
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 14 January 2020
Sec. Fungi and Their Interactions
Volume 10 - 2019 | https://doi.org/10.3389/fmicb.2019.03065
 Meihua Xie1,2,3,4
Meihua Xie1,2,3,4 Na Bai1,2,3
Na Bai1,2,3 Jiangliu Yang1,2,3
Jiangliu Yang1,2,3 Kexin Jiang1,2,3
Kexin Jiang1,2,3 Duanxu Zhou1,2,3
Duanxu Zhou1,2,3 Yining Zhao1,2,3
Yining Zhao1,2,3 Dongni Li1,2,3
Dongni Li1,2,3 Xuemei Niu1,2,3
Xuemei Niu1,2,3 Ke-Qin Zhang1,2,3
Ke-Qin Zhang1,2,3 Jinkui Yang1,2,3*
Jinkui Yang1,2,3*Inducer of meiosis 2 (Ime2), a protein kinase that has been identified in diverse fungal species, functions in the regulation of various cellular processes, such as ascospore formation, pseudohyphal growth, and sexual reproduction. In this study, AoIme2, an ortholog of Saccharomyces cerevisiae Ime2, was characterized in the nematode-trapping fungus Arthrobotrys oligospora. Disruption of the gene Aoime2 caused defective growth, with slower mycelial growth in ΔAoime2 mutants than the wild type (WT) strain, and in the mutants, the number of hyphal septa in mycelia was higher and the number of cell nuclei in mycelia and conidia was considerably lower than in the WT strain. The conidial yields of the ΔAoime2 mutants were decreased by ∼33% relative to the WT strain, and the transcription of several sporulation-related genes, including abaA, fluG, rodA, aspB, velB, and vosA, was markedly downregulated during the conidiation stage. The ΔAoime2 mutants were highly sensitive to the osmotic stressors NaCl and sorbitol, and the cell wall of partial hyphae in the mutants was deformed. Further examination revealed that the cell wall of the traps produced by ΔAoime2 mutants became loose, and that the electron-dense bodies in trap cells were also few than in the WT strain. Moreover, Aoime2 disruption caused a reduction in trap formation and serine-protease production, and most hyphal traps produced by ΔAoime2 mutants did not form an intact hyphal loop; consequently, substantially fewer nematodes were captured by the mutants than by the WT strain. In summary, an Ime2-MAPK is identified here for the first time from a nematode-trapping fungus, and the kinase is shown to be involved in the regulation of mycelial growth and development, conidiation, osmolarity, and pathogenicity in A. oligospora.
Mitogen-activated protein kinase (MAPK) cascades function as key intracellular signal transducers that use protein phosphorylation/dephosphorylation cycles to transmit information, and orthologous MAPK signaling modules in yeast and filamentous fungi have been found to be involved in regulating mating, filament growth, hyperosmotic-stress response, cell-wall integrity (CWI), and spore-wall assembly (Xu, 2000; Zhao et al., 2007; Rispail et al., 2009). MAPK is generally activated by phosphorylation at the well-conserved threonine-x-tyrosine (TXY) motif by MAPK kinase (MAPKK), which is in turn activated by MAPKK kinase (MAPKKK) (Yang et al., 2003; Jiang et al., 2018). These MAPKKK–MAPKK–MAPK cascades are evolutionarily conserved in eukaryotes (Herskowitz, 1995; Schaeffer and Weber, 1999). In Saccharomyces cerevisiae, five MAPK pathways (Kss1, Fus3, Hog1, Slt2, and Smk1 pathways) have been identified and shown to regulate mating, invasive growth, CWI, osmolarity, and ascospore formation (Jiang et al., 2018), and in filamentous fungi, three MAPK pathways corresponding to pathways in S. cerevisiae have been reported: the Fus3/Kss1-homolog, Slt2-homolog, and Hog1-homolog pathways (Jiang et al., 2018). Moreover, a fourth MAPK pathway, the inducer of meiosis 2 (Ime2)-homolog pathway, was found in several fungi, such as Aspergillus nidulans (Bayram et al., 2009), Cryptococcus neoformans (Liu and Shen, 2011), Neurospora crassa (Hutchison and Glass, 2010), and Ustilago maydis (Garrido et al., 2004). Unlike in the three classic MAPK pathways, the MAPKKK and MAPKK in the Ime2-MAPK cascade remain unidentified, and thus these kinases belong to a distinct Ime2-MAPK class (Garrido and Pérez-Martín, 2003; Schindler et al., 2003). Ime2 homologs are conserved in not only in fungi, but also all eukaryotic taxa examined (Krylov et al., 2003), and the common feature of these kinases is that their N-terminal region harbors a TXY motif, which is typically found in the activation loop of MAPKs (Payne et al., 1991).
The ime2 was first identified in S. cerevisiae as a gene that is expressed exclusively during meiosis (Smith and Mitchell, 1989; Yoshida et al., 1990), and, subsequently, was shown to be also involved in normal spore formation (Sari et al., 2008) and pseudohyphal growth (Strudwick et al., 2010). Recently, Ime2 homologs from various fungal species have been increasingly shown to function in not only the control of meiosis, but also the regulation of diverse cellular processes, including ascospore formation, pseudohyphal growth, and sexual reproduction in response to light and nutrient deprivation (Irniger, 2011). For example, Crk1, a homolog of yeast Ime2, was first identified in U. maydis to participate in the regulation of morphogenesis and plant infection (Garrido et al., 2004), and later reported to function in the negative regulation of mating in C. neoformans (Liu and Shen, 2011). The Ime2 homolog ImeB was found to be not required for meiosis in A. nidulans, but vegetative growth of the ΔimeB mutants was diminished, and fully fertile ascospores were produced in morphologically normal cleistothecia (Bayram et al., 2009). Moreover, an Ime2-like MAPK was shown to be involved in cellulase expression in Trichoderma reesei (Chen et al., 2015). Therefore, the conserved Ime2-family proteins display an unexpected diversification in their cellular functions in fungi.
Nematode-trapping (NT) fungi constitute a group of fungi that can capture nematodes; NT fungi develop specific trapping devices (traps) for nematode predation (such as adhesive networks, adhesive knobs, and constricting rings), are widely distributed in terrestrial and aquatic ecosystems, and survive mainly as saprophytes and enter a predacious phase in response to signals released by nematodes (Ahrén et al., 1998; Nordbring-Hertz et al., 2001; Su et al., 2017). The traps produced by NT fungi harbor numerous electron-dense (ED) bodies, but normal vegetative hyphae lack ED bodies (Veenhuis et al., 1985, 1989). Traps are critical tools used by NT fungi and their integrity affects the nematode-predation efficiency of the fungi. Moreover, NT fungi produce extracellular serine proteases that can degrade the nematode cuticle and thereby facilitate fungal penetration and colonization (Tunlid et al., 1994; Yang et al., 2013). Therefore, NT fungi are potential agents for controlling parasitic nematodes of plants and animals.
Arthrobotrys oligospora, a common NT-fungus species that has been isolated from diverse soil and aquatic environments, can produce adhesive traps (three-dimensional networks) to capture nematodes (Nordbring-Hertz, 2004; Yang et al., 2011). We have sequenced the genome of A. oligospora (Yang et al., 2011) and identified several signaling proteins involved in A. oligospora growth, conidiation, and pathogenicity, such as the MAPK protein AoSlt2 (Zhen et al., 2018) and the Rab protein AoRab-7A (Yang et al., 2018). However, little is known regarding the role of the Ime2-homolog MAPK in NT fungi. Here, we identified AoIme2, an ortholog of S. cerevisiae Ime2 in A. oligospora, and characterized by constructing deletion mutants of Aoime2. Our results suggest that AoIme2 plays crucial roles in regulating hyphal growth, cell nucleus development, conidiation, trap formation, and pathogenicity in A. oligospora.
Arthrobotrys oligospora Fres. (ATCC 24927) and ΔAoime2 mutants were cultured on potato dextrose agar (PDA) plates at 28°C. S. cerevisiae strain FY834 used for constructing recombinant plasmid vectors was cultured in yeast extract peptone dextrose (YPD) medium. Plasmids pRS426 and pCSN44 were maintained in Escherichia coli strain DH5α (TaKaRa, Shiga, Japan). Protoplasts of A. oligospora were regenerated on PDASS medium (PDA supplemented with 10 g/L molasses and 0.4 M saccharose) containing hygromycin (200 μg/mL). CMY (20 g/L maizena, 5 g/L yeast extract, 20 g/L agar), TG (10 g/L tryptone, 10 g/L glucose, 20 g/L agar), and TYGA (10 g/L tryptone, 5 g/L yeast extract, 10 g/L glucose, 5 g/L molasses, 20 g/L agar) media were prepared as previously described (Yang et al., 2018) and used for analyzing mycelial growth and related phenotypic traits. Caenorhabditis elegans (strain N2) worms were cultured on oatmeal medium at 26°C and then used for bioassays.
AoIme2 sequence (AOL_s00188g140) was retrieved by performing BLASTP searches1 with queries for orthologous Ime2 proteins of the model fungi S. cerevisiae, A. nidulans, and N. crassa in the NCBI database. The theoretical isoelectric point and molecular weight of AoIme2 were calculated using a pI/MW tool2. Ime2 orthologs in fungi were searched for by using BLAST algorithm, and the similarity of the orthologs from different fungi was examined by aligning them by using DNAman software package (version 5.2.2; Lynnon Biosoft, San Ramon, CA, United States) and then performing phylogenetic analysis by using MEGA 7.0 software (Kumar et al., 2016).
Aoime2 was disrupted through homologous recombination as previously described (Tunlid et al., 1999; Colot et al., 2006). All primers used in this study are described in Supplementary Table S1. The upstream and downstream fragments of the Aoime2 were, respectively, PCR-amplified from A. oligospora by using paired primers (Supplementary Table S1), and the hygromycin-resistance gene cassette (hph) was amplified using pSCN44 plasmid as the template. Next, the three DNA fragments and a linearized pRS426 vector were cotransformed into S. cerevisiae strain FY834 cells through electroporation, and after isolating the recombinant plasmid pRS426-AoIme2-hph from the yeast, the complete target sequence was amplified using primers AoIme2-5f and AoIme2-3r (Supplementary Table S1) and transformed into protoplasts of A. oligospora as described (Zhen et al., 2019). Colonies grown on PDASS medium containing 200 μg/mL hygromycin B were further verified using PCR and Southern blotting analyses. Southern blotting was performed using a North2South chemiluminescent hybridization and detection kit (Pierce, Rockford, IL, United States), according to the manufacturer’s instructions. Genomic DNA was extracted from WT and ΔAoime2 mutant strains by using a plant genomic DNA Kit (TaKaRa) and digested with KpnI for Southern blotting analysis.
Wild type (WT) and mutant strains were cultured on PDA plates at 28°C for 6 days, and then a 7-mm-diameter hyphal disk from each strain was, respectively, inoculated on PDA, TYGA, and TG plates at 28°C for 3–7 days; mycelial growth rate and colony morphology were examined and quantified at specific time intervals (Xie et al., 2019). WT and mutant strains were also incubated on CMY medium at 26°C for 15 days, and the conidial yield was determined as previously described (Liu et al., 2017). For observation of the septum in mycelia and conidia, freshly harvested conidia and hyphae of WT and mutant strains were stained with 20 μg/mL calcofluor white (CFW) (Sigma-Aldrich, St. Louis, MO, United States), and mycelial and conidial cell nuclei were visualized by staining with 20 μg/mL DAPI and 20 μg/mL CFW as described (Li et al., 2015); samples were analyzed using an inverted fluorescence microscope (Carl Zeiss, Heidenheim, Germany). Hyphal and conidial morphology was examined using scanning electron microscopy (SEM), and the ED bodies of trap cells in WT and mutant strains were examined using transmission electron microscopy (TEM). Samples were processed for SEM and TEM as previously described (Zhang et al., 2013, 2019).
Resistance to chemical stresses was evaluated by measuring the relative growth inhibition (RGI) values of fungal colonies on TG medium alone (control) or medium supplemented with reagents that induce chemical stresses, such as H2O2 and menadione for oxidative stress, NaCl and sorbitol for osmotic pressure, and SDS and Congo red for cell-wall-perturbing stress (Liu et al., 2017). The colonies were examined, and the diameter of each colony was measured. These experiments were performed in triplicate.
Freshly harvested conidia (2 × 104) of WT and mutant strains were evenly spread on WA (water agar, 20%) medium and incubated at 28°C for 3–4 days, and ∼300 nematodes were added to each WA plate to induce trap formation. Traps and captured nematodes were examined and quantified (per plate) under a microscope (Olympus, Tokyo, Japan) at 12-h intervals. The experiments were performed in triplicate.
Wild type and mutant strains were cultured in PL-4 liquid medium (Yang et al., 2011) at 28°C for 6 days, after which the fermentation broth was collected and proteolytic activity was qualitatively assayed on milk-plate medium (2% skimmed milk powder) as previously described (Zhao et al., 2004). Protease activity was also quantified as described (Wang et al., 2006). The effect of a protease inhibitor on enzyme activity and hyphal biomass was determined as described (Xie et al., 2019). Moreover, WT and mutant strains were incubated in PL-4 liquid medium at 28°C, followed by 3, 5, or 7 days incubation to collect hyphae for RNA extraction. Seven genes encoding serine proteases belonged to different subfamilies in A. oligospora were selected, and their transcriptional levels were analyzed in WT and mutant strains as described (Yang et al., 2018).
To analyze the expression of sporulation-related genes in WT and mutant strains, total RNAs were extracted from 3-, 5-, and 7-day cultures grown on TYGA by using an AxyPrep multisource RNA miniprep kit (Axygen, Jiangsu, China), and then reverse-transcribed into cDNAs by using a FastQuant RT kit with gDNase (TaKaRa). The cDNA samples of each strain were used as the template to assess the transcript level of each gene by using SYBRH Premix Ex Taq (TaKaRa) and performing RT-PCR with paired primers (Supplementary Table S2); β-tubulin served as an internal standard, and the 2–ΔΔCt method (Livak and Schmittgen, 2001) was used for quantification. The relative transcription level (RTL) of each gene was calculated as the ratio of the transcription level in the deletion mutant to the transcription level in the WT strain at a given time point.
Data are presented as means ±standard deviation (SD). SPSS program (version 16.0) (SPSS, Inc., Chicago, IL, United States) was used to analyze data from triplicate experiments, and p < 0.05 was used as the threshold for determining significant differences.
The gene Aoime2 encodes a 790-aa polypeptide featuring an isoelectric point of 10.29 and a molecular mass of 86.91 kDa. AoIme2 contains a conserved protein kinase domain (IPR000719), an active site (IPR008271), and an ATP-binding site (IPR017441). For phylogenetic analysis, we used 26 proteins orthologous to Ime2 and classic MAPK-family proteins (Kss1/Fus3, Slt2, and Hog1) from diverse fungi; the constructed neighbor-joining phylogenetic tree revealed that these MAPK-family proteins were divided into two clades (A and B) (Supplementary Figure S1). Ime2 orthologs from different fungi are clustered in Clade A, with the Ime2 orthologs from NT fungi being clustered in Subclade A-II; conversely, the classic MAPK-family members from different fungi are clustered in Clade B, which is further divided into three subclades: Fus3, Slt2, and Hog1 (Supplementary Figure S1). The similarity of orthologous Ime2 and MAPKs from different fungi was analyzed using DNAman software, and AoIme2 sequence was found to share 88.8 and 86.4% identity, respectively, with the sequences from the NT fungi Dactylellina haptotyla (Meerupati et al., 2013) and Drechslerella stenobrocha (Liu et al., 2014). The sequence identity between AoIme2 and the orthologs from other filamentous fungi reaches 49.9–51.7%, and the lowest identity is with the S. cerevisiae ortholog, 22.9%. Moreover, AoIme2 shares only 18.8–26.6% sequence similarity with fungal classic MAPKs. The Ime2 orthologs from various fungi contain two conserved motifs: the protein kinase active-site motif “-D[L/I/V]K-,” and the phosphorylation site “-TXY-” located in the activation loop. The phosphorylation-site sequences in orthologous Fus3 and Slt2 are both “-TEY-”, the sequence in Hog1 is “-TGY-”, and that in Ime2 from different fungi is “-TTY-” except in the case of ScIme2 (from S. cerevisiae) (Supplementary Figure S2).
Aoime2 was disrupted by inserting the hph cassette, and transformants were selected on PDASS medium containing hygromycin B (Tunlid et al., 1999). Genomic DNA was isolated from the WT strain and transformants and used as the template for PCR amplification with primers yz140-5f and yz140-3r (Supplementary Table S1); the sizes of the fragments amplified from the WT strain and transformants were consistent with the expected sizes, respectively, 3,044 and 2,207 bp (Supplementary Figures S3A,B). Subsequently, the genomic DNA of the WT strain and transformants was digested with the restriction enzyme KpnI and used in Southern blotting analysis (Supplementary Figure S3A), which revealed a single band, of the expected size, representing the DNA that hybridized with the Aoime2 probe in the case of both the WT strain and the transformants (Supplementary Figure S3C). Ultimately, two positive transformants (MT-1 and MT-14) were obtained based on the PCR and Southern blotting analyses.
The WT strain and the two ΔAoime2 mutants (MT-1 and MT-14) were cultured on PDA, TG, and TYGA media at 28°C. Mycelial growth rates of each fungal strain did not differ markedly on these media, but the mycelial growth rate of the ΔAoime2 mutants was significantly lower than that of the WT strain on all three media (Figures 1A,B). The WT strain produced extremely dense mycelia when grown on TYGA medium, and its aerial mycelia grew robustly. By contrast, the ΔAoime2 mutant colonies became loose and the aerial hyphae became sparse (Figures 1A,B). Moreover, the hyphae of the ΔAoime2 mutants contained 20% more hyphal septa than WT hyphae (Figure 1C), and thus the hyphal cells of the mutants were shorter than those of the WT strain (Figure 1D).
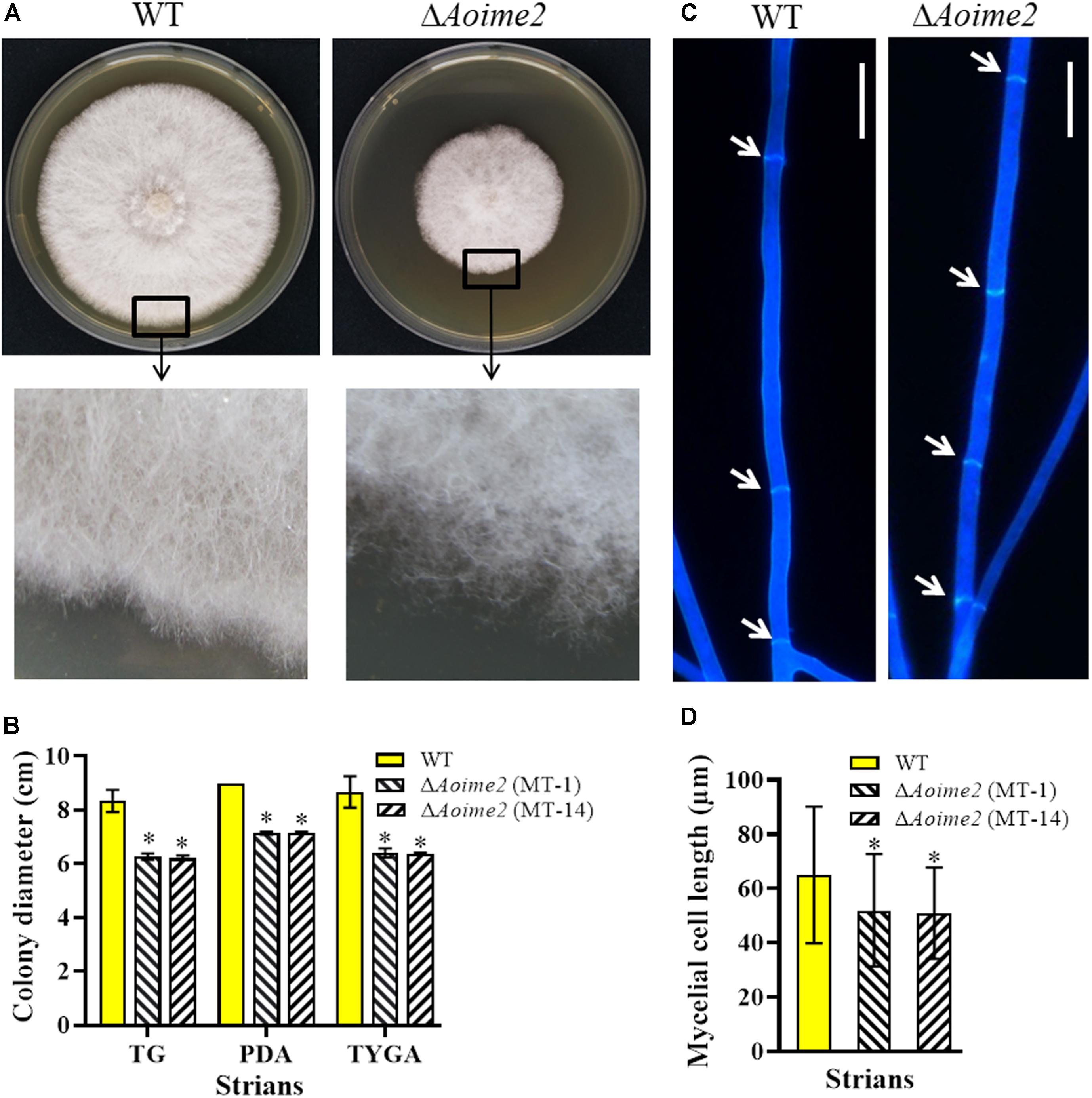
Figure 1. Comparison of mycelial growth, morphology, and septum formation between wild type (WT) and ΔAoime2 mutants. (A) Colony morphology (upper panel) and hyphae (lower panel) of WT and ΔAoime2 mutant strains incubated on TYGA medium for 5 days at 28°C. (B) Colony diameters of WT and ΔAoime2 mutants incubated on PDA, TYGA, and TG media for 7 days. (C) Hyphal septa of WT and mutants were stained with 20 μg/mL calcofluor white (CFW) after the fungal strains were incubated on CMY medium for 7 days. Arrows: hyphal septa. Bar = 10 μm. (D) Comparison of mycelial cell size of WT and mutants; 60 mycelial cells were randomly selected, and the distance between two hyphal septa was measured using ImageJ software. Error bars: SD from 60 replicates; asterisk: significant difference between mutant and WT (Tukey’s HSD, p < 0.05).
Nematode-trapping fungi are unique hyphomycetes containing multiple cell nuclei (Nordbring-Hertz, 2004). To determine whether AoIme2 regulates cell nucleus development, we used DAPI to stain cell nuclei in WT and ΔAoime2 mutant strains. Whereas the hyphal cells of the WT strain contained 6–22 nuclei, these cells in ΔAoime2 mutants contained 4–12 nuclei (Figures 2A,B). Moreover, the conidial cells of the WT strain contained 13–25 nuclei, but only 4–10 cell nuclei were present in the conidia of ΔAoime2 mutants (Figures 2C,D).
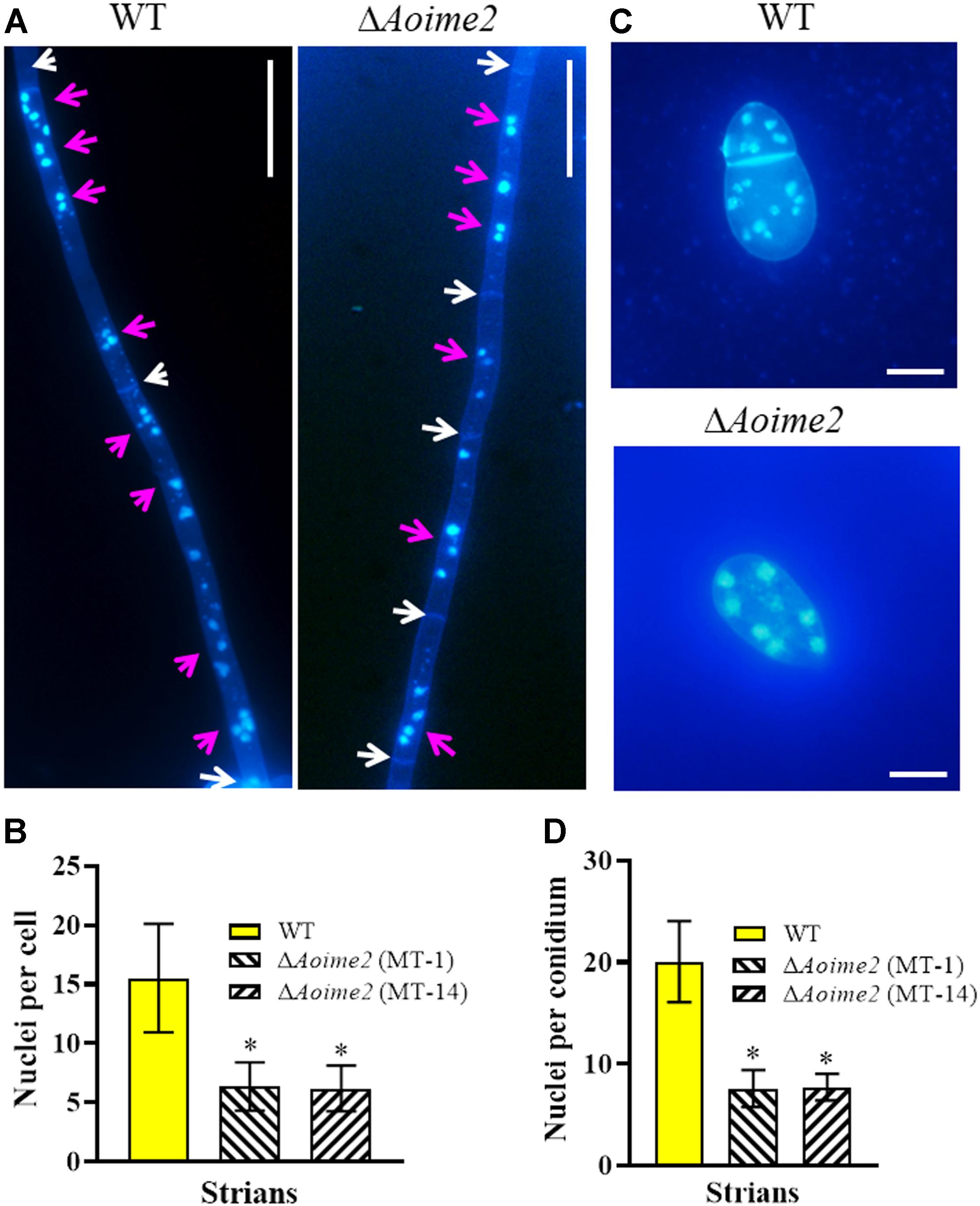
Figure 2. Mycelial and conidial cell nuclei in WT versus ΔAoime2 mutant strains. (A) Hyphae of WT and ΔAoime2 mutant strains were stained with CFW and DAPI after the fungal strains were grown for 7 days on CMY medium; samples were examined using an inverted fluorescence microscope. White arrows: septa; pink arrows: cell nuclei, the strong stained and bright points are identified as nuclei. Bar = 20 μm. (B) Comparison of mycelial cell nuclei between WT and mutant strains; 30 mycelial cells were randomly selected and the nucleus in each cell was counted separately. (C) Conidia of WT and ΔAoime2 mutant strains were stained with CFW and DAPI. Bar = 10 μm. (D) Comparison of conidial cell nuclei between WT and mutant strains; 30 conidia were randomly selected for counting cell nuclei. Error bars: SD from 30 replicates; asterisk: significant difference between mutant and WT (Tukey’s HSD, p < 0.05).
Aoime2 deletion markedly affected conidiation, with the ΔAoime2 mutants producing fewer conidiophores than the WT strain on CMY medium. Furthermore, the partial conidiophores of the Aoime2 mutant strains produced branches, but similar branches were not detected in WT conidiophores (Supplementary Figure S4). Analysis of 15-day-old cultures on CMY medium revealed that the conidial yields from the WT and ΔAoime2 mutant strains were, respectively, 6.6–7.7 × 105 and 4.3–5.2 × 105 conidia cm–2 (Figure 3A), the yields of the mutants being decreased by 33.0% relative to WT. Aoime2 deletion also noticeably altered conidial morphology. For example, the WT strain produced an obovoid spore, with one septum formed near the base of the spore; by contrast, most of the conidia (70.6%) of the ΔAoime2 mutants were morphologically abnormal, with 53.4% of the conidia lacking a septum, 11.4% of the conidial septa formed in the middle of the spore, and 5.8% of the conidia elongated in shape (Figure 3B).
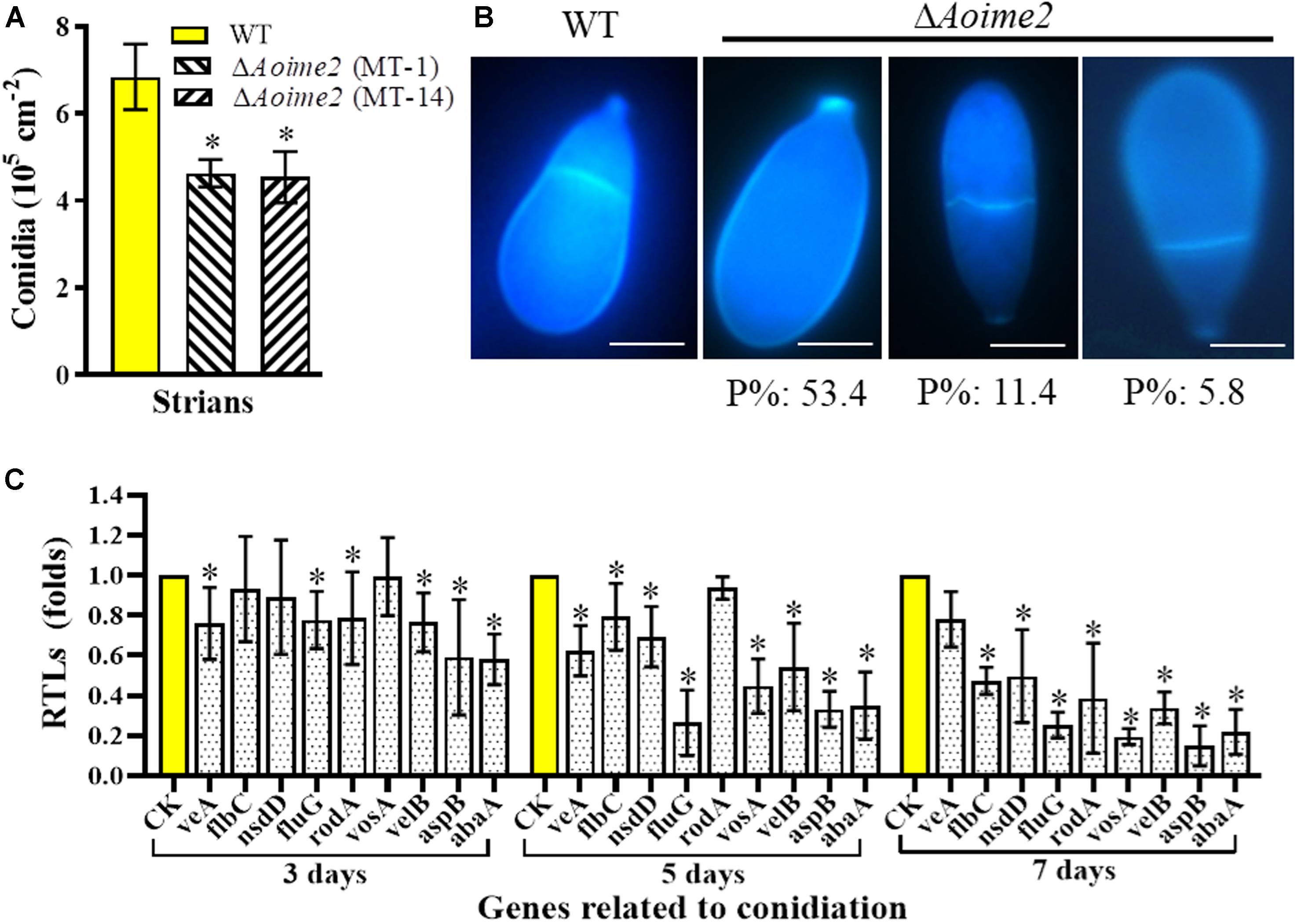
Figure 3. Comparison of conidial yields, morphology, and transcriptional levels (RTLs) of sporulation-related genes between WT and mutant strains. (A) Comparison of conidial yields between WT and ΔAoime2 mutants. Conidial yields were determined after incubation on CMY medium for 15 days. (B) Conidia were stained with CFW and examined under a microscope. Bar = 10 μm. P%: the percentage of each abnormal spore in the ΔAoime2 mutant. (C) Comparison of RTLs of sporulation-related genes between WT and ΔAoime2 mutants at different time points. CK, featuring an RTL of 1, was used as the standard for statistical analysis of the RTL of each gene in the deletion mutants to that in the WT strain under a given condition. Error bars: SD from three replicates, asterisk: significant difference between mutant and WT (Tukey’s HSD, p < 0.05).
From the A. oligospora genome, 9 sporulation-related genes were retrieved – abaA, flbC, fluG, nsdD, rodA, aspB, veA, velB, and vosA – based on their homologous genes from the model fungus A. nidulans (Krijgsheld et al., 2013), and the transcription of the genes was analyzed using RT-PCR. Most of these genes were expressed at lower levels in ΔAoime2 mutants than in the WT strain, and the transcription of the genes in the ΔAoime2 mutants was downregulated during conidiation relative to that in vegetative-growth stages. The expression levels of 8 genes, abaA, flbC, fluG, nsdD, rodA, aspB, velB, and vosA, were substantially downregulated during the conidiation stage in the ΔAoime2 mutants (Figure 3C).
To investigate whether AoIme2 regulates the biological responses to stresses in A. oligospora, we incubated the WT and ΔAoime2 mutant strains on media containing different concentrations of chemical agents and then analyzed their responses to these stressors. Mycelial growth of ΔAoime2 mutants relative to WT was strongly inhibited on TG medium supplemented with 0.5 M sorbitol or 0.2 M NaCl (Figure 4A); exposure to sorbitol and NaCl increased the RGI values of the ΔAoime2 mutants by 40.4–49.7% and 45.8–46.1%, respectively, as compared with the WT strain (Figure 4B). However, no significant difference was measured in the RGI values between WT and ΔAoime2 mutant strains on TG media supplemented with oxidants or cell-wall-perturbing agents (Supplementary Figure S5). Furthermore, the mycelial surface of the ΔAoime2 mutants was smoother than that in the WT strain and partial hyphae were deformed and shriveled (Figure 4C), and the cell wall of the trap cells of the ΔAoime2 mutants became loose, and considerably fewer trap-cell ED bodies were present in the mutants than in the WT strain (Figure 4D).
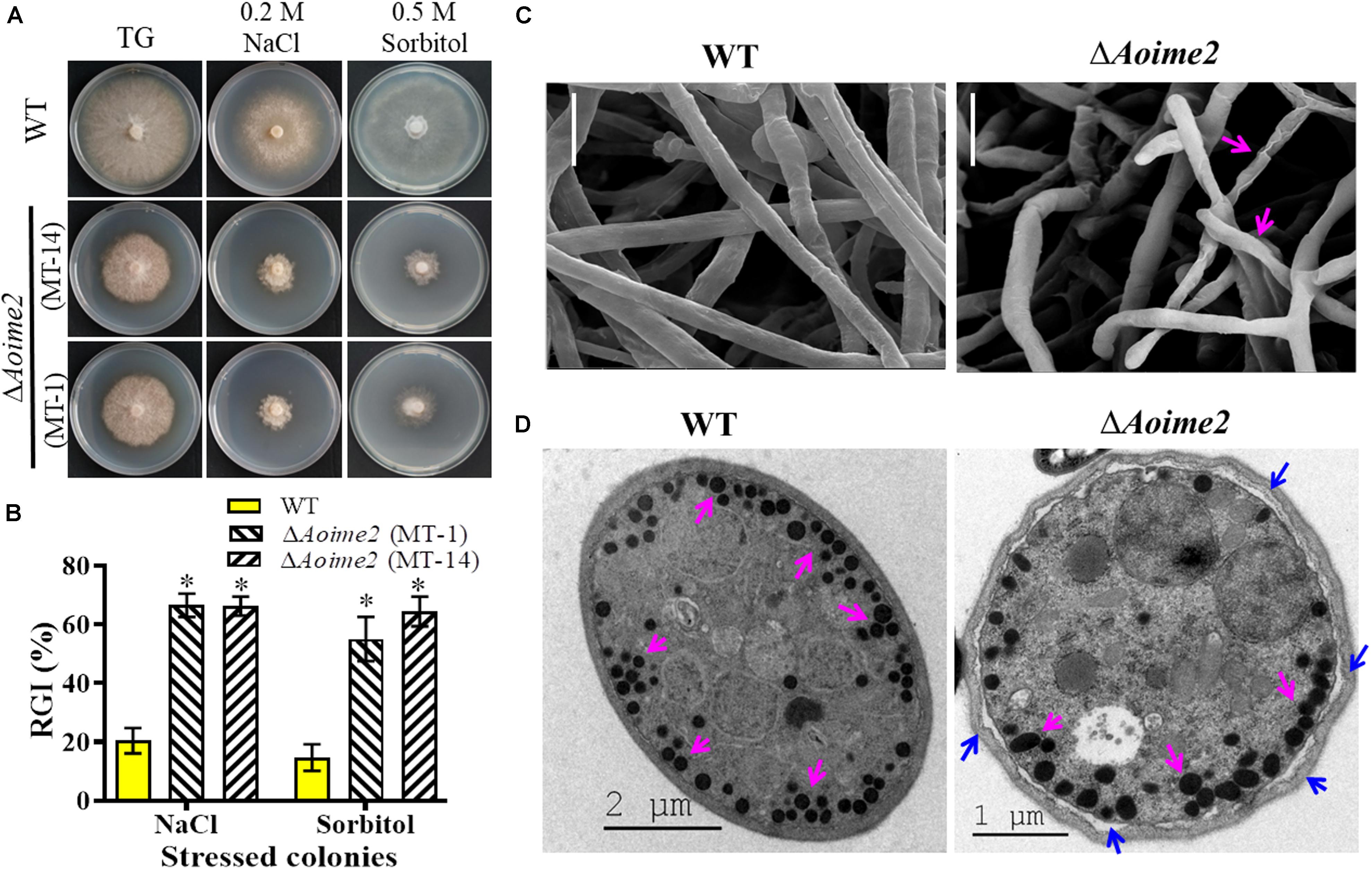
Figure 4. Tolerance to hyperosmotic stress and mycelial morphology: WT versus ΔAoime2 mutants. (A) Comparison of colonial morphology under high-osmolarity stress between WT and ΔAoime2 mutants. (B) RGI values of WT and ΔAoime2 mutants grown on TG plates supplemented with 0.2 M NaCl or 0.5 M sorbitol. Error bars: SD from three replicates, asterisk: significant difference between mutant and WT strain (Tukey’s HSD, p < 0.05). (C) Mycelia of WT and ΔAoime2 mutant strains were examined using scanning electron microscopy (SEM). Pink arrows: deformed mycelia in ΔAoime2 mutants. Bar = 10 μm. (D) Traps produced by WT and ΔAoime2 mutant strains were examined using transmission electron microscopy. Pink arrows: ED bodies in trap cells of WT and ΔAoime2 mutant strains; blue arrows: separation of cell wall and plasma membrane in ΔAoime2 mutants.
The WT and mutant strains were incubated on WA medium at 28°C for 3 days, and trap formation was examined at 12 h after adding nematodes. The WT strain began to produce fresh traps containing 1–2 hyphal loops at 12 h, and mature traps consisting of multiple hyphal loops appeared at 24 h after addition of the nematodes (Figure 5A). Most of the nematodes were captured by the WT strain at 36 h and were digested at 48 h. By comparison, diminished trap formation was observed in the ΔAoime2 mutants at all corresponding time points, and most of the hyphal loops were not closed (Figure 5A). At 24, 36, and 48 h after nematode addition, the WT strain produced 11.4, 14.1, and 17.9 traps cm–2, respectively, whereas the ΔAoime2 mutants produced only 8.1, 10.4, and 12.2 traps cm–2 (Figure 5B). Thus, 43.8, 82.5, and 90.1% of the nematodes were captured by the WT strain at 24, 36, and 48 h, respectively, but only 18.8–21.9%, 25.1–27.5%, and 44.3–46.1% were captured by the ΔAoime2 mutants at the corresponding time points (Figure 5C).
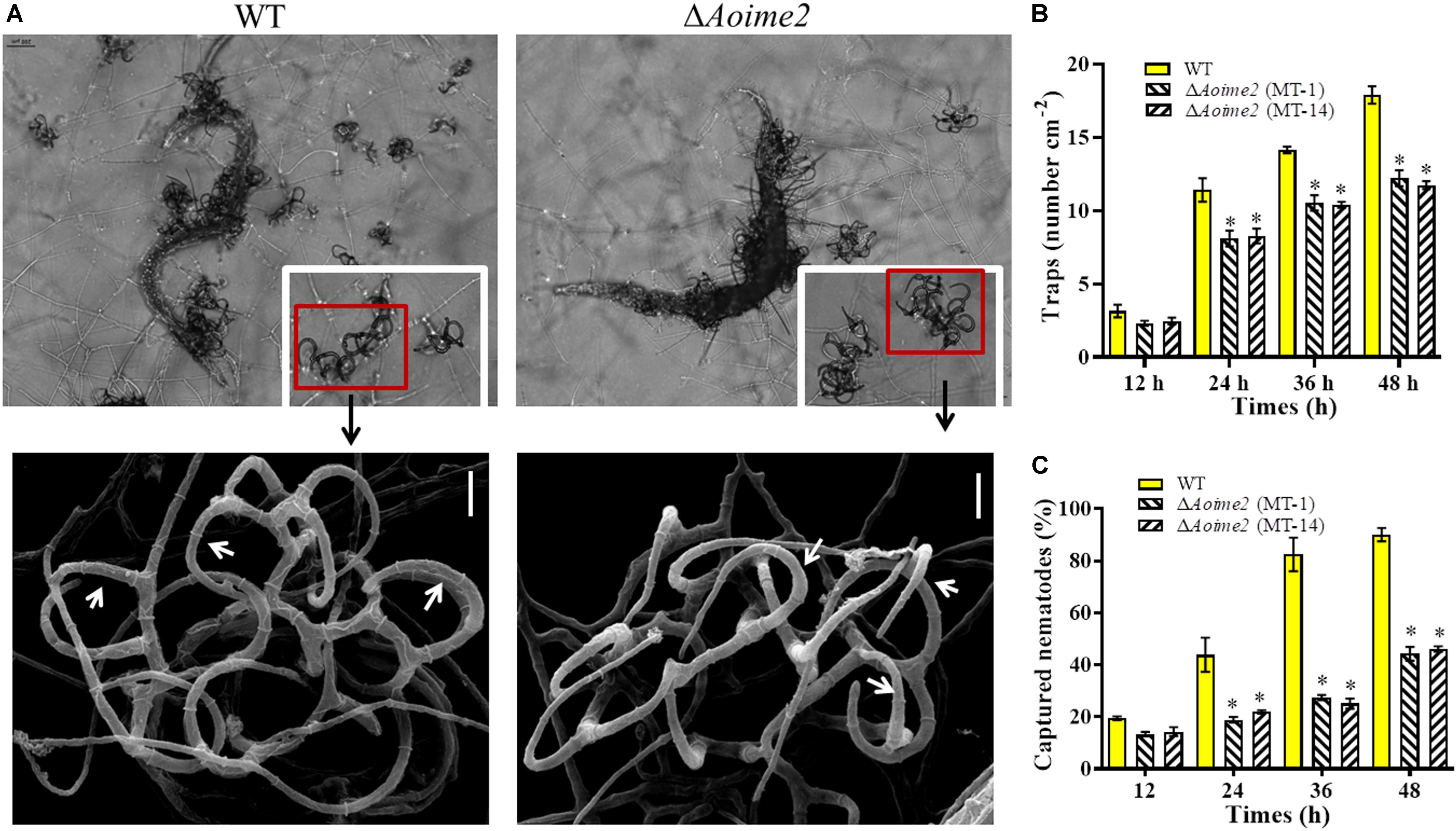
Figure 5. Trap formation, trap morphology, and nematicidal activity of WT and ΔAoime2 mutants. (A) Trap formation and morphology (upper images: light microscopy; lower images: SEM) of WT and mutants after 48-h induction of traps by nematodes. Red boxes: traps produced by WT and ΔAoime2 mutants; arrows: hyphal loops in traps. (B) Comparison of traps produced by WT and mutants at 12, 24, 36 and 48 h. (C) Percentages of nematodes captured by WT and mutant strains at different time points. Bar = 10 μm. Error bars: SD from three replicates, asterisk: significant difference between mutant and WT (Tukey’s HSD, p < 0.05).
Serine proteases are closely related to the virulence in the NT fungi and entomopathogenic fungi (Yang et al., 2013). A. oligospora can produce serine proteases to immobilize the nematodes and degrade the nematode cuticle (Tunlid et al., 1994). To determine whether AoIme2 functions in serine-protease production, we qualitatively analyzed the proteolytic activities of the WT and ΔAoime2 mutant strains by using medium containing skimmed milk (Zhao et al., 2004), which revealed that proteolytic activity of the ΔAoime2 mutants was lower than that of the WT strain (Figure 6A). Quantification of hyphal biomass from 7-day-old PL-4 cultures of the ΔAoime2 mutants further revealed 9.9–10.3% decrease relative to that from WT cultures (Figure 6B), and total activity of extracellular proteases in the mutant cultures was found to be decreased by 32.2–38.6% as compared with that in WT cultures (Figure 6C). Notably, the proteolytic activities of the WT and mutant strains were inhibited by 90% when we added the serine-protease inhibitor PMSF (phenylmethylsulfonyl fluoride, 5 mM) (Figure 6C). Lastly, the transcription of almost all tested serine-protease genes was lower in the mutants than in the WT strain, and the transcripts of PII (76g4) and PII-like protease genes (75g8 and 215g702) in particular were significantly downregulated in the ΔAoime2 mutants (Figure 6D).
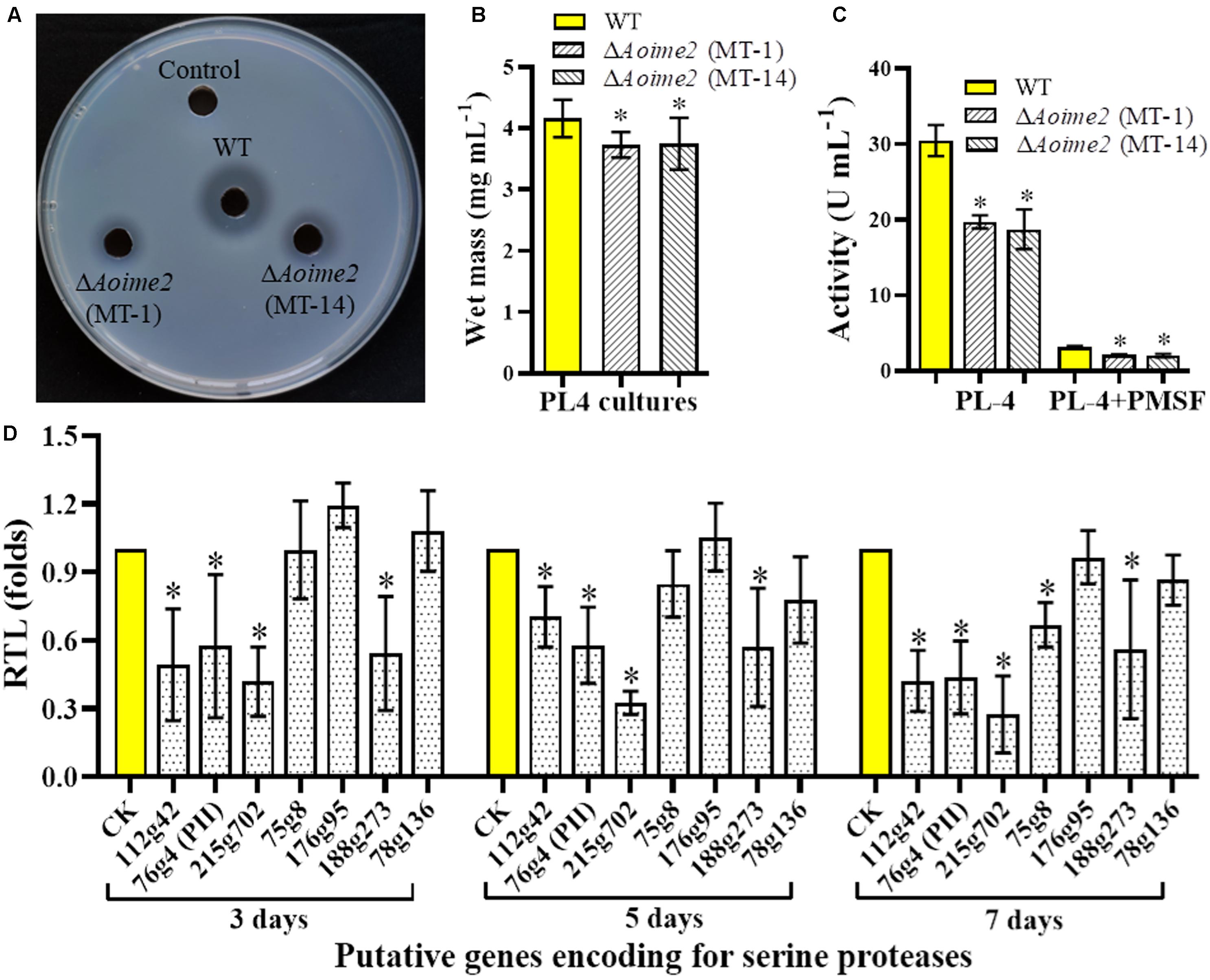
Figure 6. Comparison of extracellular proteolytic activity and transcriptional levels (RTLs) of serine-protease genes between WT and mutant strains. (A) Comparison of extracellular proteolytic activity of WT and ΔAoime2 mutants on casein plates. (B) Hyphal biomass of WT and ΔAoime2 mutants cultured in PL-4 medium for 7 days. (C) Proteolytic activity of WT and ΔAoime2 mutants quantified from 7-day-old PL-4 cultures. PMSF: serine-protease inhibitor, used at 5 mM. (D) RTLs of protease-encoding genes between WT and ΔAoime2 mutants at different time points. CK (RTL = 1) was used as the standard for statistical analysis of the RTL of each gene in the deletion mutants to that in the WT strain under a given condition. Error bars: SD from three replicates, asterisk: significant difference between mutant and WT (Tukey’s HSD, p < 0.05).
Inducer of meiosis 2, a non-classical MAPK-pathway molecule, has recently been identified in increasing numbers of fungal species and has been found to be involved in the regulation of multiple cellular processes, including vegetative growth, ascospore formation, environmental adaptation, and mating (Irniger, 2011). Here, we characterized AoIme2, an ortholog of S. cerevisiae Ime2, in the NT fungus A. oligospora. Similar to other Ime2 orthologs from yeast and filamentous fungi, AoIme2 and its orthologs share the conserved domains of MAPKs, but differ from the three classic MAPKs, Hog1-MAPK, Fus3/Kss1-MAPK, and Slt2-MAPK, in that they share differences in the phosphorylation site “-TXY-” and thus belong to the distinct Ime2-MAPK class (Garrido and Pérez-Martín, 2003; Schindler et al., 2003).
Disruption of the gene Aoime2 caused defective mycelial growth in two ΔAoime2 mutant strains, and the aerial hyphae of the mutants were sparse. Our result agrees with what has been observed with the Δime2 mutants of several filamentous fungi. In Nomuraea rileyi, deletion of Nrime2 severely affected the growth rate of the strain and hyphal growth was delayed (Li, 2018). Similarly, inactivation of crk1 (ime2) suppressed filamentous growth in U. maydis (Garrido and Pérez-Martín, 2003). Moreover, hyphal septa were increased here in ΔAoime2 mutants, as in the ΔNrime2 mutant in N. rileyi (Li, 2018), whereas mycelial and conidial cell nucleus numbers were markedly lower in the ΔAoime2 mutants than in the WT strain. Our results and the results of previous studies together suggest that Ime2 plays a critical role in hyphal growth, septum formation, and cell nucleus development in fungi.
We also recorded here a notable decrease (relative to WT) in the number of conidiophores and conidia in the ΔAoime2 mutants. Our result is similar to the result obtained after the deletion of Scime2 in S. cerevisiae, which suggested that the Ime2 plays a unique role in the activation of sporulation-specific genes (Mitchell et al., 1990). However, a different result was reported in the case of the ΔNrime2 mutant in N. rileyi: deletion of Nrime2 delayed the sporulation time but did not affect normal sporulation (Li, 2018). Furthermore, conidial morphology showed marked changes in the ΔAoime2 mutants here, and this is similar to two reported results: in Schizosaccharomyces pombe, disruption of Ime2-like protein kinase gene led to frequent production of asci containing an abnormal number of spores and spores of aberrant sizes (Abe and Shimoda, 2000); and in a budding yeast, where Ime2 is required for normal spore formation and spore-number control, abnormalities in spore formation were observed in diploid strains of the truncation mutant ime2ΔC241 (Sari et al., 2008). Corresponding to the reduction in the number of conidia, the expression of several sporulation-related genes, particularly abaA, fluG, velB, and aspB, was substantially downregulated in the ΔAoime2 mutants during the conidiation stage. These genes are necessary for conidiation in A. nidulans and other filamentous fungi (Krijgsheld et al., 2013). For example, fluG is required for the synthesis of an extracellular sporulation-inducing factor and serves as a key activator of conidiation in A. nidulans (Seo et al., 2006); aspB is expressed during vegetative growth and asexual sporulation in A. nidulans, deletion of aspB is not lethal but results in delayed septation and greatly reduced conidiation (Momany and Hamer, 1997; Hernández-Rodríguez et al., 2012). Recently, we characterized the ortholog of VelB in A. oligospora, and we found that the ΔAovelB mutant displayed severe sporulation defects (Zhang et al., 2019). These results indicate that Ime2 plays a vital role in conidiation and asexual development in A. oligospora and other fungi.
The growth of ΔAoime2 mutants was inhibited under hyperosmotic stresses but was unaffected by oxidative and cell-wall-perturbing stresses. This is similar to the decreased growth of ΔNrime2 mutant after NaCl addition in the case of N. rileyi (Li, 2018). Furthermore, the protein encoded by crk1 also plays a crucial role in environmental adaptation in U. maydis: when crk1 was disrupted in U. maydis, cells were unable to appropriately respond to environmental stimuli (Garrido and Pérez-Martín, 2003). Moreover, we noted here that the hyphae of ΔAoime2 mutants were smoother than those of the WT strain, that partial hyphae were deformed, and that the cell wall of the trap cells of the ΔAoime2 mutants became loose; this suggested that AoIme2 regulates CWI and causes the increasing sensitivity to osmotic pressure. These results show that Ime2 is critical for osmotic-pressure adaptation in A. oligospora and other fungi.
Nematode-trapping fungi break down the nematode epidermis by producing serine proteases and related hydrolytic enzymes; this enables infection and helps in the assimilation of essential nutrients from the nematodes (Yang et al., 2013). In 1994, the serine protease PII was isolated from A. oligospora; the protease can immobilize Panagrellus redivivus and hydrolyze nematode-epidermis proteins (Tunlid et al., 1994). Subsequently, 24 genes encoding putative serine proteases were identified in the A. oligospora genome, and categorized into four subtilisin families (Yang et al., 2011). We found here that extracellular proteolytic activity of the ΔAoime2 mutants was markedly diminished and further that the proteolytic activity was potently inhibited by PMSF; this suggests that the main extracellular proteases are serine proteases. Furthermore, the reduction in protease activity coincided well with the transcriptional repression of almost all tested serine-protease genes in the mutant strains relative to WT. These results indicate that AoIme2 regulates the expression of serine-protease genes, and thus plays a crucial role in serine-protease production in A. oligospora.
Traps are essential for NT fungi to capture and infect nematodes. Aoime2 disruption caused severe defects in trap formation: markedly fewer traps were present in ΔAoime2 mutants than in the WT strain, and most of the hyphal traps in the mutants did not form an intact ring; consequently, nematode capture by the ΔAoime2 mutants was substantially diminished relative to that by the WT strain. Similarly, Nrime2 deletion in N. rileyi was reported to severely affect the virality caused by infestation (Li, 2018), and crk1 in U. maydis was found to be required for pathogenicity, with the Δcrk1 mutant being unable to cause infected plants to develop tumors (Garrido et al., 2004). Moreover, examination of the ED bodies in the traps produced by WT and ΔAoime2 mutant strains revealed that considerably fewer trap-cell ED bodies were present in the ΔAoime2 mutants. These results indicate that Ime2 plays a key role in the development of infectious structures required for fungal pathogenicity.
Mitogen-activated protein kinase signaling modules have been identified in diverse fungi and have been demonstrated to be involved in the regulation of multiple biological processes, such as conidiation, cell-wall biogenesis, and stress response (Xu, 2000; Zhao et al., 2007; Rispail et al., 2009). Previously, we characterized Slt2-MAPK in two NT fungi, A. oligospora and Monacrosporium haptotylum, and reported that Slt2 disruption reduced mycelial growth, increased sensitivity to environmental stresses, and abolished the production of conidia and traps (Zhen et al., 2018). Here, we have identified an Ime2-MAPK ortholog for the first time from A. oligospora and have shown that AoIme2 is a multifunctional regulator, which may regulate the expression of phenotype-related genes, such as mycelial growth, conidiation, and trap formation genes; meanwhile, it also regulates the expression of serine-protease genes. Thus Aolme2 contributes to development and pathogenicity of A. oligospora.
We characterized the Ime2-MAPK ortholog AoIme2 from the NT fungus A. oligospora. In this fungus, AoIme2 plays a critical role in mycelial growth, conidiation, and trap formation, as well as in septum formation and cell nucleus development. Moreover, AoIme2 functions in hyperosmotic-stress response and serine-protease production. Our results provide a basis for understanding the roles of Ime2-MAPK in other NT fungi, as well as for uncovering the mechanisms underlying mycelial development, trap formation, and lifestyle switching in NT fungi.
All datasets generated for this study are included in the article/Supplementary Material.
JKY and K-QZ conceived and designed the study. MX and JKY wrote the manuscript. MX, NB, KJ, and JLY conducted the experiments. DZ, YZ, and DL analyzed the data. JKY and XN revised the manuscript. All authors read and approved the final manuscript.
This research described here is jointly supported by the National Natural Science Foundation of China (31960556, U1402265, and U1502262), the National Basic Research Program of China (2013CB127503), and the Yunnan Local Colleges Applied Basic Research Project (2017FH001-030).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.03065/full#supplementary-material
Abe, H., and Shimoda, C. (2000). Autoregulated expression of Schizosaccharomyces pombe meiosis-specific transcription factor Mei4 and a genome-wide search for its target genes. Genetics 154, 1497–1508.
Ahrén, D., Ursing, B. M., and Tunlid, A. (1998). Phylogeny of nematode-trapping fungi based on 18S rDNA sequences. FEMS Microbiol. Lett. 158, 179–184. doi: 10.1111/j.1574-6968.1998.tb12817.x
Bayram, O., Sari, F., Braus, G. H., and Irniger, S. (2009). The protein kinase ImeB is required for light-mediated inhibition of sexual development and for mycotoxin production in Aspergillus nidulans. Mol. Microbiol. 71, 1278–1295. doi: 10.1111/j.1365-2958.2009.06606.x
Chen, F., Chen, X. Z., Su, X. Y., Qin, L. N., Huang, Z. B., Tao, Y., et al. (2015). An Ime2-like mitogen-activated protein kinase is involved in cellulase expression in the filamentous fungus Trichoderma reesei. Biotechnol. Lett. 37, 2055–2062. doi: 10.1007/s10529-015-1888-z
Colot, H. V., Park, G., Turner, G. E., Ringelberg, C., Crew, C. M., Litvinkova, L., et al. (2006). A high-throughput gene knockout procedure for Neurospora reveals functions for multiple transcription factors. Proc. Natl. Acad. Sci. U.S.A. 103, 10352–10357. doi: 10.1073/pnas.0601456103
Garrido, E., and Pérez-Martín, J. (2003). The crk1 gene encodes an Ime2-related protein that is required for morphogenesis in the plant pathogen Ustilago maydis. Mol. Microbiol. 47, 729–743. doi: 10.1046/j.1365-2958.2003.03323.x
Garrido, E., Voss, U., Müller, P., Castillo-Lluva, S., Kahmann, R., and Pérez-Martín, J. (2004). The induction of sexual development and virulence in the smut fungus Ustilago maydis depends on Crk1, a novel MAPK protein. Gene Dev. 18, 3117–3130. doi: 10.1101/gad.314904
Hernández-Rodríguez, Y., Hastings, S., and Momany, M. (2012). The septin AspB in Aspergillus nidulans forms bars and filaments and plays roles in growth emergence and conidiation. Eukaryot Cell. 11, 311–323. doi: 10.1128/EC.05164-11
Herskowitz, I. (1995). MAP kinase pathways in yeast: for mating and more. Cell 80, 187–197. doi: 10.1016/0092-8674(95)90402-6
Hutchison, E. A., and Glass, N. L. (2010). Meiotic regulators Ndt80 and Ime2 have different roles in Saccharomyces and Neurospora. Genetics 185, 1271–1282. doi: 10.1534/genetics.110.117184
Irniger, S. (2011). The Ime2 protein kinase family in fungi: more duties than just meiosis. Mol. Microbiol. 80, 1–13. doi: 10.1111/j.1365-2958.2011.07575.x
Jiang, C., Zhang, X., Liu, H., and Xu, J. R. (2018). Mitogen-activated protein kinase signaling in plant pathogenic fungi. PLoS Pathog. 14:e1006875. doi: 10.1371/journal.ppat.1006875
Krijgsheld, P., Bleichrodt, R. V., Van Veluw, G. J., Wang, F., Müller, W. H., Dijksterhuis, J., et al. (2013). Development in Aspergillus. Stud Mycol. 74, 1–29. doi: 10.3114/sim0006
Krylov, D. M., Nasmyth, K., and Koonin, E. V. (2003). Evolution of eukaryotic cell cycle regulation: stepwise addition of regulatory kinases and late advent of the CDKs. Curr. Biol. 13, 173–177. doi: 10.1016/s0960-9822(03)00008-3
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Li, C. (2018). The Gene Cloning and Functional Analysis of Rim15, Ume6 and Ime2 Gene in Nomuraea Rileyi. [dissertation/master’s thesis]. Chongqing: Chongqing University.
Li, C., Melesse, M., Zhang, S., Hao, C., Wang, C., Zhang, H., et al. (2015). FgCDC14 regulates cytokinesis, morphogenesis, and pathogenesis in Fusarium graminearum. Mol. Microbiol. 98, 770–786. doi: 10.1111/mmi.13157
Liu, J., Wang, Z. K., Sun, H. H., Ying, S. H., and Feng, M. G. (2017). Characterization of the Hog1 MAPK pathway in the entomopathogenic fungus Beauveria bassiana. Environ. Microbiol. 19, 1808–1821. doi: 10.1111/1462-2920.13671
Liu, K., Zhang, W., Lai, Y., Xiang, M., Wang, X., Zhang, X., et al. (2014). Drechslerella stenobrocha genome illustrates the mechanism of constricting rings and the origin of nematode predation in fungi. BMC Genomics 15:114. doi: 10.1186/1471-2164-15-114
Liu, K. H., and Shen, W. C. (2011). Mating differentiation in Cryptococcus neoformans is negatively regulated by the Crk1 protein kinase. Fungal Genet. Biol. 48, 225–240. doi: 10.1016/j.fgb.2010.11.005
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Meerupati, T., Andersson, K. M., Friman, E., Kumar, D., Tunlid, A., and Ahrén, D. (2013). Genomic mechanisms accounting for the adaptation to parasitism in nematode-trapping fungi. PLoS Genet. 9:e1003909. doi: 10.1371/journal.pgen.1003909
Mitchell, A. P., Driscoll, S. E., and Smith, H. E. (1990). Positive control of sporulation-specific genes by the IME1 and IME2 products in Saccharomyces cerevisiae. Mol. Cell Biol. 10, 2104–2110. doi: 10.1128/mcb.10.5.2104
Momany, M., and Hamer, J. E. (1997). The Aspergillus nidulans septin encoding gene, aspB, is essential for growth. Fungal Genet. Biol. 21, 92–100. doi: 10.1006/fgbi.1997.0967
Nordbring-Hertz, B. (2004). Morphogenesis in the nematode-trapping fungus Arthrobotrys oligospora-an extensive plasticity of infection structures. Mycologist 18, 125–133. doi: 10.1017/s0269915x04003052
Nordbring-Hertz, B., Jansson, H. B., and Tunlid, A. (2001). “Nematophagous fungi,” in Encyclopedia of Life Sciences, ed. O. H. Chichester, (Hoboken, NJ: JohnWiley & Sons), 1–13.
Payne, D. M., Rossomando, A. J., Martino, P., Erickson, A. K., Her, J. H., Shabanowitz, J., et al. (1991). Identification of the regulatory phosphorylation sites in pp42/mitogen-activated protein kinase (MAP kinase). EMBO J. 10, 885–892. doi: 10.1002/j.1460-2075.1991.tb08021.x
Rispail, N., Soanes, D. M., Ant, C., Czajkowski, R., Grünler, A., Huguet, R., et al. (2009). Comparative genomics of MAP kinase and calcium-calcineurin signalling components in plant and human pathogenic fungi. Fungal Genet. Biol. 46, 287–298. doi: 10.1016/j.fgb.2009.01.002
Sari, F., Heinrich, M., Meyer, W., Braus, G. H., and Irniger, S. (2008). The C-terminal region of the meiosis-specific protein kinase Ime2 mediates protein instability and is required for normal spore formation in budding yeast. 378, 31–43. doi: 10.1016/j.fgb.2009.01.002
Schaeffer, H. J., and Weber, M. J. (1999). Mitogen-activated protein kinases: specific messages from ubiquitous messengers. Mol. Cell Biol. 19, 2435–2444. doi: 10.1128/mcb.19.4.2435
Schindler, K., Benjamin, K. R., Martin, A., Boglioli, A., Herskowitz, I., and Winter, E. (2003). The Cdk-activating kinase Cak1p promotes meiotic S phase through Ime2p. Mol. Cell Biol. 23, 8718–8728. doi: 10.1128/mcb.23.23.8718-8728.2003
Seo, J. A., Guan, Y., and Yu, J. H. (2006). FluG-dependent asexual development in Aspergillus nidulans occurs via derepression. Genetics 172, 1535–1544. doi: 10.1534/genetics.105.052258
Smith, H. E., and Mitchell, A. P. (1989). A transcriptional cascade governs entry into meiosis in Saccharomyces cerevisiae. Mol. Cell Biol. 9, 2142–2152. doi: 10.1128/mcb.9.5.2142
Strudwick, N., Brown, M., Parmar, V. M., and Schroder, M. (2010). Ime1 and Ime2 are required for pseudohyphal growth of Saccharomyces cerevisiae on nonfermentable carbon sources. Mol. Cell Biol. 30, 5514–5530. doi: 10.1128/MCB.00390-10
Su, H., Zhao, Y., Zhou, J., Feng, H., Jiang, D., Zhang, K. Q., et al. (2017). Trapping devices of nematodetrapping fungi: formation, evolution, and genomic perspectives. Biol. Rev. 92, 357–368. doi: 10.1111/brv.12233
Tunlid, A., Åhman, J., and Oliver, R. P. (1999). Transformation of the nematode-trapping fungus Arthrobotrys oligospora. FEMS Microbiol. Lett. 173, 111–116. doi: 10.1016/s0378-1097(99)00059-2
Tunlid, A., Rosen, S. E., Ek, B., and Rask, L. (1994). Purification and characterization of an extracellular serine protease from the nematode-trapping fungus Arthrobotrys oligospora. Microbiology 140, 1687–1695. doi: 10.1099/13500872-140-7-1687
Veenhuis, M., Nordbring-Hertz, B., and Harder, W. (1985). An electron-microscopical analysis of capture and initial stages of penetration of nematodes by Arthrobotrys oligospora. Antonie Van Leeuwenhoek 51, 385–398. doi: 10.1007/bf02275043
Veenhuis, M., Van Wijk, C., Wyss, U., Nordbring-Hertz, B., and Harder, W. (1989). Significance of electron dense microbodies in trap cells of the nematophagous fungus Arthrobotrys oligospora. Antonie Van Leeuwenhoek 56, 251–261. doi: 10.1007/bf00418937
Wang, M., Yang, J., and Zhang, K. Q. (2006). Characterization of an extracellular protease and its cDNA from the nematode-trapping fungus Monacrosporium microscaphoides. Can. J. Microbiol. 52, 130–139. doi: 10.1139/w05-110
Xie, M. H., Wang, Y. C., Tang, L. Y., Yang, L., Zhou, D. X., Li, Q., et al. (2019). AoStuA, an APSES transcription factor, regulates the conidiation, trap formation, stress resistance, and pathogenicity of the nematode-trapping fungus Arthrobotrys oligospora. Environ. Microbiol. 21, 4648–4661. doi: 10.1111/1462-2920.14785
Xu, J. R. (2000). MAP kinases in fungal pathogens. Fungal Genet. Biol. 31, 137–152. doi: 10.1006/fgbi.2000.1237
Yang, J. K., Liang, L. M., Li, J., and Zhang, K. Q. (2013). Nematicidal enzymes from microorganisms and their applications. Appl. Microbiol. Biotechnol. 97, 7081–7095. doi: 10.1007/s00253-013-5045-0
Yang, J. K., Wang, L., Ji, X., Feng, Y., Li, X., Zou, C., et al. (2011). Genomic and proteomic analyses of the fungus Arthrobotrys oligospora provide insights into nematode-trap formation. PLoS Pathog. 7:e1002179. doi: 10.1371/journal.ppat.1002179
Yang, S. H., Sharrocks, A. D., and Whitmarsh, A. J. (2003). Transcriptional regulation by the MAP kinase signaling cascades. Gene 320, 3–21. doi: 10.1016/s0378-1119(03)00816-3
Yang, X. W., Ma, N., Yang, L., Zheng, Y. Q., Zhen, Z. Y., Li, Q., et al. (2018). Two Rab GTPases play different roles in conidiation, trap formation, stress resistance, and virulence in the nematode-trapping fungus Arthrobotrys oligospora. Appl. Microbiol. Biotechnol. 102, 4601–4613. doi: 10.1007/s00253-018-8929-1
Yoshida, M., Kawaguchi, H., Sakata, Y., Kominami, K., Hirano, M., Shima, H., et al. (1990). Initiation of meiosis and sporulation in Saccharomyces cerevisiae requires a novel protein kinase homologue. Mol. Gen. Genet. 221, 176–186.
Zhang, G. S., Zheng, Y. Q., Ma, Y. X., Yang, L., Niu, X. M., Zhou, D. X., et al. (2019). The Velvet proteins VosA and VelB play different roles in conidiation, trap formation, and pathogenicity in the nematode-trapping fungus Arthrobotrys oligospora. Front. Microbiol. 10:1917. doi: 10.3389/fmicb.2019.01917
Zhang, L., Wang, J., Xie, X. Q., Keyhani, N. O., Feng, M. G., and Ying, S. H. (2013). The autophagy gene BbATG5, involved in the formation of the autophagosome, contributes to cell differentiation and growth but is dispensable for pathogenesis in the entomopathogenic fungus Beauveria bassiana. Microbiology 159, 243–252. doi: 10.1099/mic.0.062646-0
Zhao, M. L., Mo, M. H., and Zhang, K. Q. (2004). Characterization of a neutral serine protease and its full-length cDNA from the nematode-trapping fungus Arthrobotrys oligospora. Mycologia 96, 16–22. doi: 10.1080/15572536.2005.11832991
Zhao, X., Mehrabi, R., and Xu, J. R. (2007). Mitogen-activated protein kinase pathways and fungal pathogenesis. Eukaryot. Cell. 6, 1701–1714. doi: 10.1128/ec.00216-07
Zhen, Z. Y., Xing, X. J., Xie, M. H., Yang, L., Yang, X. W., Zheng, Y. Q., et al. (2018). MAP kinase Slt2 orthologs play similar roles in conidiation, trapformation, and pathogenicity in two nematode-trapping fungi. Fungal Genet. Biol. 116, 42–50. doi: 10.1016/j.fgb.2018.04.011
Zhen, Z. Y., Zhang, G. S., Yang, L., Ma, N., Li, Q., Ma, Y. X., et al. (2019). Characterization and functional analysis of the calcium/calmodulin-dependent protein kinases (CaMKs) in the nematode-trapping fungus Arthrobotrys oligospora. Appl. Microbiol. Biotechnol. 103, 819–832. doi: 10.1007/s00253-018-9504-5
Keywords: Arthrobotrys oligospora, inducer of meiosis 2, mycelial development, conidiation, osmolarity, pathogenicity
Citation: Xie M, Bai N, Yang J, Jiang K, Zhou D, Zhao Y, Li D, Niu X, Zhang K-Q and Yang J (2020) Protein Kinase Ime2 Is Required for Mycelial Growth, Conidiation, Osmoregulation, and Pathogenicity in Nematode-Trapping Fungus Arthrobotrys oligospora. Front. Microbiol. 10:3065. doi: 10.3389/fmicb.2019.03065
Received: 23 November 2019; Accepted: 18 December 2019;
Published: 14 January 2020.
Edited by:
Sheng-hua Ying, Zhejiang University, ChinaReviewed by:
Huiquan Liu, Northwest A&F University, ChinaCopyright © 2020 Xie, Bai, Yang, Jiang, Zhou, Zhao, Li, Niu, Zhang and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jinkui Yang, amlua3VpOTYwQHludS5lZHUuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.