- 1Department of Marine Sciences and Convergent Technology, Hanyang University, Seoul, South Korea
- 2School of Earth and Environmental Sciences and Research Institute of Oceanography, Seoul National University, Seoul, South Korea
- 3Department of Microbiology, Chungbuk National University, Cheongju, South Korea
- 4Department of Biological Sciences, Eastern Illinois University, Charleston, IL, United States
- 5Max Planck Institute for Marine Microbiology, Bremen, Germany
- 6Korea Institute of Ocean Science & Technology, Busan, South Korea
- 7Korea Polar Research Institute, Incheon, South Korea
Polynyas in the polar seas are regarded as windows through which ecosystem responses associated with global climate changes are to be noticed. However, little information is available on benthic microbial communities in the Amundsen Sea polynya (ASP), where environmental changes due to global warming are occurring rapidly, from which future climate change-induced ecosystem responses could be assessed. We performed high-throughput sequencing of 16S rRNA genes and quantitative PCR in combination with biogeochemical analyses and metabolic rate measurements to determine the composition, diversity and controls of major microbial communities in sediments of the ASP. A large fraction of the sequenced benthic microbial community (40% on average) in the polynya was uniquely affiliated with the phylum Planctomycetes, whereas Thaumarchaeota (51% on average) predominated in non-polynya areas. The relative abundance of Planctomycetes correlated significantly with organic carbon (Corg) content in the polynya sediment underlying the Phaeocystis-dominated water column. These results suggest that Planctomycetes comprise a major bacterial group utilizing relatively recalcitrant Corg produced primarily by Phaeocystis blooms. In contrast, the predominance of chemolithoautotrohic Thaumarchaeota in the sea-ice zone was attributed to low Corg supply due to low primary productivity in the ice-covered water column. The Planctomycetes-dominated microbial communities in the ASP is in stark contrast to that Proteobacteria (Delta- and Gamma-proteobacteria) occupy ecological niches as primary mineralizers of organic materials in most benthic systems in the Southern Ocean, where organic materials in the sediments mostly originate from diatom blooms. Given that microbial communities respond quickly to environmental changes, and that global climate change is proceeding rapidly in the ASP, our results suggest that any modifications in the Planctomycetes-dominated microbial communities will provide valuable insight into changes in organic matter transport to the seafloor that may result from shifts in planktonic communities from Phaeocystis to diatoms associated with climate change.
Introduction
In marine surface sediments, microorganisms occur in enormous numbers (approximately 1.7 × 1028 cells worldwide) (Whitman et al., 1998), are extremely diverse (Huber et al., 2007), and are notably involved in global carbon, nitrogen, and sulfur cycles (Canfield et al., 2005; Orcutt et al., 2011). Because microbial community composition, diversity, and metabolic activity are significantly influenced by environmental change (Bertics and Ziebis, 2009; Nguyen and Landfald, 2015; Sebastián et al., 2018; Kim et al., 2019; Richert et al., 2019), characterization of microbial distribution provides relevant information on spatial and temporal variations in environmental conditions (Fuhrman, 2009; Schauer et al., 2010; Robador et al., 2016). However, little is known about benthic microbial communities in the Southern Ocean (SO), where environmental changes due to global warming are occurring rapidly (Baldi et al., 2010; Ruff et al., 2014; Learman et al., 2016).
The SO plays a profound role in regulating global carbon cycles, accounting for approximately 20–30% of global ocean CO2 uptake (Gruber et al., 2009; Takahashi et al., 2009). As atmospheric pCO2 increases (Petit et al., 1999), the role of the SO as an atmospheric carbon sink has received more attention. During the austral summer, the coastal zone of the SO is typically characterized by the occurrence of polynyas, areas of seasonally recurring open water surrounded by sea ice (Smith and Barber, 2007; Nihashi and Oshima, 2015). Because of the combined effects of enhanced light conditions and iron supply resulting from melting sea ice, polynyas are one of the most productive marine ecosystems (Sedwick and DiTuillo, 1997; Arrigo and van Dijken, 2003; Montes-Hugo and Yuan, 2012), and thus are regarded as significant sinks for atmospheric CO2 (Miller and DiTuillo, 2007; Arrigo et al., 2008). In addition, due to the significant gas exchange (CO2, DMS, etc.) and heat flux between the sea surface and atmosphere, polynyas are regarded as windows through which we can anticipate ecosystem responses associated with climate changes (Smith and Barber, 2007).
Among the 37 known coastal polynyas around Antarctica, the Amundsen Sea polynya (ASP) is reported to be the most productive (Arrigo et al., 2008, 2012), with a primary productivity per unit area of ∼220 g C m–2 y–1 (Lee et al., 2012; Kim B.K. et al., 2014). The prymnesiophyte Phaeocystis antarctica is the main contributor to the high primary productivity in the central polynya (Figure 1B) (Yager et al., 2012; Ducklow et al., 2015; Lee et al., 2016; Williams et al., 2016; Yang et al., 2018). Due to the inflow of warm circumpolar deep water (CDW), glaciers near the Amundsen Sea (AS) are undergoing the highest rates of melting and thinning on the Antarctic continent (Rignot, 2008; Jenkins et al., 2010; Jacobs et al., 2011). Consequently, as global warming continues, the heat flux intensity of the CDW may stimulate ice melting, which results in changes in phytoplankton productivity and community composition (Deppeler and Davidson, 2017), thereby regulating the function of the ASP in carbon sequestration (Thoma et al., 2008; Lee et al., 2017). The composition and metabolic activities of benthic microbial communities are ultimately determined by the quality and quantity of the organic matter supplied from the overlying water column (Franco et al., 2007; Bienhold et al., 2012; Ruff et al., 2014; Hoffmann et al., 2017). Therefore, given that microbial communities respond quickly to environmental change (Danovaro et al., 2000; Luria et al., 2016), quantitative and qualitative information on benthic microbial communities and their metabolic activities is essential for assessing the response of these ecosystems to climate change-induced variations in primary productivity in the water column of the SO. The benthic microbial community has been studied in several regions of the Southern Ocean, including the Mertz Glacier polynyas, Ross Sea, Bellingshausen Sea, and Australian-Antarctic ridge (Bowman and McCuaig, 2003; Baldi et al., 2010; Carr et al., 2013; Learman et al., 2016). In those studies, Proteobacteria were reported as a predominant bacterial group in the sediment and are regarded as major organic carbon (Corg) oxidizers. However, there is no information on the composition and diversity of entire microbial communities in the sediment of the ASP.
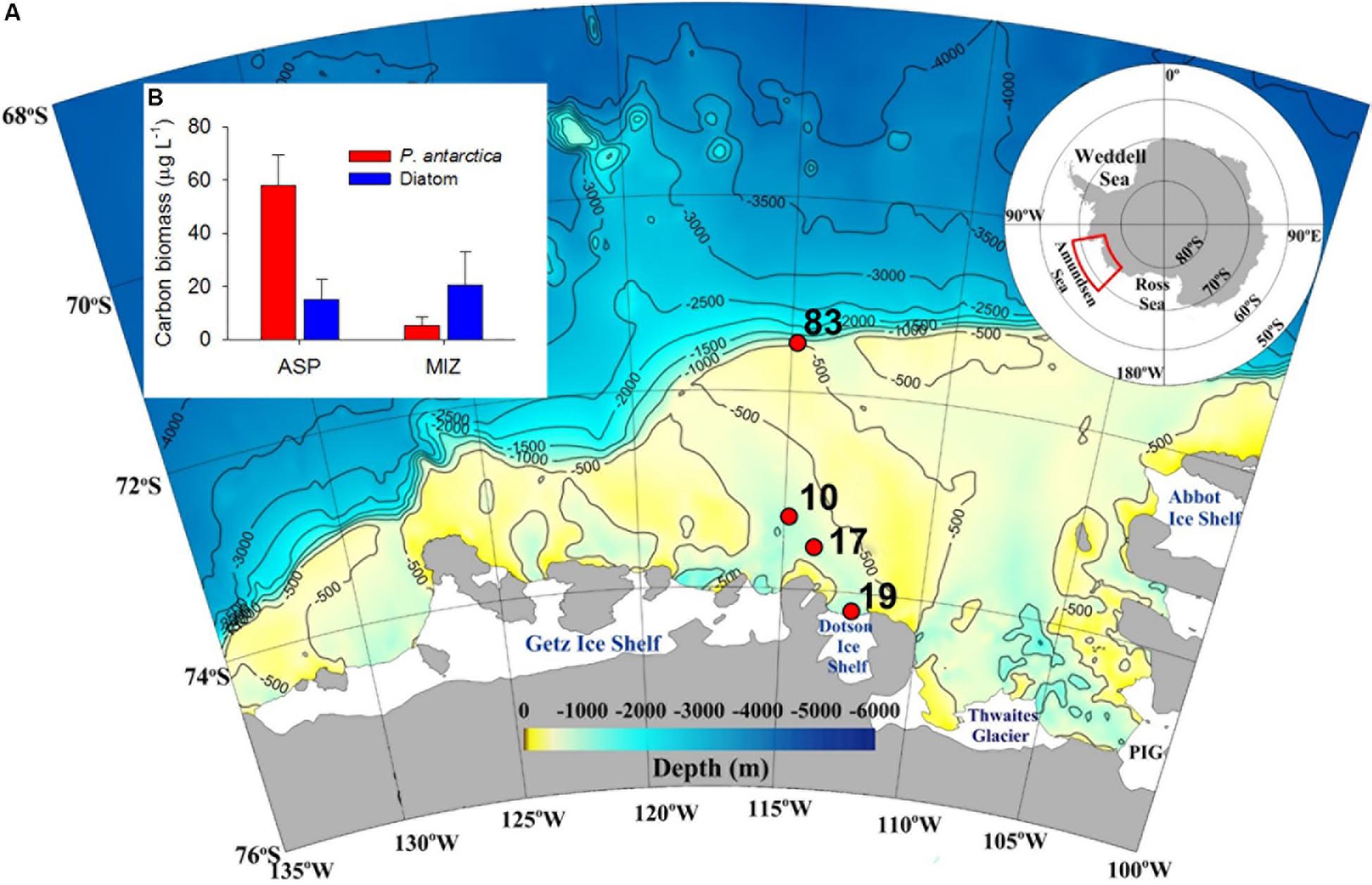
Figure 1. A map showing the sampling sites (A), and the average carbon biomass (μg L–1) of the major phytoplankton groups (B) at Amundsen Sea sites during the austral summer 2012. ASP and MIZ denote Amundsen Sea polynya and marginal ice zone (MIZ), respectively.
The objectives of this paper were: (1) to identify microbial communities in the sediment underlying the Phaeocystis-dominated water column, (2) to elucidate the major factors controlling microbial communities, with a special emphasis on the Corg content in sediment across the marginal ice zone (MIZ), polynya, and ice shelf zone, and (3) to suggest an appropriate microbial proxy for assessing the response of benthic ecosystem to ongoing climate changes in the Amundsen Sea.
Materials and Methods
Study Area
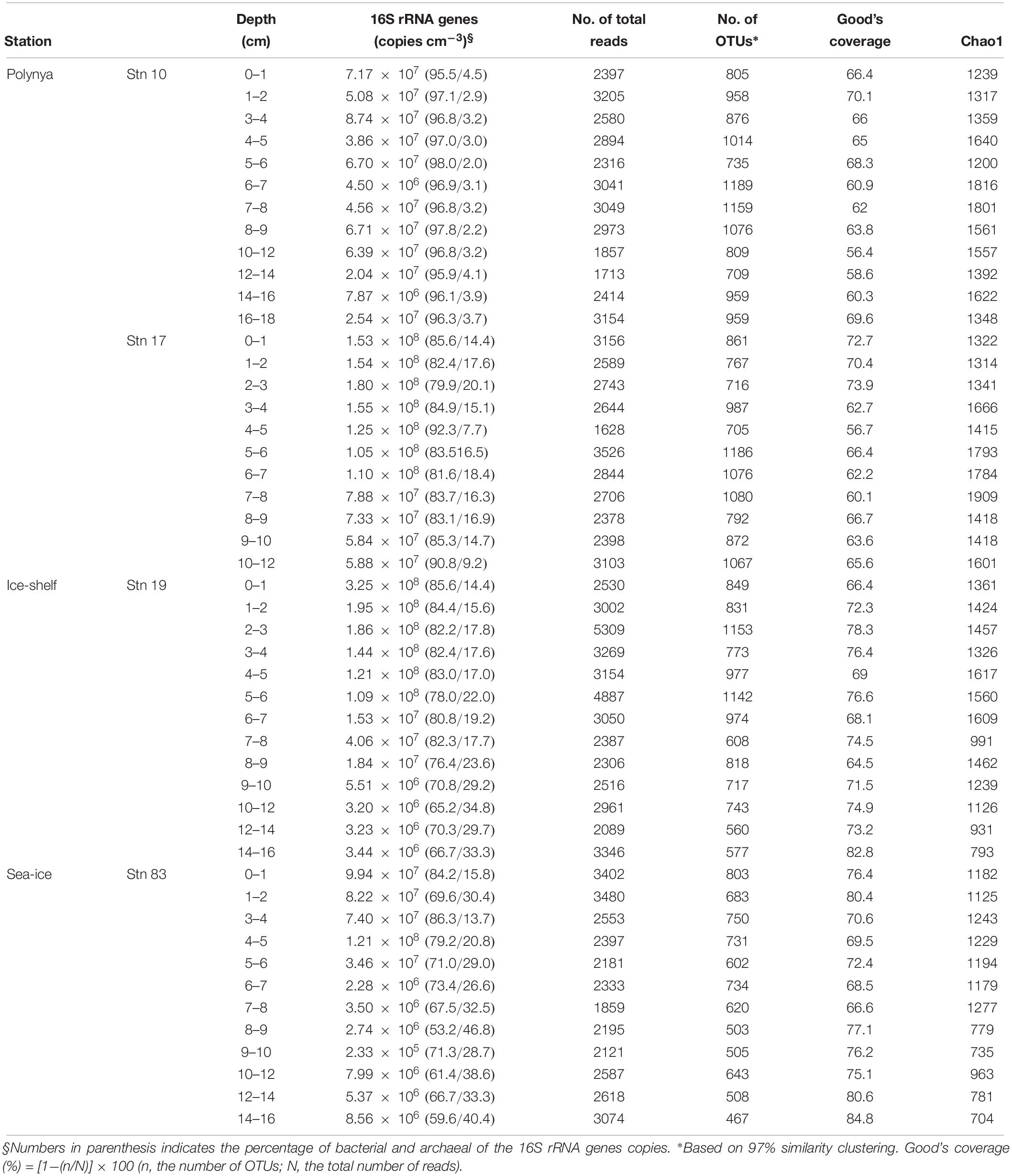
The Amundsen Sea is located in western Antarctica between the Ross Sea and Bellingshausen Sea (69°S–74°S; 100°W–135°W, Figure 1), and is characterized by a large polynya from November to February (Arrigo and van Dijken, 2003). The Korean Amundsen Sea Expedition was conducted during the austral summer, from February 9 to March 10 in 2012, aboard the Korean icebreaker research vessel RV Araon. Water depth ranged from 530 to 1,064 m, and bottom water temperature ranged from −1.8 to −1.1°C (Table 1).
Sediment samples were collected using a box corer at four stations at three contrasting sites: the MIZ (Stn 83), inside the polynya (Stns 10 and 17), and in direct proximity to the ice shelf site (Stn 19) (Table 1). P. antarctica was the major planktonic algae in this highly productive polynya area, whereas diatoms were more abundant in the relatively less-productive MIZ (Stn 83) (Figure 1B) (Yang et al., 2018). Subsamples for DNA extraction were taken from the center portion of the box corer using acryl sub-core liners (6 cm in diameter). Cores were sliced at 1-cm intervals to a depth of 11 or 18 cm, and immediately frozen at −80°C.
DNA Extraction, Quantitative PCR, and Pyrosequencing of 16S rRNA Genes
Total genomic DNA was extracted from the different sediment layers using a PowerMax DNA Isolation kit (Mo Bio Laboratories, Carlsbad, CA, United States), following the manufacturer’s instructions. Quantitative real-time polymerase chain reaction (q-PCR) was used to determine the copy number of 16S rRNA genes and archaeal ammonia monooxygenase A (amoA) genes using a TaqMan assay and a SYBR Green I assay, respectively (Supplementary Table S1). Details are described in Supplementary Material.
For each of the 48 sampled sediment layers at the polynya and non-polynya sites, we generated a 16S rRNA gene amplicon library, with one primer set covering the V5–V8 region of both bacterial and archaeal taxa (Jorgensen et al., 2012). PCR amplification of the 16S rRNA genes was performed in triplicate using a primer set of Uni787F (Roesch et al., 2007) and Uni1391R (Lane et al., 1985) according to Jorgensen et al. (2012). Resulting amplicons were sequenced by Macrogen Corporation (Korea) using the 454 GS FLX + system (Roche). The raw data have been deposited to the NCBI SRA database under accession number SRX3405376. Raw flowgrams of pyrosequencing reads were filtered and de-noised by a PyroNoise algorithm (Quince et al., 2011) implemented in MOTHUR (ver. 1.36.1) (Schloss et al., 2009). Singleton OTUs were removed prior to analysis. Chimeric sequences were identified and removed by ChimeraSlayer. A total of 132,914 pyrosequencing reads from 48 samples were qualified for further processing. The sequences were then clustered into operational taxonomic units (OTUs) that met the criteria of a 97% similarity threshold and a minimum cluster size of 2 using a QIIME pipeline (ver. 1.9.1) (Caporaso et al., 2010). Taxonomy for each OTU0.97 was assigned using the RDP classifier method (Wang et al., 2007) with the Greengenes database (ver. 13_8) (McDonald et al., 2012). To avoid the effects of different sample sizes for estimating diversity, comparison sequences were randomly subsampled to the smallest library size (Kirchman et al., 2010), which were 1,628 sequences in the present study. Chao1 estimates were created using QIIME software to assess diversity (Chao, 1984).
Geochemical Characterization
Geochemical constituents (, NOX [ + ], , and Fe2+) in the pore water, oxygen penetration depth (OPD), total oxygen uptake (TOU), sulfate reduction rates, and N2 removal rates by denitrification and anaerobic ammonia oxidation (anammox) were adopted from the results reported by Choi et al. (2016) and Kim et al. (2016). Total organic carbon (TOC) and total nitrogen (TN) were analyzed with Elemental Analyzer (Carlo Erba, NA-1500) (Verardo et al., 1990) (see Supplementary Material for detailed methods).
Statistical Analyses
Spatial difference of geochemical constituents was assessed using a Mann–Whitney U-test, with a statistical significance level of 0.05. The relative abundance of microbial populations was determined using a Kruskal–Wallis test. Tukey’s honest significance difference (HSD) test was used for multiple comparison of means at a 95% confidence interval. The OTU table from QIIME and geochemical measurements were analyzed using R (v. 3.3.2) (R Core Team, 2016) with custom scripts and several packages including vegan (v. 2.4-2) and lmtest (v. 0.9-35). Exploratory data analysis was carried out for both microbial communities and geochemical measurements data using non-metric multidimensional scaling (NMDS), diversity measures, and hierarchical clustering. Microbial community was analyzed at the OTU0.97 level as well as at the phylum and order levels. Compositional difference of microbial communities among stations was tested by multivariate analysis of variance–like non-parametric tests (analysis of similarities [ANOSIM] and permutational multivariate analysis of variance [PERMANOVA]). Ordination of microbial communities was fitted with geochemical measurements by vector fitting. Constrained ordination models were constructed by redundancy analysis (RDA) in an iterative fashion considering collinearity among constraining geochemical variables. Community structures were compared among stations, and between bacteria and archaea using a Procrustes test on RDA ordination configuration and a Mantel test.
Results
Environmental Parameters and Microbial Abundance
The distribution of geochemical constituents and community structures showed distinct spatial variation between the polynya sites and non-polynya sites of the Amundsen Sea. Contents of TOC and TN were approximately 1.25 to 2.1 times higher at the polynya than at the ice shelf (Stn 19) and sea-ice zone (Stn 83) (Figure 2 and Table 1) (P < 0.001). Accordingly, OPD in the polynya sites (1.8–2.0 cm) was shallower than in non-polynya sites (3.5–3.6 cm) (P < 0.001). Pore-water analysis revealed that the concentration of NOX at the polynya sites decreased with depth from approximately 30 μM at the top to 8 μM at 3–5 cm depth, and then remained constant down to a depth of 10–20 cm (Figure 2). In contrast, NOX concentration at the non-polynya sites (ice shelf and sea-ice zone) was high (>20 μM) at all depths. concentration was higher at the polynya sites than at the non-polynya sites (Figure 2) (P < 0.001). Concentration of Fe2+ in the pore-water was low at all sites (<10 μM), but the average concentration of dissolved Fe2+ was higher at the polynya sites (4.9 μM) than at the non-polynya sites (1.98 μM) (P = 0.003). Metabolic activities such as TOU rate and anaerobic respiration by sulfate reduction were consistently higher in the polynya sites with relatively higher Corg content compared to those measured in non-polynya sites (Figure 2 and Table 1) (P = 0.002).
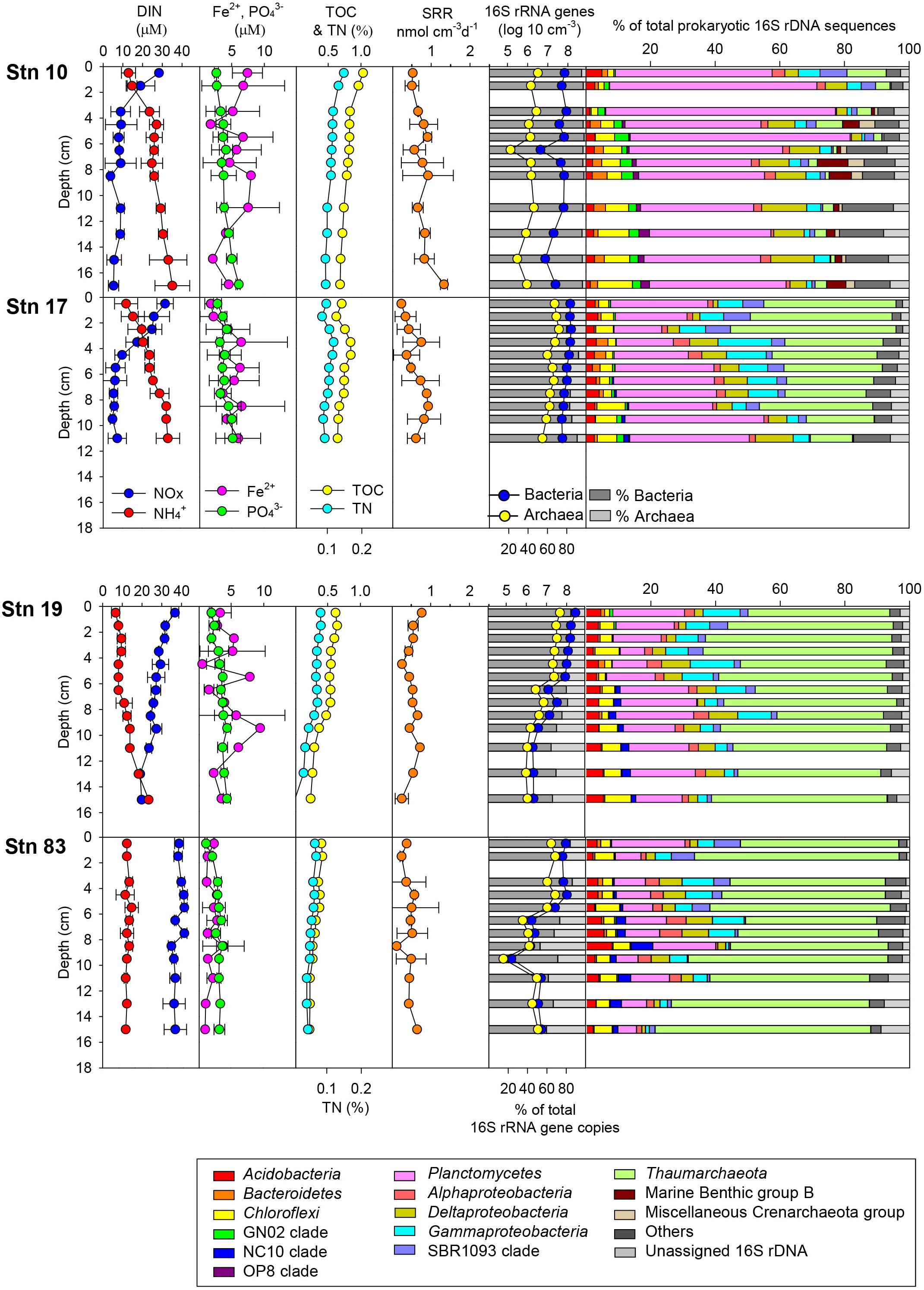
Figure 2. Distribution of geochemical constituents [dissolved inorganic nitrogen (DIN; and NOx), and Fe2+] in pore-water and contents of total organic carbon (TOC), total nitrogen (TN), sulfate reduction rate (SRR), prokaryotic cell abundance based on quantitative-PCR, and relative abundance of major phyla in the sediment of the polynya sites (Stns 10 and 17) and non-polynya sites (Stns 19 and 83) of the Amundsen Sea.
Total prokaryotic abundance determined by q-PCR of 16S rRNA genes ranged from 2.3 × 105 to 3.3 × 108 copies cm–3 per each sample (Table 2). Although the 16S rRNA gene copy numbers appeared higher at Stn 17 than those at Stn 10, the variation between polynya sites was not significant (P = 0.06). However, 16S rRNA gene copies in the sediment of Stn 17 were clearly higher than those of Stn 83 (P < 0.05). Total prokaryotic abundances were higher in the surface sediments and decreased with depth at all sites (Figure 2). The relative abundance of bacterial 16S rRNA gene copies to total prokaryotic cells was highest at Stn 10 (96.7 ± 3.3%), and then decreased to 84.8 ± 3.7% (Stn 17), 77.6 ± 7.0% (Stn 19) and 70.3 ± 9.7% on average (Stn 83). In contrast, archaeal 16S rRNA gene proportion comprised 3.3 ± 0.7% at Stn 10, and then gradually increased with distance from Stn 10 to 15.2 ± 3.7% (Stn 17), 22.4 ± 7.0% (Stn 19), and 29.7 ± 9.7% on average (Stn 83) (Table 2). The percentage of bacterial and archaeal 16S rRNA gene copies of the total 16S rRNA genes determined by q-PCR (Table 2) corresponded with the relative abundance of Bacteria versus Archaea as estimated from pyrosequencing data (Figure 2). On the other hand, copy numbers of archaeal amoA gene ranged from 2.8 × 104 to 5.7 × 106 and from 1.7 × 104 to 8.3 × 106 cm–3 for polynya sites and non-polynya sites, respectively. The depth profiles of amoA gene copy numbers in the ASP sediments showed a similar trend to the archaeal 16S rRNA gene copy numbers (Supplementary Figure S1).
Microbial Community Structure
In total of 132,914 pyrosequencing reads with an average length of 601 bp, 9,852 reads were unique. Chao1 indices are as high as an average of 1,909 in the 7–8 cm depth of Stn 17, and were lowest at 704 in the 14–16 cm depth of Stn 83 (Table 2). Chao1 and observed OTU0.97 counts of the polynya and ice-shelf sites were higher than those of the sea-ice zone. These two indices were estimated to be highest within the sub-oxic layers (3–8 cm depth) of all sites.
Non-metric multidimensional scaling of OTUs resulted in a separate clustering between polynya and non-polynya samples, and the separation was mainly driven by differences in geochemical constituents (, Fe2+, and ), TOC, TN, and sulfate reduction rate (Figure 3). Both archaeal and bacterial communities were quite distinctive between polynya sites and non-polynya sites (P < 0.001 from both PERMANOVA and ANOSIM) (Supplementary Figure S2). Similar patterns in community structure were observed at order-level resolution (Supplementary Figure S2D). From RDA analysis (Supplementary Figure S3), NOX concentrations correlated with surface microbial communities at all stations. Corg contents correlated with microbial communities in surface sediments of Stn 10 and at intermediate depths of Stns 17 and 19, respectively. Archaeal communities were more distinctive among stations, while bacterial communities were more similar overall (Supplementary Table S2). Compositional similarity at the phylum level better reflected ecological or geographic settings.
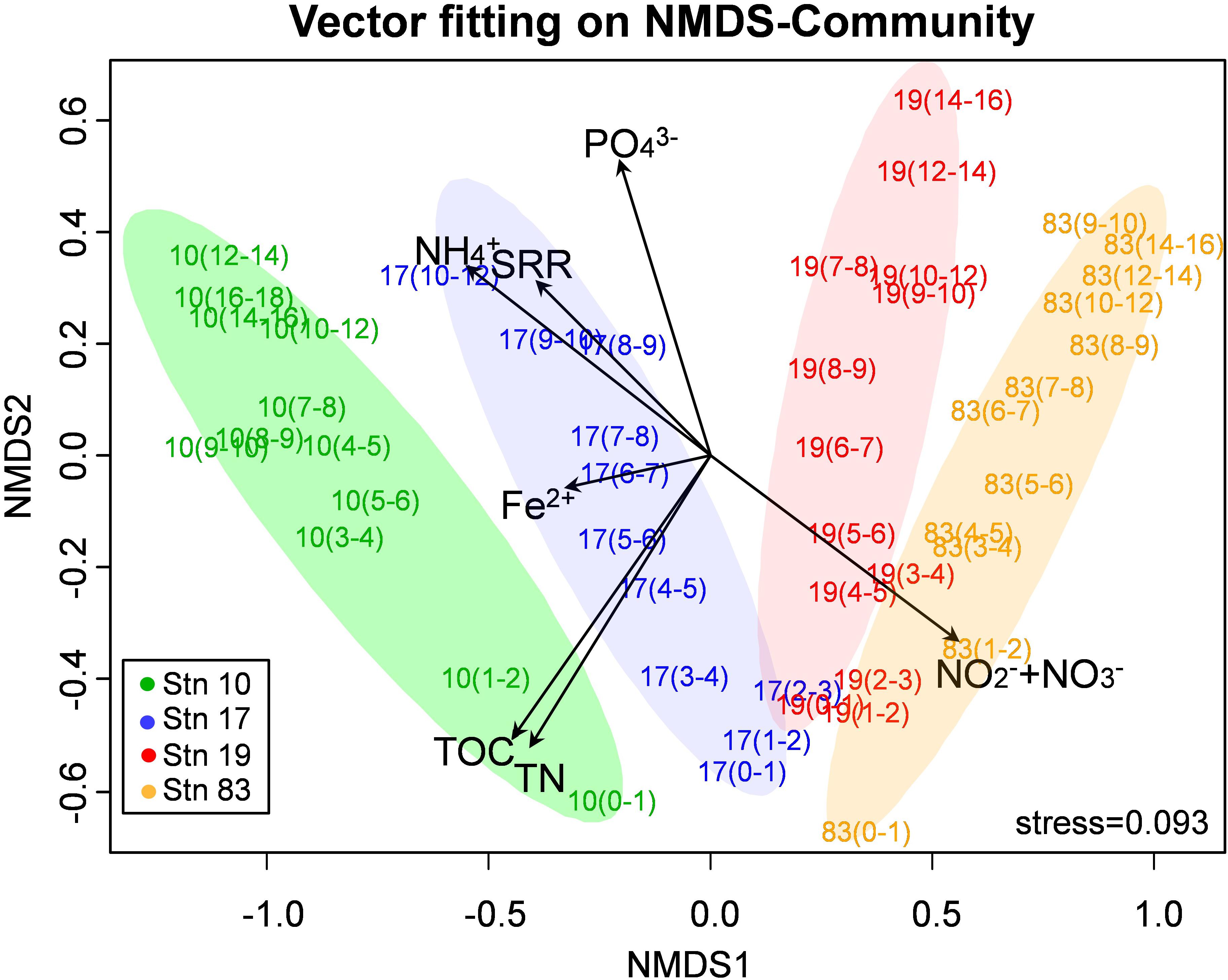
Figure 3. Microbial community by 16S rRNA gene (non-metric multidimensional scaling ordination) showing the microbial communities segregated according to the polynya and non-polynya, driven by differences in nutrients (, Fe2+, and ), contents of total organic carbon (TOC) and total nitrogen (TN), and sulfate reduction rate (SRR).
The relative contributions of each phylum were dramatically different between sites (Figures 2, 4). At the polynya site (Stn 10), the most dominant phyla were Planctomycetes (35–71%), Proteobacteria (4.8–22%), Thaumarchaeota (1.2–12.1%), and Chloroflexi (1.9–10.8%). Candidate Division GN02 (0.5–4.4%), Candidate Division SBR1093 (0.2–8.3%), Acidobacteria (1.2–5.0%), and Bacteroidetes (0.4–3.2%) were of minor abundance. At Stn17, the proportion of Planctomycetes decreased slightly (14.8–43%), whereas the percentage of Thaumarchaeota (13–51%) appeared to be more abundant than at Stn 10. Other phyla included Proteobacteria (10.6–31%), Chloroflexi (2.1–8.7%), Candidate Division SBR1093 (0.5–8.3%), and Acidobacteria (2.6–4.1%). The proportion of archaeal sequences prominently increased at Stns 19 and 83, which is consistent with the archaeal cell abundance estimated from 16S rRNA gene quantification (Figure 2 and Table 2). Relative abundance of Thaumarchaeota accounted for 41–59% and 41–67% at Stn 19 and Stn 83, respectively. Proteobacteria (4.1–27.8%), Planctomycetes (5.8–24%), Chloroflexi (1.6–8.1%), Acidobacteria (2–8%), and Candidate Division SBR1093 (0.3–8.1%) appeared in similar proportions at both non-polynya sites (Figure 2). The relative abundance of Planctomycetes in the polynya sediments showed a high positive correlation with (r = 0.64, P < 0.001), Fe2+ (r = 0.46, P < 0.001), TOC (r = 0.75, P < 0.001), TN (r = 0.73, P < 0.001), and SRR (r = 0.48, P < 0.001) (Figure 5). In contrast, the relative abundant of Thaumarchaeota in the non-polynya sediments showed a negative correlation with TOC (r = −0.75, P < 0.001) (Supplementary Figure S4).
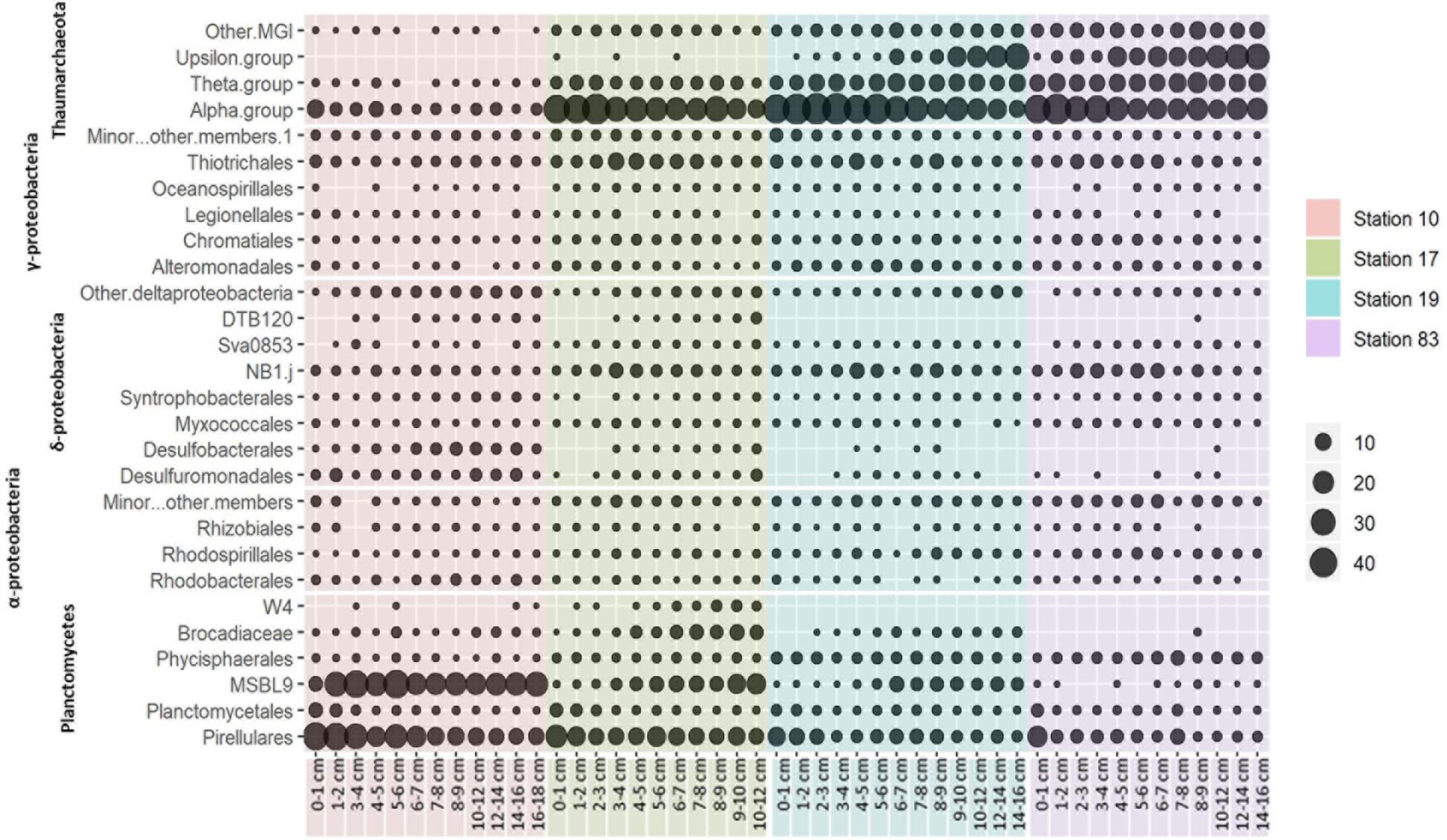
Figure 4. Phylogenetic distribution of major microbial 16S rRNA gene OTUs based on a 97% similarity cut-off. Circle size indicates the relative abundance of the clade based on order level.
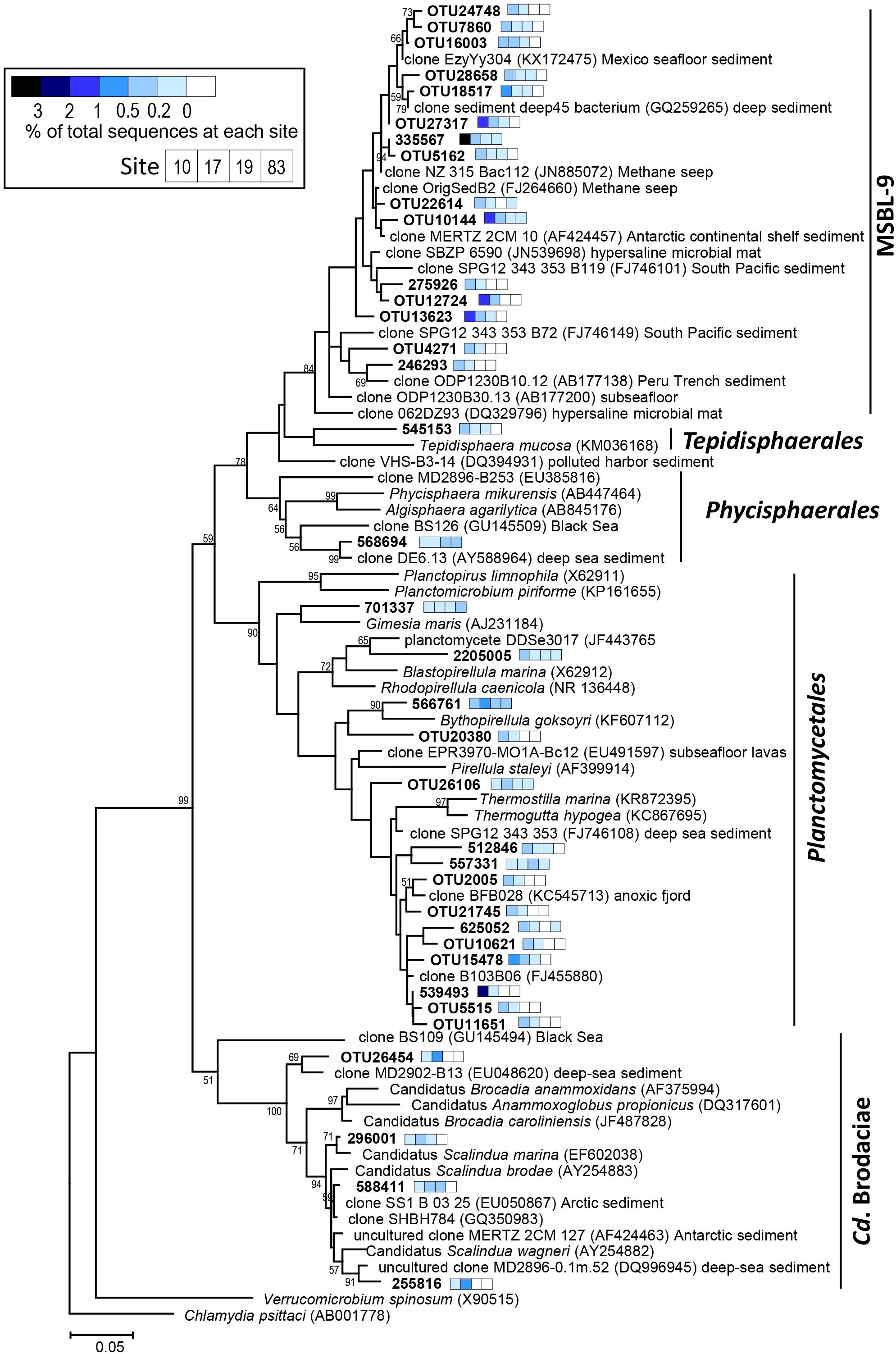
Figure 5. Phylogenetic tree showing the distribution of the 16S rRNA genes sequences retrieved from the ASP sediments within the phylum Planctomycetes. The tree was constructed using the maximum-likelihood algorithm in MEGA 7.0 (Kumar et al., 2016). The scale bar indicates the number of substitutions per nucleotide position. The color of the square bar represents the relative abundance of the sequences for each site (Stn 10, Stn 17, Stn 19, and Stn 83). Node support estimated using 1,000 bootstrap replicates. Bootstrap values above 50% are shown.
Bacterial Community Composition
The most abundant Planctomycetes in the microbial communities were divided into three clades: a Pirellula-like group, candidate order MSBL-9 (Mediterranean Sea Brine Lake-9) (Pachiadaki et al., 2014), and Candidatus Brocadiae (Supplementary Figure S5). The Pirellula-like group and the candidate order MSBL-9 were the two most dominant bacterial groups at Stn 10, representing 34% and 32% of the total 16S rRNA gene sequences, respectively (Figure 4 and Supplementary Figure S5). In contrast, at Stn 83, the relative abundance of the Pirellula-like group decreased to < 17%, and the presence of the candidate order MSBL-9 group was not discernible. The members in Candidatus Brocadiae, which are capable of anaerobic ammonium oxidation (anammox) using nitrite as an electron acceptor (Schmid et al., 2003), were most abundant at Stn 17, and comprised a maximum of 9.2% of the total prokaryotic sequences at the 9–10 cm depth (Figure 4 and Supplementary Figure S5).
The relative abundance of Proteobacteria did not exhibit significant spatial variation among sites [Kruskal–Wallis, χ2(3) = 1.375, P = 0.711]. Similarly, the spatial distribution of Gammaproteobacteira was similar between sites. Most gammaproteobacterial sequences in all sites were affiliated with the orders Thiotrichales and Chromatiales (Supplementary Figure S6). On the other hand, based on the order level, delta- and alphaproteobacterial compositions showed spatial variations (Supplementary Figure S6). Desulfobacterales and Desulfuromonadales, which are well-known as sulfate- and sulfur (S0)-reducing bacterial groups in marine sediments, appeared to be most abundant at Stn 10, but were rarely detected at Stn 83. In contrast, the deltaproteobacterial sequences in the candidate order NB1-j appeared higher at Stn 19 and Stn 83, but showed low relative abundance at Stn 10 (Figure 4 and Supplementary Figure S6). Members of the Alphaproteobacteria made up 0.4–6.9% of total reads and were divided into three major orders (Supplementary Figure S6): Rhodobacterales, Rhodospirillales, and Rhizobiales. The sequences falling into the Rhodobacteraceae appeared more frequently in Stn 10, while members of Rhodospirillacaea were detected more frequently at non-polynya sites (Stns 19 and 83).
The OTUs in Chloroflexi and Bacteroidetes appeared to vary between polynya sites and non-polynya sites (Supplementary Figure S7). The Anaerolineae were a major member of Chloroflexi at Stn 10, whereas sequences clustered in class SAR202 and TK17 were more abundant at Stns 17, 19, and 83. Members of Bacteroidetes that have been reported as major organic matter decomposers in SO sediment (Carr et al., 2013; Ruff et al., 2014; Learman et al., 2016) were substantially low with relative abundance less than 5% of total 16S rRNA gene sequences in the polynya (Figure 2).
Archaeal Community Composition
The Thaumarchaeota were a predominant microbial group in sediments from the Amundsen Sea, except at the polynya center (Stn 10) (Figure 2). Most Thaumarchaeota sequences could be classified into three subgroups termed Alpha, Theta, and Upsilon (Durbin and Teske, 2011). The Thaumarchaeota subgroup Alpha was dominant in the top layers (0–5 cm) at all sites. The community composition of the Thaumarchaeota subgroup changed gradually from the top to the bottom at the non-polynya sites (Supplementary Figure S8). The relative abundance of archaeal sequences associated with Theta and Upsilon subgroups in Thaumarchaeota increased with increasing depth, especially in the marginal sea ice zone (Stn 83). Marine Benthic Group B (MBGB) and Miscellaneous Crenarchaeotic Group (MCG), which have been proposed to be Thorarchaeota (Seitz et al., 2016) and Bathyarchaeota (Rinke et al., 2013), respectively, were only detected at Stn 10 (∼9.5% and ∼5% of total sequences, respectively). They have been reported as putative heterotrophic microorganisms (Biddle et al., 2006; Jorgensen et al., 2012; He et al., 2016). Likewise, archaeal sequences related to the methane cycle, including Methanobacteria, Methanomicrobia, and Thermoplasmata, were only detected at Stn 10 (<2% of the total sequences).
Discussion
Planctomycetes-Dominated Microbial Communities in ASP Sediments
One of the prominent features revealed by 16S rRNA genes pyrosequencing was that the members of Planctomycetes were the most abundant microbial members detected in the highly productive polynya sites, especially at Stn 10 (Figure 2). Many Planctomycetes have been found attached to sinking marine aggregates in the water column (DeLong et al., 1993; Fuchsman et al., 2012). At the ASP, however, none were detected in the water column (Delmont et al., 2014; Kim J.-G. et al., 2014). Thus, the highly abundant Planctomycetes sequences in the ASP sediment could not have originated from the water column, confirming the results reported by Probandt and coworkers who showed that Planctomycetes in subtidal, sandy sediments differed from those in the overlaying water column (Probandt et al., 2017). Pirellula members in Planctomycetes have been reported often as heterotrophic bacteria that degrade organic matter produced by algae (Glöckner et al., 2003; Morris et al., 2006; Bižić–Ionescu et al., 2015), while little information is available to speculate about the ecological role of uncultured Planctomycetes (Figure 5). Most cultivated Planctomycetes are known as aerobes or facultative aerobes (Schlesner et al., 2004). However, environmental sequences from Planctomycetes are often retrieved from anoxic zones, such as methane hydrate-bearing sediment (Inagaki et al., 2006) and subsurface seafloor sediment (Jorgensen et al., 2012). Therefore, detection of these sequences in oxygen-depleted layers is unsurprising (Figure 4).
In fine-grained marine sediment receiving high organic material input, Delta- and Gamma-proteobacteria have been reported as the predominant bacterial groups (Rooney-Varga et al., 1997; Bowman and McCuaig, 2003; Bissett et al., 2006; Liu et al., 2015). Similarly, in most benthic systems of the SO, where organic materials primarily originate from diatom blooms in the water column, proteobacterial groups such as Delta- and Gamma-proteobacteria occupy ecological niches as primary mineralizers of organic materials in the sediments (Bowman and McCuaig, 2003; Baldi et al., 2010; Ruff et al., 2014; Learman et al., 2016). Therefore, our observation of high Planctomycetes abundance, an average of 40% of total sequences (Figure 2) in ASP sediments, is intriguing. Further discussion on the dominance of Planctomycetes is presented in next section.
Organic Substrates Control Spatial Distribution of Planctomycetes
To the best of our knowledge, this is the first report suggesting the predominance of Planctomycetes in the ASP, which contrasts with the microbial communities in other benthic systems underlying diatom-dominated water columns. Major controls regulating the distribution of Planctomycetes in the sediments remain unknown, largely because no culture-based studies are available (Schlesner et al., 2004; Lee et al., 2013). However, our statistical analysis (Figure 6 and Supplementary Figure S9) revealed that relative abundance of Planctomycetes in the ASP showed a significant positive correlation with TOC contents and inorganic constituents (, , and Fe2+), presumably resulting from the benthic Corg mineralization. Most labile Corg produced by P. antarctica blooms are rapidly decomposed by heterotrophic bacteria in the water column before reaching the sediment of the ASP (Kirchman et al., 2001). The export flux of Corg formed by a Phaeocystis bloom is twofold slower than that formed by a diatom bloom (DeJong et al., 2017), and thus the contribution of P. antarctica cells to total export flux below the photic zone declines dramatically (Reigstad and Wassmann, 2007). Indeed, in the ASP, most photosynthetically-produced particulate organic carbon (POC) (>95%) exported from the surface layer is converted to suspended POC and/or dissolved carbon within the top 400 m of the water column (Ducklow et al., 2015; Lee et al., 2017). In addition, direct rate measurement conducted in this study also demonstrated that, despite high primary production (110 mmol C m–2 d–1) in the Phaeocystis-dominated water column of the ASP, benthic Corg mineralization (average, 2.1 mmol C m–2 d–1) accounted for only 1.9% of primary production, which was strikingly lower than that measured in other less productive polar regions (Kim et al., 2016). Therefore, organic materials accumulated in surface sediments of the deep ASP (>700 m) would be intrinsically recalcitrant. Meanwhile, it has been well-established that Phaeocystis colonies excrete a mucous matrix containing both carboxylated and sulfated heteropolysaccharides as main constituents (van Boekel, 1992; Alderkamp et al., 2007). Based on metagenomic information from Namibia and Oregon coastal upwelling systems, Woebken et al. (2007) revealed that all marine Planctomycete genomes, except for Candidatus Kuenenia stuttgartiensis, possess a high number of sulfatase genes, which suggests that marine Planctomycetes may be able to break down recalcitrant sulfated heteropolysaccharides (Glöckner et al., 2003; Wegner et al., 2013). Consequently, the high relative abundance of Planctomycetes at the center of the polynya (Stns 10 and 17) suggests that the members of Planctomycetes constitute a significant heterotrophic bacterial group utilizing recalcitrant organic materials produced by Phaeocystis blooms in the water column of the ASP. Recently, Probandt et al. (2017) also suggested that Planctomycetes play a key role in the degradation of high molecular weight compounds and recalcitrant materials entering surface sediment from the water column of the Wadden Sea.
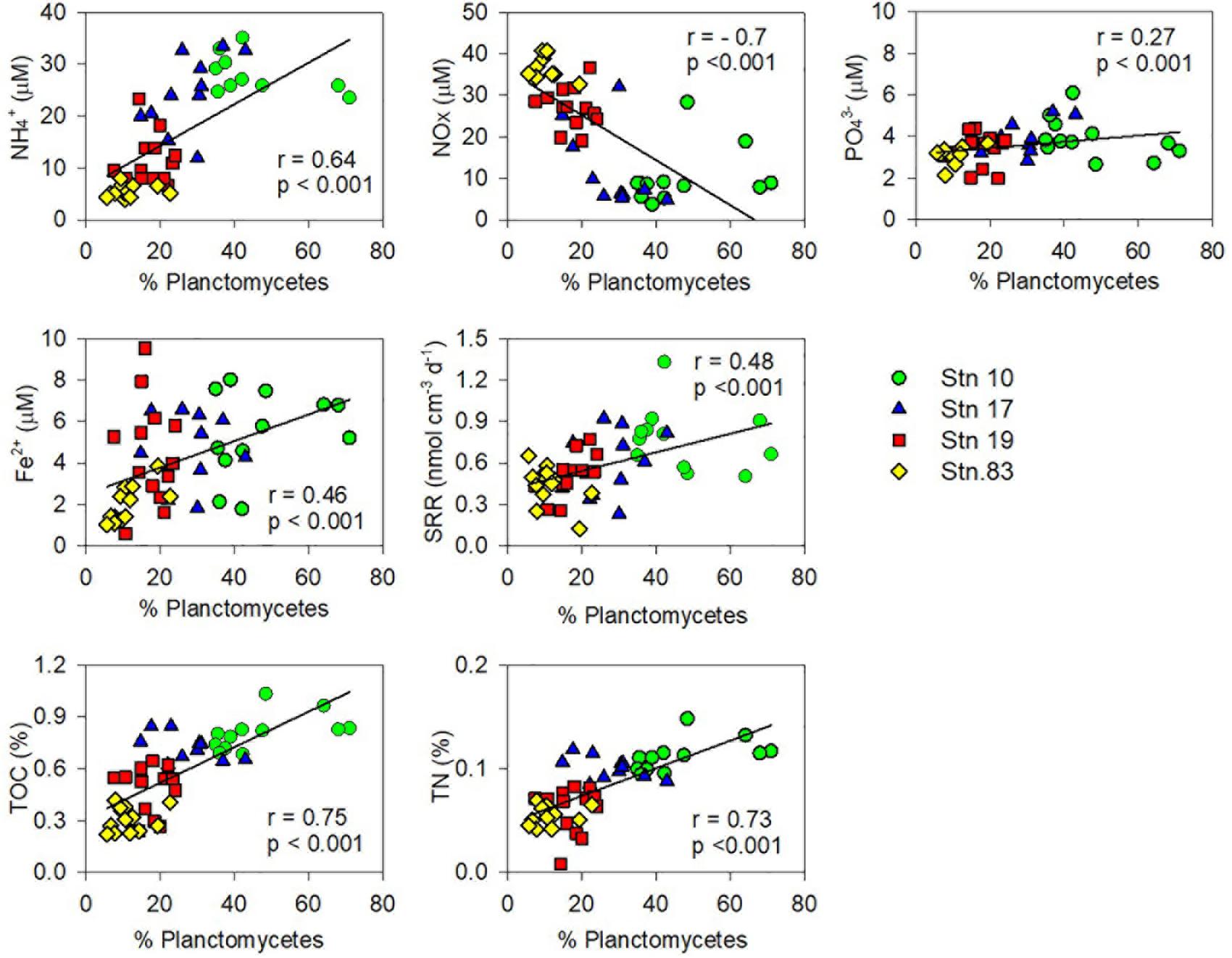
Figure 6. Correlations between the relative abundance of the planctomycetal 16S rRNA genes in total sequences and geochemical properties [the concentrations of , NOX ( + ), , and Fe2+ in pore-water, sulfate reduction rate (SRR), and contents of total organic carbon (TOC) and total nitrogen (TN)].
High Archaeal Abundance in Marginal Sea Ice Zone
In addition to the Planctomycetes-dominated bacterial communities in the sediments of the polynya sites (Stns 10 and 17), another interesting finding revealed from the quantification of 16S rRNA gene was that archaeal abundance occupied more than half (30–71%) of the total prokaryotic abundance in the MIZ (Stn 83), in which most archaeal 16S rRNA gene sequences were assigned to Thaumarchaeota (Figure 2 and Table 2). Members of Thaumarchaeota are major contributors to aerobic ammonia and nitrite oxidation in aquatic environments (Könneke et al., 2005). To examine the potential of sedimentary Thaumarchaeota to oxidize ammonia, we quantified the archaeal amoA gene, which is known for a genetic marker for the ammonia oxidation. The depth profiles of archaeal amoA and 16S rRNA gene copy numbers were very similar to each other in the samples except for some layers (Supplementary Figure S1), which implies that most archaeal members at all depths have a gene encoding ammonia monooxidase.
Both the proportion of archaeal abundance in total prokaryotic abundance (Supplementary Figure S10) and the relative abundance of Thaumarchaeota in total 16S rRNA gene sequences (Supplementary Figure S4B) showed a negative correlation with TOC contents. Previously, the environmental members of Thaumarchaeota have been reported in oligotrophic marine sediments as an important chemolithotrophic microbial assemblage (Inagaki et al., 2006; Durbin and Teske, 2011). Similarly, cultivated thaumarchaeal ammonia oxidizers have been shown to be adapted to oligotrophic conditions (Martens-Habbena et al., 2009). Recently, based on continuous succession of an active microbial group belonging to Thaumarchaeota in the absence of an external Corg supply, Sebastián et al. (2018) suggested that Thaumarchaeota play a significant role in sustaining oligotrophic bathypelagic ecosystems where the Corg supply is limited. Likewise, metaproteomic data indicate that MG-I are abundant and metabolically active at the surface water of the West Antarctic Ocean during the ice-covered winter (Williams et al., 2012), and chemoautotrophic carbon fixation by Thaumarchaeota contributes up to 9% of bacterioplankton production during the Antarctic winter (Tolar et al., 2016). Indeed, in the present study, primary production was low in the water column of the ice-covered sea-ice zone (Stn 83) (Lee et al., 2012; Hyun et al., 2016), and the Corg content in the sediment was consequently lowest (<0.5%, Table 1). Our results thus suggest that the Thaumarchaeota predominating in the sediment of non-polynya sites are a significant chemolithoautotrophic group, sustaining the benthic ecosystem where input of Corg from the water column is limited.
Planctomycetes-Dominated Communities as a Proxy for Assessing Climate Change-Induced Ecosystem Response in the ASP
The structure and function of benthic heterotrophic microbial communities are ultimately controlled by the quantity and quality of organic matter from the water column (Danovaro et al., 2000; Bissett et al., 2006; Jamieson et al., 2013; Learman et al., 2016; Probandt et al., 2017). Thus, the predominance of Planctomycetes in ASP sediment provides ecological and environmental baseline information for assessing the responses of benthic ecosystems to transitions in phytoplankton communities that may result from ongoing and future climate change in the Amundsen Sea. Due to the inflow of warm CDW, the western Antarctic Ocean including the Amundsen Sea has experienced significant surface warming and loss of sea ice (Stammerjohn et al., 2008; Thoma et al., 2008; Jenkins et al., 2010; Jacobs et al., 2011). In particular, glaciers near the Amundsen Sea are undergoing the highest rates of melting and thinning on the Antarctic continent (Rignot, 2008). Sea ice melting increases the supply of nutrients and intensifies water column stratification (Arrigo et al., 1999, 2012; Poulton and Raiswell, 2005), which consequently results in increased primary productivity (Arrigo et al., 2017; Oliver et al., 2018) and shifts in phytoplankton community structure from P. antarctica to diatoms (Arrigo and van Dijken, 2003; Arrigo et al., 2008, 2012). As discussed in previous sections, while Corg originating from Phaeocystis blooms settles slowly, the particulate organic material produced by diatoms possesses a relatively faster sinking rate and a lower C:N ratio (i.e., they are relatively more labile) than those produced by Phaeocystis (Alderkamp et al., 2007; DeJong et al., 2017). Therefore, the transition of dominant phytoplankton from Phaeocystis to diatoms would deposit more labile organic matter into the benthic system of the ASP, which ultimately results in a shift in the benthic microbial community structure from Planctomycetes to Proteobacteria. Likewise, in the marginal sea-ice zone, the loss of sea ice due to global warming may cause an increase in primary production in the water column (Arrigo et al., 2017; Oliver et al., 2018). As a result, benthic microbial communities in the MIZ can undergo a shift from Thaumarchaeota to other heterotrophic bacteria. Consequently, in the Amundsen Sea, a spatial distribution of the benthic microbial community reflecting the composition and production of dominant phytoplankton groups in the water column can act as a relevant proxy for detecting possible variation in primary productivity and phytoplankton community that is associated with sea ice melt due to the global warming.
Conclusion
Characterization of benthic microbial communities is underrepresented in the Southern Ocean, especially in the Amundsen Sea where environmental changes due to global warming are occurring rapidly. We conducted high-throughput sequencing of 16S rRNA genes and quantitative PCR in combination with biogeochemical analyses and metabolic rate measurements to determine the composition, diversity and controls of major microbial communities in sediments of the ASP. Planctomycetes predominated benthic bacterial communities in the polynya sediment underlying the Phaeocystis-dominated water column, which suggests that Planctomycetes comprise a major bacterial group utilizing relatively recalcitrant Corg produced primarily by Phaeocystis blooms. Planctomycetes-dominated microbial communities in the ASP sediment are in stark contrast to that Proteobacteria (Delta- and Gamma-proteobacteria) occupy ecological niches as primary mineralizers of organic materials in most benthic systems in the Southern Ocean, where organic materials in the sediments mostly originate from diatom blooms. Sea ice melting by global warming increases the supply of nutrients and intensifies water column stratification, which consequently results in increased primary productivity and shifts in phytoplankton community structure from P. antarctica to diatoms. Given that microbial communities respond quickly to environmental changes, and that global climate change is proceeding rapidly in the ASP, our results suggest that any modifications in the Planctomycetes-dominated microbial communities will provide valuable insight into changes in organic matter transport to the seafloor that may result from shifts in planktonic communities from Phaeocystis to diatoms associated with climate change.
Data Availability Statement
The datasets generated for this study can be found in the SRX3405376, https://www.ncbi.nlm.nih.gov/sra/SRX3405376.
Author Contributions
HC and J-HH as the first and corresponding author, designed the original experiments and conducted most writing. HC, CH, J-GK, and SK conducted molecular and statistical analysis. S-HK, AC, and J-HH participated in onboard research activities and performed biogeochemical analysis. J-GK, SK, KK, S-KR, EY, and SL participated in writing and discussion of the results. All authors read and approved the final manuscript.
Funding
Analysis for microbial communities was supported by the Korea Polar Research Institute (PE19060) and the Mid-career Researcher Program funded by the Korean Ministry of Science and ICT (No. 2018R1A2B2006340). Biogeochemical analysis and metabolic rate measurements were supported by the Korea-Long-Term Marine Ecological Research (K-LTMER) funded by the Korean Ministry of Oceans and Fisheries.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank the captain and crew of the Korean Research Icebreaker Araon, for their outstanding assistance during the cruises.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2019.00797/full#supplementary-material
References
Alderkamp, A.-C., Buma, A. G. J., and van Rijssel, M. (2007). The carbohydrates of Phaeocystis and their degradation in the microbial food web. Biogeochemistry 83, 99–118. doi: 10.1007/978-1-4020-6214-8_9
Arrigo, K. R., Lowry, K. E., and van Dijken, G. L. (2012). Annual changes in sea ice and phytoplankton in polynyas of the Amundsen Sea, Antarctica. Deep Sea Res. Part II Top. Stud. Oceanogr. 71–76, 5–15. doi: 10.1016/j.dsr2.2012.03.006
Arrigo, K. R., Robinson, D. H., Worthen, D. L., Dunbar, R. B., DiTullio, G. R., VanWoert, M., et al. (1999). Phytoplankton community structure and the drawdown of nutrients and CO2 in the Southern Ocean. Sciences 283, 365–367. doi: 10.1126/science.283.5400.365
Arrigo, K. R., and van Dijken, G. L. (2003). Phytoplankton dynamics within 37 Antarctic coastal polynya systems. J. Geophys. Res. 108:3271. doi: 10.1029/2002jc001739
Arrigo, K. R., van Dijken, G. L., Castelao, R. M., Luo, H., Rennermalm, A. K., Tedesco, M., et al. (2017). Melting glaciers stimulate large summer phytoplankton blooms in southwest Greenland waters. Geophys. Res. Lett. 44, 6278–6285. doi: 10.1002/2017GL073583
Arrigo, K. R., van Dijken, G. L., and Long, M. (2008). Coastal Southern Ocean: a strong anthropogenic CO2 sink. Geophys. Res. Lett. 35:L21302. doi: 10.1029/2008GL035624
Baldi, F., Marchetto, D., Oini, F., Fani, R., Michaud, L., Giudice, A. L., et al. (2010). Biochemical and microbial features of shallow marine sediments along the Terra Nova Bay (Ross Sea, Antarctica). Cont. Shelf Res. 30, 1614–1625. doi: 10.1016/j.csr.2010.06.009
Bertics, V., and Ziebis, W. (2009). Biodiversity of benthic microbial communities in bioturbated coastal sediments is controlled by geochemical microniches. ISME J. 3, 1269–1285. doi: 10.1038/ismej.2009.62
Biddle, J. F., Lipp, J. S., Lever, M. A., Lloyd, K. G., Sørensen, K. B., Anderson, R., et al. (2006). Heterotrophic Archaea dominate sedimentary subsurface ecosystems off Peru. Proc. Natl. Acad. Sci. U.S.A. 103, 3846–3851. doi: 10.1073/pnas.0600035103
Bienhold, C., Boetius, A., and Ramette, A. (2012). The energy-diversity relationship of complex bacterial communities in Arctic deep-sea sediments. ISME J. 6, 724–732. doi: 10.1038/ismej.2011.140
Bissett, A., Bowman, J., and Burke, C. (2006). Bacterial diversity in organically–enriched fish farm sediments. FEMS Microbiol. Ecol. 55, 48–56. doi: 10.1111/j.1574-6941.2005.00012.x
Bižić–Ionescu, M., Zeder, M., Ionescu, D., Orlić, S., Fuchs, B. M., Grossart, H.-P., et al. (2015). Comparison of bacterial communities on limnic versus coastal marine particles reveals profound differences in colonization. Environ. Microbiol. 17, 3500–3514. doi: 10.1111/1462-2920.12466
Bowman, J. P., and McCuaig, R. D. (2003). Biodiversity, community structural shifts, and biogeography of prokaryotes within Antarctic continental shelf sediment. Appl. Environ. Microbiol. 69, 2463–2483. doi: 10.1128/AEM.69.5.2463-2483.2003
Canfield, D. E., Thamdrup, B., and Kristensen, E. (eds) (2005). Aquatic Geomicrobiology. San Diego, CA: Elsevier, 711.
Caporaso, J. G., Bittinger, K., Bushman, F. D., Desantis, T. Z., Andersen, G. L., and Knight, R. (2010). PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics 26, 266–267. doi: 10.1093/bioinformatics/btp636
Carr, S. A., Vogel, S. W., Dunbar, R. B., Brandes, J., Spear, J. R., Levy, R., et al. (2013). Bacterial abundance and composition in marine sediments beneath the Ross ice shelf, Antarctica. Geology 11, 377–395. doi: 10.1111/gbi.12042
Chao, A. (1984). Non-parametric estimation of the number of classes in a population. Scand. J. Stat. 11, 265–270. doi: 10.2307/4615964
Choi, A., Cho, H., Kim, S.-H., Thamdrup, B., Lee, S. H., and Hyun, J.-H. (2016). Rates of N2 production and diversity and abundance of functional genes associated with denitrification and anaerobic ammonium oxidation in the sediment of the Amundsen Sea Polynya, Antarctica. Deep Sea Res. Part II Top. Stud. Oceanogr. 123, 113–125. doi: 10.1016/j.dsr2.2015.07.016
Danovaro, R., Marrale, D., Dell’Anno, A., Croce, N. D., Tselepides, A., and Fabiano, M. (2000). Bacterial response to seasonal changes in labile organic matter composition on the continental shelf and bathyal sediments of the Cretan Sea. Prog. Oceanogr. 46, 345–366. doi: 10.1016/s0079-6611(00)00025-2
DeJong, H. B., Dunbar, R. B., Koweek, D. A., Mucciarone, D. A., Berovici, S. K., and Hansell, D. A. (2017). Net community production and carbon export during the late summer in the Ross Sea, Antarctica. Glob. Biogeochem. Cycles 31, 473–491. doi: 10.1002/2016GB005417
Delmont, T. O., Hammar, K. M., Ducklow, H. W., Yager, P. L., and Post, A. F. (2014). Phaeocystis antarctica blooms strongly influence bacterial community structures in the Amundsen Sea polynya. Front. Microbiol. 5:646. doi: 10.3389/fmicb.2014.00646
DeLong, E. F., Franks, D. G., and Allderdge, A. L. (1993). Phylogenetic diversity of aggregate-attached vs. free-living marine bacterial assemblages. Limnol. Oceanogr. 38, 924–934. doi: 10.4319/lo.1993.38.5.0924
Deppeler, S. L., and Davidson, A. T. (2017). Southern ocean phytoplankton in a changing climate. Front. Mar. Sci. 4:40. doi: 10.3389/fmars.2017.00040
Ducklow, H. W., Wilson, S. E., Post, A. F., Stammerjohn, S. E., Erickson, M., Lee, S. H., et al. (2015). Particle flux on the continental shelf in the Amundsen Sea Polynya and Western Antarctic Peninsula. Elementa 3:46. doi: 10.12952/journal.elementa.000046
Durbin, A. M., and Teske, A. (2011). Microbial diversity and stratification of South Pacific abyssal marine sediments. Environ. Microbiol. 13, 3219–3234. doi: 10.1111/j.1462-2920.2011.02544.x
Franco, M. A., Mesel, I. D., Demba Diallo, M., Van der Gucht, K., Van Gansbeke, D., van Rijswijk, P., et al. (2007). Effect of phytoplankton bloom deposition on benthic bacterial communities in two contrasting sediments in the southern North Sea. Aquat. Microb. Ecol. 48, 241–254. doi: 10.3354/ame048241
Fuchsman, C. A., Kirkpatrick, J. B., Brazelton, W. J., Murray, J. W., and Staley, J. T. (2012). Metabolic strategies of free-living and aggregate-associated bacterial communities inferred from biologic and chemical profiles in the Black Sea suboxic zone. FEMS Microbiol. Ecol. 75, 586–603. doi: 10.1111/j.1574-6941.2011.01189.x
Fuhrman, J. A. (2009). Microbial community structure and its functional implications. Nature 459, 193–199. doi: 10.1038/nature08058
Glöckner, F. O., Kube, M., Teeling, H., Lombardot, T., Ludwig, W., Gade, D., et al. (2003). Complete genome sequence of the marine planctomycete Pirellula sp. strain 1. Proc. Natl. Acad. Sci. U.S.A. 100, 8298–8303. doi: 10.1073/pnas.1431443100
Gruber, N., Gloor, M., Fletcher, S. E. M., Doney, S. C., Dutkiewicz, S., Follows, M., et al. (2009). Oceanic sources, sinks, and transport of atmospheric CO2. Glob. Biogeochem. Cycles 23:GB1005. doi: 10.1029/2008GB003349
He, Y., Li, M., Perumal, V., Feng, X., Fang, J., Xie, J., et al. (2016). Genomic and enzymatic evidence for acetogenesis among multiple lineages of the archaeal phylum Bathyarchaeota widespread in marine sediments. Nat. Microbiol. 1:16035. doi: 10.1038/nmicrobiol.2016.35
Hoffmann, K., Hassenrück, C., Salman-Carvalho, V., Holtappels, M., and Bienhold, C. (2017). Response of bacterial communities to different detritus compositions in Arctic deep-sea sediments. Front. Microbiol. 8:266. doi: 10.3389/fmicb.2017.00266
Huber, J. A., Welch, D. B. M., Morrison, H. G., Huse, S. M., Neal, P. R., Butterfield, D. A., et al. (2007). Microbial population structures in the deep marine biosphere. Science 318, 97–100. doi: 10.1126/science.1146689
Hyun, J. H., Kim, S.-H., Yang, E. J., Choi, A., and Lee, S. H. (2016). Biomass, production, and control of heterotrophic bacterioplankton during a late phytoplankton bloom in the Amundsen Sea Polynya, Antarctica. Deep Sea Res. Part II Top. Stud. Oceanogr. 102, 102–112. doi: 10.1016/j.dsr2.2015.10.001
Inagaki, F., Nunoura, T., Nakagawa, S., Teske, A., Lever, M., Laier, A., et al. (2006). Biogeographical distribution and diversity of microbes in methane hydrate-bearing deep marine sediments on the Pacific Ocean Margin. Proc. Natl. Acad. Sci. U.S.A. 103, 2815–2820. doi: 10.1073/pnas.0511033103
Jacobs, S. S., Jenkins, A., Giulivi, C. F., and Dutrieus, R. (2011). Stronger ocean circulation and increased melting under Pine Island Glacier ice shelf. Nat. Geosci. 4, 519–523. doi: 10.1038/ngeo1188
Jamieson, R. E., Heywood, J. L., Rogers, A. D., Billett, D. S. M., and Paerce, D. A. (2013). Bacterial biodiversity in deep-sea sediments from two regions of contrasting surface water productivity near the Crozet Islands, Southern Ocean. Deep Sea Res. Part I Oceanogr. Res. Pap. 175, 67–77. doi: 10.1016/j.dsr.2012.12.012
Jenkins, A., Dutrieux, P., Jacobs, S. S., McOhail, S. D., Perrett, J. R., Webb, A. T., et al. (2010). Observations beneath Pine Island Glacier in West Antarctica and implications for its retreat. Nat. Geosci. 3, 468–472. doi: 10.1038/ngeo890
Jorgensen, S. L., Hannisdal, B., Lansén, A., Baumberger, T., Flesland, K., Fonseca, R., et al. (2012). Correlating microbial community profiles with geochemical data in highly stratified sediments from the Arctic Mid-Ocean Ridge. Proc. Natl. Acad. Sci. U.S.A. 109, 2846–2855. doi: 10.1073/pnas.1207574109
Kim, B. K., Joo, H., Song, H. J., Yang, E. J., Lee, S. H., Hahm, D., et al. (2014). Large seasonal variation in phytoplankton production in the Amundsen Sea. Polar Biol. 38, 319–331. doi: 10.1007/s00300-014-1588-5
Kim, J.-G., Park, S.-J., Quan, Z.-X., Jung, M.-Y., Cha, I.-T., Kim, S.-J., et al. (2014). Unveiling abundance and distribution of planktonic Bacteria and Archaea in a polynya in Amundsen Sea, Antarctica. Environ. Microbiol. 16, 1566–1578. doi: 10.1111/1462-2920.12287
Kim, S. H., Choi, A., Yang, E. J., Lee, S. H., and Hyun, J. H. (2016). Low benthic respiration and nutrient flux at the highly productive Amundsen Sea Polynya, Antarctica. Deep Sea Res. Part II Top. Stud. Oceanogr. 123, 92–101. doi: 10.1016/j.dsr2.2015.10.004
Kim, S.-J., Kim, J.-G., Lee, S.-H., Park, S.-J., Gwak, J.-H., Jung, M.-Y., et al. (2019). Genomic and metatranscriptomic analyses of carbon remineralization in an Antarctic polynya. Microbiome 7:29. doi: 10.1186/s40168-019-0643-4
Kirchman, D. L., Cottrell, M. T., and Lovejoy, C. (2010). The structure of bacterial communities in the western Arctic Ocean as revealed by pyrosequencing of 16S rRNA genes. Environ. Microbiol. 12, 1132–1143. doi: 10.1111/j.1462-2920.2010.02154.x
Kirchman, D. L., Meon, B., Ducklow, H. W., Carlson, C. A., Hansell, D. A., and Steward, G. F. (2001). Glucose fluxes and concentrations of dissolved combined neutral sugars (polysaccharides) in the Ross Sea and polar front zone, Antarctica. Deep Sea Res. Part II Top. Stud. Oceanogr. 48, 4179–4197. doi: 10.1016/s0967-0645(01)00085-6
Könneke, M., Bernhard, A. E., de la Torre, J. R., Walker, C. B., Waterbury, J. B., and Stahl, D. A. (2005). Isolation of an autotrophic ammonia-oxidizing marine archaeon. Nature 437, 543–546. doi: 10.1038/nature03911
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Lane, D. J., Pace, B., Olsen, G. J., Stahl, D. A., Sogin, M. L., and Pace, N. R. (1985). Rapid determination of 16S ribosomal RNA sequences for phylogenetic analyses. Proc. Natl. Acad. Sci. U.S.A. 82, 6955–6959. doi: 10.1073/pnas.82.20.6955
Learman, D. R., Henson, M. W., Thrash, J. C., Temperton, B., Brannock, P. M., Santos, S. R., et al. (2016). Biogeochemical and microbial variation across 5500 km of Antarctic surface sediment implicates organic matter as a driver of benthic community structure. Front. Microbiol. 7:284. doi: 10.3389/fmicb.2016.00284
Lee, H. W., Roh, S. W., Shin, N. R., Lee, J., Whon, T. W., Jung, M. J., et al. (2013). Blastopirellula cremea sp. nov., isolated from a dead ark clam. Int. J. Syst. Evol. Microbiol. 63, 2314–2319. doi: 10.1099/ijs.0.044099-0
Lee, S. H., Hwang, J., Ducklow, H. W., Hahm, D., Lee, S. H., Kim, D., et al. (2017). Evidence of minimal carbon sequestration in the productive Amundsen Sea polynya. Geophys. Res. Lett. 44, 7892–7899. doi: 10.1002/2017GL074646
Lee, S. H., Kim, B. K., Yun, M. S., Joo, H., Yang, E. J., Kim, Y. N., et al. (2012). Spatial distribution of phytoplankton productivity in the Amundsen Sea, Antarctica. Polar Biol. 35, 1721–1733. doi: 10.1007/s00300-012-1220-5
Lee, Y., Yang, E. J., Park, J., Jung, J., Kim, T. W., and Lee, S. H. (2016). Physical-biological coupling in the Amundsen Sea: influence of physical factors on phytoplankton community structure and biomass. Deep Sea Res. Part I Oceanogr. Res. Pap. 117, 51–60. doi: 10.1016/j.dsr.2016.10.001
Liu, J., Liu, X., Wang, M., Qiao, Y., Zheng, Y., and Zhang, X.-H. (2015). Bacterial and archaeal communities in sediments of the North Chinese Marginal Seas. Microb. Ecol. 70, 105–117. doi: 10.1007/s00248-014-0553-8
Luria, C. M., Amaral-Zettler, L. A., Ducklow, H. W., and Rich, J. J. (2016). Seasonal succession of free–living bacterial communities in coastal waters of the western Antarctic peninsula. Front. Microbiol. 7:1731. doi: 10.3389/fmicb.2016.01731
Martens-Habbena, W., Berube, P. M., Urakawa, H., de la Torre, J. R., and Stahl, D. A. (2009). Ammonia oxidation kinetics determine niche separation of nitrifying Archaea and Bacteria. Nature 461, 976–979. doi: 10.1038/nature08465
McDonald, D., Price, M. N., Goodrich, J., Nawrocki, E. P., DeSantis, T. Z., Probst, A., et al. (2012). An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and Archaea. ISME J. 6, 610–618. doi: 10.1038/ismej.2011.139
Miller, L. A., and DiTuillo, G. R. (2007). “Gas fluxes and dynamics in polynyas,” in Polynyas: Windows to the World, Vol. 74, eds W. O. Smith and D. G. Barber (Amsterdam: Elsevier), 163–191. doi: 10.1016/s0422-9894(06)74005-3
Montes-Hugo, M. A., and Yuan, X. (2012). Climate patterns and phytoplankton dynamics in Antarctic latent heat polynyas. J. Geophys. Res. 117:C05031. doi: 10.1029/2010JC006597
Morris, R. M., Longnecker, K., and Giovannoni, S. J. (2006). Pirellula and OM43 are among the dominant lineages identified in an Oregon coast diatom bloom. Environ. Microbiol. 8, 1361–1370. doi: 10.1111/j.1462-2920.2006.01029.x
Nguyen, T., and Landfald, B. (2015). Polar front associated variation in prokaryotic community structure in Arctic shelf seafloor. Front. Microbiol. 6:17. doi: 10.3389/fmicb.2015.00017
Nihashi, S., and Oshima, K. I. (2015). Circumpolar mapping of Antarctic coastal polynyas and landfast sea ice: relationship and variability. J. Clim. 28, 3650–3670. doi: 10.1175/JCLI-D-14-00369.1
Oliver, H., Luo, H., Castelao, R., van Dijken, G., Mattingly, K., Rosen, J. J., et al. (2018). Exploring the potential impact of Greenland meltwater on stratification, photosynthetically active radiation, and primary production in the Labrador Sea. J. Geophys. Res. Oceans 123, 2570–2591. doi: 10.1002/2018JC013802
Orcutt, B. N., Sylvan, J. B., Knab, N. J., and Edwards, K. J. (2011). Microbial ecology of the dark ocean above, at, and below the seafloor. Microbiol. Mol. Biol. Rev. 75, 361–422. doi: 10.1128/MMBR.00039-10
Pachiadaki, M. G., Yakimov, M. M., LaCono, V., Leadbetter, E., and Edgcomb, V. (2014). Unveiling microbial activities along the halocline of Thetis, a deep-sea hypersaline anoxic basin. ISME J. 8, 2478–2489. doi: 10.1038/ismej.2014.100
Petit, J. R., Jouzel, J., Raynaud, D., Barkov, N. I., Barnola, J.-M., Basile, I., et al. (1999). Climate and atmospheric history of the past 420,000 years from the Vostok ice core, Antarctica. Nature 399, 429–436. doi: 10.1038/20859
Poulton, S. W., and Raiswell, R. (2005). Chemical and physical characteristics of iron oxides in riverine and glacial meltwater sediments. Chem. Geol. 218, 203–221. doi: 10.1016/j.chemgeo.2005.01.007
Probandt, D., Knittel, K., Tegetmeyer, H. E., Ahmerkamp, S., Holtappels, M., and Amann, R. (2017). Permeability shapes bacterial communities in sublittoral surface sediments. Environ. Microbiol. 19, 1584–1599. doi: 10.1111/1462-2920.13676
Quince, C., Lanzen, A., Davenport, R. J., and Turnbaugh, P. J. (2011). Removing noise from pyrosequenced amplicons. BMC Bioinformatics 12:38. doi: 10.1186/1471-2105-12-38
R Core Team, (2016). R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing.
Reigstad, M., and Wassmann, P. (2007). Does Phaeocystis spp. contribute significantly to vertical export of organic carbon? Biogeochemistry 83, 217–234. doi: 10.1007/s10533-007-9093-3
Richert, I., Yager, P. L., Dinasquet, J., Logares, R., Riemann, L., Wendeberg, A., et al. (2019). Summer comes to the Southern Ocean: how phytoplankton shape bacterioplankton communities far into the deep dark sea. Ecosphere 10:e02641. doi: 10.1002/ecs2.2641
Rignot, E. (2008). Changes in West Antarctic ice stream dynamics observed with ALOS PALSAR data. Geophys. Res. Lett. 35:L12505. doi: 10.1029/2008GL033365
Rinke, C., Schwientek, P., Sczyrba, A., Ivanova, N. N., Anderson, I. J., Cheng, J. F., et al. (2013). Insights into the phylogeny and coding potential of microbial dark matter. Nature 499, 431–437. doi: 10.1038/nature12352
Robador, A., Müller, A., Sawicka, J. E., Berry, D., Hubert, C. R. J., Loy, A., et al. (2016). Activity and community structures of sulfate-reducing microorganisms in polar, temperate and tropical marine sediments. ISME J. 10, 796–809. doi: 10.1038/ismej.2015.157
Roesch, L. F., Fulthorpe, R. R., Riva, A., Casella, G., Hadwin, A. K., Kent, A. D., et al. (2007). Pyrosequencing enumerates and contrasts soil microbial diversity. ISME J. 1, 283–290. doi: 10.1038/ismej.2007.53
Rooney-Varga, J. N., Dvereux, R., Evans, R. S., and Hines, M. E. (1997). Seasonal changes in the relative abundance of uncultivated sulfate-reducing bacteria in a salt marsh sediment and in the rhizosphere of Spartina alterniflora. Appl. Environ. Microbiol. 63, 3895–3901.
Ruff, S. E., Probandt, D., Zinkann, A.-C., Iversen, M. H., Klaas, C., Würzberg, L., et al. (2014). Indications for algae-degrading benthic microbial communities in deep-sea sediments along the Antarctic Polar Front. Deep Sea Res. Part II Top. Stud. Oceanogr. 108, 6–16. doi: 10.1016/j.dsr2.2014.05.011
Schauer, R., Bienhold, C., Ramette, A., and Harder, J. (2010). Bacterial diversity and biogeography in deep-sea surface sediments of the South Atlantic Ocean. ISME J. 4, 159–170. doi: 10.1038/ismej.2009.106
Schlesner, H., Rensmann, C., Tindall, B. J., Gade, D., Rabus, R., Pfeiffer, S., et al. (2004). Taxonomic heterogeneity within the Planctomycetales as derived by DNA-DNA hybridization, description of Rhodopirellula baltica gen. nov., sp. nov., transfer of Pirellula marina to the genus Blastopirellula gen. nov. as Blastopirellula marina comb. nov. and emended description of the genus Pirellula. Int. J. Syst. Evol. Microbiol. 54, 1567–1580. doi: 10.1099/ijs.0.63113-0
Schloss, P. D., Westcott, S. L., Ryabin, T., Hall, J. R., Hartmann, M., Hollister, E. B., et al. (2009). Introducing mother: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75, 7537–7541. doi: 10.1128/aem.01541-09
Schmid, M., Walsh, K., Webb, R., Rijpstra, W. I., van de Pas-Schoonen, K., Verbruggen, M. J., et al. (2003). Candidatus “Scalindua brodae”, sp. nov., Candidatus “Scalindua wagneri”, sp. nov., two new species of anaerobic ammonium oxidizing bacteria. Syst. Appl. Micorobiol. 26, 529–538. doi: 10.1078/072320203770865837
Sebastián, M., Auguet, J.-C., Restrepo-Ortiz, C. X., Sala, M. M., Marrase, C., and Gasol, J. M. (2018). Deep ocean prokaryotic communities are remarkably malleable when facing long-term starvation. Environ. Microbiol. 20, 713–723. doi: 10.1111/1462-2920.14002
Sedwick, R. N., and DiTuillo, G. R. (1997). Regulation of algal blooms in Antarctic shelf waters by the release of iron from melting sea ice. Geophys. Res. Lett. 24, 2515–2518. doi: 10.1029/97GL0259
Seitz, K. W., Lazar, C. S., Hinrichs, K.-U., Teske, A. P., and Baker, B. J. (2016). Genomic reconstruction of a novel, deeply branched sediment archaeal phylum with pathways for acetogenesis and sulfur reduction. ISME J. 10, 1696–1705. doi: 10.1038/ismej.2015.233
Smith, W. O. Jr., and Barber, D. G. (eds) (2007). Polynyas: Windows to the World. Amsterdam: Elsevier.
Stammerjohn, S. E., Martinson, D. G., Smith, R. C., Yuan, X., and Rin, D. (2008). Trends in annual Antarctic sea ice retreat and advance and their relation to El Niño–Southern Oscillation and Southern annual mode variability. J. Geophys. Res. 113:C03S90. doi: 10.1029/2007jc004269
Takahashi, T., Sutherland, S. C., Wanninkhof, R., Sweeney, C., Feely, R. A., Chipman, D. W., et al. (2009). Climatological mean and decadal change in surface ocean p CO2, and net sea–air CO2 flux over the global oceans. Deep Sea Res. Part II Top. Stud. Oceanogr. 56, 554–577. doi: 10.1016/j.dsr2.2008.12.009
Thoma, M., Jenkins, A., Holland, D., and Jacobs, S. (2008). Modelling circumpolar deep water intrusions on the Amundsen Sea continental shelf, Antarctica. Geophys. Res. Lett. 35:L18602. doi: 10.1029/2008GL034939
Tolar, B. B., Ross, M. J., Wallsgrove, N. J., Alusihare, L. I., Popp, B. N., and Hollobaugh, J. T. (2016). Contribution of ammonia oxidation to chemoautotrophy in Antarctic coastal waters. ISME J. 10, 2605–2619. doi: 10.1038/ismej.2016.61
van Boekel, W. H. M. (1992). Phaeocystis colony mucus components and the importance of calcium ions for colony stability. Mar. Ecol. Prog. Ser. 87, 301–305. doi: 10.3354/meps087301
Verardo, D. J., Froelich, P. N., and McIntyre, A. (1990). Determination of organic carbon and nitrogen in marine sediments using the Carlo Erba Na-1500 Analyzer. Deep Sea Res. Part A Oceanogr. Res. Pap. 37, 157–165. doi: 10.1016/0198-0149(90)90034-S
Wang, Q., Garrity, G. M., Tiedje, J. M., and Cole, J. R. (2007). Naïve Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73, 5261–5267. doi: 10.1128/aem.00062-07
Wegner, C.-E., Richter-Heitmann, T., Klindworth, A., Klockow, C., Richter, M., Achstetter, T., et al. (2013). Expression of sulfatases in Rhodopirellula baltica and the diversity of sulfatases in the genus Rhodopirellula. Mar. Genomics 9, 51–61. doi: 10.1016/j.margen.2012.12.001
Whitman, W. B., Coleman, D. C., and Wiliiam, J. W. (1998). Prokaryotes: the unseen majority. Proc. Natl. Acad. Sci. U.S.A. 95, 6578–6583. doi: 10.1073/pnas.95.12.6578
Williams, C. M., Dupont, A. M., Loevenich, J., Post, A. F., Dinasquet, J., and Yager, P. L. (2016). Pelagic microbial heterotrophy in response to a highly productive bloom of Phaeocystis antarctica in the Amundsen Sea Polynya, Antarctica. Elementa 4:102. doi: 10.12952/journal.elementa.000102
Williams, T. J., Long, E., Evans, F., DeMaere, M. Z., Lauro, F. M., Raftery, M. J., et al. (2012). A metaproteomic assessment of winter and summer bacterioplankton from Antarctic Peninsula coastal surface waters. ISME J. 6, 1883–1900. doi: 10.1038/ismej.2012.28
Woebken, D., Teeling, H., Wecker, P., Dumitriu, A., Kostadinov, I., DeLong, E. F., et al. (2007). Fosmids of novel marine Planctomycetes from the Namibian and Oregon coast upwelling systems and their cross-comparison with planctomycete genomes. ISME J. 1, 419–435. doi: 10.1038/ismej.2007.63
Yager, P. L., Sherrell, R. M., Stammerjohn, S. E., Alderkamp, A.-C., Schofield, O., Abrahamsen, E. P., et al. (2012). ASPIRE: the Amundsen Sea polynya international research expedition. Oceanography 25, 40–53. doi: 10.5670/oceanog.2012.73
Keywords: Amundsen Sea polynya, Antarctica, benthic microbial community, global climate change, organic carbon mineralization, Phaesocystis antarctica, Planctomycetes, Thaumarchaeota
Citation: Cho H, Hwang CY, Kim J-G, Kang S, Knittel K, Choi A, Kim S-H, Rhee S-K, Yang EJ, Lee S and Hyun J-H (2020) A Unique Benthic Microbial Community Underlying the Phaeocystis antarctica-Dominated Amundsen Sea Polynya, Antarctica: A Proxy for Assessing the Impact of Global Changes. Front. Mar. Sci. 6:797. doi: 10.3389/fmars.2019.00797
Received: 27 September 2019; Accepted: 11 December 2019;
Published: 10 January 2020
Edited by:
David A. Walsh, Concordia University, CanadaReviewed by:
Jiwen Liu, Ocean University of China, ChinaYong Jiang, Ocean University of China, China
André M. Comeau, Dalhousie University, Canada
Copyright © 2020 Cho, Hwang, Kim, Kang, Knittel, Choi, Kim, Rhee, Yang, Lee and Hyun. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jung-Ho Hyun, aHl1bmpoQGhhbnlhbmcuYWMua3I=
 Hyeyoun Cho
Hyeyoun Cho Chung Yeon Hwang
Chung Yeon Hwang Jong-Geol Kim
Jong-Geol Kim Sanghoon Kang4
Sanghoon Kang4 Katrin Knittel
Katrin Knittel Ayeon Choi
Ayeon Choi Sung-Han Kim
Sung-Han Kim Sung-Keun Rhee
Sung-Keun Rhee Jung-Ho Hyun
Jung-Ho Hyun