- 1Department of Aquaculture, Faculty of Fisheries, Hajee Mohammad Danesh Science and Technology University, Dinajpur, Bangladesh
- 2School of Biological sciences, Universiti Sains Malaysia, George Town, Malaysia
- 3School of Applied Biosciences, College of Agriculture and Life Sciences, Kyungpook National University, Daegu, South Korea
- 4Department of Biotechnology, Yonsei University, Seoul, South Korea
- 5Department of Protein Chemistry and Technology, CSIR-CFTRI, Mysuru, India
We identified an antimicrobial peptide (AMP) from Lactobacillus acidophilus that was antagonistic to Aeromonas hydrophila. In vitro studies such as well-diffusion and field trials revealed that the AMP was active against A. hydrophila. The field trials of AMP using A. hydrophila-infected Channa striatus with a mannone oligosaccharide (MOS) prebiotic, A. hydrophila antigens, A. hydrophila-infected fish serum, L. acidophilus, and Lactobacillus cell free-supernatant (LABS-CFS) on an indicator organism further revealed that the antimicrobial agent could protect C. striatus. Other than the AMP, none of the above were able to eliminate the infectious agent A. hydrophila, and were only able to delay the death rate for 3–4 days. Thus, we conclude that the AMP is antagonistic to A. hydrophila and may be used for treatment of A. hydrophila infections. Subsequent L. acidophilus whole-genome sequence analyses enabled an understanding of the (probable) gene arrangement and its location on the chromosome. This information may be useful in the generation of recombinant peptides to produce larger quantities for treatment.
Introduction
Gut bacteria play a major role in the maintenance of human health by participating in multiple functions that are beneficial to the host (Patel and DuPont, 2015; Kristensen et al., 2016). Therefore, the current global focus has been to understand, analyze, and exploit gut microbiota for human health purposes. The World Health Organization (WHO) defined probiotics as “live microorganisms which when administered in adequate amounts confer a health benefit on the host” (WHO/FAO, 2006; Hill et al., 2014). It has also been reported that probiotics and their products influence the activity of host gut bacterial components (Scott et al., 2015). Various health benefits from the use of probiotics have been reported, beginning with the supplementation of Bifidobacterium (Yin et al., 2010; Chen et al., 2012; Plaza-Diaz et al., 2014; Reichold et al., 2014; Savcheniuk et al., 2014; Wang et al., 2015) and Lactobacillus species, which are known to reduce body weight gain and adipose tissue in experimental mice. Such findings suggest that probiotic microbes stimulate the production of adiponectin (Kim et al., 2013; Kobyliak et al., 2016).
Di Cerbo et al. (2016) reported that Lactobacilli have therapeutic effects in various pathologies (Klaenhammer et al., 2012). There have also been many reports revealing the beneficial effects of probiotics, including the mucosal immune response regulated by probiotics (Sang et al., 2010), enhancement of macrophage proliferation (Breyner et al., 2017), and the regulation of gene expression. Breyner et al. (2017) reported that probiotic supplementation influenced the host immune response by inhibiting the nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB) pathway in vitro and in vivo, further confirming the anti-inflammatory and therapeutic effects of probiotics. Nazemian et al. (2016) reported that probiotic supplementation enhanced the levels of anti-inflammatory cytokines and immunoglobulins, and increased immune cell proliferation and the production of pro-inflammatory cytokines by T cells.
The concept of monostrain and multistrain probiotics is a recent development in the field. This concept helps in catalyzing the inculcation of enhanced beneficial effects (Timmerman et al., 2004). The most advanced and recent exploitation of probiotics is fecal microbiota transplantation (FMT) (Kelly, 2013). FMT is exclusively used for C. difficile patients and is known to be very effective. FMT reduces gut dysbiosis, reinstates beneficial gut bacteria, and eliminates pathogenic bacteria.
Gut bacteria/probiotic microbes produce lipopolysaccharides (LPSs), fructooligosaccharides (FOSs), galactooligosaccharides (GOSs), and other substances that contain a non-digestible compound. The compounds selectively stimulate the growth and/or activity of indigenous bacteria and are known as prebiotics. The production of antimicrobial substances that are antagonistic to other microbes is a basic and important characteristic of a probiotic. Antimicrobial peptides (AMPs) are secondary metabolites that exhibit biological activity, but are inanimate. AMPs differ in their sequence, structure, have various nomenclatures, and are grouped as cecropins, defensins, and bacteriocins. Post-biotics mimic the beneficial effects of probiotics, while avoiding the risk of administering live microorganisms. Butyrate (Tsukahara et al., 2003; Tsilingiri and Rescigno, 2012), short chain fatty acids, small AMPs, and heat-killed probiotics are considered post-biotics and have been shown to have potent health benefits (Patel et al., 2012). Various studies support prebiotic and post-biotic use as potential alternatives to probiotic therapy. Major concerns surround probiotic therapy, however, including the creation of dysbiosis in hosts. The American Academy of Pediatrics Committee on Nutrition – Section On Gastroenterology, Hepatology, and Nutrition, and the European Society for Pediatric Gastroenterology Hepatology and Nutrition (ESPGHAN) Committee on Nutrition have suggested that large, well-designed clinical research studies be performed before probiotics are widely used (Thomas and Greer, 2010; Braegger et al., 2011). Post-biotics are the products of probiotics. Therefore, global research has begun to exploit the products of probiotics as they do not exhibit side effects such as sepsis, which is common with the use of probiotic therapy.
Motile Aeromonas hydrophila septicemia, hemorrhagic septicemia, and ulcer or red-sore diseases are caused by the Gram-negative bacterium A. hydrophila (Austin and Austin, 1999; Austin and Zhang, 2006). Aeromonas hydrophila are opportunistic pathogens and are highly infectious due to unsatisfactory water quality, such as high levels of carbon dioxide or nitrites, and low levels of dissolved oxygen (Laleh et al., 2015). Terramycin and Remet-30 are the two most extensively used FDA approved antibiotics against A. hydrophila infections in fish. There are reports of the existence of antibiotic-resistant Aeromonas spp., and therefore, one needs to consider the alternatives before administering antibiotics (Aoki et al., 1971; Pettibone et al., 1996; Son et al., 1997; Vivekanandan et al., 2002; Hemant et al., 2016). Several studies (Ansary et al., 1992; Yucel et al., 2005; Dias et al., 2012) have reported the presence of antibiotic-resistant Aeromonas spp., after which growing concern has arisen regarding resistance to ampicillin, carbenicillin, erythromycin, and streptomycin. Such resistance highlights the need to develop potent and rapid Aeromonas-eradicating drugs.
The most recent trends of the use of LPS and oral vaccines as potent/effective drug candidates have been well-received in fisheries. Baba et al. (1988) concluded that LPS induced better protection against A. hydrophila as a vaccine than a formalin-treated killed vaccine. Oral vaccines are also considered to provide good protection, but the immune response is slow, and to date, no commercialized oral vaccines have been developed against A. hydrophila (Agius et al., 1983). Attenuated and live vaccines have been shown to be successful in the fisheries industry, and in particular against bacterial infections. However, there are no reports of reliable and efficient commercial vaccines available in the industry.
The outer membrane proteins (OMPs) of A. hydrophila, such as OmpW and Aha1, are the best possible vaccine candidates (Maiti et al., 2012). Through wet laboratory experiments, Maiti et al., 2020 reported the above antigens as possible vaccine candidates. Furthermore, it was concluded that immunization with those two antigens show good immune responses. The survival of the infected A. hydrophila was considered to be in the range of 50–70%, which further suggests the protective effect of immunization with two antigens. Numerous laboratories have been engaged in the development of antigens for vaccines, but have failed to commercialize their results. This raises doubt about the efficacy of laboratory developed drug candidates.
Recent developments in the field have helped to understand and develop a new era of vaccine/drug candidates or AMPs with the following features. Candidates should (1) be manageable, with no skilled manpower required; (2) be easily produced in large quantities; (3) be easily incorporated into livestock feed; (4) not create antibiotic-resistant bacteria; (5) not be susceptible to intrinsic resistance mechanisms; (6) provide strong protection against highly active bacteria; and (7) show rapid action without any damage to the host. Based on the above criteria, the best option may be protein-based therapeutics with specific lethality to A. hydrophila that are easily manufactured and originate from GRAS (Generally Recognized As Safe) organisms. Therefore, this study focuses on post-biotic identification, isolation, characterization, and exploitation for therapy. Specifically, we aimed to identify, isolate, purify, and characterize post-biotics for A. hydrophila therapy.
Materials and Methods
Cultures, Chemicals, and Reagents
Lactobacillus acidophilus (ATCC-4356 or DSM 20079), mannone oligosaccharide (MOS), nisin, sodium dodecyl sulfate (SDS), tricine, HPLC-grade acetonitrile, and trifluoroacetic acid were purchased from Sigma-Aldrich Chemicals, Ltd. (Bengaluru, India). The indicator strain Micrococcus luteus (Microbial Type Culture Collection Centre [MTCC] Acc. No. 1739), and wild type A. hydrophila (MTCC Acc. No. 106T) was obtained from the MTCC (Chandigarh, India). De Man, Rogosa, and Sharpe (MRS), Luria Bertani (LB), and nutrient broth (NB) were purchased from Hi-Media Laboratories (Mumbai, India).
Extraction of L. acidophilus and A. hydrophila Cell-Free Supernatant (CFS)
Lactobacillus acidophilus and A. hydrophila were individually streaked (to obtain isolated single colonies) onto MRS agar plates and incubated at 37°C overnight. Subsequently, a single isolated colony was inoculated into fresh MRS broth and grown for 12–16 h, at 37°C without shaking. The resulting cultures of L. acidophilus and A. hydrophila were subcultured at 4–5% in fresh MRS medium and grown until an OD600 of 0.4 was reached. The supernatant was obtained by centrifugation at 9000 × g for 10–15 min at 4°C. Both supernatants (L. acidophilus and A. hydrophila) were individually collected, and passed through a 0.22-μm filter to remove any remaining traces of cellular debris. The resulting filtrate was filtered through a 10-kDa cutoff membrane filter. Based on the requirements to obtain highly concentrated protein, the CFSs were further concentrated using Amicon Ultra centrifugal protein concentrators (Merck-Millipore, India) at room temperature for 30 min. The resulting concentrated CFSs of L. acidophilus and A. hydrophila were used for the well-diffusion assay.
Well-Diffusion Assays
Wherever necessary, A. hydrophila and type cultures/indicators such as M. luteus and stock cultures were maintained at −80°C in 20% glycerol for long-term storage. For short-term storage, bacteria were maintained on their respective agar plates at 4°C, and new plates were streaked every 15 days. The indicator organisms M. luteus and A. hydrophila were grown in MRS broth for 10–14 h at 37°C with shaking at 200 rpm. Before cultures were harvested, the absorbance at OD600 was measured.
MRS agar (2%) was used as a solid substrate after melting the agar at 40°C for 30 s. Subsequently, M. luteus and A. hydrophila in mid-exponential growth were mixed well at a 1% concentration (100 μL of cells in 10 mL of low melting agar), poured into sterile Petri dishes under sterile conditions, and allowed to solidify. Wells of 5-mm diameter were made with a sterile aluminum bore maker (Hi-Media), and 50–100 μL of Lactobacillus cell free supernatant (LABS; 10 μg) was added to the wells. Plates were incubated at 37°C overnight. Nisin (15 μg) was used as a positive control.
Electrophoretic Separation of CFSs
Tricine-SDS-PAGE gels were used to electrophoretically separate low-molecular-weight and high-molecular-weight protein bands ranging from 1 to 100 kDa. A volume of 50–100 μL of CFS was boiled, followed by centrifugation for 15 min at 7000 × g and the supernatant was separated by tricine-SDS-PAGE. We used cathode buffer (upper tank; 0.1M Tris, 0.1M Tricine, 0.1% SDS) as the inner electrode buffer and anode buffer (lower tank; 0.2M Tris, pH 8.9) as the lateral electrode buffer. Electrophoresis was performed at room temperature without cooling measures, except for heat conduction of the surrounding air. The parameters for 16% gels (0.7 mm) included an initial voltage of 30 (45 min), followed by 200 (45 min), and an end voltage of 300, for 2–4 h. After electrophoresis, protein bands were fixed in fixing solution (0.025% Coomassie dye in 10% acetic acid) for 45 min and the gel was destained in 10% acetic acid for 15–60 min.
Purification of the Antimicrobial Substance
Lactobacillus acidophilus was grown overnight, subcultured in 1 L of MRS medium, and incubated for 72 h at 37°C with shaking (200 rpm). The cultures were harvested by centrifugation at 9000 × g for 30 min at 4°C. The collected supernatant was filtered through a 0.22-μm filter. The filtered supernatant was ammonium sulfate precipitated (70% optimum) and dialyzed against phosphate buffer using a 2-kDa cutoff membrane. Finally, the sample was purified using a Superdex-75 gel filtration column (Sigma-Aldrich). The fractions were collected and tested for antimicrobial activity. A Superdex 75 gel filtration preparation grade column (Sigma-Aldrich) with a bed volume of 120 mL, particle size of 24–44 μm, and 150 mM sodium phosphate buffer (pH 7) was used. The column was washed, equilibrated with the buffer, and the sample was injected. After ammonium sulfate precipitation and dialysis, a protein sample of 5% of the total bed volume was injected into the column. The flow rate (0.2 mL/min) and other parameters (A280) were held constant prior to fraction collection.
HPLC Analysis of the Purified Protein
The final purification to obtain 99% homogenous protein was carried out using high-performance liquid chromatography (HPLC) (SHIMADZU-PDA detector, XBridge Waters column, C18, 5 μm, 10 × 250 mm of parameters). A mobile phase of solvent A, 5 mM ammonium acetate buffer (pH 5.5) and solvent B, 100% acetonitrile was used. The gradient elution was performed as 5–45% solvent B in 40 min, 45–90% B in 12 min, and reverse 90–5% B in 8 min. The flow rate was kept at 2 mL/min. A sample of 500 μL was injected and the peaks were analyzed at 220 nm. All peak fractions were collected and assayed for bioactivity. The active peak fraction was identified and purity was determined by analytical HPLC. Antimicrobial activity of the purified compound was checked against the target organism, A. hydrophila.
Overlay Assay of the Purified Antimicrobial Substance
The chromatography-purified AMP peptide was subjected to tricine-SDS-PAGE (Schägger, 2006; Rajagopal et al., 2015). Electrophoresis was performed to separate the CFS. One part of the gel contained the molecular weight marker. The other part of the gel was subjected to fixation and washed with 100 mM sodium phosphate buffer. SDS was removed by washing with 2.5% Triton-X for 3 h. The gel was then washed with Milli-Q water to remove Triton X-100.
MRS Agar Plate Preparation/Overlay
The solid substrate MRS agar was incubated at 40°C for 30 s as described in the well-diffusion assay protocol. Aeromonas hydrophila in mid-exponential growth were mixed well at a 1% concentration (100 μL of cells in 10 mL of low melting agar), poured into sterile Petri dishes, and allowed to solidify. The antimicrobial peptide (10 μg) was electrophoretically separated using tricin-SDS-PAGE and the gel was washed with water to remove excess Triton X-100. The resulting gel was overlayed onto the Petri plates seeded with A. hydrophila, which were incubated at 37°C overnight. Nisin (15 μg) was used as a positive control.
Mass Spectrometry (MS)
Following gel filtration chromatography, the purified peptide was subjected to LC-ESI-MS (Agilent 6550 I funnel QTOF) in positive ion mode. Subsequently, the resulting mass spectrum was analyzed in the range of 400–4000 m/z. For MALDI-TOF analysis, a complex of peptide sample and α-cyano-4-hydroxycinnamic acid (CHCA) matrix was made and the mass spectrum was obtained on the MALDI-TOF mass spectrometer (AB Sciex 5800). MS/MS analysis was performed on the same instrument with the TOF-TOF analyzer. Amino acid analysis was carried out with the PICO-TAG amino analysis system (Waters) according to the manufacturer’s instructions.
The Complete Genome Sequence of Lactobacillus acidophilus DSM 20079 or ATCC-4356 (CP020620.1)
Genomic DNA was isolated from LA using the Qiagen gDNA extraction kit (Qiagen, Valencia, CA, United States) following the manufacturer’s protocol. The DNA quality was evaluated using a Qubit fluorometer (Thermo Fisher Scientific, Waltham, MA, United States) and Nanodrop spectrophotometer (Thermo Fisher Scientific). The high quality genomic DNA was fragmented using the g-TUBE (Covaris, Woburn, MA, United States) via a 20-kb template library preparation workflow and then end-repaired to prepare SMRTbell DNA template libraries according to the manufacturer’s instructions (Pacific Biosciences, Menlo Park, CA, United States). The library quality was analyzed by Qubit and average fragment size was estimated using an Agilent 2100 Bio-analyzer (Agilent, Santa Clara, CA, United States). SMRT sequencing was performed using a Pacific Biosciences RSII sequencer (Pacific Biosciences, Menlo Park, CA, United States) using one SMRT cell and P4-C2 chemistry with a PacBio RS II sequencer at 120 min movie length. The library preparation and PacBio sequencing was performed by Macrogen, Inc. (Geumcheon-gu, Seoul, South Korea).
Single molecule real-time sequencing reads were de novo assembled using the Hierarchical Genome Assembly Process (HGAP) workflow, which is a part of PacBio SMRT Analysis 2.3.0 package and subsequently polished with Quiver (Chin et al., 2013). Finally, the genome sequence was circularized using Circulator version 1.1.4 (Hunt et al., 2015). The protein coding sequences (CDSs) were predicted by carrying out with NCBI’s Prokaryotic Genome Annotation Pipeline (PGAP) version 4.2 (Tatusova et al., 2013), and Rapid Annotations using Subsystems Technology tool kit (RASTtk) was used for functional annotation of the assembled contig (Brettin et al., 2015). The mining of the bacteriocinogenic gene clusters was performed by using the BAGEL4 platform (van Heel et al., 2018). Default parameters were used for all steps of Bio-informatic analysis.
Maintenance of Fish
Before starting the animal experiments, institutional animal ethics committee (IAEC-USM-PEN-2016-11) clearance was obtained. A group of 3–4-month-old Channa striatus juveniles (55 ± 10 g) was purchased from a commercial market and maintained in canvas tanks of 4 m ± 1 m ± 1 m. The fish were provided commercial feed and acclimatized to laboratory conditions. Before commencing the experiment, fish were distributed into seven different canvas tanks of the same size, as described above (including one positive and one negative control tank). The approximate stocking density was maintained, at 5 fish m–3, and during the trials, the dissolved oxygen content was maintained at 4.50 ± 1.42 mg L–1. The pH was 6.3–6.9, and the temperature was maintained at 29 ± 2°C. Tanks were cleaned by frequent washing and nitrogenous waste was kept to a minimum.
Seven fishponds (labeled A–G) containing 10 healthy fish each were selected for the experiment. Group A was the positive control and fed a lethal dose of A. hydrophila. Group B was fed Aeromonas CFS (AMS), followed by re-challenge with A. hydrophila at 108 colony forming units (CFU) after 2 days (rechallenge at day 7). Group C was fed LAB (1010–1011) and re-challenged with A. hydrophila, similar to group B. Group D was fed LABS followed by challenge with A. hydrophila, identical to group C. Group E was fed mannone oligosaccharide (MOS, 1% quantity) prebiotic, followed by A. hydrophila infection. Group F was injected with serum collected from Aeromonas-infected fish (100 μL and re-challenged with A. hydrophila at a lethal dose of 108 CFU). Group G was the negative control group; fish were not infected and maintained in the same conditions as the experimental groups. The experiment is illustrated in the flow diagram shown in Figures 1A–G. After infection with a lethal dose of A. hydrophila, fish were maintained under observation. Fish with severe septicemia were removed and declared dead. Fully active fish lacking symptoms of A. hydrophila-infection were considered alive. To conclude the efficacy and efficiency of L. acidophilus antimicrobial substance the field experiments were performed in triplicate and repeated three times.
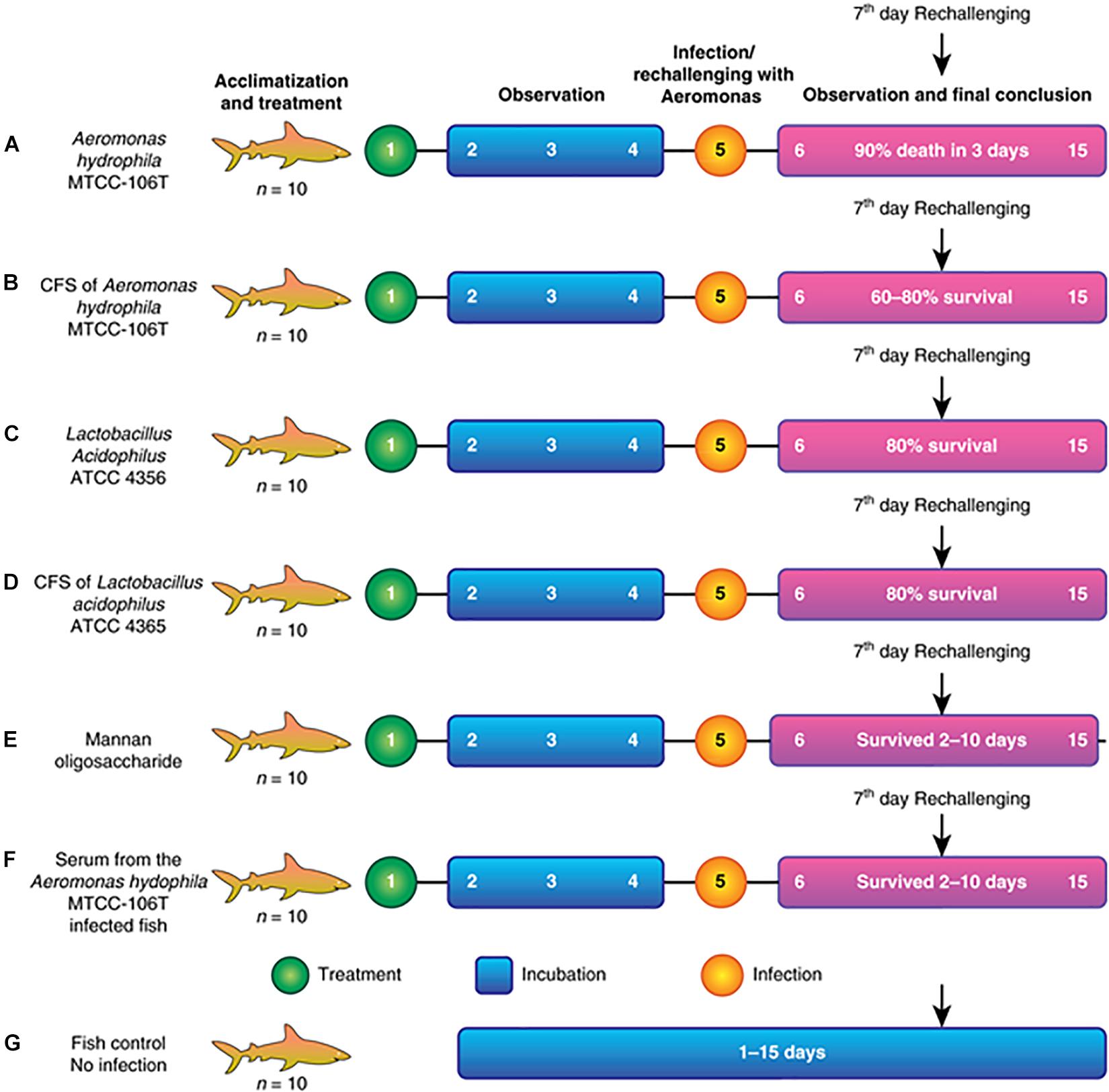
Figure 1. (A) Fish injected with a lethal dose of A. hydrophila disappeared, and fish recovered by the end. (B) Fish injected with CFS of A. hydrophila. Septicemia (C) Fish injected with L. acidophilus and re- challenged. (D) Fish injected with CFS of L. acidophilus and re- challenged severe septicemia led to death after 5 days of infection. (E) Fish injected with MOS and re- challenged with a septicemia led to death after 6 days of infection. (F) Fish injected with Aeromonas hydrophila serum of infection and severe septicemia led to death after (G) Control fish.
Statistical Evaluation of the Challenge Data
Experiment was done in triplicates and statistical significance of the data was analyzed through Two-way ANOVA (α = 0.05) with the Geisser–Greenhouse correction followed by Dunnett’s multiple comparisons test using GraphPad Prism version 8.0.0 using GraphPad Prism Version 8.0.
Results
Characterization of the Purified Antimicrobial Agent
The well-diffusion assays, tricine-SDS-PAGE, and zymography of the purified protein showed that the active anti-A. hydrophila molecule was a low molecular weight protein/peptide. This finding was verified by the corresponding protein band in the zymogram (Figure 2A, Lanes 1 and 2). The activity observed through well-diffusion assays was positive and reproducible (Figure 2A, Lane 2; Figure 2B, Inset A). Efficient and reproducible antagonistic activity was also observed with M. luteus cultures. The antimicrobial agent from L. acidophilus (10 μg) showed antagonistic activity against A. hydrophila [Figure 2B (+ ve,−ve, denotes with and without hallow zone/zone of inhibition), and 2C, Inset A]. Nisin (15 μg) was included as a control with M. luteus (data not shown), and confirmed to exhibit antagonistic activity. Thus, we consider this molecule to be an antimicrobial polypeptide with a molecular weight of 5 kDa (Figures 2B-2,C, mass spectra).
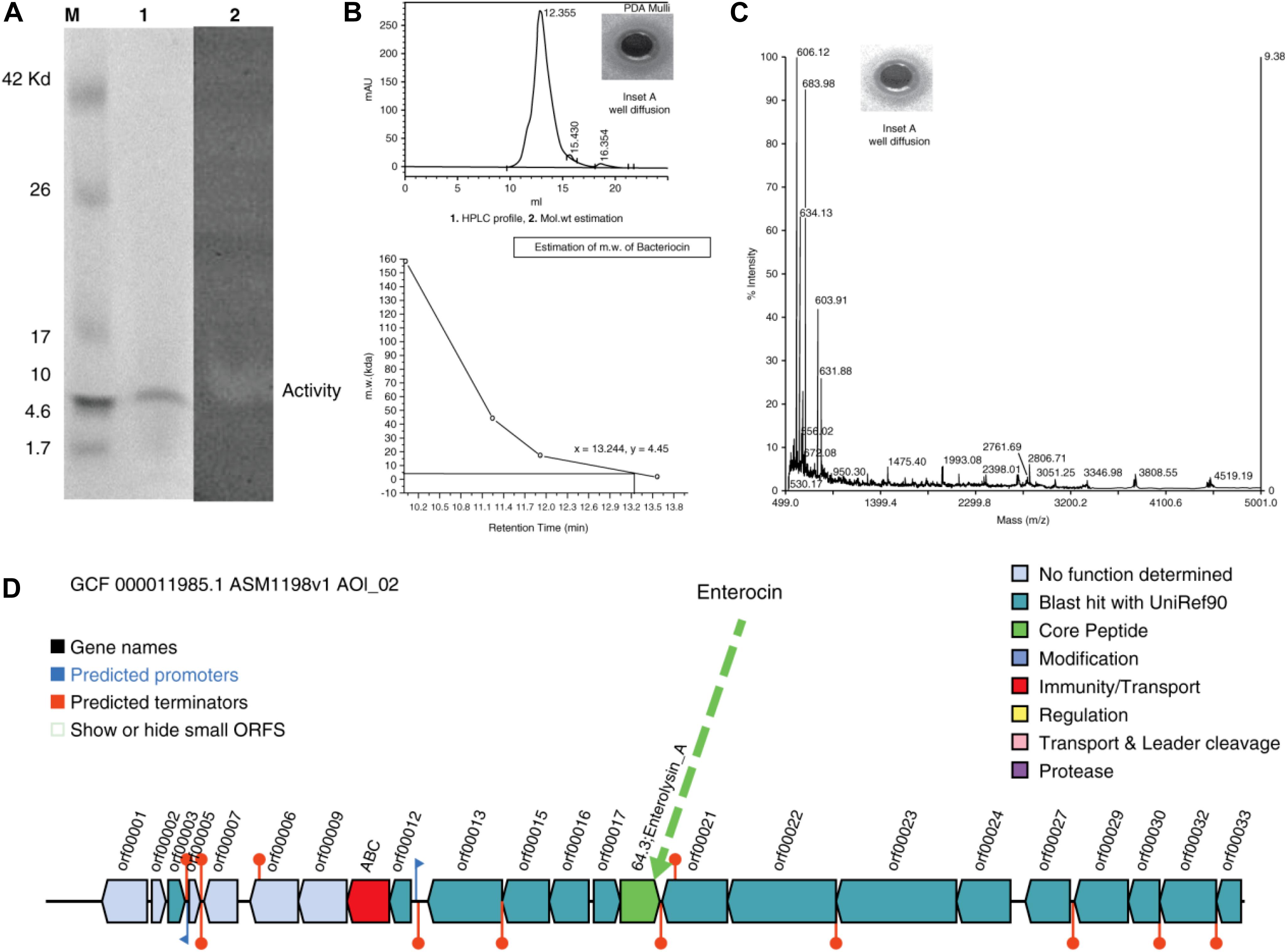
Figure 2. (A) Tricine-SDS-PAGE analysis and zymogram of L. acidophilus purified protein. M, marker; lane 1, purified protein; lane 2, arrow indicates zone of inhibition. (B) (1) HPLC profile [Inset shows the zone of inhibition (ZOI) of the peak fractions]. (2) Molecular weight estimation (+ve and –ve indicate for with and without activity). (C) Mass spectra (Inset shows the ZOIs of each peak fraction). (D) Antimicrobial biosynthetic cluster.
Gel filtration chromatography was used to obtain a single, pure, homogeneous protein. Using an automatic fraction collector, 2 mL fractions were collected and a total of 120 mL of sample was collected. The peaks were pooled and the activity was assessed against the indicator strain. Subsequently, the purified protein was subjected to HPLC for retention time analysis and revealed a single peak (Figure 2B-1), with a zone of inhibition (Figure 2B-1,+ve,−ve, Inset A). To determine the exact molecular weight of the peptide, a curve was drawn between the molecular weight and the retention time (Figure 2B-2), which confirmed the molecular weight as ∼5 kDa. The peptide was also analyzed by MS (Figure 2C, Inset A). To determine the probable corresponding genetic location and position of the predicted peptide in the genome, the complete genome sequence of L. acidophilus (ATCC-4356/DSM-20079) was annotated, assembled, and analyzed. Figure 2D shows the antimicrobial gene cluster containing the coordination of many genes for its function.
Field Trials to Understand Antimicrobial Efficiency in an Aquatic System
The present study described the isolation, identification, and characterization of a post-biotic with antagonistic activity against A. hydrophila. The post-biotic molecule was a peptide originating from L. acidophilus, which has GRAS status. Therefore, the present study evaluated the efficacy of the peptide at inhibiting A. hydrophila growth. We observed the efficacy of the molecule through different assays. Figure 1 presents the detailed study plan to develop an AMP that acts against A. hydrophila. The overall study involved 70 specimens of C. striatus that were placed in seven different canvas tanks (n = 10 for each). Assays began with the acclimatization and treatment of fish, followed by observation, infection/immunization, incubation, and observation. The experiment was ended after 15 days. A flowchart of the assay is shown in Figure 1. The results of the field trials of antimicrobial is as follows. These experiments were performed thrice as per the GLP (Good Lab Practice) standards.
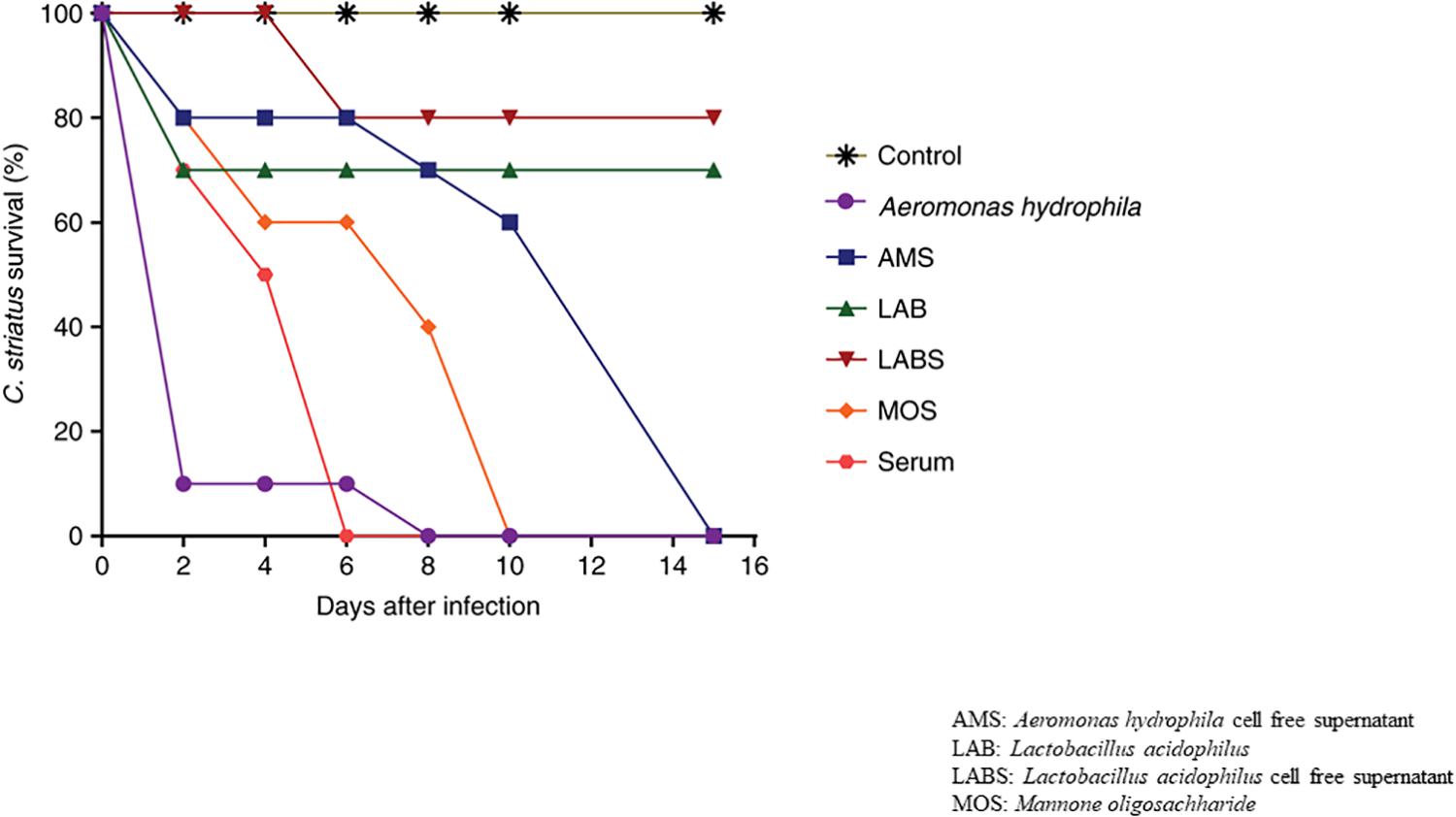
Fish infected with A. hydrophila (108 CFU, lethal dose) developed severe septicemia; 80% died within 3 days (Figures 3, 4). The remaining fish showed typical symptoms of A. hydrophila infection (Figures 3, 4), but did not die within 3 days. After 8 days, all fish were dead. Fish treated with MOS and serum survived with symptoms for 6–10 days (Figures 3, 4), but 80% of Lactobacillus-, and LABS (LA-CFS)-treated fish survived. Only 20% death occurred in the first 2 days. A mortality rate of 30% was observed on the second day in fish treated with L. acidophilus, while the remaining fish (7/10) survived for the remainder of the experiment (Figures 3, 4). A total of 80% of the fish treated with LABS survived for 3–4 days and the overall mortality rate did not exceed 20%. The remaining fish survived without symptoms after 15 days of infection. The survival curves show the growth and survival of fish treated differently, as stated in the protocol flowchart (Figure 1). Therefore, we conclude that LABS contained a compound that was active against A. hydrophila. To explore this further, we used a plate assay with the same supernatant (Figure 2B, Inset A). Fish treated with AMS and LABS exhibited 80% survival, even after 10 days. Only 20% mortality was observed during the first 4 days of treatment. The rapid killing rate initiated late on day 4 in the A. hydrophila infection group ultimately resulted in 100% mortality after 15 days. This phenomenon was unique to A. hydrophila infection and was not observed with LABS. A maximum mortality of 20% was observed with LABS. This clarifies the potency of LABS compared to AMS. The killing/lethal effect was much more pronounced in the case of Aeromonas-treated fish as the death rate was 90%, which began after 3 days of infection. Upon LABS treatment, the mortality rate was 20%, with 80% viability. The field experiments were repeated thrice and observations confirmed that the antimicrobial protein may be used to control A. hydrophila infections.
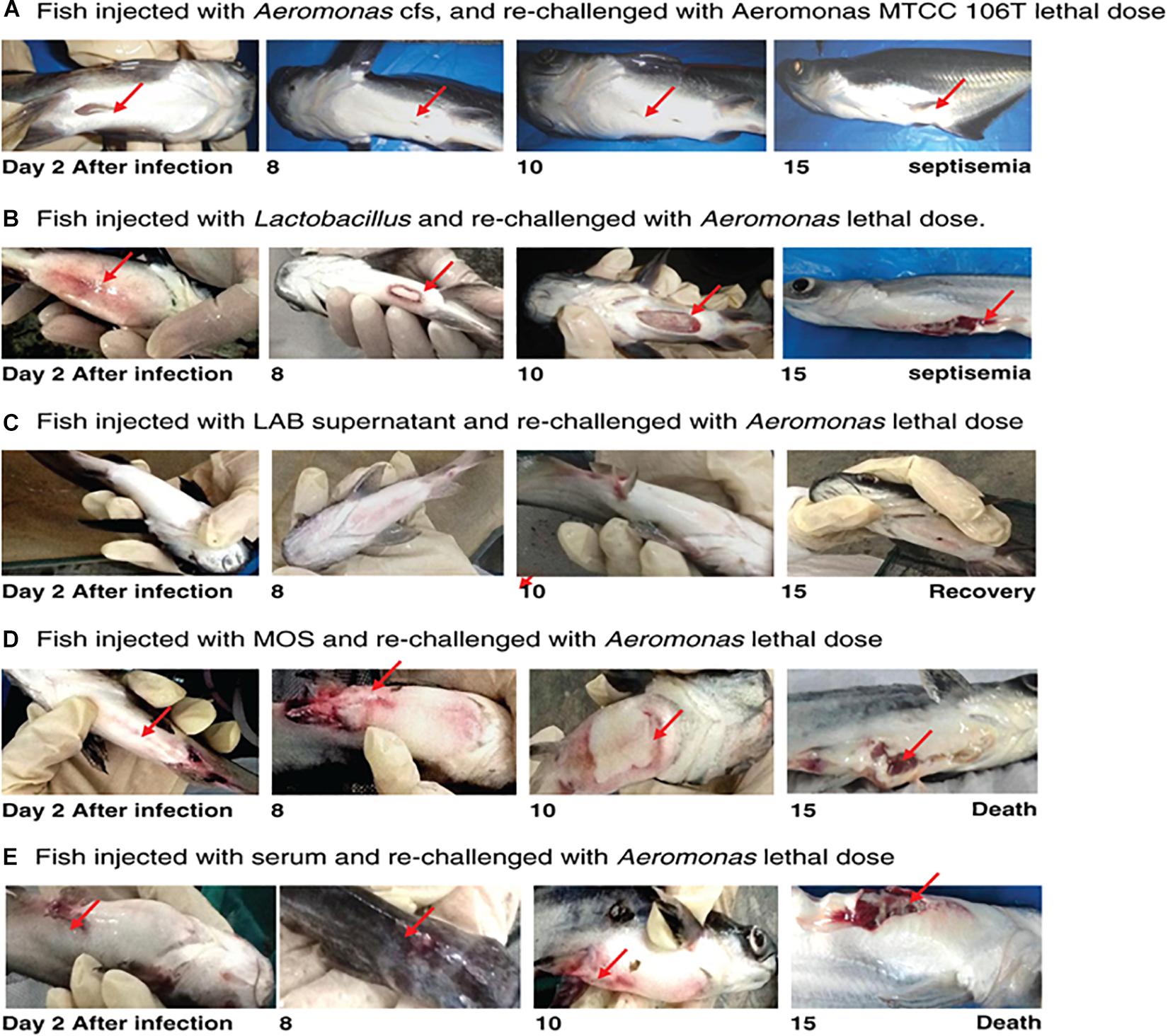
Figure 3. (A) Fish injected with Aeromonas CFS and re-challenged with a lethal dose of A. hydrophila MTCC 106T. (B) Fish injected with L. acidophilus and re-challenged with a lethal dose of A. hydrophila. (C) Fish injected with LAB supernatant and re-challenged with a lethal dose of A. hydrophila. (D) Fish injected with MOS and re-challenged with a lethal dose of A. hydrophila. (E) Fish injected with serum of A. hydrophila-infected fish and re-challenged with a lethal dose of A. hydrophila.
Statistical Analysis of the Challenge Data Through ANOVA Studies
Samples displayed extremely significant difference between groups compared to control group with p-value less than 0.01 upon all the time points observed [A. hydrophila (p = 0.0024), AMS (p = 0.0004), LAB (p = 0.0001), LABS (p = 0.0001), MOS (p < 0.0001), SERUM (p < 0.0001)]. After 15 days of infection, significant difference in survival in groups exposed to A. hydrophila (mean difference in survival of 26.0%), AMS (mean survival time of 32.6%), LAB (mean survival time of 42.8%), LABS (mean survival time of 46.2%), MOS (mean survival time of 55.8%), SERUM (mean survival time of 64.4%) was observed (Figures 3, 4).
Tricine-SDS-PAGE and Zymogram Assays
Tricine-SDS-PAGE is a suitable tool for the separation of low-molecular-weight proteins (Schägger, 2006). Based on HPLC and MS studies, we concluded that the approximate molecular weight of the antimicrobial agent was 5 kDa (Figure 2A, lane 1). To validate this observation, the same gel was overlaid on a lawn of A. hydrophila to evaluate the antimicrobial activity (Figure 2A, lane 2), and a zone of inhibition was obtained. Thus, the data confirmed that the molecular weight of the antimicrobial agent was ∼5 kDa.
HPLC Profile
The homogeneous and purified protein, as well as the standards, were subjected to HPLC to determine the retention time of the peptide. Figure 2B-1 reveals a single peak, and Inset A shows the well-diffusion assay. Once the retention time was obtained, a graph was drawn between the retention time and the molecular weight (control/standards). Figure 2B-2 confirms that the molecular weight of the protein is ∼5 kDa.
MS Analysis Through LC-ESI-MS
The approximate molecular weight of the antimicrobial agent was estimated through tricine SDS-PAGE and HPLC retention analyses. The purified protein was also subjected to LC-ESI-MS to determine its mass. Our institutional proteomic facility was used to determine the mass of the peptide, and the MS data of the purified antimicrobial compound are shown in Figure 2C. The protein was then used for a well-diffusion assay (Figure 2C, Inset A). These assays confirmed the molecular weight, and it was hypothesized that the DNA sequence would aid in the design of novel antimicrobial agents.
Whole Genome Sequence Analysis of Lactobacillus acidophilus ATCC-4356/DSM 20079
The resulting assembly consisted of a single contig with a total size of 2,009,973 bp long and the G + C content was 34.72%. The total number of predicted protein-coding genes was 1,824, with 75 predicted RNAs and 99 predicted pseudogenes. The bacteriocin genome-mining tool BAGEL-4 identified one area of interest (AOI) corresponding to bacteriocin, and the antimicrobial biosynthetic gene cluster is shown in Figure 2D, demarcating the arrangement of the bacteriocin gene (green color).
Discussion
In ancient time, it was assumed that the consumption of whole viable organisms was an essential factor to experience health benefits (Markowiak and śliżewska, 2017). However, subsequent studies on the use of probiotics for therapy resulted in doubts in their efficacy. Changes in the composition and functioning of gut bacteria due to probiotic use is not yet clear. However, some studies have reported that probiotics alter the composition of gut microbiota, which co-occurs with health-promoting effects (Zmora et al., 2019). Due to the lack of strong and reproducible experimental evidence of the beneficial effects of probiotics, it is not only difficult, but also detrimental to claim microbiome alterations are indeed beneficial (Kim et al., 2013; Zmora et al., 2019). The most frequent disadvantage of the use of whole microorganisms as probiotics is sepsis, which can be difficult to control. Therefore, researchers have begun searching for effective alternatives.
Prebiotics have been defined as “a substrate that is selectively used by host microbes conferring a health benefit” (Suez et al., 2019), and Vyas and Ranganathan (2012) concluded that prebiotics may govern microbiota compositions of a host. FOS, MOS, short chain GOS, and long chain FOS are well-studied and extensively used prebiotic substances (Giovannini et al., 2014). The most important effects observed upon the use of prebiotics are enhanced growth and activity of Bifidobacteria spp. Subsequently, to fulfill the missing functions of probiotics and prebiotics, synbiotics have evolved, which are a synergistic mixture of pro- and prebiotics. The major health benefits of the use of probiotics, prebiotics, and synbiotics largely depend on the synthesis of short chain fatty acids (SCFAs), active proteins/biomolecules, and secreted polysaccharides (Paulina et al., 2017). These observations have led to the generation of a new era of food supplements known as post-biotics.
Post-biotics are defined as non-viable substances, which confer benefits to the host upon their intake in sufficient amounts (Tsilingiri and Rescigno, 2012). The immunomodulatory properties of probiotic microorganisms shows their presence beyond their viability (Schley and Field, 2002; Patel et al., 2012; Patel and DuPont, 2015; Scott et al., 2018). Olle (2013) stated that post-biotics may impart togetherness among microbiology, food, and personalized treatment fields. The efficacy of post-biotics depends on the proteins, metabolites, fats, lipids, carbohydrates, and vitamins that are produced during the fermentation process. Post-biotic products have similar effects on human health as probiotics; however, given that sepsis is a major concern when using probiotics for therapy, their replacement with post-biotics may be efficacious. The study illustrating the useful effects of pre-, pro-, and post-biotics are explained in Supplementary Figure 1. These substances have an enormous beneficial effect in the aquatic system.
The studies such as tricine-SDS-PAGE, zymogram, well-diffusion assay, HPLC, and LC-MS concluded that the low-molecular weight antimicrobial substance/fraction (∼5 kDa) produced by L. acidophilus exhibited strong antibacterial activity against A. hydrophila. HPLC and MS confirmed the molecular weight. Whole-genome sequencing predicted that the antimicrobial compound was bacteriocin. Thus, A. hydrophila supernatants containing the secretory antigens are possible drug/vaccine candidates (discussed by Maiti et al., 2012; Khushiramani et al., 2012) because they showed maximum protection in fish infected with A. hydrophila with a high survival rate of 80%.
In general, clinical studies of probiotics have shown positive effects on intestinal and allergic diseases (Sánchez et al., 2017; Suez et al., 2019). Such studies have also proven the effectiveness of probiotics for the treatment of metabolic disorders such as obesity, insulin resistance, type 2 diabetes, and non-alcoholic fatty liver disease (Kim et al., 2019). Furthermore, those studies also proved the efficacy of probiotics in increasing immunity (immunomodulation). In terms of specific effects and/or advantages of probiotics, the modes of action of prebiotics begin with maintaining or creating an efficient epithelial barrier and subsequently inducing effective adhesion to the intestinal mucosa (Taverniti and Guglielmetti, 2011; Tsilingiri and Rescigno, 2012; Sánchez et al., 2017). Such events prevent pathogen adhesion by competitive exclusion, the production of antimicrobial substances, and modulation of the host immune system (Vyas and Ranganathan, 2012). Therefore, probiotics and their products, such as post-biotics, may be the best possible candidates to control infections. Advanced molecular biological and genetic tools are used to understand the beneficial effects of probiotics, including (1) their antagonistic activity through the production of antimicrobials (Vandenbergh, 1993; Brandao et al., 1998; Isolauri et al., 2001; Guillot, 2003); (2) competitive inhibition to kill the pathogens; and (3) immunomodulation. Newly developed probiotic formulas/food supplements that contain both probiotic strains and synergistic prebiotics enhanced probiotic effects in the small intestine and colon (Guillot, 2003; Olle, 2013; Giovannini et al., 2014; Szajewska et al., 2015). Those “enhanced” products were even more effective in controlling pathogenic infections, and superior to conventional antibiotics when each component was administered separately (Paulina et al., 2017).
Not all antigens derived from A. hydrophila may function as a vaccine candidate since a major disadvantage is the presence of trace amounts of A. hydrophila in the supernatant, which may cause severe infections. Therefore, A. hydrophila antigens may not be possible drug candidates for stressed fish in contaminated ponds as they will be more susceptible to infection. Therefore, we advise caution before using A. hydrophila antigens to treat A. hydrophila infections. Nevertheless, we consider this antimicrobial to be a possible drug candidate for A. hydrophila infections if the CFS is free of the bacterium.
Pang et al. (2015) found that an MOS-supplemented diet has been shown to enhance resistance to infection (Liu et al., 2013). Hence, we studied the MOS component in the present study. Mannone oligosaccharide was able to protect fish for 2 days, but following that, the death rate increased from 40 to 60%, and all fish died after 10 days. Hence, MOS can delay the infection rate and increase survival in the short term. Fish injected with serum showed a similar pattern to MOS; the death rate was delayed 40-60% for 3–4 days, after which all fish died. Based on the severity of infection, the most suitable and potent drug candidate for A. hydrophila can be decided. The above stated possible candidates, such as serum, MOS, LAB, LABS, AM, and AMS, are possible drug candidates as they all may be used for therapy. We conclude that the antimicrobial agent found in LABS may be considered for Aeromonas infections in aquatic systems, which exhibit high levels of protection. This molecule will require further characterization to understand its structure, amino acid sequence, and initiation of the immune response. The data obtained through field trials was subjected to statistical evaluation (ANOVA-test) and found that there is an extremely significant difference between groups compared to control group. Fifteen days infection samples show much higher significant difference in survival groups exposed to A. hydrophila. Two-way ANOVA revealed a statistically significant interaction between groups compared to control group (one-way ANOVA followed by Dunnett’s multiple comparisons test was performed using GraphPad Prism version 8.0.0 for Windows; GraphPad Software, San Diego, CA, United States)1 (McHugh, 2011).
It has yet to be shown that synthetic peptide vaccines/drugs are the best candidates to treat infections (Weidang et al., 2014). Until this is confirmed, it will be essential to exploit LABS and the extraction of antimicrobial agents for A. hydrophila infections. Antimicrobial agents from L. acidophilus may be possible drug candidates because of their rapid action, efficiency, and reproducibility. Lactobacillus-based antimicrobials are also considered to be thermostable, protease-resistant, and active (Konstantinos et al., 2016) against physiological enzymes such as trypsin and chymotrypsin. Molecular and genetic studies have reported that immunomodulation of the host occurs following consumption of probiotics (Isolauri et al., 2001). This may occur because adhesion of probiotics to epithelial cells subsequently triggers the immune system signaling, further leading to immunological modulation. Low molecular weight substances produced by probiotic microbes help inhibit pathogen replication (Oelschlaeger, 2010). Probiotics are known to influence the acquired immune system through metabolites, cellular components, and DNA. Prebiotics play a major role in increased secretion of IgA (Schley and Field, 2002). Therefore, probiotic products such as low molecular weight proteins/substances are possible drug candidates.
Liu et al. (2015) reported that A. hydrophila is virulent and pathogenic, and may lead to rapid mortality. However, the presence of Aeromonas antigens in the supernatant may trigger an immune response that protects animals. This phenomenon was observed when AMS was injected. Similarly, Lactobacillus is known to be a probiotic microbe with proven health benefits; therefore, LAB-fed animals performed much better than Aeromonas- or AMS-treated animals. Similar to AMS, LABS also provided more protection than MOS and serum. This may have been due to the presence of an antimicrobial agent active against A. hydrophila, as shown in the well-diffusion assays (Figures 2B,C, Inset). The other components, such as MOS and serum, delayed fish mortality, but they failed to protect against A. hydrophila infection. Therefore, we conclude that LABS may be the best possible drug candidate as it effectively protected fish. The prebiotics, serum, and other components may protect against A. hydrophila and delay mortality, but may not confer permanent protection. Therefore, we conclude that L. acidophilus and LABS may be suitable candidates for the long-term control of A. hydrophila infections and can be introduced into fish feed, which is the best way to administer drug candidates. Live probiotics are known to cause health problems (possibly by antibiotic resistance gene transfer); however, the products of Lactobacillus do not cause any adverse effects, such as non-specific killing (Gareau et al., 2010; do Carmo et al., 2018). The presence of excessive amounts of antimicrobial agents of L. acidophilus does not cause environmental pollution, and does not create resistant bacteria. Therefore, we conclude that antimicrobials (bacteriocins/lactocins/enterocins) of probiotic origin may be possible drug candidates for A. hydrophila infection.
We successfully isolated, identified, characterized, and validated AMP efficacy against A. hydrophila infections in aquaculture. We found that prebiotics and probiotics protect fish from A. hydrophila infections to some extent. However, an antimicrobial agent from L. acidophilus provided maximal protection against A. hydrophila infections. The other test components, including probiotics in general and LABS specifically, may provide permanent protection; prebiotics confer only temporary protection. Therefore, we consider the AMP identified from L. acidophilus may be a possible drug candidate to protect fish against A. hydrophila infections. We intend to extend this study to isolate, identify, and characterize the antimicrobial agent of L. acidophilus. Although the LABS antimicrobial agent exhibited strong antagonistic activity and protection against A. hydrophila, it was not sufficient to determine the functionality of the concept. Injectable antimicrobials may not be commercially viable, and therefore, a novel, simple, and commercially viable process for the incorporation of antimicrobials into feed is required.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: “https://www.ncbi.nlm.nih.gov/ CP020620.1”
Ethics Statement
The animal study was reviewed and approved by IAEC-USM-PEN-2013/22 FRGS- RH. Institutional Animal Ethics committee of Universiti Sains Malaysia Penang-yr 2013, Grant22 under FRGS from RH.
Author Contributions
NA: the fieldwork, bench work, and executed the hypothesis. RH: collaborator provided the laboratory space and resources at USM-Malaysia. HP: whole genome data annotation, assembly, and analysis. S-DC and D-WL: genome data compilation. J-HS: helped us to shape up the final manuscript during Brain pool fellowship and collaborator. KR: generated the idea, planned the experiments, analyzed the data, and wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
The research was financially, supported by the Strategic Initiative for Microbiomes in Agriculture and Food (Grant Number 918010-4), Ministry of Agriculture, Food and Rural Affairs, South Korea. Funding from USM, Malaysia and TWAS is highly appreciated.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to acknowledge CSIR-CFTRI, CSIR-IMT, Third World Academy of Sciences (TWAS) – University of Sains, Malaysia (TWAS – USM) for financial assistance, to KR and the KyungPook National University (KNU)-fellowship to Phaum. At-last, but not the least we thank Anagha N. a research student for her enthusiastic contributions.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.570851/full#supplementary-material
Supplementary Figure 1 | The effect of probiotics, prebiotics, and post-biotics in aquatic system.
Abbreviations
AH, Aeromonas hydrophila; AMP, Antimicrobial protein; AMS, Aeromonas supernatant; CFS, cell free supernatant; LA, Lactobacillus acidophilus; LABS, Lactobacillus acidophilus cell free- supernatant; ML, Micrococcus luteus.
Footnotes
References
Agius, C., Horne, M. T., and Ward, P. D. (1983). Immunization of rainbow trout, Salmo gairdneri Richardson, against vibriosis: comparison of an extract antigen with whole cell bacteriocins by oral and intraperitoneal routes. J. Fish Dis. 6, 129–134. doi: 10.1111/j.1365-2761.1983.tb00060.x
Ansary, A., Haneef, R. M., Torres, J. L., and Yadav, M. (1992). Plasmids and antibiotic resistance of Aeromonas hydrophila. J. Fish Biol. 15, 191–196. doi: 10.1111/j.1365-2761.1992.tb00653.x
Aoki, T., Egusa, S., Ogata, Y., and Watanabe, T. (1971). Detection of resistance factors in fish pathogen Aeromonas liquefaciens. J. Gen. Microbiol. 65, 343–349. doi: 10.1099/00221287-65-3-343
Austin, B., and Austin, D. A. (1999). Bacterial Fish Pathogens: Disease in Farmed and Wild Fish, 3rd Edn. New York, NY: Springer.
Austin, B., and Zhang, X. H. (2006). Vibrio harveyi: a significant pathogen of marine vertebrates and invertebrates. Lett. Appl. Microbiol. 43, 119–124. doi: 10.1111/j.1472-765x.2006.01989.x
Baba, T., Imamura, J., Ikeda, K., Izawa, K., and Ikeda, K. (1988). Immune protection in carp, Cyprinus carpio L., after immunization with Aeromonas hydrophila crude lipopolysaccharide. J. Fish. Dis. 11, 237–244. doi: 10.1111/j.1365-2761.1988.tb00544.x
Braegger, C., Chmielewska, A., Decsi, T., Kolacek, S., Mihatsch, W., Moreno, L., et al. (2011). Supplementation of infant formula with probiotics and/or prebiotics: a systematic review and comment by ESPGHAN committee on nutrition. J. Pediatr. Gastroenterol. Nutr. 52, 238–250. doi: 10.1097/mpg.0b013e3181fb9e80
Brandao, R. L., Castro, I. M., Bambirra, E. A., Amaral, S. C., Fietto, L. G., and Tropia, M. J. M. (1998). Intracellular signal triggered by cholera toxin in Saccharomyces boulardii and Saccharomyces cerevisiae. Appl. Environ. Microbiol. 64, 564–568. doi: 10.1128/aem.64.2.564-568.1998
Brettin, T., Davis, J. J., Disz, T., Edwards, R. A., Gerdes, S., Olsen, G. J., et al. (2015). RASTtk: a modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci. Rep. 5:8365. doi: 10.1038/srep08365
Breyner, N. M., Michon, C., de Sousa, C. S., Vilas Boas, P. B., Chain, F., Azevedo, V. A., et al. (2017). Microbial anti-inflammatory molecule (MAM) from Faecalibacterium prausnitzii show a protective effect on DNBS and DSS-induced colitis model in mice through inhibition of NF-κB pathway. Front. Microbiol. 8:114. doi: 10.3389/fmicb.2017.00114
Chen, J., Wang, R., Li, X. F., and Wang, R. L. (2012). Bifidobacterium adolescentis supplementation ameliorates visceral fat accumulation and insulin sensitivity in an experimental model of the metabolic syndrome. Br. J. Nutr. 107, 1429–1434. doi: 10.1017/s0007114511004491
Chin, C. S., Alexander, D. H., Marks, P., Klammer, A. A., Drake, J., and Heiner, C. (2013). Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat. Methods 10, 563–569. doi: 10.1038/nmeth.2474
Di Cerbo, A., Palmieri, B., Aponte, M., Morales-Medina, J. C., and Iannitti, T. J. (2016). Mechanisms and therapeutic effectiveness of lactobacilli. Clin. Pathol. 69, 187–203. doi: 10.1136/jclinpath-2015-202976
Dias, C., Mota, V., Martinez-Murcia, A., and José Saavedra, M. (2012). Antimicrobial resistance patterns of Aeromonas spp. Isolated from ornamental fish. J. Aquacult. Res. Dev. 3:131.
do Carmo, M. S., Santos, C. I. D., Araújo, M. C., Girón, J. A., Fernandes, E. S., and Monteiro-Neto, V. (2018). Probiotics, mechanisms of action, and clinical perspectives for diarrhea management in children. Food Funct. 9, 5074–5095. doi: 10.1039/c8fo00376a
Gareau, M. G., Sherman, P. M., and Walker, W. A. (2010). Probiotics and the gut microbiota in intestinal health and disease. Nat. Rev. Gastroenterol. Hepatol. 7, 503–514. doi: 10.1038/nrgastro.2010.117
Giovannini, M., Verduci, E., Gregori, D., Ballali, S., Soldi, S., Ghisleni, D., et al. (2014). Prebiotic effect of an infant formula supplemented with galacto-oligosaccharides: randomized multicenter trial. PLAGOS trial study group. J. Am. Coll. Nutr. 33, 385–393. doi: 10.1080/07315724.2013.878232
Hemant, J. P., Benet-Perelberg, A., Naor, A., Smirnov, M., Ofek, T., Nasser, A., et al. (2016). Evidence of increased antibiotic resistance in phylogenetically- diverse Aeromonas isolates from semi-intensive fish ponds treated with antibiotics. Front. Microbiol. 7:1875. doi: 10.3389/fmicb.2016.01875
Hill, C., Guarner, F., Reid, G., Gibson, G. R., Merenstein, D. J., Pot, B., et al. (2014). Expert consensus document. The International Scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat. Rev. Gastroenterol. Hepatol. 11, 506–514. doi: 10.1038/nrgastro.2014.66
Hunt, M., Silva, N. D., Otto, T. D., Parkhill, J., Keane, J. A., and Harris, S. R. (2015). Circulator: automated circularization of genome assemblies using long sequencing reads. Genome Biol. 16:294.
Isolauri, E., Sütas, Y., Kankaanpää, P., Arvilommi, H., and Salminen, S. (2001). Probiotics: effects on immunity. Am. J. Clin. Nutr. 73(Suppl. 2), 444S–450S.
Kelly, C. P. (2013). Fecal microbiota transplantation–an old therapy comes of age. N. Engl. J. Med. 368, 474–475. doi: 10.1056/nejme1214816
Khushiramani, R. M., Maiti, B., Shekar, M., Girisha, S. K., Akash, N., Deepanjali, A., et al. (2012). Recombinant Aeromonas hydrophila outer membrane protein 48 (Omp48) induces a protective immune response against Aeromonas hydrophila and Edwardsiella tarda. Res. Microbiol. 163, 286–291. doi: 10.1016/j.resmic.2012.03.001
Kim, S. K., Guevarra, R. B., Kim, Y. T., Kwon, J., Kim, H., Cho, J. H., et al. (2019). Role of probiotics in human gut microbiome-associated diseases. Microbiol. Biotechnol. 29, 1335–1340. doi: 10.4014/jmb.1906.06064
Kim, S. W., Park, K. Y., Kim, B., Kim, E., and Hyun, C. K. (2013). Lactobacillus rhamnosus GG improves insulin sensitivity and reduces adiposity in high-fat diet-fed mice through enhancement of adiponectin production. Biochem. Biophys. Res. Commun. 431, 258–263. doi: 10.1016/j.bbrc.2012.12.121
Klaenhammer, T. R., Kleerebezem, M., Kopp, M. V., and Rescigno, M. (2012). The impact of probiotics and prebiotics on the immune system. Nat. Rev. Immunol. 12, 728–734. doi: 10.1038/nri3312
Kobyliak, N., Conte, C., Cammarota, G., Haley, A. P., Styriak, I., Gaspar, L., et al. (2016). Probiotics in prevention and treatment of obesity: a critical view. Nutr. Metab. 13:14.
Konstantinos, P., Ángel, A., Bron, P. A., de Angelis, M., Gobbetti, M., Kleerebezem, M., et al. (2016). Stress physiology of lactic acid bacteria. Microbiol. Mol. Biol. Rev. 80, 837–890.
Kristensen, N. B., Bryrup, T., Allin, K. H., Nielsen, T., Hansen, T. H., and Pedersen, O. (2016). Alterations in a fecal microbiota composition by probiotic supplementation in healthy adults: a systematic review of randomized controlled trials. Genome Med. 8:52.
Laleh, Y. G., Mohammad, E. J. Z., and Milad, A. (2015). The study on effect of temperature stress on occurrence of clinical signs caused by Aeromonas hydrophila in Capoeta damascina in in vitro condition. Adv. Anim. Vet. Sci. 3, 406–412. doi: 10.14737/journal.aavs/2015/3.7.406.412
Liu, B., Xu, L., Ge, X., Xie, J., Xu, P., Zhou, Q., et al. (2013). Effects of Mannane oligosaccharide on the physiological responses, HSP70 gene expression and disease resistance of Allogynogenetic crucian carp (Carassius auratus gibelio) under Aeromonas hydrophila infection. Fish Shellfish Immunol. 34, 1395–1403. doi: 10.1016/j.fsi.2013.02.028
Liu, H., Gong, J., Chabot, D., and Wang, Q. (2015). Protection of heat-sensitive probiotic bacteria during spray-drying by sodium caseinate stabilized fat particles. Food Hydrocoll. 51, 459–467. doi: 10.1016/j.foodhyd.2015.05.015
Markowiak, P., and śliżewska, K. (2017). Effects of probiotics, prebiotics, and synbiotics on human health. Nutrients 9:1021. doi: 10.3390/nu9091021
Maiti, B., Dubey, S., Munang’andu, H. M., Karunasagar, I., Karunasagar, I., and Evensen, Ø (2020). Application of outer membrane protein-based vaccines against major bacterial fish pathogens in india. Front. Immunol. 11:1362. doi: 10.3389/fimmu.2020.01362
Maiti, B., Shetty, M., Shekar, M., and Karunasagar, I. (2012). Evaluation of two outer membrane proteins, Aha1 and OmpW of Aeromonas hydrophila as a vaccine candidate for common carp. Vet. Immunol. Immunopathol. 149, 298–301. doi: 10.1016/j.vetimm.2012.07.013
McHugh, M. L. (2011). Multiple comparison analysis testing in ANOVA. Biochemia Medica. 21, 203–209. doi: 10.11613/bm.2011.029
Nazemian, V., Shadnoush, M., Manaheji, H., and Zaringhalam, J. (2016). Probiotics and inflammatory pain: a literature review study. Middle East J. Rehabil. Health Stud. 3:e360871.
Oelschlaeger, T. A. (2010). Mechanisms of probiotic actions. Int. J. Med. Microbiol. 300, 57–62. doi: 10.1016/j.ijmm.2009.08.005
Pang, M., Jiang, J., Xie, X., Wu, Y., and Dong, Y. (2015). Novel insights into the pathogenicity of epidemic Aeromonas hydrophila ST251 clones from comparative genomics. Sci. Rep. 5:9833.
Patel, R., and DuPont, H. L. (2015). New approaches for bacteriotherapy: prebiotics, new-generation probiotics, and synbiotics. Clin. Infect. Dis. 15(60 Suppl. 2), S108–S121.
Patel, R. M., Myers, L. S., Kurundkar, A. R., Maheshwari, A., Nusrat, A., and Lin, P. W. (2012). Probiotic bacteria induce maturation of intestinal claudin 3 expression and barrier function. Am. J. Pathol. 180, 626–635.
Pettibone, G. W., Mear, J. P., and Sampsell, B. M. (1996). The incidence of antibiotic and metal resistance and plasmid carriage in Aeromonas isolated from brown null head (Ictalurus nebulosus). Lett. Appl. Microbiol. 23, 234–240. doi: 10.1111/j.1472-765x.1996.tb00073.x
Plaza-Diaz, J., Gomez-Llorente, C., Abadia-Molina, F., Saez-Lara, M. J., Campaña-Martin, L., Muñoz-Quezada, S., et al. (2014). Effects of Lactobacillus paracasei CNCM I-4034, Bifidobacterium breve CNCM I4035 and Lactobacillus rhamnosus CNCM I-4036 on hepatic steatosis in Zucker rats. PLoS One 9:e98401. doi: 10.1371/journal.pone.0098401
Rajagopal, K., Gowda, C. T., and Singh, P. K. (2015). Diffusion, frowning and smiling of low molecular weight protein bands: a simple, rapid and efficient solution. Int. J. Pept. Res. Ther. 21, 7–11. doi: 10.1007/s10989-014-9429-5
Reichold, A., Brenner, S. A., Spruss, A., Förster-Fromme, K., Bergheim, I., and Bischoff, S. C. (2014). Bifidobacterium adolescentis protects from the development of non-alcoholic steato hepatitis in a mouse model. J. Nutr. Biochem. 25, 118–125. doi: 10.1016/j.jnutbio.2013.09.011
Sánchez, B., Delgado, S., Blanco-Míguez, A., Lourenço, A., Gueimonde, M., and Margolles, A. (2017). Probiotics, gut microbiota, and their influence on host health and disease. Mol. Nutr. Food Res. 61:1600240. doi: 10.1002/mnfr.201600240
Sang, L. X., Chang, B., Zhang, W. L., Wu, X. M., Li, X. H., and Jiang, M. (2010). Remission induction and maintenance effect of probiotics on ulcerative colitis: a meta-analysis. World J. Gastroenterol. 16, 1908–1915. doi: 10.3748/wjg.v16.i15.1908
Savcheniuk, O., Kobyliak, N., Kondro, M., Virchenko, O., Falalyeyeva, T., and Beregova, T. (2014). Short-term periodic consumption of multi-probiotic from childhood improves insulin sensitivity, prevents development of non-alcoholic fatty liver disease and adiposity in adult rats with glutamate-induced obesity. BMC Complement. Altern. Med. 16:247. doi: 10.1186/1472-6882-14-247
Schley, P. D., and Field, C. J. (2002). The immune-enhancing effects of dietary fibres and prebiotics. Br. J. Nutr. 87(Suppl. 2), S221–S230.
Scott, A. J., Merrifield, C. A., Younes, J. A., and Pekelharing, E. P. (2018). Pre-, pro- and synbiotics in cancer prevention and treatment—a review of basic and clinical research. Ecancermedicalscience 12:869. doi: 10.3332/ecancer.2018.869
Scott, K. P., Antoine, J. M., Midtvedt, T., and van Hemert, S. (2015). Manipulating the gut microbiota to maintain health and treat disease. Microb. Ecol. Health Dis. 26:25877.
Son, R., Rusul, G., Sahilah, A. M., Zainuri, A., Raha, A. R., and Salmah, I. (1997). Antibiotic resistance and plasmid profile of Aeromonas hydrophila isolates from cultured fish, Tilapia (Tilapia mossambica). Lett. Appl. Microbiol. 24, 479–482. doi: 10.1046/j.1472-765x.1997.00156.x
Suez, J., Zmora, N., Segal, E., and Elinav, E. (2019). The pros, cons, and many unknowns of probiotics. Nat. Med. 25, 716–729. doi: 10.1038/s41591-019-0439-x
Szajewska, H., Skórka, A., and Pieścik-Lech, M. (2015). Fermented infant formulas without live bacteria: a systematic review. Eur. J. Pediatr. 174, 1413–1420. doi: 10.1007/s00431-015-2629-y
Tatusova, T., DiCuccio, M., Badretdin, A., Chetvernin, V., Ciufo, S., and Li, W. (2013). Prokaryotic Genome Annotation Pipeline. Bethesda, MD: National Center for Biotechnology Information.
Taverniti, V., and Guglielmetti, S. (2011). ghost probiotics: proposal of para-probiotic concept. Genes Nutr. 6, 261–274. doi: 10.1007/s12263-011-0218-x
Thomas, D. W., and Greer, F. R. (2010). Probiotics and prebiotics in pediatrics. American academy of pediatrics committee on nutrition, American academy of pediatrics section on gastroenterology, hepatology, and nutrition. Supplementation of infant formula with probiotics and/or prebiotics: a systematic review and comment by the ESPGHAN committee on nutrition. Pediatrics 126, 1217–1231. doi: 10.1542/peds.2010-2548
Timmerman, H. M., Koning, C. J., Mulder, L., Rombouts, F. M., and Beynen, A. C. (2004). Mono-strain, multi-strain and multi species probiotics–A comparison of functionality and efficacy. Int. J. Food Microbiol. 96, 219–233.
Tsukahara, T., Iwasaki, Y., Nakayama, K., and Ushida, K. J. (2003). Stimulation of butyrate production in the large intestine of weaning piglets by dietary fructo-oligosaccharides and its influence on the histological variable of the large intestinal mucosa. J. Nutr. Sci. Vitaminol. 49, 414–421. doi: 10.3177/jnsv.49.414
Vandenbergh, P. A. (1993). Lactic acid bacteria, their metabolic products and interference with microbial growth. FEMS Microbiol. Rev. 12, 221–238. doi: 10.1111/j.1574-6976.1993.tb00020.x
van Heel, A. J., Jong, A., Song, C., Viel, J. H., Kok, J., and Kuipers, O. P. (2018). BAGEL4: a user-friendly web server to thoroughly mine RiPPs and bacteriocins. Nucleic Acids Res. 46, W278–W281. doi: 10.1093/nar/gky383
Vivekanandan, G., Savithamani, K., Hatha, A. A. M., and Lakshmanaperumalsamy, P. (2002). Antibiotic resistance of Aeromonas hydrophila isolated from marketed fish and prawn of South India. Int. J. Food Microbiol. 76, 165–168. doi: 10.1016/s0168-1605(02)00009-0
Vyas, U., and Ranganathan, N. (2012). Probiotics, prebiotics, and synbiotics: gut and beyond. Gastroenterol. Res. Pract. 2012:872716.
Wang, J., Tang, H., Zhang, C., Zhao, Y., Derrien, M., Rocher, E., et al. (2015). Modulation of gut microbiota during probiotic-mediated attenuation of metabolic syndrome in high fat diet-fed mice. ISME J. 9, 1–15. doi: 10.1038/ismej.2014.99
Weidang, L., Medha, D. J., Singhania, S., Ramsey, K. H., and Murthy, A. K. (2014). Peptide vaccine: progress and challenges. Vaccines 2, 515–536. doi: 10.3390/vaccines2030515
WHO/FAO (2006). Probiotics in Food Health and Nutritional Properties and Guidelines for Evaluation. Rome: Food and Agriculture Organization of the United Nations.
Yin, Y. N., Yu, Q. F., Fu, N., Liu, X. W., and Lu, F. G. (2010). Effects of four Bifidobacteria on obesity in high-fat diet induced rats. World J. Gastroenterol. 16, 3394–3401. doi: 10.3748/wjg.v16.i27.3394
Yucel, N., Aslim, B., and Beyatli, Y. (2005). Prevalence and resistance to antibiotics, for Aeromonas species isolated from retail fish in Turkey. J. Food Qual. 28, 313–324. doi: 10.1111/j.1745-4557.2005.00037.x
Keywords: Aeromonas hydrophila, antimicrobials, antibiotics, chemotherapy, Lactobacillus acidophilus, Ocins, outer membrane protein
Citation: Akter N, Hashim R, Pham HQ, Choi S-D, Lee D-W, Shin J-H and Rajagopal K (2020) Lactobacillus acidophilus Antimicrobial Peptide Is Antagonistic to Aeromonas hydrophila. Front. Microbiol. 11:570851. doi: 10.3389/fmicb.2020.570851
Received: 09 June 2020; Accepted: 02 September 2020;
Published: 09 October 2020.
Edited by:
Ramona Iseppi, University of Modena and Reggio Emilia, ItalyReviewed by:
Terence L. Marsh, Michigan State University, United StatesHasan Cihad Tekedar, Mississippi State University, United States
Copyright © 2020 Akter, Hashim, Pham, Choi, Lee, Shin and Rajagopal. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kammara Rajagopal, cmFqYWdvcGFsa0BjZnRyaS5yZXMuaW4=; a3Jnb3BhbDIyQHJlZGlmZm1haWwuY29t; Jae-Ho Shin, amhzaGluQGtudS5hYy5rcg==
 Nahid Akter1,2
Nahid Akter1,2 Kammara Rajagopal
Kammara Rajagopal