- 1State Key Laboratory of Crop Stress Biology for Arid Areas, College of Plant Protection, Northwest A&F University, Yangling, China
- 2Hunan Provincial Key Laboratory for Biology and Control of Plant Diseases and Insect Pests, College of Plant Protection, Hunan Agricultural University, Changsha, China
Eukaryotic ribosomes are essential for proliferation, differentiation, and cell growth. RPs26 is a ribosomal subunit structural protein involved in the growth and development process. Little is known about the function of PsRPs26 in pathogenic fungi. In this study, we isolated the RPs26 gene, PsRPs26, from Puccinia striiformis f. sp. tritici (Pst). PsRPs26 contains a eukaryotic-specific Y62–K70 motif and is more than 90% identical with its ortholog gene in other fungi. PsRPs26 was found to be localized in both the nucleus and cytoplasm. Expression of PsRPs26 increased when wheat seedlings were inoculated with the Pst CYR31 isolate. Moreover, knockdown of PsRPs26 by a host-induced gene silencing system inhibited growth and limited urediospore production in Pst. Our discovery that PsRPs26 may contribute to the pathogenicity of Pst and open a new way in the pathogenic function of PsRPs26 in cereal rust fungi.
Introduction
Ribosomes are responsible for protein synthesis and essential to many organisms, ranging from bacteria to animals (Karsi et al., 2002). Ribosomal proteins (RPs) and ribosomal RNA (rRNA) are macromolecular components of the ribosome (Ferreira-Cerca et al., 2007). Mammalian ribosomes consist of 79 RPs and four rRNA species (Wool et al., 1995; Warner and Nierras, 1998). The eukaryotic 80S ribosome is composed of a small (40S) and a large subunit (60S). The 40S ribosome contains the 18S rRNA and 32 RPs, whereas the 60S ribosome is composed of three rRNAs and 47 RPs (Wool, 1979; Wool et al., 1995). As ribosomes exist in a wide spectrum of organisms, RPs have been highly conserved – 35 RP homologs exist in eubacteria, archaea, and eukarya (Verschoor et al., 1998; Belyy et al., 2016).
RPs26 is a structural protein of the ribosomal 40S subunit. In 1977, the first RPs26 was identified in rat livers (Collatz et al., 1977). Subsequently, the corresponding gene was isolated from human cDNAs (Filipenko et al., 1998). RPs26 was also cloned from the giant Panda (Hou et al., 2010). RPs26a and RPs26b are found to be 97% identical in the yeast Saccharomyces cerevisiae (Strittmatter et al., 2006). There is no significant eubacterial counterpart of RPs26; but S18 is the functional homolog of RPs26 in eubacteria, which contains a similar rRNA-contacting structural motif (Malygin and Karpova, 2009). In humans, the connection between mRNA and Rps26 was established via the Y62–K70 motif (62-YXXPKXYXK-70) conserved in eukaryotes (Sharifulin et al., 2011). Recently, RPs have been proposed as the model for understanding post-transcriptional regulation of gene expression.
It has been demonstrated that ribosome biogenesis is essential for cell growth and development. A genetic study found that depleting Sfp1, which controls the expression of numerous genes related to ribosome assembly, resulted in reduced cell size (Jorgensen et al., 2002). RPs26 is also involved in multiple growth and development processes. RPs26 genes in Diamond-Blackfan anemia patients affect the function of the proteins in rRNA processing (Doherty et al., 2010). Rps26 suppressed the splicing of its own pre-mRNA when it was expressed in Escherichia coli, suggesting some feedback mechanism might be controlling Rps26 synthesis (Malygin et al., 2003; Ivanov et al., 2005). In S. cerevisiae, Rps26B can support the growth of single yeast cells but not filamentous growth. Rps26Ap appears to be a product of the yeast translation machinery, as it is not only required for general translation but also functions in the diploid pseudohyphal growth and regulation of haploid adhesive in yeast (Strittmatter et al., 2006; Belyy et al., 2016). Additionally, Rps26 is shown to participate in endoplasmic reticulum stress in yeast (Steffen et al., 2012).
Wheat (Triticum aestivum L.) is an important food crop worldwide, the grain qualities and production of which are greatly impacted by pathogens. Wheat stripe rust, caused by the fungus pathogen Puccinia striiformis f. sp. tritici (Pst), has become the largest biotic suppression factor to the yield of wheat, causing more than 90% production losses in a field (Wang et al., 2018). Additionally, Pst rapidly evolves new virulent rust fungal isolates to adapt to most race-specific host resistance genes (Fisher et al., 2012). Pst was found to complete its sexual stage in barberry (Berberis shensiana), leading to pathogenic variation in Pst (Zheng et al., 2013).
To study the epidemiology, biology, and pathogenic factors of Pst, a complementary DNA (cDNA) library was built from wheat seedlings inoculated with Pst CYR31 isolate (Ma et al., 2009). We identified a 26S ribosomal protein gene, PsRPs26, from the library. PsRPs26 contains the eukaryotic-specific YxxPKxYxK motif. Functional characterization showed that knockdown of PsRPs26 leads to suppressed fungal growth and development and limited urediospore production of Pst.
Materials and Methods
Plant Material and Treatments
Puccinia striiformis f. sp. tritici CYR31 isolate and wheat cultivar Suwon 11 (Su11) were used. Su11 is susceptible to Pst CYR31 isolate. Fresh urediniospores of the CYR31 isolate were collected from inoculated wheat leaves. The culture, inoculation, and incubation of Su11 followed the report by Kang et al. (2002). Inoculated and control wheat leaves were harvested at 24, 48, 120, 168, and 192 h post-inoculation (hpi), and inoculated leaves of barberry at 12 days post-inoculation (dpi) were collected. The plant samples were quickly frozen and stored at -80°C.
Isolation of RNA and Quantitative Real-Time (qRT) PCR Analysis
For total RNA extraction, wheat leaves that were inoculated with Pst CYR31 isolate were extracted using TRIzolTM reagent (Invitrogen, Carlsbad, CA, United States) following the recommended protocol. First-strand cDNA was synthesized using the qRT-PCR System (Promega Corp., Madison, WI, United States). qRT-PCR primer design and reaction conditions were based on Wang et al. (2009). The Pst elongation factor PsEF was selected as the internal reference gene for the qRT-PCR analyses. The specific primers of PsRPs26 used in the qRT-PCR analysis were listed in Supplementary Table S1. All relative gene expression results were assayed using the comparative 2-ΔΔCT method (Livak and Schmittgen, 2001).
Sequence Analysis, Alignment, and Structure Prediction
The homologs are in a set of fungal genomes deposited in the National Center for Biotechnology Information (NCBI1) databases and Ensembl Fungi2. Phylogenetic trees were constructed with the neighbor-joining algorithm MEGA5 (Tamura et al., 2011). To identify intraspecific polymorphisms, we compared the coding regions of 11 Pst isolates CYR32, CYR23, CYR31, CYR33, V26, Su11-4, Yr9, PST-21, PST-78, PST-08/21, and PST-87/7. To identify nucleotide substitutions in PsRPs26, we performed PCR amplifications with the cDNA of these isolates (Wang et al., 2017). Multiple protein sequence alignments were generated using CLUSTALW (Thompson et al., 1994).
Subcellular Localization
A pCAMBIA-1302-PsRPs26-GFP fusion vector was used to verify the subcellular localization. We performed a transient expression analysis using Nicotiana benthamiana to examine the subcellular localization of PsRPs26. The reconstructed vector pCAMBIA-1302-PsRPs26-GFP and empty vector were transformed into strain GV3101 of Agrobacterium tumefaciens by electroporation. The leaves of 4-week-old N. benthamiana were transiently transformed with strains carrying the empty vector or PsRPs26-GFP. Green fluorescent protein (GFP) signals were examined with an Olympus BX-51 microscope (Olympus Corp., Tokyo).
PsRPs26 Gene Silencing Using Host-Induced Gene Silencing (HIGS)
Barley stripe mosaic virus (BSMV) γRNA-based vectors used to knockdown the expression of PsRPs26 were followed as described by Holzberg et al. (2002). The sequenced PCR product of PsRPs26 were digested with NotI and PacI and inserted into the digested BSMV:g vector. The entire second leaf was infected with virus transcripts by mildly rubbing it on the leaf surface at the two-leaf stage (Hein et al., 2005). The wheat phytoene desaturase (TaPDS) was used as a control. Three independent sets of wheat plants were used for each experiment (BSMV:TaPDS, BSMV:γ, and BSMV: PsRPs26). The fourth leaves were inoculated with fresh urediniospores of the Pst virulence race CYR31 10 days later. The disease phenotypes were recorded and photographed at 14 days post-inoculation (dpi). The fourth leaves with Pst were excised at 24, 48, and 120 hpi for RNA isolation and histological determination.
Histological Determination of Fungal Growth
Wheat samples were stained as described previously by Wang et al. (2007). Transparent leaf segments were examined using an Olympus BX-51 microscope for haustorial mother cells, hyphal lengths, and infection areas. Thirty to fifty infection sites were examined for each treatment in each biological replication. Hyphal lengths, infection areas, and haustorial mother cells were calculated using DP-BSW software. Statistical analyses were performed with SPSS software.
Results
Identification of a 26S Ribosomal Protein Gene From Puccinia striiformis
One transcript that encodes a putative 40S ribosomal protein subunit was identified by mapping the Pst genome (Zheng et al., 2013). The wheat cDNA sequence comprises of a 381-bp open reading frame (ORF) and encodes a protein of 126 aa. The corresponding protein was predicted to have a molecular weight of 13.95 kDa. To identify the subfamily of the 40S ribosomal protein subunits, a phylogenetic analysis was constructed using 31 different 40S ribosomal protein subunits of S. cerevisiae from Ensembl Fungi (see footnote 2). The results indicated that the deduced protein grouped with ScRP26S (Figure 1A). Additionally, a multi-sequence alignment was conducted with various RP26Ss obtained from Ensembl Fungi and NCBI. The predicted protein shares high similarity (99.21%) with PgRP26S in Puccinia graminis using BLASTP analysis. Further analysis confirmed its similarity to other fungal RPs26 proteins, including Fusarium oxysporum FoRPs26, Ustilago maydis UmRPs26, and Magnaporthe oryzae MoRPs26 (Figure 1B). Thus, this gene was designated as PsRPs26.
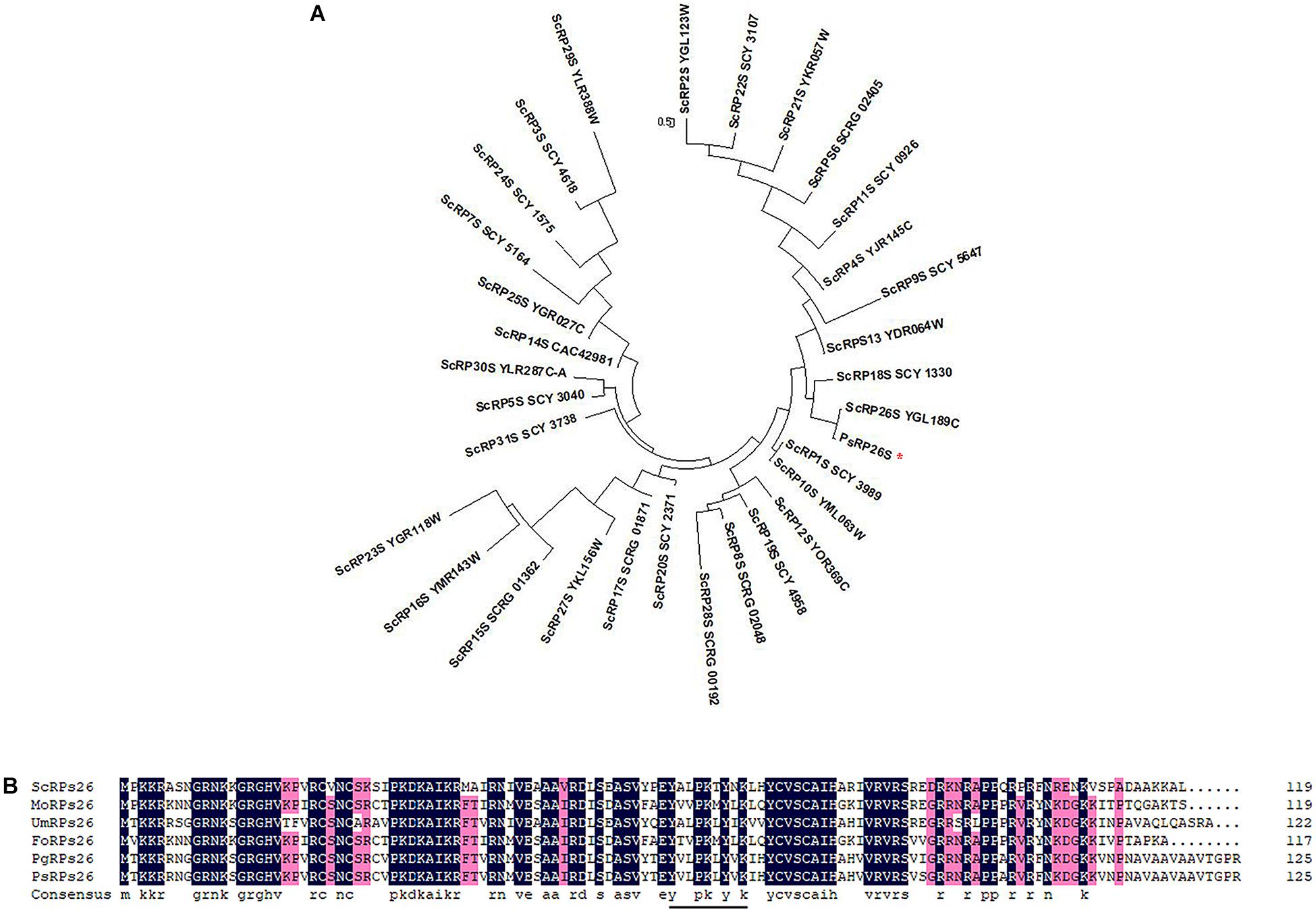
Figure 1. Phylogenetic analysis and multiple sequence alignment of PsRPs26. (A) Phylogenic analysis of PsRPs26 and Saccharomyces cerevisiae RPs family members using MEGA 5.0 software. (B) Protein multiple alignment. Identical and similar amino acid residues are shaded in black and light gray, respectively. Ps, Puccinia striiformis f. sp. tritici; Pg, Puccinia graminis f. sp. tritici; Us, Ustilago maydis; Mo, Magnaporthe oryzae; Fo, Fusarium oxysporum; and Sc, Saccharomyces cerevisiae.
Most 40S ribosomal protein subunits are highly conserved. We compared the coding regions between 11 different Pst isolates. Compared with the PsRPs26 sequence from CYR31, which is one of the predominant Pst isolates in China, only four synonymous substitutions were observed. However, no nucleotide substitutions in the YxxPKxYxK motif were found among the 11 Pst isolates (Supplementary Figure S1). These results indicated PsRPs26 is highly conserved.
PsRPs26 Localizes to the Nucleus and Cytoplasm
A fusion pCAMBIA-1302-PsRPs26-GFP vector was constructed to determine the subcellular localization of PsRPs26. The control vector and pCAMBIA-1302-PsRPs26-GFP vector were transformed into N. benthamiana leaf cells. Microscopic observation showed that the PsRPs26-GFP fusion protein was located in the nucleus and cytoplasm (Figure 2). Florescent signals were localized in the nucleus, perinuclear area, and cytoplasm in the control. These results demonstrated that PsRPs26 is located in the cytoplasm and nucleus.
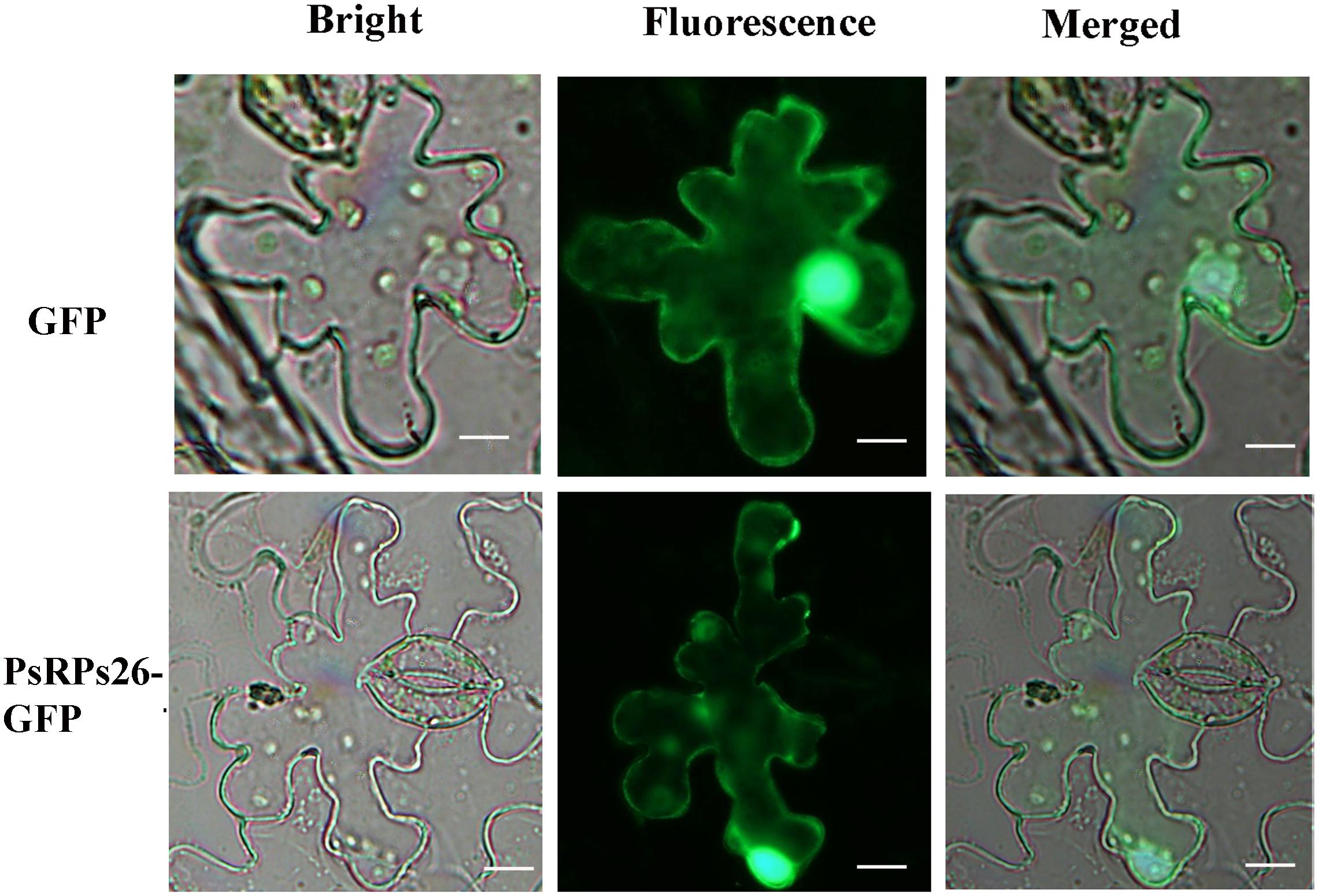
Figure 2. Subcellular localization of the PsRPs26 protein. The PsRPs26-GFP fusion protein and GFP (control) were transiently expressed in Nicotiana benthamiana. Bar = 20 μm. Similar results were obtained from three biological replicates.
PsRPs26 Is Highly Induced During the Infection Stage of Wheat and Barberry
To investigate if PsRPs26 is involved in wheat and Pst interactions, qRT-PCR was performed to assay the transcript level of PsRPs26 in different stages of Pst-host interactions. PsRPs26 transcript levels increased as early as 24 hpi in Pst-wheat interactions. At 168 hpi with the CYR31 isolate, the transcript level of PsRPs26 reached the maximum level of 22-fold compared with the control, which corresponds to the initiation of the sporulation stage. Furthermore, PsRPs26 was highly expressed in Pst-barberry interaction (sexual reproduction, Figure 3). These results strongly supported our hypothesis that the transcript level of PsRPs26 is induced during the Pst infection stage in host plants.
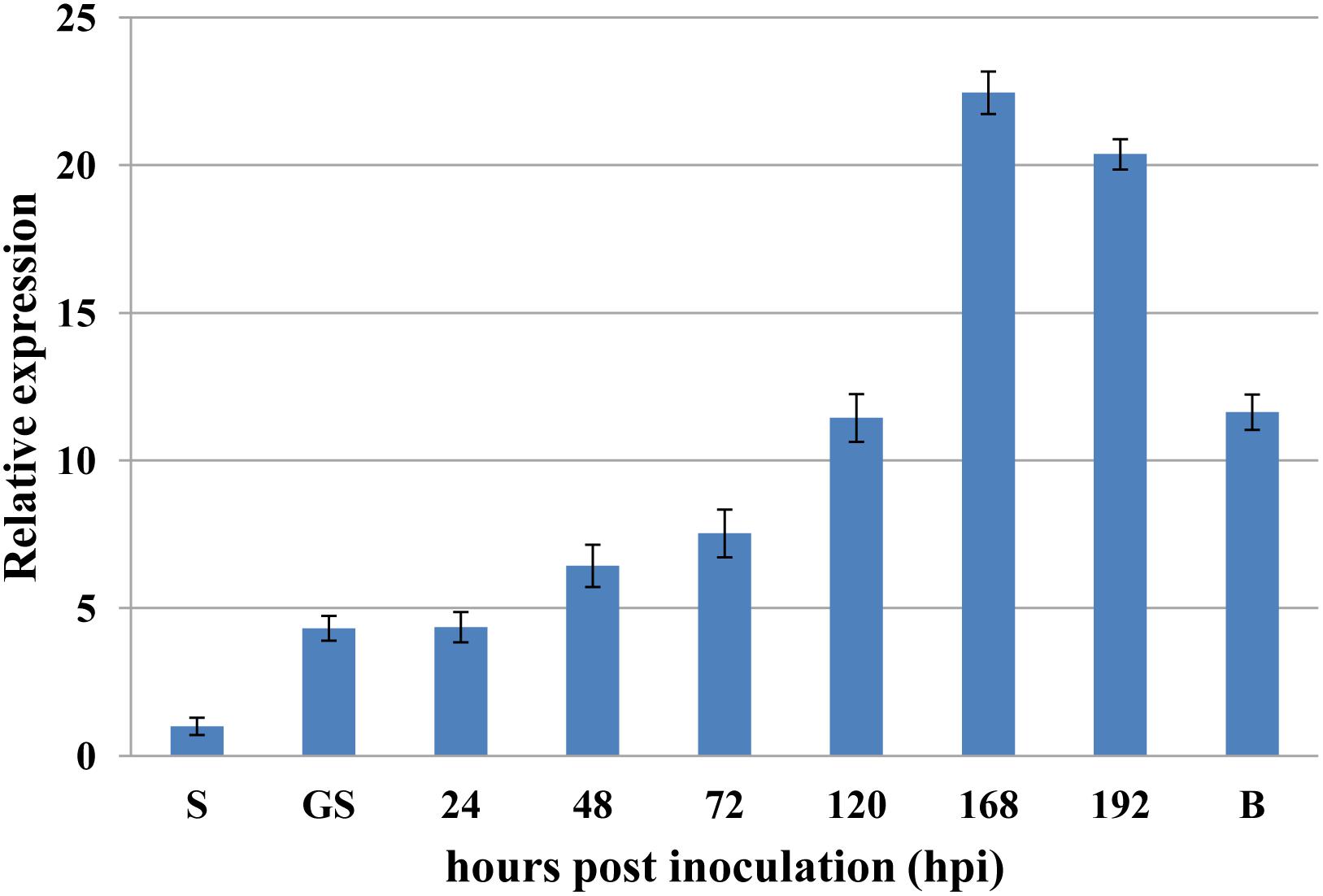
Figure 3. Expression of PsRPs26 during the different infection stages of P. striiformis. The expression levels were normalized by PsEF-1a. S, urediniospores; GS, in vitro germ tube; B, infected Berberis shensiana. Mean expression values were calculated from three independent replicates.
Silencing of PsRPs26 Attenuates Pst Growth and Development
A HIGS system with BSMV was used to further characterize the function of PsRPs26 during Pst infection. At 10-day dpi with the virus, the BSMV:γ (control) and BSMV:PsRPs26-infected leaves displayed mild chlorotic mosaic symptoms. There was no other obvious defect on leaves infected virus. Furthermore, we used the recombinant virus BSMV:TaPDS to silence the wheat PDS gene, causing severe chlorophyll photo bleaching under the same conditions. These results proved that the HIGS system was functioning successfully.
To determine how PsRPs26 participates in Pst growth and development, we observed the cytological changes in wheat seedlings with PsRPs26 knocked down and infected with Pst. The fourth leaves of wheat infected with the virus were inoculated with Pst CYR31 isolate at 10 dpi. We then assayed for the haustoria and number of haustorial mother cells, as well as the hyphal length and the number of hyphal branches. In the BSMV:PsRPs26-treated leaves, the length of the infection hyphae and number of haustoria clearly decreased. But there were no significant differences in the number of hyphal branches and haustorial mother cells (Figures 5A,B). At 48 hpi with the CYR31 isolate, hyphal growth was significantly inhibited in the PsRPs26-silenced plants (Figure 5C). Compared to the control, Pst hyphal colony size was significantly suppressed (P < 0.05) in the BSMV:PsRPs26-treated wheat leaves at 120 hpi (Figures 5A,D). These results indicated that PsRPs26 is involved in Pst growth and development during the infection stage in wheat.
Transient Silencing of PsRPs26 Significantly Limits Urediospore Production of Pst
After 14 dpi, there were masses of urediospores on the CYR31-infected wild-type seedlings and BSMV:γ infection seedlings (control). Interestingly, leaves treated with BSMV:PsRPs26 exhibited increased resistance to the CYR31 isolate, as indicated by the limited urediospore production compared with the controls (Figure 4A). We then confirmed the silencing efficiency of PsRPs26 in the HIGS system using qRT-PCR. We found that the transcript levels of PsRPs26 were reduced by 54, 67, and 63% at 24, 48, and 120 hpi, respectively (Figure 4B). The pustule density on the wheat leaves infected with BSMV:PsRPs26 decreased by 58% (Figure 4C). These results suggested that the PsRPs26 gene contributes to the pathogenicity of Pst.
Figure 4. Functional characterization of PsRPs26 during the interaction between wheat and stripe rust using host-induced gene silencing. (A) Phenotypic changes in the fourth leaves of plants pre-inoculated with the positive control vector (BSMV-PDS), FES buffer (CK), or empty BSMV vector (BSMV:γ). Phenotypes for the fourth leaves inoculated with the P. striiformis f. sp. tritici (Pst) CYR31 race at 14 dpi. (B) Relative transcript levels of PsRPs26 in the silenced leaves. BSMV:γ leaves infected with the Pst CYR31 race were used as a control. (C) Quantification of uredinial density in PsRPs26 knockdown plants 14 dpi with the CYR31 isolate. Means and standard deviations were calculated from three independent replicates. Significant differences were determined using Student’s t-test: ∗P < 0.05.
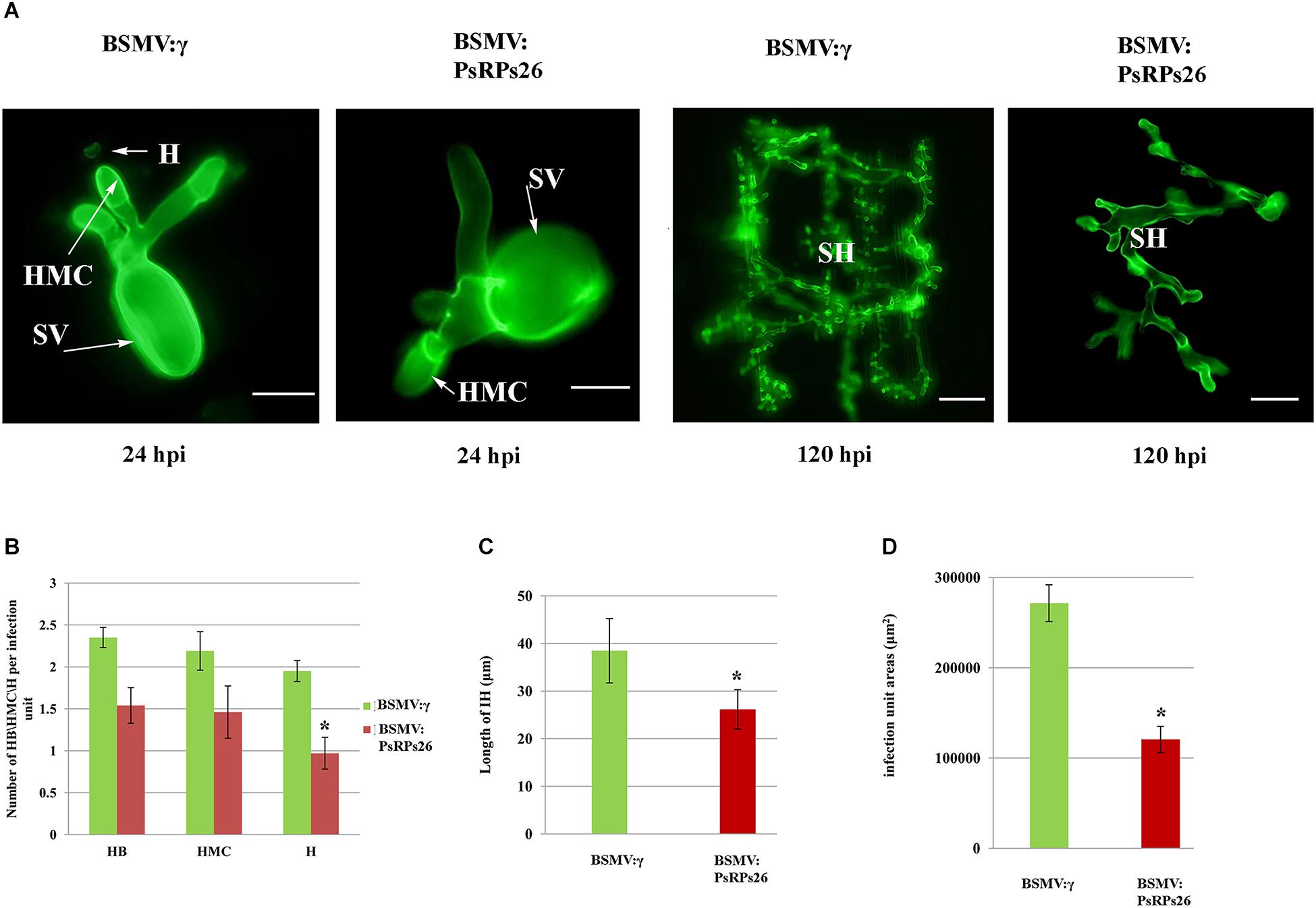
Figure 5. Histological determination of fungal growth in host cells. (A) Cytological observation of fungal development in PsRPs26 knockdown wheat leaves inoculated with the Pst CYR31 race. Leaves were sampled at 24 and 120 hpi with the CYR31 isolate. (B) The number of haustoria was significantly reduced in the PsRPs26-silenced plants at 24 hpi with the CYR31 isolate. (C) Significant decrease in the length of the infection hyphae in the PsRPs26-silenced plants at 48 hpi with the CYR31 isolate. (D) PsRPs26 -silenced plants show a significant decrease in infection unit area at 120 hpi. H, haustorium; SH, secondary hypha; HMC, haustorial mother cells; SV, substomatal vesicle (24 hpi, bar = 20 μm; 120 hpi, bar = 100 μm). Significant differences were determined using Student’s t-test: ∗P < 0.05.
Discussion
Ribosome biogenesis is essential for proliferation, cell growth, differentiation, and development (Zhou et al., 2015). RPs26 is an elemental protein of the ribosomal 40S subunit. Since the discovery of the first RPs26 in rat livers, RPs26s have been identified in diverse species, ranging from fungi to animals (Collatz et al., 1977; Hou et al., 2010; Sharifulin et al., 2011). However, little is known about the pathogenic function of RPs26 in cereal pathogenic fungi. In this study, we characterized the 40S ribosomal protein subunit, PsRPs26, from wheat leaves infected with Pst. PsRPs26 contained the conserved Y62–K70 motif typical of the eukaryotic RPs26 family. Interestingly, the PsRPs26 motif is highly conserved across different Pst isolates. These results suggested that PsRPs26 is one of the elemental structural proteins of the Pst ribosomal subunit.
Ribosomes are produced in the nucleus and are exported to the cytoplasm (Görlich and Kutay, 1999). Ribosomal subunits are organized within multiple cellular compartments: the cytoplasm, the nucleoplasm, and the nucleolus. 60S ribosomes undergo primary assembly in the cell nucleolus and are then transferred to the cytoplasm through the nucleoplasm for maturity (Nissan et al., 2002). Furthermore, RPS19 contributes to the function of ribosome biogenesis by translocating from the cytoplasm to the nucleus (Orrù et al., 2007). In this study, we determined that PsRPs26 localizes in both the nucleus and cytoplasm. These findings strongly support our hypothesis that PsRPs26 may function as a ribosomal protein subunit responsible for ribosome biogenesis.
Impairment of ribosome biogenesis can severely retard cell growth, as has been demonstrated in various eukaryotic systems. In Drosophila, RPs gene insufficiency reduces the number of ribosomes synthesized, leading to a small phenotype characterized by recessive lethality, delayed larval development, and small body size (Lambertsson, 1998; Saeboe-Larssen et al., 1998). RPL29-knockout mice displayed global skeletal growth defects that led to reduced postnatal viability in mammals (Kim-Safran et al., 2007). In S. cerevisiae, the RPS26A gene is essential for normal growth. Additionally, yeast cell is lethal when RPS26A and RPS26B genes are knocked out simultaneously (Strittmatter et al., 2006). In this study, we used the HIGS system to identify the function of PsRPs26 to Pst growth and discovered that PsRPs26 was knocked down during wheat-Pst interaction. PsRPs26 was efficiently silenced by 50–60%. The formation and development of haustoria and haustorial mother cells were restricted during the early stage of Pst infection in plants with PsRPs26 knocked down. Interestingly, PsRPs26 was upregulated during Pst-barberry interaction. Pst infection was similar to that of wheat, including haustorium formation, and fungal colonization (Cheng et al., 2015). Furthermore, silencing of PsRPs26 suppressed the formation of secondary hyphae and fungal colonization at 120 hpi with Pst, indicating that PsRPs26 is involved in fungi development during infection.
RPs26 function has been well studied in yeast and mammals. However, little is known regarding PsRPs26 function in pathogenic fungi. In this study, the observed knock-down effect of PsRPs26 during wheat-Pst interaction led to increased resistance against infection by the CYR31 isolate. However, there is little direct functional evidence to show which genes were involved in rust pathogenicity because mutagenesis and transformation protocols are lacking in Pst. There are two possible reasons that can explain how PsRPs26 contribute to Pst pathogenicity. The first proposes that PsRPs26 silencing suppresses Pst growth and development during infection, as indicated by the reduced number of uredinia on wheat leaves. Interestingly, PsRPs26 was highly upregulated at 168–196 hpi with the CYR31 isolate, which corresponded with initiation of sporulation. PsRPs26 was efficiently silenced, as characterized by limited urediospore production when compared to the controls. Thara et al. (2003) reported that a large number of fungal-like RPs, including RPs26, were characterized from susceptible wheat leaves infected with Puccinia triticina during sporulation, a stage likely to require vigorous protein synthesis. We, therefore, infer that the second reason may be silencing PsRPs26 influences protein synthesis during sporulation. All these data suggested that PsRPs26 contributes to Pst pathogenicity. However, the complex mechanism of how PsRPs26 affects growth and pathogenicity needs to be studied further.
In conclusion, PsRPs26 contained a eukaryotic-specific motif, suggesting that fungi and animals share conserved ribosomal protein components. PsRPs26 were localized in both the nucleus and cytoplasm, indicating that PsRPs26 might participate in ribosome biogenesis. PsRPs26 was induced in wheat infected with the Pst CYR31 isolate and contributed positively to the growth, development, and pathogenicity of Pst. Our findings may facilitate the design of fungicides for controlling disease in cereal crops.
Author Contributions
BW carried out most of the experiments. BW, CT, and ZK wrote the manuscript. NS and JM performed the quantitative RT-PCR and analyzed the data. YS and NW grew the plant samples. NW collected all the phenotypic data and ZK revised the manuscript. All authors read and approved the final manuscript.
Funding
This study was supported by grants from the National Natural Science Foundation of China (Grant Nos. 31801721 and 31772116).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.00968/full#supplementary-material
FIGURE S1 | Multiple sequence alignment of PsRPs26 among different Pst isolates. The black line represents the Y62–K70 motif site.
TABLE S1 | Primers used in this study.
Footnotes
References
Belyy, A., Levanova, N., Tabakova, I., Rospert, S., and Belyi, Y. (2016). Ribosomal protein Rps26 influences 80S ribosome assembly in Saccharomyces cerevisiae. mSphere 1, e109–e115. doi: 10.1128/mSphere.00109-15
Cheng, Y., Wang, X., Yao, J., Voegele, R. T., Zhang, Y., Wang, W., et al. (2015). Characterization of protein kinase PsSRPKL, a novel pathogenicity factor in the wheat stripe rust fungus. Environ. Microbiol. 17, 2601–2617. doi: 10.1111/1462-2920.12719
Collatz, E., Ulbrich, N., Tsurugi, K., Lightfoot, H. N., MacKinlay, W., Lin, A., et al. (1977). Isolation of eukaryotic ribosomal proteins. purification and characterization of the 40 S ribosomal subunit proteins Sa, Sc, S3a, S3b, S5’, S9, S10, S11, S12, S14, S15, S15’, S16, S17, S18, S19, S20, S21, S26, S27’, and S29. J. Biol. Chem. 252, 9071–9080.
Doherty, L., Sheen, M. R., Vlachos, A., Choesmel, V., O’Donohue, M. F., Clinton, C., et al. (2010). Ribosomal protein genes RPS10 and RPS26 are commonly mutated in diamond-blackfan anemia. Am. J. Hum. Genet. 86, 222–228. doi: 10.1016/j.ajhg.2009.12.015
Ferreira-Cerca, S., Pöll, G., Kühn, H., Neueder, A., Jakob, S., Tschochner, H., et al. (2007). Analysis of the in vivo assembly pathway of eukaryotic 40S ribosomal proteins. Mol. Cell 28, 446–457. doi: 10.1016/j.molcel.2007.09.029
Filipenko, M. L., Vinichenko, N. A., Karpova, G. G., Mertvetsov, N. P., and Amaldi, F. (1998). Isolation, structural analysis and mapping of the functional gene of human ribosomal protein S26. Gene 211, 287–292. doi: 10.1016/s0378-1119(98)00108-5
Fisher, M. C., Henk, D. A., Briggs, C. J., Brownstein, J. S., Madoff, L. C., McCraw, S. L., et al. (2012). Emerging fungal threats to animal, plant and ecosystem health. Nature 484, 186–194. doi: 10.1038/nature10947
Görlich, D., and Kutay, U. (1999). Transport between the cell nucleus and the cytoplasm. Annu. Rev. Cell Dev. Biol. 15, 607–660. doi: 10.1146/annurev.cellbio.15.1.607
Hein, I., Barciszewska-Pacak, M., Hrubikova, K., Williamson, S., Dinesen, M., Soenderby, I. E., et al. (2005). Virus-induced gene silencing-based functional characterization of genes associated with powdery mildew resistance in barley. Plant Physiol. 138, 2155–2164. doi: 10.1104/pp.105.062810
Holzberg, S., Brosio, P., Gross, C., and Pogue, G. P. (2002). Barley stripe mosaic virus-induced gene silencing in a monocot plant. Plant J. 30, 315–327. doi: 10.1046/j.1365-313x.2002.01291.x
Hou, Y. L., Sun, B., and Hou, W. R. (2010). cDNA cloning and sequence analysis of ribosomal protein S26 gene (rps26) from the giant panda. J. Beijing Norm Univ. Nat. Sci. 46, 177–181.
Ivanov, A. V., Malygin, A. A., and Karpova, G. G. (2005). Human ribosomal protein S26 suppresses the splicing of its pre-mRNA. Biochim. Biophys. Acta 1727, 134–140. doi: 10.1016/j.bbaexp.2004.12.011
Jorgensen, P., Nishikawa, J. L., Breitkreutz, B. J., and Tyers, M. (2002). Systematic identification of pathways that couple cell growth and division in yeast. Science 297, 395–400. doi: 10.1126/science.1070850
Kang, Z., Huang, L., and Buchenauer, H. (2002). Ultrastructural changes and localization of lignin and callose in compatible and incompatible interactions between wheat and Puccinia striiformis/Ultrastrukturelle veränderungen und lokalisierung von lignin und kailose in kompatiblen und inkompatiblen interaktionen zwischen weizen und Puccinia striiformis. J. Plant Dis. Prot. 109, 25–37.
Karsi, A., Patterson, A., Feng, J., and Liu, Z. (2002). Translational machinery of channel catfish: i. a transcriptomic approach to the analysis of 32 40S ribosomal protein genes and their expression. Gene 291, 177–186. doi: 10.1016/s0378-1119(02)00595-4
Kim-Safran, C. B., Oristian, D. S., Focht, R. J., Parker, S. G., Vivian, J. L., and Carson, D. D. (2007). Global growth deficiencies in mice lacking the ribosomal protein HIP/RPL29. Dev. Dyn. 236, 447–460. doi: 10.1002/dvdy.21046
Lambertsson, A. (1998). 3 The minute genes in Drosophila and their molecular functions. Adv. Genet. 38, 69–134. doi: 10.1016/s0065-2660(08)60142-x
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2- ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Ma, J., Huang, X., Wang, X., Chen, X., Qu, Z., Huang, L., et al. (2009). Identification of expressed genes during compatible interaction between stripe rust (Puccinia striiformis) and wheat using a cDNA library. BMC Genomics 10:586. doi: 10.1186/1471-2164-10-586
Malygin, A., Baranovskaya, O., Ivanov, A., and Karpova, G. (2003). Expression and purification of human ribosomal proteins S3, S5, S10, S19, and S26. Protein Expr. Purif. 28, 57–62. doi: 10.1016/s1046-5928(02)00652-6
Malygin, A. A., and Karpova, G. G. (2009). Structural motifs of the bacterial ribosomal proteins S20, S18 and S16 that contact rRNA present in the eukaryotic ribosomal proteins S25, S26 and S27A, respectively. Nucleic Acids Res. 38, 2089–2098. doi: 10.1093/nar/gkp1170
Nissan, T. A., Baßler, J., Petfalski, E., Tollervey, D., and Hurt, E. (2002). 60S pre-ribosome formation viewed from assembly in the nucleolus until export to the cytoplasm. EMBO J. 21, 5539–5547. doi: 10.1093/emboj/cdf547
Orrù, S., Aspesi, A., Armiraglio, M., Caterino, M., Loreni, F., Ruoppolo, M., et al. (2007). Analysis of the ribosomal protein S19 interactome. Mol. Cell. Proteomics 6, 382–393. doi: 10.1074/mcp.m600156-mcp200
Saeboe-Larssen, S., Lyamouri, M., Merriam, J., Oksvold, M. P., and Lambertsson, A. (1998). Ribosomal protein insufficiency and the minute syndrome in Drosophila: a dose-response relationship. Genetics 148, 1215–1224.
Sharifulin, D., Khairulina, Y., Ivanov, A., Meschaninova, M., Ven’yaminova, A., Graifer, D., et al. (2011). A central fragment of ribosomal protein S26 containing the eukaryote-specific motif YxxPKxYxK is a key component of the ribosomal binding site of mRNA region 5’ of the E site codon. Nucleic Acids Res. 40, 3056–3065. doi: 10.1093/nar/gkr1212
Steffen, K. K., McCormick, M. A., Pham, K. M., MacKay, V. L., Delaney, J. R., Murakami, C. J., et al. (2012). Ribosome deficiency protects against ER stress in Saccharomyces cerevisiae. Genetics 191, 107–118. doi: 10.1534/genetics.111.136549
Strittmatter, A. W., Fischer, C., Kleinschmidt, M., and Braus, G. H. (2006). FLO11 mediated filamentous growth of the yeast Saccharomyces cerevisiae depends on the expression of the ribosomal RPS26 genes. Mol. Genet. Genomics 276, 113–125. doi: 10.1007/s00438-006-0127-7
Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M., and Kumar, S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739. doi: 10.1093/molbev/msr121
Thara, V. K., Fellers, J. P., and Zhou, J. M. (2003). In planta induced genes of Puccinia triticina. Mol. Plant Pathol. 4, 51–56. doi: 10.1046/j.1364-3703.2003.00142.x
Thompson, J. D., Higgins, D. G., and Gibson, T. J. (1994). CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680. doi: 10.1093/nar/22.22.4673
Verschoor, A., Warner, J. R., Srivastava, S., Grassucci, R. A., and Frank, J. (1998). Three-dimensional structure of the yeast ribosome. Nucleic Acids Res. 26, 655–661. doi: 10.1093/nar/26.2.655
Wang, B., Song, N., Zhang, Q., Wang, N., and Kang, Z. (2018). TaMAPK4 acts as a positive regulator in defense of wheat stripe-rust infection. Front. Plant Sci. 9:152. doi: 10.3389/fpls.2018.00152
Wang, B., Sun, Y., Song, N., Zhao, M., Liu, R., Feng, H., et al. (2017). Puccinia striiformis f. sp. tritici microRNA-like RNA 1 (Pst-milR1), an important pathogenicity factor of Pst, impairs wheat resistance to Pst by suppressing the wheat pathogenesis-related 2 gene. New Phytol. 215, 338–350. doi: 10.1111/nph.14577
Wang, C. F., Huang, L. L., Buchenauer, H., Han, Q. M., Zhang, H. C., and Kang, Z. S. (2007). Histochemical studies on the accumulation of reactive oxygen species (O2- and H2O2) in the incompatible and compatible interaction of wheat-Puccinia striiformis f. sp. tritici. Physiol. Mol. Plant Pathol. 71, 230–239. doi: 10.1016/j.pmpp.2008.02.006
Wang, X., Tang, C., Zhang, G., Li, Y., Wang, C., Liu, B., et al. (2009). cDNA-AFLP analysis reveals differential gene expression in compatible interaction of wheat challenged with Puccinia striiformis f. sp. tritici. BMC Genomics 10:289. doi: 10.1186/1471-2164-10-289
Warner, J. R., and Nierras, C. R. (1998). Trapping human ribosomal protein genes. Genome Res. 8, 419–421. doi: 10.1101/gr.8.5.419
Wool, I. G. (1979). The structure and function of eukaryotic ribosomes. Annu. Rev. Biochem. 48, 719–754. doi: 10.1146/annurev.bi.48.070179.003443
Wool, I. G., Chan, Y. L., and Glück, A. (1995). Structure and evolution of mammalian ribosomal proteins. Biochem. Cell Biol. 73, 933–947. doi: 10.1139/o95-101
Zheng, W., Huang, L., Huang, J., Wang, X., Chen, X., Zhao, J., et al. (2013). High genome heterozygosity and endemic genetic recombination in the wheat stripe rust fungus. Nat. Commun. 4:2673. doi: 10.1038/ncomms3673
Keywords: ribosomal subunit, growth, Puccinia striiformis, wheat, RPs26
Citation: Wang B, Song N, Tang C, Ma J, Wang N, Sun Y and Kang Z (2019) PsRPs26, a 40S Ribosomal Protein Subunit, Regulates the Growth and Pathogenicity of Puccinia striiformis f. sp. Tritici. Front. Microbiol. 10:968. doi: 10.3389/fmicb.2019.00968
Received: 15 January 2019; Accepted: 16 April 2019;
Published: 10 May 2019.
Edited by:
Dirk Albert Balmer, Syngenta, SwitzerlandReviewed by:
Peter Michael Dracatos, The University of Sydney, AustraliaChongjing Xia, Washington State University, United States
Copyright © 2019 Wang, Song, Tang, Ma, Wang, Sun and Kang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhensheng Kang, a2FuZ3pzQG53c3VhZi5lZHUuY24=
†These authors have contributed equally to this work
 Bing Wang
Bing Wang Na Song1†
Na Song1† Jinbiao Ma
Jinbiao Ma Zhensheng Kang
Zhensheng Kang