
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Oncol. , 10 March 2020
Sec. Cancer Molecular Targets and Therapeutics
Volume 10 - 2020 | https://doi.org/10.3389/fonc.2020.00259
This article is part of the Research Topic Targeting the Microenvironment Niche in Solid Tumors View all 13 articles
Integrin, beta-like 1 (ITGBL1) protein is located in the extracellular matrix (ECM) and involved in the development and metastasis of many tumors. However, the regulatory mechanism of ITGBL1 in colorectal cancer (CRC) remains unclear. This study was to analyze the expression profile of CRC and to identify the expression change of ITGBL1 gene at different stages of CRC. Survival analysis showed that ITGBL1 was related to the metastasis of CRC, and CRC patients with a high expression of ITGBL1 had earlier metastasis. Gene Set Enrichment Analysis (GSEA) indicated the relationship between ITGBL1 expression and molecular events of CRC. The results indicated that a high expression of ITGBL1 was linked to Wnt signaling pathway, cell polarity, and tissue development, while a low expression of ITGBL1 was related to cellular respiration, electron transfer chain, and oxidative phosphorylation. With the expression profiles from interstitial and parenchyma CRC tissues, a comparison was made to determine the difference between high/low expression of ITGBL1 and Wnt signaling pathway, respectively, and further confirmed the close relation between ITGBL1 and Wnt signaling pathway. To determine the relation, an interaction network of ITGBL1 and Wnt signaling proteins was constructed. It was found that β-catenin interacted with multiple extracellular Wnt signals and could bind to ITGBL1. As a result, the regulatory mechanism of ITGBL1 in CRC is related to extracellular Wnt signals and may affect extracellular Wnt signals via β-catenin.
Integrin, beta-like 1 (ITGBL1) is a β-integrin-related extracellular matrix (ECM) protein. Recently, studies on ITGBL1 have been increasing, and it was reported that ITGBL1 could promote bone metastasis of breast cancer through transforming growth factor (TGF)-β signaling pathway (1). Studies showed that ITGBL1 could promote the invasion of ovarian cancer cell through Wnt/planar cell polarity (PCP) signaling and focal adhesion kinase (FAK)/Src pathway (2), and high expression of ITGBL1 was related to the poor prognosis and drug resistance of ovarian cancer (3). In gastric cancer, ITGBL1 was linked to epithelial–mesenchymal transition (EMT) phenotype and poor prognosis (4). Studies reported that hypermethylation of ITGBL1 was correlated with poor prognosis of acute myeloid leukemia (5). Furthermore, studies also revealed that ITGBL1 could activate nuclear factor (NF)-κB signaling pathway and promote the EMT, invasion, and migration of prostatic cancer (6). The abovementioned studies revealed that ITGBL1 was associated with the invasion and metastasis of tumors. It was reported that ITGBL1 was significantly upregulated in CRC, and its high expression was related to shortened survival of CRC patients. Additionally, knockdown of ITGBL1 suppressed CRC cell proliferation, migration, and invasion (7). Another study showed that ITGBL1 was associated with the overall survival rate (OSR) and relapse-free survival (RFS) of CRC patients, and subgroup validation demonstrated that a high ITGBL1 expression was correlated with shorter RFS in stage II patients, which suggested that ITGBL1 was a promising candidate biomarker for predicting the relapse of CRC (8). In the earlier study, a comparison of CRC expression data from normal to distant organ metastasis (normal, stage I, II, III, IV, liver, and lung metastasis) was made. There was screening of 39 genes with continuously increasing expression which contained ITGBL1 (9). These studies showed that ITGBL1 played a vital role in the development of CRC. ITGBL1 was involved in the formation of tumor microenvironment, but the molecular mechanism of ITGBL1 in CRC remained unclear. Therefore, this study aims to analyze the molecular mechanism of ITGBL1 in CRC and determine the regulatory mechanism of ITGBL1 in the metastasis of CRC.
It was found previously that the expression of ITGBL1 was continually increasing in CRC. In this study, CRC expression profiles GSE41258 (54 normal cases, 28 stage I cases, 50 stage II cases, 49 stage III cases, 58 stage IV cases, 47 liver metastasis cases, and 20 lung metastasis cases) from the Gene Expression Omnibus (GEO) database (10) which showed the expression of ITGBL1 at different stages of CRC were analyzed. Additionally, we confirmed the expression of ITGBL1 through expression profile GSE49355 and RNA-sequencing GSE50760. GSE49355 included 18 normal cases and 20 CRC cases with primary focus and 19 CRC cases with liver metastasis. GSE50760 included 18 normal cases, 18 CRC cases with primary focus, and 18 CRC cases with liver metastasis. ITGBL1 gene expression in that data was obtained and divided into different groups according to tumor progression. The comparison of the difference in each group was performed by one-way ANOVA test, and P-value was calculated by Kruskal–Wallis test.
ITGBL1 gene expression was continuously increasing in CRC, and many studies reported that ITGBL1 was associated with tumor metastasis. Therefore, a survival analysis of ITGBL1 gene expression and CRC metastasis was conducted by using expression profile GSE28722 from the GEO database. GSE28722 included 125 CRC cases with data of survival times and metastasis. ITGBL1 gene expression in those data was obtained and divided into the high ITGBL1 expression group (n = 62) and the low ITGBL1 expression group (n = 62) according to a median value (deleting median). Kaplan–Meier curve (11) was used to depict the survival curve of the two groups, and log rank test (12) was performed to analyze the statistical difference between the two groups with the P-value calculated. Considering the American Joint Committee on Cancer (AJCC) staging and the effect of patients' age (taking 60-year-olds as the dividing point) on metastasis, and Cox proportional-hazards regression was performed to further determine the effect of ITGBL1 on metastasis.
To clarify the molecular mechanism of ITGBL1 in CRC, an analysis of the dataset GSE39582 from the GEO database was carried out. GSE39582 included 566 CRC cases, which were divided into the high ITGBL1 expression group (n = 283) and the low ITGBL1 expression group (n = 283) according to the median value of ITGBL1 expression (probe ID: 205422_s_at) based on the Gene Ontology (including biological process, molecular function, and cellular component) and signaling pathway (including KEGG pathway and REACTOME pathway). Enrichment analysis was performed on the high ITGBL1 expression group and the low ITGBL1 expression group by using Gene Set Enrichment Analysis (GSEA) [false discovery rate (FDR) < 25%, nominal p < 1%], with the version of gene set as V7.0 (13). ITGBL1 was mainly located in the ECM and related to the tumor microenvironment, so GSE35602 was used to screen the ITGBL1-related differentially expressed gene (DEG) in the parenchyma and interstitial of CRC. GSE35602 included 12 cases with CRC parenchymal tissue data and 12 cases with CRC interstitial tissue data. According to the median value of ITGBL1 expression (probe ID: A_23_P408363), ITGBL1 expression data from CRC parenchymal tissue and interstitial tissue were divided in the high ITGBL1 expression group (n = 6) and the low ITGBL1 expression group (n = 6), respectively. GEO2R (14) was used to screen DEG in the high ITGBL1 expression group and the low ITGBL1 expression group with the P-value limited < 0.01 and the fold change as 4. Based on the DEG, ITGBL1-related molecular events in parenchymal and interstitial tissues were examined to elicit the differences between them.
GSE41258, GSE49355, and GSE50760 showed the expression change of ITGBL1 during the progression of CRC. The results showed a significant difference of ITGBL1 expression at different stages of CRC (P < 0.0001, P = 0.0003, and P < 0.0001). ITGBL1 expression was continuously increasing with the development of CRC (Figures 1A–C), suggesting that ITGBL1 played an important role in the development and metastasis of CRC.
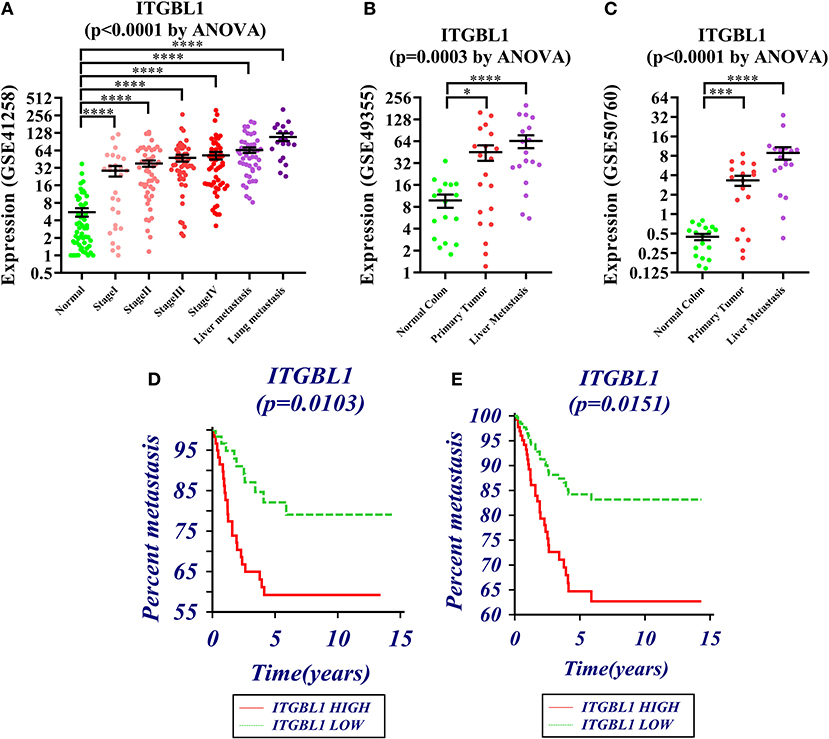
Figure 1. (A–C) Expression changes of ITGBL1 in the development and metastasis of colorectal cancer (CRC) (*P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001). (D) Survival curve of ITGBL1 and the metastasis time of CRC patients (Kaplan–Meier survival estimate and univariable survival analysis model). (E) Survival curve of ITGBL1 and the metastasis time of CRC patients (Cox proportional hazards regression model and multivariate analysis model, excluding the influence of tumor stages and ages on metastasis time).
The analysis on the association between ITGBL1 expression and the metastasis of CRC patients through dataset GSE28722 showed that ITGBL1 expression was correlated with the metastasis of CRC (log rank P = 0.0103) [hazard ratio (HR) = 0.3924 (95% CI: 0.1982–0.7772)] (Figure 1D). ITGBL1 expression was negatively related to the metastasis-free survival, and a high expression of ITGBL1 could promote the metastasis of CRC. Cox proportional-hazards regression showed that ITGBL1 was an independent factor of CRC metastasis (P = 0.0151). Patients in the high ITGBL1 expression group had a higher risk of metastasis than that of the low ITGBL1 expression group [HR = 2.5345 (95% CI: 1.2012–5.3477)] (Figure 1E).
GSEA was performed on the high ITGBL1 expression group and the low ITGBL1 expression group based on the GSE39582 dataset. It was found that in the high ITGBL1 expression group, top 20 of biological process in enrichment were mainly related to Wnt signaling pathway, cell polarity, tissue development, axon, and morphogenesis. Cellular component was mainly associated with cell matrix junction, cytoskeleton, and cell membrane. Molecular function was related to ECM binding, adhesion molecule binding, integrin binding, and FRIZZLED protein binding. Signaling pathway related to high ITGBL1 expression was mainly linked to cytoskeleton, Wnt signaling pathway, pathway in cancer, and pathway related to tumor development. In the low ITGBL1 expression group, top 20 of biological process in enrichment were mainly related to cellular respiration, electron transfer chain, and oxidative phosphorylation (see Supplementary Material). Those results demonstrated that ITGBL1 was involved in various molecular events in the progression of CRC and was mainly related to cell adhesion. Enrichment analysis showed that Wnt signaling pathway in the high ITGBL1 expression group occurred frequently in the biological process, which suggested that ITGBL1 might be closely related to Wnt signaling pathway.
DEG screening was performed on CRC parenchymal tissue and interstitial tissue based on the dataset GSE35602. In parenchymal tissue, resulting in 137 upregulated genes (SERP4 had the highest significant difference) and 17 downregulated genes in the high ITGBL1 expression group. In interstitial tissue, there were 343 upregulated genes (SERP2 and SERP4 had the highest significant difference) and 38 downregulated genes in the high ITGBL1 expression group. GSEA was used to analyze 12 cases with CRC interstitial tissue data, and the results were similar to GSE39582 analysis. This further confirmed the accuracy of the enrichment analysis. In enrichment result of molecular function, Wnt protein binding ranked first in the high ITGBL1 expression group, which proved the close association between ITGBL1 expression and Wnt signaling pathway in the interstitial tissue of CRC. Based on the GSEA analysis of GSE39582, 31 genes with enrichment function in KEGG_WNT_SIGNALING_PATHWAY were obtained. Intersection was performed between those 31 genes and upregulated genes in the high ITGBL1 expression group in CRC parenchymal tissue and interstitial tissue, respectively. In interstitial tissue, seven intersected genes (SFRP2, WNT2, FZD1, FZD7, FZD8, SFRP4, and DKK2) were attained. In parenchymal tissue, two intersected genes (SFRP4 and DKK2) were obtained. So, SFRP4 and DKK2 were significantly expressed in both parenchymal tissue and interstitial tissue. The encoded proteins of those genes were mainly located in the cell membrane or ECM as the extracellular signal protein of Wnt signaling pathway. Co-expression analysis was performed between the above seven genes and ITGBL1 using 222 microarray data of CRC obtained in The Cancer Genome Atlas (TCGA) (15) by using cBioPorta (16). It was observed that SFRP4 was the most relevant gene with ITGBL1 expression, and other genes were also correlated with ITGBL1 in varying degrees (Figure 2). PrePPI was performed to analyze the interaction relationship between ITGBL1 and the seven proteins. Cytoscape (17) was applied to construct protein–protein interaction network by selecting proteins with combining probability more than 0.8. It is found that six out of seven of those genes (proteins) could be bound to ITGBL1 indirectly, and CTNNB1 (β-catenin) was the key protein (Figure 3). Hence, those six Wnt-related genes were upregulated in the high ITGBL1 expression group and had co-expression relationship with ITGBL1. The encoded proteins of those six genes could bind to CTNNB1, and CTNNB1 could bind to ITGBL1, which suggested that CTNNB1 played an important role in the regulatory mechanism of ITGBL1 in the metastasis of CRC. According to the GSEA analysis of GSE39582, REACTOME_BETA_CATENIN_INDEPENDENT_WNT_ SIGNALING ranked first in the enrichment of REACTOME, which also suggested the important role of CTNNB1 in the relationship of ITGBL1 and Wnt signaling pathway.

Figure 2. Co-expression relationship of ITGBL1 and seven extracellular Wnt signals. Y axis in this figure represented mRNA relative expressions of these seven extracellular Wnt signals, and X axis represented mRNA relative expressions of ITGBL1. The red line indicated regression line.

Figure 3. Interaction network of ITGBL1 and extracellular Wnt signal. The red nodes (SFRP2, FZD1, FZD7, FZD8) were extracellular Wnt signals that were upregulated in the stroma. The purple nodes (SFRP4, DKK2) were extracellular Wnt signals that were upregulated in both the stroma and parenchyma. The blue nodes (FN1, CTNNB1, COL1A1) were all ITGBL1 binding proteins, and they also bound to extracellular Wnt signal. The green nodes were ITGBL1 binding proteins. The yellow node was the ITGBL1 protein.
ECM is closely related to the invasion and metastasis of tumors and involved in the formation of the tumor microenvironment. ITGBL1 is located in the ECM and related to the tumor microenvironment. This study analyzed the expression change of ITGBL1 at different stages of CRC and determined that ITGBL1 expression was associated with the metastasis of CRC. By analyzing the expression profile of CRC, it was discovered that ITGBL1 was closely related to extracellular Wnt signals (SFRP2, WNT2, FZD1, FZD7, FZD8, SFRP4, and DKK2) via CTNNB1. CTNNB1 (β-catenin) is a key protein of Wnt signals and linked to the development of tumor (18). As reported, CTNNB1 could affect autophagy in glioblastoma, and cell autophagy was related to the tumor microenvironment (19) and was also involved in adrenocortical carcinomas (20). Molecular mechanism related in CRC was correlated with CTNNB1; for instance, genetic variations of the CTNNB1 were related to the progression of CRC (21). SNX3 could inhibit the metastasis of CRC via downregulating β-catenin (22), miR-150 could suppress the metastatization process of CRC by inhibiting β-catenin (23). FOXM1 could promote the growth and metastatization process of CRC by activating β-catenin (24). In the serum of patients with CRC, the expression of β-catenin was higher than that in normal people (25). In addition to CTNNB1, this study also found that extracellular Wnt signals (SFRP2, WNT2, FZD1, FZD7, FZD8, SFRP4, and DKK2) were correlated with ITGBL1 expression. Those proteins and ITGBL1 were cell membrane or extracellular proteins and had higher relation with ITGBL1 in the interstitial tissue of CRC. Those proteins were closely related to the development of CRC. For instance, methylation could inhibit gene expression and hypermethylation of SERP2 and was negatively associated with the invasion of CRC (26). Depletion of WNT2 could inhibit CRC (27), while cancer-associated fibroblasts (CAFs)-derived WNT2 could promote the progression of CRC. Moreover, FZD8 might be a WNT2 receptor (28, 29), while miR-375 could suppress human CRC metastasis by inhibiting FZD8 (30). FZD1 is a Wnt responsive gene in colon-derived tissues which were expressed in CRC, and paracancerous normal mucosa was involved in Wnt signaling within the tumor microenvironment (31). RNA interference-mediated silencing of FZD7 inhibited invasion in CRC, and its expression was associated with the activation of Wnt signaling (32). High expression of SFRP4 was correlated with advanced CRC (33), and CRC patients with overexpressed SFRP4 had lower overall survival (34). DKK2 expression accelerates aerobic glycolysis and promotes angiogenesis in CRC (35). Therefore, those extracellular Wnt signals were related to activation of Wnt signaling pathway and the metastatization of CRC. The study established that the regulatory mechanism of ITGBL1 in the development and metastatization of CRC might be closely related to those proteins. Hence, ITGBL1 is closely associated with the metastatization of CRC and involved in the tumor microenvironment.
All datasets generated for this study are included in the article/Supplementary Material.
LQ designed and performed the study, analyzed the data, and wrote the manuscript. FS collected the data and literature. YD revised and approved the final manuscript.
This work was supported by the National Key Basic Research Program of China (973 program, 2015CB554002), the National Natural Science Foundation of China (81972754, 81773101), and the Scientific Research Starting Foundation of Southern Medical University (PY2018N003).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2020.00259/full#supplementary-material
Table S1. GSEA enrichment analysis of the high ITGBL1 expression group and the low ITGBL1 expression group. Screening results of ITGBL1-related differentially expressed genes (DEGs) in parenchyma and interstitial of CRC.
1. Li XQ, Du X, Li DM, Kong PZ, Sun Y, Liu PF, et al. ITGBL1 Is a Runx2 transcriptional target and promotes breast cancer bone metastasis by activating the TGFbeta signaling pathway. Cancer Res. (2015) 75:3302–13. doi: 10.1158/0008-5472.CAN-15-0240
2. Sun L, Wang D, Li X, Zhang L, Zhang H, Zhang Y. Extracellular matrix protein ITGBL1 promotes ovarian cancer cell migration and adhesion through Wnt/PCP signaling and FAK/SRC pathway. Biomed Pharmacother. (2016) 81:145–51. doi: 10.1016/j.biopha.2016.03.053
3. Song J, Yang P, Lu J. Upregulation of ITGBL1 predicts poor prognosis and promotes chemoresistance in ovarian cancer. Cancer Biomark. (2019) 27:51–61. doi: 10.3233/CBM-190460
4. Li R, Zhuang C, Jiang S, Du N, Zhao W, Tu L, et al. ITGBL1 predicts a poor prognosis and correlates EMT phenotype in gastric cancer. J Cancer. (2017) 8:3764–73. doi: 10.7150/jca.20900
5. Lian XY, Ma JC, Zhou JD, Zhang TJ, Wu DH, Deng ZQ, et al. Hypermethylation of ITGBL1 is associated with poor prognosis in acute myeloid leukemia. J Cell Physiol. (2019) 234:9438–46. doi: 10.1002/jcp.27629
6. Li W, Li S, Yang J, Cui C, Yu M, Zhang Y. ITGBL1 promotes EMT, invasion and migration by activating NF-kappaB signaling pathway in prostate cancer. Onco Targets Ther. (2019) 12:3753–63. doi: 10.2147/OTT.S200082
7. Qiu X, Feng JR, Qiu J, Liu L, Xie Y, Zhang YP, et al. ITGBL1 promotes migration, invasion and predicts a poor prognosis in colorectal cancer. Biomed Pharmacother. (2018) 104:172–80. doi: 10.1016/j.biopha.2018.05.033
8. Matsuyama T, Ishikawa T, Takahashi N, Yamada Y, Yasuno M, Kawano T, et al. Transcriptomic expression profiling identifies ITGBL1, an epithelial to mesenchymal transition (EMT)-associated gene, is a promising recurrence prediction biomarker in colorectal cancer. Mol Cancer. (2019) 18:19. doi: 10.1186/s12943-019-0945-y
9. Qi L, Ding Y. Construction of key signal regulatory network in metastatic colorectal cancer. Oncotarget. (2018) 9:6086–94. doi: 10.18632/oncotarget.23710
10. Clough E, Barrett T. The gene expression omnibus database. Methods Mol Biol. (2016) 1418:93–110. doi: 10.1007/978-1-4939-3578-9_5
11. Stel VS, Dekker FW, Tripepi G, Zoccali C, Jager KJ. Survival analysis I: the Kaplan-Meier method. Nephron Clin Pract. (2011) 119:c83–8. doi: 10.1159/000324758
12. Koletsi D, Pandis N. Survival analysis, part 2: Kaplan-Meier method and the log-rank test. Am J Orthod Dentofacial Orthop. (2017) 152:569–71. doi: 10.1016/j.ajodo.2017.07.008
13. Subramanian A, Kuehn H, Gould J, Tamayo P, Mesirov JP. GSEA-P: a desktop application for gene set enrichment analysis. Bioinformatics. (2007) 23:3251–3. doi: 10.1093/bioinformatics/btm369
14. Barrett T, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, et al. NCBI GEO: archive for functional genomics data sets–update. Nucleic Acids Res. (2013) 41:D991–5. doi: 10.1093/nar/gks1193
15. Zhu Y, Qiu P, Ji Y. TCGA-assembler: open-source software for retrieving and processing TCGA data. Nat Methods. (2014) 11:599–600. doi: 10.1038/nmeth.2956
16. Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. (2013) 6:l1. doi: 10.1126/scisignal.2004088
17. Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. (2003) 13:2498–504. doi: 10.1101/gr.1239303
18. Gao C, Wang Y, Broaddus R, Sun L, Xue F, Zhang W. Exon 3 mutations of CTNNB1 drive tumorigenesis: a review. Oncotarget. (2018) 9:5492–508. doi: 10.18632/oncotarget.23695
19. Nàger M, Sallán MC, Visa A, Pushparaj C, Santacana M, Macià A, et al. Inhibition of WNT-CTNNB1 signaling upregulates SQSTM1 and sensitizes glioblastoma cells to autophagy blockers. Autophagy. (2018) 14:619–36. doi: 10.1080/15548627.2017.1423439
20. Maharjan R, Backman S, Åkerström T, Hellman P, Björklund P. Comprehensive analysis of CTNNB1 in adrenocortical carcinomas: identification of novel mutations and correlation to survival. Sci Rep. (2018) 8:8610. doi: 10.1038/s41598-018-26799-2
21. Sygut A, Przybyłowska K, Ferenc T, Dziki Ł, Spychalski M, Mik M, et al. Genetic variations of the CTNNA1 and the CTNNB1 genes in sporadic colorectal cancer in Polish population. Pol Przegl Chir. (2012) 84:560–4. doi: 10.2478/v10035-012-0093-1
22. Pan B, Zhang T, Yang W, Liu Y, Chen Y, Zhou Z, et al. SNX3 suppresses the migration and invasion of colorectal cancer cells by reversing epithelial-to-mesenchymal transition via the beta-catenin pathway. Oncol Lett. (2019) 18:5332–40. doi: 10.3892/ol.2019.10860
23. He Z, Dang J, Song A, Cui X, Ma Z, Zhang Y. The involvement of miR-150/beta-catenin axis in colorectal cancer progression. Biomed Pharmacother. (2020) 121:109495. doi: 10.1016/j.biopha.2019.109495
24. Yang K, Jiang B, Lu Y, Shu Q, Zhai P, Zhi Q, et al. FOXM1 promotes the growth and metastasis of colorectal cancer via activation of beta-catenin signaling pathway. Cancer Manag Res. (2019) 11:3779–90. doi: 10.2147/CMAR.S185438
25. Li S, Huang M, Liu Q, Wang D, Wu R, Zhang X, et al. Serum expression of beta-catenin is a potential detection marker in patients with colorectal cancer. Dis Markers. (2019) 2019:5070524. doi: 10.1155/2019/5070524
26. Yang Q, Huang T, Ye G, Wang B, Zhang X. Methylation of SFRP2 gene as a promising noninvasive biomarker using feces in colorectal cancer diagnosis: a systematic meta-analysis. Sci Rep. (2016) 6:33339. doi: 10.1038/srep33339
27. Jung YS, Jun S, Lee SH, Sharma A, Park JI. Wnt2 complements Wnt/beta-catenin signaling in colorectal cancer. Oncotarget. (2015) 6:37257–68. doi: 10.18632/oncotarget.6133
28. Kramer N, Schmöllerl J, Unger C, Nivarthi H, Rudisch A, Unterleuthner D, et al. Autocrine WNT2 signaling in fibroblasts promotes colorectal cancer progression. Oncogene. (2017) 36:5460–72. doi: 10.1038/onc.2017.144
29. Aizawa T, Karasawa H, Funayama R, Shirota M, Suzuki T, Maeda S, et al. Cancer-associated fibroblasts secrete Wnt2 to promote cancer progression in colorectal cancer. Cancer Med. (2019) 8:6370–82. doi: 10.1002/cam4.2523
30. Xu L, Wen T, Liu Z, Xu F, Yang L, Liu J, et al. MicroRNA-375 suppresses human colorectal cancer metastasis by targeting Frizzled 8. Oncotarget. (2016) 7:40644–56. doi: 10.18632/oncotarget.9811
31. Planutis K, Planutiene M, Nguyen AV, Moyer MP, Holcombe RF. Invasive colon cancer, but not non-invasive adenomas induce a gradient effect of Wnt pathway receptor frizzled 1 (Fz1) expression in the tumor microenvironment. J Transl Med. (2013) 11:50. doi: 10.1186/1479-5876-11-50
32. Ueno K, Hiura M, Suehiro Y, Hazama S, Hirata H, Oka M, et al. Frizzled-7 as a potential therapeutic target in colorectal cancer. Neoplasia. (2008) 10:697–705. doi: 10.1593/neo.08320
33. Kamposioras K, Konstantara A, Kotoula V, Lakis S, Kouvatseas G, Akriviadis E, et al. The prognostic significance of WNT pathway in surgically-treated colorectal cancer: beta-catenin expression predicts for disease-free survival. Anticancer Res. (2013) 33:4573–84.
34. Nfonsam LE, Jandova J, Jecius HC, Omesiete PN, Nfonsam VN. SFRP4 expression correlates with epithelial mesenchymal transition-linked genes and poor overall survival in colon cancer patients. World J Gastrointest Oncol. (2019) 11:589–98. doi: 10.4251/wjgo.v11.i8.589
Keywords: colorectal cancer, integrin, beta-like 1 (ITGBL1), Wnt signaling pathway, tumor microenvironment
Citation: Qi L, Song F and Ding Y (2020) Regulatory Mechanism of ITGBL1 in the Metastasis of Colorectal Cancer. Front. Oncol. 10:259. doi: 10.3389/fonc.2020.00259
Received: 16 October 2019; Accepted: 14 February 2020;
Published: 10 March 2020.
Edited by:
Cirino Botta, Cosenza Hospital, ItalyReviewed by:
Daniele Caracciolo, University Magna Graecia of Catanzaro, ItalyCopyright © 2020 Qi, Song and Ding. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lu Qi, a3V1Z2E4ODhAcXEuY29t; Yanqing Ding, ZHlxQGZpbW11LmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.