needed for swimming, swarming, and twitching motilities of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U S A. 97, 4885–4890. doi: 10.1073/pnas.060030097
Rodolfo, G. C., Mariano, M. V., Norma, V. G., Alejandra, G., V., P., Takahiro, H., et al. (2013). Resistance to the quorum-quenching compounds brominated furanone C-30 and 5-fluorouracil in Pseudomonas aeruginosa clinical isolates. Patho Dis. 68 (1), 8–11. doi: 10.1111/2049-632X.12039
Schuster, M., Sexton, D. J., Diggle, S. P., Greenberg, E. P. (2013). Acyl-homoserine lactone quorum sensing: from evolution to application. Annu. Rev. Microbiol 67, 43–63. doi: 10.1146/annurev-micro-092412-155635
Shao, J., Cheng, H., Wang, C., Wang, Y. (2012). A phytoanticipin derivative, sodium houttuyfonate, induces in vitro synergistic effects with levofloxacin against biofilm formation by Pseudomonas aeruginosa. Molecules 17, 11242–11254. doi: 10.3390/molecules170911242
Shao, J., Cheng, H., Wang, C., Wu, D., Zhu, X., Zhu, L., et al. (2013). Sodium houttuyfonate, a potential phytoanticipin derivative of antibacterial agent, inhibits bacterial attachment and pyocyanine secretion of Pseudomonas aeruginosa by attenuating flagella-mediated swimming motility. World J. Microbiol Biotechnol. 29, 2373–2378. doi: 10.1007/s11274-013-1405-2
Sun, Q. L., Zhao, C. P., Wang, T. Y., Hao, X. B., Wang, X. Y., Zhang, X., et al. (2015). Expression profifile analysis of long non-coding RNA associated with vincristine resistance in colon cancer cells by next-generation sequencing. Gene 572, 79–86. doi: 10.1016/j.gene.2015.06.087
Sun, D., Zhang, W., Mou, Z., Chen, Y., Guo, F., Yang, E., et al. (2017). Transcriptome analysis reveals silver nanoparticle-decorated quercetin antibacterial molecular mechanism. ACS Appl. Mater Interfaces 9, 10047–10060. doi: 10.1021/acsami.7b02380
Szamosvári, D., Reichle, V. F., Jureschi, M., Böttcher, T. (2016). Synthetic quinolone signal analogues inhibiting the virulence factor elastase of Pseudomonas aeruginosa. Chem. Commun. 52, 13440–13443. doi: 10.1039/c6cc06295d
Van Delden, C., Iglewski, B. H. (1998). Cell-to-cell signaling and Pseudomonas aeruginosa infections. Emerg Infect. Dis. 4, 551–560. doi: 10.3201/eid0404.980405
Wang, L., Feng, Z., Wang, X., Wang, X., Zhang, X. (2010). DEGseq: an R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics. 26, 136–138. doi: 10.1093/bioinformatics/btp612
Wang, Y., Gao, L., Rao, X., Wang, J., Yu, H., Jiang, J., et al. (2018). Characterization of lasR-defcient clinical isolates of Pseudomonas aeruginosa. Sci. Rep. 8 (1), 13344. doi: 10.1038/s41598-018-30813-y
Wiegand, I., Hilpert, K., Hancock, R. E. (2008). Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat. Protoc. 3, 163–175. doi: 10.1038/nprot.2007.521
Williams, P., Camara, M. (2009). Quorum sensing and environmental adaptation in Pseudomonas aeruginosa: a tale of regulatory networks and multifunctional signal molecules. Curr. Opin. Microbiol 12, 182–191. doi: 10.1016/j.mib.2009.01.005
Wu, D., Cheng, H., Huang, W., Duan, Q. (2015). Sodium houttuyfonate in vitro inhibits the biofilm formation and alginate biosynthesis gene expression of Pseudomonas aeruginosa clinical strain. Exp.Ther. Med. 10, 753–758. doi: 10.3892/etm.2015.2562
Wu, D., Huang, W., Duan, Q., Li, F., Cheng, H. (2014). Sodium houttuyfonate affects production of N-acyl homoserine lactone and quorum sensing-regulated genes expression in Pseudomonas aeruginosa. Front. Microbiol. 5, 635. doi: 10.3389/fmicb.2014.00635
Wu, J., Wu, D., Zhao, Y., Si, Y., Mei, L., Shao, J., et al. (2020). Sodium New Houttuyfonate Inhibits Candida albicans Biofilm Formation by Inhibiting the Ras1-cAMP-Efg1 Pathway Revealed by RNA-seq. Front. Microbiol. 11, 2075. doi: 10.3389/fmicb.2020.02075
Xiaoming, Z., Jiaqing, Y., Changfeng, Z., Huijuan, C. (2012). Huanglianjiedu decoction against Pseudomonas aeruginosa biofilm and its synergistic antibacterial effect with azithromycin. Chin. J. Exp. Traditional Med. Formulae. 18, 155–158. doi: 10.13422/J.Cnki.Syfjx.2012.11.051
Yang, Z., Yefei, Z., Meixian, Z. (2016). Effects of sodium new houttuyfonate on replication and antibody growth and decline of newcastle disease virus. Heilongjiang Anim. Husbandry Veterinary Medicine. 10, P.150–P.152. doi: 10.13881/j.cnki.hljxmsy.2016.0895
Yefei, Z., Meixian, Z. (2016). Effect of sodium new houttuyfonate nanoemulsion adjuvant on immune function of broilers. J. Northwest Agriculture. 11, 1591–1596. doi: 10.7606/j.issn.1004-1389.2016.11.001
Yumei, L., Hong, H., Lu, H., Lingli, Z. (2012). Spectrographic study on structural confirmation of sodium new houttuyfonate. China Pharmacy. 23, 1777–1779. doi: CNKI:SUN:ZGYA.0.2012-19-027
Keywords: Pseudomonas aeruginosa, virulence factors, biofilm, quorum sensing, sodium new houttuyfonate, transcriptome
Citation: Zhao Y, Mei L, Si Y, Wu J, Shao J, Wang T, Yan G, Wang C and Wu D (2020) Sodium New Houttuyfonate Affects Transcriptome and Virulence Factors of Pseudomonas aeruginosa Controlled by Quorum Sensing. Front. Pharmacol. 11:572375. doi: 10.3389/fphar.2020.572375
Received: 14 June 2020; Accepted: 14 September 2020;
Published: 02 October 2020.
Edited by:
Jean Sévigny, Laval University, CanadaReviewed by:
Rodolfo García-Contreras, National Autonomous University of Mexico, MexicoJintae Lee, Yeungnam University, South Korea
Copyright © 2020 Zhao, Mei, Si, Wu, Shao, Wang, Yan, Wang and Wu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Daqiang Wu, ZGFxd3VAMTI2LmNvbQ==
†These authors have contributed equally to this work
 Yeye Zhao
Yeye Zhao Longfei Mei1†
Longfei Mei1† Jiadi Wu
Jiadi Wu Jing Shao
Jing Shao Guiming Yan
Guiming Yan Daqiang Wu
Daqiang Wu