
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Nutr. , 10 December 2019
Sec. Nutrition and Microbes
Volume 6 - 2019 | https://doi.org/10.3389/fnut.2019.00184
 Sophie Fehlbaum1
Sophie Fehlbaum1 Christophe Chassard1
Christophe Chassard1 Clarissa Schwab1
Clarissa Schwab1 Maarja Voolaid1
Maarja Voolaid1 Candice Fourmestraux2
Candice Fourmestraux2 Muriel Derrien2
Muriel Derrien2 Christophe Lacroix1*
Christophe Lacroix1*Consumption of probiotic bacteria can result in a transient colonization of the human gut and thereby in potential interactions with the commensal microbiota. In this study, we used novel PolyFermS continuous fermentation models to investigate interactions of the candidate probiotic strain Lactobacillus paracasei CNCM I-1518 (L. paracasei) with colonic microbiota from healthy elderly subjects using 16S rRNA gene amplicon sequencing and metatranscriptomics, or with microbiota in vitro-colonized with Clostridioides difficile (C. difficile NCTC 13307 and C. difficile DSM 1296)—an enteropathogen prevalent in the elderly population. Small changes in microbiota composition were detected upon daily addition of L. paracasei, including increased abundances of closely related genera Lactobacillus and Enterococcus, and of the butyrate producer Faecalibacterium. Microbiota gene expression was also modulated by L. paracasei with distinct response of the Faecalibacterium transcriptome and an increase in carbohydrate utilization. However, no inhibitory effect of L. paracasei was observed on C. difficile colonization in the intestinal models under the tested conditions. Our data suggest that, in the in vitro experimental conditions tested and independent of the host, L. paracasei has modulatory effects on both the composition and function of elderly gut microbiota without affecting C. difficile growth and toxin production.
The large intestine is the most densely populated site of the human body with over 1014 microbial cells. This diverse microbial community exerts functions that are important to maintain host health, including energy, and nutrients supply by fermentation of otherwise indigestible food components, development of a balanced immune system and the protection against pathogens, termed colonization resistance (1, 2). Different diseases have been associated with compositional changes in intestinal communities (2) and a disruption of the healthy microbial communities (also referred to dysbiosis) can result in the loss of colonization resistance and an overgrowth of pathogens, such as Clostridium difficile (3), recently renamed Clostridioides difficile (4).
Apart from disease, diet and medications are important modulators of the gut microbiota. However, the gut microbiota also changes throughout lifespan and it is suggested that the aging-associated differences in gut microbiota might be linked to the general decline in the health status (5–8). Old age was associated with a decrease in potentially beneficial bacteria, including bifidobacteria (9), and Faecalibacterium prausnitzii, and an increase in facultative anaerobes such as enterobacteria (10–12). Furthermore, the risk of C. difficile infection (CDI) is elevated in old age following antibiotic treatment (13). Decreases in short chain fatty acids (SCFA) production have also been described for the elderly (9, 14).
Probiotics are defined as “live micro-organisms which, when administered in adequate amounts, confer a health benefit on the host” (15). Specific probiotic strains have been shown to promote colonization resistance and are promising adjunct therapy for the treatment of gastrointestinal infections, such as CDI (16–18). A meta-analysis based on 6,261 subjects reported that incidence of C. difficile was lower for subjects who consumed probiotic than of controls. Notably, a better efficacy was observed when probiotics were administered closer to the first antibiotic dose (19).
Mechanisms of probiotic action include the direct interaction with the commensal gut microbiota, inhibition of enteric pathogens or their metabolites, and modulation of the immune system (20). Probiotics have been associated with improved clinical outcome in several studies (21) but the effect of probiotics on gut microbiota composition and especially on the functional activity is not always known. Lactobacillus strains are often used as probiotics due to their technological properties and the general assumption that they are safe as they have been traditionally used in fermented dairy products (22). Lactobacillus paracasei CNCM I-1518 (L. paracasei) is a candidate probiotic strain that belongs to the Lactobacillus casei group (consisting of L. casei, L. paracasei subspecies paracasei, and L. rhamnosus). This strain can survive gastrointestinal transit and to modulate immune function (23–27). Fermented milk product containing L. paracasei CNCM I-1518 was associated with a decreased duration of common gastrointestinal and respiratory infections (28), and consumption of fermented milk containing the same strain reduced the incidence of antibiotic- and C. difficile-associated diarrhea in elderly patients taking antibiotics (29). Recently, this L. paracasei strain was administered to intensive care unit patients for prevention of antibiotic associated diarrhea and CDI. The trial was of a small sample size and it was found that one patient in the probiotic group developed CDI compared to three in the control group (30).
Assessing the effect of probiotics on the gut microbiota composition and activity can be difficult due to the hindered accessibility of the gastrointestinal tract. In vitro models simulating the human colon represent a useful tool for mechanistic studies on the interactions of probiotics with the gut microbiota and pathogens independent of the host (31, 32).
We recently developed in vitro fermentation models on the novel PolyFermS platform (33) with elderly immobilized fecal microbiota for investigations of C. difficile colonization and antibiotic treatment testing (34, 35). In this study, we assessed the potential of L. paracasei to modulate the composition and function of elderly gut microbiota reproduced in these in vitro colonic continuous fermentation models of the PolyFermS platform with and without C. difficile inoculation on the composition and activity of microbial communities and their functional properties using 16S rRNA gene amplicon sequencing and metatranscriptomics, respectively. To investigate associations observed in modeled microbiota, we also performed single and co-cultures of L. paracasei, F. prausnitzii, and C. difficile.
L. paracasei CNCM I-1518 was provided by Danone Research (Palaiseau, France). Faecalibacterium prausnitzii DSM 17677 was purchased from the Deutsche Sammlung von Mikroorganismen und Zellkulturen (DSMZ, Braunschweig, Germany). C. difficile DSM 1296 (PCR ribotype 001) and C. difficile NCTC 13307 (PCR ribotype 012) were purchased from DSMZ and the National Collection of Type Cultures (NCTC, Salisbury, United Kingdom), respectively.
For inoculation of colonic fermentation studies L. paracasei and vegetative cells of C. difficile DSM 1296 were cultured from glycerol stocks (33%, −80°C) at 37°C in serum flasks flushed with N2 and CO2 at 3:1 ratio or using the anaerobic Hungate culturing technique (36) containing fermentation medium simulating human chyme as previously described (34). Spores of C. difficile DSM 1296 and NCTC 13307 were prepared according to Sorg and Dineen (37) as previously described (35).
Yeast extract-casein hydrolysate-fatty acids (YCFA) medium (38) was used to routinely culture the bacterial strains in anaerobic Hungate tubes at 37°C for co-culture studies of L. paracasei with either F. prausnitzii or C. difficile DSM 1296. YCFA was supplemented with glucose, soluble starch and cellobiose (Sigma-Aldrich Chemie GmbH, Buchs, Switzerland), each at a concentration of 2 g L−1 (YCFA-GSC).
All three continuous in vitro fermentation models investigated in this study were based on the PolyFermS design and are displayed in Figure 1. The PolyFermS model allows the parallel testing of different treatments on singular microbiota as described previously (33). Common to all three models was an inoculum reactor (IR, 37°C, retention time of 9 h, pH 5.7) inoculated with single donor fecal microbiota immobilized in gellan-xanthan beads. Model 1 and 3 were inoculated with fecal microbiota obtained from the same elderly donor (75–80 years old) with a 7-months interval. Model 2 was inoculated with fecal microbiota from a different donor (70–75 years old) (34, 35). Fecal donors did not receive antibiotic treatment for at least 3 months prior to sample collection and did not consume probiotics for at least 1 month before fecal sampling. An informed written consent was obtained from both donors. A fermentation medium simulating human chyme was used in all three models, as presented previously (35). Different models were used for the experiments as previously described in details (34, 35). In model 1 run at conditions mimicking the proximal colon (PC, 37°C, retention time 9 h, pH 5.7), the IR was connected in parallel to a control reactor (PC_CR) and test reactor treated with L. paracasei (PC_LpC). PC_CR and PC_LpC were continuously inoculated with 10% effluent from IR and 90% fresh fermentation medium. The IR of model 2 was connected in parallel to two sets of two-stage reactors, mimicking the proximal (PC, 37°C, retention time of 9 h, pH 5.7) and transverse-distal colon (DC, 37°C, retention time of 18 h, pH 6.8). The two sets consisted of a control (PC_CR + DC_CR) and L. paracasei test reactors (PC_LpC + DC_LpC). The PC reactors were continuously fed with 10% effluent from IR and 90% fresh fermentation medium while DC reactors received 100% effluent from the respective PC reactor. The IR of the model 3 was used to feed 100% one control (DC_CR) and one L. paracasei test reactor (DC_LpC) operated at transverse-distal colon conditions (37°C, retention time of 25 h, pH 6.8). During antibiotic treatment and recovery period in model 3, control and test reactors were fed with fresh fermentation medium to avoid the inflow of untreated microbiota.
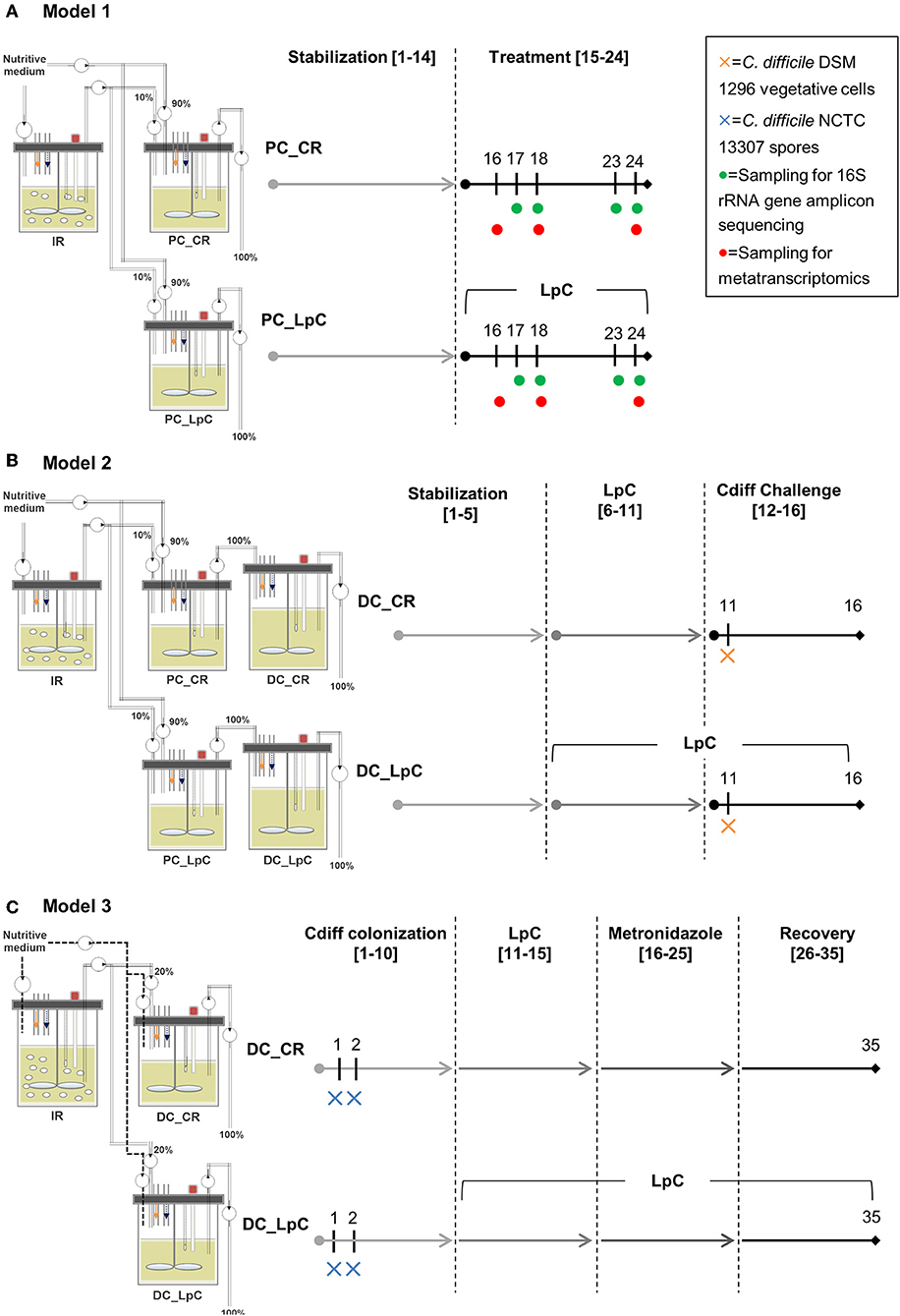
Figure 1. Experimental timeline of continuous colonic fermentation models. (A) Model 1. L. paracasei (LpC)-treated reactor mimicking the proximal colon section (PC_LpC) was inoculated with L. paracasei twice daily at days 15–24. Sampling for 16S rRNA gene amplicon sequencing was performed on days 17, 18, 23, and 24. Sampling for metatranscriptomics analysis was done on days 16, 18, and 24. (B) Model 2. L. paracasei (LpC)-treated reactor mimicking the proximal colon section (PC_LpC) was inoculated with L. paracasei twice daily at days 6–16; C. difficile DSM 1296 vegetative cells were inoculated on day 11 into DC_CR and DC_LpC mimicking the distal colon section. (C) Model 3. C. difficile NCTC 13307 spores were inoculated on day 1 and 2 into DC_CR and DC_LpC mimicking the distal colon section; DC_LpC was treated with L. paracasei twice daily at days 11–35; DC_CR and DC_LpC were treated with metronidazole (days 16–25) and recovery was observed at days 26–35. In all models sampling for qPCR and HPLC was performed daily. IR, inoculum reactor; PC, proximal colon; DC, distal colon; CR, control reactor; LpC, L. paracasei CNCM I-1518.
In model 1, the effect of L. paracasei on the healthy elderly proximal colonic microbiota was investigated. A stabilization period of 14 days was performed before PC_LpC was inoculated with L. paracasei twice daily for 10 days (Figure 1). L. paracasei was prepared from an overnight culture which was centrifuged (6,000 g, 5 min). The pellet was re-suspended in fresh fermentation medium and inoculated with a syringe to obtain final concentrations of around log10 7.5 cells mL−1 that corresponds to the approximate number of living L. paracasei cells detected in stool samples following ingestion (24). Microbiota composition was analyzed with 16S rRNA gene amplicon sequencing on four selected days at the beginning and at the end of L. paracasei treatment (days 16, 17, 23, and 24, Figure 1). qPCR was performed during the last days of stabilization period and throughout the treatment period of selected bacterial groups that were impacted by L. paracasei according to 16S rRNA amplicon sequencing or metatranscriptomics. The metatranscriptome was analyzed on 3 days corresponding to the beginning, middle, and end of L. paracasei treatment (days 16, 18, and 24). The metabolic activity was assessed with high performance liquid chromatography with refractive index detection (HPLC-RI) during the three last days of stabilization period and throughout the L. paracasei treatment period.
Model 2 was previously described for development of elderly microbiota models and for C. difficile colonization investigations (34, 35). After an initial stabilization phase of 18 days (34) and treatment periods for C. difficile investigations (35), control and test reactors were exchanged with new reactors, that were connected to IR for stabilization phase of 5 days before treatment with L. paracasei was started (Figure 1). L. paracasei was inoculated twice daily into the test system 2 (PC_LpC + DC_LpC) for 11 days (days 6–16). On day 11, DC_CR and DC_LpC were inoculated once with vegetative cells of C. difficile DSM 1296. The C. difficile cells were prepared from an overnight culture which was centrifuged (6,000 g, 5 min). The pellet was re-suspended in fresh fermentation medium and inoculated with a syringe to obtain final concentrations of approximately log10 6 cells mL−1. Reactor effluents of the DC reactors were collected 6 h post C. difficile inoculation and afterwards daily to determine cell numbers of L. paracasei and C. difficile as well as cytotoxin titers.
In model 3, the effects of L. paracasei on C. difficile NCTC 13307 were investigated in transverse-distal colon conditions before, during and after metronidazole treatment (Figure 1). The C. difficile NCTC 13307 strain was chosen due to a better colonization of reactors upon spore inoculation as described before (35). Both DC reactors were instilled with C. difficile spores at a concentration of 107 cfu, which were added once on two consecutive days at the beginning of C. difficile colonization period. L. paracasei treatment was performed in reactor DC_LpC from day 10 of fermentation with twice daily addition of L. paracasei cells throughout the remaining days of fermentation as described above for model 1. Metronidazole (Sigma-Aldrich) treatment was performed twice daily at a final concentration of 333 mg L−1 at days 16–25 in DC_CR and DC_LpC. Reactor effluents of DC's were collected daily for qPCR analysis of L. paracasei and C. difficile abundance, and for cytotoxin determination using Vero cell analysis.
The effect of ceftriaxone and metronidazole on C. difficile spore germination and colonization, respectively, as well as the general effect of these antibiotics on the gut microbiota was presented earlier (35).
Growth of F. prausnitzii was investigated in co-culture with L. paracasei because an increase in relative abundance of the genus Faecalibacterium was observed during L. paracasei treatment in PC_LpC reactor of model 1 by 16S rRNA gene amplicon sequencing. Culturing was performed in Hungate tubes containing 10 mL YCFA-GSC medium. For each measurement point, individual tubes were inoculated with 2% of three overnight cultures. Optical density (OD600nm), pH and metabolites were analyzed at 0, 8, 48, and 72 h of incubation. qPCR analysis was performed to determine cell numbers of L. paracasei and F. prausnitzii as described below. Metabolite concentrations were assessed from culture supernatants using HPLC-RI analysis. Strains were also grown individually for comparison. The co-culture test was performed four times with three replicates each time and average values of the four tests are presented.
L. paracasei and C. difficile DSM 1296 were investigated in co-cultures to assess the effect of L. paracasei on C. difficile growth and toxin production in the absence of complex microbiota. Strains were grown in Hungate tubes containing 10 mL YCFA-GSC medium and for each intended measurement time point separate tubes were inoculated with 2 or 4%, of C. difficile and L. paracasei overnight cultures, respectively. Because L. paracasei grew slower than C. difficile, L. paracasei was inoculated first and C. difficile was added after 5 h. OD, pH and metabolites were determined at 0, 5, 10, 13, and 25 h. Cell counts were additionally determined by plating on Wilkins-Chalgren agar (Oxoid AG) supplemented with cysteine-HCL and resazurin (Sigma-Aldrich). Serial dilutions were prepared in an anaerobic chamber and plates were incubated at 37°C in anaerobic jars (BioMérieux Suisse SA). Colonies of L. paracasei and C. difficile were distinguished by different colony morphology. L. paracasei had milky appearance, smooth surface, colony diameter (2–5 mm) larger than for C. difficile, round with entire margin. In contrast, C. difficile exhibited yellow ground-glass appearance, ruffled edges and smaller colonies. Toxin production was assessed after 13 and 25 h incubation in co-cultures and compared to single cultures using the Vero cell assay test. The co-culture test was performed twice with three replicates each time, and average values of the two tests are presented.
Genomic DNA was extracted from 2 mL fermentation effluent and co-cultures using the FastDNA SPIN Kit for Soil (MP Biomedicals, Illkirch, France). Abundance of specific bacterial and archaeal groups or species were measured in duplicate on an ABI PRISM 7500-PCR (Applied Biosystems, Zug, Switzerland) using a reaction volume of 25 μl as described before (39). All assays were carried out using the 2 × SYBR Green PCR Master Mix (Applied Biosystems). Specific primers were used for enumeration of different bacterial groups, including L. paracasei, F. prausnitzii, and C. difficile (Table S1). A factor of 6 and 10 was used1 to calculate the number of cells for F. prausnitzii and C. difficile, respectively, to account for several copies of 16S rRNA gene (40). Standard curves preparation and reaction conditions were described previously (41).
16S rRNA gene amplicon sequencing of effluent samples of colonic model 1 was carried out at DNAVision (Gosselies, Belgium) on a 454 Life Sciences Genome Sequencer FLX instrument (Roche Applied Science, Vilvoorde, Belgium). Amplification of the V5-V6 hypervariable 16S rRNA region was performed using primers 784F and 1061R (42). Data was analyzed using the open source software package Quantitative Insights Into Microbial Ecology (QIIME), v1.9 (43) as described before (35).
One mL effluent samples of colonic model 1 was collected directly from reactors and mixed with 1 mL 60% glycerol at −40°C, kept on ice for 20 min and centrifuged for 15 min (3,220 × g, 4°C). The supernatant was discarded, and the pellet was shock-frozen in liquid nitrogen and stored at −80°C. For total RNA isolation, pellets were re-suspended in 400 μL cold Man, Rogosa and Sharpe medium (MRS) supplemented with cysteine at 0.5 g/l and transferred to a screw cap tube containing 500 μL chlorophorm/phenol (1:1, v/v), 30 μL SDS 10% (44), 30 μL 3 M Na-acetate and 400 mg zirconium beads (0.1 mm). The mixture was disrupted in a bead beater (4 × 40 s, 5 m s−1) with cooling on ice between cycles and centrifuged for 12 min (12,000 × g, 4°C). The supernatant was added to 200 μL ice cold chloroform, centrifuged again as before and from the resulting supernatants RNA was isolated using the High Pure RNA Isolation Kit (Roche Diagnostics, Switzerland) according to the manufacturer's instructions. RNA concentrations and quality were determined on a NanoDrop 1000 spectrophotometer (Thermo Fisher Scientific, Washington, USA) and on an Agilent 2100 Bioanalyzer (Agilent, Basel, Switzerland), respectively. Paired-end RNA-seq using an Illumina HiSeq 2500 v4 was conducted at the Functional Genomics Center Zurich (ETH Zurich, Switzerland). RNA libraries were prepared using TruSeq RNA stranded library preparation kit and standard protocols supplied by Illumina.
For bioinformatics analysis, a pipeline consisting of SortMeRNA (45) for separation of rRNA and mRNA, and FLASH (46) for overlapping the paired-end sequences were used. rRNA sequences (100,000) were compared to the modified SILVA database provided by CREST (47). Putative mRNA reads were compared to the NCBI RefSeq database using MALT (http://ab.inf.uni-tuebingen.de/software/malt/) which is based on DIAMOND (48). Transcripts were taxonomically classified using MEGAN (49). Putative mRNA reads were also uploaded to MG-RAST for functional classification according to the SEED Subsystem scheme using default settings.
Acetate, butyrate, propionate, formate, and lactate were determined in fermentation effluent and co-culture samples by HPLC in duplicate (Thermo Fisher Scientific Inc. Accela, Wohlen, Switzerland). Sample supernatants were filtered into vials through a 0.45 μm nylon HPLC filter (Infochroma AG, Zug, Switzerland). The analysis was run at a flow rate of 0.4 mL min−1 using an Aminex HPX-87H (Bio-Rad Laboratories AG, Reinach, Switzerland) or Rezex ROA-Organic Acid column (Phenomenex, Basel, Switzerland), for effluent and co-culture samples, respectively and 10 mM H2SO4 as eluent. A refractive index detector was used for detection.
C. difficile cytotoxin production was monitored in effluent samples of colonic models 2 and 3 and in samples of co-culture test of L. paracasei with C. difficile DSM 1296 using a Vero cell cytotoxicity assay as described before (35).
Statistical analyses of co-culture studies were done using JMP 10.0 (SAS Institute, USA). All data are expressed as mean ± SD of several co-culture tests performed in triplicate in batch fermentation studies. Growth (log10-transformed), pH values, metabolites, and toxin production were compared between pure and co-cultures using the non-parametric Kruskal-Wallis test. Statistical analyses of metatranscriptomics data were done using a one-tailed Student's t-test for relative abundance comparisons between control reactor and L. paracasei reactor.
All 454-pyrosequencing files have been deposited to the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA) under bioproject accession number SRP144222. The mRNA reads are available at MG-RAST under project ID: IFT_antibiotics.
In a first model mimicking proximal colon (model 1), we investigated the impact of 10-days inoculation of L. paracasei on the gut microbiota composition of a healthy elderly donor using 16S rRNA gene sequencing. In the control reactor (PC_CR), Firmicutes and Bacteroidetes were the dominant phyla, while Proteobacteria and Actinobacteria represented <6% of the community. Clostridiales were the dominant bacterial order with Lachnospiraceae and Ruminococcaceae contributing the majority of reads (Figure S1), Lactobacillaceae represented between 0.9 and 2.7% of effluent microbiota; a mean relative abundance of 2.4% of Lactobacillus spp. relative to total bacterial 16S rRNA genes was determined using qPCR.
L. paracasei was not detected during stabilization phase in PC_CR and PC_LpC, and in PC_CR during treatment using strain specific qPCR (Table S1). However, in the inoculated reactor, addition of L. paracasei at log10 8 cfu mL−1 led to progressive increase of relative abundance of the strain, from 0.1 up to 0.4% after 10 days (Figure 2A). The addition of L. paracasei in proximal colon had little impact on the relative abundance of major phyla, however, analysis of days 17, 18, 23, and 24 at the beginning and end of the treatment showed an increase of Lactobacillus, Faecalibacterium, Ruminococcaceae, and Enterococcus, in PC_LpC compared to control reactor (PC_CR) (Figure 3A). At the same time, the addition of L. paracasei was associated with decreased abundance of Roseburia, Ruminococcaceae incertae sedis, Bacteroides, and Paraprevotella. The increase of Faecalibacterium and decrease in Bacteroides spp. and Roseburia spp./E. rectale after the addition of L. paracasei was confirmed by qPCR (Figure 2B).
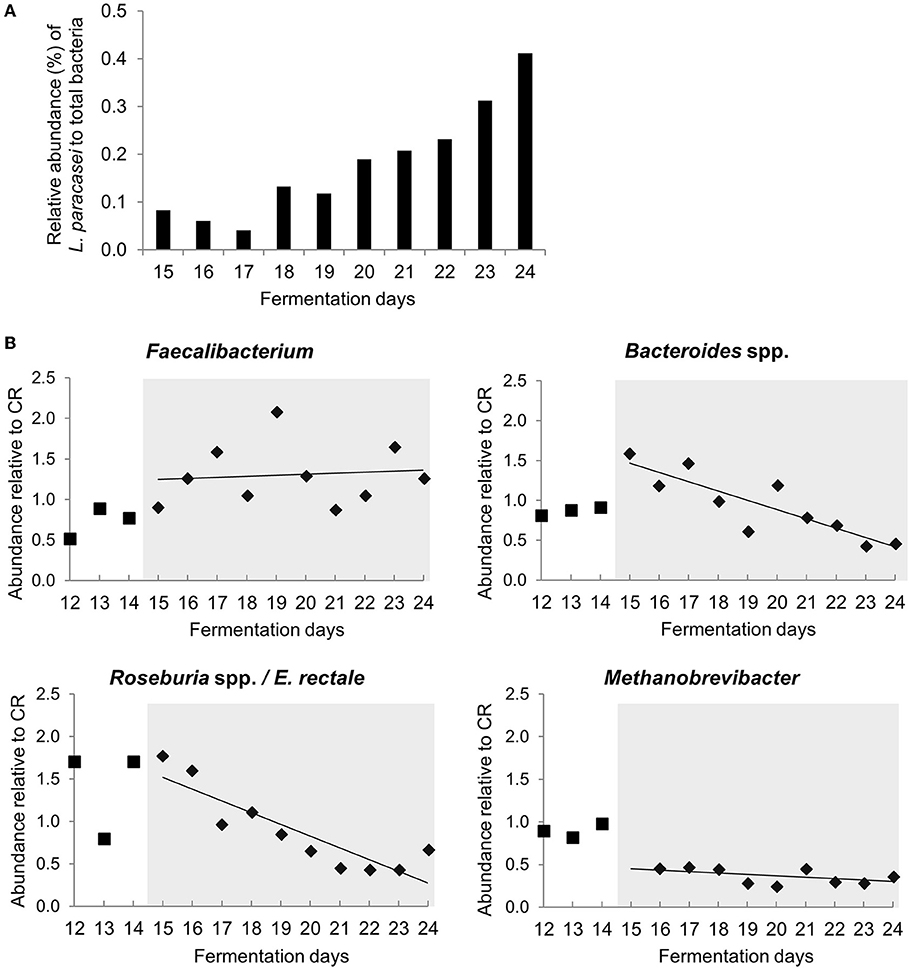
Figure 2. Abundance of selected bacterial groups in proximal colon section of model 1 assessed with qPCR (A) L. paracasei abundance compared to total bacteria in PC_LpC during treatment period. (B) Relative abundance of Faecalibacterium, Bacteroides spp., Roseburia spp. /Eubacterium rectale and Methanobrevibacter in PC_LpC (16S rRNA genes target taxon relative to 16S rRNA genes total bacteria) normalized to relative abundance in PC_CR during last 3 days of stabilization period (days 12–14,■) and during treatment period (days 15–24,♦).
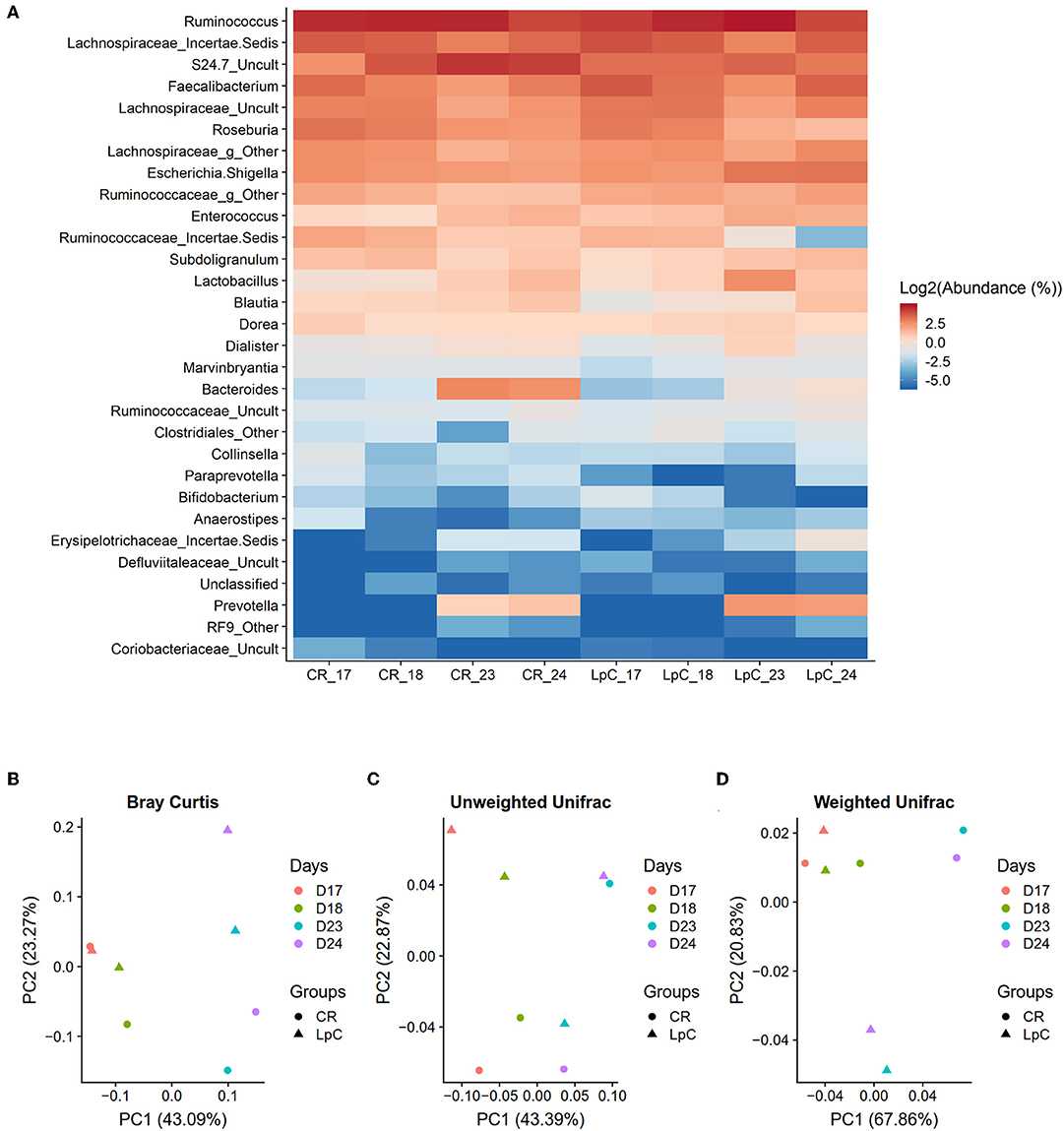
Figure 3. Effect of L. paracasei supplementation on microbiome composition in (LpC)-treated reactor mimicking the proximal colon section (PC_LpC) of model 1 assessed with 16S rRNA gene amplicon sequencing on days 17, 18, 23, and 24. (A) Heatmap showing the relative abundance at genus level of PC_LpC compared to PC_CR. (B) PCoA analysis of Bray-Curtis distances in PC_LpC and PC_CR. (C) PCoA analysis of unweighted UniFrac distances in PC_LpC and PC_CR. (D) weighted UniFrac distances in PC_LpC and PC_CR.
Beta-diversity was assessed by the Bray-curtis distances, unweighted UniFrac distances, and weighted UniFrac distances (Figures 3B–D). A clear separation of the reactor with L. paracasei at days 23 and 24 and all the other samples was observed with weighted UniFrac (Figure 3D).
Next, we evaluated the impact of L. paracasei inoculation on activity of the gut microbiota in the same experience using metatranscriptome analysis. RNA sequencing of fermentation effluents yielded between 4.5 and 12.8 million overlapped reads (average size 153 ± 31 bp), between 7.4 and 10.7% of those reads were identified as putative mRNA transcripts (225,000 and 1.57 million reads) (Table S2).
A subset of 100,000 rRNA reads per samples was used for taxonomic classification. Bacteria and archaea were detected in the effluents of the in vitro fermentation model (Figure 4 and Table S3). Clostridiales were the dominant bacterial order (78–89% of rRNA reads and 66–73% of mRNA reads, Tables S3, S4), with Ruminococcaceae (38–49%), and Lachnospiraceae (23–27%) contributing the majority of transcripts (Tables S3, S4) and confirming results obtained by 16S rRNA gene amplicon sequencing (Figure S1). Relative abundance of unclassified Veillonellaceae and Veillonella were increased in the presence of L. paracasei while Paraprevotella and Peptoniphilus decreased (Figure 4). Methanobacteriales contributed between 0.01 and 0.04% of 16S rRNA transcripts. Relative abundance of unclassified Methanobrevibacteriaceae and Methanobrevibacter also decreased in reactors to which L. paracasei had been added. This decrease after L. paracasei addition could be confirmed by qPCR (Figure 2B). Taxonomic classification of mRNA reads was consistent with the community structure revealed by rRNA analysis (Table S3).
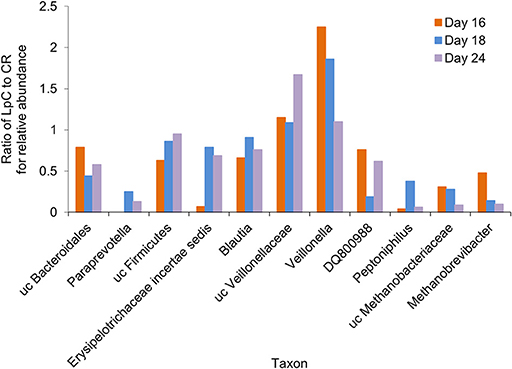
Figure 4. Ratio of the relative abundance of taxonomic groups between the L. paracasei (LpC)-treated reactor and the control reactor (CR) mimicking the proximal colon section (model 1) assessed with metatranscriptomics on days 16, 18, and 24. Only taxonomic groups that differed in abundance between PC_CR and PC_LpC are shown.
Between 74,166 and 1.7 million reads were assigned to SEED categories using MG-RAST. Almost 40% of all transcripts belonged to SEED categories “Carbohydrate metabolism” and “Protein metabolism.”
The addition of L. paracasei had modest impact on the relative abundance of most SEED categories (Table 1). However, relative abundance of transcripts assigned to SEED category “Membrane transport” was significantly (p < 0.05) increased when L. paracasei was present in the fermentation vessel due to the enhanced transcription of fructose and mannose, galactose, and sucrose specific sugar phosphotransferase systems [(PTS) Tables 1, 2]. As these categories were all related to sugar transport, we further investigated contributors to the SEED category “Carbohydrate Metabolism.” Subcategories “Di- and Oligosaccharides,” and “Fermentation” were consistently slightly enhanced on all 3 days tested when L. paracasei was added compared to controls. Within “Di- and Oligosaccharides,” sucrose phosphorylase (EC2.4.1.7), sucrose-6-phosphate hydrolase (EC3.2.1.26) and “Alpha-galactosidase” were 1.2–5 fold more abundant compared to controls. Within category “Fermentation,” “Butanol biosynthesis” (Pyruvate formate-lyase (EC 2.3.1.54), 1.2–1.8 fold) was the only noted change.
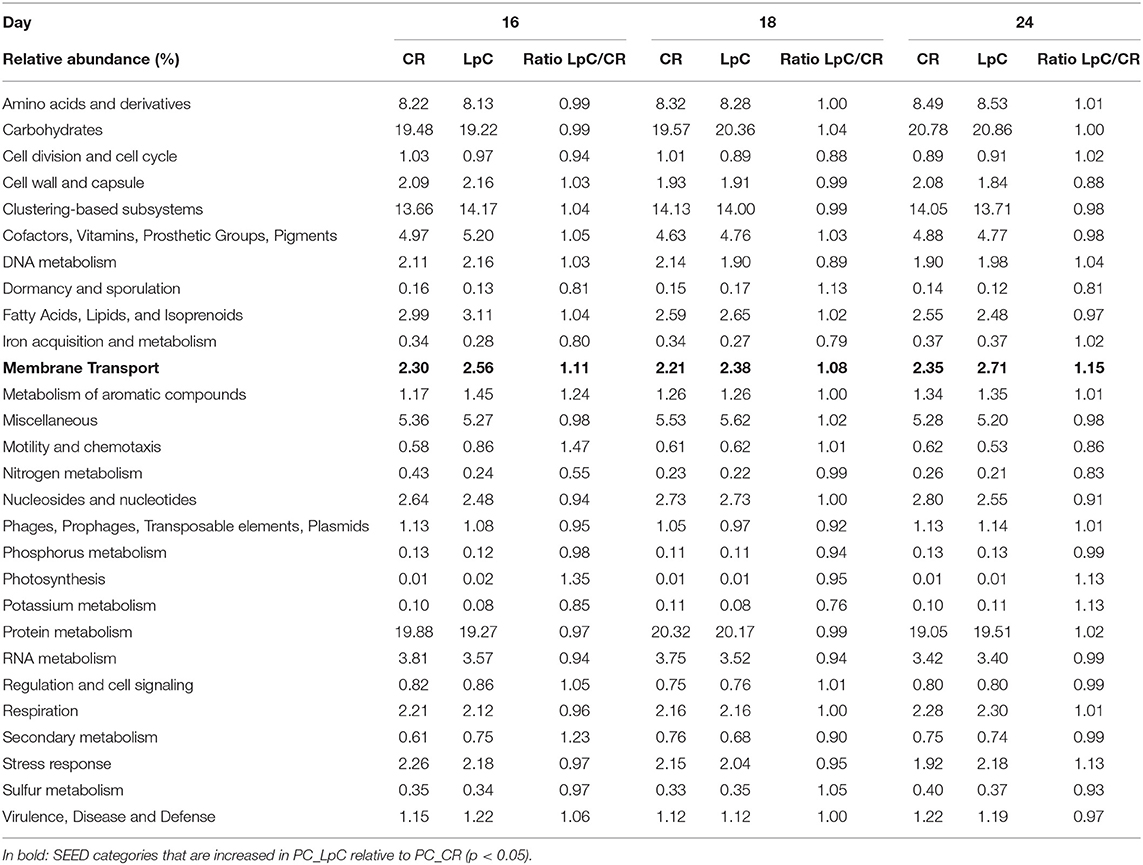
Table 1. Relative abundance of SEED categories Level 1 in control and L. paracasei (LpC)-treated reactor mimicking proximal colon section (model 1).
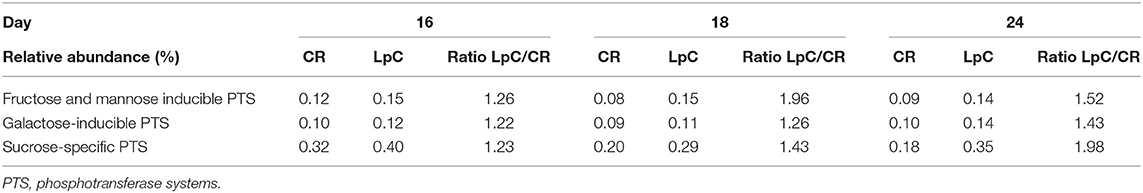
Table 2. Relative abundance of transcripts assigned to PTS systems “SEED subcategories of “Membrane Transport (SEED L1)” in control and L. paracasei (LpC)-treated reactor mimicking the proximal colon section (model 1).
As we observed an increase in relative abundance of Faecalibacterium after the addition of L. paracasei both in 16S rRNA amplicon sequencing and qPCR, we performed a targeted analysis on transcriptome from Faecalibacterium (33.000–75.000 transcripts).
Most of transcripts were assigned to “Carbohydrates” (~25%), “Protein metabolism” (~16%), and “Clustering-based subsystems” (~14%) (Table S5). Addition of L. paracasei had little impact on relative abundance of most SEED categories L1, but “Clustering- based subsystems” and “Metabolism of aromatic compounds” were significantly (p < 0.05) reduced and increased, respectively. Increase of the latter was due to increased abundance of transcripts (1.2–1.3 fold) of acetyl-CoA acetyltransferase in PC_LpC (Table S6). Acetyl-CoA acetyltransferase is involved in several pathways, therefore we also observed enhanced relative abundance of SEED subcategories level 2 “Anaerobic degradation of aromatic compounds” (subcategory level L3 “Anaerobic benzoate metabolism”), “Fermentation” (“Butanol biosynthesis”), and “Lysine, threonine, methionine, and cysteine” (“Lysine fermentation”) in PC_LpC. Also, the transcription of acetate kinase was enhanced 1.1–1.8 fold in PC_LpC. Acetate kinase is involved in pathways represented by SEED categories related to “Fermentations” (“Fermentations: Lactate,” “Fermentations: Mixed acid”), “Lysine, threonine, methionine, and cysteine” (“Lysine fermentation”), “Central carbohydrate metabolism” (“Pyruvate metabolism II: acetyl-CoA, acetogenesis from pyruvate”), and “Sugar Alcohols” (“Ethanolamine utilization”).
In contrast, 4-alpha-glucanotransferase (amylomaltase) (EC 2.4.1.25) and Oligopeptide ABC transporter, periplasmic oligopeptide-binding protein OppA (TC 3.A.1.5.1) were 1.3–1.7 fold and 1.5–1.7 fold higher expressed in control fermentations (PC_CR), respectively.
Metabolic activity at proximal colon conditions was assessed using HPLC-RI. Metabolite concentrations were stable and similar for PC_CR and PC_LpC throughout the 10 days treatment period (Figure S2). The main metabolite was acetate with concentrations around 55 mM followed by butyrate (25 mM) and propionate (8 mM).
The increase in Faecalibacterium abundance associated with L. paracasei in proximal colon conditions (model 1) led us to explore whether there are direct interactions of the two species in batch fermentations (Figure 5). In single cultures, F. prausnitzii reached maximum OD of 0.4 ± 0.1 after 8 h that decreased to 0.1 ± 0.1 after 72 h (Figure 5A). Within 8 h, log cell numbers increased by 1.3, and a decrease of F. prausnitzii cell numbers after 48 and 72 h was also observed by qPCR (Figure 5B). The pH dropped from 6.8 ± 0.04 to 6.2 ± 0.1 within 48 h and remained stable thereafter (Figure 5C).
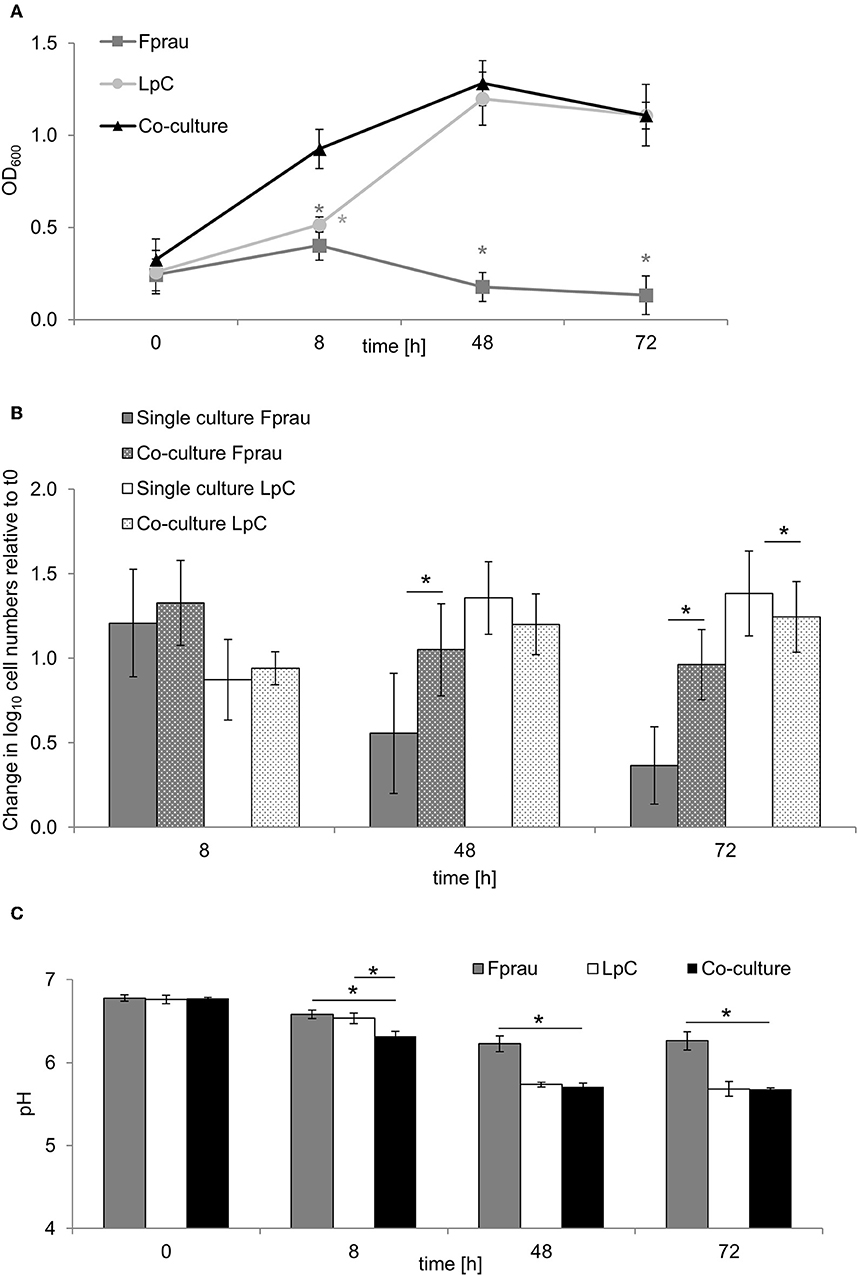
Figure 5. Single and co-culture study of L. paracasei (LpC) and F. prausnitzii (Fprau). (A) OD600 values of single and co-cultures of L. paracasei and F. prausnitzii. (B) Changes in cell numbers (log10 cell mL−1) of L. paracasei and F. prausnitzii in single and co-cultures measured with qPCR (C) pH values in single and co-cultures of L. paracasei and F. prausnitzii. Values are means ± SD of triplicate analysis of four separate experiments (n = 12), except for time point 72 h that was tested in three different experiments (n = 9); Values with an asterisk (*) indicate significant difference between single and co-cultures growth conditions (p < 0.05).
In co-cultures, maximum OD values for L. paracasei and F. prausnitzii were measured after 48 h (1.3 ± 0.1), they were similar to OD values recorded when L. paracasei was grown alone (1.2 ± 0.1). Also, pH values were similar in co-cultures and in L. paracasei single cultures and were significantly lower than of F. prausnitzii single cultures. L. paracasei cell numbers increased up to 48 h of incubation in single cultures as well as co-cultures (Figures 5A,B). In co-cultures, F. prausnitzii log cell numbers increased by 1.3 similar to single culture after 8 h incubation, however, log cell numbers decreased significantly less in co-culture thereafter. Butyrate concentrations were not significantly different in F. prausnitzii single cultures (5.3 ± 0.9 mM) compared to co-cultures (5.9 ± 2.2 mM) at all tested time points (Table S7).
To investigate the potential of L. paracasei to attenuate growth and toxin production of C. difficile, we first performed batch cultures of L. paracasei co-cultivated with C. difficile DSM 1296. Due to slower growth, L. paracasei was inoculated 5 h ahead of C. difficile to yield a balanced growth of both strains (Figure 6A). Significantly reduced viable cell numbers were measured for C. difficile after 10 and 13 h of fermentation in co-cultures (7.4 ± 0.1 and 7.3 ± 0.2 log10 cfu mL−1, respectively) compared to pure cultures (7.8 ± 0.1 and 7.9 ± 0.1 log10 cfu mL−1, respectively). After 25 h cell counts decreased to 5.3 ± 0.3 and 6.5 ± 0.2 in pure and co-cultures, respectively. Cytotoxin titers were significantly lower in co-cultures compared to single cultures after 13 and 25 h fermentation (Figure 6B). The pH was significantly lower in co-cultures compared to single cultures after 25 h (Table S8).
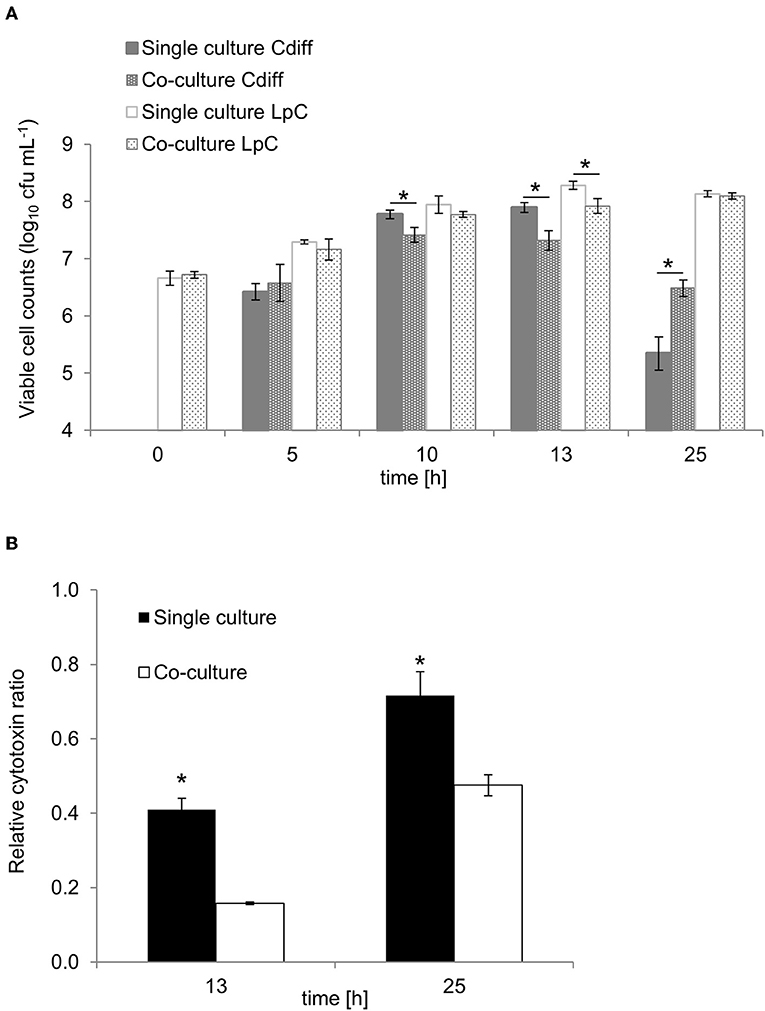
Figure 6. Growth and cytotoxin production in co-culture study of L. paracasei (LpC) with C. difficile DSM 1296 (Cdiff). (A) Viable cell counts (log10 cfu mL−1) of C. difficile and L. paracasei in single and co-cultures. (B) Cytotoxin titers produced during single and co-cultures after 13 and 25 h of incubation calculated relative to C. difficile cell counts. Values are means ± SD of triplicate analysis of two separate experiments (n = 6). Values with an asterisk (*) correspond to growth or cytotoxin titers in single cultures that are significantly different from co-cultures.
Lactate and formate formation were significantly higher in single cultures compared to co-cultures for L. paracasei and C. difficile, respectively, while acetate formation in C. difficile single cultures was similar to co-cultures (Table S9).
The interaction between L. paracasesi and C. difficile was further investigated in complex microbiota at distal colon conditions as we previously showed that C. difficile did not establish in the proximal colon section of our colonic fermentation models (35).
The impact of preventive L. paracasei inoculation on C. difficile DSM 1296 colonization was determined in model 2. In model 3, L. paracasei was inoculated after establishment of C. difficile and was tested on C. difficile NCTC 13307 colonization in combination with metronidazole, an antibiotic prescribed in case of CDI.
In the preventive approach (model 2), L. paracasei remained stable during days 6–11 in DC_LpC (log10 7.7 ± 0.3 cells mL−1 effluent, Figure 7A). However, after C. difficile inoculation cell numbers of L. paracasei increased by more than one log during the first days and remained constant thereafter, averaging log10 8.8 ± 0.1 cells mL−1 effluent. On day 11, C. difficile DSM 1296 (log10 8.5 cells) was added into both distal colon reactors of the model. During the first 24 h, C. difficile cell numbers decreased by approximately 1 log, and then steadily increased from day 13 to 16 to reach a similar concentration of log10 6.2 and 6.6 cells mL−1 in DC_CR and DC_LpC, respectively. Toxin titers were similar in control and treatment reactor throughout the fermentation period, with average titers of log10 2.9 ± 0.2 and 2.8 ± 0.2 per mL effluent in DC_CR and DC_LpC, respectively.
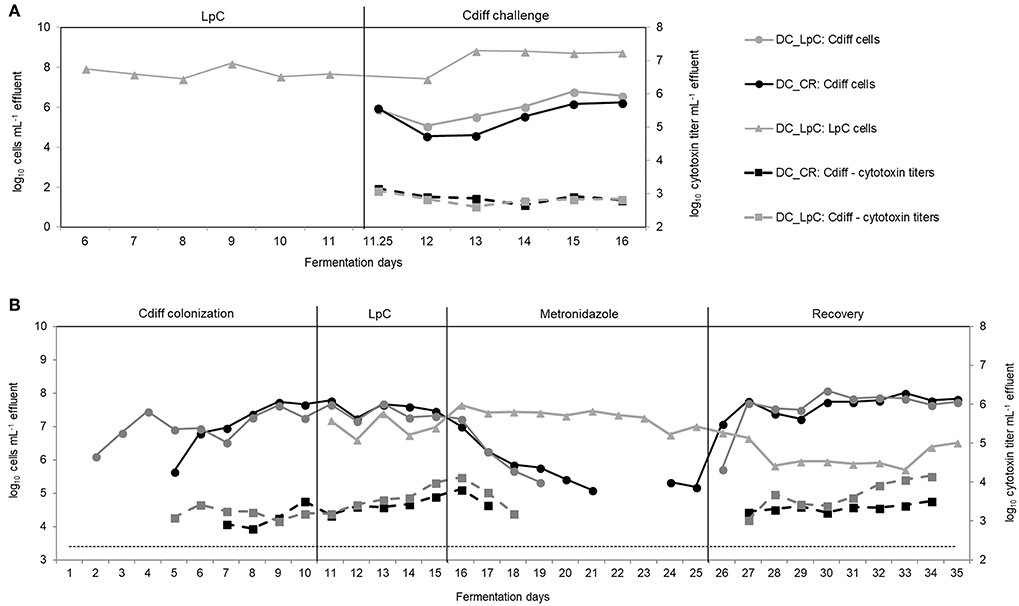
Figure 7. L. paracasei (LpC) and C. difficile (Cdiff) cell numbers (log10 cell mL−1) and cytotoxin titers in effluent samples from reactors mimicking the distal colon section. (A) Model 2. L. paracasei was inoculated twice daily from day 6 on into L. paracasei (LpC)-treated reactor mimicking the proximal colon section (PC_LpC) and on day 11 C. difficile DSM 1296 vegetative cells were inoculated into distal colon reactors of both test systems (DC_CR and DC_LpC). (B) Model 3. C. difficile NCTC 13307 was inoculated twice (day 1–2) in DC_CR and DC_LpC, and cell numbers and cytotoxin titer were monitored during different experimental periods. L. paracasei was inoculated twice daily in DC_LpC starting from day 11. Both reactors were treated with metronidazole from day 16–25 and recovery was monitored during days 26–35. (– – –) C. difficile detection limit of 3.4 log10 cell numbers mL−1.
Next, we examined the impact of L. paracasei on already established C. difficile in model 3. The effect of L. paracasei on C. difficile was assessed during 5 days after colonization of C. difficile NCTC 13307 and then during 10 days of metronidazole treatment followed by 10 days of post antibiotic recovery (Figure 7B). C. difficile cell numbers were similar in DC_CR and DC_LpC before the addition of L. paracasei with average values of log10 7.6 ± 0.2 and log10 7.4 ± 0.2 cells mL−1 effluent (day 8–10), respectively.
After addition, L. paracasei cell numbers ranged at log10 7.0 ± 0.3 per mL and slightly increased during metronidazole treatment (log10 7.3 ± 0.3 cells mL−1). During the first 3 days of recovery L. paracasei decreased by around 1 log and reached final abundance of log10 6.0 ± 0.3 cells mL−1 at day 35. C. difficile NCTC 13307 growth continuously decreased in both reactors during metronidazole treatment and reached the detection limit after 4 days in DC_LpC compared to 6 days in DC_CR. Similarly, toxin production was below detection limit shortly after start of metronidazole treatment. C. difficile reappeared at 5.3 cells mL−1 in DC_CR at days 24 and 25 of metronidazole treatment. A fast recovery of C. difficile was observed in both DC's with a slight delay for DC_LpC. After 2 day recovery, C. difficile cell numbers were stable in both reactors (log10 7.7 ± 0.2 and 7.8 ± 0.2 mL−1 in DC_CR and DC_LpC, respectively) until the end of the period. Cytotoxin titers were similar in both reactors during the recovery period.
Probiotics can transiently colonize the human colon, leading to an alteration of both the composition and activity of the commensal microbiota (50). It was suggested that these changes could enhance general homeostasis of the gut microbiota, thereby preventing overgrowth of enteric pathogens such as C. difficile (51). The hindered accessibility of the gastrointestinal tract hampers clinical studies on the effect of probiotics on gut microbiota of different colonic sections. In the current work we applied different in vitro colonic fermentation models operated with controlled conditions to test the response of the commensal elderly gut microbiota to the probiotic strain L. paracasei CNCM I-1518 and to investigate the probiotic-pathogen interaction with C. difficile, independently of host factors such as the epithelial cell layer and immune response.
Levels of L. paracasei applied in the in vitro model were in the range of fecal concentrations of the same strain assessed in vivo (24, 52). Upon daily addition, numbers of L. paracasei remained stable or even increased indicating the possibility of temporary persistence. Colonization of L. paracasei in the reactors was nevertheless transient since cessation of probiotic addition was accompanied by a rapid wash-out within 3 days (data not shown). Transient properties were ascribed to many other Lactobacillus species used as probiotics (50).
Despite low abundance relative to commensal bacteria and transient properties, probiotics impact elderly microbiota composition. Consumption of probiotics led to an increase in bifidobacteria, lactobacilli or Faecalibacterium (53–55) and decreased the abundance of opportunistic pathogens (56). Here, we showed increases in abundances of phylotypes belonging to the Lactobacillales, including Lactobacillus and Enterococcus, suggesting that L. paracasei enhances niche colonization of closely related genera (57). An increase in fecal concentrations of the Lactobacillus/Enterococcus group was previously observed upon consumption of fermented milk containing a Lactobacillus salivarius strain (58) or Lactobacillus rhamnosus (59) by healthy adults.
It was earlier reported that Bacteroidetes were more abundant in elderly compared to adults (60). Here, L. paracasei supplementation was associated with decreased abundances of phylotypes affiliated to Bacteroidetes. Compositional modifications observed after addition of L. paracasei might be related to changes in trophic interactions.
In recent studies no effect on fecal microbiota composition but community-wide transcriptional changes were observed after consumption of fermented milk products (61, 62). McNulty et al. observed that 7-weeks consumption of fermented milk product containing B. animalis subsp lactis CNCM I-2494 did not change composition based on 16S rRNA gene sequencing, but interesting changes in fecal gene expression were measured, notably related to plant polysaccharide metabolism and SCFA production, indicating an expansion of the carbohydrate metabolizing ability of the microbiota during transient colonization with the product (62). In another study, healthy elderly subjects were given Lactobacillus rhamnosus GG (61). Similarly, as McNulty et al., no modification of the microbiota composition but a clear transcriptional response was observed. Across all functional categories, increased expression of genes involved in flagellar motility, chemotaxis, and adhesion from Bifidobacterium and the dominant butyrate producers Roseburia and Eubacterium were observed during probiotic consumption. These studies highlight the interest of analyzing transcriptional activity of gut microbiota. Here we determined the impact of L. paracasei CNCM I-1518 on the proximal colon microbiota. Colonic microbiota transcriptional functional profile varied little during the test period confirming stability of the fermentation model. The high proportion of transcripts assigned to “Carbohydrates” and “Protein metabolism” indicated bacterial growth, carbohydrate fermentation and SCFA formation in the presence of high substrate concentrations, successfully mimicking the scenario in the human proximal colon as observed before (63). Despite shifts in microbiota composition, we only observed minor alterations of relative abundance of microbial functions upon addition of L. paracasei, possibly due to functional redundancy of the intestinal microbiota. For example, a shift in butyrate-producing phylotypes after addition of L. paracasei with decreased abundance of Roseburia and Peptoniphilus, and increased abundance of Faecalibacterium was observed, nevertheless, relative abundance of the SEED category “Fermentation” including transcripts related to butyrate formation was not impacted.
SCFA profiles in both control and treated reactors were highly comparable despite the presence of L. paracasei. A positive correlation between L. paracasei supplementation and Faecalibacterium was reported earlier for healthy adults (64). In co-cultures, L. paracasei enhanced survival of F. prausnitzii in the stationary growth phase. A possible reason could be the lower pH in co-cultures compared to pure F. prausnitzii cultures that may have protected cells from autolysis as it has been observed in several bacterial species (65). That there was no negative impact on growth of F. prausnitzii by L. paracasei, indicates that both strains do not compete for substrates or are inhibited by metabolites formed in the test conditions.
Probiotics were suggested as alternative treatment for gastrointestinal diseases including antibiotic-related infections, such as CDI (66). CDI often occurs after treatment with broad-spectrum antibiotics and incidences are increased in the elderly population (13). Several studies reported a reduction in C. difficile-associated diarrhea with probiotics, including Saccharomyces boulardii (67) and Lactobacillus acidophilus in combination with L. casei (68) as well as a milk drink with yogurt starter bacteria and L. paracasei (29, 30). S. boulardii acts by secreting a protease which is able to cleave toxin A and possesses enzymatic activity against C. difficile toxin B (69). However, for most probiotics the mechanism of action in CDI remains unknown. Here, we found reduced C. difficile cytotoxin titers when co-cultivated with L. paracasei compared to pure cultures, but no reduction in continuous fermentation studies with L. paracasei tested as preventive treatment (model 2) or as adjuvant therapy to metronidazole treatment (model 3). During co-cultures, C. difficile was inoculated 5 h after L. paracasei to account for the fast growth of the first, while for the modeled microbiota, C. difficile only grew in distal reactor conditions set at a pH of 6.8 which is less favorable for lactobacilli growth (35). Our data suggest that host factors that are not accounted in the in vitro colonic fermentation model, may contribute to the prevention effect of probiotics observed in vivo (19, 70).
To conclude, this is the first-time investigation of the effect of a candidate probiotic strain on the transcriptome of elderly gut microbiota using in vitro intestinal fermentation models. We showed a compositional and functional response of the microbiota on L. paracasei, with an enhancing effect on Faecalibacterium abundance and activity, a decrease in abundance of H2 and CH4 fermentative bacteria, and an increase in carbohydrate utilization, indicating a possible contribution of L. paracasei in the trophic interaction of dietary carbohydrate utilization with the commensal microbiota. We thus showed that the L. paracasei strain directly interacts with the human gut microbiota independent of the host. In contrast, no effect of L. paracasei was observed on C. difficile in complex microbiota uncoupled from the host when tested as preventive treatment or concomitantly to metronidazole, which may be partly due to the limits of in vitro microbiota models not accounting for host factors. Thus, host-microbiota interaction studies should be conducted for further investigations of the mechanism of L. paracasei in treatment or prevention of CDI.
The datasets generated for this study can be found in the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA) under bioproject accession number SRP144222.
The Ethics Committee of ETH Zurich exempted this study from review because sample collection was not in terms of intervention.
SF, CC, CS, CF, MD, and CL conceived and designed the experiments. SF and MV conducted the experiments. SF, MV, CS, and MD conducted the analysis. SF, CC, CS, MD, and CL interpreted the results and wrote the paper. All authors read and approved the final manuscript.
The authors declare that this study received funding from Danone Research (Palaiseau, France). The funder had the following involvement with the study: MD and CF were involved in study design and in the preparation of the manuscript.
MD and CF are employed by Danone Research (Palaiseau, France).
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank Lennart Opitz for help with bioinformatics analysis of metatranscriptomics data.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fnut.2019.00184/full#supplementary-material
1. Vollaard EJ, Clasener HA. Colonization resistance. Antimicrob Agents Chemother. (1994) 38:409–14. doi: 10.1128/AAC.38.3.409
2. Robles Alonso V, Guarner F. Linking the gut microbiota to human health. Br J Nutr. (2013) 109 (Suppl. 2):S21–6. doi: 10.1017/S0007114512005235
3. Kamada N, Chen GY, Inohara N, Núñez G. Control of pathogens and pathobionts by the gut microbiota. Nat Immunol. (2013) 14:685–90. doi: 10.1038/ni.2608
4. Lawson PA, Citron DM, Tyrrell KL, Finegold SM. Reclassification of Clostridium difficile as Clostridioides difficile (Hall and O'Toole 1935) Prevot 1938. Anaerobe. (2016) 40:95–9. doi: 10.1016/j.anaerobe.2016.06.008
5. O'Toole PW, Claesson MJ. Gut microbiota: changes throughout the lifespan from infancy to elderly. Int Dairy J. (2010) 20:281–91. doi: 10.1016/j.idairyj.2009.11.010
6. Duncan SH, Flint HJ. Probiotics and prebiotics and health in ageing populations. Maturitas. (2013) 75:44–50. doi: 10.1016/j.maturitas.2013.02.004
7. Jeffery IB, Lynch DB, O'Toole PW. Composition and temporal stability of the gut microbiota in older persons. ISME J. (2016) 10:170–82. doi: 10.1038/ismej.2015.88
8. Bian G, Gloor GB, Gong A, Jia C, Zhang W, Hu J, et al. The gut microbiota of healthy aged Chinese is similar to that of the healthy young. mSphere. (2017) 2:e00327–17. doi: 10.1128/mSphere.00327-17
9. Woodmansey EJ, McMurdo ME, Macfarlane GT, Macfarlane S. Comparison of compositions and metabolic activities of fecal microbiotas in young adults and in antibiotic-treated and non-antibiotic-treated elderly subjects. Appl Environ Microbiol. (2004) 70:6113–22. doi: 10.1128/AEM.70.10.6113-6122.2004
10. Mueller S, Saunier K, Hanisch C, Norin E, Alm L, Midtvedt T, et al. Differences in fecal microbiota in different European study populations in relation to age, gender, and country: a cross-sectional study. Appl Environ Microbiol. (2006) 72:1027–33. doi: 10.1128/AEM.72.2.1027-1033.2006
11. Biagi E, Nylund L, Candela M, Ostan R, Bucci L, Pini E, et al. Through ageing, and beyond: gut microbiota and inflammatory status in seniors and centenarians. PLoS ONE. (2010) 5:e10667. doi: 10.1371/journal.pone.0010667
12. Odamaki T, Kato K, Sugahara H, Hashikura N, Takahashi S, Xiao JZ, et al. Age-related changes in gut microbiota composition from newborn to centenarian: a cross-sectional study. BMC Microbiol. (2016) 16:90. doi: 10.1186/s12866-016-0708-5
13. Oldfield IEC, Johnson DA. Clinical update for the diagnosis and treatment of Clostridium difficile infection. World J Gastrointest Pharmacol Ther. (2014) 5:1–26. doi: 10.4292/wjgpt.v5.i1.1
14. Tiihonen K, Tynkkynen S, Ouwehand A, Ahlroos T, Rautonen N. The effect of ageing with and without non-steroidal anti-inflammatory drugs on gastrointestinal microbiology and immunology. Br J Nutr. (2008) 100:130–7. doi: 10.1017/S000711450888871X
16. Barker AK, Duster M, Valentine S, Hess T, Archbald-Pannone L, Guerrant R, et al. A randomized controlled trial of probiotics for Clostridium difficile infection in adults (PICO). J Antimicrob Chemother. (2017) 72:3177–80. doi: 10.1093/jac/dkx254
17. Sanders ME, Merenstein D, Merrifield CA, Hutkins R. Probiotics for human use. Nutr Bull. (2018) 43:212–25. doi: 10.1111/nbu.12334
18. Goldenberg JZ, Mertz D, Johnston BC. Probiotics to prevent Clostridium difficile infection in patients receiving antibiotics. JAMA. (2018) 320:499–500. doi: 10.1001/jama.2018.9064
19. Shen NT, Maw A, Tmanova LL, Pino A, Ancy K, Crawford CV, et al. Timely use of probiotics in hospitalized adults prevents Clostridium difficile infection: a systematic review with meta-regression analysis. Gastroenterology. (2017) 152:1889–900 e9. doi: 10.1053/j.gastro.2017.02.003
20. Oelschlaeger TA. Mechanisms of probiotic actions—A review. Int J Med Microbiol. (2010) 300:57–62. doi: 10.1016/j.ijmm.2009.08.005
21. Martinez RC, Bedani R, Saad SM. Scientific evidence for health effects attributed to the consumption of probiotics and prebiotics: an update for current perspectives and future challenges. Br J Nutr. (2015) 114:1993–2015. doi: 10.1017/S0007114515003864
22. Ouwehand AC, Salminen S, Isolauri E. Probiotics: an overview of beneficial effects. Antonie Van Leeuwenhoek. (2002) 82:279–89. doi: 10.1023/A:1020620607611
23. Rochet V, Rigottier-Gois L, Levenez F, Cadiou J, Marteau P, Bresson JL, et al. Modulation of Lactobacillus casei in ileal and fecal samples from healthy volunteers after consumption of a fermented milk containing Lactobacillus casei DN-114 001Rif. Can J Microbiol. (2008) 54:660–7. doi: 10.1139/W08-050
24. Oozeer R, Leplingard A, Mater DD, Mogenet A, Michelin R, Seksek I, et al. Survival of Lactobacillus casei in the human digestive tract after consumption of fermented milk. Appl Environ Microbiol. (2006) 72:5615–7. doi: 10.1128/AEM.00722-06
25. Tien MT, Girardin SE, Regnault B, Le Bourhis L, Dillies MA, Coppée JY, et al. Anti-inflammatory effect of Lactobacillus casei on Shigella-infected human intestinal epithelial cells. J Immunol. (2006) 176:1228–37. doi: 10.4049/jimmunol.176.2.1228
26. Borruel N, Carol M, Casellas F, Antolín M, de Lara F, Espín E, et al. Increased mucosal tumour necrosis factor alpha production in Crohn's disease can be downregulated ex vivo by probiotic bacteria. Gut. (2002) 51:659–64. doi: 10.1136/gut.51.5.659
27. Belkacem N, Serafini N, Wheeler R, Derrien M, Boucinha L, Couesnon A, et al. Lactobacillus paracasei feeding improves immune control of influenza infection in mice. PLoS ONE. (2017) 12:e0184976. doi: 10.1371/journal.pone.0184976
28. Guillemard E, Tondu F, Lacoin F, Schrezenmeir J. Consumption of a fermented dairy product containing the probiotic Lactobacillus casei DN-114001 reduces the duration of respiratory infections in the elderly in a randomised controlled trial. Br J Nutr. (2010) 103:58–68. doi: 10.1017/S0007114509991395
29. Hickson M, D'Souza AL, Muthu N, Rogers TR, Want S, Rajkumar C, et al. Use of probiotic Lactobacillus preparation to prevent diarrhoea associated with antibiotics: randomised double blind placebo controlled trial. Br Med J. (2007) 335:80. doi: 10.1136/bmj.39231.599815.55
30. Alberda C, Marcushamer S, Hewer T, Journault N, Kutsogiannis D. Feasibility of a Lactobacillus casei drink in the intensive care unit for prevention of antibiotic associated diarrhea and Clostridium difficile. Nutrients. (2018) 10:539. doi: 10.3390/nu10050539
31. Payne AN, Zihler A, Chassard C, Lacroix C. Advances and perspectives in in vitro human gut fermentation modeling. Trends Biotechnol. (2012) 30:17–25. doi: 10.1016/j.tibtech.2011.06.011
32. Lacroix C, de Wouters T, Chassard C. Integrated multi-scale strategies to investigate nutritional compounds and their effect on the gut microbiota. Curr Opin Biotechnol. (2015) 32C:149–55. doi: 10.1016/j.copbio.2014.12.009
33. Zihler Berner A, Fuentes S, Dostal A, Payne AN, Vazquez Gutierrez P, Chassard C, et al. Novel Polyfermentor intestinal model (PolyFermS) for controlled ecological studies: validation and effect of pH. PLoS ONE. (2013) 8:e77772. doi: 10.1371/journal.pone.0077772
34. Fehlbaum S, Chassard C, Haug MC, Fourmestraux C, Derrien M, Lacroix C. Design and investigation of PolyFermS in vitro continuous fermentation models inoculated with immobilized fecal microbiota mimicking the elderly colon. PLoS ONE. (2015) 10:e0142793. doi: 10.1371/journal.pone.0142793
35. Fehlbaum S, Chassard C, Poeker SA, Derrien M, Fourmestraux C, Lacroix C. Clostridium difficile colonization and antibiotics response in PolyFermS continuous model mimicking elderly intestinal fermentation. Gut Pathog. (2016) 8:63. doi: 10.1186/s13099-016-0144-y
36. Bryant MP. Commentary on the Hungate technique for culture of anaerobic bacteria. Am J Clin Nutr. (1972) 25:1324–8. doi: 10.1093/ajcn/25.12.1324
37. Sorg JA, Dineen SS. Laboratory maintenance of Clostridium difficile. Curr Protoc Microbiol. (2009) Chapter 9:Unit9A.1. doi: 10.1002/9780471729259.mc09a01s12
38. Duncan SH, Hold GL, Barcenilla A, Stewart CS, Flint HJ. Roseburia intestinalis sp. nov., a novel saccharolytic, butyrate-producing bacterium from human faeces. Int J Syst Evol Microbiol. (2002) 52(Pt 5):1615–20. doi: 10.1099/ijs.0.02143-0
39. Zihler A, Gagnon M, Chassard C, Hegland A, Stevens MJ, Braegger CP, et al. Unexpected consequences of administering bacteriocinogenic probiotic strains for Salmonella populations, revealed by an in vitro colonic model of the child gut. Microbiology. (2010) 156(Pt 11):3342–53. doi: 10.1099/mic.0.042036-0
40. Větrovský T, Baldrian P. The variability of the 16S rRNA gene in bacterial genomes and its consequences for bacterial community analyses. PLoS ONE. (2013) 8:e57923. doi: 10.1371/journal.pone.0057923
41. Dostal A, Fehlbaum S, Chassard C, Zimmermann MB, Lacroix C. Low iron availability in continuous in vitro colonic fermentations induces strong dysbiosis of the child gut microbial consortium and a decrease in main metabolites. FEMS Microbiol Ecol. (2013) 83:161–75. doi: 10.1111/j.1574-6941.2012.01461.x
42. Andersson AF, Lindberg M, Jakobsson H, Bäckhed F, Nyrén P, Engstrand L. Comparative analysis of human gut microbiota by barcoded pyrosequencing. PLoS ONE. (2008) 3:e2836. doi: 10.1371/journal.pone.0002836
43. Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. (2010) 7:335–6. doi: 10.1038/nmeth.f.303
44. Stevens MJ, Wiersma A, de Vos WM, Kuipers OP, Smid EJ, Molenaar D, et al. Improvement of Lactobacillus plantarum aerobic growth as directed by comprehensive transcriptome analysis. Appl Environ Microbiol. (2008) 74:4776–8. doi: 10.1128/AEM.00136-08
45. Kopylova E, Noé L, Touzet H. SortMeRNA: fast and accurate filtering of ribosomal RNAs in metatranscriptomic data. Bioinformatics. (2012) 28:3211–7. doi: 10.1093/bioinformatics/bts611
46. Magoč T, Salzberg SL. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics. (2011) 27:2957–63. doi: 10.1093/bioinformatics/btr507
47. Lanzén A, Jørgensen SL, Huson DH, Gorfer M, Grindhaug SH, Jonassen I, et al. CREST–classification resources for environmental sequence tags. PLoS ONE. (2012) 7:e49334. doi: 10.1371/journal.pone.0049334
48. Buchfink B, Xie C, Huson DH. Fast and sensitive protein alignment using DIAMON. Nat Methods. (2015) 12:59–60. doi: 10.1038/nmeth.3176
49. Huson DH, Auch AF, Qi J, Schuster SC. MEGAN analysis of metagenomic data. Genome Res. (2007) 17:377–86. doi: 10.1101/gr.5969107
50. Derrien M, van Hylckama Vlieg JE. Fate, activity, and impact of ingested bacteria within the human gut microbiota. Trends Microbiol. (2015) 23:354–66. doi: 10.1016/j.tim.2015.03.002
51. Hell M, Bernhofer C, Stalzer P, Kern JM, Claassen E. Probiotics in Clostridium difficile infection: reviewing the need for a multistrain probiotic. Benef Microbes. (2013) 4:39–51. doi: 10.3920/BM2012.0049
52. Collins JW, Chervaux C, Raymond B, Derrien M, Brazeilles R, Kosta A, et al. Fermented dairy products modulate Citrobacter rodentium-induced colonic hyperplasia. J Infect Dis. (2014) 210:1029–41. doi: 10.1093/infdis/jiu205
53. Lahtinen SJ, Forssten S, Aakko J, Granlund L, Rautonen N, Salminen S, et al. Probiotic cheese containing Lactobacillus rhamnosus HN001 and Lactobacillus acidophilus NCFM(R) modifies subpopulations of fecal lactobacilli and Clostridium difficile in the elderly. Age. (2012) 34:133–43. doi: 10.1007/s11357-011-9208-6
54. Ouwehand AC, Bergsma N, Parhiala R, Lahtinen S, Gueimonde M, Finne-Soveri H, et al. Bifidobacterium microbiota and parameters of immune function in elderly subjects. FEMS Immunol Med Microbiol. (2008) 53:18–25. doi: 10.1111/j.1574-695X.2008.00392.x
55. Gao R, Zhang X, Huang L, Shen R, Qin H. Gut microbiota alteration after long-term consumption of probiotics in the elderly. Probiotics Antimicrob Proteins. (2019) 11:655–66. doi: 10.1007/s12602-018-9403-1
56. Rampelli S, Candela M, Severgnini M, Biagi E, Turroni S, Roselli M, et al. A probiotics-containing biscuit modulates the intestinal microbiota in the elderly. J Nutr Health Aging. (2013) 17:166–72. doi: 10.1007/s12603-012-0372-x
57. Stecher B, Chaffron S, Käppeli R, Hapfelmeier S, Freedrich S, Weber TC, et al. Like will to like: abundances of closely related species can predict susceptibility to intestinal colonization by pathogenic and commensal bacteria. PLoS Pathog. (2010) 6:e1000711. doi: 10.1371/journal.ppat.1000711
58. Collins JK, Dunne C, Murphy L, Morrissey D, O'Mahony L, O'Sullivan E, et al. A randomised controlled trial of a probiotic Lactobacillus strain in healthy adults: assessment of its delivery, transit and influence on microbial flora and enteric immunity. Microb Ecol Health Dis. (2002) 14:81–9. doi: 10.1080/08910600260081720
59. Tannock GW, Munro K, Harmsen HJ, Welling GW, Smart J, Gopal PK. Analysis of the fecal microflora of human subjects consuming a probiotic product containing Lactobacillus rhamnosus DR20. Appl Environ Microbiol. (2000) 66:2578–88. doi: 10.1128/AEM.66.6.2578-2588.2000
60. Claesson MJ, Cusack S, O'Sullivan O, Greene-Diniz R, de Weerd H, Flannery E, et al. Composition, variability, and temporal stability of the intestinal microbiota of the elderly. Proc Natl Acad Sci USA. (2011) 108 (Suppl. 1):4586–91. doi: 10.1073/pnas.1000097107
61. Eloe-Fadrosh EA, Brady A, Crabtree J, Drabek EF, Ma B, Mahurkar A, et al. Functional dynamics of the gut microbiome in elderly people during probiotic consumption. MBio. (2015) 6:e00231–15. doi: 10.1128/mBio.00231-15
62. McNulty NP, Yatsunenko T, Hsiao A, Faith JJ, Muegge BD, Goodman AL, et al. The impact of a consortium of fermented milk strains on the gut microbiome of gnotobiotic mice and monozygotic twins. Sci Transl Med. (2011) 3:106ra106. doi: 10.1126/scitranslmed.3002701
63. Doo EH, Chassard C, Schwab C, Lacroix C. Effect of dietary nucleosides and yeast extracts on composition and metabolic activity of infant gut microbiota in PolyFermS colonic fermentation models. FEMS Microbiol Ecol. (2017) 93:fix088. doi: 10.1093/femsec/fix088
64. Zhang J, Wang L, Guo Z, Sun Z, Gesudu Q, Kwok L, et al. 454 pyrosequencing reveals changes in the faecal microbiota of adults consuming Lactobacillus casei Zhang. FEMS Microbiol Ecol. (2014) 88:612–22. doi: 10.1111/1574-6941.12328
65. Rice KC, Bayles KW. Molecular control of bacterial death and lysis. Microbiol Mol Biol Rev. (2008) 72:85–109. doi: 10.1128/MMBR.00030-07
66. Sanders ME, Guarner F, Guerrant R, Holt PR, Quigley EM, Sartor RB, et al. An update on the use and investigation of probiotics in health and disease. Gut. (2013) 62:787–96. doi: 10.1136/gutjnl-2012-302504
67. Pothoulakis C. Review article: anti-inflammatory mechanisms of action of Saccharomyces boulardii. Aliment Pharmacol Ther. (2009) 30:826–33. doi: 10.1111/j.1365-2036.2009.04102.x
68. Gao XW, Mubasher M, Fang CY, Reifer C, Miller LE. Dose-response efficacy of a proprietary probiotic formula of Lactobacillus acidophilus CL1285 and Lactobacillus casei LBC80R for antibiotic-associated diarrhea and Clostridium difficile-associated diarrhea prophylaxis in adult patients. Am J Gastroenterol. (2010) 105:1636–41. doi: 10.1038/ajg.2010.11
69. Castagliuolo I, Riegler MF, Valenick L, LaMont JT, Pothoulakis C. Saccharomyces boulardii protease inhibits the effects of Clostridium difficile toxins A and B in human colonic mucosa. Infect Immun. (1999) 67:302–7.
Keywords: Lactobacillus paracasei CNCM I-1518, Clostridioides difficile, gut microbiota, intestinal model, metataxonomics, metatranscriptomics, Faecalibacterium, elderly
Citation: Fehlbaum S, Chassard C, Schwab C, Voolaid M, Fourmestraux C, Derrien M and Lacroix C (2019) In vitro Study of Lactobacillus paracasei CNCM I-1518 in Healthy and Clostridioides difficile Colonized Elderly Gut Microbiota. Front. Nutr. 6:184. doi: 10.3389/fnut.2019.00184
Received: 24 September 2019; Accepted: 22 November 2019;
Published: 10 December 2019.
Edited by:
Clara G. De Los Reyes-Gavilan, Spanish National Research Council (CSIC), SpainReviewed by:
Susana Delgado, Institute of Dairy Products of Asturias (IPLA), SpainCopyright © 2019 Fehlbaum, Chassard, Schwab, Voolaid, Fourmestraux, Derrien and Lacroix. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Christophe Lacroix, Y2hyaXN0b3BoZS5sYWNyb2l4QGhlc3QuZXRoei5jaA==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.