- 1Key Laboratory of Quality and Safety Risk Assessment for Aquatic Products on Storage and Preservation (Shanghai), China Ministry of Agriculture, College of Food Science and Technology, Shanghai Ocean University, Shanghai, China
- 2Department of Microbiology, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA, United States
- 3Department of Epidemiology and Biostatistics, Faculty of Public Health, Kuwait University, Safat, Kuwait
Vibrio cholerae is a leading waterborne pathogenic bacterium worldwide. It can cause human cholera that is still pandemic in developing nations. Detection of V. cholerae contamination in drinking water and aquatic products is imperative for assuring food safety. In this study, a simple, sensitive, specific, and visualized method was developed based on loop-mediated isothermal amplification (LAMP) (designated sssvLAMP) to detect virulence-associated (ctxA, tcpA, hapA, mshA, pilA, and tlh) and species-specific (lolB) genes of V. cholerae. Three pairs of oligonucleotide primers (inner, outer, and loop primers) were designed and or synthesized to target each of these genes. The optimal conditions of the sssvLAMP method was determined, and one-step sssvLAMP reaction was performed at 65°C for 40 min. Positive results were simply read by the naked eye via color change (from orange to light green) under the visible light, or by the production of green fluorescence under the UV light (260 nm). The sssvLAMP method was more efficient in detecting 6.50 × 101–6.45 × 104-fold low number of V. cholerae cells, and more sensitive in V. cholerae genomic DNA (1.36 × 10–2-4.42 × 10–6 ng/reaction) than polymerase chain reaction (PCR) method. Among 52 strains of V. cholerae and 50 strains of non-target species (e.g., other Vibrios and common pathogens) examined, the sensitivity and specificity of the sssvLAMP method were 100% for all the target genes. Similar high efficiency of the method was observed when tested with spiked samples of water and aquatic products, as well as human stool specimens. Water from various sources and commonly consumed fish samples were promptly screened by this simple and efficient visualized method and diversified variation in the occurrence of the target genes was observed. V. cholerae strains could be mostly detected by the presence of hapA and tlh alone or in combination with other genes, indicating a variable risk of potentially pathogenic non-O1/O139 strains in edible food products. This novel LAMP method can be a promising tool to address the increasing need of food safety control of aquatic products.
Introduction
Vibrio cholerae can cause cholera, a severe human diarrhoeal disease that can be quickly fatal if untreated and is typically transmitted via contaminated water and person-to-person contact (Baker-Austin et al., 2018). It was estimated that cholera caused roughly 2.9 million cases and 95,000 deaths annually worldwide between 2008 and 2012 (Ali et al., 2015). Cholera is still pandemic in developing nations in recent years such as in Zambia, Tanzania, Mozambique, Somalia, South Sudan, Kenya, and Congo (Kinshasa) in 2018, as well as in Mozambique and Somalia in 2019 (World Health Organization)1. V. cholerae is reported to harbor a highly conserved species-specific gene lolB (Lalitha et al., 2008). Previous studies have indicated that cholerae toxin (CTX) and toxin coregulated pilus (TCP) are major toxic factors of epidemic V. cholerae strains of serotypes O1 and O139. Nevertheless, some non-O1/O139 strains lacking the ctx and tcp genes have been reported to cause sporadic episodes of diarrhea and gastroenteritis (Austin, 2010; Ceccarelli et al., 2015), indicating that other virulence factors exist. Thus, detection of the potential pathogenic non-O1/O139 V. cholerae contamination in food is also imperative for assuring food safety.
Previous studies have revealed virulence-associated genes that are present in V. cholerae chromosomes (Hacker et al., 1997). For instance, a hlyA gene encodes an extracellular pore-forming toxin, produced by biotype El Tor of serogroup O1 and most of the non-O1/O139 strains. The HlyA is known to be associated with multiple virulence-related traits, including the hemolytic activity, lethality, cardiotoxicity, cytotoxicity and enterotoxicity (Benitez and Silva, 2016; Gao et al., 2018). V. cholerae produces at least three morphologically distinct types of pili (Hall et al., 1988), including the TCP, mannose-sensitive hemagglutination (MSHA) pilus (Chiavelli et al., 2001; Moorthy and Watnick, 2004), and Type IV-A pilus (Fullner and Mekalanos, 1999; Aagesen and Hase, 2012), which all play important roles in the adaptability and pathogenicity of the bacterium. The mshBACD gene cluster is responsible for the structure of MSHA, which is reported to act not only as a receptor of a widespread filamentous bacteriophage facilitating transfer of virulence genes in V. cholerae O139 strain (Jouravleva et al., 1998), but also aiding bacterial association with aquatic plankton to support environmental adaptation in non O1/O139 strains (Chiavelli et al., 2001; Moorthy and Watnick, 2004; Gong et al., 2019). The third type of pilus is essential for the colonization of Vibrio species in the environment and/or host tissues (Fullner and Mekalanos, 1999). It is encoded by a 5.4-kb pilABCD gene cluster, in which the pilA gene encodes one of the major subunits of this Type IV-A pilus (Aagesen and Hase, 2012). Among other virulence-associated factors in V. cholerae include the tlh (Fiore et al., 1997), and hapA genes (Datta-Roy et al., 1986), the former encodes a thermolabile hemolysin with phospholipase and lecithinase activities (Fiore et al., 1997), and the latter encodes a hemagglutinin protease involved in V. cholerae interaction with aquatic hosts (Halpern et al., 2003).
To date, many methods have been developed for effective detection of O1/O139 V. cholerae contamination in food. Compared with the conventional culture-based microbiological detection assays, molecular biology-based methods are more rapid and sensitive, such as PCR (Kumar et al., 2010; Zago et al., 2017), real-time PCR (Garrido-Maestu et al., 2015; Casasola-Rodriguez et al., 2018), multiplex PCR (Bwire et al., 2018; Vu et al., 2018), oligonucleotide array hybridization (Nasrabadi et al., 2017), strand displacement amplification (SDA) (Phillips et al., 2018), rolling circle amplification (RCA) (Osterberg et al., 2014), cross-priming amplification (CPA) (Zhang et al., 2015), and nucleic acid sequence-based amplification (NASBA) (Fykse et al., 2012). Nevertheless, these methods require expensive equipments, which limit their wide application, particularly in on-site testing and large-scale survey. On the other hand, the loop-mediated isothermal amplification (LAMP) technique is an alternative promising tool because of its simplicity, rapidness and suitability for on-site large-scale screening (Notomi et al., 2000; Soli et al., 2013; Engku Nur Syafirah et al., 2018).
The LAMP technique, originally developed by Notomi et al. (2000), can amplify nucleic acids from a single copy to 109 copies at a constant temperature (typically 60–70°C) (Notomi et al., 2000). This one-step reaction method requires just a simple equipment such as a water bath or temperature block. The LAMP-positive amplicons can be confirmed by the gel electrophoresis analysis with fluorescent dsDNA intercalating dyes, e.g., the ethidium bromide (EB) (Almasi et al., 2012) and Synergy Brands (SYBR) Green (Yu et al., 2014; Yang et al., 2018). Utilization of metal indicators, such as hydroxynaphthol blue (HNB) (Soli et al., 2013; Zhong et al., 2018), and magnesium pyrophosphate (Notomi et al., 2000), allows the observation of results with the naked eye. However, the post-amplification detection requires opening of reaction tubes and therefore significantly increases the risk of carry-over contamination (Zanoli and Spoto, 2013). Although the HNB dye can be added prior to the reaction, positive amplicons with color change from violet to sky blue can be variable and imperfect for different observers (Wastling et al., 2010). The drawback of the magnesium pyrophosphate-based detection method is time-dependent and instable. The turbidity generated as a by-product (magnesium pyrophosphate) of DNA amplification of positive samples is stable but just for a short time, which needs to be judged soon after taking out the samples from the water bath (Mori et al., 2001). On the other hand, the use of MnCl2-calcein dye has been a more recent approach to circumvent the instability problem of other dyes applied in LAMP-based detection (Fang et al., 2018; Sayad et al., 2018) to develop the sssvLAMP system. Calcein is a metal indicator that yields strong fluorescence by forming complexes with divalent metallic ions, such as Ca2+ and Mg2+ (Toffaletti and Kirvan, 1980; Fiedoruk-Pogrebniak and Koncki, 2015). As LAMP reaction proceeds in the presence of target DNA, calcein is deprived of Mn2+ by newly generated pyrophosphate ion, and instead combines with residual Mg2+, producing green fluorescence (Tomita et al., 2008). In contrast, if no amplification occurred, no color change and green fluorescence are observed.
To date, only few studies have been conducted to detect V. cholerae genes by the LAMP, i.g., the ctxA (Okada et al., 2010), hlyA (He et al., 2009), ompW (Srisuk et al., 2010), rtxA (Tourlousse et al., 2012), toxR (Zhang et al., 2015), and lolB (Liew et al., 2015). Development of a simple, rapid, specific and sensitive LAMP method for V. cholerae detection is very important for food safety control, particularly, as a highly efficient tool for large-scale screening of the bacterial contamination in water and aquatic products.
Materials and Methods
Bacterial Strains and Culture Conditions
Bacterial strains and culture media used in this study are listed in Table 1. The thiosulfate citrate bile salt sucrose (TSBS), Luria-Bertani (LB), Tryptic Soy Broth (TSB), and de Man Rogosa Sharpe (MRS) media were purchased from Beijing Land Bridge Technology Co., Ltd., Beijing, China. The Enterobacteria Enrichment (EE), and Brain Heart Infusion (BHI) media were purchased from Qingdao Hope Bio-Technology Co., Ltd., Qingdao, China, while the Marine 2216 was from Becton, Dickinson and Company, United States. Vibrio and non-Vibrio strains were individually inoculated from laboratory stock at −80°C onto corresponding agar plates, respectively, incubated at 37°C for 16–18 h. Single colonies of each strain was individually streaked into corresponding broth supplemented with 3.0% (pH 8.4–8.5, Vibrio strains) and 0.5/1.0% NaCl (pH 7.0–7.2, non-Vibrio strains), respectively, and incubated at 37°C for 16–18 h for further analysis.
Genomic DNA Preparation
Bacterial genomic DNA was prepared using the TaKaRa MiniBEST Bacterial Genomic DNA Extraction Kit Ver. 3.0 (TaKaRa Biomedical Technology Co., Ltd., Beijing, China) following the manufacturer’s instructions. Extracted DNA samples were analyzed by agarose gel electrophoresis, visualized under short-wavelength UV light (260 nm), and imaged using the UVP EC3 Imaging system (UVP LLC, Upland, CA, United States) as described previously (He et al., 2015). The DNA concentration and purity (A260/A280) were determined using a multimode microplate reader (Synergy 2, Vermont, United States).
Bacterial genomic DNA was also extracted by a thermal lysis method as described previously (Okada et al., 2010) with minor modifications. Briefly, 1 mL of bacterial cell culture was centrifugated at 12,000 rpm for 5 min, and the cell pellet was resuspended with 1 mL of sterile 1 × phosphate buffer saline (PBS, pH 7.4–7.6, Shanghai Sangon Biological Engineering Technology and Services Co., Ltd., Shanghai, China). The resuspension was then 10-fold serially diluted. Bacterial cells were enumerated via plating appropriate dilutions of cell suspension onto the LB agar plates as described previously (Sun et al., 2014). In parallel, a 1 mL of appropriately diluted resuspension was centrifugated at 12,000 rpm for 5 min, and the cell pellet was heated in 200 μL of sterile ultrapure water at 95°C for 10 min, and then transferred on ice for cooling. After centrifugation at 12,000 rpm for 5 min, the resulting lysis solution was used as DNA template for the detection of V. cholerae cells in water.
Designing LAMP Primers
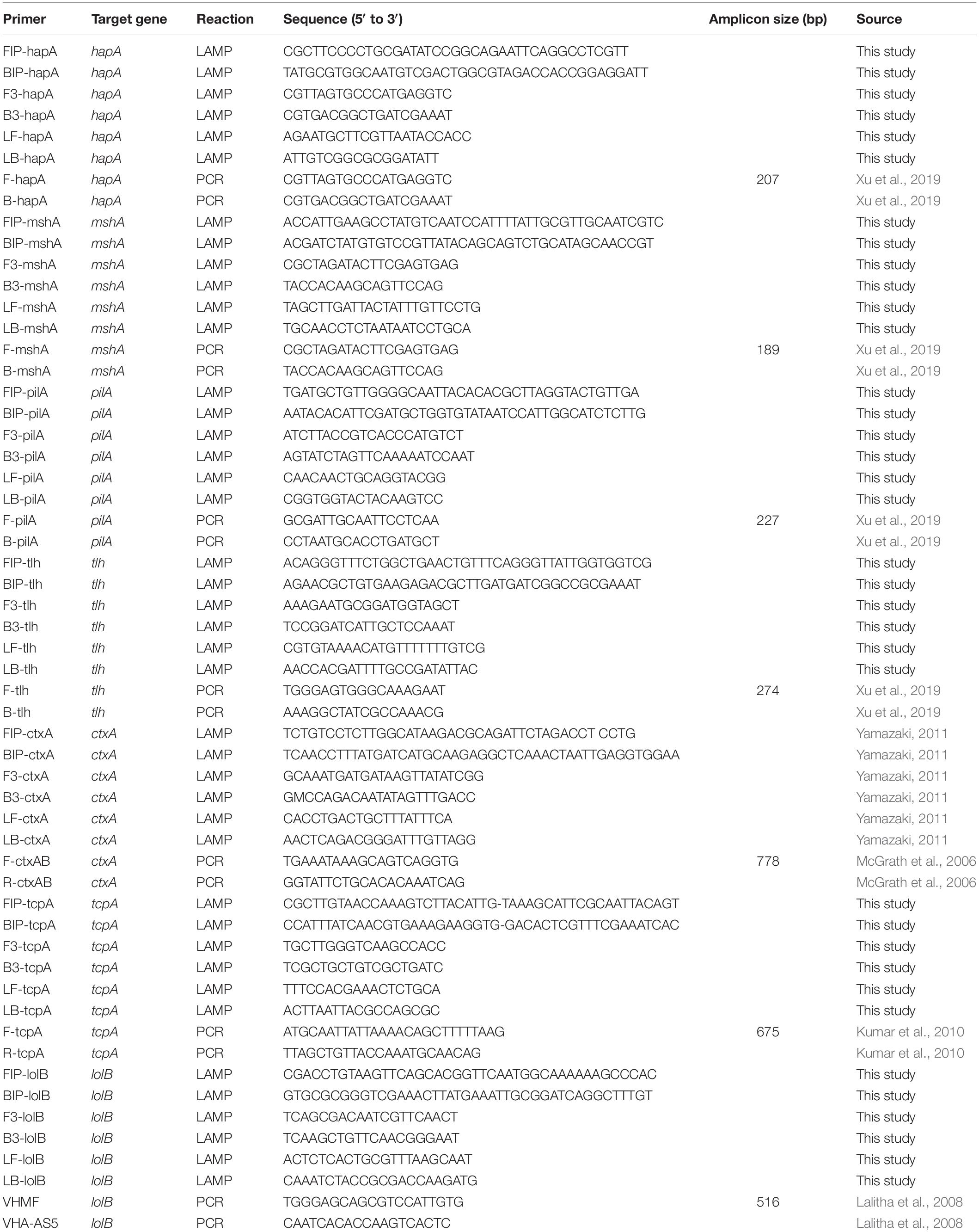
The primers used in this study were listed in Table 2. The sequences of the target genes of V. cholerae were downloaded from the National Center for Biotechnology Information (NCBI) GenBank database2, including the tcpA, hapA, mshA, pilA, tlh, and lolB genes, with GenBank accession Nos. listed in Supplementary Figure S1. Two pairs of inner and outer primers (FIP and BIP, F3 and B3) targeting conserved sequences of each gene was designed using the Primer Explorer Version 5 software3 with default parameters by importing DNA sequence of each target gene in a FASTA format, while the pair of the loop primers (LF and LB) were designed using the SnapGene Viewer version 4.1.4 software (GSL Biotech LLC, Chicago, IL, United States) by choosing loop primer sequences based on proper annealing temperatures and sequence locations required by inner and outer primers. The locations of each pair of the newly designed primers were marked in Supplementary Figure S1. The ctxA gene was detected with the primers described previously (Yamazaki, 2011). All primers were synthesized by the Sangon (Shanghai, China). Comparative sequence alignments between the newly designed LAMP primers versus target gene sequences were performed using the BioEdit software (Hall, 1999) (Supplementary Figure S1). The virulence-associated genes were amplified from some representative V. cholerae strains by PCR reactions, and DNA sequencing was carried out by the Sangon (Shanghai, China). The sequences were submitted to the GenBank with accession Nos. listed in Supplementary Figure S1.
Preparation of MnCl2-Calcein Dye Stock Solution
Calcein (Sigma, St. Louis, MO, United States) was first dissolved with 1 M NaOH (Analytical Reagent, Sinopharm Chemical Reagent Co., Ltd, Shanghai, China), and 6.5 mM calcein solution was prepared with ultrapure water. MnCl2⋅4H2O (Sangon, Shanghai, China) was used to prepare 130 mM MnCl2 solution. Then a stock solution consisting of 1.30 mM calcein and 15.60 mM MnCl2 was prepared and stored at −20°C.
Optimization of Reaction Parameters of the sssvLAMP Method
The initial LAMP reaction solution was prepared according to the method described previously (Srisuk et al., 2010; Yamazaki, 2011) with minor modifications. A 25 μL of LAMP reaction solution contained 1.6 μM of each of the inner primers (FIP and BIP), 0.2 μM of each of the outer primers (F3 and B3), and 0.8 μM of each of the loop primers (LF and LB), 1× Thermopol buffer (pH 8.8, contains 20 mM Tris-HCl, 10 mM (NH4)2SO4, 10 mM KCl, 2 mM MgSO4, 0.10% Triton® X-100, New England Biolabs, Beverly, MA, United States), 0.8 M betaine (Sigma, St. Louis, MO, United States), 1.4 mM of each dNTP (TaKaRa, China), 6 mM MgSO4 (New England Biolabs), 12 U Bst DNA polymerase (New England Biolabs), and 2 μL of DNA template. Finally, 1 μL of the MnCl2-calcein stock solution was added to the reaction solution, in which final concentrations of Mn2+ and calcein were 600 and 50 μM, respectively. A negative control was prepared using DNase/RNase-free deionized water (Tiangen Biotech Co., Ltd., Beijing, China) instead of bacterial culture or DNA template.
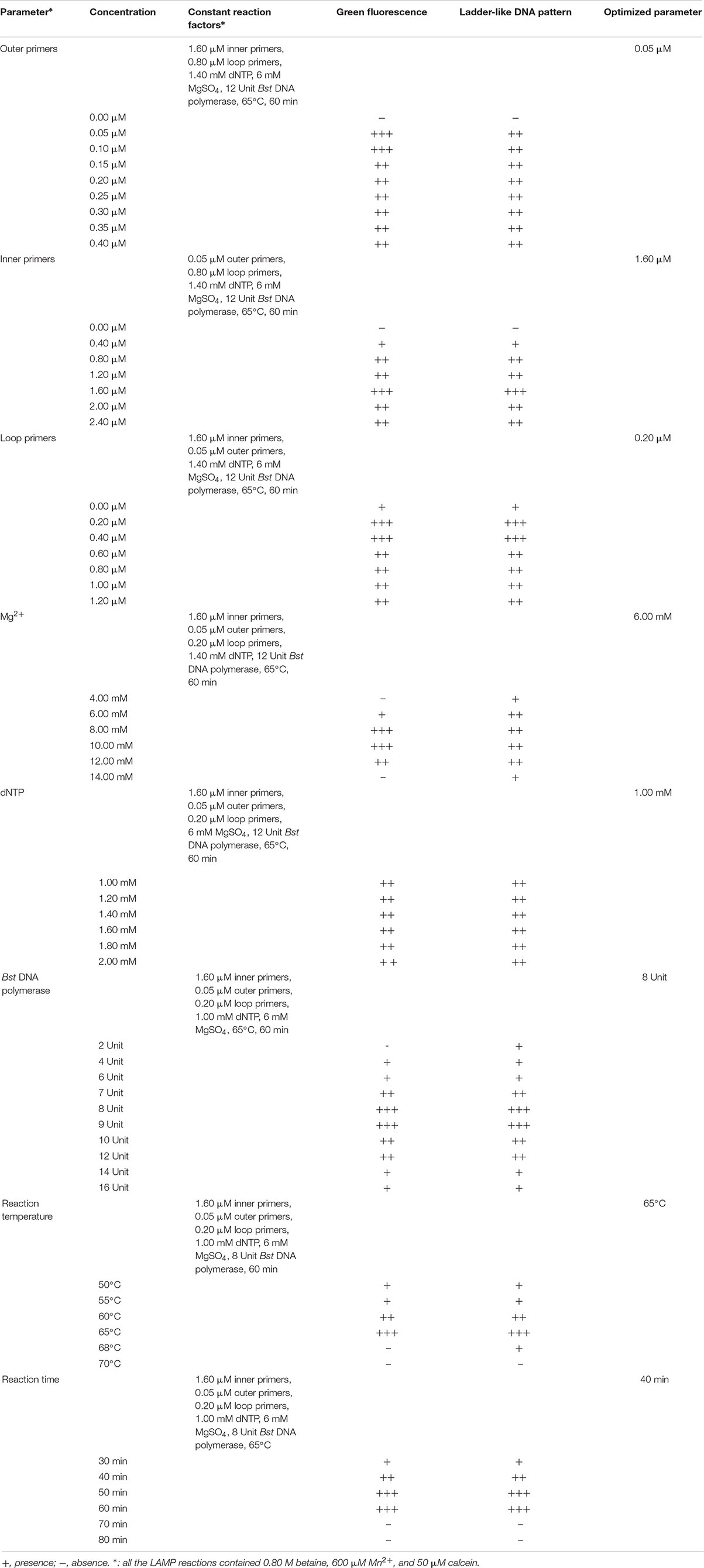
The reaction parameters were optimized, including different concentrations of the outer primers (0.05–0.40 μM), inner primers (0.4–2.4 μM), loop primers (0.2–1.2 μM), Mg2+ (4.0–14.0 mM), dNTP (1.0–2.0 mM), and Bst DNA polymerase (2–16 Unit) per reaction, as well as different reaction temperature (50–70°C) and reaction time (30–80 min) (Table 3). The LAMP was terminated at 80°C for 10 min. Additionally, the function of loop primers in the reaction system was also investigated. Some target genes were used in the optimization of reaction parameters.
Confirmation of the LAMP Results
Positive and negative results were confirmed by observing the color change of the sssvLAMP reaction solutions with the naked eye under the visible light, or under the UV transilluminator (302 nm) on a blue background, if needed. A light green color or bright fluorescence is typical characteristics of all positive reactions, while an original orange color or no fluorescence for negative results. Also, the LAMP products were verified by 2.0% agarose gel electrophoresis analysis, which forms ladder-like DNA patterns (Tomita et al., 2008). The gels were visualized and recorded using a UVP EC3 Imaging system as described above.
Determination of Specificity and Sensitivity of the sssvLAMP Method
The inclusivity (determined as 100% of positive detection of target strains) of the sssvLAMP method was examined with 52 V. cholerae strains (Table 1), including pandemic (i.g., ATCC39315/N16961), and non-pandemic strains. Among these strains, V. cholerae GIM 1.449 is widely used as a reference non-O1/O139 strain in China. Virulence-associated gene types of non-pandemic strains were confirmed by PCR assays (Xu et al., 2019). The exclusivity (determined as 100% of negative detection of non-target strains) of the method was tested with 50 bacterial strains, including closely related Vibrio species (n = 16), and non-Vibrio species (n = 34). It contained common bacterial pathogens, e.g., Vibrio alginolyticus, Vibrio fluvialis, Vibrio harvey, Vibrio metschnikovii, Vibrio mimicus, Vibrio parahemolyticus, Vibrio vulnificus, as well as Aeromonas hydrophila, Escherichia coli, Enterobacter cloacae, Enterobacter sakazakii, Klebsiella pneumoniae, Listeria monocytogenes, Salmonella paratyphi, and Staphylococcus aureus (Table 1). Genomic DNA was individually extracted from these bacteria as described above and diluted serially with the DNase/RNase-free deionized water as DNA templates.
The sensitivity of the sssvLAMP method was evaluated for each targeted gene in the 52 V. cholerae strains (Table 1). Cell culture and genomic DNA samples of these strains were used in the evaluation. For the detection of V. cholerae cells, overnight cell cultures of V. cholerae strains were individually inoculated (1%, v/v) into fresh media (Table 1) and incubated at 37°C, and bacterial cells grown to log-phase were harvested by centrifugation, resuspended, diluted and enumerated as described above. For instance, 9.3 × 107–9.3 colony forming units (CFU)/mL, and 1.44 × 106–1.44 CFU/mL of V. cholerae GIM 1.449 cells were used for the detection of the hapA, and tlh genes, respectively. The 1.09 × 108–1.09 CFU/mL of V. cholerae Q07-21 cells were tested for the mshA gene, while 1.28 × 108–12.8 CFU/mL of V. cholerae L10-36 cells were for the pilA gene. For the detection of V. cholerae genomic DNA, for instance, 1.67 × 101–1.67 × 10–9 ng/μL of V. cholerae GIM 1.449 genomic DNA samples was used for the detection of the hapA gene, while 1.41 × 101–1.41 × 10–9 ng/μL of V. cholerae Q07-21, 1.31 × 101–1.31 × 10–9 ng/μL of V. cholerae L10-36, and 1.48 × 101–1.48 × 10–9 ng/μL of V. cholerae GIM 1.449 genomic DNA samples were tested for the target genes mshA, pilA, and tlh by the sssvLAMP method, respectively. The last dilution of genomic DNA or cell culture samples, which was tested positive for the target gene by the sssvLAMP method, was considered as the limit of detection (LOD) in this study.
Determination of Sensitivity of the sssvLAMP Method for Spiked Fish, Crustaceans and Shellfish Samples, as well as Human Stool Specimens
Fresh fish (Parabramis pekinensis), crustaceans (Litopenaeus vannamei), and shellfish (Perna viridis) were sampled from a local food market (30°53′7.16′′N, 121°54′48.39′′E) in Shanghai, China. Fresh meat of the aquatic products without skin was collected with sterile surgical scalpels, and then homogenated (Yamazaki, 2011). The resulting homogenates were plated onto the TCBS agar plates. Only the sample homogenates that were detected negative for V. cholerae and the virulence-associated genes were used in the following spiked experiments.
The spiked fish, crustaceans and shellfish samples, as well as human stool specimens were prepared according to the method described previously (Yamazaki, 2011) with minor modifications. Briefly, 1 g (wet weight) of fresh sample was individually added into 9 mL alkaline peptone water (APW, pH 8.5, 3% NaCl, Land Bridge, Beijing, China) and homogenized thoroughly. V. cholerae strains GIM1.449, Q07-21, and L10-36 were individually inoculated into 5 mL TSB (pH 8.5, 3% NaCl) broth and incubated at 37°C for 16–18 h. Serial 10-fold dilutions of V. cholerae culture were prepared, and bacterial cells were calculated by plating counting method as described above, and meanwhile, a 100 μL of each dilution was spiked into 900 μL of the fresh homogenate and mixed well. Then, two microliters of 10-fold dilution of the mixture was used for the sssvLAMP method targeting the hapA, mshA, pilA, and tlh genes, while one microliter was used for the PCR assay.
PCR Assay
Oligonucleotide primers used for the PCR assay were listed in Table 2. A 20 μL of the PCR reaction solution contained 8 μL of DNase/RNase-Free Deionized Water (Tiangen Beijing, China), 10 μL of 2 × Taq Master Mix (Novoprotein Technology Co., Ltd., Shanghai, China), 0.5 μL of each primer and 1 μL of bacterial culture or genomic DNA template. PCR reaction was performed for 30 cycles, each of which consisted of denaturation at 94°C for 1 min, annealing at 52–62°C for 1 min and extension at 72°C for 1 min. The annealing temperatures and elongation times were based on melting temperatures of primer pairs and the predicted lengths of PCR products. PCR reactions were performed in a Mastercycler® pro PCR thermal cycler (Eppendorf, Hamburg, Germany). Amplicons were analyzed by agarose gel electrophoresis, then visualized and recorded as described above.
Sample Collection and Analysis by the sssvLAMP Method
Water samples were collected from various sources in June of 2019 in Shanghai, China, including mineral water, spring water, tap water, river water, lake water, and sea water along the East China Sea (Table 4). The latter three types of water samples (n = 3 per type) were collected from the surface water layers (<30 cm) as described previously (He et al., 2015). The commonly consumed mineral water (n = 3), and spring water (n = 3) samples were purchased from local food markets in Shanghai, while the tap water samples (n = 3) were collected from the tap water system available throughout the city. Commonly consumed freshwater fish (Aristichthys nobilis, Carassius auratus, Ctenopharyngodon idellus, and P. pekinensis) samples (n = 3 per fish species, >500 g/sample) were collected from the local food markets as described above. Health huaman stool samples were provided by M. Yang in Shanghai Ocean University. All samples were maintained at 4°C and analyzed immediately after transported to the laboratory in Shanghai Ocean University, Shanghai, China. Bacterial cells of each 500 mL of water samples (>1.5 L/sample) were filtered through polycarbonate membranes with 0.22-μm pore size (47 mm diameter, Millipore, Corcaigh, Ireland). Consequently, each membrane was washed with 10 mL of sterile 1 × PBS. After centrifugation, bacterial cell pellet was resuspended with 1 mL of the 1 × PBS, and 2 μL of which was used as the DNA template for the sssvLAMP as described above. Fish meat and intestinal samples, as well as human stool specimens were homogenated according to the methods described above. All tests in this study were conducted in triplicate.
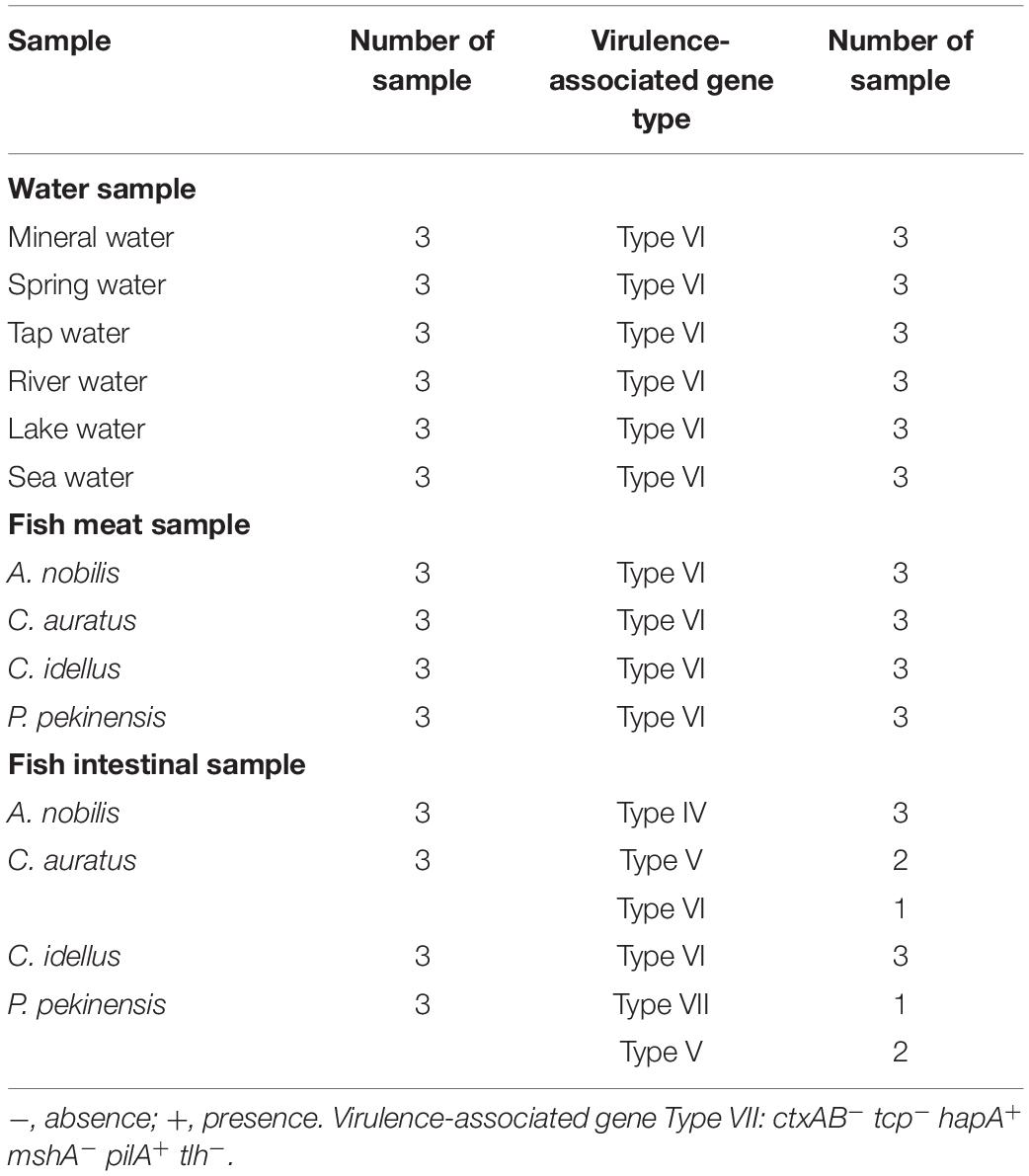
Table 4. The genetic diversity of virulence-associated genes of V. cholerae in water and fish samples by the sssvLAMP method.
Results
Reaction Parameters Optimized for the sssvLAMP Method
Optimal Concentration of Outer Primers
To determine the optimal concentration of outer primers for the sssvLAMP method, different concentrations of the F3 and B3 primers (0.05–0.40 μM) were evaluated, while 1.6 μM of inner primers, and 0.8 μM of loop primers were set in reaction systems according to previous reports. The results are illustrated in Figure 1 and Table 3. The color of all the reaction solutions containing 0.05–0.40 μM of the outer primers were observed to change from the original orange to light green by the naked eye under the visible light (Figure 1A, r1: tubes 2–9), and also to display bright green fluorescence under the UV light (Figure 1B, r1: tubes 2–9), which indicated positive amplicons of the target gene (mshA) in these reaction tubes. Moreover, no obvious difference in color or fluorescence was found among these positive reactions. These results were confirmed by the agarose gel electrophoresis analysis, which showed characteristic ladder-like DNA patterns (Figure 1C, lanes 2–9). In contrast, the reaction solution without the outer primers was in orange (Figure 1A, tube 1), and showed no green fluorescence under the UV light (Figure 1B, tube 1), indicating no amplification of the target gene. The negative result was also confirmed by the electrophoresis analysis, on which no characteristic ladder-like DNA pattern was observed (Figure 1C, lane 1). Thus, for a cost-effective purpose, the 0.05 μM of the outer primers were chosen for the sssvLAMP method in the further analysis.
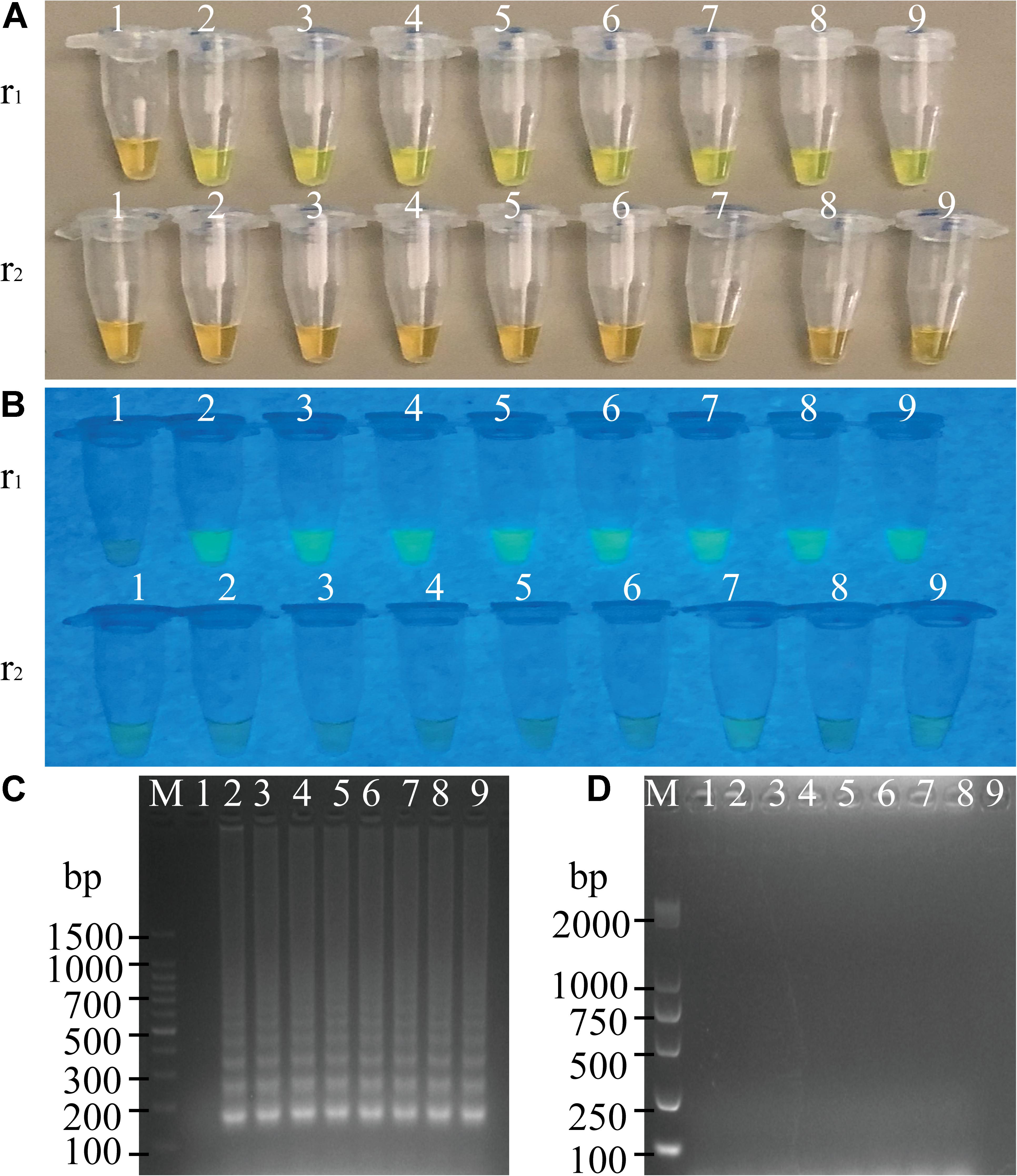
Figure 1. Optimization of the outer primers (F3 and B3) of the sssvLAMP method. (A–D) The results observed by the naked eye under the visible light (A) and the UV light (302 nm) (B), and verified by 2% agarose gel electrophoresis analysis (C: positive results; D: negative results). r1 and r2: with and without DNA templates, respectively. Lane M: DNA molecular weight marker (100 bp). Tubes/Lanes 1–9: containing 0.00, 0.05, 0.10, 0.15, 0.20, 0.25, 0.30, 0.35, and 0.40 μM of the F3 and B3 primers, respectively.
Optimal Concentration of Inner Primers
Different concentrations (0.4, 0.8, 1.2, 1.6, 2.0, and 2.4 μM) of the inner primers were examined in the sssvLAMP reaction system (Table 3). The target gene was amplified from all reaction solutions supplemented with the FIP and BIP primers, based on the observation of the color change and green fluorescence under the visible light and UV light, respectively (Figures not shown). Nevertheless, the amplification with 0.4 μM of the inner primers yielded weaker ladder-like DNA patterns than those with the other concentrations, as confirmed by the agarose gel electrophoresis analysis (Figures not shown). Moreover, the concentration of 1.6 μM primers produced the strongest DNA bands (Figures not shown). Therefore, the 1.6 μM of the inner primers was chosen for the sssvLAMP method in the further analysis.
Optimal Concentration of Loop Primers
As summarized in Table 3, different concentrations (0.2–1.2 μM) of the loop primers were evaluated for the sssvLAMP method, in which 0.05 μM of the outer primers and 1.6 μM of the inner primers were involved. The target gene was amplified from all the reaction tubes containing the 0.2–1.2 μM of the LF and LB primers, showing the color change, production of bright green fluorescence and characteristic ladder-like DNA patterns (Figures not shown). Moreover, no observed difference was found among the positive reactions. Thus, the 0.2 μM of outer primers were chosen for the sssvLAMP method. The optimal proportion among the outer, inner, and loop primers was 1:32:4 (μM/μM/μM).
Optimal Concentration of Mg2+
The optimal concentration of Mg2+ for the sssvLAMP method was determined, and the results were shown in Table 3. The color of reaction solutions containing 6.0–12.0 mM of Mg2+ was all observed to change from orange to light green, which indicated positive amplification of the target gene (Figures not shown). These results were confirmed by the agarose gel electrophoresis analysis, which showed characteristic ladder-like DNA patterns (Figures not shown). In contrast, no color change was observed in the reaction solutions containing 4.0 and 14.0 mM of Mg2+, respectively, while only weak ladder-like DNA pattern was in the tube with 14.0 mM Mg2+ (Figures not shown). Thus, the concentration of 6 mM Mg2+ was chosen for the sssvLAMP method.
Optimal Concentration of dNTP
As presented in Table 3, the optimal concentration of dNTP was examined for the sssvLAMP method. All the concentrations of dNTP (1.0–2.0 mM) led to the same color change typical for positive results (Figures not shown), consistent with the results yielded from the electrophoresis analysis (Figures not shown). Thus, the minimum concentration of dNTP (1.0 mM) was chosen in the further analysis.
Optimal Unit of Bst DNA Polymerase
As shown in Table 3, Bst DNA polymerase was also tested in the sssvLAMP system. A weak amplification of the target gene was observed in the reaction solution containing 2 Unit of Bst DNA polymerase (Figures not shown). All the other reaction solutions with 4–16 Unit enzyme were in green, indicating positive amplicons in these reaction tubes (Figures not shown). Moreover, the reaction tube with 8 U enzyme showed the strongest ladder-like DNA pattern (Figures not shown). Therefore, the 8 U of Bst DNA polymerase was chosen for the sssvLAMP method.
Optimal Reaction Temperature
To determine the optimal reaction temperature for the sssvLAMP method, different temperatures (50–70°C) were tested, and the results were presented in Table 3. The typical color change was observed in the reaction tubes when incubated at 50, 55, 60, and 65°C, respectively, whereas no color change was found in the tube incubated at 68 and 70°C (Figures not shown). The results were consistant with those by agarose gel electrophoresis analysis, except that only weak ladder-like DNA pattern was observed in the reaction tube at 68°C (Figures not shown). The sssvLAMP appeared to be completely inhibited at 70°C. The reaction temperature of 65°C was chosen for the further analysis, given the manufacturer’s suggestion for the Bst DNA polymerase.
Optimal Reaction Time and Utility of the Loop Primers
Variation of reaction times (30–80 min) was also tested, and the results were shown in Table 3. Without the loop primers, a light green color was observed in the tube after the LAMP reaction was performed for 30 min (Figures not shown). However, the sssvLAMP produced bright green fluorescence of good intensity for positive reaction tubes after incubated for 40, 50, 60, 70, and 80 min, respectively (Figures not shown), in congruence with the visualization of strong bands of DNA amplicons by gel electrophoresis was observed, and there was no obvious difference of fluorescence intensity due to variable reaction time (40–80 min).
The usefulness of reaction time reduction by addition of the loop primers was observed. As illustrated in Figure 2, the reaction solution without the loop primers turned light green in color after the reaction was performed for 50 min (Figures 2A,B, tube 5), but those containing the LF and LB primers (0.2 μM) turned light green at 40 min (Figures 2A,B, tube 4). The results by the electrophoresis analysis indicated that there was a weak amplification in the reaction tube with the loop primers at 30 min (Figure 2C, lane 2). These results indicated that the loop primers increased the amplification efficiency and shorten reaction time in the sssvLAMP method.
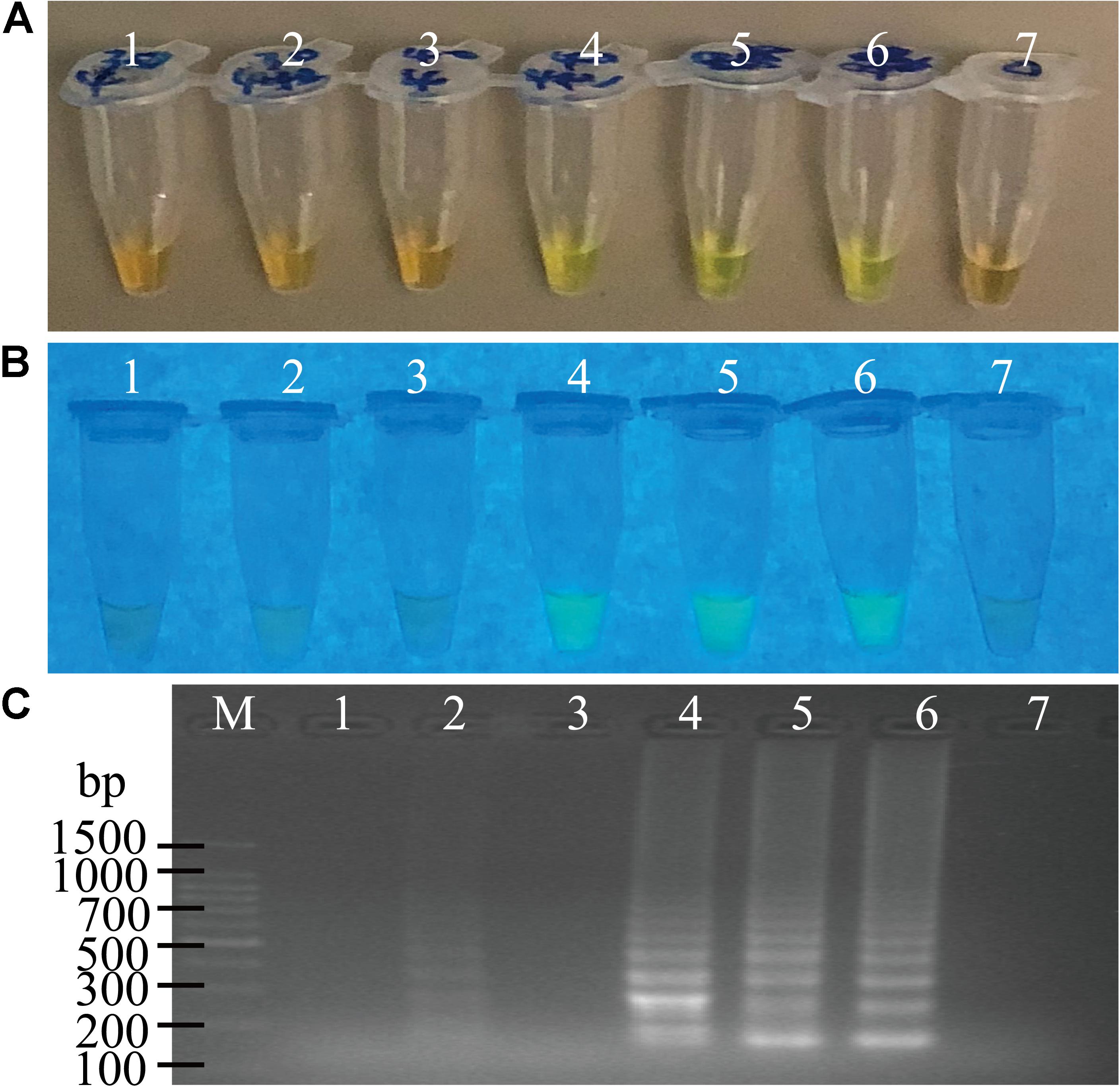
Figure 2. The function of the loop primers (LF and LB) in the sssvLAMP method. (A–C) The results observed by the naked eye under the visible light (A) and the UV light (302 nm) (B), and verified by 2% agarose gel electrophoresis analysis (C). Lane M: DNA molecular weight Marker (100 bp). Tubes/Lanes 2, 4, 6: reaction mixture with the loop primers at 65°C for 30, 40, 50 min, respectively, while Tubes/Lanes 1, 3, 5: reaction mixture without the loop primers. Tube/Lane 7: negative control.
Taken together, the optimal parameters of the sssvLAMP method included 1.6 μM of FIP and BIP primers, 0.05 μM of F3 and B3 primers, 0.20 μM of LF and LB primers, 6 mM Mg2+, 1.0 mM dNTP, and 8 U of Bst DNA polymerase. The LAMP reaction was performed at 65°C for 40 min with 0.2 μM of the loop primers (Table 3).
Specificity of the sssvLAMP Method
To evaluate specificity of the sssvLAMP method developed in this study, we firstly examined the inclusivity of the method with the 52 V. cholerae strains (Table 1). The results indicated that all V. cholerae strains were detected positive for the species-specific lolB gene. Nevertheless, diversified variation in the occurrence of virulence-associated genes (Types I to VII) was observed. V. cholerae strains were mostly detected by the presence of hapA and tlh alone or in combination with other genes (mshA, pilA), but the absence of the ctxA and tcpA genes, except that V. cholerae ATCC39315 (N16961) was detected positive for the latter two toxigenic genes (Figures not shown) (Table 1). Consequentially, the exclusivity of the sssvLAMP method was tested with the 50 bacterial strains (Table 1). No positive amplification targeting any of the target genes was recorded in the reaction tubes containing DNA templates of the closely related Vibrio species tested (n = 16), including V. fluvialis, V. harvey, V. metschnikovii, V. mimicus, V. parahemolyticus, and V. vulnificus, based on the observed origin color, no green fluorescence and no characteristic ladder-like DNA patterns (e.g., Figures 3, 7). Also, no positive amplification was observed from non-Vibrio species (n = 34), including the common bacterial pathogens tested, indicating high specificity of the sssvLAMP method targeting the virulence-associated genes of V. cholerae.
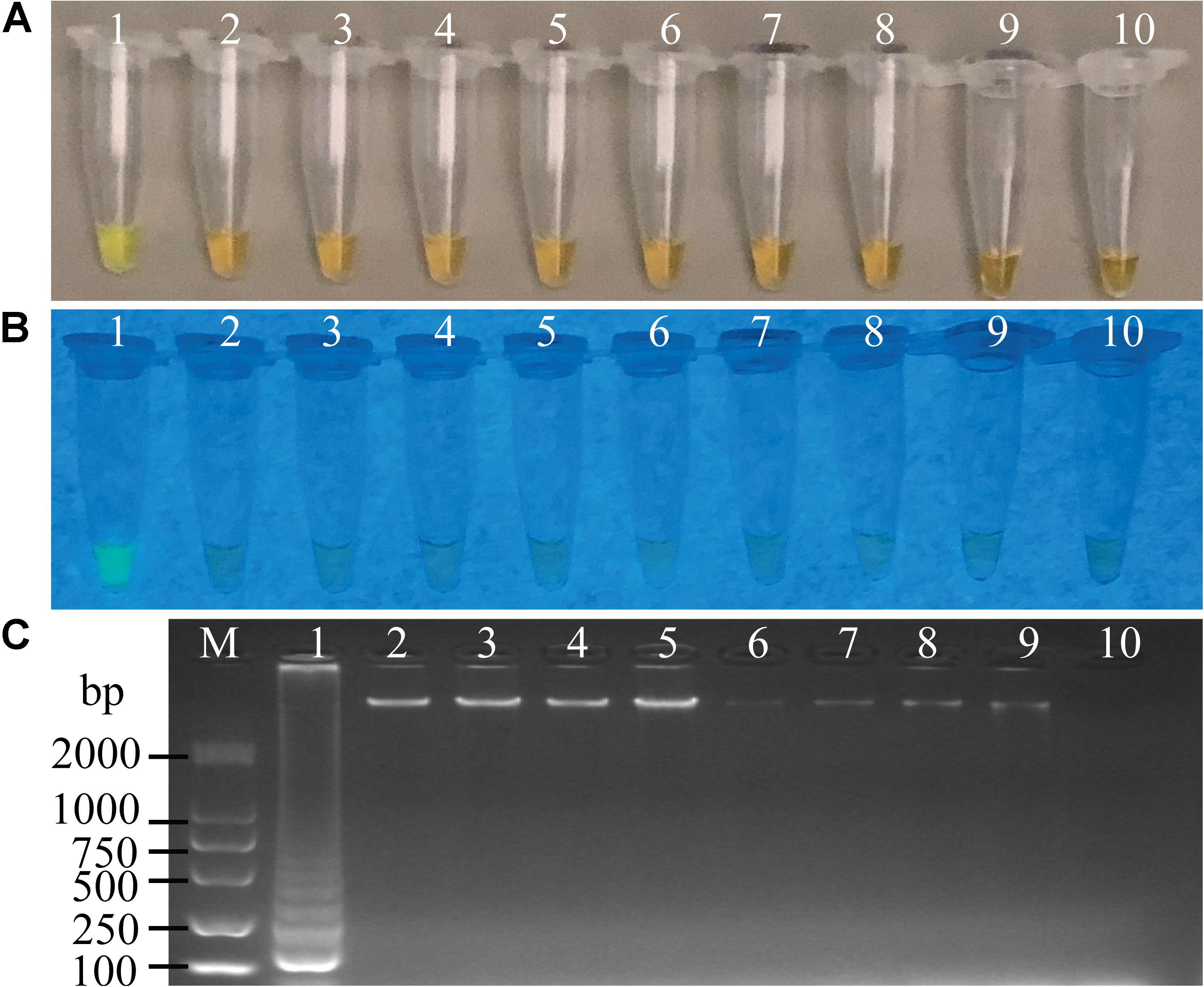
Figure 3. Specificity of the sssvLAMP method for the detection of the hapA gene in some bacterial strains. (A–C) The results observed by the naked eye under the visible light (A) and the UV light (302 nm) (B), and verified by 2% agarose gel electrophoresis analysis (C). Lane M: DNA molecular weight Marker (100 bp). Tubes/Lanes 1–10: containing genomic DNA of V. cholerae GIM 1.449, 216.79 ng/μL; V. parahemolyticus ATCC 33847, 226.93 ng/μL; V. vulnificus, 421.14 ng/μL; V. alginolyticus, 225.90 ng/μL; E. coli ATCC25922, 94.45 ng/μL; S. aureus, 94.09 ng/μL; A. hydrophila, 10.18 ng/μL; Salmonella spp., 78.54 ng/μL; E. cloacae, 132.28 ng/μL; negative control, respectively.
LOD of the sssvLAMP Method for Cell Culture and Genomic DNA of V. cholerae Strains
Sensitivity of the sssvLAMP method was determined for the detection of the 52 V. cholerae strains (Table 5). For instance, for the target gene hapA, V. cholerae GIM 1.449 cells ranged from 9.3 × 107 to 9.3 CFU/mL were tested in the reaction tubes. Cell cultures (5.6 × 108–1.25 CFU/mL) of the other 23 V. cholerae strains carrying the hapA genes were examined as well. The positive results with the typical color change, production of the bright green fluorescence, and forming of characteristic ladder-like DNA pattern showed that the sensitivity ranged from 1.3 × 10–1 to 92 CFU/reaction for 91.3% of the tested V. cholerae strains (e.g., Figure 4). Similarly, for the target gene mshA, the 3 V. cholerae strains (mshA+) were tested with 1.09 × 108–1.17 CFU/mL cells, and the results indicated that the LOD ranged from 1.5 × 10–1 to 28 CFU/reaction (Figures not shown) (Table 5). Likewise, the LOD values for the pilA and tlh genes ranged from 1.1 × 10–1 to 33 CFU/reaction (100% strains tested) and from 1.7 × 10–1 to 70 CFU/reaction (87% strains tested), respectively, when cell cultures ranged from 1.28 × 108 to 1.04 CFU/mL of 3 V. cholerae (pilA+) and from 3.5 × 107 to 1.01 CFU/mL of 23 V. cholerae (tlh+) strains were tested, respectively (Figures not shown) (Table 5).
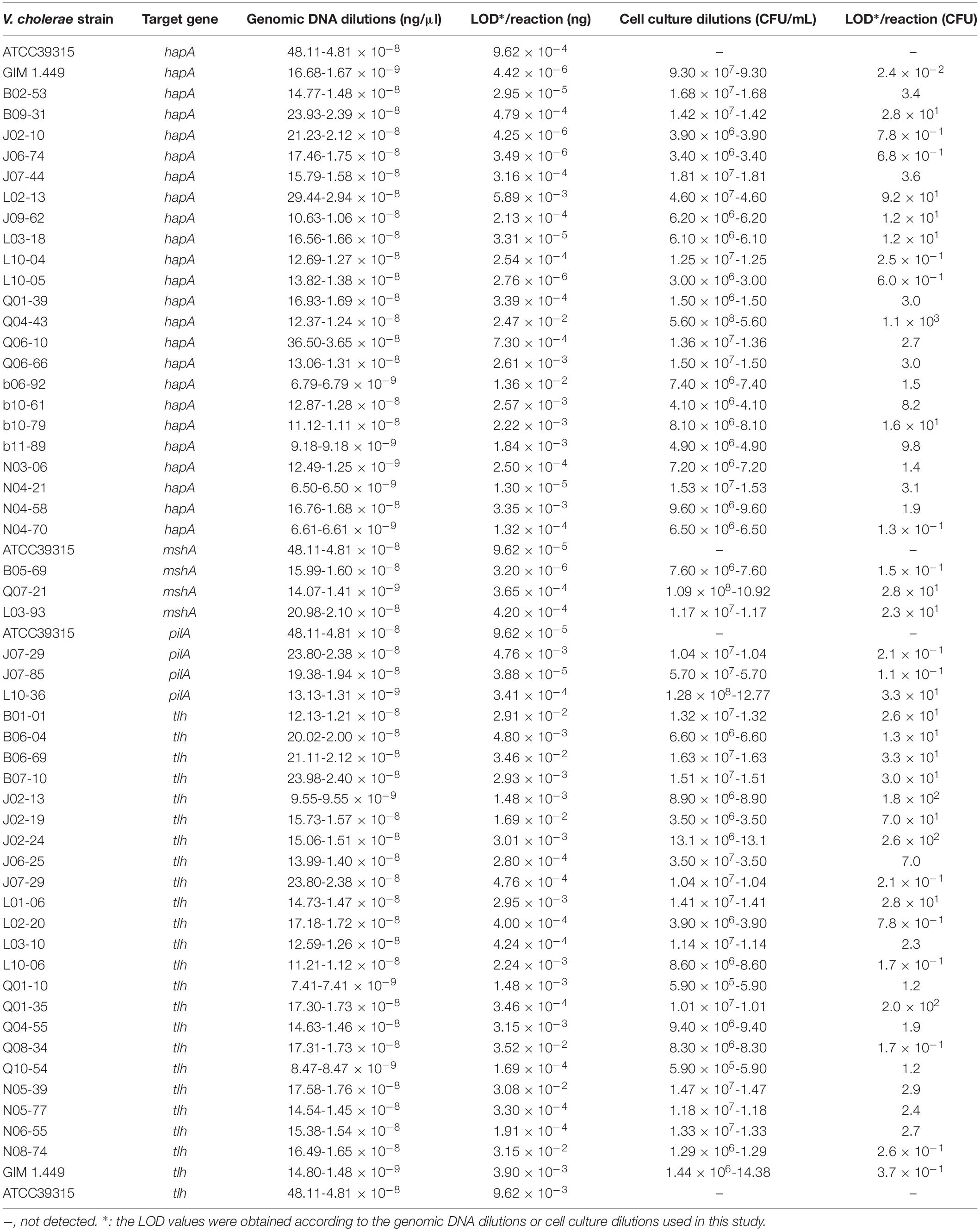
Table 5. Sensitivity of the sssvLAMP method for the detection of genomic DNA and cell cultures of V. cholerae strains.
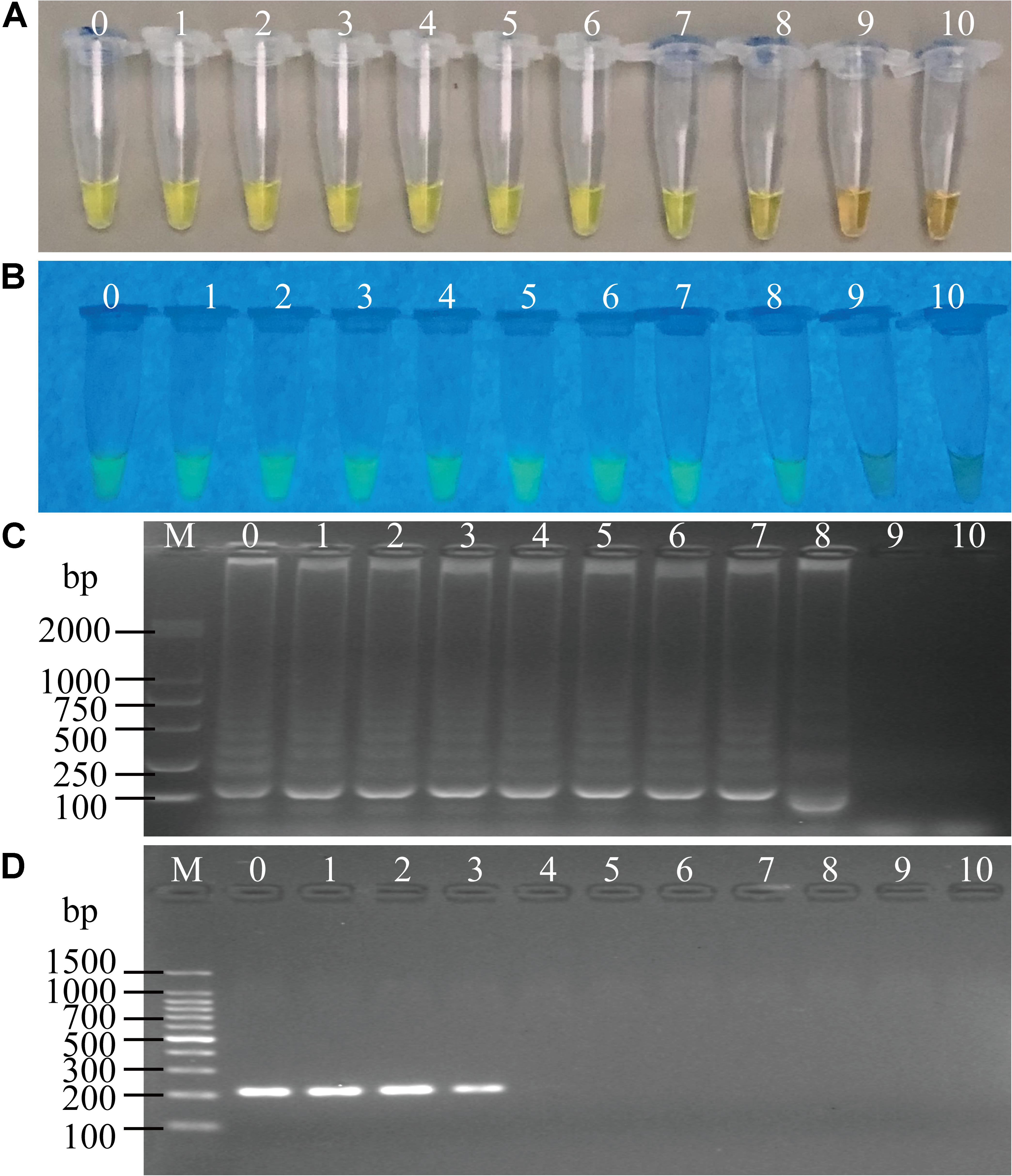
Figure 4. Sensitivity of the sssvLAMP method for the detection of the hapA gene of V. cholerae cell culture. (A–C) The results observed by the naked eye under the visible light (A) and the UV light (302 nm) (B), and verified by 2% agarose gel electrophoresis analysis (C) by the sssvLAMP method. D: the results by the PCR assay. Lane M: DNA molecular weight Marker (100 bp). Tubes/Lanes 0–9: containing 9.3 × 107–9.3 × 10–2 CFU/mL of V. cholerae GIM 1.449 cells, respectively; 10: negative control.
The LOD of the sssvLAMP method for genomic DNA of the 52 V. cholerae strains was also determined. For instance, for the target gene hapA, genomic DNA ranged from 6.50 to 9.18 × 10–9 ng/μL of 24 V. cholerae (hapA+) strains was tested, and the results indicated that the sensitivity of the sssvLAMP method ranged from 1.36 × 10–2 to 4.42 × 10–6 ng/reaction (Figures not shown). Similarly, for the target genes mshA, pilA, and tlh, the LOD values ranged from 3.65 × 10–4 to 3.20 × 10–6 ng/reaction, from 4.76 × 10–3 to 9.62 × 10–5 ng/reaction, and from 1.69 × 10–2 to 4.76 × 10–4 ng/reaction, respectively, when 3.20-1.41 × 10–9 ng/μL, 13.13-4.81 × 10–8 ng/μL, and 8.47-9.55 × 10–9 ng/μL genomic DNA samples of V. cholerae strains (mshA+, pilA+, and tlh+) were tested in the sssvLAMP method, respectively (e.g., Figure 5) (Table 5). These results also indicated strain-dependent variation in LOD (CFU versus genomic DNA of V. cholerae).
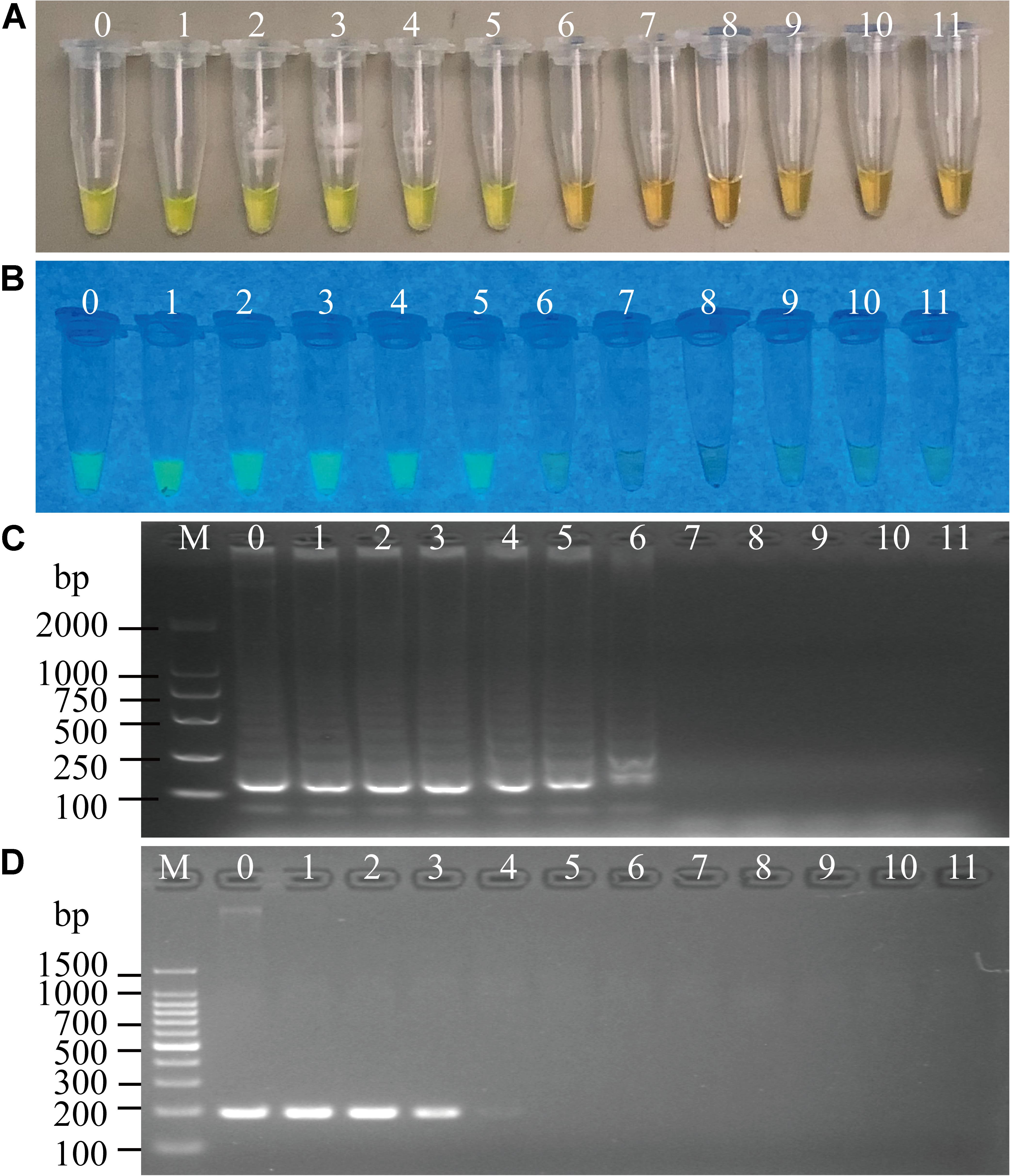
Figure 5. Sensitivity of the sssvLAMP method for the detection of the mshA gene of V. cholerae genomic DNA. (A–C) The results observed by the naked eye under the visible light (A) and the UV light (302 nm) (B), and verified by 2% agarose gel electrophoresis analysis (C) by the sssvLAMP method. (D) The results by the PCR assay. Lane M: DNA molecular weight Marker (100 bp). Tubes/Lanes 0–10: containing 14.07 ng/μL –14.07 × 10–10 ng/μL of V. cholerae Q07-21 genomic DNA; 11: negative control.
Comparison of the sensitivity by the routine PCR assay. For example, as shown in Figure 4D, the LOD of hapA gene by the PCR assay was 6.0 × 104CFU/mL and 1.1 × 10–3 ng DNA/μL for the detection of V. cholerae GIM 1.449 cells and genomic DNA samples, respectively, which indicated that the sssvLAMP method was 6.45 × 104-, and 6.50 × 103-fold more sensitive than the PCR assay. Similarly, for the target gene mshA in V. cholerae Q07-21, our data showed the LOD of 7.10 × 105 CFU/mL, and 9.15 ng DNA/μL by the PCR, which were 650.18, and 650.32-fold lower than the sssvLAMP method (Figures not shown). Likewise, the LOD of the pilA gene in V. cholerae L10-36 by the PCR assay was 8.30 × 104 CFU/mL, and 8.53 × 10–1 ng DNA/μL, respectively, which were 65-fold lower than LAMP assays, while those of the tlh gene in V. cholerae GIM 1.449 were 9.35 × 104CFU/mL, and 9.62 × 10–1 ng DNA/μL respectively, indicating that the sssvLAMP method was 6.50 × 103, and 6.41 × 103-fold more sensitive than the PCR assay, respectively (e.g., Figure 5).
Taken together, for the detection of V. cholerae cells and genomic DNA samples in water, the sensitivity of the sssvLAMP method for the virulence-associated genes were 6.50 × 101-6.45 × 104, and 6.50 × 101-6.50 × 103-fold sensitive than the PCR assay for these tested V. cholerae strains, respectively.
Sensitivity of the sssvLAMP Method for Spiked Fish, Shrimp and Shellfish Samples, as Well as Human Stool Specimens
Cell cultures of V. cholerae strains GIM1.449, Q07-21 and L10-36 were added into P. pekinensis homogenates, respectively, and the sensitivity of the sssvLAMP Method was determined. The observed LOD values ranged from 0.2 to 250 CFU/reaction of V. cholerae, which were 6.5 × 103, 6.5 × 102, 11.50, and 6.5 × 102-fold higher than those of the PCR assay for targeting the hapA, mshA, pilA, and tlh genes, respectively (e.g., Figure 6). Additionally, each of the three strains was spiked into L. vannamei, and P. viridis homogenates, respectively, and LOD values for the four target genes were evaluated. The resulting data showed that the sssvLAMP Method was more sensitive for the L. vannamei matrix (LOD: 0.59–8.7 CFU/reaction) than for the P. viridis matrix (LOD: 8.7–590 CFU/reaction) (Table 6). Additionally, health human stool specimens were spiked with each of the V. cholerae GIM1.449, Q07-21 and L10-36 strains, and the sensitivity of the sssvLAMP method was also tested. As shown in Table 6, the LOD values ranged from 0.59 to 87 CFU/reaction targeting the four virulence-associated genes tested, suggesting application potential in the detection of human clinical diarrhea patient samples in the future.
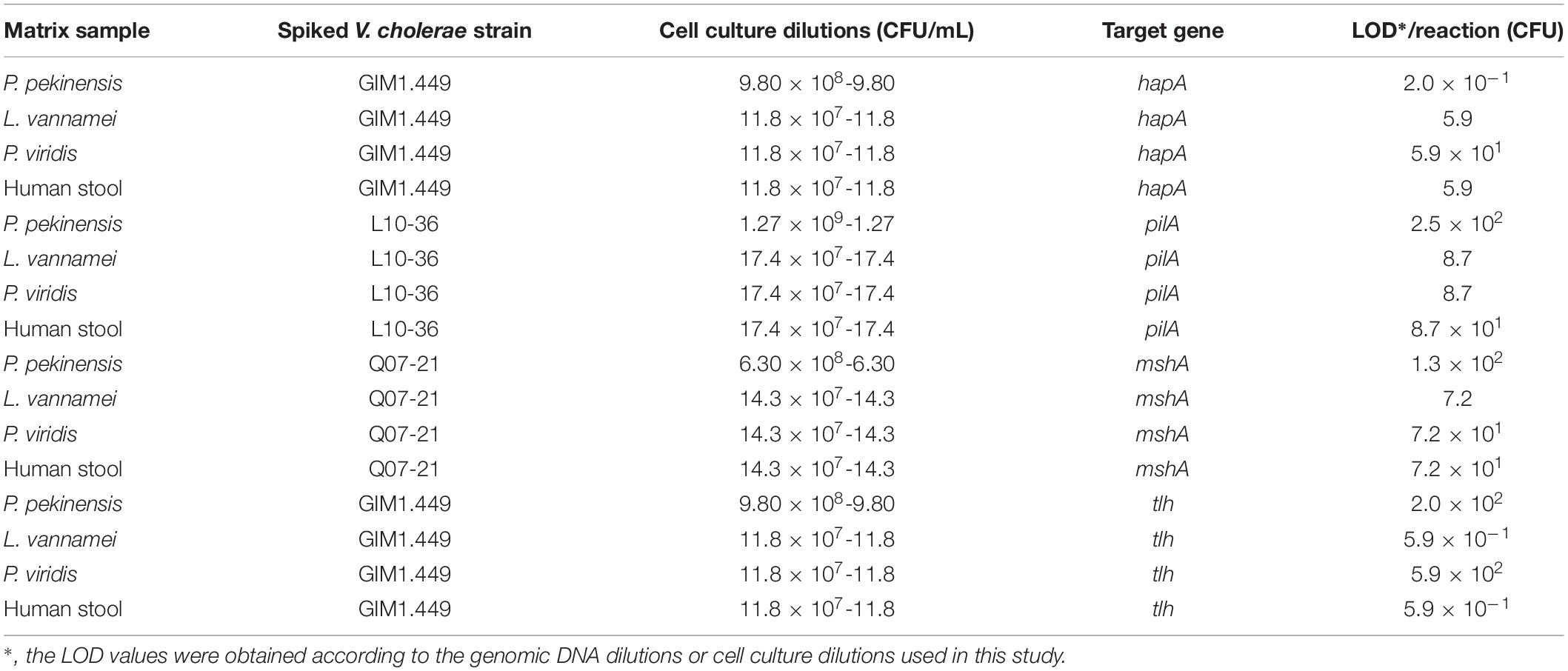
Table 6. Sensitivity of the sssvLAMP method for spiked aquatic product samples and human stool specimens.
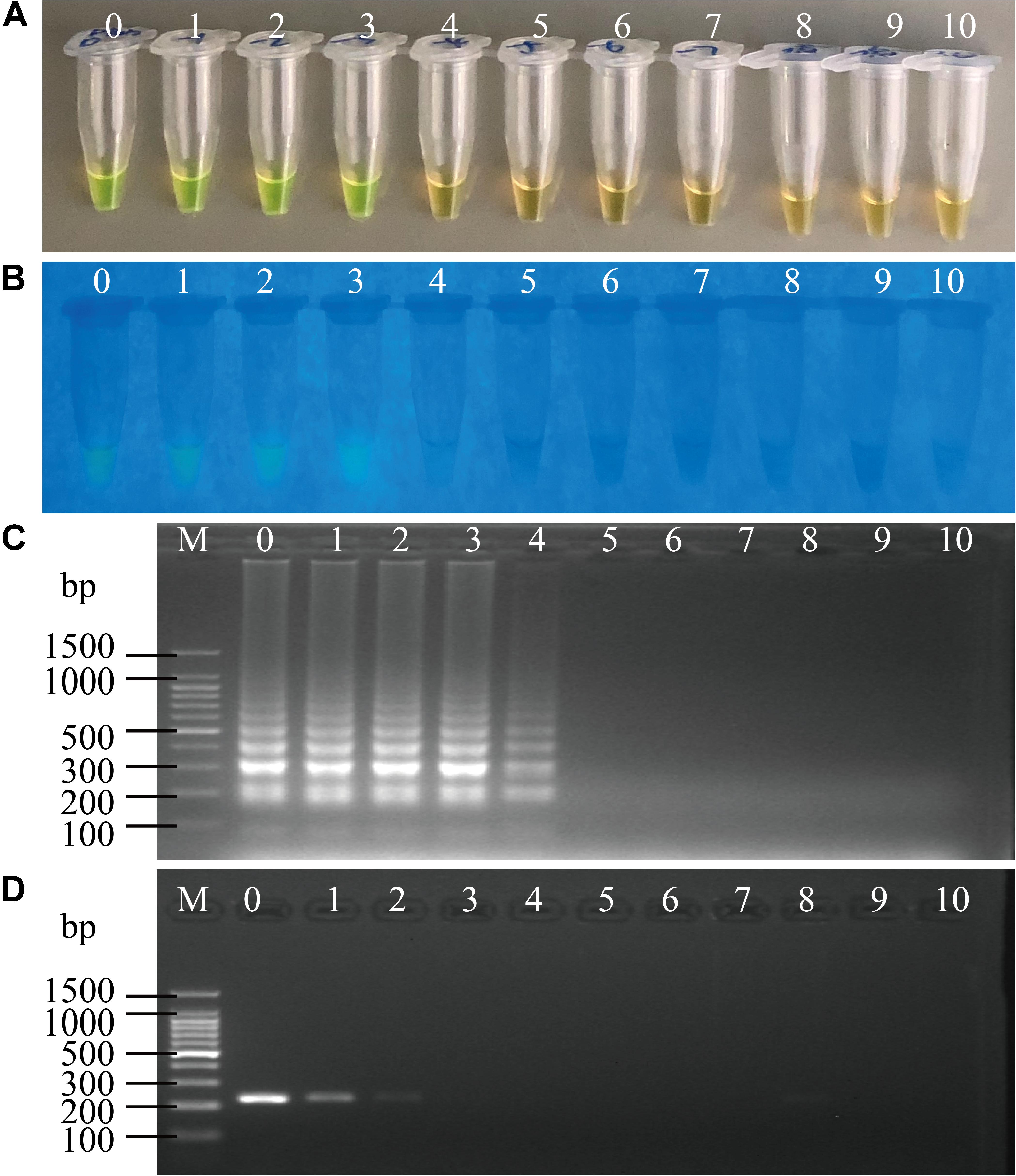
Figure 6. Sensitivity of the sssvLAMP method for the detection of the pilA gene in the spiked fish matrix samples. (A–C) the results observed by the naked eye under the visible light (A) and the UV light (302 nm) (B), and verified by 2% agarose gel electrophoresis analysis (C) by the sssvLAMP method. (D) The results by the PCR assay. Lane M: DNA molecular weight Marker (100 bp). Tubes/Lanes 0–9: 4.85 × 106-4.85 × 10–3 CFU/Ml of Vibrio cholerae L10-36 cells; 10: negative control.
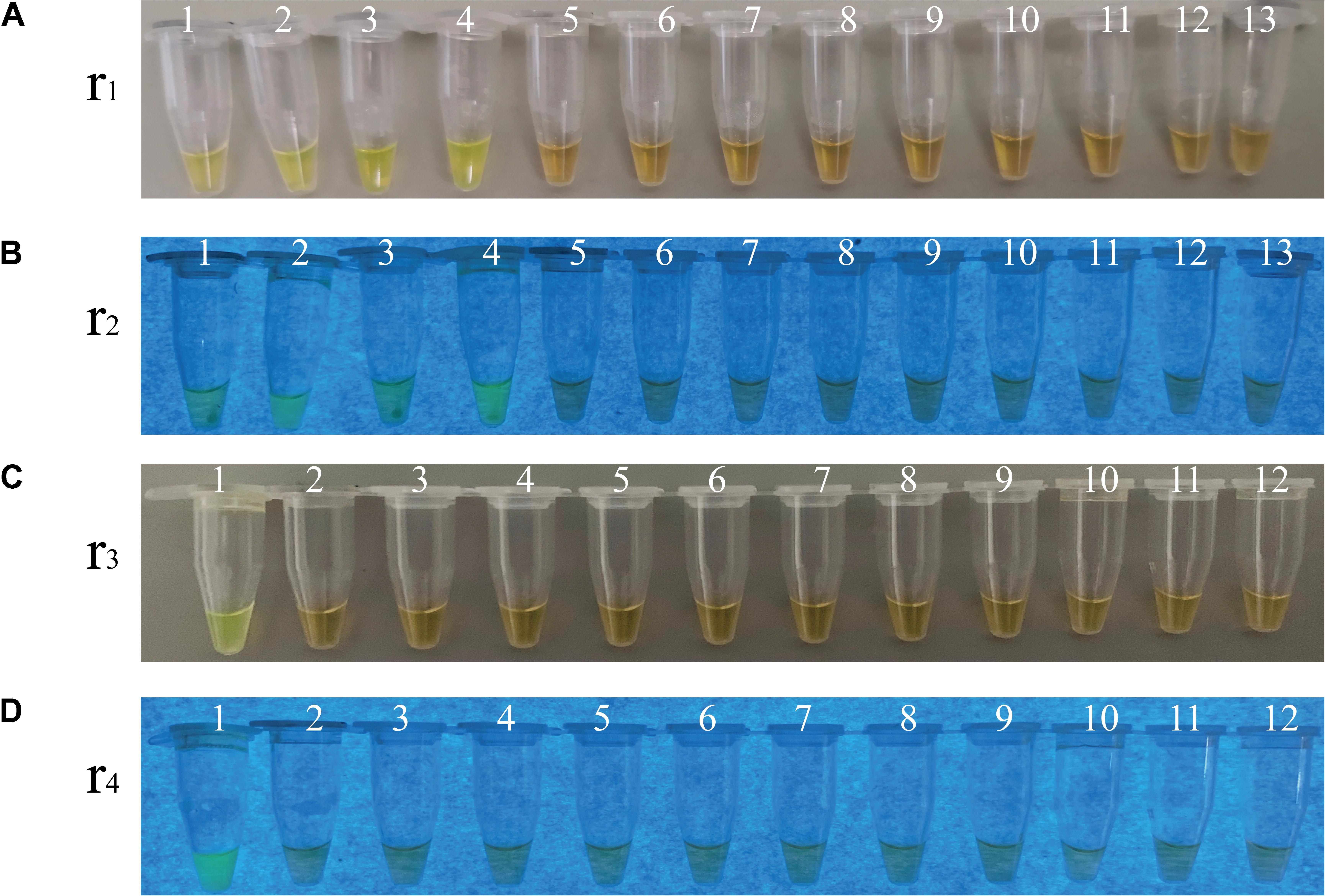
Figure 7. Specificity of the sssvLAMP method for the detection of the ctxA, tcpA, lolB, and hapA genes in some bacterial strains. (A–D) The results observed by the naked eye under the visible light (A,C) and the UV light (302 nm) (B,D). Tubes 1–13 (A,B): containing genomic DNA of V. cholerae ATCC39315 (N16961); V. cholerae ATCC39315 (N16961); V. cholerae ATCC39315 (N16961); V. cholerae GIM1.449; V. fluvialis ATCC33809; V. harvey ATCC BAA-1117; V. harveyi ATCC33842; V. mimicus bio-56759; V. metschnikovii ATCC 700040; V. parahemolyticus ATCC17802; V. parahemolyticus ATCC33847; V. vulnificus ATCC27562; and negative control, respectively. Tubes 1-12 (C,D): containing genomic DNA of V. cholerae GIM1.449 (lolB); A. hydrophila ATCC35654; E. cloacae ATCC13047; E. coli ATCC8739; E. sakazakii CMCC45401; L. monocytogenes ATCC19115; P. aeruginosa ATCC9027; S. enterica subsp. Enterica Leminor et popoff ATCC13312; S. typhimurium ATCC15611; S. flexneri ATCC12022; V. alginolyticus ATCC33787; and negative control, respectively. All the tubes contained the LAMP primers targeting the hapA gene, except the tubes 1 and 2 (A,B), and tube 1 (C,D) targeting the ctxA, tcpA, and lolB genes, respectively. The highest genomic DNA concentrations given in Table 5 were used in the specificity assays.
Reproducibility of the sssvLAMP Method
For all target genes, all positive results were repeated in all the experiments performed not only for V. cholerae cells or genomic DNA samples, but also for the spiked fish samples within the LOD ranges, indicating high reproductivity (100%) of the sssvLAMP method developed in this study (Figures not shown).
Detection of Water and Fish Samples by the sssvLAMP Method
Eighteen water samples collected from various sources in June of 2019 in Shanghai, China were examined by the sssvLAMP method for the target genes of V. cholerae. As shown in Table 4, all the water samples were negative for the targeted genes, indicating the drinking water and environmental water samples tested were negative for potential pathogenic V. cholerae. Additionally, four commonly consumed fish species samples (A. nobilis, C. auratus, C. idellus, and P. pekinensis) were also examined by the sssvLAMP method. The results showed that all the fish meat samples were negative for the targeted genes. However, among the twelve fish intestinal samples, seven were positive for hapA, pilA, and/or tlh genes. Although only a small number of samples were detected, variable virulence-associated gene profiles were observed among intestinal samples of the fish species (Table 4). The hapA and tlh genes were detected in the A. nobilis intestinal samples, while the former gene also existed in the C. auratus and P. pekinensis intestines. The pilA gene was only present in P. pekinensis intestines, whereas all the virulence-associated genes were absent from C. idellus intestinal samples. The results were confirmed by the PCR assay and the conventional isolation and identification assay of V. cholerae from the fish intestinal samples.
Discussion
The development of rapid, sensitive, and cost-effective methods is needed not only for the quick and appropriate treatment of patients infected with pathogenic Vibrio strains, but also for the prompt control and prevention of the diseases (Okada et al., 2010). To date, only few studies have been conducted on the detection of V. cholerae genes by the LAMP technique (He et al., 2009; Okada et al., 2010; Srisuk et al., 2010; Tourlousse et al., 2012; Liew et al., 2015; Zhang et al., 2015). In this study, an sssvLAMP method was successfully developed for the detection of virulence-associated genes ctxA, tcpA, hapA, mshA, pilA, and tlh of V. cholerae, as well as species-specific gene lolB of the bacterium.
The magnesium is required for the LAMP reaction, however, the lower or higher concentration of Mg2+ strongly influenced the LAMP reaction and its product. For instance, the excessive Mg2+ stabilizes the mispriming of primers that decrease the specificity of the assay, and also prevents the denaturation of the DNA double strands by stabilizing the duplex structure (Markoulatos et al., 2002). In contrast, lower concentrations of Mg2+ fail to amplify the target genes. In this study, the optimal concentration of Mg2+ was 6.0 mM for the sssvLAMP method, which was consistent with previous studies on the detection of ctxA gene of V. cholerae (Engku Nur Syafirah et al., 2018), dengue viruses (Kim et al., 2018), and Mycoplasma hyopneumoniae (Liu et al., 2015), but less than those (8 mM) for the detection of ctxA and ompW genes of V. cholerae (Okada et al., 2010; Srisuk et al., 2010).
The dNTP is essential for the synthesis of double-stranded DNAs in the nucleic acid amplification in vitro. In this study, the LAMP amplification was observed in all reaction tubes containing 1.0–2.0 mM dNTP, and the concentration of 1.0 mM was chosen for the sssvLAMP method, which was less than some previous reports (Goto et al., 2009; Srisuk et al., 2010). The minimum usage of dNTP reduces the cost of the method, particularly when a large number of samples need to be analyzed.
In the LAMP system, the autocycling and strand displacement of DNA synthesis were mediated by the Bst DNA polymerase and four specially designed primers (Notomi et al., 2000). It has been reported that the LAMP reaction could be accelerated by the addition of the loop primers (Nagamine et al., 2002; Lim et al., 2013). In this study, our results indicated that positive amplicons were initially detected at a reaction time of 30 min with the loop primers, which was 20-min faster than those without the loop primers. The significance of optimal concentrations of each pair of primers in the LAMP system has been pointed out in previous studies (Okada et al., 2010; Srisuk et al., 2010; Lau et al., 2011; Kakizaki et al., 2018), in which a 1.6 μM of the inner primers, 0.2 μM of the outer primers, and 0.8 μM of the loop primers was often used with an optimal ratio 8:1:4 (μM/μM/μM) of the three primers. In this study, our data showed that 1.60 μM of inner primers, 0.05 μM of the outer primers, and 0.2 μM of the loop primers were optimum for the sssvLAMP method with a final ratio of 32:1:4 (μM/μM/μM), showing fourfold less amount of the outer and loop primers. The elongation reactions in the LAMP system are sequentially repeated by Bst DNA polymerase-mediated strand-displacement synthesis (Notomi et al., 2000, 2015). In this study, an 8 U concentration of Bst DNA polymerase was used in the sssvLAMP reaction, which yielded positive amplicons with the distinct color change, consistent with some previous studies (Priya et al., 2018; Huang et al., 2019; Zhang and Gleason, 2019), but higher than the other assays (Srisuk et al., 2010; Yamazaki, 2011).
In this study, all positive reactions can be easily judged by the naked eye under the visible light, or under the UV light by the use of MnCl2-calcein dye in the LAMP system prior to amplification. Moreover, the sssvLAMP method did not require opening of reaction tubes with no probable cross-contamination, which usually arise from opened tubes after amplification (Sayad et al., 2018). Furthermore, the sssvLAMP method was carried out in a one-step reaction at 65°C for 40–50 min, which was faster than the PCR assay and more suitable for low-equipment setting laboratories.
Sensitivity is particularly important in the detection of foodborne pathogenic bacteria (Yuan et al., 2018). It has been reported that 10 CFU per reaction targeting bacterial resistance integrons (intI1, intI2, and intI3) (Yu et al., 2014), 8 and 0.54 CFU per reaction targeting the ompW (Srisuk et al., 2010) and ctxA (Okada et al., 2010) genes of V. cholerae, respectively, could be detected by LAMP methods. In this study, for the target genes hapA, mshA, pilA, and tlh, the LOD values ranged from 9.2 × 101 to 1.1 × 10–1CFU/reaction for the most of 52 V. cholerae strains in the inclusivity tests. Given the LOD values below 1 CFU/reaction, one possibility could be non-culturable Vibrio cells in cell dilutions. Another possibility could be the released genomic DNA from cracked V. cholerae cells during cell incubation, or from thermal lysis reactions before the sssvLAMP assays. These DNA in dilutions could be detected by the sssvLAMP method. Tourlousse et al. (2012) reported that detection and quantification of 10–100 genomes per μL could be performed in a polymer microfluidic chip by a real-time fluorogenic LAMP for targeting rtxA and toxR genes of V. cholerae. Different sensitivities of the LAMP technique were also reported, e.g., 6.2 pg DNA per tube for the hlyA (He et al., 2009) and 5 fg DNA per reaction for the ctxA (Okada et al., 2010) genes of V. cholerae, 100 pg DNA per μL for resistance gene vanA of Enterococcus faecium (Huang et al., 2019), and 10 pg DNA per tube for bacterial resistance integrons (intI1, intI2, and intI3) (Yu et al., 2014), all of which were less sensitive than those of the hapA (4.42 × 10–6 ng DNA/reaction) and tlh (3.90 × 10–3 ng DNA/reaction) genes of V. cholerae GIM1.449 by the sssvLAMP method developed in this study (Table 5). Variable LOD for the same or different targeted genes in different strains of V. cholerae may be related to the amplification efficiency of designed primers, or the multiple occurrence of a toxigenic gene in the genome of a particular stain, which may be different than that of another strain harboring the same gene. Additionally, variable sensitivity of the sssvLAMP method was observed when it was used to detect aquatic product samples and human stool specimens, which may be resulted from the influence of different matrix in homogenate samples tested.
The specificity data of the sssvLAMP method indicated that the method were applicable only in the existence of both the inner and outer primers. Moreover, no false positive amplification of the targeted genes of V. cholerae was observed in the closely related Vibrio species, as well as in non-targeting pathogenic bacteria tested, indicating high specificity of the designed primers targeting virulence-associated genes of V. cholerae. High occurrence of V. cholerae isolates carrying target genes has been reported (Meena et al., 2019; Xu et al., 2019), e.g., hapA (95.0%), and tlh (76.0%) in the fish species (Xu et al., 2019). Since the target genes can be present in many non-pathogenic strains of V. cholerae, cytotoxicity or enterotoxicity experiments may be performed to further justify pathogenesis of target genes-positive V. cholerae strains.
In this study, similar high efficiency of the sssvLAMP method was observed when tested with spiked samples of water and aquatic products, as well as human stool specimens. The results showed none of the V. cholerae contamination in all drinking water and environmental water samples, as well as human stool specimens tested. However, virulence-associated gene Types IV, V, and VII were found from the intestinal samples from three of the four commonly consumed fish species in China, including A. nobilis, C. auratus, and P. pekinensis, in which hapA, tlh, and pilA genes were detected positive. High occurrence of the hapA gene was also detected in the presence of V. cholerae strains isolated from the fish intestines (Xu et al., 2019), which may be related to its function involved in V. cholerae interaction with aquatic hosts (Halpern et al., 2003). In contrast, the mshA gene was absent from the fish intestinal samples, and low percentage was present in the 52 V. cholerae strains tested in this study, suggesting missing or truncation of the mshA gene in the bacterium, although the MSHA gene cluster is reported to exist and aid bacterial association with aquatic plankton to support environmental adaptation in many non-O1/O139 strains (Chiavelli et al., 2001; Moorthy and Watnick, 2004; Gong et al., 2019). The finding in this study, coupled with the previous research enhanced need for regular monitoring of V. cholerae contamination in these aquatic products for ensuring food safety.
Overall, the sssvLAMP method developed in this study was simple, rapid, and visible to the naked eye, showing greater advantages when compared with the routine PCR assay. This method was successfully employed to detect virulence-associated genes hapA, mshA, pilA and tlh, toxigenic gene ctxA and tcpA, and species-specific gene lolB of V. cholerae. In the future research, the sssvLAMP method should detect other important virulence factors, e.g., Type III secretion system, non-agglutinable heat-stable enterotoxin (NAG-ST), and cholix toxin, which have been shown to induce entertoxicity or cytotoxicity. It should support the field or clinical diagnosis where rapid and reliable detection of virulence-associated genes of V. cholerae is urgently required.
Data Availability Statement
The datasets generated for this study are available on request to the corresponding author.
Author Contributions
MX, ZS, JZ, WA, and LC participated in the design and or discussion of the study. MX, HF, and DC carried out the experiments. MX, HF, and LC analyzed the data. MX and LC wrote the manuscript. WA revised the manuscript. All authors read and approved the final version to be published.
Funding
This work was supported by the grants from the Shanghai Municipal Science and Technology Commission (No. 17050502200) and the National Natural Science Foundation of China (No. 31671946).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank M. Yang in Shanghai Ocean University for providing us human stool specimens, and Drs. Weili Liang, and Biao Kan in the National Institute for Communicable Disease Control and Prevention, Chinese Center for Disease Control and Prevention for providing us genomic DNA of Vibrio cholerae ATCC39315 (N16961) in this study.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.02899/full#supplementary-material
FIGURE S1 | Comparative sequence alignments between the newly designed LAMP primers versus the targeted genes hapA (A), lolB (B), mshA (C), pilA (D), tlh (E), and tcpA (F) amplified in representative strains by PCR reactions with GenBank accession numbers MN708522 to MN708551.
Footnotes
- ^ http://www.who.int/
- ^ https://www.ncbi.nlm.nih.gov/genbank
- ^ http://primerexplorer.jp/lampv5e/index.html
References
Aagesen, A. M., and Hase, C. C. (2012). Sequence analyses of type IV pili from Vibrio cholerae, Vibrio parahaemolyticus, and Vibrio vulnificus. Microb. Ecol. 64, 509–524. doi: 10.1007/s00248-012-0021-2
Ali, M., Nelson, A. R., Lopez, A. L., and Sack, D. A. (2015). Updated global burden of cholera in endemic countries. PLoS Negl. Trop. Dis. 9:e0003832. doi: 10.1371/journal.pntd.0003832
Almasi, M. A., Moradi, A., Nasiri, J., Karami, S., and Nasiri, M. (2012). Assessment of performance ability of three diagnostic methods for detection of Potato Leafroll virus (PLRV) using different visualizing systems. Appl. Biochem. Biotechnol. 168, 770–784. doi: 10.1007/s12010-012-9818-1
Austin, B. (2010). Vibrios as causal agents of zoonoses. Vet. Microbiol. 140, 310–317. doi: 10.1016/j.vetmic.2009.03.015
Baker-Austin, C., Oliver, J. D., Alam, M., Ali, A., Waldor, M. K., Qadri, F., et al. (2018). Vibrio spp. infections. Nat. Rev. Dis. Primers. 4:8. doi: 10.1038/s41572-018-0005-8
Benitez, J. A., and Silva, A. J. (2016). Vibrio cholerae hemagglutinin (HA)/protease: an extracellular metalloprotease with multiple pathogenic activities. Toxicon 115, 55–62. doi: 10.1016/j.toxicon.2016.03.003
Bwire, G., Debes, A. K., Orach, C. G., Kagirita, A., Ram, M., Komakech, H., et al. (2018). Environmental surveillance of Vibrio cholerae O1/O139 in the five African great lakes and other major surface water sources in Uganda. Front. Microbiol. 9:1560. doi: 10.3389/fmicb.2018.01560
Casasola-Rodriguez, B., Ruiz-Palacios, G. M., Pilar, R. C., Losano, L., Ignacio, M. R., and Orta de Velasquez, M. T. (2018). Detection of VBNC Vibrio cholerae by RT-real time PCR based on differential gene expression analysis. FEMS Microbiol. Lett. 365:fny126. doi: 10.1093/femsle/fny156
Ceccarelli, D., Chen, A., Hasan, N. A., Rashed, S. M., Huq, A., and Colwell, R. R. (2015). Non-O1/non-O139 Vibrio cholerae carrying multiple virulence factors and V. cholerae O1 in the Chesapeake Bay. Maryland. Appl. Environ. Microbiol. 81, 1909–1918. doi: 10.1128/AEM.03540-14
Chiavelli, D. A., Marsh, J. W., and Taylor, R. K. (2001). The mannose-sensitive hemagglutinin of Vibrio cholerae promotes adherence to zooplankton. Appl. Environ. Microbiol. 67, 3220–3225. doi: 10.1128/AEM.67.7.3220-3225.2001
Datta-Roy, K., Banerjee, K., De, S. P., and Ghose, A. C. (1986). Comparative study of expression of hemagglutinins, hemolysins, and enterotoxins by clinical and environmental isolates of non-O1 Vibrio cholerae in relation to their enteropathogenicity. Appl. Environ. Microbiol. 52, 875–879.
Engku Nur Syafirah, E. A. R., Nurul Najian, A. B., Foo, P. C., Mohd Ali, M. R., Mohamed, M., and Yean, C. Y. (2018). An ambient temperature stable and ready-to-use loop-mediated isothermal amplification assay for detection of toxigenic Vibrio cholerae in outbreak settings. Acta Trop. 182, 223–231. doi: 10.1016/j.actatropica.2018.03.004
Fang, J., Wu, Y., Qu, D., Ma, B., Yu, X., Zhang, M., et al. (2018). Propidium monoazide real-time loop-mediated isothermal amplification for specific visualization of viable Salmonella in food. Lett. Appl. Microbiol. 67, 79–88. doi: 10.1111/lam.12992
Fiedoruk-Pogrebniak, M., and Koncki, R. (2015). Multicommutated flow analysis system based on fluorescence microdetectors for simultaneous determination of phosphate and calcium ions in human serum. Talanta 144, 184–188. doi: 10.1016/j.talanta.2015.06.001
Fiore, A. E., Michalski, J. M., Russell, R. G., Sears, C. L., and Kaper, J. B. (1997). Cloning, characterization, and chromosomal mapping of a phospholipase (lecithinase) produced by Vibrio cholerae. Infect. Immun. 65, 3112–3117. doi: 10.0000/PMID9234762
Fullner, K. J., and Mekalanos, J. J. (1999). Genetic characterization of a new type IV-A pilus gene cluster found in both classical and El Tor biotypes of Vibrio cholerae. Infect. Immun. 67, 1393–1404. doi: 10.1007/BF02560516
Fykse, E. M., Nilsen, T., Nielsen, A. D., Tryland, I., Delacroix, S., and Blatny, J. M. (2012). Real-time PCR and NASBA for rapid and sensitive detection of Vibrio cholerae in ballast water. Mar. Pollut. Bull. 64, 200–206. doi: 10.1016/j.marpolbul.2011.12.007
Gao, H., Xu, J., Lu, X., Li, J., Lou, J., Zhao, H., et al. (2018). Expression of hemolysin is regulated under the collective actions of HapR, Fur, and HlyU in Vibrio cholerae El Tor serogroup O1. Front. Microbiol. 9:1310. doi: 10.3389/fmicb.2018.01310
Garrido-Maestu, A., Chapela, M. J., Vieites, J. M., and Cabado, A. G. (2015). lolB gene, a valid alternative for qPCR detection of Vibrio cholerae in food and environmental samples. Food Microbiol. 46, 535–540. doi: 10.1016/j.fm.2014.09.012
Gong, L., Yu, P., Zheng, H., Gu, W., He, W., Tang, Y., et al. (2019). Comparative genomics for non-O1/O139 Vibrio cholerae isolates recovered from the Yangtze River Estuary versus V. cholerae representative isolates from serogroup O1. Mol. Genet. Genomics 294, 417–430. doi: 10.1007/s00438-018-1514-6
Goto, M., Honda, E., Ogura, A., Nomoto, A., and Hanaki, K. (2009). Colorimetric detection of loop-mediated isothermal amplification reaction by using hydroxy naphthol blue. Biotechniques 46, 167–172. doi: 10.2144/000113072
Hacker, J., Blum-Oehler, G., Muhldorfer, I., and Tschape, H. (1997). Pathogenicity islands of virulent bacteria: structure, function and impact on microbial evolution. Mol. Microbiol. 23, 1089–1097. doi: 10.1046/j.1365-2958.1997.3101672.x
Hall, R. H., Vial, P. A., Kaper, J. B., Mekalanos, J. J., and Levine, M. M. (1988). Morphological studies on fimbriae expressed by Vibrio cholerae 01. Microb. Pathog. 4, 257–265. doi: 10.1016/0882-4010(88)90086-1
Hall, T. A. (1999). BioEdit, A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41, 95–98. doi: 10.1016/j.micpath.2019.04.039
Halpern, M., Gancz, H., Broza, M., and Kashi, Y. (2003). Vibrio cholerae hemagglutinin/protease degrades chironomid egg masses. Appl. Environ. Microbiol. 69, 4200–4204. doi: 10.1128/aem.69.7.4200-4204.2003
He, N., Lei, Z., Liang, C., Wang, D., Liu, Y., Jia, J., et al. (2009). Detection of Vibrio cholerae by LAMP assay. China Tropical Med. 9, 23–26.
He, Y., Tang, Y., Sun, F., and Chen, L. (2015). Detection and characterization of integrative and conjugative elements (ICEs)-positive Vibrio cholerae isolates from aquacultured shrimp and the environment in Shanghai. China. Mar. Pollut. Bull. 101, 526–532. doi: 10.1016/j.marpolbul.2015.10.062
Huang, Q. Q., Liu, B. B., Zhu, H. F., Ma, J. J., Tsoi, M., Yao, B. Q., et al. (2019). Rapid and sensitive detection of resistance gene vanA from clinical Enterococcus faecium and E. faecalis isolates by loop-mediated isothermal amplification. J. Glob. Antimicrob. Resist. 16, 262–265. doi: 10.1016/j.jgar.2018.10.012
Jouravleva, E. A., McDonald, G. A., Marsh, J. W., Taylor, R. K., Boesman-Finkelstein, M., and Finkelstein, R. A. (1998). The Vibrio cholerae mannose-sensitive hemagglutinin is the receptor for a filamentous bacteriophage from V. cholerae O139. Infect. Immun. 66, 2535–2539.
Kakizaki, E., Sonoda, A., Sakai, M., and Yukawa, N. (2018). Simple detection of bacterioplankton using a loop-mediated isothermal amplification (LAMP) assay: first practical approach to 72 cases of suspected drowning. Forensic. Sci. Int. 289, 289–303. doi: 10.1016/j.forsciint.2018.05.035
Kim, J. G., Baek, S. H., Kim, S., Kim, H. I., Lee, S. W., Phan, L. M. T., et al. (2018). Rapid discriminative detection of dengue viruses via loop mediated isothermal amplification. Talanta 190, 391–396. doi: 10.1016/j.talanta.2018.08.019
Kumar, P., Peter, W. A., and Thomas, S. (2010). Rapid detection of virulence-associated genes in environmental strains of Vibrio cholerae by multiplex PCR. Curr. Microbiol. 60, 199–202. doi: 10.1007/s00284-009-9524-6
Lalitha, P., Siti Suraiya, M. N., Lim, K. L., Lee, S. Y., Nur Haslindawaty, A. R., Chan, Y. Y., et al. (2008). Analysis of lolB gene sequence and its use in the development of a PCR assay for the detection of Vibrio cholerae. J. Microbiol. Methods 75, 142–144. doi: 10.1016/j.mimet.2008.05.001
Lau, Y. L., Fong, M. Y., Mahmud, R., Chang, P. Y., Palaeya, V., Cheong, F. W., et al. (2011). Specific, sensitive and rapid detection of human plasmodium knowlesi infection by loop-mediated isothermal amplification (LAMP) in blood samples. Malar. J. 10, 197. doi: 10.1186/1475-2875-10-197
Liew, P. S., Lertanantawong, B., Lee, S. Y., Manickam, R., Lee, Y. H., and Surareungchai, W. (2015). Electrochemical genosensor assay using lyophilized gold nanoparticles/latex microsphere label for detection of Vibrio cholerae. Talanta 139, 167–173. doi: 10.1016/j.talanta.2015.02.054
Lim, K. T., Teh, C. S., and Thong, K. L. (2013). Loop-mediated isothermal amplification assay for the rapid detection of Staphylococcus aureus. Biomed. Res. Int. 2013, 895816. doi: 10.1155/2013/895816
Liu, M. J., Du, G. M., Bai, F. F., Wu, Y. Z., Xiong, Q. Y., Feng, Z. X., et al. (2015). A rapid and sensitive loop-mediated isothermal amplification procedure (LAMP) for Mycoplasma hyopneumoniae detection based on the p36 gene. Genet. Mol. Res. 14, 4677–4686. doi: 10.4238/2015.May.4.27
Markoulatos, P., Siafakas, N., and Moncany, M. (2002). Multiplex polymerase chain reaction: a practical approach. J. Clin. Lab. Anal. 16, 47–51. doi: 10.1002/jcla.2058
McGrath, B. M., O’Halloran, J. A., Piterina, A. V., and Pembroke, J. T. (2006). Molecular tools to detect the IncJ elements: a family of integrating, antibiotic resistant mobile genetic elements. J. Microbiol. Methods 66, 32–42. doi: 10.1016/j.mimet.2005.10.004
Meena, B., Anburajan, L., Sathish, T., Das, A. K., Vinithkumar, N. V., Kirubagaran, R., et al. (2019). Studies on diversity of Vibrio sp. and the prevalence of hapA, tcpI, st, rtxA&C, acfB, hlyA, ctxA, ompU and toxR genes in environmental strains of Vibrio cholerae from Port Blair bays of South Andaman. India. Mar Pollut Bull. 144, 105–116. doi: 10.1016/j.marpolbul.2019.05.011
Moorthy, S., and Watnick, P. I. (2004). Genetic evidence that the Vibrio cholerae monolayer is a distinct stage in biofilm development. Mol. Microbiol. 52, 573–587. doi: 10.1111/j.1365-2958.2004.04000.x
Mori, Y., Nagamine, K., Tomita, N., and Notomi, T. (2001). Detection of loop-mediated isothermal amplification reaction by turbidity derived from magnesium pyrophosphate formation. Biochem. Biophys. Res. Commun. 289, 150–154. doi: 10.1111/j.1365-2958.2004.04000.x
Nagamine, K., Hase, T., and Notomi, T. (2002). Accelerated reaction by loop-mediated isothermal amplification using loop primers. Mol. Cell Probes. 16, 223–229. doi: 10.1006/mcpr.2002.0415
Nasrabadi, Z., Ranjbar, R., Poorali, F., and Sarshar, M. (2017). Detection of eight foodborne bacterial pathogens by oligonucleotide array hybridization. Electron. Physician. 9, 4405–4411. doi: 10.19082/4405
Notomi, T., Mori, Y., Tomita, N., and Kanda, H. (2015). Loop-mediated isothermal amplification (LAMP): principle, features, and future prospects. J. Microbiol. 53, 1–5. doi: 10.1007/s12275-015-4656-9
Notomi, T., Okayama, H., Masubuchi, H., Yonekawa, T., Watanabe, K., Amino, N., et al. (2000). Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 28, e63–e63. doi: 10.1093/nar/28.12.e63
Okada, K., Chantaroj, S., Taniguchi, T., Suzuki, Y., Roobthaisong, A., Puiprom, O., et al. (2010). A rapid, simple, and sensitive loop-mediated isothermal amplification method to detect toxigenic Vibrio cholerae in rectal swab samples. Diagn. Microbiol. Infect. Dis. 66, 135–139. doi: 10.1016/j.diagmicrobio.2009.09.004
Osterberg, F. W., Rizzi, G., Donolato, M., Bejhed, R. S., Mezger, A., Stromberg, M., et al. (2014). On-chip detection of rolling circle amplified DNA molecules from Bacillus globigii spores and Vibrio cholerae. Small 10, 2877–2882. doi: 10.1002/smll.201303325
Phillips, E. A., Moehling, T. J., Bhadra, S., Ellington, A. D., and Linnes, J. C. (2018). Strand displacement probes combined with isothermal nucleic acid amplification for instrument-free detection from complex samples. Anal Chem. 90, 6580–6586. doi: 10.1021/acs.analchem.8b00269
Priya, G. B., Agrawal, R. K., Milton, A. A. P., Mishra, M., Mendiratta, S. K., Agarwal, R. K., et al. (2018). Development and evaluation of isothermal amplification assay for the rapid and sensitive detection of Clostridium perfringens from chevon. Anaerobe 54, 178–187. doi: 10.1016/j.anaerobe.2018.09.005
Sayad, A., Ibrahim, F., Mukim Uddin, S., Cho, J., Madou, M., and Thong, K. L. (2018). A microdevice for rapid, monoplex and colorimetric detection of foodborne pathogens using a centrifugal microfluidic platform. Biosens. Bioelectron. 100, 96–104. doi: 10.1016/j.bios.2017.08.060
Soli, K. W., Kas, M., Maure, T., Umezaki, M., Morita, A., Siba, P. M., et al. (2013). Evaluation of colorimetric detection methods for Shigella, Salmonella, and Vibrio cholerae by loop-mediated isothermal amplification. Diagn Microbiol Infect. Dis. 77, 321–323. doi: 10.1016/j.diagmicrobio.2013.09.009
Srisuk, C., Chaivisuthangkura, P., Rukpratanporn, S., Longyant, S., Sridulyakul, P., and Sithigorngul, P. (2010). Rapid and sensitive detection of Vibrio cholerae by loop-mediated isothermal amplification targeted to the gene of outer membrane protein ompW. Lett. Appl. Microbiol. 50, 36–42. doi: 10.1111/j.1472-765X.2009.02749.x
Sun, X., Liu, T., Peng, X., and Chen, L. (2014). Insights into Vibrio parahaemolyticus CHN25 response to artificial gastric fluid stress by transcriptomic analysis. Int. J. Mol. Sci. 15, 22539–22562. doi: 10.3390/ijms151222539
Toffaletti, J., and Kirvan, K. (1980). Spectrophotometric micro method for measurement of dialyzable calcium by use of cresolphthalein complexone and continuous-flow analysis. Clin. Chem. 26, 1562–1565.
Tomita, N., Mori, Y., Kanda, H., and Notomi, T. (2008). Loop-mediated isothermal amplification (LAMP) of gene sequences and simple visual detection of products. Nat. Protoc. 3, 877–882. doi: 10.1038/nprot.2008.57
Tourlousse, D. M., Ahmad, F., Stedtfeld, R. D., Seyrig, G., Tiedje, J. M., and Hashsham, S. A. (2012). A polymer microfluidic chip for quantitative detection of multiple water- and foodborne pathogens using real-time fluorogenic loop-mediated isothermal amplification. Biomed. Microdevices. 14, 769–778. doi: 10.1007/s10544-012-9658-3
Vu, T. T. T., Alter, T., and Huehn, S. (2018). Prevalence of Vibrio spp. in retail seafood in Berlin. Germany. J. Food Prot. 81, 593–597. doi: 10.4315/0362-028X.JFP-17-366
Wastling, S. L., Picozzi, K., Kakembo, A. S., and Welburn, S. C. (2010). LAMP for human African trypanosomiasis: a comparative study of detection formats. PLoS Negl. Trop. Dis. 4:e865. doi: 10.1371/journal.pntd.0000865
Xu, M., Wu, J., and Chen, L. (2019). Virulence, antimicrobial and heavy metal tolerance, and genetic diversity of Vibrio cholerae recovered from commonly consumed freshwater fish. Environ. Sci. Pollut. Res. Int. 26, 27338–27352. doi: 10.1007/s11356-019-05287-8
Yamazaki, W. (2011). Sensitive and rapid detection of cholera toxin-producing Vibrio cholerae using loop-mediated isothermal amplification. Methods Mol. Biol. 739, 13–22. doi: 10.1007/978-1-61779-102-4_2
Yang, X., Al-Attala, M. N., Zhang, Y., Zhang, A. F., Zang, H. Y., Gu, C. Y., et al. (2018). Rapid detection of Ustilaginoidea virens from rice using loop-mediated isothermal amplification assay. Plant Dis. 102, 1741–1747. doi: 10.1094/PDIS-01-18-0065-RE
Yu, G., Chen, L., Lin, C. W., Li, B., Cui, H., Chen, S., et al. (2014). Loop-mediated isothermal amplification assays for screening of bacterial integrons. Biol. Res. 47:53. doi: 10.1186/0717-6287-47-53
Yuan, H., Chao, Y., Li, S., Tang, M. Y. H., Huang, Y., Che, Y., et al. (2018). Picoinjection-enabled multitarget loop-mediated isothermal amplification for detection of foodborne pathogens. Anal. Chem. 90, 13173–13177. doi: 10.1021/acs.analchem.8b03673
Zago, V., Zambon, M., Civettini, M., Zaltum, O., and Manfrin, A. (2017). Virulence-associated factors in Vibrio cholerae non-O1/non-O139 and V. mimicus strains isolated in ornamental fish species. J. Fish Dis. 40, 1857–1868. doi: 10.1111/jfd.12659
Zanoli, L. M., and Spoto, G. (2013). Isothermal amplification methods for the detection of nucleic acids in microfluidic devices. Biosensors 3, 18–43. doi: 10.3390/bios3010018
Zhang, L., and Gleason, C. (2019). Loop-mediated isothermal amplification for the diagnostic detection of Meloidogyne chitwoodi and M. fallax. Plant Dis. 103, 12–18. doi: 10.1094/PDIS-01-18-0093-RE
Zhang, X., Du, X. J., Guan, C., Li, P., Zheng, W. J., and Wang, S. (2015). Detection of Vibrio cholerae by isothermal cross-priming amplification combined with nucleic acid detection strip analysis. Mol. Cell Probes 29, 208–214. doi: 10.1016/j.mcp.2015.05.001
Keywords: Vibrio cholerae, virulence-associated genes, loop-mediated isothermal amplification, water, aquatic product
Citation: Xu M, Fu H, Chen D, Shao Z, Zhu J, Alali WQ and Chen L (2019) Simple Visualized Detection Method of Virulence-Associated Genes of Vibrio cholerae by Loop-Mediated Isothermal Amplification. Front. Microbiol. 10:2899. doi: 10.3389/fmicb.2019.02899
Received: 05 July 2019; Accepted: 02 December 2019;
Published: 20 December 2019.
Edited by:
Abd El-Latif Hesham, Assiut University, EgyptReviewed by:
Sucharit Basu Neogi, International Centre for Diarrhoeal Disease Research (ICDDR), BangladeshEckhard Strauch, Federal Institute for Risk Assessment (BfR), Germany
Copyright © 2019 Xu, Fu, Chen, Shao, Zhu, Alali and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lanming Chen, bG1jaGVuQHNob3UuZWR1LmNu
†These authors have contributed equally to this work
 Mengjie Xu
Mengjie Xu Huiyu Fu
Huiyu Fu Dailing Chen
Dailing Chen Zehuai Shao1
Zehuai Shao1 Walid Q. Alali
Walid Q. Alali Lanming Chen
Lanming Chen