
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Pediatr. , 10 July 2020
Sec. Pediatric Hematology and Hematological Malignancies
Volume 8 - 2020 | https://doi.org/10.3389/fped.2020.00278
This article is part of the Research Topic New Perspectives on Pediatric Acute Leukemia View all 9 articles
 Teofila Ksiazek1,2
Teofila Ksiazek1,2 Malgorzata Czogala3,4
Malgorzata Czogala3,4 Przemyslaw Kaczowka2,3
Przemyslaw Kaczowka2,3 Beata Sadowska2
Beata Sadowska2 Katarzyna Pawinska-Wasikowska3,4
Katarzyna Pawinska-Wasikowska3,4 Mirosław Bik-Multanowski1
Mirosław Bik-Multanowski1 Barbara Sikorska-Fic5
Barbara Sikorska-Fic5 Michał Matysiak5
Michał Matysiak5 Jolanta Skalska-Sadowska6
Jolanta Skalska-Sadowska6 Jacek Wachowiak6
Jacek Wachowiak6 Anna Rodziewicz-Konarska7
Anna Rodziewicz-Konarska7 Alicja Chybicka7
Alicja Chybicka7 Katarzyna Muszynska-Rosłan8
Katarzyna Muszynska-Rosłan8 Maryna Krawczuk-Rybak8
Maryna Krawczuk-Rybak8 Dominik Grabowski9
Dominik Grabowski9 Jerzy Kowalczyk9
Jerzy Kowalczyk9 Lucyna Maciejka-Kemblowska10
Lucyna Maciejka-Kemblowska10 Elzbieta Adamkiewicz-Drozynska10
Elzbieta Adamkiewicz-Drozynska10 Wojciech Mlynarski11
Wojciech Mlynarski11 Renata Tomaszewska12
Renata Tomaszewska12 Tomasz Szczepanski12
Tomasz Szczepanski12 Joanna Pohorecka13
Joanna Pohorecka13 Grazyna Karolczyk13
Grazyna Karolczyk13 Agnieszka Mizia-Malarz14
Agnieszka Mizia-Malarz14 Katarzyna Mycko15
Katarzyna Mycko15 Wanda Badowska15
Wanda Badowska15 Karolina Zielezinska16
Karolina Zielezinska16 Tomasz Urasinski16
Tomasz Urasinski16 Irena Karpinska-Derda17
Irena Karpinska-Derda17 Mariola Woszczyk17
Mariola Woszczyk17 Małgorzata Ciebiera18
Małgorzata Ciebiera18 Monika Lejman19
Monika Lejman19 Szymon Skoczen3,4*
Szymon Skoczen3,4* Walentyna Balwierz3,4
Walentyna Balwierz3,411q23/MLL rearrangements are frequently detected in pediatric acute myeloid leukemia. The analysis of their clinical significance is difficult because of the multitude of translocation fusion partners and their low frequency. The presence of t(10;11)(p12;q23) translocation was previously identified in pediatric acute myelogenous leukemia (AML). It is considered as the second most common translocation detected in pediatric 11q23/MLL-rearranged (present KMT2A) AML, after t(9;11)(p22;q23). The presence of the above translocation was previously identified as an unfavorable prognostic factor. Since June 2015, the Polish Pediatric Leukemia/Lymphoma Study Group has applied the therapeutic protocol requiring extensive diagnostics of genetic changes in pediatric AML. Until November 2019, molecular genetic studies were performed in 195 children with diagnosed AML to identify carriers of fusion gene transcripts for 28 most common chromosomal translocations in acute leukemia. The fusion gene transcript for translocation t(10;11)(p12;q23) involving MLL gene was detected with unexpectedly high frequency (8.9%) in our research. It was the highest frequency of all detected MLL rearrangements, as well as other detected fusion gene transcripts from chromosomal aberrations characteristic for AML. It seems that chromosomal aberration between chromosomes 10 and 11 can be relatively frequent in some populations. Paying attention to this fact and ensuring proper genetic diagnosis seem to be important for appropriate allocation of patients to risk groups of pediatric AML treatment protocols.
Acute myelogenous leukemia (AML) is a heterogeneous group of hematologic malignancies, characterized by unregulated, clonal proliferation of abnormal myeloid progenitor cells. Despite major improvements in outcome over the past decades, it remains a life-threatening malignancy in children. Several constitutive genetic variants and acquired chromosomal abnormalities have been identified as prognostic markers in leukemia (1, 2). In AML in children, genetic diagnostics play an extremely important role in stratifying the risk of treatment failure. Identified genetic markers together with the response to induction treatment are the most important factors that allow selection of optimal therapy for the patient. Among the most relevant factors are chromosomal translocations and gene mutations, which recently have become crucial for risk stratification in pediatric AML (3, 4).
Characteristics for AML genetic changes include structural chromosomal aberrations (translocations, inversions), leading to the formation of gene fusions. The fusion genes present in leukemia cells undergo expression that cause malfunctioning of their protein products (4–6). Common genetic abnormalities in AML with high frequency and well-established favorable prognostic significance include t(8;21)(q22;q22) (RUNX1-RUNX1T1), inv(16) (p13;q22) (CBFB-MYH11), and t(15;17)(q24;q21) (PML-RARA). A basic diagnostic tool for detecting these changes is cytogenetic analysis using the karyotyping technique, supported by fluorescence in situ hybridization (FISH). However, nowadays, molecular biology methods are increasingly used to detect transcripts of specific fusion genes in patient's leukemia cells as an important part of the diagnostic process. Molecular techniques have found wide diagnostic application in the detection of not only the best known AML-related fusion genes but also the detection of fusion partners, for example, after the MLL (present KMT2A) gene rearrangement (7).
A variety of recurrent chromosomal rearrangements involving 11q23/MLL gene have been reported in adult and pediatric acute leukemias. So far, more than 135 different fusion partners of MLL gene have been identified in acute leukemias (8). Most common for pediatric AML are translocations t(9;11)(p21.3;q23.3) (MLL-MLLT3), t(10;11)(p12;q23.3) (MLL-MLLT10), t(11;19)(q23.3;p13.1) (MLL-ELL), and t(6;11)(q27;q23.3) (MLL-AFDN) (7–9). The prognostic impact of MLL rearrangement depends on detected fusion partner but is not always clearly defined. However, the presence of translocation t(10;11)(p12;q23) is usually associated with a poor prognosis (3, 7, 9).
Since June 2015, the Polish Pediatric Leukemia/Lymphoma Study Group has applied the therapeutic protocol for pediatric AML requiring extensive diagnostics of genetic changes in leukemic cells. For all patients, conventional and molecular cytogenetics analyses were recommended—classical karyotype and FISH analyses for the most common translocation in AML: t(8; 21)(q22; q22)/RUNX1-RUNX1T1, inv(16)(p13; q22)/CBFB-MYH11, t(15;17)(q24; q21)/PML-RARA, and MLL rearrangements by break-apart FISH probes. Molecular analyses were also performed to screen for further, less common transcripts of fusion genes founded in leukemia, as well as mutation in WT1, FLT3, NPM1, CEBPA, and GATA1 genes. Until November 2019, molecular genetic studies were performed in 195 children with de novo diagnosed AML. This represents nearly all pediatric patients with AML in Poland during this period.
One of the performed molecular analyses was genotyping for carriers of fusion gene transcripts for 28 most common chromosomal translocations with prognostic significance in leukemia by a CE-marked (approved for use in Europe) in vitro diagnostic test based on multiplex reverse transcription–polymerase chain reaction (RT-PCR) screening assay, HemaVision-28N RT-PCR (DNA Diagnostic A/S, Risskov, Denmark). The detailed information about 28 leukemia-causing translocations detected by HemaVision-28N is presented in Supplement Table 1. Diagnostic tests were conducted on mRNA samples isolated from mononuclear cells from bone marrow collected before the treatment. For this purpose, bone marrow samples were centrifuged in a density gradient (Histopaque-1077; Sigma-Aldrich, St. Louis, MO, USA), and the obtained bone marrow mononuclear cells were lysed in TRI Reagent Solution (Thermo Fisher Scientific Baltics, Vilnius, Lithuania). The isolation of total RNA was performed according to the manufacturer's instructions. Next, 2 μg of non-degraded RNA was used as a template for synthesis of cDNA in RT reaction (SuperScript™ II Reverse Transcriptase; Thermo Fisher Scientific). HemaVision RT-PCR assay used cDNA as a template for multiplex PCR amplification reactions, followed by nested PCR reactions. All PCR reactions were performed with primer mixes from the HemaVision HV01-28N kit and Multiplex PCR Master Mix (Eurx, Gdansk, Poland). The final PCR products were analyzed by agarose gel electrophoresis.
All children were treated according to the same therapeutic protocol—AML BFM 2012/2019. The genetic tests and individual persons' data used in publication were obtained and processed according to written informed consent obtained from guardians of all patients in accordance with the Declaration of Helsinki. The study was approved by the Ethical Committee of the Jagiellonian University in Krakow (KBET 122.6120.17.2015, dated January 29, 2015). The data that support the findings of the study are available on request from the corresponding author.
In the performed molecular genetic studies, the presence of fusion genes transcripts was revealed in 46.7% (91/195 patients) (Figure 1). The fusion gene transcript for translocation t(10;11)(p12;q23) (MLL-MLLT10) was detected in 8.9% (17/195) of cases with surprisingly the highest frequency among all other marked chromosomal abnormalities determined in HemaVision-28N. Slightly lower frequency was observed for fusion transcripts generated by chromosomal translocation t(8;21) (q22;q22) (RUNX1-RUNX1T1) (8.2%; 16 patients) and t(9;11)(p22:q23) (MLL-MLLT3) (7.7%; 15 patients). Furthermore, other AML-characteristic transcripts related to 11q23/MLL rearrangements, such as t(11;19)(q23;p13.1), t(6;11)(q27;q23), and t(4;11)(q21;q23), were also found. Together, five different fusion gene transcripts for 11q23/MLL rearrangements were detected in 43 patients (22.1%).
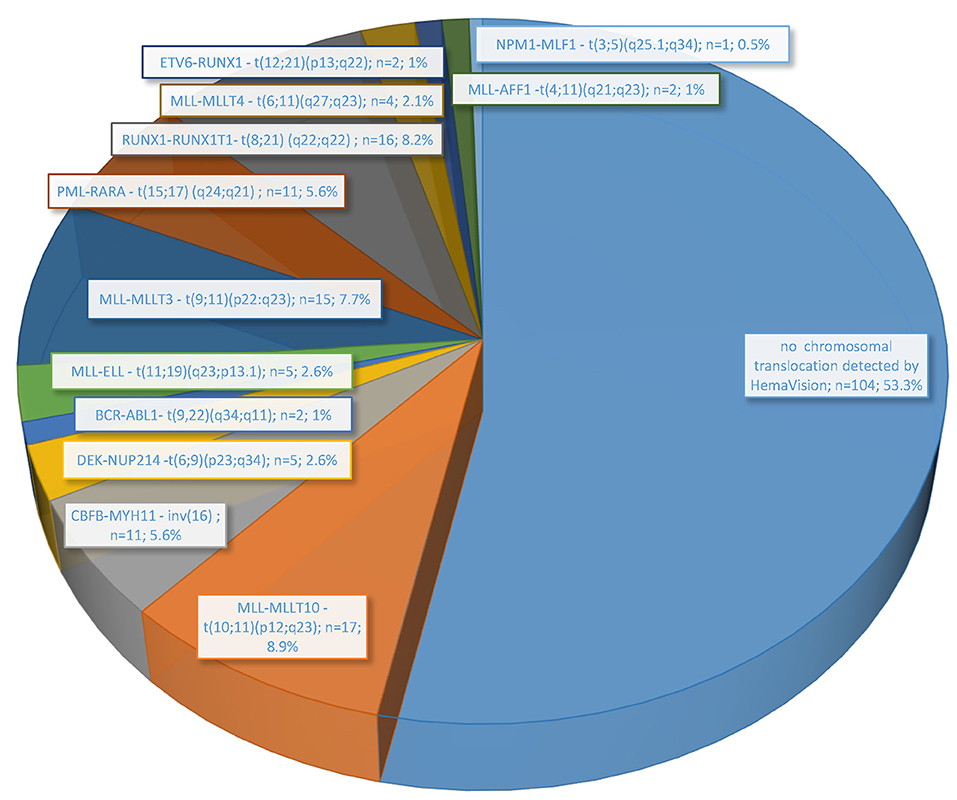
Figure 1. Frequency of chromosomal translocations associated with leukemia detected by molecular methods (HemaVision RT-PCR; DNA Diagnostic A/S) in 195 pediatric AML patients treated in Poland from June 2015 to November 2019.
For 14 of 17 children with molecularly confirmed of MLL-MLLT10 fusion transcript, the results of karyotype and FISH analysis were obtained, and a summary is shown in Table 1. In some cases, the karyotype result indicated other position of chromosome breaks than expected due to molecular result, especially in the case of chromosome 10 (p11, p13, p?). Pediatric karyotypes always pose a great diagnostic challenge due to achieved resolution and G-band quality. Obtained cytogenetic results (karyotype, FISH) were performed in local laboratories, and molecular verification was carried out in a reference laboratory. No correction of chromosome breakpoints assessed in the classical karyotype after molecular analysis results was performed.
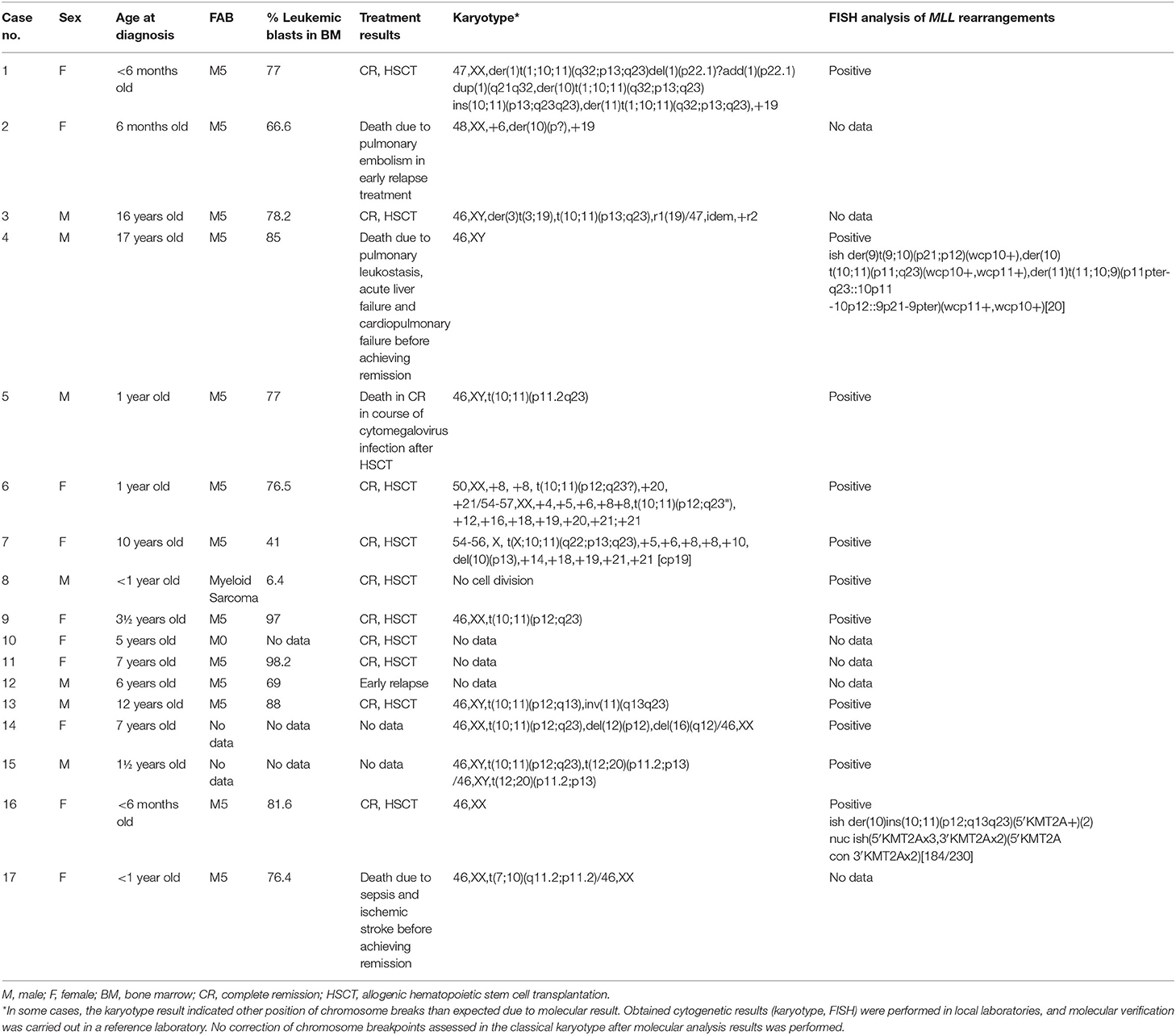
Table 1. Clinical and genetic characteristics of 17 children with diagnosed AML and confirmed t(10;11)(p12;q23) fusion gene transcript in the molecular genetic study.
In four of those 14 patients, only rearrangements in chromosomes 10 and 11 were observed (cases 5, 9, 8, and 16, for whom rearrangements of MLL gene were confirmed only by FISH). For the next seven cases, additional chromosomal abnormalities or complex karyotype was detected (cases 1, 3, 6, 7, 13, 14, and 15; the results in detail in Table 1). For patient 4, the karyotyping result was normal, but in a metaphase FISH, the variant translocation with chromosomes 11, 10, and 9 was confirmed (cryptic rearrangement). Moreover, variant translocations were also observed in two other cases—patient 1: t(1;10;11) and patient 7: p t(X;10:11). It should be emphasized that in all these cases the fusion gene transcript for MLL-MLLT10 was detected in molecular studies. For the last two children (cases 2 and 17), the presence of translocation t(10;11)(p12;q23) was not simply confirmed; however, abnormalities in chromosome 10 were indicated in both cases. Unfortunately, no data from FISH analysis were available for them. In the case of low G-band quality and resolution in the karyotype analysis for children with leukemia, such results do not exclude rearrangement of chromosomes 10 and 11 for patient 2 or cryptic variant translocation of chromosomes 7, 10, and 11 for patient 17.
The clinical characteristics of 17 presented children with diagnosed AML and molecular confirmation of MLL-MLLT10 fusion gene transcript are presented in Table 1. All of the children were treated according to the AML-BFM 2012/2019−10 girls and 7 boys with median age of 3.6 years (range, 4 months to 16.2 years). In four of them, hyperleukocytosis (91–245 × 109/L) was observed at diagnosis. Ten patients achieved complete remission (CR) and underwent allogenic hematopoietic stem cell transplantation (HSCT). Two patients died early before achieving remission—the first because of sepsis and ischemic stroke 1.4 months after diagnosis and the second one due to pulmonary leukostasis, acute liver failure, and cardiopulmonary failure 13 days after final diagnosis of AML. Two further patients had an early relapse; one of them died because of pulmonary embolism. One patient died in CR in course of cytomegalovirus infection 2.6 months after HSCT. Median observation time was 11.5 months (range, 0.4–49.7 months). For two patients, no follow–up data were available.
The presence of t(10;11)(p12;q23) translocation was previously identified in pediatric AML and considered as an unfavorable prognostic factor, but it was observed rarely (1, 3, 8–11). This rearrangement is described in the latest large cohort studies as the second most common translocation detected in pediatric 11q23/MLL-rearranged AML, after t(9;11)(p22;q23) (8). Its occurrence is being estimated at 3% (3). Surprisingly, in our research, the fusion gene transcript resulting from t(10;11)(p12;q23) frequency (8.9%) was not only the highest in the case of all detected MLL rearrangements but also in relation to other common chromosomal translocation in AML, among others t(8;21)(q22;q22) (RUNX1-RUNX1T1)-−8.2% (16/195 patients). Similar results were presented by Stasevich et al. (12), who confirmed the presence of the t(10;11)(p12;q23) at a very high level in children treated for AML in 1 year at Minsk Oncology Center. In the mentioned article, five children with of t(10;11)(p12;q23) translocation were being described among 18 pediatric patients. Worth noting, all of five children do not have the simple reciprocal translocation; in all cases, MLL-MLLT10 fusion was created by several rearrangements. Also, in our research, in 3 of 13 children with karyotype result, variant translocation of 11q23/MLL was revealed [t(1;10;11), t(11;10;9), t(X;10;11)]. Therefore, it seems that in some cases the ambiguity and complexity of changes detected by cytogenetic analysis do not always allow for a correct and rapid assessment of the presence of rearrangement of chromosomes 10 and 11 in leukemic blasts. Hence, the molecular genetic studies seem to be a more appropriate technique to unequivocal assessment of the presence of MLL-MLLT10 fusion transcript, especially in a case of possible missing of this chromosomal aberration during karyotyping (two children with normal karyotype reported) or when no karyotyping result is obtained.
In the presented group of 17 patients with positive molecular results for MLL-MLLT10, in three patients (patients 10–12 in Table 1), there is no cytogenetic confirmation of the results because of the lack of access to archival data. For the next two patients (patients 2 and 17), only the karyotype analysis was performed, with the results not indicating the presence of MLL rearrangement. For this reason, no FISH analysis was performed in these cases. However, in both these cases, chromosome breakage in 10p was observed [48,XX,+6,der(10)(p?),+19 and 46,XX,t(7;10)(q11.2;p11.2)/46,XX]. In the context of a positive molecular result for the MLL-MLLT10 fusion gene transcript, the presence of more complex chromosomal rearrangements, including chromosome 11 rearrangement, may be possible in these patients (cryptic or variant translocation). This also indicates the need for obligatory FISH for MLL rearrangements in all AML patients.
Therefore, because of the large amount of molecular diagnostic tests performed with the HemaVision-28N RT-PCR IVD assay (for 195 children diagnosed with AML) and cytogenetic confirmation of the obtained result in the 12 presented cases, also the molecular results for this group of five patients without direct cytogenetic confirmation were considered reliable and included in the analysis.
According to the recommendations of an international expert panel, the patients with translocation t(10;11)(p12;q23) should be allocated to the high-risk group of pediatric AML treatment protocols (3). The MLL gene encodes a histone methyltransferase that positively regulates homeobox (HOX) gene expression (13). The presence of MLL-MLLT10 fusion protein observed in patients with t(10;11)(p12;q23) in leukemic blasts affects the histone methylation ability and dysregulation of HOXA and MEIS1 expression, finally leading to MLL rearrangement-dependent leukemia (14). It cannot be excluded that the presence of the MLL-MLLT10 fusion transcript detected by molecular methods in the case of variant chromosomal rearrangements or complex karyotype does not have an equally significant negative effect on cell function. However, further research is required in this matter. To sum up, it seems that fusion gene transcript resulting from t(10;11)(p12;q23) can be relatively frequent in some populations, or its frequency may be underestimated because of the difficulty of its cytogenetic assessment. Paying attention to this fact and ensuring proper genetic diagnosis seem to be important for appropriate allocation of patients to risk groups of pediatric AML treatment protocols.
The datasets generated for this study are available on request to the corresponding author.
The studies involving human participants were reviewed and approved by Ethical Committee of the Jagiellonian University in Krakow. Written informed consent to participate in this study was provided by the participants' legal guardian/next of kin.
TK, MCz, and WBal: study concept and design, acquisition of data, analysis and interpretation of data, drafting of the manuscript, and critical revision of the manuscript for important intellectual content. PK, BS, and ML: analysis and interpretation of data and critical revision of the manuscript for important intellectual content. KP-W: analysis and interpretation of data assistance, acquisition and accumulation of data, and critical revision of the manuscript for important intellectual content. MB-M and SS: critical revision of the manuscript for important intellectual content. BS-F, MM, JS-S, JW, AR-K, AC, KM-R, MK-R, DG, JK, LM-K, EA-D, WM, RT, TS, JP, GK, AM-M, KM, WBad, KZ, TU, IK-D, MW, and MCi: acquisition of data. All authors contributed to the article and approved the submitted version.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors wish to thank the colleagues in the pediatric oncology centers from Polish Pediatric Leukemia/Lymphoma Study Group (PPLLSG) for their participation in providing the clinical data and the biological material for genetic research.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fped.2020.00278/full#supplementary-material
1. von Neuhoff C, Reinhardt D, Sander A, Zimmermann M, Bradtke J, Betts DR, et al. Prognostic impact of specific chromosomal aberrations in a large group of pediatric patients with acute myeloid leukemia treated uniformly according to trial AML-BFM 98. J Clin Oncol. (2010) 28:2682–9. doi: 10.1200/JCO.2009.25.6321
2. Meshinchi S, Arceci RJ. Prognostic factors and risk-based therapy in pediatric acute myeloid leukemia. Oncologist. (2007) 12:341–55. doi: 10.1634/theoncologist.12-3-341
3. Creutzig U, van den Heuvel-Eibrink MM, Gibson B, Dworzak MN, Adachi S, de Bont E, et al. Diagnosis and management of acute myeloid leukemia in children and adolescents: recommendations from an international expert panel. Blood. (2012) 120:3187–205. doi: 10.1182/blood-2012-03-362608
4. Pui CH, Carroll WL, Meshinchi S, Arceci RJ. Biology, risk stratification, and therapy of pediatric acute leukemias: an update. J Clin Oncol. (2011) 29:551–65. doi: 10.1200/JCO.2010.30.7405
5. Manola KN. Cytogenetics of pediatric acute myeloid leukemia. Eur J Haematol. (2009) 83:391–405. doi: 10.1111/j.1600-0609.2009.01308.x
6. Braoudaki M, Tzortzatou-Stathopoulou F. Clinical cytogenetics in pediatric acute leukemia: an update. Clin Lymphoma Myeloma Leuk. (2012) 12:230–7. doi: 10.1016/j.clml.2012.04.004
7. Balgobind BV, Zwaan CM, Pieters R, Van den Heuvel-Eibrink MM. The heterogeneity of pediatric MLL-rearranged acute myeloid leukemia. Leukemia. (2011) 25:1239–48. doi: 10.1038/leu.2011.90
8. Meyer C, Burmeister T, Gröger D, Tsaur G, Fechina L, Renneville A, et al. The MLL recombinome of acute leukemias in 2017. Leukemia. (2018) 32:273–84.
9. Balgobind BV, Raimondi SC, Harbott J, Zimmermann M, Alonzo TA, Auvrignon A, et al. Novel prognostic subgroups in childhood 11q23/MLL-rearranged acute myeloid leukemia: results of an international retrospective study. Blood. (2009) 114:2489–96. doi: 10.1182/blood-2009-04-215152
10. Meyer C, Hofmann J, Burmeister T, Gröger D, Park TS, Emerenciano M, et al. The MLL recombinome of acute leukemias in 2013. Leukemia. (2013) 27:2165–76.
11. DiNardo CD, Tang G, Pemmaraju N, Wang SA, Pike A, Garcia-Manero G, et al. Acute myeloid leukemia with t(10;11): a pathological entity with distinct clinical presentation. Clin Lymphoma Myeloma Leuk. (2015) 15:47–51. doi: 10.1016/j.clml.2014.06.022
12. Stasevich I, Utskevich R, Kustanovich A, Litvinko N, Savitskaya T, Chernyavskaya S, et al. Translocation (10;11)(p12;q23) in childhood acute myeloid leukemia: incidence and complex mechanism. Cancer Genet Cytogenet. (2006) 169:1140. doi: 10.1016/j.cancergencyto.2006.03.011
13. Benjamin DY, Hess JL, Horning SE, Brown GA, Korsmeyer SJ. Altered Hox expression and segmental identity in Mll-mutant mice. Nature. (1995) 378:505–8. doi: 10.1038/378505a0
Keywords: acute myeloid leukemia, 11q23/KMT2A rearrangements, MLL rearrangements, children, risk stratification, treatment results
Citation: Ksiazek T, Czogala M, Kaczowka P, Sadowska B, Pawinska-Wasikowska K, Bik-Multanowski M, Sikorska-Fic B, Matysiak M, Skalska-Sadowska J, Wachowiak J, Rodziewicz-Konarska A, Chybicka A, Muszynska-Rosłan K, Krawczuk-Rybak M, Grabowski D, Kowalczyk J, Maciejka-Kemblowska L, Adamkiewicz-Drozynska E, Mlynarski W, Tomaszewska R, Szczepanski T, Pohorecka J, Karolczyk G, Mizia-Malarz A, Mycko K, Badowska W, Zielezinska K, Urasinski T, Karpinska-Derda I, Woszczyk M, Ciebiera M, Lejman M, Skoczen S and Balwierz W (2020) High Frequency of Fusion Gene Transcript Resulting From t(10;11)(p12;q23) Translocation in Pediatric Acute Myeloid Leukemia in Poland. Front. Pediatr. 8:278. doi: 10.3389/fped.2020.00278
Received: 15 January 2020; Accepted: 04 May 2020;
Published: 10 July 2020.
Edited by:
Daniele Zama, Sant'Orsola-Malpighi Polyclinic, ItalyReviewed by:
Yana Pikman, Dana–Farber Cancer Institute, United StatesCopyright © 2020 Ksiazek, Czogala, Kaczowka, Sadowska, Pawinska-Wasikowska, Bik-Multanowski, Sikorska-Fic, Matysiak, Skalska-Sadowska, Wachowiak, Rodziewicz-Konarska, Chybicka, Muszynska-Rosłan, Krawczuk-Rybak, Grabowski, Kowalczyk, Maciejka-Kemblowska, Adamkiewicz-Drozynska, Mlynarski, Tomaszewska, Szczepanski, Pohorecka, Karolczyk, Mizia-Malarz, Mycko, Badowska, Zielezinska, Urasinski, Karpinska-Derda, Woszczyk, Ciebiera, Lejman, Skoczen and Balwierz. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Szymon Skoczen, c3p5bW9uLnNrb2N6ZW5AdWouZWR1LnBs
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.