- 1Department of Breast and Medical Research Office, Affiliated Cancer Hospital of Zhengzhou University, Zhengzhou, China
- 2College of Public Health, Zhengzhou University, Zhengzhou, China
- 3Medical Record Statistics Office, Affiliated Hospital of Hebei University of Engineering, Handan, China
- 4Department of Infectious Disease, People’s Hospital of Zhengzhou University, Henan Provincial People’s Hospital, Zhengzhou, China
- 5Department of Nosocomial Infection Management, The First Affiliated Hospital of Zhengzhou University, Zhengzhou, China
Background: Numerous studies have demonstrated the presence of microRNA-124 abnormalities involving gene expression, methylation, and single nucleotide polymorphism (SNP) in multiple and diverse cancers, but the prognostic value of these abnormalities in cancer remains inconclusive.
Objective: The aim of this study is to determine the prognostic value of miR-124 in cancer.
Methods: We scrutinized the electronic databases and estimate the association between miR-124 expression, methylation and single nucleotide polymorphisms (SNPs), and prognosis in cancers. The pooled hazard ratios with 95% confidence intervals (CIs) for overall survival (OS), and disease-free survival/recurrence-free survival (RFS)/progression-free survival (PFS) were calculated to estimate the effects of miR-124 expression, methylation, and SNPs on cancer prognosis. The Quality in Prognosis Studies and Newcastle-Ottawa Scale were utilized to assess the quality of included studies.
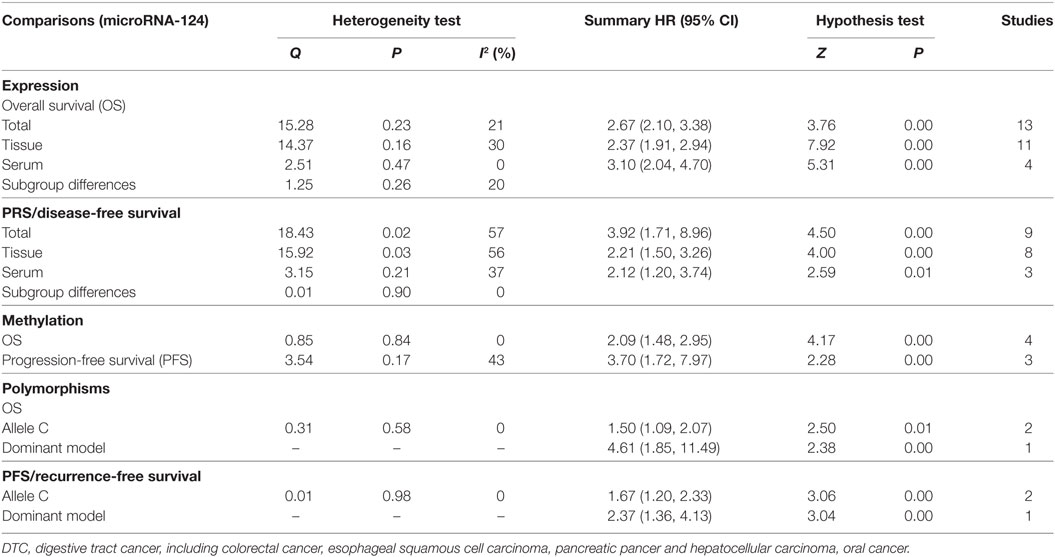
Results: A total of 20 studies involving 3,574 participants were analyzed in evidence synthesis. Our findings showed that the low expression of miR-124 was significantly associated with poor OS (HR = 2.37, 95% CI: 1.91–2.94, P = 0.00; HR = 3.10, 95% CI: 2.04–4.70, P = 0.00) and PFS/RFS (HR = 2.21, 95% CI: 1.50–3.26, P = 0.00; HR = 2.12, 95% CI: 1.20–3.74, P = 0.00). The hyper-methylation of miR-124 was associated with poor OS (HR = 2.09, 95% CI: 1.48–2.95, P = 0.00) and PFS (HR = 3.70, 95% CI: 1.72–7.97, P = 0.00) (Table 3). The patients carrying with Allele C of miR-124 rs5315649 had a worse OS (HR = 1.50, 95% CI: 1.09–2.07, P = 0.00) and PFS (HR = 1.67, 95% CI: 1.20–2.33, P = 0.00) than the carriers with Allele G.
Conclusion: The low expression and hyper-methylation of miR-124 was strongly associated with poor prognosis, and genetic variations of miR-124 rs531564 affected prognosis in cancer patients.
Introduction
MicroRNAs (miRNAs) are small, non-protein-coding RNA molecules involved in RNA silencing and posttranscriptional regulation of gene expression (1, 2). Numerous studies have proved that abnormalities of miRNAs are involved in various cancers, which play important roles in many aspects of carcinogenesis and act as oncogenes or tumor suppressors, including cell differentiation, proliferation, angiogenesis, and metastasis (3–6).
MicroRNAs regulate genes expression by binding to the 3′-untranslated region of target mRNAs (7, 8). Given the high stability of miRNAs in formalin-fixed, paraffin-embedded tissue and circulation, they are increasingly considered as biomarkers for predicting cancer prognosis and treatment response (9–11). Previous studies have demonstrated that miRNAs are aberrantly expressed in various types of cancer and involved in different biological processes, such as differentiation, cell growth, migration, and apoptosis (12).
Human microRNA-124 (miR-124) is encoded by three loci: miR-124-1 (8p23.1), miR-124-2 (8q12.3), and miR-124-3 (20q13.33) (13). MiR-124 is significantly downregulated in various tissues and cell lines of cancer. Overexpression of miR-124 suppresses migration, cell proliferation, and invasion and induces apoptosis by regulating Rac1, indicating that miR-124 plays a tumor suppressive role in various cancer (14–16). It had been demonstrated in diverse cancer types, such as non-small cell lung cancer (NSCLC) (16), hepatocellular carcinoma (17), glioblastoma multiforme (18), gastric cancer (19), ovarian cancer (20), breast cancer (21–24), and colorectal cancer (25). However, little is known about the association between the cancer prognosis and expression levels of miR-124 in tissues or serum.
Aberrant DNA methylation of promoter CpG islands permanently inactivates tumor suppressor genes and is profoundly involved in carcinogenesis, similar to chromosomal abnormalities and mutations (26). Downregulation of miR-124 by promoter methylation has been observed in gastric cancer (27), colorectal cancer (28), prostate cancer (29), cervical and pancreatic cancers (13). Methylation-mediated downregulation of MiR-124 can be observed in 85% of lung cancer patients (30). As a novel risk marker for cancer, the methylation levels of miR-124 and the epidemiological risk of cancer patients need to be specified.
Common genetic polymorphisms in miRNAs and miRNA-processing pathway genes are well established in tumor development and progression (31). Single nucleotide polymorphisms (SNPs) in miRNA-processing pathway genes or miRNAs may alter the transcription and expression of miRNAs and are, therefore, associated with the risks and outcomes of various cancers (32). Since SNPs associated with the risk of cancer may affect prognosis, analysis of relevant SNPs in miRNAs may help to find novel cancer therapeutic targets and prognostic biomarkers (33).
To date, there is no available information on system-based evidence-based medicine for the prognostic value of miR-124. Furthermore, the role of miR-124 in cellular proliferation and invasion of cancer is not fully understood. Numerously, previous studies have few new or insightful arguments in their reports that contributed significantly to the field of cancer biology. Therefore, the prognostic data of miR-124 need to be assimilated from different studies to draw the conclusion. In this study, we used quantitative synthesis to precisely quantify the expression, methylation levels, and SNP (rs5315649) of miR-124 to assess the prognostic significance in cancer patients.
Materials and Methods
Search Strategy
This study was executed in accordance with criteria of Meta-analysis of Observational Studies in Epidemiology group (MOOSE) (34) and the Preferred Reporting Items for Systematic Reviews and Meta-analysis (PRISMA) (35). The protocol of this meta-analysis has not been published or registered to any databases.
We scrutinized the following electronic databases until December 2017: PubMed, Web of Science, Google Scholar, Embase, Wanfang medicine online, and Chinese National Knowledge Infrastructure (CNKI). The search strategy was set up using the key words: “carcinoma” or “cancer” or “tumor” and “microRNA-124” or “miR-124” and “Methylation” and “polymorphisms” and “prognosis” or “survival” or “outcome” in humans. We also manually searched reference lists of relevant articles to further identify potential studies that not retrieved by databases exploration.
Subsequently, citations selected from initial search were screened for eligibility by two authors independently (Fujiao Duan and Zhenxing Yang). Articles that met all selection criteria were retrieved.
Inclusion and Exclusion Criteria
The including criteria were: (i) cohort studies that investigated the relationship between miR-124 and prognostic indicators including overall survival (OS) and/or progression-free survival (PFS)/recurrence-free survival (RFS)/disease-free survival (DFS) of cancer patients; (ii) the expression levels of miR-124 was measured in cancer tissue or serum; (iii) hazard ratios (HRs) and corresponding 95% CIs for survival analysis were reported in studies or could be computed from given data; (iv) available in Chinese or English language.
The exclusion criteria were: (i) studies that were not conducted in cancer patients; (ii) neither Chinese nor English language; (iii) review articles, case reports, or letters; (iv) with insufficient data to calculate the HRs and their 95% CIs, or the Kaplan–Meier curve unable to calculate HRs and 95% CI parameters.
Duplicate publications were eliminated through the Mendeley software (36). If a study had overlapping data with other published literatures, we selected the study with a larger sample size or the latest published article. All targeted articles were then evaluated and screened for eligibility by two reviewers (Zhen Peng and Weigang Liu) independently, and conflicts were finalized after consultation with third author.
Methodological Quality Assessment
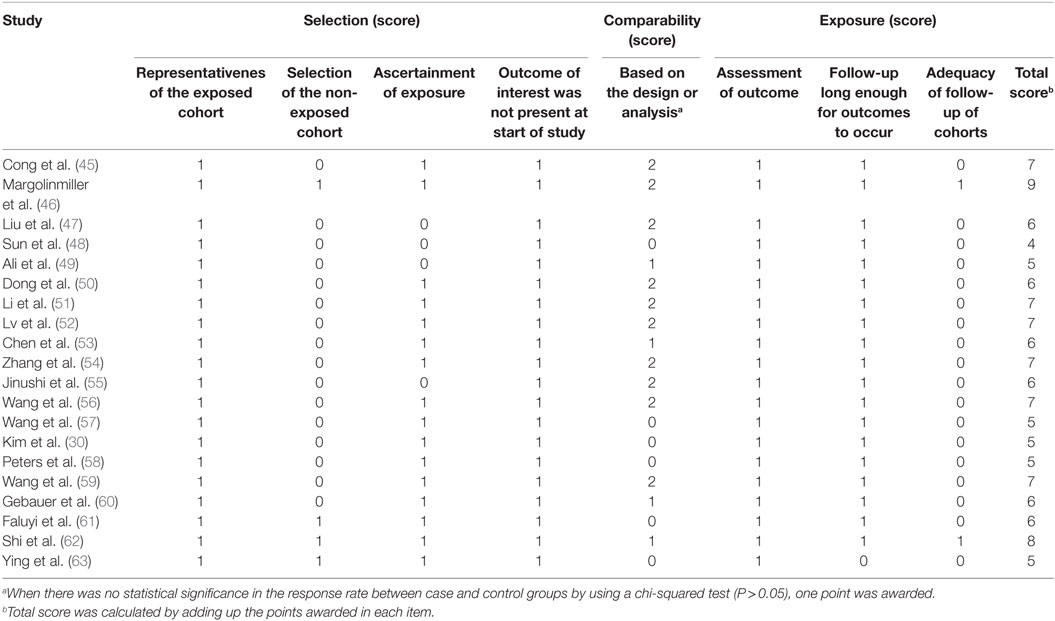
When the prognostic result was reported only as the Kaplan–Meier curves in some studies, the Engauge Digitizer 4.1 was then used to obtain the survival data, and Tierney’s method to calculate the HRs and their 95% CIs (37). The quality of the enrolled studies was assessed by the Newcastle-Ottawa Scale (NOS). The NOS consists of three quality parameters with a total of 9 points. Studies with a NOS score greater than 6 were considered as high-quality.
The specific Quality in Prognosis Studies (QUIPS) for specific biases of prognosis was appraised based on the approach of Hayden et al. (38). Estimation of the potential bias of the items included study participation, study attrition, prognostic factor measurement, outcome measurement, study confounding, statistical analysis, and reporting.
Two reviewers (Fujiao Duan and Zhenxing Yang) performed the quality assessments separately and, in case of any inconsistency, the final decision was reached with consensus.
Statistical Analysis
The HR with 95% CI was used to evaluate the impact of miR-124 expression on of cancer patients. Inter-study heterogeneity was quantified using Q-tests and the I-squared (I2) test (39). In the absence of significant heterogeneity (Pheterogeneity > 0.10 or I2 < 50%), a fixed-effects model (Mantel–Haenszel method) (40) was appropriately used to calculate the pooled effect, otherwise, the random-effects model (DerSimonian and Laird method) (41) was employed, and meta-regression was further utilized to explore sources of heterogeneity (42).
Begg’s funnel plot (rank correlation test) (43) and Egger’s test (44) determined the potential publication bias among included studies. One-way sensitivity analyses were performed, and then by omitting each study in turn to examine the stability of the pooled results.
All statistical analyses were performed with RevMan (Version 5.3.5 for Windows, Cochrane Collaboration, Oxford, UK) and Stata 13.1 MP (Stata Corporation, College Station, TX, USA). A two-tailed value of P < 0.05 was considered statistically significant.
Results
Study Identification
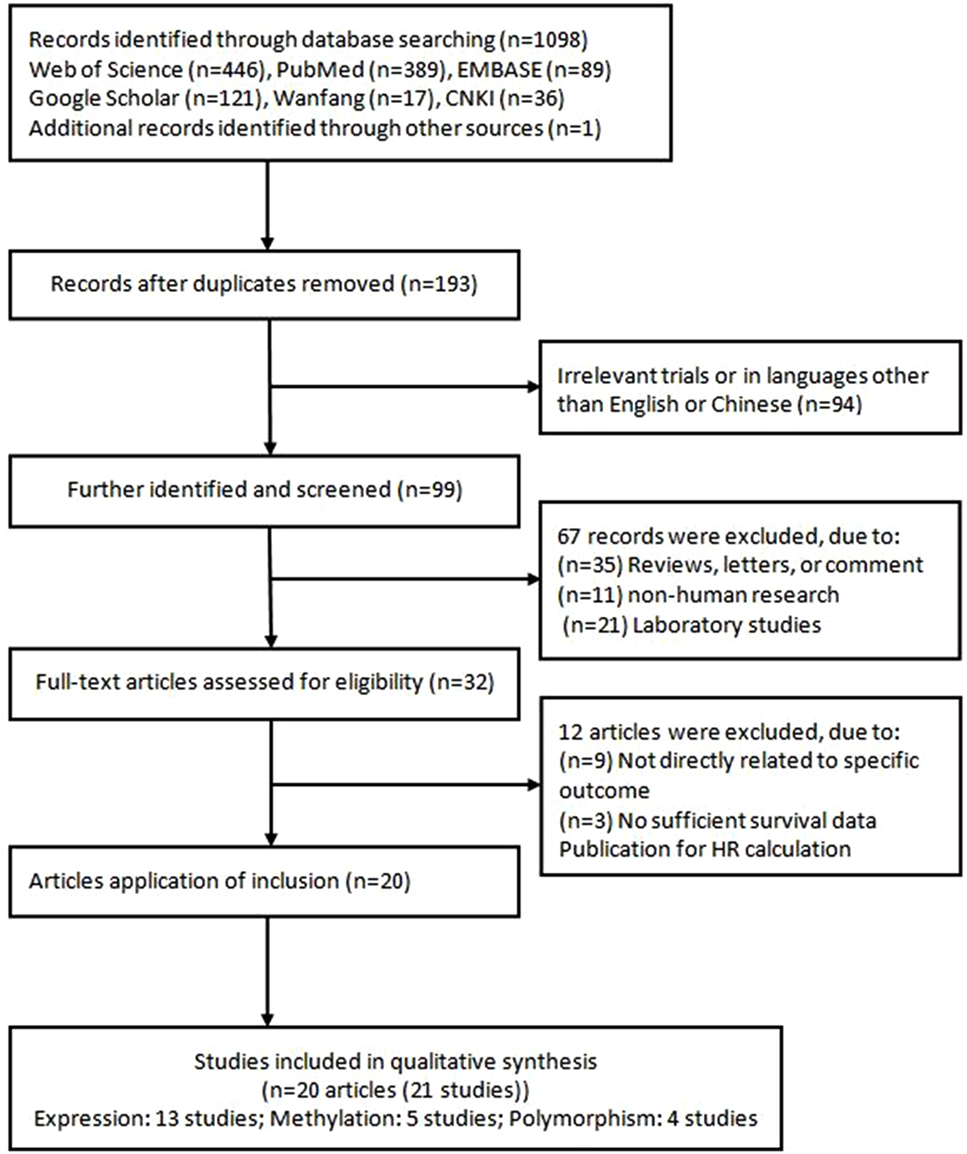
The systematic search returned 1,098 publications based on the search strategy (Figure 1). According to the exclusion criteria, the abstracts of 193 studies were reviewed. Of them, 94 were excluded because of irrelevant trials or in languages other than English or Chinese; 68 were excluded because they were reviews, letters, comments, non-human research, or laboratory studies. Eventually, 32 articles were eligible for further analysis. However, 12 articles were excluded as they were not directly related to specific outcome or they had insufficient survival data published for a HR calculation. Therefore, 20 articles (30, 45–63) (21 studies) were finally included in the meta-analysis. One of the articles (61) performed two cohorts in different populations, and we considered it as two studies.
Baseline Characteristics of Included Studies
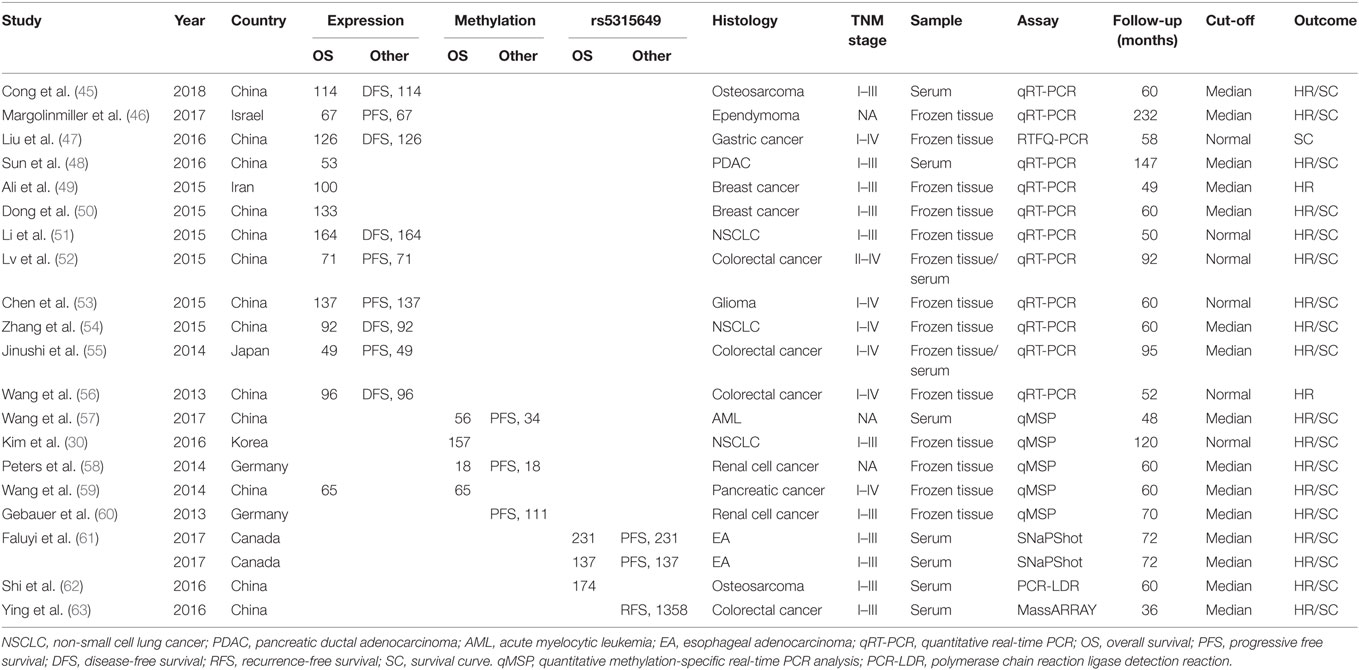
The major characteristics of eligible studies are summarized in Table 1. The studies were published from 2013 to 2017 and included a total of 3,574 patients from China, Iran, Japan Korea, Germany, and Canada. The patients were classified as Asian or Caucasian according to their ethnic background. The types of cancer included colorectal cancer, gastric cancer, osteosarcoma, pancreatic ductal adenocarcinoma (PDAC), breast cancer, NSCLC, glioma, renal cell cancer, acute myelocytic leukemia, pancreatic cancer, and esophageal adenocarcinoma. The method of miR-124 detection was quantitative real-time polymerase chain reaction (qRT-PCR), quantitative methylation-specific real-time PCR analysis (qMSP), and polymerase chain reaction ligase detection reaction (PCR-LDR) in 21 studies. MiR-124 expression, methylation levels, and rs5315649 for OS and/or DFS/RFS/PFS were measured in tissue or serum. The cutoff values of miR-124 were different in the studies, with most taken as the median.
Qualitative Assessment
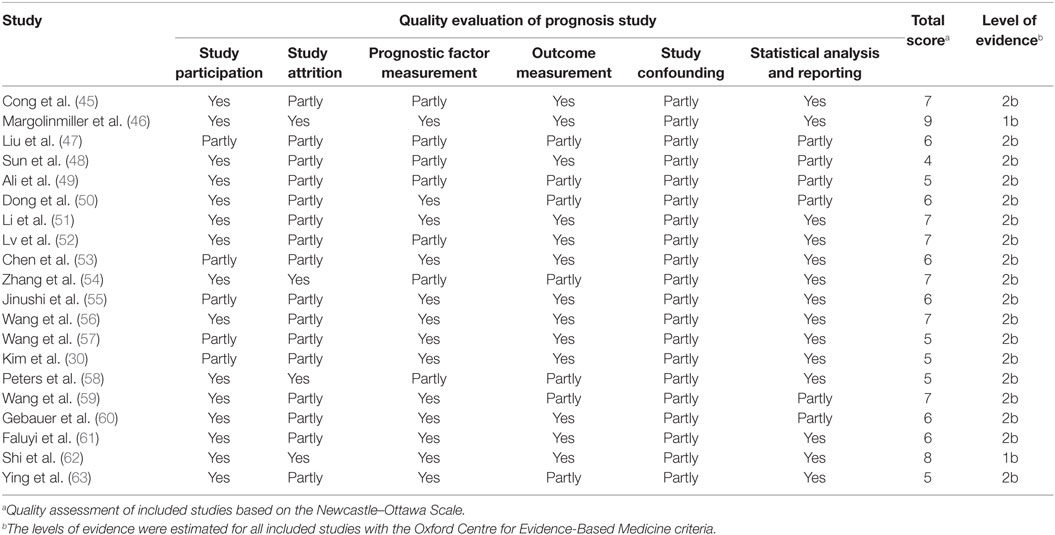
The result of quality assessment of the included studies based on QUIPS was summarized in Table 2. The bias domains of estimated items include participation, attrition, measurement of prognostic factor, confounding measurement and account, outcome measurement, and analysis and reporting. The risks of bias legend were presented in Figures 2 and 3. Based on the NOS (Table A1 in Appendix), 70 percent (14/20) of the enrolled studies were high-quality (quality score ≥ 6).
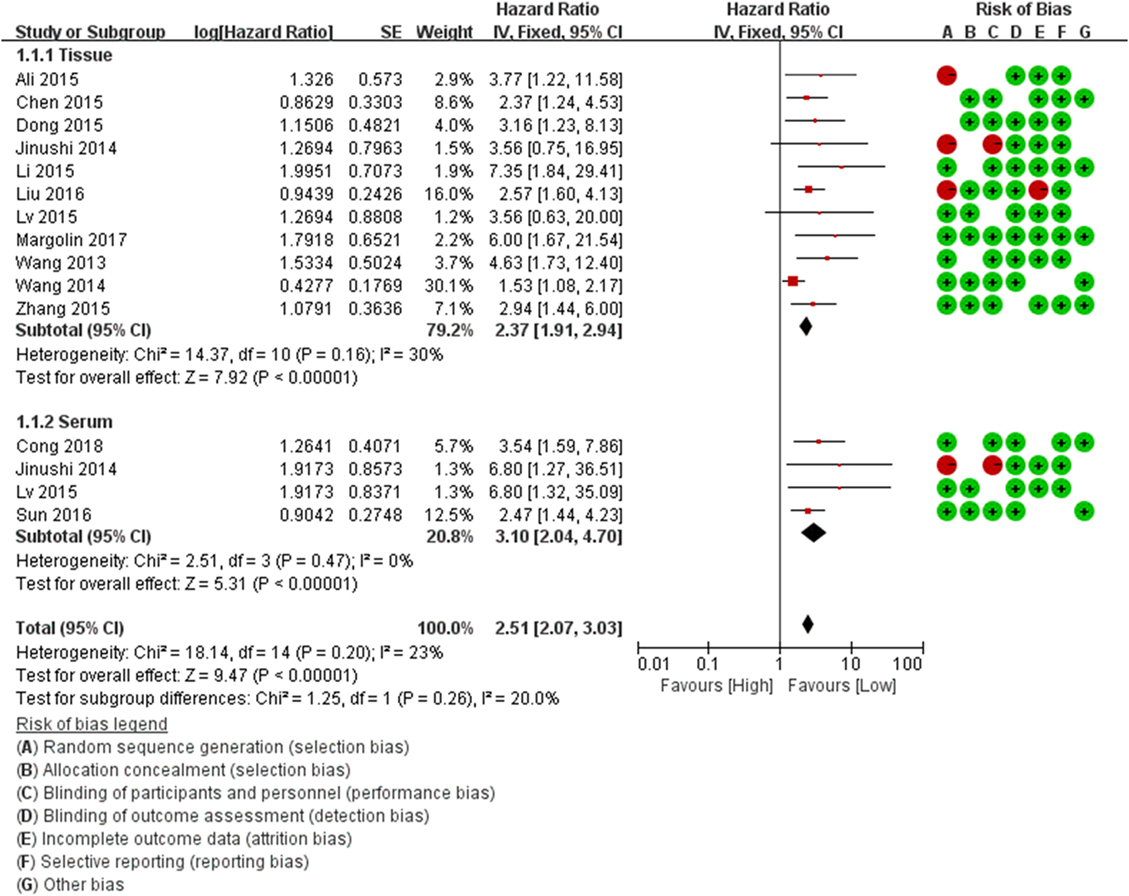
Figure 2. Forest plots of studies evaluating the hazard ratios of miR-124 expression (tissue and serum) with respect to overall survival.
Meta-Analysis Findings
Relationship Between the Expression of mir-124 and Patients’ Survival
For the OS, HRs were provided in 13 studies, and a significant association was observed between low miR-124 level and poor OS in patients (HR = 2.67, 95% CI: 2.10–2.38, P = 0.00). We conduct stratified analysis based on different sources, and the results showed that low expression of miR-124 in both serum (HR = 2.37, 95% CI: 1.91–2.94, P = 0.00) and cancer tissue (HR = 3.10, 95% CI: 2.04–4.70, P = 0.00) was significantly associated with poor OS (HR = 2.37, 95% CI: 1.91–2.94, P = 0.00; HR = 3.10, 95% CI: 2.04–4.70, P = 0.00). The test results showed that there was no heterogeneity between subgroups (I2 = 20%, P = 0.26) (Table 3; Figure 2).
Our analysis revealed a negative correlation between miR-124 level and PFS/RFS (HR = 3.92, 95% CI: 1.71–8.96, P = 0.00). Meanwhile, stratified analysis of different sources showed the low expression of miR-124 in serum (HR = 2.21, 95% CI: 1.50–3.26, P = 0.00) and cancer tissue (HR = 2.12, 95% CI: 1.20–3.74, P = 0.00) was statistically significant with the poor OS respectively. In tests for subgroup differences, the results showed that there was no heterogeneity between subgroups (I2 = 0%, P = 0.90) (Table 3).
Relationship Between the Methylation of mir-124 and Patients’ Survival
The results showed that hyper-methylation of miR-124 was associated with poor OS (HR = 2.09, 95% CI: 1.48–2.95, P = 0.00) and PFS (HR = 3.70, 95% CI: 1.72–7.97, P = 0.00) (Table 3).
Relationship Between the SNP of mir-124 and Patients’ Survival
The patients carrying with Allele C of miR-124 rs5315649 had a worse OS than the carriers with Allele G (HR = 1.50, 95% CI: 1.09–2.07, P = 0.00). Compared with the carriers with CG + GG genotype of miR-124 rs531564, for the OS, patients with CC showed significant association (HR = 4.61, 95% CI: 1.85–11.49, P = 0.00). Patients carrying with Allele C and CC genotype were associated with a poor PFS (HR = 1.67, 95% CI: 1.20–2.33, P = 0.00; HR = 2.37, 95% CI: 1.36–4.13, P = 0.00) (Table 3).
Test of Heterogeneity
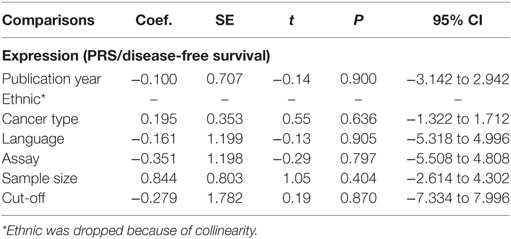
The results of heterogeneity tests were presented in Table 3. There was no significant heterogeneity between the miR-124 expression (OS, I2= 21%, P = 0.23), methylation (OS, I2 = 0%, P = 0.84; PFS, I2 = 43%, P = 0.17), and polymorphisms (OS, allele, I2 = 0%, P = 0.58; PFS/RFS, allele, I2 = 0%, P = 0.98) and the risk of tumorigenesis, except the expression for PRS/DFS (I2 = 57%, P = 0.02). Therefore, the fixed effects were applied to calculate the pooled HR for miR-124. Meanwhile, meta-regression was applied to investigate sources of heterogeneity for PRS/DFS of expression (Table 4).
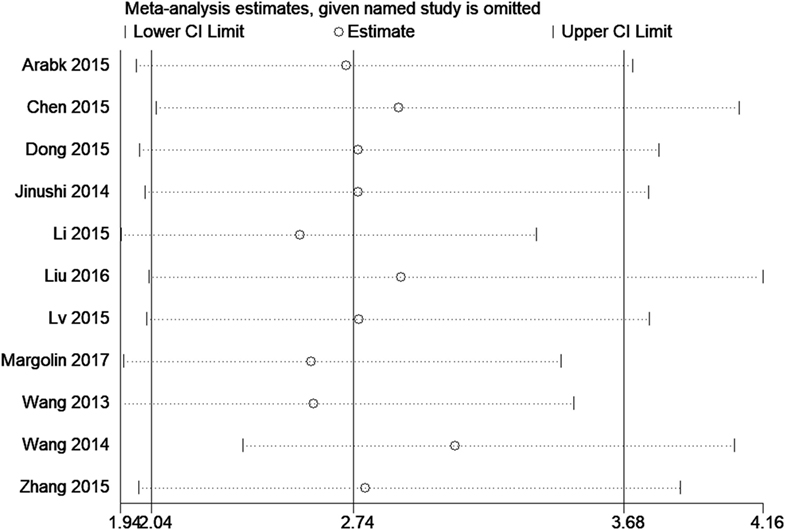
Sensitivity Analyses
Sensitivity analyses were carried out to assess the contribution of each study to the pooled estimate. Omitting individual dataset in each comparisons and recalculating did not substantially change the pooled HR, indicating that pooled HRs were quite stable (Figure 3).
Publication Bias
Begg’s and Egger’s test were used to evaluate the publication bias. The results suggested no evidence of publication bias (Table 5). Meanwhile, the shape of the funnel plots revealed no visual evidence of the asymmetry (Figures 4A,B).
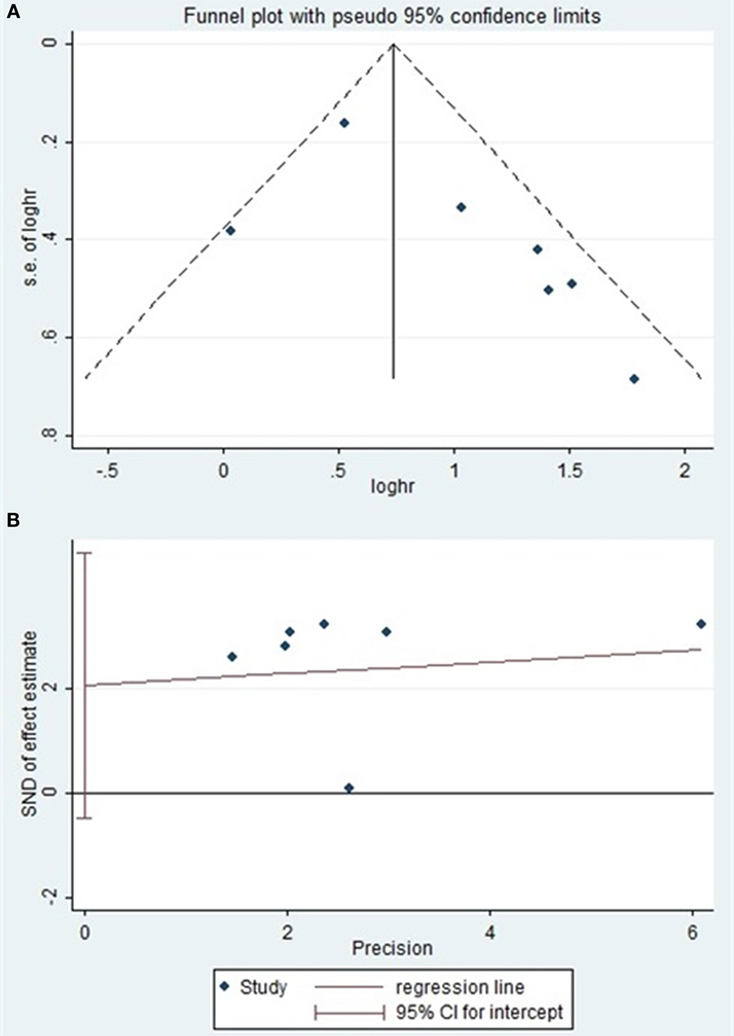
Figure 4. (A) Begg’s funnel plot of publication bias on the relationship between miR-124 expression and PRS/disease-free survival (DFS). (B) Egger’s funnel plot of publication bias on the relationship between miR-124 expression and PRS/DFS.
Discussion
Emerging studies have indicated that miRNAs could act as oncogenes or tumor suppressors and played key roles in proliferation, differentiation, metastasis, and cell apoptosis of cancer cells (64–66). Therefore, exploring the profiles of miRNAs related to tumorigenesis may promote the understanding of potential mechanisms of cancer development and progression and provide valuable insights for early diagnosis and prognosis of cancer (67, 68).
Several studies have indicated that miR-124 inhibits the epithelial–mesenchymal transition, proliferation, invasion, migration, and angiogenesis of cancer cells (69). However, the association between miR-124 expression, methylation, and genetic variants and cancer survival is still unknown. Therefore, it is very important to address why miR-124 as a prognostic indicator is valuable for judging prognosis and guiding treatment.
In the present study, we revealed that the low expression levels of miR-124 in serum and tissue were significantly associated with poor OS (HR = 2.37, 95% CI: 1.91–2.94, P = 0.00 and HR = 3.10, 95% CI: 2.04–4.70, P = 0.00) and PFS/RFS (HR = 2.21, 95% CI: 1.50–3.26, P = 0.00 and HR = 2.12, 95% CI: 1.20–3.74, P = 0.00). We also analyzed the correlation between different methylation levels (OS, HR = 2.09, 95% CI: 1.48–2.95, P = 0.00; PFS, HR = 3.70, 95% CI: 1.72–7.97, P = 0.00) and SNP (rs5315649) (Allele G: OS, HR = 1.50, 95% CI: 1.09–2.07, P = 0.00; Allele G: PFS, HR = 1.67, 95% CI: 1.20–2.33, P = 0.00) to evaluate their prognostic significance in cancer patients.
Downregulation of miR-124 has also been observed in various malignancies, including both solid tumors and hematological malignancies (70, 71). It is strictly conservative in both primary sequences and spatial expression patterns, which are limited to the nervous system of different metazoan, including aplysia, nematodes, flies, and all vertebrates studied. This protective effect indicates that miR124 plays an important role in controlling the expression of neural genes (72). Functional studies have linked vertebrate miR-124 to diverse aspects of neural specification or differentiation (73). Dysregulated miRNA expression can be induced by abnormal DNA methylation and contributes to the development and progression of multiple human cancers, including pancreatic cancer (59).
DNA hyper-methylation of miR-124 in pancreatic cancer is mediated by at least part of epigenetic mechanisms (74). Reduced expression of miRNA-124 can be found in pancreatic cancer tissues, and its downregulation was significantly associated with poor OS of PDAC patients. Rac1 as a direct target of miR-124, it has a fundamental role in tumorigenesis and invasion of cancer cells (59).
Epigenetic modifications have been proved to be essential for mammalian development, and epigenetic changes are related to different cancers (75). In cancer cells, some tumor suppressive miRNAs are silenced by the abnormal DNA methylation of CpG islands (76, 77). Therefore, to some extent, aberrant DNA methylation contributes to carcinogenesis and cancer progression.
Polymorphisms of miRNAs can create or destroy miRNA-binding sites and modulate miRNA–mRNA interaction potentially, while those in processing genes can achieve miRNA transcription by altering processing, transcription, or maturation (32). Hsa-mir-124 rs531564 is a relatively consistent predictor of OS, where mutation of each allele can reduce mortality by 30–40% (61). It is a SNP that has been previously found to be associated with the development of cervical cancer, colorectal cancer, and esophageal squamous cell carcinoma (78). Our study bears out this result. In the present study, systematic evaluation was analyzed to precisely quantify the miR-124 expression, methylation levels, and genetic variants. Although our results are robust, following several limitations are worth noting. First, due to not all the included studies reported adjusted HRs and theirs 95% CI, in this case, some data were extracted from survival curves, which could result in several tiny errors. Second, although no evidence of publication bias was found, included studies were mostly in Chinese region, which may generate publication bias. Third, the cut-off values (median, normal mean) were applied to evaluate the different miR-124 expression, methylation levels, and rs531564, which may lead to the deviations of actual values due to different algorithms. Finally, for DFS/PFS, the included studies were not stratified because of the limited availability of eligible studies.
In summary, this is the first study to evaluate the prognostic effects of miR-124 expression, methylation levels, and polymorphisms in different cancer patients. This study showed that low expression and hyper-methylation of miR-124 was strongly associated with poor prognosis, and genetic variations of miR-124 rs531564 affected prognosis in cancer patients. Given its limitations, the results of the study should be interpreted with caution. Future studies are needed to validate these results in prospective studies and evaluate their prognostic role in clinical practice.
Author Contributions
FD, YS and WL: Conceived and designed the study; YF and JS: Performed the dataset; ZP and LD: Analyzed the data; KW: Contributed analysis and tools material; DF: Wrote the manuscript; WL and YF Reference collection and data management; FD, WL, ZY, and KW: Statistical analyses and paper writing; FD: Study design; YS, revised manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer HK and handling Editor declared their shared affiliation.
Acknowledgments
This study was supported by the National Natural Science Foundation of China (No: 81701536).
References
2. Arora S, Rana R, Chhabra A, Jaiswal A, Rani V. miRNA-transcription factor interactions: a combinatorial regulation of gene expression. Mol Genet Genomics (2013) 288:77–87. doi:10.1007/s00438-013-0734-z
3. Winbanks CE, Ooi JY, Nguyen SS, Mcmullen JR, Bernardo BC. MicroRNAs differentially regulated in cardiac and skeletal muscle in health and disease: potential drug targets? Clin Exp Pharmacol Physiol (2014) 41:727–37. doi:10.1111/1440-1681.12281
4. Henry WRF, Claudia V, Robert W, Jeremias W, Christian CD, Werner SK, et al. microRNAs are differentially regulated between MDM2-positive and negative malignant pleural mesothelioma. Oncotarget (2016) 7:18713–21. doi:10.18632/oncotarget.7666
5. Lin CW, Lin PY, Yang PC. Noncoding RNAs in tumor epithelial-to-mesenchymal transition. Stem Cells Int (2016) 2016:1–13. doi:10.1155/2016/2732705
6. Vorvis C, Koutsioumpa M, Iliopoulos D. Developments in miRNA gene signaling pathways in pancreatic cancer. Future Oncol (2016) 12:1135–50. doi:10.2217/fon-2015-0050
7. He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet (2004) 5:522–31. doi:10.1038/nrg1379
8. Ha M, Kim VN. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol (2014) 15:509–24. doi:10.1038/nrm3838
9. Gomes BC, Santos B, Rueff J, Rodrigues AS. Methods for studying microRNA expression and their targets in formalin-fixed, paraffin-embedded (FFPE) breast cancer tissues. Methods Mol Biol (2016) 1395:189. doi:10.1007/978-1-4939-3347-1_11
10. Treece AL, Duncan DL, Tang W, Elmore S, Morgan DR, Dominguez RL, et al. Gastric adenocarcinoma microRNA profiles in fixed tissue and in plasma reveal cancer-associated and Epstein-Barr virus-related expression patterns. Lab Invest (2016) 96:661–71. doi:10.1038/labinvest.2016.33
11. Wen J, Feng Y, He X, Li S, Huang X, Xiao X, et al. Development and validation of a prognostic nomogram based on the log odds of positive lymph nodes (LODDS) for breast cancer. Oncotarget (2016) 7:21046–53. doi:10.18632/oncotarget.8091
12. Jones KB, Salah Z, Sara DM, Galasso M, Gaudio E, Nuovo GJ, et al. MicroRNA signatures associate with pathogenesis and progression of osteosarcoma. Cancer Res (2012) 72:1865–77. doi:10.1158/0008-5472.CAN-11-2663
13. Wilting SM, Boerdonk RAV, Henken FE, Meijer CJ, Diosdado B, Meijer GA, et al. Methylation-mediated silencing and tumour suppressive function of hsa-miR-124 in cervical cancer. Cancer Genet Cytogenet (2010) 203:167. doi:10.1186/1476-4598-9-167
14. Geng S, Zhang X, Chen J, Liu X, Zhang H, Xu X, et al. The tumor suppressor role of miR-124 in osteosarcoma. PLoS One (2014) 9:e91566. doi:10.1371/journal.pone.0091566
15. Deng D, Wang L, Chen Y, Li B, Xue L, Shao N, et al. MicroRNA-124-3p regulates cell proliferation, invasion, apoptosis, and bioenergetics by targeting PIM1 in astrocytoma. Cancer Sci (2016) 107:899. doi:10.1111/cas.12946
16. Lin J, Xu K, Wei J, Heimberger AB, Roth JA, Ji L. MicroRNA-124 suppresses tumor cell proliferation and invasion by targeting CD164 signaling pathway in non-small cell lung cancer. J Gene Ther (2016) 2:6.
17. Lang Q, Ling C. MiR-124 suppresses cell proliferation in hepatocellular carcinoma by targeting PIK3CA. Biochem Biophys Res Commun (2012) 426:247–52. doi:10.1016/j.bbrc.2012.08.075
18. Lv Z, Yang L. MiR-124 inhibits the growth of glioblastoma through the downregulation of SOS1. Mol Med Rep (2013) 8:345–9. doi:10.3892/mmr.2013.1561
19. Xie L, Zhang Z, Tan Z, He R, Zeng X, Xie Y, et al. microRNA-124 inhibits proliferation and induces apoptosis by directly repressing EZH2 in gastric cancer. Mol Cell Biochem (2014) 392:153. doi:10.1007/s11010-014-2028-0
20. Zhang H, Wang Q, Zhao Q, Di W. MiR-124 inhibits the migration and invasion of ovarian cancer cells by targeting SphK1. J Ovarian Res (2013) 6:84. doi:10.1186/1757-2215-6-84
21. Li W, Zang W, Liu P, Wang Y, Du Y, Chen X, et al. MicroRNA-124 inhibits cellular proliferation and invasion by targeting Ets-1 in breast cancer. Tumor Biol (2014) 35:10897–904. doi:10.1007/s13277-014-2402-2
22. Feng T, Xu D, Tu C, Li W, Ning Y, Ding J, et al. MiR-124 inhibits cell proliferation in breast cancer through downregulation of CDK4. Tumor Biol (2015) 36:1–11. doi:10.1007/s13277-015-3275-8
23. Du S, Li H, Sun X, Li D, Yang Y, Tao Z, et al. MicroRNA-124 inhibits cell proliferation and migration by regulating SNAI2 in breast cancer. Oncol Rep (2016) 36:3259–66. doi:10.3892/or.2016.5163
24. Wang Y, Chen L, Wu Z, Wang M, Jin F, Wang N, et al. miR-124-3p functions as a tumor suppressor in breast cancer by targeting CBL. BMC Cancer (2016) 16:826. doi:10.1186/s12885-016-2862-4
25. Zhou L, Xu Z, Ren X, Chen K, Xin S. MicroRNA-124 (MiR-124) inhibits cell proliferation, metastasis and invasion in colorectal cancer by downregulating Rho-associated protein kinase 1(ROCK1). Cell Physiol Biochem (2016) 38:1785–95. doi:10.1159/000443117
26. Peter AJ, Stephen BB. The epigenomics of cancer. Cell (2007) 128:683–92. doi:10.1016/j.cell.2007.01.029
27. Ando T, Yoshida T, Enomoto S, Asada K, Tatematsu M, Ichinose M, et al. DNA methylation of microRNA genes in gastric mucosae of gastric cancer patients: its possible involvement in the formation of epigenetic field defect. Int J Cancer (2009) 124:2367–74. doi:10.1002/ijc.24219
28. Ueda Y, Ando T, Nanjo S, Ushijima T, Sugiyama T. DNA methylation of microRNA-124a is a potential risk marker of colitis-associated cancer in patients with ulcerative colitis. Dig Dis Sci (2014) 59:2444–51. doi:10.1007/s10620-014-3193-4
29. Shi XB, Xue L, Ma AH, Tepper CG, Gandouredwards R, Kung HJ, et al. Tumor suppressive miR-124 targets androgen receptor and inhibits proliferation of prostate cancer cells. Oncogene (2013) 32:4130–8. doi:10.1038/onc.2012.425
30. Kim YH, Lee WK, Lee EB, Son JW, Kim DS, Park JY. Combined effect of metastasis-related microRNA, miR-34 and miR-124 family, methylation on prognosis of non–small-cell lung cancer. Clin Lung Cancer (2017) 18:e13–20. doi:10.1016/j.cllc.2016.06.005
31. Sung H, Jeon S, Lee KM, Han S, Song M, Choi JY, et al. Common genetic polymorphisms of microRNA biogenesis pathway genes and breast cancer survival. BMC Cancer (2012) 12:195. doi:10.1186/1471-2407-12-195
32. Ryan BM, Robles AI, Harris CC. Genetic variation in microRNA networks: the implications for cancer research. Nat Rev Cancer (2010) 10:389–402. doi:10.1038/nrc2867
33. Hartman M, Loy EY, Ku CS, Chia KS. Molecular epidemiology and its current clinical use in cancer management. Lancet Oncol (2010) 11:383–90. doi:10.1016/S1470-2045(10)70005-X
34. Stroup DF, Berlin JA, Morton SC, Olkin I, Williamson GD, Rennie D, et al. Meta-analysis of observational studies in epidemiology: a proposal for reporting. Meta-analysis Of Observational Studies in Epidemiology (MOOSE) group. J Am Med Assoc (2000) 283:2008–12. doi:10.1001/jama.283.15.2008
35. Moher D, Liberati A, Tetzlaff J, Altman DG, The PRISMA Group. Prisma statement-preferred reporting items for systematic reviews and meta-analyses. J Chin Integr Med (2009) 7:889–96. doi:10.3736/jcim20090918
36. Jatinder S. Mendeley: a free research management tool for desktop and web. J Pharmacol Pharmacother (2010) 1:62–3. doi:10.4103/0976-500X.64539
37. Tierney JF, Stewart LA, Ghersi D, Burdett S, Sydes MR. Practical methods for incorporating summary time-to-event data into meta-analysis. Trials (2007) 8:16. doi:10.1186/1745-6215-8-16
38. Hayden JA, Van Der Windt DA, Cartwright JL, Côté P, Bombardier C. Assessing bias in studies of prognostic factors. Ann Intern Med (2013) 158:280–6. doi:10.7326/0003-4819-158-4-201302190-00009
39. Higgins JPT, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. Br Med J (2003) 327:557–60. doi:10.1136/bmj.327.7414.557
40. Mantel N. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst (1959) 22:719–48.
41. DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials (1986) 7:177. doi:10.1016/0197-2456(86)90046-2
42. Thompson SG, Higgins JPT. How should meta-regression analyses be undertaken and interpreted? Stat Med (2002) 21:1559–73. doi:10.1002/sim.1187
43. Begg CB, Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics (1994) 50:1088. doi:10.2307/2533446
44. Stuck AE, Rubenstein LZ, Wieland D, Egger M, Davey Smith G, Schneider M, et al. Bias in meta-analysis detected by a simple, graphical test. BMJ (1998) 315(7109):629–34.
45. Cong C, Wang W, Tian J, Gao T, Zheng W, Zhou C. Identification of serum miR-124 as a biomarker for diagnosis and prognosis in osteosarcoma. Cancer Biomark (2018) 21:449–54. doi:10.3233/CBM-170672
46. Margolinmiller Y, Yanichkin N, Shichrur K, Toledano H, Ohali A, Tzaridis T, et al. Prognostic relevance of miR-124-3p and its target TP53INP1 in pediatric ependymoma. Genes Chromosomes Cancer (2017) 56:639–50. doi:10.1002/gcc.22467
47. Liu F, Xie LM, Zhang ZW, Tang HL, Wu LX. Expression of miR-124 in gastric cancer and its clinical signifcance. China Oncol (2016) 26:215–30. doi:10.3969/j.issn.1671-7171.2012.03.002
48. Sun B, Liu X, Gao Y, Li L, Dong Z. Downregulation of miR-124 predicts poor prognosis in pancreatic ductal adenocarcinoma patients. Br J Biomed Sci (2016) 73:152. doi:10.1080/09674845.2016.1220706
49. Ali A, Aghdas S, Mehri S, Emad Y, Masoumeh G. Down-regulated microRNA-124 expression as predictive biomarker and its prognostic singifcance with clinicopathological features in breast cancer patients. Diagn Pathol (2015) 10:178. doi:10.1186/s13000-015-0391-0
50. Dong LL, Chen LM, Wang WM, Zhang LM. Decreased expression of microRNA-124 is an independent unfavorable prognostic factor for patients with breast cancer. Diagn Pathol (2015) 10:45. doi:10.1186/s13000-015-0257-5
51. Li X, Yu Z, Li Y, Liu S, Gao C, Hou X, et al. The tumor suppressor miR-124 inhibits cell proliferation by targeting STAT3 and functions as a prognostic marker for postoperative NSCLC patients. Int J Oncol (2015) 46:798–808. doi:10.3892/ijo.2014.2786
52. Lv M, Huang X, Li QW, Wang YY. Expression levels of microRNA-124 in plasma and tissure correlated with prognosis in patients with colorectal cancer. Chin J Diffic Compl Cas (2015) 14:1125–8. doi:10.3969/j.issn.1671-6450.2015.11.008
53. Chen T, Wang XY, Li C, Xu SJ. Downregulation of microRNA-124 predicts poor prognosis in glioma patients. Neurol Sci (2015) 36:131–5. doi:10.1007/s10072-014-1895-1
54. Zhang Y, Li H, Han J, Zhang Y. Down-regulation of microRNA-124 is correlated with tumor metastasis and poor prognosis in patients with lung cancer. Int J Clin Exp Pathol (2015) 8:1967.
55. Jinushi T, Shibayama Y, Kinoshita I, Oizumi S, Jinushi M, Aota T, et al. Low expression levels of microRNA-124-5p correlated with poor prognosis in colorectal cancer via targeting of SMC4. Cancer Med (2014) 3:1544–52. doi:10.1002/cam4.309
56. Wang MJ, Li Y, Wang R, Wang C, Yu YY, Yang L, et al. Downregulation of microRNA-124 is an independent prognostic factor in patients with colorectal cancer. Int J Colorectal Dis (2013) 28:183–9. doi:10.1007/s00384-012-1550-3
57. Wang H, Zhang TT, Jin S, Liu H, Zhang X, Ruan CG, et al. Pyrosequencing quantified methylation level of miR-124 predicts shorter survival for patients with myelodysplastic syndrome. Clin Epigenetics (2017) 9:91. doi:10.1186/s13148-017-0388-5
58. Peters I, Dubrowinskaja N, Abbas M, Seidel C, Kogosov M, Scherer R, et al. DNA methylation biomarkers predict progression-free and overall survival of metastatic renal cell cancer (mRCC) treated with antiangiogenic therapies. PLoS One (2014) 9:e91440. doi:10.1371/journal.pone.0091440
59. Wang P, Chen L, Zhang J, Chen H, Fan J, Wang K, et al. Methylation-mediated silencing of the miR-124 genes facilitates pancreatic cancer progression and metastasis by targeting Rac1. Oncogene (2014) 33:514. doi:10.1038/onc.2012.598
60. Gebauer K, Peters I, Dubrowinskaja N, Hennenlotter J, Abbas M, Scherer R, et al. Hsa-mir-124-3CpG island methylation is associated with advanced tumours and disease recurrence of patients with clear cell renal cell carcinoma. Br J Cancer (2013) 108:131. doi:10.1038/bjc.2012.537
61. Faluyi OO, Eng L, Xin Q, Che J, Zhang Q, Cheng D, et al. Validation of microRNA pathway polymorphisms in esophageal adenocarcinoma survival. Cancer Med (2017) 6:361. doi:10.1002/cam4.989
62. Shi ZW, Wang JL, Zhao N, Guan Y, He W. Single nucleotide polymorphism of hsa-miR-124a affects risk and prognosis of osteosarcoma. Cancer Biomark (2016) 17:249. doi:10.3233/CBM-160637
63. Ying HQ, Peng HX, He BS, Pan YQ, Wang F, Sun HL, et al. MiR-608, pre-miR-124-1 and pre-miR26a-1 polymorphisms modify susceptibility and recurrence-free survival in surgically resected CRC individuals. Oncotarget (2016) 7:75865–73. doi:10.18632/oncotarget.12422
64. Cho WC. OncomiRs: the discovery and progress of microRNAs in cancers. Mol Cancer (2007) 6:60. doi:10.1186/1476-4598-6-1
65. Cho WC. MicroRNAs: potential biomarkers for cancer diagnosis, prognosis and targets for therapy. Int J Biochem Cell Biol (2010) 42:1273. doi:10.1016/j.biocel.2009.12.014
66. Duan F, Wang K, Dai L, Zhao X, Feng Y, Song C, et al. Prognostic significance of low microRNA-218 expression in patients with different types of cancer: evidence from published studies. Medicine (2016) 95:e4773. doi:10.1097/MD.0000000000004773
67. Bryant RJ, Pawlowski T, Catto JWF, Marsden G, Vessella RL, Rhees B, et al. Changes in circulating microRNA levels associated with prostate cancer. Br J Cancer (2012) 106:768. doi:10.1038/bjc.2011.595
68. Browne G, Taipaleenmäki H, Stein GS, Stein JL, Lian JB. MicroRNAs in the control of metastatic bone disease. Trends Endocrinol Metab (2014) 25:320–7. doi:10.1016/j.tem.2014.03.014
69. Babashah S, editor. MicroRNAs: Key Regulators of Oncogenesis. Switzerland: Springer International Publishing (2014). p. 433. doi:10.1007/978-3-319-03725-7
70. Cai WL, Huang WD, Li B, Chen TR, Li ZX, Zhao CL, et al. microRNA-124 inhibits bone metastasis of breast cancer by repressing Interleukin-11. Mol Cancer (2018) 17:9. doi:10.1186/s12943-017-0746-0
71. Han ZB, Yang Z, Chi Y, Zhang L, Wang Y, Ji Y, et al. MicroRNA-124 suppresses breast cancer cell growth and motility by targeting CD151. Cell Physiol Biochem (2013) 31:823. doi:10.1159/000350100
72. Sun K, Westholm JO, Tsurudome K, Hagen JW, Lu Y, Kohwi M, et al. Neurophysiological defects and neuronal gene deregulation in Drosophila mir-124 mutants. PLoS Genet (2012) 8:e1002515. doi:10.1371/journal.pgen.1002515
73. Abernathy DG, Yoo AS. MicroRNA-dependent genetic networks during neural development. Cell Tissue Res (2015) 359:179–85. doi:10.1007/s00441-014-1899-4
74. Inês PE, De CSGP, Marcelo C, Nogueira DSNC, Guerreiro IDC, Carvalho AL, et al. Mechanisms and role of microRNA deregulation in cancer onset and progression. Genet Mol Biol (2011) 34:363–70. doi:10.1590/S1415-47572011000300001
76. Kubo T, Toyooka S, Tsukuda K, Sakaguchi M, Fukazawa T, Soh J, et al. Epigenetic silencing of microRNA-34b/c plays an important role in the pathogenesis of malignant pleural mesothelioma. Clin Cancer Res (2011) 17:4965–74. doi:10.1158/1078-0432.CCR-10-3040
77. Kong KL, Kwong DLW, Chan HM, Law YK, Chen L, Li Y, et al. MicroRNA-375 inhibits tumour growth and metastasis in oesophageal squamous cell carcinoma through repressing insulin-like growth factor 1 receptor. Gut (2012) 61:33–42. doi:10.1136/gutjnl-2011-300178
78. Gao XR, Wang HP, Zhang SL, Wang MX, Zhu ZS. Pri-miR-124 rs531564 polymorphism and colorectal cancer risk. Sci Rep (2015) 5:14818. doi:10.1038/srep14818
Appendix
Keywords: miR-124, prognosis, cancer, risk factor, comprehensive assessment
Citation: Sun Y, Duan F, Liu W, Peng Z, Dai L, Feng Y, Yang Z, Shang J and Wang K (2018) Comprehensive Assessment of the Relationship Between MicroRNA-124 and the Prognostic Significance of Cancer. Front. Oncol. 8:252. doi: 10.3389/fonc.2018.00252
Received: 26 April 2018; Accepted: 21 June 2018;
Published: 16 July 2018
Edited by:
Imtiaz Ahmad Siddiqui, University of Wisconsin-Madison, United StatesReviewed by:
Ritika Das, New York University, United StatesHamidullah Khan, University of Wisconsin-Madison, United States
Nidhi Jain, Cedars-Sinai Medical Center, United States
Copyright: © 2018 Sun, Duan, Liu, Peng, Dai, Feng, Yang, Shang and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Fujiao Duan, ZmpkdWFuQDEyNi5jb20=
 Yadong Sun1
Yadong Sun1 Fujiao Duan
Fujiao Duan Kaijuan Wang
Kaijuan Wang