- 1Life and Health Sciences Research Institute (ICVS), School of Medicine, Braga, Portugal
- 2ICVS/3B’s - PT Government Associate Laboratory, University of Minho, Braga, Portugal
- 3Biotechnology Unit, Yucatan’s Center for Scientific Research (CICY), Merida, Mexico
- 4Laboratory of Cellular Mycology and Proteomics, Biology Institute, University of Rio de Janeiro State, Rio de Janeiro, Brazil
- 5Centre of Molecular and Environmental Biology (CBMA), Department of Biology, University of Minho, Braga, Portugal
- 6Genomic Science and Biotechnology, Catholic University of Brasília, Brasília, Brazil
- 7Department of Cellular Biology, University of Brasília, Brasília, Brazil
- 8Laboratório de Bioinformática, Laboratório Nacional de Computação Científica, Petrópolis, Brazil
- 9Faculdade de Ciências Farmacêuticas de Ribeirão Preto, Universidade de São Paulo, São Paulo, Brazil
- 10Faculdade de Ciências Farmacêuticas, Universidade de São Paulo, São Paulo, Brazil
The pathogenic clade of the Sporothrix genus comprises the etiological agents of sporotrichosis, a worldwide emergent disease. Despite the growing understanding of their successful pathogen traits, there is little information on genome sizes and ploidy within the genus. Therefore, in this work, we evaluated the ploidy of four species of the Sporothrix genus, specifically Sporothrix brasiliensis, Sporothrix schenckii, Sporothrix globosa, and Sporothrix pallida. Through cell cycle analysis of the yeast-phase cells, we showed that the DNA content of G0/G1 cells was similar to the genome size determined by whole genome sequencing. Moreover, ploidy of S. schenckii, S. brasiliensis, and S. pallida that was determined by allele composition using next-generation sequencing (NGS) data is consistent with monomorphic positions at each allele. These data show that the analyzed strains of Sporothrix are haploid, or at least aneuploid, thereby laying the foundation for the development of a molecular toolbox for Sporothrix spp.
Introduction
Fungi of the genus Sporothrix comprise four species of a pathogenic clade that are the etiological agents of sporotrichosis, a chronic subcutaneous infection that affects humans and other mammals (Teixeira et al., 2014; de Beer et al., 2016). Sporotrichosis is the most prevalent subcutaneous mycosis worldwide, despite being frequently neglected (Queiroz-Telles et al., 2017). Although this mycosis is mainly attributed to Sporothrix schenckii, new clinically relevant species are emerging, namely, Sporothrix globosa, Sporothrix lurei, and Sporothrix brasiliensis, the latter being the most virulent species (Marimon et al., 2007; Arrillaga-Moncrieff et al., 2009; Castro et al., 2013; Zhang et al., 2015).
Infection usually results from the direct inoculation of fungal cells into traumatized cutaneous lesions arising via plant- or animal-invoked injuries (Rodrigues et al., 2016). Subsequently, in the majority of cases, nodules develop in the infection site leading to ulceration (Kauffman, 1999). Poorly controlled sporotrichosis can disseminate to distant anatomical sites, including bones, lungs, and central nervous system (Ferreira et al., 2016; Hassan et al., 2016; Rodrigues et al., 2016; Mialski et al., 2018). Apart from being a global health problem, sporotrichosis is hyperendemic in Brazil, mainly due to the zoonotic and crossed transmission of S. brasiliensis from infected felines (Montenegro et al., 2014; Sanchotene et al., 2015; Gremião et al., 2017). This mycosis is also endemic in Asia, where regions with high incidence of S. globosa infection are reported (Zhang et al., 2015). Species of the Sporothrix genus exhibit thermal dimorphism, with a yeast phase at 37°C, while at 25°C, the filamentous form is predominant (Kauffman, 1999). Similar to other human pathogens, this dimorphic behavior is an important factor for virulence (Lopes-Bezerra et al., 2006; Téllez et al., 2014). Accordingly, not only yeast cells can be mediators of zoonotic transmission (Gremião et al., 2017), but also mycelium can be present in the environment and transmitted by cat scratches.
The genome structure of S. schenckii, S. brasiliensis, and S. globosa is similar with a total genome length of 32.4, 33.2, and 33.4 Mb, respectively (Teixeira et al., 2014; Huang et al., 2016). On the other hand, S. pallida has a genome length of 37.8 Mb, while S. mexicana genome size is still unknown (D’Alessandro et al., 2016). Moreover, S. schenckii is reported to have high genetic diversity, compared to S. brasiliensis and S. globosa (Rangel-Gamboa et al., 2016, 2018). Analysis of sequence data from S. globosa revealed lack of genetic heterozygosity (Huang et al., 2016). Fungi of the S. schenckii complex are highly polymorphic regarding chromosome number and size (Marimon et al., 2007; Sasaki et al., 2014). Additionally, S. schenckii is thought to be a diploid organism, whereas the ploidy of other species of the complex has not been described so far (Torres-Guerrero, 1999).
Therefore, in the present work, we developed a flow cytometry (FCM) protocol for cell cycle analysis (Almeida et al., 2007) in order to determine the DNA content per cell (DNAC) of S. brasiliensis, S. schenckii, S. globosa, and S. pallida yeast cells. Ploidy of the analyzed strains was determined from the comparison of the DNAC with the previously reported genome size, and further validated for S. schenckii, S. brasiliensis, and S. pallida with the analysis of the distribution of base frequencies at variable sites in the genome using the next-generation sequencing (NGS) data.
Materials and Methods
Microorganisms and Culture Media
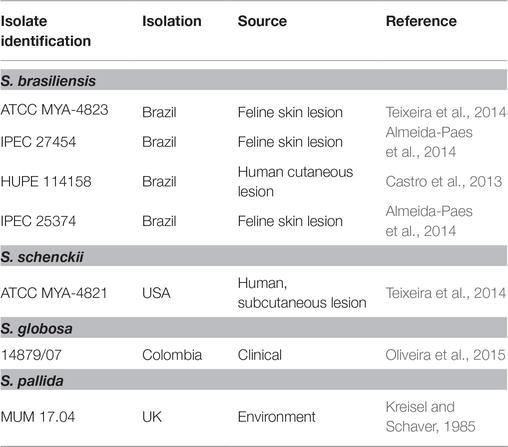
The strains of the Sporothrix genus used in this study are listed in Table 1. Yeast cells were maintained at 37°C in yeast extract peptone dextrose (YPD) solid medium (2% glucose, 1% peptone, 0.5% yeast extract, and 1.5% agar; pH = 7.8). For the subsequent assay, yeast cells were cultured in YPD liquid medium at 37°C with aeration on a mechanical shaker (150 rpm). Conidia were obtained after incubation in YPD liquid medium at 25°C with mechanical aeration (150 rpm) for 3 days. Conidia were recovered through successive gaze filtration. Aspergillus nidulans A4 and R21/R153 (dos Reis et al., 2018), haploid and diploid strains, respectively, were maintained in YPD solid medium at 30°C. For subsequent analysis, conidia were collected and washed with phosphate buffered saline (PBS) (1×) (8 g NaCl, 0.2 g KCl, 1.44 g Na2HPO4, 0.24 g KH2PO4 per liter of sterilized water).
Measurements of DNA Content per Cell
The DNA content of isolated cells was determined accordingly to the protocol described in Almeida et al. (2007) with modifications. Exponentially grown yeast cells and conidia were collected, centrifuged (13,000 rpm for 3 min), and washed with PBS (1×). To obtain an uniform single-cell suspension, collected cells were filtered through sterile gauze and fixed overnight with ethanol 70% (vol/vol) at 4°C. Afterwards, cells were harvested, washed, and resuspended in 850 μl of sodium citrate buffer (50 mM; pH = 7.5). Briefly sonicated Sporothrix spp. cells were treated at 50°C for 4 h with RNase A (0.50 mg/ml) (GRiSP, Porto, Portugal) and for 2 h with proteinase K (1 mg/ml) (GRiSP). A. nidulans conidia, after a brief sonication, were treated at 50°C for 2 h with RNase A (0.50 mg/ml) (GRiSP) and for 2 h with proteinase K (1 mg/ml) (GRiSP). A volume of 50 μl of treated cells was stained overnight with SYBR Green I (10,000×) (Invitrogen, CA, USA) at 4°C. For the yeast cells, a concentration of SYBR Green I of 2% (vol/vol) was used from a 10-fold diluted working solution in Tris-EDTA (pH 8.0) (Sigma-Aldrich). For the case of conidia, a final concentration of 12% (vol/vol) of SYBR Green I (10,000×) was used. Prior to flow cytometry (FCM) analysis, 750 μl of sodium citrate buffer (50 mM; pH 7.5) with 0.25% (vol/vol) of Triton X-100 (Sigma-Aldrich) was added to the stained cells.
Flow Cytometry
Stained cells were analyzed in a FACS LSRII flow cytometer (Becton Dickinson, NJ, USA) with a 488 nm excitation laser. Signals from a minimum of 30,000 cells per sample were captured in FITC channel (530 nm ± 30 nm) at low flow rate of about 1,000 cells/s and an acquisition protocol was defined to measure forward scatter (FSC) and side scatter (SSC) on a four-decade logarithmic scale and mean green fluorescence intensity of SYBR Green I (MFISGI) on a linear scale. FACS Diva was used as the acquisition software. Results were analyzed with FlowJo (Tree Star, OR, USA) software, version 10, and the coefficients of variation (CV), as well as MFISGI, were estimated using Modfit LT software (Verity Software House, ME, USA).
Fluorescence Microscopy
Yeast cells of genome strains, S. schenckii ATCC MYA-4821 and S. brasiliensis ATCC MYA-4823, and S. pallida MUM 17.04 labeled with SYBR Green I were analyzed using a fluorescence microscope (DP71 Olympus). The images (1,036 × 1,024 pixels) were acquired with an oil immersion objective (100×/1.40) and analyzed with cellSens imaging software (Olympus Life Science, Tokyo, Japan).
Ploidy Estimation by Next-Generation Sequencing Data
Raw sequencing data for S. schenckii (SRX342487) and S. pallida (SRX550176) were downloaded, in FASTQ format, from NCBI’s Sequence Read Archive through the SRA-toolkit (Leinonen et al., 2011). The original NGS sequencing files for S. brasiliensis were used (Teixeira et al., 2014). As controls, the raw sequencing data for Aspergillus flavus NRRL 3357 (haploid, SRX2124714) and Candida albicans SC 5314 (diploid, SRX2250255) were also obtained from NCBI-SRA.
Short raw reads for each species were aligned to the corresponding assembled genome (S. schenckii 1,099–18 GCF_000961545.1, S. brasiliensis 5110 GCA_000820605.1, S. pallida SPA8 GCA_000710705.2, A. flavus NRRL357 GCF_000006275.2, and C. albicans SC5314 GCF_000182965.3) with Bowtie2 (Leinonen et al., 2011) using the “very-sensitive” option. If raw reads were paired, “no-mixed, no-discordant, no-overlap, no-contain” parameters were also used. For each genome, the resulting SAM files were converted to BAM files and sorted with samtools (Li et al., 2009), and then split by chromosomes/scaffolds using bamtools (Barnett et al., 2011).
Allele frequencies at every position along each chromosome/scaffold for all genomes were calculated using the “ploidyNGS” algorithm (Corrêa dos Santos et al., 2017), with default parameters (max_allele_freq 0.95, max_depth 100). Chromosome/scaffold frequency files for each genome were then merged. Ploidy was estimated by constructing histograms from the frequency files and comparing to the haploid and diploid controls.
Statistical Analysis
Data are reported as the mean ± standard deviation (SD) of at least three independent assays. Data were analyzed using MS Excel 2016 (Office, Microsoft®, WA, USA) and GraphPad Prism Software, version 7.0 (GraphPad Software Inc., CA, USA). A linear regression was calculated to predict the MFISGI based on genome size (Mb).
Results and Discussion
The DNA Content of Sporothrix spp. Yeast Cells
Measuring the DNA content is a well-established method to monitor cell proliferation, cell cycle, and cell ploidy. Despite the relevance of Sporothrix spp. in human and animal diseases, ploidy determination in species of this genus is scarce. Indeed, the diploid status of S. schenckii has been reported (Torres-Guerrero, 1999). To determine the DNA content of the yeast cells of different species of the Sporothrix genus, a protocol for cell cycle analysis using FCM (Fortuna et al., 2000; Almeida et al., 2007) was applied with modifications to obtain lower coefficients of variation. The CV is a measurement of the peak width of DNA from cells in G0/G1 phase of the cell cycle and therefore, low CV values are critical for the accuracy of cellular ploidy determination (Fortuna et al., 2000; Almeida et al., 2007). A minimum of cellular debris and clumps was achieved by using exponentially growing cells and gauze filtration (data not shown). Cell separation and the absence of filamentous forms were confirmed by optical microscopy. The RNase A and proteinase K treatments were optimized to achieve CVs lower than 7, as described elsewhere (Table 2; Rodrigues et al., 2003; Almeida et al., 2007). Additionally, different SYBR Green I concentrations – 0.2, 2, 4, and 10% – were used for DNA staining, ensuring that the dye was not limiting (data not shown). Fluorescence microscopy analysis revealed specific nuclear staining, as expected for SYBR Green I, for the analyzed strains (Figure 1).
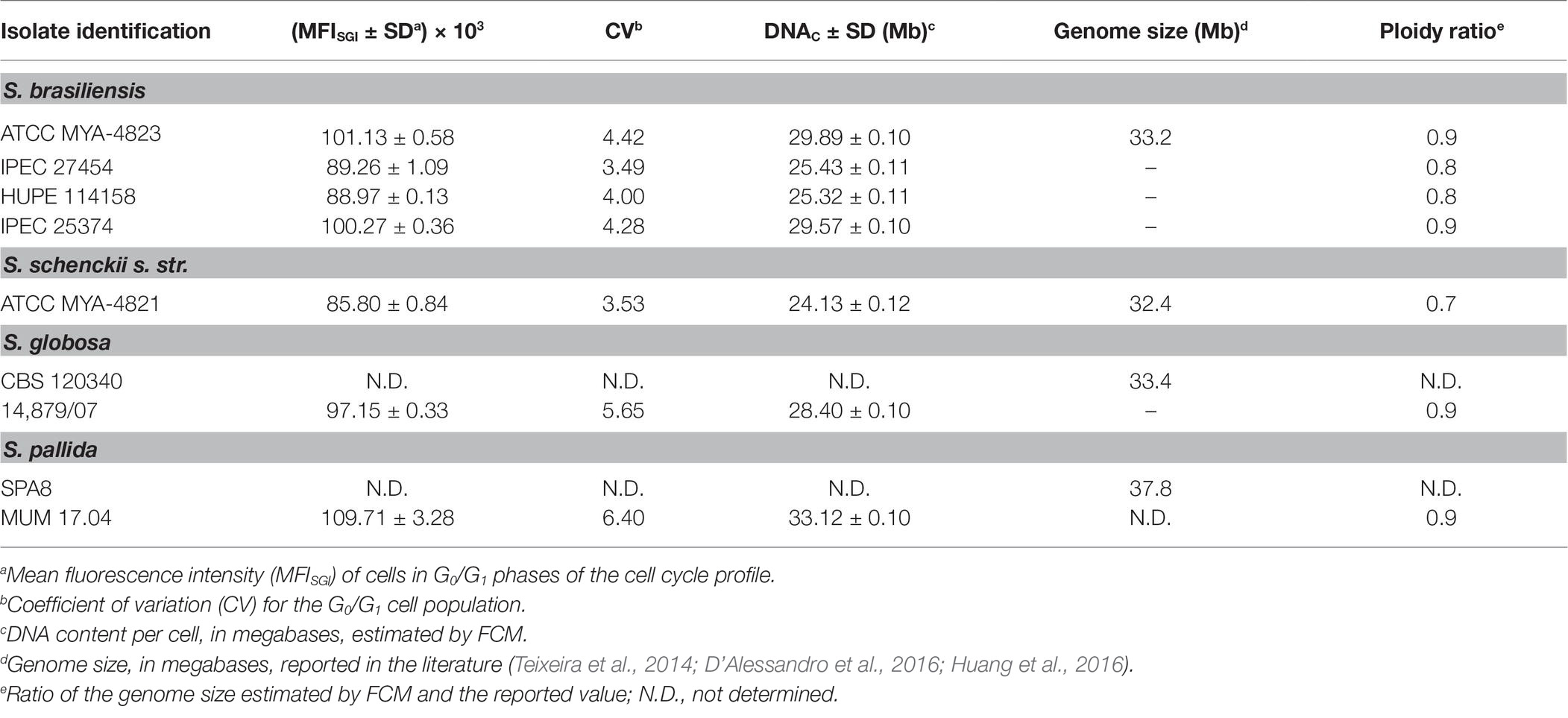
Table 2. Mean fluorescent intensity and corresponding calculated DNA content per cell (DNAC) (Mb) of Sporothrix genus strains estimated by flow cytometry (FCM) of SYBR Green I stained cells.
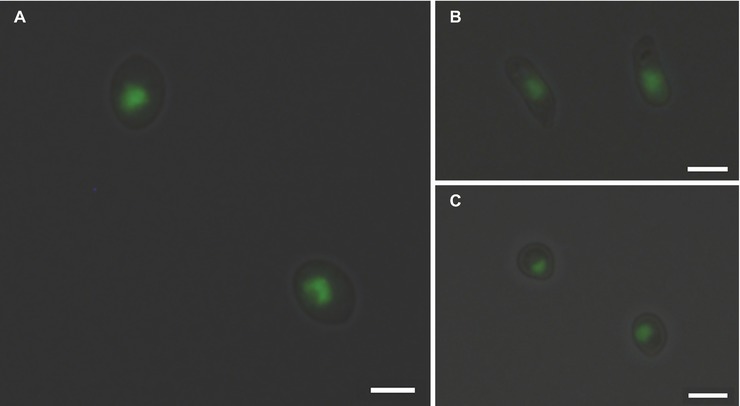
Figure 1. SYBR Green I nuclear staining of Sporothrix spp. Fluorescence microscopy analysis of (A) S. brasiliensis ATCC MYA-4823, (B) S. schenckii ATCC MYA-4821, and (C) S. pallida MUM 17.04 stained with SYBR Green I reveals the presence of mononucleated yeast cells. Images (1,036 × 1,024 pixels) were acquired in a fluorescence microscope (DP71 Olympus) with an oil immersion objective (100×/1.4) and cropped using the cellSens imaging software (Olympus Life Science). Scale bar equals 3 μm.
FCM allows the establishment of a direct correlation between MFISGI and the DNAC (Rodrigues et al., 2003). As a first approach, the genome size of Sporothrix spp. was estimated using a correlation based in the MFISGI and the DNAC of haploid and diploid strains of Saccharomyces cerevisiae (data not shown). The utilization of this well-established DNA content cell standard would allow the analysis of cell populations with n, 2n, and 4n of DNA content. However, SYBR Green I exhibits a preferential binding to GC-rich sequences (Gudnason et al., 2007), with S. cerevisiae and Sporothrix spp. presenting different GC content. In fact, whereas for S. cerevisiae, the percentage of GC content is around 38%, for S. brasiliensis and S. schenckii, the values are of 62%, being for S. globosa and S. pallida of 54 and 52%, respectively (Teixeira et al., 2014; D’Alessandro et al., 2016; Huang et al., 2016). Thus, for the analysis of the cell DNA content of Sporothrix spp., we took advantage of haploid and diploid strains of A. nidulans as reference, which presents a GC content of 50% (Galagan et al., 2005). As such, from the described haploid genome size of A. nidulans (30.07 Mb) (Galagan et al., 2005) and the MFISGI obtained for each cellular DNA content (n and 2n, corresponding to G0/G1 phase of the cell cycle of haploid and diploid A. nidulans conidia) (Figure 2A1), a linear regression was established – MFISGI = (2,659 ± 141.6)*DNAC + (21,647 ± 6,726) (Figure 2A2; R 2 = 0.9888).
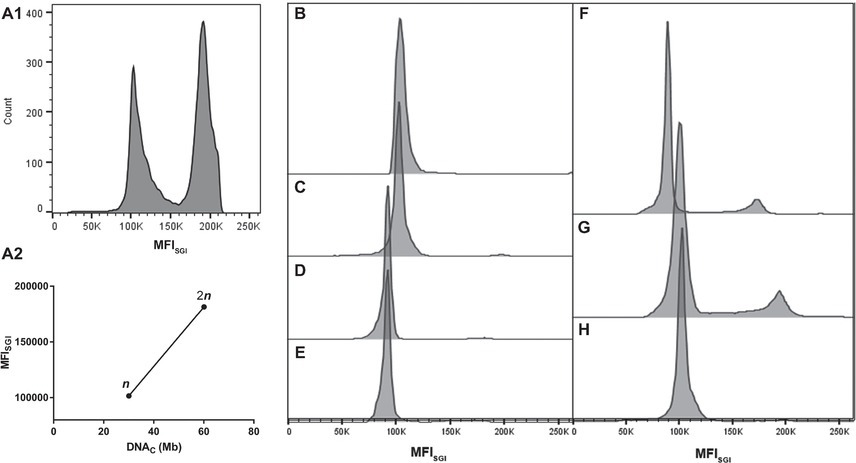
Figure 2. Representative histograms of cell cycle analysis of the analyzed microorganisms. (A) Analyses of A. nidulans strains: (A1) mixed populations of A. nidulans haploid and diploid strains and (A2) graph showing a typical standard curve relating MFISGI of n and 2n peaks of A. nidulans strains and the theoretical amount of DNA per cell (DNAC). Histograms for: S. brasiliensis (B) ATCC MYA-4823, (C) IPEC 25374, (D) IPEC 27454, and (E) HUPE 114158; (F) S. schenckii ATCC MYA-4821; (G) S. globosa 14879/07; and (H) S. pallida MUM 17.04.
Representative histograms for S. brasiliensis, S. schenckii, S. globosa, and S. pallida obtained by FCM are shown in Figures 2B–H, respectively. This figure includes the histograms obtained for the fully sequenced strains: S. brasiliensis ATCC MYA-4823 and S. schenckii ATCC MYA-4821 (Figures 2B,F, respectively) (Teixeira et al., 2014). The average of the DNAC estimated according to the cells in G0/G1 phase of the cell cycle for the different strains of S. brasiliensis analyzed ranged between 25.32 and 29.89 Mb, and was of 24.13, 28.40, and 33.12 Mb for S. schenckii, S. globosa, and S. pallida, respectively (Table 2). Moreover, S. brasiliensis ATCC MYA-4823 and S. schenckii ATCC MYA-4821 conidia presented identical values of MFISGI to those presented for the yeast form (Figure 3). Overall, these data suggest that there are no variations in ploidy in the morphological transition from conidia to yeast cells of these Sporothrix spp.
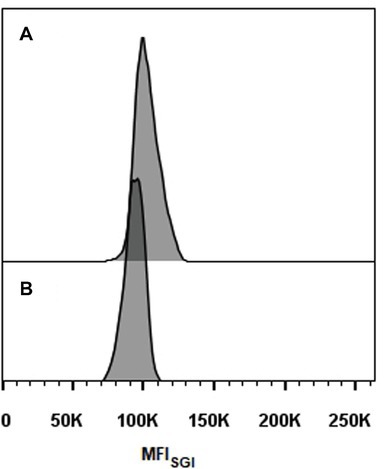
Figure 3. Histograms of cell cycle analysis of Sporothrix spp. conidia. (A) S. brasiliensis ATCC MYA-4823 and (B) S. schenckii ATCC MYA-4821.
Ploidy Estimation of Sporothrix spp. by Flow Cytometry and Next-Generation Sequencing Data
To determine the ploidy of the analyzed organisms, a comparison was performed between the DNAC of cells in G0/G1 phase of the cell cycle, determined by FCM (Table 2), and the genome length reported for each organism from the full sequence analysis. The ploidy state of the studied organisms was inferred by the ratio obtained from these two parameters (designed as ploidy ratio) (Almeida et al., 2007). S. schenckii presented a ploidy ratio of 0.7, suggestive of a haploid profile, although aneuploidy cannot be excluded (Table 2). In what regards S. brasiliensis, the ploidy ratio ranged from 0.9, for the sequenced strain, to 0.8 in other isolates (Table 2). These data point for a haploid profile in these strains. For the analyzed strains of S. globosa and S. pallida, a ploidy ratio of 0.9 was obtained, suggestive also of a haploid profile. Additionally, a DNAC for S. mexicana MUM 17.07 identical to the other isolates tested was obtained suggesting a haploid status, considering a genome length similar to the other Sporothrix spp. (data not shown).
Ploidy can be determined indirectly – without measuring cellular DNAC – from the analysis of the short-read sequencing data generated by NGS experiments. Therefore, to strengthen our data, this methodology was also applied to determine the ploidy of Sporothrix spp. Briefly, a typical NGS run produce millions of reads coming from every piece of DNA present on the original sample. After mapping, the NGS reads to an assembled genome, the frequency of variations at every position along the genome can be interpreted to be supporting different alleles at each position and used to infer the ploidy level. In the case of a haploid organism, a single allele for the vast majority of reads it is expected (monomorphic positions), whereas for diploids, polymorphic positions are expected where half of the readings support one allele and the other half support an alternative allele. As controls, we analyzed the allele frequency of NGS data from haploid and diploid fungus, A. flavus and C. albicans, respectively (Figures 4A,B).
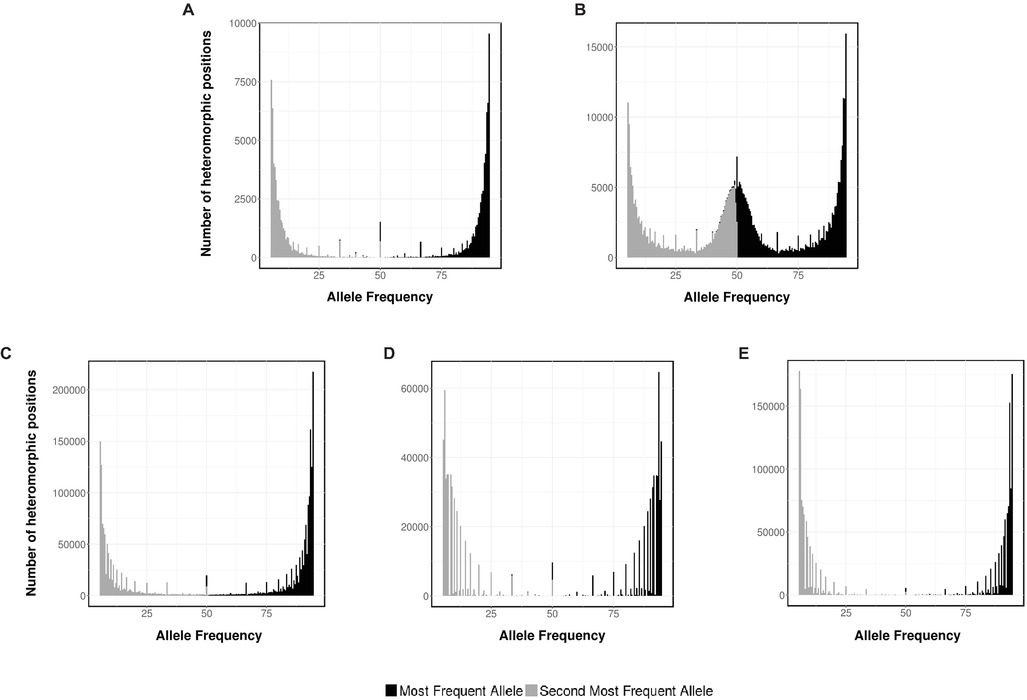
Figure 4. Ploidy estimation by NGS data. Distribution of the two most frequent putative alleles for (A) A. flavus NRRL 3357, (B) C. albicans SC 5314, (C) S. schenckii ATCC MYA-4821, (D) S. brasiliensis ATCC MYA-4823, and (E) S. pallida CBS 120340.
As expected, for A. flavus the most abundant allele is close to 95% (monomorphic positions). Conversely, for C. albicans, the histogram displays monomorphic positions, with a frequency close to 95%, and heterozygous positions, with frequencies close to 50% for the first and second more frequent base. The allele frequency plots for S. schenckii, S. brasiliensis, and S. pallida (Figures 4C–E, respectively) present a single peak for both the most abundant allele (close to 95%) and the second most abundant one (close to 5%), supporting the hypothesis that these three strains of Sporothrix spp. are haploid organisms. For S. globosa, a low heterozygosity was also previously described (Huang et al., 2016). The differences on the densities of the histograms can be accounted due to the difference on depth coverage and NGS sequencing technology used in each case (S. schenckii Illumina HiSeq 2000, S. brasiliensis 454 GS FLX, and S. pallida Ion Torrent PGM).
Conclusion
Our results exclude a diploid DNA content for these organisms and propose a haploid or at least a near haploid profile for S. schenckii, S. brasiliensis, S. globosa, and S. pallida.
The construction of whole-genome knockout collections (e.g., by random insertion) are invaluable tools for connecting gene sequence to function. The application of such methodologies to identify and characterize virulence traits from Sporothrix spp. has not been received significant experimental attention, most likely because S. schenckii was thought to be diploid (Torres-Guerrero, 1999). The novel information presented herein represents a timely and practical advance that may now be exploited using molecular techniques like Agrobacterium-based transformation methods.
Apart from the biological significance of the ploidy state in the vegetative growth and sexual cycle, this knowledge, coupled to the power of modern molecular technologies, as CRISPR-mediated gene disruption, may open new avenues for the identification of virulence traits of these pathogens.
Data Availability
The datasets analyzed for this study can be found in the NCBI-SRA (https://www.ncbi.nlm.nih.gov/sra/?term=SRX342487+OR+SRX550176+OR+SRX2124714+OR+SRX2250255) with the following accession codes: SRX342487 for Sporothrix schenckii, SRX550176 for S. pallida, SRX2124714 for Aspergillus flavus NRRL 3357, and SRX2250255 for Candida albicans SC 5314. Data for S. brasiliensis are available upon request from MF.
Author Contributions
LL-B and FR conceived the study. BF, JR-P, and GN performed the experiments. BF, JR-P, and FR analyzed the data. BF, JR-P, and ET drafted the manuscript. GG, AC, CC, PS, MF, and AV contributed reagents, materials, and analysis tools. All the authors reviewed the manuscript critically.
Funding
This work was supported by the Northern Portugal Regional Operational Programme (NORTE 2020), under the Portugal 2020 Partnership Agreement, through the European Regional Development Fund (FEDER) (NORTE-01-0145-FEDER-000013), and to Fundação para a Ciência e Tecnologia (FCT) (IF/00735/2014 to AC, SFRH/BPD/96176/2013 to CC, and IF/01390/2014 to ET). LL-B and GN were research fellows of FAPERJ (Fundação Carlos Chagas Filho de Amparo a Pesquisa do Estado do Rio de Janeiro). LL-B is a research fellow of Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq). JR-P thanks CONACYT grant INFR-2016-01-269833 for bioinformatics infrastructure.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors thank Dr. Hector Mora-Montes, Universidad Guanajuato; Dr. Ricardo Silvestre and Dr. Célia Pais, University of Minho, for the suggestions and contributions in the discussion. The authors also thank Dr. Rosane Orofino Costa from Hospital Universitário Pedro Ernesto for the HUPE 114158 isolate.
References
Almeida-Paes, R., de Oliveira, M., Freitas, D., do Valle, A., Zancopé-Oliveira, R., and Gutierrez-Galhardo, M. (2014). Sporotrichosis in Rio de Janeiro, Brazil: Sporothrix brasiliensis is associated with atypical clinical presentations. PLoS Negl. Trop. Dis. 8:e3094. doi: 10.1371/journal.pntd.0003094
Almeida, A., Carmona, J., Cunha, C., Carvalho, A., Rappleye, C., Ludovico, P., et al. (2007). Towards a molecular genetic system for the pathogenic fungus Paracoccidioides brasiliensis. Fungal Genet. Biol. 44, 1387–1398. doi: 10.1016/j.fgb.2007.04.004
Arrillaga-Moncrieff, I., Capilla, J., Mayayo, E., Marimon, R., Mariné, M., Gené, J., et al. (2009). Different virulence levels of the species of Sporothrix in a murine model. Clin. Microbiol. Infect. 15, 651–655. doi: 10.1111/j.1469-0691.2009.02824.x
Barnett, D. W., Garrison, E. K., Quinlan, A. R., Strömberg, M. P., and Marth, G. T. (2011). BamTools: a C++ API and toolkit for analyzing and managing BAM files. Bioinformatics 27, 1691–1692. doi: 10.1093/bioinformatics/btr174
Castro, R. A., Kubitschek-Barreira, P. H., Teixeira, P. A. C., Sanches, G. F., Teixeira, M. M., Quintella, L. P., et al. (2013). Differences in cell morphometry, cell wall topography and gp70 expression correlate with the virulence of Sporothrix brasiliensis clinical isolates. PLoS One 8:e75656. doi: 10.1371/journal.pone.0075656
Corrêa dos Santos, A. R., Goldman, G. H., and Riaño-Pachón, D. M. (2017). ploidyNGS: visually exploring ploidy with next generation sequencing data. Bioinformatics 33, 2575–2576. doi: 10.1093/bioinformatics/btx204
D’Alessandro, E., Giosa, D., Huang, L., Zhang, J., Gao, W., Brankovics, B., et al. (2016). Draft genome sequence of the dimorphic fungus Sporothrix pallida, a nonpathogenic species belonging to Sporothrix, a genus containing agents of human and feline sporotrichosis. Genome Announc. 4, e00184–e00916. doi: 10.1128/genomeA.00184-16
de Beer, Z. W., Duong, T. A., and Wingfield, M. J. (2016). The divorce of Sporothrix and Ophiostoma: solution to a problematic relationship. Stud. Mycol. 83, 165–191. doi: 10.1016/j.simyco.2016.07.001
dos Reis, T. F., Silva, L. P., de Castro, P. A., Almeida de Lima, P. B., do Carmo, R. A., Marini, M. M., et al. (2018). The influence of genetic stability on Aspergillus fumigatus virulence and azole resistance. G3 8, 265–278. doi: 10.1534/g3.117.300265
Ferreira, L. C., Barroso, P. F., Tonomura, E., Akiti, T., Rodrigues, K. M. de P., Ferreira, L. C., et al. (2016). Osteomyelitis caused by Sporothrix schenckii in an immunocompetent patient. Rev. Soc. Bras. Med. Trop. 49, 527–529. doi: 10.1590/0037-8682-0354-2015
Fortuna, M., Sousa, M. J., Côrte-Real, M., Leão, C., Salvador, A., and Sansonetty, F. (2000). “Cell cycle analysis of yeasts” in Current Protocols in Cytometry. eds. Robinson, J. P. Darzynkiewicz, Z. Dean, P. New York, USA: John Wiley & Sons, Inc. 11.13.1–11.13.9.
Galagan, J. E., Calvo, S. E., Cuomo, C., Ma, L. -J., Wortman, J. R., Batzoglou, S., et al. (2005). Sequencing of Aspergillus nidulans and comparative analysis with A. fumigatus and A. oryzae. Nature 438, 1105–1115. doi: 10.1038/nature04341
Gremião, I., Miranda, L., Reis, E. G., Rodrigues, A. M., and Pereira, S. A. (2017). Zoonotic epidemic of sporotrichosis: cat to human transmission. PLoS Pathog. 13:e1006077. doi: 10.1371/journal.ppat.1006077
Gudnason, H., Dufva, M., Bang, D. D., and Wolff, A. (2007). Comparison of multiple DNA dyes for real-time PCR: effects of dye concentration and sequence composition on DNA amplification and melting temperature. Nucleic Acids Res. 35:e127. doi: 10.1093/nar/gkm671
Hassan, K., Turker, T., and Zangeneh, T. (2016). Disseminated sporotrichosis in an immunocompetent patient. Case Reports Plast. Surg. Hand Surg. 3, 44–47. doi: 10.3109/23320885.2016.1168703
Huang, L., Gao, W., Giosa, D., Criseo, G., Zhang, J., He, T., et al. (2016). Whole-genome sequencing and in silico analysis of two strains of Sporothrix globosa. Genome Biol. Evol. 8, 3292–3296. doi: 10.1093/gbe/evw230
Leinonen, R., Sugawara, H., and Shumway, M. International Nucleotide Sequence Database Collaboration (2011). The sequence read archive. Nucleic Acids Res. 39, D19–D21. doi: 10.1093/nar/gkq1019
Li, H., Handsaker, B., Wysoker, A., Fennell, T., Ruan, J., Homer, N., et al. (2009). The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079. doi: 10.1093/bioinformatics/btp352
Lopes-Bezerra, L. M., Schubach, A., and Costa, R. O. (2006). Sporothrix schenckii and sporotrichosis. An. Acad. Bras. Cienc. 78, 293–308. doi: 10.1590/S0001-37652006000200009
Marimon, R., Cano, J., Gené, J., Sutton, D. A., Kawasaki, M., and Guarro, J. (2007). Sporothrix brasiliensis, S. globosa, and S. mexicana, three new Sporothrix species of clinical interest. J. Clin. Microbiol. 45, 3198–3206. doi: 10.1128/JCM.00808-07
Mialski, R., de Oliveira, J. N., da Silva, L. H., Kono, A., Pinheiro, R. L., Teixeira, M. J., et al. (2018). Chronic meningitis and hydrocephalus due to Sporothrix brasiliensis in immunocompetent adults: a challenging entity. Open Forum Infect. Dis. 5, ofy081. doi: 10.1093/ofid/ofy081
Montenegro, H., Rodrigues, A. M., Dias, M. A. G., da Silva, E. A., Bernardi, F., and de Camargo, Z. P. (2014). Feline sporotrichosis due to Sporothrix brasiliensis: an emerging animal infection in São Paulo, Brazil. BMC Vet. Res. 10:269. doi: 10.1186/s12917-014-0269-5
Oliveira, M. M., Santos, C., Sampaio, P., Romeo, O., Almeida-Paes, R., Pais, C., et al. (2015). Development and optimization of a new MALDI-TOF protocol for identification of the Sporothrix species complex. Res. Microbiol. 166, 102–110. doi: 10.1016/J.RESMIC.2014.12.008
Queiroz-Telles, F., Fahal, A. H., Falci, D. R., Caceres, D. H., Chiller, T., and Pasqualotto, A. C. (2017). Neglected endemic mycoses. Lancet Infect. Dis. 17, e367–e377. doi: 10.1016/S1473-3099(17)30306-7
Rangel-Gamboa, L., Martínez-Hernandez, F., Maravilla, P., Arenas-Guzmán, R., and Flisser, A. (2016). Update of phylogenetic and genetic diversity of Sporothrix schenckii sensu lato. Med. Mycol. 54, 248–255. doi: 10.1093/mmy/myv096
Rangel-Gamboa, L., Martinez-Hernandez, F., Maravilla, P., and Flisser, A. (2018). A population genetics analysis in clinical isolates of Sporothrix schenckii based on calmodulin and calcium/calmodulin-dependent kinase partial gene sequences. Mycoses 61, 383–392. doi: 10.1111/myc.12751
Rodrigues, A. M., de Hoog, G. S., and de Camargo, Z. P. (2016). Sporothrix species causing outbreaks in animals and humans driven by animal-animal transmission. PLoS Pathog. 12, 1–7. doi: 10.1371/journal.ppat.1005638
Rodrigues, F., Ludovico, P., Sousa, M. J., Steensma, H. Y., Côrte-Real, M., and Leão, C. (2003). The spoilage yeast Zygosaccharomyces bailii forms mitotic spores: a screening method for haploidization. Appl. Environ. Microbiol. 69, 649–653. doi: 10.1128/AEM.69.1.649-653.2003
Sanchotene, K. O., Madrid, I. M., Klafke, G. B., Bergamashi, M., Terra, P. P. Della, Rodrigues, A. M., et al. (2015). Sporothrix brasiliensis outbreaks and the rapid emergence of feline sporotrichosis. Mycoses 58, 652–658. doi: 10.1111/myc.12414
Sasaki, A. A., Fernandes, G. F., Rodrigues, A. M., Lima, F. M., Marini, M. M., Dos S. Feitosa, L., et al. (2014). Chromosomal polymorphism in the Sporothrix schenckii complex. PLoS One 9:e86819. doi: 10.1371/journal.pone.0086819
Teixeira, M., de Almeida, L. G. P., Kubitschek-Barreira, P., Alves, F. L., Kioshima, E. S., Abadio, A. K. R., et al. (2014). Comparative genomics of the major fungal agents of human and animal Sporotrichosis: Sporothrix schenckii and Sporothrix brasiliensis. BMC Genomics 15:943. doi: 10.1186/1471-2164-15-943
Téllez, M. D., Batista-Duharte, A., Portuondo, D., Quinello, C., Bonne-Hernández, R., and Carlos, I. Z. (2014). Sporothrix schenckii complex biology: environment and fungal pathogenicity. Microbiology 160, 2352–2365. doi: 10.1099/mic.0.081794-0
Torres-Guerrero, H. (1999). Ploidy study in Sporothrix schenkii. Fungal Genet. Biol. 27, 49–54. doi: 10.1006/fgbi.1999.1128
Keywords: sporotrichosis, Sporothrix schenckii complex, S. brasiliensis, S. schenckii, S. globosa, S. pallida, ploidy, yeast phase
Citation: Ferreira BH, Ramírez-Prado JH, Neves GWP, Torrado E, Sampaio P, Felipe MSS, Vasconcelos AT, Goldman GH, Carvalho A, Cunha C, Lopes-Bezerra LM and Rodrigues F (2019) Ploidy Determination in the Pathogenic Fungus Sporothrix spp. Front. Microbiol. 10:284. doi: 10.3389/fmicb.2019.00284
Edited by:
Orazio Romeo, Università degli Studi di Messina, ItalyReviewed by:
Shanshan Li, First Affiliated Hospital of Jilin University, ChinaChristina A. Cuomo, Broad Institute, United States
Copyright © 2019 Ferreira, Ramirez-Prado, Neves, Torrado, Sampaio, Felipe, Vasconcelos, Goldman, Carvalho, Cunha, Lopes-Bezerra and Rodrigues. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Fernando Rodrigues, ZnJvZHJpZ3Vlc0BtZWQudW1pbmhvLnB0
†These authors have contributed equally to this work
 Beatriz H. Ferreira
Beatriz H. Ferreira Jorge H. Ramírez-Prado
Jorge H. Ramírez-Prado Gabriela W. P. Neves
Gabriela W. P. Neves Egídio Torrado
Egídio Torrado Paula Sampaio
Paula Sampaio Maria Sueli S. Felipe6,7
Maria Sueli S. Felipe6,7 Ana Tereza Vasconcelos
Ana Tereza Vasconcelos Gustavo H. Goldman
Gustavo H. Goldman Agostinho Carvalho
Agostinho Carvalho Leila M. Lopes-Bezerra
Leila M. Lopes-Bezerra Fernando Rodrigues
Fernando Rodrigues