- 1Institute of Food Sciences, National Research Council (CNR-ISA), Avellino, Italy
- 2Institute of Biomembranes, Bioenergetics and Molecular Biotechnologies, National Research Council (CNR-IBIOM), Bari, Italy
Probiotics are commensal microorganisms that are present in the intestinal tract and in many fermented foods and positively affect human health, promoting digestion and uptake of dietary nutrients, strengthening intestinal barrier function, modulating immune response, and enhancing antagonism toward pathogens. The proteosurfaceome, i.e., the complex set of proteins present on the bacterial surface, is directly involved as leading actor in the dynamic communication between bacteria and host. In the last decade, the biological relevance of surface-exposed proteins prompted research activities exploiting the potentiality of proteomics to define the complex network of proteins that are involved in the molecular mechanisms at the basis of the adaptation to gastrointestinal environment and the probiotic effects. These studies also took advantages of the recent technological improvements in proteomics, mass spectrometry and bioinformatics that triggered the development of ad hoc designed innovative strategies to characterize the bacterial proteosurfaceome. This mini-review is aimed at describing the key role of proteomics in depicting the cell wall protein architecture and the involvement of surface-exposed proteins in the intimate and dynamic molecular dialogue between probiotics and intestinal epithelial and immune cells.
Introduction
Probiotics are commensal microorganisms that are present in the intestinal tract and in many fermented foods, and are defined as “live microorganisms that, when administered in adequate amounts, confer a health benefit on the host” (1). Most probiotics are Gram-positive bacteria, mainly lactic acid bacteria (LAB), and Bifidobacteria (2). More recently, Propionibacterium freudenreichii, a beneficial bacterium traditionally used as a cheese ripening starter, has been recognized to exhibit probiotic abilities, some of these due to the production of nutraceuticals and beneficial metabolites (3). The mechanisms by which probiotics positively affect human health include promotion of digestion and uptake of dietary nutrients, strengthening of intestinal barrier function, modulation of immune response and enhancement of antagonism toward pathogens, either by producing antimicrobial compounds or through competition for mucosal binding sites (4, 5).
Surface-exposed proteins constitute the first-line of contact between bacteria and host being leading actors in this complex interplay as they directly interact with epithelial and immune cells (6). This interaction could lead to the inhibition or activation of key signaling pathways in the intestinal cells involving nuclear factor-κB (NFκB) and mitogen-activated protein kinases (MAPKs), and influencing the regulation of downstream pathways such as the secretion of cytokines (chemokines and interleukins) responsible for the immunomodulation or antibacterial peptides (defensins). The activation of these cascades prompts physiological modifications (increase of mucin secretion, changes in the surface properties, rearrangement of the tight junctions, etc.) and changes in genetic expression that could affect cell proliferation and survival (7–9).
The in-depth analysis of the proteosurfaceome, defined as “the proteinaceous subset of the surfaceome found at the cell wall and totally or partially exposed on the external side of the cell membrane,” represents a crucial point to elucidate molecular mechanisms underlying host/probiotic crosstalk (10, 11). In general, sorting of bacterial proteins to the cell surface is governed by the presence or absence of signal peptides which direct them to the protein export machinery thus allowing their migration to bacterial surface, and surface-retention domains responsible for their anchoring to cell wall or cytoplasmic membrane (12). Surface proteins can be mainly divided in four groups: (i) proteins anchored to the cytoplasmic membrane by hydrophobic transmembrane domain(s) (integral membrane proteins, IMP), (ii) lipoproteins which are covalently attached to membrane lipids after cleavage of a signal peptide by signal peptidase II, (iii) proteins containing C-terminal LPXTG-like motif and covalently attached to peptidoglycan by sortases, and (iv) non-covalently bound proteins which are associated to the cell wall through weak interactions (van der Waals forces, hydrogen or ion bonds) taking advantage of conserved structural domains (LysM proteins, WXL proteins, GW proteins, proteins with choline binding domains). Some bacteria can also show supramolecular structures, formed by the assembly of specific protein subunits on the cell envelope, such as flagella, mainly involved in cell motility, and pili or fimbriae, mainly involved in cell adhesion, aggregation, and immunomodulation (10, 11, 13). Furthermore, in several bacterial species (Lactobacillus acidophilus, Lactobacillus helveticus, P. freudenreichii, etc.) the outermost constituent of the cell wall is represented by a S-layer that is a semiporous proteinaceous crystalline array composed of self-assembling (glyco)protein subunits called S-layer proteins (SLPs) which act as a scaffold for non-covalently attached secreted proteins (also named S-layer associated proteins, SLAPs). SLPs are involved in several processes including maintaining cell shape, acting as molecular sieves, serving as binding sites, protecting against environmental stresses and mediating bacterial adhesion and gut immune response (14, 15). Noteworthy, cytoplasmic housekeeping proteins (metabolic enzymes, molecular chaperones, translational elongation factors, ribosomal proteins, etc.) have been identified in bacterial surface proteomes. Such proteins, defined as anchorless proteins or moonlighting proteins, display different, seemingly unrelated, functions in different cell locations, and lack any extra-cytoplasmic sorting sequence or binding domain, so that non-canonical secretion pathways have been hypothesized (16, 17).
Subcellular localization of bacterial proteins has been postulated by several bioinformatics tools mainly based on predictive algorithms. The most widely used tools are PSORTb v3.0 server that allows to obtain subcellular localization prediction on the basis of a multi-component approach including different modules such as SCL-BLAST for homology-based prediction, HMMTOP transmembrane helix prediction tool and a signal peptide identification tool (18, 19) and SignalP server, that predicts the presence and location of signal peptide cleavage sites in amino acid sequences from prokaryotes and eukaryotes (20). Prediction of transmembrane-spanning domains (helices) in IMP could be carried out by TMHMM v2.0 (21). Prediction of non-classically secreted proteins (such as moonlighting proteins) could be achieved by SecretomeP server (22) and MoonProt (23).
In the last decades, proteomics significantly contributed to depict an overall picture of the proteosurfeoceome through the identification of hundreds of proteins in a single analysis. In fact, this approach provided direct information on protein localization and topology, thus corroborating the bioinformatics prediction with experimental evidence, and allowed the identification of moonlighthing proteins, “unexpectedly” present on cell surface. More importantly, as the proteosurfaceome is a highly dynamic entity, tightly modulated by the host/bacteria molecular dialogue, proteomics represent the most suitable tool to monitor the wide reorganization of the surface proteins induced by either gastro intestinal tract (GIT) environment or other growth conditions, that could modulate the probiotic functionalities.
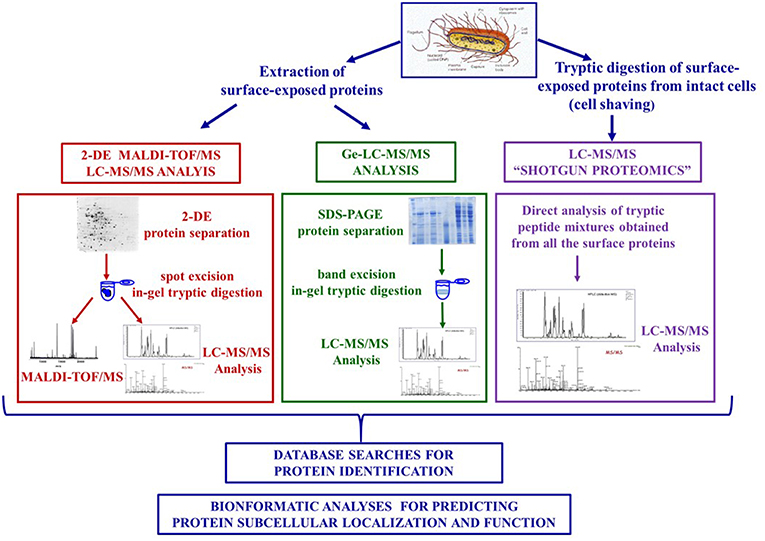
However, the proteomic analysis of these proteins proved to be a challenging task due to their low abundance and hydrophobicity. First generation proteomic strategies integrated protein extraction from subcellular fractions, two dimensional electrophoresis (2-DE) and mass spectrometry, i.e., Matrix Assisted Laser Desorption Ionization—Time of Flight Mass Spectrometry (MALDI-TOF-MS) or nano-liquid chromatography coupled to tandem mass spectrometry (LC-MS/MS) and usually led to the identification of a limited number of surface proteins as these are hardly amenable to 2-DE analysis. The need to overcome these experimental drawbacks successively prompted the design of sophisticated strategies that selectively targeted surface proteins either by proteases digestion (shaving procedures) or biotin labeling (protein biotinylation procedures) of intact cells (24). These approaches exploited technological advancements in mass spectrometry and proteomics that allowed the development of gel-free proteomic strategies (also named shotgun proteomics) based on the direct analysis of peptides obtained from the tryptic digestion of the entire proteome by LC-MS/MS and Ge-LC-MS/MS approaches that include a preliminary SDS-PAGE fractionation step of the extracted proteins (25) (Figure 1).
This mini-review is aimed at describing both methods applied in the last years to define the bacterial proteosurfaceome and proteomic studies that significantly contributed in depicting the cell wall protein architecture and the involvement of surface-exposed proteins in the intimate and dynamic molecular dialogue between probiotics and host.
Proteomic Methods for the Analyis of Surface-Exposed Proteins
Protocols for Surface Protein Extraction
The analysis of surface proteins of Gram positive bacteria takes advantage of the cell envelope structure consisting of a cross-linked peptidoglycan layer. Cell wall degradation of intact cells is carried out for a short time (30–60 min) using enzymes such as lysozyme, mutanolysin (glycosidases), and/or lysostaphin (endopeptidase) in a buffer containing protease inhibitors and high sugar concentrations to create an osmotic pressure in order to maintain protoplast integrity and prevent contamination of cytoplasmic proteins. Surface proteins contained in the supernatants are recovered by centrifugation, precipitated, separated by 2-DE, and identified by mass spectrometry (26–29) (Figure 1). Denaturating agents (urea, guanidine-HCl, SDS buffers) have been also used to extract non-covalently bound surface proteins. Different protocols tailored to specific bacterial features have been proposed to ameliorate protein recovery and mass spectrometric analyses (26, 30, 31).
Surface proteins from S-layer forming bacteria are usually extracted using 5 M lithium chloride (LiCl) solutions thus breaking the hydrogen bonds that stabilize these supramolecular structures (32). More recently, a modified LiCl extraction protocol has been set up by Johnson et al. (33) to specifically separate the highly abundant SLPs from the less abundant SLAPs, exploiting their different solubility in 1 M LiCl solution, and applied to study SLAPs isolated from different probiotic bacteria (34–36).
As to Gram negative bacteria, classical methods for the extraction of surface exposed proteins take into account the peculiar features of their cell-envelope and include the subfractionation of outer membrane, cytoplasmic membrane and periplasmic proteins. Commonly, inner and outer membrane vesicles are prepared by lysozyme-EDTA lysis or French press lysis of bacteria, separated by density centrifugation using a sucrose gradient and analyzed by proteomics (37, 38).
Cell Shaving Strategy
The concept underlying this innovative strategy is that, regardless of anchoring mechanisms, surface exposed protein fragments can be selectively released through a limited proteolytic digestion step carried out on intact bacterial cells. The “shaved” peptides can be then separated from the whole cells by centrifugation and analyzed by shotgun proteomics thus achieving protein identification (39). Trypsin is the proteolytic enzyme more often used in these experiments, as lysine and arginine residues are widely represented in protein sequences and often easily accessible to the enzyme (Figures 1,2A).
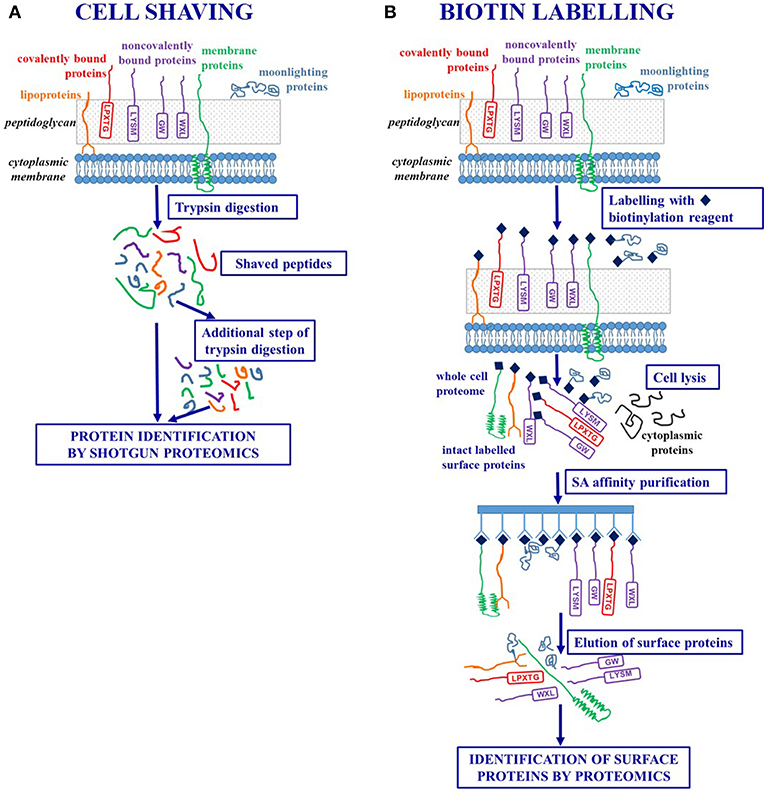
Figure 2. Workflows of ad hoc designed strategies for selectively targeting surface-exposed proteins. (A) Cell shaving strategies based on a limited proteolytic digestion step of intact bacterial cells in order to release surface exposed protein fragments. (B) Labeling strategies based on biotinylation of surface-exposed proteins of intact bacterial cells in order to achieve their purification by affinity chromatography.
In order to rule out osmotic shock and contamination of cytoplasmic proteins, bacterial cells are suspended in an isotonic buffer (obtained through the addition of sucrose or arabinose) and protease digestion is carried out for a short time (15–30 min) to preserve cell integrity. However, the limited proteolysis could produce high molecular weight protein fragments which are not amenable for LC-MS/MS analyses and a further extensive tryptic digestion step (18 hs) carried out on the shaved peptides has been included in optimized protocols (40, 41).
The cell shaving approach allows a comprehensive description of the bacterial proteosurfaceome and, as trypsin molecules could diffuse into the cell wall and interact with membrane and cell wall embedded proteins, a single experiment can provide the simultaneous identification of IMP, lipoproteins, LPXTG-proteins, non-covalently bound proteins, moonlighting proteins (40). Moreover, as the procedure selectively identifies peptides generated from surface-exposed domains, this methodology can contribute to refine the protein topology derived from bioinformatics by matching prediction and experimental data (39).
The method was conceived to identify potential targets of drugs and novel candidates for vaccines, and applied to study Streptococcus pyogenes, leading to the identification of 68 surface-associated proteins, one of which was validated in mice as a potential vaccine candidate (39, 42).
This protocol is particularly suitable for the analysis of Gram positive bacteria, due to their thick and rigid cell wall that is less prone to lysis during treatments, and comprehensive proteomic studies have been performed on several probiotics including Lactococcus lactis (43), Bifidobacterium animalis (44), Lactobacillus rhamnosus (45), P. freudenreichii (46, 47), and food pathogens like Staphylococcus aureus (48–50) and Listeria monocytogenes (51, 52). On the other hand, the cell shaving method has been applied for the analysis of surface proteins of Gram negative bacteria such as Escherichia coli (53) and Salmonella enterica (54), even if their cell envelope can be more easily damaged thus causing cytoplasmic protein contamination.
Labeling of Surface-Exposed Proteins
These approaches rely on labeling strategies that specifically target surface proteins of intact bacterial cells. A first approach is based on the labeling of surface-exposed proteins with fluorescent CyDye reagents. After the labeling step, a complete cell lysis is carried out and the proteins are separated by 2-DE. Labeled surface proteins are highlighted by fluorescence imaging and identified using mass spectrometry (46, 55) (Figure 1).
A more sophisticated approach uses ad hoc designed biotin containing reagents to target surface proteins and suitably exploits the highly specific and stable non-covalent interaction between avidin [or the bacterial protein streptavidin (SA)] and biotin (56). As the biotinylation reagents can penetrate the peptidoglycan structure, proteins that are buried within the cell wall can be labeled. After a cell lysis step, labeled surface proteins are separated from non-labeled cytoplasmic proteins by SA affinity chromatography. The intact surface proteins thus purified could be identified using both classical proteomic approaches (including 2-DE) and Ge-LC-MS/MS or shotgun proteomics (27, 57–59) (Figures 1,2B).
The chemical features of the labeling reagent have a crucial role in this protocol. The biotin moiety is at the basis of the affinity purification step and several spacers can be linked to the valeric acid carboxylic group of biotin and carry functional groups (such as N-hydroxysuccinimide, NHS) that covalently link to primary amino groups of proteins (ε-amino group of lysine residues and N-terminal α-amine group) (60).
The elution of biotinylated proteins from SA-coated resin could be severely hampered by the high stability of the biotin-SA complex. To overcome this problem, cleavable spacers that include a disulphide bridge in their structure have been introduced thus allowing to easily and completely elute the biotinylated proteins by using reducing reagents such as dithiotreitol. Sulpho-NHS-SS-biotin is nowadays the most commonly used reagent in proteomic studies. The negative charge of sulphonate group present on the NHS ring makes this molecule water soluble and unable to cross the cell membrane thus drastically reducing the contamination of cytoplasmic proteins (60).
This method is suitable to study the proteosurfaceome of both Gram-positive and Gram-negative bacteria but, unfortunately, up to now, it has been mainly applied to analyze the surface proteins of pathogens such as S. aureus (27, 57, 61), L. monocytogenes (52), E. coli (62). We can foresee that in the near future, this strategy will be also applied to the characterization of the proteosurfaceome of probiotics thus contributing to the advancement of knowledge in this field.
Proteomics for Studying Surface-Exposed Proteins In Probiotics
The high biological relevance of surface-exposed proteins in the dynamic crosstalk between bacteria and their environment prompted the design of dedicated proteomic strategies useful for investigating the molecular mechanisms of adaptation to GIT environment, adhesion, colonization, and immunomodulation, key features for the probiotic action (63–65). First studies on this topic, integrating proteomics and biochemical assays, confirmed the presence of moonlighting proteins on the surface of Lactobacilli and Bifidobacteria and their ability to bind extracellular matrix components (plasminogen, fibronectin, mucin) and adhere to GIT (28, 66–72). More recently, a comparative proteomic study performed on potentially probiotic strains of Lactobacillus pentosus led to identify moonlighting proteins in the cell-wall proteome and to correlate their abundance level to strain specific adhesion capacities to mucus (73).
Importantly, the adaptation to GIT can enhanced probiotic features as, for instance, bile stress can be a signal of gut entry for bacteria, which triggers the re-organization of the surface protein pattern resulting in improved adhesion ability in the new niche. In fact, the abundance of surface-exposed Clp proteins (ClpB, ClpE), chaperone (DnaK), and enzymes involved in carbohydrate metabolism was enhanced in L. rhamnosus GG after exposure to bile stress. The abundance of some of these enzymes increased in the proteosurfaceome but not in the proteome, thus suggesting a sort of protein relocalization triggered by bile (55). Changes in the expression profile of moonlighting proteins (in particular ribosomal proteins and glycolytic enzymes) promoted by bile were also observed in B. animalis subsp. lactis and B. longum (29, 74).
Several factors supplied with diets such as plant polyphenols (resveratrol, ferulic acid, etc.) and prebiotic carbohydrates (raffinose, fructooligosaccharides, etc.) could influence the surface protein profile of probiotics. These molecules improved the adhesive capabilities to mucin and HT-29 cells of L. acidophilus NCFM probably by modulating biosynthesis and/or secretion of moonlighting proteins (75–77).
Proteomics also contributed to assess that the surface protein pattern is strain specific. In fact, a trypsin shaving approach applied to compare the proteosurfaceomes of L. rhamnosus GG and the closely related dairy strain Lc705, led to the identification of 102 and 198 proteins anchored to the surface trough different mechanisms (IMP, LPXTG proteins, lipoproteins, C-term and N-Term anchored proteins and moonlighting proteins). Strain specific differences were mainly associated to moonlighting proteins and could be related to the adaptation to their ecological niches and probiotic functions (response to bile, hydrolysis of casein, immunostimulation, pathogen exclusion) (45). In addition, SpaC and SpaA proteins, involved in the assembly of pilus structures, were identified only in the proteome of GG and related to adhesion properties and prolonged residence in the GIT of this strain compared to Lc705 (78).
Similarly, a pilin-like protein was identified in a L. lactis strain able to adhere to Caco-2 cells and the synthesis of pili was confirmed by immunoblotting detection and electron and atomic force microscopy observations (43). Several proteins potentially involved in adhesion, including the pilus structure proteins FimA and FimB, were also identified in a B. animalis ssp lactis strain (44).
S-layer proteins (SLPs) and S-layer associated proteins (SLAPs) have a key role in GIT adaptation and immunomodulation processes, as highlighted by functional studies integrated by proteomics. First studies performed on L. acidophilus NCFM, a S-layer forming microorganism, showed the involvement of the main SLP (SlpA) in adhesion to Caco2 cells (79) and dendritic cells (DCs) immunomodulation (80). Recently, using an optimized extraction protocol, 37 SLAPs of L. acidophilus NCFM were identified and most of them were predicted to be extracellular proteins while only four proteins were potentially moonlighting proteins. The protein SLAP LBA1029 contributed to a pro-inflammatory response in murine DCs (inducing the expression of TNF-α), thus indicating that SLAPs may impart immunological properties to microbes (33). The surface-exposed proteins of L. acidophilus NCFM were also characterized by Celebioglu and Svensson (35), leading to the identification of a higher number of moonlighting proteins. In addition, a quantitative proteomic study performed using more sophisticated mass spectrometric approaches, defined a detailed catalog of the L. acidophilus NCFM surface proteins containing 276 SLAPs and demonstrated that the cell surface proteome was modulated by growth phase. This feature could be exploited to optimize probiotic actions and enhance their delivery, persistence, and general efficacy (36).
The presence or absence of the S-layer has a clear impact on the composition and complexity of proteosurfaceome of different lactobacilli. In fact, a proteomic study led to identify numerous SLAPs in proteosurfaceome of S-layer-forming strains of L. acidophilus, L. helveticus, L. crispatus, L. amylovorus, and L. gallinarum. On the other hand, the few proteins isolated with LiCl treatment of the non-S-layer forming strains of L. delbrueckii subsp. bulgaricus and L. casei were mostly intracellular proteins, likely presented extracellularly due to cell lysis in stationary phase. These findings also confirmed the role of SLAPs as integral components of S-layer (34).
Recently surface proteins from P. freudenreichii, a S-layer forming bacterium, have been characterized by using three complementary proteomic methods (guanidine hydrochloride extraction, cell shaving and fluorescent labeling with CyDye coupled to mass spectrometric analyses) and their ability in enhancing the production of the anti-inflammatory cytokines IL6 and IL10 by human immune cells was assessed (46). Furthermore, a study combining comparative genomics, transcriptomics and surface proteomics, coupled with gene inactivation, was performed on 23 strains of P. freudenreichii with different anti-inflammatory potential. Results evidenced the involvement of SlpB and SlpE in the release of IL10 by human immune cells and demonstrated that different combinations of surface and cytoplasmic proteins, depending on the strain, exerted a pleiotropic effect on the anti-inflammatory properties (47). Finally, do Carmo et al. demonstrated a direct role of SlpB in the adhesion to HT-29 cells and reported that the inactivation of slpB gene caused profound modifications in whole cell and surface proteomes as well as bacterial stress tolerance (81, 82).
The reported achievements clearly assess the ability of proteomics in investigating the structural features of different classes of surface proteins and its role in elucidating the mechanisms of bacteria/host interaction.
Concluding Remarks
In the past few years, proteomics proved to be the method of choice to investigate surface architecture of bacteria cells, contributing to define protein location and topology, and to deal with the extremely dynamic and constantly renewing nature of proteosurfaceome that is deeply affected by the environment. The exploitation of technological innovations in proteomics and mass spectrometry has improved the knowledge in the probiotics field. This review displays the essential role of proteomics in the elucidation of the molecular mechanisms of the probiotic action and the identification of key actors of the probiotics/host molecular dialogue, thus potentially providing new tools for selecting strains with specific healthy promoting functions. Future research perspectives should include the analysis of post-translational modifications of surface proteins (glycoproteomics, phosphoproteomics, etc.), and the investigation of their effects in the interaction with host epithelial cells.
Author Contributions
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1. Hill C, Guarner F, Reid G, Gibson GR, Merenstein DJ, Pot B, et al. Expert consensus document. The International Scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat Rev Gastroenterol Hepatol. (2014) 11:506–14. doi: 10.1038/nrgastro.2014.66
2. Marco ML, Pavan S, Kleerebezem M. Towards understanding molecular modes of probiotic action. Curr Opin Biotechnol. (2006) 17:204–10. doi: 10.1016/j.copbio.2006.02.005
3. Rabah H, do Carmo FLR, Jan G. Dairy propionibacteria: versatile probiotics. Microorganisms. (2017) 5:24. doi: 10.3390/microorganisms5020024
4. Bron PA, van Baarlen P, Kleerebezem M. Emerging molecular insights into the interaction between probiotics and the host intestinal mucosa. Nat Rev Microbiol. (2012) 10:66–78. doi: 10.1038/nrmicro2690
5. Lebeer S, Bron PA, Marco ML, Van Pijkeren JP, O'Connell Motherway M, Hill C, et al. Identification of probiotic effector molecules: present state and future perspectives. Curr Opin Biotechnol. (2018) 49:217–23. doi: 10.1016/j.copbio.2017.10.007
6. Ruiz L, Hevia A, Bernardo D, Margolles A, Sánchez B. Extracellular molecular effectors mediating probiotic attributes. FEMS Microbiol Lett. (2014) 359:1–11. doi: 10.1111/1574-6968.12576
7. Lebeer S, Vanderleyden J, De Keersmaecker SC. Host interactions of probiotic bacterial surface molecules: comparison with commensals and pathogens. Nat Rev Microbiol. (2010) 8:171–84. doi: 10.1038/nrmicro2297
8. Sánchez B, Urdaci MC, Margolles A. Extracellular proteins secreted by probiotic bacteria as mediators of effects that promote mucosa-bacteria interactions. Microbiology. (2010) 156:3232–42. doi: 10.1099/mic.0.044057-0
9. Hevia A, Delgado S, Sánchez B, Margolles A. Molecular players involved in the interaction between beneficial bacteria and the immune system. Front Microbiol. (2015) 6:1285. doi: 10.3389/fmicb.2015.01285
10. Desvaux M, Dumas E, Chafsey I, Hébraud M. Protein cell surface display in Gram-positive bacteria: from single protein to macromolecular protein structure. FEMS Microbiol Lett. (2006) 256:1–15. doi: 10.1111/j.1574-6968.2006.00122.x
11. Desvaux M, Candela T, Serror P. Surfaceome and proteosurfaceome in parietal monoderm bacteria: focus on protein cell-surface display. Front Microbiol. (2018) 9:100. doi: 10.3389/fmicb.2018.00100
12. Scott JR, Barnett TC. Surface proteins of gram-positive bacteria and how they get there. Annu Rev Microbiol. (2006) 60:397–423. doi: 10.1146/annurev.micro.60.080805.142256
13. Kleerebezem M, Hols P, Bernard E, Rolain T, Zhou M, Siezen RJ, et al. The extracellular biology of the lactobacilli. FEMS Microbiol Rev. (2010) 34:199–230. doi: 10.1111/j.1574-6976.2010.00208.x
14. Hynönen U, Palva A. Lactobacillus surface layer proteins: structure, function and applications. Appl Microbiol Biotechnol. (2013) 97:5225–43. doi: 10.1007/s00253-013-4962-2
15. do Carmo FLR, Rabah H, De Oliveira Carvalho RD, Gaucher F, Cordeiro BF, da Silva SH, et al. Extractable bacterial surface proteins in probiotic-host interaction. Front. Microbiol. (2018) 9:645. doi: 10.3389/fmicb.2018.00645
16. Wang W, Jeffery CJ. An analysis of surface proteomics results reveals novel candidates for intracellular/surface moonlighting proteins in bacteria. Mol Biosyst. (2016) 12:1420–31. doi: 10.1039/c5mb00550g
17. Jeffery CJ. Protein moonlighting: what is it, and why is it important? Philos Trans R Soc Lond B Biol Sci. (2018) 373:20160523. doi: 10.1098/rstb.2016.0523
18. Gardy JL, Brinkman FS. Methods for predicting bacterial protein subcellular localization. Nat Rev Microbiol. (2006) 4:741–51. doi: 10.1038/nrmicro1494
19. Yu NY, Wagner JR, Laird MR, Melli G, Rey S, Lo R, et al. PSORTb 3.0: improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes. Bioinformatics. (2010) 26:1608–15. doi: 10.1093/bioinformatics/btq249
20. Petersen TN, Brunak S, von Heijne G, Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods. (2011) 8:785–6. doi: 10.1038/nmeth.1701
21. Krogh A, Larsson B, von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol. (2001) 305:567–80. doi: 10.1006/jmbi.2000.4315
22. Bendtsen JD, Kiemer L, Fausbøll A, Brunak S. Non-classical protein secretion in bacteria. BMC Microbiol. (2005) 5:58. doi: 10.1186/1471-2180-5-58
23. Chen C, Zabad S, Liu H, Wang W, Jeffery C. MoonProt 2.0: an expansion and update of the moonlighting proteins database. Nucleic Acids Res. (2018) 46:D640–4. doi: 10.1093/nar/gkx1043
24. Solis N, Cordwell SJ. Current methodologies for proteomics of bacterial surface-exposed and cell envelope proteins. Proteomics. (2011) 11:3169–89. doi: 10.1002/pmic.201000808
25. Aebersold R, Mann M. Mass spectrometry-based proteomics. Nature. (2003) 422:198–207. doi: 10.1038/nature01511
26. Nandakumar R, Nandakumar MP, Marten MR, Ross JM. Proteome analysis of membrane and cell wall associated proteins from Staphylococcus aureus. J Proteome Res. (2005) 4:250–7. doi: 10.1021/pr049866k
27. Gatlin CL, Pieper R, Huang ST, Mongodin E, Gebregeorgis E, Parmar PP, et al. Proteomic profiling of cell envelope-associated proteins from Staphylococcus aureus. Proteomics. (2006) 6:1530–49. doi: 10.1002/pmic.200500253
28. Izquierdo E, Horvatovich P, Marchioni E, Aoude-Werner D, Sanz Y, Ennahar S. 2-DE and MS analysis of key proteins in the adhesion of Lactobacillus plantarum, a first step toward early selection of probiotics based on bacterial biomarkers. Electrophoresis. (2009) 30:949–56. doi: 10.1002/elps.200800399
29. Ruiz L, Couté Y, Sánchez B, de los Reyes-Gavilán CG, Sanchez JC, Margolles A. The cell-envelope proteome of Bifidobacterium longum in an in vitro bile environment. Microbiology. (2009) 155:957–67. doi: 10.1099/mic.0.024273-0
30. Sánchez B, Bressollier P, Chaignepain S, Schmitter JM, Urdaci MC. Identification of surface-associated proteins in the probiotic bacterium Lactobacillus rhamnosus GG. Int Dairy J. (2009) 19:85–8. doi: 10.1016/j.idairyj.2008.09.005
31. Tiong HK, Hartson S, Muriana PM. Comparison of five methods for direct extraction of surface proteins from Listeria monocytogenes for proteomic analysis by orbitrap mass spectrometry. J Microbiol Methods. (2015) 110:54–60. doi: 10.1016/j.mimet.2015.01.004
32. Sleytr UB, Schuster B, Egelseer EM, Pum D. S-layers: principles and applications. FEMS Microbiol Rev. (2014) 38:823–64. doi: 10.1111/1574-6976.12063
33. Johnson B, Selle K, O'Flaherty S, Goh YJ, Klaenhammer T. Identification of extracellular surface-layer associated proteins in Lactobacillus acidophilus NCFM. Microbiology. (2013) 159:2269–82. doi: 10.1099/mic.0.070755-0
34. Johnson BR, Hymes J, Sanozky-Dawes R, Henriksen ED, Barrangou R, Klaenhammer TR. Conserved S-layer-associated proteins revealed by exoproteomic survey of S-layer-forming lactobacilli. Appl Environ Microbiol. (2016) 82:134–45. doi: 10.1128/AEM.01968-15
35. Celebioglu HU, Svensson B. Exo- and surface proteomes of the probiotic bacterium Lactobacillus acidophilus NCFM. Proteomics. (2017) 17:1700019. doi: 10.1002/pmic.201700019
36. Klotz C, O'Flaherty S, Goh YJ, Barrangou R. Investigating the effect of growth phase on the surface-layer associated proteome of Lactobacillus acidophilus using quantitative proteomics. Front Microbiol. (2017) 8:2174. doi: 10.3389/fmicb.2017.02174
37. Weiner JH, Li L. Proteome of the Escherichia coli envelope and technological challenges in membrane proteome analysis. Biochim Biophys Acta. (2008) 1778:1698–713. doi: 10.1016/j.bbamem.2007.07.020
38. Thein M, Sauer G, Paramasivam N, Grin I, Linke D. Efficient subfractionation of gram-negative bacteria for proteomics studies. J Proteome Res. (2010) 9:6135–47. doi: 10.1021/pr1002438
39. Rodríguez-Ortega MJ, Norais N, Bensi G, Liberatori S, Capo S, Mora M, et al. Characterization and identification of vaccine candidate proteins through analysis of the group A Streptococcus surface proteome. Nat Biotechnol. (2006) 24:191–7. doi: 10.1038/nbt1179
40. Tjalsma H, Lambooy L, Hermans PW, Swinkels DW. Shedding and shaving: disclosure of proteomic expressions on a bacterial face. Proteomics. (2008) 8:1415–28. doi: 10.1002/pmic.200700550
41. Olaya-Abril A, Gómez-Gascón L, Jiménez-Munguía I, Obando I, Rodríguez-Ortega MJ. Another turn of the screw in shaving Gram-positive bacteria: optimization of proteomics surface protein identification in Streptococcus pneumoniae. J Proteomics. (2012) 75:3733–46. doi: 10.1016/j.jprot.2012.04.037
42. Doro F, Liberatori S, Rodríguez-Ortega MJ, Rinaudo CD, Rosini R, Mora M, et al. Surfome analysis as a fast track to vaccine discovery: identification of a novel protective antigen for Group B Streptococcus hypervirulent strain COH1. Mol Cell Proteomics. (2009) 8:1728–37. doi: 10.1074/mcp.M800486-MCP200
43. Meyrand M, Guillot A, Goin M, Furlan S, Armalyte J, Kulakauskas S, et al. Surface proteome analysis of a natural isolate of Lactococcus lactis reveals the presence of pili able to bind human intestinal epithelial cells. Mol Cell Proteomics. (2013) 12:3935–47. doi: 10.1074/mcp.M113.029066
44. Zhu D, Sun Y, Liu F, Li A, Yang L, Meng XC. Identification of surface-associated proteins of Bifidobacterium animalis ssp. lactis KLDS 2.0603 by enzymatic shaving. J Dairy Sci. (2016) 99:5155–72. doi: 10.3168/jds.2015-10581
45. Espino E, Koskenniemi K, Mato-Rodriguez L, Nyman TA, Reunanen J, Koponen J, et al. Uncovering surface-exposed antigens of Lactobacillus rhamnosus by cell shaving proteomics and two-dimensional immunoblotting. J Proteome Res. (2015) 14:1010–24. doi: 10.1021/pr501041a
46. Le Maréchal C, Peton V, Plé C, Vroland C, Jardin J, Briard-Bion V, et al. Surface proteins of Propionibacterium freudenreichii are involved in its anti-inflammatory properties. J Proteomics. (2015) 113:447–61. doi: 10.1016/j.jprot.2014.07.018
47. Deutsch SM, Mariadassou M, Nicolas P, Parayre S, Le Guellec R, Chuat V, et al. Identification of proteins involved in the anti-inflammatory properties of Propionibacterium freudenreichii by means of a multi-strain study. Sci Rep. (2017) 7:46409. doi: 10.1038/srep46409
48. Solis N, Larsen MR, Cordwell SJ. Improved accuracy of cell surface shaving proteomics in Staphylococcus aureus using a false-positive control. Proteomics. (2010) 10:2037–49. doi: 10.1002/pmic.200900564
49. Solis N, Parker BL, Kwong SM, Robinson G, Firth N, Cordwell SJ. Staphylococcus aureus surface proteins involved in adaptation to oxacillin identified using a novel cell shaving approach. J Proteome Res. (2014) 13:2954–72. doi: 10.1021/pr500107p
50. Monteiro R, Hébraud M, Chafsey I, Chambon C, Viala D, Torres C, et al. Surfaceome and exoproteome of a clinical sequence type 398 methicillin resistant Staphylococcus aureus strain. Biochem Biophys Rep. (2015) 3:7–13. doi: 10.1016/j.bbrep.2015.07.004
51. Zhang CX, Creskey MC, Cyr TD, Brooks B, Huang H, Pagotto F, et al. Proteomic identification of Listeria monocytogenes surface-associated proteins. Proteomics. (2013) 13:3040–5. doi: 10.1002/pmic.201200449
52. Esbelin J, Santos T, Ribière C, Desvaux M, Viala D, Chambon C, et al. Comparison of three methods for cell surface proteome extraction of Listeria monocytogenes biofilms. OMICS. (2018) 22:779–87. doi: 10.1089/omi.2018.0144
53. Walters MS, Mobley HL. Identification of uropathogenic Escherichia coli surface proteins by shotgun proteomics. J Microbiol Methods. (2009) 78:131–5. doi: 10.1016/j.mimet.2009.04.013
54. Fagerquist CK, Zaragoza WJ. Proteolytic surface-shaving and serotype-dependent expression of SPI-1 invasion proteins in Salmonella enterica subspecies enterica. Front Nutr. (2018) 5:124. doi: 10.3389/fnut.2018.00124
55. Koskenniemi K, Laakso K, Koponen J, Kankainen M, Greco D, Auvinen P, et al. Proteomics and transcriptomics characterization of bile stress response in probiotic Lactobacillus rhamnosus GG. Mol. Cell Proteomics. (2011) 10:M110.002741. doi: 10.1074/mcp.M110.002741
56. Scheurer SB, Roesli C, Neri D, Elia G. A comparison of different biotinylation reagents, tryptic digestion procedures, and mass spectrometric techniques for 2-D peptide mapping of membrane proteins. Proteomics. (2005) 5:3035–9. doi: 10.1002/pmic.200402069
57. Hempel K, Pané-Farré J, Otto A, Sievers S, Hecker M, Becher D. Quantitative cell surface proteome profiling for SigB-dependent protein expression in the human pathogen Staphylococcus aureus via biotinylation approach. J. Proteome Res. (2010) 9:1579–90. doi: 10.1021/pr901143a
58. Bonn F, Maaß S, van Dijl JM. Enrichment of cell surface-associated proteins in Gram-positive bacteria by biotinylation or trypsin shaving for mass spectrometry analysis. Methods Mol Biol. (2018) 1841:35–43. doi: 10.1007/978-1-4939-8695-8_4
59. Elia G. Cell surface protein biotinylation for SDS-PAGE analysis. Methods Mol Biol. (2019) 1855:449–59. doi: 10.1007/978-1-4939-8793-1_37
60. Elia G. Biotinylation reagents for the study of cell surface proteins. Proteomics. (2008) 8:4012–24. doi: 10.1002/pmic.200800097
61. Moche M, Schlüter R, Bernhardt J, Plate K, Riedel K, Hecker M, et al. Time-resolved analysis of cytosolic and surface-associated proteins of Staphylococcus aureus HG001 under planktonic and biofilm conditions. J Proteome Res. (2015) 14:3804–22. doi: 10.1021/acs.jproteome.5b00148
62. Monteiro R, Chafsey I, Leroy S, Chambon C, Hébraud M, Livrelli V, et al. Differential biotin labelling of the cell envelope proteins in lipopolysaccharidic diderm bacteria: exploring the proteosurfaceome of Escherichia coli using sulfo-NHS-SS-biotin and sulfo-NHS-PEG4-bismannose-SS-biotin. J Proteomics. (2018) 181:16–23. doi: 10.1016/j.jprot.2018.03.026.
63. Siciliano RA, Mazzeo MF. Molecular mechanisms of probiotic action: a proteomic perspective. Curr Opin Microbiol. (2012) 15:390–6. doi: 10.1016/j.mib.2012.03.006
64. De Angelis M, Calasso M, Cavallo N, Di Cagno R, Gobbetti M. Functional proteomics within the genus Lactobacillus. Proteomics. (2016) 16:946–62. doi: 10.1002/pmic.201500117
65. Ruiz L, Hidalgo C, Blanco-Míguez A, Lourenço A, Sánchez B, Margolles A. Tackling probiotic and gut microbiota functionality through proteomics. J Proteomics. (2016) 147:28–39. doi: 10.1016/j.jprot.2016.03.023
66. Granato D, Bergonzelli GE, Pridmore RD, Marvin L, Rouvet M, Corthésy-Theulaz IE. Cell surface-associated elongation factor Tu mediates the attachment of Lactobacillus johnsonii NCC533 (La1) to human intestinal cells and mucins. Infect Immun. (2004) 72:2160–9. doi: 10.1128/IAI.72.4.2160-2169.2004
67. Bergonzelli GE, Granato D, Pridmore RD, Marvin-Guy LF, Donnicola D, Corthésy-Theulaz IE. GroEL of Lactobacillus johnsonii La1 (NCC 533) is cell surface associated: potential role in interactions with the host and the gastric pathogen Helicobacter pylori. Infect Immun. (2006) 74:425–34. doi: 10.1128/IAI.74.1.425-434.2006
68. Candela M, Bergmann S, Vici M, Vitali B, Turroni S, Eikmanns BJ, et al. Binding of human plasminogen to Bifidobacterium. J Bacteriol. (2007) 189:5929–36. doi: 10.1128/JB.00159-07
69. Siciliano RA, Cacace G, Mazzeo MF, Morelli L, Elli M, Rossi M, et al. Proteomic investigation of the aggregation phenomenon in Lactobacillus crispatus. Biochim Biophys Acta. (2008) 1784:335–42. doi: 10.1016/j.bbapap.2007.11.007
70. Beck HC, Madsen SM, Glenting J, Petersen J, Israelsen H, Nørrelykke MR, et al. Proteomic analysis of cell surface-associated proteins from probiotic Lactobacillus plantarum. FEMS Microbiol Lett. (2009) 297:61–6. doi: 10.1111/j.1574-6968.2009.01662.x
71. Castaldo C, Vastano V, Siciliano RA, Candela M, Vici M, Muscariello L, et al. Surface displaced alfa-enolase of Lactobacillus plantarum is a fibronectin binding protein. Microb Cell Fact. (2009) 8:14. doi: 10.1186/1475-2859-8-14
72. González-Rodríguez I, Sánchez B, Ruiz L, Turroni F, Ventura M, Ruas-Madiedo P, et al. Role of extracellular transaldolase from Bifidobacterium bifidum in mucin adhesion and aggregation. Appl Environ Microbiol. (2012) 78:3992–8. doi: 10.1128/AEM.08024-11
73. Pérez Montoro B, Benomar N, Caballero Gómez N, Ennahar S, Horvatovich P, Knapp CW, et al. Proteomic analysis of Lactobacillus pentosus for the identification of potential markers of adhesion and other probiotic features. Food Res Int. (2018) 111:58–66. doi: 10.1016/j.foodres.2018.04.072
74. Candela M, Centanni M, Fiori J, Biagi E, Turroni S, Orrico C, et al. DnaK from Bifidobacterium animalis subsp. lactis is a surface-exposed human plasminogen receptor upregulated in response to bile salts. Microbiology. (2010) 156:1609–18. doi: 10.1099/mic.0.038307-0
75. Celebioglu HU, Ejby M, Majumder A, Købler C, Goh YJ, Thorsen K, et al. Differential proteome and cellular adhesion analyses of the probiotic bacterium Lactobacillus acidophilus NCFM grown on raffinose - an emerging prebiotic. Proteomics. (2016) 16:1361–75. doi: 10.1002/pmic.201500212
76. Celebioglu HU, Olesen SV, Prehn K, Lahtinen SJ, Brix S, Abou Hachem M, et al. Mucin- and carbohydrate-stimulated adhesion and subproteome changes of the probiotic bacterium Lactobacillus acidophilus NCFM. J Proteomics. (2017) 163:102–10. doi: 10.1016/j.jprot.2017.05.015
77. Celebioglu HU, Delsoglio M, Brix S, Pessione E, Svensson B. Plant polyphenols stimulate adhesion to intestinal mucosa and induce proteome changes in the probiotic Lactobacillus acidophilus NCFM. Mol Nutr Food Res. (2018) 62:1700638. doi: 10.1002/mnfr.201700638
78. Savijoki K, Lietzén N, Kankainen M, Alatossava T, Koskenniemi K, Varmanen P, et al. Comparative proteome cataloging of Lactobacillus rhamnosus strains GG and Lc705. J Proteome Res. (2011) 10:3460–73. doi: 10.1021/pr2000896
79. Ashida N, Yanagihara S, Shinoda T, Yamamoto N. Characterization of adhesive molecule with affinity to Caco-2 cells in Lactobacillus acidophilus by proteome analysis. J Biosci Bioeng. (2011) 112:333–7. doi: 10.1016/j.jbiosc.2011.06.001
80. Konstantinov SR, Smidt H, de Vos WM, Bruijns SC, Singh SK, Valence F, et al. S layer protein A of Lactobacillus acidophilus NCFM regulates immature dendritic cell and T cell functions. Proc Natl Acad Sci USA. (2008) 105:19474–9. doi: 10.1073/pnas.0810305105
81. do Carmo FLR, Rabah H, Huang S, Gaucher F, Deplanche M, Dutertre S, et al. Propionibacterium freudenreichii surface protein SlpB is involved in adhesion to intestinal HT-29 cells. Front Microbiol. (2017) 8:1033. doi: 10.3389/fmicb.2017.01033
Keywords: proteomics, probiotics, proteosurfaceome, surface-exposed proteins, S-layer proteins, moonlighting proteins, immunomodulation
Citation: Siciliano RA, Lippolis R and Mazzeo MF (2019) Proteomics for the Investigation of Surface-Exposed Proteins in Probiotics. Front. Nutr. 6:52. doi: 10.3389/fnut.2019.00052
Received: 25 February 2019; Accepted: 05 April 2019;
Published: 24 April 2019.
Edited by:
Fernanda Mozzi, CONICET Centro de Referencia para Lactobacilos (CERELA), ArgentinaReviewed by:
Silvina Graciela Fadda, CONICET Centro de Referencia para Lactobacilos (CERELA), ArgentinaMaria de los Angeles Serradell, National Council for Scientific and Technical Research (CONICET), Argentina
Copyright © 2019 Siciliano, Lippolis and Mazzeo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rosa Anna Siciliano, cnNpY2lsaWFub0Bpc2EuY25yLml0
 Rosa Anna Siciliano
Rosa Anna Siciliano Rosa Lippolis2
Rosa Lippolis2 Maria Fiorella Mazzeo
Maria Fiorella Mazzeo